Abstract
Trinucleotide repeat (TNR) instability is associated with over 42 neurodegenerative diseases and cancer, for which the molecular mechanisms remain to be elucidated. We have shown that the DNA base excision repair (BER) pathway and its central component, DNA polymerase β (pol β), in particular, its polymerase activity plays an active role in regulating somatic TNR instability. Herein, we revealed a unique role of the pol β dRP lyase in preventing somatic TNR instability. We found that deficiency of pol β deoxyribose phosphate (dRP) lyase activity locked the pol β dRP lyase domain to a dRP group, and this ‘tethered’ pol β to its template forcing the polymerase to perform a processive DNA synthesis. This subsequently promoted DNA strand slippage allowing pol β to skip over a template loop and causing TNR deletion. We showed that the effects were eliminated by complementation of the dRP lyase deficiency with wild-type pol β protein. The results indicate that pol β dRP lyase activity restrained the pol β-dRP interaction to suppress a pol β processive DNA synthesis, thereby preventing TNR deletion. This further implicates a potential of pol β dRP lyase inhibition as a novel treatment of TNR-expansion diseases.
INTRODUCTION
Expansion of trinucleotide repeats (TNRs) in the human genome is responsible for the development of over 42 human neurodegenerative and neuromuscular diseases including Huntington's disease (HD), myotonic dystrophy type 1 (DM1), fragile X syndrome (FXS) and Friedreich's ataxia (FRDA) (1,2). TNRs associated with the neurodegenerative diseases include CAG, CTG, CGG and GAA repeats, which are located at different loci of the related disease-causing genes (2). On the other hand, deletions of CAG repeats in the androgen receptor are associated with prostate and ovarian cancers (3,4). Thus, TNR instability, i.e. TNR expansion and deletion, is involved in the initiation and progression of both neurodegeneration and cancer. Extensive studies have shown that DNA replication, repair, recombination, and gene transcription underlie TNR instability (5–7). Among them, the role of DNA repair in mediating TNR somatic instability is implicated due to its association with age-dependent somatic TNR expansions in non-proliferating neuronal tissues of human (8). Major DNA repair pathways including DNA base excision repair (BER) (9), mismatch repair (MMR) (10) and transcription coupled-nucleotide excision repair (TC-NER) (11) are all involved in modulating TNR instability. An age-dependent somatic TNR expansion induced by oxidative DNA damage highlights a crucial role of the BER pathway in modulating somatic TNR instability (12). This is because TNRs are tandem repeats containing stretches of guanines, cytosines, and adenines, which form hotspots of DNA base damage induced by endogenous and environmental stresses. These include oxidized and alkylated bases that are subject to BER (13). We have previously shown that BER of oxidized and alkylated DNA base lesions in tracts of TNRs can cause both TNR deletions and expansions (14–16) upon the location of a base lesion in a duplex TNR tract (17). We further demonstrate that BER of a base lesion located in a TNR hairpin loop results in the removal of the hairpin preventing TNR expansion (16,18). This indicates that DNA base lesions exhibit a position effect on TNR instability during BER. The effect is mediated by the formation of secondary structures that occur in a TNR tract, which subsequently regulates the efficiency of synthesis and removal of TNRs by DNA polymerase β (pol β) and flap endonuclease 1 (FEN1). If pol β synthesize more repeats than those removed by FEN1, repeat expansion occurs. However, if pol β inserts fewer repeats than those removed by FEN1, repeat deletion occurs (17). Thus, the balance between the activities of pol β DNA synthesis and FEN1 flap cleavage in processing TNRs determines whether TNRs are expanded or deleted. Moreover, the balance can be altered by the crosstalk between pol β and the mismatch repair protein, MSH2-MSH3, which stimulates pol β synthesis of TNRs and promotes TNR expansion (19). These results have demonstrated the importance of the polymerase activity of pol β in mediating TNR instability during BER.
However, as a bifunctional DNA polymerase, pol β also contains an 8-kD N-terminal lyase domain (20), which has the deoxyribose phosphate (dRP) lyase activity that removes a 5′-dRP group generated from the 5′-incision of an abasic site by AP endonuclease 1 (APE1) (21) through β-elimination (22,23). It has been shown that it is the pol β dRP lyase activity that protects cells from DNA damage-induced cytotoxicity rather than its DNA polymerase activity (24,25). Thus, the dRP lyase activity of pol β plays an essential role in mammalian embryonic and cellular survival (26,27), whereas its polymerase activity can be compensated by DNA replication polymerases and other DNA repair polymerases. It has also been shown that the dRP lyase activity of pol β is much faster than its DNA synthesis activity (20). A recent study from the Wilson group has also demonstrated that pol β dRP lyase domain performs a processive search for a 1 nt gap (28,29). This indicates that pol β dRP lyase initially searches for the 1 nt gap with a dRP group and removes the dRP group leaving the 1 nt gap (22,23,30) that is subsequently filled in by pol β gap-filling DNA synthesis via its polymerase activity. This results in a nick that is ligated by DNA ligases (20). Thus, the dRP lyase and polymerase activities of pol β are sequentially coordinated to ensure the efficient single-nucleotide/short-patch BER that removes AP sites and dRP groups preventing cell death and genome instability. However, it remains a puzzle how pol β dRP lyase activity is involved in the cellular maintenance of genome stability including repeat sequence stability.
Since TNRs form hotspots of DNA base lesions and their lengths undergo dynamic changes during BER, BER-mediated TNR instability can be used as an ideal system to test the importance of both the polymerase and dRP lyase activities of pol β in maintaining genome stability. Furthermore, since the removal of 5′-dRP residue by the pol β dRP lyase is mediated by β-elimination during which a transient covalent bond is formed between Lys 72 of the pol β dRP lyase domain and a dRP group (31), it is possible that during BER of a base lesion in a TNR tract, the interaction between the pol β dRP lyase domain and a dRP group may prevent DNA slippage and the formation of DNA secondary structures, thereby suppressing TNR instability. Also, since the lyase domain of pol β cooperates with the N-terminal subdomain of its polymerase domain to form a doughnut-shaped structure to wrap around double-strand DNA (20), pol β dRP lyase activity may also cooperate with its polymerase activity to prevent TNR instability.
In this study, employing pol β mutational analysis, a fluorescence-based DNA slippage assay that was newly developed by our group along with the sodium boric hydride crosslink trapping assay, we explored a unique role of the pol β dRP lyase activity in modulating TNR instability during BER. Surprisingly, we found that pol β dRP lyase activity prevented TNR deletions by suppressing the processive synthesis of TNRs by pol β as well as inhibiting DNA strand slippage and the formation of TNR secondary structures on the template strand.
MATERIALS AND METHODS
Materials
DNA oligonucleotides were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). Deoxynucleotide 5′-triphosphates (dNTPs) were obtained from Sigma-Aldrich (St. Louis, MO, USA). T4 polynucleotide kinase and terminal deoxynucleotidyl transferase were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Radionucleotides [γ-32P] ATP (3000 Ci/mmol) and Cordycepin 5′-triphosphate 3′-[α-32P] (5000 Ci/mmol) were purchased from Perkin Elmer Inc. (Boston, MA, USA). Micro Bio-Spin 6 chromatography columns were acquired from Bio-Rad Laboratories (Hercules, CA, USA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), l(+)-glutamine, and 0.25% trypsin-EDTA were purchased from Thermo Fisher Scientific (Waltham, MA, USA). All standard chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Thermo Fisher Scientific (Waltham, MA, USA). Escherichia coli uracil-DNA glycosylase (UDG) was purchased from New England BioLabs Inc. (Ipswich, MA, USA). Purified recombinant pol β dRP lyase mutant (K72A and H34G) and pol β 8 kD domain proteins were generous gifts from Dr Samuel H. Wilson at the National Institute of Environmental Health Sciences/National Institutes of Health. Purified recombinant human APE1, wild-type (WT) pol β, FEN1 and DNA ligase I (LIG I) were expressed and purified as described previously (14,18,32).
Oligonucleotide substrates
DNA oligonucleotide substrates containing uracil (U), were designed to mimic a base lesion that occurs in a (GAA)20 or (CAG)20 repeat tract. The guanine in the tenth repeat unit of (GAA)20 or (CAG)20-containing substrates was substituted with U. Substrates were constructed by annealing a U-containing oligonucleotide to its template strand at a molar ratio of 1:2. The U-containing substrates were incubated with E. coli UDG to create an abasic site with a native sugar residue before each experiment. The 5′-dRP lyase substrates were subsequently generated upon treatment with APE1. The sequences of the oligonucleotides are listed in Supplementary Table S1.
In vitro BER in mouse embryonic fibroblast (MEF) cell extracts
Pol β null (pol β−/−) and wild-type (pol β+/+) MEFs were grown to near confluence. Cells were washed twice with phosphate buffered saline (PBS), harvested by cell scrapers, and collected by centrifugation at 3000 rpm for 15 min. Cell extracts were made as described previously (33) and dialyzed into BER reaction buffer containing 30 mM 4-(2-hydroxyethyl)-1-piperazineethane-sulfonic acid (HEPES), 50 mM KCl, 0.1 mM EDTA, 0.1 mg/ml bovine serum albumin, 1 mM dithiothreitol (DTT), and 0.01% Nonidet P-40, pH 7.5. Substrates were pre-incubated with UDG at 37°C for 30 min. This allowed all uracils in the substrates to be converted into native abasic sites that were completely cleaved by APE1. In vitro BER of a native abasic site in pol β−/− and pol β+/+ cell extracts was performed by incubating a (GAA)20 or (CAG)20 repeat substrate containing a native abasic site (25 nM) with 60 μg cell extracts and 25 nM APE1 in a 25 μl reaction mixture that contained BER reaction buffer, 10% (v/v) glycerol, 0.25% (w/v) inositol, 50 μM dNTPs, 5 mM Mg2+ and 2 mM ATP. BER in pol β−/− cell extracts that were complemented with purified pol β protein, was examined in the presence of 10 nM pol β K72A or H34G or WT or pol β 8 kD domain protein under the same experimental condition. Reaction mixture was assembled on ice and incubated at 37°C for 30 min. The template strand of the substrates was biotinylated at the 5′-end for isolation of repaired products. Repaired products were incubated with avidin agarose beads (Pierce-Thermo Scientific, Rockford, IL, USA) in the binding buffer that contained 0.1 M phosphate, 0.15 M NaCl, pH 7.2 and 1% (v/v) Nonidet P-40 at 4°C for 2 h with rotation. Agarose beads were centrifuged at 5000 rpm for 1 min and washed three times with binding buffer. Repaired strands were then separated from their template strands by incubation with 0.15 M NaOH for 15 min with rotation at room temperature. This allowed the repaired strands to be released into the supernatant. Repaired strands were then collected from the supernatant by centrifugation at 5000 rpm for 2 min and precipitated with ethanol. Repaired strands were dissolved in TE buffer and stored at –20°C for subsequent repeat sizing analysis.
BER reconstituted with purified enzymes
BER of a native abasic site was reconstituted by incubating 25 nM purified APE1, 10 nM pol β K72A or H34G or WT protein along with 25 nM FEN1 and 25 nM LIG I, with 25 nM (GAA)20 or (CAG)20 repeat-containing substrates with a native abasic site. Reaction mixture (20 μl) contained BER reaction buffer with 50 μM dNTPs, 5 mM Mg2+, 2 mM ATP, and indicated concentrations of BER enzymes and substrates. Reaction mixture was assembled on ice and incubated at 37°C for 30 min. Reactions were terminated by heating at 95°C for 10 min in stopping buffer containing 95% formamide and 2 mM EDTA. Repair intermediates and products were separated by 18% urea-denaturing polyacrylamide gel electrophoresis (PAGE) and detected by the Pharos FX Plus PhosphorImager from Bio-Rad Laboratory (Hercules, CA, USA). To isolate repaired products, the template strand of the substrates was biotinylated at the 5′-end. Repaired strands were separated from the template strand with 0.15 M NaOH and precipitated with ethanol, dissolved in TE buffer, and stored at –20°C for subsequent repeat sizing analysis.
Sizing analysis of TNR length
Repaired products resulting from BER reconstituted with MEF cell extracts or purified enzymes in the context of (GAA)20 and (CAG)20 repeats were amplified by PCR with a forward primer (5′-CGA GTC ATC TAG CAT CCG TA-3′) and a reverse primer tagged by a 6-carboxyfluorescein (6-FAM) (5′-6-FAM-CA ATG AGT AAG TCT ACG TA-3′). PCR amplification was performed under the following conditions: 95°C for 10 min, 1 cycle; 95°C for 30 s, 50°C for 30 s and 72°C for 1.5 min, 35 cycles; 72°C for 1 h. The 6-FAM-labeled PCR products were then subjected to capillary electrophoresis using an ABI 3130XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) (Florida International University DNA Sequencing Core Facility). The size of repaired products was determined by DNA fragment analysis with GeneMapper version 5.0 software (Applied Biosystems, Foster City, CA). Size standards, MapMarker 1000 (Bioventures, Murfreesboro, TN) were run in parallel with PCR-amplified repaired products. For all the experiments, only repaired strands were able to be amplified by PCR with AmpliTaq Gold 360 DNA polymerase (Thermo Fisher Scientific, Waltham, MA, USA) used in our experiments.
Formation of TNR loops probed by S1 nuclease digestion
Formation of TNR hairpin or loop structures in the template strand of the substrates was probed by incubating 18 U S1 nuclease with 25 nM substrates that contained (GAA)20 and (CAG)20 repeats. Substrates containing a native abasic site were incubated with 25 nM APE1, 10 nM pol β K72A or H34G or WT, and 25 nM LIG I in the absence or presence of 25 nM FEN1 at 37°C for 30 min allowing the completion of BER. Subsequently, the reaction mixture was subject to S1 nuclease digestion. The reaction mixture of S1 nuclease digestion (20 μl) was assembled in reaction buffer containing 50 mM sodium acetate (pH 4.5), 280 mM NaCl and 4.5 mM ZnSO4, and was incubated at 37°C for 3, 5, 10 and 15 min, and subsequently subjected to protease K digestion at 55°C for 30 min. Reaction mixture was then incubated at 95°C for 10 min for denaturing DNA. Substrates and nuclease digestion products were separated by 18% urea-denaturing PAGE and detected by a PhosphorImager. Synthesized DNA size markers were used to indicate the size of nuclease cleavage products.
Measurement of TNR strand slippage with a fluorescence-based DNA slippage assay
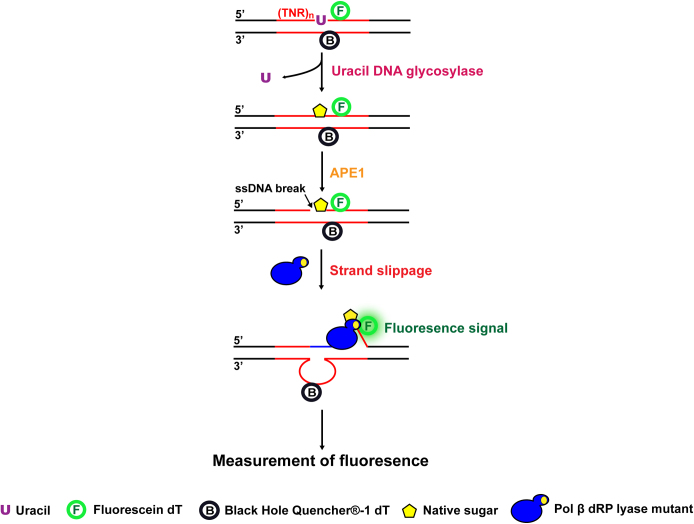
To further determine the ability for the dRP lyase domain of pol β mutants or WT to interact and hold/grab the downstream strand of the damaged strand for preventing DNA slippage after APE1 5′-incision of an abasic site, a new fluorescence-based DNA slippage assay was developed (Figure 1). For this assay, a 6-fluorescein (6-FAM)-tagged dT was inserted downstream of the uracil (U) at the damaged strand of the (GAA)20 and (CAG)20 repeat-containing substrates or random DNA sequence-containing substrates, while a Black Hole Quencher-1 (BHQ-1)-tagged dT was inserted in the template strand in proximity to the 6-FAM-dT. For (GAA)20-repeat-containing substrates, 6-FAM-dT was inserted immediately downstream of the U, whereas BHQ-1-dT was inserted in the template strand opposite to the A that was four nucleotides upstream of the U on the damaged strand (Supplementary Table S1). For (CAG)20 repeat-containing substrates, 6-FAM-dT was inserted 1 nt downstream of the U, while BHQ-1-dT was inserted opposite to the A that was four nucleotides upstream of the dU on the damaged strand (Supplementary Table S1). For the random DNA sequence-containing substrate, 6-FAM-dT was inserted immediately downstream of the U, whereas BHQ-1-dT was inserted in the template strand opposite to the A that was five nucleotides upstream of the U on the damaged strand (Supplementary Table S1). The (GAA)20 or (CAG)20 repeat or random DNA sequence-containing substrates with a native abasic site was generated by incubating the substrates with UDG at 37°C for 30 min. Subsequently, 50 nM substrates containing a native abasic site were incubated with 25 nM APE1, 10 nM pol β K72A or H34G or WT at 37°C in the BER reaction buffer using a CFX Connect Real-Time PCR Detection System from Bio-Rad Laboratories (Hercules, CA). Fluorescence signal was monitored over a 2-min incubation period. The relative fluorescence units (Intensity of fluorescence, IF) were recorded and used to illustrate the level of DNA slippage resulting from the dissociation of the downstream damaged strand from its template strand with the BHQ-1 quencher.
Figure 1.
The schematic diagram of the fluorescence-based DNA slippage assay.
Detection of the crosslink between pol β dRP lyase domain and 5′-dRP by a NaBH4 trapping assay
The crosslink between pol β dRP lyase domain and 5′-dRP group in TNR tracts was captured by incubating 100 nM of pol β K72A or H34G or WT protein with 2.5 nM (GAA)20 or (CAG)20 substrates containing a native sugar in BER reaction buffer with 10 mM MgCl2, 50 μM dNTP and 1 mM 2-mercaptoethanol in the absence or presence of 50 mM NaBH4. The substrates mimic the intermediates resulting from APE1 5′-incision of a native abasic site. This excluded a possibility that APE1 may also crosslink with the native sugar. The substrates were constructed by annealing the 32P-labeled downstream primer with a 5′-phosphorylated uracil and an upstream primer with the template strand at a molar ratio of 1:2:3 (Supplementary Table S1). The 5′-dRP was generated by incubating the U-containing substrates with UDG (5 U) for 30 min. Wild-type and mutant pol β proteins were incubated with the substrates at 37°C at the time intervals of 30 s, 1 min, 5 min, 10 min or 15 min to allow DNA synthesis. The 10 μl reaction mixture was subsequently incubated for 1 h at 37°C in the presence of 50 mM NaBH4 to allow the formation of crosslink, and the reaction was terminated with 4 μl 4× SDS loading buffer containing 100 mM Tris–HCl, pH 6.8, 4% (w/v) SDS, 20% (v/v) glycerol, 0.2% (w/v) bromophenol blue, and 200 mM DTT. All samples were heated at 95°C for 10 min, and pol β-dRP crosslink complexes were separated from the substrates with 10% SDS-PAGE. Substrates and crosslink complexes were detected and analyzed using the Pharos FX Plus PhosphorImager.
RESULTS
The dRP lyase activity of pol β prevents GAA and CAG repeat deletions during BER
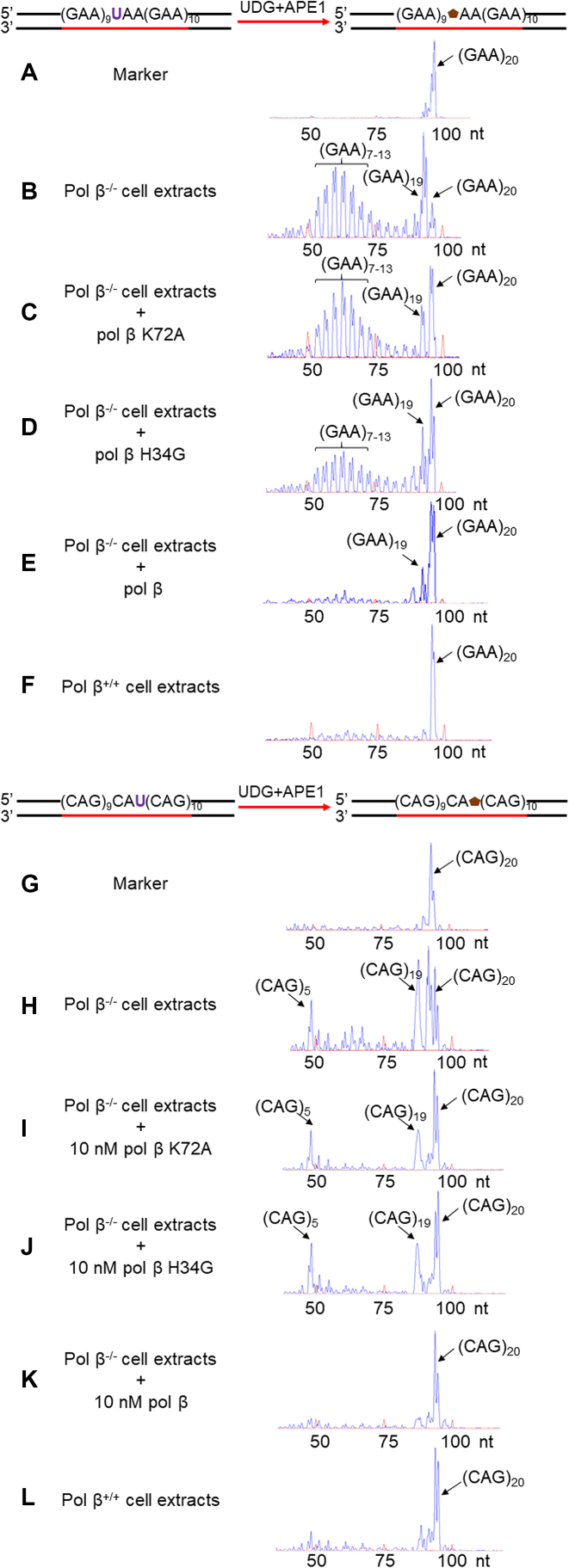
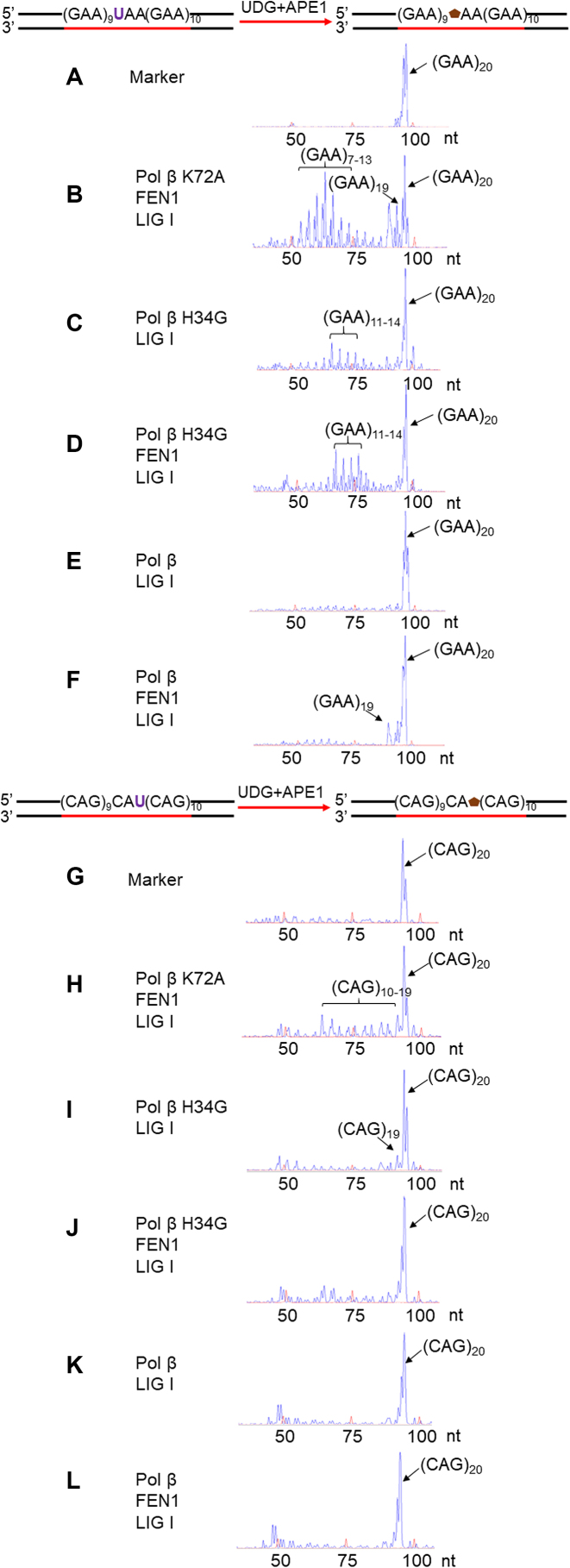
To determine whether the dRP lyase activity of pol β is involved in the modulation of TNR instability during BER, we initially examined GAA and CAG repeat instability during BER of a native abasic site in a repeat tract in the absence and presence of pol β using the cell extracts of pol β−/- or pol β+/+ MEFs and a (GAA)20 or (CAG)20 repeat substrate with a native sugar generated from a uracil (Figure 2) (Supplementary Table S1). The results showed that BER in pol β−/− cell extracts resulted in (GAA)19 and (GAA)7–13 deletion products as well as (CAG)19 and (CAG)5 deletion products (Figure 2, panels B and H). These appeared to be generated by the pol β-independent long-patch BER pathway mediated by replicative DNA polymerases suggesting that the pol β-independent long-patch BER pathway also resulted in TNR instability. Complementation of pol β−/− cell extracts with purified pol β K72A mutant protein having less than 10% wild-type level of dRP lyase activity (34) failed to affect repeat deletions (Figure 2, panels C and I) during BER, whereas complementation of pol β−/− cell extracts with pol β H34G mutant protein having ∼50% wild-type level of dRP lyase activity (34) significantly reduced GAA repeat deletions (Figure 2, panel D), but failed to affect CAG repeat deletion (Figure 2, panel J). Complementation of pol β−/− cell extracts with pol β WT eliminated the GAA and CAG repeat deletion products (Figure 2, panels E and K), whereas BER in pol β+/+ cell extracts did not result in any deletion products from the repeats (Figure 2, panels F and L). In addition, complementation of pol β−/− extracts with the pol β 8 kD domain resulted in the similar sizes of GAA and CAG repeat deletion products as the ones generated by pol β−/− extracts mediated-BER (compare Supplementary Figure S1, panels A and B to Figure 2, panels B and H), indicating that pol β dRP lyase domain alone was insufficient to prevent TNR instability during BER. Similarly, BER reconstituted with purified pol β K72A mutant protein resulted in the repeat deletion products containing (GAA)7–19 and (CAG)10–19 (Figure 3, panels B and H). BER reconstituted with pol β H34G mutant protein resulted in deletion products with (GAA)11–14 independent of FEN1 (Figure 3, panels C and D). In contrast, BER reconstituted with pol β H34G mutant led to one CAG repeat deletion in the absence of FEN1 (Figure 3, panel I), but failed to affect repeat instability in the presence of FEN1 (Figure 3, panel J). Consistent with the results from the cell extracts, BER reconstituted with pol β WT proteins did not produce GAA and CAG repeat deletion products in the absence of FEN1 (Figure 3, panels E and K). In the presence of FEN1, BER reconstituted with pol β WT protein only resulted in a (GAA)19 product with one GAA deletion (Figure 3, panel F), but did not cause CAG repeat deletion (Figure 3, panel L). Thus, the results indicate that the dRP lyase activity of pol β prevented TNR deletions during BER.
Figure 2.
Pol β dRP lyase activity prevents TNR deletions during BER mediated by MEFs cell extracts. The effect of pol β dRP lyase activity on TNR instability was examined by reconstituting BER with (GAA)20 or (CAG)20 repeat substrates containing a native abasic site using pol β−/− or pol β+/+ MEF extracts as described in the Materials and Methods. (A) and (G) The DNA fragment analysis results of a DNA marker containing (GAA)20 or (CAG)20 repeats without DNA damage. (B) and (H) The DNA fragment analysis results of the repaired products from BER mediated by pol β−/− MEF extracts with the (GAA)20 or (CAG)20 substrate. (C) and (I) The DNA fragment analysis results of the repaired products from BER mediated by pol β−/−MEFs extracts complemented with pol β K72A mutant protein with the (GAA)20 or (CAG)20 substrate. (D) and (J) The DNA fragment analysis results of the repaired products from BER mediated by pol β−/− MEF extracts complemented with pol β H34G mutant protein with the (GAA)20 or (CAG)20 substrate. (E) and (K) The DNA fragment analysis results of repaired products from BER mediated by pol β−/−MEF extracts complemented with wild-type pol β protein with the (GAA)20 or (CAG)20 substrate. (F) and (L) The DNA fragment analysis result of the repaired products from BER mediated by pol β+/+ MEF extracts with the (GAA)20 or (CAG)20 substrate. The schemes for generating the substrates were illustrated above the graphs. The template strand of the substrate was biotinylated at the 5′-end. Major repair products are indicated with black arrows. The red peaks are DNA size marks. The sizes of DNA fragments are illustrated as nucleotide, nt. The experiment was conducted in triplicate, and only representative results are illustrated in the figures.
Figure 3.
Pol β dRP lyase activity prevents TNR deletions during BER reconstituted with purified BER enzymes. The effect of pol β dRP lyase activity on TNR instability was examined by reconstituting BER with purified BER enzymes and the (GAA)20 or (CAG)20 repeat substrate containing a native abasic site as described in the Materials and Methods. (A) and (G) The DNA fragment analysis results of a DNA marker containing (GAA)20 or (CAG)20 repeats without a base lesion. (B) and (H) The DNA fragment analysis results of the repair products from BER reconstituted with the (GAA)20 or (CAG)20 repeat substrate in the presence of pol β K72A and FEN1. (C) and (I) The DNA fragment analysis results of the repair products from BER reconstituted with the (GAA)20 or (CAG)20 repeat substrate in the presence of pol β H34G. (D) and (J) The DNA fragment analysis results of the repair products from BER reconstituted with the substrate containing (GAA)20 or (CAG)20 repeats in the presence of pol β H34G and FEN1. (E) and (K) The DNA fragment analysis results of the repair products from BER reconstituted with the substrate containing (GAA)20 or (CAG)20 repeats in the presence of wild-type pol β. (F) and (L) The DNA fragment analysis results of the repair products from BER reconstituted with the substrate containing (GAA)20 or (CAG)20 repeats in the presence of wild-type pol β and FEN1. The schemes for generating the substrates were illustrated above the graphs. The template strand of the substrate was biotinylated at the 5′-end. Major repair products are indicated with black arrows. The red peaks are DNA size markers. The sizes of DNA fragments are illustrated as ‘nt’. Each experiment was conducted in triplicate, and only representative results are illustrated in the figures.
The dRP lyase activity of pol β suppresses a processive DNA synthesis during BER in TNR tracts
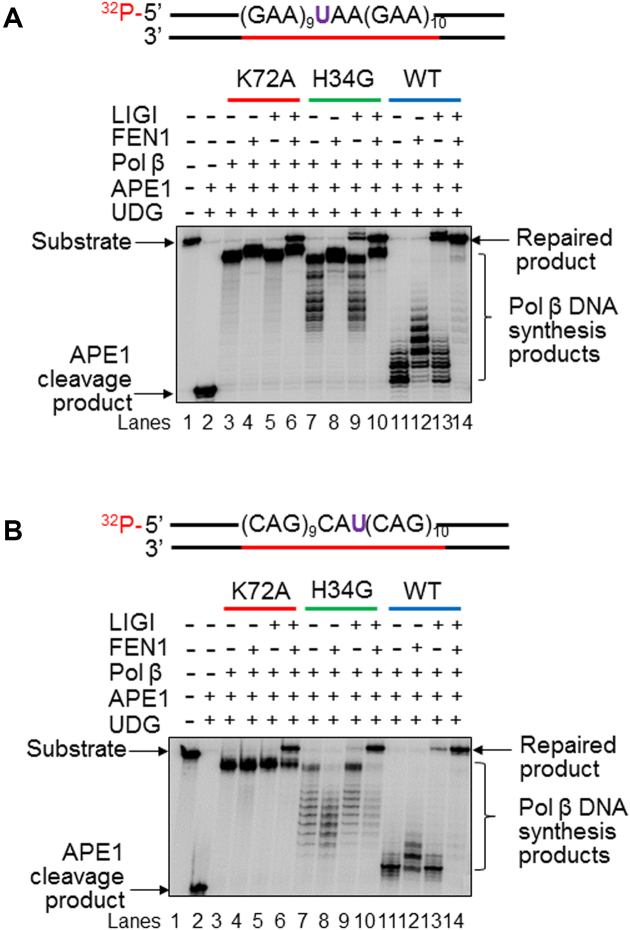
Because the DNA polymerase activity of pol β plays a crucial role in mediating TNR instability, we further asked whether it is possible that the pol β dRP lyase modulated TNR instability by altering its DNA polymerase activity. To address this, we characterized pol β DNA synthesis of the pol β dRP lyase deficient mutants during the repair of a native sugar in a GAA and CAG repeat tract. The results revealed that pol β K72A mutant at 10 nM exhibited a highly processive DNA synthesis on both the (GAA)20 and (CAG)20 repeat substrates, and this was independent of FEN1 (Figure 4, panels A and B, lanes 3–4). Pol β H34G mutant at the same concentration also exhibited a significantly improved processive DNA synthesis on the substrates (Figure 4, panels A and B, lanes 7–8). However, pol β WT only exhibited a distributive DNA synthesis on the substrates (Figure 4, panels A and B, lanes 11–12). The results showed that the processive DNA synthesis by the pol β dRP lyase mutants could not reach to the end of the substrates because the synthesis products were shorter than the full length of the substrates (Figure 4, panels A and B, compare the size of the pol β synthesis products in lanes 3–4 and 7–8 to the size of the substrates in lane 1). Thus, the repaired products were generated only through FEN1 cleavage and ligation by LIG I. This was further supported by the fact that BER reconstituted with pol β K72A mutant protein resulted in the repaired product only in the presence of both FEN1 and LIG I (Figure 4, panels A and B, lanes 5 and 6), whereas BER reconstituted with pol β H34G mutant or pol β WT led to the repaired product with the presence of LIG I (Figure 4, panels A and B, lanes 9–10 and 13–14). The presence of FEN1 significantly increased the amounts of the repaired product (Figure 4, panels A and B, compare lane 10 to lane 9 and lanes 14 to 13). The results indicate that deficiency of pol β dRP lyase activity significantly promoted the pol β processive DNA synthesis activity during BER in TNR tracts. This further indicated that pol β dRP lyase activity suppressed the polymerase activity suggesting that efficient removal of the 5′-dRP group by pol β dRP lyase domain restrained its ability to further synthesize DNA. This was further supported by the results showing that the sustainment of a reduced sugar that is refractory to the pol β dRP lyase also led to large GAA and CAG repeat deletions during BER (Supplementary Figure S2) by facilitating a processive pol β DNA synthesis of GAA and CAG repeats by pol β H34G and pol β WT (Supplementary Figure S3, panels A and B, lanes 6 and 9).
Figure 4.
DNA synthesis of pol β WT, pol β K72A, and pol β H34G during BER of a native abasic site in TNR tracts. The DNA synthesis activities of pol β WT and pol β dRP lyase mutant, K72A and H34G during BER were characterized with the (GAA)20 (A) or (CAG)20 (B) repeat substrate containing a native abasic site as described in the Materials and Methods. Lane 1 represents the substrate only. Lane 2 indicates the reaction mixture with 1 U UDG and 25 nM APE1. Lanes 3 and 4 correspond to the reaction mixture with 10 nM pol β K72A in the absence and presence of 25 nM FEN1. Lanes 5 and 6 correspond to the reaction mixture with 10 nM pol β K72A and 25 nM LIG I in the absence and presence of 25 nM FEN1. Lanes 7 and 8 correspond to the reaction mixture with 10 nM pol β H34G in the absence and presence of 25 nM FEN1. Lanes 9 and 10 correspond to the reaction mixture with 10 nM pol β H34G and 25 nM LIG I in the absence and presence of 25 nM FEN1. Lanes 11 and 12 correspond to the reaction mixture with 10 nM pol β WT in the absence and presence of 25 nM FEN1. Lanes 13 and 14 correspond to the reaction mixture with 10 nM pol β WT and 25 nM LIG I in the absence and presence of 25 nM FEN1. Substrates were 32P-labeled at the 5′-end of the damaged strand and are illustrated above each gel. The experiments were repeated at least three times. Representative gels are illustrated.
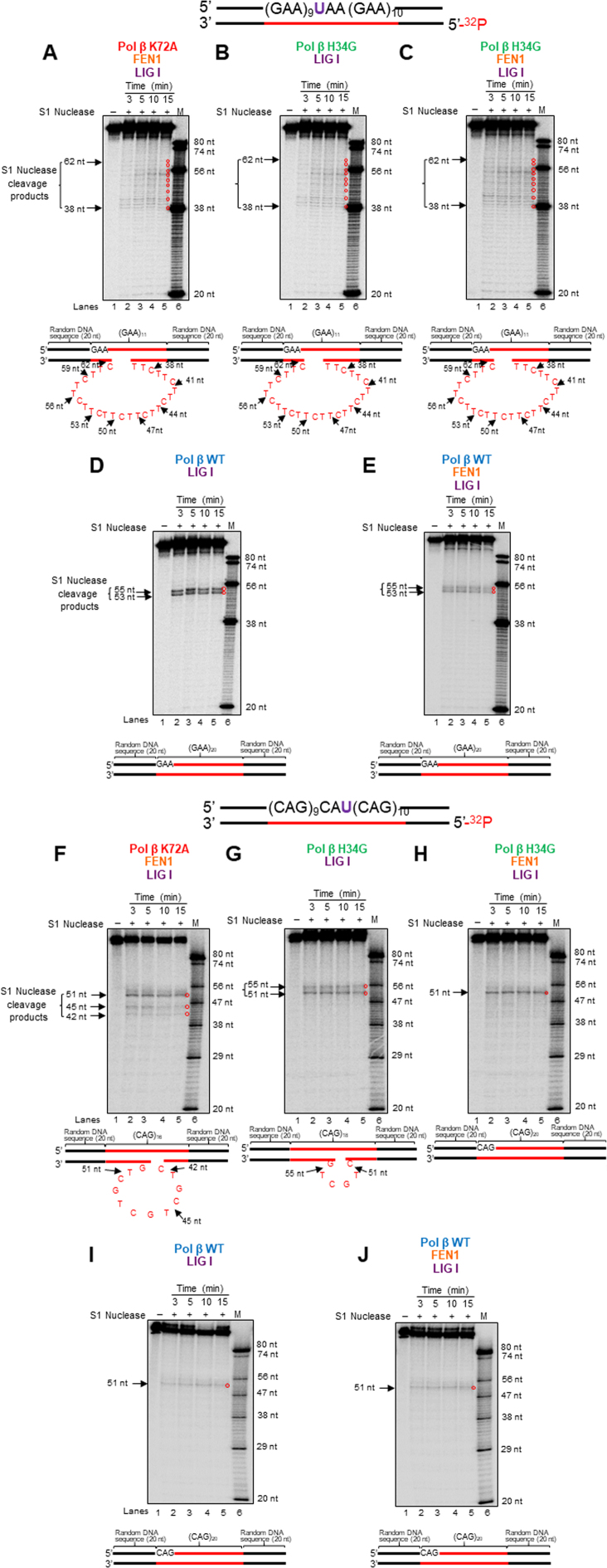
The dRP lyase activity of pol β prevents the formation of loop structures on the template strand in TNR tracts during BER
Since the formation of hairpins and loops on the template strand of TNR tracts underlies repeat deletion (15,17,19,35), it is possible that during BER, deficiency of pol β dRP lyase activity facilitates the formation of these structures on the template strand to promote repeat deletion, whereas proficient dRP lyase of pol β prevents these events and repeat deletion. We tested this possibility by detecting the formation of TNR loop structures on the template strands of the repaired products resulting from BER with the (GAA)20 and (CAG)20 substrates using S1 nuclease that specifically cleaves the single-stranded regions of TNR hairpins and loops (36). We found that S1 nuclease cleavage on the repaired product from BER with pol β K72A mutant protein and FEN1 resulted in the cleavage products containing 38 to 62 nucleotides on the template strand of the (GAA)20 substrate (Figure 5, panel A, lanes 2–5). S1 cleavage products with the same sizes were produced through BER with pol β H34G mutant protein in the absence and presence of FEN1 (Figure 5, panels B and C, lanes 2–5). The results indicate that BER mediated by the pol β mutant proteins resulted in the formation of a (TTC)9 loop on the template strand of the repaired product. However, S1 nuclease cleavage on the repaired product from BER with pol β WT generated only 53 nt and 55 nt products independent of FEN1 (Figure 5, panels D and E, lanes 2–5) indicating no repeat loops formed on the template strand of the repaired product. Similarly, for the (CAG)20 substrate, S1 nuclease cleavage on the template strand of the repaired product with pol β K72A mutant in the presence of FEN1 or pol β H34G mutant in the absence of FEN1 resulted in the cleavage products containing 42–51 nucleotides (Figure 5, panel F, lanes 2–5) and 51 nt and 55 nt products (Figure 5, panel G, lanes 2–5). However, S1 nuclease cleavage on the template strand of the repaired product with pol β H34G mutant in the presence of FEN1 led to the cleavage product containing 51 nt only. The results indicate that the mutant promoted the formation of a (CTG)4 and (CTG)2 loop on the template strand of the repaired product. In contrast, S1 nuclease cleavage on the repaired product resulting from pol β WT only generated the 51 nt product (Figure 5, panels I and J, lanes 2–5). This indicates that no secondary structures formed on the template strand of the repaired products from the CAG repeat substrate during BER mediated by pol β WT. Thus, the results indicate that the proficient dRP lyase activity of pol β WT efficiently suppressed the formation of TNR secondary structures on the template strand of TNR tracts, thereby preventing TNR deletion during BER. It should be noted that to detect the repaired strand of the products with the template loops shown in Figure 3, the repaired strands were isolated and amplified by PCR. This made the signal of the repaired strands shown in Figure 3 looked much more than the ones shown in Figure 5 (compare Figure 5, panel A to Figure 3, panel B; Figure 5, panel B to Figure 3, panel C; Figure 5, panel C to Figure 3, panel D).
Figure 5.
The formation of loop structures on the template strand of TNR tracts during BER of a native abasic site. The formation of loop structures on the template strand of the repaired products resulting from BER with the (GAA)20 and (CAG)20 repeat substrates containing a native abasic site was probed by S1 nuclease as described in the Materials and Methods. (A) and (F) The results of S1 nuclease cleavage on the template of the repaired products generated from pol β K72A-mediated BER in the presence of FEN1 with the (GAA)20 or (CAG)20 repeat substrate. (B) and (G) The results of S1 nuclease cleavage on the template of the repaired products generated from pol β H34G-mediated BER with the (GAA)20 or (CAG)20 repeat substrate. (C) and (H) The results of S1 nuclease cleavage on the template of the repaired products generated from pol β H34G-mediated BER in the presence of FEN1 with the (GAA)20 or (CAG)20 repeat substrate. (D) and (I) The results of S1 nuclease cleavage on the template of the repaired products generated from BER mediated by pol β WT with the (GAA)20 or (CAG)20 repeat substrate. (E) and (J) The results of S1 nuclease cleavage on the template of the repaired products generated from BER mediated by pol β WT in the presence of FEN1 with the (GAA)20 or (CAG)20 repeat substrate. Lane 1 represents substrate alone. Lanes 2–5 represent the S1 nuclease cleavage products generated at various time intervals. Lane 6 represents synthesized size markers (M). Substrates were 32P-labeled at the 5′-end of the template strand and are illustrated above each gel. S1 nuclease digestion sites are indicated. S1 nuclease cleavage sites and loop structures formed on the template strands of the repaired products are illustrated below the gels. Experiments were repeated in triplicate.
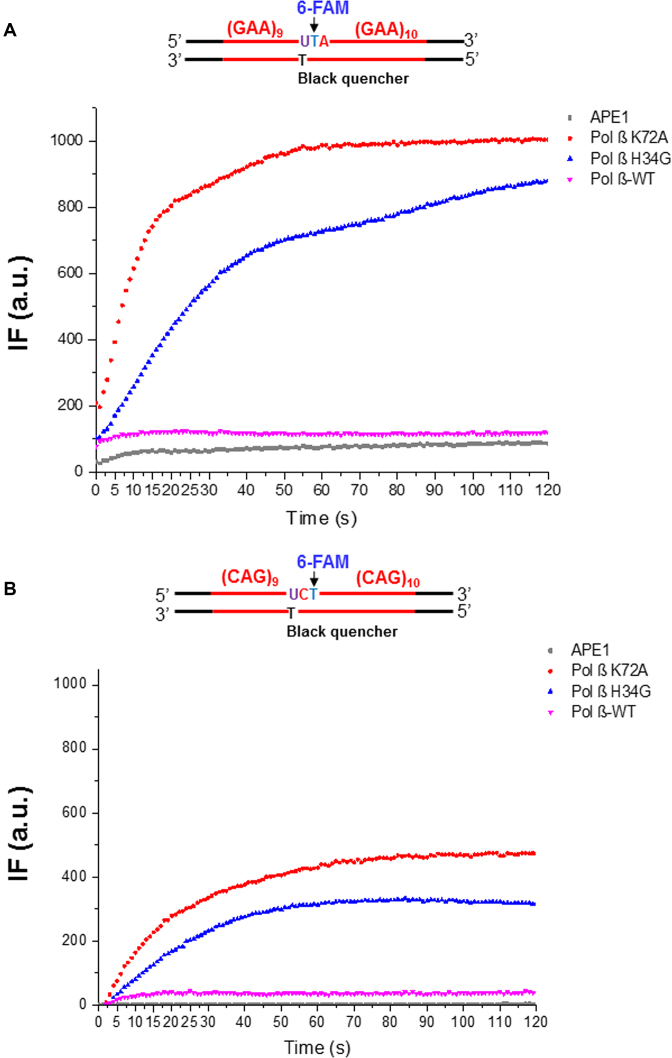
The dRP lyase activity of pol β prevents TNR strand slippage during BER of a native sugar
Since the pol β mutant proteins facilitated TNR deletion by promoting the formation of loop structures on the template strand of the repeat-containing substrates, we reason that the mutant proteins stimulate TNR strand slippage leading to the formation of TNR loop structures and deletions. To test this, we developed a fluorescence-based DNA slippage assay (Figure 1) to measure the dynamics of repeat strand slippage of the substrates in the presence of pol β WT and mutant proteins during BER. During BER, a single-stranded break is generated in the damaged strand in TNR tracts by incubation with UDG and APE1, and the downstream of the damaged strand slips upon DNA synthesis of pol β. This allows the 6-FAM group on the damaged strand of the substrates to be dissociated from the template strand and positioned far away from the BHQ-1 leading to a detectable level of the fluorescence signal. Our results revealed that during BER at different time intervals, pol β K72A mutant resulted in the highest level of fluorescence in both GAA and CAG repeats (red), and pol β H34G mutant resulted in the second highest level of fluorescence signal (blue) (Figure 6, panels A and B). However, pol β WT only resulted in a low background level of fluorescence (magenta) that is comparable to the one with APE1 only (grey). It should be noted that under the condition without the repeat strand slippage, i.e. in the presence of APE1 with the substrate alone (grey), only a low background fluorescence signal was detected. This indicates that no spontaneous separation between the fluorophore and quencher in the substrates in the absence of pol β DNA synthesis. Similarly, pol β WT with weak DNA synthesis activity also led to a low fluorescence signal (magenta) demonstrating that limited DNA strand slippage induced by the limited DNA synthesis of pol β WT only produced a low level of fluorescence signal (Figure 6, panels A and B, compared ‘magenta’ to ‘grey’). In contrast, the presence of a processive DNA synthesis by pol β H34G (blue) and K72A (red) mutants (compare Figure 6 to Figure 4) produced a high level of fluorescence signal (Figure 6, panels A and B, compare ‘blue’ and ‘red’ to ‘grey’), indicating the dissociation of 6-FAM-dT on the damaged strand from BHQ-1-dT on the template strand resulted from DNA strand slippage caused by the pol β dRP lyase mutants. A linear regression model was then used to quantify the rate of the changes of fluorescence intensities. The parameters calculated from the model were summarized in Table 1. Based on this model, the slope of the curve, k, was obtained, and the rate of strand slippage (the separation of the 6-FAM group in the downstream damaged strand from the BHQ-1 group in the template strand) was calculated according to the equation: The rate of strand slippage = [Total substrate amount (1000 fmol) × percentage of the products (>1 nt insertion)/IF2min] × k. The percentage of the products (> 1 nt insertion) was determined by conducting the gel-based experiments using the 32P-labeled substrates in parallel (data not shown). The results showed that during BER in GAA repeats, pol β K72A mutant caused the strand slippage 2-fold faster than pol β H34G mutant (26.80 fmol × s−1 vs. 11.20 fmol × s−1), and 14.9-fold faster than pol β WT (1.80 fmol × s−1) (Table 1). Similarly, for BER with CAG repeat substrate, pol β K72A mutant also caused strand slippage (17.20 fmol × s−1) 1.7-fold faster than pol β H34G mutant (10.00 fmol × s−1), and 8-fold faster than wild-type pol β (2.00 fmol × s−1) (Table 1). The results indicate that deficiency of pol β dRP lyase activity significantly promoted TNR strand slippage, whereas efficient dRP lyase activity of pol β WT suppressed repeat strand slippage. This further suggests that failure in removal of the dRP group led to a persistent interaction between pol β dRP lyase domain and the 5′-dRP group promoting TNR slippage during BER. In addition, for the random DNA sequence-containing substrate, pol β H34G (blue) and WT (magenta) only led to a low background level of fluorescence that is comparable to the one with APE1 only (grey) (Supplementary Figure S4). Pol β K72A mutant produced a lower level of fluorescence signal in the context of random DNA sequence (red) (Supplementary Figure S4) than that in the context of GAA and CAG repeats (compare Figure 6, panels A and B, ‘red’ to Supplementary Figure S4, ‘red’). This suggests that the strand-displacement DNA synthesis by pol β K72A led to the dissociation of the 6-FAM-dT from the BHQ-1-dT in the template strand. Interestingly, the fluorescence signal generated by pol β K72A mutant (red) was reduced during later time intervals (20–120 s) (Supplementary Figure S4) suggesting the reannealing of the displaced 5′-flaps to the template strand. This further suggests that no secondary structureswere formed in the 5′-flaps with random DNA sequence.
Figure 6.
The strand slippage of TNRs during BER of a native abasic site. The strand slippage during BER of a native abasic site in the context of (GAA)20 (A) or (CAG)20 (B) repeats, was examined as described in the Materials and Methods. The intensity of fluorescence (IF) generated from each reaction condition was recorded by the Bio-Rad CFX Connect Real-Time PCR Detection System and plotted against time (s). The IF from each reaction condition was indicated with different color. A schematic diagram of (GAA)20 or (CAG)20 substrate containing a native abasic site, BHQ-1 dT, and 6-FAM-dT was illustrated above the graph. Experiments were repeated in triplicate, and the representative results are shown.
Table 1.
DNA slippage rates of Pol β
| (GAA)20 | (CAG)20 | |||||
|---|---|---|---|---|---|---|
| Parameters | Pol β K72A | Pol β H34G | Pol β WT | Pol β K72A | Pol β H34G | Pol β WT |
| k (s-1) | 39.99 | 17.43 | 2.30 | 17.51 | 10.28 | 2.87 |
| Strand slippage rate (fmol·s-1) | 26.80 | 11.20 | 1.80 | 17.20 | 10.00 | 2.00 |
| R 2 | 0.98 | 0.99 | 0.84 | 0.98 | 0.99 | 0.87 |
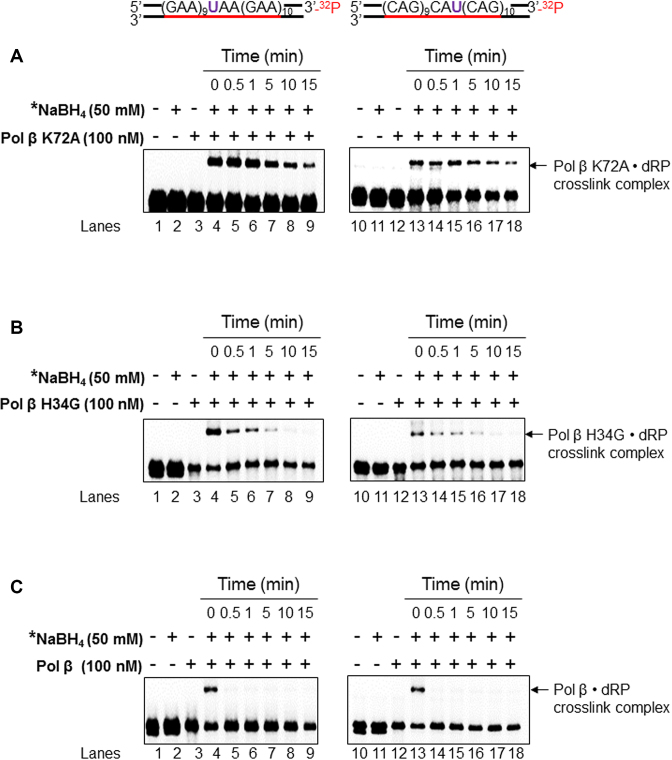
Efficient pol β dRP lyase activity restrains the interaction between the dRP lyase domain and a dRP group during BER
We have previously shown that the pol β dRP lyase domain can interact with both a native and oxidized sugar during BER in duplex TNR tracts and hairpins (32). Thus, we reason that the dRP lyase mutant proteins, pol β K72A, and H34G may persistently interact with 5′-dRP due to lack of the ability of removing the sugar, and this then keeps the mutant polymerases staying on the template and continuously synthesizing DNA without a constraint, thereby leading to a processive DNA synthesis. To test this possibility, we examined the formation of a pol β-dRP complex via the Schiff base covalent linkage in TNR tracts at different time intervals (0–15 min) using NaHB4 crosslinking trapping assay (Figure 7). The results showed that the formation of the pol β K72A-dRP crosslink complex in both (GAA)20 and (CAG)20 repeat tracts, was detected even over 15 min (Figure 7, panel A, lanes 4–9 and lanes 13–18), whereas pol β H34G-dRP crosslink complex in the repeat tracts was only detected between 0 min to 5 min (Figure 7, panel B, lanes 4–7 and lanes 13–16). However, the pol β WT-dRP crosslink complex was detected only within the 0–0.5 min (Figure 7, panel C, lanes 4 and 13). This indicates that deficiency of dRP lyase activity of pol β through K72A and H34G mutations allowed the dRP lyase domain to persistently interact with the dRP group. This is further supported by the results from the experiment during which the processivity of pol β H34G and pol β WT was significantly stimulated during BER of a reduced sugar, (tetrahydrofuran) in both (GAA)20 and (CAG)20 repeat tracts (Supplementary Figure S3). All of the results suggest that deficiency of pol β dRP lyase forced the dRP lyase domain to interact with the dRP group allowing the polymerase to stay on the DNA template and perform a processive DNA synthesis.
Figure 7.
The crosslink between the dRP lyase domain of wild-type and mutant pol β proteins and a dRP group. The formation of the pol β K72A•dRP crosslink complex (A), pol β H34G• dRP crosslink complex (B), and pol β• dRP crosslink complex (C) in TNRs, respectively was measured using the (GAA)20 (left panel) or (CAG)20 (right panel) substrates containing 5′- dRP as described in the Materials and Methods. Lane 1 indicates the uracil-containing substrate that was pre-incubated with UDG. Lane 2 indicates the pre-cut substrate in the presence of 50 mM NaBH4. Lanes 3 and 4 represent the reaction mixtures containing the pre-cut substrates and pol β K72A or H34G or pol β (wild-type) (100 nM) in the absence and presence of 50 mM NaBH4. Lanes 5 to 9 represent reaction mixtures that contained the substrates and pol β K72A or H34G or pol β (100 nM) in the presence of 50 mM NaBH4 with pre-incubation at the time points of 30 s, 1 min, 5 min, 10 min and 15 min. ‘*’ denotes 50 mM NaBH4 was added after pre-incubation with pol β K72A or H34G or pol β at various time intervals. Substrates were 32P-labeled at the 3′-end of the downstream damaged strand and are illustrated above each gel. Each experiment was repeated in triplicate. Only the representative gels are shown.
DISCUSSION
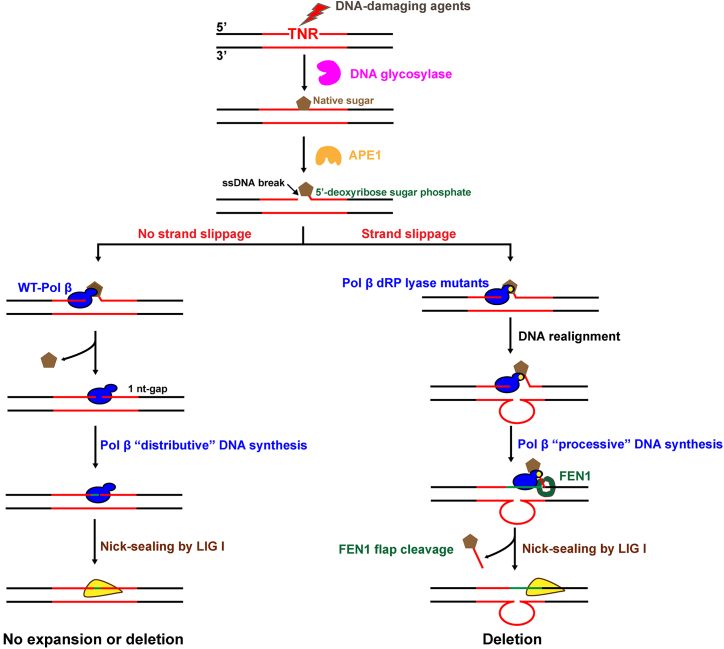
In this study, we explored a critical role of pol β dRP lyase activity in sustaining TNR stability during BER. We found that deficiency of pol β dRP lyase activity failed to suppress GAA and CAG repeat deletions (Figures 2 and 3). However, complementation of the dRP lyase deficiency with pol β WT eliminated the repeat deletions (Figures 2 and 3). We demonstrated that pol β dRP lyase activity efficiently prevented a processive DNA synthesis (Figure 4), the formation of loop structures on the template (Figure 5), and repeat strand slippage (Figure 6), thereby suppressing repeat deletions. We further demonstrated that deficiency of pol β dRP lyase activity led to sustainment of a dRP group, which subsequently ‘tethered’ pol β protein on the TNR tracts through the pol β-dRP interaction (Figure 7) forcing the polymerase to switch its DNA synthesis from a distributive mode to a processive mode (Figure 4 and Supplementary Figure S3). The results allowed us to propose a model that illustrates the role of pol β dRP lyase activity in maintaining TNR instability during BER (Figure 8). During BER of a base lesion in a TNR tract, the base lesion is removed by a damage-specific DNA glycosylase. This results in an abasic site that is 5′-incised by APE1, leaving a 1 nt gap with a 5′-dRP group. Proficient pol β dRP lyase activity removes the dRP group and fills in the 1 nt gap allowing pol β to fall off the DNA template. This suppresses pol β processive DNA synthesis, TNR strand slippage, and the formation of loop structures on the template, thereby preventing TNR deletions (Figure 8, sub-pathway 1). Deficiency of pol β dRP lyase results in the sustainment of the dRP group, which ‘tethers’ pol β on the TNR template allowing the polymerase to perform a highly processive DNA synthesis. This subsequently promotes repeat strand slippage and the formation of loop structures on the template strand and allows the enzyme to skip over the loop structures causing repeat deletions (Figure 8, sub-pathway 2).
Figure 8.
The dRP lyase of pol β prevents TNR deletion by suppressing a processive DNA synthesis and repeat strand slippage during BER. A DNA base lesion that occurs in the TNR tract is removed by a damage-specific DNA glycosylase. This results in an abasic site that is subsequently 5′-incised by APE1, leaving a 1 nt gap with a 5′-dRP moiety. If the dRP lyase activity of pol β is proficient, the Lys72 of pol β forms the Schiff base with the 5′-dRP group preventing DNA strand slippage. The dRP lyase then removes the dRP group leaving a 1 nt gap. Pol β then fills in the gap generating nick and dissociates from the product, and the nick is sealed by LIG I. This allows pol β to adopt a distributive DNA synthesis mode, thereby preventing repeat deletion (sub-pathway 1). On the other hand, deficiency of pol β dRP lyase caused by dRP lyase mutations locks the dRP lyase domain to the dRP residue. This ‘tethers’ pol β to its template allowing it to adopt a processive DNA synthesis mode. This results in DNA slippage and promotes the formation of loop structures on the template and pol β skip-over of the loop structures leading to repeat deletions (sub-pathway 2).
Here, we also revealed different DNA synthesis modes adopted by pol β and the mechanisms that govern the modes of DNA synthesis during BER in TNR tracts. Our results support a model that pol β dRP lyase domain persistently interacts with the dRP group through the lysines at its lysine pocket (34) for its substrate binding and subsequent dRP removal via β-elimination in TNRs (Figure 7) (30). In a scenario where dRP lyase activity is deficient or the dRP group is refractory to the dRP lyase activity of pol β via β-elimination (i.e. reduced sugar such as THF), pol β persistently interacts with the dRP group, and this “tethers” the polymerase on its DNA template forcing the polymerase to perform a processive synthesis of TNRs (Figure 4 and Supplementary Figure S3). The Wilson group has shown that pol β can perform a processive DNA synthesis in a short gap (5–6 nt) by interacting with the 5′-phosphate of the dRP group through Lys 35 of the lyase domain (34,37). Our results further indicate the pivotal role of pol β-dRP interaction in promoting the mode of processive DNA synthesis (compare lanes 7–8 of Figure 4 to lanes 6–7 of Supplementary Figure S3 and lanes 11–12 of Figure 4 to lanes 9–10 of Supplementary Figure S3). Thus, our results suggest that upon the presence of the dRP group, pol β is ‘tethered’ to its template, adopting a processive mode of DNA synthesis until the dRP group is ultimately removed. Further, our results indicated that the deficiency of pol β dRP lyase activity by the point mutation at K72 resulted in the failure of the removal of the dRP group leading to a persistent interaction of pol β K72A with its substrate through the other lysine residues in the ‘lysine pocket’ of the dRP lyase domain (Figure 7). This in turn ‘tethered’ the polymerase on its DNA template forcing the polymerase to perform a processive synthesis of TNRs (Figure 4). Thus, our results suggest that proficient pol β dRP lyase activity efficiently removes a dRP group to prevent pol β from performing a processive DNA synthesis. This is critically important for preventing DNA strand slippage, the formation of TNR loops and TNR instability, and mutations mediated by pol β. Although K72 has been identified as the major lysine residue, it is not the only one that can form a Schiff base with a native sugar (31,34,38,39). Our results further suggest that other lysines including K84 in the ‘lysine pocket’ of the pol β lyase domain can also form a Schiff base with a native sugar (31,34,38,39) to tether the polymerase on its template (Figure 7). Thus, it is conceivable that in a scenario where dRP lyase residue/s is mutated in the dRP lyase domain of pol β, the BER pathway is forced to switch from the SN-BER pathway to the long-patch BER pathway promoting TNR instability (14,15,17).
Our results showed that the DNA synthesis activity of pol β was inversely correlated with its dRP lyase activity (Figure 4) suggesting that there is functional cooperation between the dRP lyase domain of pol β and its polymerase domain that suppresses the processive DNA synthesis of pol β. This is supported by our results showing that the pol β 8 kD domain alone failed to rescue TNR stability during BER in pol β−/− cell extracts (Supplementary Figure S1). This is further supported by the fact that although these domains can function independent of each other, the catalytic efficiency of the isolated polymerase and dRP lyase domains is significantly reduced compared with the intact pol β protein. This indicates that the two domains of pol β synergize to fulfill efficient enzymatic activities of the intact pol β (38,39). The synergistic effect appears to be also critically important to restrain pol β DNA synthesis activity, thereby ensuring the efficient short-patch BER that in turn prevents DNA strand slippage and TNR instability. This is also supported by our results showing that in the absence of pol β polymerase domain, pol β lyase domain alone failed to coordinate with replicative DNA polymerase to prevent TNR instability (Supplementary Figure S1), further demonstrating a necessity of the cooperation between the activities of pol β dRP lyase and its polymerase in sustaining TNR stability. A recent study from the Wilson group has shown that pol β uses positively charged lysine residues within the lyase domain to processively search for DNA damage, i.e. a 1 nt gap through hopping and intersegmental transfer along DNA (28,29). Once a gapped DNA is encountered, the lyase domain makes specific interactions with the gap to allow the lesion recognition. Moreover, the interaction between the lyase domain and nucleotides in the gapped DNA strengthens its affinity of binding to DNA, which subsequently facilitates its DNA polymerase domain to engage in the gap and initiate the DNA synthesis. This sheds light on the cooperation between the pol β dRP lyase domain and its polymerase domain in damage search. Our results suggest that pol β dRP lyase domain with a proficient dRP lyase can also serve as the ‘guard’ to prevent the processive DNA synthesis mode of pol β. The underlying structural base is implicated by an early study from the Wilson group showing that pol β adopts a ‘closed thumb’ as an active conformation and ‘open thumb’ as an inactive conformation for nucleotidyl transfer through an ‘induced-fit’ mechanism (40). Since the active conformation of pol β DNA synthesis, the ‘closed thumb’ conformation is mediated by the contact between the C-terminal thumb subdomain and the dRP lyase domain, we suggest that dRP lyase deficiency causes the failure of the removal of the dRP group resulting in the persistent pol β-dRP interaction. This then forces pol β to keep ‘the closed thumb conformation’ that in turn ‘tethers’ the polymerase on its template leading to a processive DNA synthesis. On the other hand, a dRP group is removed efficiently by pol β WT allowing the polymerase to adopt the ‘open thumb’ conformation. This then results in the dissociation of pol β from its product leading to distributive DNA synthesis. Thus, our results suggest that malfunction of pol β dRP lyase promotes the processive DNA synthesis by locking the enzyme in the ‘closed thumb’ conformation through an interaction between the dRP lyase domain and a dRP group.
Removal of the dRP group by the pol β dRP lyase determines if BER undergoes the single-nucleotide BER sub-pathway or long-patch BER sub-pathway (31,41). Since the long-patch BER involves multiple-nucleotide synthesis by pol β or other DNA polymerases, which allows the addition or removal of repeats, the long-patch BER mediates TNR instability (14,15,17). Our results further suggest that the choice of the BER sub-pathways and modes of pol β DNA synthesis can be regulated through inhibition of pol β dRP lyase activity. This may preferentially lead to deletions of expanded TNR tracts. Also, it has been reported that pol β can be acetylated by acetyltransferase p300 at Lys 72 both in vitro and in vivo (42). Acetylated pol β was less efficient in reconstituted BER due to the reduced dRP lyase activity (42). This suggests that pol β acetylation can act as a switch to govern the modes of pol β DNA synthesis and the BER sub-pathway to regulate TNR instability. The role of pol β dRP lyase inhibition in mediating TNR instability warrants further studies. This will help to pave the way for developing new strategies for the treatment of TNR expansion diseases by targeting pol β dRP lyase activity.
In conclusion, we provide the primary evidence that pol β dRP lyase activity synergizes with its polymerase activity to prevent TNR deletions during BER. We demonstrated that this effect was achieved through the prevention of a processive synthesis of TNRs by pol β, repeat strand slippage, and the formation of secondary structures in TNR tracts.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Samuel H. Wilson, Laboratory of Genome Integrity and Structural Biology, National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH) for generously providing plasmids for expressing BER enzymes and pol β K72A and H34G mutant proteins.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [ES023569 to Y.L.]; NSF CAREER Award [DMR-1555361 to Y.W.]. Funding for open access charge: National Institutes of Health [ES023569 to Y.L.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Paulson H.L., Fischbeck K.H.. Trinucleotide repeats in neurogenetic disorders. Annu. Rev. Neurosci. 1996; 19:79–107. [DOI] [PubMed] [Google Scholar]
- 2. McMurray C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010; 11:786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nelson K.A., Witte J.S.. Androgen receptor CAG repeats and prostate cancer. Am. J. Epidemiol. 2002; 155:883–890. [DOI] [PubMed] [Google Scholar]
- 4. Schildkraut J.M., Murphy S.K., Palmieri R.T., Iversen E., Moorman P.G., Huang Z., Halabi S., Calingaert B., Gusberg A., Marks J.R. et al. Trinucleotide repeat polymorphisms in the androgen receptor gene and risk of ovarian cancer. Cancer Epidemiol. Biomark. 2007; 16:473–480. [DOI] [PubMed] [Google Scholar]
- 5. Mirkin S.M. Expandable DNA repeats and human disease. Nature. 2007; 447:932–940. [DOI] [PubMed] [Google Scholar]
- 6. Usdin K., House N.C., Freudenreich C.H.. Repeat instability during DNA repair: insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015; 50:142–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jakupciak J.P., Wells R.D.. Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life. 2000; 50:355–359. [DOI] [PubMed] [Google Scholar]
- 8. De Biase I., Rasmussen A., Endres D., Al-Mahdawi S., Monticelli A., Cocozza S., Pook M., Bidichandani S.I.. Progressive GAA expansions in dorsal root ganglia of Friedreich's ataxia patients. Ann. Neurol. 2007; 61:55–60. [DOI] [PubMed] [Google Scholar]
- 9. Liu Y., Wilson S.H.. DNA base excision repair: a mechanism of trinucleotide repeat expansion. Trends Biochem. Sci. 2012; 37:162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iyer R.R., Pluciennik A., Napierala M., Wells R.D.. DNA triplet repeat expansion and mismatch repair. Annu. Rev. Biochem. 2015; 84:199–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pearson C.E., Nichol Edamura K., Cleary J.D.. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005; 6:729–742. [DOI] [PubMed] [Google Scholar]
- 12. Kovtun I.V., Liu Y., Bjoras M., Klungland A., Wilson S.H., McMurray C.T.. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007; 447:447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wallace S.S. Base excision repair: a critical player in many games. DNA Repair. 2014; 19:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu Y., Prasad R., Beard W.A., Hou E.W., Horton J.K., McMurray C.T., Wilson S.H.. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 2009; 284:28352–28366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu M., Gabison J., Liu Y.. Trinucleotide repeat deletion via a unique hairpin bypass by DNA polymerase beta and alternate flap cleavage by flap endonuclease 1. Nucleic Acids Res. 2013; 41:1684–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu M., Lai Y., Torner J., Zhang Y., Zhang Z., Liu Y.. Base excision repair of oxidative DNA damage coupled with removal of a CAG repeat hairpin attenuates trinucleotide repeat expansion. Nucleic Acids Res. 2014; 42:3675–3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lai Y., Xu M., Zhang Z., Liu Y.. Instability of CTG repeats is governed by the position of a DNA base lesion through base excision repair. PLoS One. 2013; 8:e56960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beaver J.M., Lai Y., Xu M., Casin A.H., Laverde E.E., Liu Y.. AP endonuclease 1 prevents trinucleotide repeat expansion via a novel mechanism during base excision repair. Nucleic Acids Res. 2015; 43:5948–5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lai Y., Budworth H., Beaver J.M., Chan N.L., Zhang Z., McMurray C.T., Liu Y.. Crosstalk between MSH2-MSH3 and pol beta promotes trinucleotide repeat expansion during base excision repair. Nat. Commun. 2016; 7:12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beard W.A., Wilson S.H.. Structure and mechanism of DNA polymerase beta. Biochemistry. 2014; 53:2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Doetsch P.W., Cunningham R.P.. The enzymology of apurinic/apyrimidinic endonucleases. Mutat. Res. 1990; 236:173–201. [DOI] [PubMed] [Google Scholar]
- 22. Matsumoto Y., Kim K.. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995; 269:699–702. [DOI] [PubMed] [Google Scholar]
- 23. Piersen C.E., Prasad R., Wilson S.H., Lloyd R.S.. Evidence for an imino intermediate in the DNA polymerase beta deoxyribose phosphate excision reaction. J. Biol. Chem. 1996; 271:17811–17815. [DOI] [PubMed] [Google Scholar]
- 24. Sobol R.W., Prasad R., Evenski A., Baker A., Yang X.P., Horton J.K., Wilson S.H.. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000; 405:807–810. [DOI] [PubMed] [Google Scholar]
- 25. Sobol R.W., Horton J.K., Kuhn R., Gu H., Singhal R.K., Prasad R., Rajewsky K., Wilson S.H.. Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature. 1996; 379:183–186. [DOI] [PubMed] [Google Scholar]
- 26. Gu H., Marth J.D., Orban P.C., Mossmann H., Rajewsky K.. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994; 265:103–106. [DOI] [PubMed] [Google Scholar]
- 27. Sugo N., Aratani Y., Nagashima Y., Kubota Y., Koyama H.. Neonatal lethality with abnormal neurogenesis in mice deficient in DNA polymerase beta. EMBO J. 2000; 19:1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howard M.J., Rodriguez Y., Wilson S.H.. DNA polymerase beta uses its lyase domain in a processive search for DNA damage. Nucleic Acids Res. 2017; 45:3822–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Howard M.J., Wilson S.H.. Processive searching ability varies among members of the gap-filling DNA polymerase X family. J. Biol. Chem. 2017; 292:17473–17481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prasad R., Beard W.A., Strauss P.R., Wilson S.H.. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J. Biol. Chem. 1998; 273:15263–15270. [DOI] [PubMed] [Google Scholar]
- 31. Prasad R., Batra V.K., Yang X.P., Krahn J.M., Pedersen L.C., Beard W.A., Wilson S.H.. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair. 2005; 4:1347–1357. [DOI] [PubMed] [Google Scholar]
- 32. Beaver J.M., Lai Y., Rolle S.J., Weng L., Greenberg M.M., Liu Y.. An oxidized abasic lesion inhibits base excision repair leading to DNA strand breaks in a trinucleotide repeat tract. PLoS One. 2018; 13:e0192148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biade S., Sobol R.W., Wilson S.H., Matsumoto Y.. Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J. Biol. Chem. 1998; 273:898–902. [DOI] [PubMed] [Google Scholar]
- 34. Prasad R., Beard W.A., Chyan J.Y., Maciejewski M.W., Mullen G.P., Wilson S.H.. Functional analysis of the amino-terminal 8-kDa domain of DNA polymerase beta as revealed by site-directed mutagenesis. DNA binding and 5′-deoxyribose phosphate lyase activities. J. Biol. Chem. 1998; 273:11121–11126. [DOI] [PubMed] [Google Scholar]
- 35. Lai Y., Beaver J.M., Lorente K., Melo J., Ramjagsingh S., Agoulnik I.U., Zhang Z., Liu Y.. Base excision repair of chemotherapeutically-induced alkylated DNA damage predominantly causes contractions of expanded GAA repeats associated with Friedreich's ataxia. PLoS One. 2014; 9:e93464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wiegand R.C., Godson G.N., Radding C.M.. Specificity of the S1 nuclease from Aspergillus oryzae. J. Biol. Chem. 1975; 250:8848–8855. [PubMed] [Google Scholar]
- 37. Singhal R.K., Wilson S.H.. Short gap-filling synthesis by DNA polymerase beta is processive. J. Biol. Chem. 1993; 268:15906–15911. [PubMed] [Google Scholar]
- 38. Beard W.A., Wilson S.H.. Structure and mechanism of DNA polymerase Beta. Chem. Rev. 2006; 106:361–382. [DOI] [PubMed] [Google Scholar]
- 39. Prasad R., Kumar A., Widen S.G., Casas-Finet J.R., Wilson S.H.. Identification of residues in the single-stranded DNA-binding site of the 8-kDa domain of rat DNA polymerase beta by UV cross-linking. J. Biol. Chem. 1993; 268:22746–22755. [PubMed] [Google Scholar]
- 40. Sawaya M.R., Prasad R., Wilson S.H., Kraut J., Pelletier H.. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997; 36:11205–11215. [DOI] [PubMed] [Google Scholar]
- 41. Prasad R., Dianov G.L., Bohr V.A., Wilson S.H.. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J. Biol. Chem. 2000; 275:4460–4466. [DOI] [PubMed] [Google Scholar]
- 42. Hasan S., El-Andaloussi N., Hardeland U., Hassa P.O., Burki C., Imhof R., Schar P., Hottiger M.O.. Acetylation regulates the DNA end-trimming activity of DNA polymerase beta. Mol. Cell. 2002; 10:1213–1222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.