Abstract
Detection of microsatellite-instability in colonoscopy-obtained polyps, as well as in plasma-circulating DNA, is frequently confounded by sensitivity issues due to co-existing excessive amounts of wild-type DNA. While also an issue for point mutations, this is particularly problematic for microsatellite changes, due to the high false-positive artifacts generated by polymerase slippage (stutter-bands). Here, we describe a nuclease-based approach, NaME-PrO, that uses overlapping oligonucleotides to eliminate unaltered micro-satellites at the genomic DNA level, prior to PCR. By appropriate design of the overlapping oligonucleotides, NaME-PrO eliminates WT alleles in long single-base homopolymers ranging from 10 to 27 nucleotides in length, while sparing targets containing variable-length indels at any position within the homopolymer. We evaluated 5 MSI targets individually or simultaneously, NR27, NR21, NR24, BAT25 and BAT26 using DNA from cell-lines, biopsies and circulating-DNA from colorectal cancer patients. NaME-PrO enriched altered microsatellites and detected alterations down to 0.01% allelic-frequency using high-resolution-melting, improving detection sensitivity by 500–1000-fold relative to current HRM approaches. Capillary-electrophoresis also demonstrated enhanced sensitivity and enrichment of indels 1–16 bases long. We anticipate application of this highly-multiplex-able method either with standard 5-plex reactions in conjunction with HRM/capillary electrophoresis or massively-parallel-sequencing-based detection of MSI on numerous targets for sensitive MSI-detection.
INTRODUCTION
High levels of microsatellite instability (MSI) are predictive for colorectal cancer (CRC) therapy outcome in chemotherapy and immunotherapy and has been associated with distinct characteristics and favorable results including better prognosis, a higher 5-year survival, and lesser metastasis (1,2). Most CRCs derive from colonic adenomas which are the main precursor for CRC (3) and the current gold standard for adenoma screening is colonoscopy. Early diagnosis of the condition followed by surveillance can reduce the risk of developing CRC (4), especially in Lynch syndrome (LS) where MSI is found in 90% of cases. Adenocarcinoma development in LS often occurs within 1–3 years compared to sporadic cases 8–12 years, indicating that polyps without appropriate diagnosis can become malignant fast (5,6). Since MSI takes place at early stage of adenoma formation (7–10), MSI screening of polyps can be an early indicator of progression, especially in LS patients. In the therapy setting, while tumor testing is the gold standard, a convenient approach to screen for MSI longitudinally during cancer treatment is via circulating DNA (cfDNA, liquid biopsy) using a blood draw, thereby interrogating systemic MSI reflecting primary or secondary (occult) tumor status at the time of blood collection. Furthermore, chemotherapy can induce secondary MSI not present in the primary tumor (11–13) and systemic MSI can be predictive for immunotherapy response (2). Accordingly, real time monitoring of MSI in plasma can be clinically valuable for predictive applications or as a marker for assessment of residual tumor load.
However, detection of MSI in colonoscopy-obtained polyps (8,10), as well as in cfDNA (14), is frequently confounded by sensitivity issues due to co-existing excessive amounts of wild-type DNA (15). Improvements in sensitivity of MSI detection include utilization of long nucleotide repeats that display increased instability as compared to shorter repeats (10). Further, COLD-PCR technology (16–21) has recently been adapted to enrich altered microsatellites and suppress wild-type (WT) alleles for sensitive detection of single microsatellite sequences in the HSP110 microsatellite (22). Despite improvements, the enrichment of mutations via PCR is ultimately limited by polymerase-introduced errors (stutter bands) (23,24) that introduce WT allele changes indistinguishable from genuine indels. Thus, small indels comprising few nucleotide changes unavoidably fall within stutter bands that confound interpretation when capillary electrophoresis is employed for endpoint detection. High resolution melting-based MSI detection enables convenient assessment of MSI (25), but is also PCR-based and liable to stutter artifacts. Similarly, NGS is error-prone when it comes to identifying changes in homopolymers (26). Recent studies have illustrated that mononucleotides are highly mutable with 92.4% and 93% of MSI events in CRC and EC respectively, to be due to MSI in mononucleotide repeats (27,28). Thus, although di-nucleotide and tri-nucleotide repeats are also common in coding sequences, mononucleotides are often preferred as sensitive markers of instability (1). However, the limitations of the current technology for accurate length determination of the homopolymers including mononucleotide repeats are known and well documented (29,30). While bioinformatic approaches significantly reduce polymerase and sequencing errors (31), detection of small indels within large homopolymers remains a problem that limits the otherwise highly promising NGS-based detection of MSI.
Here, we present a new approach for enrichment of altered micro-satellites at the genomic DNA level prior to DNA-amplification, thereby reducing or eliminating the universal influence of stutter artifacts in MSI detection. We recently developed nuclease-assisted minor-allele enrichment with probe-overlap, NaME-PrO (32), a single-step approach that removes WT-DNA at the genomic DNA level and enriches single point mutation-containing alleles. As originally described, NaME-PrO employs a double-strand-DNA-specific nuclease (33) and overlapping oligonucleotide-probes interrogating multiple point mutation targets. Here we demonstrate that, by appropriate design of the overlapping oligonucleotides, NaME-PrO eliminates WT alleles in long single base homopolymers of at least 27 nucleotides length, while sparing targets containing variable-length indels at any position within the homopolymer, Figure 1A. Following DNA denaturation and cooling at ∼60–65°C, the probes form double-stranded regions with their targets, thereby guiding nuclease digestion to the selected sites. Microsatellite indels create ‘bulges’ that inhibit digestion, thus subsequent amplification yields DNA with microsatellite alterations enhanced at multiple targets. The assay is applied at the genomic or circulating-DNA level prior to amplification, thereby avoiding polymerase-introduced ‘stutter bands’ arising from WT DNA. Since NaME-PrO can be highly multiplexed to enrich hundreds (34) of targets simultaneously, it can be used to enrich complete panels of markers and holds unique potential for improved MSI analysis via massively parallel sequencing.
Figure 1.
Concept and workflow of Multiplex-NaME PrO for MSI screening. (A) Following DNA denaturation, NaME-PrO probes bind to their microsatellite targets on multiple sites simultaneously. DSN then digests wild type alleles whereas indel-containing sites remain substantially undigested and can be amplified. (B) NaME-PrO sense and antisense probe design. Poly A/T homopolymers contribute relatively less to the probe melting temperature Tm as compared to other nucleotides. Hence despite the large overlap between sense and antisense probes the Tm of the flanking, non-overlapping nucleotides raises the overall probe Tm and prevents excessive probe-probe binding. An example for microsatellite NR27 is depicted; due to the low Tm of the polyA homopolymer there is a 5°C difference between the probe-probe heterodimer and the probe-DNA target duplex.
MATERIALS AND METHODS
Cell lines and clinical samples
Human genomic DNA from human colon adenocarcinoma cell lines HCT-15, LS174T (Sigma-Aldrich) and Human genomic DNA (Promega) were used as mutant and WT control DNA respectively and serial dilutions of mutant into WT DNA were prepared. Plasma samples obtained from colon cancer patients under consent and Institutional Review Board approval were provided by the Dana Farber Cancer Institute GI Bank. Cell-free circulating DNA (cfDNA) was isolated from plasma using the QIAamp Circulating Nucleic Acid Kit (Qiagen), and the concentration of cfDNA was measured on a Qubit 3.0 fluorometer (Thermo Fisher Scientific) using a dsDNA HS assay (Q32854). Cancer and corresponding normal tissues biopsies were anonymously obtained from the Massachusetts General Hospital Tumor Bank under IRB approval and DNA was extracted using the Blood and Tissue kit (Qiagen).
NaME-PrO protocol
Single-plex NaME-PrO was performed on genomic DNA using serial dilution mixtures of WT and mutant DNA using ∼30–50 ng input per reaction or, alternatively on cfDNA using 10 ng input per reaction, or on tissue DNA from normal colon and colon cancer with DNA input of 30 ng per reaction. The 10-μl NaME-PrO reaction contained genomic DNA, 0.75× DSN buffer, 0.375× GoTaq buffer, and 100 nmol/l each overlapping probe (TGFBR2, HSP110, BAT25, BAT26, NR21, NR24, NR27 listed in Table S1, online Data Supplement). The initial mix in the absence of DSN was prepared on ice and mixed well. Denaturation at 98°C for 2 min was then performed on a mastercycler™ (Eppendorf Nexus). The program was then paused to remove the samples. Tubes were opened, then 0.5 units DSN (Evrogen) were applied on the tube wall, centrifuged for 2 s and immediately placed back on the thermocycler where the program resumed as follows: incubation at 61°C for NR21, NR24, NR27, BAT25 and BAT26 for 20 min followed by 2 min at 95°C for DSN inactivation. Following DSN inactivation, NaME-PrO products were kept on ice for up to 2 h, until used without purification in same-day amplification reactions as described below. No-DSN control samples (no-DSN enzyme) were included and assessed in parallel. For multiplexed NaME-PrO reactions using NR21, NR24, NR27, BAT25 and BAT26 the respective probes were all mixed together and the reactions was performed as described above. Multiplex NaME-PrO was followed by multiplex PCR amplification.
PCR amplification
PCR reactions targeting TGFRB2, HSP110, NR21, NR24, NR27, BAT25 and BAT26 were prepared in a final volume of 25 μl (2 μl of NaME-PrO sample, 1× GoTaq buffer (Promega), 200 nmol/l of each primer, 200 μmol/l of each of the four deoxynucleotide triphosphates (BioLine), 1.25 U of GoTaq Polymerase (Promega), and 0.8× LC Green). The primer sequences for TGFRB2, HSP110,, NR21, NR24, NR27, BAT25 and BAT26 are depicted in Table S2 in the Data Supplement. PCR protocol on a CFX Connect™ real-time PCR machine (Biorad) included an initial denaturation step at 98°C for 2 min followed by 50 cycles of denaturation at 98°C for 10 s, annealing at 55°C (for NR21, NR24, NR27, BAT25 and BAT26) or 58°C (for TGFRB2 and HSP110) for 20 s, and elongation at 72°C for 10 s. The final step included melting curve analysis (0.2°C step increments, 2 or 4 s hold before each acquisition) from 65 to 98°C.
Multiplex PCR following addition of 200 nmol/l of each primer, was performed at Ta = 55°C using the same conditions as described above.
High resolution melting (HRM) analysis
Ten-microliters of PCR product were transferred to a 96-well plate, and 20 μl of mineral oil were added to each well. HRM was performed on a 96-well LightScanner® system (Idaho Technology). The software sensitivity level was set as 1.2 for computing DNA variant groups. All experiments were replicated at least three independent times for assessing the reproducibility of the results. Single-plex and multiplex HRM were performed under the same HRM protocol.
Capillary electrophoresis for microsatellite analysis
PCR reactions targeting TGFRB2, HSP110, NR21, NR24, NR27, BAT25 and BAT26 were prepared in a final volume of 25 μl [2μl of 1 in 100 diluted post-NaME-PrO PCR sample, 1× GoTaq buffer (Promega), 200 nmol/l of each primer (1:1 ratio FAM labeled forward: unlabeled reverse), 200 μmol/l of each of the four deoxynucleotide triphosphates (BioLine), 1.25 U of GoTaq Polymerase (Promega)] using the primers listed in Table S1 in Data Supplement. The reaction was performed on a CFX Connect™ real-time PCR machine. PCR protocol included an initial denaturation step at 98°C for 2 min followed by 25 cycles of denaturation at 98°C for 10 s, annealing at 55°C (for NR21, NR24, NR27, BAT25 and BAT26) 58°C (for TGFRB2 and HSP110) for 20 s, and elongation at 72°C for 10 s. The fluorescent labeled PCR products were subjected to fragment analysis on an Applied Biosystems, Inc. 3130xl Genetic Analyzer. The fragments were separated by capillary electrophoresis on a 36cm capillary array with POP7 polymer. Each sample contained an Internal Lane Standard 600 (ILS 600, Promega, Inc.) composed of 22 carboxy-X-rhodamine-labeled DNA fragments for size determination. The data were analyzed with the assistance of GeneMapper v4.0 software. The panel and bin files were created based on the sizes of the wild type microsatellite marker-containing PCR amplicons.
RESULTS
NaME-PrO probe design for enrichment of microsatellite changes
In the original application of NaME-PrO (32) probes targeting single point mutations were designed to overlap each other by no more than up to 10–15 bases, in order to avoid probe-probe interactions. In contrast, for MSI detection the entire poly-A/T homopolymer is part of both sense and anti-sense probes and the non-overlapping portion of the probes only comprises 3–5 bases on either side. Despite the large sense-antisense probe overlap, the few non-overlapping bases make a significant contribution to the overall probe melting temperature (Tm) since the poly-A portion contributes relatively less to the Tm (Figure 1B). Thereby the design takes advantage of the low-Tm of the A-homopolymer portion, and at NaME-PrO reaction temperatures that approach probe Tm the probes are anticipated to bind preferably to their respective targets as opposed to each other.
BAT25, TGFRB2, HSP110 and BAT26 microsatellite testing: serial dilutions
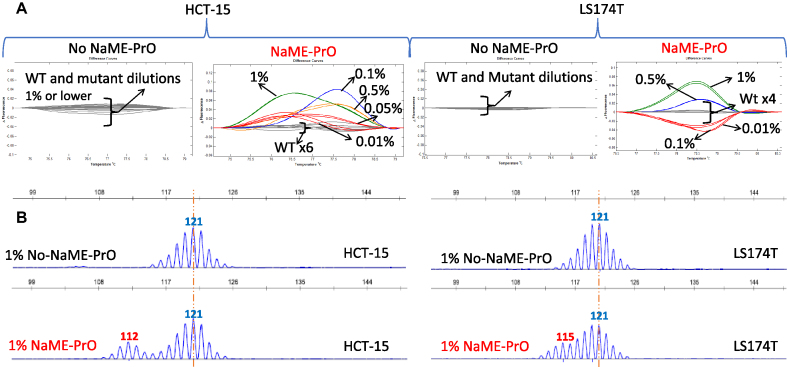
To examine NaME-PrO efficiency in removing WT DNA and enriching indels in microsatellites, we first tested marker BAT25 using NaME-PrO-PCR-HRM in a serial dilution experiment (mutated DNA abundance of 1%, 0.5%, 0.1%, 0.05%, and 0.01%), depicted in Figure 2A. NaME-PrO was performed on ∼50 ng input genomic DNA and assessed on DNA from different cell lines HCT-15, LS174T that are MSI-H and harbor distinct 5–9bp deletions. When NaME-PrO was applied, HRM could identify indels down to 0.01% mutation abundance, Figure 2A. The deletion size in BAT25 for HCT-15 and LS174T was confirmed using capillary electrophoresis, Figure 2B. In contrast, the HRM sensitivity limit without application of NaME-PrO was about ∼10% mutation abundance as shown in Supplementary Figure S1 indicating that NaME-PrO provides an almost 3-order of magnitude improvement in HRM sensitivity. Similarly, NaME-PrO enrichment of homopolymer changes in 10-mer or 17-mer microsatellites was assessed in TGFBR2 and HSP110 respectively, for HCT15 and HCT-116 DNA harboring 2–6 bp deletions in these targets, and HRM sensitivity improvements of 500–100-fold were observed Supplementary Figure S2. Following NaME-PrO, HRM could detect deletions down to 0.01% mutation abundance while the lowest detectable level without NaME-PrO was about 5%. Serial dilutions of cell line to WT DNA were also assessed for BAT26, Supplementary Figure S3, and HRM-based detection down to 0.01% was demonstrated for NaME-PrO-treated samples.
Figure 2.
NaME-PrO application on BAT25 microsatellite in serial dilutions of DNA from MSI-H cell lines. (A). Comparison between NaME-PrO-treated and untreated DNA for BAT25 microsatellites on DNA from colon cancer cell lines HCT15 (left) and LS174T (right) serially diluted into WT DNA, detected via high resolution melting. (B) Capillary electrophoresis analysis for 1% DNA abundance FOR HCT-15 DNA (left) and LS174T DNA (right) (red = variant, blue = WT).
Colon cancer circulating-DNA and cancer testing, BAT25
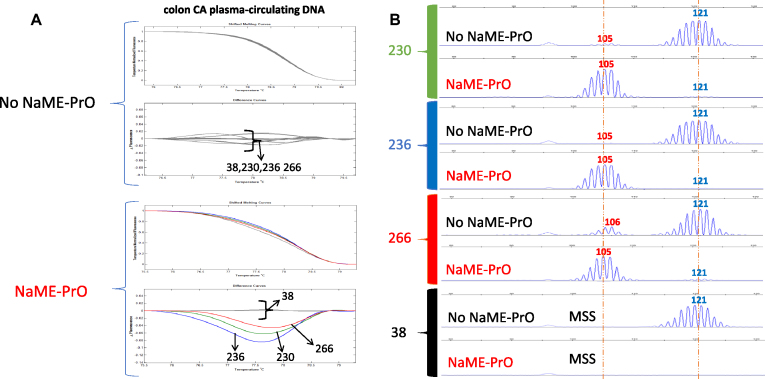
The presence of indels in BAT25 was investigated in clinical samples using plasma circulating DNA cfDNA (10ng input) using blood donated by colon cancer patients. Screening BAT-25 microsatellite marker revealed indels in cfDNA from 4/33 (12%) samples. Three representative samples with deletion (#230, 236 and 266) and a sample from a normal volunteer (#38) are shown on Figure 3. Indel enrichment was identified initially with HRM and capillary electrophoresis confirmed the 16bp deletions. The No-NaME-PrO treated samples display either absence or a minor trace of the deletion in capillary electrophoresis, Figure 3. The data indicate that NaME-PrO increases the ability to detect MSI in plasma.
Figure 3.
NaME-PrO application on BAT25 microsatellite using circulating DNA from colon cancer samples. (A) Comparison between NaME-PrO-treated and untreated screening for BAT25 microsatellites on plasma circulating DNA following HRM analysis: examples from three positive samples (#230, 236, 266) plus cfDNA from a healthy control patient (#38) are shown. (B) Capillary electrophoresis analysis for the same samples (red = variant, blue = WT).
Next, a series of DNA samples from matched colon cancer-normal tissues was similarly tested for BAT25 changes. 5/10 cancer samples demonstrated BAT25 changes, which were not present in their normal counterpart. These changes were evident both in the NaME-PrO-treated and the No-NaME-PrO samples, indicating a high level of MSI and cancer purity in these samples, Supplementary Figure S4. The remaining 5 samples did not demonstrate BAT25 changes with either method (not shown).
Finally, BAT25 was also tested in DNA from a series of advanced colon cancer patients with low cancer content, and 7/16 (43%) demonstrated BAT25 indels evident on 9 NaME-PrO-treated samples, while no-NaME-PrO samples demonstrated no indels (Supplementary Figure S5, samples #259, 235 and 301). Samples that demonstrated 1–6bp indels (#256, 313, 307 and 280) are similarly shown in Supplementary Figure S6, indicating the ability of pre-PCR enrichment via NaME-PrO to reveal single base changes despite polymerase slippage (stutter). The BAT25 screening of clinical samples is summarized in Table 1.
Table 1. Summary of BAT25 screening of clinical patient samples.
| Clinical Samples | No. of Patients | No NaME-PrO HRM positive | NaME-PrO HRM positive | No NaME-PrO Capillary Electrophoresis positive | NaME-PrO Capillary Electrophoresis positive |
|---|---|---|---|---|---|
| Circulating Plasma DNA | 33 | 0 | 4 | 0 | 4 |
| Matched normal tissue and colon tumor pairs | 10 | 5 (tumor) | 5 (tumor) | 5 (tumor) | 5 (tumor) |
| Advanced Colon Cancer Genomic DNA | 16 | 0 | 9 | 0 | 7 |
Multiplexed NaME-PrO, HRM and capillary electrophoresis on colon cancer samples
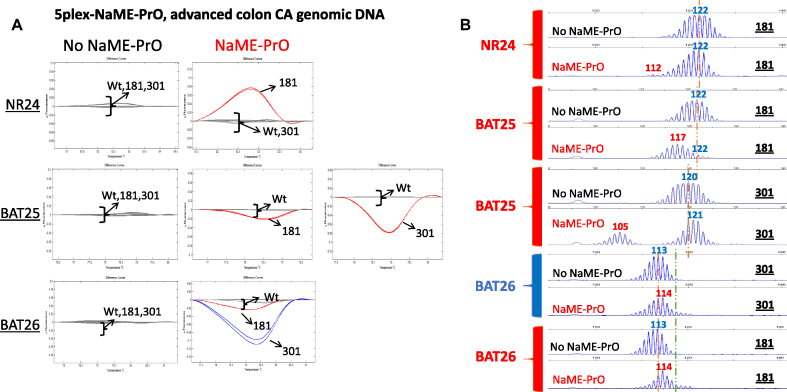
NaME-PrO is inherently multiplex-able since multiple probes can be used to target diverse sites simultaneously (32,34). To apply multiplexed NaME-PrO for enriching multiple altered microsatellites, probes for 5 markers were employed, NR21, NR24, NR27, BAT25 and BAT26. Samples found to be positive for BAT25 deletion were selected and re-screened using multiplex NaME-PrO. Supplementary Figures S7 and S8 depict HRM and capillary electrophoresis, respectively, for matched normal-cancer CN1/CT1 samples. Following multiplex NaME-PrO, the product was amplified via simplex PCR and assessed via HRM, followed by capillary electrophoresis. Indel enrichment was evident for all 5 targets following multiplexed NaME-PrO. Figure 4 depicts 5-plex screening of DNA from advanced colon cancer patients where deletions were only evident for NaME-PrO-treated samples. For sample #181 deletions were found in NR24 (10bp), BAT25 (5bp) and an insertion for BAT26 (1bp). For sample #301 a deletion was found in BAT25 (16bp) and insertion in BAT26 (1bp).
Figure 4.
Multiplexed NaME-PrO application on cancer tumor DNA from advanced colon cancer patients. (A). Comparison between multiplexed NaME-PrO-treated and untreated screening for NR21, NR24, NR27, BAT25 and BAT26 markers on advanced colon cancer patients using HRM: an example for patients 181 and 301 is shown. NaME-PrO treated samples display variant melting curves for NR24, BAT25 and BAT26. HRM for NR21 and NR27 markers did not identify a variant (not shown). No-NaME-PrO samples did not identify any variant via HRM. (B) Capillary electrophoresis was conducted for the HRM positive patient samples 181 and 301. Sample-181 carried deletions for NR24 (10 bp), BAT25(5bp) and insertion for BAT26 (1 bp). On the other hand, 301 carried a deletion in BAT25 (16 bp) and insertion in BAT26 (1 bp). No-NaME-PrO samples did not identify indels in these samples (red = variant, blue = WT).
Finally, multiplexed NaME-PrO was followed by a multiplexed PCR reaction and HRM was applied to all five amplicons simultaneously, as previously described for point mutations (35). Supplementary Figure S9 indicates a clear demonstration of the presence of at least one positive target for NaME-PrO treated samples. In contrast, no-NaME-PrO samples indicated no indel present.
DISCUSSION
Colorectal cancer (CRC) is classified into three categories, the hereditary form referred to as Hereditary Nonpolyposis Colorectal Cancer (HNPCC) or Lynch Syndrome (LS), the sporadic form and the colitis-associated CRC (36). Microsatellite instability (MSI) is present in 15% of CRC, 3% Lynch syndrome and 12% sporadic (37), and colon tumors are traditionally classified into three distinct groups according to the Bethesda guidelines, MSI-high (MSI-H), MSI-low (MSI-L) or MSI stable (MSS) (38). MSI-H compared to MSI-S is predictive for therapy outcome in chemotherapy and immunotherapy and has been associated with distinct characteristics and favorable results including better prognosis, a higher 5-year survival, and less metastatic potential (1,2). Despite classification in three distinct groups, there is evidence that colorectal cancers have inherent instability and if enough markers are tested a majority of colorectal cancers have some degree of MSI (39) illustrating a continuous phenotype, as demonstrated recently via massively parallel sequencing, MPS (26,31,40,41). A range of other cancers such as endometrial, rectal and stomach cancer also present MSI phenotype, reflecting mismatch repair deficiency and highlighting the potential role of MSI beyond just colon cancer (40,42).
Furthermore, there are compelling data indicating that immune checkpoint inhibitor therapy is more likely to be effective in patients with MSI-H colorectal cancer underlined by mismatch repair deficiency (2,42). Inaccurate assessment of MSI could cause patients to be missed who would otherwise have benefitted from immunotherapy and possibly have had a robust, durable tumor response. Therefore, improving detection sensitivity for MSI is of great clinical importance. Although one may infer mismatch repair deficiency by detecting mutations via targeted re-sequencing (43), this approach is relatively expensive since a broad genome coverage is needed to provide accurate assessment (44). In contrast to screening multi-gene panels for detecting single point mutations, microsatellites have a higher chance of harboring indels due to mismatch repair deficiency, hence surveying MSI in small set of pre-defined markers maybe a more effective and low-cost approach to use for clinical classification.
Given the renewed interest in sensitive, multi-target screening of clinical samples for varying degrees of MSI, either using established markers and approaches or using new markers and massively parallel sequencing, the current approach using NaME-PrO for enrichment of altered microsatellite markers provides a powerful new tool. The combination of NaME-PrO with HRM provides the most sensitive approach to identify positive samples, as Figures 2–4 demonstrate. Therefore, as we recently proposed for point mutations (35) it may be possible to employ HRM in multiplexed format for pre-screening for the presence of MSI-positive samples, while HRM-negative samples may not need further examination and can be classified as MSI-stable.
Given that according to Shagin et al. (33) the DSN nuclease requires a minimum of 10–12 bases of perfect base-pairing for digestion, it was somewhat surprising that WT mononucleotide strings of at least 27bp were digested to provide highly effective indel enrichment. Our current hypothesis is that in view of the poly-A sequence the ‘bulge’ created between an indel and NaME-PrO probes is de-localized, ie at any moment it can be anywhere along the poly-A, thus conferring temporal instability along the entire poly-A string. Effectively, it is like the bulge is constantly travelling ‘back and forth’ along the poly-A target thus preventing DSN from finding a stable 10–12 bp sequence to digest. Potentially even longer targets might also be enriched with NaME-PrO, although current studies were restricted to the sequences most commonly recommended for assessing MSI. About potential application of the method with dinucleotide repeats, one could anticipate similar behavior as with mono-nucleotides, at least with shorter di-nucleotide repeats and provided that the NaME-PrO probes designed for the WT sequence retain a Tm in the range of 60–70°C where DSN digestion is prevalent. However further studies must verify the behavior of the system with di-tri-nucleotide repeats.
As regards to population-level heterogeneity in micro-satellite length, the microsatellite markers selected for the present study are known to be either monomorphic or quasi-monomorphic therefore a single probe design applies for almost all cases. If a DNA target is polymorphic in the population, then NaME-PrO probes that match all the polymorphic versions may need to be used simultaneously in the reaction, as we previously demonstrated for SNPs (32,35).
In summary, we adapted NaME-PrO technology for enrichment of altered microsatellites and demonstrated that combination with high resolution melting and/or capillary electrophoresis provides a highly sensitive approach to detect MSI changes in clinical samples. The highly multiplexed format of NaME-PrO, combined with pre-PCR enrichment which circumvents problems associated with polymerase slippage provides a powerful enhancement in the ability to assess MSI in biological samples.
Supplementary Material
ACKNOWLEDGEMENTS
We gratefully acknowledge the helpful advice and discussions with Richard Cawthon about TGFRB2 genetic instability in cancer and aging. In addition, the assistance of Sigitas Verselis of the Dana Farber Cancer Institute Sequencing Core Facility with capillary electrophoresis is gratefully acknowledged. The contents of this manuscript do not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R33 CA217652 to G.M.M.] (in part). Funding for open access charge: Departmental funds.
Conflict of interest statement. None declared.
REFERENCES
- 1. Vilar E., Gruber S.B.. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010; 7:153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Le D.T., Uram J.N., Wang H., Bartlett B.R., Kemberling H., Eyring A.D., Skora A.D., Luber B.S., Azad N.S., Laheru D. et al. PD-1 blockade in tumors with mismatch-repair deficiency. N. Engl. J. Med. 2015; 372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yurgelun M.B., Goel A., Hornick J.L., Sen A., Turgeon D.K., Ruffin M.T., Marcon N.E., Baron J.A., Bresalier R.S., Syngal S. et al. Microsatellite instability and DNA mismatch repair protein deficiency in Lynch syndrome colorectal polyps. Cancer Prev. Res. (Phila.). 2012; 5:574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vasen H.F., Abdirahman M., Brohet R., Langers A.M., Kleibeuker J.H., van Kouwen M., Koornstra J.J., Boot H., Cats A., Dekker E. et al. One to 2-year surveillance intervals reduce risk of colorectal cancer in families with Lynch syndrome. Gastroenterology. 2010; 138:2300–2306. [DOI] [PubMed] [Google Scholar]
- 5. Sehgal R., Sheahan K., O’Connell P.R., Hanly A.M., Martin S.T., Winter D.C.. Lynch syndrome: an updated review. Genes (Basel). 2014; 5:497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. De Jong A.E., Morreau H., Van Puijenbroek M., Eilers P.H., Wijnen J., Nagengast F.M., Griffioen G., Cats A., Menko F.H., Kleibeuker J.H. et al. The role of mismatch repair gene defects in the development of adenomas in patients with HNPCC. Gastroenterology. 2004; 126:42–48. [DOI] [PubMed] [Google Scholar]
- 7. Heinen C.D., Shivapurkar N., Tang Z., Groden J., Alabaster O.. Microsatellite instability in aberrant crypt foci from human colons. Cancer Res. 1996; 56:5339–5341. [PubMed] [Google Scholar]
- 8. Beggs A.D., Domingo E., Abulafi M., Hodgson S.V., Tomlinson I.P.. A study of genomic instability in early preneoplastic colonic lesions. Oncogene. 2013; 32:5333–5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kloor M., Huth C., Voigt A.Y., Benner A., Schirmacher P., von Knebel Doeberitz M., Bläker H.. Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: a pathological study. Lancet Oncol. 2012; 13:598–606. [DOI] [PubMed] [Google Scholar]
- 10. Bacher J.W., Sievers C.K., Albrecht D.M., Grimes I.C., Weiss J.M., Matkowskyj K.A., Agni R.M., Vyazunova I., Clipson L., Storts D.R. et al. Improved detection of microsatellite instability in early colorectal lesions. PLoS One. 2015; 10:e0132727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kamat N., Khidhir M.A., Hussain S., Alashari M.M., Rannug U.. Chemotherapy induced microsatellite instability and loss of heterozygosity in chromosomes 2, 5, 10, and 17 in solid tumor patients. Cancer Cell Int. 2014; 14:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pinto J.L., Fonseca F.L., Marsicano S.R., Delgado P.O., Sant’anna A.V., Coelho P.G., Maeda P., Del Giglio A.. Systemic chemotherapy-induced microsatellite instability in the mononuclear cell fraction of women with breast cancer can be reproduced in vitro and abrogated by amifostine. J. Pharm. Pharmacol. 2010; 62:931–934. [DOI] [PubMed] [Google Scholar]
- 13. Fonseca F.L., Sant Ana A.V., Bendit I., Arias V., Costa L.J., Pinhal A.A., del Giglio A.. Systemic chemotherapy induces microsatellite instability in the peripheral blood mononuclear cells of breast cancer patients. Breast Cancer Res. 2005; 7:R28–R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shaw J.A., Smith B.M., Walsh T., Johnson S., Primrose L., Slade M.J., Walker R.A., Coombes R.C.. Microsatellite alterations plasma DNA of primary breast cancer patients. Clin. Cancer Res. 2000; 6:1119–1124. [PubMed] [Google Scholar]
- 15. Buhard O., Lagrange A., Guilloux A., Colas C., Chouchène M., Wanherdrick K., Coulet F., Guillerm E., Dorard C., Marisa L. et al. HSP110 T17 simplifies and improves the microsatellite instability testing in patients with colorectal cancer. J. Med. Genet. 2016; 53:377–384. [DOI] [PubMed] [Google Scholar]
- 16. Li J., Wang L., Mamon H., Kulke M.H., Berbeco R., Makrigiorgos G.M.. Replacing PCR with COLD-PCR enriches variant DNA sequences and redefines the sensitivity of genetic testing. Nat. Med. 2008; 14:579–584. [DOI] [PubMed] [Google Scholar]
- 17. Milbury C.A., Correll M., Quackenbush J., Rubio R., Makrigiorgos G.M.. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin. Chem. 2012; 58:580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. How Kit A., Mazaleyrat N., Daunay A., Nielsen H.M., Terris B., Tost J.. Sensitive detection of KRAS mutations using enhanced-ice-COLD-PCR mutation enrichment and direct sequence identification. Hum. Mutat. 2013; 34:1568–1580. [DOI] [PubMed] [Google Scholar]
- 19. Galbiati S., Brisci A., Lalatta F., Seia M., Makrigiorgos G.M., Ferrari M., Cremonesi L.. Full COLD-PCR protocol for noninvasive prenatal diagnosis of genetic diseases. Clin. Chem. 2011; 57:136–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murphy D.M., Bejar R., Stevenson K., Neuberg D., Shi Y., Cubrich C., Richardson K., Eastlake P., Garcia-Manero G., Kantarjian H. et al. NRAS mutations with low allele burden have independent prognostic significance for patients with lower risk myelodysplastic syndromes. Leukemia. 2013; 27:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Milbury C.A., Li J., Liu P., Makrigiorgos G.M.. COLD-PCR: improving the sensitivity of molecular diagnostics assays. Expert Rev. Mol. Diagn. 2011; 11:159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. How-Kit A., Daunay A., Buhard O., Meiller C., Sahbatou M., Collura A., Duval A., Deleuze J.F.. Major improvement in the detection of microsatellite instability in colorectal cancer using HSP110 T17 E-ice-COLD-PCR. Hum. Mutat. 2017; 3:441–453. [DOI] [PubMed] [Google Scholar]
- 23. Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004; 5:435–445. [DOI] [PubMed] [Google Scholar]
- 24. Milbury C.A., Li J., Makrigiorgos G.M.. PCR-based methods for the enrichment of minority alleles and mutations. Clin. Chem. 2009; 55:632–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Janavicius R., Matiukaite D., Jakubauskas A., Griskevicius L.. Microsatellite instability detection by high-resolution melting analysis. Clin. Chem. 2010; 56:1750–1757. [DOI] [PubMed] [Google Scholar]
- 26. Gan C., Love C., Beshay V., Macrae F., Fox S., Waring P., Taylor G.. Applicability of next generation sequencing technology in microsatellite instability testing. Genes (Basel). 2015; 6:46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Walker C.J., Miranda M.A., O’Hern M.J., Blachly J.S., Moyer C.L., Ivanovich J., Kroll K.W., Eisfeld A.K., Sapp C.E., Mutch D.G. et al. MonoSeq variant caller reveals novel mononucleotide run indel mutations in tumors with defective DNA mismatch repair. Hum. Mutat. 2016; 37:1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim T.M., Laird P.W., Park P.J.. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013; 155:858–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balzer S., Malde K., Jonassen I.. Systematic exploration of error sources in pyrosequencing flowgram data. Bioinformatics. 2011; 27:i304–i309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van den Hoecke S., Verhelst J., Vuylsteke M., Saelens X.. Analysis of the genetic diversity of influenza A viruses using next-generation DNA sequencing. BMC Genomics. 2015; 16:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Maruvka Y.E., Mouw K.W., Karlic R., Parasuraman P., Kamburov A., Polak P., Haradhvala N.J., Hess J.M., Rheinbay E., Brody Y. et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat. Biotechnol. 2017; 35:951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Song C., Liu Y., Fontana R., Makrigiorgos A., Mamon H., Kulke M.H., Makrigiorgos G.M.. Elimination of unaltered DNA in mixed clinical samples via nuclease-assisted minor-allele enrichment. Nucleic Acids Res. 2016; 44:e146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shagin D.A., Rebrikov D.V., Kozhemyako V.B., Altshuler I.M., Shcheglov A.S., Zhulidov P.A., Bogdanova E.A., Staroverov D.B., Rasskazov V.A., Lukyanov S.. A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res. 2002; 12:1935–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu Y., Song C., Ladas I., Fitarelli-Kiehl M., Makrigiorgos G.M.. Methylation-sensitive enrichment of minor DNA alleles using a double-strand DNA-specific nuclease. Nucleic Acids Res. 2017; 45:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ladas I., Fitarelli-Kiehl M., Song C., Adalsteinsson V.A., Parsons H.A., Lin N.U., Wagle N., Makrigiorgos G.M.. Multiplexed elimination of Wild-Type DNA and High-Resolution melting prior to targeted resequencing of liquid biopsies. Clin. Chem. 2017; 63:1605–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang D., Dubois R.N.. The role of COX-2 in intestinal inflammation and colorectal cancer. Oncogene. 2010; 29:781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang X., Li J.. Era of universal testing of microsatellite instability in colorectal cancer. World J. Gastrointest. Oncol. 2013; 5:12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Umar A., Boland C.R., Terdiman J.P., Syngal S., de la Chapelle A., Ruschoff J., Fishel R., Lindor N.M., Burgart L.J., Hamelin R. et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J. Natl. Cancer Inst. 2004; 96:261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Laiho P., Launonen V., Lahermo P., Esteller M., Guo M., Herman J.G., Mecklin J.P., Jarvinen H., Sistonen P., Kim K.M. et al. Low-level microsatellite instability in most colorectal carcinomas. Cancer Res. 2002; 62:1166–1170. [PubMed] [Google Scholar]
- 40. Hause R.J., Pritchard C.C., Shendure J., Salipante S.J.. Classification and characterization of microsatellite instability across 18 cancer types. Nat. Med. 2016; 22:1342–1350. [DOI] [PubMed] [Google Scholar]
- 41. Salipante S.J., Scroggins S.M., Hampel H.L., Turner E.H., Pritchard C.C.. Microsatellite instability detection by next generation sequencing. Clin. Chem. 2014; 60:1192–1199. [DOI] [PubMed] [Google Scholar]
- 42. Lee V., Murphy A., Le D.T., Diaz L.A. Jr. Mismatch repair deficiency and response to immune checkpoint blockade. Oncologist. 2016; 21:1200–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stadler Z.K., Battaglin F., Middha S., Hechtman J.F., Tran C., Cercek A., Yaeger R., Segal N.H., Varghese A.M., Reidy-Lagunes D.L. et al. Reliable detection of mismatch repair deficiency in colorectal cancers using mutational load in Next-Generation sequencing panels. J. Clin. Oncol. 2016; 34:2141–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ngeow J., Eng C.. Mismatch repair deficiency in colorectal cancers: is somatic genomic testing the Grab-Bag for all answers. J. Clin. Oncol. 2016; 34:2085–2087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.