Abstract
Organoid cultures in 3D matrices are relevant models to mimic the complex in vivo environment that supports cell physiological and pathological behaviors. For instance, 3D epithelial organoids recapitulate numerous features of glandular tissues including the development of fully differentiated acini that maintain apico-basal polarity with hollow lumen. Effective genetic engineering in organoids would bring new insights in organogenesis and carcinogenesis. However, direct 3D transfection on already formed organoids remains challenging. One limitation is that organoids are embedded in extracellular matrix and grow into compact structures that hinder transfection using traditional techniques. To address this issue, we developed an innovative approach for transgene expression in 3D organoids by combining single-cell encapsulation in Matrigel microbeads using a microfluidic device and electroporation. We demonstrate that direct electroporation of encapsulated organoids reaches up to 80% of transfection efficiency. Using this technique and a morphological read-out that recapitulate the different stages of tumor development, we further validate the role of p63 and PTEN as key genes in acinar development in breast and prostate tissues. We believe that the combination of controlled organoid generation and efficient 3D transfection developed here opens new perspectives for flow-based high-throughput genetic screening and functional genomic applications.
INTRODUCTION
Tissues and organs are multicellular structures that self-organize in three dimensions (3D). Cells within a tissue interact with neighboring cells and with extracellular matrix (ECM) through biochemical and mechanical cues that maintain specificity and homeostasis of biological tissues. While traditional 2D cultures on rigid surfaces fail to reproduce in vivo cell behavior, 3D matrices are becoming increasingly popular supports for cell cultures because they allow mimicking the complex environment that supports cell physiological functions (1) to better predict in vivo responses (2,3) and thus to limit the need for animal models (4). For example, epithelial organoid culture in Matrigel recapitulates numerous features of glandular tissues including the development of fully differentiated acini that maintain apico-basal polarity by enclosing a central lumen (5). Therefore, deciphering the key genetic networks underlying epithelial differentiation and polarity in organoids brings new insights in organogenesis and allows us to better understand how they may be disrupted in disease states such as cancer. RNA interference (RNAi) and plasmid transfection have been widely used as powerful tools to alter the expression of specific genes and to observe resulting phenotypic changes (6). While nucleic acid transfection is highly effective in the majority of mammalian cells cultured under standard 2D conditions, additional obstacles are encountered for transfection of solid tissues or 3D models. Indeed, one limitation is that organoids are embedded in ECM, which constitutes a barrier for efficient transfection. Moreover, organoids grow into dense and compact structures that impede diffusion, penetration and cellular accumulation of genetic material, which makes transfection via traditional techniques difficult (7–9). In addition, quiescent cells that are located at the center of 3D structures are often refractory to transfection (10). Thus, direct 3D transfection on already formed organoids remains challenging.
Gene delivery methods are usually divided between viral and non-viral vectors. Viral vectors provide the highest transfection efficiency but have serious limitations such as the size of DNA carried in the vector, intrinsic biosafety issues, concern for viral insertion mutagenesis (11) and an inability to diffuse through ECM (12). To overcome these limitations, a common strategy is to dissociate organoids into single cells or small group of cells before transfecting them and subsequently re-embedding them into Matrigel (13–15). Hence, viral transduction is limited to multi-cellular tumor spheroids (MCTS) or dissociated organoids devoid of ECM with heterogeneous efficiency and the need to further select transduced structures (16). However, under these conditions, the natural self-organization of organoids is lost along with their spatial architecture and polarity, ultimately going back to a 2D cell transfection. Among non-viral-based approaches, lipofection and electroporation are widely used in biological research and usually allow more than 80% of transfection efficiency in 2D cultures. However, they have been proven to be relatively inefficient in transfecting 3D cultures with transfection efficiencies lower than 5 and 20%, respectively (7–9). A common strategy to circumvent this issue is to transfect cells that are grown in 2D and to transfer them into 3D culture, which limits the biological issues that we can address (15,17). However, progeny cells that colonize the matrix will not be transfected and will gradually lose the desired effect. In addition, this method becomes highly challenging when performing high-throughput assays because it requires sequentially transfecting cells, detaching them and seeding them onto ECM. Gene-activated matrices that combine scaffolds or hydrogels and lipid-based gene delivery reagents were developed to allow direct in situ transfection of cells that grow in 3D matrices (18,19). While this system maintains a permanent transfection with 60–90% of siRNA-mediated gene knock-down during 3D organoid development, it does not allow transfection at different time points. To identify efficient non-viral gene delivery methods that are compatible with high-throughput analysis of 3D models in vitro, we systematically evaluate the methods for siRNA transfer into 3D organoids cultured within Matrigel microbeads, which are produced via flow-focusing-based microfluidics (20). Droplet or digital microfluidic aims at forming water-in-oil or oil-in-water emulsions by using microfabricated chips. It ensures high-throughput formation of droplets of controlled size, shape and monodispersity. In this study, we adapted this method to produce Matrigel-in-oil emulsion, leading to a high-throughput production of Matrigel microbeads were each single microbead can be considered as a single ‘bioreactor’ for 3D epithelial cell culture. Here, we took advantage of the reduced amount of ECM that surrounds organoids and the ease of microbead manipulation to develop a method for ex vivo gene function studies. In particular, we demonstrate the efficiency of gene delivery using electroporation in combination with microencapsulated 3D organoids.
MATERIALS AND METHODS
Cell lines and 2D cell culture
The RWPE-1 cell line was obtained from ATCC (CRL-11609). This cell line was derived from non-neoplastic human prostate epithelial cells via immortalization with human papillomavirus. RWPE-1 cells were previously used as a model for normal prostate epithelial cell behavior, as characterized by a polarized acinar morphology in 3D cultures (20). RWPE-1 cells were maintained in KSFM (Gibco Life Technologies, Courtaboeuf, France) supplemented with 5 ng/ml Epidermal Growth Factor (Gibco), 50 μg/ml Bovine Pituitary Extract (Gibco) and 1% Penicillin-Streptomycin (Gibco). Cells were cultured up to 70% confluence and seeded in T25 Flasks at a density of 2 × 104 viable cells/cm2.The MCF10A cell line was obtained from ATCC (CRL-10317). This cell line was derived from spontaneously immortalized non-neoplastic human breast epithelial cells. MCF10A cells were used as a model for normal breast epithelial cell behavior as characterized by a polarized acinar morphology in 3D cultures. MCF10A cells were maintained in DMEM/F12 (Gibco) supplemented with 5% Horse serum (Eurobio, AbCys, Courtaboeuf, France), 20 ng/ml Epidermal Growth Factor (Life Technologies), 10 μg/ml Insulin (Sigma-Aldrich, St Quentin Fallavier, France), 0.5 μg/ml Hydrocortisone (Sigma-Aldrich), 100 ng/ml Cholera toxin (Sigma-Aldrich) and 1% Penicillin-Streptomycin (Gibco). Cells were maintained in culture until ∼80% confluence and seeded in T25 Flasks at a density of 3 × 104 viable cells/cm2. RWPE-1 and MCF10A cells that stably express GFP (RWPE-1-GFP (engineered in our lab) and MCF10A-GFP, (kind gifts of Dr Odile Filhol-Cochet, CEA Grenoble)) were obtained via transduction using a lentiviral vector that encodes the enhanced green fluorescent protein cDNA (GFP). GFP+ cells were sorted using a MoFlo cell sorter (Beckman Coulter, Villepinte, France). Cell number and viability were measured via trypan blue dye exclusion staining using an EVE™ Automatic Cell Counter (NanoEnTek, VWR, Strasbourg, France).
Matrigel microbead production
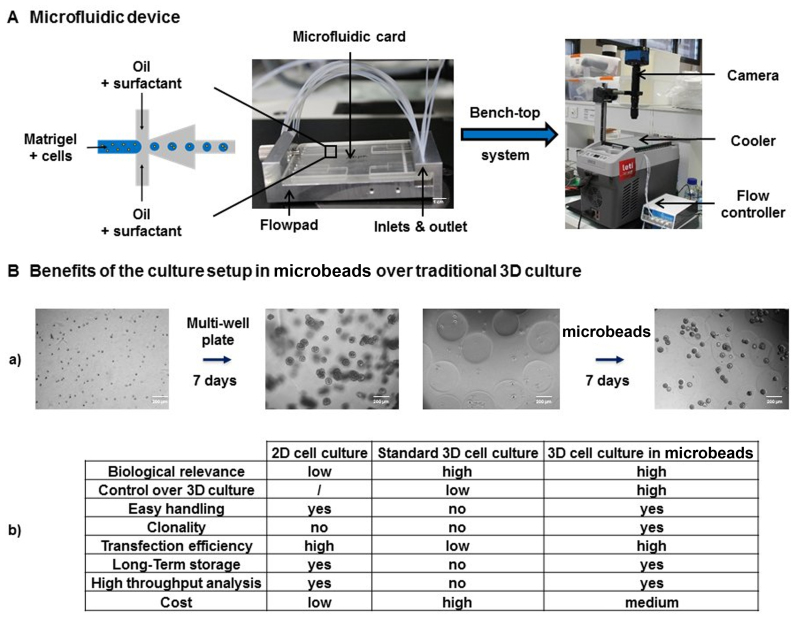
A Cyclic Olefin Copolymer microfluidic chip with a flow focusing geometry connected to a FlowPad microfluidic platform was used within a compact bench-top cooler system (Figure 1A). Two different microfluidic chips were designed, with channel square cross-sections of 200 × 200 and 300 × 300 μm to produce microbeads 250 and 450 μm in diameter, respectively. Perfluorinated oil HFE-7500 (3M, Cergy-Pontoise, France) was used with a 1.5% w/v PFPE-PEG-PFPE surfactant (Chemipan R&D laboratories, Warsaw, Poland) as a continuous phase and Matrigel (Corning, Brumath, France) premixed with cells as a dispersed phase. All experiments were performed at 4°C to prevent Matrigel polymerization inside microfluidic channels. Cells were mixed with Matrigel and injected in a reservoir in a Fluiwell-4C device (Fluigent, Villejuif, France). The flow inside the microfluidic device was induced and controlled using a microfluidic flow control system (Fluigent). A pressure of 40 mbar was applied to the cell/Matrigel mix as well as to the oil/surfactant mix. The cell/Matrigel mix was pushed into the flow-focusing junction and cut by perfluorinated oil flow, which led to the formation of Matrigel droplets. Matrigel droplets were collected in an Eppendorf tube for ∼15 min and subsequently incubated at 37°C for 20 min to promote Matrigel droplet polymerization into microbeads. Transfer of microbeads from oil into culture media was achieved by mixing microbeads, culture media and 1H, 1H, 2H, 2H-Perfluoro-1-octanol (Sigma-Aldrich). Real-time Matrigel droplet formation in perfluorinated oil was controlled using a camera (Basler, i2c, Pessac, France, acA800–510 μm). Microbeads were imaged after the transfer from oil into culture media with an inverted microscope (Olympus KX41, Rungis, France).
Figure 1.
High-throughput Matrigel microbead production for 3D organoid culture. (A) Scheme of the microfluidic platform that was used for Matrigel microbead production. The cell/Matrigel mix was pushed into the flow-focusing junction and cut via perfluorinated oil flow, which led to the formation of Matrigel droplets. Flow inside the microfluidic device was induced by a Fuigent flow control system. Then, droplets were allowed to polymerize at 37°C for 20 min. Real-time Matrigel droplet formation in perfluorinated oil was controlled using an acA800–560 μm camera. (B) (a) Single cells dispersed within a gel generate acini that are overlapping and heterogeneous in size and shape (left panel), which is in contrast to single cells microencapsulated within a gel (right panel). (b) Table that recapitulates the benefits of the 3D culture setup in microbeads over traditional 2D and 3D culture.
3D culture in Matrigel microbeads
For the acinar morphogenesis assay, RWPE-1 cells in Matrigel microbeads were cultured in KSFM supplemented with 50 ng/ml epidermal growth factor, 2% fetal bovine serum (PAA Clone, Les Mureaux, France), 1% Matrigel and 1% Penicillin-Streptomycin in multiwell plates. MCF10A cells in Matrigel microbeads were cultured in DMEM/F12 supplemented with 5% Horse serum, 20 ng/ml Epidermal Growth Factor, 10 μg/ml Insulin (Sigma-Aldrich), 0.5 μg/ml Hydrocortisone (Sigma-Aldrich), 100 ng/ml Cholera toxin (Sigma-Aldrich), 2% Matrigel and 1% Penicillin-Streptomycin in multiwell plates. Cell culture medium was changed every two days. All cells were routinely cultured in a humidified atmosphere with 5% CO2 at 37°C. For long-term organoid storage, Matrigel microbeads containing organoids at different developmental stages were frozen using 10% cryoprotective dimethylsulfoxide (DMSO, Sigma-Aldrich) in media according to standard procedures (21).
RNAi and plasmid transfection
To assess the transfection efficiency, RWPE-1-GFP and MCF10A-GFP cells, which were seeded in Matrigel microbeads, were allowed to develop into organoids for 4 and 11 days, respectively, and were subsequently transfected with siRNA (siAllStars, siAllStars AF546, siGFP, Qiagen, Les Ulis, France) or plasmid (pMAX-GFP, Lonza, Bazel, Switzerland) at a final concentration of 20 nM. This selected dose is used in routine in our lab (22) and is consistent with other works from the literature working with MCF10A (23) or RWPE1 (24). As a negative control, a siRNA without homology to any known mammalian gene was used (AllStars negative control, Qiagen, Valencia, CA, USA). AF546 stands for siRNA AllStars labeled at the 3′ end of the sense strand with an Alexa Fluor 546 nm fluorescent dye. For each experiment, cell viability was measured in controls (untreated cells, cells treated with siRNA alone, cells transfected without siRNA and cells transfected with siRNA AllStars) as well as in cells transfected with the different siRNA used. No significant differences should be observed between the different conditions of paired experiments to ensure a robust data analysis (see section “direct transfection of organoids in Matrigel microbeads”). Different transfection reagents were tested including Lipofectamine RNAiMax (Invitrogen, Life Technologies), Oligofectamine (Invitrogen), 3DFect, 3DFectIN (OZ Biosciences, Marseille, France) or TransIT2020 (Mirus, Euromedex, Mundolsheim, France) according to the manufacturer’s instructions. Electroporation was assessed using an AMAXA Transfection System (Lonza, program X-001) by resuspending microbeads in an electroporation buffer (HEPES 21 mM, NaCl 137 mM, KCl 5 mM, Na2HPO4–7H2O 0.7 mM, Dextrose 6 mM, pH 7.15). GFP was used as a surrogate RNAi target in determining siRNA knock-down efficiency with the variety of methods. After 3 days, the cells were extracted from microbeads, and GFP intensity was analyzed using flow cytometry. Mean fluorescence intensities (MFI) were used to quantify siRNA extinction efficiency as follows: extinction efficiency = 100 − (MFI siGFP/MFI siAllStars) × 100. For the functional transfection assays, RWPE-1 and MCF10A organoids in Matrigel microbeads were transfected at different time points with siRNA targeting p63 (sip63, Qiagen) or PTEN (siPTEN, Qiagen) genes and siRNA AllStars (Qiagen) at a final concentration of 20 nM.
Lentiviral transduction
RWPE-1 cells in Matrigel microbeads were allowed to develop into organoids for 4 days in media without antibiotics. After addition of 8 μg/ml Polybrene (Sigma-Aldrich), organoids were infected with lentiviruses diluted in assay media for 24 h. Then, half of the media was replaced with fresh media. After 2 days, 0.2 μg/ml Puromycin (Sigma-Aldrich) was added to select transduced cells. After 3 days of Puromycin selection, cells were extracted from organoids and analyzed via flow cytometry. Lentiviral transduction was performed using a Multiplicity of Infection (MOI) ranging from 0 to 20. MOI was derived as the number of virus particles against the average number of cells per mature organoid.
Flow cytometry
After transfection, microbeads were dispatched in 48-well plates or 24-well-plates. Superposing microbeads should be avoided as it could induce microbead aggregation during long-term culture experiments and be critical in transfection experiments. Adding Matrigel in 3D culture medium (see section “3D culture in Matrigel microbeads”) is essential to prevent cells from escaping microbeads and growing in 2D at the bottom of the plates. Single cells were extracted from Matrigel microbeads by combining prolonged trypsin incubation and several vortex steps (Supplementary Figure S1). Matrigel microbeads were pelleted by quick centrifugation (30 s), and rinsed twice with phosphate-buffered saline (PBS) 1× before addition of 500 μl of Trypsin-ethylenediaminetetraacetic acid 1 × 0.05% (Gibco, Ref 25300–054). Samples were incubated for 30 min at 37°C and vortexed for about 20 s every 5 min. Cells were then pelleted by centrifugation (5 min at 150 G) and rinsed twice in PBS 1× before flow cytometry analysis. In all experiments, gate was performed on 10 000 viable cells per condition. Cells were analyzed via flow cytometry using an LSR II flow cytometer (Becton Dickinson) gated on 10 000 viable cells per condition.
Apoptosis assay
CellEvent (Invitrogen) staining was performed to detect activated Caspase-3/7 and to quantify apoptosis in organoids within Matrigel microbeads. Briefly, Matrigel microbeads were incubated 30 min at 37°C with 10 μM of Cell Event reagent. As a positive cell death control, Matrigel microbeads were transfected with siRNA Cell Death (AllStars Hs Cell Death siRNA from Qiagen, 20 nM). After Cell Event staining, Hoechst staining was performed and images were acquired using a High Content CellInsight Screening Platform (ThermoFischer Scientific) for all conditions. Analysis was performed using R software and the percentage of dead organoids was evaluated based on negative control levels (Supplementary Figure S2).
Morphological analysis
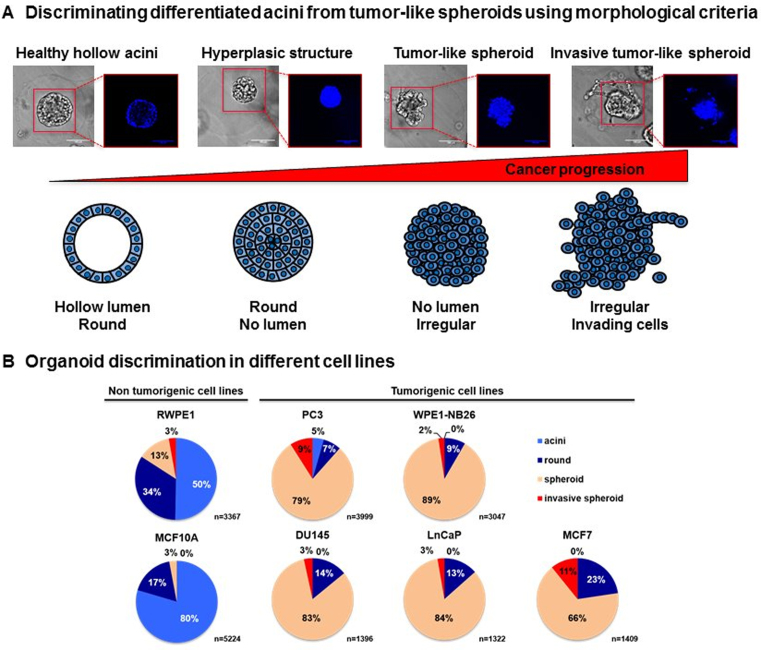
Morphological analysis of 3D structures was performed by eye counting under a phase microscope. While this method might appear long and laborious, it requires less time and efforts compared to immunofluorescence staining and confocal imaging that remain the gold standard to analyze organoid fate in 3D cultures (25,26). Organoid structures were classified into four different categories according to (27): round and regular polarized organoids with a hollow lumen resembling acinar structures of glandular tissues (acini), round and regular full structures (round), disorganized and irregular full spheroid structures (spheroid), loose, disorganized irregular full structures with invading cells migrating outside the organoid (invasive spheroid) (Figure 5A). This morphology-based classification allows the quantification of the tumorigenic potential of different cell lines and the quick visualization of different conditions.
Figure 5.
Discriminating differentiated acini from tumor-like spheroids based on morphological criteria. (A) Encapsulated RWPE-1 cells were allowed to develop into organoids for 7 days. Organoid structure was then discriminated into four different categories: round and regular polarized organoids with a hollow lumen resembling acinar structures of glandular tissues (acini), round and regular full structures (round), disorganized and irregular full spheroid structures (spheroid), and loose, disorganized irregular full structures with invading cells migrating outside the organoid (invasive spheroid). (B) Morphological classification of organoids, which were obtained from five prostate cell lines (RWPE-1, PC3, DU145, LnCaP and WPE1-NB26) and two breast cell lines (MCF10A and MCF7) that originated from benign or tumor tissues.
Western blot analysis
Cells were extracted from Matrigel microbeads by combining prolonged trypsin incubation and several vortex steps (Supplementary Figure S1). Cell recovery (Corning® Cell Recovery Solution, 100 ml (Product #354253) was further used to remove all traces of Matrigel. This solution enables the recovery of cells cultured on Corning® Matrigel® Basement Membrane Matrix for subsequent biochemical analyses. It has been tested for its ability to depolymerize a 1 mm thick layer of gelled Matrigel Matrix after 1 h at 2–8°C. Briefly, after trypsin digestion of Matrigel microbeads, cells and remaining traces of Matrigel were pelleted by centrifugation (5 min at 150 g), supernatant was discarded and 1 ml of cell recovery was added. Samples were mixed and incubated for 30 min at 4°C. Cells were pelleted by centrifugation (5 min at 150 g) and rinsed twice in PBS 1× before protein extraction for western blot analysis. Cellular proteins were extracted using a RIPA lysis buffer (Sigma-Aldrich) that was complemented with a protease inhibitor cocktail (Roche Diagnostics, Cat. No. 11697498001, Sigma-Aldrich). According to manufacturer’s instructions, one tablet was used extemporaneously to prepare 50 ml of protein extraction solution. After quantification with a Pierce BCA protein assay kit (Thermo Fisher Scientific), equal amounts of total protein (typically 10 ng of protein per sample) were separated via electrophoresis on a NuPAGE™ Novex™ 4–12% Bis-Tris Protein Gel (Life Technologies) in MES buffer and then transferred onto a nitrocellulose membrane (D. Dutscher, Brumath, France). The membranes were blocked in 5% non-fat milk in Tris-buffered saline, 0.1% Tween 20 (TBS-T, Sigma) for 1 h at room temperature (RT) and incubated with primary antibodies diluted at 1:1000 in 5% non-fat milk in TBS-T overnight at 4°C. Rabbit polyclonal antibodies directed against p63 (Cell Signaling Technology, Ozyme, St Quentin Fallavier, France), PTEN (Cell Signaling Technology) and GAPDH (Santa Cruz Biotechnology, Heidelberg, Germany) were used. Membranes were washed three times for 10 min in TBS-T and incubated with secondary horseradish peroxidase-conjugated antibodies (anti-rabbit, eBiosciences, Paris, France) at a dilution of 1:10 000 in TBS-T for 50 min. Membranes were washed three times for 10 min in TBS-T and developed using the Amersham ECL™ Prime Western Blotting System (D. Dutscher). Chemiluminescence was analyzed using a ChemiDoc Touch Imaging System (BioRad, Marne-la-Coquette, France), and quantification of the intensity of the bands was performed using the ImageLab™ Touch Software (BioRad).
Immunofluorescence staining
Matrigel microbeads were immobilized at the bottom of wells using Celltak coating (7 μg/cm2 of surface area, Corning) followed by a quick centrifugation (30 s at 200 g). Then, microbeads were fixed using 2% paraformaldehyde for 20 min and permeabilized with Trisbuffered saline (TBS) 0.5% Triton X-100 (Sigma-Aldrich) for 10 min at RT. To prevent non-specific adsorption of antibodies, microbeads were incubated with TBS-T 0.1% BSA 10% goat serum for 1 h at RT. Primary antibodies (rabbit anti-giantin 1:400) were added in TBS-T 0.1% BSA 10% goat serum overnight at 4°C. Primary antibodies were washed away four times with TBS-T for 15 min, and secondary antibodies (Rabbit-Cy3 1:400, Sigma-Aldrich) were added in TBS-T 0.1% BSA 10% goat serum for 1 h at RT. F-actin was labeled with 500-fold diluted FITC-phalloidin (Sigma-Aldrich), and nuclei were counterstained with 1 μg/ml Hoechst 33258 (Molecular Probes, Life Technologies) in TBS-T 0.1% BSA 5% goat serum for 30 min. Finally, microbeads were washed twice with TBS-T, and fluorescence images were taken using an AxioObserver.Z1 inverted microscope (Zeiss, Marly-le-Roi, France) mounted with an AxioCam 503 monochrome digital camera or a confocal spinning-disk inverted microscope (Nikon TI-E Eclipse) equipped with an Evolve EM-CCD camera. Hoechst, FITC and Cyanine 3 (Cy3) fluorophores were excited and collected sequentially (400 Hz line by line). Controls for auto-fluorescence or non-specific signal were performed by incubating microbeads without antibodies or with fluorochrome-matching isotype controls.
Statistical analysis
All data are represented as a mean of at least three separate experiments. Statistical significance was determined using the Mann–Whitney-Wilcoxon test using R software. Significant P-values are indicated by an asterisk (*P < 0.05; **P < 0.01; ***P < 0.0001).
RESULTS
Automatic generation of microencapsulated organoids
The process of microencapsulation was previously described (20). Because the process of droplet formation is highly repeatable under laminar flow conditions, it is possible to produce thousands of identical bioreactors. As previously demonstrated, our approach enables and improves acini culture homogeneity, recovery of 3D structures for further analysis such as fundamental genomics, throughput of the 3D culture preparation and most importantly high-throughput analysis, as we have shown by applying large-particle FACS. Now that feasibility has been demonstrated, progress has been made in our group on process automation. To do so, plastic chips with flow-focusing geometry connected to a FlowPad microfluidic platform were integrated within a bench-top cooler system (Figure 1A) (‘Materials and Methods’ section). This setup allows the automatic generation of homogeneous organoids that are embedded within microbeads of controlled size, composition and cell distribution (Figure 1B–a) (20). The table in Figure 1B recapitulates the benefits of this setup compared with 2D culture or standard 3D culture in Lab-Tek chambers. In each microbead, one single cell will give rise to an organoid derived from clonal origin. One single cell was encapsulated initially and acinar/spheroid morphology was assessed on ∼400 cells for RWPE1 following 7 days of growth. For breast acinar formation, following 14 days, the final number of cells was difficult to determine since it could, in theory, reach several thousands of cells per acini. This setup would be compatible with high-throughput screening applications and is less expensive than traditional 3D culture because it requires two to three times less Matrigel to obtain the same amount of organoids. For example, 350 μl of Matrigel for one well of a Lab-Tek 4-chambers slide allows to produce 42802 microbeads with a diameter of 250 μm (Figure 1B–b). Finally, these organoids are easy to handle, allowing us to store them for long periods by using cryopreservation procedures.
Direct transfection of organoids in Matrigel microbeads
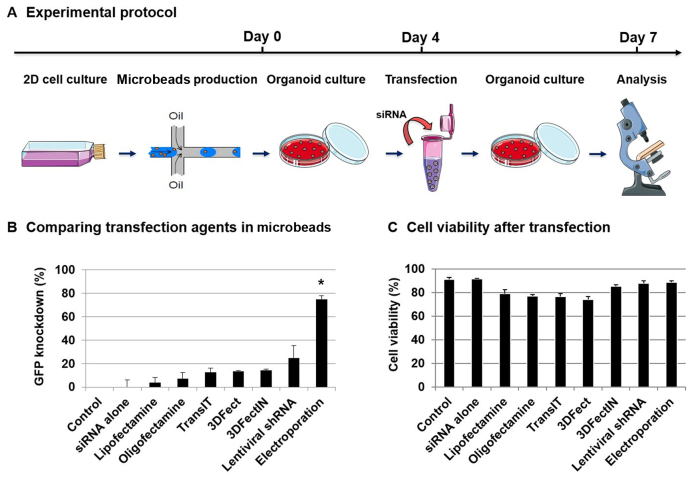
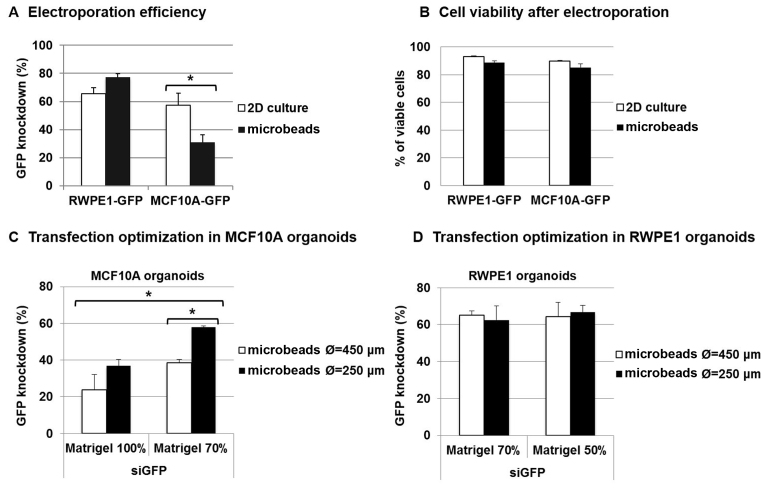
To evaluate the potential of micro-encapsulation for siRNA transfections into 3D organoids, we compared the transfection efficiency of the most common methods, namely, liposome/polymer-mediated transfection (Lipofectamin and Oligofectamin), electroporation (AMAXA system) and virus-mediated delivery (Lentiviral vector). We also evaluated compounds that are specifically designed to improve 3D culture transfections (TransIT, 3DFect and 3DFectIn). We used green fluorescent protein (GFP) to evaluate the extinction efficiency in 3D cultures. To this end, a prostate RWPE-1 cell line that stably expresses GFP was engineered using lentiviral vectors. RWPE-1-GFP was first used as a fast growing and well-characterized cellular model for normal epithelial acinar morphology in 3D cultures. Single RWPE-1-GFP cells were encapsulated and allowed to develop into organoids for 4 days before being transfected using siRNA directed against the GFP gene or siRNA AllStars as a control. After 3 days of culture, transfection efficiency was quantified by analyzing the GFP fluorescence intensity in cells that were extracted from organoids via flow cytometry (Figure 2A; Supplementary Figures S3 and 4). Actually, we measured the siRNA transfection efficiency because, at the same time, we monitored the siRNA transfection and target gene (GFP) knock-down. As expected, siRNA alone without any vectors was not sufficient to transfect organoids. Lipofectamin and Oligofectamin demonstrated low extinction efficiencies (≈5%), while TransIT, 3DFect and 3DFectIn reached up to 15% extinction efficiency (Figure 2B). Then, we used lentiviruses containing GFP shRNA expressing constructs in the presence of polybrene with or without Puromycin selection. The best results were obtained with an MOI (estimated number of virus per cell) of 10, which led to a 25% decrease in GFP fluorescent signal (Supplementary Figure S5). Finally, the highest results were obtained using electroporation, with GFP silencing efficiencies of almost 80% (P = 0.029). To ensure that the reduction of fluorescent signal was due to GFP silencing and not to the transfection protocol, cell death was evaluated using trypan blue dye exclusion staining and Caspase-3/7 detection. A small but not significant (ns) decrease in cell viability (from 91 to 88%) (P = 0.1) was observed in cells subjected to electroporation or to lentiviral transduction (Figure 2C). In other experiments, a not significant decrease in cell viability was also observed (from 85 to 74%) (ns, P = 0.1) (Supplementary Figure S6). These results highlight the efficiency and inocuity of electroporation to transfect RWPE-1 organoids in microbeads compared with other techniques. The versatility of this method was also showed by successfully transfecting tumorigenic cell lines which form full tumor-like spheroids in the beads (Supplementary Figure S7).
Figure 2.
Direct transfection of organoids in Matrigel microbeads. (A) Experimental protocol for direct transfection of encapsulated RWPE-1-GFP organoids. Encapsulated RWPE-1-GFP cells were allowed to develop into organoids for 4 days and were then transfected with siRNA (control siAllStars and siGFP, 20 nM) for 3 days before analysis. (B) After 3 days of transfection by comparing different techniques, cells were extracted from microbeads and analyzed via flow cytometry. GFP knock-down was determined using the GFP MFI measurement as follows: GFP knock-down = 100 − (MFI siGFP/MFI siAllStars) × 100. Results are expressed as the percentage of GFP signal extinction and represent the mean value ± SEM of three experiments (*P < 0.05). (C) Cell viability was measured via trypan blue dye exclusion staining.
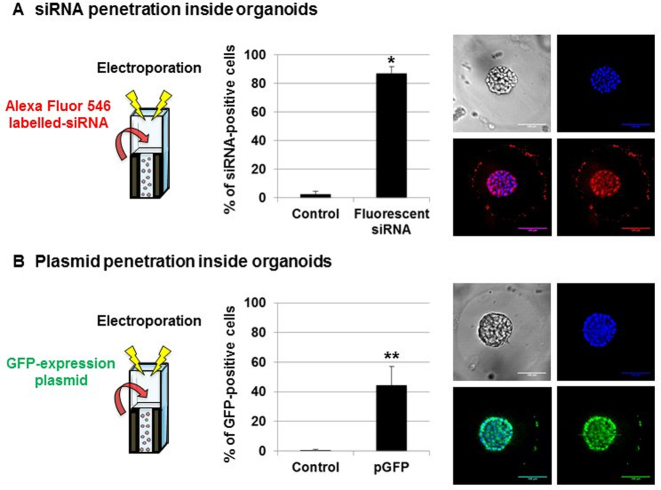
siRNA transfection can be achieved at the center of organoids in microbeads
Even in spheroid models produced by cell aggregation and devoid of ECM, direct 3D transfection is usually limited to the outer layer of cells (28), which reduces its relevance and leads to heterogeneous results. From this perspective, we decided to assess siRNA penetration inside organoids using electroporation of Alexa Fluor 546-labeled siRNA. RWPE-1 organoids were allowed to develop for 4 days in Matrigel microbeads before being transfected. After 3 days of culture, cell nuclei were stained with Hoechst, and fluorescence images were taken using a spinning disk confocal microscope. We purposely display an organoid without hollow lumen (full of cells) to show the extent of siRNA penetration in cells that are located at the center of 3D structures (Figure 3A). We observed a homogeneous distribution of siRNA in organoids, even in the cells that are located at the center of 3D structures. We further used flow cytometry analysis on cells that were extracted from transfected organoids, which demonstrated that almost 90% of RWPE-1 cells have integrated the labeled siRNA (P = 0.05) (Supplementary Figure S8A). These results are consistent with the measured transfection efficiency, as shown in Figure 2B. Given the efficiency of this technique regarding siRNA transfection, we wondered whether this can also be used for bigger nucleic acids such as expression vectors. We then assessed plasmid penetration inside organoids using electroporation of GFP-expressing plasmid. A dose-response curve was performed with p-GFP (Supplementary Figure S9). RWPE-1 organoids were allowed to develop for 4 days in Matrigel microbeads before being transfected. After 3 days of culture, cell nuclei were stained with Hoechst and fluorescence images were taken using a spinning disk confocal microscope. We observed a homogeneous distribution of GFP-expressing cells in organoids, even in cells that are located at the center of 3D structures (Figure 3B). We further used flow cytometry analysis on cells that were extracted from transfected organoids and demonstrated that more than 40% of RWPE-1 cells were expressing the GFP protein (P = 0.004) (Supplementary Figure S8B). Despite these promising results, a high heterogeneity was observed between experiments when using plasmids. Indeed, most of the cells were expressing GFP in some organoids while only a few cells were expressing the GFP in others. This is probably due to the size of the plasmid (∼5000 bp) when compared to siRNA (20 bp), as previously described (6). This differential GFP expression may also be related to the fact that large molecules such as plasmid DNA face physical barriers (nuclear envelope in addition to cytoplasmic membrane), before reaching the nucleus, while small molecules like siRNA freely cross the electropermeabilized membrane and have a free access to the cytoplasm. Moreover, this process may be emphasized by a slower diffusion of plasmids through the matrix and may contribute to reduce transfection efficiency (29). However, these results clearly demonstrate the general applicability of microbead electroporation to transfect siRNA and to a lower extent plasmids, even into cells that are located at the center of organoids.
Figure 3.
siRNA versus plasmid penetration into organoids that are grown in Matrigel microbeads. Fluorescent siRNA penetration (A) or GFP plasmid expression (B) inside encapsulated RWPE-1 organoids was assessed using flow cytometry and spinning disk confocal microscopy (40× magnification, scale bar 100 μm) 3 days after electroporation with 20 nM fluorescent siRNA or GFP plasmid (*P < 0.05; **P < 0.01). False color imaging is used here for better visualization.
Improving transfection efficiency by modulating microbead size and Matrigel concentration
To validate the extensive applicability of 3D organoid electroporation, we compared the extinction efficiency of 2D-cultured cells and 3D-organoids that are derived from prostate RWPE-1 and breast MFC10A cell lines. As previously observed, electroporation was found to be very effective for transfecting RWPE-1 cells. Interestingly, extinction efficiency was even higher in encapsulated organoids compared with 2D-cultured cells (77 versus 65%) (ns, P = 0.114) (Figure 4A). In contrast, electroporation efficiency was found to be lower in MCF10A cells compared with RWPE-1 cells under 2D culture conditions (from 65 to 57%) (ns, P = 0.5) and further decreased when performing direct 3D organoid transfection (from 57% in 2D to 31% in microbeads) (P = 0.024). Cell viability was found to be more than 85% under all conditions (Figure 4B). A small but not significant decrease (∼5%) in cell viability was observed after 3D transfection compared with 2D transfection in both RWPE-1 and MCF10A cells. However, this is probably due to the cell extraction process, which combines prolonged trypsin incubation and several vortex steps (‘Materials and Methods’ section) that are needed to extract cells from Matrigel microbeads before assessing their viability. Because in some cases ECM seems to be the major obstacle to siRNA transfection in 3D organoids, we decided to modulate Matrigel quantity around organoids to optimize transfection efficiency, particularly for MCF10A organoids. To do so, we adjusted two parameters: microbead size (cf diameter Ø) and Matrigel concentration. In RWPE-1 cells, no significant difference was observed when varying microbead size (from 450 to 250 μm) or Matrigel concentration (from 70 to 50%) (Figure 4C). This is probably because the extinction efficiency was already high. In contrast, transfection efficiency increased from 23 to 36% when decreasing microbead size (from 450 to 250 μm) and from 23 to 38% when decreasing Matrigel concentration (from 100 to 70%) in MCF10A organoids. Finally, we were able to reach up to 57% extinction efficiency when decreasing both microbead size and Matrigel concentration in MCF10A organoids (P = 0.05). This extinction efficiency is equal to the one we previously observed in 2D-cultured MCF10A cells (Figure 4A). Altogether, these results demonstrate the possibility of increasing extinction efficiency in organoid refractory for electroporation by optimizing parameters such as microbead size and Matrigel concentration to decrease Matrigel quantity around 3D structures. By doing so, we obtained extinction efficiencies that are comparable between 2D cultures and 3D organoids.
Figure 4.
Improving transfection efficiency by modulating microbead size and Matrigel concentration. RWPE-1-GFP and MCF10A-GFP-encapsulated organoids or 2D cell cultures were electroporated with siRNA (control siAllStars and siGFP, 20 nM). After 3 days, transfection efficiency (A) was determined using flow cytometry via GFP MFI measurement, as follows: transfection efficiency = 100 − (MFI siGFP/MFI siAllStars) × 100 and cell viability (B) was measured via trypan blue dye exclusion staining. The results represent the mean value ± SEM of three experiments (*P < 0.05). (C) Transfection optimization in MCF10A organoids: MCF10A-GFP microbeads of different sizes (450 and 250 μm in diameter Ø) and Matrigel concentrations (70 and 100%) were produced and electroporated with siRNA (control siAllStars and siGFP, 20 nM). After 3 days, extinction efficiency was determined using flow cytometry, as previously described. (D) Transfection optimization in RWPE1 organoids: RWPE-1-GFP microbeads of different sizes (450 and 250 μm in diameter Ø) and Matrigel concentrations (50 and 70%) were produced and electroporated with siRNA (control siAllStars and siGFP, 20 nM). After 3 days, extinction efficiency was determined using flow cytometry, as previously described.
Discriminating differentiated organoids from tumor-like spheroids based on morphological criteria
Current analysis of organoids uses confocal microscopy and is thus limited by time-consuming acquisitions and the need for human assistance, which hampers their applicability for pharmaceutical studies. Here, we used morphological classification as a quick and reliable read-out to discriminate organoid features and gene functions, as previously described (27). During glandular tumor development, the cells first begin to proliferate at the center of differentiated acini leading to the formation of polarized but hyperplasic structures. Cell polarization will then progressively be lost, which leads to the formation of disorganized and irregular tumor-like spheroids. Finally, cells or groups of cells will begin to migrate outside the tumor mass to invade the surrounding tissues, which ultimately leads to the formation of metastasis. Following this reasoning, organoid structures were discriminated into four different categories: round and regular polarized organoids with a hollow lumen that resembles acinar structures of glandular tissues (acini), round and regular full structures (round), disorganized and irregular full tumor-like structures (spheroid), and loose, disorganized irregular full structures with cells migrating outside the spheroid (invasive spheroid) (Figure 5A). We believe that this classification reflects the different stages of tumor development and can thus be used to quantify tumorigenic potential of the cells. To further validate its relevance for distinguishing normal from tumor-like phenotypes, we performed morphological classification of organoids, which were obtained from epithelial cells lines (Supplementary Figure S10): five prostate cell lines (RWPE-1, PC3, DU145, LNCaP and WPE1-NB26) and two breast cell lines (MCF10A and MCF7) that correspond to different stages of the disease (Figure 5B). As expected, we observed an increased proportion of spheroids and invasive spheroids in all tumor cell lines (PC3, DU145, LNCaP, WPE1-NB26 and MCF7) compared with the control cell lines (RWPE-1 and MCF10A) (30). MCF10A cells displayed a higher acini-forming capability compared with RWPE-1 cells (respectively 80 and 50%) (P = 0.004). This was accompanied by a lower percentage of round structures (17 versus 34%) (P = 0.048) as well as spheroids (3 versus 13%) (P = 0.004) and invasive spheroids (0 versus 3%) (P = 0.004). Interestingly, 5% of the PC3-derived organoids were able to differentiate into hollow acini, while this phenomenon was rarely observed for other cancer cell lines. These results are consistent with a previous study, which demonstrates that PC3 cells probably retain some differentiation capabilities (31). Among the cancer cell lines we studied, PC3 and MCF7-derived organoids demonstrated the highest invasive capabilities, as supported by high percentages of invasive spheroids (respectively 9 and 11%). Altogether, these results support the use of morphological criteria to systematically discriminate glandular organoids from tumor-like spheroids.
Gene silencing in Matrigel microbeads validates p63 and PTEN as key genes in mammary and prostatic acinar development
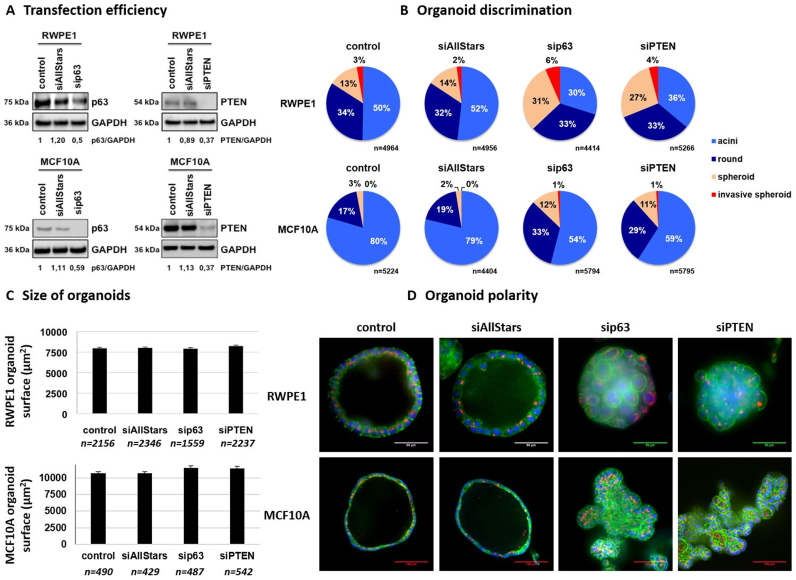
To further validate the versatility of direct organoid electroporation to identify tumor-suppressor genes during acinar development, we inhibited p63 and PTEN expression, two genes that are described as tumor-suppressors that play a key role in breast and prostate tissue development (32–35). To do so, prostate RWPE-1 and breast MCF10A organoids were allowed to develop in microbeads for 4 and 10 days, respectively, before performing transfection. Organoids were then cultured for 3 days before being analyzed. Cells were extracted from microbeads, and western blot analysis was performed on total protein extracts after the complete Matrigel removal (‘Materials and Methods’ section). We used antibodies directed against p63, PTEN and GAPDH as an internal control (Figure 6A). The results demonstrate no difference between untreated controls and organoids that are transfected with siRNA AllStars, while p63 and PTEN protein levels were effectively decreased by ∼50% in both RWPE-1 and MCF10A models. We then performed morphological classification of organoids as previously described (Figure 5A). We observed no difference in the proportion of different morphological categories between untreated organoids or organoids transfected with siRNA AllStars (Figure 6B). This demonstrates that the transfection method does not affect acini development in either RWPE-1 or MCF10A models. However, when transfecting RWPE-1 organoids with siRNA directed against p63, we observed an increase in both spheroids (from 14% to 31%) (P = 0.008) and invasive spheroids (from 2% to 6%) (P = 0.056) at the expense of acini (from 52 to 30%) (P = 0.008) compared with transfection with siRNA AllStars. When performing transfection with siRNA directed against the PTEN gene, we also observed an increase in spheroids (from 14 to 27%) (P = 0.008) and invasive spheroids (from 2 to 4%) (P = 0.095) at the expense of acini (from 52 to 36%) (P = 0.016). Similar results were obtained using MCF10A organoids, with an increase in round structures (from 19 to 33% for p63 inhibition (P = 0.008) and 19 to 29% for PTEN inhibition (P = 0.008)), spheroids (from 3 to 7% for p63 inhibition (P = 0.008) and 3 to 6% for PTEN inhibition (P = 0.008)) and invasive spheroids (from 0.07 to 0.18% for p63 inhibition (P = 0.016) and 0.07 to 0.24% for PTEN inhibition (P = 0.016)). This was accompanied by a decrease in the percentage of acini from 78 to 60% for p63 inhibition (P = 0.008) and from 78 to 65% for PTEN inhibition (P = 0.008). This marks a failure of cell polarization and lumen clearance, which leads to the generation of spheroids that consist of loosely associated disordered cells in the absence of either p63 or PTEN expression during organoid development. Interestingly, we observed no significant difference when comparing the size of organoids between the different conditions in RWPE-1 and MCF10A models (Figure 6C), which shows the relevance of morphological classification used here. Finally, encapsulated organoids were immobilized using Celltak coating and fixed before performing immunofluorescence staining for nucleus (Hoechst), F-Actin (Phalloidin) and Golgi apparatus (anti-Giantin antibody) (Figure 6D). RWPE-1 and MCF10A cell lines gave rise to polarized hollow acini in both controls (untreated and siRNA AllStars), as shown by the absence of cells at the center of acini and by the apical localization of the Golgi apparatus. In contrast, organoids transfected with siRNA that targeted p63 or PTEN failed to develop into fully differentiated acini, which demonstrates a tumor-like phenotype, as evidenced by the loss of polarity and the presence of cells at the center of 3D structures. Altogether, these results demonstrate that electroporation of organoids that are grown in Matrigel microbeads is efficient in decreasing p63 and PTEN expression and leads to the failure of acini development accompanied by the transition toward a tumor-like phenotype.
Figure 6.
Gene silencing in Matrigel microbeads validates p63 and PTEN as common tumor suppressors during mammary and prostatic acinar development. Encapsulated RWPE-1 or MCF10A cells were allowed to develop into organoids for respectively 4 and 11 days and were then transfected with indicated siRNA at 20 nM for 3 days before western blot analysis (A), morphological discrimination (B), size measurement (C) or immunostaining (D) for F-actin (Phalloidin-FITC, green), Golgi (Anti-Giantin Cy3, red) and nuclei (Hoechst, blue). Images were acquired using a spinning disk confocal microscope (20 × magnification, scale bar 50 μm).
DISCUSSION
Even though 3D cultures are widely used as relevant models to study physiological and pathological processes, direct 3D transfection remains challenging and limits high-throughput genetic screening studies in 3D cultures. Despite the wide use of siRNA, large efforts are still being made to develop more effective and safe methods to deliver siRNAs into cells, particularly in 3D cultures, and because of the great clinical potential of RNAi to treat diseases.
To date, current 3D culture systems rely on multiwell plates where cells that are embedded in ECM scaffolds develop into heterogeneous and overlapping organoids, limiting experiment reproducibility, analysis and gene delivery. Thus, there is a need for new culture formats to produce homogeneous and easy-to-handle organoids for high-throughput applications. Building on our previous microfluidic chip which delivers highly controlled conditions for the seeding and growth of polarized cellular acini within Matrigel microbeads (20), we designed an automatic and robust microfluidic platform that provides incremental improvements toward a high-throughput production of well-controlled organoids that are embedded within microbeads (∼2000 microbeads/min).
In contrast to static 3D cultures in Lab-Tek chambers, organoids that are grown in Matrigel microbeads are easy to handle and can be manipulated at will to perform electroporation. In addition, the reduced amount of Matrigel surrounding organoids constitutes a permissive 3D environment that facilitates transfection. Taking advantage of these characteristics, we demonstrate that direct electroporation of encapsulated organoids reach up to 80% transfection efficiency in RWPE-1 organoids. Despite reaching lower transfection efficiencies in MCF10A organoids, we also showed that gene silencing can be improved by tuning parameters such as microbead size or Matrigel concentration. In contrast with a previous study demonstrating low plasmid transfection efficiency (about 6%) in 3D cultures (30), we managed to reach plasmid extinction efficiency of more than 40%. Moreover, in a recent study (36) demonstrating the benefits of a novel gas plasma-facilitated cellular transfection method, siRNA extinction efficiency of about 20% (as compared to more than 80% in our system) and plasmid transfection efficiency of less than 5% (as compared to more than 40% in our system) were reached in a 3D culture system. Strikingly, we managed for the first time to reach siRNA transfection efficiencies that are comparable between 2D cultures and 3D organoids. While viral transduction systems usually allow higher transfection efficiencies, it has been demonstrated that ECM impedes virus-mediated gene delivery since its digestion dramatically improves extinction efficiency (12), which explains the limited effect on Matrigel microbeads. Direct organoid electroporation is particularly useful for transfecting long-term cultured organoids and at different development stages. For example, the siRNA effect would be totally lost if performing 2D transfection before 3D culture using MCF10A cells because they need a minimum of 14 days to fully develop into differentiated acini. Thus far, different methods have also been successfully developed to achieve efficient transgene expression in 3D models. For example, retroviral infection allowed the sustained expression of GFP in more than 90% of cells from organoid fragments that were removed from Matrigel (12). In other studies, researchers managed to achieve high transfection efficiencies using polyplex nanomicelles (37), flexible nanocarriers (38) or polyamine nanoparticles (39) on MCTS. However, these studies were performed on incomplete 3D models, cell aggregates such as MCTS or dissociated organoids devoid of any exogenous ECM (40). A viable strategy to transfect ECM-embedded organoids is to use inducible systems (41). Nonetheless, while ensuring a controlled and efficient transfection, this system is hardly compatible with high-throughput analysis. In another study, MCTS from mouse sarcoma were transfected via electroporation in the presence of propidium iodide. The method was highly efficient since practically the entire cell population was permeabilized, as measured by propidium iodide uptake (28). In addition, no difference was detected between cells at the periphery and the cells located deeper within the spheroid's core. However, the uptake efficiency of macromolecules such as plasmid DNA was found to be inefficient. Indeed, only a few cells from colon cancer MCTS were transfected using a plasmid that codes for the GFP. A striking observation is that transfected cells were heterogeneously distributed and appeared to be only localized in the outer layers of the spheroids. These results suggest that limited access to central regions of MCTS remains a significant barrier to gene delivery. Indeed, close contacts between cells may act as physical barriers that limit the diffusion of plasmid DNA and therefore its access to cells present in the core of the spheroid. In contrast, our results demonstrate high levels of siRNA molecule penetration through ECM and increased efficacy in quiescent cells at the center of organoids afforded by electroporation, which makes it a promising strategy for gene silencing studies. Thus, the combination of organoid microencapsulation and efficient siRNA delivery methods developed here encompasses existing techniques, and allows the transfection of physiological organoid cultures embedded in ECM that mimic complex tissue development steps.
Using this system, we further explored the effects of p63 and PTEN gene silencing on acini formation. To do so, we used morphological classification as a read-out to distinguish healthy organoids from tumor spheroids. We believe that this classification contains better criteria compared with 3D structure size to discriminate healthy organoids from tumor spheroids. In this study, morphological analysis of 3D structures was performed by eye counting under a phase microscope, which remains time consuming and laborious, although more rapid that the conventional immunostaining and confocal z-stack analysis. However, an automatic image analysis software is currently under development and will be applied to discriminate the different categories of organoids to meet the need of high-throughput screening applications. Indeed, we observed no significant difference between the sizes of organoids after p63 or PTEN inhibition, while we observed a significant increase in the proportion of spheroids at the expense of fully differentiated acini. This difference could be explained by an increase in cell proliferation and/or a loss of apoptosis, which occurs at the center of acini, leading to an increase in cell number progressively filling the lumen. This phenomenon would likely not be reflected at first by an increase in the overall size of organoids because of cell proliferation that occurs at the center of 3D structures such as for in situ carcinoma. We observed that down-regulation of p63 and PTEN led to the failure of acinar development as evidenced by the lack of cell polarization, lumen clearance and the generation of spheroids that consist of loosely associated disordered cells. These results validate the roles of p63 and PTEN as tumor-suppressor genes that play a key role in acinar development in both breast and prostate tissues.
Our results highlight the potential of microencapsulated organoid electroporation to study tissue development and tumorigenesis and provide valuable tools that should find versatile applications in fundamental research and organoid/spheroid-based drug assays. Being able to perform organoids versus tumor-like spheroids discrimination using label-free and real time flow-based strategies would prove very convenient for the future High Content Screening in 3D models. We believe that in addition to high-throughput screening for functional genomics studies, this approach will also be useful in identifying potential RNAi therapeutics and in probing their effectiveness in a 3D culture environment that will more accurately predict in vivo cell response.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Odile Filhol-Cochet of CEA Grenoble for the production of GFP-positive MCF10A cell line. We thank ‘live microscopy facility’ of BIG (μLife, funded by CEA Nanobio and Labex Gral) for access to the confocal microscope.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Commissariat à l’Energie Atomique (CEA) [Plan de couplage DSV-DRT]; Agence Nationale de la Recherche (ANR); Investissements d’avenir [ANR-11-NANB-0002]; Grenoble Alps Metropole (Proof of Concept program of Canceropole CLARA (PROscan3D project)). Funding for open access charge: CEA; ANR.
Conflict of interest statement. None declared.
REFERENCES
- 1. Edmondson R., Broglie J.J., Adcock A.F., Yang L.. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014; 12:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edmondson R., Adcock A.F., Yang L.. Influence of matrices on 3D-cultured prostate cancer cells’ drug response and expression of drug-action associated proteins. PLoS One. 2016; 11:e0158116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Weigelt B., Ghajar C.M., Bissell M.J.. The need for complex 3D culture models to unravel novel pathways and identify accurate biomarkers in breast cancer. Adv. Drug Deliv. Rev. 2014; 69–70:42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dutta R.C., Dutta A.K.. Human-organoid models: accomplishments to salvage test-animals. J. Biomed. Eng. Med. Devices. 2016; 1:1–8. [Google Scholar]
- 5. O’Brien L.E., Zegers M.M.P., Mostov K.E.. Opinion: building epithelial architecture: insights from three-dimensional culture models. Nat. Rev. Mol. Cell Biol. 2002; 3:531–537. [DOI] [PubMed] [Google Scholar]
- 6. Kim T.K., Eberwine J.H.. Mammalian cell transfection: the present and the future. Anal. Bioanal. Chem. 2010; 397:3173–3178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kostarelos K., Emfietzoglou D., Papakostas A., Yang W.-H., Ballangrud A., Sgouros G.. Binding and interstitial penetration of liposomes within avascular tumor spheroids. Int. J. Cancer. 2004; 112:713–721. [DOI] [PubMed] [Google Scholar]
- 8. Canatella P.J., Black M.M., Bonnichsen D.M., McKenna C., Prausnitz M.R.. Tissue electroporation: quantification and analysis of heterogeneous transport in multicellular environments. Biophys. J. 2004; 86:3260–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang H., Lee M.-Y., Hogg M.G., Dordick J.S., Sharfstein S.T.. High-throughput transfection of interfering RNA into a 3D cell-culture chip. Small Weinh. Bergstr. Ger. 2012; 8:2091–2098. [DOI] [PubMed] [Google Scholar]
- 10. Mellor H.R., Davies L.A., Caspar H., Pringle C.R., Hyde S.C., Gill D.R., Callaghan R.. Optimising non-viral gene delivery in a tumour spheroid model. J. Gene Med. 2006; 8:1160–1170. [DOI] [PubMed] [Google Scholar]
- 11. Salimzadeh L., Jaberipour M., Hosseini A., Ghaderi A.. Non-viral transfection methods optimized for gene delivery to a lung cancer cell line. Avicenna J. Med. Biotechnol. 2013; 5:68–77. [PMC free article] [PubMed] [Google Scholar]
- 12. Koo B.-K., Stange D.E., Sato T., Karthaus W., Farin H.F., Huch M., van Es J.H., Clevers H.. Controlled gene expression in primary Lgr5 organoid cultures. Nat. Methods. 2011; 9:81–83. [DOI] [PubMed] [Google Scholar]
- 13. Schwank G., Andersson-Rolf A., Koo B.-K., Sasaki N., Clevers H.. Generation of BAC transgenic epithelial organoids. PLoS One. 2013; 8:e76871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang N., Zhang H., Zhang B.-Q., Liu W., Zhang Z., Qiao M., Zhang H., Deng F., Wu N., Chen X. et al. Adenovirus-mediated efficient gene transfer into cultured three-dimensional organoids. PloS One. 2014; 9:e93608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Broutier L., Andersson-Rolf A., Hindley C.J., Boj S.F., Clevers H., Koo B.-K., Huch M.. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016; 11:1724–1743. [DOI] [PubMed] [Google Scholar]
- 16. Miyoshi H., Stappenbeck T.S.. In vitro expansion and genetic modification of gastrointestinal stem cells in spheroid culture. Nat. Protoc. 2013; 8:2471–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong Q., Wu G., Han L., Zhang Z., Du J., Sun W., Cao L.. A transfection method of PS-asODNs targeting ANGPTL4 in multicellular structures of hepatocarcinoma cell line. Cancer Gene Ther. 2015; 22:285–290. [DOI] [PubMed] [Google Scholar]
- 18. Zoldan J., Lytton-Jean A.K.R., Karagiannis E.D., Deiorio-Haggar K., Bellan L.M., Langer R., Anderson D.G.. Directing human embryonic stem cell differentiation by non-viral delivery of siRNA in 3D culture. Biomaterials. 2011; 32:7793–7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sapet C., Formosa C., Sicard F., Bertosio E., Zelphati O., Laurent N.. 3D-fection: cell transfection within 3D scaffolds and hydrogels. Ther. Deliv. 2013; 4:673–685. [DOI] [PubMed] [Google Scholar]
- 20. Dolega M.E., Abeille F., Picollet-D’hahan N., Gidrol X.. Controlled 3D culture in Matrigel microbeads to analyze clonal acinar development. Biomaterials. 2015; 52:347–357. [DOI] [PubMed] [Google Scholar]
- 21. Yokoyama W.M., Thompson M.L., Ehrhardt R.O.. Cryopreservation and thawing of cells. Curr. Protoc. Immunol. 2012; Appendix 3:3G. [DOI] [PubMed] [Google Scholar]
- 22. Benzina S., Pitaval A., Lemercier C., Lustremant C., Frouin V., Wu N., Papine A., Soussaline F., Romeo P.-H., Gidrol X.. A kinome-targeted RNAi-based screen links FGF signaling to H2AX phosphorylation in response to radiation. Cell. Mol. Life Sci. 2015; 72:3559–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Martin S.E., Jones T.L., Thomas C.L., Lorenzi P.L., Nguyen D.A., Runfola T., Gunsior M., Weinstein J.N., Goldsmith P.K., Lader E. et al. Multiplexing siRNAs to compress RNAi-based screen size in human cells. Nucleic Acids Res. 2007; 35:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ezponda T., Popovic R., Shah M.Y., Martinez-Garcia E., Zheng Y., Min D.-J., Will C., Neri A., Kelleher N.L., Yu J. et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013; 32:2882–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee G.Y., Kenny P.A., Lee E.H., Bissell M.J.. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods. 2007; 4:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fessart D., Begueret H., Delom F.. Three-dimensional culture model to distinguish normal from malignant human bronchial epithelial cells. Eur. Respir. J. 2013; 42:1345–1356. [DOI] [PubMed] [Google Scholar]
- 27. Taubenberger A.V., Bray L.J., Haller B., Shaposhnykov A., Binner M., Freudenberg U., Guck J., Werner C.. 3D extracellular matrix interactions modulate tumour cell growth, invasion and angiogenesis in engineered tumour microenvironments. Acta Biomater. 2016; 36:73–85. [DOI] [PubMed] [Google Scholar]
- 28. Wasungu L., Escoffre J.-M., Valette A., Teissie J., Rols M.-P.. A 3D in vitro spheroid model as a way to study the mechanisms of electroporation. Int. J. Pharm. 2009; 379:278–284. [DOI] [PubMed] [Google Scholar]
- 29. Gibot L., Golzio M., Rols M.-P.. How imaging membrane and cell processes involved in electropermeabilization can improve its development in cell biology and in clinics. Adv. Anat. Embryol. Cell Biol. 2017; 227:107–118. [DOI] [PubMed] [Google Scholar]
- 30. Härmä V., Schukov H.-P., Happonen A., Ahonen I., Virtanen J., Siitari H., Åkerfelt M., Lötjönen J., Nees M.. Quantification of dynamic morphological drug responses in 3D organotypic cell cultures by automated image analysis. PLoS One. 2014; 9:e96426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Härmä V., Virtanen J., Mäkelä R., Happonen A., Mpindi J.-P., Knuuttila M., Kohonen P., Lötjönen J., Kallioniemi O., Nees M.. A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One. 2010; 5:e10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shore A.N., Chang C.-H., Kwon O.-J., Weston M.C., Zhang M., Xin L., Rosen J.M.. PTEN is required to maintain luminal epithelial homeostasis and integrity in the adult mammary gland. Dev. Biol. 2016; 409:202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang S., Gao J., Lei Q., Rozengurt N., Pritchard C., Jiao J., Thomas G.V., Li G., Roy-Burman P., Nelson P.S. et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003; 4:209–221. [DOI] [PubMed] [Google Scholar]
- 34. Tucci P., Agostini M., Grespi F., Markert E.K., Terrinoni A., Vousden K.H., Muller P.A.J., Dötsch V., Kehrloesser S., Sayan B.S. et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:15312–15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoh K.E., Regunath K., Guzman A., Lee S.-M., Pfister N.T., Akanni O., Kaufman L.J., Prives C., Prywes R.. Repression of p63 and induction of EMT by mutant Ras in mammary epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E6107–E6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xu D., Wang B., Xu Y., Chen Z., Cui Q., Yang Y., Chen H., Kong M.G.. Intracellular ROS mediates gas plasma-facilitated cellular transfection in 2D and 3D cultures. Sci. Rep. 2016; 6:27872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Han M., Bae Y., Nishiyama N., Miyata K., Oba M., Kataoka K.. Transfection study using multicellular tumor spheroids for screening non-viral polymeric gene vectors with low cytotoxicity and high transfection efficiencies. J. Control. Release. 2007; 121:38–48. [DOI] [PubMed] [Google Scholar]
- 38. Jung H., Shimatani Y., Hasan M., Uno K., Hama S., Kogure K.. Development of flexible nanocarriers for siRNA delivery into tumor tissue. Int. J. Pharm. 2017; 516:258–265. [DOI] [PubMed] [Google Scholar]
- 39. Zamora G., Wang F., Sun C.-H., Trinidad A., Kwon Y.J., Cho S.K., Berg K., Madsen S.J., Hirschberg H.. Photochemical internalization-mediated nonviral gene transfection: polyamine core-shell nanoparticles as gene carrier. J. Biomed. Opt. 2014; 19:105009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Picollet-D’hahan N., Dolega M.E., Liguori L., Marquette C., Le Gac S., Gidrol X., Martin D.K.. A 3D toolbox to enhance physiological relevance of human tissue models. Trends Biotechnol. 2016; 34:757–769. [DOI] [PubMed] [Google Scholar]
- 41. Herr R., Wöhrle F.U., Danke C., Berens C., Brummer T.. A novel MCF-10A line allowing conditional oncogene expression in 3D culture. Cell Commun. Signal. 2011; 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.