Abstract
Sin3a is a core component of histone-deacetylation-activity-associated transcriptional repressor complex, playing important roles in early embryo development. Here, we reported that down-regulation of Sin3a led to the loss of embryonic stem cell (ESC) self-renewal and skewed differentiation into mesendoderm lineage. We found that Sin3a functioned as a transcriptional coactivator of the critical Nodal antagonist Lefty1 through interacting with Tet1 to de-methylate the Lefty1 promoter. Further studies showed that two amino acid residues (Phe147, Phe182) in the PAH1 domain of Sin3a are essential for Sin3a–Tet1 interaction and its activity in regulating pluripotency. Furthermore, genome-wide analyses of Sin3a, Tet1 and Pol II ChIP-seq and of 5mC MeDIP-seq revealed that Sin3a acted with Tet1 to facilitate the transcription of a set of their co-target genes. These results link Sin3a to epigenetic DNA modifications in transcriptional activation and have implications for understanding mechanisms underlying versatile functions of Sin3a in mouse ESCs.
INTRODUCTION
Embryonic stem cells (ESCs), isolated from the inner cell mass (ICM) of blastocysts, are self-renewing cells that maintain pluripotency to differentiate into all types of cells (1–4). When given the appropriate cellular signals, ESCs are subjected to constant cell fate choices between self-replication and differentiation (4). Thus, an exact understanding of the molecular mechanisms underlying the transition between these cell fates is pivotal for the developmental studies and the application of ESCs in regenerative medicine. Multiple signaling pathways (e.g., LIF/Stat3, Wnt/β-Catenin, MAPK and BMP/TGF-β) are involved in the regulation of ESC self-renewal and differentiation potential (5–7). The Nodal-Smad2 pathway, which belongs to the TGF-β signaling pathway, participates in a series of key events, including early embryonic development (8,9) and ESC mesendoderm specification (7,10). In addition, maintaining the unique identity of ESCs requires an intricate network of transcription factors (e.g. Oct4, Sox2, Nanog, Esrrb, Tbx3) (11–13), epigenetic modification molecules (e.g. Tet1, MOF, Wdr5) (14–17), nucleosome remodelers, cofactors, miRNAs and ncRNAs, etc (13). Previous studies have indicated that Sin3a, a well-known master scaffold protein and transcriptional corepressor, is ubiquitously expressed at the blastocyst stage and is essential for early embryonic development (18). The Sin3a–HDAC complex has also been found to be required for ESC proliferation under self-renewing conditions (19). However, the role of Sin3a in ESC self-renewal and differentiation potential has not yet been fully elucidated.
Among cofactors that participate in the regulation of transcription, Sin3a can interact with a variety of transcriptional factors through its six conserved domains that include four paired amphipathic helices (PAH1–4), one histone deacetylase interaction domain (HID) and one highly conserved region (HCR). Each of these domains has been reported to possess innate specificity for recruiting certain transcriptional repressors, thereby permitting the complex to modulate transcriptional output by altering chromatin structure (20–22). PAH1 and PAH2 can specifically identify transcription factors, while PAH3 and PAH4 tend to play supporting roles in the complex. The HID domain of Sin3a has been generally reported to be responsible for recruiting histone deacetylases (HDACs) to form an HDAC-associated transcriptional repressor complex (20,23–25). Previous studies have indicated that the multi-subunit nature of Sin3a provides unique contact surfaces for interaction with particular accessory proteins to repress the transcription of specific genes (26). For example, the Sin3a–HDAC complex has been reported to interact with Fam60a, decreasing histone acetylation at the cyclin D1 promoter and thus repressing the expression of cyclin D1 (27). Actions of Sin3a and estrogen receptor alpha (ERα) at the proximal promoter of ESR1 can overcome other activating signals at distal or proximal sites and ultimately decrease the expression levels of ESR1 (28). However, the important role of Sin3a in transcriptional activation is often understated. In fact, the molecular mechanisms of how Sin3a promotes gene transcription remain to be unveiled.
Tet1 has been shown to be capable of erasing DNA methylation by reiterative oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (29), thus playing an important role in transcriptional activation of target genes (30). Although Tet1 possesses an N-terminal CXXC structure typical for DNA binding, this domain alone fails to recognize specific target gene sequences for Tet1 (31), implying that other proteins are involved in the regulatory mechanism by which Tet1 promotes the conversion of 5mC to 5hmC at specific DNA sites. Moreover, Sin3a and other epigenetic modifiers such as MeCP2, HDAC1, EZH2 and LSD1 have been identified as Tet1-interacting proteins using proximity ligation in situ assays (32). However, it is challenging to link these particular interactions, including the Sin3a–Tet1 interaction, to specific biological functions.
In our study, we found that knockdown of Sin3a not only impaired ESC self-renewal but also skewed the differentiation of ESCs toward mesendoderm lineage. Mechanistically, Sin3a regulated Nodal signaling and promoted Lefty1 transcription via Tet1-mediated DNA de-methylation. Moreover, we found that Sin3a interacted with Tet1 to facilitate the transcription of a set of their co-target genes across the mouse genome, suggesting that activation of gene expression is a universal function of Sin3a–Tet1 interaction in mouse ESCs.
MATERIALS AND METHODS
Cell culture and in vitro differentiation of ESCs
The mouse ESC line E14 was provided by the Cell Bank of Shanghai Institute for Biological Sciences, Chinese Academy of Sciences. Cells were maintained on plates coated with 0.1% gelatin in high glucose Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 15% fetal bovine serum (Gibco), 1% penicillin and streptomycin (Gibco), 2 mM l-glutamine (Thermo), 100 mM nonessential amino acids (NEAA; Thermo), 100 μM β-mercaptoethanol (Gibco) and leukemia inhibitory factor (1000 U/ml LIF; Millipore). For retinoic acid (RA)-induced differentiation, cells were maintained in culture media without LIF, and 100 nM RA (Sigma) added.
Real-time quantitative PCR
Total RNA was isolated from the cells using TRIzol reagent (Invitrogen). 1 μg of total RNA was reverse transcribed using PrimeScriptTM RT reagent kit (TaKaRa). Real-time quantitative PCR (RT-qPCR) was performed using SYBR Green reagent (Bio-Rad) in the Stratagene Mx3000 QPCR system (Stratagene). The results were normalized to the Gapdh gene. The primers used for RT-qPCR are listed in Supplementary Table S1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (33). Briefly, Sin3a knockdown and control ESCs were dissociated into single cells and cross-linked with 1% formaldehyde for 10 min at room temperature, followed by quenching with 0.125 M glycine. Samples were lysed after two washes with PBS and sonicated to generate DNA fragments of 100–1000 bp. Then, the chromatin fragments were immunoprecipitated overnight with specific antibodies at 4°C. After dissociation from the immunocomplexes, the immunoprecipitated DNA was quantified by qPCR and normalized against the genomic DNA input prepared before immunoprecipitation. The primers used in ChIP-qPCR are listed in Supplementary Table S2.
Bisulfite amplicon sequencing analysis
Promoter CpG methylation was analyzed by bisulfite PCR as previously described (34). Briefly, genomic DNA was treated with RNase A (NEB) and extracted using a genomic DNA extraction kit (TIANGEN). A total of 1 μg genomic DNA was then used for DNA methylation analysis using the bisulfite PCR primers listed in Supplementary Table S3. Nested PCR products were cloned into the pMD19-T vector (TaKaRa) and then sequenced. The percent methylation at each CpG site was derived from sequencing 10–15 clones.
Western blotting
For western blotting, cells were harvested and lysed with protease inhibitor-containing RIPA buffer. Protein concentrations were standardized using a Pierce BCA Protein Assay Kit (Thermo Scientific). 15–30 μg protein was resolved by SDS-PAGE, transferred to PVDF membranes (Perkin Elmer Life Sciences) and probed with primary antibodies. Gapdh was used as a loading control. After incubation with the appropriate secondary antibodies, signals were visualized by enhanced chemiluminescence (ECL) (ImageQuant LAS 4000 mini).
Immunoprecipitation
Immunoprecipitation assays were performed as described previously (35). Briefly, ESCs and HEK293FT cells were collected and lysed with lysis buffer (1% Triton X-100 in 50 mM Tris–HCl at pH 7.4 containing 150 mM NaCl, 2 mM Na3VO4, 100 mM NaF and protease inhibitors). For endogenous immunoprecipitation of ESCs, 1 mg cell lysate was incubated overnight with specific antibodies at 4°C followed by 4 h of incubation with a 1:1 mixture of Ezview Red Protein A Affinity Gel (Sigma, P6486) and Ezview Red Protein G Affinity Gel (Sigma, E3403). For exogenous immunoprecipitation of HEK293FT cells, 1 mg cell lysate was incubated with Ezview Red Anti-FLAG M2 affinity gel (Sigma, F2426) or Ezview Red Anti-HA affinity gel (Sigma, E6779) at 4°C for 4 h. Immunoprecipitations were submitted to western blotting after adequate washing.
Statistical analysis
GraphPad Prism 4.0 (GraphPad) was used for the data analysis. All statistical data were presented as the mean ± S.E.M. of at least three independent experiments. Statistical significance was determined using unpaired two-tailed Student's t-tests. P < 0.05, P < 0.01 or P < 0.001 was considered to be statistically significant. No statistical methods were used to predetermine the sample size. The investigators were not blinded to allocation and experimental outcome.
RESULTS
Knockdown of Sin3a impairs ESC self-renewal and skews differentiation toward mesendoderm lineage
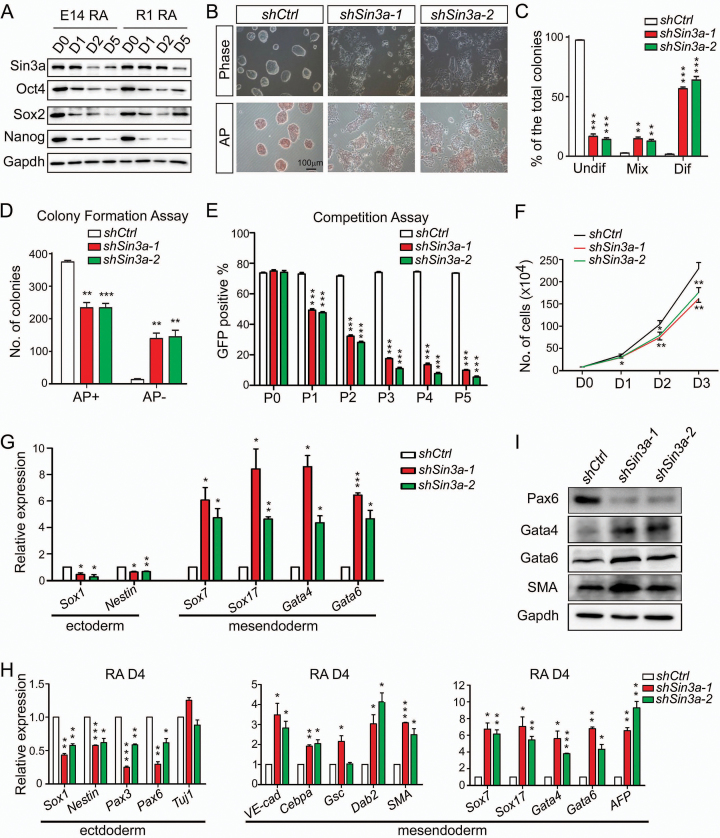
During RA-induced (100 nM) ESC differentiation, Sin3a protein levels showed marked reduction (Figure 1A). To explore a potential role of Sin3a in mouse ESCs, we designed two shRNAs against Sin3a. Compared to cells infected with lentivirus expressing non-targeting shRNA (shCtrl) as control, the two Sin3a knockdown ESC lines (shSin3a-1 and shSin3a-2) showed more than a 70% decrease in both Sin3a mRNA and protein levels (Supplementary Figure S1A and S1B). Knockdown of Sin3a resulted in obvious cell morphology changes and, more importantly, decreased alkaline phosphatase (AP) activity, indicative of ESC differentiation (Figure 1B and C). Furthermore, ESC colony formation and fluorescence-based competition assays were used to evaluate ESC self-renewal capability (17,36). We found that knockdown of Sin3a markedly reduced the capacity of ESC colony formation (Figure 1D). In addition, we co-cultured shCtrl, shSin3a-1 and shSin3a-2 transduced GFP-positive (GFP+) ESCs with wild-type ESCs (GFP-) in a 4:1 ratio in the presence of LIF. The percentage of GFP+ cells was measured at every passage ending with passage five. Two Sin3a knockdown ESCs led to a decrease in the expansion of ESCs and lower GFP levels than that of control ESCs (Figure 1E). Mouse ESCs derived from Sin3a-/- blastocysts were previously reported to undergo cell cycle arrest at G2/M phase followed by apoptosis (37). In contrast, we observed a decreased proliferation ability following Sin3a knockdown (Figure 1F) accompanied by an extended G1 phase and a reduced S phase (Supplementary Figure S1C and D), with no significant increase in apoptosis (Supplementary Figure S1E–G), which was in agreement with a recently published study (38). These differences could potentially be explained by incomplete inactivation of Sin3a in the present study. Sin3a knockdown ESCs still expressed some ESC markers, such as Oct4, Sox2, Nanog, Tbx3 and Esrrb (Supplementary Figure S1H–J), indicating that they maintained at least some aspects of an undifferentiated state. To better understand the pattern for marker gene expression in Sin3a knockdown cells, we used RT-qPCR to measure changes in early differentiation marker expression, and found that knockdown of Sin3a resulted in selective up-regulation of mesendoderm lineage marker genes under self-renewing conditions (Figure 1G). Taken together, these results suggested that Sin3a normally functions to maintain ESC self-renewal ability. To further characterize the effect of prolonged sh-Sin3a on mouse ESC developmental potential, we cultured Sin3a knockdown and control ESCs under RA-induced differentiation conditions. RT-qPCR and western blot analyses revealed that mesendoderm marker genes were generally up-regulated, while the great majority of ectodermal marker genes were down-regulated at day 4 following RA treatment after Sin3a knockdown (Figure 1H and I). To investigate the effect of Sin3a knockdown on ESC differentiation potential in vivo, we injected control and two Sin3a knockdown ESC lines subcutaneously into immune-deficient mice (NOD-SCID) and evaluated the subsequent teratoma formation. Consistent with the results obtained from the cell culture experiments, Sin3a knockdown teratomas also exhibited skewed differentiation toward mesendoderm lineage, compared to control teratomas (Supplementary Figure S2A and B). Collectively, our data suggest that Sin3a is required for ESC self-renewal and that down-regulation of Sin3a skewed differentiation toward mesendoderm.
Figure 1.
Knockdown of Sin3a impairs ESC self-renewal and skews differentiation toward mesendodermal fate. (A) Western blot analysis for Sin3a, Oct4, Sox2 and Nanog expression changes during 4 days of RA-induced ESC differentiation. (B) Cell morphology changes and AP staining after 4 days of Sin3a knockdown. (C) Statistical assay for the percentages of undifferentiated (Undif), mixed (Mix) and differentiated (Dif) ESCs described in (B). (D) Colony formation assay with AP staining following Sin3a knockdown. (E) Competition assay with a FACS plot showing the percentages of GFP positive (GFP+) ESCs that were respectively infected with lentivirus expressing shSin3a-1, shSin3a-2 or control shRNA. (F) Growth curve of ESCs following Sin3a knockdown. (G) RT-qPCR analysis of several early differentiation markers after 4 days of Sin3a knockdown under self-renewing conditions. (H, I) RT-qPCR (H) and western blot (I) analyses of various lineage markers at day 4 of RA-induced differentiation from Sin3a knockdown ESCs. Ctrl: control (and similarly hereafter). Data are representative of one experiment with at least three independent biological replicates. Data in (C–H) represent mean ± S.E.M. *P< 0.05, **P< 0.01, ***P< 0.001 versus the control, Student's t-test (n = 3).
Sin3a regulates Nodal signaling and Lefty1 serves as a direct target of Sin3a in mouse ESCs
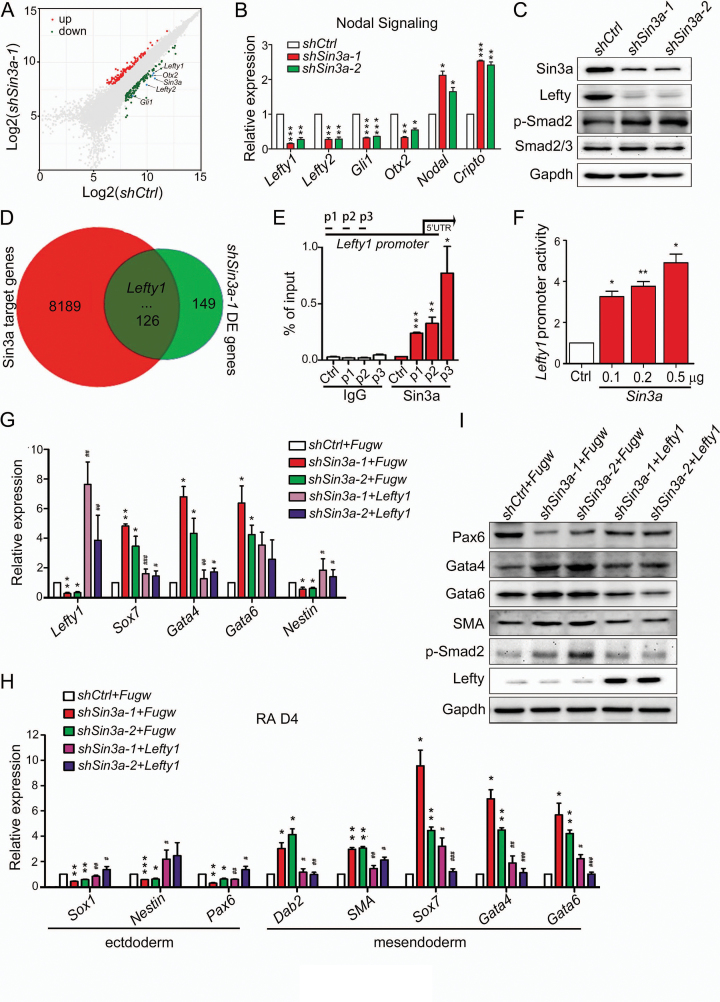
To investigate how Sin3a regulates ESC pluripotency, we compared gene expression differences between shSin3a-1 and control ESCs with mouse gene expression microarrays. Scatterplot analysis of differentially expressed genes (DE genes) revealed that key components of the Nodal pathway (Lefty1, Lefty2, Gli1 and Otx2) were remarkably down-regulated in shSin3a-1 ESCs (Figure 2A and Supplementary Table S4). Changes in the expression levels of key Nodal signaling genes were verified by RT-qPCR and western blot experiments (Figure 2B and C). Specifically, Sin3a knockdown caused up-regulation of Smad2 phosphorylation (Figure 2C and Supplementary Figure S3A), suggesting that decreasing Sin3a levels activated Nodal signaling in mouse ESCs. The Nodal signaling pathway is known to play important roles in the maintenance of ESC pluripotency, especially subsequent mesendodermal lineage specification (7,10). We thus hypothesized that Sin3a knockdown ESCs displayed impaired self-renewal and skewed differentiation toward mesendoderm due to Nodal signaling pathway activation. To test this hypothesis, Sin3a knockdown ESCs were treated with the Nodal signaling inhibitor (SB431542, 20μM) under self-renewing conditions. As expected, we found that SB431542 treatment reversed the effect of Sin3a knockdown, including weaker AP activity (Supplementary Figure S3B and S3C) and increased expression of mesendodermal lineage genes (Supplementary Figure S3D). Thus, inhibition of Sin3a promoted increased Nodal signaling in mouse ESCs. To identify which Nodal pathway components are directly regulated by Sin3a, we analyzed publicly available Sin3a ChIP-seq data of ESCs (30) and differentially expressed genes in shSin3a-1 ESCs (shSin3a-1 DE genes). Among the overlapped genes from two datasets, Lefty1 stands out as a strong candidate for direct downstream effector of Sin3a (Figure 2D and Supplementary Table S5). To confirm this, we performed chromatin immunoprecipitation quantitative PCR (ChIP-qPCR) assay with a Sin3a antibody and observed direct occupancy of the Sin3a protein at the promoter of Lefty1 (Figure 2E). Furthermore, we carried out dual-luciferase reporter (TLR) assay with the vector containing the promoter region of Lefty1 spanning Sin3a binding site identified through Sin3a ChIP-seq data. As expected, overexpression of Sin3a promoted TLR signal-induced luciferase activity of the Lefty1 promoter (Figure 2F). We further performed rescue experiments by introducing exogenous Lefty1 in Sin3a knockdown ESCs (Figure 2G and Supplementary Figure S3E). Although the decreased AP activity of Sin3a knockdown ESCs could not be rescued by expressing exogenous Lefty1 (Supplementary Figure S3F), increased mesendoderm marker (Sox7, Gata4) and decreased ectoderm marker (Nestin) expression were partially rescued in Sin3a knockdown ESCs post Lefty1 overexpression day 4 under self-renewing conditions (Figure 2G). Following RA treatment for 4 days, Sin3a knockdown ESCs with ectopic Lefty1 displayed down-regulation of Smad2 phosphorylation accompanied by decreased mesendoderm marker expression and up-regulation of ectoderm markers (Figure 2H and I). Together, our results demonstrated that the Nodal antagonist Lefty1 served as a direct target of Sin3a in maintaining ESC pluripotency.
Figure 2.
The Nodal antagonist Lefty1 serves as a direct target of Sin3a. (A) Dot plots representing the mRNA expression changes following Sin3a knockdown relative to control. The genes Lefty1, Lefty 2, Gli1, Otx2 and Sin3a are highlighted. (B) RT-qPCR analysis of key Nodal signaling genes following Sin3a knockdown. (C) Western blot analysis of Sin3a, Lefty, p-Smad2 and Smad2/3 after 4 days of Sin3a knockdown. The Lefty antiserum recognizes both Lefty1 and Lefty2. (D) Venn diagram of the overlap between Sin3a target genes and shSin3a-1 DE genes. Lefty1 is highlighted. (E) ChIP-qPCR analysis of Sin3a enrichment at the promoter of Lefty1. The primers (p1, p2 and p3) used were designed at the promoter region of Lefty1 spanning the Sin3a binding site identified from the Sin3a ChIP-seq data as shown in (Figure 3E), and the primer to sequences that are adjacent (∼0.5 kb) to Sin3a binding regions served as a negative control. (F) Luciferase activities in the lysates of ESCs transfected with the Lefty1 promoter reporter (0.2 μg Lefty1-luc) and the indicated plasmid (0.1 μg, 0.2 μg or 0.5 μg Fugw-Sin3a). The blank Fugw vector was co-transfected with Lefty1-luc as control. (G) RT-qPCR analysis of Lefty1 and several early differentiation markers upon expressing ectopic Lefty1 in Sin3a knockdown ESCs under self-renewing conditions. (H) RT-qPCR analysis of lineage markers at day 4 of RA-induced differentiation from Sin3a knockdown ESCs expressing ectopic Lefty1. (I) Western blot analysis for Pax6, Gata4, Gata6, SMA, p-Smad2 and Lefty expression changes in cells described in (H). Data in (B) and (E–H) represent mean ± S.E.M. *P< 0.05, **P< 0.01, ***P< 0.001 versus the control; #P< 0.05, ##P< 0.01, ###P< 0.001 versus the Sin3a-inhibited Fugw blank vector group. Student's t-test (n = 3).
Sin3a coordinates with Tet1 to de-methylate the Lefty1 promoter
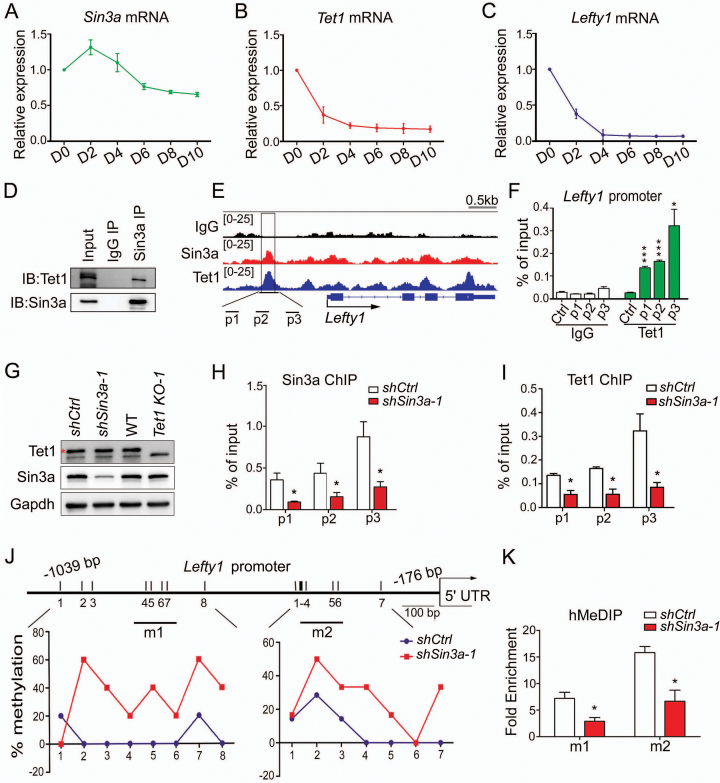
Previous studies have reported that down-regulation of Tet1 in mouse ESCs resulted in diminished expression of Lefty1 and hyperactive Nodal signaling, consequently skewing differentiation toward mesendodermal fates (34). We utilized an efficient paired gRNA-guided CRISPR/Cas9 knockout (paired-KO) strategy (39) to deplete Tet1 in ESCs (Supplementary Figure S4A). We confirmed the biallelic genomic DNA deletion by genomic PCR (Tet1 KO-1 and Tet1 KO-2) and the complete depletion of Tet1 protein by western blot (Supplementary Figure S4B). Consistent with previous studies (14,34), loss of Tet1 did not decrease AP activity of ESCs maintained in LIF, nor affected the expression of key pluripotency factors Oct4 and Nanog (Supplementary Figure S4C and E). RT-qPCR analysis of several early differentiation markers demonstrated that knocking out Tet1 resulted in selective up-regulation of mesendoderm lineage markers in ESC culture conditions (Supplementary Figure S4D). Furthermore, we observed the deregulated genes in Tet1 knockout ESCs encoded key members of the Nodal signaling pathway, including reduced expression levels of Lefty1/Lefty2 and up-regulation of Nodal (Supplementary Figure S4E and F). Collectively, loss of Tet1 resembles the sh-Sin3a phenotype in terms of Nodal pathway gene regulation and mesendoderm specification. Notably, Tet1 exhibited a similar expression pattern relative to Lefty1 during ESC differentiation by embryoid body (EB) formation, while the expression pattern of Sin3a differed from that of Lefty1 (Figure 3A–C). Consistent results were obtained from RT-qPCR and western blot experiments on undifferentiated ES and differentiated MEF cells (Supplementary Figure S4G and H). These results raised the possibility that the regulation of Lefty1 by Sin3a might depend on Tet1 in the context of ESCs. Consistent with this co-operation mode, the Tet1 protein was previously reported to interact with Sin3a (30). We confirmed this by showing that an antibody against endogenous Sin3a could precipitate Tet1 in mouse ESCs (Figure 3D). To explore how Sin3a interacts with Tet1 to regulate Lefty1, we examined Sin3a and Tet1 ChIP-seq data of ESCs from public databases (30) and observed co-occupancy of Sin3a and Tet1 at the Lefty1 promoter (Figure 3E). ChIP-qPCR analysis confirmed that the Sin3a binding DNA regions at the Lefty1 promoter perfectly overlapped with that of the Tet1 binding (Figures 2E and 3F). The functional significance of Tet1 binding at the Lefty1 promoter may be through promoter methylation. Accordingly, previous studies have reported that the Lefty1 promoter is hypo-methylated in ESCs and hyper-methylated in differentiated cells and that Tet1 maintains the Lefty1 promoter in a hypo-methylated state by facilitating DNA de-methylation (34,40). Since Tet1 does not recognize specific target gene sequences (31), we hypothesized that Sin3a, a known scaffold protein, might help Tet1 binding to the Lefty1 promoter. Western blot analysis suggested that Tet1 and Sin3a did not affect each other's expression (Figure 3G). ChIP-qPCR against Sin3a and Tet1 in Sin3a knockdown ESCs revealed that Sin3a down-regulation coincided with loss of both Sin3a and Tet1 binding at the Lefty1 promoter (Figure 3H and I). This was not due to down-regulation of Tet1 expression levels in Sin3a knockdown ESCs (Figure 3G). As expected, Sin3a knockdown ESCs showed an increase in CpG methylation levels at specific regions of the 1.0 kb Lefty1 promoter region, compared to control ESCs in which the locus was hypo-methylated (Figure 3J). Since conventional bisulfite sequencing failed to distinguish 5mC and 5hmC (41,42), hydroxyl-methylated DNA immunoprecipitation (hMeDIP) was further employed to show that knockdown of Sin3a resulted in decreased 5hmC levels at the Lefty1 promoter (Figure 3K). In conclusion, Sin3a–Tet1 interaction is required for Sin3a-dependent Lefty1 expression.
Figure 3.
Sin3a coordinates with Tet1 to de-methylate the Lefty1 promoter. (A–C) RT-qPCR analysis of the mRNA expression patterns of Sin3a (A), Tet1 (B) and Lefty1 (C) during EB formation. (D) Endogenous interaction between Sin3a and Tet1 in ESCs under self-renewing conditions. (E) Genome browser view of ChIP-seq peaks for Sin3a and Tet1 at the promoter of Lefty1 in ESCs. The black box indicates the Sin3a and Tet1 co-localization peaks. (F) ChIP-qPCR analysis of Tet1 enrichment at the promoter of Lefty1. (G) Western blot analysis of Sin3a and Tet1 expression levels following Sin3a knockdown or Tet1 depletion in ESCs. The red asterisk indicates specific bands. (H, I) The effect of Sin3a knockdown on Sin3a (H) and Tet1 (I) occupancies at the promoter of Lefty1. (J) Bisulfite sequencing analysis of CpG methylation status at the Lefty1 promoter following Sin3a knockdown. The average percentage methylation at each CpG site is derived from sequencing 10–15 clones. The black lines (m1 and m2) indicate hMeDIP-qPCR amplicons for (K). (K) hMeDIP-qPCR analysis of 5hmC level changes at the promoter of Lefty1 after Sin3a knockdown. The fold enrichments are relative to IgG controls after normalizing to the input. Data in (F, H, I and K) represent mean ± S.E.M. *P< 0.05, ***P< 0.001 versus the control, Student's t-test (n = 3).
Identification of the domain(s) and key amino acid residues of Sin3a for Tet1 binding
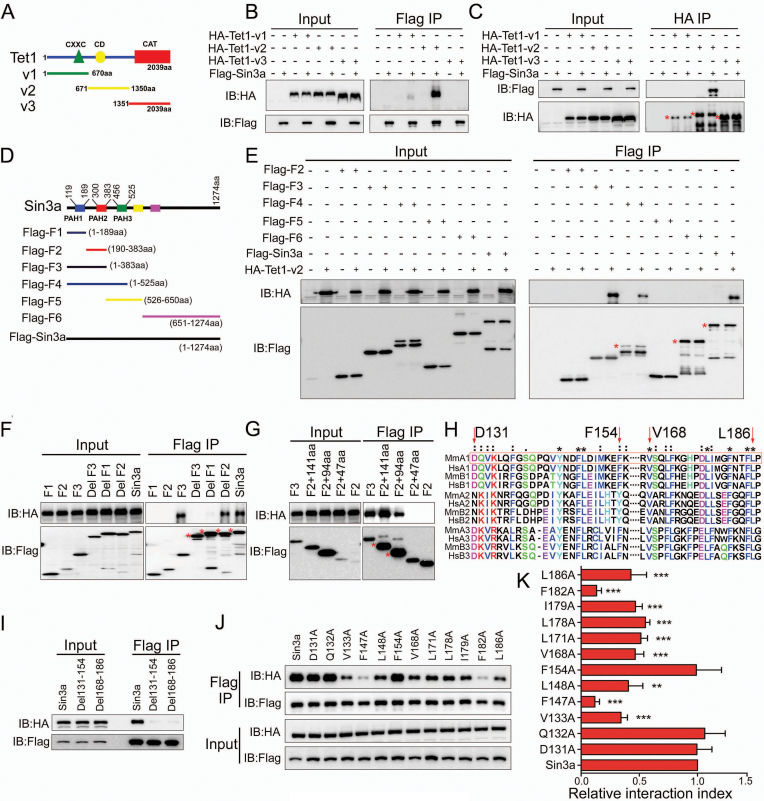
To test whether Sin3a–Tet1 interaction is direct and identify protein domains involved in binding, we performed immunoprecipitations (IP) of Flag-tagged Sin3a mutants and HA-tagged Tet1 truncations in HEK293FT cells. We first constructed three fragment plasmids of Tet1, described as HA-Tet1-v1, HA-Tet1-v2 and HA-Tet1-v3, which respectively contained CXXC domain (CXXC), cysteine-rich domain (CD) or catalytic domain (Cat) (Figure 4A). The reciprocal IP (Flag IP and HA IP) of wild-type Sin3a and truncated Tet1 mutations showed that wild-type Sin3a only interacts with HA-Tet1-v2 (Figure 4B and C). In order to identify what regions of Sin3a interact with Tet1, serial truncations of Sin3a were generated according to the predicted structure of Sin3a (Figure 4D) to evaluate their capability of interaction with HA-Tet1-v2. We found that the Flag-F3 (1–383 aa) truncation of Sin3a can interact with HA-Tet1-v2 (Figure 4E). The F1 (1–189 aa) deletion mutant (DelF1) greatly impaired Sin3a interaction with HA-Tet1-v2, while the F2 (189–383 aa) deletion mutant (DelF2) did not, suggesting that the F1 fragment, which contains the PAH1 domain of Sin3a, is critical for HA-Tet1-v2 binding (Figure 4F). Additional IP experiments demonstrated that the C-terminal 94 amino acid residues of the F1 fragment (96–189 aa) which spans the PAH1 domain (119–189 aa) was responsible for its interaction with HA-Tet1-v2 (Figure 4G). As the entire PAH1 domain is conserved among Sin3a orthologues from different species (26,43), we aligned the PAH domains of mouse and human Sin3 homologues (Sin3a and Sin3b) using ClustalX and focused on two relatively conserved motifs (Asp131-Phe154, Val168-Leu186) in the mouse PAH1 domain (Figure 4H). Further IP analysis showed that Sin3a mutants with either Asp131-Phe154 (Del131–154) or Val168-Leu186 (Del168–186) amino acid residues deleted were unable to precipitate HA-Tet1-v2 (Figure 4I). These results implied that the two relatively conserved regions described above are both essential for the interaction between Sin3a and HA-Tet1-v2. To identify the essential amino acid resides in Sin3a for such interaction, we examined the roles of some fully and semi conserved amino acid residues in the Asp131-Phe154 and Val168-Leu186 motifs (Figure 4H) by mutational analysis. We first constructed Flag-tagged versions of mutant Sin3a proteins carrying individual point mutations, which mutated these fully conserved or semi conserved individual amino acids to the polar uncharged amino acid Ala. After co-expression of these individual point mutations with HA-Tet1-v2 in HEK293FT cells, we found that some of the mutations (V133A, F147A, L148A, V168A, L171A, L178A, I179A, F182A and L186A) impaired the interaction between Sin3a and HA-Tet1-v2. In particular, two fully conserved amino acid mutations (F147A, F182A) significantly interrupted the Sin3a–Tet1 interaction (Figure 4J and K). In summary, we identified the PAH1 domain of Sin3a and two essential amino acid residues (Phe147, Phe182) that are critical for Tet1 binding.
Figure 4.
Identification of the domain(s) and key amino acid residues of Sin3a for Tet1 binding. (A) Schematic of the different truncated Tet1 mutants used in this study. (B, C) The reciprocal IP was performed using either Flag beads (B) or HA beads (C) in HEK293FT cells. HEK293FT cells were co-transfected with Flag-tagged wild-type Sin3a and the different Tet1 mutants described in (A). (D) Schematic of the different truncated Sin3a mutants used in this study. (E–G) IP of the Flag-tagged wild-type Sin3a and truncated Sin3a mutants. (H) ClustalX generated alignments of the PAH domains of putative Sin3 homologues (prefix abbreviations: Mm: M. musculus; Hs: H. sapiens; A1/2/3: Sin3a PAH1/2/3; B1/2/3: Sin3b PAH1/2/3). The red box denotes the PAH1 domain of mouse Sin3a. (*) represents completely conservative, and (:) represents semi conservative. (I) IP of the Flag-tagged wild-type Sin3a, Del131–154 and Del168–186. (J) IP of the Flag-tagged wild-type Sin3a and the individual point mutations in Sin3a. (K) Quantification of the relative HA protein levels normalized to Flag expression described in the Flag IP group of (J) using AlphaView-Fluor Chem FC3 software. **P< 0.01, ***P< 0.001 versus Sin3a, Student's t-test (mean ± S.E.M., n = 3). All Sin3a mutants were Flag-tagged, and all truncated Tet1 mutants were HA-tagged. The different Sin3a mutants in (E–G, I and J) were co-transfected with HA-Tet1-v2 in HEK293FT cells. Membranes in (B, C, E-G, I and J) were immunoblotted for Flag and HA. The red asterisks indicate specific bands. At least three independent experiments were conducted for the immunoprecipitations described above.
Sin3a–Tet1 protein interaction is required for Sin3a to maintain mouse ESC pluripotency
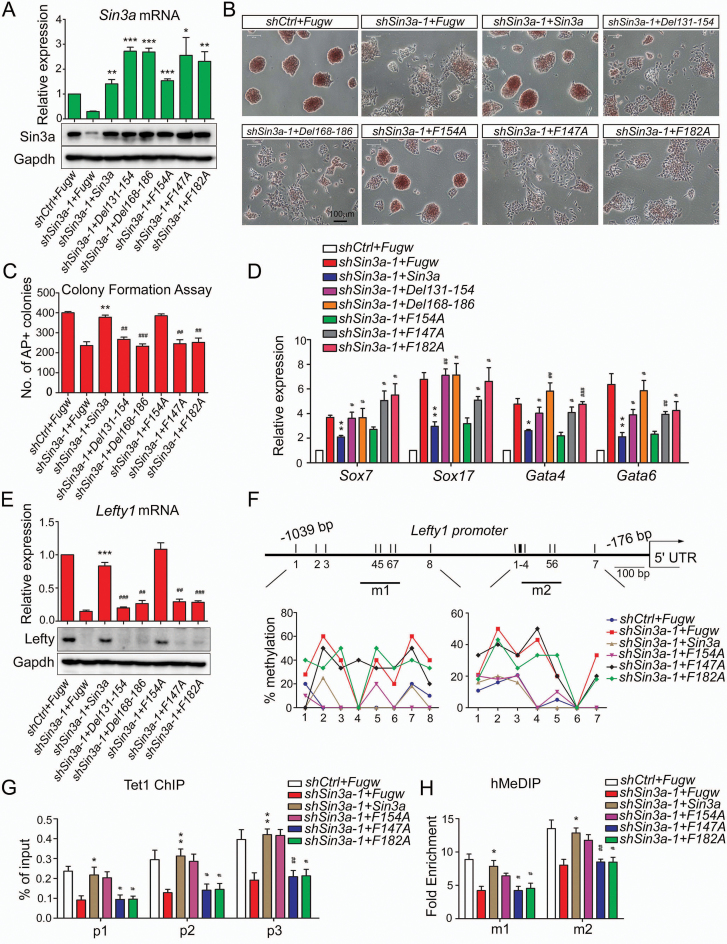
To determine whether the Sin3a–Tet1 interaction is required for Sin3a function in mouse ESCs, we performed rescue experiments using wild-type Sin3a and a series of Sin3a mutants. RT-qPCR and western blot experiments first confirmed that exogenous wild-type Sin3a and a series of Sin3a mutants (Del131–154, Del168–186, F154A, F147A and F182A) were expressed in shSin3a-1 ESCs at comparable levels (Figure 5A and Supplementary Figure S5A). As expected, wild-type Sin3a and the F154A mutant could both rescue the Sin3a knockdown phenotype, including weaker AP activity, decreased proliferation/colony formation ability and up-regulation of mesendoderm marker genes, while a series of Sin3a mutants (Del131–154, Del168–186, F147A and F182A) impairing Sin3a–Tet1 interaction failed (Figure 5B–D and Supplementary Figure S5B). We next examined the effects of ectopic expression of wild-type Sin3a and these mutant Sin3a proteins on its direct target Lefty1 in shSin3a-1 ESCs. Wild-type Sin3a and the F154A mutant both restore the sh-Sin3a effect, including reduced Lefty1 expression, increased CpG methylation levels, decreased Tet1 enrichment and 5hmC levels at the Lefty1 promoter, while a series of Sin3a mutants impairing Sin3a–Tet1 interaction did not (Figure 5E–H). Consistent results were obtained from the rescue experiments using a Tet-inducible shRNA system (44), as shown by decreased Sin3a expression, weaker AP activity, decreased colony formation ability and down-regulation of Lefty1/2 (Supplementary Figure S5C–I). Taken together, these results further confirmed that Sin3a–Tet1 interaction is required for Sin3a activity in ESCs.
Figure 5.
Sin3a–Tet1 protein interaction is required for Sin3a to maintain ESC pluripotency. (A) RT-qPCR and western blot analyses of ectopic expression of wild-type Sin3a and its mutants (Del131–154, Del168–186, F154A, F147A and F182A).The shSin3a-1-treated ESCs were co-infected with virus of Fugw, wild-type Sin3a or its mutants under self-renewing conditions. Wild-type Sin3a and all the mutants we used here were introduced as nonsense mutations against shSin3a-1 target. (B–D) The effects of expressing ectopic wild-type Sin3a or its mutants described in (A) on Sin3a knockdown phenotype. AP staining (B), colony formation assay (C) and RT-qPCR analysis of several early differentiation markers (D) were performed under self-renewing conditions. (E) The effects of ectopic expression of wild-type Sin3a and its mutants on the expression of Lefty1 in shSin3a-1 ESCs. (F–H) The effects of ectopic expression of wild-type Sin3a and its mutants (F154A, F147A and F182A) on CpG methylation levels (F), Tet1 occupancy (G) and 5hmC level changes (H) at the Lefty1 promoter in shSin3a-1 ESCs. The fold enrichments in (H) are relative to IgG controls after normalizing to the input. The shSin3a-1-treated ESCs that were co-infected with Fugw virus were used as control. *P< 0.05, **P< 0.01, ***P< 0.001 versus the control; #P< 0.05, ##P< 0.01, ###P< 0.001 versus the Sin3a-overexpressed shSin3a-1 group. Student's t-test (mean ± S.E.M., n = 3).
Genome-wide analysis of the activation of Sin3a–Tet1 interaction on gene transcription
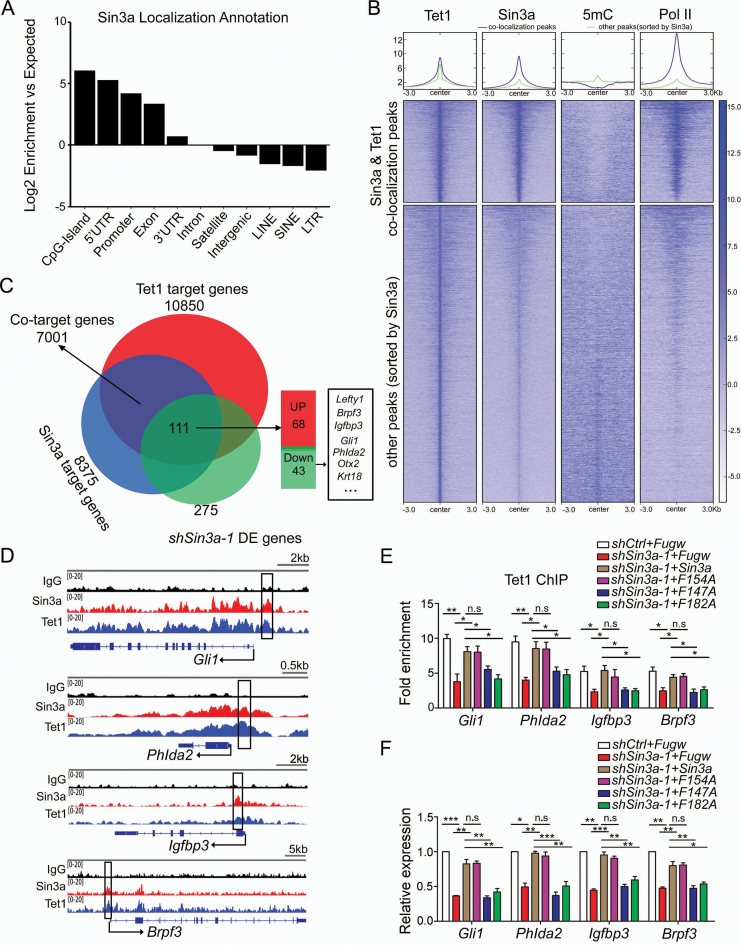
Previous studies have shown that Sin3a, a scaffolding protein, serves as a core component of the Sin3a–HDAC corepressor complex (20,24,25). In this study, we found that Sin3a also interacted with Tet1 to maintain gene (Lefty1) expression via DNA de-methylation in ESCs. Similarly, we confirmed the co-occupancy of Sin3a and Tet1 at an intragenic region of Lefty2 (Supplementary Figure S6A and B), in agreement with previous studies showing that Tet1 is enriched at the transcriptional start sites of CpG rich promoters and gene bodies in ESCs (30,45). ChIP-qPCR and bisulfite amplicon sequencing analyses further revealed that inhibition of Sin3a resulted in loss of Tet1 binding concomitant with an increase in methylation levels at a CpG-rich island of Lefty2 (Supplementary Figure S6C and D), indicating that Sin3a–Tet1 interaction similarly facilitated Lefty2 transcription. To investigate whether the chromosomal occupancy of Sin3a–Tet1 interaction is a general mechanism for promoting gene transcription in ESCs, we performed genome-wide analyses of Sin3a, Tet1 and Pol II ChIP-seq and of 5mC MeDIP-seq in mouse ESCs. Localization of Sin3a and co-localization of Sin3a and Tet1 were enriched at CpG islands, 5′UTR and Promoter, followed by Exon (Figure 6A and Supplementary Figure S6E). Activation of Sin3a–Tet1 interaction on transcription initiation events can be a consequence of lower 5mC and increased entry of Pol II into Sin3a and Tet1 co-binding regions, which are enriched at CpG islands, 5′UTR and promoters. To test this, we categorized Tet1 peaks into two groups according to Sin3a co-localization and ranked them by Sin3a peak intensity. It is noteworthy that lower levels of 5mC and significant enrichment of Pol II were detected around sites of Sin3a and Tet1 co-localization than around other peaks (Figure 6B). These results implied that Sin3a may act together with Tet1 to modulate cytosine methylation dynamics of target DNA regions and facilitate target gene transcription. Venn diagram analysis of shSin3a-1 DE genes overlapping with Sin3a and Tet1 co-target genes revealed that 40% (111) of the shSin3a-1 DE genes (275) were among the co-target genes. We found that knockdown of Sin3a in ESCs led to an approximately equal distribution of up- (68) and down-regulated (43) co-target genes (Figure 6C and Supplementary Table S6). To study how Sin3a–Tet1 interaction promotes the transcription of specific genes, we focused on the 43 down-regulated genes described above (Figure 6C and Supplementary Table S6). To further validate these bioinformatic analyses of the cooperativity between Sin3a and Tet1, we selectively tested several putative target genes with RT-qPCR and confirmed that they were also down-regulated in Tet1 knockout ESCs (Supplementary Figure S6F). As shown in the representative Sin3a and Tet1 binding map, Sin3a peaks were co-localized with Tet1 peaks at the promoter regions of these genes (Figure 6D and Supplementary Figure S6G). ChIP-qPCR analysis further demonstrated that enrichment of Tet1 at the promoter regions of these genes decreased upon Sin3a knockdown (Figure 6E and Supplementary Figure S6H). Finally, we showed that ectopic expression of wild-type Sin3a and the F154A mutant rescued the sh-Sin3a effect, including decreased Tet1 enrichment (Figure 6E) and reduced gene expression (Figure 6F) as shown in the examples provided (Gli1, Phlda2, Igfbp3 and Brpf3), while the Sin3a mutants (F147A and F182A) impairing Sin3a–Tet1 interaction failed. Collectively, these results link Sin3a to Tet1-mediated cytosine methylation modifications in transcriptional activation in mouse ESCs.
Figure 6.
Genome-wide analysis of the activation of Sin3a–Tet1 complex on gene transcription. (A) Annotations of Sin3a ChIP-seq peaks to various genomic features were indicated based on their log2 (fold enrichment) over expected values. (B) Heatmap analysis of published ChIP-seq datasets for Tet1, Sin3a and Pol II and 5mC MeDIP-seq datasets in mouse ESCs. Regions between −3kb and 3kb define boundaries of Tet1 binding sites. (C) Venn diagram of the overlap between Sin3a and Tet1 co-target genes and shSin3a-1 DE genes. (D) Genome browser view of ChIP-seq peaks for Sin3a and Tet1 at the promoters of Gli1, Phlda2, Igfbp3 and Brpf3 genes in ESCs. The black boxes indicate ChIP-qPCR amplicons for (E). (E) ChIP-qPCR analysis of Tet1 enrichment at the promoters of genes (Gli1, Phlda2, Igfbp3 and Brpf3) in shSin3a-1 ESCs expressing ectopic wild-type Sin3a and its mutants. The fold enrichments relative to IgG controls are shown after normalizing to the input. (F) The effects of ectopic expression of wild-type Sin3a and its mutants (F154A, F147A and F182A) on the expression levels of Gli1, Phlda2, Igfbp3 and Brpf3 in shSin3a-1 ESCs. *P< 0.05, **P< 0.01, ***P< 0.001, Student's t-test (mean ± S.E.M., n = 3).
DISCUSSION
Previous studies have reported that the Sin3a–HDAC complex regulates Nanog expression during ESC proliferation (19). Our work showed that knockdown of Sin3a did not affect the expression of the pluripotency genes, such as Oct4, Sox2 and Nanog, but significantly impaired ESC self-renewal and resulted in pronounced mesendoderm skewing during ESC differentiation. Mesoderm is induced from the posterior primitive streak in response to Wnt or low levels of TGF-β/Nodal/Activin signaling, whereas definitive endoderm arises in response to high, sustained Nodal/Activin signaling from ‘mesendoderm’ progenitors in the anterior posterior streak (46). Our study suggests that Sin3a regulates ESC self-renewal and balances differentiation through inhibition of Nodal-Smad2 signaling, as evidenced by reversal of the Sin3a knockdown phenotype by Nodal-Smad2 signaling inhibitor. Lefty1, a well-known Nodal antagonist, blocks Nodal signaling by binding Nodal and its EGF-CFC co-receptors, such as TDGF-1/Cripto (47). Previous studies have found that knockdown of Lefty1 caused up-regulation of Smad2 phosphorylation in ESCs and skewed differentiation towards mesendoderm under differentiating conditions (48). In the present study, we showed that Lefty1 is likely involved in Sin3a regulation, as evidenced by reversal of sh-Sin3a phenotype in terms of mesendoderm skewing by Lefty1 overexpression. However, ectopic expression of Lefty1 failed to restore the sh-Sin3a effect in terms of ESC self-renewing, in line with the normal self-renewal ability of Lefty1 knockdown ESCs (48). The sh-Sin3a phenotype might be deconstructed into the involvement of multiple Nodal signaling genes in addition to Lefty1, such as Lefty2, Gli1 and Otx2. Although the Nodal antagonist Lefty1, highly expressed in the ICM and undifferentiated ESCs (49,50), was identified as an important target of Sin3a, it is more complicated than we understand how Sin3a modulates the intensity of Nodal signaling at the crossroads between self-renewal and differentiation of mESC.
Sin3a is a scaffold protein that regulates downstream genes through interactions with transcription factors and epigenetic modification molecules (26). For example, Sin3a was shown to interact with HDACs, particularly HDAC1, to repress gene transcription (25,26). Interestingly, HDAC1 is a known transcriptional repressor of Lefty1 in ESCs (51). However, our results revealed that Sin3a mainly functions as a transcriptional coactivator for Lefty1, suggesting that the regulation of Sin3a on Lefty1 expression might be independent of HDAC1. Previous studies have indicated that Tet1 may maintain Lefty1 expression by facilitating DNA de-methylation in mouse ESCs, and that knockdown of Tet1 skewed ESC differentiation into mesendoderm lineage, resembling the sh-Sin3a phenotype in terms of mesendoderm specification (34,40). Our results confirmed that Tet1 did promote the transcription of Lefty1 in ESCs. The co-occupancy of Sin3a and Tet1 at the Lefty1 promoter was detected through analysis of public Sin3a and Tet1 ChIP-seq data (30), suggesting that Sin3a may serve as a transcriptional coactivator of the Lefty1 gene by acting with Tet1. Conventional bisulfite sequencing and hMeDIP-qPCR analysis demonstrated that knockdown of Sin3a resulted in increased DNA methylation and decreased DNA hydroxyl-methylation at the promoter of Lefty1. These data suggested that Sin3a promoted hydroxyl-methylation at the Lefty1 promoter by acting together with Tet1, hence facilitating Lefty1 transcription. However, the role and mechanism of Sin3a over transcriptional activation via epigenetic DNA modifications are poorly understood. Our results further indicated that knockdown of Sin3a led to an approximately equal distribution of up- and down-regulation of its direct target genes in ESCs. Previous studies have reported that Sin3a shares a highly overlapping binding profile with Tet1 on a genome-wide scale (30,52) and that Tet1 plays an important role in the regulation of gene activation by converting 5mC to 5hmC at CpG islands (30). One interesting discovery from our study is that lower levels of 5mC and significant enrichment of Pol II are detected around Sin3a and Tet1 co-localization than around other Tet1 peaks sorted by Sin3a. Furthermore, our study indicated that inhibition of Sin3a led to a loss of Tet1 recruitment at target genomic regions and reduced transcription of a set of their co-target genes (e.g. Gli1, Phlda2, Igfbp3 and Brpf3). Accumulating evidence suggested that Sin3a associates with Tet1 to de-methylate adjacent genomic regions and facilitates gene transcription. Our work thus revealed a novel regulatory mechanism of how Sin3a functions as a transcriptional coactivator via epigenetic DNA modifications. Like many previously identified Tet1-interacting proteins, such as Mbd3, PcG and OGT (53), Sin3a lacks a specific DNA-recognition capacity, implying that it is more likely to be a ‘functional’ partner than ‘recruiter’ for Tet1. It would be interesting to further explore the associated ‘recruiters’ for Sin3a–Tet1 interaction.
Another important contribution of this study was that we identified the PAH1 domain in Sin3a responsible for Tet1 binding, and our work for the first time systematically studied a serious of individual point mutations (V133A, F147A, L148A, V168A, L171A, L178A, I179A, F182A and L186A) in the PAH1 domain on their role in the interaction between Sin3a and Tet1. We identified two important amino acid residues when mutated (F147A and F182A) that can significantly interrupt Sin3a–Tet1 interaction. The PAH1 domain is highly conserved among Sin3a orthologues from yeast to mammals (26,43), implying the importance of the PAH1 domain in Sin3a during evolution. Structural studies have revealed that PAH1 and PAH2 of mammalian Sin3a share a high degree of similarity, and specifically identify transcription factors (54,55). Although the PAH2 domain has been reported to exhibit conformational heterogeneity that enables Sin3a to interact with diverse proteins (e.g. Mad1, HBP1, Mnt/Rox, Pf1) (23,56,57), the innate ability of the PAH1 domain to recruit specific proteins has been poorly studied. The PAH domains of Sin3a has been generally reported to possess innate specificity for recruiting particular transcriptional repressors (20–22). Meanwhile, the amphipathic helix structural motif has been previously noted in the activation domains of transcriptional activators when bound to their cellular targets (58). This is in line with our result that Sin3a serves as a coactivator dependent on the interaction between its PAH1 domain and Tet1. The structure of PAH1 domain in Sin3a provides a basis for specificity in binding with Tet1 and, ultimately, facilitates gene transcription. Our findings help greatly in dissecting the role of PAH1 domain in the interaction between Sin3a and Tet1. The rescue experiments using these Sin3a mutants further demonstrated that the cooperativity between Sin3a and Tet1 was required for Sin3a function in ESCs, including its regulation on ESC pluripotency and expression of a set of their co-target genes (e.g., Lefty1, Gli1, Phlda2, Igfbp3 and Brpf3). However, we cannot exclude the presence of additional protein interactions that are important for Sin3a activity in ESCs. Recently, researchers found that Sin3a binds together with Fam60a, Tet1 and OGT at H3K4me3-positive promoters in ESCs, and that Fam60a is an important subunit of a variant Sin3a complex that is required to promote ESC pluripotency (38). It should be emphasized that models of Sin3a-Fam60a interaction and Sin3a–Tet1 cooperation are not mutually exclusive. A fundamental question remains on how these cooperative interactions spatially and temporally work together to orchestrate general transcriptional activation in the context of ESCs.
In summary, the roles of Sin3a in regulating mouse ESC self-renewal and mesendodermal specification are being gradually uncovered. Our findings reveal a novel molecular mechanism by which Sin3a interacts with Tet1 to promote gene transcription and ESC pluripotency. The involvement of Sin3a in epigenetic DNA modifications sheds new light on understanding the role of Sin3a–Tet1 interaction in the regulation of diverse biological processes.
DATA AVAILABILITY
The microarray data sets have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE104192.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Prof. Gang Pei for the kind gift of the mouse ESC line R1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology [2016YFA0101300]; National Natural Science Foundation of China [81530042, 31701110, 41476120, 31721003, 31571529, 31571519, 31471250, 31571390, 31771506, 81600675, 31671533]; Ministry of Education Grants [IRT_15R51]; Science and Technology Commission of Shanghai Municipality [15JC1403201]; Fundamental Research Funds for the Central Universities [1500219106, 20002310002, 1515219039, 1515219040]; Shanghai Municipal Medical and Health Discipline Construction Projects [2017ZZ02015]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Evans M.J., Kaufman M.H.. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981; 292:154–156. [DOI] [PubMed] [Google Scholar]
- 2. Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ying Q.L., Nichols J., Chambers I., Smith A.. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003; 115:281–292. [DOI] [PubMed] [Google Scholar]
- 4. Rossant J. Stem cells and early lineage development. Cell. 2008; 132:527–531. [DOI] [PubMed] [Google Scholar]
- 5. Niwa H., Ogawa K., Shimosato D., Adachi K.. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009; 460:118–122. [DOI] [PubMed] [Google Scholar]
- 6. Ng H.H., Surani M.A.. The transcriptional and signalling networks of pluripotency. Nat. Cell. Biol. 2011; 13:490–496. [DOI] [PubMed] [Google Scholar]
- 7. Fei T., Zhu S., Xia K., Zhang J., Li Z., Han J.D., Chen Y.G.. Smad2 mediates Activin/Nodal signaling in mesendoderm differentiation of mouse embryonic stem cells. Cell Res. 2010; 20:1306–1318. [DOI] [PubMed] [Google Scholar]
- 8. Nomura M., Li E.. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998; 393:786–790. [DOI] [PubMed] [Google Scholar]
- 9. Moustakas A., Heldin C.H.. The regulation of TGFbeta signal transduction. Development. 2009; 136:3699–3714. [DOI] [PubMed] [Google Scholar]
- 10. Lee K.L., Lim S.K., Orlov Y.L., Yit le Y., Yang H., Ang L.T., Poellinger L., Lim B.. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet. 2011; 7:e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I.R.. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006; 442:533–538. [DOI] [PubMed] [Google Scholar]
- 12. Kim J., Chu J., Shen X., Wang J., Orkin S.H.. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008; 132:1049–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young R.A. Control of the embryonic stem cell state. Cell. 2011; 144:940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dawlaty M.M., Ganz K., Powell B.E., Hu Y.C., Markoulaki S., Cheng A.W., Gao Q., Kim J., Choi S.W., Page D.C. et al. . Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell. 2011; 9:166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y.. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010; 466:1129–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li X., Li L., Pandey R., Byun J.S., Gardner K., Qin Z., Dou Y.. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012; 11:163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ang Y.S., Tsai S.Y., Lee D.F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B. et al. . Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011; 145:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dannenberg J.H., David G., Zhong S., van der Torre J., Wong W.H., Depinho R.A.. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 2005; 19:1581–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baltus G.A., Kowalski M.P., Tutter A.V., Kadam S.. A positive regulatory role for the mSin3A-HDAC complex in pluripotency through Nanog and Sox2. J. Biol. Chem. 2009; 284:6998–7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ayer D.E., Lawrence Q.A., Eisenman R.N.. Mad-Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995; 80:767–776. [DOI] [PubMed] [Google Scholar]
- 21. Schreiber-Agus N., Chin L., Chen K., Torres R., Rao G., Guida P., Skoultchi A.I., DePinho R.A.. An amino-terminal domain of Mxi1 mediates anti-Myc oncogenic activity and interacts with a homolog of the yeast transcriptional repressor SIN3. Cell. 1995; 80:777–786. [DOI] [PubMed] [Google Scholar]
- 22. Wang H., Clark I., Nicholson P.R., Herskowitz I., Stillman D.J.. The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol. Cell. Biol. 1990; 10:5927–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brubaker K., Cowley S.M., Huang K., Loo L., Yochum G.S., Ayer D.E., Eisenman R.N., Radhakrishnan I.. Solution structure of the interacting domains of the Mad-Sin3 complex: implications for recruitment of a chromatin-modifying complex. Cell. 2000; 103:655–665. [DOI] [PubMed] [Google Scholar]
- 24. Cowley S.M., Kang R.S., Frangioni J.V., Yada J.J., DeGrand A.M., Radhakrishnan I., Eisenman R.N.. Functional analysis of the Mad1-mSin3A repressor-corepressor interaction reveals determinants of specificity, affinity, and transcriptional response. Mol. Cell. Biol. 2004; 24:2698–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Silverstein R.A., Ekwall K.. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 2005; 47:1–17. [DOI] [PubMed] [Google Scholar]
- 26. Kadamb R., Mittal S., Bansal N., Batra H., Saluja D.. Sin3: insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013; 92:237–246. [DOI] [PubMed] [Google Scholar]
- 27. Munoz I.M., MacArtney T., Sanchez-Pulido L., Ponting C.P., Rocha S., Rouse J.. Family with sequence similarity 60A (FAM60A) protein is a cell cycle-fluctuating regulator of the SIN3-HDAC1 histone deacetylase complex. J. Biol. Chem. 2012; 287:32346–32353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ellison-Zelski S.J., Solodin N.M., Alarid E.T.. Repression of ESR1 through actions of estrogen receptor alpha and Sin3A at the proximal promoter. Mol. Cell. Biol. 2009; 29:4949–4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L. et al. . Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009; 324:930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williams K., Christensen J., Pedersen M.T., Johansen J.V., Cloos P.A., Rappsilber J., Helin K.. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011; 473:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Frauer C., Rottach A., Meilinger D., Bultmann S., Fellinger K., Hasenoder S., Wang M., Qin W., Soding J., Spada F. et al. . Different binding properties and function of CXXC zinc finger domains in Dnmt1 and Tet1. PLoS One. 2011; 6:e16627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cartron P.F., Nadaradjane A., Lepape F., Lalier L., Gardie B., Vallette F.M.. Identification of TET1 partners that control Its DNA-demethylating function. Genes Cancer. 2013; 4:235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wei T., Chen W., Wang X., Zhang M., Chen J., Zhu S., Chen L., Yang D., Wang G., Jia W. et al. . An HDAC2-TET1 switch at distinct chromatin regions significantly promotes the maturation of pre-iPS to iPS cells. Nucleic Acids Res. 2015; 43:5409–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G. et al. . Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011; 8:200–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu S., Jing R., Yang Y., Huang Y., Wang X., Leng Y., Xi J., Wang G., Jia W., Kang J.. A motif from Lys216 to Lys222 in human BUB3 protein is a nuclear localization signal and critical for BUB3 function in mitotic checkpoint. J. Biol. Chem. 2015; 290:11282–11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lee D.F., Su J., Sevilla A., Gingold J., Schaniel C., Lemischka I.R.. Combining competition assays with genetic complementation strategies to dissect mouse embryonic stem cell self-renewal and pluripotency. Nat. Protoc. 2012; 7:729–748. [DOI] [PubMed] [Google Scholar]
- 37. McDonel P., Demmers J., Tan D.W., Watt F., Hendrich B.D.. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 2012; 363:62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Streubel G., Fitzpatrick D.J., Oliviero G., Scelfo A., Moran B.. Fam60a defines a variant Sin3a-Hdac complex in embryonic stem cells required for self-renewal. EMBO J. 2017; 36:2216–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Z., Hui Y., Shi L., Chen Z., Xu X., Chi L., Fan B., Fang Y., Liu Y., Ma L. et al. . Efficient CRISPR/Cas9-Mediated versatile, predictable, and Donor-Free gene knockout in human pluripotent stem cells. Stem Cell Rep. 2016; 7:496–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai H.Q., Wang B.A., Yang L., Chen J.J., Zhu G.C., Sun M.L., Ge H., Wang R., Chapman D.L., Tang F. et al. . TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling. Nature. 2016; 538:528–532. [DOI] [PubMed] [Google Scholar]
- 41. Nestor C., Ruzov A., Meehan R., Dunican D.. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. Biotechniques. 2010; 48:317–319. [DOI] [PubMed] [Google Scholar]
- 42. Jin S.G., Kadam S., Pfeifer G.P.. Examination of the specificity of DNA methylation profiling techniques towards 5-methylcytosine and 5-hydroxymethylcytosine. Nucleic Acids Res. 2010; 38:e125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Ingen H., Lasonder E., Jansen J.F., Kaan A.M., Spronk C.A., Stunnenberg H.G., Vuister G.W.. Extension of the binding motif of the Sin3 interacting domain of the Mad family proteins. Biochemistry. 2004; 43:46–54. [DOI] [PubMed] [Google Scholar]
- 44. Wiederschain D., Wee S., Chen L., Loo A., Yang G., Huang A., Chen Y., Caponigro G., Yao Y.M., Lengauer C. et al. . Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle (Georgetown, Tex.). 2009; 8:498–504. [DOI] [PubMed] [Google Scholar]
- 45. Wu H., D’Alessio A.C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y.E., Zhang Y.. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011; 473:389–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schier A.F., Shen M.M.. Nodal signalling in vertebrate development. Nature. 2000; 403:385–389. [DOI] [PubMed] [Google Scholar]
- 47. Cheng S.K., Olale F., Brivanlou A.H., Schier A.F.. Lefty blocks a subset of TGFbeta signals by antagonizing EGF-CFC coreceptors. PLoS Biol. 2004; 2:E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim D.K., Cha Y., Ahn H.J., Kim G., Park K.S.. Lefty1 and lefty2 control the balance between self-renewal and pluripotent differentiation of mouse embryonic stem cells. Stem Cells Dev. 2014; 23:457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adjaye J., Huntriss J., Herwig R., BenKahla A., Brink T.C., Wierling C., Hultschig C., Groth D., Yaspo M.L., Picton H.M. et al. . Primary differentiation in the human blastocyst: comparative molecular portraits of inner cell mass and trophectoderm cells. Stem Cells. 2005; 23:1514–1525. [DOI] [PubMed] [Google Scholar]
- 50. Sperger J.M., Chen X., Draper J.S., Antosiewicz J.E., Chon C.H., Jones S.B., Brooks J.D., Andrews P.W., Brown P.O., Thomson J.A.. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:13350–13355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu P., Dou X., Liu C., Wang L., Xing C., Peng G.. Histone deacetylation promotes mouse neural induction by restricting Nodal-dependent mesendoderm fate. Nat. Commun. 2015; 6:6830. [DOI] [PubMed] [Google Scholar]
- 52. Zhong J., Li X., Cai W., Wang Y., Dong S., Yang J., Zhang J., Wu N., Li Y., Mao F. et al. . TET1 modulates H4K16 acetylation by controlling auto-acetylation of hMOF to affect gene regulation and DNA repair function. Nucleic Acids Res. 2017; 45:672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu H., Zhang Y.. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014; 156:45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Le Guezennec X., Vermeulen M., Stunnenberg H.G.. Molecular characterization of Sin3 PAH-domain interactor specificity and identification of PAH partners. Nucleic Acids Res. 2006; 34:3929–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sahu S.C., Swanson K.A., Kang R.S., Huang K., Brubaker K., Ratcliff K., Radhakrishnan I.. Conserved themes in target recognition by the PAH1 and PAH2 domains of the Sin3 transcriptional corepressor. J. Mol. Biol. 2008; 375:1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swanson K.A., Knoepfler P.S., Huang K., Kang R.S., Cowley S.M., Laherty C.D., Eisenman R.N., Radhakrishnan I.. HBP1 and Mad1 repressors bind the Sin3 corepressor PAH2 domain with opposite helical orientations. Nat. Struct. Mol. Biol. 2004; 11:738–746. [DOI] [PubMed] [Google Scholar]
- 57. Zhang Y., Zhang Z., Demeler B., Radhakrishnan I.. Coupled unfolding and dimerization by the PAH2 domain of the mammalian Sin3A corepressor. J. Mol. Biol. 2006; 360:7–14. [DOI] [PubMed] [Google Scholar]
- 58. Berk A.J. Activation of RNA polymerase II transcription. Curr. Opin. Cell Biol. 1999; 11:330–335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data sets have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE104192.