Abstract
In Eukaryotes, DNA is wound around the histone octamer forming the basic chromatin unit, the nucleosome. Atomic structures have been obtained from crystallography and single particle cryo-electron microscopy (cryoEM) of identical engineered particles. But native nucleosomes are dynamical entities with diverse DNA sequence and histone content, and little is known about their conformational variability, especially in the cellular context. Using cryoEM and tomography of vitreous sections we analyse native nucleosomes, both in vitro, using purified particles solubilized at physiologically relevant concentrations (25–50%), and in situ, within interphase nuclei. We visualize individual nucleosomes at a level of detail that allows us to measure the distance between the DNA gyres wrapped around. In concentrated solutions, we demonstrate a salt-dependent transition, with a high salt compact conformation resembling the canonical nucleosome and an open low salt one, closer to nuclear nucleosomes. Although further particle characterization and cartography are needed to understand the relationship between this conformational variability and chromatin functional states, this work opens a route to chromatin exploration in situ.
INTRODUCTION
The nucleosome is the basic packaging unit of eukaryotic chromatin and consists in 145–147 bp DNA wrapped into 1.65 turn of a left-handed superhelix around a histone octamer (two copies of H2A, H2B, H3 and H4). Atomic resolution structures are available since 1997 from X-ray crystallography (1–4) and more recently from cryo-electron microscopy (cryoEM) (5). A canonical structure of the particle has thus emerged from high resolution structures of identical, symmetric and highly stable engineered particles assembled from recombinant histones and strongly positioning DNA sequences.
There are now increasing evidences that there is not a unique nucleosome entity but a whole family of structures and conformations (6–8). Nucleosomes do not only play a role in DNA packaging but are also involved in the regulation of many nuclear processes from gene expression to DNA replication and repair (9,10). To serve these functions, they are chemically modified (epigenetic modifications, histone post-translational modifications and DNA methylation), but also unwrapped, remodelled, disassembled. They show remarkable dynamical properties (11–18), with conformational changes, such as DNA breathing (11,12), H2B/H2A dimer splitting (16) or nucleosome gaping (17,18). The nucleosome conformation may also depend on the environment, e.g. local ionic conditions or macromolecular crowding, but to our knowledge, such as effects have little been investigated (18,19). In all cases, this variability is mainly documented in vitro or in silico. Even more interesting would be the possibility to explore nucleosome conformational variations in their nuclear context, but this domain remains terra incognita.
Using cryoEM and tomography of vitreous sections, we analyse the conformation of purified isolated nucleosomes (with their native chemical heterogeneity) under physiological relevant concentrations (25–50% w:v). In the presence of monovalent ions, above a concentration threshold, nucleosomes stack into columns that further order the ones relative to the others to form multiple liquid crystalline phases (LC) at higher concentrations (20–24). These phases constitute ideal objects to explore the influence of ionic environment or nucleosome concentration, as well as sample preparation methods. We visualize nucleosomes in LC phases and analyse the conformation of the nucleosomal DNA. In particular, we access the distance P between the DNA gyres wrapped around the particle. We observe variations of P and analyse these variations by comparison with the crystallographic structure (1,2). We show that the nucleosome conformations are on average more open in low salt conditions whilst compact conformations, closer to the canonical structure, are found at higher salt.
We then question the occurrence of open conformations in situ, investigating nucleosomal DNA in interphase cell nuclei from different cell types, namely two-cultured human cell lines, and Drosophila embryos. We obtain both 2D images and cryo-tomograms of vitreous sections where nucleosomes can be recognized individually and analysed at a high enough level of detail to visualize DNA wrapped around the particle and measure P. On average, nucleosomes in situ are more open than the canonical crystallographic structure, with P-value closer to that of native purified particles in low salt solution. Yet the particle’s variability is more important, probably reflecting the different functional states found in the cell nucleus.
MATERIALS AND METHODS
Nucleosome purification and preparation of concentrated solutions
Native H1-depleted nucleosomes (histone octamer associated with 167 ± 10 bp DNA) and native nucleosome core particles (NCP) (146 ± 3 bp DNA) were respectively prepared from calf thymus and chicken erythrocyte chromatin. Chromatin was extracted in a low ionic strength buffer, followed by histone H1 removal. Nucleosome particles with different DNA associated lengths were obtained by controlled digestion with micrococcal nuclease, and purified by gel filtration (Sephacryl S300 HR, GE Healthcare). The integrity of all core histones, and the absence of contaminating di- and oligo-nucleosomes were checked by gel electrophoresis. Purified particles were extensively dialysed at a concentration of 1 mg/ml against a TE buffer (10 mM Tris pH 7.6, 1 mM ethylenediaminetetraacetic acid) supplemented with either 5 mM NaCl (low salt) or 150 mM (high salt). The solutions were concentrated by ultrafiltration through a cellulose membrane (Ultracell YM100, Millipore) in a pressurized cell up to about 250 mg/ml (25% w:v). Higher concentrations were reached by osmotic pressure, induced either by extensive dialysis against PolyEthyleneGlycol (PEG, MW20000) dissolved in the same buffer at concentrations ranging from 10 to 22% (w:v), or by progressive mixing with the PEG solution, as described in (21).
Cell culture and preparation
Cell suspensions of the human cell lines HT29 (25) and KE37 (26) were pelleted by centrifugation (3000 g, 3 min), and gently re-suspended in an equal volume of dextran (PM 40 000, Sigma-Aldrich) dissolved at a concentration of 40% (w:w) in phosphate-buffered saline (PBS). Drosophila melanogaster (Bloomington Stock number 30564) were maintained in a standard Bloomington medium. Embryos were collected on apple agar plates, dechorionated in 50% (v:v) bleach (Sigma-Aldrich) and washed with distilled water. Developmental stages 14–15 were selected by visual inspection under a stereomicroscope (Leica M165 FC) and transferred into gold-plated copper carriers with a 0.1 mm indentation (B-type, Engineering Office M. Wohlwend GmbH) filled with 25% dextran in PBS and processed for high-pressure freezing.
Bulk sample vitrification and cryo-ultramicrotomy
Nucleosome LC solutions and cell pellets were frozen by slam-freezing against a copper block cooled to liquid helium temperature (Cryovacublock, Reichert) as described in (27). Drosophila embryos were frozen by high pressure freezing using an HPM 010 machine (ABRA Fluid AG). Frozen specimens were then transferred into a Leica FC6/UC6 ultramicrotome (Leica Microsystems) installed in a controlled environment with a relative humidity maintained below 20%. Nucleosome solutions and cell suspensions were sectioned at −160°C, Drosophila embryos at −145°C, using a 25° or a 35° diamond knife (Diatome). The cutting feed was set at 40 nm for sections dedicated to 2D imaging, in order to minimize superimposition effects. A cutting feed of 75 nm was used for thicker sections dedicated to tomography. Sections were respectively collected on Quantifoil S7/2 grids coated by a continuous thin carbon film (2 D imaging), or directly on C-flat CF-2/1 grids (Electron Microscopy Sciences) for tomography. They were pressed against the grid using a mechanical press (28) or an electrostatic press (Crion (29), Leica).
Thin film vitrification
Drops of LC nucleosome solutions were deposited onto an electron microscopy grid covered with a holy carbon film. On account of their high viscosity, drops were strongly blotted manually between two filter papers, and immediately vitrified by immersion in liquid ethane using a home-made plunging device. To avoid water evaporation, the procedure was performed under humidified atmosphere (30).
Cryo-electron microscopy
For 2D imaging, grids were mounted in a Gatan 626 cryoholder (Gatan, Pleasanton, USA) and transferred to a JEOL 2010 Field Emission Gun (FEG) transmission electron microscope (TEM) operated at 200 kV or a FEI CM12 TEM operated at 80 kV. The vitreous state of the specimens was checked by electron diffraction. Images were recorded on Kodak SO163 films, under low-dose conditions at direct magnification of 25 000× to 45 000×, with defocus in the range of 1.5 μm (200kV) and 900 nm (80 kV). Films were developed in full-strength Kodak D19 for 12 min. In these imaging conditions, the signal of interest (typically in the range of 2.5–3.5 nm, lies far below the first zero of the contrast transfer function (CTF) (1.9 nm for both imaging conditions). We checked that CTF correction is not necessary for image interpretation (see Supplementary Figure S1). Images presented in Figures 1–3 were denoized by wavelet filtration using the ImageJ ‘A trous wavelet filter’ plugin (with k1 = 20, k2 = k3 = k4 = k5 = 0).
Figure 1.
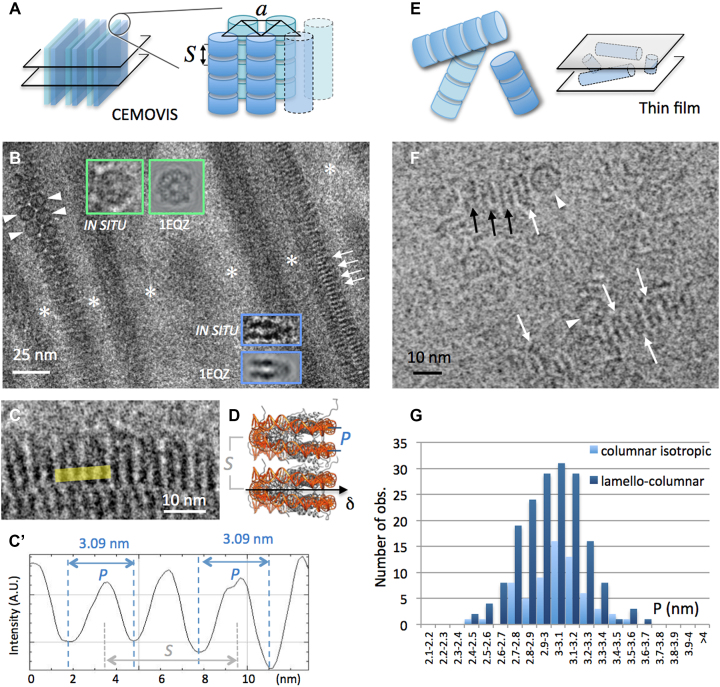
CryoEM of NCP forming columnar phases in low salt solutions (15 mM [+]). (A) The lamello-columnar phase is composed of stacks of nucleosomes (columns) that align in parallel into a triangular lattice of parameter a, forming bilayers. Bulk phase (long range ordered bilayers) can be sectioned (e.g. transversally, left) and analysed by CEMOVIS. (B and C) CEMOVIS imaging of the lamello-columnar phase. Bilayers in transverse section are separated by particle-free layers of solvent (white stars). Columns of nucleosomes are seen in top (arrowheads), side (arrows) or oblique views. Top and side views are compared to the corresponding projections of the crystallographic structure of the NCP (from PDB ID: 1EQZ) in the inserts. (C and C’) Measurement of P-values on columns in side view. (D) In the columns of the lamello-columnar phase, nucleosomes stack on top of each other with parallel alignment of their dyad axes (δ). P corresponds to the distance between the DNA gyres measured at the front side of the particle. S corresponds to the stacking repeat. (F) In the columnar isotropic phase observed by thin film cryoEM, columns of nucleosomes form a random network sketched in (E). Nucleosomes are recognized in side (arrows) and top (arrowheads) views. (B and F) White arrows indicate the orientation of the nucleosome's dyad axis δ where it lies in the observation plane. Black arrows point to side view particles with their dyad axis out of the observation plane. (G) Distribution of P-values measured in the lamello-columnar and columnar isotropic phases.
Figure 3.
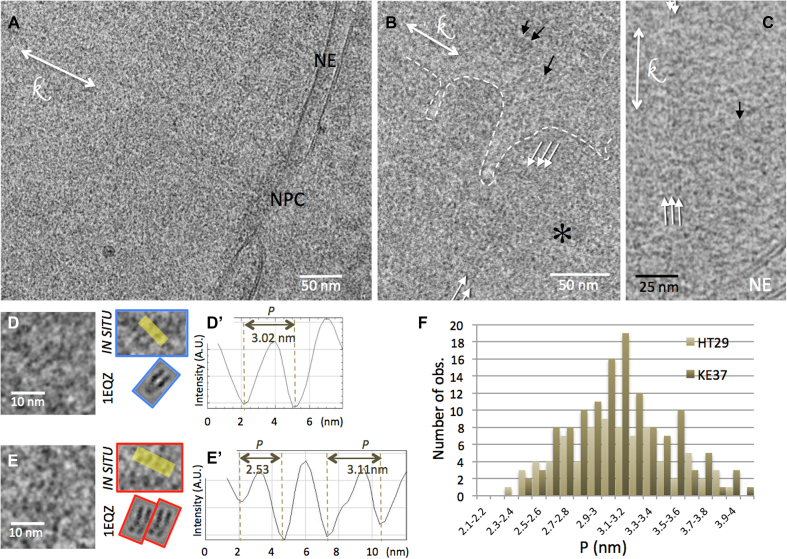
CEMOVIS of interphase nuclei of the human cell lines KE37 (A, B and D) and HT29 (C and E). (A) Local view of a nucleus limited by the NE. An NPC is visible. (B) A chromatin-dense region (*), delineated by the white dotted line, coexists with a less dense region. (B and C) Striations are visible, either locally (black arrows) or extending over longer distances (white arrows). (D and E) Nucleosomes visualized in side views. The patterns are compared with CTF-modulated projections of the crystallographic structure of the particle (from PDB ID: 1 EQZ). (D’ and E’) The distance between the DNA gyres P, is measured on line profiles indicated in yellow in (D) and (E). (F) Distribution of P for the two cell types.
For cryo-electron tomography grids were mounted into Autogrid rings (FEI, Thermofisher, Eindhoven, The Netherlands) and transferred into a Titan Krios (FEI, Thermofischer, Eindhoven, The Netherlands) operated at 300 kV equipped with a GATAN GIF Quantum SE post-column energy filter and K2 Summit direct electron detector (Gatan, Pleasanton, USA). Tilt series were recorded using Serial EM software (bio3d.colorado.edu/SerialEM (31)) at a nominal magnification of 64000 × (2.2 Å/pixel). The dose-symmetric recording scheme macro (32), was run within an angular range from −60° to +60°, with a starting angle 0° and an angular increment of 2°. The electron dose was set to 1.5 e–/A2 for individual tilt images, corresponding to the total dose 91.5 e–/A2 for the complete tilt series. Contrast of tilt images recorded with imaging conditions used for 2D images (CTF first zero at 1.9 nm) was not sufficient for fiducial-free alignment. A reliable alignment was achieved when the target defocus was set to −3.5 μm, corresponding to the CTF first zero at 2.4 nm. Since this value is relatively close to the signal of interest (P sampling range, see ‘Results’ section), CTF correction was performed for all tomographic data. Generation of fiducial-free markers was done by patch tracking algorithm of IMOD package (33). Tilt series alignment based on marker positions generated by IMOD, CTF correction and tomogram reconstruction were performed using customs scripts (34,35). The reconstructed volumes were denoized by filtering by 3D non-linear anisotropic diffusion (k = 1, 10 iterations) (36). 5 nm tomographic slices through the reconstructed section volumes were generated in IMOD.
Generation of CTF-modulated projections and tomographic slices from the canonical NCP
For simulation of 2D nucleosome reference images, a 3D electron density map was generated from PDB ID: 1EQZ map using the pdb2em function of the EMAN software package (37) with a resolution of 1 nm and pixel size 0.025 nm. All subsequent calculations were done with using custom scripts running under the Matlab platform (MathWorks, Natick, USA). The nucleosome electron density map was rotated in space with all three rotational angles assigned to random numbers, and a projection of the complete volume was generated. The CTF was applied to the projection image with defocus assigned as a random number within the defocus range of real 2D images of nucleosomes acquired at 200 kV (−1.5 ± 0.5 μm) and at 80 kV (900 nm). This procedure was repeated to generate a set of reference images (Supplementary Figure S2). Similar patterns were obtained for both imaging conditions. To simulate 3D reference images, the 1EQZ nucleosome electron density map was binned down to 64 × 64 × 64 pixel volumes with the final voxel of 0.2 nm3. We generated a 75-nm thick virtual chromatin section as a volume of 1024 × 1024 × 375 pixels where 750 randomly oriented nucleosomes were placed into random positions, without density overlapping. A series of tilt images was generated by rotation and projection of each volume, using the same angle range increment as for real tomograms. We found that in our tomograms of vitreous sections, an average defocus corresponded to −3.7 μm at 0° tilt, whilst it fluctuated from −3 to −4.5 μm at higher angles. Accordingly, 0° tilt images of the generated volume changes were modulated with CTF for the defocus of −3.7 μm, whereas defocus values for other tilt angles were generated as a list of random numbers within the aforementioned range. The generated tilt series were then aligned, CTF corrected and reconstructed with the same scripts as used for the real tomogram reconstructions. Similarly, 5 nm thick tomographic slices were generated in IMOD.
P-measurements and statistical analyses
Line profiles measurements were performed using the ImageJ software (38). The Shapiro–Wilk (SW) test was used to assess normality of P distance measurements distributions. All data samples were distributed normally with the exception of high-salt solution measurements. The two-sample t-test was used to assess difference between normally distributed data samples. The Kolmogorov–Smirnov test was used to assess the differences between high salt and other data samples. All tests were performed at an alpha level of 0.05. Statistical analysis was done using the XLSTAT software (Addinsoft, New York, USA).
RESULTS
CryoEM of nucleosomes in concentrated solutions
LC phases of nucleosomes were prepared at two different concentrations of added monovalent salt: 15 mM (low salt) and 160 mM (high salt), in order to question the effect of salt concentration on nucleosome conformation. We analysed both H1-depleted nucleosomes and NCP (DNA lengths of 167 ± 10 and 145 ± 3 bp, respectively, see ‘Materials and Methods’ section). Within the range of concentration explored here (25–50% w:v), we found no difference, both in terms of LC organization and particle conformation, between the two sets of particles. They will hereafter simply be referred as ‘NCP’.
Low salt solutions: evidence for a variable nucleosome conformation, on average more open than the canonical crystallographic structure
At low added salt concentration (≤40 mM), and at NCP concentrations between 28 and 32% (w:v), columns of NCP order with a local triangular lattice, into a lamello-columnar phase (Figure 1A). This phase was previously analysed by cryoEM of vitreous sections (CEMOVIS), providing a molecular scale understanding of nucleosome order and interactions (24). Figure 1B, shows a transverse section of the lamello-columnar phase, with bilayers of columns in apical and side views. In both cases, the orientation of the NCP can be determined by comparison with an image gallery generated from the PDB ID: 1EQZ, applying the CTF corresponding to our imaging conditions. The overall shape of the NCP is that of a flat cylinder, about 11.5 nm in width and 6 nm in height, with DNA wrapped into 1.65 turn on its lateral surface. Multiple patterns are expected (Supplementary Figure S2). Both disc-shaped top views, and highly contrasted striated side views, characterized by a pair of stripes drawn by the nearly two gyres of DNA wrapped around the particle, are unambiguously identified (Figure 1B inserts and pointed by arrowheads and arrows, respectively). Top views reveal the triangular lattice formed by columns. Side views present long-range periodic striations corresponding to DNA of stacked particles. The period S corresponds to the stacking repeat of the NCPs (Figure 1C, sketched in 1D). The distance P characterizes the distance between the two gyres of DNA wrapped around a given particle. We systematically measured P on intensity line profiles using ImageJ (Figure 1C’), wherever NCP are oriented in side views. The P distribution is given in Figure 1G (dark blue): it varies from 2.5 to 3.67 nm, and follows a normal distribution (SW test p = 0.399), with an average value of 2.99 ± 0.23 nm.
CEMOVIS-associated compression does not affect nucleosome structure
The major artefact associated to the CEMOVIS approach is the compression induced during the cutting process (39–41). Compression occurs in the cutting direction k and is coupled to an increase of the section thickness. It has been reported to induce severe deformations, up to 20–30%, of long-range structures e.g. entire bacterial or yeast cells, or organelles. Supramolecular assemblies, such as microtubules, were also reported to deform. In contrast, at the molecular level, deformations have been shown to be negligible or even inexistent: the structure of ribosomes, as well as the local packing of DNA in sperm cells or crystalline protein lattices were shown to be un- or little affected (42–45). To determine whether or not compression modifies the P parameter of the NCP, we sorted out a subset of measurements obtained on the lamello-columnar phase in vitro. It appears that most of the P-values were recorded on side view NCP oriented between 0 and 45° to the cutting direction k. Only few were observed oriented perpendicular, suggesting that long-range compression may perturb chromatin visualization, possibly inducing local reorientations. Nonetheless, where clearly visible side view NCP were oriented perpendicular to k, P did not show any variation. We also compared P for particles oriented at 0(±5)°, 45(±5)° and 90(±5)° to k, and this analysis confirms that there is no compression effect on the P-value (Supplementary Figure S3).
CEMOVIS versus thin film cryoEM: the sample preparation procedure does not influence nucleosome structure
Single bilayers of columns can also be trapped within thin films (Supplementary Figure S4). Bilayers are then always parallel to the observation plane, and the columns of the two layers superimpose within the film thickness. Side views of individual nucleosomes are recognized in some places, and P-values measured on line profiles as described above. P also follows a normal distribution (SW test p = 0.863), and varies from 2.41 to 3.51 nm (3 ± 0.21 nm). The comparison with CEMOVIS measurements (Student test p = 0.519; see Supplementary Figure S4) shows that there is here no influence of the sample preparation method.
The inter-distance and relative orientation of stacked particles do not influence the nucleosome conformation
The stacking distance S between particles varies with the particle concentration, from 5.9 to 6.2 nm in the lamello-columnar phase (45), and from 5.9 to about 7 nm within the less concentrated columnar isotropic phase (25–28% w:v). The local order between neighbouring particles also differs in both phases (24). NCP always stack in parallel in the lamello-columnar phase (with their dyad axis oriented inward the bilayer, Figure 1B, white arrows and arrowheads). In contrast, there is no preferred orientation of NCP dyad axes within columns of the isotropic phase, where parallel as well as anti-parallel and intermediate orientations are found. This is observed in Figure 1F, which shows a local view of the columnar isotropic phase trapped in a thin film. P varies from 2.48 to 3.51 nm in the columnar isotropic phase (3.03 ± 0.2 nm). The P distribution is normal (SW test p = 0.606) and can be compared to that found for the lamello-columnar phase (Figure 1G). No significant difference can be found (t-test p = 0.197), which suggests that both the local stacking density and the relative orientation of neighbours do not significantly influence the conformation of stacked NCP.
NCP in low salt columnar solutions present a range of conformations. The global set of P data obtained from the different low salt phases follows a normal distribution (SW test p = 0.551), with an average value of 3 ± 0.22 nm. In order to compare these data with the canonical crystallographic structure, we measured P on the different striated projection patterns generated from the PDB file (Supplementary Figures S2 and 5), exactly as it was done on EM images. This also results in a normal distribution (SW test p = 0.142), but with an average value of 2.79 ± 0.14 nm (Supplementary Figure S5), which is significantly different from P in low salt solutions (t-test p = 9.4 × 10−15). This comparison reveals that native NCP in low salt solution markedly differ from the particle’s canonical PDB structure: they present a whole range of conformations rather than a single one, and are on average more open than synthetic particles in crystals.
High salt solutions: nucleosomes adopt more compact conformations resembling the canonical structure
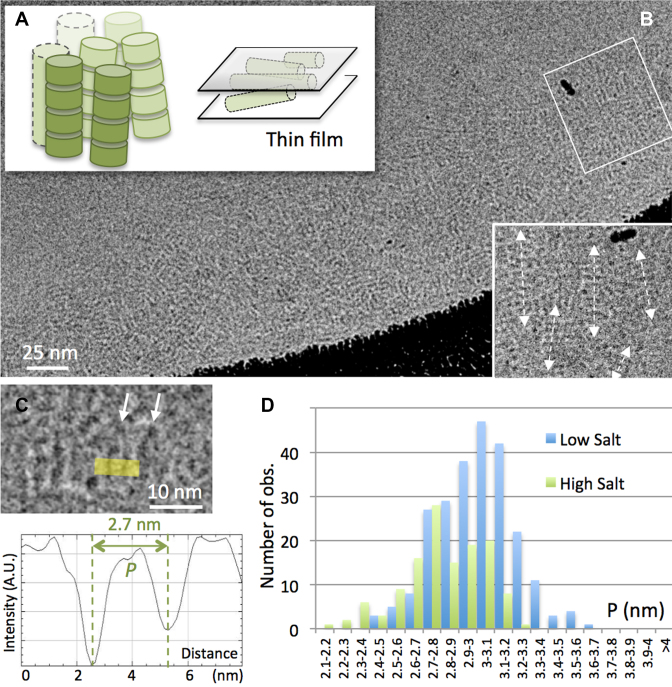
Above 100 mM added salt concentration, dense columnar nematic and columnar hexagonal phases form, at NCP concentrations higher than 45 and 50% (w:v), respectively (21,22,45). Both vitreous thin films (Figure 2A) and sections (not shown) were obtained from hereafter so-called ‘high salt’ (160 mM) columnar solutions, and imaged by cryoEM. In both phases, striated patterns typical of stacked side view particles are observed. In thin films, columns are oriented parallel to the film and superimpose; a local nematic order can be detected locally (Figure 2B). Individual NCP are recognized and P measured on line profiles (Figure 2C). Measurements obtained from thin film cryoEM (mean 2.81 nm, s.d. 0.218 nm) and CEMOVIS (mean 2.78 nm, s.d. 0.22 nm) do not show a significant difference (Kolmogorov–Smirnov test p = 0.588, α = 0.05). They were therefore combined into a single data sample, whose distribution is given in Figure 2D (in green), and compared with low salt data (in blue). The SW test (p = 0.032) does not allow us to hypothesize a normal distribution for P at high salt. To compare the high salt and low salt populations, we therefore use a Komolgorov–Smirnov test, resulting in a p value < 0.0001 (α = 0.05). Whilst in both cases P presents a broad distribution, it is larger in low salt than in high salt solutions. We also compared the high salt data with the simulated measurements from PDB. The mean P-value at high salt (2.79 nm) is the same as that simulated from PDB, thus supporting a similarity between native NCP in high salt solution and synthetic particles in crystals. Nevertheless the distribution of the high salt data is significantly different from the simulated measurements (Komolgorov–Smirnov test p = 0.022; α = 0.05). It is broader (s.d. 0.23 versus 0.14 nm), and enriched in larger P-distances, typical of the low salt particles (Supplementary Figure S6).
Figure 2.
CryoEM of NCP forming columnar phases in high salt solution and comparison of P-values measured in low versus high salt conditions. (A) The columnar phase formed in high salt (160 mM [+]) is trapped within a thin film and observed by cryoEM in (B). Columns align parallel to the film and superimpose within its thickness, leading to complex striated patterns. A local nematic order is observed in an enlargement (white rectangle). (C) Side view nucleosomes are recognized locally and P is measured on line profiles. (D) Comparison of the distributions of P-values recorded in the columnar hexagonal phase (160mM [+], green) and in the lamello-columnar phase (15 mM [+], blue, see Figure 1).
Altogether, these observations reveal that nucleosome undergo a salt-dependent conformational change at physiologically relevant nucleosome concentrations. The canonical particle resembles native nucleosomes in high salt solutions, whilst more open conformations are found in low salt environment.
By comparing thin film cryoEM and CEMOVIS approaches, we demonstrate that vitreous cryo-sectioning does not alter the NCP conformation. Note that, although it is not the focus of the present paper, CEMOVIS approaches also do not disturb significantly inter-particle interactions and ordering, as already shown in vitro (21). Having thus shown that CEMOVIS is well adapted to access nucleosome conformation in crowded environments, we carried out a CEMOVIS analysis of nucleosomes in their nuclear context. We prepared vitreous sections of interphase nuclei of different cell types, two human cell lines (HT29 and KE 37) and D. melanogaster embryonic brain cells. HT29 and KE37 were imaged in 2D, and cryo-tomograms were recorded from Drosophila embryos.
Cryo-electron microscopy and tomography of vitreous sections (CEMOVIS and CETOVIS) of interphase chromatin
Nucleosomes can be visualized and their conformation analysed in the interphase nucleus
Figure 3A shows a local view of the nucleus of a KE37 cell, with its nuclear envelope (NE) and a nuclear pore complex (NPC). Chromatin presents a homogeneous grainy texture resembling that already described (46–48). Yet, this texture, observed in the two cell lines, is here finer than classically reported in CEMOVIS studies, on account of our choice of lower underfocus imaging. Dense chromatin rich regions are interspaced by less dense regions, yet with no well defined frontiers (Figure 3B). As already described in mammal cells, dense chromatin regions are always found in the vicinity of NE or nucleoli (49,50). In addition, we observe striated patterns (Figure 3B and C, arrows). These striations are ubiquitous in the nucleus, and are found both in dense and less dense regions. They may remain very local, corresponding to a single pair of stripes (Figure 3B and C, black arrows), or propagate over larger distances (Figure 3B and C, white arrows) either in the streak direction (up to 60–100 nm) or perpendicularly (typically 10–15 nm and up to 40–45 nm in a few cases). Nucleosomes being the major chromatin component, we postulate that these striped patterns correspond to side views of nucleosomes, twice more contrasted than apical and oblique views (see Supplementary Figure S2), and therefore expected to dominate other contributions.
To avoid superimposition effects of 2D imaging and access the conformation of variously oriented nucleosomes, we recorded cryo-tomograms of vitreous sections. We found that acquisition of tilt series is much more demanding; therefore obtaining the highest density of nuclei within the section is essential to find locations suitable for tomography. Since organ/tissue samples better fulfil this criterion than cell suspensions, we chose the developing Drosophila brain. It shows both a high cell density and high nuclei/cytoplasm ratio, contributing to the yield of suitable chromatin areas (51). In addition, Drosophila embryos can be efficiently vitrified (52) and are surrounded by the liquid-impermeable vitelline membrane (53,54), which remarkably preserves the native extracellular environment during sample preparation and freezing.
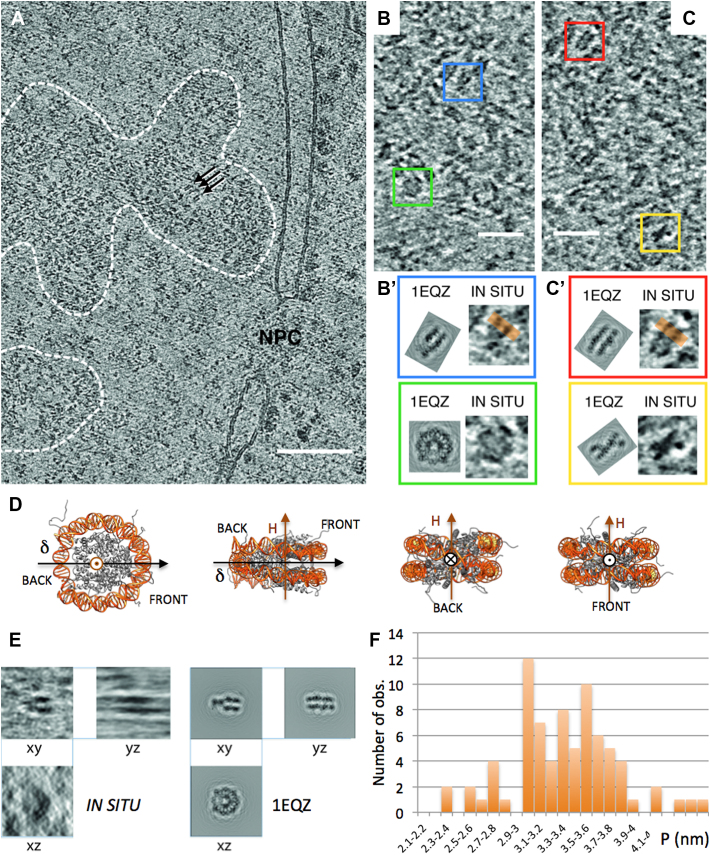
Chromatin of embryonic Drosophila brain cells is organized into two main regions: the highly compact chromocenters which accumulate constitutive heterochromatin regions of all chromosomes attached to NE (not shown), and less compact domains containing facultative heterochromatin and euchromatin. These are distributed within nucleoplasm and proved more accessible for cryo-tomography analyses. Figure 4A shows a typical 5-nm thick section extracted from a tomogram. The NE with its NPC is recognized. Within the nucleus, disperse chromatin-dense regions are identified (Figure 4A, dotted lines). As in 2D images, striations are locally observed (arrows). Chromatin rich regions are surrounded by the low density nucleoplasm. In Figure 4B and C, the local observed patterns are compared with the gallery of 5-nm thick projections generated from PDB maps of the NCP (PDB ID: 1EQZ) applying the CTF-modulation and correction of the tomograms (Supplementary Figure S7). Our analysis unambiguously reveals not only striated patterns, which are representative of side views (and are the only recognizable ones in 2D images), but also all other characteristic patterns, including top views (Figure 4B, green). Amongst the variety of side views, it is now possible to fully determine the particle’s orientation (as sketched in Figure 4D), and in particular to distinguish back and front views when the nucleosome dyad axis lies perpendicular to the section plane. Figure 4C shows examples of back (red) and front (yellow) views showing the entry/exit DNA sites. We checked the compatibility of the xy view of each particle, with its xz and yz views (Figure 4E), which unambiguously proves that the particles do indeed correspond to a nucleosome. In addition, these 3D data allowed us, in a few cases, to determine the handedness of the DNA superhelix wrapped around the histone octamer (Supplementary Figure S8). Twelve particles were analysed, all showing a left-handed superhelix, as in the canonical structure.
Figure 4.
CETOVIS of interphase nuclei of embryonic fly brain. (A) A 5-nm thick slice through a tomographic reconstruction showing an area of nucleus (on the right) with a segment of NE containing an NPC. Dashed lines delineate two chromatin areas, in which individual nucleosomes are recognizable as dispersed 11 nm densities. (B and C) Enlargements of the chromatin areas show four characteristic patterns of 5 nm thick nucleosome sections, outlined with coloured square frames. Those are a round top view (green), and a family of side views, with a ‘v-shaped’ section through the pseudo-dyad symmetry axis (blue), a striated front view (red) and an ‘x-shape’ back view showing the entry/exit DNA regions (yellow). These patterns are compared with the corresponding CTF-modulated sections generated from the crystallographic structure of nucleosome (from PDB ID: 1 EQZ). (D) Sketch of different orientations of the NCP (1EQZ) with its superhelical (H) and dyad (δ) axes. (E) Three perpendicular views of a nucleosome selected from the reconstructed volume are compared with the corresponding views of CTF-modulated crystallographic structure. The horizontal smearing visible in the YZ view is a missing wedge artefact. (F) Distribution of P.
Measurement of the distance P between DNA gyres: the nucleosome conformation in situ is variable and more open than the canonical crystallographic structure
On 2D images, wherever striated nucleosomes patterns were recognized (e.g. Figure 3D, D’, E and E’), P was systematically measured on intensity line profiles (Figure 3D’ and E’). Within HT29 and KE37 cell nuclei, P varies from 2.35 to 4.1 nm, with an average value of 3.13 ± 0.36 nm. The distributions of the P-values are shown in Figure 3F for the two types of cells. In both cases the distribution is normal (SW test p = 0.399 and 0.168 respectively) and no significant difference was observed between the two cell lines (Student’s t-test p = 0.518). For Drosophila embryonic brain cells P measurements were obtained from line profiles on computational slices of the reconstructions (orange lines in Figure 4B’ and C’). P also shows a normal distribution (SW test p = 0.45) with an average value 3.37 nm (s.d. 0.45 nm) (Figure 4F).
As we did above for nucleosomes in solution, we compare these data with projections of the canonical crystallographic structure mimicking our imaging conditions (Supplementary Figures S5 and 7 for 2D and 3D data, respectively). Relative to the canonical PDB structure, in situ measurements show a much broader distribution and a shift toward high P-values: 3.13 ± 0.36 nm versus 2.79 ± 0.14 nm for 2D data, and 3.37 ± 0.45 nm versus 2.98 ± 0.24 for 3D ones. Nucleosomes observed in situ in frozen hydrated interphase nuclei on average significantly differ from the particle’s canonical structure. As we found for purified NCP in low salt solutions, they present a whole range of conformations and are more open than the canonical structure.
Note that the measured P-values depend on the imaging conditions (voltage and underfocus values), as analysed in Supplementary Figure S9. CTF correction does not correct for this effect (3D data are CTF-corrected; for 2D data, we checked that CTF-correction does not change measured P-values, see Supplementary Figure S1). Since imaging conditions differ for 2D and tomographic data acquisitions, it is not possible to directly compare, nor simply normalize, the 2D and 3D datasets. Yet, the ratio of the mean P-value of nuclear nucleosomes to that of the corresponding PDB reference is the same for the two datasets (1.12 and 1.13, respectively). This suggests that the different nuclear nucleosome populations are similar whatever the considered cell type.
DISCUSSION
We are able to visualize, recognize and analyse nucleosomes in their native nuclear environment by using cryoEM and tomography of vitreous sections (CEMOVIS and CETOVIS). We detect in interphase nuclei of very different cell types—from human cell lines to embryonic insect cells—the occurrence of multiple conformations of the nucleosomes. P distances measured between DNA gyres wrapped around the particle are on average larger than in the PDB canonical structure, and this parameter varies of more than 0.5 nm around the average. We show that isolated NCP in solution also exhibit variations of this parameter P, and demonstrate the influence of the ionic environment. Compact conformations of the nucleosome are found at high ionic strength (with an average P-value close to that of the PDB at 160 mM), and extended ones in low salt solutions (larger P-values at 15 mM).
A first main step here has been to visualize individual nucleosomes at sufficient resolution to allow the analysis nucleosomal DNA. Conventional EM of chemically fixed or freeze-substituted and stained cells long failed to evidence nucleosomes until Kizilyaprak et al. (55) identified 11 nm fibres and electron dense particles in euchromatin regions. Then, phosphorus-rich DNA periphery and phosphorus-poor core histone octamer were distinguished using electron spectroscopic tomography, but the DNA gyres remained unsolved (56). With the development of cryoEM that keeps the native state of hydrated samples, new perspective were open but so far chromatin was described by CEMOVIS as a homogeneous grainy compartment (46–48) and nucleosomes were only detected as an 11-nm signal in FFTs (46,57). Cryo-electron tomography studies went further, with nucleosomes visualized as ellipsoidal densities ∼10 nm-wide and 6 nm-thick (58,59). We use ultrathin cryo-sections, optimized for nucleosome imaging (40 and 75 nm thick for 2D and 3D imaging, respectively). The comparison of data recorded on vitreous sections and thin films preparations (nucleosomes in solution) shows that cryo-sectioning does not alter nucleosome conformation, making CEMOVIS a method of choice for nucleosome imaging, not only in vitro in concentrated solutions, but also in the cellular context. We identify here individual nucleosomes and resolve DNA gyres wrapped around, which allows their comparison with PDB structures, and opens the way to new questions.
CryoEM provides snapshots of macromolecules and may reflect either static or dynamic variability of nucleosomes. The purified particles analysed here are chemically diverse (in terms of DNA sequence, histone variant content and post-translational modifications). But crystallography and high resolution cryoEM have shown no conformational changes related to the variability of DNA sequence (60), histone tail acetylation or DNA methylation (61–63), and histone variants (64–66), that could be detected at our level of resolution. Moreover, the salt-dependence of the P-distribution in vitro supports the hypothesis of a dynamical conformational transition rather than chemically distinct particle populations.
The conformational changes evidenced here occur along the direction of the DNA superhelical axis, perpendicular to the nucleosome plane (or ‘out of plane’). The two turns of nucleosomal DNA open up in low salt environment. It is different from the well documented partial uncoiling of DNA, or ‘breathing’ that involves breaking of histone–DNA contacts at the entry/exit sites of the DNA (11,12,15,67). We may hypothesize that this ‘out of plane’ phenomenon results from the electrostatic repulsion between the negatively charged DNA gyres, which would be screened at high salt. Indeed, the Debye length in low salt solution is in the range of P (3 nm at 10 mM), whilst it falls below 1 nm above 100 mM. Note that the increase of DNA stiffness in low salt environment could also be involved.
The variations of P could describe different modes of out of plane nucleosome opening, discussed below: (i) a global variation of the DNA superhelical pitch; or (ii) an edge opening/closing, or ‘gaping’, of the particle.
An increase of the DNA superhelical pitch, was first proposed by Marky and Manning (68), as the result of DNA elastic instability. Global out of plane motions in the direction of the superhelical axis were also found in molecular dynamics simulations (69), but with small amplitudes (about 0.1 nm compared to 0.5 nm here). Experimentally, extended conformations with increase of the DNA superhelical pitch in low salt environment, are reported in two EM studies of dried and freeze-dried particles (19,70), but the dimensions of the particles can hardly be reconciled with our cryoEM measurements probably on account of dehydration artefacts associated with freeze-drying.
A gaping transition, corresponding to an edge opening of the nucleosome (like a gaping oyster), was proposed by Mozziconacci et al. (17,71). Interestingly, a Fluorescence Resonance Energy Transfert (FRET) analysis of synthetic 147 bp, 601 particles in dilute solution has reported a change in the distance between DNA gyres on the front side of the particle, i.e. a gaping movement (18). It occurs through about 0.5 to 1 nm, an opening range in good agreement with our measurements. The transition is described as a slow switch between two discrete states: a compact state (with smaller distance between DNA gyres corresponding to the canonical crystallographic structure), and a gaped open state. The authors report a salt dependence: increasing the ionic strength increases the dwell time in compact conformation, and could also increase the compactness of the two states (18). Our P distribution in high salt solutions (Figure 2D), with one peak around 2.7–2.8 nm, and the other around 3 nm, could be indicative of such a switch mechanism. Compact canonical conformations would disappear with the increase of Debye length in low salt solutions.
Although gaping is more probable than a global increase of the superhelical pitch, sorting back and front P-distances would be necessary to tell apart these two possibilities. But it would require a much larger sampling to obtain statistically reliable information. Note that both may exist. More generally, further studies are necessary to characterize this conformational change, for example simulations of cryoEM images obtained from gaped and extended atomic models, or hybrid approaches, such as the combination of FRET and cryoEM (72). The use of single particle cryoEM methods developed to access continuous conformational changes (73,74) would also be needed to characterize the conformational modes of the nucleosome.
Interestingly, open conformations that predominate in situ are found in vitro in low salt solutions, whilst high salt conditions (in the range of 150 mM usually referred as ‘physiological’) favour compact canonical-like structures. The net negative charge of the NCP is about 150 (in the absence of post-translational modification and binding of additional basic proteins). At particle concentrations ranging in our solutions from 25 to 50% (i.e. 1.2–2.4 mM), the counter-ion concentration reaches 180–360 mM. As a result, low salt concentrated solutions may present a total cation concentration closer to the nuclear physiological value, whilst high salt ones may be well above. Although counterintuitive, low added salt solutions may therefore be more relevant chromatin models.
By comparison with low salt concentrated solutions, in situ, the P distribution is significantly broader and shifted toward higher values. This is not surprising: in situ nucleosomes interact with many other nuclear components (histone H1, remodellers, enzymes). The topological constraints introduced by the continuity of the chromatin fibre could also play a role, as also the more complex ionic environment (for example the presence of divalent cations).
Ionic effects play a role at all levels of chromatin organization: histone tails that condense or extend (75), inter-particle interactions (76,77), order-disorder transitions and local tuning of chromatin concentrations (21,77,78), and, at large scale, coil-globule transitions of chromosome domains (79–81). Local changes within the nuclear ionic environment (for example a local increase in salt concentration induced by release of counter-ions upon binding of basic proteins), could trigger conformational changes of the nucleosome and regulate its accessibility. Opening/closing the DNA gyres could also contribute to the capability of chromatin to accommodate various levels of supercoiling (‘topological buffer’(82)). However, the interplay between the conformation of the particles, their stacking/unstacking, and the chain topology remains to be understood.
Our observations are likely to describe only part of the conformational variability of nucleosomal DNA in situ. We did identify here only those particles that resemble closely the canonical structure. In particular, we did not search for more exotic particles, such as hemisomes, hexasomes, reversomes (6,83,84). To progress in this direction, CETOVIS, allowing us to follow the helical path of the DNA wrapped on single nucleosomes, is a very promising tool. Lastly, to question the physiological significance of nucleosome opening it is necessary to target in situ measurements. Correlative light-electron microscopy (85) approaches are promising tools to map nucleosomes and investigate possible conformational differences between particles found in hetero- and euchromatin regions, in telomeres or centromeres, or at transcription, replication and repair foci for example.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J.J. Lacapère and L. Issop (Faculté X. Bichat, Paris) and A.M. Tassin (I2BC, Gif-sur-Yvette) for providing HT29 and KE37 cells, respectively. We thank G. Pehau-Arnaudet for his support at the federative FEG-cryo-electron microscope (Institut Pasteur, Paris) and J. Dubochet for access and support at the LAU (Uni. Lausanne, Switzerland). We thank H. Gnaegi (Diatome) for providing diamond knives for tomographic experiments and M. Kunz (Univ. Frankfurt) for help with tilt series reconstruction. We also thank an anonymous referee for contributing to the discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale de la Recherche [ANR-06-PCVI-0029]; Investissements d’Avenir’ LabEx PALM [ANR-10-LABX-0039-PALM]; German Research Foundation [EL 861/1-1 to M.E., D.G.]. Funding for open access charge: Laboratoire de Physique des Solides; CNRS UMR 8502 Deutsche Forschungsgemeinschaft [EL 861/1-1].
Conflict of interest statement. None declared.
REFERENCES
- 1. Luger K., Mäder A.W., Richmond R.K., Sargent D.F., Richmond T.J.. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997; 389:251–260. [DOI] [PubMed] [Google Scholar]
- 2. Harp J.M., Hanson B.L., Timm D.E., Bunick G.J.. Asymmetries in the nucleosome core particle at 2.5 Å resolution. Acta Crystal. D. 2000; 56:1513–1534. [DOI] [PubMed] [Google Scholar]
- 3. White C.L., Suto R.K., Luger K.. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001; 20:5207–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Davey C.A., Sargent D.F., Luger K., Maeder A.W., Richmond T.J.. X-ray structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 2002; 319:1097–1113. [DOI] [PubMed] [Google Scholar]
- 5. Chua E.Y., Vogirala V.K., Inian O., Wong A.S., Nordenskiöld L., Plitzko J.M., Sandin S.. 3.9 Å structure of the nucleosome core particle determined by phase-plate cryo-EM. Nucleic Acids Res. 2016; 44:8013–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zlatanova J., Bishop T.C., Victor J.M., Jackson V., van Holde K.. The nucleosome family: dynamic and growing. Structure. 2009; 17:160–171. [DOI] [PubMed] [Google Scholar]
- 7. Andrew A.J., Luger K.. Nucleosome structure(s) and stability: variation on a theme. Annu. Rev. Biophys. 2011; 40:99–117. [DOI] [PubMed] [Google Scholar]
- 8. Tan S., Davey C.A.. Nucleosome structural studies. Curr. Opin. Struct. Biol. 2011; 21:128–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ramachandran S., Henikoff S.. Nucleosome dynamics during chromatin remodeling in vivo. Nucleus. 2016; 7:20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Polach K.J., Widom J.. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 1995; 254:130–149. [DOI] [PubMed] [Google Scholar]
- 11. Blossey R., Schiessel H.. The dynamics of the nucleosome: thermal effects, external forces and ATP. FEBS J. 2011; 278:3619–3632. [DOI] [PubMed] [Google Scholar]
- 12. Koopmans W.J.A., Brehm A., Logie C., Schmidt T., Van Noort J.. Single-pair FRET microscopy reveals mononucleosome dynamics. J. Fluoresc. 2007; 17:785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li G., Widom J.. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004; 11:763–769. [DOI] [PubMed] [Google Scholar]
- 14. Ordu O., Lusser A., Dekker N.H.. Recent insights from in vitro single molecule studies into nucleosome structure and dynamics. Biophys. Rev. 2016; 20168:S33–S49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tomschik M., Zheng H., van Holde K., Zlatanova J., Leuba S.H.. Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:3278–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Böhm V., Hieb A.R., Andrews A.J., Gansen A., Rocker A., Toth K., Luger K., Langowski J.. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2011; 39:3093–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mozziconacci J., Victor J.M.. Nucleosome gaping supports a functional structure for the 30nm chromatin fiber. J. Struct. Biol. 2003; 143:72–76. [DOI] [PubMed] [Google Scholar]
- 18. Ngo T.M., Ha T.. Nucleosomes undergo slow spontaneous gaping. Nucleic Acids Res. 2015; 43:3964–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Czarnota G., Ottensmeyer F.P.. Structural states of the nucleosome. J. Biol. Chem. 1996; 271:3677–3683. [DOI] [PubMed] [Google Scholar]
- 20. Leforestier A., Fudaley S., Livolant F.. Spermidine-induced aggregation of nucleosome core particles: evidence for multiple liquid crystalline phases. J. Mol. Biol. 1999; 290:481–494. [DOI] [PubMed] [Google Scholar]
- 21. Mangenot S., Leforestier A., Durand D., Livolant F.. Phase diagram of nucleosome core particles. J. Mol. Biol. 2003; 333:907–916. [DOI] [PubMed] [Google Scholar]
- 22. Livolant F., Mangenot S., Leforestier A., Bertin A., De Frutos M., Raspaud E., Durand D.. Are liquid crystalline properties of nucleosomes involved in chromosome structure and dynamics. Philos. Trans. A Math. Phys. Eng. Sci. 2006; 364:2615–2633. [DOI] [PubMed] [Google Scholar]
- 23. Bertin A., Mangenot S., Renouard M., Durand D., Livolant F.. Structure and phase diagram of nucleosome core particles aggregated by multivalent cations. Biophys. J. 2007; 93:3652–3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leforestier A., Dubochet J., Livolant F.. Bilayers of nucleosome core particles. Biophys. J. 2001; 81:2414–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Issop L., Ostuni M.A., Lee S., Laforge M., Péranzi G., Rustin P., Lacapère J.J.. Translocator protein-mediated stabilization of mitochondrial architecture during inflammation stress in colonic cells. PLoS One. 2016; 11:e0152919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gogendeau D., Guichard P., Tassin A.M.. Purification of centrosomes from mammalian cell lines. Methods Cell Biol. 2015; 129:171–189. [DOI] [PubMed] [Google Scholar]
- 27. Leforestier A., Livolant F.. Cholesteric liquid crystalline DNA; a comparative analysis of cryofixation methods. Biol. Cell. 1991; 71:115–122. [DOI] [PubMed] [Google Scholar]
- 28. Hsieh C.-E., Leith A., Mannella C.A., Frank J., Marko M.. Towards high-resolution three-dimensional imaging of native mammalian tissue: electron tomography of frozen-hydrated rat liver sections. J. Struct. Biol. 2006; 153:1–13. [DOI] [PubMed] [Google Scholar]
- 29. Pierson J., Fernández J.J., Bos E., Amini S., Gnaegi H., Vos M., Peters P.J.. Improving the technique of vitreous cryo-sectioning for cryo-electron tomography: electrostatic charging for section attachment and implementation of an anti-contamination glove box. J. Struct. Biol. 2010; 169:219–225. [DOI] [PubMed] [Google Scholar]
- 30. Wade R., Chrétien D.. Lattice defects in microtubules: protofilament numbers vary within individual microtubules. J. Cell Biol. 1992; 117:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005; 152:36–51. [DOI] [PubMed] [Google Scholar]
- 32. Hagen W.J.H., Wan W., Briggs J.A.C.. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 2017; 197:191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kremer J.R., Mastronarde D.N., McIntosh J.R.. Computer visualisation of three-dimensional image data using IMOD. J. Struct. Biol. 1996; 116:71–76. [DOI] [PubMed] [Google Scholar]
- 34. Kunz M., Frangakis A.S.. Super-sampling SART with ordered subsets. J. Struct. Biol. 2014; 188:107–115. [DOI] [PubMed] [Google Scholar]
- 35. Kunz M., Frangakis A.S.. Three-dimensional CTF correction improves the resolution of electron tomograms. J. Struct. Biol. 2017; 197:114–122. [DOI] [PubMed] [Google Scholar]
- 36. Frangakis A., Hegerl R.. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 2001; 135:239–205. [DOI] [PubMed] [Google Scholar]
- 37. Ludtke S.J., Baldwin P.R., Chiu W.. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 1999; 128:82–97. [DOI] [PubMed] [Google Scholar]
- 38. Collins T.J. ‘ImageJ for microscopy’. Biotechniques. 2007; 43:25–30. [DOI] [PubMed] [Google Scholar]
- 39. Al-Amoudi A., Studer D., Dubochet J.. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J. Struct. Biol. 2005; 150:109–121. [DOI] [PubMed] [Google Scholar]
- 40. Han H.M., Zuber B., Dubochet J.. Compression and crevasses in vitreous sections under different cutting conditions. J. Microsc. 2008; 230:167–171. [DOI] [PubMed] [Google Scholar]
- 41. Dubochet J., Zuber B., Eltsov M., Bouchet‐Marquis C., Al‐Amoudi A., Livolant F.. How to ‘read’ a vitreous section. Methods Cell Biol. 2007; 79:385–406. [DOI] [PubMed] [Google Scholar]
- 42. Sartori Blanc N., Senn A., Leforestier A., Livolant F., Dubochet J.. DNA in human and stallion spermatozoa forms local hexagonal packing with twist and many defects. J. Struct. Biol. 2001; 134:76–81. [DOI] [PubMed] [Google Scholar]
- 43. Sader K., Studer D., Zuber B., Gnaegi H., Trinick J.. Preservation of high resolution protein structure by cryo-electron microscopy of vitreous sections. Ultramicroscopy. 2009; 110:43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pierson J., Ziese U., Sani M., Peters P.J.. Exploring vitreous cryo-section-induced compression at the macromolecular level using electron cryo-tomography; 80S yeast ribosomes appear unaffected. J. Struct. Biol. 2011; 173:345–349. [DOI] [PubMed] [Google Scholar]
- 45. Mangenot S., Leforestier A., Durand D., Livolant F.. X-ray diffraction characterization of the dense phases formed by nucleosome core particles. Biophys. J. 2003; 84:2570–2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. McDowall A.W., Smith J.M., Dubochet J.. Cryo-electron microscopy of vitrified chromosomes in situ. EMBO J. 1986; 5:1395–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dubochet J., Blanc N.S.. The cell in absence of aggregation artifacts. Micron. 2001; 32:91–99. [DOI] [PubMed] [Google Scholar]
- 48. Bouchet-Marquis C., Dubochet J., Fakan S.. Cryoelectron microscopy of vitrified sections: a new challenge for the analysis of functional nuclear architecture. Histochem. Cell Biol. 2006; 125:43–51. [DOI] [PubMed] [Google Scholar]
- 49. Kalverda B., Röling M.D., Fornerod M.. Chromatin organization in relation to the nuclear periphery. FEBS Lett. 2008; 582:2017–2022. [DOI] [PubMed] [Google Scholar]
- 50. Padeken J., Heun P.. Nucleolus and nuclear periphery: velcro for heterochromatin. Curr. Opin. Cell Biol. 2014; 28:54–60. [DOI] [PubMed] [Google Scholar]
- 51. Therianos S., Leuzinger S., Hirth F., Goodman C.S., Reinchert H.. Embryonic development of the Drosophila brain: formation of commissural and descending pathway. Development. 1995; 121:3849–3860. [DOI] [PubMed] [Google Scholar]
- 52. Eltsov M., Dubé N., Yu Z., Pasakarnis L., Haselmann-Weiss U., Brunner D., Frangakis A.S.. Quantitative analysis of cytoskeletal reorganization during epithelial tissue sealing by large-volume electron tomography. Nat. Cell Biol. 2015; 17:605–614. [DOI] [PubMed] [Google Scholar]
- 53. Mahovald A.P. Fixation problems for electron microscopy of Drosophila embryos. Dros. Inf. Serv. 1962; 36:130–131. [Google Scholar]
- 54. Limbourg B., Zalokar M.. Permeabilization of Drosophila eggs. Dev. Biol. 1973; 35:382–387. [DOI] [PubMed] [Google Scholar]
- 55. Kizilyaprak C., Spehner D., Devys D., Schultz P.. In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS One. 2010; 5:e11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fussner E., Strauss M., Djuric U., Li R., Ahmed K., Hart M., Ellis J., Bzett-Jones D.P.. Open and closed domains in the mouse genome are configured as 10-nm chromatin fibres. EMBO Rep. 2012; 13:992–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gan L., Ladinsky M.S., Jensen G.J.. Chromatin in a marine picoeukaryote is a disordered assemblage of nucleosomes. Chromosoma. 2013; 122:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scheffer M.P., Eltsov M., Frangakis A.S.. Evidence for short-range helical order in the 30-nm chromatin fibers of erythrocyte nuclei. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:16992–16997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mahamid J., Pfeffer S., Schaffer M., Villa E., Danev R., Cuellar L.K., Baumeister W.. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science. 2016; 351:969–972. [DOI] [PubMed] [Google Scholar]
- 60. Frouws T.D., Duda S.C., Richmond T.J.. X ray structure of the MNTV-A nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:1214–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watanabe S., Resch M., Lilyestrom W., Clark N., Hansen J.C., Luger K.. Structural characterization of H3K56Q nucleosomes and nucleosomal arrays. Biochim. Biophys. Acta. 2010; 1799:480–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wakamori M., Fujii Y., Suka N., Shirouzu M., Sakamoto K., Umehara T., Yokoyama S.. Intra-and inter-nucleosomal interactions of the histone H4 tail revealed with a human nucleosome core particle with genetically-incorporated H4 tetra-acetylation. Sci. Rep. 2015; 5:17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fujii Y., Wakamori M., Umehara T., Yokoyama S.. Crystal structure of human nucleosome core particle containing enzymatically introduced CpG methylation. FEBS Open Bio. 2016; 6:498–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tachiwana H., Osakabe A., Shiga T., Miya Y., Kimura H., Kawaga W., Kurumizaka H.. Structures of human nucleosomes containing major histone H3 variants. Acta Cristallogr. D Biol. Crystallogr. 2011; 67:578–583. [DOI] [PubMed] [Google Scholar]
- 65. Suto R.K., Clarkson M.J., Tremethick D.J., Luger K.. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000; 7:1121–1124. [DOI] [PubMed] [Google Scholar]
- 66. Roulland Y., Ouararhni K., Naidenov M., Ramos L., Shuaib M., Syed S.H., Tachiwana H.. The flexible ends of CENP-A nucleosome are required for mitotic fidelity. Mol. Cell. 2016; 63:674–685. [DOI] [PubMed] [Google Scholar]
- 67. Li G., Levitus M., Bustamante C., Widom J.. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 2005; 12:46–53. [DOI] [PubMed] [Google Scholar]
- 68. Marky N.L., Manning G.S.. The elastic resilience of DNA can induce all-or-none structural transitions in the nucleosome core particle. Biopolymers. 1991; 31:1543–1557. [DOI] [PubMed] [Google Scholar]
- 69. Roccatano D., Barthel A., Zacharias M.. Structural flexibility of the nucleosome core particle at atomic resolution studied by molecular dynamics simulation. Biopolymers. 2007; 85:407–421. [DOI] [PubMed] [Google Scholar]
- 70. Oliva R., Bazett-Jones D.P., Locklear L., Dixon G.H.. Histone hyperacetylation can induce unfolding of the nucleosome core particle. Nucleic Acids Res. 1990; 18:2739–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mozziconacci J., Lavelle C., Barbi M., Lesne A., Victor J.M.. A physical model for the condensation and decondensation of eukaryotic chromosomes. FEBS Lett. 2006; 580:368–372. [DOI] [PubMed] [Google Scholar]
- 72. Komar A.A. Unravelling co-translational protein folding: concept and methods. Methods. 2017; S1046-2023:30273–30276. [Google Scholar]
- 73. Jin Q., Sorzano C.O., de la Rosa-Trevin J.M., Bilbao-Castro J.R., Nunez-Ramirez R., Llorca O., Tama F., Jonic S.. Iterative elastic 3D-to-2D alignment method using normal modes for studying structural dynamics of large macromolecular complexes. Structure. 2014; 22:496–506. [DOI] [PubMed] [Google Scholar]
- 74. Dashti A., Schwander P., Langlois R., Fung R., Li W., Hosseinizadeh A., Liao H.Y., Pallesen J., Sharma G., Stupina V.A. et al. Trajectories of the ribosome as a Brownian nanomachine. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:17492–17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mangenot S., Leforestier A., Vachette P., Durand D., Livolant F.. Salt-induced conformation and interaction changes of nucleosome core particles. Biophys. J. 2002; 82:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Korolev N., Allahverdi A., Lyubartsev A.P., Nordenskiöld L.. The polyelectrolyte properties of chromatin. Soft Matter. 2012; 8:9322–9333. [Google Scholar]
- 77. Cui Y., Bustamante C.. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Widom J. Physicochemical studies of the folding of the 100 Å nucleosome filament into the 300 Å filament: cation dependence. J. Mol. Biol. 1986; 190:411–424. [DOI] [PubMed] [Google Scholar]
- 79. Emanuel M., Radja N.H., Henriksson A., Schiessel H.. The physics behind the larger scale organization of DNA in eukaryotes. Phys. Biol. 2009; 6:025008. [DOI] [PubMed] [Google Scholar]
- 80. Chiariello A.M., Annunziatella C., Bianco S., Esposito A., Nicodemi M.. Polymer physics of chromosome large-scale 3D organisation. Sci. Rep. 2016; 6:29775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cortini R., Barbi M., Caré B.R., Lavelle C., Lesne A., Mozziconacci J., Victor J.M.. The physics of epigenetics. Rev. Mod. Phys. 2016; 88:025002. [Google Scholar]
- 82. Bancaud A., e Silva N.C., Barbi M., Wagner G., Allemand J.F., Mozziconacci J., Viovy J.L.. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat. Struct. Mol. Biol. 2006; 13:444–450. [DOI] [PubMed] [Google Scholar]
- 83. Kato D., Osakabe A., Arimura Y., Mizukami Y., Horikoshi N., Saikusa K., Akashi S., Nishimura Y., Park S.-Y., Nogami J. et al. Crystal structure of the overlapping dinucleosome composed of hexasome and octasome. Science. 2017; 356:205–208. [DOI] [PubMed] [Google Scholar]
- 84. Vlijm R., Lee M., Lipfert J., Lusser A., Dekker C., Dekker N.. Nucleosome assembly dynamics involve spontaneous fluctuations in the handedness of tetrasomes. Cell Rep. 2015; 10:216–225. [DOI] [PubMed] [Google Scholar]
- 85. Schorb M., Gaechter L., Avinoam O., Sieckmann F., Clarke M., Bebeacua C., Briggs J.A.. New hardware and workflows for semi-automated correlative cryo-fluorescence and cryo-electron microscopy/tomography. J. Struct. Biol. 2017; 197:83–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.