Abstract
In all cells, initiation of chromosome replication depends on the activity of AAA+ initiator proteins that form complexes with replication origin DNA. In bacteria, the conserved, adenosine triphosphate (ATP)-regulated initiator protein, DnaA, forms a complex with the origin, oriC, that mediates DNA strand separation and recruitment of replication machinery. Complex assembly and origin activation requires DnaA-ATP, which differs from DnaA-ADP in its ability to cooperatively bind specific low affinity sites and also to oligomerize into helical filaments. The degree to which each of these activities contributes to the DnaA-ATP requirement for initiation is not known. In this study, we compared the DnaA-ATP dependence of initiation from wild-type Escherichia coli oriC and a synthetic origin (oriCallADP), whose multiple low affinity DnaA sites bind DnaA-ATP and DnaA-ADP similarly. OriCallADP was fully occupied and unwound by DnaA-ADP in vitro, and, in vivo, oriCallADP suppressed lethality of DnaA mutants defective in ATP binding and ATP-specific oligomerization. However, loss of preferential DnaA-ATP binding caused over-initiation and increased sensitivity to replicative stress. The findings indicate both DnaA-ATP and DnaA-ADP can perform most of the mechanical functions needed for origin activation, and suggest that a key reason for ATP-regulation of DnaA is to control replication initiation frequency.
INTRODUCTION
In all organisms, initiation of chromosome replication must be precisely timed during the cell cycle, since delayed or under-initiation leads to eventual chromosome loss, while re-initiation from the same origin can result in replication fork collapse and genome instability (1). Although diverse mechanisms exist among different domains of life to regulate the onset of chromosome replication (2), much of the control focuses on formation, activation and/or inactivation of initiator protein complexes that bind to and unwind replication origin DNA, recruit replicative helicase and help load replication proteins (3).
In bacteria, the complex that triggers initiation of chromosome replication is termed the orisome, and its assembly is best characterized in the model organism, Escherichia coli (4). Escherichia coli orisomes assemble in stages, as monomers of the highly conserved initiator protein, DnaA, bind first to widely spaced high affinity (Kd = 4–20 nM) (5) recognition sites in the replication origin, oriC, and then to arrayed low affinity (Kd > 200 nM) sites positioned in gap regions between strong sites (Figure 1A) (4).
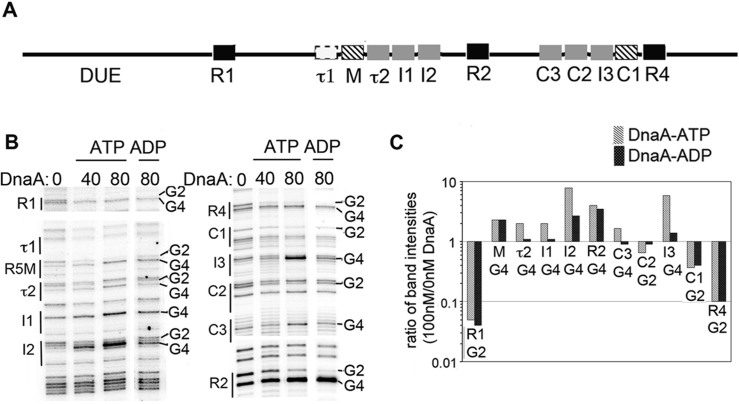
Figure 1.
Characterization of low affinity DnaA binding sites in Escherichia coli oriC. (A) Map of oriC, showing low affinity DnaA recognition sites that preferentially bind DnaA-ATP (gray boxes) and high and low affinity sites that bind both nucleotide forms (black and hatched boxes, respectively). The putative τ1 site is marked with a dashed outline. The DUE is indicated. (B) DMS footprint comparing in vitro binding of purified DnaA-ATP and DnaA-ADP to oriC. Binding sites and positions of the G2 and G4 residues diagnostic of DnaA binding are marked. (C) Quantitation of G2 or G4 band intensities for each site when bound to 80 nM DnaA-ATP (gray) or DnaA-ADP (black), relative to band intensity of site in the absence of DnaA.
DnaA contains four domains, each with distinct functions. Targeted mutations in Domains I, III and IV have revealed key roles for these domains in DnaA recruitment (I), binding (IV), oligomerization (I, III) and helicase loading (I, III), reviewed in (6,7). Further, as a member of the AAA+ family of adenosine triphosphatases (ATPases), DnaA has structural similarity to archaeal and eukaryotic initiators (8) and tightly binds both ATP and adenosine diphosphate (ADP) in domain III (9). While either form of DnaA can occupy high affinity recognition sites in oriC, (10), a subset of lower affinity sites preferentially bind DnaA-ATP (11,12). In vivo, DnaA is bound to high affinity sites throughout the cell cycle (13), but the origin is unwound, and initiation triggered, only when initiator binding extends to the low affinity sites (14–16). Low affinity site occupation requires cooperative binding, which is mediated by bound DnaA recruiting another DnaA molecule and positioning it so that it can bind the adjacent recognition sequence (17,18). Not all of the low affinity sites are essential for origin activation, but at least some of the DnaA-ATP preferring sites must be occupied, particularly in the left region of oriC (19). Thus, the long recognized DnaA-ATP requirement for in vitro unwinding (9) depends, at least in part, on this subset of origin binding sites.
DnaA-ATP is also uniquely capable of oligomerizing via interactions between AAA+ domains (domain III). The solved oligomeric structure is a helical filament having single-stranded (ss) DNA binding activity (20). A number of roles for the ssDNA-binding oligomer in orisome function have been suggested, including stabilization of the unwound complex (21), DNA strand separation by DNA stretching (22) and recruitment of the helicase loader (23). An alternative form of the DnaA-ATP oligomer is purported to associate with double-stranded (ds)DNA recognition sites in oriC, altering oriC topology as part of the unwinding mechanism (24,25).
Because origin binding by DnaA-ATP is a prerequisite for subsequent events in origin activation, it has not been possible to rigorously examine the requirement for this initiator form in all stages of orisome assembly. We have addressed this issue by designing a version of oriC (oriCallADP) that binds to both nucleotide forms of DnaA equivalently at all recognition sites, and using oriCallADP to examine the need for DnaA-ATP in origin unwinding and in vivo origin activation. We report that origin recognition, but not cooperative binding between low affinity sites, requires DnaA-ATP. Once bound, DnaA-ADP and DnaA-ATP have similar unwinding activities in vitro. Further, when harbored as the sole chromosomal replication origin, oriCallADP suppressed lethality of DnaA mutants with defects in ATP binding and ATP-dependent oligomer formation. These observations are consistent with DnaA-ATP being the active form of the initiator primarily because this form is required for occupation of key dsDNA low affinity binding sites in oriC. If allowed equivalent access to oriC, both DnaA-ADP and DnaA-ATP are capable of performing the mechanical functions required to trigger initiation. However, loss of preferential DnaA-ATP binding caused over-initiation and increased sensitivity to replicative stress, demonstrating that an important consequence of ATP-dependent DnaA binding is controlled replication initiation frequency.
MATERIALS AND METHODS
Chemicals, proteins, oligonucleotides and enzymes
Supplementary Material contains a description of the reagents used.
Bacterial strains and plasmids
The E. coli strains and plasmids used in this study, including details of their construction, are described in Supplementary Material.
Recombineering
OriC allADP was introduced into ACL402 by recombineering (26) (described in more detail in Supplementary Materials and Methods and Figure S1A). Replacement was verified by sequence analysis, using sequencing primers shown in Supplementary Table S2.
Dimethyl sulfate (DMS) footprinting
DnaA or DnaA(R285A) at the concentrations indicated in the figures was pre-incubated with 5 mM ATP or 5 mM ADP for 5 min before adding DNA, followed by dimethyl sulfate (DMS) modification of DNA and primer extension, and analysis as described (16). Primer sequences were 5′-GATCGCACGATCTGTATAC, for revealing R1-I2, and 5′-GGATCATTAACTGTGAATG, for revealing R4-R2. Representative scans are shown in the figures. Images were analyzed by using Bio-Rad Quantity One software. Intensities of bands in binding sites (relative to the total intensity of all selected bands) were calculated. Deviations in band intensities among experiments were <10%.
EMSA
Electrophoretic mobility shift assay (EMSA) was done as described previously (17) using end-labeled fragment and DnaA-ATP, DnaA-ADP or DnaA(R285A)-ATP. To generate the fragments, complementary single-stranded oligonucleotides: 5′-CAGTCATTGGTCATTCACAGCT CATTCACAGAGTTATCCACAGTAGATCGCA and its complement, or 5′-CAGTCATTGGTCATTCACAGCT CATTCACAGAGGATATAGTTGTAGATCGCA and its complement were annealed as described (17). Representative scans (from triplicate experiments) are shown, with some intervening lanes removed.
P1 endonuclease digestions
Supercoiled plasmid (10 fmol) in 20 μl of 40 mM HEPES-KOH, pH 7.6, 8 mm MgCl2, 30% glycerol, 0.32 mg/ml bovine serum albumin (BSA) and 20 ng hydroxyurea (HU) protein was incubated for 10 min at 38°C with varying mixtures of ATP and ADP (total concentration of nucleotide was 5 mM). DnaA-ATP and/or DnaA-ADP at the concentrations indicated in Figure 3, and 1.2 units of P1 endonuclease were then added, and reactions were incubated for 10–15 s. Reactions were stopped and samples processed as described in (16). Images were analyzed by using Bio-Rad Quantity One software. Triplicate experiments were done, with representative scans shown in Figure 3.
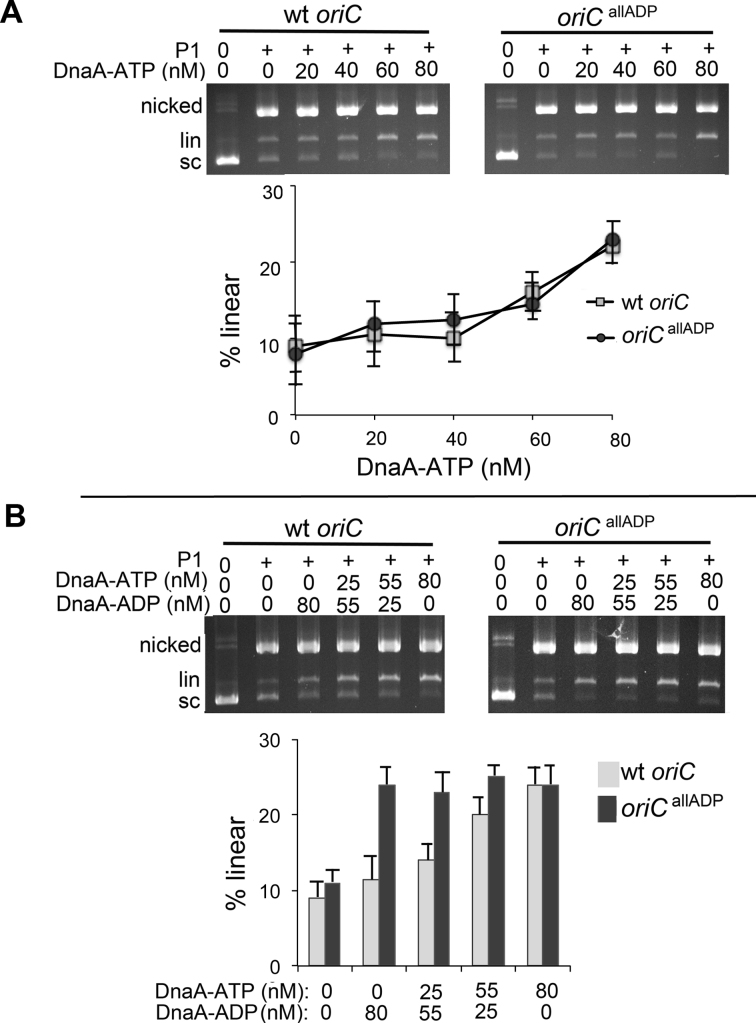
Figure 3.
DnaA-ADP is active in unwinding oriCallADP, but not wild-type oriC. Purified supercoiled plasmid was incubated with the indicated concentrations of DnaA-ATP (A), or a mixture of DnaA-ATP and DnaA-ADP at the concentrations indicated in (B), and treated with P1 nuclease. Topological forms were separated by electrophoresis through agarose gels, followed by staining with ethidium bromide. The positions of uncut supercoiled, nicked and linearized forms are shown. The percentage of DNA converted to linear form was quantified.
Protein crosslinking
Reactions were set up as for DMS modification, except that no BSA was added to the reaction. After incubation of DNA and DnaA for 5 min, the samples were incubated with 3,3′-dithiobis(sulfosuccinimidylpropionate) (DTSSP; 100 mM) for 5 min at 37°C. Reactions were stopped by the addition of lysine (50 mM). Loading buffer lacking reducing agent was then added to the samples, and the samples were resolved on 8% sodium dodecyl sulphate-polyacrylamide gel electrophoresis gels. The proteins were transferred to nitrocellulose, and DnaA on the blots was revealed using an anti-DnaA antibody, detected by chemiluminescence (Bio-Rad ImunoStar kit). Images were analyzed by using Bio-Rad Quantity One software. Assays were done in triplicate, with representative scans shown in Figure 4.
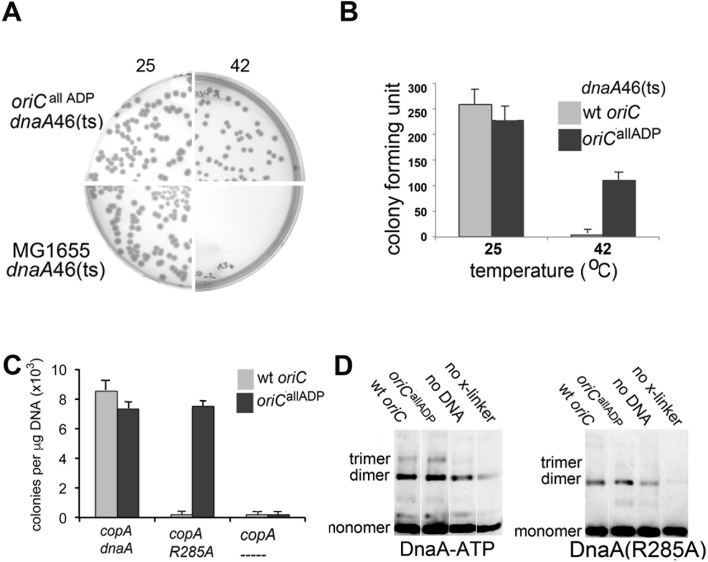
Figure 4.
Initiation from oriCallADP has a reduced requirement for DnaA-ATP in vivo. (A) Cells from a single colony of MG1655(dnaA46 ts)(bottom) or MG1655(oriCallADP, dnaA46 ts) (top) were diluted, plated and incubated at either 25°C (left) or 42°C (right). Colonies were counted after 24 or 18 h, respectively. Representative plates are shown. (B) Quantitation of the plating study shown in (A). Colony counts from two separate experiments, with triplicate plates for each temperature in each experiment were run. Error bars are SD of mean. (C) DnaA null cells, carrying both a plasmid R1 origin and either oriC (gray bars) or oriCallADP (black bars), were transformed with a plasmid expressing the plasmid R1 origin inhibitor copA or a plasmid co-expressing either wild-type DnaA or DnaA(R285A) and copA. Transformation efficiency was calculated after 18 h of growth. Two separate experiments, with triplicate plates were run. Error bars are SD of mean. (D) Oligomeric DnaA forms after DTSSP treatment of purified supercoiled plasmids incubated with 0 or 100 nM DnaA-ATP or DnaA(R285)-ATP. Three separate experiments were performed. Quantitation is shown in Supplementary Figure S3.
Flow cytometry
MG1655 or MG1655(oriCallADP) were grown in LB media or minimal salts media (27) supplemented with 0.2% glucose, 0.2% uracil and 0.2% casamino acids to an OD450 of 0.2, and then incubated with 300 μg/ml rifampicin and 15 μg/ml cephalexin at 37°C for 4 h. A total of 1 ml of treated cells were fixed with 9 ml of 70% ethanol and stored at 4°C. Prior to flow cytometric analysis, 1 ml of the fixed cell suspension was pelleted, washed with 50 mM Tris–Cl, pH 7.5, 150 mM NaCl (TBS) and resuspended in 1 ml of TBS containing 0.5 μl Vybrant DyeCycle Green (Thermofisher). Stained cells (3000–5000 cells/ml) were analyzed using an Accuri C6 personal flow cytometer, and data from 10 000 cells were collected. The threshold was set to prevent counting of non-fluorescing particles, and gating was used to exclude non-bacterial particles from the histogram images. Forward scatter was used for cell mass measurement.
Drug sensitivity assays
An overnight culture of MG1655 asnB::tet or MG1655(oriCallADP, asnB::tet) was grown in LB-media supplemented with tetracycline (12.5 μg/ml) at 37°C. The cultures were serially diluted in LB media and different dilutions were plated on LB plates containing HU at 0, 5, 10, 15 or 20 mM or azidothymidine (AZT) at 0, 0.5, 1, 1.5 and 2 μM, followed by incubation for 24 h at 37°C. Duplicate plates were used for each concentration. The total cell viability and surviving fraction was calculated as described in (28). Three individual assays were performed.
RESULTS
Escherichia coli oriC contains six low affinity sites that preferentially bind DnaA-ATP
Previous studies established that the I1, I2, I3 and τ2 recognition sites preferentially bind DnaA-ATP, while R1, R2, R4 and R5M bind DnaA-ATP and DnaA-ADP equivalently (11,12). Binding attributes have not been reported for three recently identified low affinity sites (C1, C2 and C3) (17). To complete characterization of DnaA-ATP and DnaA-ADP binding to oriC, DMS footprinting was used to reveal the DnaA binding patterns on supercoiled templates (Figure 1B and C). DnaA was pre-incubated with either ATP or ADP, and then bound to supercoiled oriC plasmid prior to DMS treatment and primer extension. Sites that are occupied by DnaA are revealed by distinctive changes to the DMS modification pattern resolved on sequencing gels. Specifically, the G4 of the 9 mer consensus 5′-TGTGGATAA (or variations of this sequence) becomes hypersensitive, and the G2 less sensitive to DMS (Figure 1B), causing darker and lighter bands, respectively. It should be noted that not all binding sites have guanosines at both positions. Based on the footprints, C1 and R5M bind DnaA-ATP and DnaA-ADP equally, as do R1, R2 and R4. In contrast, C2 and C3 preferentially bind DnaA-ATP. We are unable to detect any DnaA binding to the putative τ1 site, located to the left of R5M, by either DMS footprinting (Figure 1B) or EMSA (17). Thus, we conclude that in E. coli oriC, each gap region between strong sites contains four low affinity sites: one non-discriminatory site (C1 or R5M), and three sites that preferentially bind DnaA-ATP.
DnaA-ATP is not required for the cooperative binding needed to fill low affinity DnaA sites
For low affinity sites to become occupied, DnaA must not only recognize the nucleotide sequence of the site to be filled, but also must be recruited and positioned (cooperative binding) by previously bound DnaA (17,18). There is evidence that both site recognition and recruitment aspects of cooperative binding require DnaA-ATP. Single nucleotide changes in the I2 and I3 sites allow them to recognize and bind DnaA-ADP (11), while mutation of a critical arginine (R285) in DnaA results in loss of both AAA+-dependent interactions and low affinity site binding (12). However, DnaA bound to R1 or R4 can use its N-terminal domain (domain I) to recruit DnaA-ADP for binding to R5M or C1, respectively (29), and similar domain I interactions might promote cooperative DnaA-ADP binding between the remaining low affinity sites, as long as the recruited DnaA could recognize the 9 mer sequences. To examine this possibility, the ability of DnaA-ADP and DnaA(R285A) to extend from an anchor R4 site to occupy two flanking R5M sites was measured by EMSA (Figure 2A). End-labeled probe was incubated with two concentrations of DnaA-ATP, DnaA-ADP or DnaA(R285A)-ATP, and the resulting complexes were resolved on polyacrylamide gels. At the higher DnaA concentration, all forms of DnaA were capable of filling all three sites on the oligonucleotide probe, including the two low affinity sites, to form complexes containing three bound DnaA molecules (Figure 2A). Binding to the two closely spaced R5M sites required the high affinity anchor site (Figure 2A, first panel), as has been reported previously (5,17).
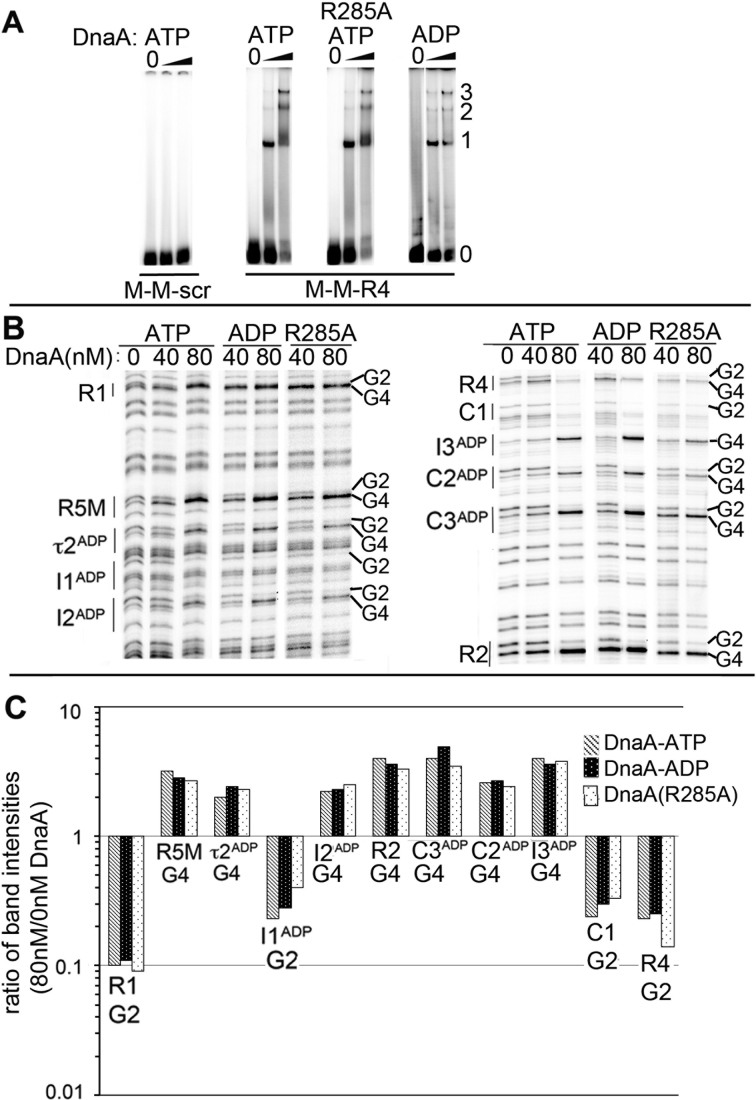
Figure 2.
DnaA-ATP oligomerization is not required for cooperative binding between low affinity sites. (A) EMSA of wild-type DnaA-ATP, DnaA(R285A)-ATP or DnaA-ADP binding to probe carrying the closely spaced sites shown below the panels. DnaA:DNA molar ratios were 0:1, 2.5:1 and 10:1. Positions of unbound probe (0), and probe bound to 1, 2 or 3 molecules of DnaA are marked. (B) DMS footprints of DnaA-ATP, DnaA-ADP and DnaA(R285A)-ATP bound to oriCallADP. Binding sites and diagnostic G2 and G4 residues are marked. (C) Quantitation of G2 or G4 band intensities for each site when bound to 80 nM DnaA-ATP (gray), DnaA-ADP (black) or DnaA(R285A)-ATP (stippled) relative to intensities at 0 nM DnaA.
We next designed a version of oriC (termed oriCallADP) that can become fully occupied with DnaA-ADP. To ensure that orisome assembly on the oriCallADP template remained staged, the DnaA-ATP sites were converted to sequences similar (or identical) to the low affinity R5M and C1 sites. The sequences of the converted sites are shown in Supplementary Table S1. Staged binding of DnaA-ADP and DnaA(R285A) to oriCallADP was verified using DMS footprinting (Figure 2B and C); the binding pattern is similar to the ordered binding of DnaA-ATP to wild-type oriC (Figure 1B). Combined, these results indicate that site recognition, more than ATP-dependent recruitment, contributes to the requirement for DnaA-ATP in oriC binding.
DnaA-ADP is capable of unwinding oriCallADP plasmids
Binding of DnaA to low affinity sites in E. coli oriC is a prerequisite for DNA strand separation (30). DnaA-ATP is required to form the unwound complex (9), but mixed complexes of DnaA-ATP and DnaA-ADP are also active (31), and it remains unclear how much unwinding activity DnaA-ADP might possess if this form could access the low affinity sites in the origin. To address this, oriCallADP, was used to compare unwinding activities of DnaA-ATP and DnaA-ADP. First, we verified that the site mutations in oriCallADP did not increase or decrease the unwinding activity of DnaA-ATP. Supercoiled wild-type oriC and oriCallADP plasmids were incubated with increasing concentrations of DnaA-ATP, treated with single-strand specific P1 endonuclease, and the topological forms were separated on agarose gels (Figure 3A). When the plasmid DNA unwinds, P1 can make opposing nicks on the ssDNA, producing a linear form. In this assay, wild-type and mutant plasmids showed indistinguishable unwinding patterns with DnaA-ATP. This is the expected result if oriCallADP and wild-type oriC have similar overall affinities for DnaA-ATP, since DnaA would unwind oriCallADP at a lower DnaA concentration if the converted sites had higher affinity than their wild-type counterparts. Next, to determine if the mutations in oriCallADP reduced the requirement for DnaA-ATP in plasmid origin unwinding, the wild-type or mutant plasmids were incubated with varying ratios of DnaA-ATP/DnaA-ADP, while keeping the total DnaA concentration constant (at 80 nM) (Figure 3B). We observed that oriCallADP was unwound in reactions containing only DnaA-ADP. In contrast, although DnaA-ADP could supplement DnaA-ATP in unwinding the wild-type oriC plasmid, the majority of DnaA in the reaction mixture had to be in the ATP form, similar to what has been reported previously (31). These results are consistent with earlier work that suggested that I2 and I3 play a role in determining the DnaA-ATP dependence of unwinding. The data demonstrate that both DnaA-ADP and DnaA-ATP are mechanically capable of unwinding oriC DNA if they can bind to the origin.
DnaA mutants defective in ATP binding can activate oriCallADPin vivo
The ability of DnaA-ADP to unwind the oriCallADP plasmid suggests that the binding site mutations might also decrease the initiation requirement for DnaA-ATP in vivo. Since it is not possible to generate cells containing only DnaA-ADP, we chose to examine oriCallADP activity in cells carrying the dnaA46(ts) allele, which has two amino acid substitutions (A184Vand H252Y) in the AAA+ domain. The A184V mutation disrupts nucleotide binding and is responsible for the temperature sensitive growth (permissive at 25°C, and non-permissive at 42°C) (32). The H252Y mutation reduces interactions between AAA+ domains and ATP-dependent filament assembly, as well as affinity for single stranded DNA (20). If oriCallADP does have a decreased requirement for DnaA-ATP in origin activation, then as the sole chromosomal replication origin, oriCallADP should suppress the dnaA46 mutation and allow growth at non-permissive temperature. To test this, recombineering (homologous recombination-mediated genetic engineering)(26) was used to delete the chromosomal copy of wild-type oriC, and replace it with oriCallADP at the native location (Supplementary Figure S1A shows a scheme of procedure). The mutant chromosomal origin was then transferred from the recombineering strain into MG1655(dnaA46) by transduction using P1 phage. Individual colonies were selected after growth in selective media at 25°C. The selected colonies were then diluted in LB, plated and tested for growth at 25°C and 42°C (Figure 4A and B). As expected, no MG1655(dnaA46) colonies appeared at non-permissive temperature (Figure 4A, bottom right, 4B). In contrast, oriCallADP was able to suppress the DnaA46 mutation and form colonies at 42°C (Figure 4A, top right and B). Only partial suppression was observed; at non-permissive temperature, MG1655(oriCallADP,dnaA46) have half the colony forming unit activity as MG1655(oriCallADP, wild-type dnaA) (Figure 4B), possibly because DnaA46 has lower activity and stability even at permissive temperature (33). To ensure that the ability of MG1655(oriCallADP, dnaA46) to grow at 42°C was not caused by activation of a cryptic origin, P1 transduction was used to replace the origins in MG1655(dnaA46) and MG1655(oriCallADP, dnaA46) with ΔoriC::pKN1562(kan), asnA::cat; colonies were selected after growth at 25°C. This transduction inactivates oriC, and inserts a plasmid R1 replication origin at the native oriC location. The transduced cells were then transformed with a plasmid expressing copA or dnaA. CopA is an antisense RNA that prevents activation of the integrated R1 plasmid origin (34), and is lethal to the cells unless an alternative origin can trigger initiation. No colonies were formed in cells derived from MG1655(wt oriC, dnaA46) or MG1655(oriCallADP, dnaA46) after transformation with the plasmid expressing copA, while the plasmid expressing DnaA transformed the cells with high efficiency (Supplementary Figure S2). Combined, these results suggest that even DnaA that is defective in nucleotide binding and has reduced oligomerization capacity can trigger chromosome replication from oriCallADP.
OriC allADP can be activated by DnaA(R285A) in vivo
The suppression of DnaA46 temperature sensitivity by oriCallADP supports a model of E. coli replication initiation in which the ability of DnaA to occupy low affinity sites in oriC is key to determining whether that DnaA can trigger initiation, and suggests that ATP-dependent oligomerization may be less critical in origin activation. However, although the H252Y mutation in DnaA46 reduces interactions between AAA+ domains, it does not eliminate them (20). Therefore, to test the requirement for ATP-dependent DnaA oligomerization further, we examined oriCallADP activation by the more robust assembly mutant, DnaA(R285A), which is similar to DnaA-ADP in its inability to oligomerize and to bind ssDNA (20). DnaA(R285A) is also unable to activate wild-type oriC in vitro and in vivo (12). However, since R285 is not essential for cooperative binding at low affinity DnaA-ADP recognition sites (Figure 2), it seemed possible that DnaA(R285A) might trigger initiation from oriCallADP. This idea was tested using the oriCallADP recombineered cells, which, in addition to the transplanted origin, carry a deletion in the chromosomal dnaA gene and must initiate replication from an integrated plasmid R1 replication origin (Supplementary Figure S1A). Cells containing either wild-type oriC or oriCallADP at the native position were transformed with plasmids expressing: copA; copA and wild-type dnaA; or copA and dnaA(R285A) (Figure 4C and Supplementary Figure S1B). CopA is lethal to the cells unless the chromosomal oriC or oriCallADP can be activated by co-expressed DnaA. Accordingly, in the absence of exogenous DnaA, no colonies appeared after transformation of either the strain carrying transplanted wild-type oriC or oriCallADP with the copA expression plasmid (Figure 4C). This result also verifies that the cells do not harbor any other alternative origins that function in the absence of DnaA. Further, as expected with cells that harbor functional versions of oriC, both strains were successfully transformed with the plasmid co-expressing wild-type DnaA and copA (Figure 4C). In contrast, only the cells carrying transplanted oriCallADP could be transformed with the plasmid co-expressing DnaA(R285A) and copA (Figure 4C). These results suggest that when DnaA can access all low affinity oriC recognition sites, the requirement for arginine finger/ATP interactions and thus for robust ATP-dependent oligomerization and ssDNA binding, can be suppressed.
Although the assembly defect caused by the R285A mutation has been well characterized (20), DnaA(R285A) binds ATP similarly to wild-type DnaA and presumably DnaA(R285A)-ATP and wild-type DnaA-ATP monomers have comparable conformations. It is possible that the close positioning of low affinity sites in oriCallADP promoted some interactions between the AAA+ domains of adjacent DnaA(R285A) monomers even without the R285 arginine finger, and in this way allowed DnaA(R285A) to activate oriCallADP. To investigate this possibility, protein crosslinking experiments were performed to evaluate oligomerization of DnaA(R285A) on wild-type oriC and oriCallADP (Figure 4D). Supercoiled plasmids carrying oriC or oriCallADP were incubated with DnaA-ATP or DnaA(R285A)-ATP at levels slightly higher than required for full origin occupation and unwinding (100 nM). The complexes were then treated with the amine specific crosslinker DTSSP, separated on denaturing protein gels in the absence of reducing agent and oligomeric species were detected by immunoblotting. DnaA-ATP formed dimers and trimers on both origins (Figure 4D and Supplementary Figure S3), with formation of multimers being stimulated by the oriC template; these results are consistent with previous studies (20,29). In contrast, DnaA(R285A)-ATP was impaired in formation of any higher order species on either origin (Figure 4D and Supplementary Figure S3), and the very low level of DnaA(R285A) trimer formation on oriC and oriCallADP (<1% of total species) was the same as that seen without any template (Supplementary Figure S3). Additionally, the crosslinking assays suggest that DnaA(R285A) is either not oligomerizing on the ssDNA in the unwound region, or it is forming only very unstable filaments that could not be detected in this assay. Combined, these results suggest that formation of stable DnaA oligomers longer than dimers is not essential for oriCallADP activation.
Cells with chromosomal oriCallADP initiate more than once per cell division cycle
Over a range of growth rates, a key determinant of cell initiation age is the level of available DnaA-ATP (35), which fluctuates during the cell cycle (36). Logically, since occupation of the low affinity DnaA-ATP sites in oriC requires the elevated DnaA-ATP levels attained just prior to initiation, these sites should play a role in setting the time of initiation in the cell cycle (17). Thus, if oriCallADP can be activated by any available form of DnaA, then initiation timing should no longer be tightly coupled to DnaA-ATP levels, but rather to available levels of either form of DnaA. To generate cells where oriCallADP is the sole replication origin, the mutant origin or wild-type oriC was transferred from the recombineered strains into the native oriC location in MG1655(ΔoriC::pKN1562(kan), asnA::cat, asnB::tet) by transduction with P1 phage, replacing the integrated plasmid origin. The resulting colonies, regardless of whether their cells contained wild-type oriC or oriCallADP as the sole chromosomal replication origin, appeared at the same time interval after transduction. This result is not at all similar to the extremely small, slow to appear colonies observed after an hda null mutation is transduced into cells (37) and suggests that survival and growth of oriCallADP cells does not require generation of suppressor mutations.
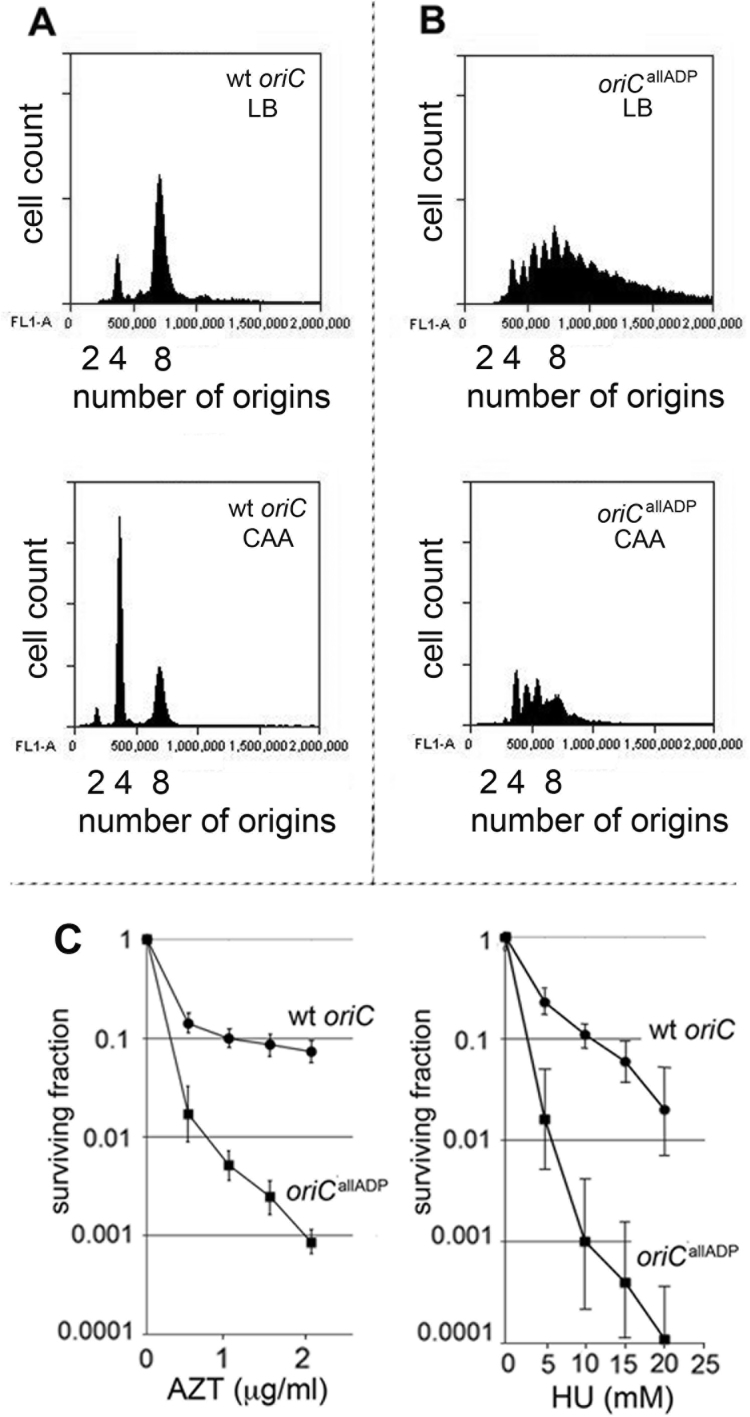
To evaluate timing of initiation, MG1655 or MG1655(oriCallADP) cells growing exponentially in LB (generation time of 22± 2 min for both strains) or minimal media supplemented with glucose, casamino acids and uracil (generation time of 33± 2 min), were treated with rifampicin to inhibit new rounds of chromosome replication and cephalexin, which prevents cell division, followed by incubation to allow completion of ongoing rounds of replication. After staining cellular DNA, the number of chromosomes in the cell, which reflects the number of oriC copies present at the time of drug addition, was detected by flow cytometry. Under rapid growth conditions, the E. coli generation time is less than the time needed to complete chromosome replication (38), so exponentially growing cells normally contain more than one copy of oriC, and all origin copies initiate chromosome replication synchronously, once per cell cycle, on partially duplicated chromosomes (39). The origin number doubles at the time of initiation, and only 2n origins should be in the cells. Accordingly, cultures of MG1655 cells contained 4 or 8 origins when grown in LB (Figure 5A, top panel), or 2, 4 or 8 origins when grown in minimal media supplemented with glucose and casamino acids (Figure 5A, bottom panel). In contrast, initiation timing in MG1655(oriCallADP) cells was perturbed, since cells containing odd numbers of origins were detected (Figure 5B, top and bottom panels). The histograms are consistent with over-initiation, since the number of origins per cell is higher than observed in wild-type cells; this is expected if origin activation is less tightly coupled to DnaA-ATP levels.
Figure 5.
Over-initiation in oriCallADP cells leads to increased sensitivity to replicative stress. (A and B) DNA histograms of wild-type oriC (A) or oriCallADP (B) cells growing in LB (top panels), or minimal media supplemented with glucose, casamino acids and uracil (bottom panels), treated with cephalexin and rifampicin, and analyzed by flow cytometry. The number of origins in the cells at the time of drug treatment is shown. (C) Survival fraction of cells incubated in the presence of AZT or HU are plotted after 18 h of growth. Two separate experiments, with triplicate plates were run. Error bars are SD of mean.
To verify that the MG1655(oriCallADP) cells were over-initiating and not simply resistant to rifampicin-induced inhibition of initiation, we measured the DNA content of cells growing in minimal salts media at the time of drug addition, and then at 20 min intervals after drug addition. Exponentially growing MG1655(oriCallADP) cells had a higher DNA content than MG1655 carrying wild-type oriC, as is expected from cells that over-initiate, as long as the forks can progress (Supplementary Figure S4A, top panel). In contrast, cells that have rifampicin-resistant initiations, such as cells carrying a deletion of the datA locus, have the same DNA content as their wild-type parental strain when growing exponentially (40). After addition of rifampicin and cephalexin, DNA content increased in both MG1655(oriCallADP) and MG1655 for approximately 50 minutes (Supplementary Figure S4), which is consistent with the time required to complete all rounds of ongoing chromosome replication. In contrast, rifampicin-resistant datA cells continue to increase DNA content about twice as long as wild-type cells (40). At last, the total DNA content in both MG1655 and MG1655(oriCallADP) increased to a similar extent (∼1.5-fold) (Supplementary Figure S4A and B), unlike rifampicin-resistant cells which have a greater increase in DNA content (e.g. 2-fold for datA cells) (40). The over-initiation also does not cause MG1655(oriCallADP) cells to be discernably bigger than MG1655 cells, as measured by either light scatter or microscopy (Supplementary Figure S4C).
Although the rate of appearance of the transduced colonies did not support the idea that generation of suppressor mutations was required for survival of MG1655(oriCallADP) cells, some over-initiating cells, such as cells lacking hda, do generate suppressors (37) (41). Therefore, we examined MG1655(oriCallADP) cells more thoroughly for suppressors that might affect initiation. Origins in MG1655 or MG1655(oriCallADP) were replaced with ΔoriC::pKN1562(kan), asnA::cat using transduction. Wild-type oriC was then re-introduced into both strains, creating MG1655 (wt oriC to wt oriC) and MG1655(oriCallADP to wt oriC). Initiation timing in both strains was examined by treating exponentially growing cells with rifampicin and cephalexin, and analyzing DNA content after replication run out using flow cytometry (Supplementary Figure S4D). DNA histograms of both MG1655 (wt oriC to wt oriC) and MG1655(oriCallADP to wt oriC) were very similar, and both looked like MG1655 (compare FigS4A, bottom, with S4D). These data indicate that although MG1655(oriCallADP) cells over-initiate, this does not appear to result in generation of suppressor mutations that would decrease initiation frequency.
Loss of ATP-dependent DnaA binding sites in oriC increases sensitivity to inhibitors of replication fork movement
Loss of once per cycle initiation regulation raises the probability of head-to-tail (co-directional) fork collisions (1), since over-initiation can cause forks to be closer together. Although under unstressed growth conditions, over-initiation did not cause measurable growth defects in MG1655(oriCallADP), over-initiation could make the cells more susceptible to agents that impair replication fork movement, since these would further increase the odds of collision between closely spaced replication forks (28). To test this, we measured the sensitivity of MG1655 and MG1655(oriCallADP) to HU and to AZT, both of which slow replication forks (42,43). Exponentially growing cells were plated onto LB-agar plates containing varying concentrations of drug, and the number of colony forming units was counted after overnight growth at 37°C (Figure 5B). MG1655(oriCallADP) was at least 3-fold more susceptible to both drugs, indicating that loss of ATP-dependent initiator binding to dsDNA resulted in increased sensitivity to replication stress. This behavior emphasizes the need for an ‘on-off’ switch to regulate replication origin firing and maintain genome stability, a switch that is provided by coupling origin-initiator protein interactions to ATP binding and hydrolysis.
DISCUSSION
In all forms of life, the ATP-bound form of DNA replication initiators is required to license replication origins (44). This requirement was first identified in E. coli, based on studies showing that only DnaA-ATP has activity in in vitro unwinding and replication assays (9). Various explanations are proposed for the dependence on DnaA-ATP in E. coli DNA replication; however, because origin binding is ATP-dependent, it has been difficult to rigorously test the DnaA-ATP requirement during all stages of the initiation process. In this study, we addressed this issue by creating a version of oriC (oriCallADP) in which DnaA binding to oriC is independent of bound nucleotide. Using this origin, we found that DnaA-ATP and DnaA-ADP were both competent to unwind oriCallADP plasmids and that bound nucleotide and robust intermolecular interactions of AAA+ domains were not essential for activation of oriCallADPin vivo. The data suggest that after binding to low affinity sites, DnaA-ATP is not unique in its ability to perform the mechanical processes used in origin activation.
This and previous studies (11,12) identified low affinity sites that bind DnaA-ATP and DnaA-ADP equivalently, as well as sites that preferentially bind DnaA-ATP. It is not known why some specific binding sites allow both DnaA’s nucleotide forms to bind, and others do not, but, logically, the answer lies in conformational differences between DnaA-ATP and DnaA-ADP. In the ATP-bound form, the DNA binding domain IV bends toward the AAA+ domain III, placing the two domains in proximity (24). Alternatively, the bent structure of DnaA-ATP could allow amino acid side chains from both domains III and IV to participate in site binding, similar to the ‘lobster claw’ mode of interaction suggested for dsDNA binding of archaeal and eukaryotic initiator proteins (45,46). Since ‘lobster claw’ binding is stabilized by AAA+ interactions (46), it is possible that in wild-type oriC, the AAA+ domains of DnaA molecules occupying DnaA-ATP sites are linked, thus coupling DnaA-ATP oligomerization to site occupation. This would explain the inability of DnaA(R285A) to stably occupy DnaA-ATP sites, and also why low affinity DnaA binding in wild-type oriC requires that sites be arrayed in the same orientation, and separated by 2 bp (17). In contrast, the low affinity R5M and C1 sites may be able to make more base-specific contacts with domain IV, which would alleviate the need for the contribution from domain III in binding to these sites. Consistent with this idea, the A in position 7 in the consensus sequence 5′- TTA/TTNCACA, which aids sequence-specific binding to domain IV (47), is a common feature among 9 mer sequences that bind DnaA-ADP. It is also possible that some conformational changes in DnaA, observed at physiological ATP concentrations (5 mM) (48), contribute to the cooperative binding that extends DnaA between low affinity DnaA-ATP sites. It should also be noted that DnaA-ATP sites have not been identified in many bacterial origins, and some bacterial DnaAs do not contain a domain 1 that is capable of DnaA–DnaA interactions (49). In these origins, it is possible that low affinity sites can be filled only using DnaA-ATP oligomerization. This may be the case inBacillus subtilus, where one mode of initiation regulation utilizes anti-cooperativity factors that prevent DnaA-ATP oligomerization (50,51).
The observations that E. coli oriCallADP can be unwound by DnaA-ADP, and activated by DnaA46 and DnaA(R285A) raise questions about the role of DnaA-ATP oligomers in origin activation. Our data are not compatible with models for E. coli origin unwinding that invoke a requirement for an oligomeric DnaA-ATP filament that extends from R1 into the DNA unwinding element (DUE) (21), stretches DUE DNA (22) or creates right-handed supertwists when bound to the dsDNA DnaA-ATP sites (24,25). The data may be more consistent with models that invoke engagement of DUE DNA with key amino acids in the central channel of a DnaA-ATP oligomer associated with low affinity sites (52), but only if a similar channel can be formed on oriCallADP by close spacing of any form of DnaA molecules, or if only one DnaA molecule bound to R5M is needed to unwind, as has been recently suggested (53). However, if a DnaA-ATP filament is not essential, the question remains how DnaA in any form is able to separate origin DNA strands. Perhaps the inherent DNA bending activity of bound DnaA is sufficient. DnaA produces a 30–40° bend in the DNA of each site to which it binds (5), similar to the DNA distortions caused by binding of archaeal and eukaryotic initiator proteins (45,46). Concerted DNA bending by DnaA bound at multiple arrayed sites may generate sufficient torsional stress to unwind the AT-rich DUE. In this model, the mechanical activity for unwinding results from domain IV interacting with DNA, while domains I and III, mediating cooperative binding to DnaA-ATP sites, are needed for assembly of the unwinding complex. While it is not known how many arrayed sites might be required, there is evidence that not all the low affinity sites in E. coli oriC are essential (19,53,54). Additionally, although we can detect oriCallADP activation in the absence of robust DnaA oligomers, it remains likely that in any cells containing wild-type DnaA, a DnaA-ATP filament bound to ssDNA plays a role in initiation, by providing a docking site for the helicase loader (DnaC) or by expanding the unwound region (22,23). While the location of ssDNA DnaA binding in E. coli is not yet determined, the recently identified DnaA-trio elements located in the right 13 mer region of the DUE (55) are candidate sites. It is also possible that bacteria are capable of unwinding oriC using different mechanisms, depending on the origin configurations. Future studies are required to elucidate this critical step in origin activation.
Our data also reveal a critical role for preferential DnaA-ATP binding sites in regulating initiation timing, since oriCallADP initiated more than once per cell cycle. It is perhaps less obvious why the over-initiation activity of oriCallADP remained compatible with host growth. Over-initiation is most lethal when closely spaced replication forks are combined with some problem, such as a DNA lesion, that results in stalled forks, because this increases the frequency of co-directional collisions (28,56). To avoid this, cells maintain a minimal inter-initiation interval, termed the eclipse period, which ensures some spacing between replication forks. In E. coli, the majority of the eclipse period is caused by the SeqA protein (57,58), which prevents re-initiation by blocking low affinity DnaA binding sites in oriC immediately after replication begins (15,59). SeqA also aids repair of collapsed forks and replication restart (60). SeqA null mutants over-initiate, and because the closely spaced forks encounter unrepaired collapsed forks, growth of SeqA null cells is severely inhibited. Cells that carry too much DnaA-ATP (such as hda null cells) also over-initiate, and this is further complicated by DnaA-ATP-induced repression of nrdAB (61,62), resulting in lower than normal nucleotide levels. Insufficient nucleotides could slow fork movement directly, or could make it more difficult to bypass DNA lesions caused by events such as oxidative stress (56,63). In contrast oriCallADP cells retain functional SeqA, and the mechanisms controlling cellular DnaA-ATP levels should be operating normally. Thus, despite over-initiating, the SeqA protein should maintain the eclipse period and fork stability in oriCallADP cells, and the over-initiation should not be not exasperated by slowed or stalled forks caused by low nucleotide levels. However, since DnaA-ATP hydrolysis will be ineffective at preventing extra initiations after the end of the eclipse period, the oriCallADP cells are more susceptible to drug-induced fork slowing, probably because the extra forks arising from extra-initiation events can collide under these conditions (28). Thus, requiring DnaA-ATP for origin recognition links orisome assembly to its inactivation switch, and contributes to the once-per-cycle timing mechanism that helps maintain genome stability. Based on the findings of this paper, however, it appears that this ATP-dependent regulatory mechanism is distinct from DnaA’s mechanical activities that cause the DNA conformational changes required for origin activation.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM54042 to A.C.L.]; Florida Institutes of Technology Holzer Lequear Fund for Molecular Genetics (to J.E.G.); Funding for open access charge: Open Access Subvention Fund from the Florida Tech Libraries (in part).
Conflict of interest statement. None declared.
REFERENCES
- 1. Simmons L.A., Breier A.M., Cozzarelli N.R., Kaguni J.M.. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 2004; 51:349–358. [DOI] [PubMed] [Google Scholar]
- 2. Costa A., Hood I.V., Berger J.M.. Mechanisms for initiating cellular DNA replication. Annu. Rev. Biochem. 2013; 82:25–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Leonard A.C., Grimwade J.E.. Building a bacterial orisome: emergence of new regulatory features for replication origin unwinding. Mol. Microbiol. 2005; 55:978–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leonard A.C., Grimwade J.E.. The orisome: structure and function. Front. Microbiol. 2015; 6:545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schaper S., Messer W.. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J. Biol. Chem. 1995; 270:17622–17626. [DOI] [PubMed] [Google Scholar]
- 6. Kaguni J.M. DnaA: controlling the initiation of bacterial DNA replication and more. Annu. Rev. Microbiol. 2006; 60:351–375. [DOI] [PubMed] [Google Scholar]
- 7. Katayama T., Kasho K., Kawakami H.. The DnaA cycle. Front. Microbiol. 2017; 8:2496–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Erzberger J.P., Pirruccello M.M., Berger J.M.. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002; 21:4763–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sekimizu K., Bramhill D., Kornberg A.. ATP activates dnaA protein in initiating replication of plasmids bearing the origin of the E. coli chromosome. Cell. 1987; 50:259–265. [DOI] [PubMed] [Google Scholar]
- 10. Noguchi Y., Sakiyama Y., Kawakami H., Katayama T.. The arg fingers of key DnaA protomers are oriented inward within the replication origin oriC and stimulate DnaA subcomplexes in the initiation complex. J. Biol. Chem. 2015; 290:20295–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McGarry K.C., Ryan V.T., Grimwade J.E., Leonard A.C.. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2811–2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawakami H., Keyamura K., Katayama T.. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005; 280:27420–27430. [DOI] [PubMed] [Google Scholar]
- 13. Samitt C.E., Hansen F.G., Miller J.F., Schaechter M.. In vivo studies of DnaA binding to the origin of replication of Escherichia coli. EMBO J. 1989; 8:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cassler M.R., Grimwade J.E., Leonard A.C.. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995; 14:5833–5841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nievera C., Torgue J.J., Grimwade J.E., Leonard A.C.. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol. Cell. 2006; 24:581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ryan V.T., Grimwade J.E., Camara J.E., Crooke E., Leonard A.C.. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol. Microbiol. 2004; 51:1347–1359. [DOI] [PubMed] [Google Scholar]
- 17. Rozgaja T.A., Grimwade J.E., Iqbal M., Czerwonka C., Vora M., Leonard A.C.. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 2011; 82:475–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Speck C., Weigel C., Messer W.. ATP- and ADP-dnaA protein, a molecular switch in gene regulation. EMBO J. 1999; 18:6169–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stepankiw N., Kaidow A., Boye E., Bates D.. The right half of the Escherichia coli replication origin is not essential for viability, but facilitates multi-forked replication. Mol. Microbiol. 2009; 74:467–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Duderstadt K.E., Mott M.L., Crisona N.J., Chuang K., Yang H., Berger J.M.. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010; 285:28229–28239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Speck C., Messer W.. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. EMBO J. 2001; 20:1469–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Duderstadt K.E., Chuang K., Berger J.M.. DNA stretching by bacterial initiators promotes replication origin opening. Nature. 2011; 478:209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mott M.L., Erzberger J.P., Coons M.M., Berger J.M.. Structural synergy and molecular crosstalk between bacterial helicase loaders and replication initiators. Cell. 2008; 135:623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Erzberger J.P., Mott M.L., Berger J.M.. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006; 13:676–683. [DOI] [PubMed] [Google Scholar]
- 25. Zorman S., Seitz H., Sclavi B., Strick T.R.. Topological characterization of the DnaA-oriC complex using single-molecule nanomanipuation. Nucleic Acids Res. 2012; 40:7375–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L.. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009; 4:206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helmstetter C.E., Krajewski C.A.. Initiation of chromosome replication in dnaA and dnaC mutants of Escherichia coli B/r F. J. Bacteriol. 1982; 149:685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sutera V.A., Lovett S.T.. The role of replication initiation control in promoting survival of replication fork damage. Mol. Microbiol. 2006; 60:229–239. [DOI] [PubMed] [Google Scholar]
- 29. Miller D.T., Grimwade J.E., Betteridge T., Rozgaja T., Torgue J.J., Leonard A.C.. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:18479–18484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leonard A.C., Grimwade J.E.. Regulation of DnaA assembly and activity: taking directions from the genome. Annu. Rev. Microbiol. 2011; 65:19–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yung B.Y., Crooke E., Kornberg A.. Fate of the DnaA initiator protein in replication at the origin of the Escherichia coli chromosome in vitro. J. Biol. Chem. 1990; 265:1282–1285. [PubMed] [Google Scholar]
- 32. Carr K.M., Kaguni J.M.. The A184V missense mutation of the dnaA5 and dnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 1996; 20:1307–1318. [DOI] [PubMed] [Google Scholar]
- 33. Boye E., Løbner-Olesen A., Skarstad K.. Timing of chromosomal replication in Escherichia coli. Biochim. Biophys. Acta. 1988; 951:359–364. [DOI] [PubMed] [Google Scholar]
- 34. Molin S., Nordström K.. Control of plasmid R1 replication: functions involved in replication, copy number control, incompatibility, and switch-off of replication. J. Bacteriol. 1980; 141:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Løbner-Olesen A., Skarstad K., Hansen F.G., von Meyenburg K., Boye E.. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989; 57:881–889. [DOI] [PubMed] [Google Scholar]
- 36. Kurokawa K., Nishida S., Emoto A., Sekimizu K., Katayama T.. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999; 18:6642–6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Riber L., Fujimitsu K., Katayama T., Løbner-Olesen A.. Loss of Hda activity stimulates replication initiation from I-box, but not R4 mutant origins in Escherichia coli. Mol. Microbiol. 2009; 71:107–122. [DOI] [PubMed] [Google Scholar]
- 38. Cooper S., Helmstetter C.E.. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968; 31:519–540. [DOI] [PubMed] [Google Scholar]
- 39. Skarstad K., Boye E., Steen H.B.. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986; 5:1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Morigen, Molina F., Skarstad K.. Deletion of the datA site does not affect once-per-cell-cycle timing but induces rifampin-resistant replication. J. Bacteriol. 2005; 187:3913–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Charbon G., Riber L., Cohen M., Skovgaard O., Fujimitsu K., Katayama T., Lobner-Olesen A.. Suppressors of DnaA(ATP) imposed overinitiation in Escherichia coli. Mol. Microbiol. 2011; 79:914–928. [DOI] [PubMed] [Google Scholar]
- 42. Elwell L.P., Ferone R., Freeman G.A., Fyfe J.A., Hill J.A., Ray P.H., Richards C.A., Singer S.C., Knick V.B., Rideout J.L.. Antibacterial activity and mechanism of action of 3′-azido-3′-deoxythymidine (BW A509U). Antimicrob Agents Chemother. 1987; 31:274–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Timson J. Hydroxyurea. Mutat. Res. 1975; 32:115–132. [DOI] [PubMed] [Google Scholar]
- 44. Lee D.G., Bell S.P.. ATPase switches controlling DNA replication initiation. Curr. Opin. Cell Biol. 2000; 12:280–285. [DOI] [PubMed] [Google Scholar]
- 45. Sun J., Kawakami H., Zech J., Speck C., Stillman B., Li H.. Cdc6-induced conformational changes in ORC bound to origin DNA revealed by cryo-electron microscopy. Structure. 2012; 20:534–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dueber E.L., Corn J.E., Bell S.D., Berger J.M.. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007; 317:1210–1213. [DOI] [PubMed] [Google Scholar]
- 47. Sutton M.D., Kaguni J.M.. Threonine 435 of Escherichia coli DnaA protein confers sequence-specific DNA binding activity. J. Biol. Chem. 1997; 272:23017–23024. [DOI] [PubMed] [Google Scholar]
- 48. Saxena R., Vasudevan S., Patil D., Ashoura N., Grimwade J.E., Crooke E.. Nucleotide-Induced conformational changes in escherichia coli DnaA protein are required for bacterial ORC to Pre-RC conversion at the chromosomal origin. Int. J. Mol. Sci. 2015; 16:27897–27911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zawilak-Pawlik A., Nowaczyk M., Zakrzewska-Czerwińska J.. The role of the N-Terminal domains of bacterial initiator DnaA in the assembly and regulation of the bacterial replication initiation complex. Genes (Basel). 2017; 8:136–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Merrikh H., Grossman A.D.. Control of the replication initiator DnaA by an anti-cooperativity factor. Mol. Microbiol. 2011; 82:434–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scholefield G., Murray H.. YabA and DnaD inhibit helix assembly of the DNA replication initiation protein DnaA. Mol. Microbiol. 2013; 90:147–159. [DOI] [PubMed] [Google Scholar]
- 52. Ozaki S., Kawakami H., Nakamura K., Fujikawa N., Kagawa W., Park S.Y., Yokoyama S., Kurumizaka H., Katayama T.. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008; 283:8351–8362. [DOI] [PubMed] [Google Scholar]
- 53. Sakiyama Y., Kasho K., Noguchi Y., Kawakami H., Katayama T.. Regulatory dynamics in the ternary DnaA complex for initiation of chromosomal replication in Escherichia coli. Nucleic Acids Res. 2017; 45:12354–12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ozaki S., Katayama T.. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012; 40:1648–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richardson T.T., Harran O., Murray H.. The bacterial DnaA-trio replication origin element specifies single-stranded DNA initiator binding. Nature. 2016; 534:412–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Charbon G., Riber L., Løbner-Olesen A.. Countermeasures to survive excessive chromosome replication in Escherichia coli. Curr. Genet. 2018; 64:71–79. [DOI] [PubMed] [Google Scholar]
- 57. Olsson J., Dasgupta S., Berg O.G., Nordström K.. Eclipse period without sequestration in Escherichia coli. Mol. Microbiol. 2002; 44:1429–1440. [DOI] [PubMed] [Google Scholar]
- 58. von Freiesleben U., Krekling M.A., Hansen F.G., Løbner-Olesen A.. The eclipse period of Escherichia coli. EMBO J. 2000; 19:6240–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lu M., Campbell J.L., Boye E., Kleckner N.. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994; 77:413–426. [DOI] [PubMed] [Google Scholar]
- 60. Pedersen I.B., Helgesen E., Flåtten I., Fossum-Raunehaug S., Skarstad K.. SeqA structures behind Escherichia coli replication forks affect replication elongation and restart mechanisms. Nucleic Acids Res. 2017; 45:6471–6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Babu V.M.P., Itsko M., Baxter J.C., Schaaper R.M., Sutton M.D.. Insufficient levels of the nrdAB-encoded ribonucleotide reductase underlie the severe growth defect of the Δhda E. coli strain. Mol. Microbiol. 2017; 104:377–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gon S., Camara J.E., Klungsøyr H.K., Crooke E., Skarstad K., Beckwith J.. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006; 25:1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Charbon G., Bjørn L., Mendoza-Chamizo B., Frimodt-Møller J., Løbner-Olesen A.. Oxidative DNA damage is instrumental in hyperreplication stress-induced inviability of Escherichia coli. Nucleic Acids Res. 2014; 42:13228–13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.