Abstract
Fragile X syndrome (FXS), the most common form of inherited intellectual disability, is due to the functional deficiency of the fragile X mental retardation protein (FMRP), an RNA-binding protein involved in translational regulation of many messenger RNAs, playing key roles in synaptic morphology and plasticity. To date, no effective treatment for FXS is available. We searched for FMRP targets by HITS-CLIP during early development of multiple mouse brain regions (hippocampus, cortex and cerebellum) at a time of brain development when FMRP is most highly expressed and synaptogenesis reaches a peak. We identified the largest dataset of mRNA targets of FMRP available in brain and we defined their cellular origin. We confirmed the G-quadruplex containing structure as an enriched motif in FMRP RNA targets. In addition to four less represented motifs, our study points out that, in the brain, CTGKA is the prominent motif bound by FMRP, which recognizes it when not engaged in Watson–Crick pairing. All of these motifs negatively modulated the expression level of a reporter protein. While the repertoire of FMRP RNA targets in cerebellum is quite divergent, the ones of cortex and hippocampus are vastly overlapping. In these two brain regions, the Phosphodiesterase 2a (Pde2a) mRNA is a prominent target of FMRP, which modulates its translation and intracellular transport. This enzyme regulates the homeostasis of cAMP and cGMP and represents a novel and attractive therapeutic target to treat FXS.
INTRODUCTION
Fragile X mental retardation protein (FMRP) is an RNA binding protein (RBP) encoded by the fragile X mental retardation 1 (FMR1) gene whose silencing causes the fragile X syndrome (FXS), the most common form of intellectual disability and a leading genetic cause of autism. FMRP is involved in different steps of RNA metabolism, ranging from nuclear export to transport of mRNA along neurites and translational control in the soma as well as at synapses (1). FMRP deficiency impacts the size of brain regions, synaptogenesis and alters the morphology of dendritic spines as well as some forms of synaptic plasticity (2,3). To date, no specific and effective therapy is available for FXS. Current clinical approaches are focused on behavioral therapy and off-label medications that only mitigate a limited set of symptoms, such as hyperactivity, seizures or anxiety (4). Recent clinical trials based on antagonist of metabotropic glutamate Receptor 5 (mGlu5) and antagonist of GABA-B receptor were discontinued (5,6). The understanding of the physiopathology of FXS and the development of a specific therapy are intimately linked to the understanding of FMRP function and then to the identification of FMRP mRNA targets. For this reason, the search for molecular interactors (proteins and mRNAs) of FMRP has been very active (1). By using high-throughput sequencing-crosslinking immunoprecipitation (HITS-CLIP) on mRNAs associated with polyribosomes in whole brains of 11–25 day-old mice, 842 mRNAs were identified as targets of FMRP, likely during their translation (7). A phospho-activatable ribonucleoside CLIP (PAR-CLIP) analysis was also performed in a non-neuronal cell line (HEK) co-expressing FMRP endogenously and its inducible flag-tagged form that was immunoprecipitated together with 6.000 mRNAs (8). More recently, a CLIP study using microarray performed in 8-day in vitro cultured neurons resulted in the identification of one predominant mRNA target of FMRP (9). Nevertheless, it is not clear if most of these targets have a critical role in the physiopathology of FXS and in which cells they interact with FMRP.
Here, we used HITS-CLIP to identify FMRP targets at an early mouse developmental stage [postnatal day (PND) 13], when FMRP is most highly expressed (10,11) and synaptogenesis peaks (12). Our analysis resulted in the identification of the largest set of brain mRNA targets of FMRP to date. This allowed us to dissect the role of FMRP in different brain regions and cell types. On the basis of these findings we were able to identify a predominant motif bound by FMRP in brain and a prominent mRNA target that is a promising druggable pathway for this disorder.
MATERIALS AND METHODS
HITS-CLIP (high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation)
The protocol was optimized following previously described published methods (13–16). Briefly, to isolate mRNAs associated with FMRP in vivo, UV-crosslinking and immunoprecipitations were performed on 100 mg of tissues grinded in liquid nitrogen. Powders were spread onto 60 mm dishes floating onto liquid nitrogen. RNAs and protein were crosslinked through three rounds of 254 nm UV irradiation (400 mJ each). IP was carried out as described previously with minor modifications (13–16). Crosslinked material was lysed in NP40 buffer [50 mM Hepes pH 7.6, 150 mM NaCl, 0.5% NP40, 1 mM NaF, 0.5 mM Dithiothreitol (DTT), proteases/phosphatases inhibitors (pierce), RNaseOut (1:1000)] as described in Spitzer et al. (15). Debris were removed by centrifugation (10 min, 20 000 g, 4°C) and supernatants were incubated with RNaseI (0.01 u/μl; Ambion) for 15 min at 22°C. RNase digestion was stopped by the addition of SUPERase In RNAse inhibitor (0.2 u/μl). For each assay, 15 μl of Rb11 polyclonal anti-FMRP antibody were bound to 50 μl of protein A dynabeads (Invitrogen) for each mg of protein lysate in the input. Salt concentration in the lysates was adjusted to 400 mM for 5 min then brought back to 150 mM. IP was carried out at 4°C for 4 h and ∼2% of the input lysate and 10% of the immunoprecipitated material were saved for western blot analysis to check IP quality. Beads were washed once in NP40 buffer, washed once in high salt buffer (50 mM Hepes pH 7.6, 0,1% sodium dodecyl sulphate, 1 M NaCl, 1 mM ethylenediaminetetraacetic acid, 1% NP40) then twice again in NP40 buffer. Co-precipitated RNAs were dephosphorylated in PNK buffer (50 mM Tris pH 8, 100 mM NaCl, 10 mM MgCl2, 1 mM DTT) without ATP, then DTT was adjusted to 5mM and RNAs were labeled with gamma-32P-ATP and T4 Polynucleotide Kinase (PNK) (NEB). Beads were washed 5 times in PNK buffer without DTT complemented with 0.2% Tween 20. IP materials were resuspended in LDS (Invitrogen), heat-denatured for 10 min at 70°C and separated on NuPage 4–12% gel, run in 3-(N-morpholino)propanesulfonic acid (MOPS) buffer. Ribonucleoprotein complex were transferred to nitrocellulose membrane with NuPage neutral transfer buffer (Invitrogen), washed with RNAse free water (Ambion) and exposed to phosphorimager screens over night at −20°C. The area containing the specific radioactive signal corresponding to FMRP ribonucleoprotein complexes (100 to 130 kDa) was cut out of the membrane, sliced and digested with proteinase K (1 mg/ml) for 30 min at 56°C (Ambion) then urea concentration was adjusted to 3 M and the samples were further incubated for 10 min at 56°C (16). After purificarion, RNAs were cloned with the CleanTag Ligation Kit for Small RNA Library Prep (Trilink; adapter diluted 1/4) according to the manufacturer’s instructions, reverse transcribed with superscript II (Invitrogen) and amplified through 16–20 cycles of polymerase chain reaction (PCR) (NEB next).
CLIP sequencing
CLIP samples have been sequenced on Ion torrent Proton and SOLiD WildFire. Due to the RNase digestion, fragments should fit the FMRP footprint and may contain adapter sequences at the 3′ end of the reads. Using cutapdapt (v1.2.1), reads were trimmed for the 5′ adapter sequence present due to the Proton library construction protocol used (–front = TTCTACAGTCCGACGATC). Cutadapt was also used to trim for potential 3′ adapter sequence (–adapter = TGGAATTCTCGGGTGCCAAGG, for Proton and –adapter = 33020103 for SOLiD reads). Trimmed reads with a length under 20 nt were discarded from further analysis to avoid ambiguous mapping. Remaining reads were aligned with bowtie (version 0.12.7) directly to refseq transcriptome (release 20161019) with ‘–best –strata –norc’. Custom Java class, based on Picard Java API, was used to discard reads mapping to more than one unique gene. Alignment files were filtered to discard PCR duplicates by keeping only one read per alignment start position taking into account possible stretch of soft clipped bases at the start of the reads. Resulting reads are called fragments and represent the total signal of CLIP experiments aiming at the detection of the FMRP binding sites. The CLIP signal per sample is presented in Supplementary Table S1. Results of the various filtering steps are presented in Supplementary Table S2.
Peak calling and motif detection
FMRP binding sites were identified with Pyicoclip from Pyicoteo Tools Suite (17), that was applied independently for all the samples. Bam files with no multi gene hit reads filtered for PCR duplicates were converted to Bed file with bedtools2 BamToBed and then provided to pyicoclip for peak calling on the whole mouse transcriptome described in refseq (release 19 October 2016). We started from the unfiltered list of peaks detected by pyicoclip, processed the file to filter peaks with <2 fragments counts and removed the peaks present in mir, RNU, Sno Rny, mitochondrial and riken sequences. Given the large amount of splice variants described in refseq for each gene we generated a method to remove peaks called in different splice variants of the same gene and identifying the same genomic region using a custom perl script. We finally called a total of 18.124 peaks across all samples. We gathered all peaks in a unique file and removed peaks identifying the same genomic region with the same custom perl script previously used. We finally identified a total of 18.124 peaks (8962, 5082, 4080 peaks for CB, HC and CX, respectively) that we used to feed to the motif detection software DREME (Meme release 4.10.1) using ‘–norc –m 5’ parameter in order to identify enriched motifs within our sets of peaks. Genebank annotations were used to annotate the coding sequence (cds) and the untranslated regions for each mRNA to map G-4 regions and FMRP binding sites.
Motifs fold analysis
In vivo DMS-mediated RT stop scores were computed as previously described (18) for each base of transcripts. We then mapped the 14.376 peaks (FMRP binding sites) and computed unpairing scores for motifs contained in FMRP binding sites or for the same motif in the same transcript outside of an FMRP binding site. Only transcripts for which we detected 100% base coverage for all motifs in a given transcript in the DMS_vitro_95C condition were processed for folding analysis in the three replicates of mESC_DMS_vivo conditions.
Codon analysis
Codon composition of FMRP binding site in the cds regions was analyzed according to the following formula PauseScore = (Readscodon/orf/ReadsORF)/(Nbr codon/ORF/LengthORF). In which Readscodon/orf = number of reads covering a given codon for a given ORF, ReadsORF = number of reads covering the ORF, Nbr codon/ORF: number of a given codon in a given ORF and LengthORF: the length of the ORF.
G-quadruplex (G-4) mapping
Presence of a G-4 structure in the various RNAs was assessed by reverse transcriptase-mediated primer extension based on a previously described protocol (19) with some modifications. The DNA sequence of interest was PCR amplified and subcloned into pGEM-T easy vector. The insertion was verified by sequencing and the sequence of interest was PCR amplified with the following primers (T7invitroT; 5′-GACTGACTTAATACGACTCACTATAGGG-3′; M13Rev; 5′- CACACAGGAAACAGCTATGAC -3′). Electropherograms were generated and analyzed with the QuShape software (20).
Single cell dataset analysis
Mouse cortex and hippocampus single cell RNAseq data from Zeisel et al. (21) was downloaded from their website (http://linnarssonlab.org/cortex/) and analyzed using the R package Seurat. Raw RNA molecule counts (i.e. unique molecule identifier counts, UMI) were downloaded from http://linnarssonlab.org/cortex/. Count data were normalized to the median count, then log2 transformed. For each organ, among all FMRP targets identified, the most variable genes between all cell types were selected using a coefficient of variation cut-off of 0.7 and an average expression value >0.2. This resulted in the selection of 74 FMRP target genes for hippocampus and 58 FMRP target genes for cortex. These genes were then used as input for unsupervised hierarchical analysis. Hierarchical clustering and heatmaps of gene expression were generated using the R package pheatmap. Selected genes were clustered using the Ward.D clustering method and the Pearson correlation as distance. Cells were ordered by cell types using the level 1 classification provided with the original data. For each cell type, a list of genes differentially expressed was determined using the FindMarkers function from the R package Seurat, with parameters thresh.use = 1 and min.pct = 0.25. For each cell type, the 30 genes with highest P-values were used for enrichment analysis. Enrichment in FMRP targets by cell types was assessed using hypergeometric test. All P-values were adjusted for multiple testing using the Benjamini–Hochberg correction.
RT-qPCR
quantitative PCR (qPCR) was performed on a Light Cycler 480 (Roche) with MasterMix SYBRGreen (Roche) following the manufacturer's instructions and according to the MIQE guidelines (22). Primer sequences are reported in Supplementary Table S3.
Synaptosomes purification
Synaptosomes fractions were prepared by centrifugation on sucrose density gradients. Mouse PND13 cortices were homogenized in sucrose buffer (0.32 M sucrose, 10 mM Tris–HCl, pH 7.4) in a 7× weight/volume ratio. The homogenates were centrifuged at 1000 g for 5 min at 4°C. The supernatants were brought to a final volume of 2 ml with sucrose buffer and carefully placed on sucrose-percoll 2–6–10–20% non-continuous density gradients. The gradients were ultracentrifuged at 18 000 rpm for 10 min at 4°C with slow brake, and the crude synaptosomal fractions, placed between 10 and 20% sucrose-percoll, were carefully recovered with a glass pipette pre-coated with sucrose buffer. The crude synaptosomal fractions were diluted to 10 ml with Hepes buffer (140 mM NaCl, 3 mM KCl, 1.2 mM MgSO4, 1.2 mM CaCl2, 1 mM NaH2PO4, 5 mM NaHCO3, 10 mM glucose, 5 mM Hepes, pH 7.4 at 37°C) and centrifuged at 10 000 g for 30 min at 4°C with full brake. The pellets were resuspended in NP40 lysis buffer (1 M Tris–HCl, 3 M NaCl, 12 mM MgCl2, 0,1 M DTT, 1% NP40), centrifuged full speed for 10 min at 4°C and then resuspended in sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer.
Polyribosome fractionation
Polyribosome fractionation was performed as described previously (23) on 20–50% (w/w) continuous sucrose gradients. Fractions were separated on a BR-188 Density Gradient Fractionation System (Brandel). Fold changes in Tbp and Pde2a mRNA levels were assessed by RT-qPCR and were calculated for individual fraction 8 to 14 according to the 2−∂Cp formula for which ∂Cp is (Cp genex KO fractionn – Cp genex WT fractionn). Results from fractions 6 to 8 (Light), 9 to 11 (Medium) and 12–14 (Heavy) were pooled and analyzed together.
Protein extraction and western blot analysis
Immunoblot was performed as follows: cells or grinded tissues were homogenized in NP40 buffer, debris were removed by centrifugation (20 000 g, 10 min, 4°C). Protein content in the supernatant was measured using the Bradford assay (Biorad) and samples were separated on NuPage bis-tris 4–12% gels in MOPS buffer. Separated proteins were transferred to nitrocellulose membranes (Biorad). Membranes were blocked with phosphate-buffered saline-Tween (0.05%) and milk (5%). Primary antibodies were incubated overnight. Antibody Rb11 was generated by immunization of rabbits with a truncated FMRP protein corresponding to its C-terminal region, as previously described (10). Rb11 was used at the dilution of 1:500 for Western blot. Monoclonal 1C3 anti-FMRP antibody (24) was used at the dilution 1:1000; mouse polyclonal anti-FMRP 1R was used at the dilution 1:1000 (11). Anti PDE2A (Fabgennix, # PD2A-101AP), PSD95 (Millipore, AB9708), Anti-rpS6 (Cell signaling Technology, 54D2) anti ß-Actin (Sigma, clone AC-74).
Cloning of cDNA fragments corresponding to FMRP binding sites
We cloned fragments bound by FMRP. These fragments were cloned by PCR from mouse genomic DNA and inserted in the PmeI site of pSIcheck2 (Promega). The sequences of the various constructs were verified by Sanger sequencing on an Applied Biosystems 3130XL and are provided in Supplementary Table S3.
Luciferase assays
STEK cells, expressing or not FMRP were seeded in 96-wells plates (20 000 cells/well) and were immediately transfected with pSi-check2-derived constructs using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Renilla and firefly activities were measured with the Dual-glo luciferase assay (Promega) using the Glomax 96-wells plate luminometer (Promega).
Probes, smFISH and image analysis
smFISH primary probes and FLAPs (secondary fluorescent probes) were produced and purchased from Integrated DNA Technologies (IDT) as decribed (25). Three-dimensional image stacks were captured on a wide-field microscope (Zeiss Axioimager Z1/Apotome) equipped with a 63× objective and a CCD camera (Axiocam MRm 4), controlled with Metamorph (Molecular Devices). Spots in the Cy3 channels were detected in 3D stacks with Imaris (Bitplane) and categorized as being labeled. Spots were split into different branching surfaces drawn by Imaris software (filament) or into cell bodies surfaces.
Statistical analysis
Results are expressed as mean±SEM. All statistical analyses were based on biological replicates. Appropriate statistical tests used for each experiment are described in the corresponding figure legends or in the ‘Materials and Methods’ section for HITS-CLIP. All statistical analyses were carried out using the GraphPad Prism Version 6.0e.
RESULTS
FMRP target RNAs in different mouse brain areas
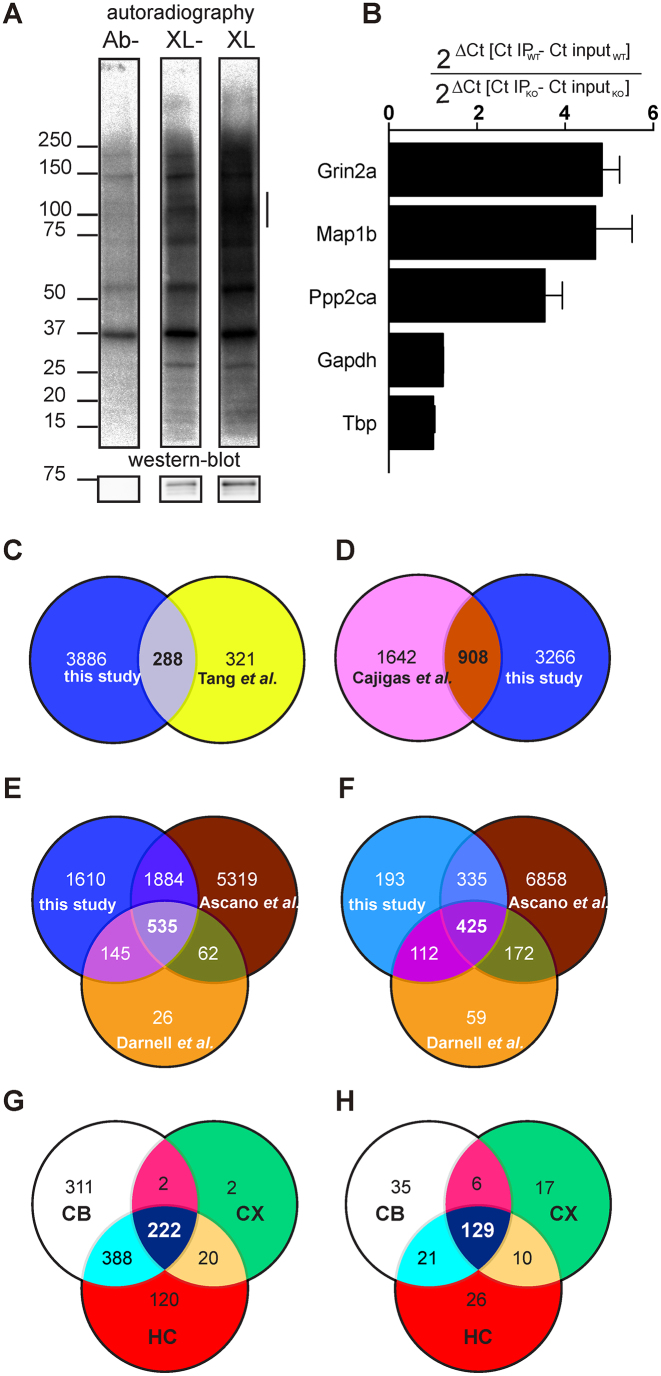
We purified a new polyclonal antibody, Rb#11 and showed that it efficiently pulls down FMRP and recognizes FMRP in Western blot (Supplementary Figure S1A and B). We then set up the CLIP conditions adapting a procedure from previous HITS-CLIP, PAR-CLIP and iCLIP assays (8,13,16). Specific brain regions (cortex, hippocampus and cerebellum) from Post Natal Day (PND) 13 male mice were grinded in liquid nitrogen into fine powder, UV crosslinked (lanes Ab- and XL), and proteins solubilized in NP40 buffer (Figure 1A). After RNaseI treatment, FMRP was immunoprecipitated and bound RNAs were radioactively labeled. Ribonucleoprotein complexes (RNPs) were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Radiolabeled RNPs were detected by autoradiography (Figure 1A, upper panel). The same membrane was subsequently analyzed by Western blot to detect FMRP (Figure 1A, bottom panel). We observed that an increased amount of FMRP/fragmented RNA complex is immunoprecipitated after cross-link (XL), compared to the no XL (XL-) condition (Figure 1A)—an example using immunoprecipitated FMRP from cortex is shown), while no FMRP and FMRP-containing RNPs were detected in the control lane Ab- (without antibody). We selected a set of previously described target mRNAs of FMRP (Grin2a, Map1b and Ppp2ca) and some control RNAs (Gapdh and Tbp). In the absence of RNase treatment, we analyzed their relative enrichment by RT-qPCR in immunoprecipitated mRNAs from wild-type (WT) and compared them with immunoprecipitated mRNAs from Fmr1-KO cortices (Figure 1B), confirming the specificity of co-immunoprecipitated target mRNAs of FMRP. We then performed HITS-CLIP, sequenced co-immunoprecipitated cross-linked fragments. The size distribution of the mapped reads was consistent with a protection from RNAse degradation since we could observe reads longer than 40 nt (see an example Supplementary Figure S1C). We repeated this assay twice for cerebellum, three times for cortex and four times for hippocampus. The correlation in the fragment counts per gene from replicate experiments showed the robustness of the experimental protocol (Supplementary Figure S1D). We mapped 712 000 unique reads to the latest ref seq assembly (NCBI ftp web server ref10/2016). We used pyico clip (17) to identify FMRP binding sites, referred thereafter as peaks, resulting in the identification of 14 376 peaks corresponding to 4.174 RNA targets (Supplementary Table S1). A total of 288 of our hits overlap with deregulated synaptic proteins identified in the cortex of young Fmr1-null mice (26) (Figure 1C). Approximately one-fourth of the FMRP targets we identified were previously described as distantly transported RNAs in hippocampus neuropils of rats (Figure 1D) (27), which is consistent with the role of FMRP in RNA transport in specialized cells. Our target mRNA repertoire clearly shows some similarity with datasets obtained by other groups (Figure 1E and Supplementary Table S1) as it shares 680 hits with the list of FMRP target mRNAs associated with brain polyribosomes (13) and 2419 hits with the FMRP target list in Human Embryonic Kidney (HEK) 293 cells (8) (Figure 1E). Taking in consideration only those having at least 50 fragments (counts) co-immunoprecipitated with FMRP, we found a total of 1065 targets (Supplementary Table S4 and Figure 1F), among them 872 are overlapping with previous studies (Figure 1F) (8,13). Considering our most stringent analysis, we identified 192 new RNA targets of FMRP. Further strengthening the interest of a tissue-specific CLIP approach, 80 these new targets were found specifically in the cerebellum and 36 in the hippocampus. To identify signaling pathways regulated by FMRP in the brain, we performed Gene Ontology analysis of its target RNAs in the cortex, hippocampus and cerebellum using Ingenuity Pathway Analysis (IPA) by considering the most restricted number of targets (Supplementary Table S5). Even if most of these mRNAs are common to the various brain areas (Figure 1G), specific pathways are regulated in each brain region (Figure 1H and Supplementary Table S5). We also took advantage of the recent publication of single-cell transcriptomic datasets and compared our dataset with the recent single cell transcriptomic analysis in the somatosensory cortex and the hippocampus (21) and observed that FMRP target RNAs are mostly expressed in neuronal cells (interneurons and pyramidal cells) in the cortex and the hippocampus (Supplementary Figure S2A and B). Cumulative Distribution analysis shows that mRNA targets carrying a G-4 forming motif, or encoded by autism-related genes (autismdb; https://gene.sfari.org/autdb/Welcome.do) are amongst the best targets of FMRP (Figure 2A and Supplementary Table S1). Target mRNAs harboring at least one peak are also enriched in the immuno-precipitated material compared to those devoid of any FMRP binding site (Figure 2A).
Figure 1.
Identification of mRNA targets of FMRP. (A) Upper panel: audioradiography of immunoprecipitated UV-crosslinked RNPs from 13 PND male mouse brain cortex extracts after separation on a SDS-PAGE gel and then transferred onto a nitrocellulose membrane. Ab-: immunoprecipitation in the absence of the polyclonal anti-FMRP antibody Rb#11; XL-: Immunoprecipitation in the presence of Rb#11, but without UV-crosslink; XL: Immunoprecipitation in the presence of both Rb#11 and UV cross-link. Bottom panel: the presence of FMRP on the nitrocellulose membrane was revealed by immunoblot using the monoclonal anti-FMRP 1R antibody. On the left, the apparent molecular weights (kDa) of proteins and RNPs are indicated. On the right, the presence of RNPs coimmunoprecipitated with FMRP in a dose-dependent manner but absent in the control lane (Ab-) is indicated by a vertical line. (B) Levels of RNA targets co-immunoprecipitated with FMRP were measured by qRT-PCR. Grin2a, Map1b, Ppp2ca, were the positive controls of this CLIP and showed a significant enrichment according to one sample t-test. Gapdh and Tbp, two RNA that are not targeted by FMRP were used as negative controls. (C–E) Schematic representation of the shared identified FMRP targets between our HITS-CLIP dataset and (C) proteins that were previously described to be differentially localized in cortical synaptosomes between WT and Fmr1-KO mice and (D) mRNAs localized at the neuropil of rat CA1. (E) FMRP targets previously identified by CLIP analysis using polyribosome-associated RNAs and RNAs obtained by HEK-293 cells expressing an inducible and tagged FMRP. (F) Schematic representation of the shared identified FMRP targets between our HITS-CLIP restricted (only 50 counts) dataset and FMRP targets previously identified by CLIP analysis using polyribosome-associated RNAs and RNAs obtained by HEK-293 cells expressing an inducible and tagged FMRP. (G) Schematic representation of shared FMRP targets (restricted dataset) in the cortex (CX), hippocampus (HC) and cerebellum (CB). (H) Schematic representation of shared regulated pathways identified by Ingenuity Pathways analysis of FMRP targets in the cortex (CX), hippocampus (HC) and cerebellum (CB). See Supplementary Table S5 for details.
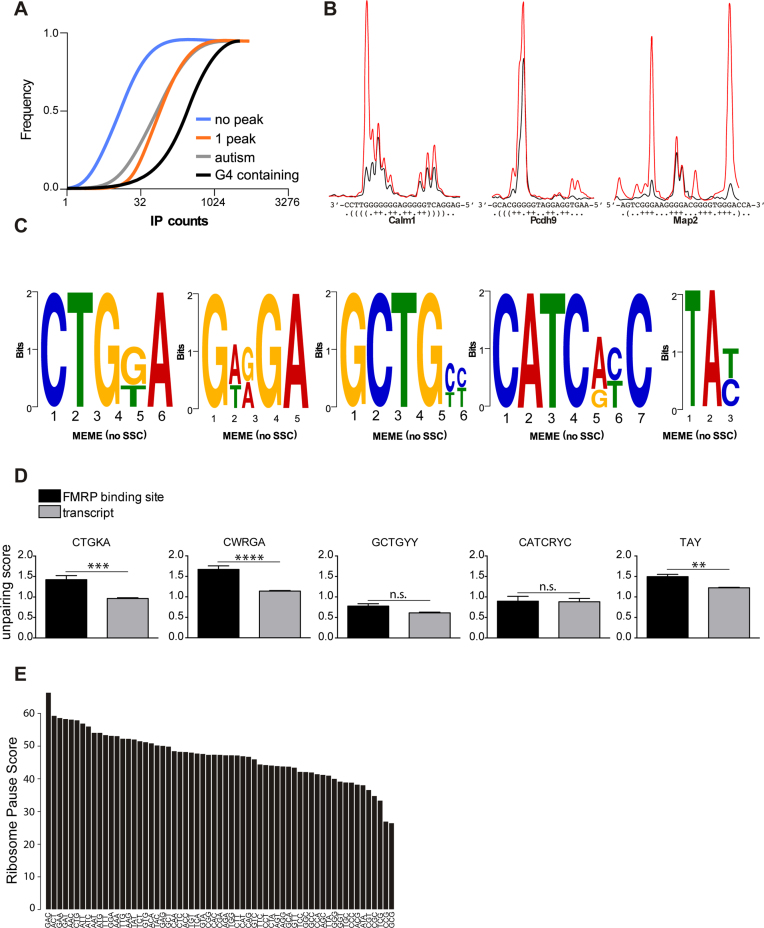
Figure 2.
Preferred motifs bound by FMRP. (A) Cumulative distribution of counts in immuno-precipitated transcripts. RNAs harboring one binding FMRP binding site, RNAs from SFARI database (07/2016) and predicted G-4-containing transcripts are enriched in the immuno-precipitated material. Genes that are not conserved across species and the RNA that were not detected in the IP were removed from the analysis. (B) RNAs from FMRP bound regions in Calm1, Pscdh9 and Map2 were produced and folded in vitro in the presence of K+ (red line) or Li+ ions (blue line). G-4 forming structures were mapped by fluorescent primer extension, fragments were separated by capillary electrophoresis and signals processed with QuShape (20). The sharp increase of the red over the blue signal highlights strong RT-stops in the presence of K+ ion. (C) DREME analysis of FMRP binding sites in its target mRNAs shows a significant enrichment for the CTGKA (E-value = 7.7e−40); GCTGYY (E-value = 9.4e−28); GWRGA (E-value = 2.4e−25); CATCRYC (E-value = 1.9e−17) and TAY (E-value = 2.0e−16) motifs. (D) FMRP recognizes unpaired motifs presented in stem-loop structures. Each nucleotide of a given transcript containing an FMRP binding site was attributed a Watson–Crick pairing score according to the Guo and Bartel dataset (18). Scores were computed for each motif when present in a FMRP binding site or outside in the corresponding transcript. Results are presented as mean ± SEM. Statistical significance was assessed by the Wilcoxon matched-pairs signed rank test; n.s.: not significant; **P < 0.005; ***P < 0.0005; ****P < 0.0001. (E) FMRP binds preferentially to coding sequences enriched in GAC codon. The ribosome pause score is computed based on the read coverage of each codon and normalized to its frequency in the ORF considering the expression and length of the ORF.
Motifs bound by FMRP
Consistent with previous studies we found that FMRP mostly binds cds regions of mRNA (59–68% of our identified FMRP binding sites) and 25–37% of FMRP binding sites are present in the 3′UTRs (Supplementary Figure S3A). Interestingly, one of the peaks overlaps with the Fmr1 G-4-forming structure, a site that has been described previously as bound by FMRP (19). We thus analyzed the folding of the peaks sequences with RNA fold (28) and identified 208 peaks overlapping strong RT-stop regions described in Guo et al., (18). We could also identify 149 G-4-forming structures, according to RNAfold (28), that are strikingly enriched in the CLIPed RNA (Figure 2A). These RNA structures were predominantly located in cds sequences (49.7%) of FMRP target mRNA, but G-4 are strikingly more abundant in 5′UTR compared to the other motifs we identified (23.5 versus 4.5–13%) (Supplementary Figure S3B). We validated in vitro the presence of G-4 forming structure in three of them (Calm1, Pcdh9 and Map2) by fluorescent primer extension (Figure 2B).
We studied the peak sequences of the three analyzed tissues with DREME (29) and we identified a consensus CTGKA-based motif (Figure 2C), and other less prominent motifs (Figure 2C). Taking advantage of the recent structure-seq dataset in mouse embryonic stem cells (18) we could assess whether the motifs that we identified were engaged in Watson-crick pairing in vivo. Our analysis clearly shows that the CTGKA, the GWRGA and the TAY motifs present in FMRP binding sites are more accessible to DMS modification in vivo (Figure 2D) and in vitro (not shown) than the unbound cognate motifs present in the same transcripts. This shows that FMRP recognizes motifs that are presented in single stranded regions or loop sequences of stem loop structures, suggesting a combinatorial RNA binding modality by FMRP through both structure and sequence recognition (Figure 2D).
It was proposed that FMRP could stall ribosomes onto mRNA thereby reducing the translation of its targets (13). We reasoned that FMRP could achieve such an effect by binding to specific mRNA regions that contain sequences or structures that slow down ribosome progression. We computed a ribosome pause score based on the read coverage of each codon, normalized to its frequency in the ORF also considering the expression and length of the ORF. Our approach therefore computes FMRP binding on codons based on the context sequence and expression of the different ORFs. Our results show that while GC rich codons are underrepresented from these sequences the GAC codon is strikingly enriched in FMRP binding sites (Figure 2E). This latter observation is consistent with a recent report (30). However, to our knowledge, the GAC codon has not been shown to interfere with ribosome elongation speed.
Pde2a mRNA is a prominent target of FMRP
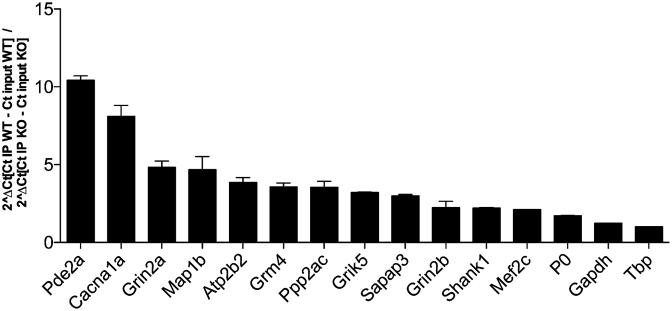
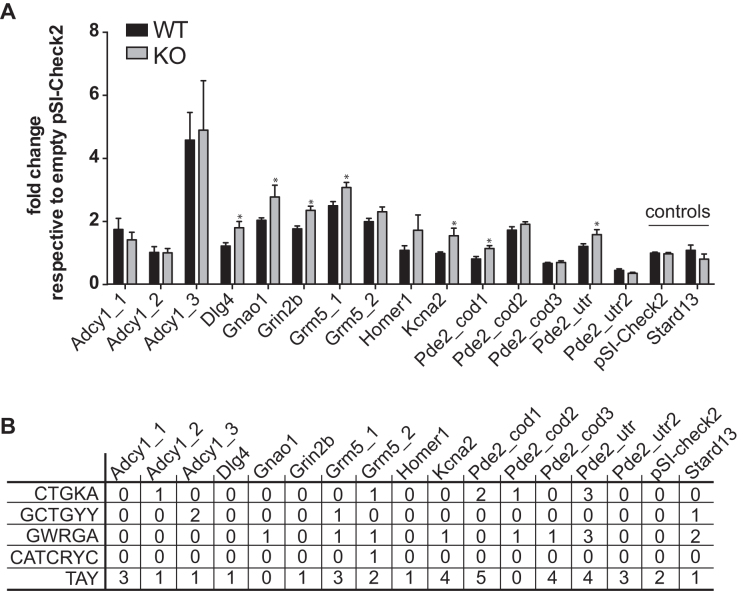
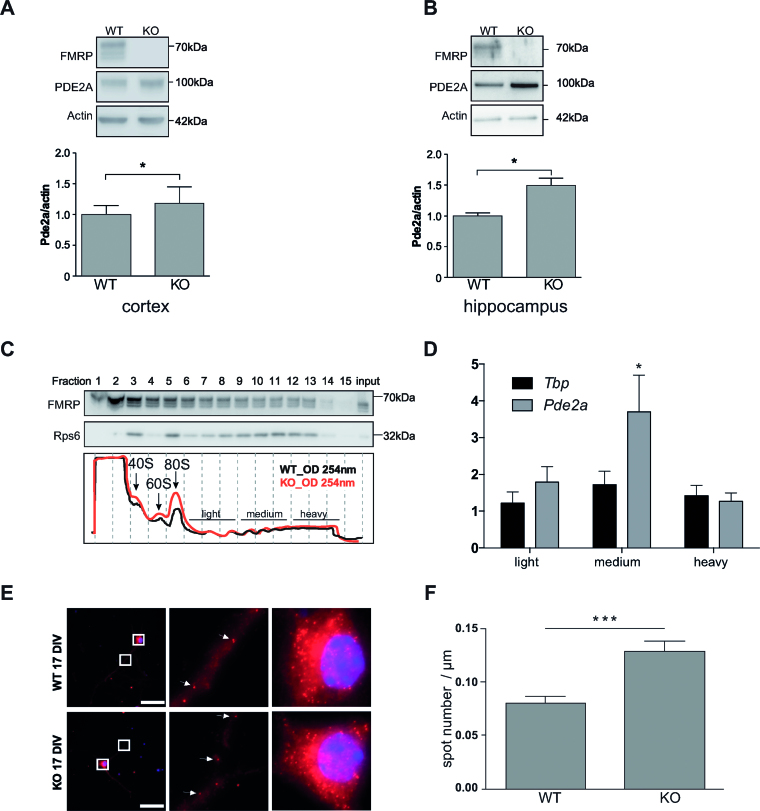
We tested by RT-qPCR the enrichment of prominent CLIPed RNAs (Pde2a, Cacna1a, Atp2b2, Grm4, Grik5, Shank1 and Mef2c) in addition to four positive controls (Grin2a, Map1b, Ppp2ca and Sapap3: RNAs that have been previously indicated as targets of FMRP) and three negative controls (P0, Gapdh and Tbp) (Figure 3) and we confirmed the results of CLIP. We sub-cloned in the 3′UTR of the luciferase reporter gene the peak sequence bound by FMRP in some of the best targets we identified and we transfected cell lines expressing or not Fmr1 mRNA. As shown in Figure 4A and B, all the best motifs bound by FMRP behave by repressing the translation in the presence of FMRP. Indeed, in the absence of FMRP the activity of luciferase is increased when its mRNA is bound to one of the motifs we identified (Figure 4A). We could also verify by RT-qPCR that the relative levels of expression of the Renilla luciferase mRNA of each construct were identical in the presence and in the absence of FMRP (not shown) when compared to the levels of the Firefly luciferase mRNA. The results we found using a reporter protein are consistent with the data of synaptic expression of the protein considered (26). For instance, PDE2A is overexpressed in the Fmr1-null cortical synapses of young mice [(26) and Figure 5] while the levels of Adenylate cyclase 1 are not different in the presence or in the absence of FMRP in the same extracts, even if its coding mRNA is bound by this protein. Suggesting that FMRP may modulate the metabolism of this mRNA in a step other than translation. We found that the Pde2a mRNA was the most enriched among the mRNAs co-immunoprecipitated with FMRP in cortex (Figure 3) and hippocampus (not shown). PDE2A is a phosphodiesterase involved in cAMP and cGMP degradation in various tissues including the brain (31–33). Interestingly, many of the pathways most represented in our IPA analysis are directly connected to cAMP/cGMP signaling and involve PDE2A (Supplementary Table S5). These findings suggest that the Pde2a mRNA is a crucial target of FMRP and may be involved in the physiopathology of FXS. While findings obtained using various approaches (34–37) highlight the presynaptic localization of PDE2A, we found that PDE2A is also post-synaptically located and partially co-localizes with PSD95 in dendritic spines of 17-day in vitro cultured cortical neurons (Supplementary Figure S4A). Furthermore, levels of Pde2a mRNA in cultured primary cortical neurons are increased in vitro upon neuron development (Supplementary Figure S4B). High-throughput sequencing showed that Pde2a mRNA levels are similar in the cortex of 13-day-old Fmr1-KO and WT mice (Supplementary Figure S4C) and, even if other PDEs are expressed in this brain area, Pde2a mRNA levels are the highest (Supplementary Figure S4C). An increased abundance of PDE2A was reported in synaptosomes prepared from the brain of mice at 17 days of age, but not at the adult age (26). Consistent with these results, we found that PDE2A protein levels are increased in highly purified synaptosomal fractions obtained from cortices and hippocampi of 13-day-old Fmr1-null mice compared with WT animals (Figure 5A, B). These data, together with the findings reported in Figure 4, suggested that FMRP negatively modulates the translation of the Pde2a mRNA. We thus carried out polyribosome fractionation on cortical extracts from 13-day-old WT and Fmr1-KO (Figure 5C) and measured Pde2a mRNA levels by RT-qPCR in pooled fractions (Figure 5D). We showed that, compared to WT cortical tissue, Pde2a mRNA is more abundantly associated with polyribosomes obtained from Fmr1-KO tissue. This result is consistent with an upregulated translation of this mRNA in the absence of FMRP. We assumed that a similar mechanism of FMRP-dependent regulation occurs in the hippocampus, since PDE2A levels are increased in Fmr1-KO hippocampi. To assess the implication of FMRP in the dendritic transport, the level of the Pde2a mRNA was measured by smFISH (38) both in WT and Fmr1-KO primary cortical neurons at 17 days in vitro. Compared to WT, the abundance of Pde2a mRNA was higher in neurites of Fmr1-KO neurons (Figure 5E–F) while no difference was observed in the soma (not shown). Collectively, these results strongly suggest that FMRP negatively modulates the translation of the Pde2a mRNA as well as its axonal/dendritic transport. An increased level of PDE2A is predicted to impact on the intracellular amount of cAMP and cGMP both in cortex and hippocampus.
Figure 3.
Validation of target mRNAs of FMRP. FMRP was immunoprecipitated from two independent UV-crosslinked assays. FMRP-associated RNAs were quantified by RT-qPCR. All RNAs except P0, Gapdh and Tbp (these mRNAs are not a target of FMRP) showed a significant enrichment according to one sample t-test.
Figure 4.
Role of RNA motifs bound by FMRP in translational regulation. (A) Influence of FMRP on luciferase reporter gene assay bearing motifs bound by FMRP or an unrelated control. pSI-CHECK-2 plasmids expressing Renilla carrying the various peak sequences in its’ 3′-UTR were transfected in STEK cells expressing or not FMRP. At least three-independent experiments with two biological replicates, for each transfection were quantified. For each transfection Renilla luciferase activity was normalized with Firefly luciferase activity. Ratios for all conditions were divided by the mean of the ratio measured for the empty vector (pSI-CHECK-2). Results are presented as the mean ±SEM (t-test, *P < 0.05). Empty vector and pSI-CHECK-2 carrying a sequence not bound by FMRP are named ‘controls’. (B) Summary Table reporting the motifs present in the various sequence analyzed.
Figure 5.
Characterization of Pde2A mRNA as predominant target of FMRP. (A) Synaptoneurosomes from 13 days old WT and Fmr1-KO cortices and (B) hippocampi were purified and proteins extracted as described in the ‘Material and Methods’ section. The presence of FMRP and PDE2A was revealed by western blot. Upper panels/representative western-blot of FMRP, PDE2A and β-Actin protein levels in synaptosomal extracts from cortex (A) and hippocampus (B). Densitometric quantification of immunoblots reveal that the absence of FMRP (KO) leads to a significant increase in PDE2A protein levels relative to controls (WT). Data are presented as mean ± SEM of n = 4 independent samples (Mann–Whitney test * P < 0.05). (C) Cortical protein extracts from PND 13 WT and Fmr1-KO male mice were analyzed by sedimentation velocity through a 20–50% sucrose gradient. Fifteen fractions were generated. The integrity and distribution of polyribosomes were based on the 254 nm UV profile (in the lower panel representative profiles of two sucrose gradients preparations from WT and Fmr1-KO cortices, respectively, are shown). The distribution of polyribosomes was also verified by the presence of ribosomal protein S6 (rpS6), a core protein of the small ribosomal subunit. The presence of rpS6 and FMRP in the various fractions was detected by immunoblot (top panels) using specific antibodies for these two proteins. The name of the protein is indicated on the left while the molecular weight is indicated on the right of each immunoblot. RNAs purified from the indicated fractions were pooled and the abundance of Pde2a mRNA measured by qRT-PCR. (D) Fold changes in Tbp or Pde2a mRNA levels in the pooled fractions were measured as described in the ‘Materials and Methods’ section. Mean ± SEM from three independent experiments (three WT and three Fmr1-KO brain were used) are shown. One sample t-test was performed for each mRNA in the pooled fractions, *P < 0.05 (E) Left panels: representative pictures of cells expressing the Pde2a mRNA detected by smFISH in WT and Fmr1-KO cultured cortical neurons. Scale bar: 50 μm. Images of cell bodies and dendrites boxed in this panel are enlarged (zoom 900×) in middle and right panels, respectively. Each dot corresponds to a single RNA molecule. White arrows indicate examples of individual mRNA molecules. (F) Number of transported mRNAs (spots) localized in the distal segment of dendrites of 17 days in vitro WT (n = 44) or Fmr1-KO (n = 60) neurons. Mean ± SEM is shown. Statistical significance was calculated using the unpaired t-test, ***P < 0.0005.
DISCUSSION
Despite a wealth of research on FXS, the mechanistic action of FMRP (e.g. the RNA motifs that are targeted and the functional role of FMRP/RNA interaction) remains unclear and a specific and effective therapy for FXS is not yet available. With the purpose of addressing both of these issues, we define here a set of mRNA targets of FMRP at a critical step of postnatal development of the brain. We performed HITS-CLIP in mouse cerebellum, hippocampus and cortex at PND 13, when FMRP is most highly expressed (10,11) and the number of synapses reaches the highest level (12). We identified the largest set of FMRP mRNA targets in the mouse brain to date, including 1610 new targets, 57 of which have been previously linked to autism (SFARI list). Considering a restricted list of targets harboring at least 50 counts in the immunoprecipitated material from each tissue, we identified 193 new targets, 80 of which were found in the cerebellum IP and 36 in the hippocampus further illustrating the need to perform tissue-restricted CLIP analysis to identify a comprehensive set of FMRP RNA targets.
Indeed, another study considered only FMRP targets associated to polyribosomes (13), while we immunoprecipitated FMRP from various subcellular localizations. This confirms that FMRP is involved in functions other than translational regulation and suggests that this protein interacts with different RNAs at different steps of their metabolism. Indeed, we show here that FMRP negatively modulates both translation and transport of the Pde2a mRNA at the synapse. Overall, we identified fewer RNA targets than Ascano et al. (8) but their study used overexpression of FMRP in a cell line also expressing endogenous FMRP (8), overexpression that could have biased the mRNA binding repertoire of FMRP. Furthermore, even if HEK share a large proportion of their transcriptome with the brain tissue, the authors have probably identified RNA targets of FMRP that are not neuronally expressed.
Molecular bases of FMRP/RNA interaction
The ACUK/WGGA motif (8) was previously shown to be necessary (at least in HEK cells) but not sufficient to mediate FMRP/RNA interaction (1,39,40). Our dataset shows an enrichment of G-4-forming structures in FMRP-bound RNA fragments, which further confirms the role of G-4 in mediating FMRP/RNA interaction. However, as now clearly established, not all FMRP mRNA targets harbor this motif (41). Indeed, we identified here a novel and very significantly enriched consensus motif (CTGKA) bound by FMRP, suggesting that we defined the prominent RNA motif driving FMRP-dependent translation modulation in brain. Among the less prominent motifs we found, two of them (TAY and GWRGA) are very similar to those identified by another study (30) combining results from the two previous CLIP assays (8,13). However, the other motifs we found appear specific to our study, suggesting that our approach (in different brain areas), combined with an antibody efficiently precipitating FMRP, allowed the identification not only of new targets (see later) but also of new binding-motifs. All the motifs (alone or in combination) are able to negatively modulate FMRP-driven translational regulation consistent with the well-established function of FMRP as a repressor of translation, even if some exceptions have been described (9,23,42). The fact that we found that some RNA regions bound by FMRP do not have an impact on the FMRP-translational modulation (Figure 4A) suggests that their interaction with FMRP mediate a function other than translation regulation. Alternatively, additional co-factors are needed to achieve such a regulation in a given cell type or in a spatio/temporal control. In other cases, the reasons could be more complexes and suggest an implication of FMRP in large number of regulation of RNA metabolism, not only including intracellular transport but possibly an implication in RNA folding and/or RNPs assembly. Also, the sequences we—and others—found are relatively short and this strongly indicate that their location in a special structural context is crucial for FMRP recognition, as we have already suggested (39). This suggestion is now validated by showing that three of the motifs bound by FMRP in brain, (CTGKA, GWRGA, TAY) are recognized in the context of a single strand RNA region (likely a loop structure). Interestingly, Taliaferro et al. recently showed that the structural context in which a given sequence motif is crucial for the specificity of binding of the cognate RNA-binding protein (43). Similar studies will be needed to decipher the determinants of this structural context that modulate FMRP binding onto its target sequences. The combinatorial recognition of its target sequence by FMRP may be explained by the presence of multiple RNA binding domains in this protein. On one hand, the RGG-box was already shown to recognize several structures and a combined sequence/structure (7,15,23,44) and, on the other hand, the KH2 domain was shown to recognize a complex structure, the kissing complex, (45). Even if a pathological mutation in the KH2 (e.g. I304N) does not prevent the ability of this protein to bind RNA (7), this mutant FMRP is not associated to polyribosomes, is localized in small RNAse-resistant RNPs (46) and impacts the size and number of RNA granules (47,48). Furthermore, deletion of the RGG box resulted in a less efficient association to polyribosomes (49). Collectively, these findings suggest that, even if these two RNA binding domains can act in a modular manner, the precise recognition of the mRNA targets and their integration of functional RNPs depends on the integrity of both RNA-binding domains.
By analyzing the codon composition of FMRP binding sites in coding sequences we showed that, intriguingly, the GAC codon is overrepresented and GC-rich codons are underrepresented in these sequences. The GAC codon is not particularly rare and its corresponding tRNA has an average level of expression. We believe that this further argues in favor of the requirement of a structural motif (i.e. a sequence presented in a structure context) bound by FMRP. In addition to the presence of RNA motifs and its surrounding sequences, the specificity of binding of this protein might imply the action of FMRP-interacting proteins that are components of FMRP-containing RNPs and that can interfere (compete/cooperate/prevent) with the function of FMRP. In this context, we can only argue that to assess the functional significance of the FMRP/target interaction and the molecular role of RNA target encoded proteins in the physiopathology of FXS, the critical targets of FMRP must be studied individually in the appropriate cellular context. Indeed, FMRP may play crucial regulatory roles in the metabolism of cell-specific transcripts even in cell types where it is not highly expressed. For this reason, we studied the cell origin of FMRP targets. Some of these previously overlooked targets include Slc1a2 in astrocytes/ependymal cells, Mef2c in pyramidal cells from CA1, Scd2 and Marcks in oligodendrocytes, and Rapgef4 in endothelial mural cells. These results also suggest the functional importance of FMRP in glia, even if in these cells its expression is lower than in pyramidal neurons. Moreover, it is interesting that some FMRP RNA targets are particularly abundant in a given tissue, such as Grm4, Fat2 or Unc5C that are specific to the cerebellum, Camk2a, Kif5a and Agap2 that are more enriched in immunoprecipitations from hippocampi, or Calm1 and Enc1 that are more enriched in IPs from cortex. Pde2a is only bound by FMRP in cortex and hippocampus (Supplementary Table S1). Considering that the role of FMRP in the various steps of RNA metabolism is modulated through the presence of its interacting partners, as we have previously shown (23,43), we strongly believe that our results are critical to unravel cell-dependent FMRP regulations. In conclusion, we can predict that the functional relevance of each target can be studied individually. A similar approach was used to understand the role of the RNA G-4 in the molecular bases of FXS. This motif has been shown to modulate the FMRP-dependent translation (39,41) but not FMRP-dependent dendritic transport (50). Conversely, the complex with FXR1P (paralog of FMRP)/G-4 has also been involved in the control of RNA stability (51).
Identification of mRNAs target of FMRP critical for FXS physiopathology
Our approach allowed us to define the role of FMRP in various brain areas by investigating its implication in the regulation of mRNAs expressed in specific brain regions. A large overlap is observed between FMRP targets in cortex and in hippocampus, while targets from cerebellum appear more divergent. However, specific targets for each analyzed area have been found and these identifications could be possible only by the ‘regionalized’ approach. Gene Ontology analysis allowed to point out that FMRP targets several pathways in the brain like CREB signaling in neurons (Supplementary Table S5), axonal guidance, RAR activation, Rac signaling, regulation of p70s6k signaling and synaptic long-term depression and long-term potentiation. Of particular interest is the specificity of the Gai signaling in the hippocampus as well as the iNos and Tgf-ß signaling in the cortex. In addition, our analysis also pointed out that cAMP/cGMP is one the most prominent deregulated pathways in the Fmr1-KO mouse brain and that the Pde2a mRNA is one prominent target of FMRP. We show here that in the absence of FMRP the level of PDE2A is elevated both in cortex and hippocampus, implying a reduced level of cAMP and cGMP in those brain areas in Fmr1-null mice. Indeed, the PDE2A enzyme is involved in cGMP-dependent degradation of both cAMP and cGMP, two second messengers at the crossroad of many signaling pathways which modulate a large array of intracellular processes in neurons strongly impacting memory and cognition (52). In neurons, PDE2A is mainly synaptic and exerts both pre- and post-synaptic functions [this study and (37)] being involved in synaptogenesis (53) and synaptic plasticity (37,54). All these considerations suggest that this protein is a putative therapeutic target for an effective treatment of FXS.
We focused here on Pde2a mRNA, a mRNA target of FMRP in hippocampus and cortex, two brain areas whose dysfunction is most likely causative to cognitive and behavior deficits in FXS. However, we would like to underline that it is known that altered cerebellum volume and connectivity are associated with etiology of Autism Spectrum Disorder (ASD) (55,56). However, specific cerebellar pathways involved in those pathophysiological mechanisms are poorly described. Thus, our analysis of the FMRP targets in this tissue may help in revealing them. For instance, the implication and regulation of (Estrogen Receptor) Erb2 and Erb3 by FMRP is very intriguing and, if extended to ASD, could provide a molecular clue to explain the 1 to 4 ratio between female and male ASD patients (57). In addition, for instance, one of the pathways specifically modulated by FMRP in cerebellum is the GDNF (Glial Cell Line-derived Neurotrophic Factor) that has been recently associated to normal cerebellar motor learning (58). In conclusion, all these pathways may contribute to cellular and/or behavioral phenotypes of FXS.
DATA AVAILABILITY
Sequencing data have been submitted to GEO: GSE104269.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Lalli and M. Capovilla for critical reading of the manuscript, M. Drozd for discussion and N. Durand for technical help.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Université Côte d’Azur; INSERM; CNRS; Agence Nationale de Recherches [ANR-11-LABX-0028-01, ANR-12-BSV4-0020, ANR-12-SVSE8-0022, ANR-15-CE16-0015]; Fondation Jeéroôme Lejeune; Monaco Against Autism Foundation; FRM [DEQ20140329490 to B.B, FRM-ING20140129004 to B.B., T.M.]; FRAXA Foundation (to T.M.); ‘Signalife Program’ International PhD Fellowship (to S.C.). Funding for open access charge: FRM [DEQ20140329490].
Conflict of interest statement. None declared.
REFERENCES
- 1. Maurin T., Zongaro S., Bardoni B.. Fragile X syndrome: from molecular pathology to therapy. Neurosci. Biobehav. Rev. 2014; 46:242–255. [DOI] [PubMed] [Google Scholar]
- 2. Bassell G.J., Warren S.T.. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008; 60:201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sidorov M.S., Auerbach B.D., Bear M.F.. Fragile X mental retardation protein and synaptic plasticity. Mol. Brain. 2013; 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Erickson C.A., Davenport M.H., Schaefer T.L., Wink L.K., Pedapati E.V., Sweeney J.A., Fitzpatrick S.E., Brown W.T., Budimirovic D., Hagerman R.J. et al. Fragile X targeted pharmacotherapy: lessons learned and future directions. J. Neurodev. Disord. 2017; 9:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. van Karnebeek C.D., Bowden K., Berry-Kravis E.. Treatment of neurogenetic developmental conditions: from 2016 into the future. Pediatr. Neurol. 2016; 65:1–13. [DOI] [PubMed] [Google Scholar]
- 6. Castagnola S., Bardoni B., Maurin T.. The search for an effective therapy to treat fragile X syndrome: dream or reality?. Front. Synaptic Neurosci. 2017; 9:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darnell J.C., Jensen K.B., Jin P., Brown V., Warren S.T., Darnell R.B.. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001; 107:489–499. [DOI] [PubMed] [Google Scholar]
- 8. Ascano M. Jr, Mukherjee N., Bandaru P., Miller J.B., Nusbaum J.D., Corcoran D.L., Langlois C., Munschauer M., Dewell S., Hafner M. et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012; 492:382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tabet R., Moutin E., Becker J.A., Heintz D., Fouillen L., Flatter E., Krezel W., Alunni V., Koebel P., Dembele D. et al. Fragile X mental retardation protein (FMRP) controls diacylglycerol kinase activity in neurons. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E3619–E3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davidovic L., Navratil V., Bonaccorso C.M., Catania M.V., Bardoni B., Dumas M.E.. A metabolomic and systems biology perspective on the brain of the fragile X syndrome mouse model. Genome Res. 2011; 21:2190–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bonaccorso C.M., Spatuzza M., Di Marco B., Gloria A., Barrancotto G., Cupo A., Musumeci S.A., D’Antoni S., Bardoni B, Catania M.V.. Fragile X mental retardation protein (FMRP) interacting proteins exhibit different expression patterns during development. Int. J. Dev Neurosci. 2015; 42:15–23. [DOI] [PubMed] [Google Scholar]
- 12. Semple B.D., Blomgren K., Gimlin K., Ferriero D.M., Noble-Haeusslein L.J.. Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013; 106–107:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Darnell J.C., Van Driesche S.J., Zhang C., Hung K.Y., Mele A., Fraser C.E., Stone E.F., Chen C., Fak J.J., Chi S.W. et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011; 146:247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr, Jungkamp A.C., Munschauer M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010; 141:129–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Spitzer J., Hafner M., Landthaler M., Ascano M., Farazi T., Wardle G., Nusbaum J., Khorshid M., Burger L., Zavolan M. et al. PAR-CLIP (Photoactivatable Ribonucleoside-Enhanced Crosslinking and Immunoprecipitation): a step-by-step protocol to the transcriptome-wide identification of binding sites of RNA-binding proteins. Methods Enzymol. 2014; 539:113–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Konig J., Zarnack K., Rot G., Curk T., Kayikci M., Zupan B., Turner D.J., Luscombe N.M., Ule J.. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat. Struct. Mol. Biol. 2010; 17:909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Althammer S., Gonzalez-Vallinas J., Ballare C., Beato M., Eyras E.. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics. 2011; 27:3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schaeffer C., Bardoni B., Mandel J.L., Ehresmann B., Ehresmann C., Moine H.. The fragile X mental retardation protein binds specifically to its mRNA via a purine quartet motif. EMBO J. 2001; 20:4803–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Karabiber F., McGinnis J.L., Favorov O.V., Weeks K.M.. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA. 2013; 19:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zeisel A., Munoz-Manchado A.B., Codeluppi S., Lonnerberg P., La Manno G., Jureus A., Marques S., Munguba H., He L., Betsholtz C. et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015; 347:1138–1142. [DOI] [PubMed] [Google Scholar]
- 22. Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009; 55:611–622. [DOI] [PubMed] [Google Scholar]
- 23. Bechara E.G., Didiot M.C., Melko M., Davidovic L., Bensaid M., Martin P., Castets M., Pognonec P., Khandjian E.W., Moine H. et al. A novel function for fragile X mental retardation protein in translational activation. PLoS Biol. 2009; 7:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Castets M., Schaeffer C., Bechara E., Schenck A., Khandjian E.W., Luche S., Moine H., Rabilloud T., Mandel J.L., Bardoni B.. FMRP interferes with the Rac1 pathway and controls actin cytoskeleton dynamics in murine fibroblasts. Hum. Mol. Genet. 2005; 14:835–844. [DOI] [PubMed] [Google Scholar]
- 25. Tsanov N., Samacoits A., Chouaib R., Traboulsi A.M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. et al. smiFISHand FISH-quant—a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016; 44:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tang B., Wang T., Wan H., Han L., Qin X., Zhang Y., Wang J., Yu C., Berton F., Francesconi W. et al. Fmr1 deficiency promotes age-dependent alterations in the cortical synaptic proteome. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4697–E4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cajigas I.J., Tushev G., Will T.J., tom Dieck S., Fuerst N., Schuman E.M.. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012; 74:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey T.L. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011; 27:1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Anderson B.R., Chopra P., Suhl J.A., Warren S.T., Bassell G.J.. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016; 44:6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaccolo M., Movsesian M.A.. cAMP and cGMP signaling cross-talk: role of phosphodiesterases and implications for cardiac pathophysiology. Circ. Res. 2007; 100:1569–1578. [DOI] [PubMed] [Google Scholar]
- 32. Averaimo S., Nicol X.. Intermingled cAMP, cGMP and calcium spatiotemporal dynamics in developing neuronal circuits. Front. Cell Neurosci. 2014; 8:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maurice D.H., Ke H., Ahmad F., Wang Y., Chung J., Manganiello V.C.. Advances in targeting cyclic nucleotide phosphodiesterases. Nat. Rev. Drug Discov. 2014; 13:290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Russwurm C., Zoidl G., Koesling D., Russwurm M.. Dual acylation of PDE2A splice variant 3: targeting to synaptic membranes. J. Biol. Chem. 2009; 284:25782–25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu F., Ren J., Zhang J.E., Zhong W., Luo M.. Natriuretic peptides block synaptic transmission by activating phosphodiesterase 2A and reducing presynaptic PKA activity. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:17681–17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boyken J., Gronborg M., Riedel D., Urlaub H., Jahn R., Chua J.J.. Molecular profiling of synaptic vesicle docking sites reveals novel proteins but few differences between glutamatergic and GABAergic synapses. Neuron. 2013; 78:285–297. [DOI] [PubMed] [Google Scholar]
- 37. Fernandez-Fernandez D., Rosenbrock H., Kroker K.S.. Inhibition of PDE2A, but not PDE9A, modulates presynaptic short-term plasticity measured by paired-pulse facilitation in the CA1 region of the hippocampus. Synapse. 2015; 69:484–496. [DOI] [PubMed] [Google Scholar]
- 38. Tsanov N., Samacoits A., Chouaib R., Traboulsi A.M., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Peter M. et al. smiFISH and FISH-quant—a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. 2016; 44:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maurin T., Melko M., Abekhoukh S., Khalfallah O., Davidovic L., Jarjat M., D’Antoni S., Catania M.V., Moine H., Bechara E. et al. The FMRP/GRK4 mRNA interaction uncovers a new mode of binding of the Fragile X mental retardation protein in cerebellum. Nucleic Acids Res. 2015; 43:8540–8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Suhl J.A., Chopra P., Anderson B.R., Bassell G.J., Warren S.T.. Analysis of FMRP mRNA target datasets reveals highly associated mRNAs mediated by G-quadruplex structures formed via clustered WGGA sequences. Hum. Mol. Genet. 2014; 23:5479–5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Melko M., Bardoni B.. The role of G-quadruplex in RNA metabolism: involvement of FMRP and FMR2P. Biochimie. 2010; 92:919–926. [DOI] [PubMed] [Google Scholar]
- 42. Gross C., Yao X., Pong D.L., Jeromin A., Bassell G.J.. Fragile X mental retardation protein regulates protein expression and mRNA translation of the potassium channel Kv4.2. J. Neurosci. 2011; 31:5693–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Taliaferro J.M., Lambert N.J., Sudmant P.H., Dominguez D., Merkin J.J., Alexis M.S., Bazile C., Burge C.B.. RNA sequence context effects measured in vitro predict in vivo protein binding and regulation. Mol. Cell. 2016; 64:294–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bechara E., Davidovic L., Melko M., Bensaid M., Tremblay S., Grosgeorge J., Khandjian E.W., Lalli E., Bardoni B.. Fragile X related protein 1 isoforms differentially modulate the affinity of fragile X mental retardation protein for G-quartet RNA structure. Nucleic Acids Res. 2007; 35:299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Darnell J.C., Fraser C.E., Mostovetsky O., Stefani G., Jones T.A., Eddy S.R., Darnell R.B.. Kissing complex RNAs mediate interaction between the fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005; 19:903–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zang J.B., Nosyreva E.D., Spencer C.M., Volk L.J., Musunuru K., Zhong R., Stone E.F., Yuva-Paylor L.A., Huber K.M., Paylor R. et al. A mouse model of the human fragile X syndrome I304N mutation. PLoS Genet. 2009; 5:e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schrier M., Severijnen L.A., Reis S., Rife M., van’t Padje S., van Cappellen G., Oostra B.A., Willemsen R.. Transport kinetics of FMRP containing the I304N mutation of severe fragile X syndrome in neurites of living rat PC12 cells. Exp. Neurol. 2004; 189:343–353. [DOI] [PubMed] [Google Scholar]
- 48. Levenga J., Buijsen R.A., Rifé M., Moine H., Nelson D.L., Oostra B.A., Willemsen R., de Vrij F.M.. Ultrastructural analysis of the functional domains in FMRP using primary hippocampal mouse neurons. Neurobiol. Dis. 2009; 35:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mazroui R., Huot M.E., Tremblay S., Boilard N., Labelle Y., Khandjian E.W.. Fragile X Mental Retardation protein determinants required for its association with polyribosomal mRNPs. Hum. Mol. Genet. 2003; 12:3087–3096. [DOI] [PubMed] [Google Scholar]
- 50. Subramanian M., Rage F., Tabet R., Flatter E., Mandel J.L., Moine H.. G-quadruplex RNA structure as a signal for neurite mRNA targeting. EMBO Rep. 2011; 12:697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davidovic L., Durand N., Khalfallah O., Tabet R., Barbry P., Mari B., Sacconi S., Moine H., Bardoni B.. A novel role for the RNA-binding protein FXR1P in myoblasts cell-cycle progression by modulating p21/Cdkn1a/Cip1/Waf1 mRNA stability. PLoS Genet. 2013; 9:e1003367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gomez L., Breitenbucher J.G.. PDE2 inhibition: potential for the treatment of cognitive disorders. Bioorg. Med. Chem. Lett. 2013; 23:6522–6527. [DOI] [PubMed] [Google Scholar]
- 53. Boess F.G., Hendrix M., van der Staay F.J., Erb C., Schreiber R., van Staveren W., de Vente J., Prickaerts J., Blokland A., Koenig G.. Inhibition of phosphodiesterase 2 increases neuronal cGMP, synaptic plasticity and memory performance. Neuropharmacology. 2004; 47:1081–1092. [DOI] [PubMed] [Google Scholar]
- 54. Domek-Lopacinska K., Strosznajder J.B.. The effect of selective inhibition of cyclic GMP hydrolyzing phosphodiesterases 2 and 5 on learning and memory processes and nitric oxide synthase activity in brain during aging. Brain Res. 2008; 1216:68–77. [DOI] [PubMed] [Google Scholar]
- 55. Traut N., Beggiato A., Bourgeron T., Delorme R., Rondi-Reig L., Paradis A.L., Toro R.. Cerebellar volume in autism: literature meta-analysis and analysis of the autism brain imaging data exchange cohort. Biol. Psychiatry. 2018; 83:579–588. [DOI] [PubMed] [Google Scholar]
- 56. Stoodley C.J., D’Mello A.M., Ellegood J., Jakkamsetti V., Liu P., Nebel M.B., Gibson J.M., Kelly E., Meng F., Cano C.A. et al. Altered cerebellar connectivity in autism and cerebellar-mediated rescue of autism-related behaviors in mice. Nat. Neurosci. 2017; 20:1744–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Loomes R., Hull L., Mandy W.P.L.. What is the male-to-female ratio in autism spectrum disorder? A systematic review and Meta-Analysis. J. Am. Acad. Child Adolesc. Psychiatry. 2017; 56:466–474. [DOI] [PubMed] [Google Scholar]
- 58. Sergaki M.C., López-Ramos J.C., Stagkourakis S., Gruart A., Broberger C., Delgado-García J.M., Ibáñez C.F.. Compromised survival of cerebellar molecular layer interneurons lacking GDNF receptors GFRα1 or RET impairs normal cerebellar motor learning. Cell Rep. 2017; 19:1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been submitted to GEO: GSE104269.