Abstract
The development of red blood cell (RBC) alloantibodies and autoantibodies complicates transfusion therapy in sickle cell disease (SCD) patients. In an effort to reduce the risk of alloimmunization, some strategies have been used to provide antigen-matched RBC transfusions to patients with SCD in Brazil, including molecular matching in 3 levels: RH and K matching; extended matching (RH, KEL, FY, JK, MNS, DI), and extended matching including RHD and RHCE variant alleles. Molecular matching has shown clinical benefits to the patients with SCD, contributing significantly to reduce the rates of alloimmunization. Improvements in the clinical outcomes of the patients have also been observed as shown by an increase in their hemoglobin levels and reduction in their percentage of hemoglobin S as well as better in vivo RBC survival and diminished frequency of transfusions. However, prevention of RBC alloimmunization still remains a challenge in Brazil due to the difficulty to fulfill all transfusion requests of the patients with antigen-matching units, inaccuracy of RBC phenotyping, RBC transfusions outside the institution where the patient is treated, advanced age of some patients, the RBC antigen discrepancy between donors and recipients, and the presence of RH variants.
Keywords: Sickle cell disease, Alloimmunization, Antigen matching, Red cell genotyping
Introduction
Sickle cell disease (SCD) is the most prevalent hereditary disease in Brazil and affects around 30,000 patients in the whole country. The incidence of SCD in newborn babies varies among the states in Brazil, reflecting the ethnic heterogeneity of the Brazilian population. SCD is more prevalent in the state of Bahia with 1 in 650 patients affected, followed by Rio de Janeiro with 1 in 1,200 patients affected [1]. The Africans brought to Brazil belonged to two major groups: the West-African and the Bantu people (Angola, Congo, and Mozambique). The West-African people were sent to Bahia and the Bantus were sent to the Southeast and Northeast Brazil. In a study reporting the genomic ancestry of individuals from different regions of Brazil, it was demonstrated that the African proportion is highest in the Northeast (29%) followed by the Southeast (17%), but most SCD patients in Brazil share European and African genetic ancestries, and many have also a significant proportion of Amerindian ancestry [2].
Nationwide, in 2016, 1,071 newborn babies had SCD, and >60,000 were heterozygotes for the βS allele [3]. Around 50% of the patients are chronically transfused, and RBC alloimmunization is a serious complication in these patients with severe clinical consequences, including delay in obtaining matched blood as well as potentially life-threatening delayed hemolytic transfusion reactions, generation of autoantibodies, and hyperhemolysis syndrome.
The incidence of alloimmunization in patients with SCD in Brazil ranges from 10 to 60% depending on the region and on the protocol of antigen matching used [4,5,6,7,8]. Even though our population is very mixed, we have observed that patients and donors exhibit differences in their genotype frequencies what could explain the high rates of alloimmunization in some regions [4,7].
Antigen-Matched in Sickle Cell Disease Patients: Chances
In an effort to reduce the risk of alloimmunization, some strategies have been used to provide antigen-matched RBC transfusions to patients with SCD in Brazil. The most common strategy is provision of prophylactic matching for C, E, and K antigens. But phenotype matching for antigens in the Kidd, Duffy and MNS systems has also been a practice. Molecular genotyping was introduced in Brazil in 2002 for the management of multiply transfused patients with SCD in order to improve transfusion therapy by allowing the determination of the true blood group genotype, by assisting in the identification of suspected alloantibodies, and by helping to decrease the risk of transfusion reactions, especially delayed transfusion reactions to existing alloantibodies, and to prevent alloimmunization [9].
With the evolution of the molecular methods to genotype RBC antigens, we started to use the DNA array platforms to genotype SCD patients and to find the most compatible blood for transfusion. In a study performed in 2009 [10], we observed that in alloimmunized patients, who had been in need for transfusions each week, the transfusion interval could be increased to 30–45 days when 2–3 units of genotype-matched RBCs were given. After that, we also provided evidence that molecular matching of RBCs is superior to serological matching in SCD patients by showing that patients who previously had been received serologically matching units benefited from being transfused with molecularly matched units in terms of better in vivo RBC survival [11]. Therefore, according to our experience, matching at DNA level can provide an added level of safety and efficacy for chronically transfused patients with SCD and can be used to decrease the risk of alloimmunization.
However, despite provision of extended antigen-matched RBCs using molecular matching, the patients still produced Rh antibodies, and in many cases the antibodies were considered autoantibodies because the patient‘s own RBC type was serologically and molecularly positive for the corresponding antigen [12]. As we identified altered RH alleles in our SCD patients with Rh antibodies and showed that most of the antibodies produced by patients with RH variants were involved in delayed hemolytic transfusion reactions or decreased survival of transfused RBCs, matching patients based on RH variants started to be considered to improve matching in the Rh system [13].
In order to provide more highly matched blood and to be cost-effective for the treatment of chronically transfused patients with SCD, we have implemented molecular matching in 3 levels: RH and K matching (level 1); extended matching (RH, KEL, FY, JK, MNS, DI) (level 2) and extended matching including RHD and RHCE variant alleles (level 3). Extended genotyping is performed in patients with a recent transfusion history (chronically transfused patients), in patients with positive direct antiglobulin test, and in those with RH variants predicting partial antigens and/or lack of high prevalence antigens. Although molecular matching is not yet a routine in our institution, it has shown clinical benefits to the patients with SCD, contributing significantly to reduce the rates of alloimmunization to 5–10% with level 1 of matching and to <1% with level 2 of matching. Improvements in the clinical outcomes of the patients have also been observed as shown by an increase in their hemoglobin (Hb) levels, a reduction in their percentage of HbS, a better in vivo RBC survival, and a diminished frequency of transfusions.
Considering the total of RBC units requested for each patient and a number of 2 donations/year for the compatible donors, we are fulfilling the needs of patients in level 1 of matching and for most of patients in level 2, but when we considered level 3 of matching, it has been much more difficult to find compatible donors for patients with SCD. Currently, alloimmunized patients with clinically significant Rh antibodies are receiving level 3-matching units if possible as the number of RH-genotyped donors is limited at present, especially for the SCD patient's needs. As we are not fulfilling all transfusion requests with level 3-matching units, we are concerned because prophylaxis of maintaining higher Hb levels in those patients is restricted and the number of transfusions in such patients is lower. Moreover, knowing well clinical risk, incompatible blood is being transfused. In addition, we are also observing that in some patients more closely matched donors are necessary. As the number of RH-genotyped donors available is limited to meet the needs of patients with variants and confirmed alloantibodies, we are developing some strategies to optimize the identification of donors with RH variant alleles by screening donors self-declared as Blacks, donors with known African genetic ancestry markers, donors with partial D, and donors with the r‘s phenotype, but donor selection for SCD patients in a country with increased miscegenation is a real problem [14].
Besides of the challenges to fulfill all requests connected with level 3 matching units, the cost effectiveness of those practices to minimize the risk of alloimmunization remains controversial, partly because not all patients develop alloantibodies despite extensive exposure to donor RBC antigens [15].
Several factors are known to influence the recipient‘s immune system to react to alloantigens, e.g., the dose, the immunogenicity of the antigen as well as genetic or acquired patient-related factors [16,17,18,19,20,21]. However, possible differences in the immune makeup of transfused patients who produce alloantibodies (responders) and those who do not (non-responders) are not known. By identifying such markers, it may be possible to predict responders and non-responders, thereby avoiding the use of costly antigen-matched units for non-responders and only selecting phenotyped/genotyped-matched units for responders in order to reduce alloimmunization-associated morbidity and mortality.
In previous studies, we found that TNFA-308A, IL1B-511T, RHAG 808G>A and HLA-DRB1*15 alleles may predict a good responder phenotype [8] and that RHAG 808G>A and HLA-DRB*15 alleles are closely linked to RH alloimmunization [8,22]. These findings contributed to the development of a transfusion strategy for non-alloimmunized SCD patients as typing for these polymorphisms could potentially help in the classification of responder and non-responder SCD patients, allowing blood with high level of compatibility to be transfused to responders in order to avoid production of RBC antibodies and the negative consequences of alloimmunization. We searched for TNFA-308A, IL1B-511T polymorphisms in non-alloimmunized SCD patients in order to select responders and non-responders for transfusion with level 2-matching RBC units and for the presence of HLA-DRB1*15 allele and RHAG 808G>A polymorphism to select responders and non-responders SCD patients for transfusion with level 3-matching RBC units [23].
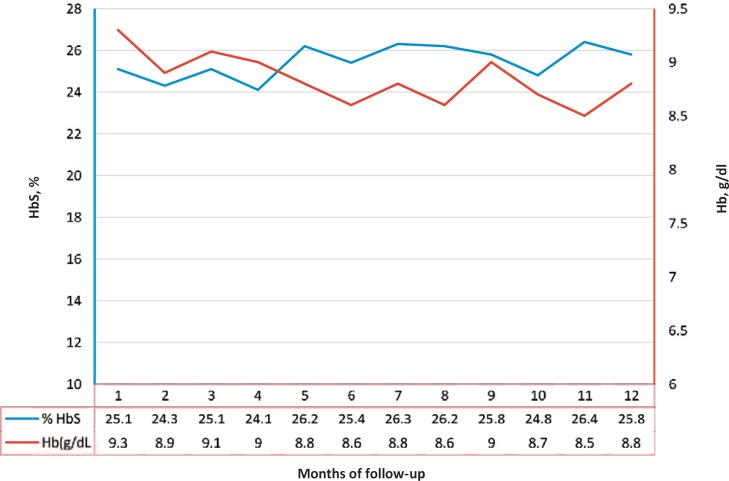
We selected 96 non-alloimmunized patients on chronic transfusion receiving at least level 1-matching units. 57 patients had minimum of 5 negative antigens and are in the level 2 matching protocol while 21 patients had RH variants and we are trying to provide level 3-matched units. 13 of the 57 patients with multiple negative antigens have polymorphisms associated with risk of alloimmunization and are being transfused with level 2 of matching while the other 44 patients with no genetic risk factors associated with RBC alloimmunization are being transfused with level 1 of matching. Only 4 of the 21 patients with clinically relevant RHD-CE genotypes have the polymorphisms associated with risk of RH alloimmunization and are being transfused with level 3 of matching. This practice has improved the ability to find antigen-matched components for transfusion support in non-alloimmunized patients and has been of benefit to patients, as observed by the lack in the development of alloantibodies. Transfusion follow-up during 1 year showed that this transfusion strategy was efficient to keep the patient's Hb baseline levels and the percentage of HbS to less than 30% (fig. 1).
Fig. 1.
Transfusion follow-up during 12 months of a SCD patient receiving antigen-matched blood.
Antigen-Matching in Sickle Cell Disease Patients: Challenges
Even though prospective antigen-matched transfusions are a reality for many Brazilian transfusion centers which take care of SCD patients, the alloimmunization rates observed in our country are still above that described in the literature [7]. There are multiple factors underlying the higher risk of post-transfusion alloantibody formation observed among Brazilian SCD patients, some of them already depicted as potential pitfalls by multiple studies [24,25] and some reflecting the heterogeneity of transfusion and laboratory policies throughout the country.
We have recently studied 80 SCD patients who were under chronic transfusion treatment receiving level 1-matched transfusions [7]. The first surprising factor was that 46% of the individuals were already alloimmunized (mainly against Rh antigens) at the beginning of the antigen-matched transfusion protocol. As the studied transfusion center of reference only serves adult patients, this high rate of alloimmunization presented by the individuals joining the service probably reflected heterogeneity of efficacy of the alloimmunization prophylaxis protocols applied by the pediatric transfusion services. While there were transfusion centers providing level 2-matched transfusions to the SCD patients, others did not have any specific transfusion policy to meet the needs of this patient population. Among the non-alloimmunized group of patients, 33.7% developed alloantibodies during the transfusion therapy, most of which directed against RH and K antigens that were included in the antigen-matching protocol. When evaluating the possible causes underlying the prophylaxis failure, it was demonstrated that the occurrence of transfusions outside the institution of reference and the presence of RH variants were the most important factors. Even though less than 25% of the alloimmunized patients received transfusions in external hospitals, they were registered in other services and received non-matched RBC units when admitted to the emergency department. It has been previously demonstrated that the occurrence of alloimmunization is intrinsically correlated to the patients' inflammation background and that the risk of alloantibody formation following antigen-mismatched transfusion is higher when SCD individuals are acutely ill [26]. This partly explains why our SCD patients easily developed alloantibodies when receiving antigen-mismatched transfusions in other hospitals. This problem can be overcome by providing the patients with their complete RBC phenotype and genotype, what is not complex, but most importantly by providing the external hospitals with phenotype-compatible RBC units when needed, what is far more complicated.
The presence of RH variants was detected in 32% of the Brazilian SCD patients who developed Rh alloantibodies, irrespective of receiving level 1-matched transfusions [7]. Recently, we have evaluated 35 SCD patients with unexplained Rh antibody, meaning they exhibited a certain Rh antibody in the sera but phenotyped as antigen-positive, using a NGS-based strategy in the Ion Torrent system (Thermo Fisher Scientific, Waltham, MA, USA) [27]. RHD*weak D 4.2.2 was the most common RHD variant allele (50%), followed by RHD*weak partial D 4.0 (20%), RHD*DVII (10%), and RHD*DAU0.01 (10%); and the most prevalent RHCE-altered alleles were RHCE*ce48C (40%), RHCE*ceAR (20%), and RHCE*ce48C,733G (8%). These alleles were also found in other cohorts of Afro-American and Afro-Caribbean SCD patients [28,29]. Identifying RH variants at the beginning of the transfusion protocol could have prevented RH alloimmunization by providing antigen-negative or haplotype-compatible RBC units, but RH genotyping is not currently available for all Brazilian centers that treat SCD patients. Also, even though the RH alleles identified in this study were associated with alloantibody development, if RH genotyping was performed for all SCD recipients under transfusion therapy, other variant RH alleles with low risk of alloantibody formation would be identified and their inclusion in the antigen-matched protocol would be questionable [29]. One important data derived from this study evaluating RH variants among sensitized Brazilian SCD patients refers to the inaccuracy of serology to distinguish Rh alloantibodies from Rh autoantibodies, possibly due to the high transfusion rates in the studied population and further exacerbated by the fact that transfusions occurring outside the reference hospital may be omitted to the laboratory team, as previously discussed [7]. Of 42 cases of abnormal Rh antibodies, 38% were correctly classified by serological methods, and 62% were considered inaccurate. Of the inaccurate results, 73.1% were considered as clinically relevant, as they were associated with shortage of RBC units available for transfusion, as in the case of patients receiving D-negative-only units due to unconfirmed partial-D diagnosis or due to delayed hemolytic transfusion reactions / poor transfusion increment in the case of patients with anti-e alloantibodies classified as autoantibodies [7]. As so, molecular confirmation of RH variants is strongly suggested for SCD patients with unexplained Rh antibodies, especially when serology is not totally trustworthy.
One of the factors that underlie alloantibody development among Brazilian SCD patients is the RBC antigen discrepancy between donors and recipients. Even though the Brazilian population is mixed, mainly of African, European and Amerindian ancestry, the prevailing logic that the donors' RBC phenotype would be similar to that of SCD patients has proved wrong [7]. It has been demonstrated, for example, that the RHCE*Ce allele, frequent among Europeans, is much more prevalent among blood donors in comparison to patients [7]. Also, considering the FYBES allele (FYB*-67T>C) in homozygosis as an ancestry marker of African descent, the frequency of this genotype is significantly higher among SCD patients in comparison to blood donors (25 vs. 8.3%), reflecting the racial disparity between these groups [7]. Thus, antigen discrepancies between donors and recipients are also true for Brazilian population, irrespective of its intense miscegenation.
Conclusion
In order to reduce the high alloimmunization rates and, in parallel, the drawbacks associated to this transfusion complication, such as delayed hemolytic transfusion reactions and hemolytic disease of the fetus and newborn, Brazilian transfusion centers should prefer the molecular matching between donors and recipients, aiming to overcome potential serological limitation. In order to assure that, genotyping of blood group systems, including RH, should be accessible throughout the country. Also, the smaller transfusion centers that frequently transfuse SCD patients originally under primary antigen-matching transfusion program should have access to both the patients' genotype data as well to compatible phenotyped units, which can be provided by larger transfusion centers.
According to our experience with molecular matching and RBC alloimmunization in SCD patients, our future plans are to look for more clinical and laboratory evidences on the benefits of molecular matching, select responders and non-responders, and find good approaches for screening more donors with homozygous genotypes and RH variants to have enough compatible donor units to fulfill the patient's needs. With the advance of next-generation sequencing, it will be possible to obtain even more accurate blood group phenotype predictions in both blood donors and patients, thus allowing patient-specific transfusion therapy.
Disclosure Statement
The authors have disclosed no conflicts of interest.
References
- 1.Silva WS, de Oliveira RF, Ribeiro SB, da Silva IB, de Araújo EM, Baptista AF. Screening for structural hemoglobin variants in Bahia, Brazil. Int J Environ Res Publ Health. 2016;13:225. doi: 10.3390/ijerph13020225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pena SDJ, Di Pietro G, Fuchshuber-Moraes M, Genro JP, Hutz MH, Kehdy Fde S, Kohlrausch F, Magno LA, Montenegro RC, Moraes MO, de Moraes ME, de Moraes MR, Ojopi EB, Perini JA, Racciopi C, Ribeiro-Dos-Santos AK, Rios-Santos F, Romano-Silva MA, Sortica VA, Suarez-Kurtz G. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS ONE. 2011;6:e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lobo CL, Ballas SK, Domingos AC, Moura PG, do Nascimento EM, Cardoso GP, de Carvalho SM. Newborn screening program for hemoglobinopathies in Rio de Janeiro, Brazil: Pediatr Blood Cancer. 2014;61:34–39. doi: 10.1002/pbc.24711. [DOI] [PubMed] [Google Scholar]
- 4.Moreira Junior G, Bordin JO, Kuroda A, Kerbauy J. Red blood cell alloimmunization in sickle cell disease: the influence of racial and antigenic pattern differences between donors and recipients in Brazil. Am J Hematol. 1996;52:197–200. doi: 10.1002/(SICI)1096-8652(199607)52:3<197::AID-AJH11>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 5.Zanette AMD, Gonçalves MS, Schettini LV, Aguiar LM, Bahia RCS, Nogueira LAV, Brandão CJF, Azevedo CAN, Aragão LR, Arruda SM. Alloimmunization and clinical profile of sickle cell disease patients from Salvador-Brazil. Ethn Dis. 2010;20:136–141. [PubMed] [Google Scholar]
- 6.Murao M, Viana MB. Risk factors for alloimmunization by patients with sickle cell disease. Braz J Med Biol Res. 2005;38:675–682. doi: 10.1590/s0100-879x2005000500004. [DOI] [PubMed] [Google Scholar]
- 7.Dezan MR, Oliveira VB, Bianchi JVS, Rofrigues V, Solano JH, Gomes FC, Bonifácio SL, Guallandro SFM, Krieger JE, Pereira AC, Sabina EC, Mendrone-Junior A, Dinardo CL. Effectiveness of a red cell antigen matching transfusion protocol in sickle cell disease patients. ISBT Sci Ser. 2016;11:132–139. [Google Scholar]
- 8.Sippert EA, Visentainer JEL, Alves HV, Rodrigues C, Gilli SCO, Addas-Carvalho M, Saad STO, Costa FF, Castilho L. Red blood cell alloimmunization in patients with sickle cell disease: correlation with HLA and cytokine gene polymorphisms. Transfusion. 2017;57:379–389. doi: 10.1111/trf.13920. [DOI] [PubMed] [Google Scholar]
- 9.Castilho L, Rios M, Bianco C, Pellegrino J, Jr, Alberto FL, Saad STO, Costa FF. DNA based typing of blood groups for the management of polytransfused sickle cell disease patients. Transfusion. 2002;42:232–238. doi: 10.1046/j.1537-2995.2002.00029.x. [DOI] [PubMed] [Google Scholar]
- 10.Ribeiro KR, Guarnieri MH, Costa DC, Costa FF, Pellegrino J, Jr, Castilho L. DNA array analysis for red blood cell antigens facilitates the transfusion support with antigen-matched blood in patients with sickle cell disease. Vox Sang. 2009;97:147–152. doi: 10.1111/j.1423-0410.2009.01185.x. [DOI] [PubMed] [Google Scholar]
- 11.da Costa DC, Pellegrino J, Jr, Guelsin GA, Ribeiro KA, Gilli SC, Castilho L. Molecular matching of red blood cells is superior to serological matching in sickle cell disease patients. Rev Bras Hematol Hemoter. 2013;35:35–38. doi: 10.5581/1516-8484.20130012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sippert E, Fujita CR, Machado D, Guelsin G, Gaspardi A, Gilli SCO, Saad STO, Castilho L. Variant RH alleles and Rh immunization in patients with sickle cell disease. blood Transfusion. 2015;13:72–77. doi: 10.2450/2014.0324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaspardi AC, Sippert EA, De Macedo MD, Pellegrino J, Jr, Costa FF, Castilho L. Clinically relevant RHD-CE genotypes in patients with sickle cell disease and in African Brazilian donors. Blood Transfus. 2016;14:449–454. doi: 10.2450/2016.0275-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prisco Arnoni C, Guilhem Muniz J, de Paula Vendrame TA, de Medeiros Person R, Roche Moreira Latini F, Castilho L. RHCE variants inherited with altered RHD alleles in Brazilian blood donors. Transfus Med. 2016;26:285–290. doi: 10.1111/tme.12309. [DOI] [PubMed] [Google Scholar]
- 15.Kacker S, Ness PM, Savage WJ, Frick KD, Shirey RS, King KE, Tobian AA. Cost-effectiveness of prospective red blood cell antigen matching to prevent alloimmunization among sickle cell patients. Transfusion. 2014;54:86–97. doi: 10.1111/trf.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodrigues C, Sell AM, Guelsin GAS, Higa TT, Pagliarini E Silva S, Macedo LC, Sippert EÂ, de Alencar JB, Zanette Â, Acorsi CRL, Castilho L, Visentainer JEL. HLA polymorphisms and risk of red blood cell alloimmunisation in polytransfused patients with sickle cell anaemia. Transfus Med. 2017;27:437–443. doi: 10.1111/tme.12459. [DOI] [PubMed] [Google Scholar]
- 17.Higgins JM, Sloan SR. Stochastic modeling of human RBC alloimmunization: evidence for a distinct population of immunologic responders. Blood. 2008;112:2546–2553. doi: 10.1182/blood-2008-03-146415. [DOI] [PubMed] [Google Scholar]
- 18.Milton JN, Ashley-Koch AE, Garrett ME, Soldano KL, Orringer EP, Sebastiani P, Dworkis DA, Quillen K, Steinberg MH, Telen MJ. Genes associated with alloimmunization to blood group antigens in sickle cell disease. Blood. 2014;124:762. [Google Scholar]
- 19.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: pathophysiology, risk factors, and transfusion management. Blood. 2012;120:528–537. doi: 10.1182/blood-2011-11-327361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baleotti W, Ruiz MO, Fabron A, Castilho L, Giuliatti S, Donadi EA. HLA-DRB1*07:01 allele is primarily associated with the Diego a alloimmunization in a Brazilian population. Transfusion. 2014;54:2468–2476. doi: 10.1111/trf.12652. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira VB, Dezan MR, Gomes FCA, Menosi Guallandro SF, Krieger JE, Pereira AC, Levi JE, Rocha V, Mendrone-Junior A, Sabino EC, Dinardo CL. -318C/T polymorphism of the CTLA-4 gene is an independent risk facotor for RBC alloimmunization among sickle cell disease. Int J Immunogenet. 2017;44:219–224. doi: 10.1111/iji.12334. [DOI] [PubMed] [Google Scholar]
- 22.Sippert EA, Botelho MA, Gaspardi AC, Castilho L. RHAG GZMB and PRKCQ genes and alloimmunization to red blood cell antigens in patients with sickle cell disease and β-thalassemia. Transfusion. 2015;55((suppl 3)):29A. [Google Scholar]
- 23.Santos TD, Sippert EA, Macedo MD, Menegati SFP, Castilho L. Using genetic markers to select responders and non-responders sickle cell disease (SCD) patients for transfusion with RH haplotype matching red blood cell (RBC) units. Transfusion. 2017;57((suppl 3)):63A. [Google Scholar]
- 24.O'Suoji C, Liem RI, Mack AK, Kingsberry P, Ramsey G, Thompson AA. Alloimmunization in sickle cell anemia in the era of extended red cell typing. Pediatr Blood Cancer. 2013;60:1487–1491. doi: 10.1002/pbc.24530. [DOI] [PubMed] [Google Scholar]
- 25.Telen MJ, Afenyi-Annan A, Garrett ME, Combs MR, Orringer EP, Ashley-Koch AE. Alloimmunization in sickle cell disease: changing antibody specificities and association with chronic pain and decreased survival. Transfusion. 2015;55:1378–1387. doi: 10.1111/trf.12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fasano RM, Booth GS, Miles M, Du L, Koyama T, Meier ER, Luban NL. Red blood cell alloimmunization is influenced by recipient inflammatory state at time of transfusion in patients with sickle cell disease. Br J Haematol. 2015;168:291–300. doi: 10.1111/bjh.13123. [DOI] [PubMed] [Google Scholar]
- 27.Dezan MR, Ribeiro IH, Oliveira VB, Vieira JB, Gomes FC, Franco LAM, Varuzza L, Ribeiro R, Chinoca KZ, Levi JE, Krieger JE, Pereira AC, Gualandro SFM, Rocha VG, Mendrone-Junior A, Sabino EC, Dinardo CL. RHD and RHCE genotyping by next-generation sequencing is an effective strategy to identify molecular variants within sickle cell disease patients. Blood Cells Mol Dis. 2017;65:8–15. doi: 10.1016/j.bcmd.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Reid ME, Halter Hipsky C, Hue-Roye K, Hoppe C. Genomic analyses of RH alleles to improve transfusion therapy in patients with sickle cell disease. Blood Cells Mol Dis. 2014;52:195–202. doi: 10.1016/j.bcmd.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noizat-Pirenne F, Tournamille C. Relevance of RH variants in transfusion of sickle cell patients. Transfus Clin Biol. 2011;18:527–535. doi: 10.1016/j.tracli.2011.09.001. [DOI] [PubMed] [Google Scholar]