Abstract
Background
High-frequency blood group antigens (HFA) are present in >90% of the human population, according to some reports even in >99% of individuals. Therefore, patients lacking HFA may become challenging for transfusion support because compatible blood is hardly found, and if the patient carries alloantibodies, the cross-match will be positive with virtual every red cell unit tested.
Methods
In this study, we applied high-throughput blood group SNP genotyping on >37,000 Swiss blood donors, intending to identify homozygous carriers of low-frequency blood group antigens (LFA).
Results
326 such individuals were identified and made available to transfusion specialists for future support of patients in need of rare blood products.
Conclusion
Thorough comparison of minor allele frequencies using population genetics revealed heterogeneity of allele distributions among Swiss blood donors which may be explained by the topographical and cultural peculiarities of Switzerland. Moreover, geographically localized donor subpopulations are described which contain above-average numbers of individuals carrying rare blood group genotypes.
Keywords: Blood groups, Low-frequency antigen, High-frequency antigen, Rare donor panel/program, Rare/molecular blood group, Blood group allele, Population genetics, Switzerland
Introduction
One of the challenges in transfusion medicine is to provide compatible blood for patients negative for a high-frequency blood group antigen (HFA) and who have an alloantibody against the antigen [1]. Such HFA negativity may either be caused by a complete lack of the protein serving as carrier of a certain blood group system with concomitant negativity for all its associated antigens, or by the lack of only one specific antigen, because of the presence of an identical variant blood group protein inherited on both parental haplotypes. Low-frequency blood group antigens (LFA) are the less frequent antithetic variants of HFAs. LFAs do not create a major transfusion problem from the aspect of finding compatible donors. However, a potentially dangerous antibody to an LFA could remain undetected if a full cross-match analysis was not performed [2].
The Kell blood group system is an exemplary model for the above-mentioned model. The system is highly polymorphic and expresses 36 antigens, all encoded on a type II glycoprotein of 732 amino acids encoded by the KEL gene and its allelic variants [3,4]. Some of the Kell antigens have been classified into antithetical pairs, each represented by one HFA and its correspondent LFA, e.g. K and k or Kpa and Kpb, others being independently expressed or having unknown antithetical partners. KEL2 or k+, formerly also named ‘Cellano’, is a HFA and its antithetical variant is the LFA: KEL1 or K+. As a result, K+ k- homozygous individuals are encountered only rarely, e.g. at an exemplary frequency of 1 per 1,371 Swiss [5]. While, K+ k- individuals still express other Kell antigens, e.g. Kpb or KEL11, none of the Kell antigens are expressed on cells of the Kell-null phenotype, K₀, which arise from homozygous or compound heterozygous KEL-inactivating mutations [6,7,8]. K₀ individuals are exceedingly rare and may only be found at frequency fewer than 1 individual per 1 million Austrians [6].
Some blood types are extremely scarce worldwide, and requests for transfusion are particularly difficult to fulfill. On the other side of the frequency spectrum, e.g. where prevalence of certain antigens is shifting towards ‘public antigen frequency', a generally accepted numerical definition for ‘rarity’ is lacking. For instance, D- blood is common in Caucasians (approximately 15% of the population), but it is rare in Asia (less than 1%) [2]. In some nations, a blood type with a prevalence of 1 in 100 is considered rare, whereas in other countries, the same status requires antigen prevalence of less than 1 in 5,000. Moreover, in some programs, rare donors are exclusively determined by being negative for single HFAs, whereas in other programs, donors negative for a combination of several public (common) red cell antigens are also recognized as being rare. As a consequence, the definition of a ‘rare donor’ is widely different, as vividly demonstrated by the existent variety of national rare donor programs [9].
Traditionally, red cell antigens have been identified by serology. Recent advances in molecular biology made it possible to genotype most of the blood group antigens employing high-throughput technology platforms [10]. Also, today, blood group antigens without suitable anti-sera - such as Scianna and Dombrock - are screened for using high-throughput genotyping methods [11,12].
The presented project was conducted for the main purpose to ensure the supply of rare blood units to the Swiss population. MALDI-TOF MS was adapted and used to genotype 37,253 Swiss blood donors by a customized ‘RARE module’ covering 26 blood group single nucleotide polymorphisms (SNPs) including 22 antithetical HFA/LFA pairs (table 1) [13].
Table 1.
Specificities included in the matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry (MALDI-TOF MS) based ‘RARE module’
| Blood group system | ISBT # | Blood group | Gene (HGNC) | Chromosome | Allele name 1 | Allele name 2 | CDS§ position | CDS on mRNA accession number | nt 1 | nt 2 | Amino acid exchange | dbSNP rs number | Antigens | HFA/LFA | Allele ct. | SNP ct. | Plex W1 | Plex W2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABO | 001 | ABO A vs O1 | ABO | 9q34.2 | ABO*A(wt) | ABO*O.01 | 261 | NM_020469.2 | G | del G | fsThr88Pro | rs8176719 | 2 | – | 2 | 1 | – | W2 |
| ABO | 001 | ABO A vs O2 | ABO | 9q34.2 | ABO*A(wt) | ABO*O.02 | 802 | NM_020469.2 | G | A | Gly268Arg | rs41302905 | – | – | 1 | 1 | – | W2 |
| ABO | 001 | ABO A vs B | ABO | 9q34.2 | ABO*A(wt) | ABO*B | 803 | NM_020469.2 | G | C | Gly268Ala | rs8176747 | 1 | – | 1 | 1 | W1 | – |
| Lutheran | 005 | Lua / Lub | BCAM | 19q13.32 | LU*01 | LU*02 (wt) | 230 | NM_005581.4 | A | G | His77Arg | rs28399653 | 2 | 1 | 2 | 1 | W1 | – |
| Lutheran | 005 | Lu8 / Lu14 | BCAM | 19q13.32 | LU*02.14 | LU*02 (wt) | 611 | NM_005581.4 | A | T | Lys204Met | rs28399656 | 2 | 1 | 1 | 1 | W1 | – |
| Lutheran | 005 | Aua / Aub | BCAM | 19q13.32 | LU*02.19 | LU*02 (wt) | 1615 | NM_005581.4 | G | A | Ala539Thr | rs1135062 | 2 | – | 1 | 1 | – | W2 |
| Kell | 006 | K / k | KEL | 7q34 | KEL*01 | KEL*02 (wt) | 578 | NM_000420.2 | T | C | Met193Thr | rs8176058 | 2 | 1 | 2 | 1 | W1 | – |
| Kell | 006 | Kpa / Kpb | KEL | 7q34 | KEL*02.03 | KEL*02 (wt) | 841 | NM_000420.2 | T | C | Trp281Arg | rs8176059 | 2 | 1 | 1 | 1 | – | W2 |
| Kell | 006 | K11 / K17 | KEL | 7q34 | KEL*02.17 | KEL*02 (wt) | 905 | NM_000420.2 | C | T | Ala302Val | rs61729034 | 2 | 1 | 1 | 1 | W1 | – |
| Kell | 006 | Jsa / Jsb | KEL | 7q34 | KEL*02.06 | KEL*02 (wt) | 1790 | NM_000420.2 | C | T | Pro597Leu | rs8176038 | 2 | 1 | 1 | 1 | W1 | – |
| Diego | 010 | Dia / Dib | SLC4A1 | 17q21.31 | DI*01 | DI*02 (wt) | 2561 | NM_000342.3 | T | C | Leu854Pro | rs2285644 | 2 | 1 | 2 | 1 | W1 | – |
| Wright | 010 | Wra / Wrb | SLC4A1 | 17q21.31 | DI*02.03 | DI*02 (wt) | 1972 | NM_000342.3 | A | G | Glu658Lys | rs75731670 | 2 | 1 | 1 | 1 | – | W2 |
| Cartwright | 011 | Yta / Ytb | ACHE | 7q22.1 | YT*01 (wt) | YT*02 | 1057 | NM_001302621.1 | C | A | His353Asn | rs1799805 | 2 | 1 | 2 | 1 | W1 | – |
| Scianna | 013 | SC1, SC2 | ERMAP | 1p34.2 | SC*01 (wt) | SC*02 | 169 | NM_001017922.1 | G | A | Gly57Arg | rs56025238 | 2 | 1 | 2 | 1 | – | W2 |
| Dombrock | 014 | Doa / Dob | ART4 | 12p12.3 | DO*01 | DO*02 (wt) | 793 | NM_021071.2 | A | G | Asn265Asp | rs11276 | 2 | – | 2 | 1 | – | W2 |
| Dombrock | 014 | Hy+ / Hy- | ART4 | 12p12.3 | DO*02.–04 | DO*02 (wt) | 323 | NM_021071.2 | T | G | Val108Gly | rs28362797 | 2 | 1 | 1 | 1 | W1 | – |
| Dombrock | 014 | Jo(a+) / Jo(a–) | ART4 | 12p12.3 | DO*01.–05 | DO*02 (wt) | 350 | NM_021071.2 | T | C | Ile117Thr | rs28362798 | 2 | 1 | 1 | 1 | – | W2 |
| Colton | 015 | Coa / Cob | AQP1 | 7p14.3 | CO*01.01 (wt) | CO*02 | 134 | NM_198098.2 | C | T | Ala45Val | rs28362692 | 2 | 1 | 2 | 1 | W1 | – |
| Landst.-Wien. | 016 | LWa / LWb | ICAM-4 | 19p13.2 | LW*05 (wt) | LW*07 | 299 | NM_001544.4 | A | G | Gln100Arg | rs77493670 | 2 | 1 | 2 | 1 | W1 | – |
| Cromer | 021 | Cra / Cra- | CD55 | 1q32.2 | CROM*–01 | CROM*01 (wt) | 679 | NM_000574.3 | C | G | Pro227Ala | rs60822373 | 2 | 1 | 2 | 1 | W1 | – |
| Cromer | 021 | Tca / Tcb | CD55 | 1q32.2 | CROM*01.03 | CROM*01 (wt) | 155 | NM_000574.3 | T | G | Leu52Arg | rs28371588 | 2 | 1 | 1 | 1 | W1 | – |
| Cromer | 021 | Tca / Tcc | CD55 | 1q32.2 | CROM*01.04 | CROM*01 (wt) | 155 | NM_000574.3 | C | G | Pro52Arg | rs28371588 | 1 | 1 | 1 | – | W1 | – |
| Knops | 022 | Kna / Knb | CR1 | 1q32.2 | KN*01 (wt) | KN*02 | 4681 | NM_000573.3 | G | A | Val1561Met | rs41274768 | 2 | 1 | 2 | 1 | W1 | – |
| Knops | 022 | McCa / McCb | CR1 | 1q32.2 | KN*01.06 | KN*01 (wt) | 4768 | NM_000573.3 | G | A | Glu1590Lys | rs17047660 | 2 | 1 | 1 | 1 | W1 | – |
| Knops | 022 | Vil+ / Vil– | CR1 | 1q32.2 | KN*01.07 | KN*01 (wt) | 4801 | NM_000573.3 | G | A | Gly1601Arg | rs17047661 | 2 | 1 | 1 | 1 | – | W2 |
| Indian | 023 | Ina / Inb | CD44 | 11p13 | IN*01 | IN*02 (wt) | 137 | NM_001001391.1 | C | G | Pro46Arg | rs369473842 | 2 | 1 | 2 | 1 | W1 | – |
| Vel | 034 | Vel+ / Vel- | SMIM1 | 1p36.32 | VEL*–01 | VEL*01 | c.64–80 del | NM_001163724.2 | del 17 bp | - | Ser22Glnfs* | rs566629828 | 2 | 1 | 2 | 1 | – | W2 |
| Gender | n.a. | female/male | GYG2/paralog | Xp22.33/Yp11.2 | GYG2*Xfemale | GYGpar*Ymale | i2+3291 | NG_021257.1 | ||||||||||
| (female)+ | ||||||||||||||||||
| AC002992.1 (male)+ | C | A | – | no rs | – | – | 2 | 1 | W1 | – | ||||||||
| Gender | n.a. | female/male | AMEL/paralog | Xp22.2/Yp11.2 | AMEL*Xfemale | AMEL*Ymale | i3–7/i4–140 | NG_012040.1 | ||||||||||
| (female)+ | ||||||||||||||||||
| NG_008011.1 (male)+ | T | C | – | no rs | – | – | 2 | 1 | – | W2 | ||||||||
| SUM excluding SNPs for gender determination | 50 | 22 | 40 | 26 | ||||||||||||||
CDS =coding sequence, + genomic sequences.
Material and Methods
Samples
Between 2012 and 2014, samples from 37,253 blood donors were collected at different sites throughout Switzerland. Donor samples were provided by 11 regional blood transfusion services (BTSs) headed in Geneva (n = 1,348), Lausanne (n = 1,526), Neuchâtel (n = 1,029), Sion (n = 760), Basel (n = 1,222), Aarau (n = 855), Luzern (n = 2,770), Zurich (n = 24,058), Lugano (n = 768), St. Gallen (n = 1,476), and Chur (n = 1,441). In 2012, blood donations were collected by 13 independent BTSs, most of them covering more than their administrative-regional area, also known as ‘cantons’. As a consequence, respective inhabitant numbers and deduced ‘coverage’ of every BTS given need to be considered as approximate (table 2). The ethical approval of the study was waived by the ethical committee of the Canton of Zurich, and all donors explicitly permitted genetic laboratory investigations by written consent.
Table 2.
Geographic organisation of the 13 Blood Transfusion Services (BTSs) of Switzerland in 2015. Demographic data on Swiss cantons [23]; their BTSs and headquarter locations; origin and number of blood donor samples, sampling coverage of Swiss population
| Canton Name | Canton | BTS | Number of | Coverage per | Donor | % of Swiss | % of coverage |
|---|---|---|---|---|---|---|---|
| (local language) | abbreviation | headquarter | Inhabitants | BTS | tested | ||
| Genève | GE | Geneva | 484,736 | 484,736 | 1,348 | 5.82 | 6.96 |
| Vaud | VD | Lausanne | 773,407 | 773,407 | 1,5260 | 9.29 | 11.10 |
| Neuenburg/Neuchâtel | NE | Neuchâtel | 178,107 | 250,889 | 1,0290 | 3.01 | 3.60 |
| Jura | JU | 72,782 | |||||
| Wallis/Valais | VS | Sion | 335,696 | 335,696 | 760 | 4.03 | 4.82 |
| Basel-Stadt | BS | Basel | 191,817 | 475,048 | 1,222 | 5.70 | 6.82 |
| Basel-Landschaft | BL | 283,231 | |||||
| Aargau | AG | Aarau | 653,675 | 920,093 | 855 | 11.05 | 13.21 |
| Solothurn | SO | 266,418 | |||||
| Luzern | LU | Luzern | 398,762 | 600,392 | 2,770 | 7.21 | 8.62 |
| Obwalden | OW | 37,076 | |||||
| Nidwalden | NW | 42,420 | |||||
| Zug | ZG | 122,134 | |||||
| Zürich | ZH | Zurich | 1,466,424 | 1,967,782 | 24,058 | 23.63 | 28.25 |
| Schwyz | SZ | 154,093 | |||||
| Schaffhausen | SH | 79,836 | |||||
| Thurgau | TG | 267,429 | |||||
| Ticino | TI | Lugano | 351,946 | 351,946 | 768 | 4.23 | 5.05 |
| St. Gallen | SG | St. Gallen | 499,065 | 569,582 | 1,476 | 6.84 | 8.18 |
| Appenzell Ausserrhoden | AR | 54,543 | |||||
| Appenzell Innerrhoden | AI | 15,974 | |||||
| Graubünden | GR | Chur | 196,610 | 236,638 | 1,441 | 2.84 | 3.40 |
| Glarus | GL | 40,028 | |||||
| Swiss areas covered | 6,966,209 | 6,966,209 | 37,253 | 83.66 | 100.00 | ||
| Freiburg/Fribourg | FR | 307,461 | 307,461 | 0 | 3.69 | ||
| Bern | BE | 1,017,483 | 1,053,456 | 0 | 12.22 | ||
| Uri | UR | 35,973 | 0 | 0.43 | |||
| Swiss areas uncovered | 1,360,917 | 1,360,917 | 0 | 16.34% | |||
| Total Switzerland | 8,327,126 | 8,327,126 | 37,253 | 100.00% | |||
Blood Group Polymorphisms, SNPs, Analyzed by MALDI-TOF MS ‘RARE Module'
For automated DNA extraction, magnetic bead technology was used (Chemagen; Perkin Elmer, Baesweiler, Germany). Assay design for all SNPs (table 1), quality control of the primer mixes, and MALDI-TOF MS-based genotyping was done as described previously [5].
Prior to implementation of the ‘RARE module’ into routine use, molecular typing performance was validated by assessing a panel of 95 natural and artificial, reference DNAs representing all blood group specificities of the module. Additionally, every individual typing batch was controlled for specificity, using the identical reference DNA panel. Reference DNAs representing the blood group phenotypes, Lu(a+b-), KK, Kp(a+b-) and Yt(a-b+), and heterozygous phenotypes representative of Lu(08+14+), Au(a+b+), Js(a+b+), KEL(11+ 7+) and Do(a+b+) were provided by BTS Zurich [5,14,15,16]. Sample material of individuals with phenotypes Di(a+b+), Wr(a+b+), Co(a-b+), and Kn(a-b+) was given by Susanne Kilga-Nogler (Zentralinstitut für Bluttransfusion und Immunologische Abteilung, Innsbruck, Austria). Indian, In(a+b-), reference DNAs were provided by Joyce Pool (International Blood Group Reference Laboratory, NHS Blood and Transplant, Bristol, UK). Sample material of Vel- individuals was provided by Christof Jungbauer (Austrian Red Cross, Blood Service for Vienna, Lower Austria and Burgenland, Vienna, Austria).
Artificially synthesized control DNAs were used in cases where natural blood group DNA was not available and were generated by standard PCRs using one mutated and one regular amplification primer each, in order to cover the respective polymorphic SNPs. Artificial DNA fragments were generated for the following LFAs and their antithetical HFA partners: SC2 (SC*02), LWb (LW*07), Hy- (DO*02.-04), Jo(a-) (DO*01.-05), McCb (KN*01.06), Vil+ (KN*01.07), and all Cromer antigens [17,18,19,20,21]. Before use, artificial DNA fragments were titrated to equimolar copy concentrations as found in genomic DNA extracts from donor samples before validation.
Provision of Diagnostic Anti-Sera
Rare diagnostic anti-sera for serotyping by standard techniques, such as anti-Dib, anti-Coa, anti-LWa, and anti-Jsa, were made available by exchange programs or were provided by sources as described previously [5,22].
Data Sources, Statistical Methods, and Allele Frequency Calculation
Population data of Switzerland was taken from the report published annually by the Schweizerische Eidgenossenschaft, ‘Bundesamt für Statistik’ (table 2) [23].
Absolute allele frequencies were calculated by direct allele counting (table 3) according to Hardy-Weinberg proportions for all samples originating from the greater area covered by BTS Zurich and are given as ‘minor allele frequencies’ (maf) [24]. Allele frequencies of individual BTSs were calculated by direct allele counting as described above and, in order to give an averaged ‘Swiss allele frequency’, statistically corrected (weighted) according to the number of inhabitants of the areas covered by the respective BTSs (tables 2, 4). Additionally, for each allele, the mean, standard deviation (SD) and coefficient of correlation (CV) across all cantons is given (supplementary table 1; available at http://content.karger.com/ProdukteDB/produkte.asp?doi=490714). The coefficient of correlation is calculated by division of the SD of the mean and is a measure of dispersion that is independent of the scale. We also checked for correlation between sample size and MAF using the rank correlation coefficient by Spearman [25].
Table 3.
Genotyping results for ISBT 005 (Lutheran) to ISBT 013 (Scianna) and for ISBT 014 (Dombrock) to ISBT 034 (Vel)a
| Blood group system |
|||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lutheran | Lutheran | Lutheran | Kell | Kell | Kell | Kell | Diego | Wright | Cartwright | Scianna | Dombrock | Dombrock | Dombrock | Colton | Landst.-Wiener | Cromer | Cromer | Cromer | Knops | Knops | Knops | Indian | Vel | ||||||||||||||||||||||||
| ISBT number | 005 | 005 | 005 | 006 | 006 | 006 | 006 | 010 | 010 | 011 | 013 | 014 | 014 | 014 | 015 | 016 | 021 | 021 | 021 | 022 | 022 | 022 | 023 | 034 | |||||||||||||||||||||||
| Frequent | Lub | Lu8 | Aub | k | Kpb | K11 | Jsb | Dib | Wrb | Yta | SC:1 | Dob | Hy+ | Jo(a+) | Coa | LWa | Cra | Tca | Kna | McCa | Vil- | Inb | Vel+ | ||||||||||||||||||||||||
| antigen | |||||||||||||||||||||||||||||||||||||||||||||||
| Rare antigen | Lua | Lu14 | Aua | K | Kpa | K17 | Jsa | Dia | Wra | Ytb | SC:2 | Doa | Hy- | Jo(a-) | Cob | LWb | Cra- | Tcb | Tcc | Knb | McCb | Vil+ | Ina | Vel- | |||||||||||||||||||||||
| Geneva | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 1,237 | 1,304 | 623 | 1,212 | 1,313 | 1,313 | 1,313 | 1,318 | 1,294 | 1,163 | 1,319 | 507 | 1,323 | 1,318 | 1,239 | 1,318 | 1,302 | 1,318 | 1,268 | 1,311 | 1,296 | 1,307 | 1,297 | ||||||||||||||||||||||||
| Aa | 78 | 16 | 575 | 106 | 16 | 6 | 6 | 3 | 3 | 151 | 12 | 623 | 2 | 3 | 80 | 3 | 1 | 1 | 2 | 53 | 8 | 29 | 0 | 33 | |||||||||||||||||||||||
| aa | 1 | 0 | 132 | 1 | 1 | 0 | 0 | 0 | 0 | 7 | 0 | 201 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 6 | 0 | 1 | |||||||||||||||||||||||
| invalid | 32 | 28 | 18 | 29 | 18 | 29 | 29 | 27 | 51 | 27 | 17 | 17 | 23 | 27 | 28 | 27 | 45 | 27 | 27 | 27 | 17 | 41 | 17 | ||||||||||||||||||||||||
| Lausanne | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 1,397 | 1,451 | 721 | 1,392 | 1,478 | 1,487 | 1,486 | 1,493 | 1,502 | 1,324 | 1,495 | 533 | 1,510 | 1,478 | 1,390 | 1,491 | 1,407 | 1,489 | 1,421 | 1,471 | 1,487 | 1,490 | 1,459 | ||||||||||||||||||||||||
| Aa | 83 | 40 | 649 | 101 | 32 | 6 | 6 | 0 | 0 | 162 | 15 | 735 | 1 | 2 | 100 | 2 | 2 | 0 | 3 | 70 | 8 | 21 | 2 | 52 | |||||||||||||||||||||||
| aa | 1 | 0 | 138 | 0 | 1 | 0 | 0 | 0 | 0 | 5 | 0 | 242 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 0 | |||||||||||||||||||||||
| invalid | 45 | 35 | 18 | 33 | 15 | 33 | 34 | 33 | 24 | 35 | 16 | 16 | 15 | 46 | 33 | 33 | 117 | 34 | 34 | 47 | 15 | 34 | 15 | ||||||||||||||||||||||||
| Neuchâtel | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 957 | 1,002 | 502 | 957 | 1,002 | 1,026 | 1,026 | 1,028 | 999 | 909 | 1,020 | 361 | 1,028 | 1,027 | 956 | 1,024 | 1,027 | 1,026 | 972 | 1,024 | 1,018 | 1,023 | 1,002 | ||||||||||||||||||||||||
| Aa | 69 | 25 | 416 | 69 | 27 | 2 | 2 | 0 | 0 | 116 | 8 | 499 | 1 | 0 | 68 | 4 | 0 | 0 | 1 | 55 | 4 | 10 | 1 | 27 | |||||||||||||||||||||||
| aa | 0 | 105 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 162 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | ||||||||||||||||||||||||
| invalid | 3 | 1 | 6 | 1 | 0 | 1 | 1 | 1 | 30 | 3 | 1 | 7 | 0 | 2 | 1 | 1 | 2 | 2 | 1 | 1 | 0 | 5 | 0 | ||||||||||||||||||||||||
| Sion | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 685 | 732 | 376 | 692 | 742 | 745 | 744 | 746 | 755 | 659 | 748 | 275 | 755 | 742 | 699 | 741 | 716 | 744 | 723 | 742 | 752 | 746 | 755 | ||||||||||||||||||||||||
| Aa | 53 | 13 | 313 | 50 | 13 | 1 | 2 | 0 | 0 | 85 | 7 | 375 | 0 | 0 | 46 | 5 | 0 | 0 | 1 | 23 | 0 | 3 | 0 | 0 | |||||||||||||||||||||||
| aa | 1 | 1 | 66 | 2 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 104 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 21 | 14 | 5 | 16 | 5 | 14 | 14 | 14 | 5 | 14 | 5 | 6 | 5 | 18 | 14 | 14 | 44 | 15 | 14 | 18 | 5 | 14 | 5 | ||||||||||||||||||||||||
| Basel | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 982 | 1,057 | 516 | 997 | 1,033 | 1,064 | 1,060 | 1,080 | 1,027 | 955 | 482 | 379 | 1,047 | 1,064 | 1,004 | 1,073 | 1,007 | 1,073 | 1,024 | 1,063 | 1,047 | 1,079 | 475 | ||||||||||||||||||||||||
| Aa | 82 | 22 | 426 | 69 | 18 | 3 | 1 | 0 | 0 | 122 | 1 | 522 | 0 | 0 | 75 | 7 | 0 | 0 | 4 | 56 | 2 | 9 | 0 | 9 | |||||||||||||||||||||||
| aa | 0 | 0 | 100 | 3 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 154 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 158 | 143 | 180 | 153 | 171 | 155 | 161 | 142 | 195 | 142 | 739 | 167 | 175 | 158 | 142 | 142 | 215 | 145 | 142 | 157 | 166 | 143 | 738 | ||||||||||||||||||||||||
| Aarau | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 790 | 841 | 411 | 781 | 833 | 844 | 844 | 849 | 854 | 758 | 854 | 288 | 855 | 850 | 776 | 843 | 850 | 849 | 792 | 849 | 854 | 846 | 817 | ||||||||||||||||||||||||
| Aa | 60 | 9 | 368 | 68 | 22 | 6 | 6 | 1 | 1 | 91 | 1 | 417 | 0 | 0 | 69 | 7 | 0 | 0 | 1 | 57 | 1 | 1 | 0 | 38 | |||||||||||||||||||||||
| aa | 0 | 0 | 76 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 150 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 5 | 5 | 0 | 5 | 0 | 5 | 5 | 5 | 0 | 4 | 0 | 0 | 0 | 5 | 8 | 5 | 5 | 5 | 5 | 5 | 0 | 9 | 0 | ||||||||||||||||||||||||
| Luzern | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 2,514 | 2,661 | 1,333 | 2,495 | 2,664 | 2,697 | 2,712 | 2,716 | 2,710 | 2,446 | 898 | 975 | 2,724 | 2,719 | 2,494 | 2,701 | 2,715 | 2,717 | 2,535 | 2,719 | 2,728 | 2,708 | 876 | ||||||||||||||||||||||||
| Aa | 197 | 58 | 1,147 | 222 | 72 | 24 | 8 | 1 | 2 | 261 | 4 | 1,306 | 4 | 1 | 219 | 19 | 0 | 0 | 0 | 184 | 1 | 8 | 1 | 26 | |||||||||||||||||||||||
| aa | 6 | 0 | 252 | 4 | 1 | 0 | 0 | 0 | 0 | 12 | 0 | 456 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 53 | 51 | 38 | 49 | 33 | 49 | 50 | 53 | 58 | 51 | 1,868 | 33 | 42 | 50 | 54 | 50 | 55 | 53 | 50 | 50 | 34 | 61 | 1,868 | ||||||||||||||||||||||||
| Zurich | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 22,013 | 23,287 | 11,766 | 21,953 | 23,237 | 23,628 | 23,591 | 23,698 | 23,120 | 21,075 | 14,681 | 8,451 | 23,717 | 23,713 | 21,971 | 23,536 | 23,630 | 23,677 | 22,459 | 23,684 | 23,680 | 23,702 | 14,753 | ||||||||||||||||||||||||
| Aa | 1,678 | 440 | 9,773 | 1,693 | 527 | 66 | 74 | 36 | 9 | 2,567 | 98 | 11,427 | 7 | 13 | 1,705 | 201 | 14 | 4 | 44 | 1,259 | 36 | 115 | 3 | 26 | |||||||||||||||||||||||
| aa | 29 | 5 | 2,207 | 50 | 4 | 0 | 0 | 1 | 0 | 92 | 0 | 3,911 | 0 | 0 | 31 | 1 | 0 | 0 | 0 | 18 | 2 | 6 | 0 | 3 | |||||||||||||||||||||||
| invalid | 338 | 326 | 312 | 362 | 290 | 364 | 393 | 323 | 929 | 324 | 9279 | 269 | 334 | 332 | 351 | 320 | 414 | 333 | 322 | 336 | 257 | 353 | 9,276 | ||||||||||||||||||||||||
| Lugano | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 718 | 745 | 322 | 698 | 749 | 758 | 754 | 753 | 728 | 674 | 380 | 269 | 762 | 760 | 691 | 757 | 753 | 756 | 726 | 759 | 759 | 758 | 382 | ||||||||||||||||||||||||
| Aa | 43 | 12 | 335 | 56 | 12 | 0 | 1 | 0 | 1 | 80 | 3 | 382 | 0 | 0 | 61 | 4 | 0 | 0 | 0 | 35 | 1 | 4 | 0 | 0 | |||||||||||||||||||||||
| aa | 0 | 0 | 105 | 4 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 112 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |||||||||||||||||||||||
| invalid | 7 | 11 | 6 | 10 | 7 | 10 | 13 | 15 | 39 | 8 | 385 | 5 | 6 | 8 | 14 | 7 | 15 | 12 | 7 | 8 | 5 | 10 | 385 | ||||||||||||||||||||||||
| St. Gallen | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 1,361 | 1,435 | 745 | 1,349 | 1,444 | 1,460 | 1,453 | 1,460 | 1,435 | 1,297 | 1,458 | 491 | 1,447 | 1,462 | 1,347 | 1,450 | 1,461 | 1,460 | 1,395 | 1,462 | 1,458 | 1,454 | 1,404 | ||||||||||||||||||||||||
| Aa | 99 | 27 | 596 | 113 | 22 | 1 | 8 | 1 | 0 | 158 | 7 | 724 | 0 | 0 | 113 | 12 | 1 | 0 | 1 | 65 | 0 | 7 | 0 | 60 | |||||||||||||||||||||||
| aa | 2 | 0 | 124 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 249 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 14 | 14 | 11 | 14 | 10 | 15 | 15 | 15 | 41 | 15 | 11 | 12 | 29 | 14 | 14 | 14 | 14 | 15 | 15 | 14 | 11 | 22 | 12 | ||||||||||||||||||||||||
| Chur | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 1,343 | 1,398 | 636 | 1,301 | 1,396 | 1,411 | 1,419 | 1,422 | 1,421 | 1,249 | 393 | 516 | 1,426 | 1,422 | 1,318 | 1,415 | 1,420 | 1,417 | 1,360 | 1,421 | 1,421 | 1,421 | 395 | ||||||||||||||||||||||||
| Aa | 76 | 24 | 628 | 119 | 33 | 12 | 3 | 0 | 0 | 166 | 2 | 665 | 3 | 0 | 100 | 7 | 1 | 2 | 1 | 60 | 0 | 9 | 0 | 0 | |||||||||||||||||||||||
| aa | 2 | 0 | 161 | 1 | 0 | 0 | 0 | 0 | 0 | 7 | 0 | 247 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | |||||||||||||||||||||||
| invalid | 20 | 19 | 16 | 20 | 12 | 18 | 19 | 19 | 20 | 19 | 1,046 | 13 | 12 | 19 | 21 | 19 | 20 | 21 | 19 | 20 | 11 | 20 | 1,046 | ||||||||||||||||||||||||
| Total | |||||||||||||||||||||||||||||||||||||||||||||||
| AA | 33,997 | 35,913 | 17,951 | 33,827 | 35,891 | 36,433 | 36,402 | 36,563 | 35,845 | 32,509 | 23,728 | 13,045 | 36,594 | 36,555 | 33,885 | 36,349 | 36,288 | 36,526 | 34,675 | 36,505 | 36,500 | 36,534 | 23,615 | ||||||||||||||||||||||||
| Aa | 2,518 | 686 | 15,226 | 2,666 | 794 | 127 | 117 | 42 | 16 | 3,959 | 158 | 17,675 | 18 | 19 | 2,636 | 271 | 19 | 7 | 58 | 1,917 | 61 | 216 | 7 | 271 | |||||||||||||||||||||||
| aa | 42 | 7 | 3466 | 68 | 7 | 0 | 0 | 1 | 0 | 143 | 0 | 5988 | 0 | 0 | 52 | 1 | 0 | 0 | 0 | 25 | 4 | 16 | 0 | 5 | |||||||||||||||||||||||
| invalid | 696 | 647 | 610 | 692 | 561 | 693 | 734 | 647 | 1,392 | 642 | 13,367 | 545 | 641 | 679 | 680 | 632 | 946 | 662 | 636 | 683 | 521 | 712 | 13,362 | ||||||||||||||||||||||||
| % Invalid | 1.87 | 1.74 | 1.64% | 1.86 | 1.51 | 1.86 | 1.97 | 1.74% | 3.74 | 1.72 | n.a. | 1.46 | 1.72 | 1.82 | 1.83 | 1.70 | 2.54 | 1.78 | 1.71 | 1.83 | 1.4% | 1.9% | n.a. | ||||||||||||||||||||||||
| Overall | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | 37,253 | ||||||||||||||||||||||||
Top to bottom: Numbers of frequent homo- (AA), hetero- (Aa), and rare homozygotes(aa) and invalid typings for each BTS, identified by their respective head quarter locations, ordered according to geographical latitude from West to East. Left to right: antigens tested, ordered according to the ISBT number of the respective blood group system. Donors with rare blood group phenotypes are underlayed in gray (excluding blood group system Knops) and are summing up to a total of 326 (line ‘rare homo’ of total numbers). Only 23,886 and 23,848 blood donors, were analyzed for Scianna rs56025238 and Vel rs566629828, respectively. Donor samples not tested for these antigens are indicated as n.t.
Table 4.
Allele frequency data given for the minor allele of each SNP as minor allele frequency (MAF) for the greater area of Zurich or Switzerlanda
| Blood group system |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lutheran | Lutheran | Lutheran | Kell | Kell | Kell | Kell | Diego | Wright | Cartwright | Scianna | Dombrock | |
| Rare allele | LU*01 | LU*02.14 | LU*02.19 | KEL*01 | KEL*02.03 | KEL*02.17 | KEL*02.06 | DI*01 | DI*02.03 | YT*02 | SC* 02 | DO*01 |
| c.nt position | 230 | 611 | 1615 | 578 | 841 | 905 | 1790 | 2561 | 1972 | 1057 | 169 | 793 |
| Rare nt. | A | A | G | T | T | C | C | T | A | A | A | A |
| rs number | rs28399653 | rs28399656 | rs1135062 | rs8176058 | rs8176059 | rs61729034 | rs8176038 | rs2285644 | rs75731670 | rs1799805 | rs56025238 | rs11276 |
| MAF Zurich only | 0.03659 | 0.00948 | 0.29872 | 0.03783 | 0.01125 | 0.00139 | 0.00156 | 0.00080 | 0.00019 | 0.05795 | 0.00332 | 0.40458 |
| MAF Swiss covered (weighted) | 0.03451 | 0.00930 | 0.30523 | 0.03853 | 0.01064 | 0.00196 | 0.00184 | 0.00043 | 0.00028 | 0.05796 | 0.00298 | 0.40412 |
| Delta % ZH vs. Swiss | 6 | 2 | –3 | –2 | 6 | –29 | –15 | 88 | –30 | 0 | 11 | 0 |
| Blood group system |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dombrock | Dombrock | Colton | Landst. Wiener | Cromer | Cromer | Cromer | Knops | Knops | Knops | Indian | Vel | |
| Rare allele | DO*02.–04 | DO*01.–05 | CO*02 | LW*07 | CROM*–01 | CROM*01.03 | CROM*01.04 | KN*02 | KN*01.06 | KN*01.07 | IN*01 | VEL*–01 |
| c.nt position | 323 | 350 | 134 | 299 | 679 | 155 | 155 | 4681 | 4768 | 4801 | 137 | c.64–80del |
| Rare nt. | T | T | T | G | C | T | C | A | G | G | G | del 17 bp |
| rs number | rs28362798 | n.a. | rs28362692 | rs77493670 | rs60822373 | rs28371588 | rs28371588 | rs41274768 | rs17047660 | rs17047661 | rs369473842 | rs566629828 |
| MAF Zurich only | 0.00023 | 0.00027 | 0.03727 | 0.00428 | 0.00030 | 0.00008 | 0.00093 | 0.02728 | 0.00084 | 0.00267 | 0.00006 | 0.00108 |
| MAF Swiss covered (weighted) | 0.00025 | 0.00025 | 0.03790 | 0.00321 | 0.00023 | 0.00007 | 0.00072 | 0.02662 | 0.00112 | 0.00405 | 0.00013 | 0.01022 |
| Delta % ZH vs. Swiss | –6 | 11% | -2% | 33% | 29% | 28% | 29% | 2% | -25% | -34% | -50% | -89% |
Left to right: antigens investigated, ordered according to the ISBT number of the respective blood group system.
To visualize overall similarities in relative blood group frequencies among BTSs, we performed a principal coordinate analysis (PCoA) on the relative blood frequencies using the R-package ape 5.0. Cantons with comparable blood group frequency will cluster closely together in this analysis, whereas those with unequal blood group frequency will spread apart. In population genetics, a measure of population structure is the fixation index (here FST[26]) that is usually calculated on SNPs or microsatellite data. FST can take a value between 0 and 1. In simplified terms, the smaller the FST the more similar the genetic background and vice versa. Here we use FST to elucidate population substructures based on blood group antigens. The PCoA was performed on the minor blood group alleles having a frequency of >0.1% across all cantons, using an Eucledian distance measure, and on FST using it as distance measure. Since we used the FST as distance measure for the second PCoA, we calculated the variance explained by the first two PCoA components across only the positive eigenvalues. To visualize pairwise (comparison of each canton to the rest) FST values, we plotted a heatmap using the gplots package 3.0.1 of R. In short, a heatmap plots the differences of the input values using different color intensities. Dendrograms are constructed using hierarchical clustering to depict overall similarities of features using the complete linkage algorithm [27]. We also performed pairwise Fisher's exact tests for each blood group on the contingency tables listing the absolute frequencies of the respective homozygous and heterozygous antigen counts for the analyzed cantons. We adjusted for the sample size of the different panels as described (supplementary table 2; available at http://content.karger.com/ProdukteDB/produkte.asp?doi=490714) [28]. Moreover, we adjusted for multiple testing using the correction proposed by Benjamini and Hochberg [29].
Results
MALDI-TOF MS-Based ‘RARE Module’
The ‘RARE module’ consisted of two multiplex reactions, comprising a total of 26 biallelic or triallelic SNP assays for the simultaneous analysis of 13 blood group genes and 40 of their alleles, representing a total of 50 blood group antigens, most of them described as HFAs or LFAs (table 1). Genotyping for Scianna (SC1/SC2, rs56025238) and Vel (Vel+/Vel-, rs566629828) only became available in a later revised version of the RARE module and after the description of the genetic background of Vel negativity [30]. Therefore, only 23,886 and 23,848 blood donors were analyzed for Scianna rs56025238 and Vel rs566629828, respectively. All other SNPs were analyzed on 37,253 DNA samples. Approximately two-thirds of the analyzed blood donors (n = 24,058) were from the greater area of Zurich, whereas the other third (n = 13,195) was provided by the other 10 BTSs (fig. 1, table 2). Approximately 84% of the Swiss population, e.g. 6.966 of the total 8.327 million inhabitants are covered by all 11 BTSs. Therefore, roughly 1 of every 200, or 37,253 of 8.327 million Swiss individuals were assessed by our approach.
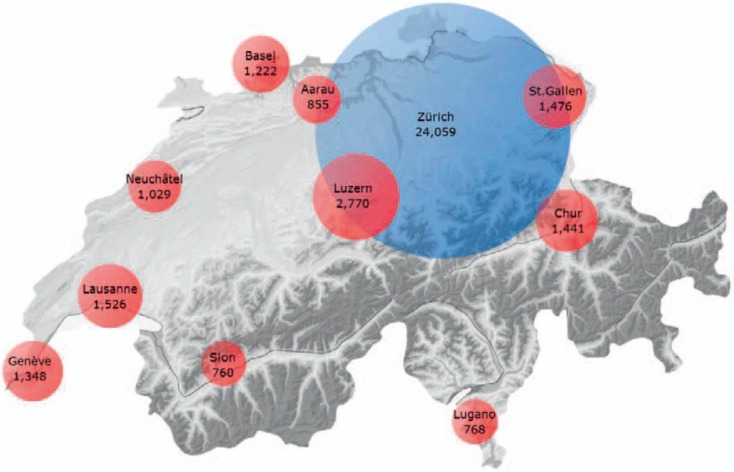
Fig. 1.
Origin of blood donor samples and BTSs, identified by their headquarter locations (local language), participating in this project. The respective head quarter locations are given with their approximate geographical location and in their local languages. The area of each circle is correspondent to the number of samples investigated for each BTS (also see table 2). Approximately two-thirds of the analyzed blood donors (n = 24,058) were from the greater area of Zurich (blue circle), whereas the other third (n = 13,195) were from another 10 different blood transfusion services distributed throughout Switzerland (red circles), summing up to a total of 37,253 individual blood donor samples investigated in the course of this project. Topographically, Swiss Alps are shown in dark grey. Lugano and Sion are located south and Chur within the Swiss Alps.
Both multiplexes included additional assays, one each for gender determination, and three additional assays for the specific detection of ABO SNPs, located at coding nucleotide positions 261, 802 and 803. The respective assays served as quality control measure, e.g. to link DNA samples to their available donor phenotype data, thereby allowing for the exclusion of serial sample mistake. Comparison of ABO geno- and serotyping will be published elsewhere (manuscript in preparation). Calling failures, caused by samples with either a negative result for all SNPs or for only a single SNP assay failure, were excluded from the finally analyzed data set and ranged between 1.40% and 3.73% per assay (average 1.87%, median 1.78%; table 3, bottom line).
Rare Blood Donors Negative for HFAs and Homozygous Positivity for LFAs
The identified individuals with rare blood group antigen constellations had genotypes, known to encode for the blood group phenotypes Lu(a+b-) (n = 42), Lu(8–14+) (n = 7), K+k- (n = 68), Kp(a+b-) (n = 7), Di(a+b-) (n = 1), Yt(a-b+) (n = 143), Co(a-b+) (n = 52), LW(a-b+) (n = 1), Vel- (n = 5), summing up to a total of n = 326 (table 3). Knops blood group antigens are defined by clinically insignificant antibodies, but are notoriously difficult to identify [2]. Therefore, individuals with predicted phenotypes Kn(a-b+) (n = 25), McC(a-b+) (n = 4), heterozygous for Vil+/- (n = 216), and homozygous for Vil+ (n = 16) are listed separately from the above. All donors beside those showing rare Knops phenotypes were reported to the Swiss Rare Donor File [31]. Successive samples of rare genotype carriers were used for serological confirmation of genotype. So far, the reinvestigated individuals with the following phenotypes were: Lu(a+b-) (n = 15 of 42), K+k- (n = 26 of 68), Kp(a+b-) (n = 2 of 7), Di(a+b-) (n = 1 of 1), Yt(a-b+) (n = 52 of 143), Co(a-b+) (n = 18 of 52).
Blood Group Allele Frequencies
Genotyping allowed for the identification of frequent and rare homozygous and heterozygous genotypes, the later in some cases not detectable by serologic testing. For instance, using standard serological methods, Vel positivity is undistinguishable in between VEL*01/ VEL*-01 heterozygotes and VEL*01/ VEL*01 homozygotes. On a molecular and statistical basis however, heterozygotes will be recognized and are much more frequent as compared to rare homozygotes. Therefore, genotyping data provided exact blood group allele frequency estimates. The MAF of all blood group SNPs from all BTSs are shown in supplementary table 3 (available at http://content.karger.com/ProdukteDB/produkte.asp? doi=490714). MAF data, separately calculated for the donor panel from the greater area of Zurich, and an average Swiss MAF, averaging data of all BTSs according to the number of inhabitants covered by the respective BTSs, are given in table 4.
The most common alleles are encoding the two public antigens of Doa and Dob (mean MAF of 0.40 (0.39, 0.42)) and Aua and Aub (MAF of 0.31 (0.29, 0.36)), followed by Ytb (MAF 0.059 (0.052, 0.063)) with borderline frequency and by K with a clear LFA value (MAF of 0.039 (0.0338, 0.043)),. Alleles with very low frequency are encoding Ina (MAF 0.0001, (0.0007)) and Cr(a-) (MAF 0.0002 (0.0007)). In general, differences in allele frequencies can be observed for all analyzed blood group SNPs. These differences are more pronounced for some blood groups than for others.
Looking at the very rare alleles (MAF < 0.1%), a comparably high variability of (CV > 0.8) across cantons can be observed which may be linked to the fact, that for these even a small changes of MAF has a higher impact on overall frequency. All of the following predicted antigens, Jo(a-), McCb, Wra, Hy-, Vel-, Tcc, Dia, are observed in some but not all cantons (table 3). For Vel+/Vel- we saw a correlation of R2 < 0.7 between sample size and MAF, which explains the observed frequency differences across the cantons by differences in sample sizes. To some extent, this is also true for KEL11/17 with a Spearman correlation coefficient of 0.6. The highest variability across the more frequent rare blood group antigens (MAF > 1%) is seen for Kpa and Knb followed by Lua.
Analysis of Inter-Cantonal Blood Group Variability
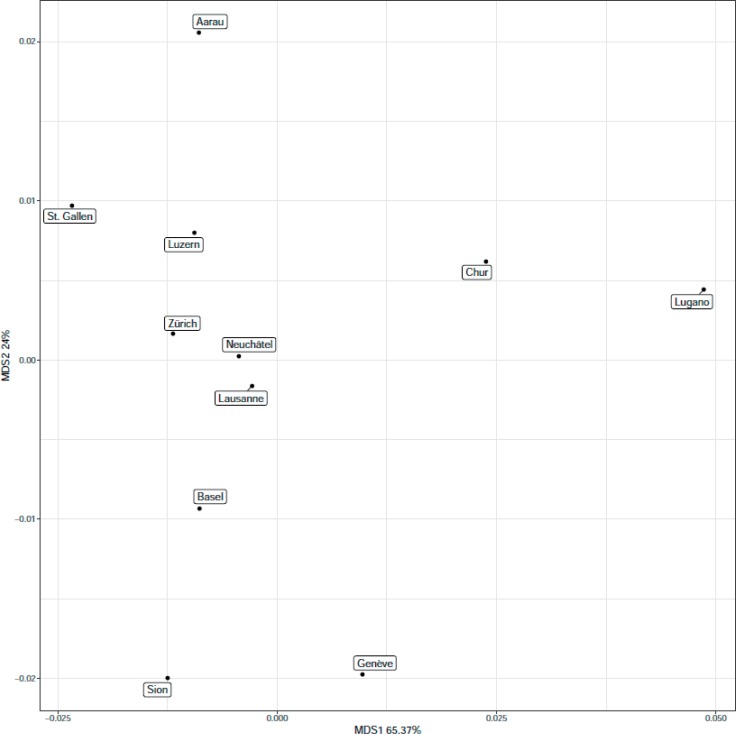
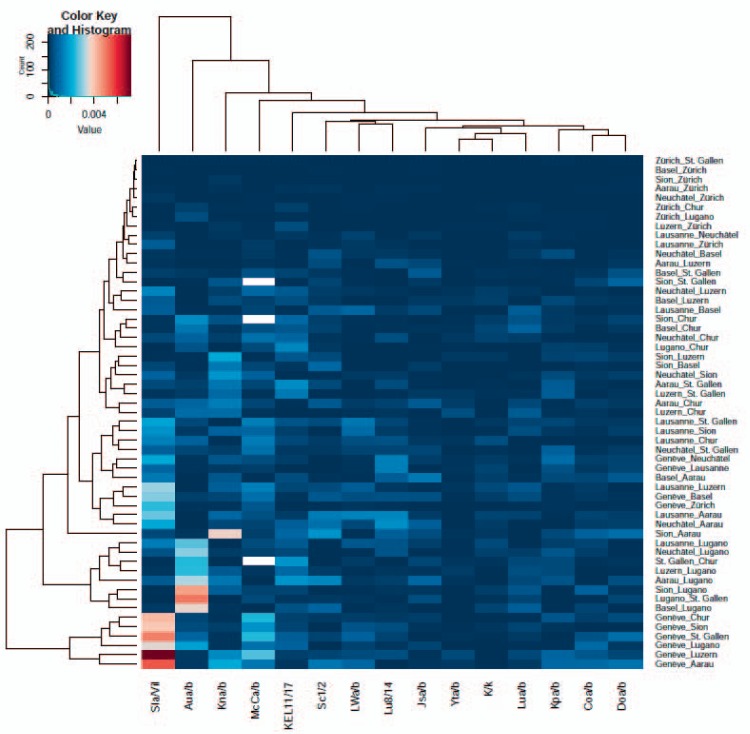
The PCoA analyses, both on the relative frequencies of the blood group antigens (fig. 2) and the FST, were performed to visualize frequency differences in the different cantons to facilitate their investigation (supplementary tables 4 and 5; available at http://content.karger.com/ProdukteDB/produkte.asp?doi=490714). The FST is used to determine population differences because of genetic differentiation and is usually applied to SNP data. Figure 2 shows the four outlying cantons TI (headed in Lugano), GE (Geneva), VS (Sion), and AG (Aarau) (for abbreviations of canton names, their areas covered and location of headquarters see table 2). It was performed on 13 blood group antigens with mean MAF of 0.1% across the cohorts, e.g. Lua, Lu14 and Aub, of the Lutheran system, K, Kpa, Jsa and Kel17 of the Kell system, Knb, McCb and Vil+ of the Knops system as well as on Ytb, Cob and Doa (Doa and Dob antigens have ‘public’ allele frequency). Figure 3 shows the values of the F-Statistics in a heatmap for the blood groups with rare antigen frequencies of >0.1%. Overall FST were very low, ranging from 0.79 × 10−5 for the comparison between BL and BS (Basel) and AG (Aarau) to 80.8 × 10−5 for the comparison between VS (Sion) and TI (Lugano), showing that genetic population structures, as was to be expected, are very similar between the cantons. ZH (Zurich), NE (Neuchâtel), and VD (Lausanne) clustered together with NE at the center, with mean FST of 4.8 × 10−5 and 10.7 × 10−5. On the outposts are Lugano with a mean FST of 58.42 × 10−5, Sion, Aarau, Chur and Geneva.
Fig. 2.
PcoA analysis of the MAF using only blood group antigens with a minimal allele frequency of 0.1% or higher, across all head quarter locations of the participating BTS. Genetic blood group profiles of samples collected from BTSs headquartered in Sion, Lugano, Aarau, Geneva, and Chur cluster far away from the other Swiss regions investigated. Except for Aarau, this overlaps well with local languages spoken and geographical profiles of the other headquarters representative of the respective cantons (table 2). All but Sion, Geneva and Lugano are located in a region with a German language profile (beside some areas of Graubünden, with its capital Chur, and its inhabitants, still cultivating Raetho-Romanic language). Additionally, Lugano, Chur and Sion are clearly separated from the other areas investigated by the Swiss Alps, reaching maximal altitudes of up to 4,634 m above sea level in Switzerland (fig. 1).
Fig. 3.
Heatmap of the pairwise F-Statistics FST across all cantons. Larger values indicate a higher degree of genetic variation within the comparison. The row dendrogram shows two main clusters with comparisons for Geneva and Lugano and a third broader cluster comprised of comparisons made to Geneva, Aarau, Chur, and Sion. The main differences are driven by Aua/b antigens, member of the Lutheran, and the Vil+/-, Kna/b and McCa/b antigens of the Knops blood group system.
The differences between the cantons based on the antigen frequencies (here always the rarer of the alleles is analyzed) were mainly driven by Aub (Geneva and Lugano), Knb, Vil+ (Geneva), and McCb (fig. 3). After sample size correction using pairwise Fisher Exact Test, Aub remains significantly associated with differences between the cantons (see supplementary table 6; available at http://content.karger.com/ProdukteDB/produkte.asp?doi=490714). For the Zurich samples, we had access to the ZIP codes of all donors. With this, we were able to classify them into probands living in extremely elevated (>1,500 m) and extremely low (<300 m) regions of the greater cantonal area of Zurich. For these samples, we tested for differences in antigen frequencies according to the altitude above sea level of the region of living with no significant results (data not shown). Of note, out of the total 24,058 samples analyzed, only 40 belonged to the ‘extreme height’ set.
Discussion
This country-wide search for rare blood group antigens was conducted to increase the pool of the Swiss rare blood donors and thus to improve the supply of rare blood units. In total 326 Swiss blood donors with rare or extremely rare blood group antigen constellations were identified. Thanks to this pool of rare blood donors, it will be possible to provide rare donor blood on demand, without the need for frozen stocks of respective erythrocytes. Once, the expected recognition of molecular specificities had been validated, the ‘mission’ could be accomplished without the need of commercially available typing sera, e.g. diagnostic antibodies, some of them directed against Aub, Dia, Cob, Doa, Jsa and SC2 notoriously being unobtainable [11,12].
Molecular blood group typing also allowed for correct identification of heterozygotes for all antigens investigated, thereby delivering exact frequency data. Frequencies of certain antigens differed pronouncedly among different regions of Switzerland. Based on these results, targeted searches for certain rare phenotypes focused on regions with expected higher occurrence of the respective rare antigens are possible in the future. For instance, individuals negative for Vel were to be expected once among 2,025 inhabitants covered by BTS of Aarau, in comparison to one Vel- among 146,689 inhabitants, expected by Hardy-Weinberg proportions, of the Italian part of Switzerland and represented by the BTS headed in Lugano [24]. Of course, it is expected, that the same local antigen frequencies will be observed in the local patients. Thereby, such frequencies have important clinical implications, on one hand with respect to the expected occurrence of the respective antigens among patients and on the other hand, and as an indirect result, with respect to the prevalence of respective antibodies directed against them.
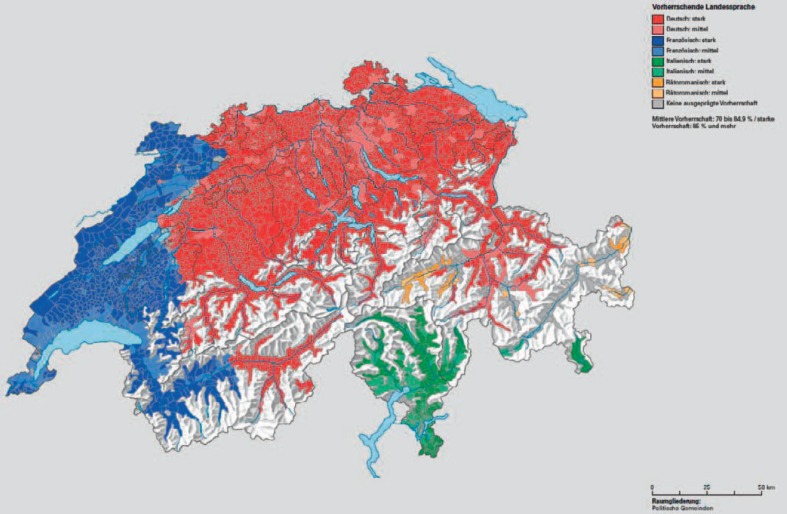
Previous genetic studies on the European population have revealed a substructure within Switzerland [32,33]. Using a genome-wide set of common markers, the existing ‘language clusters’ (fig. 4) were re-identified within Switzerland. These studies showed (again) that genetic distance varies with geographic distance and that language is an important barrier for reproduction. Geographically, TI (Lugano), and VS (Sion) are located south, whereas GR (Chur) lies within the massive mountain wall of the Swiss Alps (fig. 1). This ‘wall’ reaches maximal altitudes of up to 4634 m (peak Dufourspitze) above sea level. First inspections with respect to VEL*-01 allele prevalence, seemed to show that it is much rarer south than in the north of the Alps. However, the greater cantonal area of ZH (Zurich) north of the Alps proved this hypothesis statistically wrong, or alternatively, and purely speculative, suggested this region having predominantly been populated from the southern part of the Alps. Additionally, Switzerland also has a very distinct language profile with four spoken languages, French, German, Italian and Rhaeto-Romanic, also known as ‘Romansch’ (fig. 4) [34]. Of the analyzed cantons, GE, VD, NE, and some parts of JU and VS belong to the French, TI and some parts of GR to the Italian, and some parts mainly located in GR and partially in TI to the Rhaeto-Romanic speaking regions, whereas all other cantons are in regions where the generally spoken language is German. This language profile may also be reflected in the ethnic and genetic background of the local blood donors investigated. The cantons BS and BL with their capitol of Basel, for instance, are both located close to France but have a German language profile and cluster between the cantons with German and French language profiles. GR with its capitol Chur and TI with its capitol Lugano both share Italian and Rhaeto-Romanic influences and cluster closest together.
Fig. 4.
Languages of Switzerland [34].
However, frequency data need to be interpreted with caution. The rarer certain alleles are observed and the smaller respective donor panels are, the less reliable the frequency estimates might be calculated. For instance, the only Vel- allele carrier identified in the Italian-speaking part of Switzerland also typed phenotypically Vel- and was homozygous for VEL*-01/ VEL*-01. Thereby this individual represented a highly significant statistical outlier, without any further VEL*01/VEL*-01 heterozygotes among 383 other individuals investigated and donating blood in the area of Lugano. There is a chance that this individual is a retired Swiss citizen and ‘refugee’ from the cold winter climate of Aarau. Similarly, the only Di(a+b-) individual identified within all 37,235 donors investigated, turned out to be an immigrant from Peru.
Heterozygous SNP carriers may also represent an important resource for further scientific analysis of blood group antigen genetics. In heterozygotes, each mutation affecting the allelic expression would become directly visible on the phenotypic level. Previously, the underlying principle had been used to identify K₀ alleles among apparently KEL*01/KEL*02 heterozygotes, but with a discrepant K+k- phenotype [6]. Accordingly, among the total of 2,518 apparently LU*A/LU*B heterozygotes identified in the course of this study, 500 were reinvestigated by serology. All showed a congruent Lu(a+b+) phenotype, suggesting a low frequency of ill expressed Lutheran alleles within Switzerland (data not shown, manuscript in preparation). In addition, the data set allowed for new observations with respect to allelic Lutheran haplotypes, For example, among the samples investigated, six LU*A homozygous samples were identified which also were proven to be homozygous for LU*19. Sequence analysis proved the existence of this new LU allele, now being recognized by the ISBT as LU*01.19 [4,16].
The present study is an example for the magnitude of information delivered by applying high-throughput blood group genotyping. Gathered data provided new scientific insights into blood group genetics, completed allele frequency data for practical use, and delivered newly identified blood donors with rare and very rare antigen constellations, now available for the provision of rare donor blood.
Declaration of Financial Support
Financial support for this project was granted by the Humanitarian Foundation of the Swiss Red Cross (SRC, support: 47%), the Blood Transfusion Service Zurich, SRC (support: 33%), Switzerland, and with respect to funding, to smaller extents, by the Swiss blood transfusion umbrella organization Blutspende Schweiz, SRC, Bern, Switzerland, and Agena Bioscience GmbH, Hamburg, Germany. The presented technological approach represents a joint collaborative effort of the Blood Transfusion Service Zurich, and the company Agena Bioscience GmbH.
Author Contributions
N.T., K.N., Y.M., C.P., and S.S., performed experiments.
C.G., S.M., C.V., F.D., M.K., C.E, A.K. and J.G., performed experiments and analyzed data.
S. AelD., S.WA., C.T., JD.T., SM.M., A.S., JD.B., M.S., L.I., A. B., J.S., B.W., D.C., MC.B, J.T., S.H. and T.S., contributed essential material and collected data.
C.G., S.M., C.V., BM.F, F.D., M.K., A.F. discussed the results and commented on the manuscript.
C.G., S.M., C.V. and BM.F. designed the study.
C.G. and S.M. supervised the study.
C.G., F.D. and BM.F. wrote the manuscript.
C.G. and F.D made the tables and figures.
All authors revised and edited the manuscript.
Disclosure Statement
Christoph Gassner is an employee of the Blood Transfusion Service Zurich, SRC, and acts as a consultant for inno-train GmbH, Kronberg i.T., Germany. Caren Vollmert is employed at Agena Bioscience GmbH, Hamburg, Germany. All other authors do not disclose any competing interests.
Supplementary Material
Supplementary data
Christoph Gassner, Frauke Degenhardt, and Stefan Meyer contributed equally to this work.
References
- 1.Woodfield G, Poole J, Nance ST, Daniels G. A review of the ISBT rare blood donor program. Immunohematology. 2004;20:244–248. [PubMed] [Google Scholar]
- 2.Daniels G. 3rd ed. Oxford: Blackwell; 2013. Human Blood Groups. [Google Scholar]
- 3.Storry JR, Casthilho L, Chen Q, Daniels G, Denomme G, Flegel WA, et al. International society of blood transfusion working party on red cell immunogenetics and terminology: report of the Seoul and London meetings. ISBT Sci Ser. 2016;11:118–122. doi: 10.1111/voxs.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Society of Blood Transfusion Committee on Terminology for RBC Surface Antigens. 2018 www.isbtweb.org/working-parties/red-cell-immunogenetics-and-blood-group-terminology/blood-group-terminology/blood-group-allele-terminology/. (last accessed June 27, 2018) [Google Scholar]
- 5.Meyer S, Vollmert C, Trost N, Bronnimann C, Gottschalk J, Buser A, et al. High-throughput Kell, Kidd, and Duffy matrix-assisted laser desorption/ionization, time-of-flight mass spectrometry-based blood group genotyping of 4000 donors shows close to full concordance with serotyping and detects new alleles. Transfusion. 2014;54:3198–3207. doi: 10.1111/trf.12715. [DOI] [PubMed] [Google Scholar]
- 6.Kormoczi GF, Wagner T, Jungbauer C, Vadon M, Ahrens N, Moll W, et al. Genetic diversity of KELnull and KELel: a nationwide Austrian survey. Transfusion. 2007;47:703–714. doi: 10.1111/j.1537-2995.2007.01174.x. [DOI] [PubMed] [Google Scholar]
- 7.Onodera T, Kawai M, Obara K, Enomoto T, Sasaki K, Osabe T, et al. Silent KEL alleles identified from Japanese individuals with the Ko phenotype. Vox Sang. 2018;113:290–296. doi: 10.1111/vox.12628. [DOI] [PubMed] [Google Scholar]
- 8.Boturao-Neto E, Yamamoto M, Chiba AK, Kimura EY, de Oliveira Mdo C, do Monte Barretto CL, et al. Molecular basis of KELnull phenotype in Brazilians. Transfus Med Hemother. 2015;42:52–58. doi: 10.1159/000370232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nance S, Scharberg EA, Thornton N, Yahalom V, Sareneva I, Lomas-Francis C. International rare donor panels: a review. Vox Sang. 2016;110:209–218. doi: 10.1111/vox.12357. [DOI] [PubMed] [Google Scholar]
- 10.Svensson AM, Delaney M. Considerations of red blood cell molecular testing in transfusion medicine. Expert Rev Mol Diagn. 2015;15:1455–1464. doi: 10.1586/14737159.2015.1086646. [DOI] [PubMed] [Google Scholar]
- 11.Jungbauer C, Hobel CM, Schwartz DW, Mayr WR. High-throughput multiplex PCR genotyping for 35 red blood cell antigens in blood donors. Vox Sang. 2012;102:234–242. doi: 10.1111/j.1423-0410.2011.01542.x. [DOI] [PubMed] [Google Scholar]
- 12.van der Schoot CE, de Haas M, Engelfriet CP, Reesink HW, Panzer S, Jungbauer C, et al. Genotyping for red blood cell polymorphisms. Vox Sang. 2009;96:167–179. doi: 10.1111/j.1423-0410.2008.01131.x. [DOI] [PubMed] [Google Scholar]
- 13.Gassner C, Meyer S, Frey BM, Vollmert C. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry-based blood group genotyping - the alternative approach. Transfus Med Rev. 2013;27:2–9. doi: 10.1016/j.tmrv.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Meyer S, Sigurdardottir S, Engström C, Zorbas-Nikos A, Frey B, Gassner C. A rare KEL*02.17|KEL*02N.06 (IVS3 + 1g>a) compound heterozygous individual, prone to anti-KEL11 immunization. Transfus Med Hemother. 2012;39((suppl 1)):65. [Google Scholar]
- 15.Hegemann I, Jaquet P, Furrer M, Frey BM. Genotypic and Phenotypic characterization of Dombrock (Do) blood group antigens in blood donors. Transfus Med Hemother. 2006;33((suppl 1)):46. [Google Scholar]
- 16.Trost N, Meyer S, Vollmert C, Gottschalk J, Ries J, Markovic A, et al. MALDI-TOF MS based BCAM genotyping on 37,234 Swiss proves two new Lutheran blood group alleles, both positive for Aub specific 1,615G. Vox Sang. 2016;111((suppl 1)) 5A-S30-05 (abstract) [Google Scholar]
- 17.Wagner FF, Poole J, Flegel WA. Scianna antigens including Rd are expressed by ERMAP. Blood. 2003;101:752–757. doi: 10.1182/blood-2002-07-2064. [DOI] [PubMed] [Google Scholar]
- 18.Hermand P, Gane P, Mattei MG, Sistonen P, Cartron JP, Bailly P. Molecular basis and expression of the LWa/LWb blood group polymorphism. Blood. 1995;86:1590–1594. [PubMed] [Google Scholar]
- 19.Moulds JM, Zimmerman PA, Doumbo OK, Kassambara L, Sagara I, Diallo DA, et al. Molecular identification of Knops blood group polymorphisms found in long homologous region D of complement receptor 1. Blood. 2001;97:2879–2885. doi: 10.1182/blood.v97.9.2879. [DOI] [PubMed] [Google Scholar]
- 20.Agre P, Smith BL, Baumgarten R, Preston GM, Pressman E, Wilson P, et al. Human red cell Aquaporin CHIP. II. Expression during normal fetal development and in a novel form of congenital dyserythropoietic anemia. investigation Clin Invest. 1994;94:1050–1058. doi: 10.1172/JCI117419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Telen MJ, Rao N, Udani M, Thompson ES, Kaufman RM, Lublin DM. Molecular mapping of the Cromer blood group Cra and Tca epitopes of decay accelerating factor: toward the use of recombinant antigens in immunohematology. Blood. 1994;84:3205–3211. [PubMed] [Google Scholar]
- 22.SCARF: Serum, Cells & Rare Fluids Exchange 2018 http://scarfex.jove.prohosting.com/exchange.html (last accessed June 27, 2018) [Google Scholar]
- 23.Schweizerische Eidgenossenschaft. Bundesamt-für-Statistik Die Bevölkerung der Schweiz 2015. 2018 www.bfs.admin.ch/bfs/de/home/statistiken/kataloge-datenbanken/publikationen.assetdetail.1401562.html (last accessed June 27, 2018) [Google Scholar]
- 24.Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28:49–50. doi: 10.1126/science.28.706.49. [DOI] [PubMed] [Google Scholar]
- 25.Spearman C. The proof and measurement of association between two things. By C. Spearman, 1904. Am J Psychol. 1987;100:441–471. [PubMed] [Google Scholar]
- 26.Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 27.Defays S. An efficient algorithm for a complete link method. Computer J. 1977;20:364–366. [Google Scholar]
- 28.Moskvina V, Smith M, Ivanov D, Blackwood D, StClair D, Hultman C, et al . Genetic differences between five European populations. Hum Hered. 2010;70:141–149. doi: 10.1159/000313854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995:289–300. Series B. [Google Scholar]
- 30.Storry JR, Joud M, Christophersen MK, Thuresson B, Akerstrom B, Sojka BN, et al. Homozygosity for a null allele of SMIM1 defines the Vel-negative blood group phenotype. Nat Genet. 2013;45:537–541. doi: 10.1038/ng.2600. [DOI] [PubMed] [Google Scholar]
- 31.Hustinx H, Blutspende Schweiz. Rare Donors - Seltene Spender - Rare donneurs. 2018 www.iblutspende.ch/en/rare-donors/rare-donors.html (last accessed June 27, 2018) [Google Scholar]
- 32.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lao O, Lu TT, Nothnagel M, Junge O, Freitag-Wolf S, Caliebe A, et al. Correlation between genetic and geographic structure in Europe. Curr Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 34.Schweizerische Eidgenossenschaft. Statistischer Atlas der Schweiz, Vorherrschende Landessprachen in den Gemeinden, 2000. 2018 www.atlas.bfs.admin.ch/maps/13/de/12957_3072_104_70/21290.html (last accessed June 27, 2018) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data