Abstract
Long intergenic non-coding RNAs (lincRNAs) are non-coding transcripts >200 nucleotides long that do not overlap protein-coding sequences. Importantly, such elements are known to be tissue-specifically expressed and to play a widespread role in gene regulation across thousands of genomic loci. However, very little is known of the mechanisms for the evolutionary biogenesis of these RNA elements, especially given their poor conservation across species. It has been proposed that lincRNAs might arise from pseudogenes. To test this systematically, we developed a novel method that searches for remnants of protein-coding sequences within lincRNA transcripts; the hypothesis is that we can trace back their biogenesis from protein-coding genes or posterior transposon/retrotransposon insertions. Applying this method, we found 203 human lincRNA genes with regions significantly similar to protein-coding sequences. Our method provides a visualization tool to trace the evolutionary biogenesis of lincRNAs with respect to protein-coding genes by sequence divergence. Subsequently, we show the expression correlation between lincRNAs and their identified parental protein-coding genes using public RNA-seq repositories, hinting at novel gene regulatory relationships. In summary, we developed a novel computational methodology to study non-coding gene sequences, which can be applied to identify the evolutionary biogenesis and function of lincRNAs.
INTRODUCTION
In eukaryotic cells, long intergenic non-coding RNAs (lincRNAs) are the largest class of long non-coding RNA (lncRNA) molecules. They retain exon-intron structure and are encoded in highly regulated regions of the genome, which are transcribed from thousands of genomic loci in mammals. Increasing evidence suggests that these resulting RNA molecules play a functional and critical role in gene regulation (1,2). LincRNAs show tissue-specific expression patterns, form structured RNAs, and participate in events such as transcript 5′ capping, polyadenylation and splicing. All of these explains the role of lincRNAs in the maintenance of cell integrity, and their involvement in disease and development (3,4).
The biogenesis of lincRNAs is not well understood and its study is challenged by the fact that lincRNAs are less conserved across species than protein-coding genes (5). On the other hand, evolution from pseudogenes, more specifically from decayed protein-coding genes, has been suggested as a lincRNA biogenesis mechanism (6,7). This hypothesis is lacking support by a systematic analysis of the sequence relationship of these entities with their protein-coding counterparts. In relation to this, there have been investigations to identify certain features that are recognized by lincRNAs in the query genes they regulate, which mainly include primary sequence, secondary structure and genomic positioning of the lincRNA effector transcripts (8,9). This led to an implication that transcripts arising from pseudogenes could regulate by complementarity their parental genes (10–13) and evolution from pseudogenes was suggested as a mechanism of antisense ncRNA biogenesis (14,15). However, these hypotheses could gain further support by a systematic analysis of the sequence relationship of lincRNAs with their protein-coding counterparts. In this direction, an attempt has been recently made, by means of synteny and sequence analysis, trying to understand the evolutionary relationships between human lincRNAs and their corresponding homologous protein-coding genes in nine species (16).
Toward addressing these questions, we developed a novel algorithm that aligns protein-coding sequences against lincRNA transcript sequences to detect remnants of coding sequences within lincRNAs and to associate the lincRNAs with their protein-coding counterparts. Additionally, we developed a platform to visualize these alignments to ease the comprehension of the biogenesis of individual lincRNAs.
We employed this approach to detect significant alignments with protein-coding regions for 203 lincRNA genes, for which we are able to trace mutational signatures leading to their biogenesis. Additionally, by means of transcriptomic analysis, we demonstrate that the expression signatures of these lincRNAs correlate to their corresponding parental protein-coding genes indicating a possible regulatory relation. Taken together, this study provides a tool to study non-coding sequence similarity with coding sequences, opening a research line to study lincRNA evolution and function.
MATERIALS AND METHODS
Generation of a scoring matrix for the alignment of protein-coding genes against non-coding elements
In order to generate a proper scoring matrix for the alignment of protein-coding sequences with lincRNAs, we simulate point mutations according to the neutral theory of molecular evolution between human and chimpanzees (17). For each amino acid, we select randomly one of its different codon codifications and then one random nucleotide from the triplet is mutated accordingly to the probabilities of transitions and transversions. For example, TTT (coding for Phenylalanine) could be mutated to TTA (coding for Leucine). To obtain robust results, we mutated each amino acid 107 times. Note that a point mutation in codons for some amino acids can lead to a stop-codon. We took then the frequencies of changes from each amino acid to some other amino acid or stop codon after a point mutation (see Table 1). Note that it is important to define the protein-coding genes as query and the lincRNAs as targets. Finally, a scoring matrix was calculated adding 1 to the values of an amino acid not changing, to favor conservation, and penalizing zero-probability events by assigning them a score of –4.
Table 1.
The probabilities of transition for the different amino acids after receiving a point mutation. Rows and columns are the query and target amino acids, respectively
| A | C | D | E | F | G | H | I | K | L | M | N | P | Q | R | S | T | V | W | Y | * | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | 0.33 | 0 | 0.03 | 0.03 | 0 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0.06 | 0.22 | 0.22 | 0 | 0 | 0 |
| C | 0 | 0.22 | 0 | 0 | 0.06 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.22 | 0.11 | 0 | 0 | 0.06 | 0.22 | 0.06 |
| D | 0.06 | 0 | 0.22 | 0.11 | 0 | 0.22 | 0.06 | 0 | 0 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0.06 | 0 |
| E | 0.06 | 0 | 0.11 | 0.22 | 0 | 0.22 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0.06 |
| F | 0 | 0.06 | 0 | 0 | 0.22 | 0 | 0 | 0.06 | 0 | 0.33 | 0 | 0 | 0 | 0 | 0 | 0.22 | 0 | 0.06 | 0 | 0.06 | 0 |
| G | 0.06 | 0.03 | 0.11 | 0.11 | 0 | 0.33 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 | 0.11 | 0 | 0.06 | 0.01 | 0 | 0.01 |
| H | 0 | 0 | 0.06 | 0 | 0 | 0 | 0.22 | 0 | 0 | 0.06 | 0 | 0.06 | 0.06 | 0.11 | 0.22 | 0 | 0 | 0 | 0 | 0.22 | 0 |
| I | 0 | 0 | 0 | 0 | 0.04 | 0 | 0 | 0.22 | 0.02 | 0.07 | 0.11 | 0.04 | 0 | 0 | 0.02 | 0.04 | 0.22 | 0.22 | 0 | 0 | 0 |
| K | 0 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0.03 | 0.22 | 0 | 0.03 | 0.11 | 0 | 0.06 | 0.22 | 0 | 0.06 | 0 | 0 | 0 | 0.06 |
| L | 0 | 0 | 0 | 0 | 0.11 | 0 | 0.02 | 0.04 | 0 | 0.44 | 0.02 | 0 | 0.15 | 0.02 | 0.04 | 0.07 | 0 | 0.06 | 0.01 | 0 | 0.03 |
| M | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.33 | 0.06 | 0.11 | 2 | 0 | 0 | 0 | 0.06 | 0 | 0.22 | 0.22 | 0 | 0 | 0 |
| N | 0 | 0 | 0.22 | 0 | 0 | 0 | 0.06 | 0.06 | 0.11 | 0 | 0 | 0.22 | 0 | 0 | 0 | 0.22 | 0.06 | 0 | 0 | 0.06 | 0 |
| P | 0.06 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0.22 | 0 | 0 | 0.33 | 0.03 | 0.06 | 0.22 | 0.06 | 0 | 0 | 0 | 0 |
| Q | 0 | 0 | 0 | 0.06 | 0 | 0 | 0.11 | 0 | 0.06 | 0.06 | 0 | 0 | 0.06 | 0.22 | 0.22 | 0 | 0 | 0 | 0 | 0 | 0.22 |
| R | 0 | 0.07 | 0 | 0 | 0 | 0.11 | 0.07 | 0.01 | 0.07 | 0.04 | 0.01 | 0 | 0.04 | 0.07 | 0.33 | 0.06 | 0.02 | 0 | 0.05 | 0 | 0.05 |
| S | 0.04 | 0.04 | 0 | 0 | 0.07 | 0.07 | 0 | 0.02 | 0 | 0.07 | 0 | 0.07 | 0.15 | 0 | 0.06 | 0.3 | 0.06 | 0 | 0.01 | 0.02 | 0.03 |
| T | 0.22 | 0 | 0 | 0 | 0 | 0 | 0 | 0.17 | 0.03 | 0 | 0.06 | 0.03 | 0.06 | 0 | 0.03 | 0.08 | 0.33 | 0 | 0 | 0 | 0 |
| V | 0.22 | 0 | 0.03 | 0.03 | 0.03 | 0.06 | 0 | 0.17 | 0 | 0.08 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0.33 | 0 | 0 | 0 |
| W | 0 | 0.11 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0 | 0.28 | 0.06 | 0 | 0 | 2 | 0 | 0.44 |
| Y | 0 | 0.22 | 0.06 | 0 | 0.06 | 0 | 0.22 | 0 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0.06 | 0 | 0 | 0 | 0.22 | 0.11 |
Input sequences to the aligner
The aligner uses protein-coding gene sequences as query and lincRNA sequences as target. The DNA sequences are translated into the three different frames corresponding to the direction of the transcription of the lincRNA.
The longest common sub-string algorithm to find out blocks of pairwise alignments
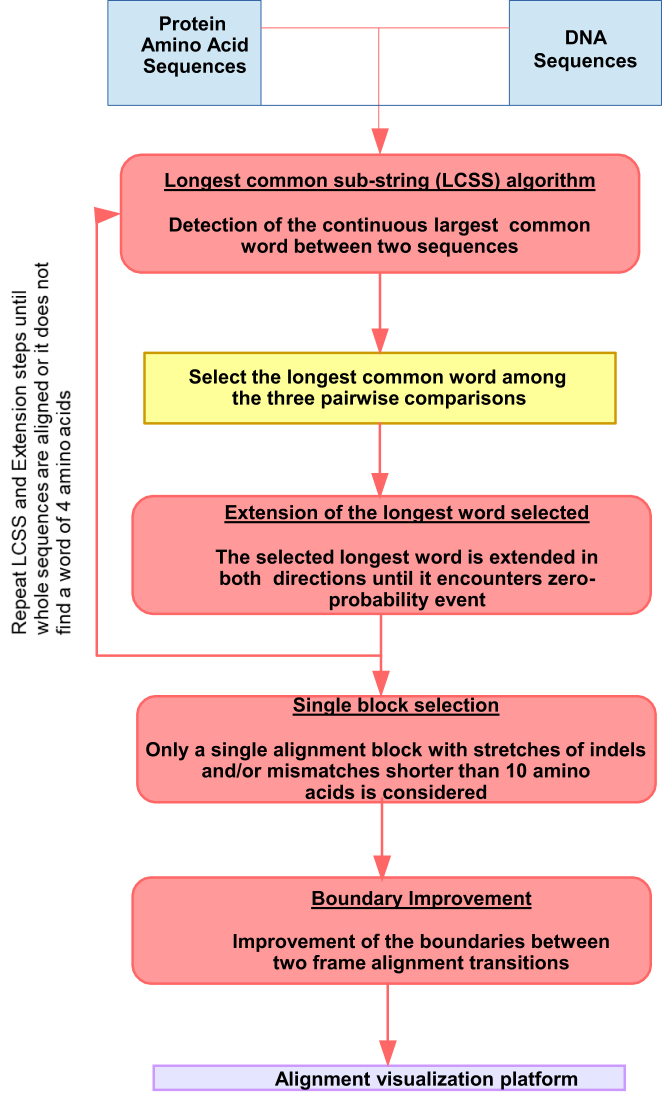
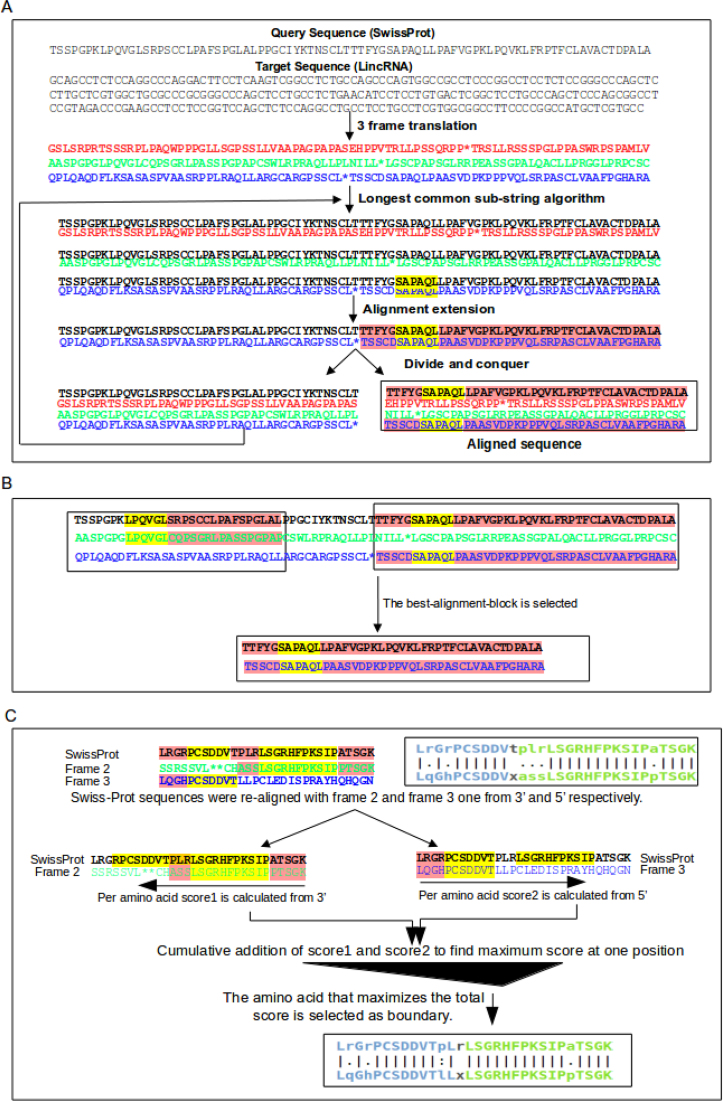
As an initial step, we use the Longest Common Sub-String (LCSS) algorithm to find the longest identical residue pairwise alignment between the protein-coding sequence and the three frame translated lincRNA sequences. The minimum pairwise alignment length allowed is four amino acids. After, the longest alignment is selected (LCSS-word). Further, the LCSS-word is extended in both directions until a zero-probability event is found. Then, we proceed with a divide and conquer strategy (Figure 1). The unaligned sequences of the protein-coding sequence and the three lincRNA open reading frames (ORFs) are identified and aligned in an iterative process (Figure 2A). The loop ends when LCSS-words of four or more amino acids cannot be found.
Figure 1.
Methodology for finding remnants of protein-coding sequences within lincRNA. Flow chart for the alignment of protein-coding genes (amino acid sequences) with non-coding genes (DNA sequence) with the following steps: determination of the longest common sub-string, followed by extension in both directions until a zero-probability event is found. The process iterates on unaligned regions. Then, a single block is selected with stretches of indels and/or mismatches shorter than 10 amino acids. Finally, the boundaries between different frames are optimized, maximizing the alignment score is defined according to our scoring matrix (see Materials and Methods for details).
Figure 2.
Illustrative example of the procedure to align a protein sequence to a lincRNA. The alignment algorithm is divided in three major steps: (A) Flowchart representation of an alignment that includes longest common sub-string (LCSS) and extension. The longest common word between the two sequences is highlighted in yellow color in the first step and extended in both directions, highlighted in red color. Then, the sequences are divided and the unaligned sequences are aligned using, recursively, the same procedure described above. (B) An alignment-block has less than 10 consecutive amino-acids composed by indels and/or mismatches. The alignment-block with the best score is selected. (C) The alignment between different open reading frames is optimized by maximizing the score. See that in the figure, the final alignment is shifted two amino acids towards the blue frame (frame 3) as PL has a higher score where P→ I are conserved and L→ L are identical residues compared to the green frame (frame 2) where P→ A and I→ S were semi conserved. Note that conservation is defined according to our scoring matrix (see Methods for details).
Selection of the best alignment-block
When the alignment of the protein-coding gene against the three lincRNA ORFs is completed, we retrieve the longest alignment-block. An alignment-block contains no more than 10 consecutive unaligned amino acids (indels and/or mismatches). The one with the highest alignment score is chosen (see Figure 2B). This strategy reduces the number of false positives.
Optimization of the boundary-alignment between different frames
However, in an alignment-block there can be alignments from different ORFs because of frame-shift mutations. We optimized the detection of the boundaries between frames, selecting in each case, the boundary that maximizes the alignment score (see Figure 2C).
Finding remnants of protein-coding genes within lincRNA sequences
We applied the described methodology to detect the presence of protein-coding gene signatures within lincRNAs and to trace their biogenesis and sequence-divergence. A total of 20 239 protein sequences were obtained from Swiss-Prot (release of September 2017) (18). The lincRNA sequences were obtained from GENCODE v19 (19), a total of 11 904 lincRNA transcripts corresponding to 7256 different lincRNA genes. LincRNA low complexity regions were masked based on RepeatMasker annotations from the UCSC genome browser (hg19). Finally, 20 239 Swiss-Prot protein-coding sequences were aligned against the set of 11 904 lincRNA low-complexity-masked sequences.
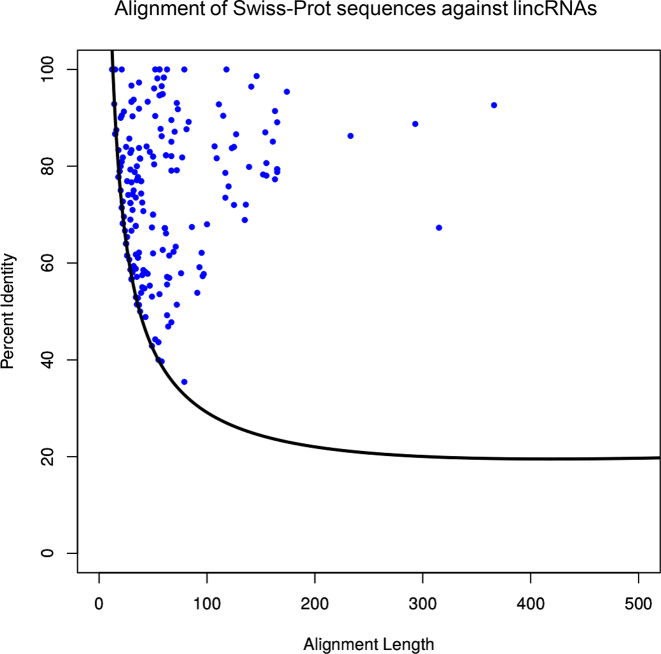
The Rost-curve is currently used to determine if two proteins are homologous (20,21). Here, we used the Rost-curve both to find homologous between protein-coding genes and lincRNAs, also to select the best hit for each lincRNA gene (Figure 3). The percent of identity is represented in the ordinates and the alignment length in the abscissas. Alignments located above the Rost-curve are selected as significant. If a lincRNA has more than one alignment above the curve, the one with the largest Euclidean distance to the Rost-curve is selected (see Supplementary Table S1).
Figure 3.
Filtering the alignments. Percent identity versus alignment length of alignments between 164 human protein-coding genes against 203 lincRNA genes. The depicted Rost-curve was used to select the best alignment if multiple proteins were aligning to one lincRNA (see Methods for details).
Development of an alignment visualization platform for the alignments
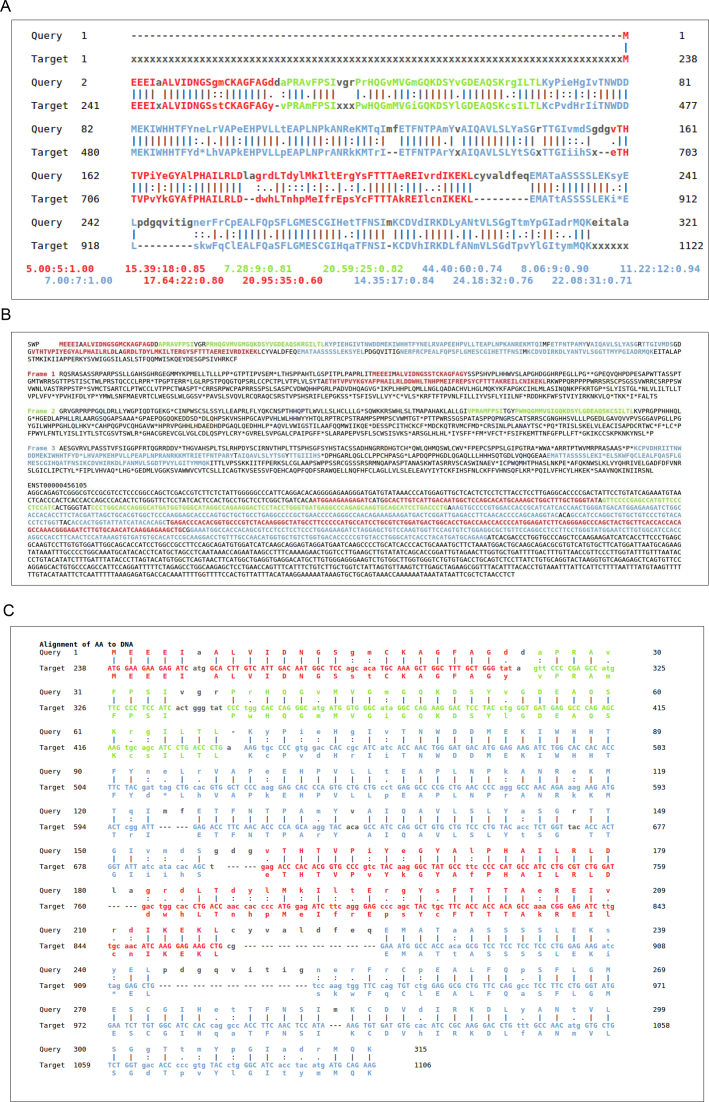
We developed a sequence alignment visualization platform that allows the analysis of the sequence divergence following the lincRNA biogenesis (see Figure 4A–C). The visualization is divided into three main sections. First, the protein sequence against the three-frame-translated lincRNA sequence; highlighted colors are used for the different translated frames. Second, the protein sequence against the three different ORFs of the three translated lincRNA sequences; one alignment for each frame. Third, Alignment of the protein sequence against the lincRNA DNA sequence, where the divergence can be observed with a resolution of one nt.
Figure 4.
Visualization of the alignment. Alignment of human Actin, cytoplasmic 2 (Swiss-Prot AC P63261) against lincRNA gene ENSG00000234996 (ENST00000456105). The alignment visualization platform is divided in three main sections: (A) Alignment of the protein sequence against the three frame lincRNA-translated sequences. The alignments for the different frames are colored differently (red: frame 1, green: frame 2, and blue: frame 3). The alignment scores are depicted below, colored for each alignment-block in the following format: ‘Alignment score: Alignment length: ratio Alignment score / Alignment Length’. (B) The protein sequence is aligned against the three translated frames. (C) Alignment of the protein sequence to the lincRNA sequence. The non-coding translated-frame sequences are also represented. Highlights apart from color include; ‘|’ for perfect match, ‘:’ for conserved residue, ‘.’ for semi-conserved residue, ‘-’ for a gap, and residues in black indicate mismatches.
The alignments are highlighted with three different colors: red, green and blue for frames 1, 2 and 3, respectively. Furthermore, standard symbols have been used to represent the alignment quality: ‘|’ for identity, ‘:’ for conserved (probability value ≥ 0.2) and ‘.’ for semi-conserved (probability < 0.2) (Figure 4A–C). Note that here, conservation is defined by the probability of amino acid substitutions we calculated above.
Expression analysis of lincRNAs and protein-coding genes
BAM files pertaining to RNA-seq datasets for human tissues of ectodermal (pre-frontal cortex), mesodermal (heart) and endodermal (liver) origin were downloaded from ENCODE (22) (see Supplementary Table S2). Reads falling in subject lincRNAs and protein-coding genes were quantified using QuasR (23). Only the subset of lincRNAs and protein-coding genes that are located in the 21 autosomal chromosomes were retained for further analysis. Library size normalization across RNA-seq datasets was done using DESeq (24). Normalized read counts were then log-scaled (base 2) for performing further correlative analysis.
RESULTS
Identification of lincRNA genes containing remnants of transcribed protein-coding sequences
To study the evolutionary biogenesis of lincRNA genes by sequence-divergence from protein-coding genes, we aligned protein-coding sequences against the lincRNA transcribed sequences, translated in the three ORFs that are determined by the direction of the transcription. To challenge the lack of an aligner sensitive enough for this purpose, we developed a novel method (Figures 1 and 2A–C; Supplementary Figure S1; see Materials and Methods for details).
The aligner was optimized for aligning protein-coding sequences against non-coding sequences, considering (i) a stochastic model of sequence evolution within non-coding sequences, (ii) analysis of all ORFs, and (iii) detection of frame-shift mutations. The aligner works as follows (Figure 1): (i) Find the longest LCSS-word in the three ORFs. (ii) Extend the LCSS-word allowing conserved and semiconserved residue alignments. (iii) Select the best alignment-block. (iv) Select the boundaries that optimize the alignment between different frames. See Materials and Methods for details.
Substitution matrices provide a scoring platform for pairwise and multiple sequence alignments of amino acid sequences. BLOSUM (25) is a paradigm of such matrices, and it is the BLAST default substitution matrix. However, BLOSUM was derived from the BLOCKS database (26), which contains information for protein sequences conserved across protein families. This makes the use of this scoring matrix unreliable for non-coding genomic sequences, especially considering that non-coding elements do not evolve like protein-coding genes (27). In contrast, non-coding sequences show enhanced random-mutation patterns and lower selection pressure. Thus, in order to address this, we generated a new substitution matrix based on probability calculations, where the probability of amino-acid substitutions was calculated from stochastic point mutations in triplet-codons as explained in Methods.
For illustration, we compared our alignment methodology to tBLASTn (28) for the alignment of lincRNA gene ENSG00000234277 (ENST00000445817) with the amino-acid sequence of the human 40S ribosomal protein R18 (Swiss-Prot AC P62269) (Supplementary Figure S2A-B). One major difference is that our method captures similarity to the C-terminal of the protein (amino acids 132–152), which is missing by tBLASTn. This can be attributed to the new substitution matrix and also because the algorithm provides an optimized alignment of the query and target sequence, with the ability to merge alignments from all the ORFs that correspond to the direction of the transcription, unlike the highest scoring pairs (HSPs) from tBLASTn.
Next, we created a visualization platform (see Materials and Methods for details) to ease the analysis of evolutionary sequence-divergence in non-coding gene sequences. This method provides three different views, allowing to observe frame-shifts, indels and other point mutations at the nucleotide resolution level. The exemplary visualization of human Actin, cytoplasmic 2 (Swiss-Prot AC P63261) alignment against lincRNA gene ENSG00000234996 (ENST00000456105) is shown in Figure 4A–C. TBLASTn finds 8 HSPs for this particular lincRNA, but misses a significant part of the alignment obtained with our method (from amino acid positions 159–216; frame +1, red color).
In order to validate the proposed method, we have performed the following analysis: From the 8,715 processed pseudogenes annotated at psiDR (29) we have removed those whose sequences are composed exclusively by low complexity regions, remaining a total of 7988 processed pseudogenes. We aligned those, with the method proposed, against human proteins. Interestingly, 96.84% (7735 out of 7988) of the processed pseudogenes are captured by our method as non-coding gene sequences having protein coding remnants within and validating our approach.
Then, we applied our method to find remnants of protein-coding genes within human lincRNA genes in order to explain their origin and function. A total of 413 lincRNA genes with alignments against Swiss-Prot protein sequences were detected. We used the Rost-curve to find 6876 reliable alignments from these 413 lincRNAs genes against 340 protein-coding genes (see Supplementary File S1). If a lincRNA aligns to multiple proteins, we computed the Euclidean distance of each alignment to the Rost-curve, selecting from those the alignment that maximizes the distance to the curve (see Methods for details).
We noted that some of the 413 lincRNA genes overlap protein-coding genes. To handle this, we first identified 336 parental proteins that are aligning to the complete set. Then, we retrieved from UniProt their annotated protein-coding genes (identified by their Ensemble IDs, GeneIDs and UCSC identifiers). We successfully mapped the genomic coordinates of 208 out of the 336 proteins using three different identifiers. To achieve this, we used a multi-step approach. First, we retrieved genomic coordinates using ENSEMBL Ids from GENCODE v19, UCSC Gene ids (retrieving coordinates from the UCSC Table browser (30) and GeneIDs to RefSeq (their genomic coordinates were retrieved from the UCSC Table Browser). Most of the genomic coordinates were retrieved for the hg19 genome and some remaining were retrieved from hg38, which were further converted into hg19 using the liftOver tool of UCSC for batch conversions (31).
The 208 Swiss-Prot proteins that we could map to genomic coordinates are parental to 254 lincRNA genes, of which 203 lincRNAs do not overlap with the parental protein-coding genes (see Supplementary File S2). We consider this a, stringently filtered, set of lincRNAs that we will use for a subsequent analysis (see below). Of note, from the remaining set of proteins that we could not map to genomic coordinates (128 out of 336), 41 are annotated as lincRNAs in Swiss-Prot.
Thus from this stringent filtering with the aim of maximizing specificity, we identify 203 lincRNA genes to have protein-coding remnants within them. Those remnants correspond to 164 different protein-coding genes.
Comparison of 413 lincRNA genes, with protein-coding remnants, with pseudogenic regions
We expect that many of the sequences we identified should be part of known pseudogenes. To investigate the overlap of the 413 lincRNAs with protein-coding remnants with regions identified to be pseudogenic, we took the 11 216 pseudogenes from the psiDR resource (29). When we compare these with the list of 413 lincRNA genes that we identify in our study, we see an overlap of 76 lincRNAs at the transcribed regions. This suggests a link between pseudogenes and lincRNA.
To try to add more evidence about the relation of our lincRNAs to pseudogenes, we obtained a list of 193 human lincRNAs identified by a complementary approach to find human lincRNAs that arised by drift from coding sequences (16). While these have a minor overlap to the 11 216 psiDR pseudogenes (17 sequences), their agreement with our set of 413 lincRNAs is much larger (56 sequences), further supporting that the lincRNAs we detected were generated by pseudogenization. The comparison to proteins in nine vertebrate species used by Liu et al. explains why their dataset contains sequences that we could not identify; on the other hand, alignment to human protein coding genes using an alternative to BLAST explains the larger number of lincRNAs identified by our method.
Transcriptome analysis shows expression correlations between lincRNAs and their corresponding protein-coding genes
To further study the possible functional relevance of our findings of protein-coding remnants in lincRNA sequences, we employed RNA-seq analysis to examine expression patterns of lincRNAs and related protein-coding genes.
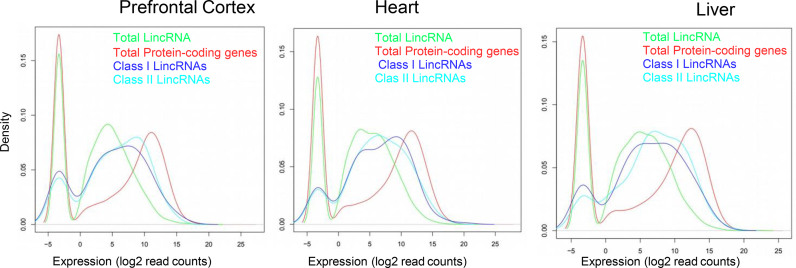
On this line, we compared the expression of (i) total lincRNAs (n = 7256), (ii) total protein-coding genes (n = 20 239), and our set of selected 413 lincRNAs with significant similarity to protein-coding genes distinguishing: (iii) Class I lincRNAs (n = 203) are those lincRNAs that are aligning to human protein-coding genes above the Rost-curve and do not overlap with genomic coordinates of protein-coding genes and (iv) Class II lincRNAs (n = 210) are those lincRNAs that are aligning to human protein-coding genes above the Rost-curve and overlap with the genomic coordinates of protein coding genes or the genomic coordinates of the Swiss-Prot proteins cannot be obtained by means of Ensembl, Gene Id or UCSC identifier. The RNA-seq data was analysed across three tissues of different germline origin (ectoderm: pre-frontal cortex, endoderm: liver and mesoderm: heart).
The expression of Class I lincRNAs (n = 181 from 203, on autosomal chromosomes) and Class II lincRNAs (n = 182 from 210) is significantly higher than that of all lincRNAs (Figure 5), suggesting functional impact of protein-coding remnants within these lincRNA sequences. This implies that lincRNAs having protein-coding sequence remnants are more capable to be expressed pointing out to functionality.
Figure 5.
LincRNAs that align to protein-coding genes are highly expressed. Density plots depicting log-normalized expression of total lincRNAs (green), total protein-coding genes (red), Class I lincRNAs (blue) and Class II lincRNAs (cyan), in the tissues indicated (prefrontal cortex, heart and liver) (see text for details).
To test if the set of 181 Class I lincRNAs could have any impact on the regulation of their parental protein-coding genes because of their sequence-similarity (10), we compared their expression with that of their respective parental genes.
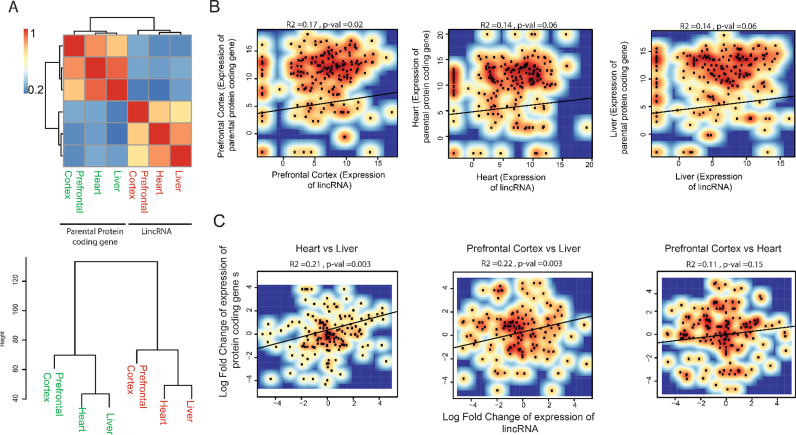
As it could be expected, higher correlation values (Pearson scores) are obtained when comparing the expression of either lincRNAs or the parental genes between tissues, than when comparing lincRNAs with protein-coding genes (Figure 6A). Interestingly, the lowest values of correlation correspond to comparisons across different tissues (e.g. lowest correlation is between protein-coding gene expression in heart and lincRNA expression in liver).
Figure 6.
The expression of lincRNAs and their respective parental protein-coding genes are positively correlated. (A) Heatmap of the Pearson correlation scores of log2-normalized expression values across three human tissues (prefrontal cortex, heart and liver) for 181 lincRNA genes and their 181 respective parental protein-coding genes. (B) Correlation scatter plot between lincRNAs and parental protein-coding genes for each of the selected tissues: prefrontal cortex (left), liver (middle) and heart (right). (C) Correlation scatter plot depicting log fold change in the expression of protein-coding genes and the corresponding lincRNA genes across two tissue types: heart versus liver (left), prefrontal cortex versus liver (middle) and prefrontal cortex versus heart (right).
Focusing on the comparisons between lincRNAs and parental protein-coding genes, positive correlation (R2 ranging from 0.14 to 0.17) was observed in the three tissues analysed (Figure 6B). Fold changes of expression are more meaningful than counts in terms of defining a functional effect. Thus, we next decided to compare the expression changes of lincRNA and parental protein-coding genes between tissues and found stronger correlation (R ranging from 0.11 to 0.22) (Figure 6C).
DISCUSSION
LincRNAs are more tissue specifically expressed compared to protein-coding genes, their corresponding transcripts also differ in sub-cellular locations, metabolic profiles and epigenetic regulation (32). Recent investigations have established lincRNAs to be very important in gene regulatory events underlying cell identity and function (8,33,34). They participate in a number of molecular mechanisms, many of which depend on their primary sequence and secondary structure. Using these features, lincRNAs interfere and compete with the cellular translation process, thereby, selectively targeting the synthesis of certain proteins (8).
The sequences of lincRNAs hold information about their evolutionary biogenesis, which potentially could help us to understand sequence divergence patterns during lincRNA evolution. Additionally, the sequence relationship between lincRNAs and their template protein-coding genes could potentially allow us to study their function. For instance, in relation to their target selection in the gene regulatory cascade, to ultimately understand their role in cell identity and function. However, the sequence-divergence events that contribute to their biogenesis have been poorly understood.
The major reason for this is the unavailability of a proper alignment method to compare efficiently the remnants of protein-coding gene sequences within non-coding elements. Given the aim to find functional relationships of lincRNAs, it makes sense to align the amino acid sequences of functional proteins against lincRNA sequences. In this work, we presented a novel method that finds protein-coding gene sequence remnants within lincRNA sequences. A visualization platform for the alignments in order to analyse their sequence divergence events is also provided (Figure 4). Our approach includes a new substitution matrix, and merges in an optimized way the alignments from multiple ORFs. Hence, we overcome a major limitation in studying the evolution of lincRNA gene sequences.
We used the Rost-curve to detect homology between protein-coding and lincRNA genes, obtaining 203 lincRNAs (corresponding to 164 different protein-coding genes). Our results suggest that many lincRNAs evolved from sequence divergence events from protein-coding gene sequences. For example: lincRNA gene ENSG00000223396 (transcript ENST00000441932.1) from chromosome 1 is aligning to protein-coding gene P46783 (40S ribosomal protein S10) on chromosome 6 (the sequence alignment can be visualized in Supplementary File S3). We provide similar alignments for the complementary dataset of lincRNAs as Supplementary File S4.
While, inherently, these lincRNAs might have arisen from an event of pseudogenization of a coding region, and some are currently annotated as pseudogenes, the definition of a pseudogene is functional and therefore, as evidence of the transcription and function of each of these lincRNAs accumulates they should not be considered pseudogenes. But the processes generating pseudogenes from coding regions are very prevalent (15). Using our approach, we could validate most of the psiDR processed pseudogenes supporting the concept that pseudogenization could lead to lincRNAs showing remnants of protein coding sequences.
It is very likely that for many lincRNAs we could not find any associated protein-coding gene sequences due to sequence divergence making them undetectable by sequence alignment analysis. Alternatively, the parental protein-coding gene could have been lost during evolution in humans. In these cases, we could be detecting homologs of the lost genes.
From the set of 203 Class I lincRNAs there are 15 lincRNA genes that, while not overlapping to the parental protein-coding gene, are located at a close genomic distance in the genome. This shows that these lincRNAs are the product of small tandem genomic duplications (Supplementary File S5).
Importantly, we observed that the lincRNAs with remnants of protein sequences within them are more expressed than the average lincRNA (Figure 5), hinting at a possible functionality, at least in the tissues considered. We also observed that lincRNA expression and the expression of their associated protein-coding gene positively correlates across three tissues, even more if fold changes between tissues are considered (Figure 6). This result suggests that the relations we found between lincRNAs and proteins might have post-transcriptional regulatory consequences, with a mild selection pressure to keep complementarity to the parental gene. This hypothesis is supported in a recent study (16) where lincRNAs from human have been aligned to nine different species using blastn finding 193 lincRNA human orthologues with a potential to regulate their paralogs as competing endogenous RNAs.
Collectively, our approach helps the study of non-coding RNA sequences to predict their functional relationships with protein-coding genes. Future work should aim to validate the tissue specific functions of these lincRNAs in relation to their parental protein-coding genes.
CONCLUSION
LincRNAs are the most abundant transcribed non-coding RNAs in mammalian cells. Previous evidence have established their functional role in gene regulation. However, their sequence relationship with protein-coding genes for analysing their biogenesis, as well as their regulatory targets remained under-explored. We developed an optimized methodology for the alignment of protein-coding sequence remnants within lincRNA genes. We complemented this approach with a visualization platform to analyse the sequence divergence. Collectively, this approach can be implemented to study the genetic events leading to lincRNA biogenesis and post-transcriptional target regulatory selection. By implementing this approach, we found 203 lincRNA genes that show significant alignment with protein sequences, pointing out to a functional association with those. Moreover, these hits show overall high expression levels and correlated expression with the aligning protein-coding genes, establishing their importance and inter-dependence in gene regulation. Our study provides a novel tool for analyzing non-coding RNA sequences and gives unprecedented insights into the gene regulatory function of hundreds of human lincRNAs.
DATA AVAILABILITY
The open software for obtaining the lincRNA set predictions (Class I and II, 413 lincRNAs) and for aligning each lincRNA against their corresponding protein-coding gene is available in the next GitHub repository (https://github.com/swttalyan/protsInLincRNA).
Supplementary Material
ACKNOWLEDGEMENTS
Authors Contributions: All authors contributed to the work presented in this paper. E.M. conceived the whole study. E.M. designed the study except the expression analysis (ST). E.M. and M.A. supervised ST. S.T. performed all data acquisition, programming and code execution, except the calculation of the probabilities for amino acid substitutions (E.M.). S.T., M.A. and E.M. analysed the data. S.T. wrote the first draft of the manuscript. M.A. and E.M. made extensive, critical and substantial contributions to the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [AN735/3-2 to M.A.A.]. Funding for open access charge: Johannes Gutenberg University of Mainz.
Conflict of interest statement. None declared.
REFERENCES
- 1. Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S.. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016; 539:452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hon C.C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J., Gough J., Denisenko E., Schmeier S., Poulsen T.M., Severin J. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017; 543:199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Calin G.A., Liu C.G., Ferracin M., Hyslop T., Spizzo R., Sevignani C., Fabbri M., Cimmino A., Lee E.J., Wojcik S.E. et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007; 12:215–229. [DOI] [PubMed] [Google Scholar]
- 4. Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D., Khalil A.M., Zuk O., Amit I., Rabani M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010; 142:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ulitsky I., Shkumatava A., Jan C.H., Sive H., Bartel D.P.. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011; 147:1537–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hezroni H., Ben-Tov Perry R., Meir Z., Housman G., Lubelsky Y., Ulitsky I.. A subset of conserved mammalian long non-coding RNAs are fossils of ancestral protein-coding genes. Genome Biol. 2017; 18:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milligan M.J., Lipovich L.. Pseudogene-derived lncRNAs: emerging regulators of gene expression. Front. Genet. 2014; 5:476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu W., Alvarez-Dominguez J.R., Lodish H.F.. Regulation of mammalian cell differentiation by long non-coding RNAs. EMBO Rep. 2012; 13:971–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 10. Muro E.M., Mah N., Andrade-Navarro M.A.. Functional evidence of post-transcriptional regulation by pseudogenes. Biochimie. 2011; 93:1916–1921. [DOI] [PubMed] [Google Scholar]
- 11. Poliseno L., Salmena L., Zhang J., Carver B., Haveman W.J., Pandolfi P.P.. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010; 465:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tam O.H., Aravin A.A., Stein P., Girard A., Murchison E.P., Cheloufi S., Hodges E., Anger M., Sachidanandam R., Schultz R.M. et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008; 453:534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Watanabe T., Totoki Y., Toyoda A., Kaneda M., Kuramochi-Miyagawa S., Obata Y., Chiba H., Kohara Y., Kono T., Nakano T. et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008; 453:539–543. [DOI] [PubMed] [Google Scholar]
- 14. Hawkins P.G., Morris K.V.. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription. 2010; 1:165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Muro E.M., Andrade-Navarro M.A.. Pseudogenes as an alternative source of natural antisense transcripts. BMC Evol. Biol. 2010; 10:338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu W.H., Tsai Z.T., Tsai H.K.. Comparative genomic analyses highlight the contribution of pseudogenized protein-coding genes to human lincRNAs. BMC Genomics. 2017; 18:786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ebersberger I., Metzler D., Schwarz C., Paabo S.. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002; 70:1490–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu C.H., Apweiler R., Bairoch A., Natale D.A., Barker W.C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R. et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006; 34:D187–D191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999; 12:85–94. [DOI] [PubMed] [Google Scholar]
- 21. Sander C., Schneider R.. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991; 9:56–68. [DOI] [PubMed] [Google Scholar]
- 22. Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gaidatzis D., Lerch A., Hahne F., Stadler M.B.. QuasR: quantification and annotation of short reads in R. Bioinformatics. 2015; 31:1130–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Anders S., Huber W.. Differential expression analysis for sequence count data. Genome Biol. 2010; 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Henikoff S., Henikoff J.G.. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henikoff J.G., Henikoff S.. Blocks database and its applications. Methods Enzymol. 1996; 266:88–105. [DOI] [PubMed] [Google Scholar]
- 27. Johnsson P., Lipovich L., Grander D., Morris K.V.. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim. Biophys. Acta. 2014; 1840:1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 29. Pei B., Sisu C., Frankish A., Howald C., Habegger L., Mu X.J., Harte R., Balasubramanian S., Tanzer A., Diekhans M. et al. The GENCODE pseudogene resource. Genome Biol. 2012; 13:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Speir M.L., Zweig A.S., Rosenbloom K.R., Raney B.J., Paten B., Nejad P., Lee B.T., Learned K., Karolchik D., Hinrichs A.S. et al. The UCSC genome browser database: 2016 update. Nucleic Acids Res. 2016; 44:D717–D725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ransohoff J.D., Wei Y., Khavari P.A.. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018; 19:143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Procedures Online. 2014; 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quan Z., Zheng D., Qing H.. Regulatory roles of long non-coding RNAs in the central nervous system and associated neurodegenerative diseases. Front. Cell. Neurosci. 2017; 11:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The open software for obtaining the lincRNA set predictions (Class I and II, 413 lincRNAs) and for aligning each lincRNA against their corresponding protein-coding gene is available in the next GitHub repository (https://github.com/swttalyan/protsInLincRNA).