Abstract
Background
PCR with sequence-specific priming using allele-specific fluorescent primers and analysis on a capillary sequencer is a standard technique for DNA typing. We aimed to adapt this method for donor typing in a medium-throughput setting.
Methods
Using the Extract-N-Amp PCR system, we devised a set of eight multiplex allele-specific PCR with fluorescent primers for Fya/Fyb, Jka/Jkb, M/N, and S/s. The alleles of a gene were discriminated by the fluorescent color; donor and polymorphism investigated were encoded by product length. Time, cost, and routine performance were collated. Discrepant samples were investigated by sequencing. The association of new alleles with the phenotype was evaluated by a step-wise statistical analysis.
Results
On validation using 312 samples, for 1.1% of antigens (in 5.4% of samples) no prediction was obtained. 99.96% of predictions were correct. Consumable cost per donor were below EUR 2.00. In routine use, 92.2% of samples were successfully predicted. Discrepancies were most frequently due to technical reasons. A total of 11 previously unknown alleles were detected in the Kell, Lutheran, and Colton blood group systems. In 2017, more than 20% of the RBC units prepared by our institution were from donors with predicted antigen status. A steady supply of Yt(a-), Co(a-) and Lu(b-) RBC units was ensured.
Conclusions
Pooled capillary electrophoresis offers a suitable alternative to other methods for extended donor DNA typing. Establishing a large cohort of typed donors improved transfusion support for patients.
Keywords: Blood donor typing, Colton blood group, Immunohematology, Kell blood group, Lutheran blood group, Molecular blood group typing, Null allele
Introduction
In the past decade, molecular phenotype prediction has increasingly been replacing serologic antigen determination. Systematic donor typing allows to supply units much better matched to the recipient than conventional approaches starting with antigen determination only if there is an acute need for typed units [1,2]. A large number of methods have been developed that allow high-throughput typing of blood donors for many antigens [3,4,5,6,7,8,9,10,11,12]. These methods differ in the antigens predicted, the number of rare alleles covered, and the cost per antigen and per donor. Attempted high-accuracy prediction of a donor's phenotype [3,11] contrasts with approaches that expect a serologic confirmation on release [8].
The driving force for large-scale molecular typing differs between populations. In countries with a large number of patients of African descent, the correct identification of ‘African’ RHCE alleles and Dombrock phenotypes is a major challenge and convincing argument to switch to molecular typing [12,13], because serologic typing is unsatisfactory. In some European countries like Germany, such alleles are nearly absent, and compatible units can usually be identified by serologic methods. In these countries, the major challenge is the provision of blood for patients with antibodies to the high-prevalence antigens Kpb, Lub, Yta or Vel [14], and for patients with multiple alloantibodies [15,16] to the clinical relevant antigens outside ABO and RH. Most of these antigens may be easily determined by serology; hence, any molecular method has to compete with serologic approaches.
A decade ago, we developed a simple multiplex PCR approach that allowed to screen for donors who are negative for high-frequency antigens at less than USD 3.00 per donor [17]. In order to extend this method to the more polymorphic antigens of the Duffy and Kidd blood group system, we adapted our approach to capillary gel electrophoresis with fluorescent dye-labeled sequence- specific primers. Here we present our experience with this improved method.
Material and Methods
Blood Samples
EDTA blood samples were collected for blood group determination from blood donors donating at the German Red Cross Blood Donation Service NSTOB (North-central Germany). The donors consented for scientific use of their samples.
DNA Isolation
DNA was isolated from EDTA blood samples using the Extract-N-Amp Blood PCR kit (Sigma-Aldrich, Taufkirchen, Germany) with slight modifications as reported previously [17].
Multiplex PCR
PCR amplification was performed using the Extract-N-Amp Blood PCR mix with primers and concentrations shown in table 1. Eight distinct primer sets were used that resulted in slightly different product sizes. In addition to primers, MgCl2 was added to the PCR mixture to a final extra of 1.2 mmol/l. Reactions were initially performed in a volume of 5 µl including 0.5 µl DNA preparation; during routine use reaction size was reduced to a total volume of 3.9 µl. PCR consisted of 6 min initial denaturation at 94 °C, 10 cycles of 15 s at 94 °C and 45 s at 65 °C followed by 29 cycles of 20 s at 94 °C, 30 s at 65 °C, and 1 min at 72° C and a final 15 min at 72 °C.
Table 1.
PCR primers used
| Name | Sequence* | Product size, bp | Use | Typical concentration, µmol/l |
|---|---|---|---|---|
| Lua2 | NED-cctgcatctcagccgaggctaTgt | Lua specific | 0.05 | |
| Lub2 | ROX-cctgcatctcagccgaggcAagAc | Lub specific | 0.04 | |
| 118b | aaggacccagagagagagagactga | 120 | Lu reverse set 1 | 0.04 |
| 122b | gagaaaggacccagagagagagagactga | 124 | Lu reverse set 2 | 0.04 |
| 126c | cagagagaaaggacccagagagagagagactga | 128 | Lu reverse set 3 | 0.05 |
| 130 | gctgcagagagaaaggacccag | 132 | Lu reverse set 4 | 0.05 |
| 134 | aggagctgcagagagaaaggacc | 136 | Lu reverse set 5 | 0.05 |
| 138 | agccaggagctgcagagagaaag | 140 | Lu reverse set 6 | 0.05 |
| 142 | gcagagccaggagctgcagag | 144 | Lu reverse set 7 | 0.05 |
| 146 | agaggcagagccaggagctgcag | 148 | Lu reverse set 8 | 0.05 |
| M3F | FAM-gaagaAgttgaagtgtgcattgccacac | M specific | 0.10 | |
| N3-H2 | HEX-gaagaAgttgaagtgtgcattgccacct | N specific | 0.04 | |
| mnbv6 | gggatgtgagAggaatttgtcttttgca | 154 | GYPA reverse set 1 | 0.09 |
| mnbv604 | tTTTgggatgtgagAggaatttgtcttttgca | 158 | GYPA reverse set 2 | 0.14 |
| mnbv608 | TTTTtTTTgggatgtgagAggaatttgtcttttgca | 162 | GYPA reverse set 3 | 0.14 |
| mnbv612 | TTTTttTTctcagggatgtgagAggaatttgtcttttgca | 166 | GYPA reverse set 4 | 0.14 |
| mnbv616b | TTTCTTTctCTTctcagggatgtgagAggaatttgtcttttgca | 170 | GYPA reverse set 5 | 0.14 |
| mnbv620b | TTTTTCTTTcTcttagctcagggatgtgagAggaatttgtcttttgca | 174 | GYPA reverse set 6 | 0.14 |
| mnbv624b | TtTCTCTTTCTTTcTcttagctcagggatgtgagAggaatttgtcttttgca | 178 | GYPA reverse set 7 | 0.14 |
| mnbv628c | tCTTTtTCTCTTTCTTTcTcttagctcagggatgtgagAggaatttgtcttttgca | 182 | GYPA reverse set 8 | 0.14 |
| coa5 | FAM-TgacaccttcacgttgtcctggaTcg | Coa specific | 0.03 | |
| cob5 | HEX-cgacaccttcacgttgtccttgacca | Cob specific | 0.03 | |
| Co186 | agcggtctcaggccaagccc | 188 | Co reverse set 1 | 0.04 |
| Co190 | TTTcagcggtctcaggccaagccc | 190 | Co reverse set 2 | 0.07 |
| Co194n | TATcTTTcagcggtctcaggccaagccc | 194 | Co reverse set 3 | 0.07 |
| Co198 | tgagcacccggcagcggtc | 198 | Co reverse set 4 | 0.07 |
| Co202 | gaattgagcacccggcagcg | 202 | Co reverse set 5 | 0.07 |
| Co206 | gagggaattgagcacccggcag | 206 | Co reverse set 6 | 0.09 |
| Co210 | ctcagagggaattgagcacccgg | 210 | Co reverse set 7 | 0.12 |
| Co214 | agctctcagagggaattgagcaccc | 214 | Co reverse set 8 | 0.18 |
| gs4 | FAM-gtacagtgaaacgatggacaagttgtcAca | S specific set 1,3,5,7 | 0.24 | |
| kS4-H2 | HEX-gtacagtgaaacgatggacaagttgtcTcg | s specific set 1,3,5,7 | 0.19 | |
| gSl | FAM-TctTgtacagtgaaacgatggacaagttgtcAca | S specific set 2,4,6,8 | 0.36 | |
| kSl-H2 | HEX-TctTgtacagtgaaacgatggacaagttgtcTcg | s specific set 2,4,6,8 | 0.29 | |
| S212 | attactaatggtaagactgacacattacctcataaatgtt | 220 | GYPB reverse set 1,2 | 0.34 |
| 224 | ||||
| S220 | atactgaaattactaatggtaagactgacacattacct | 230 | GYPB reverse set 3,4 | 0.23 |
| 235 | ||||
| S228b | aaaagtagatactgaaattactaatggtaagactgacacattac | 239 | GYPB reverse set 5,6 | 0.39 |
| 243 | ||||
| S236 | gggtcattaaaagtagatactgaaattactaatggtaaga | 248 | GYPB reverse set 7,8 | 0.23 |
| 252 | ||||
| YtaF | FAM-ccactagttacctgcaggTTgtg | Yta specific | 0.32 | |
| YtbH | HEX-ccactagttacctgcaggcTgtt | Ytb specific | 0.08 | |
| Yt260 | agaggcTcgtcgcagggcc | 261 | Yt reverse set 1 | 0.25 |
| Yt264.2 | tgTgagaggcTcgtcgcagg | 258 | Yt reverse set 2 | 0.33 |
| Yt268 | ggcatgTgagaggcTcgtcgc | 262 | Yt reverse set 3 | 0.33 |
| Yt272c | TgtgggcatgTgagaggcTcgtcgc | 265 | Yt reverse set 4 | 0.4 |
| Yt272c(-2) | TTTTTgtgggcatgTgagaggcTcgtcgc | 270 | Yt reverse set 5 | 0.4 |
| Yt 280c | tgTgccacggtgTgcatgggagag | 273 | Yt reverse set 6 | 0.33 |
| yt284b | acTctgTgccacggtgggcatgg | 277 | Yt reverse set 7 | 0.33 |
| Yt288b.2 | TTtTacTctgTgccacggtgggcatgg | 281 | Yt reverse set 8 | 0.33 |
| Fya6 | FAM-ggTagctgcttccaggttggcTc | Fya specific | 0.43 | |
| Fyb6 | HEX-ggcagctgcttccaggAtggcat | Fyb specific | 0.10 | |
| fy297 | cttcaggctccctgctttgtc | 298 | Fy reverse set 1 | 0.53 |
| fy301 | ggctcttcaggctccctg | 298 | Fy reverse set 2 | 0.53 |
| fy305 | ctccggctcttcaggctc | 302 | Fy reverse set 3 | 0.53 |
| fy309 | cctgctccggctcttcag | 308 | Fy reverse set 4 | 0.53 |
| fy313 | tgttcctgctccggctcttca | 311 | Fy reverse set 5 | 0.53 |
| fy317 | ggactgttcctgctccgg | 316 | Fy reverse set 6 | 0.53 |
| fy321 | cccaggactgttcctgctc | 320 | Fy reverse set 7 | 0.44 |
| fy326b | cttccccaggactgttcctgctc | 325 | Fy reverse set 8 | 0.44 |
| jkfa4 | FAM-gttgaaaccccagagtccTaagtagatgtc | Jka specific | 0.05 | |
| jkfb4-H2 | HEX-gttgaaaccccagagtccaTagtagatgtt | Jkb specific | 0.06 | |
| jk333b | actctcctcTccactcatgtgcctg | 335 | JK reverse set 1 | 0.07 |
| jk339 | tcctactctcctcTccactcatgtg | 339 | JK reverse set 2 | 0.10 |
| jk343 | gccatcctactctcctcTccact | 343 | JK reverse set 3 | 0.07 |
| jk347 | gTgagccatcctactctcctcc | 347 | JK reverse set 4 | 0.07 |
| jk351 | tcctgTgagccatcctactctcc | 351 | JK reverse set 5 | 0.10 |
| jk355 | attttcctgTgagccatcctactctcc | 355 | JK reverse set 6 | 0.08 |
| jk359 | gTgcattttcctgTgagccatcctac | 359 | JK reverse set 7 | 0.07 |
| jk363b | caggTggcattttcctgTgagccatcctac | 363 | JK reverse set 8 | 0.07 |
| kpa1107F | FAM-ctcttccttgtcaatctccatcactAcat | Kpa specific | 0.18 | |
| kpb | HEX-ctcttccttgtcaatctccatcacttTac | Kpb specific | 0.17 | |
| kp372b | gcaggacaaccagtcgatggcg | 372 | Kp reverse set 1 | 0.29 |
| kp 376b | gcaagcaggacaaccagtcgatgg | 376 | Kp reverse set 2 | 0.29 |
| kp380 | gcttgcaagcaggacaaccagtcg | 380 | Kp reverse set 3 | 0.29 |
| kp384b | tgtcgcttgcaagcaggacaaccag | 384 | Kp reverse set 4 | 0.35 |
| kp388b | tgaatgtcgcttgcaagcaggacaacc | 388 | Kp reverse set 5 | 0.35 |
| kp392b | ggtgtgaatgtcgcttgcaagcagg | 392 | Kp reverse set 6 | 0.29 |
| kp396b | catcggtgtgaatgtcgcttgcaagc | 396 | Kp reverse set 7 | 0.29 |
| Kp400b | gggacatcggtgtgaatgtcgcttg | 400 | Kp reverse set 8 | 0.35 |
Big letters denote mismatched nucleotides.
Analysis of Pooled Samples by Capillary Electrophoresis
16 PCR products, each obtained with a different primer set, were mixed and cleansed using Nucleo Spin Gel- and PCR Clean up Kits (Macheray-Nagel, Düren, Germany). 5 µl of amplificate were analyzed using an ABI 310 capillary sequencer (Thermo-Fisher, Darmstadt, Germany). Applied Biosystems GeneScan 500 LIZ dye size standard (Thermo-Fisher) was used as size marker. Donor-specific antigen status was derived from the ratio of blue and green fluorescence or red and yellow fluorescence (for Lu antigens) at the reaction- and gene-specific product sizes. In our hands, the exact position of the amplicons could differ from the calculated by a few base pairs (bp) and even varied from run to run. Therefore, peak positions calculated by the Gene mapper 4.0 Software were exported to an Excel sheet that included formulas for an advanced identification of the peaks based on the expected peak position and the position of neighboring peaks. In short, for each SNP, first the position of the whole group was identified allowing for a few bp offset, and then within each group the peaks were assigned to the respective samples. Analysis included warnings for low signal intensity, off-scale peaks, and borderline fluorescence ratios. Automated analysis was checked by visual inspection of the electropherograms.
IT Solution for Unit Release
An IT solution to use the obtained data as blood unit sorting algorithm was established: The data were linked to the donor registry. For each donation, it was checked on release whether historical data indicated the probable presence of an useful phenotype (e.g. negative for Kpb or ccD.EE and negative for Fya and Jka). These units were diverted to a ‘laboratory inventory’ and kept for 1–4 weeks depending on the rarity of the assumed phenotype. If units of this phenotype were demanded, they were confirmed by serology or used without confirmation for preventive matching. If unused, they were rescheduled to general purpose transfusion when the hold phase was finished.
Validation
After establishing the reaction conditions in preliminary experiments, 312 samples were tested and the predicted results compared to results obtained with routine serologic methods. Discrepant results were further investigated by repeat serology, repeat pooled capillary electrophoresis, and, if the discrepancy persisted, sequencing.
Sequencing
Sanger sequencing was done using BigDye sequencing kit (Thermo-Fisher). The standard approach for sequencing from genomic DNA was chosen. One or several exons were amplified from genomic DNA and sequenced using the primers also used for amplification or internal sequencing primers. For KEL, the primers of Lee et al. [18] were used.
Statistical Analysis of Mutations in Discrepant Samples
A possible causative role of a polymorphism detected in a sample displaying a discrepant phenotype was evaluated as follows:
- Polymorphisms in known null alleles and alleles with destroyed start codon, stop codons in the coding sequence, or frameshift mutations were considered to be causative due to their structure, if a predicted antigen was not present.
- A mutation interfering with primer binding was considered to be causative due to its structure, if the discrepancy was caused by a false-negative antigen prediction.
- Polymorphisms known to be highly associated with the polymorphism investigated were considered unlikely to be causing a discrepancy. The only example was c.586G>A in BCAM that is highly associated with the Lua-expressing LU*01 allele (c.230 G>A) [19].
- The residual samples were counted and the observed mutations evaluated in a sequential manner with correction for multiple testing according to Bonferoni-Holm [20] considering the known allele distribution found in large databases.
For each polymorphism observed, its frequency among non-Finnish Europeans was extracted from the ExAC database (exac.broadinstitute.org) [21]. The cumulative frequency of the encountered and all less frequent polymorphisms was calculated. This cumulative frequency was used to calculate the probability that any polymorphism at least as rare was observed among the collated samples (2n alleles) by chance. If the probability was less than 0.01, the polymorphism was considered most likely associated with the discrepant phenotype, if it was less than 0.05, it was considered likely associated or causative. The samples explained by such polymorphism were removed from the set of samples under consideration, and the calculation was repeated with the residual samples until no sample or no polymorphism with likely association was left.
Hands-On Time and Consumable Cost
Hands-on time and consumable cost were collated and estimated according to the technicians' experience.
Snapshot of Units Available in the Blood Service
Snapshots of the number of available units in our Blood Service were taken from our SAP-based inventory management system (transaction BSD/MM_Chargenbest).
Results
Antigen Prediction by Pooled Capillary Electrophoresis
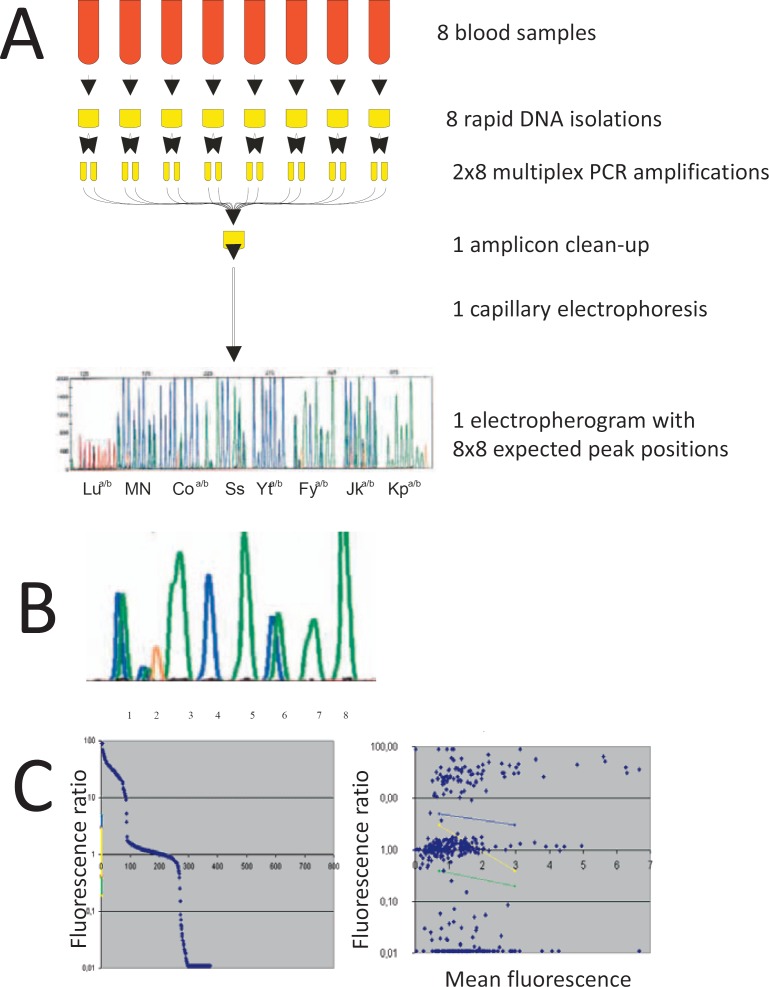
A medium-throughput donor typing method was developed based on PCR amplification of crude DNA with fluorescence-labeled allele-specific primers, product length-based barcoding, and analysis by capillary electrophoresis. DNA amplification was performed in two multiplex reactions, one containing primers to predict Coa/Cob, Jka/Jkb, Lua/Lub, and M/N, the other those for Fya/Fyb, Kpa/Kpb, S/s, and Yta/Ytb. Each reaction contained sequence-specific primers for both alleles that were 5′ labeled with different fluorescence dyes (usually FAM and HEX with the exception of Lu using NED and ROX). Eight variations of these multiplex PCR reactions were developed that yielded PCR products differing by a size of 4 bp from the next shorter version. Thus, eight donor samples were amplified in parallel using eight different versions of the multiplex PCR. All products were pooled, purified, and analyzed in a single capillary electrophoresis run (fig. 1). Obtained fluorescence signals were assigned to sample and polymorphisms according to the product size, and allele status was deduced from the ratio of fluorescences that corresponded to the different alleles. The pooling prior to capillary run increased analysis throughput by a factor of 8. Considering a run time of 50 min, the maximal throughput using a single capillary machine was limited to 230 samples per day.
Fig. 1.
Outline of the workflow. Panel A Schematic workflow of the pooled capillary electrophoresis: From eight donors, a crude DNA preparation was prepared using Extract-N-Amp system. For each sample, two multiplex PCR were performed, eight different variants of the multiplex PCR were used that yielded slightly different product sizes. PCR amplicons of these 2 × 8 multiplex PCR were pooled and analyzed by capillary electrophoresis. Panel B For each sample and polymorphism, a specific product size was expected that could show fluorescence for one, the other or both alleles. The fluorescence ratio of the two allele-specific colors was analyzed. Panel C Visualization of the fluorescence ration distribution (left panel) and the fluorescence ratio versus mean fluorescence (right panel) allowed to define the regions for heterozygosity (yellow), and borderline results (blue and green) used for the automated analysis of the fluorescence ratio.
Validation
A comparison of prediction and serologic typing was attempted for312 donor samples representing 4,992 antigens predicted by pooled capillary electrophoresis. As no suitable anti-Ytb was available, serologic typing was restricted to Coa, Cob, Fya, Fyb, Jka, Jkb, Kpa, Kpb, Lua, Lub, M, N, S, s, and Yta (table 2), resulting in a set of 4,680 serologic antigen typings. 23 allele pairs could not be predicted in 12 samples due to low signal, in addition 8 antigens were not predicted because of borderline fluorescence ratios in 7 samples. Two samples yielded both failures due to low signal and borderline ratio, resulting in a total of 54 antigens (including 3 Ytb) that could not predicted in 17 samples, corresponding to a rate of 1.1% failed antigen predictions and 5.4% of samples with at least one failed antigen prediction.
Table 2.
Validation of antigen prediction
| System | Molecular analysis |
Serologic analysis |
||
|---|---|---|---|---|
| prediction* | n | confirmed | discrepant serology | |
| JK | Jk(a+b−) | 88 | 88 | |
| Jk(a+b+) | 151 | 151 | ||
| Jk(a−b+) | 72 | 72 | ||
| no call | 1 | Jk(a+b−): 1 | ||
| FY | Fy(a+b−) | 54 | 54 | |
| Fy(a+b+) | 154 | 149 | Fy(a+b−): 5§§ | |
| Fy(a−b+) | 98 | 98 | ||
| Fy(a?b+) | 2 | Fy(a−b+): 1; Fy(a+b+): 1 | ||
| no call | 4 | Fy(a−b+): 2; Fy(a+b+): 2 | ||
| MN | M+N− | 99 | 99 | |
| M+N+ | 151 | 150 | M+N−:1$ | |
| M−N+ | 58 | 58 | ||
| M+N? | 1 | M+N+: 1 | ||
| no call | 3 | M+N−: 2; M+N+: 1 | ||
| Ss | S+s− | 28 | 28 | |
| S+s+ | 148 | 148 | ||
| S−s+ | 129 | 128 | S+s+: 1‡ | |
| S?s+ | 1 | S−s+: 1 | ||
| no call | 6 | S+s+: 4; S−s+: 2 | ||
| Co | Co(a+b−) | 284 | 284 | |
| Co(a+b+) | 24 | 24 | ||
| Co(a−b+) | 0 | 0 | ||
| Co(a+b?) | 2 | Co(a+b−): 2 | ||
| no call | 2 | Co(a+b+): 1; Co(a+b−): 1 | ||
| LU | Lu(a+b−) | 0 | 0 | |
| Lu(a+b+) | 24 | 24 | ||
| Lu(a−b+) | 286 | 286 | ||
| Lu(a?b+) | 1 | Lu(a−b+): 1 | ||
| no call | 1 | Lu(a−b+): 1 | ||
| YT† | Yt(a+b−) | 285 | 285 | |
| Yt(a+b+) | 23 | 23 | ||
| Yt(a−b+) | 1 | 1 | ||
| No call | 3 | Yt(a+): 3 | ||
| Kp | Kp(a+b−) | 0 | 0 | |
| Kp(a+b+) | 6 | 6 | ||
| Kp(a−b+) | 302 | 302 | ||
| Kp(a?b+) | 1 | Kp(a−b+): 1 | ||
| no call | 3 | Kp(a−b+): 3 |
One antigen not predicted is indicated by an ‘?’, both antigens reported as ‘no call’.
Antigen Ytb was not checked by serology.
Fyb antigen missed by serology in FYx allele.
Non-systematic misprediction: repeat molecular prediction correct.
GYPB*04/GYPB*24 sample.
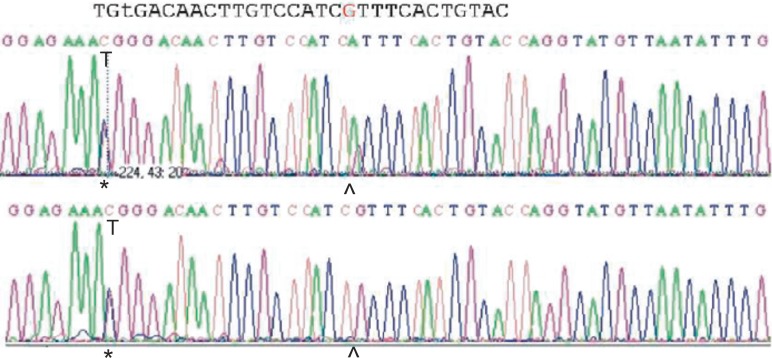
Among the evaluable 4,629 antigen predictions, 4,622 (99.8%) were identical to the serologic result. There were 7 discrepancies: In 5 cases, antigen Fyb was predicted positive, but determined negative by routine serology. These 5 cases were found to be due to the presence of a FYx allele known to express a weak Fyb antigen and considered serologic failures. In one sample, the S antigen was missed in a GYPB*24 allele (Mit-positive allele), because the additional polymorphism in GYPB*24 interfered with primer binding (fig. 2). Finally, 1 sample was predicted as N-positive but found to be N-negative in serology. Repeat molecular testing yielded the correct prediction, and the discrepancy was categorized as random failure due to technical causes. In conclusion, 4,627 of the 4,629 antigen predictions were correct (99.96%), which corresponded to 98.87% of all antigens attempted to predict.
Fig. 2.
Molecular basis of a false-negative antigen S prediction in the Mit allele. The electropherogram of the Mit+ sample (upper sequence) and a control Ss sample (lower sequence) is shown in a region encompassing the primer binding site. The asterisks indicate the 143C/T polymorphism discriminating S and s, the caret the 161G/A polymorphism characterizing the Mit allele. The A of the Mit allele introduces an additional mismatch of the primer which likely prevents efficient primer binding.
Hands-On Time and Consumable Cost
Hands-on time andconsumable cost was collated (table 3, hands-on time) and compared to a previously developed molecular typing solution based on agarose gel electrophoresis [17]. While the hands-on time per donor for the set-up was slightly lower than in the gel method, visual proofreading of the results incurred an increase of total hand-on time by 49%. Likewise, product cleanse-up and capillary electrophoresis was more expensive than agarose gel electrophoresis resulting in a 25% increase of cost per donor. Considering that the capillary electrophoresis yielded 16 predictions, while the gel method only yielded 4, the cost per antigen was about a third of the old method.
Table 3.
Hands-on time and price estimate: comparison to agarose electrophoresis
| Capillary electrophoresis |
Agarose electrophoresis |
||
|---|---|---|---|
| process | hands-on time, min | process | hands-on time, min |
| DNA Extraction | 20 | DNA Extraction (including additional | 28 |
| mixing step) | |||
| PCR set-up and start | 20 | PCR set-up and start | 21 |
| Post-PCR workup | 45 | ||
| Electrophoresis set-up | 5 | electrophoresis set-up | 37 |
| Electrophoresis maintenance | 5 | ||
| Documentation | 5 | documentation | 14 |
| Time per 96 donors | 160 | time per 91 donors | 102 |
| Time per donor | 1.04 | time per donor | 1.12 |
| Proofreading of results | 60 | ||
| Time per donor (including proofreading) | 1.67* | ||
| Time per antigen | 0.10 | time per antigen | 0.28 |
| Consumable/chemical | Cost per test, EUR | Consumable/chemical | Cost per test, EUR |
|---|---|---|---|
| Extract-N-Amp Blood Kit (quarter standard | 0.76 | Extract-N-Amp Red Blood Kit | 0.76 |
| volume) | (half standard volume) | ||
| Oligonucleotides | 0.03 | oligonucleotides | 0.04 |
| Product cleanse-up | 0.26 | ||
| Capillary electrophoresis† | 0.20 | agarose electrophoresis | 0.05 |
| Tubes, plates and tips | 0.38 | tubes and filter tips | 0.44 |
| Total | 1.63 | total | 1.3 |
| Antigen predictions per test | 16 | antigen predictions per test | 4 |
| Cost per antigen | 0.10 | cost per antigen | 0.33 |
The time for the preparation of the PCR mixes and for collecting and preparing the samples is not included.
Routine Use of Pooled Capillary Electrophoresis
Until December 2017, a total of 136,120 donor samples representative of more than 100,000 donors were tested. The system was modified several times, and Co testing was temporarily dropped to allow the inclusion of other systems. A total of 287 Yt(a-), 145 Co(a-), 96 Lu(b-) and 12 Kp(b-) donors were identified. Data predicted by pooled capillary electrophoresis were pooled with historic serologic data and used to identify units with an ‘expected’ suitable antigen pattern that were diverted to a special ‘laboratory inventory’ and kept for 1–4 weeks (fig. 3).
Fig. 3.
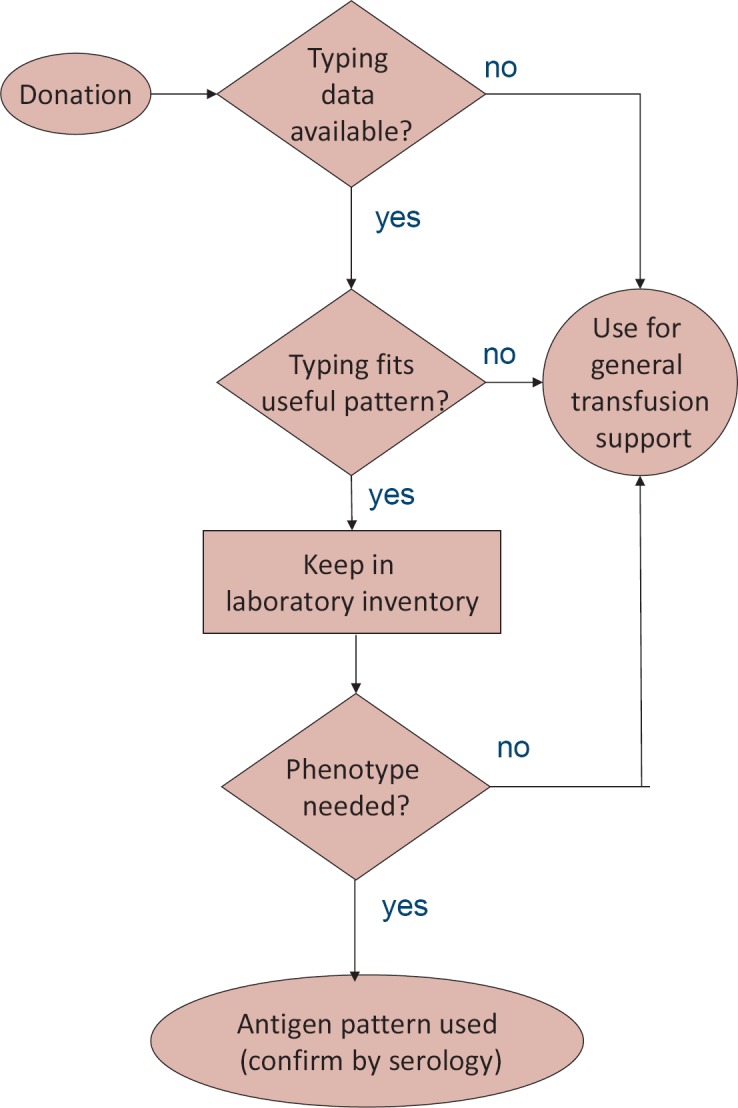
Inventory management. Use of historical typing data for patient care. On release, an algorithm checks whether antigens historically determined by serology or predicted by pooled capillary electrophoresis are known. If such antigens are known, the antigen pattern is compared to ‘suitable’ antigen patterns. Units with such patterns are kept in a laboratory inventory for 1–4 weeks depending on the rarity of the expected phenotype. If used for indications necessitating antigen negative units, the predicted antigen status is confirmed by serology. If no matching patient is found, units are used for general purpose transfusions.
The prediction rate was re-evaluated after 34,768 samples (table 4): It dropped to 92.9%, Fy and Yt being the most unstable systems. The main cause of failure was low signal, followed by a borderline fluorescence ratio. Rarely, off-scale peaks (fluorescence too strong for correct color compensation) prevented analysis. For practical purposes, a donor was considered successfully typed if a prediction of Fya and Fyb was present.
Table 4.
Antigen prediction rate in the first 34,768 samples and causes of failure
| Predicted, % | Failed to predict |
||||
|---|---|---|---|---|---|
| Low signal, % | Offscale, % | Ratio, % | |||
| Jka | 95.4 | 4.3 | 0.0 | 0.3 | |
| Jkb | 95.1 | 4.3 | 0.0 | 0.6 | |
| Fya | 86.4 | 11.5 | 0.0 | 2.1 | |
| Fyb | 87.7 | 11.5 | 0.0 | 0.8 | |
| M | 95.1 | 3.8 | 0.6 | 0.5 | |
| N | 94.1 | 3.8 | 0.6 | 1.5 | |
| S | 91.6 | 7.3 | 0.2 | 0.9 | |
| s | 91.6 | 7.3 | 0.2 | 0.9 | |
| Coa | 97.3 | 2.5 | 0.2 | 0.0 | |
| Cob | 96.5 | 2.5 | 0.2 | 0.9 | |
| Lua | 97.8 | 1.6 | 0.0 | 0.6 | |
| Lub | 98.3 | 1.6 | 0.0 | 0.1 | |
| Yta | 86.7 | 13.0 | 0.0 | 0.3 | |
| Ytb | 86.2 | 13.0 | 0.0 | 0.7 | |
| Kpa | 92.8 | 6.7 | 0.0 | 0.5 | |
| Kpb | 93.3 | 6.7 | 0.0 | 0.0 | |
| Mean | 92.9 | 6.3 | 0.1 | 0.7 | |
From January 1, 2017 to December 31, 2017, a total of 20,861 donor samples were successfully typed, and the ‘success rate’ was 91% (based on successful Fy typing). During this year, 188,309 (28%) of 670,799 RBC units were from typed donors, 42,731 (23% of typed units, 6% of all units) of these fitted a ‘rule’ and were stored in the laboratory inventories. This number represents a slight overestimate of the efficiency of the typing project, because some units were selected due to historical serologic typing information. As a result of the computer-controlled guidance of unit use, we were able to maintain a stock of about 10 Lu(b-), 25 Yt(a-) and 15 Co(a-) non-cryoconserved units in our blood service (table 5).
Table 5.
Availability of rare types
| Phenotype | Snapshots |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| April 20, 2009 | September 8, 2015 | February 22, 2016 | September 29, 2017 | March 13, 2018 | |||||
| Vel- | |||||||||
| All | 1 | 2 | 2 | - | 3 | ||||
| O RhD− | - | - | - | - | 1 | ||||
| Kp(b−) | |||||||||
| All | - | 1 | - | 1 | 2 | ||||
| O RhD− | - | - | - | - | 1 | ||||
| Lu(b−) | |||||||||
| All | 2 | 12 | 11 | 10 | 12 | ||||
| O RhD− | - | 1 | 3 | 4 | 3 | ||||
| Yt(a−) | |||||||||
| All | - | 20 | 21 | 37 | 25 | ||||
| O RhD− | - | 2 | 1 | 6 | 2 | ||||
| Co(a−) | |||||||||
| All | - | 29 | 14 | 18 | 14 | ||||
| O RhD− | - | 3 | 3 | 1 | 2 | ||||
| k− | |||||||||
| All | - | 11 | 19 | 21 | 32 | ||||
| O RhD− | - | 3 | 4 | 5 | 4 | ||||
Analysis of Mispredictions
In 2017, 62 antigen predictions turned out to be incorrect (2009–2017: 433 predictions). While this number represents only 0.14% of units scheduled for the laboratory inventory, a realistic rate of discrepancies cannot be derived from the data, because many units were used for general-purpose transfusion and not checked by serology. Most mispredictions were due to low-quality runs not identified by antigen calling (2017: 65%; all years: 49%) or an attempted manual interpretation of a result that could not be automatically called (2017: 27%; all years: 35%).
Systematic Serologic Typing of Donors Predicted to Be Kp(a+b+), Co(a+b+) or Lu(a+b+)
In order to detect null alleles, samples predicted to be Kp(a+b+), Co(a+b+), or Lu(a+b+) were checked by serology yielding 8,355 Lua typings (Lub 8,965; Kpa 2,468; Kpb 2,565; Coa 1,497; Cob 1,520). A discrepant result reproduced on re-testing was obtained for Lua in 1 proband (plus 1 proband detected during routine re-typing; Lub 10; Kpa 7, Kpb 2, Coa 2, Cob 2). These numbers compared to 10 Kp(b-), 96 Lu(b-) and 145 Co(a-) donors correctly predicted as Kp(a+b-), Lu(a+-b-) and Co(a-b+), respectively.
Molecular Causes of Discrepant or Weakened Antigen Expression
Rare genotypes detected in donors with discrepant or weakened antigen expression are collated in table 6. Several new alleles with weakened or absent antigen expression were found: 1 for Kell, 2 for Colton, and 8 for Lutheran. Adsorption/elution was done in selected cases only and found to be unreliable: 1 sample with a known KEL mod allele was found negative for Kpa on adsorption/elution. False-negative predictions due to mutations interfering with primer binding were usually detected during routine use and alleles with weak or absent antigen expression by systematic testing of predicted heterozygotes.
Table 6.
Molecular structures identified in samples with reproducible discrepancy between prediction and serology
| Samples (n) | Phenotype |
Donor |
ExAC frequency* |
||||||
|---|---|---|---|---|---|---|---|---|---|
| predicted | found | polymorphisms | protein | antigens | snp number | non-Finnish Europeans | deduced genotype | evidence for causality / association | |
| Glycophorin B | |||||||||
| False-negative antigen prediction | |||||||||
| 3 | S−s+ | S+s+ | c.143C/T; c.161G/A | 48 Met / Thr | S / s | rs7683365 | 0.3802; | GYPB*04/GYPB*24 | disrupted primer |
| 54 Arg / His | Mit+ | rs370332485 | 0.0002869 | binding | |||||
| 1 | S−s+ | S+s+ | c.143C/T; c.161G/C | 48 Met / Thr | S / s | rs7683365 | 0.3802; | GYPB*04/GYPB*03. | disrupted primer |
| 54 Arg / Pro | unknown | absent | (c.161C)† | binding | |||||
| 1 | S+s− | S+s+ | c.143C/T; c.163T/C; | 48 Met / Thr; | S / s | rs7683365 | 0.3802; | GYPB*04/GYPB*03. | disrupted primer |
| c.251 G/C | 55 Phe / Leu | unknown | rs755129621 | 0.00008228 | (c.163C,251C) or | binding | |||
| 84 Ser/ Thr | unknown | rs1132783 | 0.232 | GYPB*04(c.251C)/ | |||||
| GYPB*03(c.163C) | |||||||||
| Lutheran | |||||||||
| Lub− (including Lub mod) | |||||||||
| 2 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19.(c.586A)/ | no cause identified§: |
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | Lu*02 | ||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.(c.586A)/ | no cause identified§: |
| Elution pos | c.586 G/A | 196 Val / Ile | unknown | rs28399654 | 0.03308; | Lu*02 | |||
| 1: | Lu(a+b+) | Lu(a+b−) | c.-4036_-4029del (het) | Non-coding; | unknown; | - | NC; | LU*01.19.(c.586A)/ | the deletion is 63 bp 5′ |
| c.230 G/A; | 77 Arg / His; | Lub+/Lua+; | rs28399653 | 0.0382; | Lu*02(-4036_ | of the BCAM promoter | |||
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | -4029del)† | ||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19.(c.586A)/ | p < 0.01 |
| elution neg | c.529 G/C; | 177 Gly / Arg | unknown; | rs777617369 | W: 0.0001225 | Lu*02.(c.529C)† | |||
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | |||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01 / LU*02.c | p < 0.05 |
| c.711 C/T | (silent) | -; | rs3810140 | 0.06594; | (711T, 714T, 733A)† | ||||
| c.714 C/T | (silent) | -; | rs3810140 | 0.06590 | |||||
| c.733 G/A | 245 Glu / Lys | unknown | rs762681823 | 0.000015603 | |||||
| 2 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.(c.586A)/ | p < 0.01 |
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | Lu*02.-13.(c.997A)† | ||||
| c.997 G/A; | 333 Gly / Arg; | unknown; | rs147593462 | W:0.00002477 | |||||
| c.1671 C/T | (silent) | -; | rs28399658 | 0.04170; | |||||
| c.1742 A/T | 581 Gln / Leu | Lu:13 / −13 | rs28399659 | 0.04632 | |||||
| 1 | Lu(a+b+) | Lu(a+b−) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19/Lu*02. | unsure: p < 0.05 |
| c.1612 G/A; | 538 Gly / Ser; | unknown; | rs145626518 | 0.0005108; | (c.1612A)† or | ||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | LU*01/ LU*02.10. | ||||
| (c.1612A)† | |||||||||
| 1 | Lu(a+b+) | Lu(a+b−) | c.100_105del‡; | 34_35delArgLeu‡ | LU:12/LU:-12; | - | 0.001679; | LU*01.19.(c.586A)/ | unsure: p < 0.05 |
| elution pos | c.230 G/A; | 77 Arg / His; | Lub+/Lua+; | rs28399653 | 0.0382; | Lu*02.-12.1 | |||
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | |||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| Lub weak | |||||||||
| 1 | Lu(a+b+) | Lu(a+b+w) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19.(c.586A)/ | no cause identified§: |
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | Lu*02 | ||||
| c.1615 A/G | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a+b+w) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19 / | p < 0.05 |
| c.1183 C/T; | 395 Arg / Cys; | unknown; | rs761656868 | 0.00003004; | Lu*02.(c.1183T) † | ||||
| c.1615 T/A | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a+b+w) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19./(c.529C) / | unsure: p < 0.05 |
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | Lu*02.-26 | ||||
| c.1495 C / T | 499 Arg / Trp | LU:26/-26 | rs148391498 | 0.0007812 | |||||
| 1 | Lu(a+b+) | Lu(a−b+) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19.(c.937T) †/ | p < 0.01 |
| c.586 G/A; | 196 Val / Ile; | unknown; | rs28399654 | 0.03308; | Lu*02 | ||||
| c.937 G/C | 313 Val / Leu | unknown; | rs199762962 | W:0.000008293 | |||||
| c.1615 T/A | 539 Thr / Ala | Aua+/Aub+ | rs1135062 | 0.29932 | |||||
| 1 | Lu(a+b+) | Lu(a−b+) | c.230 G/A; | 77 Arg / His | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.(c.586A). | p < 0.01 |
| c.1722 A/T | 574 Lys / Asn | unknown; | - | absent | (c.1722T) †/ | ||||
| c.1742 A/T | 581 Gln / Leu | Lu:13 / −13 | rs28399659 | 0.04632 | Lu*02.-13 | ||||
| False-negative antigen prediction | |||||||||
| 1 | Lu(a+b−) | Lu(a+b+) | c.230 G/A; | 77 Arg / His; | Lub+/Lua+; | rs28399653 | 0.0382; | LU*01.19 / | disrupted primer |
| c.240 G/A | (silent) | - | rs779970171 | W:0.00002868 | LU*02.(c.240A) | binding | |||
| Kell | |||||||||
| Kpb- (including mod phenotype) | |||||||||
| 1 | Kp(a+b+) | Kp(a+b−) | c.841 C/T; | 281 Arg / Trp; | Kpb+ / Kpa+; | rs8176059 | 0.010544; | KEL*02.03 / | p < 0.01 |
| c.1658 C/T | 553 Pro / Leu | unknown | rs1431256906 | absent | KEL*01.01. | ||||
| (c.1658T) † | |||||||||
| 1 | Kp(a+b+) | Kp(a+b−) | c.841 C/T; | 281 Arg / Trp; | Kpb+ / Kpa+; | rs8176059 | 0.010544; | KEL*02.03 / | known null allele |
| c.1546 C/T | 516 Arg / Stop | K null | rs8176034 | 0.00007494 | KEL*02N.17 | ||||
| Kpa- (including mod phenotype) | |||||||||
| 6 | Kp(a+b+) | Kp(a−b+) | c.257 G/A; | 86 Arg / Gln; | K mod; | rs777011308 | 0.00001501; | KEL*02M.13/ | known mod allele |
| Elution | c.841 C/T | 281 Arg / Trp | Kpb+ / Kpa+ | rs8176059 | 0.010544 | KEL*01.01 | |||
| negative | |||||||||
| 1 | Kp(a+b+) | Kp(a−b+) | c.257 G/A; | 86 Arg / Gln; | K mod; | rs777011308 | 0.00001501; | KEL*02M.13/ | known mod allele |
| c.841 C/T; | 281 Arg / Trp; | Kpb+ / Kpa+ | rs8176059 | 0.010544; | KEL*01.01. | ||||
| c.1680 A/C | (silent) | - | rs8176036 | 0.04956 | (c.1680C) | ||||
| Colton | |||||||||
| Coa-(including mod phenotype) | |||||||||
| 1 | Co(a+b+) | Co(a−b+) | c.-101 C/G; | Non-coding; | -; | NC; | CO*01. | promoter, possibly transcription factor binding inhibited | |
| c.134 C/T | 45 Ala / Val | Coa+ / Cob+; | rs28362692 | 0.03976 | (c.-101G)† / | ||||
| CO*02 | |||||||||
| 1 | Co(a+b+) | Co(a−b+) | c.134 C/T; | 45 Ala / Val; | Coa+ / Cob+; | rs28362692 | 0.03976 | CO*01.(c.494A) / | no cause identified§: |
| c.494 G/A | 165 Gly / Asp | unknown | rs28362731 | 0.04309 | CO*02 | ||||
| Cob- (including mod phenotypes) | |||||||||
| 1 | Co(a+b+) | Co(a+b−) | c.134 C/T | 45 Ala / Val | Coa+ / Cob+ | rs28362692 | 0.03976 | CO*01 / CO*02 | no cause identified |
| Elution pos | |||||||||
| Cob weak | |||||||||
| 1 | Co(a+b+) | Co(a+wb+) | c.134 C/T | 45 Ala / Val | Coa+ / Cob+ | rs28362692 | 0.03976 | CO*01 / CO*02 | no cause identified |
| 1 | Co(a+b+) | Co(a+b+w) | c.134 C/T; | 45 Ala / Val; | Coa+ / Cob+; | rs28362692 | 0.03976 | CO*01 / CO*02. | p < 0.01 |
| Elution pos | c.502 G/A | 168 Ala / Thr | unknown | - | absent. | (c.502A)† | |||
| False-negative antigen prediction | |||||||||
| 1 | Co(a−b+) | Co(a+) | c.134 C/T; | 45 Ala / Val; | Coa+ / Cob+; | rs28362692 | 0.03976 | CO*01.(c.135A)† / | yes: c.135G>A interferes |
| c.135 G/A | (silent) | - | rs781356338 | 0.00004542 | CO*02 | with primer binding | |||
NC = Not covered (polymorphism outside the scope of ExAC, e.g. in the promoter); absent = no single case worldwide observed in ExAC despite coverage of the region.
Frequency of the mutation among non-Finnish Europeans according to the ExAC database (http://exac.broadinstitute.org). If the mutation was absent among non-Finnish Europeans but present wordwide, the worldwide frequency was given with a prefixed W. The mutations possibly causing the discrepancy are indicated by bold letters.
Previously unknown allele.
The shown polymorphism are frequent and unlikely to cause the discrepant phenotype.
For KEL, 2 negative serologic Kpb results were caused by the KEL*02N.17 and by the previously unknown KEL*02N/M.(1658C>T) allele. No adsorption/elution result was available for that donor; hence the allele could represent a KEL null or KEL mod allele. Furthermore, 8 false-positive Kpa predictions were caused by the KEL*02M.13 allele [22].
Ten donors predicted to be Lua/Lub were Lu(b-) by serology. We identified four previously unknown alleles with missense mutations: LU*02.(c.529C), LU*02.-13.(c.997A), LU*02.(c.711T, 714T, 733A), and LU*02.(c.1612A). In addition, we detected two silent LU*01-derived alleles, and one allele with weak expression of Lub antigen. No cause could be identified in 3 donors and possibly associated alterations in 2 donors. Most alleles identified in these donors were previously unknown and carried missense mutations, suggesting that they could represent LU mod rather than LU null alleles. Adsorption/elution was done for two alleles only and was positive in one.
For Coa, 2 donors predicted to be Coa/Cob were Co(a-). One carried a mutation in the promoter; in the other one no cause could be identified. In addition, 3 donors had weak or almost absent Cob expression, but a probably causative mutation was found in only 1 of them.
Discussion
We developed a low-cost molecular typing system to screen our donor population for rare antigen constellations. Key features included a simplified DNA isolation approach, simultaneous analysis of several samples by pooled capillary electrophoresis, and the deliberate allowance of a relevant failure rate as well as the exclusion of the complicated blood group systems ABO and RH. Implementation of our typing approach on a routine basis resulted in major improvements of RBC supply for patients with anti-Yta or anti-Lub.
Current approaches for high-throughput molecular blood group antigen prediction in blood donors [3,4,5,6,7,8,9,10,11] differ relevantly with respect to typing effort, cost, and quality of the results. On one hand, sophisticated approaches attempted a highly reliable determination of the blood type even in many rare phenotypes [3,11]. Other approaches were optimized for high throughput [8] or low cost [23] and relied on serologic confirmation on release [8,9]. Our approach may represent the extreme version of low-cost, low-quality testing: we deliberately allowed for a 10% ‘no call’ rate and did not bother with rare alleles in order to be able to screen a large donor cohort with limited financial resources.
The antigen prediction was focused on the antigens clinically most relevant in our population, excluding ABO, Rh phenotype and K that had to be determined for unit release. Therefore, we complemented predictions for Fya/Fyb, Jka/Jkb, S/s, and M/N by a screening approach covering the rare phenotypes most relevant for our population: Kp(b-), Lu(b-), Yt(a-) [14], and Co(a-). We could not include a prediction of the Vel- phenotype that is caused by a 17 bp deletion [24], because no information on its molecular basis was available at the start of our typing project. The antigens Cob, Lua, Ytb, and Kpa were included for technical reasons to balance for Coa, Lub, Yta, and Kpb.
Already in 2008, we showed that molecular typing from crude DNA preparations was useful to screen donors for rare phenotypes. The pooled capillary electrophoresis extended this concept, keeping the simplified DNA isolation method based on the Extract-N-Amp system but replacing agarose gel by capillary electrophoresis. Pooling of 8 samples in a single capillary run resulted in a cost reduction by about 66% and increased throughput by up to 8-fold. In contrast to other donor typing approaches using pooling [25], product length encoding of the sample number obviated the need for a separate pool resolution. Compared to the agarose gel approach, the number of predicted antigens was quadrupled, resulting in a lower cost per antigen of EUR 0.10 (USD 0.13). The semi-automated analysis of the data allowed the inclusion of all antigen predictions into our donor database and paved the way for an IT-controlled unit guidance system.
The simplistic approach came at a cost: the major antigens (evaluated as successful Fya/Fyb prediction) were successfully predicted in only about 90% of donors (table 4), despite the results being checked manually. Without the manual check, the prediction rate would have been even worse. Furthermore, there was a certain rate of mispredictions resulting in antigen discrepancies on serologic confirmation. These problems often occurred in samples with generally low signals, with technical problems during data interpretation being the main cause of error. Systematic mistyping of alleles was a rarity (table 6) and negligible compared to technical failures; hence we did not perceive it necessary to include additional polymorphisms for antigen prediction. Considering that the observed discrepancies are accumulated from 4 laboratories serving an area with approximately 8 million inhabitants, each laboratory encountered such a problem one to two times a month. Most technicians did not consider this discrepancy rate a practical problem and rather valued the wide availability of units for which a 99% correct antigen status was known.
It should be realized that our approach is critically dependent on the availability of the commercial ‘Extract-N-Amp’ kit. While there are other enzymes suitable for direct PCR, a change of the DNA isolation and PCR chemistry would incur relevant redesign of the method.
In very rare phenotypes like Kp(b-), compound heterozygotes with one null allele may underlie a relevant proportion of the antigen-negative donors [11]. We tried to verify this proposal by systematic serologic testing of samples predicted to be heterozygous for Kpa/Kpb, Lua/Lub, or Coa/Cob. The yield was limited: In addition to 11 Kp(b-) donors predicted by single polymorphism typing, we found 2 serologically Kp(b-) donors among 2,565 predicted to be Kpa/Kpb. For Lub, in addition to 96 donors predicted to be Lu(b-), 10 serologically Lu(b-) donors were found among 8,965 predicted Lua/Lub. For Coa, 145 Co(a-) donors found by single polymorphism prediction compared to a single additional Co(a-) donor detected by serologically typing of 1,497 predicted heterozygous donors. A total of 11 previously unknown alleles was found, many of which carried missense mutations suggesting that they may represent mod rather than null alleles. Even if many of them were seen in a single sample only, the statistical significance of the association could usually be proven by a combination of SNP frequency data taken from the ExAC database [21] with a sequential statistical testing approach [20.] For Lu and Co, in a relevant number of samples no cause for the diminished or absent antigen expression was detected, suggesting that regions outside the coding region are of relevance for Co and Lu expression. While serologic typing of predicted heterozygous donors was an interesting tool to find possible null alleles, it had minor impact on the number of identified antigen-negative donors.
Our large-scale typing program was complemented by an IT solution that allowed to select suitable donors and to guide units with predicted interesting phenotypes into the ‘laboratory inventory’ by a check of the predicted antigen pattern to a set of ‘rules’. The importance of a suitable IT integration of the typing process has been stressed by Perreault et al. [26] and becomes obvious considering the number of available k- units: For many years before the start of the project, RhD- K+ donors were systematically tested for antigen k, but without a working IT solution, we were unable to establish a stock useful in emergency situations.
In conclusion, our strategy allowed us to apply this approach to 20% of our RBC units (i.e. more than 120,000 units/year) and obviated the need for several rounds of serologic antigen screening when a patient with multiple antibodies needed blood: For a patient needing 4 Jk(a-) Fy(a-) units, in older times 64 units were tested for Jka, the negative units for Fya, and the double-negative units were cross-matched. Nowadays, 4 units are cross-matched and tested in parallel. The demand for Yt(a-), Co(a-), and Lu(b-) RBC units can now be fulfilled in the majority of cases. The typing project thus decisively improved the transfusion support for alloimmunized patients.
Authors Contributions
FFW invented the approach, did the initial primer design, established the data extraction, and analyzed the data. AD performed PCE during the development phase and analyzed discrepant samples. RB performed PCE, monitored the efficiency of the method during routine use, optimized PCR conditions and designed PCR primers in the later phase of the project. THM decided to start the typing project and continuously supported its further development, including the demand for a useful IT integration.
Disclosure Statement
Nothing to disclose.
Acknowledgment
We thank Oliver Herrmann for providing an IT solution for RBC unit guidance, Dr. Silke Beermann for helpful discussions, and Christof Weinstock, Red Cross Blood Service Baden-Württemberg - Hesse, for suggesting reference 16. The authors thank Jana Albes, Dr. Silke Beermann, Sabine Brockmann, Sindy Burau, Katrin Haase-Volm Polina Schneider, Susanne Steinhauer, Katja von Söhnen, and Nina Wilhelm for their expert technical assistance.
References
- 1.Flegel WA, Gottschall JL, Denomme GA. Integration of red cell genotyping into the blood supply chain: a population-based study. Lancet Haematol. 2015;2:e282–e288. doi: 10.1016/S2352-3026(15)00090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flegel WA, Gottschall JL, Denomme GA. Implementing mass-scale red cell genotyping at a blood center. Transfusion. 2015;55:2610–2615. doi: 10.1111/trf.13168. quiz 2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avent ND, Martinez A, Flegel WA, Olsson ML, Scott ML, Nogues N, Pisacka M, Daniels GL, Muniz-Diaz E, Madgett TE, Storry JR, Beiboer S, Maaskant-van Wijk PM, von Zabern I, Jimenez E, Tejedor D, Lopez M, Camacho E, Cheroutre G, Hacker A, Jinoch P, Svobodova I, van der Schoot E, de Haas., M The Bloodgen Project of the European Union, 2003–2009. Transfus Med Hemother. 2009;36:162–167. doi: 10.1159/000218192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paris S, Rigal D, Barlet V, Verdier M, Coudurier N, Bailly P, Bres JC. Flexible automated platform for blood group genotyping on DNA microarrays. J Mol Diagn. 2014;16:335–342. doi: 10.1016/j.jmoldx.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Lopez M, Apraiz I, Rubia M, Piedrabuena M, Azkarate M, Veldhuisen B, Vesga MA, Van Der Schoot E, Puente F, Tejedor D. Performance evaluation study of ID CORE XT, a high throughput blood group genotyping platform. Blood Transfus. 2018;16:193–199. doi: 10.2450/2016.0146-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hashmi G, Shariff T, Zhang Y, Cristobal J, Chau C, Seul M, Vissavajjhala P, Baldwin C, Hue-Roye K, Charles-Pierre D, Lomas-Francis C, Reid ME. Determination of 24 minor red blood cell antigens for more than 2000 blood donors by high-throughput DNA analysis. Transfusion. 2007;47:736–747. doi: 10.1111/j.1537-2995.2007.01178.x. [DOI] [PubMed] [Google Scholar]
- 7.Denomme GA, Van Oene., M High-throughput multiplex single-nucleotide polymorphism analysis for red cell and platelet antigen genotypes. Transfusion. 2005;45:660–666. doi: 10.1111/j.1537-2995.2005.04365.x. [DOI] [PubMed] [Google Scholar]
- 8.Montpetit A, Phillips MS, Mongrain I, Lemieux R, St-Louis M. High-throughput molecular profiling of blood donors for minor red blood cell and platelet antigens. Transfusion. 2006;46:841–848. doi: 10.1111/j.1537-2995.2006.00805.x. [DOI] [PubMed] [Google Scholar]
- 9.Di Cristofaro J, Silvy M, Chiaroni J, Bailly P. Single PCR multiplex SNaPshot reaction for detection of eleven blood group nucleotide polymorphisms: optimization, validation, and one year of routine clinical use. J Mol Diagn. 2010;12:453–460. doi: 10.2353/jmoldx.2010.090222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palacajornsuk P, Halter C, Isakova V, Tarnawski M, Farmar J, Reid ME, Chaudhuri A. Detection of blood group genes using multiplex SNaPshot method. Transfusion. 2009;49:740–749. doi: 10.1111/j.1537-2995.2008.02053.x. [DOI] [PubMed] [Google Scholar]
- 11.Meyer S, Vollmert C, Trost N, Bronnimann C, Gottschalk J, Buser A, Frey BM, Gassner C. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014;54:3198–3207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 12.Silvy M, Di Cristofaro J, Beley S, Papa K, Rits M, Richard P, Chiaroni J, Bailly P. Identification of RHCE and KEL alleles in large cohorts of Afro-Caribbean and Comorian donors by multiplex SNaPshot and fragment assays: a transfusion support for sickle cell disease patients. Br J Haematol. 2011;154:260–270. doi: 10.1111/j.1365-2141.2011.08691.x. [DOI] [PubMed] [Google Scholar]
- 13.Ribeiro KR, Guarnieri MH, da Costa DC, Costa FF, Pellegrino J, Jr, Castilho L. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang. 2009;97:147–152. doi: 10.1111/j.1423-0410.2009.01185.x. [DOI] [PubMed] [Google Scholar]
- 14.Seltsam A, Wagner FF, Salama A, Flegel WA. Antibodies to high-frequency antigens may decrease the quality of transfusion support: an observational study. Transfusion. 2003;43:1563–1566. doi: 10.1046/j.1537-2995.2003.00565.x. [DOI] [PubMed] [Google Scholar]
- 15.Reesink HW, Engelfriet CP, Schennach H, Gassner C, Wendel S, Fontao-Wendel R, de Brito MA, Sistonen P, Matilainen J, Peyrard T, Pham BN, Rouger P, Le Pennec PY, Flegel WA, von Zabern I, Lin CK, Tsoi WC, Hoffer I, Barotine-Toth K, Joshi , SR, Vasantha K, Yahalom V, Asher O, Levene C, Villa MA, Revelli N, Greppi N, Marconi M, Tani Y, Folman CC, de Haas M, Koopman MM, Beckers E, Gounder DS, Flanagan P, Wall L, Aranburu Urtasun E, Hustinx H, Niederhauser C, Flickinger C, Nance SJ, Meny GM. Donors with a rare pheno (geno) type. Vox Sang. 2008;95:236–253. doi: 10.1111/j.1423-0410.2008.01084.x. [DOI] [PubMed] [Google Scholar]
- 16.Flickinger C. In search of red blood cells for alloimmunized patients with sickle cell disease. Immunohematology. 2006;22:6. [PubMed] [Google Scholar]
- 17.Wagner FF, Bittner R, Petershofen EK, Doescher A, Muller TH. Cost-efficient sequence-specific priming-polymerase chain reaction screening for blood donors with rare phenotypes. Transfusion. 2008;48:1169–1173. doi: 10.1111/j.1537-2995.2008.01682.x. [DOI] [PubMed] [Google Scholar]
- 18.Lee S, Naime DS, Reid ME, Redman CM. Molecular basis for the high-incidence antigens of the Kell blood group system. Transfusion. 1997;37:1117–1122. doi: 10.1046/j.1537-2995.1997.37111298088039.x. [DOI] [PubMed] [Google Scholar]
- 19.Moller M, Joud M, Storry JR, Olsson ML. Erythrogene: a database for in-depth analysis of the extensive variation in 36 blood group systems in the 1000 Genomes Project. Blood Adv. 2016;1:240–249. doi: 10.1182/bloodadvances.2016001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm S. A simple sequentially rejective multiple test procedure. Scand J Statist. 1979;6:5. [Google Scholar]
- 21.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji Y, Veldhuisen B, Ligthart P, Haer-Wigman L, Jongerius J, Boujnan M, Ait Soussan A, Luo G, Fu Y, van der Schoot CE, de Haas., M Novel alleles at the Kell blood group locus that lead to Kell variant phenotype in the Dutch population. Transfusion. 2015;55:413–421. doi: 10.1111/trf.12838. [DOI] [PubMed] [Google Scholar]
- 23.Jungbauer C, Hobel CM, Schwartz DW, Mayr WR. High-throughput multiplex PCR genotyping for 35 red blood cell antigens in blood donors. Vox Sang. 2012;102:234–242. doi: 10.1111/j.1423-0410.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- 24.Ballif BA, Helias V, Peyrard T, Menanteau C, Saison C, Lucien N, Bourgouin S, Le Gall M, Cartron JP, Arnaud L. Disruption of SMIM1 causes the Vel- blood type. EMBO Mol Med. 2013;5:751–761. doi: 10.1002/emmm.201302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He YL, Gao HH, Ye LY, Guo ZH, Wang P, Zhu ZY. Multiplex polymerase chain reaction with DNA pooling: a cost-effective strategy of genotyping rare blood types. Transfus Med. 2013;23:42–47. doi: 10.1111/j.1365-3148.2012.01198.x. [DOI] [PubMed] [Google Scholar]
- 26.Perreault J, Lavoie J, Painchaud P, Cote M, Constanzo-Yanez J, Cote R, Delage G, Gendron F, Dubuc S, Caron B, Lemieux R, St-Louis M. Set-up and routine use of a database of 10,555 genotyped blood donors to facilitate the screening of compatible blood components for alloimmunized patients. Vox Sang. 2009;97:61–68. doi: 10.1111/j.1423-0410.2009.01177.x. [DOI] [PubMed] [Google Scholar]