Abstract
This review presents the French strategy for blood group genotyping in high-responder and newly diagnosed sickle cell disease (SCD) patients. In addition to FY, JK, and MNS genotyping, the RH blood group system is now explored in SCD patients in France. Molecular typing has been used for the deduction of partial RH2 (C) antigens since 2010, and the gradual implementation of systematic RHD and RHCE genotyping nationwide was initiated in late 2014. In our laboratory, 962 RH:2 (C-positive) SCD patients have been tested since 2010, and 1,148 SCD patients of all RH phenotypes have been genotyped for clinically relevant alleles of RHD and RHCE since late 2014.
Keywords: Sickle cell disease, Transfusion medicine, Blood group genotyping, RH antigens, Partial antigens
Part I: Overview and Presentation of the French Strategy
Introduction
Sickle cell disease (SCD) is a major concern in transfusion medicine in France. It is one of the most frequent genetic disorders, affecting approximately 14,000 patients nationwide, mostly of African descent. Patient care is often based on packed red blood cell (pRBC) transfusions to sustain patients and limit complications.
All patients in France are typed for ABO blood group, and for the main antigens of the RH (D, C, E, c, e) and KEL (K) systems. SCD patients are also typed for the main antigens of the FY (Fya, Fyb), JK (Jka, Jkb), and MNS (M, N, S, s) systems. Typing for these antigens is part of the initial workup for SCD patients and is generally performed before the first transfusion.
Molecular variants for several blood group systems have been described in individuals of African descent and are responsible for rare phenotypes. Different alleles in the MNS system are responsible for the S-s-U-, Uvar, and Henshaw phenotypes [1,2]. Their detection in patients is particularly important in situations in which the pool of donors is not predominantly of African descent, as in mainland France.
Many partial antigens and rare blood types have been described in the RH system [1,3,4,5], including several found in patients in France [6,7]. RH variants are considered relevant in SCD patients [8], due to the risk of allo-antibody formation. Caution is often exercised for the rarer RH variants because a lack of evidence for antibody formation or of previous clinical consequences associated with a rare variant cannot be considered to rule out the risk.
The stakes associated with rare phenotypes and molecular variants are particularly high for SCD patients, who often undergo transfusion and are at high risk of alloimmunization [9,10]. There is evidence to suggest that individuals may be high or low ‘responders’, depending on their propensity to produce antibodies [11,12], but the responder status of a patient cannot be predicted until a positive result is obtained in an antibody screening test. Delayed hemolytic transfusion reactions (DHTRs) are a major concern in SCD patient care, and are generally associated with the production of antibodies [13,14]. Alloantibodies associated with both partial RH antigens and rare RH blood types are associated with a risk of DHTR [15,16] and must be taken into account in SCD patients.
Blood Group Genotyping
Blood group genotyping has been used for more than 15 years in France and is becoming increasingly important in transfusion medicine. Specialists make particular use of this technique in the following common situations: i) positive results in a direct antiglobulin test (DAT), ii) recent transfusion episodes precluding blood group phenotyping, iii) typing for blood group systems or antigens for which there are no commercially available reagents, and iv) the detection of molecular variants, especially for the RH blood group system. Due to a lack of commercially available reagents, most molecular variants can be detected only by molecular typing.
French transfusion specialists sometimes request FY, JK, or MNS genotyping for SCD patients, if prior typing data are not available. This may be the case for patients arriving in France after transfusion in other countries or for first-time patients who have never been typed despite French guidelines. When patients are typed S-s-, U- or Uvar phenotype is deduced from genotype analysis [2]. There are no commercially available reagents for Uvar phenotype detection.
The prevalence of partial RH antigens and variant RH alleles in SCD patients has recently been investigated in France [11,17] and elsewhere [15,18,19]. SCD patients with variant alleles have been shown to have a higher risk of alloimmunization, in both these and other studies [20].
These studies have shown that partial RH antigens have a high prevalence and are clinically relevant in SCD populations. In the French studies [11,17], 20.8–27.7% of RH:2 (C-positive) SCD patients were found to carry partial RH2 antigens without conventional RH2 antigens, and 8.4% of RH:1 (D-positive) patients were found to carry partial RH1 antigens. These partial antigens are associated with high rates of alloimmunization in SCD patients: anti-RH2 antibodies are detected in 14.3–30% of all patients with partial RH2 antigens [11,17], and anti-RH1 antibodies are detected in 17.6% of patients with partial RH1 antigens [11].
The French Strategy for the Detection of RH Variants
Following the French studies cited above, two important decisions were taken concerning the care of SCD patients in France. Based on the prevalence of variant RH alleles and the clinical consequences of alloimmunization, systematic RH molecular typing was considered to be warranted for SCD patients, except those known to be low responders (no antibody formation despite multiple transfusions), to limit costly genotyping analyses. All considerations and recommendations below relate to high-responder SCD patients (alloimmunized) or SCD patients of unknown responder status (patients who have received few pRBC transfusions or newly diagnosed patients).
Firstly, since 2010, RH:2 SCD patients with normal or weak RH2 antigen expression have received RH:-2 (C negative) pRBCs until exclusion of the most frequent partial RH2 antigens, deduced from molecular typing: RN (RHD*01-RHCE*02.10.01) and CceS(RHD*03N.01-RHCE*01.20.03 and RHD*01N.06-RHCE*01.20.03) haplotypes [17], and DIVa(C)-(RHD*04.01-RHCE*CE- DIVa.2(2–3)-CE-D(5)-CE), following its description in 2012 [21] (samples from 2010 and 2011 were tested retrospectively).
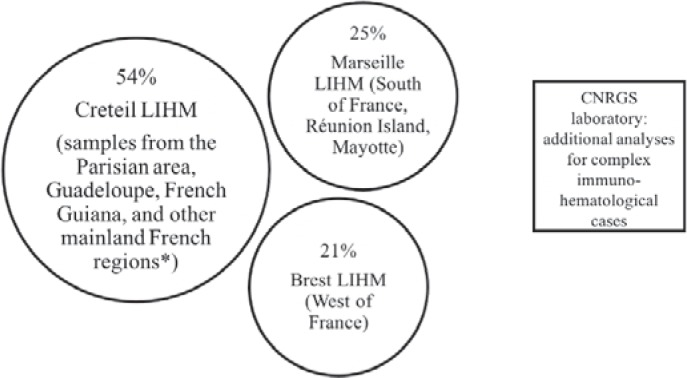
Secondly, systematic RH molecular typing for SCD patients has gradually been introduced, nationwide, since December 2014. Three molecular immunohematology laboratories (LIHM) perform RH molecular typing in France (fig. 1). The CNRGS laboratory deals with complex cases of immunization. The Créteil LIHM in the Parisian area (Ile-de-France) received 54% of all samples requiring blood group genotyping in 2017, regardless of indication, and 65% of the SCD samples tested in France, because the Parisian area has the highest SCD patient density in France.
Fig. 1.
Organization of the molecular immunohematology laboratories (LIHM) in France. 3 LIHMs receive samples from all over France for blood group genotyping (% of samples analyzed at each laboratory in 2017, for SCD and all other indications), and the CNRGS laboratory deals with complex immunization cases. *The other mainland French regions covered by the Creteil LIHM are the North, East, and Central areas of France.
We describe here the results for the Créteil LIHM. In this study, 57% of the SCD samples received originated from the Parisian area (mostly from the Mondor, Necker and Avicenne hospitals), and 39% originated from Guadeloupe (in the French Caribbean) and French Guiana (in South America). The regions of mainland France covered by the Créteil LIHM are the North, East, and Central areas of France (most prescriptions for testing come from Lille and Dijon). For data analysis, these regions were considered together under the heading ‘Other regions’.
Part II: Review of the French Strategy at LIHM Créteil between 2010 and 2017
2010–2016: RH2 Partial Antigen Detection
From 2010 to early 2016, 572 SCD patients were genotyped for the deduction of partial RH2 antigens in the absence of conventional RH2: 509 patients with an unambiguous RH:2 phenotype (fig. 2), and 63 with ambiguous RH2 antigen expression or anti-RH2 antibodies. Overall, 205 (35.8%) of those tested were deduced to have partial RH2 antigens, including 149 (72.7%) whose partial RH2 antigen would not have been detected without systematic genotyping, because RH2 antigen expression was unambiguous.
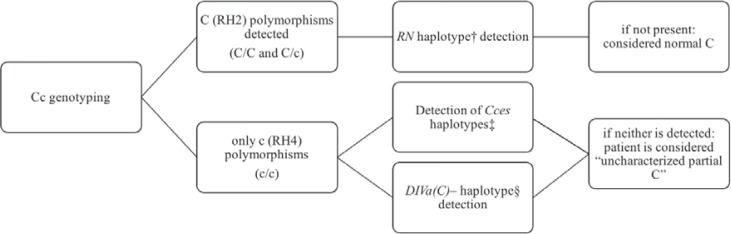
Fig. 2.
Flow chart for molecular detection of the most prevalent partial RH2 antigens in SCD patients with unambiguous RH2 antigen expression. †RHD*01-RHCE*02.10.01. ‡RHD *03N.01-RHCE*01.20.03 and RHD*01N.06-RHCE *01.20.03. §RHD*04.01-RHCE*CE- DIVa.2(2-3)-CE-D(5)-CE).†, ‡Detected by real-time polymerase chain reaction (PCR) assays, as previously described [17]. §Initially detected by Sanger sequencing analysis, and subsequently with a specific real-time PCR assay.
Variant RH2 antigen detection by serological methods seems to be more common for RN than for CceS: ambiguous RH2 antigen expression was observed for half the RN samples, but fewer than 1 in 8 of the CceS samples. This finding is of particular importance, because partial RH2 CceS seems to be associated with the highest risk of anti-RH2 immunization in SCD patients, as reported in previous studies [17] and in this cohort. One of the 8 patients with anti-RH2 antibodies had RN, and 5 had CceS.
The DIVa(C)- haplotype was found to be extremely rare in SCD patients in France, as it was detected in only 3 patients (0.5% of RH:2 patients). Weak RH2 antigen expression was due to the RZ(RHD*01 associated with RHCE*04) haplotype in some cases. In such cases, the antigen is normal, but there are fewer RH2 antigens per RBC [1].
2014–2017: Gradual Implementation of Full RH Analysis for SCD Patients
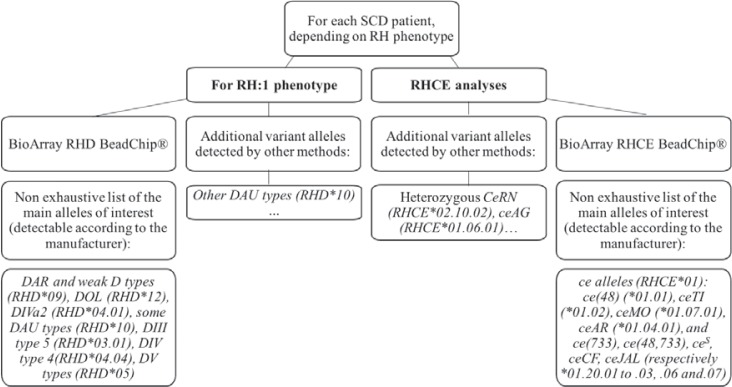
In late 2014, after the study by Silvy and coworkers [11], the detection of partial RH1, RH3, RH4, and RH5 antigens was gradually introduced for SCD patients in France. Progressive testing was initiated at the Créteil LIHM, beginning with SCD patients in Guadeloupe, French Guiana, and several of the main hospitals in the Parisian area. Over the last 3 years, 1,148 SCD patients have been typed for most of the clinically relevant RH alleles (fig. 3). This typing is based on the RHD and RHCE BeadChip DNA array (BioArray/Immucor®, Warren NJ, USA), together with several additional assays performed in our laboratory to detect alleles of interest. The c.1136C>T substitution in the DAU0 allele (RHD*10.00) is predicted to be neutral [22] and was not routinely detected at our laboratory.
Fig. 3.
Flow chart: outline of the strategy for detecting clinically relevant RHD and RHCE alleles in SCD patients. Other analyses, including Sanger sequencing for one or both RH genes, were performed on a case-by-case basis, especially if expression was ambiguous for one or more RH antigens or if RH antibodies were present.
Transfusion specialists agree on the partial status of the RH antigens produced by most of the altered RHD and RHCE alleles common in individuals of African descent (see table 1 for main alleles of interest and references). However, the partial status of the RH4 and RH5 antigens produced by alleles ce(733) (RHCE*01.20.01) and ce(48,733) (RHCE*01.20.02) remains a matter of debate. It is now agreed that no partial antigen is produced for ce(48) (RHCE*01.01) allele [19]. In France, the ce(733) (RHCE*01.20.01) and ce(48,733) (RHCE*01.20.02) alleles are not considered to produce partial antigens either. In our experience, the anti-RH5 antibodies associated with these molecular backgrounds are auto-antibodies, and patients are therefore no more prone to the formation of anti-RH4 or anti-RH5 antibodies when exposed to non-genotype-matched pRBCs. The status of the RH1 antigen associated with weak D type 4.0 (RHD*09.03) is also under scrutiny [41,42,43].
Table 1.
Main alleles of interest in SCD patients of Afro-Caribbean descent responsible for partial RH antigensa
| Common or short name | ISBT allele reference* | References |
|---|---|---|
| Haplotypes (at least partial RH2 antigen) | ||
| RN | RHD*01 associated with RHCE*02.10.01 | Le Pennec et al. [23] |
| Rouillac et al. [7] | ||
| Tournamille et al. [17] | ||
| CceS | RHD*03N.01 or *01N.06 associated with RHCE*01.20.03 | Westhoff et al. [24, 25] |
| Tournamille et al. [17] | ||
| DIVa(C)– | RHD*04.01 associated with RHCE*CE-DIVa.2(2–3)-CE-D(5)-CE | Hipsky et al.[21] |
| RHD alleles (partial RH1 antigen) | ||
| DAU alleles | RHD*10 | Srivastava et al. [22] |
| DAR alleles | RHD*09.01, .02, .06 | Hemker et al. [26] |
| Wagner et al. [27] | ||
| DIII type 5 | RHD*03.01 | Westhoff et al. [25] |
| DIV type 4 | RHD*04.04 | Rouillac et al. [28] |
| von Zabern et al.[29] | ||
| DV alleles | RHD*05 | Rouillac et al. [28] |
| Weak D type 4.0† | RHD*09.03 | Wagner et al. [27] |
| DOL alleles | RHD*12 | Flegel et al. [3] |
| RHCE*ce alleles (partial RH5 and/or RH4 antigens) | ||
| ceS | RHCE*01.20.03 | Pham et al. [30] |
| ceAG | RHCE*01.06.01 | Westhoff et al. [31] |
| ceMO | RHCE*01.07.01 | Noizat-Pirenne et al. [32] |
| Westhoff et al. [4] | ||
| ceAR | RHCE*01.04.01 | Noizat-Pirenne et al. [6] |
| Peyrard et al. [33] | ||
| Hipsky et al. [34] | ||
| ceEK | RHCE*01.05.01 | Noizat-Pirenne et al. [6] |
| Peyrard et al. [33] | ||
| ceBI | RHCE*01.08 | Reid et al. [35] |
| ceSM | RHCE*01.09 | Reid et al. [35] |
| ceJAL | RHCE*01.20.07 | Lomas-Francis et al. [36] |
| Ong et al. [37] | ||
| ceTI type alleles | RHCE*01.02, *01.03, *01.20.04 | Westhoff et al. [38] |
| ceCF‡ | RHCE*01.20.06 | Hipsky et al. [39] |
Haplotypes and alleles of interest detected in SCD patients of Afro-Caribbean descent in France that have been demonstrated to express one or more partial RH antigens. The table contains references for partial antigen status and RH10 and RH20 antigen expression (see also Daniels et al. [40]). For clarity, the main haplotypes producing partial RH2 antigens are grouped at the beginning of this table.
Except for RHCE*CE-DIVa.2(2–3)-CE-D(5)-CE (the name used in the initial report) for which no ISBT reference is available.
The status of RH1 antigen associated with weak D type 4.0 is controversial [43].
ceCF also produces some RH1 epitopes.
Of the 1,148 SCD patients tested for altered alleles of both RHD and RHCE (table 2), at least one partial RH antigen was deduced from genotyping results for 253 (22.0%). The prevalence of partial antigens was similar for SCD patients from the Parisian area and for those from other regions of mainland France. The slightly higher numbers obtained simply reflect the larger numbers of samples sent to our laboratory for testing due to serological discrepancies or antibodies against RH antigens in antigen-positive patients.
Table 2.
Exploration of RHD and RHCE in 1,148 patients from late 2014 to 2017a
| Parisian area | Guadeloupe | French Guiana | Other mainland regions | All regions | |
|---|---|---|---|---|---|
| Patients with ≥ 1 partial RH antigen / | 123 / 466 | 69 / 394 | 41 / 221 | 20 / 70 | 253 / 1,148 |
| number of patients (%) | (26.5%) | (17.5%) | (18.6%) | (29.4%) | (22.0%) |
| Number of partial RH1 / all RH:1 | 47 / 450 | 46 / 375 | 32 / 213 | 6 / 66 | 131 / 1,104 |
| patients (%) | (10.4%) | (12.3%) | (15.0%) | (9.1%) | (11.9%) |
| Number of partial RH2 / all RH:2 | 70 / 185 | 21 / 126 | 8 / 60 | 8 / 19 | 107 / 390 |
| patients (%)* | (37.8%) | (16.7%) | (13.3%) | (42.1%) | (27.4%) |
| Number of RH:3 patients | 77 | 86 | 36 | 9 | 208 |
| Number of partial RH4 / all RH:4 | 7 / 451 | 2 / 388 | 3 / 220 | 3 / 68 | 15 / 1,126 |
| patients (%) | (1.6%) | (0.5%) | (1.4%) | (4.4%) | (1.3%) |
| Number of partial RH5 / all RH:5 | 26 / 462 | 25 / 392 | 13 / 220 | 7 / 68 | 71 / 1,141 |
| patients (%) | (5.6%) | (6.4%) | (5.9%) | (10.3%) | (6.2%) |
Detection of partial RH antigens following implementation of the French strategy. No partial RH3 antigen was detected. RH antigens for which weak expression could not be explained after RH genotyping with our strategy are included, as this weak antigen expression could be due to rare undetected RH alleles.
See table 3 for further details concerning RH2 alleles.
Rare RH phenotypes due to homozygously altered alleles were deduced by molecular analysis for 8 patients (0.7%): 1 RH:-31,-34, 1 RH:-18 (homozygous for the ceAR allele RHCE*01.04.01), 3 homozygous for the ceAG allele (RHCE*01.06.01), and 3 homozygous for the ceTI allele (RHCE*01.02.01). In addition, 19 patients (1.7%) were heterozygous for two altered RHCE alleles, a situation associated with an unknown risk of alloimmunization. RH10 (V) and/or RH20 (VS) expression could be deduced for 517 of the 1,148 patients (45.0%).
The prevalence of haplotypes leading to partial RH2 antigen expression varies in the French regions covered by the Créteil LIHM (table 3). The difference in RN and CceS haplotype prevalence for all RH:2 SCD patients genotyped from 2010 to 2017 is particularly striking. These differences undoubtedly stem from the specific features of each geographic region, with differences in gene flow and genetic drift. The Parisian area (Ile-de-France) has higher levels of gene flow due to immigration. French Guiana, which is located in South America, has a different mixture. Guadeloupe is an island in the French Caribbean and is, thus, more susceptible to genetic drift.
Table 3.
Comparison of the prevalence of the RN and Cces» haplotypes leading to partial RH2 antigen expression, for all SCD patient samples tested from 2010 to 2017a
| RHCE allele | Parisian area | Guadeloupe | French Guiana | All regions |
|---|---|---|---|---|
| Number of RH:2 patients tested | 701 | 176 | 61 | 963 |
| RN haplotype† | 83 (11.8%) | 2 (1.1%) | 1 (1.6%) | 90 (9.3%) |
| CceS haplotypes‡ | 151 (21.5%) | 33 (18.8%) | 7 (11.5%) | 195 (20.2%) |
Percentages represent the number of partial RH antigens, in the absence of conventional RH2 antigens, deduced from the molecular analysis, relative to the number of RH:2 patients tested.
RHD*01-RHCE*02.10.01.
RHD*03N.01-RHCE*01.20.03 and RHD*01N.06-RHCE*01.20.03.
Overview of Genotyping for the FY, JK, and MNS Blood Group Systems from 2015 to 2017
As stated above, extended genotyping is rarely necessary for SCD patients in France because extended phenotyping is included in the initial workup. Only a small proportion of the total number of SCD samples tested in our laboratory underwent FY, JK and MNS genotyping (fig. 4). A shift from phenotyping to genotyping would not be cost-effective in France.
Fig. 4.
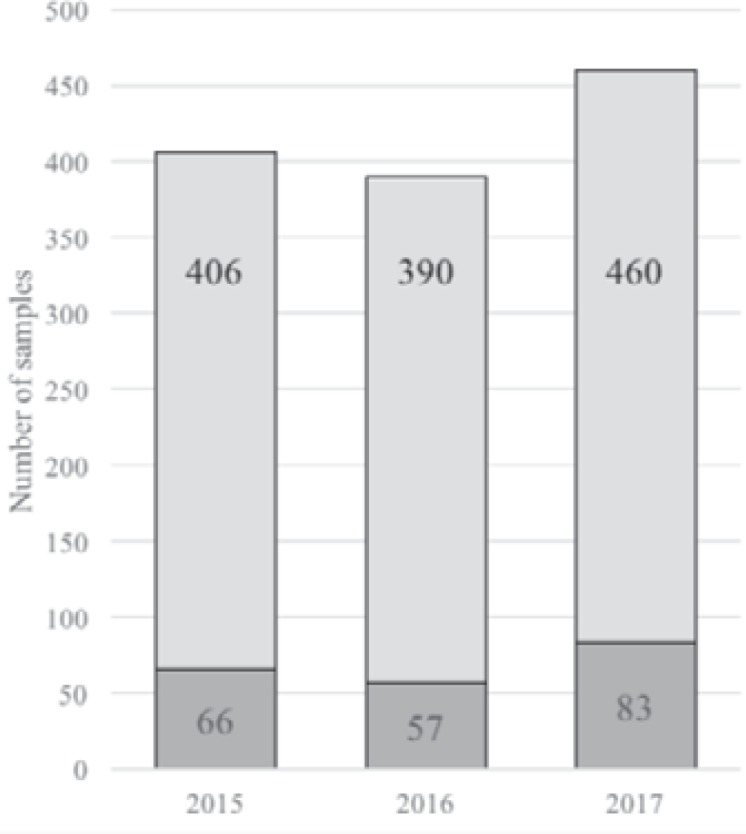
SCD samples genotyped each year from 2015 to 2017. Genotyping performed for SCD patients at the Créteil LIHM: number of samples typed for FY, JK and/or MNS blood group systems (darker gray) each year, relative to all SCD samples genotyped.
Concluding Remarks
This review of current French genotyping practices for SCD patients confirms the results of the previous studies [11,17] on a larger scale, and reveals differences between the SCD patients of different French regions. Overall, for almost 1 in 4 SCD patients in France at least one partial RH antigen can be deduced from molecular typing. Most of these cases are not picked up by phenotyping and require genotyping for detection.
Several studies have reported allele prevalences for other populations [18,19,44,45,46]. Partial RH2 antigens appear to be particularly prevalent in mainland France, whereas partial RH1 antigens are more prevalent in Guadeloupe and French Guiana. Conversely, the pattern of variation in RH alleles does not seem to differ across the US [47]. The specific features of French regions overseas may account for this difference.
The RHD and RHCE genotyping strategy is being implemented gradually, and is not yet being applied by all French transfusion specialists. Thus, samples with ambiguous antigen expression for one or more RH antigens are still more likely to be sent to our laboratory for further testing. This may affect the allele and haplotype prevalences reported here. The results summarized in this review promote generalized systematic RHD and RHCE genotyping for SCD patients, by showing it would be beneficial for SCD patients.
The large number of partial RH antigens in SCD patients of African descent highlights the importance of detecting these antigens before the occurrence of alloimmunization. The strategy presented here would be improved by advances in our knowledge of the risk of alloimmunization associated with RH variants, particularly for patients heterozygous for two altered RHCE alleles. Furthermore, these analyses would be unnecessary for many patients if it became possible to determine the high- or low-responder status of the individual before the occurrence of alloimmunization. Unfortunately, this is not yet possible, and RH genotyping remains essential for high-responder and newly diagnosed SCD patients.
Disclosure Statement
The authors have no competing interests to declare.
Acknowledgments
We would like to thank Prof. Jacques Chiaroni (EFS Marseille) for providing insight into genetic flow and genetic drift, Mr. Stephane Goumard (EFS Ile-de-France) for valuable assistance with data extraction, and Mrs. Julie Haddou (LIHM Créteil) for assistance with data collection. We would also like to thank the transfusion specialists who collaborate with LIHM Créteil daily, to ensure that SCD patients in France receive the best possible care.
References
- 1.Reid M, Lomas-Francis C, Olsson M. 3rd ed. London: Academic Press; 2012. The Blood Group Antigen FactsBook. [Google Scholar]
- 2.Storry JR, Reid ME, Fetics S, Huang C-H. Mutations in GYPB exon 5 drive the S-s-U+(var) phenotype in persons of African descent: implications for transfusion. Transfusion. 2003;43:1738–1747. doi: 10.1046/j.0041-1132.2003.00585.x. [DOI] [PubMed] [Google Scholar]
- 3.Flegel WA, von Zabern I, Doescher A, Wagner FF, Strathmann KP, Geisen C, Palfi M, Písacka M, Poole J, Polin H, Gabriel C, Avent ND. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion. 2009;49:1059–1069. doi: 10.1111/j.1537-2995.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Westhoff CM, Vege S, Horn T, Hue-Roye K, Halter Hipsky C, Lomas-Francis C, Reid ME. RHCE*ceMO is frequently in cis to RHD*DAU0 and encodes a hr(S)-, hr(B)-, RH:-61 phenotype in black persons: clinical significance. Transfusion. 2013;53:2983–2989. doi: 10.1111/trf.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid ME, Hipsky CH, Velliquette RW, Lomas-Francis C, Larimore K, Olsen C. Molecular background of RH in Bastiaan, the RH:-31,-34 index case, and two novel RHD alleles. Immunohematology. 2012;28:97–103. [PubMed] [Google Scholar]
- 6.Noizat-Pirenne F, Lee K, Pennec P-YL, Simon P, Kazup P, Bachir D, Rouzaud AM, Roussel M, Juszczak G, Ménanteau C, Rouger P, Kotb R, Cartron JP, Ansart-Pirenne H. Rare RHCE phenotypes in black individuals of Afro-Caribbean origin: identification and transfusion safety. Blood. 2002;100:4223–4231. doi: 10.1182/blood-2002-01-0229. [DOI] [PubMed] [Google Scholar]
- 7.Rouillac C, Gane P, Cartron J, Le Pennec PY, Cartron J, Colin Y. Molecular basis of the altered antigenic expression of RhD in weak D(Du) and RhC/e in RN phenotypes. Blood. 1996;87:4853–4861. [PubMed] [Google Scholar]
- 8.Noizat-Pirenne F, Tournamille C. Relevance of RH variants in transfusion of sickle cell patients. Transfus Clin Biol. 2011;18:527–535. doi: 10.1016/j.tracli.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood. 2012;120:528–537. doi: 10.1182/blood-2011-11-327361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fasano RM, Booth GS, Miles M, Du L, Koyama T, Meier ER, Luban NL. Red blood cell alloimmunization is influenced by recipient inflammatory state at time of transfusion in patients with sickle cell disease. Br J Haematol. 2015;168:291–300. doi: 10.1111/bjh.13123. [DOI] [PubMed] [Google Scholar]
- 11.Silvy M, Tournamille C, Babinet J, Pakdaman S, Cohen S, Chiaroni J, Galactéros F, Bierling P, Bailly P, Noizat-Pirenne F. Red blood cell immunization in sickle cell disease: evidence of a large responder group and a low rate of anti-Rh linked to partial Rh phenotype. Haematologica. 2014;99:e115–117. doi: 10.3324/haematol.2014.104703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sins JWR, Biemond BJ, van den Bersselaar SM, Heijboer H, Rijneveld AW, Cnossen MH, Kerkhoffs JL, van Meurs AH, von Ronnen FB, Zalpuri S, de Rijke YB, Ellen van der Schoot C, de Haas M, van der Bom JG, Fijnvandraat K. Early occurrence of red blood cell alloimmunization in patients with sickle cell disease. Am J Hematol. 2016;91:763–769. doi: 10.1002/ajh.24397. [DOI] [PubMed] [Google Scholar]
- 13.Habibi A, Mekontso-Dessap A, Guillaud C, Michel M, Razazi K, Khellaf M, Chami B, Bachir D, Rieux C, Melica G, Godeau B, Galacteros F, Bartolucci P, Pirenne F. Delayed hemolytic transfusion reaction in adult sickle-cell disease: presentations, outcomes, and treatments of 99 referral center episodes. Am J Hematol. 2016;91:989–994. doi: 10.1002/ajh.24460. [DOI] [PubMed] [Google Scholar]
- 14.de Montalembert M, Dumont M-D, Heilbronner C, Brousse V, Charrara O, Pellegrino B, Piguet C, Soussan V, Noizat-Pirenne F. Delayed hemolytic transfusion reaction in children with sickle cell disease. Haematologica. 2011;96:801–807. doi: 10.3324/haematol.2010.038307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chou ST, Jackson T, Vege S, Smith-Whitley K, Friedman DF, Westhoff CM. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood. 2013;122:1062–1071. doi: 10.1182/blood-2013-03-490623. [DOI] [PubMed] [Google Scholar]
- 16.Noizat-Pirenne F. Relevance of alloimmunization in haemolytic transfusion reaction in sickle cell disease. Transfus Clin Biol. 2012;19:132–138. doi: 10.1016/j.tracli.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 17.Tournamille C, Meunier-Costes N, Costes B, Martret J, Barrault A, Gauthier P, Galactéros F, Nzouékou R, Bierling P, Noizat-Pirenne F. Partial C antigen in sickle cell disease patients: clinical relevance and prevention of alloimmunization. Transfusion. 2010;50:13–19. doi: 10.1111/j.1537-2995.2009.02382.x. [DOI] [PubMed] [Google Scholar]
- 18.Gaspardi AC, Sippert EA, De Macedo MD, Pellegrino J, Costa FF, Castilho L. Clinically relevant RHD-CE genotypes in patients with sickle cell disease and in African Brazilian donors. Blood Transfus Trasfus Sangue. 2016;14:449–454. doi: 10.2450/2016.0275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou ST, Flanagan JM, Vege S, Luban NLC, Brown RC, Ware RE, Westhoff CM. Whole-exome sequencing for RH genotyping and alloimmunization risk in children with sickle cell anemia. Blood Adv. 2017;1:1414–1422. doi: 10.1182/bloodadvances.2017007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sippert E, Fujita CR, Machado D, Guelsin G, Gaspardi AC, Pellegrino J, Gilli S, Saad SS, Castilho L. Variant RH alleles and Rh immunisation in patients with sickle cell disease. Blood Transfus Trasfus Sangue. 2015;13:72–77. doi: 10.2450/2014.0324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hipsky CH, Hue-Roye K, Lomas-Francis C, Huang C-H, Reid ME. Molecular basis of the rare gene complex, DIVa(C)-, which encodes four low-prevalence antigens in the Rh blood group system. Vox Sang. 2012;102:167–170. doi: 10.1111/j.1423-0410.2011.01519.x. [DOI] [PubMed] [Google Scholar]
- 22.Srivastava K, Polin H, Sheldon SL, Wagner FF, Grabmer C, Gabriel C, Denomme GA, Flegel WA. The DAU cluster: a comparative analysis of 18 RHD alleles, some forming partial D antigens. Transfusion. 2016;56:2520–2531. doi: 10.1111/trf.13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Pennec PY, Rouger P, Klein MT, Kornprobst M, Brossard Y, Boizard B, Salmon C. A serologic study of red cells and sera from 18 Rh:32,-46 (RN/RN) persons. Transfusion. 1989;29:798–802. doi: 10.1046/j.1537-2995.1989.29990070185.x. [DOI] [PubMed] [Google Scholar]
- 24.Westhoff CM, Vege S. RH gene polymorphisms are risk factor for allo-immunization in patients with sickle cell disease. Blood. 2007;110:445. [Google Scholar]
- 25.Westhoff CM, Vege S, Halter-Hipsky C, Whorley T, Hue-Roye K, Lomas-Francis C, Reid ME. DIIIa and DIII type 5 are encoded by the same allele and are associated with altered RHCE*ce alleles: clinical implications. Transfusion. 2010;50:1303–1311. doi: 10.1111/j.1537-2995.2009.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemker MB, Ligthart PC, Berger L, van Rhenen DJ, van der Schoot CE, Wijk PA. DAR, a new RhD variant involving exons 4, 5, and 7, often in linkage with ceAR, a new Rhce variant frequently found in African blacks. Blood. 1999;94:4337–4342. [PubMed] [Google Scholar]
- 27.Wagner FF, Frohmajer A, Ladewig B, Eicher NI, Lonicer CB, Müller TH, Siegel MH, Flegel WA. Weak D alleles express distinct phenotypes. Blood. 2000;95:2699–2708. [PubMed] [Google Scholar]
- 28.Rouillac C, Colin Y, Hughes-Jones NC, Beolet M, D'Ambrosio AM, Cartron JP, Le Van., Kim C Transcript analysis of D category phenotypes predicts hybrid Rh D-CE-D proteins associated with alteration of D epitopes. Blood. 1995;85:2937–2944. [PubMed] [Google Scholar]
- 29.von Zabern I, Wagner FF, Moulds JM, Moulds JJ, Flegel WA. D category IV: a group of clinically relevant and phylogenetically diverse partial D. Transfusion. 2013;53:2960–2973. doi: 10.1111/trf.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pham B-N, Peyrard T, Tourret S, Beolet M, Many H, Juszczak G, Roussel M, Kappler-Gratias S, Rouger P, Le Pennec PY. Anti-HrB and anti-hrb revisited. Transfusion. 2009;49:2400–2405. doi: 10.1111/j.1537-2995.2009.02307.x. [DOI] [PubMed] [Google Scholar]
- 31.Westhoff CM, Vege S, Hipsky CH, Horn T, Hue-Roye K, Keller J, Velliquette R, Lomas-Francis C, Chou ST, Reid ME. RHCE*ceAG (254C>G, Ala85Gly) is prevalent in blacks, encodes a partial ce-phenotype, and is associated with discordant RHD zygosity. Transfusion. 2015;55:2624–2632. doi: 10.1111/trf.13225. [DOI] [PubMed] [Google Scholar]
- 32.Noizat-Pirenne F, Mouro I, Le Pennec PY, Ansart-Pirenne H, Juszczak G, Patereau C, Verdier M, Babinet J, Roussel M, Rouger P, Cartron JP. Two new alleles of the RHCE gene in Black individuals: the RHce allele ceMO and the RHcE allele cEMI. Br J Haematol. 2001;113:672–679. doi: 10.1046/j.1365-2141.2001.02802.x. [DOI] [PubMed] [Google Scholar]
- 33.Peyrard T, Pham B-N, Poupel S, Martin-Blanc S, Auxerre C, Kappler-Gratias S, Bonin P, Rouger P, Le Pennec., PY Alloanti-c/ce in a c+ceAR/Ce patient suggests that the rare RHCE ceAR allele (ceAR) encodes a partial c antigen. Transfusion. 2009;49:2406–2411. doi: 10.1111/j.1537-2995.2009.02308.x. [DOI] [PubMed] [Google Scholar]
- 34.Hipsky CH, Lomas-Francis C, Fuchisawa A, Reid ME. RHCE*ceAR encodes a partial c (RH4) antigen. Immunohematology. 2010;26:57–59. [PubMed] [Google Scholar]
- 35.Reid ME, Halter Hipsky C, Hue-Roye K, Coghlan G, Olsen C, Lomas-Francis C. The low-prevalence Rh antigen STEM (RH49) is encoded by two different RHCE*ce818T alleles that are often in cis to RHD*DOL. Transfusion. 2013;53:539–544. doi: 10.1111/j.1537-2995.2012.03754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lomas-Francis C, Alcantara D, Westhoff C, Uehlinger J, Valvasori M, Castilho L, Reid ME. JAL (RH48) blood group antigen: serologic observations. Transfusion. 2009;49:719–724. doi: 10.1111/j.1537-2995.2008.02025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong J, Walker PS, Schmulbach E, Storry JR, Vege S, Westhoff C, Lomas-Francis C, Reid ME. Alloanti-c in a c-positive, JAL-positive patient. Vox Sang. 2009;96:240–243. doi: 10.1111/j.1423-0410.2008.01135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westhoff CM, Vege S, Halter Hipsky C, Hue-Roye K, Copeland T, Velliquette RW, Horn T, Lomas-Francis C, Reid ME. RHCE*ceTI encodes partial c and partial e and is often in cis to RHD*DIVa. Transfusion. 2013;53:741–746. doi: 10.1111/j.1537-2995.2012.03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hipsky CH, Lomas-Francis C, Fuchisawa A, Reid ME, Moulds M, Christensen J, Nickle P, Vege S, Westhoff C. RHCE*ceCF encodes partial c and partial e but not CELO, an antigen antithetical to Crawford. Transfusion. 2011;51:25–31. doi: 10.1111/j.1537-2995.2010.02764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniels GL, Faas BH, Green CA, Smart E, Maaskant-van Wijk PA, Avent ND, Zondervan HA, von dem Borne AE, van der., Schoot CE The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion. 1998;38:951–958. doi: 10.1046/j.1537-2995.1998.381098440860.x. [DOI] [PubMed] [Google Scholar]
- 41.Sandler SG, Chen LN, Flegel WA. Serological weak D phenotypes: a review and guidance for interpreting the RhD blood type using the RHD genotype. Br J Haematol. 2017;179:10–19. doi: 10.1111/bjh.14757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ouchari M, Srivastava K, Romdhane H, Jemni Yacoub S, Flegel WA. Transfusion strategy for weak D Type 4.0 based on RHD alleles and RH haplotypes in Tunisia. Transfusion. 2018;58:306–312. doi: 10.1111/trf.14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flegel WA, Peyrard T, Chiaroni J, Tournamille C, Jamet D, Pirenne F. A proposal for a rational transfusion strategy in patients of European and North African descent with weak D type 4.0 and 4.1 phenotypes. Blood Transfus Trasfus Sangue. 2018:1–2. doi: 10.2450/2018.0259-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chou ST, Westhoff CM. Application of genomics for transfusion therapy in sickle cell anemia. Blood Cells Mol Dis. 2017;67:148–154. doi: 10.1016/j.bcmd.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silvy M, Di Cristofaro J, Beley S, Papa K, Rits M, Richard P, Chiaroni J, Bailly P. Identification of RHCE and KEL alleles in large cohorts of Afro-Caribbean and Comorian donors by multiplex SNaPshot and fragment assays: a transfusion support for sickle cell disease patients. Br J Haematol. 2011;154:260–270. doi: 10.1111/j.1365-2141.2011.08691.x. [DOI] [PubMed] [Google Scholar]
- 46.Granier T, Beley S, Chiaroni J, Bailly P, Silvy M. A comprehensive survey of both RHD and RHCE allele frequencies in sub-Saharan Africa. Transfusion. 2013;53:3009–3017. doi: 10.1111/trf.12409. [DOI] [PubMed] [Google Scholar]
- 47.Reid ME, Halter Hipsky C, Hue-Roye K, Hoppe C. Genomic analyses of RH alleles to improve transfusion therapy in patients with sickle cell disease. Blood Cells Mol Dis. 2014;52:195–202. doi: 10.1016/j.bcmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]