Abstract
Trypanosoma brucei live in mammals as bloodstream forms and in the Tsetse midgut as procyclic forms. Differentiation from one form to the other proceeds via a growth-arrested stumpy form with low messenger RNA (mRNA) content and translation. The parasites have six eIF4Es and five eIF4Gs. EIF4E1 pairs with the mRNA-binding protein 4EIP but not with any EIF4G. EIF4E1 and 4EIP each inhibit expression when tethered to a reporter mRNA, but while tethered EIF4E1 suppresses only when 4EIP is present, suppression by tethered 4EIP does not require the interaction with EIF4E1. In growing bloodstream forms, 4EIP is preferentially associated with unstable mRNAs. Bloodstream- or procyclic-form trypanosomes lacking 4EIP have only a marginal growth disadvantage. Bloodstream forms without 4EIP are, however, defective in translation suppression during stumpy-form differentiation and cannot subsequently convert to growing procyclic forms. Intriguingly, the differentiation defect can be complemented by a truncated 4EIP that does not interact with EIF4E1. In contrast, bloodstream forms lacking EIF4E1 have a growth defect, stumpy formation seems normal, but they appear unable to grow as procyclic forms. We suggest that 4EIP and EIF4E1 fine-tune mRNA levels in growing cells, and that 4EIP contributes to translation suppression during differentiation to the stumpy form.
INTRODUCTION
The amounts of protein synthesized from a messenger RNA (mRNA) are determined by the rates of translation initiation (1,2) and elongation (3,4). Most eukaryotic translation is initiated by binding of eIF4E to the cap structure. eIF4E recruits eIF4G, which in turn recruits the helicase eIF4A and, via other initiation factors, the 43S complex. The 43S complex, which includes the 40S subunit with charged methionyl transfer RNA as well as various additional translation factors, then scans toward the start codon (5). Most eukaryotic species examined have more than one eIF4E homolog (6). The roles of different eIF4Es have been most extensively investigated in metazoa; in general, there are one or two eIF4Es that are responsible for constitutive cap-dependent translation and the remainder have specialized roles at particular developmental stages or are involved in processes other than translation (6).
In animal cells, one mechanism for the regulation of translation initiation involves 4E-binding proteins (4E-BPs) (7). The 4E-BP–eIF4E interaction is partially mediated by a canonical binding motif, YXXXXLØ, which is found near 4E-BP N-termini (8). Since eIF4G binds eIF4E using the same motif, 4E-BP and eIF4G binding is mutually exclusive. 4E-BP–eIF4E binding is however strengthened by additional interactions (9–11).
The ‘tethering’ assay is a method that measures the effects of attachment of a protein to a reporter mRNA. The protein of interest is expressed as a fusion with a protein or peptide that has the ability to bind a short RNA sequence with high affinity and specificity, such as the lambdaN peptide. Activity is monitored using a reporter mRNA that includes the cognate RNA sequence—for the lambdaN peptide, boxB. Tethering of various 4E-BPs results in translation repression but curiously, this does not require the YXXXXLØ motif. For example, the abilities of tethered 4E-T or Drosophila Cup to suppress a target reporter mRNA does not depend on YXXXXLØ (12,13). In both cases, the authors suggested that the tethered protein was able to recruit the CAF1–NOT deadenylase complex (12,13). In contrast to the tethering results, exogenous expression of mammalian 4E-T resulted in indiscriminate translational suppression, and this was dependent on the YXXXXLØ motif (13), suggesting recruitment of 4E-T via eIF4E. It is not known whether tethered 4E-BPs without YXXXXLØ can bind to eIF4E in vivo using their additional binding surface (9–11), or whether they act independently of eIF4E.
Another mechanism by which translation can be inhibited in animals and plants depends on eIF4E-like cap-binding proteins that cannot recruit eIF4G. These include mammalian 4EHP/eIF4E2 (14), Drosophila d4EHP/EIF4E-8 (15,16) and Arabidopsis nCBP (17). Mammalian and Drosophila 4EHPs are required for correct translation control during development (18–21). Interactions with partner proteins such as Bicoid (16) or GIGYF2 ( = GIF2) (22–24), and with the RNA-induced silencing complex (25,26), have been implicated in induction of mRNA decay and/or translation suppression by 4EHPs. GIGYF2 appears to be able to act both in conjunction with (24), and independently of, 4EHP (27), and GIGYF2 interactions with both Ago2 (28) and the CAF1–NOT complex (27) have been documented.
Trypanosoma brucei is a unicellular eukaryotic parasite. The bloodstream form lives extracellularly in the blood and tissue fluids of mammals, evading the immune response by antigenic variation of a variant surface glycoprotein coat (29). The parasites multiply as long slender bloodstream forms, which depend on glycolysis for their energy metabolism (30). When parasitemia reaches a critical level, non-dividing ‘stumpy forms’ start to dominate the population (31,32). Although these cells have very low overall mRNA and translation levels (33,34), they do express a variety of new proteins, including the surface protein PAD1 (35) and some proteins of mitochondrial energy metabolism (31). Stumpy forms are pre-adapted for development in the definitive host, the Tsetse fly, which becomes infected by blood feeding. Within the Tsetse midgut the parasites differentiate to procyclic forms, which have EP and GPEET procyclins on their surface (36) and a mitochondrial energy metabolism that depends on amino acid substrates (37–39). After a few weeks, with additional differentiation steps, mammal-infective parasites reappear in the salivary glands and can be transmitted at the next blood meal (40).
Cellular differentiation requires substantial changes in gene expression, which usually depend on regulatory transcription factors. Trypanosomes and related organisms, however, lack transcriptional control of individual genes. Instead, transcription is polycistronic: individual mRNAs are excised by processing (41). The 5′-end of each mRNA is formed by trans splicing of a 39-nt leader sequence (42), which bears a highly methylated cap called cap4 (43,44). The 3′-end of each mRNA is, as usual, created by cleavage and polyadenylation (45,46). As a consequence of this arrangement, regulation of gene expression in trypanosomes is almost exclusively post-transcriptional: there are big differences between mRNAs in their processing efficiencies, decay rates and translation (47–49). Various RNA-binding proteins have been shown to regulate these steps in specific trypanosome mRNA subsets (50–52). With one exception, however (53), the molecular mechanisms by which these proteins act are not yet known.
This paper concerns control of translation in T. brucei. Trypanosoma brucei and the closely related parasite Leishmania have six eIF4Es and five eIF4Gs (54). Results from studies of protein–protein interactions, migration in sucrose gradients, intracellular location and the effects of RNAi in T. brucei suggest that the eIF4E4–eIF4G3 and EIF4E3–EIF4G4 complexes are implicated in general translation initiation (54). EIF4E3 and EIF4E4 are present in a slight molar excess relative to mRNA (55,56). RNA interference (RNAi) experiments showed that EIF4E3 is essential in both forms whereas for EIF4E4, a growth defect was seen only in bloodstream forms (57). The functions of T. brucei EIF4E5–EIF4G2, EIF4E5–EIF4G1 (58) and EIF4E6–EIF4G5 (59) complexes, and of eIF4E2 (60), are not yet known.
EIF4E1 has been studied most extensively in Leishmania major. It binds cap4 and m7GTP with a relatively low affinity which is, however, only three times worse than that of Leishmania EIF4E4 (61,62). Leishmania major EIF4E1 does not interact with any of the five eIF4G homologs, either by pull-down or by yeast two-hybrid analysis (63). In contrast, it does interact with another protein, 4EIP (63) and also directly with EIF3A (64). The latter interaction could result in EIF4G-independent translation initiation. Because of life-cycle-dependent differences in both the EIF4E1–4EIP interaction, and in expression of other EIF4Es, it was suggested the EIF4E1 might act as an unconventional initiation factor in the intracellular amastigote life-cycle stage. Further, it was hypothesized that the initiation activity of EIF4E1 is suppressed, in the promastigote stage, by interaction with 4EIP (63). The results of yeast two-hybrid assays indicated that a YXXXXLØ motif at the N-terminus of Leishmania 4EIP was required for interaction with EIF4E1 (63). The first 52 residues of 4EIP form two α-helices that interact with EIF4E1; the consensus motif is in the N-terminal helix (65). In vitro evidence suggested that binding of m7GTP by EIF4E1 was inhibited by addition of the N-terminal 4EIP fragment, probably as a consequence of a conformational change within the cap-binding pocket (65).
Trypanosoma brucei EIF4E1 (Tb927.11.2260) is found in the nucleus and cytosol (55). Since there are three times fewer EIF4E1 molecules per cell than there are mRNAs (55,66), it cannot be a major general translation initiation factor. Depletion of T. brucei eIF4E1 by RNAi halted growth of bloodstream forms and slowed growth of procyclic forms (57). EIF4E1 and 4EIP (Tb927.9.11050) were found to be extremely strong repressors when tethered to reporter mRNAs (67,68). We therefore decided to investigate the function of 4EIP in T. brucei. We were particularly interested in knowing whether 4EIP is recruited to specific mRNAs via RNA-binding proteins, and also in knowing the role of 4EIP during differentiation.
MATERIALS AND METHODS
Trypanosomes and plasmids
All experiments apart from those concerning differentiation were done using Lister 427 strain bloodstream forms or procyclic forms. For differentiation experiments, the EATRO1125 strain was used. The EATRO1125 bloodstream forms were routinely grown in the presence of 1.1% methyl cellulose (69) to retain differentiation capability. Otherwise, transfection and growth conditions were as described (70,71). Plasmids and oligonucleotides are listed in Supplementary Table S4. All growth and differentiation experiments were done in the absence of selecting drugs, and with at least a 24-h prior pre-incubation without selection. Double knockout lines were routinely grown without blasticidin and puromycin.
EATRO1125 cells were grown at 37°C, 5% carbon dioxide, in HMI-9 media containing 1.1 % methylcellulose. To obtain high density EATRO1125 stumpy cells, 5 × 105 cells were allowed to grow to maximum density. Collection was as described in (69): cultures were diluted five times with phosphate buffered saline (PBS), filtered through 615 ¼ MN filters (Macherey-Nagel, Germany) and centrifuged at 1218 × g for 10 min. This collection procedure lasted up to 20 min.
Polysomes, RNA, RNASeq and northern blotting
Polysome analysis and northern blotting were done as described in (72). For RNA preparation from differentiating cells, ∼5 × 106 cells (0 h time point), ∼3 × 107 cells (36 h time point) and ∼5 × 107 cells (48 h time point) were collected. This yield at least 1.5 μg of RNA, but the RNA from the 36 and 48 h was degraded, so could not be evaluated.
To find mRNAs that were bound to 4EIP, lysates from cells inducibly expressing 4EIP-TAP (not irradiated) were bound to IgG beads and washed. RNA was released from the beads by protease treatment and purified. Input and unbound fractions were ribosomal RNA-depleted using oligonucleotides and RNaseH, all fractions were subjected to RNASeq (E-MTAB-6240), and the data were analyzed, as described previously (71,73,74).
Immunoprecipitation, western blotting, tethering and interactome analysis
Immunoprecipitation, yeast two-hybrid screening, tethering assays and tandem affinity purification were all done as described in (53). Western blots were detected as described in (71). Apart from commercial antibodies to myc and V5, which came from various sources, antibodies were to ribosomal protein S9 (this lab), PAD1 (Keith Matthews, University of Edinburgh) and trypanothione reductase (Luise Krauth-Siegel, Heidelberg University).
Pulse labeling
For each time point, ∼3 × 107 cells were collected, washed twice with PBS and re-suspended in 350 μl labeling medium (Dulbecco’s Modified Eagle Medium (gibco) lacking l-methionine). These were grown in the conditions mentioned above for 1 h before addition of 35S- methionine for 20 min. Thereafter the cells were collected by centrifugation at 3500 × g for 2 min, washed twice with PBS and re-suspended in Laemmli buffer.
RESULTS
4EIP sequence comparisons
To obtain preliminary indications concerning 4EIP functional conservation, we searched for similar sequences and found potential homologs in many kinetoplastid genomes, including that of the free-living Bodo caudatus. The N-terminal 4E interaction domain, YxxxxLØ, is found in all complete sequences (Figure 1A and Supplementary Figure S1). All but one of the hydrophobic residues in the second EIF4E1-interacting helix (65) are also conserved, suggesting that they are important for 4EIP function. A second, highly charged, conserved region (Figure 1A and Supplementary Figure S1) is separated from the EIF4E1 interaction domain by disordered linker. In B. caudatus, L. major, Trypanosoma theileri and Trypanosoma grayi, the linker includes polyglycine tracts (Supplementary Figure S1). The remainder of the protein is much less conserved, except that the C-termini are proline- and glutamine-rich, and highly basic. The last 183 residues of T. brucei 4EIP have 21% glutamine and 17% proline and a predicted pI of 10.8. Values for the equivalent regions from B. caudatus and Endotrypanum are, respectively, 39% and 21% glutamine, 11% and 20% proline, and pIs of 11.5 and 12.0. All of the 4EIPs listed in Figure 1 have polyglutamine repeats at the far C-terminus, with the exception of those from the salivarian trypanosomes (congolense, brucei and vivax). The complete sequences terminate with (Q)1-17RR, except in Bodo which has (Q)11R. There is no indication from the sequences of which part might be implicated in mRNA binding, although the C-termini may bind via charge alone. Trypanosoma brucei 4EIP is predicted by Phyre (75) to be predominantly disordered (Supplementary Figure S1, pages 2 and 3).
Figure 1.
Trypanosoma brucei 4EIP interacts with EIF4E1. (A) Conserved regions in kinetoplastid 4EIPs. Amino acids highlighted in black are identical in all aligned sequences. Residues highlighted in gray are partially conserved—either identical or chemically similar in a majority of sequences. Sequences are (in vertical order): Trypanosoma grayi DQ04_01981000; Trypanosoma theileri ORC92063.1; Trypanosoma cruzi TcCLB.508461.290; Trypanosoma congolense TcIL3000_9_4530 Tryanosoma brucei Tb927.9.11050; Trypanosoma vivax TvY486_0905070; Endotrypanum_monterogeii EMOLV88_350043200; Leishmania major LmjF.35.3980; Blechomonas_ayalai Baya_047_0070; Bodo caudatus CUG36708.1. (B) The photos show growth of yeast on plates with stringent selection for the interaction between bait and prey plasmids. The identities of the plasmids used are shown next to the streaked yeast. Full results with controls are shown in Supplementary Figure S1A. (C) Extracts were made from bloodstream-form trypanosomes with or without a V5- in situ tagged eIF4E1 and myc-tagged 4EIP. Anti-myc immunoprecipitates were subjected to sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting. In: input extract from 5 × 106 cells; U: unbound fraction from 5 × 106 cells; E: eluate from immunoprecipitating beads, from 1 × 108 cells. Upper panel: anti-myc pull-down, with cells lacking a myc-tagged protein as control. Lower panel: anti-V5 pull-down, with cells lacking V5-tagged protein as the control. (D) As in (C) but with anti-V5 as the precipitating antibody. (E) As in (C) but with procyclic-form trypanosomes. (F) Western blot showing relative amounts of V5-tagged 4EIP and EIF4E1 in bloodstream forms. The only band from 4EIP migrated at ∼37 kDa. The control is ribosomal protein S9.
T. brucei 4EIP interacts with eIF4E1
Leishmania major 4EIP was identified as a specific interaction partner of L. major EIF4E1 (63). In a two-hybrid assay, T. brucei 4EIP also, as expected, interacted with T. brucei EIF4E1, but not with EIF4E2, EIF4E3 or EIF4E4 (Figure 1B and Supplementary Figure S2A). To investigate the interaction inside trypanosomes, we made bloodstream-form T. brucei in which one EIF4E1 open reading frame was N-terminally V5-tagged (V5-4E1). In the same cells, we expressed 4EIP with a C-terminal myc tag (4EIP-myc) (Figure 2B). Since 4EIP-myc was expressed from an RNA polymerase I promoter, it was probably overexpressed relative to V5-4E1. Anti-myc beads pulled down V5-4E1 from extracts expressing 4EIP-myc, but not from extracts from cells that did not express myc-tagged protein (Figure 1C, upper panel). Similarly, the pull-down of 4EIP-myc by anti-V5 beads was dependent on co-expression of V5-4E1 (Figure 1D). These results confirmed the interaction in T. brucei.
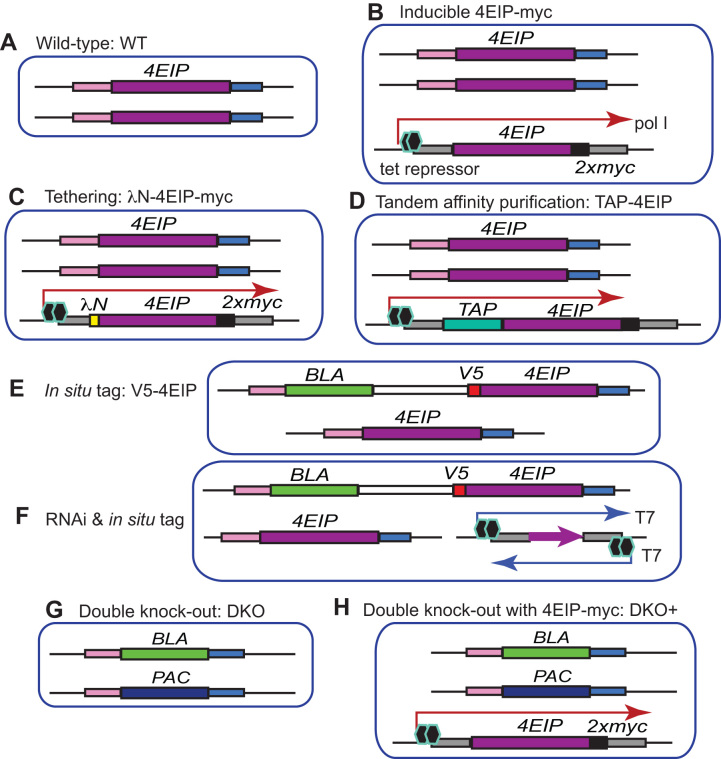
Figure 2.
Cell lines. Diagrams are not to scale. Each picture represents the relevant gene loci. The different DNA segments are labeled on the figure.
An 18-mer L. major peptide that included the YxxxxLØ motif was shown by nuclear magnetic resonance to interact with purified Leishmania EIF4E1 in vitro (63). We therefore tested whether the equivalent sequence in T. brucei 4EIP was required for its interaction with T. brucei eIF4E1. Indeed, 4EIP that lacked the first 13 amino acids failed to interact with EIF4E1 both by two-hybrid assay (Figure 1B and Supplementary Figure S2A, ΔN4EIP) and in vivo (Figure 1C). Similar results were obtained in procyclic-form trypanosomes: the EIF4E1–4EIP interaction was present and dependent on the N-terminal peptide (Figure 1E). It is however impossible to rule out the possibility that inside cells, N-terminally truncated but RNA-bound 4EIP is able to interact with cap-bound EIF4E1 in a more transient fashion via the second binding surface, with some stabilization from the linking RNA.
To assess the relative amounts of EIF4E1 and 4EIP in bloodstream forms, we compared the signals of the V5-tagged versions. The predominant band from V5-4EIP migrated at ∼37 kDa (Figure 1F), although the predicted molecular weight of the tagged protein is 56 kDa. Proteolysis of V5-4EIP might have happened during centrifugation and cell lysis, but since the cells were boiled in sample buffer immediately after harvest, we suspect that the protein is subject to cleavage in vivo, with loss of ∼200 residues from the C-terminus. Although the N-terminal V5 tag might affect protein turnover, the signal intensities suggest that EIF4E1 and 4EIP (or its N-terminal fragment) are present at roughly similar levels in bloodstream forms.
Suppression of expression by 4EIP does not require the N-terminal EIF4E1-interacting motif or the conserved acidic domain
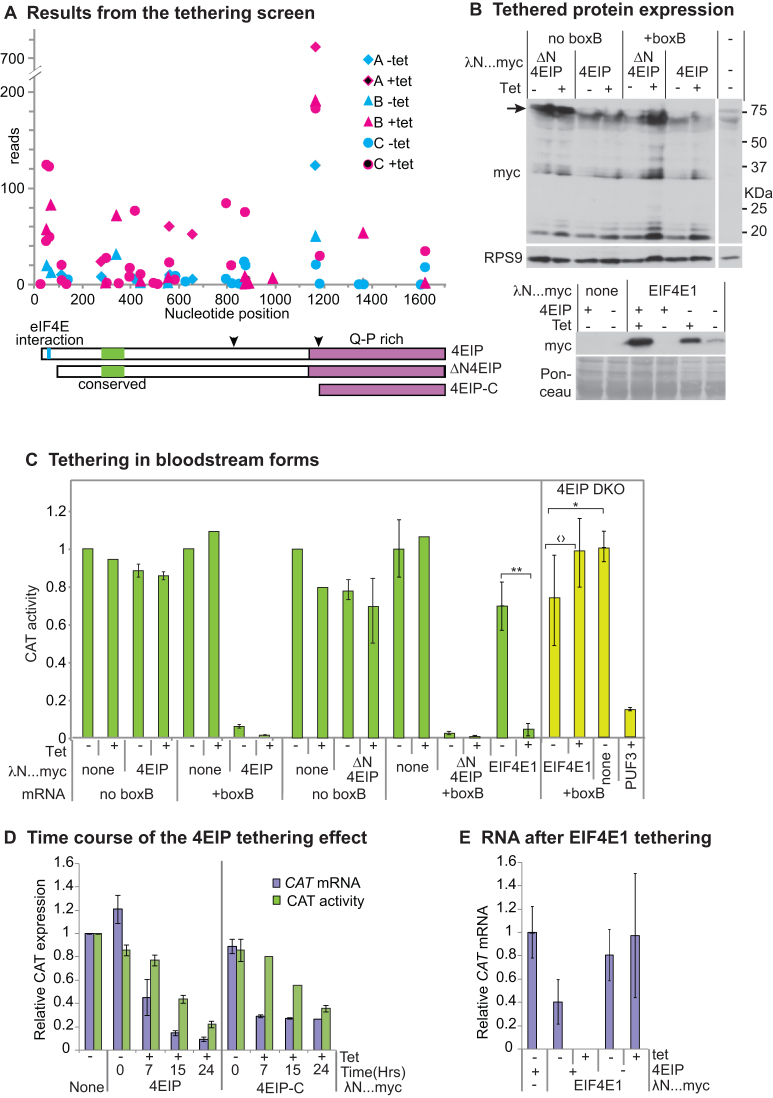
We next asked which parts of 4EIP are required to suppress expression in the tethering assay. First, we scrutinized existing data from our previous high-throughput screen. In that screen, we expressed a library of protein fragments bearing the lambdaN peptide in cells expressing a boxB reporter mRNA that encoded a toxic enzyme (67). Results for 4EIP from the screen are plotted in Figure 3A. Briefly, each symbol marks the position of the N-terminus of a lambdaN fusion. If the fusion protein suppressed expression, the magenta symbols show higher read counts than the corresponding cyan symbols. The results (67) suggested that protein fragments including the C-terminal 150 residues of 4EIP were sufficient to suppress reporter expression. This region has 20% glutamine and 18% proline, with a predicted pI of 10.4.
Figure 3.
Tethering of 4EIP to an mRNA suppresses expression. (A) Results for 4EIP from the published tethering screen (67). The trypanosomes used inducibly expressed an mRNA encoding the glycolytic enzyme PGKB, which is toxic in bloodstream forms (86). They were transfected with a library for inducible expression of lambdaN fusion proteins created from randomly sheared trypanosome DNA. After induction of both PGKB-boxB and the lamdaN fusions, cells were grown and the polymerase chain reaction products from the integrated lambdaN fusion proteins were sequenced (67). Results from three independent selections (A, B and C) are shown using different symbols. Counts that are higher in the presence of the inducer, tetracycline (magenta symbols), than without tetracycline (cyan symbols) indicate suppression of PGKB expression. Cartoons of 4EIP showing the two conserved regions (cyan and green) and the PQ-rich C-terminal region are below the x-axis. The two downward arrowheads indicate the cleavage positions that would generate the tagged proteins that were observed by western blotting in panel (B) and Figure 1F. (B) Expression of lambdaN…myc fusion proteins for the experiments in panels (C) and (E). The full-length lambdaN-4EIP (upper panel, arrow) was always found to be degraded; the two C-terminal pieces (30 and 20 kDa) will lack the lambda-N portion, so will be inactive in the assay. (C) Tethering assay using CAT reporter. Bloodstream-form trypanosomes expressing a CAT mRNA with five copies of boxB in the 3′-untranslated region (3′-UTR) were used; cells in which the CAT mRNA lacked boxB served as control. In the left-hand panel, cells were transfected with plasmids designed for tetracycline-inducible expression of either LambdaN-4EIP-myc or LambdaN-4EIP-myc with an N-terminal deletion that eliminated the interaction with eIF4E1 (ΔN4EIP). CAT activity was measured in the presence and absence of tetracycline. Results are expressed as arithmetic mean ± standard deviation of at least three measurements. Right-hand panel (with yellow bars in online version) — cells lacking 4EIP were used to test the activity of tethered EIF4E1, with PUF3 as a control. For the EIF4E1, tests were done six times. Results of Student’s t-tests were *P = 0.04; 〈?P = 0.07; **P = 0.009. Expression of myc-tagged proteins is shown on the right. (D) Time course of the tethering effect in bloodstream forms, using either 4EIP or its C-terminal domain. Results are mean and standard deviation of three measurements. (E) Effect on the CAT reporter mRNA of tethering EIF4E1 in the presence or absence of 4EIP. Results are mean and standard deviation of three measurements. In the paired bars, mRNA measurements are on the left and CAT activity on the right.
To confirm the screening results, we used bloodstream forms that contained a constitutively transcribed CAT-boxB mRNA. We first expressed full-length lambdaN-4EIP-myc from a tetracycline-inducible promoter (Figures 2C and 3B). Even in the absence of tetracycline, CAT mRNA was not detectable and chloramphenicol acetyltransferase (CAT) activity was suppressed by ∼95% (Figure 3C). As previously seen for N-terminally V5-tagged 4EIP, lambdaN-4EIP-myc appeared to be susceptible to degradation or processing. Although myc-tagged protein migrating at the expected size (∼60 kDa) was sometimes detectable, bands at ∼32 and 17 kDa were more reliably detected (Figure 3A and B). Expression of lambdaN-4EIP-myc was readily detectable in the absence of tetracycline, which explains why CAT-boxB expression was also suppressed without tetracycline (Figure 3B). The same result was obtained upon tethering of ΔN4EIP (Figure 3B). For both proteins, expression of a CAT mRNA without boxB was unaffected (Figure 3C). Thus tethered 4EIP acts independently of the EIF4E1 interaction motif, as was previously seen in Opisthokonts.
We then, using new lines, examined the time course of the tethering effect, comparing the full-length protein with the C-terminal 150 residues (Figure 3A). The kinetics of RNA decrease was similar for the full-length protein and for the C-terminal domain (Figure 3D). CAT activity declined more slowly than RNA as expected, because we are here also observing degradation of CAT protein that was synthesized prior to tetracycline addition (Figure 3D). These results confirmed that as predicted from the screen, the C-terminal domain of 4EIP is sufficient for suppression of gene expression by tethered 4EIP. This means that in the original tethering assay, all activity must have come from intact lamdaN-4EIP-myc rather than proteolytic fragments.
Results so far indicated that the activity of 4EIP in the tethering assay was independent of the EIF4E1 interaction. We therefore asked the complementary question—whether the action of EIF4E1 depended on 4EIP. EIF4E1 had highly suppressive activity in the tethering assay, both in high-throughput screens and when tested individually (67) (Figure 3B and C). To find out whether this activity required 4EIP, we repeated the tethering assays in cells lacking 4EIP (these ‘DKO’ cells are described later in this paper). A control tethered protein, PUF3, was able to suppress reporter expression in Δ4EIP cells (Figure 3C, far right). LambdaN-EIF4E1 was readily detected without tetracycline; after tetracycline addition, the amount increased 2–5-fold (Figure 3B). CAT activity was slightly higher in the presence of tetracycline than in its absence (Figure 3C), but the difference was not significant and the level in the presence of tetracycline was similar to that for the control cell line lacking tethered protein. Tethering of EIF4E1 in the presence of 4EIP rendered CAT mRNA undetectable, whereas without 4EIP there was no significant effect (Figure 3E). In the absence of 4EIP, therefore, tethered EIF4E1 is unable to repress translation and probably cannot stimulate it either. These results are exactly the same as those previously seen for mammalian 4EHP and GIGYF2: the suppressive activity of tethered 4EHP is absent in mutant mammalian cells that lack GIGYF2, but tethered GIGYF2 represses in the absence of 4EHP binding (24).
4EIP tethering inhibits translation initiation and causes mRNA decay
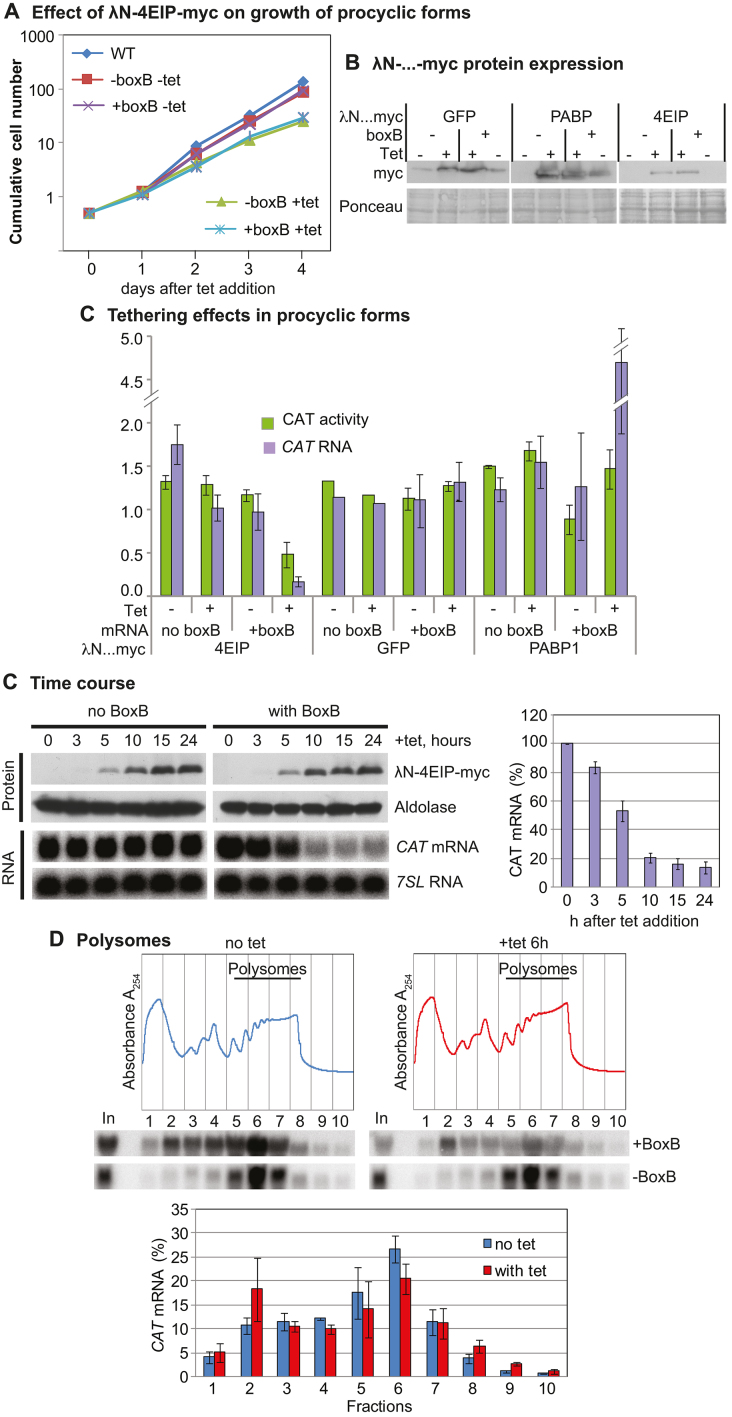
We next repeated the 4EIP tethering experiment in procyclic forms. Expression of lambdaN-4EIP-myc in procyclic forms resulted in mild growth inhibition (Figure 4A). Induction of lambdaN-4EIP-myc expression for 24 h (Figure 4B) resulted in a 10-fold decrease in CAT-boxB mRNA, and a more moderate decrease in CAT activity: CAT protein probably persisted from the pre-induction period. LambdaN-GFP-myc had no effect, while tethering of poly(A)-binding protein (PABP) 1 increased expression, as expected (76) (Figure 4B and C). In procyclic forms, full-length lamdaN-4EIP-myc was readily detected.
Figure 4.
4EIP tethering suppresses translation and induces mRNA destruction in procyclic forms. (A) Effect of LambdaN-4EIP-myc expression on proliferation. A cumulative growth curve is shown. Results are arithmetic mean with error bars showing standard deviation from three assays. (B) Expression of LambdaN-…-myc fusion proteins. (B) Effects of different tethered reporters on CAT activity (left bars, green in online version) and mRNA (right bars, mauve in online version). Results are arithmetic mean with error bars showing standard deviation. (C) Time course of LambdaN-4EIP-myc expression in cells expressing CAT mRNA with or without boxB. A northern blot showing the effect on CAT mRNA is shown below, and quantitation of replicates is to the right. (D) Polysomes were separated on sucrose gradients, either without tetracycline (blue) or 6 h after tetracycline addition (red). Typical gradient profiles and northern blots are shown at the top, and quantitation of three replicates beneath with “+tet” on the right.
To find out whether 4EIP inhibits translation, we needed to find a time point at which expression of LambdaN-4EIP-myc was evident, but CAT-boxB mRNA was still detectable. This occurred between 5 and 10 h after induction (Figure 4C). Analysis of sucrose gradient fractions revealed that even without induction, much more of the CAT-boxB mRNA was in the low density fractions than was the case for the mRNA without boxB. After 6-h induction, the amount of CAT-boxB mRNA in the polysomal fraction was preferentially decreased relative to the amount toward the top of the gradient (Figure 4D). There are two possible interpretations of this result. One is that attachment of 4EIP to the reporter mRNA represses its translation, with subsequent degradation. The alternative possibility is that 4EIP stimulates deadenylation or decapping, with a decrease in translation as a secondary effect. There was, however, no evidence for mRNA shortening from the northern blots (Figure 4D) and we would expect decapped mRNA to be destroyed very rapidly.
4EIP interactions with other proteins
In order to find out how 4EIP causes mRNA decay or translation repression, we looked for interaction partners beyond EIF4E1 either by tandem affinity purification and mass spectrometry (Supplementary Table S1 and Supplementary Figure S3A) or by yeast two-hybrid screening of a protein fragment library (53,77) (Supplementary Table S2). As expected, EIF4E1 consistently co-purified with 4EIP (Supplementary Table S1); in the yeast two-hybrid screen, the only interacting EIF4E1 fragment began at the start codon (Supplementary Table S2) and other potential interactors could not be confirmed (Supplementary Figure S4). None of the experiments provided any evidence for association of 4EIP with RNA-binding proteins that suppress expression, and no interactions were detected with known components of the mRNA degradation machinery.
mRNAs associated with 4EIP are relatively unstable
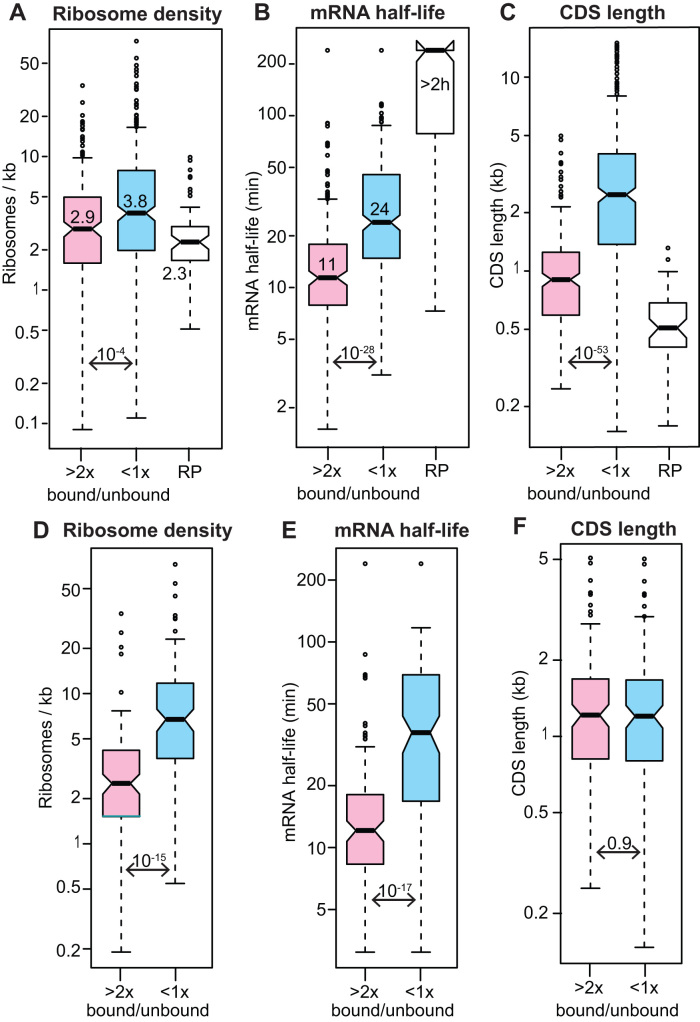
To find out which mRNAs are associated with 4EIP, we performed two independent single-step purifications of TAP-tagged 4EIP (Supplementary Figure S3B). We sequenced the 4EIP-associated RNAs and compared them with those in the unbound fraction (Supplementary Table S3, sheet 2). We classified 219 mRNAs that were at least 2-fold enriched in both pull-downs as ‘bound RNAs’. These were compared with 399 mRNAs that were reproducibly less abundant in the bound than in the unbound fractions (‘unbound RNAs’) (Supplementary Table S3, sheet 1). The 4EIP-bound mRNAs had slightly lower ribosome densities than the unbound ones (Figure 5A), but more dramatically, their median half-life was half that of the unbound mRNAs (Figure 5B). These results are consistent with a suppressive role for 4EIP.
Figure 5.
RNAs bound to 4EIP are relatively unstable mRNAs and were classified as ‘bound’ if the reads per million in the fraction bound to TAP-4EIP were at least twice those in the flow-though in both biological replicates, and the minimum count number was 10. ‘Unbound’ mRNAs has a ration of <1 in both replicates. The mRNAs encoding ribosomal proteins (RP) were mostly not bound but were considered separately. All results are for bloodstream forms. (A) Ribosome density for all ‘bound’ and ‘unbound’ mRNAs, as judged by ribosome profiling (49). Medians are indicated, as are the Student’s t-test values. (B) mRNA half-lives for all ‘bound’ and ‘unbound’ mRNAs (47). (C) Open reading frame or coding sequence (CDS) lengths of all ‘bound’ and ‘unbound’ mRNAs. (D) As (A), but for a subset of length-matched mRNAs (Supplementary Table S3, sheet 4). (E) As (B), but for the subset of length-matched mRNAs. (F) As (C), but for the subset of length-matched mRNAs.
Strangely, the average coding region length for 4EIP-bound mRNAs was less than half that of unbound mRNAs (Figure 5C). We therefore worried that the relative instability of bound mRNAs was an artifact that was in some way linked to short coding region length. To check this, we re-analyzed a size-matched subset of bound and unbound mRNAs (Figure 5F and Supplementary Table S3, sheet 4). The differences in ribosome density and half-life persisted, showing that this is an intrinsic property of 4EIP-bound mRNAs (Figure 5D and E). There was no evidence for specific association with mRNAs that are down-regulated in procyclic forms (e.g. (71)). The annotated 5′-untranslated region lengths of bound and unbound mRNAs were similar (medians of 84 and 94 nt, respectively). No enriched sequence motifs were found in either the 5′ or 3′-untranslated regions of the bound mRNAs.
No functional protein category was enriched in the bound fraction. However, some functional groups were significantly enriched in the unbound fraction: these included glucose and glycerol metabolism (Bonferroni-corrected Fisher test P-value 0.02) and lysosomal proteins (corrected P-value 0.05). In addition, the ‘unbound’ list included mRNAs encoding four of the eight T-complex protein-1 ring complex components, and, among the cytoskeletal proteins, many flagellar structure components (including tubulin) but only three motor proteins. More dramatically, more than half of all ribosomal protein mRNAs were in the ‘unbound’ fraction (P-value 10−30). The mRNAs encoding ribosomal proteins behave differently from most other mRNAs in trypanosomes. They are poorly translated (49,72), extremely stable (47), and always excluded from stress granules (72,78). Their lack of association with 4EIP shows that their poor translation is not caused by 4EIP.
We also statistically analyzed the results using DeSeq2 (79) using a custom script (https://zenodo.org/record/165132#.W2B2FS2B32I) (Supplementary Table S3, sheet 2 and Supplementary Figure S5). Although this program is not designed for pull-down experiments, the results were generally similar to those calculated from read ratios (Supplementary Figure S5).
4EIP is not essential in bloodstream-form or procyclic-form trypanosomes
To investigate the role of 4EIP in trypanosomes, we first targeted it by RNAi in monomorphic bloodstream forms that expressed in situ V5-tagged 4EIP (Figure 2F). RNAi was effective, although a small amount of tagged protein persisted (Figure 6A and Supplementary Figure S6A). There was a small but reproducible increase in the division time (Figure 6A). For unknown reasons, we were unable to obtain procyclic form clones with effective RNAi.
Figure 6.
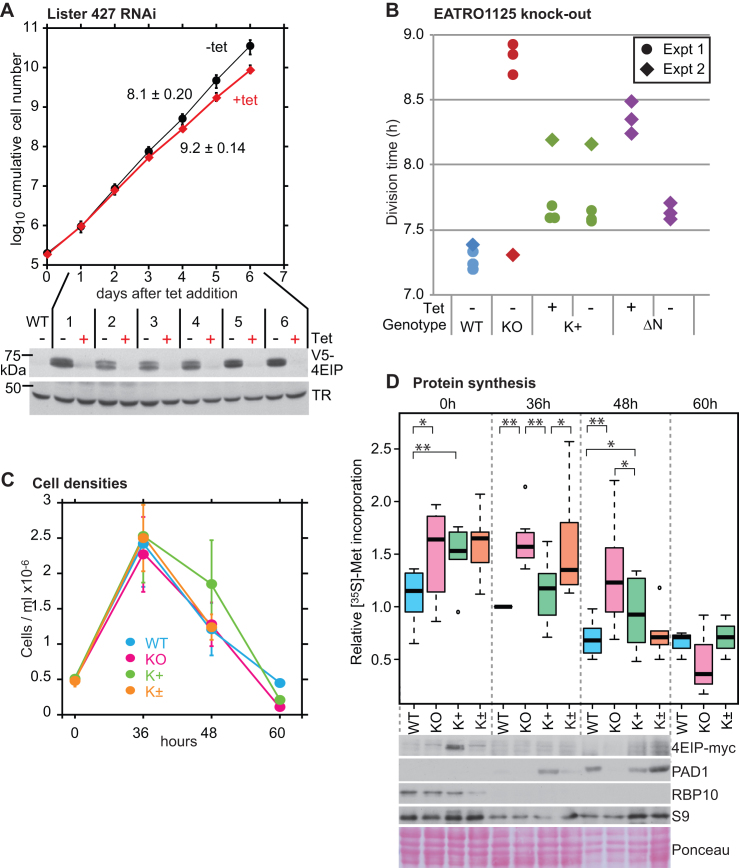
Cells without 4EIP differentiate poorly into stumpy forms. (A) Depletion of 4EIP in monomorphic bloodstream forms (Lister 427) results in a very mild growth defect. Cumulative growth of cultures with tetracycline-inducible RNAi is shown, with mean and standard deviation for three different cell lines. Expression of V5-4EIP in one of the three different RNAi lines, analyzed by western blotting, is shown below. TR is trypanothione reductase. Results for the other two cell lines are in Supplementary Figure S7A. (B) Division times of different EATRO1125 cell lines. The division times of the same cell lines were measured in two independent experiments. Results for the individual replicates are shown. KO: no 4EIP; K+: knockout line with inducible 4EIP-myc, with or without tetracycline; ΔN—knockout line with inducible lambdaN-ΔN-4EIP-myc, with or without tetracycline. Tetracycline was added 48 h prior to the start of measurements. In the second experiment (diamonds), the knockout line appeared to have adapted to the lack of 4EIP during continuous culture. (C) Growth of different EATRO1125 cell lines allowed to reach high density. Cells with a starting density of 5 × 105/ml were allowed to grow in HMI-9 media containing 1.1% methylcellulose for 60 h. Results are mean and standard deviation of three replicates. K+ are the complemented cells and K± are the K+ cells grown without tetracycline. (D) [35S]-Methionine incorporation into proteins during differentiation of EATRO1125 at high density. Trypanosomes were cultured as shown in (B), washed and pre-incubated for 1 h in labeling medium at 37°C. Methionine (35S) was then added for 20 min and proteins were analyzed by SDS-PAGE (Supplementary Figure S8B). Results for seven independent experiments are shown; four of the experiments included time = 0 h and three included time = 60 h. The color code is a paler version of (B). Student’s t-test results are: ** P < 0.01, * P < 0.05, both as paired or unpaired tests. Expression of various proteins, by western blotting, is shown under the box plot. For 4EIP-myc, the full-length protein is shown; degradation products are shown in Supplementary Figure S6F. The PAD1 signals from WT and K± are comparable.
To find out whether 4EIP was essential for survival, we deleted the genes in both Lister 427 and pleomorphic EATRO1125 bloodstream forms and procyclic forms. The deletion was successful in all cases. For Lister 427 cells (Supplementary Figure S6C), the division time increased from 8 h (wild-type and single knockout) to 10.4 h. Bloodstream-form EATRO1125 Δ4eip/Δ4eip knockout cells (Supplementary Figure S6D) had a mild growth defect (Figure 6B, ‘KO’), but after several months of continuous culture growth was normal, suggesting that the cells had adapted to life without 4EIP (Figure 6B, diamond symbols). Procyclic EATRO1125 Δ4eip/Δ4eip knockout cells (Supplementary Figure S6E) had a division time of ∼20 h, compared with 14 h for cells with both genes intact.
We introduced a tetracycline-inducible 4EIP-myc gene into the EATRO1125 Δ4eip/Δ4eip knockout cells (Figure 2H). The full-length tagged protein was detectable (migrating slightly slower than expected) but degradation products were, as before, at least as abundant (Supplementary Figure S6F). As expected from results with YFP-tagged 4EIP (see http://tryptag.org/?query=Tb927.9.11050) (80), the tagged protein was in the cytosol (Supplementary Figure S6G). These cells grew normally even in the absence of tetracycline (Figure 6B, K+). We do not know how much 4EIP-myc is expressed relative to the endogenous amount. However, we do know that RNA polymerase I transcription is 5–10 times more active than transcription by RNA polymerase II (81). Since the level of 4EIP-myc in the absence of tetracycline was 5 ± 3% of the induced level, it is quite possible that this ‘uninduced’ amount was sufficient for complementation.
Bloodstream forms with inducible expression of 4EIP with the N-terminal 11-amino-acid deletion (ΔN) grew normally in the absence of tetracycline but with tetracycline, growth was decreased (Figure 6B), suggesting that excess expression of the deleted version was toxic.
4EIP is required for normal translation arrest during stumpy form differentiation
We now attempted to induce stumpy-form differentiation in the EATRO1125 cells. The protocols used were as previously described (71). Cultures in methyl cellulose were started at 5 × 105/ml and allowed to grow for 48–60 h. Cell numbers peaked within 36 h then started to decline (Figure 6C). The complemented knockout cells, which were probably overexpressing 4EIP, consistently grew to slightly higher maximum densities and survived for longer before numbers started to decline (Figure 6C; Supplementary Figure S7A and B). After 36 h, we examined the cells by phase-contrast and fluorescence microscopy with DNA stain; all of the cultures consisted predominantly of G1-arrested cells (not shown). There was also no significant difference between the knockout and the complemented trypanosomes in the distance between the nucleus and kinetoplast (Supplementary Figure S7C). However, the stumpy-form marker PAD1 was usually not detectable in cells lacking 4EIP (Figures 6D and 7A). In several experiments, at 36 h the PAD1 signal from cells with inducible 4EIP-myc were higher than those from wild-type or uninduced cells (Figure 6D). This, together with the higher cell density at 48 h, suggests that 4EIP may enhance stumpy differentiation. All of the cultures lost expression of RBP10, a bloodstream-form regulator (71), within 36 h (Figure 6D).
Figure 7.
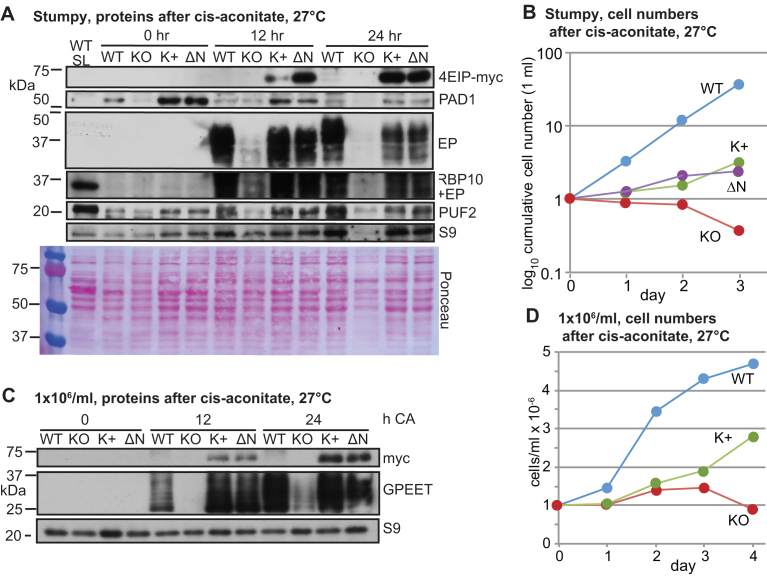
Cells without 4EIP cannot differentiate into growing procyclic forms. (A) Expression of various proteins in EATRO1125 cells cultured to high density for 60 h, then treated with cis-aconitate (CA) and transferred to 27°C. WT SL is long slender bloodstream forms; KO: no 4EIP; K+: knockout line with inducible 4EIP-myc and tetracycline; ΔN—knockout line with inducible lambdaN-ΔN-4EIP-myc, with tetracycline. In this experiment, full-length 4EIP-myc was not detected at time = 0. (B) Cell counts for cells treated as in (A) (to the end of day 1), then transferred to procyclic medium. (C) Expression of various proteins in cells cultured to 1 × 106/ml, then treated with cis-aconitate and transferred to 27°C. Replicates are in Supplementary Figure S8C. (D) Cell counts for cells from (C) transferred to procyclic medium on day 1.
Stumpy formation is accompanied by a general decrease in protein synthesis (34). To find out whether the lack of 4EIP affected this, we measured incorporation of [35S]-methionine into proteins. After 36 h, the knockout cells consistently showed higher protein synthesis than either the wild-type or the complemented controls (Figure 6D), but the difference was less marked after 48 h and had disappeared at 60 h. Examination of the protein profiles did not reveal any highly synthesized proteins that were consistently different between the cell lines (examples are in Supplementary Figure S8D).
From these experiments, we concluded that loss of 4EIP results in a delay in the inhibition of gene expression that accompanies stumpy formation. We attempted to compare mRNAs from the different time points but were unfortunately unable to obtain intact RNA from the 36 and 48 h cultures. The most likely explanation is that the cell pellets contained very high levels of RNase, either from dead cells or from the stumpy forms. Nevertheless, from the other results it seems likely that the major role of 4EIP in stumpy formation is in suppressing translation.
4EIP is required for normal differentiation to procyclic forms
Next, we investigated differentiation of the bloodstream forms into procyclic forms. Forty-eight hours after the cells attained a density of 1 × 106/ml, cis-aconitate (final concentration 6 mM) was added and the temperature was reduced to 27°C. At this point (time = 0 in Figure 7), the myc-tagged complementing proteins were not detected, presumably because stumpy-form cells have extremely low transcription and translation (33). However, within 12 h of cis-aconitate addition, the complementing proteins were again seen. After 12 h cis-aconitate, expression of the procyclic surface protein EP procyclin was clearly visible in wild-type cells, but barely detectable in the Δ4EIP knockout (Figure 7A and Supplementary Figure S8A), and the same was true after 24 h. Expression of either 4EIP-myc or ΔN4EIP-myc in the Δ4EIP cells restored EP procyclin expression. Expression of RBP10, a bloodstream-form-specific RNA-binding protein, was lost within 24 h (Supplementary Figure S8A). After 24 h in cis-aconitate, the cells were transferred to procyclic medium at 27°C. The wild-type cells grew, as previously described (71). In contrast, the Δ4EIP knockout cells lingered under procyclic-form conditions for up to 2 months, but appeared unable to divide. The complemented cells had intermediate behavior, showing that at least some of the differentiation defect in the Δ4EIP knockout cells was due to the lack of 4EIP (Figure 7B). The difference between the complemented cells and wild-type is most likely due to loss of other aspects of differentiation competence during the three rounds of clonal selection (see also Supplementary Figure S8B).
Results so far indicated that 4EIP is required for formation of stumpy forms that are competent for full procyclic-form differentiation. We therefore wondered whether the cells would be able to differentiate if they had not been left at high density for a prolonged period. To test this, the trypanosomes were grown to 1 × 106/ml, then cis-aconitate was added and the temperature was shifted to 27°C. Once again, a delay in EP or GPEET procyclin expression was specifically observed in the Δ4EIP knockout cells (Figure 7C and Supplementary Figure S8B). After 24 h, the medium was changed to procyclic form medium. The knockout cells were again unable to resume replication (Figure 7D), in contrast to the complemented cells.
Stumpy-form differentiation in the above experiments was caused by density-dependent signaling, but it can also be induced by cell-permeable, hydrolyzable cyclic AMP analogs (82). Preliminary results suggested that cells lacking 4EIP were also defective in this response (Supplementary Figure S8D).
These results showed that 4EIP is required for differentiation of bloodstream to procyclic forms. Since 4EIP is not essential in established procyclic forms (Supplementary Figure S6), we suggest that the differentiation defect is caused by an inability of the cells to suppress expression of a subset of proteins during the actual differentiation process. It was notable that the defect could be complemented by N-terminally deleted 4EIP, since this implies that the essential function of 4EIP during differentiation does not require it to interact with EIF4E1.
EIF4E1 is not essential in bloodstream forms
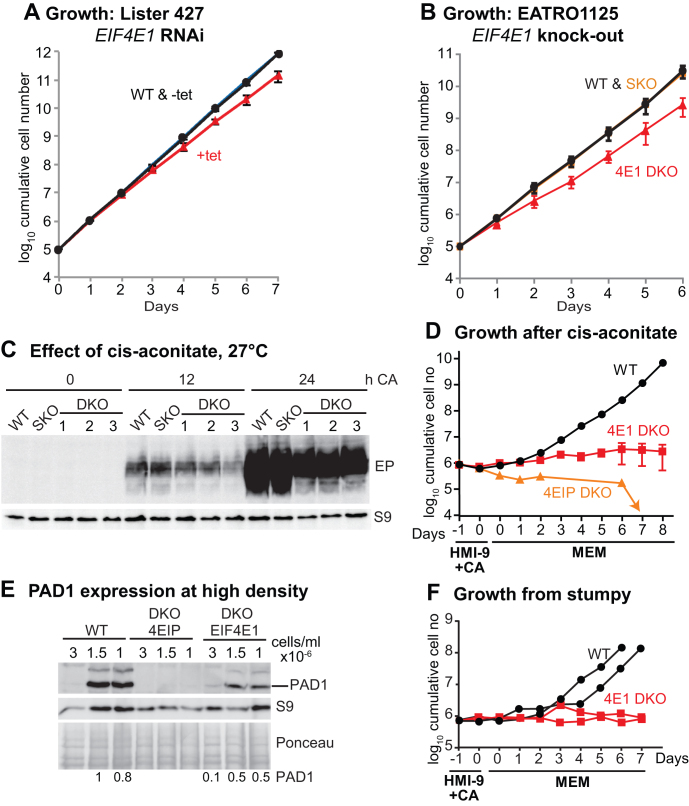
It was previously reported that while procyclic forms with RNAi targeting EIF4E1 showed only mildly slower growth (55), bloodstream-form trypanosomes with depleted EIF4E1 were unable to grow (55). Essentiality of EIF4E1 in bloodstream forms would suggest that it has a function that is independent of 4EIP. We, however, observed only a slight growth defect after RNAi (Figure 8A). Moreover, using differentiation-competent EATRO1125 bloodstream forms, both EIF4E1 gene copies were readily deleted (Supplementary Figure S9A and B), with only a mild growth defect (Figure 8B). We do not understand why the published RNAi was so deleterious: although the double-stranded RNA that was used comprises the complete open reading frame, it has no predicted off-target matches.
Figure 8.
Bloodstream forms without EIF4E1 cannot differentiate into growing procyclic forms. (A) Depletion of EIF4E1 in Lister 427 bloodstream forms results in a very mild growth defect. Cumulative growth of cultures with tetracycline-inducible RNAi is shown, with mean and standard deviation for three independent experiments. (B) Cumulative growth of different EATRO1125 cell lines. For WT and SKO (single replacement), the division times of the same cell lines were measured in two independent experiments. For DKO (both genes replaced), results for three different lines are shown. (C) Effect of adding cis-aconitate (CA) and shifting the temperature to 27°C (EATRO1125). The starting cell density was 1 × 106/ml. EP procyclin expression was measured by western blotting, with ribosomal protein S9 as the loading control. Although it is not evident from the image, the last three lanes are somewhat under-loaded, and quantitation revealed no significant differences between DKO lines and the SKO or WT. (D) Cell numbers in cultures that were treated as in (C) (HMI-9 +CA, 27°C), then transferred to medium suitable for procyclic forms (MEM, 27°C). The results shown are mean and standard deviation for three EIF4E1 cultures. Controls included two wild-type cultures (mean shown) and a 4EIP culture. (E) Expression of PAD1 after incubation at high density. Cells were incubated until they attained 3 × 106/ml, then the first samples were taken. To ensure complete comparability, subsequent samples were taken when the cell numbers had dropped to precisely 1.5 × 106 and 1.0 × 106/ml. Amounts of PAD1 relative to the Ponceau red stain are indicated. (F) Cells were left at high density for 48 h, then treated as in (D). Cumulative cell counts are shown.
Since the function of 4EIP in differentiation appeared to be independent of interaction with EIF4E1, we next tested whether the EIF4E1 knockout bloodstream forms were able to differentiate. After growth to 1 × 106/ml, followed by cis-aconitate addition, the kinetics of EP procyclin expression was similar to wild-type, although the amount was somewhat reduced (Figure 8C). Upon transfer to procyclic medium the cells lacking EIF4E1 initially appeared to recover, and expressed EP procyclin, but proliferation could not be sustained (Figure 8D and Supplementary Figure S9C). In addition, the surviving cells were abnormally long (Supplementary Figure S9D). Two attempts to knockout both EIF4E1 genes in established procyclic forms failed, although deletion of one copy was possible. To find out whether the EIF4E1 cells were defective in stumpy differentiation, we followed PAD1 expression during incubation of the cells at high density. PAD1 expression was reproducibly seen, although the level was somewhat lower than normal (Figure 8E and Supplementary Figure S9E). These cells were however once again not able to differentiate into growing procyclic forms (Figure 8F).
These results indicate that EIF4E1 and 4EIP have different roles during differentiation. While 4EIP is required for during stumpy-form differentiation, EIF4E1 is likely to be essential in growing procyclic forms.
DISCUSSION
Our results indicate that tethering of T. brucei 4EIP to an mRNA suppresses its translation and promotes RNA degradation. In bloodstream forms, 4EIP is preferentially bound to mRNAs with relatively short half-lives and lower ribosome densities, suggesting that natural binding has a similar effect. For 4EIP, we do not know which of the two effects on mRNA is the primary activity: they could be linked, since poor translation initiation might result in mRNA decay but deadenylation or decapping can prevent translation. No interactions of 4EIP with the mRNA degradation machinery were detected. The mRNAs bound to yeast 4E-BPs also had lower than average ribosome densities, shorter poly(A) tails and less PABP binding (83).
Although 4EIP appears—from ultraviolet cross-linking results—to bind mRNA directly, it lacks a known RNA-binding motif and is not clear how the protein is recruited to specific mRNAs: we found no common sequence motifs in the bound transcripts. In mammalian cells, 4EHP can be recruited to target mRNAs via specific RNA-binding proteins (22,23). Only one putative RNA-binding protein, ZC3H14, interacted with 4EIP in a yeast two-hybrid screen. ZC3H14 can indeed suppress expression in the bloodstream-form tethering assay, but there is as yet no evidence from either ribosome profiling (48,49) or proteomics (69,84) that ZC3H14 protein is expressed (Might ZC3H14 be specific to life-cycle stages that have not yet been molecularly characterized?). The mechanism(s) by which 4EIP recognizes its targets thus remains to be discovered.
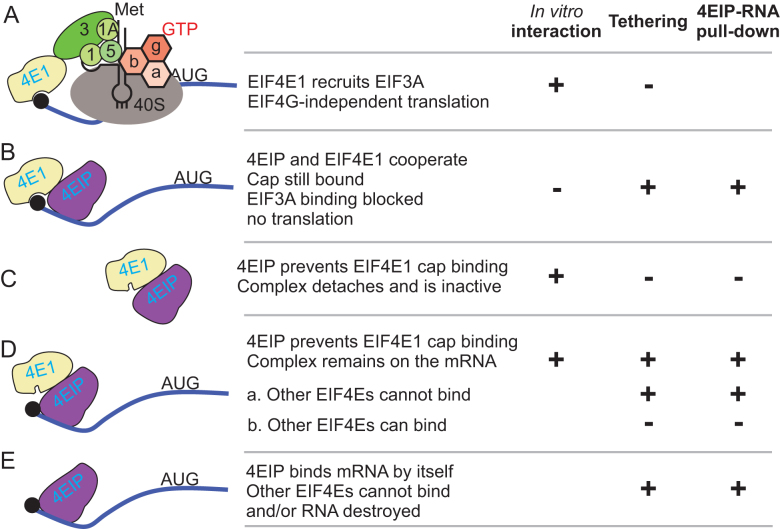
The only 4EIP partner protein that emerged reliably from our analyses was EIF4E1. There are various possible mechanisms by which 4EIP and EIF4E1 might act. One possibility is that without 4EIP, EIF4E1 can act as a translation initiation factor (Figure 9A). Leishmania major EIF4E1 can interact with EIF3 in an in vitro pull-down, and the interaction was also detected in vivo, although overexpression of EIF4E1 was required (63,64). An argument against this idea is that when EIF4E1 was tethered in the absence of 4EIP, there was no significant increase in reporter expression, whereas tethered EIF4E3/G4 and EIF4E4/G3 enhance expression considerably (67,68). However, the presence of the tags, or the fact that the protein was tethered to the 3′-UTR, could have affected EIF4E1 function. In vivo, EIF4E1 has lower abundance and cap-binding affinity than EIF4E3 and EIF4E4 (55,63), so EIF4E1 probably could not compete with EIF4E3 or EIF4E4 for cap binding by itself. Under the model in Figure 9A, 4EIP would prevent the interaction of EIF4E1 with EIF3A, in partial analogy to Opisthokont 4E-BPs. An alternative is that EIF4E1–4EIP complex is functionally similar to 4EHP-GYF2, and that EIF4E1 acts as a constitutively inhibitory translation factor (Figure 9B), with mRNA binding enhanced by cooperative interactions with 4EIP.
Figure 9.
Models for 4EIP function. For details see the ‘Discussion’ section.
The interaction of Leishmania 4EIP inhibits EIF4E1 cap binding (65). It is possible that this could result in complete dissociation of EIF4E1 from the RNA (Figure 9C). Since, however, 4EIP could be cross-linked to mRNA (68), it is also possible that the complex remains bound, but detachment of EIF4E1 from the cap exposes the latter for decapping (Figure 9D). The models in Figure 9B and D imply that the EIF4E1–4EIP complex prevents binding of active cap-binding translation initiation complexes. This is very likely to be the major effect in growing intact cells. However, several results indicated that the suppressive activity of 4EIP does not depend on its interaction with EIF4E1. First, both ΔN4EIP, which does not detectably interact with EIF4E1, and a C-terminal fragment, which lacks all possible interaction surfaces, were sufficient to repress expression when tethered to a reporter mRNA. Stronger evidence comes from the ability of ΔN4EIP to complement the differentiation defect of cells lacking full-length 4EIP (Figure 7C). This suggests that during stumpy formation, 4EIP acts independently of EIF4E1 (Figure 9E).
Our interaction studies provided no clear insights into the mechanism by which 4EIP suppresses translation and/or causes mRNA destruction. Apart from the 4E interaction motif, there is a second highly conserved charged domain, which might be implicated in protein–protein or protein–RNA interactions. However when tethered, the highly unstructured and proline-glutamine-rich C-terminus was sufficient for activity. 4EIPs from several other kinetoplastids have polyglutamine repeats (Supplementary Figure S1). Such repeats are present in many regulatory RNA-binding proteins, both from trypanosomes (68), and other species (for example, they are found in Bicoid and GIGYF2). These and other low complexity regions have been implicated in protein aggregation and in formation of hydrogels and RNA–protein granules (85). EIF4E1 is found in starvation granules (78) (http://tryptag.org/?query=EIF4E1), but it is distributed throughout the cytosol and nucleus under normal conditions (55).
Both the absence of 4EIP in bloodstream or procyclic forms, and its overexpression (as a fusion protein) in procyclic forms, resulted in a slight growth defect. We therefore suggest that in growing cells, 4EIP may help to suppress expression of proteins that are needed in only modest amounts. Overexpression of 4EIP might cause excessive suppression of genuine targets, as well as binding of 4EIP may bind to mRNAs with which it does not normally associate. It was notable that on western blots, most 4EIP appeared to be degraded into smaller species: an N-terminal part that contains the EIF4E1 interaction domain, but not the region active in the tethering assay and the Q-P rich C-terminus that was active in the tethering assay but which could not interact with EIF4E1. If this cleavage occurs in vivo, it could be a mechanism to regulate EIF4E1-dependent translational suppression by 4EIP.
When pleomorphic trypanosomes were incubated for prolonged periods at high density, the level of 4EIP became critical. Overexpression of 4EIP enhanced parasite survival and increased PAD1 expression, suggesting that translational shut-down is protective. In contrast, cells that lacked 4EIP showed a minor delay in suppression of protein synthesis. We therefore suggest that 4EIP is required in order to suppress translation in the early stages of stumpy formation, before mRNAs are lost because of transcription arrest. The inability of these cells to differentiate further could be the result of a disrupted developmental program.
DATA AVAILABILITY
The ArrayExpress accession number for the 4EIP-TAP pull-down is E-MTAB-6240.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Thomas Ruppert and the Mass Spectrometry facility at the ZMBH for mass spectrometry analysis, and David Ibbersson (Bioquant, Heidelberg) for building RNASeq libraries and taking them for sequencing. We acknowledge contributions from MSc or BSc students during lab practicals, especially Jacob Borold who did preliminary polysome analysis after tethering. We thank Keith Matthews for antibody to PAD1 and Luise Krauth-Siegel for antibody to trypanothione reductase. We also thank Michal Shapira, Osvaldo de Melo Neto and Eden Freire for useful discussions and for communicating unpublished results.
Author contributions: All authors except C.C. contributed data, mostly with accompanying text. M.T. performed 4EIP-TAP experiments (Supplementary Figure S3 and Supplementary Table S1), RNA pull-down including data analysis (Figure 5 and Supplementary Table S3) and the two-hybrid screen (Supplementary Table S2). K.K.M. performed differentiation experiments, add-backs and tethering of EIF4E1 in 4EIP knockout; Figures 1E and F, 3, 6 and 7; Supplementary Figures S7, S8 and S9. D.D.: Figure 1A–D; Supplementary Figures S2 and S3. I.M.: Figure 4. E.M.: Figure 3 and RNAi in procyclics. F.E.: EIF4E1 DKO, Figure 8 and Supplementary Figure S9. J.B.: 4EIP KO in Lister 427 and EATRO1125, Figure 6 and Supplementary Figure S7. C.C. conducted data analysis, funding acquisition, writing and editing manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [Cl112-17-1, Cl112-17-2, Cl112-24, Cl112-28]; Deutsches Akademisches Austauschdienst (DAAD) (to K.K.M.); HBIGS (Heidelberg Biosciences International Graduate School) (to F.E.). Funding for open access charge: Institutional funds and DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1. Firczuk H., Kannambath S., Pahle J., Claydon A., Beynon R., Duncan J., Westerhoff H., Mendes P., McCarthy J.E.. An in vivo control map for the eukaryotic mRNA translation machinery. Mol. Syst. Biol. 2013; 9:635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B.. Rate-limiting steps in yeast protein translation. Cell. 2013; 153:1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richter J.D., Coller J.. Pausing on Polyribosomes: Make way for elongation in translational control. Cell. 2015; 163:292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanson G., Coller J.. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018; 19:20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pestova T.V., Kolupaeva V.G., Lomakin I.B., Pilipenko E.V., Shatsky I.N., Agol V.I., Hellen C.U.. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hernandez G., Proud C.G., Preiss T., Parsyan A.. On the diversification of the translation apparatus across eukaryotes. Comp. Funct. Genomics. 2012; 2012:256848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kamenska A., Simpson C., Standart N.. eIF4E-binding proteins: new factors, new locations, new roles. Biochem. Soc. Trans. 2014; 42:1238–1245. [DOI] [PubMed] [Google Scholar]
- 8. Mader S., Lee H., Pause A., Sonenberg N.. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4gamma and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995; 15:4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kinkelin K., Veith K., Grunwald M., Bono F.. Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA. 2012; 18:1624–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Igreja C., Peter D., Weiler C., Izaurralde E.. 4E-BPs require non-canonical 4E-binding motifs and a lateral surface of eIF4E to repress translation. Nat. Commun. 2014; 5:4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Peter D., Igreja C., Weber R., Wohlbold L., Weiler C., Ebertsch L., Weichenrieder O., Izaurralde E.. Molecular architecture of 4E-BP translational inhibitors bound to eIF4E. Mol. Cell. 2015; 57:1074–1087. [DOI] [PubMed] [Google Scholar]
- 12. Igreja C., Izaurralde E.. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev. 2011; 25:1955–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamenska A., Lu W.T., Kubacka D., Broomhead H., Minshall N., Bushell M., Standart N.. Human 4E-T represses translation of bound mRNAs and enhances microRNA-mediated silencing. Nucleic Acids Res. 2014; 42:3298–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rom E., Kim H.C., Gingras A.C., Marcotrigiano J., Favre D., Olsen H., Burley S.K., Sonenberg N.. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem. 1998; 273:13104–13109. [DOI] [PubMed] [Google Scholar]
- 15. Hernandez G., Altmann M., Sierra J.M., Urlaub H., Diez del Corral R., Schwartz P., Rivera-Pomar R.. Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila. Mech. Dev. 2005; 122:529–543. [DOI] [PubMed] [Google Scholar]
- 16. Cho P.F., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N.. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005; 121:411–423. [DOI] [PubMed] [Google Scholar]
- 17. Kropiwnicka A., Kuchta K., Lukaszewicz M., Kowalska J., Jemielity J., Ginalski K., Darzynkiewicz E., Zuberek J.. Five eIF4E isoforms from Arabidopsis thaliana are characterized by distinct features of cap analogs binding. Biochem. Biophys. Res. Commun. 2015; 456:47–52. [DOI] [PubMed] [Google Scholar]
- 18. Cho P.F., Gamberi C., Cho-Park Y.A., Cho-Park I.B., Lasko P., Sonenberg N.. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol. 2006; 16:2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vardy L., Orr-Weaver T.L.. Regulating translation of maternal messages: multiple repression mechanisms. Trends Cell Biol. 2007; 17:547–554. [DOI] [PubMed] [Google Scholar]
- 20. Lasko P. Posttranscriptional regulation in Drosophila oocytes and early embryos. Wiley Interdiscip. Rev. RNA. 2011; 2:408–416. [DOI] [PubMed] [Google Scholar]
- 21. Valzania L., Ono H., Ignesti M., Cavaliere V., Bernardi F., Gamberi C., Lasko P., Gargiulo G.. Drosophila 4EHP is essential for the larval-pupal transition and required in the prothoracic gland for ecdysone biosynthesis. Dev. Biol. 2016; 410:14–23. [DOI] [PubMed] [Google Scholar]
- 22. Morita M., Ler L.W., Fabian M.R., Siddiqui N., Mullin M., Henderson V.C., Alain T., Fonseca B.D., Karashchuk G., Bennett C.F. et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol. Cell. Biol. 2012; 32:3585–3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu R., Olsen M.T., Webb K., Bennett E.J., Lykke-Andersen J.. Recruitment of the 4EHP-GYF2 cap-binding complex to tetraproline motifs of tristetraprolin promotes repression and degradation of mRNAs with AU-rich elements. RNA. 2016; 22:373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peter D., Weber R., Sandmeir F., Wohlbold L., Helms S., Bawankar P., Valkov E., Igreja C., Izaurralde E.. GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression. Genes Dev. 2017; 31:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jafarnejad S.M., Chapat C., Matta-Camacho E., Gelbart I.A., Hesketh G.G., Arguello M., Garzia A., Kim S.H., Attig J., Shapiro M. et al. Translational control of ERK signaling through miRNA/4EHP-directed silencing. Elife. 2018; 7:e35034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chapat C., Jafarnejad S.M., Matta-Camacho E., Hesketh G.G., Gelbart I.A., Attig J., Gkogkas C.G., Alain T., Stern-Ginossar N., Fabian M.R. et al. Cap-binding protein 4EHP effects translation silencing by microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5425–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amaya Ramirez C.C., Hubbe P., Mandel N., Bethune J.. 4EHP-independent repression of endogenous mRNAs by the RNA-binding protein GIGYF2. Nucleic Acids Res. 2018; 46:5792–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schopp I.M., Amaya Ramirez C.C., Debeljak J., Kreibich E., Skribbe M., Wild K., Béhune J.. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017; 8:15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Horn D. Antigenic variation in African trypanosomes. Mol. Biochem. Parasitol. 2014; 195:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hannaert V., Bringaud F., Opperdoes F.R., Michels P.A.M.. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid Biol. Dis. 2003; 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rico E., Rojas F., Mony B.M., Szoor B., Macgregor P., Matthews K.R.. Bloodstream form pre-adaptation to the tsetse fly in Trypanosoma brucei. Front. Cell. Infect. Microbiol. 2013; 3:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matthews K.R., McCulloch R., Morrison L.J.. The within-host dynamics of African trypanosome infections. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015; 370:20140288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pays E., Hanocq-Quertier J., Hanocq F., Van Assel S., Nolan D., Rolin S.. Abrupt RNA changes precede the first cell division during the differentiation of Trypanosoma brucei bloodstream forms into procyclic forms in vitro. Mol. Biochem. Parasitol. 1993; 61:107–114. [DOI] [PubMed] [Google Scholar]
- 34. Brecht M., Parsons M.. Changes in polysome profiles accompany trypanosome development. Mol. Biochem. Parasitol. 1998; 97:189–198. [DOI] [PubMed] [Google Scholar]
- 35. Dean S., Marchetti R., Kirk K., Matthews K.. A surface transporter family conveys the trypanosome differentiation signal. Nature. 2009; 459:213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roditi I., Liniger M.. Dressed for success: the surface coats of insect-borne protozoan parasites. Trends Microbiol. 2002; 10:128–134. [DOI] [PubMed] [Google Scholar]
- 37. Bringaud F., Riviere L., Coustou V.. Energy metabolism of trypanosomatids: adaptation to available carbon sources. Mol. Biochem. Parasitol. 2006; 149:1–9. [DOI] [PubMed] [Google Scholar]
- 38. Mantilla B.S., Marchese L., Casas-Sanchez A., Dyer N.A., Ejeh N., Biran M., Bringaud F., Lehane M.J., Acosta-Serrano A., Silber A.M.. Proline metabolism is essential for Trypanosoma brucei brucei survival in the Tsetse Vector. PLoS Pathog. 2017; 13:e1006158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Savage A.F., Kolev N.G., Franklin J.B., Vigneron A., Aksoy S., Tschudi C.. Transcriptome profiling of Trypanosoma brucei development in the Tsetse fly vector Glossina morsitans. PLoS One. 2016; 11:e0168877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rotureau B., Van Den Abbeele J.. Through the dark continent: African trypanosome development in the tsetse fly. Front. Cell. Infect. Microbiol. 2013; 3:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Clayton C. Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 2016; 32:46–51. [DOI] [PubMed] [Google Scholar]
- 42. Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011; 6:459–474. [DOI] [PubMed] [Google Scholar]
- 43. Perry K., Watkins K.P., Agabian N.. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc. Natl. Acad. Sci. U.S.A. 1987; 84:8190–8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Freistadt M., Cross G., Branch A., Robertson H.. Direct analysis of the mini-exon donor RNA of Trypanosoma brucei: detection of a novel cap structure also present in messenger RNA. Nucleic Acids Res. 1987; 15:9861–9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clayton C., Michaeli S.. 3′ processing in protists. Wiley Interdiscip. Rev. RNA. 2011; 2:247–255. [DOI] [PubMed] [Google Scholar]
- 46. Koch H., Raabe M., Urlaub H., Bindereif A., Preusser C.. The polyadenylation complex of Trypanosoma brucei: characterization of the functional poly(A) polymerase. RNA Biol. 2016; 13:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fadda A., Ryten M., Droll D., Rojas F., Färber V., Haanstra J., Bakker B., Matthews K., Clayton C.. Transcriptome-wide analysis of mRNA decay reveals complex degradation kinetics and suggests a role for co-transcriptional degradation in determining mRNA levels. Mol. Microbiol. 2014; 94:307–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jensen B.C., Ramasamy G., Vasconcelos E.J., Ingolia N.T., Myler P.J., Parsons M.. Extensive stage-regulation of translation revealed by ribosome profiling of Trypanosoma brucei. BMC Genomics. 2014; 15:911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Antwi E., Haanstra J., Ramasamy G., Jensen B., Droll D., Rojas F., Minia I., Terrao M., Mercé C., Matthews K. et al. Integrative analysis of the Trypanosoma brucei gene expression cascade predicts differential regulation of mRNA processing and unusual control of ribosomal protein expression. BMC Genomics. 2016; 17:306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kolev N.G., Ullu E., Tschudi C.. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell. Microbiol. 2014; 16:482–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gupta S.K., Chikne V., Eliaz D., Tkacz I.D., Naboishchikov I., Carmi S., Waldman Ben-Asher H., Michaeli S.. Two splicing factors carrying serine-arginine motifs, TSR1 and TSR1IP, regulate splicing, mRNA stability, and rRNA processing in Trypanosoma brucei. RNA Biol. 2014; 11:715–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gupta S.K., Kosti I., Plaut G., Pivko A., Tkacz I.D., Cohen-Chalamish S., Biswas D.K., Wachtel C., Waldman Ben-Asher H., Carmi S. et al. The hnRNP F/H homologue of Trypanosoma brucei is differentially expressed in the two life cycle stages of the parasite and regulates splicing and mRNA stability. Nucleic Acids Res. 2013; 41:6577–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Singh A., Minia I., Droll D., Fadda A., Clayton C., Erben E.. Trypanosome MKT1 and the RNA-binding protein ZC3H11: interactions and potential roles in post-transcriptional regulatory networks. Nucleic Acids Res. 2014; 42:4652–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Freire E.R., Sturm N.R., Campbell D.A., de Melo Neto O.P.. The role of cytoplasmic mRNA cap-binding protein complexes in Trypanosoma brucei and other trypanosomatids. Pathogens. 2017; 6:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Freire E., Dhalia R., Mouraa D., da Costa Lima T., Lima R., Reisa C., Hughes K., Figueiredo R., Standart N., Carrington M. et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol. Biochem. Parasitol. 2011; 176:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klein C., Terrao M., Inchaustegui Gil D., Clayton C.. Polysomes of Trypanosoma brucei: association with initiation factors and RNA-binding proteins. PLoS One. 2015; 10:e0135973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moura D.M., Reis C.R., Xavier C.C., da Costa Lima T.D., Lima R.P., Carrington M., de Melo Neto O.P.. Two related trypanosomatid eIF4G homologues have functional differences compatible with distinct roles during translation initiation. RNA Biol. 2015; 12:305–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Freire E.R., Vashisht A.A., Malvezzi A.M., Zuberek J., Langousis G., Saada E.A., Nascimento J.D., Stepinski J., Darzynkiewicz E., Hill K. et al. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA. 2014; 20:1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Freire E., Malvezzi A., Vashisht A., Zuberek J., Saada E., Langousis G., de F., Nascimento J., Moura D., Darzynkiewicz E., Hill K. et al. Trypanosoma brucei translation-initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot. Cell. 2014; 13:896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Freire E.R., Moura D.M.N., Bezerra M.J.R., Xavier C.C., Morais-Sobral M.C., Vashisht A.A., Rezende A.M., Wohlschlegel J.A., Sturm N.R., de Melo Neto O.P. et al. Trypanosoma brucei EIF4E2 cap-binding protein binds a homolog of the histone-mRNA stem-loop-binding protein. Curr. Genet. 2018; 64:821–839. [DOI] [PubMed] [Google Scholar]
- 61. Yoffe Y., Zuberek J., Lewdorowicz M., Zeira Z., Keasar C., Orr-Dahan I., Jankowska-Anyszka M., Stepinski J., Darzynkiewicz E., Shapira M.. Cap-binding activity of an eIF4E homolog from Leishmania. RNA. 2004; 10:1764–1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yoffe Y., Zuberek J., Lerer A., Lewdorowicz M., Stepinski J., Altmann M., Darzynkiewicz E., Shapira M.. Binding specificities and potential roles of isoforms of eukaryotic initiation factor 4E in Leishmania. Eukaryot. Cell. 2006; 5:1969–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zinoviev A., Leger M., Wagner G., Shapira M.. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res. 2011; 39:8404–8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meleppattu S., Kamus-Elimeleh D., Zinoviev A., Cohen-Mor S., Orr I., Shapira M.. The eIF3 complex of Leishmania-subunit composition and mode of recruitment to different cap-binding complexes. Nucleic Acids Res. 2015; 43:6222–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meleppattu S., Arthanari H., Zinoviev A., Boeszoermenyi A., Wagner G., Shapira M., Leger-Abraham M.. Structural basis for LeishIF4E-1 modulation by an interacting protein in the human parasite Leishmania major. Nucleic Acids Res. 2018; 46:3791–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haanstra J., Stewart M., Luu V.-D., van Tuijl A., Westerhoff H., Clayton C., Bakker B.. Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J. Biol. Chem. 2008; 283:2495–2507. [DOI] [PubMed] [Google Scholar]
- 67. Erben E., Fadda A., Lueong S., Hoheisel J., Clayton C.. Genome-wide discovery of post-transcriptional regulators in Trypanosoma brucei. PLoS Pathog. 2014; 10:e1004178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lueong S., Merce C., Fischer B., Hoheisel J., Erben E.. Gene expression regulatory networks in Trypanosoma brucei: insights into the role of the mRNA-binding proteome. Mol. Microbiol. 2016; 100:457–471. [DOI] [PubMed] [Google Scholar]
- 69. Dejung M., Subota I., Bucerius F., Dindar G., Freiwald A., Engstler M., Boshart M., Butter F., Janzen C.J.. Quantitative proteomics uncovers novel factors involved in developmental dfferentiation of Trypanosoma brucei. PLoS Pathog. 2016; 12:e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Alibu V.P., Storm L., Haile S., Clayton C., Horn D.. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 2004; 139:75–82. [DOI] [PubMed] [Google Scholar]
- 71. Mugo E., Clayton C.. Expression of the RNA-binding protein RBP10 promotes the bloodstream-form differentiation state in Trypanosoma brucei. PLoS Pathog. 2017; 13:e1006560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Minia I., Merce C., Terrao M., Clayton C.. Translation regulation and RNA granule formation after heat shock of procyclic form Trypanosoma brucei: many heat-induced mRNAs are increased during differentiation to mammalian-infective forms. PLoS Negl. Trop. Dis. 2016; 10:e0004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Droll D., Minia I., Fadda A., Singh A., Stewart M., Queiroz R., Clayton C.. Post-transcriptional regulation of the trypanosome heat shock response by a zinc finger protein. PLoS Pathog. 2013; 9:e1003286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Klein C., Terrao M., Clayton C.. The role of the zinc finger protein ZC3H32 in bloodstream-form Trypanosoma brucei. PLoS One. 2017; 12:e0177901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Delhi P., Queiroz R., Inchaustegui D., Carrington M., Clayton C.. Is there a classical nonsense-mediated decay pathway in trypanosomes. PLoS One. 2011; 6:e25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chakraborty C., Fadda F., Erben E., Mugo E., Lueong S., Hoheisel J., Clayton C.. Interactions of CAF1-NOT complex components from Trypanosoma brucei. F1000 Res. 2017; 6:858. [Google Scholar]
- 78. Fritz M., Vanselow J., Sauer N., Lamer S., Goos C., Siegel T., Subota I., Schlosser A., Carrington M., Kramer S.. Novel insights into RNP granules by employing the trypanosome's microtubule skeleton as a molecular sieve. Nucleic Acids Res. 2015; 43:8013–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Love M., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Dean S., Sunter J., Wheeler R.. TrypTag.org: A trypanosome genome-wide protein localisation resource. Trends Parasitol. 2016; 33:80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Biebinger S., Rettenmaier S., Flaspohler J., Hartmann C., Peña-Diaz J., Wirtz L.E., Hotz H.R., Barry J.D., Clayton C.E.. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 1996; 24:1202–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Laxman S., Riechers A., Sadilek M., Schwede F., Beavo J.. Hydrolysis products of cAMP analogs cause transformation of Trypanosoma brucei from slender to stumpy-like forms. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:19194–19199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Costello J., Castelli L.M., Rowe W., Kershaw C.J., Talavera D., Mohammad-Qureshi S.S., Sims P.F., Grant C.M., Pavitt G.D., Hubbard S.J. et al. Global mRNA selection mechanisms for translation initiation. Genome Biol. 2015; 16:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Christiano R., Kolev N., Shi H., Ullu E., Walther T., Tschudi C.. The proteome and transcriptome of the infectious metacyclic form of Trypanosoma brucei define quiescent cells primed for mammalian invasion. Mol. Microbiol. 2017; 106:74–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Castilla-Llorente V., Ramos A.. PolyQ-mediated regulation of mRNA granules assembly. Biochem. Soc. Trans. 2014; 42:1246–1250. [DOI] [PubMed] [Google Scholar]
- 86. Blattner J., Helfert S., Michels P., Clayton C.E.. Compartmentation of phosphoglycerate kinase in Trypanosoma brucei plays a critical role in parasite energy metabolism. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:11596–11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The ArrayExpress accession number for the 4EIP-TAP pull-down is E-MTAB-6240.