Abstract
The majority of variants identified by genome-wide association studies (GWAS) reside in the noncoding genome, affecting regulatory elements including transcriptional enhancers. However, characterizing their effects requires the integration of GWAS results with context-specific regulatory activity and linkage disequilibrium annotations to identify causal variants underlying noncoding association signals and the regulatory elements, tissue contexts, and target genes they affect. We propose INFERNO, a novel method which integrates hundreds of functional genomics datasets spanning enhancer activity, transcription factor binding sites, and expression quantitative trait loci with GWAS summary statistics. INFERNO includes novel statistical methods to quantify empirical enrichments of tissue-specific enhancer overlap and to identify co-regulatory networks of dysregulated long noncoding RNAs (lncRNAs). We applied INFERNO to two large GWAS studies. For schizophrenia (36,989 cases, 113,075 controls), INFERNO identified putatively causal variants affecting brain enhancers for known schizophrenia-related genes. For inflammatory bowel disease (IBD) (12,882 cases, 21,770 controls), INFERNO found enrichments of immune and digestive enhancers and lncRNAs involved in regulation of the adaptive immune response. In summary, INFERNO comprehensively infers the molecular mechanisms of causal noncoding variants, providing a sensitive hypothesis generation method for post-GWAS analysis. The software is available as an open source pipeline and a web server.
INTRODUCTION
Genome-wide association studies (GWAS) have successfully identified over 50,000 genetic variants associated with more than 2,300 human diseases and phenotypes (1,2), but interpretation of these signals remains difficult. First, each GWAS-identified variant tags linkage disequilibrium (LD) blocks of potentially functional variants that are inherited together, and the causal variant underlying the association signal may not be genotyped on the platform (3). Second, 90% or more of GWAS variants are in the noncoding genome where they do not directly affect coding sequences of messenger RNAs (mRNA) (4); rather, they may affect regulatory elements that modulate mRNA transcription levels such as enhancers (5). Enhancers are context-specific and annotations are incomplete, so information must be integrated across tissue contexts and data sources to identify variants affecting enhancer function (6). Finally, to translate GWAS findings into a deeper understanding of pathology, enabling the development of novel therapeutics, it is crucial to identify the tissue-specific target genes of enhancers affected by regulatory variation.
Recent large-scale efforts focused on identifying active regulatory regions within the noncoding genome (7–9), but the field lacks a comprehensive method for identifying not only causal noncoding variants and the regulatory elements they disrupt but also the relevant tissue contexts, target genes, and downstream biological processes affected by these variants. A straightforward approach for investigating noncoding genetic signals is to derive a score measuring the regulatory potential of individual variants; examples include RegulomeDB, GWAVA, CADD and GenoCanyon (10–14). However, these score-based methods do not identify both the specific affected regulatory elements and the affected target genes and mostly ignore the reality that variants may have different impacts on different traits. HaploReg (15) is the best known tool that addresses this scientific need, which expands GWAS tag variants into haplotype blocks and overlaps them with chromatin state annotations and expression quantitative trait loci (eQTL), variants whose alleles are correlated with changes in the expression level of a target gene, to identify specific regulatory loci and target genes. However, it offers no way to integrate the enhancer and eQTL overlap results to characterize the affected tissue contexts, and performs direct eQTL overlap, which is biased by LD structure and may yield both false positives and negatives.
To address these issues, we introduce INFERNO (INFERring the molecular mechanisms of NOncoding genetic variants), a new method that integrates hundreds of diverse functional genomics data sources across tissues and cell lines with GWAS summary statistics to identify sets of putatively causal noncoding variants underlying an association signal and comprehensively characterize the downstream regulatory effects of these variants including the tissue contexts, specific regulatory elements, and target genes that they affect. INFERNO includes a tissue classification scheme that integrates information across diverse functional genomics data sources to characterize the relevant tissue context of functional variants in a hypothesis-free manner. INFERNO also introduces a novel statistical model for quantifying the enrichment of enhancer overlaps in specific tissue categories for any GWAS data.
To identify tissue-specific affected target genes, INFERNO integrates eQTL data from the GTEx consortium (16) with GWAS summary statistics using a Bayesian co-localization model (17). This allows the method to avoid the biases of directly overlapping GWAS variants with eQTL measurements and to identify eQTL signals that are strongly co-localized with association signals. Furthermore, it identifies functional variants that underlie co-localized signals and overlap regulatory elements in the matching tissue category. Many eQTL signals affect long noncoding RNAs (lncRNAs) which in turn can regulate protein-coding gene expression. INFERNO identifies lncRNA co-regulatory networks and downstream biological processes using GTEx RNA sequencing data (16), both across all tissues and using a novel principal components-adjusted method to identify tissue-specific regulatory networks. In summary, INFERNO provides a powerful hypothesis generation approach for identifying putatively causal regulatory signals to guide post-GWAS research.
To demonstrate the utility of INFERNO, we first applied the method to a large GWAS for schizophrenia (36,989 cases, 113,075 controls (18)). INFERNO uncovered functionally supported variants underlying eQTL signals targeting known schizophrenia genes and novel candidates related to transmembrane cellular signaling and significant enhancer enrichments in neuron-related tissue categories. We also identified downstream effects of lncRNAs on several known schizophrenia-related pathways including MAPK signaling, splicing, and Herpes simplex infection which is a known risk factor for schizophrenia (19). We then applied INFERNO to a large GWAS for inflammatory bowel disease (IBD) (12,882 cases and 21,770 controls) (20). INFERNO identified enhancer enrichments in immune and digestive tissues and effects on IBD-related pathways including adaptive immune response and leukocyte activation. INFERNO is available as an open source software package, and users can analyze top GWAS variants using the web server at http://inferno.lisanwanglab.org/.
MATERIALS AND METHODS
INFERNO pipeline implementation
The open source INFERNO pipeline is implemented using Python v2.7.9, R v3.2.3, and bash, and is available at https://bitbucket.org/wanglab-upenn/INFERNO. The pipeline can run any or all of the analysis steps spanning annotation, enhancer enrichment analysis, eQTL co-localization, lncRNA co-regulatory network identification, and pathway analysis and implements user-definable parameters for each step, and can be run on bsub-based servers or directly in a bash shell.
P-value and LD expansion of top GWAS variants
INFERNO starts with GWAS summary statistics and a set of user-defined top tagging variants as input. First, each top variant is expanded into a set of putative causal variants for further examination. In the p-value expansion mode, INFERNO computes a set of all variants i within 500 kb of each tagging variant such that pi ≤ m * pt where pi is the p-value for variant i, pt is the p-value of the tagging variant, and m is the user-defined multiplicative constant, 10 by default for one order of magnitude. These sets are pruned by LD using PLINK v1.90b2i 64-bit (21) with ‘–indep-pairwise 500 kb 1 0.7′ (within 500 000 bp and meeting a correlation threshold of r2 > = 0.7, which are user-definable parameters). LD structure is estimated using phase 3 version 1 (11 May 2011) of the 1000 Genomes Project (22). For the GWAS analyses in this manuscript, data from the European (EUR) population was used, but INFERNO users can also select LD structure data from 1000 Genomes for African (AFR), Asian (ASN), and South American (AMR) populations. Then, variants are re-expanded by LD structure using the same distance and r2 parameters. INFERNO also implements a direct expansion mode where input tag variants are directly expanded by LD structure.
Genomic partition analysis
Variants were categorized into different functional categories using the UCSC knownGene and RepeatMasker annotations for the hg19 genome build. Only chr1-22, X and Y are used in INFERNO. The 5′ UTR exons and introns, 3′ UTR exons and introns, and exons and introns were extracted from the knownGene annotation for each protein-coding gene, and all overlapping exons were merged together. Promoter annotations were defined as 1000 bp upstream of the first exon in the transcript, either coding or in the UTR. Variants were then assigned to mutually exclusive genomic element annotations using the hierarchy: 5′ UTR exon > 5′ UTR intron > 3′ UTR exon > 3′ UTR intron > promoter > mRNA exon > mRNA intron > repeat. A variant not overlapping with any class of elements above was classified as intergenic.
Functional annotation data download and pre-processing
FANTOM5 enhancer facet-level expression BED files, Roadmap ChromHMM BED files for the five core Roadmap marks (H3K4me3, H3K4me1, H3K36me3, H3K27me3, H3K9me3), HOMER TFBS annotations, and GTEx v6p eQTL and RNAseq data were downloaded from their respective servers and further processed using the bedtools suite (23) and custom awk and Python scripts.
Dataset classification into tissue categories
Building off existing categorization of Roadmap samples and informed by the UBERON and CL ontologies (24,25) used in the FANTOM5 facet-level classification and in GTEx, the different tissues and cell types from each data source were grouped into 32 major classes, with some data sources further grouped into 58 secondary and 15 tertiary sub-classes (Supplementary Tables S1–S3).
Quantification of enhancer enrichments
10,000 random sets of background variants matched to the input set of variants (the LD pruned set for p-value expansion or the input for direct expansion) by distance to the nearest gene, minor allele frequency, and the number of variants in each tagged LD block are sampled. The variants from each background set are then expanded into their corresponding LD blocks to match the number of variants in the LD expanded input set and overlapped with the same sets of functional annotations. Then, the empirical p-value for the significance of the overlap of the input data with each functional annotation or combination of annotations a (e.g. eRNA enhancer overlap, or both eRNA enhancer overlap and eQTL overlap) in a given tissue category t is defined as pa,t = (1 + ba,t) / (1 + 10 000), where ba,t is the number of background samples that include at least as many variant counts overlapping the annotation a in the tissue context t as the input dataset. These counts are calculated so that each LD block only contributes one effective count for annotation overlap in order to correct for LD structure by default, but INFERNO also reports results counting each variant in an LD block separately. These empirical p-values are corrected for multiple testing using the Benjamini–Hochberg procedure (26). INFERNO reports empirical p-values both within and across tag regions.
eQTL co-localization analysis
INFERNO uses the COLOC R package (17) to perform co-localization of the eQTL signals tested in GTEx v6p in 44 tissues against GWAS summary statistics. For each tag region and GTEx tissue, the script identifies all the genes tested for eQTL with the tagging variant in the region, reads in the eQTL data for each gene, and performs co-localization analysis using all the GWAS variants 500,000 bp on either side of the tag variant that are also found in the eQTL data. Minor allele frequencies (MAFs) can be defined by the user or can be extracted from 1000 genomes data using a custom preprocessing script. Then, the MAF and p-values of variants in the GWAS and eQTL datasets are used for co-localization analysis, including a user-defined sample size and case/control ratio for the GWAS of interest. Only variants with MAF ≥ 1% are included in the eQTL data by design of the GTEx consortium.
lncRNA cross-tissue and tissue-specific correlation analysis
To characterize co-regulatory networks of lncRNAs with eQTLs, reads per kilobase per million (RPKM) values across all RNA-sequenced samples in GTEx are used. GENCODE annotations are used to identify GTEx target genes that are categorized as lncRNAs (27), and correlations of the lncRNA RPKM vectors against all genes are computed using corr.test from the psych R package. Genes not expressed in any sample are excluded. Two correlation measures are computed: the Pearson correlation, which measures the linear relationship between two variables, and the Spearman correlation, which is a rank-based test that does not assume a linear relationship. User-defined parameters on the absolute values of the Spearman and Pearson values, 0.5 by default, are used to identify lncRNA target genes.
For tissue-specific correlation analysis, to avoid spurious intra-tissue correlations due to sample characteristics such as read depth, INFERNO performs a principal components-based correction analogous to PEER correction in eQTL scans or population stratification in GWAS studies. We generate a matrix of gene expression (in RPKM) Gc of size nc × kc, where nc is the number of samples in tissue category c and kc is the number of genes that are expressed in at least one sample in category c. Then, the tissue-specific sample correlation matrix Cc of size nc x nc is defined as Gc × G’c. Principal components (PC) analysis using centering and scaling was performed with the prcomp function in R, and the top 10 PCs for each sample were saved. To perform tissue-specific correlation analysis, the gene expression vectors for each lncRNA of interest and gene expressed in category c are modeled by linear regression against a user-defined number of PCs, 10 by default, and correlation is performed on the regression residuals.
WebGestalt pathway analysis
The pathway analysis script uses the WebGestaltR package (R ≥ 3.3) (28–30) to query pathway annotations over the Internet and performs pathway analysis of co-localized eQTL target genes, eQTL target genes within tissue categories, cross-tissue lncRNA targets, tissue-specific lncRNA targets, and cross-tissue and tissue-specific targets of individual lncRNAs against the KEGG, Gene Ontology Biological Process (BP), Gene Ontology Cellular Component (CC), and Gene Ontology Molecular Function (MF) pathway databases. For all three Gene Ontology categories, the ‘no_redundant’ annotations were used to reduce redundant genes in each given pathway of interest. Pathway analysis on targets of individual lncRNAs is also performed if the number of genes targeted by that lncRNA exceeds the minimum pathway overlap threshold.
MetaXcan analysis
INFERNO implements MetaXcan analysis (31–33) using GTEx v7 models from the PredictDB database and calls MetaMany.py from MetaXcan to analyze GWAS summary statistics.
LD score regression
INFERNO implements partitioned heritability LD score regression analysis using data and scripts provided at http://data.broadinstitute.org/alkesgroup/LDSCORE/ (34,35). INFERNO uses 1000 Genomes Phase 1 baseline model LD scores, regression weights, and allele frequencies, and the munge_sumstats.py and ldsc.py scripts.
Schizophrenia GWAS analysis
The full summary statistics file (scz2.snp.results.txt) and 128 top variants (scz2.rep.128.txt) for the schizophrenia analysis were obtained from the Psychiatric Genomics Consortium website. Top variants were parsed to remove variants on sex chromosomes or indels and converted into INFERNO input format using awk scripts. Summary statistics were annotated with minor allele frequencies from the 1000 genomes data using a custom script, annotate_input_variants.R, in the data_preprocessing/ section of the INFERNO code. These parsed files were then used as input to INFERNO with no p-value expansion.
IBD GWAS analysis
The full summary statistics file for the European population IBD analysis (EUR.IBD.gwas_info03_filtered.assoc) was obtained from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) website and annotated with 1000 Genomes minor allele frequencies. We selected 60 genome-wide significant variants by filtering the IBD GWAS p-value to 5 × 10−8 in the ‘All loci - Eur.’ tab of Supplementary Table S1 from Liu et al. (20). These parsed files were then used as input to INFERNO with no p-value expansion using the EUR 1000 Genomes population.
RESULTS
Overview of INFERNO pipeline
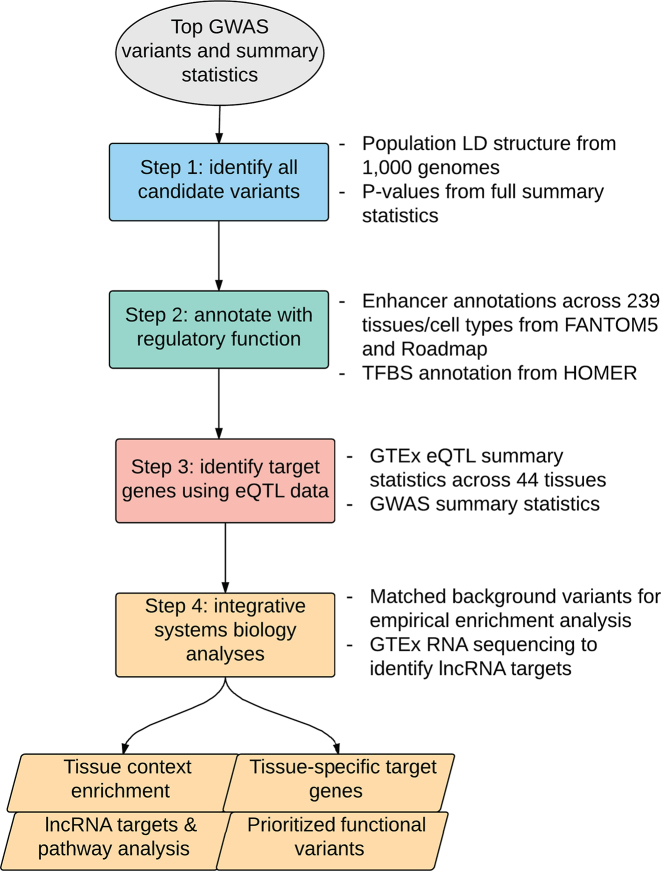
INFERNO consists of four stages (Figure 1): (i) Define sets of all potentially causal variants underlying each top GWAS signal. (ii) Characterize these variants by overlapping with various functional genomics data sources including epigenomic states, enhancer annotations, and overlap with messenger RNA (mRNA) and repeat elements. (iii) Identify tissue-specific effects on target genes using Bayesian co-localization of GWAS and eQTL data. (iv) Integrate information from all previous steps using a tissue categorization framework to identify functional variants with concordant annotation support, the tissue contexts of enhancer-gene interactions, target genes with strong functional support, significant tissue-specific enrichments of enhancer overlaps, and the downstream biological processes affected by disruption of target genes and long noncoding RNAs.
Figure 1.
Outline of INFERNO pipeline approach.
Defining comprehensive sets of potentially causal variants from GWAS findings
Given a user-defined list of top variants and summary statistics from a given GWAS, INFERNO provides two approaches for defining comprehensive sets of putatively causal variants underlying each top association signal (Methods). For high powered GWAS datasets with dense association signals, or where summary statistics are not available, INFERNO uses data from the 1000 Genomes Project (22) in a user-defined population (European, African, East Asian, or admixed American) to define blocks of variants in linkage disequilibrium (LD) with user-specified top variants using parameters for the threshold on r2 and maximum size of each LD block (0.7 and 500Kbp by default, respectively). For lower-powered GWAS datasets or those with sparse association signals, INFERNO uses GWAS summary statistics to identify all significant or almost significant variants within a user-defined window around each tagging variant (p-value expansion). This set is pruned by LD structure using PLINK (21), where representative subsets of variants within each LD block are chosen such that subjecting them to LD expansion recaptures the other variants in the LD block. These LD block-tagging variants are re-expanded into full LD blocks using PLINK. For lower-powered GWAS data, the p-value expansion enables the analysis of nominally significant signals near genome-wide significant signals by taking the full GWAS summary statistics as well as LD structure into account, potentially capturing causal variants underlying association signals. For GWAS datasets with larger sample size and denser association signals such as the two datasets we analyzed in this study, we recommend direct LD expansion from the tagging variants to avoid confounding separate genome-wide significant signals that are close to each other.
Annotation of expanded variant sets with transcriptional regulatory elements
To identify noncoding genetic variants, INFERNO quantifies the proportion of variants overlapping messenger RNA (mRNA) promoters, exons (i.e. coding variants), introns, 5′ untranslated region (UTR) exons and introns, 3′ UTR exons and introns, and RepeatMasker genomic repeats including LINEs and SINEs. Any variant outside all of these regions is classified as intergenic. All variants are subjected to annotation overlap regardless of genomic partition.
Next, each variant is overlapped with two complementary enhancer data sources. The first are sites of enhancer RNA (eRNA) transcription, which reflects enhancer activity (36), as assayed by cap analysis of gene expression (CAGE-seq) data generated by the FANTOM5 consortium across 112 tissue and cell type groupings (7). The second is enhancer states defined by ChromHMM (37) using combinatorial epigenomic states measured by chromatin immunoprecipitation and sequencing (ChIP-seq) of five histone modifications, which mark active enhancers in a stereotypical pattern (38,39), across 127 tissues and cell types generated by the Roadmap Epigenomics Project (8) and by the Encyclopedia of DNA Elements (ENCODE) project (9,40). In the Roadmap analysis, variants are overlapped with a total of 15 ChromHMM states including three types of enhancer states, sites of genic transcription, repressed regions, and active promoters, another type of transcriptional regulatory element that may harbor causal variants underlying an association signal. Enhancer overlap in all tissues and cell types from FANTOM5 and/or Roadmap is reported for each variant in the full expanded set.
In addition to overlapping variants with functional genomics annotations across tissues, INFERNO includes a sequence-based analysis to find variants affecting transcription factor binding sites (TFBSs) identified by the HOMER tool (41) (see Materials and Methods). INFERNO uses positional weight matrices (PWMs) to compute the difference in the log-odds binding probability of each affected TFBS between the reference and alternate alleles of the overlapping genetic variant (ΔPWM score) in order to identify genetic variants that either increase or decrease TFBS strength.
Tissue categorization of annotations and integrative analysis
Combining information across complementary sources of functional genomics data enables the comparison of evidence from independent experiments to improve sensitivity and robustness. However, it is not possible to directly compare results across the three consortia analyzed by INFERNO (FANTOM5, Roadmap, and GTEx) because each assayed different tissue types and cell lines at different levels of biological complexity. To integrate evidence from these disparate data sources, we designed a tissue classification scheme that grouped individual samples from all 3 data sources into one of 32 tissue categories (Supplementary Tables S1–S3, Supplementary Figure S1, Materials and Methods). This categorization approach provides additional power over using the individual enhancer data sources on their own, as an average of only 38% of FANTOM5 enhancers and 1.2% of Roadmap enhancers are shared between the two data sources within tissue categories (Supplementary Figure S2). This integrative categorization provides a high-level view of the affected tissue contexts across consortia, allowing for easy identification of the tissue contexts harboring noncoding elements affected by variants within each GWAS tag region.
Background sampling for enrichment of enhancer overlaps
Due to the widespread regulatory activity in the noncoding genome (9), selecting a large set of genetic variants in an LD block and overlapping them with hundreds of functional measurements may yield many overlaps simply by chance. To quantify the significance of enhancer overlap enrichment, INFERNO includes a statistical sampling approach to define empirical p-values for the enrichment of overlaps for each pair of annotations a and tissue category t within individual GWAS tag regions as well as across all tag regions (e.g. a = FANTOM5 enhancers, t = brain, see Materials and Methods). This provides statistical evidence of enhancer dysregulation aggregated across all genome-wide significant loci as well as enrichments within individual loci. INFERNO provides results from two modes of sampling: (i) LD-collapsed, where any number of variants that overlap a given annotation but are in LD with each other contribute just one count to that annotation-tissue category combination; (ii) direct, where each variant overlapping an annotation in a tissue category contributes one to the observed counts, regardless of other overlaps in the same LD block. The LD-collapsed approach is consistent with other genomic enrichment tools including GREGOR (42), while the direct approach is more sensitive in detecting large regions of regulatory annotations within LD blocks. As an alternate approach for quantifying the genome-wide enrichment of GWAS signals in various functional annotations, INFERNO can perform stratified LD score regression (34,35) against the 53 functional annotations from the full baseline model. We emphasize that the INFERNO enrichment analysis is distinct from LD score regression in that it focuses on identifying enhancer enrichments within significant loci rather than genome-wide enrichment, which could occur even without any genome-wide significant signal.
eQTL co-localization analysis
Current noncoding genetic variation annotation methods can identify functional variants and affected regulatory elements, and in some cases provide a hypothesis-free characterization of the relevant tissue context (14), but do not fully characterize the affected target genes. These methods either assume that the nearest gene is the affected transcript or directly overlap variants with eQTLs, including HaploReg, which uses eQTL signals from 14 sources. However, the closest gene is typically not the target of transcriptional regulatory elements (5), and direct overlap of variants with eQTL signals is biased by genomic LD structures, where an eQTL association signal may be spread across a haplotype block so that the measured variant is not the causal regulatory variant.
When summary statistics are not available, INFERNO performs direct overlap with eQTL signals across 44 tissues from the Genotype-Tissue Expression (GTEx) project (16). If summary statistics are available, INFERNO performs co-localization analysis using the COLOC method to control for bias from LD structures (17). This model uses a Bayesian statistical model to calculate posterior probabilities for different causality relationships between GWAS and tissue-specific eQTL signals. The most relevant hypothesis from the COLOC output for INFERNO is H4, that there is a shared causal variant underlying both the eQTL signal and the GWAS disease signal. INFERNO performs co-localization analysis comparing all GWAS signals within 500kb of each tag variant with eQTL signals across all 44 GTEx tissues (Methods). Strongly co-localized signals are defined as a user-definable threshold on P(H4), 0.5 by default representing a higher chance of a co-localized signal than any other hypothesis. COLOC also reports the probability of any individual variant being the shared causal variant, measured by the Approximate Bayes Factor (ABF). For further analysis of putatively causal variants, INFERNO defines sets of variants accounting for at least half of the cumulative ABF distribution at each strongly co-localized signal. This allows for the sensitive detection of truly co-localized signals to identify causal variants, the target genes they affect, and the tissue context of the regulation.
As an alternate approach for identifying affected genes, INFERNO also provides an option to perform MetaXcan analysis (31–33) using GTEx data as the reference transcriptome dataset to perform gene-based association mapping given GWAS summary statistics. MetaXcan identifies tissue-specific effects on genes but does not prioritize individual variants, so only COLOC results can be integrated with the variant-based analyses in the rest of the INFERNO pipeline.
Integrative analysis of co-localized eQTLs with annotations
To integrate the results between the enhancer and eQTL analyses, INFERNO uses the tissue categorizations of the FANTOM5, Roadmap, and GTEx datasets to filter variants in the ABF-expanded sets underlying a co-localized signal to only those overlapping an enhancer from the concordant tissue class. Those variants are then prioritized based on whether they overlap a TFBS and/or have a high individual ABF value (Materials and Methods). Prioritization by ABF enables the identification of highly confident single variants, but single variants with high ABF may be difficult to detect in complex LD regions, so prioritization by TFBS can distinguish potentially causal variants from sets of variants with low ABF. This integration of diverse data types spanning epigenomic marks, enhancer activity, transcription factor motifs, and eQTL signals provides a useful tool to identify causal variants, affected target genes, and relevant tissue contexts in an unbiased fashion. Furthermore, if users have an a priori assumption about which tissue categories might be relevant for their trait of interest, INFERNO can further prioritize variants from those specific categories.
Identification of co-regulated networks mediated by lncRNAs
Finding a GTEx eQTL signal supported by concordant enhancer support may be only the first step to understanding the affected regulatory mechanism underlying a genetic association signal because the eQTL may target a long noncoding RNA (lncRNA), which can in turn act as a transcriptional regulatory element for other genes (43). Although tools for lncRNA target prediction in specific contexts exist, lncRNA targeting mechanisms are not fully characterized, so INFERNO takes an unbiased approach to find genes that may be co-regulated with or directly regulated by lncRNAs: RNA sequencing-based expression vectors of all expressed genes in the genome across all samples and tissues from GTEx are correlated with the expression vector of a lncRNA of interest. Then, genes with correlation values meeting user-specified thresholds on the absolute value of Spearman and/or Pearson correlations across all tissues (0.5 by default, Methods) are considered to be co-regulated with the lncRNA of interest, in line with previous approaches to lncRNA target identification (44). We analyzed 1,417,168,941 pairs of transcripts expressed in GTEx and 6,007,249 met both correlation thresholds, indicating that the default settings roughly correspond to the top 0.5% of interactions transcriptome-wide (Supplementary Figure S3A). To identify tissue-specific lncRNA co-regulatory networks, INFERNO also performs a principal components-corrected intra-tissue category correlation analysis (Methods, Supplementary Figure S3B). These approaches provide lists of co-regulated genes based on expression correlation, which are potential lncRNA targets, but cannot characterize the direction of causality for lncRNA–mRNA relationships. Thus, INFERNO identifies putative co-regulatory networks affected by genetic variants across tissues and within tissue categories. This analysis is done automatically within INFERNO after the co-localization analysis is complete.
Tissue-specific pathway analysis of co-localized eQTL target genes and lncRNA targets
To characterize the biological processes affected by co-localized eQTL target genes as well as potential targets of co-localized lncRNAs, INFERNO uses the WebGestaltR package (28–30) to perform pathway analysis (Materials and Methods). This analysis is performed on all co-localized eQTL target genes, eQTL target genes within tissue categories, cross-tissue lncRNA targets, tissue-specific lncRNA targets, and cross-tissue and tissue-specific targets of individual lncRNAs.
Application to schizophrenia GWAS
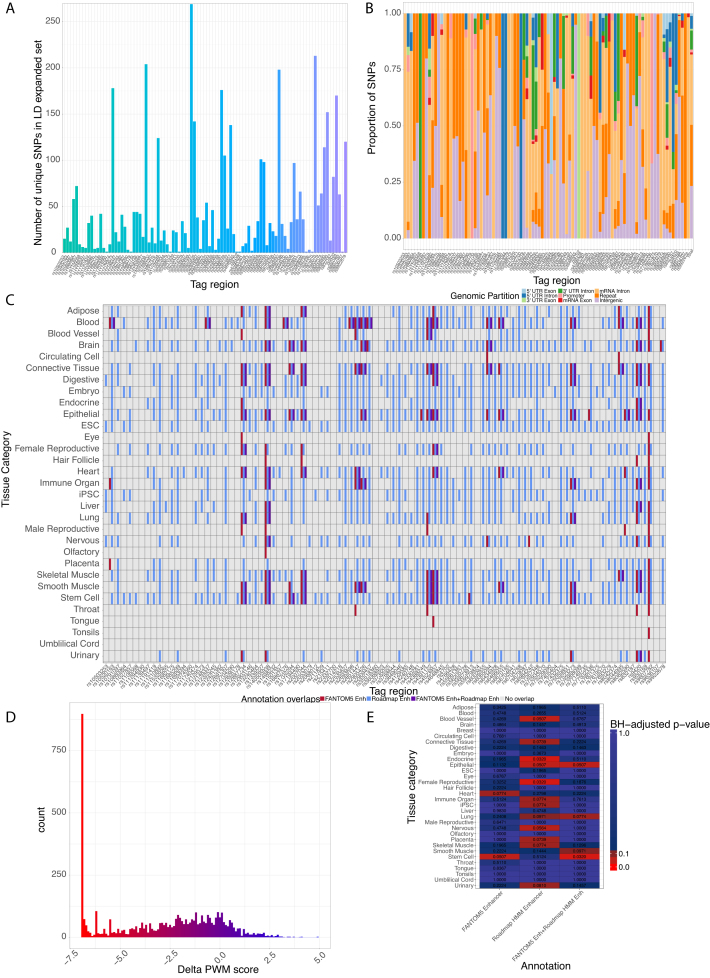
To demonstrate INFERNO’s utility, we analyzed a GWAS dataset for schizophrenia from the Psychiatric Genomics Consortium with 94 LD-independent signals (n = 36,989 cases, 113,075 controls, (18), skipping variants on chromosome X due to lack of annotations for indels). Due to the density of genome-wide significant association signals in this dataset, we performed this analysis using direct LD expansion from the tagging variants. LD expansion with a threshold of r2 > = 0.7 in the European population of 1000 genomes yielded 4,329 unique variants (Figure 2A). Only 40 of these were located in messenger RNA (mRNA) exons. The majority were in mRNA introns (1,354), repeat elements (1,175), or outside of any annotations (1,010) (Figure 2B), suggesting potential noncoding impact. Overlapping these variants with enhancer annotations found widespread enhancer signals in the Roadmap data, with 2,215 (51%) variants overlapping a ChromHMM enhancer state in at least one tissue (Figure 2C). The FANTOM5 overlaps were limited due to the more conservative nature of the eRNA measurements, with 103 (2.4%) variants overlapping a FANTOM5 enhancer in at least one tissue. Finally, overlap with HOMER TFBSs found that 2,013 (46%) unique variants overlapped TFBSs for 227 unique transcription factors for a total of 4,869 variant—TFBS overlaps. The majority (4,073) of these overlaps lowered the predicted binding strength (Figure 2D).
Figure 2.
Characteristics of expanded variant sets for schizophrenia analysis. (A) Number of variants after LD expansion. (B) Genomic partitions of expanded set variants across tag regions. (C) Summary of tissue category FANTOM5 and Roadmap enhancer overlaps across tag regions. (D) Distribution of ΔPWM scores for variants overlapping HOMER TFBSs. (E) Empirical enrichment of variants overlapping enhancers from FANTOM5 and/or Roadmap in specific tissue categories.
The INFERNO LD-collapsed sampling procedure identified significant enrichments of enhancer overlaps in 2 FANTOM5 tissue categories, 12 Roadmap tissue categories and 4 categories for concordant FANTOM5+Roadmap overlap (Figure 2E). This wide range of tissue categories reflects the low tissue-specificity of the regulatory elements affected by individual schizophrenia-associated genetic variants. There was no enrichment for brain annotations after multiple testing correction, with the lowest p-value (unadjusted empirical p-value = 0.0344) for Roadmap overlaps alone. The neuron datasets from both FANTOM5 and Roadmap are grouped into the ‘nervous’ category (adjusted p-value = 0.0564 for Roadmap) rather than brain, suggesting that neuronal regulatory mechanisms may be more genetically affected than higher-level signals measured from homogenized brain region samples in the brain category containing mixtures of various cell types, only a fraction of which may mediate schizophrenia predisposition.
Specific enriched categories with previously described relevance to schizophrenia include: blood vessel (45); endocrine, specifically thyroid hormones (46); female reproductive, given the later average onset of schizophrenia and reduced incidence rate in women (47); heart (48); immune organ, given the strong epidemiological and molecular genetic evidence of immune dysfunction and gastroenterological issues in schizophrenia (49–51); developmental categories such as stem cell and iPSCs (52,53); lung (54); placenta (55); and nervous (56). No individual tag regions were found to have LD-collapsed significant enrichments.
Comparison of the LD-collapsed enrichment counts with the direct count-based enrichment counts found fewer cross-tag region enrichments, including in the immune organ and epithelial categories, but did not identify an enrichment in stem cell (Supplementary Figure S4) or in brain. However, this approach yielded significant enrichments within 46 individual tag regions, including for brain in 4 tag regions (Supplementary Table S4).
Next, 103,016 co-localization tests for 2,928 genes (COLOC) were performed across the 94 tag regions, identifying 942 unique tissue-target gene eQTL signals spanning all 44 GTEx tissues and 286 unique genes (including 53 lncRNAs from 34 tissues) that were strongly co-localized with schizophrenia GWAS signals (Supplementary Table S5, Supplementary Figure S5). Pathway analysis on the 286 co-localized target genes found no significant enrichments. We cross-referenced these genes with several schizophrenia differential gene expression datasets (57–63), but found that only 14 co-localized genes were significantly differentially expressed across these datasets.
To prioritize the strongest signals from this analysis, we identified five regions harboring co-localized eQTL and GWAS signals supported by variants with individually high ABFs as well as enhancer overlaps from the same tissue category: rs4766428 (12q24.11), rs12826178 (12q13.3), rs56205728 (15q15.1), rs4702 (15q26.1), and rs6002655 (22q13.2) (Figure 3A, Supplementary Table S5, Table 1). In all these regions, the prioritized variant was also the tag variant, but this is not necessarily always the case.
Figure 3.
Results of GTEx co-localization analysis with schizophrenia GWAS. (A) Top results from co-localization analysis integrated with annotation overlaps. Counts in barplots refer to individual variants underlying an eQTL signal in a given tag region, including all variants in the ABF-expanded sets. (B) UCSC Genome Browser view of locus around rs4766428. In ChromHMM tracks, yellow = enhancer, green-yellow = genic enhancer, green = transcription, red = active transcription start site. Track highlighted with red box is dorsolateral prefrontal cortex.
Table 1.
Summary of INFERNO region prioritizations for schizophrenia. Strong ABF refers to signals where one variant had an ABF of 0.50 or higher for a co-localized eQTL signal
| Prioritized variant | Prioritization approach | Tissue and target gene |
|---|---|---|
| rs4766428 (12q24.11) | Strong ABF + TFBS + concordant enhancer | 12 signals including C12orf76 and VPS29 in brain and TCTN1 in nerve |
| rs12826178 (12q13.3) | Strong ABF + concordant enhancer | TSPAN31 in blood |
| rs56205728 (15q15.1) | Strong ABF + concordant enhancer | BUB1B in brain, PAK6 and PLCB2 in skeletal muscle |
| rs4702 (15q26.1) | Strong ABF + concordant enhancer | FES in subcutaneous adipose and pancreas, SLCO3A1 in transformed fibroblasts, and FURIN in esophagus mucosa |
| rs6002655 (22q13.2) | Strong ABF + concordant enhancer | NDUFAF6, RANGAP1, RP4-756G23.5 in blood |
We focused on the 12q24 region around rs4766428, the only high ABF signal with TFBS overlap. rs4766428 itself was prioritized as having high ABF underlying 12 distinct eQTL signals including for C12orf76 and VPS29 in the brain category and TCTN1 in the nerve tissue category (Figure 3B). Note that this variant lies in an intron of ATP2A2 but is not co-localized with an eQTL for that gene. These three genes are all involved in transmembrane cellular processes: C12orf76 is an unannotated transcript associated with the ‘ion channel activity’ GO pathway (64), VPS29 is part of a group of vacuolar sorting proteins (65), and TCTN1 encodes a family of secreted transmembrane proteins involved in ciliopathies and several cancer types (66). This variant also disrupts binding sites for ERRA, PPARg and RXR (ΔPWM = −1.95, −1.84, −2.06, respectively). Of particular relevance is the ChromHMM enhancer overlap in brain dorsolateral prefrontal cortex (highlighted with red box in Figure 3B), although several other brain regions have also been implicated in schizophrenia (67).
Using the top 94 LD-independent signals as input, HaploReg detected almost none of the top results identified by INFERNO. In the rs4766428 region, HaploReg did not identify any brain signals for VPS29 and did not identify any eQTL signals at all for C12orf76 and TCTN1. In the rs1286178 region, HaploReg did not detect any eQTL signal for rs1286178. In the rs56205728 region, HaploReg did not identify the brain signals for BUB1B or the PAK6 signal. In the rs4702 region, HaploReg detected the FES signals in pancreas but not subcutaneous adipose, detected the FURIN signal in esophagus mucosa, and missed the SLCO3A1 fibroblast signal, although it identified additional FES signals in fibroblasts and thyroid that INFERNO did not identify as strongly co-localized signals (P(H4) = 0.08 and 0.40, respectively). In the rs6002655 region, HaploReg detected a signal for NDUFA6-AS1, but not in whole blood, and did not identify any eQTLs for RANGAP1 or RP4-756G23.5. These discrepancies suggest that the comprehensive use of functional annotations and the Bayesian co-localization approach integrating eQTL data with GWAS summary statistics in INFERNO provides a more sensitive approach than the generic annotations in HaploReg.
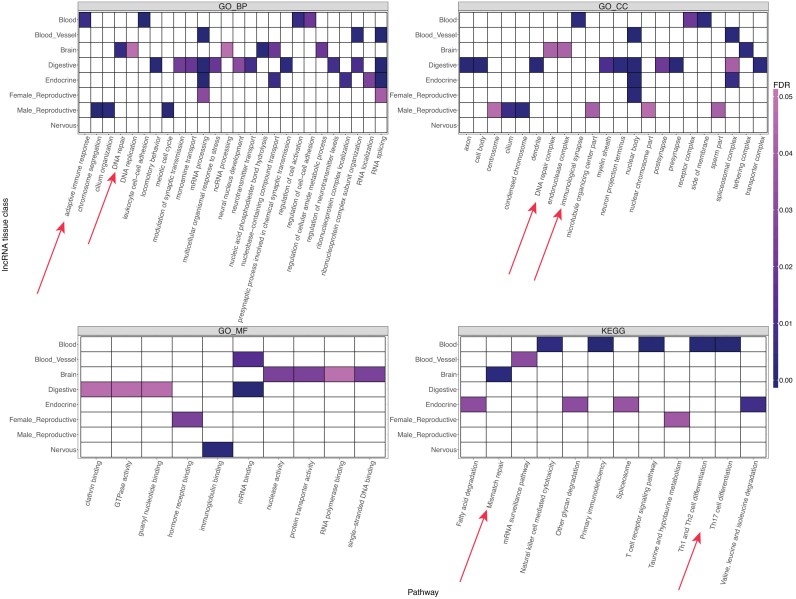
Next, we performed correlation-based target identification for the lncRNAs reported by INFERNO. INFERNO identified 5,893 unique genes co-regulated with 42 unique lncRNAs from 33 tissues and 15 tissue classes (Supplementary Figure S6). We first performed pathway analysis on all 5,893 genes across 4 functional annotation databases. This found significant enrichments in 107 pathways (Supplementary Table S6) including previously reported schizophrenia-related pathways such as RNA splicing (FDR = 2.04 × 10−11 (68)), phosphatidylinositol signaling (FDR = 0.0011 (69)), MAPK signaling pathway (FDR = 0.044) (70), Th1 and Th2 cell differentiation (FDR = 0.013 (71)), T cell receptor signaling (FDR = 0.013 (72)), spliceosome (FDR = 0.00045 (68)) and RNA transport (FDR = 0.023 (73)). One intriguing finding was the enrichment of the Herpes simplex infection (hsa05168, FDR = 0.025). Maternal Herpes simplex virus (HSV) infection may lead to increased risk of schizophrenia in their offspring (19) and HSV exposure may exacerbate cognitive function impairment in schizophrenic patients (74). We used PC-adjusted tissue-specific correlation to find putative tissue-specific lncRNA targets, which identified 2,938 unique genes targeted by 22 lncRNAs across 14 tissue categories, 1,438 of which were also identified by the cross-tissue approach. Tissue-specific pathway analysis of these 2,938 genes identified 84 enriched pathways across eight tissue categories (Figure 4, Supplementary Table S6). In brain, there were several enrichments for DNA repair-related pathways (75), and in blood, several enrichments were observed for immune-related pathways.
Figure 4.
Pathway enrichments for tissue-specific lncRNA targets in schizophrenia. Results are split by the tissue category of the lncRNA eQTL signal and pathway annotation. Red arrows denote brain and blood schizophrenia-related pathways discussed in the main text.
Application to inflammatory bowel disease GWAS
Due to the relative undersampling of functional datasets in the brain category for the schizophrenia analysis, we also applied INFERNO to an Inflammatory Bowel Disease (IBD) GWAS in Europeans (n = 12,882 cases and 21,770 controls) (20) with 60 genome-wide significant loci (Materials and Methods). Using the same parameters as in the schizophrenia analysis, INFERNO identified 2,649 potentially causal variants. Only 56 overlapped with mRNA exons. 1,638 (61%) variants overlapped with a ChromHMM enhancer state, while 181 (6.8%) overlapped with FANTOM5 enhancers. 1,259 (47%) overlapped with TFBSs for 58 unique TFs, for a total of 2,925 variant-TFBS overlaps, with the majority (2,449) of these lowering the predicted binding strength (Supplementary Figure S7A–D).
LD-collapsed sampling found significant enrichments of enhancer overlaps in eight FANTOM5 tissue categories, 16 Roadmap tissue categories, and four categories for concordant FANTOM5+Roadmap overlap (Supplementary Figure S7E). Several of these enrichments are as expected from an immune-related trait such as IBD, including the blood category (which includes all the T- and B-cell line datasets and was enriched for FANTOM5, Roadmap, and concordant overlaps), the immune organ category (which was enriched for Roadmap overlap), and the digestive category (which was enriched for all three types of overlaps).
Next, 85,609 co-localization tests were performed for 2,408 genes across the 60 tag regions, identifying 647 unique tissue-target gene co-localized eQTL signals spanning all 44 GTEx tissues and 202 unique genes (including 40 lncRNAs in 34 tissues) that were strongly co-localized with IBD GWAS signals (Supplementary Figure S8, Supplementary Table S7). Pathway analysis returned the IBD KEGG pathway (FDR = 0.0005) and found additional enrichments in leukocyte differentiation (FDR = 0.0098), leukocyte cell-cell adhesion (FDR = 0.0098), and the vacuolar cellular component (FDR = 0.0096), supporting their relevance to the trait.
Co-regulatory network analysis found 4,750 unique cross-tissue genes for 34 out of 40 lncRNAs from the co-localization analysis. These genes were enriched for 140 pathways including dozens of immune-related pathways such as the adaptive immune response (FDR = 0), leukocyte-mediated immunity (FDR = 0), and B cell activation (FDR = 6.93 × 10−11), and the IBD KEGG pathway as well (FDR = 9.303 × 10−7) (Supplementary Table S8). The enrichment of leukocyte-related pathways in both the COLOC genes and lncRNA targets supports their relevance to IBD, which is characterized by infiltration of circulating leukocytes into inflamed intestinal mucosa (76). The tissue-specific co-expression approach identified 959 unique genes targeted by 21 lncRNAs across 10 tissue categories, 344 of which were also found in the cross-tissue approach. Tissue-specific pathway analysis identified 58 enriched pathways across four tissue categories (Supplementary Table S8), including immunological synapse (FDR = 0.0089) and the Ras signaling pathway (FDR = 0.0462) in blood.
Web server and tool availability
We provide a web server (http://inferno.lisanwanglab.org) for INFERNO that accepts the top variants from any given GWAS, expands them by LD, and performs the annotation overlap analysis including directly overlapping variants with GTEx eQTL data. To run the computationally intensive enhancer sampling, eQTL co-localization, lncRNA correlation, and pathway enrichment analyses, INFERNO is also available as an open source pipeline (https://bitbucket.org/wanglab-upenn/INFERNO). INFERNO includes a master script that can be customized by the user to run any or all of the individual analysis steps (Materials and Methods).
DISCUSSION
INFERNO provides a sensitive and comprehensive hypothesis generation method for identifying functional genetic variants underlying genetic association signals and characterizing their tissue-specific effects on regulatory elements, target genes, and downstream biological processes. Analysis of the schizophrenia and IBD datasets demonstrated that INFERNO picks up many signals that converge to common tissue contexts and pathways when sufficient genetic loci are available. These two disease applications show that INFERNO can identify putatively causal variants, affected tissue contexts, regulatory elements, and target genes relevant to any type of GWAS trait, and that the lncRNA target identification can identify both cross-tissue and tissue-specific biologically relevant genes and pathways downstream of lncRNA perturbations by genetic variants. However, while the diversity of functional genomic data and tissue contexts analyzed by INFERNO allows it to characterize the potential mechanisms underlying GWAS association signals, this broad range of data sources also means that our algorithm may pick up more general regulatory mechanisms not directly related to the phenotype of interest, and these ‘hitchhikers’ could obfuscate the truly causal processes. This nonspecificity is likely to be a characteristic of complex trait genetics in general and could be a reason why most complex trait variants have very small effect sizes. Another factor that affects the specificity of INFERNO results is the currently limited availability of functional genomics annotation data, which are measured in normal tissues that do not necessarily reflect the disease state for a given GWAS signal and may not be exact matches for the relevant tissue context for a given trait. Thus, INFERNO is best used to prioritize biological processes and tissue contexts in an unbiased and systematic fashion for functional follow-up studies to prove the causality of the prioritized signals and their relevance to the phenotype of interest.
INFERNO improves on existing noncoding annotation methods for GWAS signals, the most comparable of which is HaploReg (15). HaploReg expands GWAS variants by LD structure only, missing many of the candidate variants INFERNO can identify using summary statistic-based expansion, and reports direct annotation overlaps with Roadmap but not FANTOM5 enhancer annotations. Additionally, it lacks a tissue classification framework to integrate information across disparate annotation sources. HaploReg provides an enhancer enrichment score by calculating the background frequencies of enhancer overlap in each cell type for all unique GWAS loci and all 1000 Genomes common variants and comparing these frequencies to those for a query list of variants using a binomial test. This approach ignores LD structure and does not match variants by any characteristics. INFERNO provides a more sensitive statistical method for quantifying the tissue-specific significance of annotation overlaps in a GWAS signal accounting for LD structure and other genomic characteristics. Furthermore, INFERNO allows for the calculation of enrichments both within and across tag regions, and the tissue classification approach enables the scoring of enrichments supported by disparate data sources. INFERNO also performs a more sensitive eQTL analysis by applying a Bayesian model to identify truly co-localized signals between GWAS and eQTL data, and additionally performs co-regulatory network identification for lncRNAs identified by this algorithm.
Several other approaches for characterizing noncoding genetic variants have been proposed such as RegulomeDB, GWAVA, EIGEN, LINSIGHT and CADD (10,11,13,77,78). These methods are designed for different purposes than INFERNO, as they generate phenotype-agnostic scores to prioritize regulatory variants without any information about specific regulatory mechanisms, tissue context, or target genes. INFERNO has at least two fundamental differences from these methods. First, INFERNO uses a tissue categorization framework to characterize the relevant tissue contexts of regulatory elements and target genes affected by noncoding variants comprehensively and unbiasedly. Second, rather than assigning a single score for each variant generically, INFERNO is designed to incorporate GWAS signals with functional genomics data so that the results are more specific to a phenotype of interest.
Application of INFERNO to schizophrenia and IBD GWAS data identified significant overlaps of enhancers in relevant tissue categories to each trait and eQTL signals from the same categories targeting disease-related genes. The lack of schizophrenia enrichment in the brain category may reflect the cell type- as opposed to brain region-specificity of schizophrenia genetic predisposition, as a range of regions are included in the category, each of which are generated from homogenized samples containing a mix of cell types. INFERNO also identified both cross-tissue and tissue-specific lncRNA signals targeting several biological processes known to be related to each phenotype including the MAPK signaling pathway, spliceosome, and Herpes simplex infection for schizophrenia and adaptive immunity and leukocyte differentiation for IBD. The lack of overlap with differentially expressed schizophrenia genes may be due to the fact that genetic perturbations of regulatory mechanisms may not manifest as differentially expressed genes between cases and controls. The wide range of tissue contexts and biological processes identified by INFERNO reflects the complexity of these polygenic and complex traits, although the IBD enrichments were more specific to immune-related tissues than the heterogeneous schizophrenia enrichments. The prior literature supporting these regulatory signals supports the utility and sensitivity of INFERNO for interpreting GWAS results and guiding post-GWAS followup studies.
INFERNO is limited by the amount of currently available functional genomics datasets, as certain tissue categories are overrepresented relative to others (Supplementary Figures S1 and S2). The statistical power of INFERNO will be further improved as functional datasets are generated in more tissues and cell types and as genomics technology continues to be refined and improved. Other co-localization approaches that account for different causality models but are more computationally intensive have been proposed such as eCAVIAR (79), and their incorporation into INFERNO will also improve its power to characterize noncoding association signals. Another future direction is to extend INFERNO to identify structural variation and copy number variants that contribute to traits through expression regulation.
DATA AVAILABILITY
The processed datasets supporting the conclusions of this article are available in the full INFERNO annotation file available at http://inferno.lisanwanglab.org/full_INFERNO_annotations.tar.gz. The web server is available at http://inferno.lisanwanglab.org/. The INFERNO software is open source and available at https://bitbucket.org/wanglab-upenn/INFERNO/. The INFERNO pipeline runs on Linux, is implemented using bash, Python 2.7 and R, and runs either on LSF-based cluster computing systems or linearly on a Linux machine. Further software versions, specific annotation sources and pre-processing scripts, and package requirements are documented in the Bitbucket repository and web server. INFERNO is freely available under the MIT license. The schizophrenia GWAS data are available from the Psychiatric Genomics Consortium, the IBD GWAS data are available from the International IBD Genetics Consortium.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge Chris Stoeckert for guidance on the tissue categorization, and Adam Naj and all members of the Wang lab for valuable feedback.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health, National Institute on Aging [U01-AG032984 to G.D.S., UF1-AG047133 to G.D.S., U54-AG052427 to G.D.S. and L.S.W., U24-AG041689 to L.S.W., R01-GM099962 to L.S.W., P30-AG010124, RF1-AG055477 to C.D.B., U54-NS100693 to G.D.S. and T32-AG00255 to A.A.W.]. Funding for open access charge: National Institutes of Health [U24-AG041689].
Conflict of interest statement. None declared.
REFERENCES
- 1. Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L. et al. . The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014; 42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al. . The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Evangelou E., Ioannidis J.P.A.. Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 2013; 14:379–389. [DOI] [PubMed] [Google Scholar]
- 4. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al. . Systematic localization of common Disease-Associated variation in regulatory DNA. Science. 2012; 337:1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corradin O., Scacheri P.C.. Enhancer variants: evaluating functions in common disease. Genome Med. 2014; 6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shlyueva D., Stampfel G., Stark A.. Transcriptional enhancers: from properties to genome-wide predictions. Nat. Rev. Genet. 2014; 15:272–286. [DOI] [PubMed] [Google Scholar]
- 7. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al. . An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Consortium R.E., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al. . Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bernstein B.E., Birney E., Dunham I., Green E.D., Gunter C., Snyder M.. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012; 489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S. et al. . Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ritchie G.R.S., Dunham I., Zeggini E., Flicek P.. Functional annotation of noncoding sequence variants. Nat. Methods. 2014; 11:294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lu Q., Hu Y., Sun J., Cheng Y., Cheung K.-H., Zhao H.. A statistical framework to predict functional Non-Coding regions in the human genome through integrated analysis of annotation data. Sci. Rep. 2015; 5:10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu Q., Powles R.L., Abdallah S., Ou D., Wang Q., Hu Y., Lu Y., Liu W., Li B., Mukherjee S. et al. . Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer's disease. PLoS Genet. 2017; 13:e1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2015; 44:gkv1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ardlie K.G., Deluca D.S., Segre A. V., Sullivan T.J., Young T.R., Gelfand E.T., Trowbridge C.A., Maller J.B., Tukiainen T., Lek M. et al. . The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015; 348:648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V.. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLos Genet. 2014; 10:e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ripke S., Neale B.M., Corvin A., Walters J.T.R., Farh K.-H., Holmans P.A., Lee P., Bulik-Sullivan B., Collier D.A., Huang H. et al. . Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014; 511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yolken R. Viruses and schizophrenia: a focus on herpes simplex virus. Herpes. 2004; 11:83A–88A. [PubMed] [Google Scholar]
- 20. Liu J.Z., Van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T. et al. . Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat. Genet. 2015; 47:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J. et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007; 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Abecasis G.R., Bentley D.R., Chakravarti A., Clark A.G., Donnelly P., Eichler E.E. et al. . A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mungall C.J., Torniai C., Gkoutos G. V, Lewis S.E., Haendel M.A.. Uberon, an integrative multi-species anatomy ontology. Genome Biol. 2012; 13:R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bard J., Rhee S.Y., Ashburner M.. An ontology for cell types. Genome Biol. 2005; 6:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Benjamini Y., Hochberg Y.. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Statistical Methodol. 1995; 57:289–300. [Google Scholar]
- 27. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G. et al. . The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J., Vasaikar S., Shi Z., Greer M., Zhang B.. WebGestalt 2017: A more comprehensive, powerful, flexible and interactive gene set enrichment analysis toolkit. Nucleic Acids Res. 2017; 45:W130–W137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J., Duncan D., Shi Z., Zhang B.. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013; 41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B., Kirov S., Snoddy J.. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005; 33:W741–W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L. et al. . Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018; 9:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S. V, Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Nicolae D.L., Cox N.J. et al. . A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015; 47:1091–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wheeler H.E., Shah K.P., Brenner J., Garcia T., Aquino-Michaels K., Cox N.J., Nicolae D.L., Im H.K.. Survey of the heritability and sparse architecture of gene expression traits across human tissues. PLos Genet. 2016; 12:e1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M.. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015; 47:291–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K. et al. . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015; 47:1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim T.-K., Hemberg M., Gray J.M., Costa A.M., Bear D.M., Wu J., Harmin D. a, Laptewicz M., Barbara-Haley K., Kuersten S. et al. . Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010; 465:182–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zentner G.E., Scacheri P.C.. The chromatin fingerprint of gene enhancer elements. J. Biol. Chem. 2012; 287:30888–30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ernst J., Kheradpour P., Mikkelsen T.S., Shoresh N., Ward L.D., Epstein C.B., Zhang X., Wang L., Issner R., Coyne M. et al. . Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011; 473:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple Combinations of Lineage-Determining transcription factors prime cis-Regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J.. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015; 31:2601–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Engreitz J.M., Ollikainen N., Guttman M.. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016; 17:756–770. [DOI] [PubMed] [Google Scholar]
- 44. Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., Huarte M., Zuk O., Carey B.W., Cassady J.P. et al. . Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009; 458:223–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Greene C., Kealy J., Humphries M.M., Gong Y., Hou J., Hudson N., Cassidy L.M., Martiniano R., Shashi V., Hooper S.R. et al. . Dose-dependent expression of claudin-5 is a modifying factor in schizophrenia. Mol. Psychiatry. 2017; doi:10.1038/mp.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Santos N.C., Costa P., Ruano D., MacEdo A., Soares M.J., Valente J., Pereira A.T., Azevedo M.H., Palha J.A.. Revisiting thyroid hormones in schizophrenia. J. Thyroid Res. 2012; 2012:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gogos A., Sbisa A.M., Sun J., Gibbons A., Udawela M., Dean B.. A role for estrogen in Schizophrenia: clinical and preclinical findings. Int. J. Endocrinol. 2015; 2015:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ringen P.A., Engh J.A., Birkenaes A.B., Dieset I., Andreassen O.A.. Increased mortality in schizophrenia due to cardiovascular disease - a non-systematic review of epidemiology, possible causes and interventions. Front. Psychiatry. 2014; 5:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khandaker G.M., Cousins L., Deakin J., Lennox B.R., Yolken R., Jones P.B.. Inflammation and immunity in schizophrenia: Implications for pathophysiology and treatment. Lancet Psychiatry. 2015; 2:258–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sekar A., Bialas A.R., de Rivera H., Davis A., Hammond T.R., Kamitaki N., Tooley K., Presumey J., Baum M., Van Doren V. et al. . Schizophrenia risk from complex variation of complement component 4. Nature. 2016; 530:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Severance E.G., Prandovszky E., Castiglione J., Yolken R.H.. Gastroenterology issues in Schizophrenia: why the gut matters. Curr. Psychiatry Rep. 2015; 17:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iannitelli A., Quartini A., Tirassa P., Bersani G.. Schizophrenia and neurogenesis: a stem cell approach. Neurosci. Biobehav. Rev. 2017; 80:414–442. [DOI] [PubMed] [Google Scholar]
- 53. Reif A., Fritzen S., Finger M., Strobel A., Lauer M., Schmitt A., Lesch K.P.. Neural stem cell proliferation is decreased in schizophrenia, but not in depression. Mol. Psychiatry. 2006; 11:514–522. [DOI] [PubMed] [Google Scholar]
- 54. Zuber V., Jönsson E.G., Frei O., Witoelar A., Thompson W.K., Schork A.J., Bettella F., Wang Y., Djurovic S., Smeland O.B. et al. . Identification of shared genetic variants between schizophrenia and lung cancer. Sci. Rep. 2018; 8:674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fatemi S.H., Folsom T.D., Rooney R.J., Mori S., Kornfield T.E., Reutiman T.J., Kneeland R.E., Liesch S.B., Hua K., Hsu J. et al. . The viral theory of schizophrenia revisited: Abnormal placental gene expression and structural changes with lack of evidence for H1N1 viral presence in placentae of infected mice or brains of exposed offspring. Neuropharmacology. 2012; 62:1290–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deicken R.F., Zhou L., Schuff N., Fein G., Weiner M.W.. Hippocampal neuronal dysfunction in schizophrenia as measured by proton magnetic resonance spectroscopy. Biol. Psychiatry. 1998; 43:483–488. [DOI] [PubMed] [Google Scholar]
- 57. Chang X., Liu Y., Hahn C.-G., Gur R.E., Sleiman P.M.A., Hakonarson H.. RNA-seq analysis of amygdala tissue reveals characteristic expression profiles in schizophrenia. Transl. Psychiatry. 2017; 7:e1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fillman S.G., Cloonan N., Catts V.S., Miller L.C., Wong J., Mccrossin T., Cairns M., Weickert C.S.. Increased inflammatory markers identified in the dorsolateral prefrontal cortex of individuals with schizophrenia. Mol. Psychiatry. 2013; 18:206–214. [DOI] [PubMed] [Google Scholar]
- 59. Hwang Y., Kim J., Shin J.Y., Kim J.I.I., Seo J.S., Webster M.J., Lee D., Kim S.. Gene expression profiling by mRNA sequencing reveals increased expression of immune/inflammation-related genes in the hippocampus of individuals with schizophrenia. Transl. Psychiatry. 2013; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kohen R., Dobra A., Tracy J.H., Haugen E.. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl. Psychiatry. 2014; 4:e366–e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sainz J., Mata I., Barrera J., Perez-Iglesias R., Varela I., Arranz M.J., Rodriguez M.C., Crespo-Facorro B.. Inflammatory and immune response genes have significantly altered expression in schizophrenia. Mol. Psychiatry. 2013; 18:1056–1057. [DOI] [PubMed] [Google Scholar]
- 62. Wu J.Q., Wang X., Beveridge N.J., Tooney P.A., Scott R.J., Carr V.J., Cairns M.J.. Transcriptome sequencing revealed significant alteration of cortical promoter usage and splicing in schizophrenia. PLoS One. 2012; 7:e36351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhao Z., Xu J., Chen J., Kim S., Reimers M., Bacanu S.A., Yu H., Liu C., Sun J., Wang Q. et al. . Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. Mol. Psychiatry. 2015; 20:563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Finn R.D., Attwood T.K., Babbitt P.C., Bateman A., Bork P., Bridge A.J., Chang H.Y., Dosztanyi Z., El-Gebali S., Fraser M. et al. . InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017; 45:D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Haft C.R., Sierra M.D.L.L., Bafford R., Lesniak M.A., Barr V.A., Taylor S.I.. Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: Assembly into multimeric complexes. Mol. Biol. Cell. 2000; 11:4105–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Dai X., Dong M., Yu H., Xie Y., Yu Y., Cao Y., Kong Z., Zhou B., Xu Y., Yang T. et al. . Knockdown of TCTN1 strongly decreases growth of human colon cancer cells. Med. Sci. Monit. 2017; 23:452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Knable M.B., Weinberger D.R.. Dopamine, the prefrontal cortex and schizophrenia. J. Psychopharmacol. 1997; 11:123–131. [DOI] [PubMed] [Google Scholar]
- 68. Chabot B., Shkreta L.. Defective control of pre-messenger RNA splicing in human disease. J. Cell Biol. 2016; 212:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yao J.K., Yasaei P., van Kammen D.P.. Increased turnover of platelet phosphatidylinositol in schizophrenia. Prostaglandins, Leukot. Essent. Fat. Acids. 1992; 46:39–46. [DOI] [PubMed] [Google Scholar]
- 70. Funk A.J., McCullumsmith R.E., Haroutunian V., Meador-Woodruff J.H.. Abnormal activity of the MAPK- and cAMP-associated signaling pathways in frontal cortical areas in postmortem Brain in Schizophrenia. Neuropsychopharmacology. 2012; 37:896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Avgustin B., Wraber B., Tavcar R.. Increased Th1 and Th2 immune reactivity with relative Th2 dominance in patients with acute exacerbation of schizophrenia. Croat. Med. J. 2005; 46:268–274. [PubMed] [Google Scholar]
- 72. Craddock R.M., Lockstone H.E., Rider D.A., Wayland M.T., Harris L.J.W., McKenna P.J., Bahn S.. Altered T-cell function in schizophrenia: a cellular model to investigate molecular disease mechanisms. PLoS One. 2007; 2:e692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tsuboi D., Kuroda K., Tanaka M., Namba T., Iizuka Y., Taya S., Shinoda T., Hikita T., Muraoka S., Iizuka M. et al. . Disrupted-in-schizophrenia 1 regulates transport of ITPR1 mRNA for synaptic plasticity. Nat. Neurosci. 2015; 18:698–707. [DOI] [PubMed] [Google Scholar]
- 74. Prasad K.M., Watson A.M.M., Dickerson F.B., Yolken R.H., Nimgaonkar V.L.. Exposure to herpes simplex virus type 1 and cognitive impairments in individuals with schizophrenia. Schizophr. Bull. 2012; 38:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Markkanen E., Meyer U., Dianov G.L.. Dna damage and repair in schizophrenia and autism: implications for cancer comorbidity and beyond. Int. J. Mol. Sci. 2016; 17:E856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arseneau K.O., Cominelli F.. Targeting leukocyte trafficking for the treatment of inflammatory bowel disease. Clin. Pharmacol. Ther. 2015; 97:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huang Y.F., Gulko B., Siepel A.. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017; 49:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D.. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016; 48:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hormozdiari F., van de Bunt M., Segrè A. V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E.. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 2016; 99:1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The processed datasets supporting the conclusions of this article are available in the full INFERNO annotation file available at http://inferno.lisanwanglab.org/full_INFERNO_annotations.tar.gz. The web server is available at http://inferno.lisanwanglab.org/. The INFERNO software is open source and available at https://bitbucket.org/wanglab-upenn/INFERNO/. The INFERNO pipeline runs on Linux, is implemented using bash, Python 2.7 and R, and runs either on LSF-based cluster computing systems or linearly on a Linux machine. Further software versions, specific annotation sources and pre-processing scripts, and package requirements are documented in the Bitbucket repository and web server. INFERNO is freely available under the MIT license. The schizophrenia GWAS data are available from the Psychiatric Genomics Consortium, the IBD GWAS data are available from the International IBD Genetics Consortium.