Abstract
The role of Runt-related transcription factor 3 (RUNX3) gene in breast cancer remains not fully understood. We studied the correlation between RUNX3 gene promoter methylation and estrogen receptor (ER) expression status in breast cancer. Three breast cancer cell lines and 113 formalin-fixed, paraffin-embedded breast cancer tissue samples were analyzed for RUNX3 expression. Methylation-specific polymerase chain reaction was used to analyze RUNX3 methylation on the samples. Migration and invasion ability were evaluated in MCF7 cell line (RUNX3 methylated) treated with methylation inhibitor 5-Aza-2′-deoxycytidine (5-Aza-CdR) to study the effect of RUNX3 methylation status. Our data showed that the expression of RUNX3 was high in MCF10A but not in MCF7 and SKBR3 cell lines, while the RUNX3 promoter showed hypermethylation in MCF7 but not in MCF10A and SKBR3. In tissues samples, Immunohistochemical (IHC) expression of RUNX3 protein was higher in ER-negative samples than in ER-positive cases, and it was negatively correlated with the methylation status of the RUNX3 gene promoter. Proliferation, migration, and invasion of MCF7 were suppressed when 5-Aza-CdR treated. Also, the hypermethylation status of RUNX3 gene promoter was reversed and RUNX3 expression was increased. In summary, our data suggest that hypermethylation of the RUNX3 gene promoter may play an important role in ER-positive breast tumor progression.
Keywords: breast cancer, estrogen receptor, RUNX3 methylation
Introduction
Breast cancer is associated with numerous genomic alterations including the dysregulation of tumor suppressor genes via epigenetic alterations, hemizygous deletion, and gene mutation.1 Hypermethylation of the promoter region of tumor suppressor genes is believed to be correlated with the downregulation of gene expression and has been causally linked with carcinogenesis in many tumors, including breast cancer.2–6 Runt-related transcription factor 3 (RUNX3), encoded by the RUNX3 gene, was first recognized as a tumor suppressor gene in gastric cancer owing to the causal relationship between the loss of RUNX3 and gastric carcinogenesis and progression.7 RUNX3 is located at 1p36, a region of frequent genomic loss in various human cancers, including stomach, breast, and lung carcinoma.8 Several studies have found that hypermethylation of the RUNX3 gene promoter region is the main mechanism of this downregulation.9,10
Previous research has focused on the role of RUNX3 promoter methylation in carcinogenesis; however, the correlation of RUNX3 promoter methylation with breast cancer and the subtypes of breast cancer is still not completely understood.11–14 Considering that breast cancer is highly heterogeneous, different mechanisms may be involved in the tumorigenesis of different subtypes of breast cancers. Former studies have shown that RUNX3 plays a role as a tumor suppressor gene in breast cancer by targeting the estrogen receptor (ER), which induces the proteasome-mediated degradation of ER and thus reducing its activity, affecting the expression of downstream genes.15 However, the correlation between the hypermethylation status of the RUNX3 gene promoter and ER status in breast carcinoma is still not fully clear.
Since promoter hypermethylation is a major factor in the inactivation of RUNX3 in breast cancer and since RUNX3 may work as a tumor suppressor by targeting ER, we hypothesized that the function of RUNX3 promoter hypermethylation may be associated mainly with ER-positive breast cancer. To investigate whether RUNX3 promoter methylation is related to ER status in breast cancer, we examined the methylation status of the RUNX3 gene promoter and RUNX3 protein expression in cell lines and in ER-positive and ER-negative breast cancer tissue samples, and examined RUNX3 mRNA expression in cell lines. Also, we investigated the effects of DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-Aza-CdR) on RUNX3 expression and gene promoter methylation in the ER-positive breast cancer cell line MCF7. Furthermore, we studied the relationship between RUNX3 promoter hypermethylation and clinicopathologic features in ER-positive and ER-negative human breast cancer specimens.
Materials and Methods
Cell Lines and Culture Conditions
The human breast cancer cell lines MCF7 and SKBR3 and the human normal breast cell line MCF10A were obtained from the Cell Bank at the Chinese Academy of Sciences (Shanghai, China), and were authenticated by morphology test under microscope. All cell lines were maintained in Roswell Park Memorial Institute-1640 medium (Gibco-BRL, Tokyo, Japan) supplemented with 10% fetal bovine serum at 37C in a humidified atmosphere of 5% carbon dioxide and 95% air.
Human Tissue Specimens and Patient Information
Formalin-fixed, paraffin-embedded (FFPE) archival tissue samples of 113 invasive breast carcinomas were obtained from the Department of Pathology at the Affiliated Hospital of Xuzhou Medical College (Xuzhou, Jiangsu, China). The 113 cases were from years 2008 and 2011, and included 68 ER-positive and 45 ER-negative cases. Clinicopathologic information was acquired from the pathology medical records. All cases were reviewed and confirmed independently by two pathologists using the World Health Organization system. The study was approved by the Ethics Committee of the First Affiliated Hospital of Xuzhou Medical College, informed consent was waived for this study.
Immunocytochemistry (ICC) Analysis
Cells were grown on microscope coverslips and were treated with 4% paraformaldehyde for 15 min, washed with phosphate-buffered saline solution, fixed on the slide with neutral gum, and treated with 1% Triton X-100. We then followed the streptavidin-peroxidase method using a standard kit (SP9000, Zhongshan Biotech, Beijing, China). The cells on the microscope coverslips were incubated overnight at 4C with anti-ER monoclonal antibody (1:250; ZA-0102, Zhongshan Biotech). Diaminobenzidine (ZLI-9018, Zhongshan Biotech) was used to visualize the immunoreaction. Immunoreactivity was evaluated blindly by two independent pathologists using light microscopy. The percentage of positive cells was calculated by counting the number of positive cells among 100 cells.
Western Blot Analysis
Whole-cell proteins were extracted from complete cell lysates using radioimmunoprecipitation assay lysis buffer (P0013B, Beyotime Institute of Biotechnology, Shanghai, China), and the protein concentrations of the lysates were determined with an Enhanced bicinchoninic acid (BCA) Protein Assay kit (P0010, Beyotime Institute of Biotechnology). Then, we boiled the samples in sodium dodecyl sulfate sample buffer for 5 min before subjecting them to sodium dodecyl sulfate polyacrylamide gel electrophoresis. After blocking the blot in 3% bovine serum albumin for 2 hr at room temperature, we blotted the polyvinylidene difluoride membrane with anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) monoclonal antibody (1:10,000; MB001, Bioworld Technology, St Louis Park, MN) and anti-RUNX3 monoclonal antibody (1:5,000; D234-3, R3-5G4, Medical and Biological Laboratories Co. Ltd, Nagoya, Japan). Secondary antibody IRDye 800CW goat antimouse immunoglobulin G (1:10,000; 926-32210, LI-COR, Lincoln, NE) was incubated with the blot for 2 hr in the dark at room temperature. After washing the blot extensively, we obtained images using the Odyssey infrared fluorescence imaging system (LI-COR).
Reverse-transcriptase Polymerase Chain Reaction Analysis (RT-PCR)
Total cellular RNA was extracted from the three cell lines (MCF10A, MCF7, and SKBR3) with TRIzol reagent (15596-026, Life Technologies, NY), and we performed reverse transcription (RT) on cDNA from 2 µg of total RNA in 20-µL reactions using a TIANscript RT kit (KR104, TIANGEN, Beijing, China), following the manufacturer’s instructions. Primer sequences to amplify RUNX3 and β-actin were as follows: sense for RUNX3, 5-GAGTTTCACCCTGACCATCACTGTG-3 (869 base pairs); antisense for RUNX3, 5-GCCCATCACTGGTCTTGAAGGTTGT-3 (869 base pairs)16; sense for β-actin, 5-GAGCTACGAGCTGCCTGACG-3 (466 base pairs); and antisense for β-actin, 5-CCTAGAAGCATTTGCGGTGG-3 (466 base pairs)17. These primer sequences were identical to the endogenous human target genes as confirmed by a Basic Local Alignment Search Tool search. The PCR, containing templates (2 µL), sense and antisense primer (1 µL each), double-distilled water (8.5 µL), and Taq polymerase (12.5 µL), were performed in a thermal cycler (GeneAmp RCR System 9600; Life Technologies, NY). Initial denaturation was performed at 94C for 3 min and was followed by 35 amplification cycles (30 sec at 94C and 58C, 1 min at 72C). Final elongation was performed at 72C for 5 min, and β-actin was used as a control. PCR products were analyzed on 2% agarose gels.
Methylation-specific PCR Assay (MSP)
Genomic DNA was obtained from cell lines and FFPE tissues using a TIANamp Genomic DNA kit (DP304, TIANGEN) and a QIAamp DNA FFPE Tissue kit (56404, Qiagen, Dusseldorf, Germany), respectively, according to the manufacturers’ instructions. To evaluate demethylation levels of the RUNX3 gene promoter we used MSP to analyze DNA extracted from different cells based on sequence differences between methylated and unmethylated DNA after sodium bisulfite modification (EZ DNA Methylation-Gold kit, D5005, Zymo Research, Irvine, CA), according to the manufacturer’s instructions. The primer pairs specific for methylated (M) and unmethylated (U) RUNX3 DNA were as follows13: M-sense primer 5-ATAATAGCGGTCGTTAGGGCGTCG-3; M-antisense 5-GCTTCTACTTTCCCGCTTCTCGCG-3; U-sense primer 5-ATAATAGTGGTTGTTAGGGTGTTG-3; and U-antisense 5-ACTTCTACTTTCCCACTTCTCACA-3. The size of the PCR amplification product for both the methylated and the unmethylated reactions was 115 base pairs, according to analysis using ZymoTaqPreMix (E2003, Zymo Research). DNA from normal lymphocytes treated with or without CpG methyltransferase (Sigma-Aldrich, St. Louis, MO) was used as the positive control for methylated and unmethylated alleles, respectively. PCR products were separated on a 2% agarose gel and visualized by ultraviolet transillumination. The experiment was repeated three times.
Immunohistochemical (IHC) Analysis
Four-microns FFPE tissue sections were treated for heat epitope retrieval for 2 min in citrate buffer, pH 6.0 (ZLI-9065, ZSGB-BIO, Beijing, China). RUNX3 monoclonal antibody was used for IHC reaction (1:300, clone R3-6E9, Medical and Biological Laboratories Co., Ltd, Nagoya, Japan), using the streptavidin-peroxidase method (kit SP9000, ZSGB-BIO). 3,3′-diaminobenzidine-based detection (ZLI-9019, ZSGB-BIO) was used to detect the immunoreaction in the sections.18 A positive reaction was indicated by a reddish-brown precipitate in the nucleus or cytoplasm for RUNX3. RUNX3 immunoreactivity was classified as negative when the sample had less than 10% positive cells and as positive when the sample had 10% or more positive cells.12,19 Two pathologists independently scored the slides for IHC analysis.
Cell Proliferation Assay
Cellular proliferation was analyzed using a cell counting kit (CCK-8; Beyotime Institute of Biotechnology). MCF7 cells were trypsinized and seeded in 96-well plates at a density of 2,000 cells per well and were cultured for 24 hr. Then, the cells were treated with 0.5 μmol/L, 1.0 μmol/L, 2.0 μmol/L, 4.0 μmol/L, or 8.0 μmol/L 5-Aza-CdR. Each concentration was applied to six identical wells. Untreated tumor cells (no 5-Aza-CdR) were used as a control group. The plates were incubated at 37C in 5% carbon dioxide for 24 hr, 48 hr, or 72 hr each, and the remaining procedures were performed following the manufacturer’s protocol. Cell proliferation was measured by an enzyme-linked immunosorbent assay and the results determined by optical density (OD) measured at 495 nm. The growth inhibition rate was calculated using the following formula: Inhibition rate (%) = (Average OD value of experimental group – Average OD value of control group) / Average OD value of experimental group × 100%. Each experiment was repeated three times.
Migration and Invasion Assays
Migration and invasion assays were performed in a transwell chamber (8-μm pore size, Corning Incorporated, Corning, NY), according to the manufacturer’s instructions. Cell culture inserts for the invasion assay were precoated with matrigel (BD Biosciences, Bedford, MA) for 4 hr at 37C. Cells, treated with 2.0 μmol/L and 4.0 μmol/L concentrations of 5-Aza-CdR, respectively, were seeded into the upper chamber containing serum-free medium (200 µL) at a density of 1.5×105 cells/well, while 500 µL of complete medium was added to the lower chamber as a chemotaxin. After culture for 24 hr, non-invading cells were removed with a cotton swab. Cells that migrated to the lower surface of the membrane were fixed in 4% paraformaldehyde for 30 min and then were stained with crystal violet. Five random fields were selected for cell counting under a light microscope at 200× magnification. The migration assay procedure was similar except that matrigel was not used. The experiments were performed in triplicate.
Effect of 5-Aza-CdR on RUNX3 Methylation Status, Protein and mRNA Expression, and Proliferation, Migration, and Invasion in MCF7 Cells
MCF7 cells were treated with various concentrations of 5-Aza-CdR (0.5 μmol, 1.0 μmol, 2.0 μmol, 4.0 μmol, and 8.0 μmol) for 48 hr, then RUNX3 gene promoter methylation was identified by MSP assay, RUNX3 protein and mRNA expression were detected by western blot and RT-PCR.
MCF7 cells were treated with different concentrations of 5-Aza-CdR (0.5 μmol/L, 1.0 μmol/L, 2.0 μmol/L, 4.0 μmol/L, or 8.0 μmol/L) for 24 hr, 48 hr, and 72 hr, and then CCK-8 assay and migration and invasion assays were performed to detect the effect of 5-Aza-CdR on proliferation, and invasion on MCF7 cells. This assay was done in triplicate.
Statistical Analysis
The results were expressed as means ± standard deviations. Statistical analyses were performed with one-way analysis of variance or with the χ2 test, the phi coefficient was used as measure for correlation, using the SPSS 16.0 software package. The p values less than 0.05 were considered statistically significant.
Results
Expression of ER and RUNX3 in Breast Cancer Cell Lines
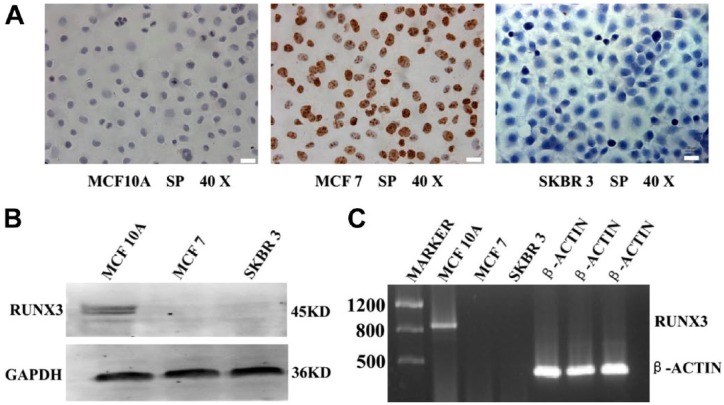
As shown in Fig. 1A, the expression of ER was undetectable in MCF10A and SKBR3 cells but was detected in MCF7 cells via ICC analysis. The expression of RUNX3 protein was detected in only normal breast epithelial cells (MCF10A) by western Blot (Fig. 1B). As expected, the mRNA of RUNX3 was detected, by RT-PCR, only in MCF10A cells (Fig. 1C). The results confirmed that the MCF7 cell line was ER-positive and SKBR3 cell line was ER-negative. RUNX3 protein and mRNA expressed in the cell line of normal breast epithelium (MCF10A) but not breast cancer cells (MCF7, SKBR3), and the RUNX3 protein level was corresponded with RUNX3 mRNA levels in both normal and neoplastic breast cell lines.
Figure 1.
ER and RUNX3 expression in normal and breast cancer cells. (A) ER was positive in MCF7 cell line, negative in MCF10A and SKBR3 cells (Immunocytochemistry, ×400). (B) RUNX3 protein was detected in MCF10A cell line but not in MCF7 and SKBR3 cells (western blot, GAPDH acted as an internal control). (C) RUNX3 mRNA was positive in MCF10A cell line but not MCF7 and SKBR3 cells (RT-PCR, β-actin as an internal control).Scale bars, 50 μm. Abbreviations: ER, estrogen receptor; RUNX3, Runt-related transcription factor 3; GAPDH, anti-glyceraldehyde 3-phosphate dehydrogenase; RT-PCR, reverse-transcriptase polymerase chain reaction analysis; SP, streptavidin-perosidase assay.
RUNX3 Expression in ER-positive and ER-negative Breast Cancer Tissue Samples
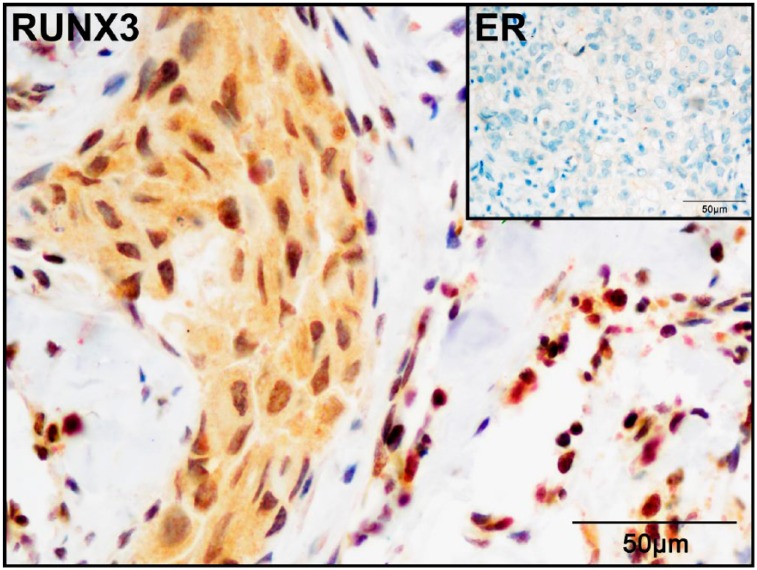
We used IHC analysis to evaluate RUNX3 expression in 113 human breast cancer tissue samples, including 68 ER-positive and 45 ER-negative cases. RUNX3 showed exclusive nucleus-enriched staining in the normal breast duct epithelial cells, but presented a cytoplasmic or a combined cytoplasmic and nuclear staining pattern in the tumor cells (Fig. 2). Forty-eight out of the 113 samples were positive for RUNX3, including 18/68 (27%) ER-positive and 30/45 (67%) ER-negative cases. The positive rate of RUNX3 expression was significantly lower in the ER-positive tissues than in the ER-negative cases (p<0.01; Table 1).
Figure 2.
RUNX3 expression in ER-negative breast cancer. RUNX3 expressed in both nucleus and cytoplasm of tumor cells in ER-negative breast cancer, while lymphocytes showed nuclear positivity (used as internal positive control; Immunocytochemistry, ×400). Abbreviations: RUNX3, Runt-related transcription factor 3; ER, estrogen receptor.
Table 1.
Relationship of RUNX3 Protein Expression and RUNX3 Gene Promoter Methylation to ER in 113 Breast Cancer Samples.
| Group | n | RUNX3 Protein |
p |
RUNX3 Hypermethylation |
p | ||
|---|---|---|---|---|---|---|---|
| Positive (%) | Negative | Methylation (%) | No-methylation | ||||
| ER-positive | 68 | 18 (27) | 50 | <0.01a | 56 (82) | 12 | <0.01a |
| ER-negative | 45 | 30 (67) | 15 | 10 (22) | 35 | ||
| Total | 113 | 48 (42) | 65 | 66 (58) | 47 | ||
Abbreviations: RUNX3, Runt-related transcription factor 3; ER, estrogen receptor.
p value compared with ER-negative group.
Hypermethylation of the RUNX3 Gene Promoter in Cell Lines and Breast Cancer Tissues
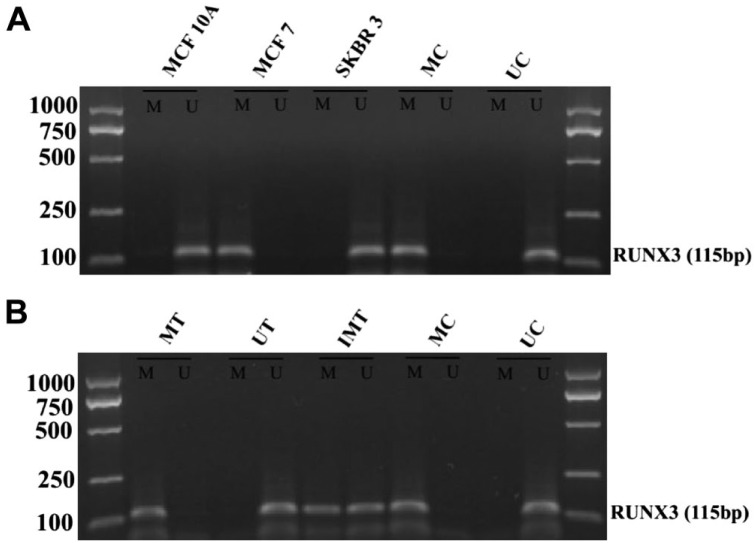
To investigate whether the silencing of the RUNX3 gene in breast cancer cell lines was associated with hypermethylation of the RUNX3 gene promoter, we performed MSP analysis on all three cell lines. We found that hypermethylation of the RUNX3 gene promoter occurred in MCF7 cells, but not in MCF10A and SKBR3 cells (Fig. 3A).
Figure 3.
Methylation status of RUNX3 gene promoter in breast cell lines and breast cancer tissues. (A) MSP analysis of the RUNX3 gene promoter in normal and breast cancer cell lines. Methylated products were found in MCF7 cells, but not in MCF10A and SKBR3 cells. (B) MSP analysis of the RUNX3 gene promoter in breast cancer tissues. RUNX3 methylated in sample 1, unmethylated in sample 2, incomplete methylation was seen in sample 3. Abbreviations: RUNX3, Runt-related transcription factor 3; MSP, Methylation-specific Polymerase Chain Reaction; MC, denotes methylated control; UC, denotes unmethylated control; MT, methylated tissue; UT, unmethylated tissue; IMT, incomplete methylated tissue.
Also, we performed an MSP assay on all 113 breast cancer tissue samples, to determine whether the RUNX3 hypermethylation in breast cancer tissue was consistent with that in the breast cancer cell lines. The results showed that 66 out of the 113 (58%) cases had RUNX3 hypermethylation, including 56/68 (82%) ER-positive cases, and 10/45 (22%) ER-negative cases. A significant difference of RUNX3 gene promoter hypermethylation rate was found between ER-positive and ER-negative groups (p<0.01; Fig. 3B, Table 1).
In the RUNX3-positive cases, 31% (15/48) presented RUNX3 promoter hypermethylation, while 79% (51/65) of the RUNX3-negative cases showed RUNX3 promoter hypermethylation. Negative correlation was found between RUNX3 protein expression and RUNX3 gene promoter hypermethylation (Phi = −.673, p<0.001; Table 2).
Table 2.
Relationship Between RUNX3 Protein Expression and Promoter Methylation in 113 Breast Cancer Samples.
| RUNX3 Protein | n | Methylation (%) | No-methylation | p Value |
|---|---|---|---|---|
| Positive | 48 | 15 (31) | 33 | <0.001a |
| Negative | 65 | 51 (79) | 14 | |
| Total | 113 | 66 (58) | 47 |
Abbreviation: RUNX3, Runt-related transcription factor 3
p value compared with RUNX3-negative group.
Correlation Between Hypermethylation of the RUNX3 Gene Promoter and Clinicopathologic Parameters
We analyzed the relationship between hypermethylation of the RUNX3 gene and the clinicopathologic parameters of the 113 breast cancer tissue samples (Table 3). In ER-positive tumors, RUNX3 promoter hypermethylation was associated with the presence of lymph node metastasis and clinical stage; while in ER-negative cases RUNX3 methylation was associated only with clinical stage. In ER-positive samples, the tumors with metastasis in less than 3 lymph nodes had a higher positive rate of hypermethylation than tumors with more than 3 lymph nodes metastasis (p<0.05), and tumors with an early clinical stage (I and II) also had a higher positive rate of RUNX3 hypermethylation than advanced-stage tumors (III) (p<0.05).
Table 3.
Relationship Between RUNX3 Promoter Methylation Status and Clinicopathological Parameters in 113 Breast Cancer Samples.
| Variables | n | ER-positive Breast Cancer |
χ2 | p | n | ER-negative Breast Cancer |
χ2 | p | ||
|---|---|---|---|---|---|---|---|---|---|---|
| M | UM | M | UM | |||||||
| Age (years) | ||||||||||
| <56 | 38 | 29 | 9 | 2.16 | 0.14 | 25 | 4 | 21 | 1.26 | 0.26 |
| ≥56 | 30 | 27 | 3 | 20 | 6 | 14 | ||||
| Tumor size (cm) | ||||||||||
| ≤2 | 27 | 22 | 5 | 1.11 | 0.66 | 22 | 6 | 16 | 2.71 | 0.19 |
| 2–5 | 34 | 29 | 5 | 18 | 2 | 16 | ||||
| >5 | 7 | 5 | 2 | 5 | 2 | 3 | ||||
| Axillary lymph nodes affected | ||||||||||
| 0–3 | 57 | 50 | 7 | 4.89 | 0.03 | 9 | 4 | 5 | 1.81 | 0.18 |
| >3 | 11 | 6 | 5 | 36 | 6 | 30 | ||||
| Histological grade | ||||||||||
| I | 7 | 5 | 2 | 1.31 | 0.54 | 6 | 2 | 4 | 0.82 | 0.79 |
| II | 31 | 25 | 6 | 21 | 4 | 17 | ||||
| III | 30 | 26 | 4 | 18 | 4 | 14 | ||||
| Clinical stage | ||||||||||
| I + II | 40 | 37 | 3 | 5.29 | 0.02 | 30 | 4 | 26 | 2.72 | 0.01 |
| III | 28 | 19 | 9 | 15 | 6 | 9 | ||||
Abbreviations: RUNX3, Runt-related transcription factor 3; ER, estrogen receptor; M, cases with RUNX3 promoter hypermethylated; U, cases with RUNX3 promoter nonmethylated.
5-Aza-CdR on the Methylation Status of RUNX3 in MCF7 Cells
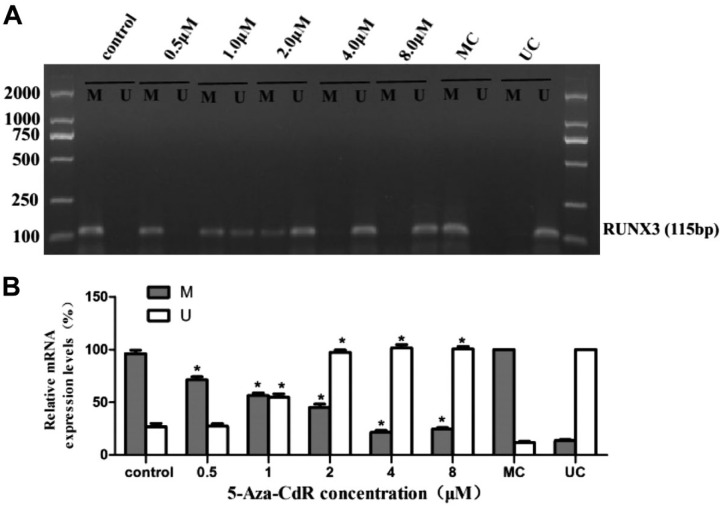
To confirm whether the hypermethylation status of the RUNX3 gene promoter could be reversed, we performed an MSP assay on MCF7 cells treated with different concentrations of 5-Aza-CdR. The experimental data revealed that the promoter regions of RUNX3 were heavily methylated in MCF7 cells. However, after 5-Aza-CdR treatment, the methylation levels of the RUNX3 gene promoter decreased gradually as doses of 5-Aza-CdR increased, and meanwhile the demethylated levels increased (Fig. 4; p<0.05).
Figure 4.
Effect of 5-Aza-CdR on RUNX3 methylation in MCF7 cell line for 48 hr. The hypermethylation of RUNX3 reduced gradually in MCF7 cell line treated with 0.5, 1.0, 2.0, 4.0, and 8.0 μM 5-Aza-CdR, respectively (MSP assay). Abbreviations: RUNX3, Runt-related transcription factor 3; 5-Aza-CdR, 5-Aza-2′-deoxycytidine; MSP, Methylation-specific Polymerase Chain Reaction; MC, denotes methylated control; UC, denotes unmethylated control. *p<0.05 (compared with control group).
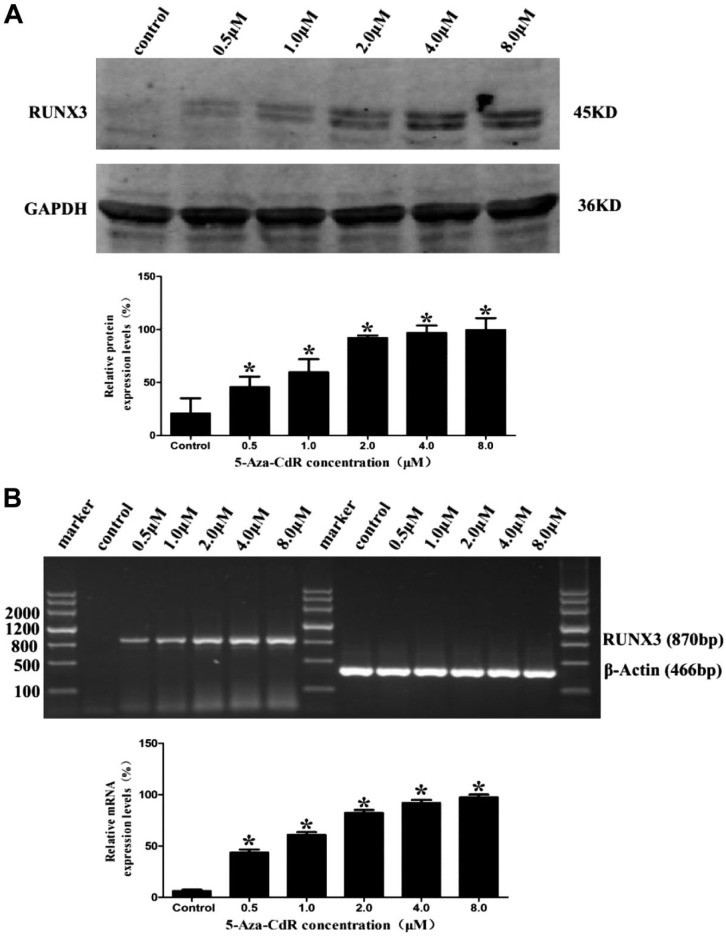
5-Aza-CdR Increased RUNX3 Protein and mRNA Expression Levels in MCF7 Cells
To examine the effect of 5-Aza-CdR on the expression of RUNX3, we tested the protein and mRNA expression with western blot and RT-PCR on the MCF7 cells after treatment with 5-Aza-CdR. Compared with the control group, RUNX3 protein expression significantly increased with the increase of 5-Aza-CdR concentration (Fig. 5A; p<0.05). Consistently, RT-PCR analysis results showed that RUNX3 mRNA expression also significantly increased with increased 5-Aza-CdR concentrations (Fig. 5B; p<0.05).
Figure 5.
Effect of 5-Aza-CdR on RUNX3 expression in MCF7 cell line for 48 hr. (A) RUNX3 protein expression increased gradually along with the increase of 5-Aza-CdR concentration (western blot, GAPDH as an internal control). (B) RUNX3 mRNA expression increased gradually along with the increase of 5-Aza-CdR concentration (RT-PCR, β-actin was an internal control). Abbreviations: RUNX3, Runt-related transcription factor 3; 5-Aza-CdR, 5-Aza-2′-deoxycytidine; GAPDH, anti-glyceraldehyde 3-phosphate dehydrogenase; RT-PCR, reverse-transcriptase polymerase chain reaction analysis. *p<0.05 (compared with control group).
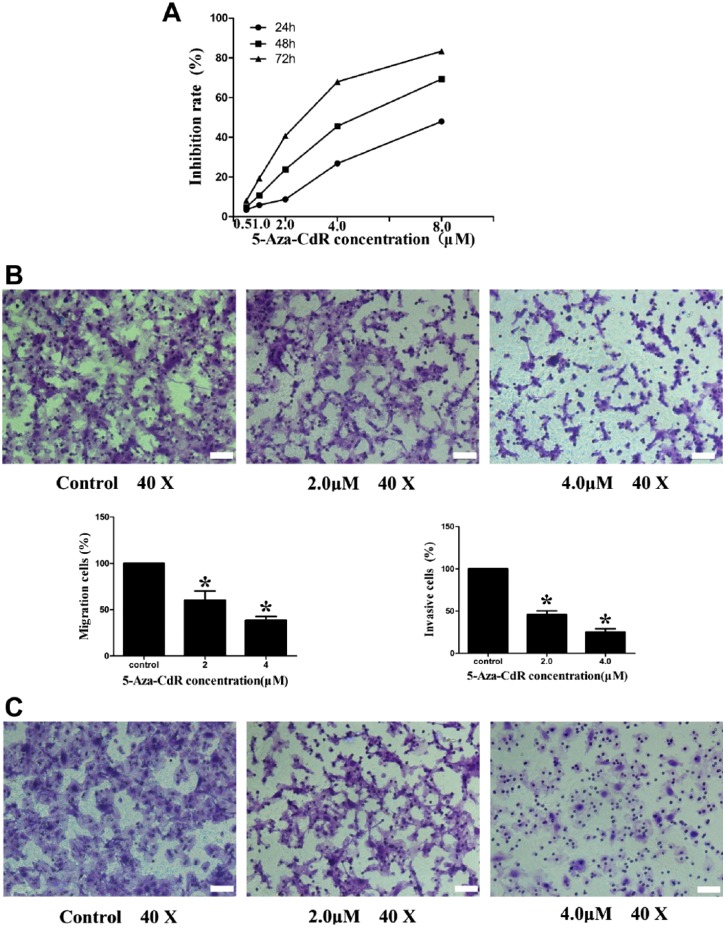
5-Aza-CdR Suppressed Proliferation, Migration, and Invasion in MCF7 Cells
To investigate the influence of demethylation of the RUNX3 gene promoter on breast cancer, we treated ER-positive MCF7 cells with different concentrations of 5-Aza-CdR and detected the effects of 5-Aza-CdR on MCF7 cell growth with a CCK-8 assay. The results revealed that 5-Aza-CdR significantly suppressed the proliferation of MCF7 cells in a dose- and time-dependent manner (Fig. 6A, Table 4). We then determined the effect of 5-Aza-CdR on MCF7 cell migration and invasion. The migration assay revealed that 5-Aza-CdR inhibited the motility potential of MCF7 cells by 40% (with the 2.0 μmol/L concentration) and 61% (with the 4.0 μmol/L concentration). The 5-Aza-CdR inhibited the invasive ability of MCF7 cells by 54% (with the 2.0 μmol/L concentration) and 75% (with the 4.0 μmol/L concentration) (Fig. 6B).
Figure 6.
The 5-Aza-CdR inhibits proliferation, migration, and invasion of MCF7 cell line. (A) Growth-inhibiting rate increased gradually along with the increase of 5-Aza-CdR concentration and time (CCK-8 analysis). (B) The migration ability of MCF7 cell line reduced gradually along with the increase of 5-Aza-CdR concentration (Transwell migration analysis). (C) The invasion ability of MCF7 cell line reduced gradually along with the increase of 5-Aza-CdR concentration (Transwell invasion analysis). Scale bars, 50 μm. Abbreviation: 5-Aza-CdR, 5-Aza-2′-deoxycytidine. *p<0.05 (compared with control group).
Table 4.
The Inhibition Rate of Proliferation of MCF7 Cells Treated With 5-Aza-CdR ( ± s, %).
| 5-Aza-CdR Concentration | Exposure Time |
||
|---|---|---|---|
| 24 hr | 48 hr | 72 hr | |
| 0.5 μmol/L | 3.45 ± 0.51* | 4.79 ±1.48* | 8.04 ± 0.94* |
| 1.0 μmol/L | 5.86 ± 0.87* | 10.83 ± 2.83* | 19.35 ± 1.56* |
| 2.0 μmol/L | 8.74 ± 0.99* | 23.77 ± 2.24* | 40.67 ± 1.62* |
| 4.0 μmol/L | 26.77 ± 0.87* | 45.61 ± 1.43* | 67.94 ± 1.47* |
| 8.0 μmol/L | 47.97 ± 1.31* | 69.34 ± 1.96* | 83.36 ± 1.76* |
| IC50 | 8.42 | 4.52 | 2.54 |
Abbreviations: 5-Aza-CdR: 5-Aza-2′-deoxycytidine; IC50, half maximal inhibitory concentration.
p<0.05 (compared with control group [0 μM/L]).
Discussion
In this study, we analyzed RUNX3 gene promoter methylation status, mRNA expression and protein expression on breast cell lines and breast cancer tissues. In addition, we correlated the results with ER expression and clinicopathological features on the breast cancer tissues. Our results showed that RUNX3 protein and mRNA expressed on the normal breast cell line MCF10A, but not on the breast cancer cell lines MCF7 or SKBR3. RUNX3 hypermethylation was detected in ER-positive breast cancer cell line MCF7 but not in ER-negative breast cell line SKBR3 or in normal breast cell line MCF10A. On the tissue samples, RUNX3 expression was significantly higher in ER-negative breast cancers than that in ER-positive group, while RUNX3 hypermethylation was detected in most of the ER-positive tumors but only in a small subset of the ER-negative tumors (p<0.05). We noted that although RUNX3 promoter hypermethylation was not detected in ER-negative SKBR3, this cell line showed RUNX3-negative, suggesting that the correlation of RUNX3 expression and promoter hypermethylation was cellular context-dependent. These results suggest that RUNX3 hypermethylation, which was consistent with RUNX3 downregulation, occurs mainly in ER-positive breast cancer. Epigenetic RUNX3 downregulation has been found in various types of solid tumor,8,9,16 and plays an important role in the inactivation of RUNX3 in breast cancer.14,16–18 The hypermethylation of RUNX3 is known as an early event in breast carcinogenesis.13 The study presented here tested the hypothesis that RUNX3 hypermethylation is associated with breast cancer mainly in ER-positive subtype. Our findings that RUNX3 protein and mRNA can be detected in MCF10A but not in MCF7 or SKBR3 cells suggest that RUNX3 is indeed inactive in breast cancer cells. Hypermethylation of RUNX3 gene promoter found in MCF7 cells but not in SKBR3 cells, furthermore, suggests that gene promoter methylation is not the only mechanism that inactivates RUNX3 in breast cancer and that, in ER-negative breast cancer, other mechanisms such as deletion or mutation could be responsible for the downregulation of RUNX3. Moreover, our results suggest that, in ER-positive breast cancer, gene promoter methylation could be a mechanism involved in RUNX3 inactivation.
To confirm the above results in human breast cancer tissues, we analyzed 113 breast cancer tissue specimens. Our results showed that RUNX3 protein was expressed in 48/113 (42%) breast cancers, and RUNX3 hypermethylation was found in 66/113 (58%) breast cancers. Our results are similar to Lau’s17 and Hwang’s16 findings, which showed a rate of hypermethylation of the RUNX3 gene promoter approximately 50% in both tissue and cell lines, also showing a higher methylation rate than observed by Kim et al.,19 which found only 25% of RUNX3 methylation in breast cancer. Also, we found that RUNX3 protein expression was significantly lower in the ER-positive samples (18/68, 27%) than in the ER-negative samples (30/45, 67%; p<0.01), and RUNX3 promoter methylation was significantly more frequent in the ER-positive cases (56/68, 82%) than the ER-negative samples (10/45, 22%; p<0.01). RUNX3 promoter methylation showed a negative correlation with RUNX3 protein expression in the tissue specimens (p<0.05). Interestingly, we found RUNX3 was mainly positive in nuclei in normal breast tissue while positive in both nuclei and cytoplasm in neoplastic tissues, suggesting that not only promoter methylation but also cytoplasm translocation might play critical roles in RUNX3 regulation. This phenomenon has been reported by other groups.2,13,17 Overall, these data confirm that RUNX3 promoter methylation is a key mechanism of RUNX3 downregulation in ER-positive breast cancer in tissue level.
The analysis of clinicopathologic parameters indicated that RUNX3 methylation was associated with clinical stage in both ER-positive and ER-negative breast cancers and with lymph node metastasis in ER-positive breast cancers. In ER-positive breast cancer, RUNX3 promoter hypermethylation was associated with more favorable prognostic factors such as lower clinical stage and fewer affected lymph nodes. This implies that detection of the methylation status of the RUNX3 gene promoter may be of prognostic value for patients with ER-positive breast cancer. However, this hypothesis must be verified with a large cohort study.
As an epigenetic mechanism of tumor suppressor gene silencing, DNA hypermethylation can be reversed by methylation inhibitors.20 5-Aza-CdR, a nucleoside analog and specific DNA methyltransferase inhibitor, has shown antitumoral activity in patients with leukemia and myelodysplastic syndrome.21,22 Although much has been learned about 5-Aza-CdR and antimethylation, few reports have been published about either RUNX3 inactivation owing to promoter hypermethylation in breast cancer or the potential reversal of hypermethylation to restore the biological function of RUNX3. Given the heterogeneous subtypes of breast cancer differ greatly in pathogenesis, clinical and pathologic features, treatment response, and prognosis, further research is necessary to profile RUNX3 epigenetic dysregulation in different subtypes of breast cancer. Our results indicate that methylation inhibitor 5-Aza-CdR reverse the downregulation of the RUNX3 gene by gene promoter methylation in a dose-dependent fashion, and suppressed proliferation, migration and invasion in MCF7 cells. This implicates that gene promoter methylation is responsible for the RUNX3 downregulation observed in ER-positive breast cancer, and hints that reversion of RUNX3 gene methylation in ER-positive breast cancers may be a new potential therapeutic target. Further studies are needed to confirm this hypothesis.
Our data also showed that hypermethylation of RUNX3 gene promoter was associated with prognostic parameters, like stage and lymph node metastasis. Overall, our study strongly suggest that RUNX3 epigenetic dysregulation may play an important role in mainly ER-positive breast carcinogenesis. Also, our in vitro data and clinicopathologic data showed a correlation between RUNX3 gene promoter methylation and the invasiveness of breast cancer tumors. Our findings suggest that RUNX3 epigenetic alterations could be a prognostic marker for breast cancer and a potential therapeutic target that could reverse the malignant potential of ER-positive breast cancer. Further research on larger cohorts of patients to explore the role of RUNX3 in both ER-positive and triple-negative breast cancer is needed to understand the clinical and mechanistic role of RUNX3 in breast carcinogenesis.
Footnotes
Authors’ Note: Abstract presented at the 2016 Annual Meeting of the United States and Canadian Academy of Pathology (USCAP), Washington State Convention Center, Seattle, WA.
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: HL designed the study and drafted the manuscript. ZY, QY, KC, and YWei performed the immunohistochemistry and the molecular genetics studies. JR-C and DM revised the manuscript. YWu designed the study. All authors have read and approved the final manuscript.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Xuzhou Science and Technology Project (KC15SH043): “RUNX3 gene promoter methylation and its reversion in breast cancer,” and by the Special Foundation of the President of Xuzhou Medical University (09KJZ05): “The clinicopathological and molecular biological study on basal-like breast carcinoma.”
Contributor Information
Hui Liu, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Zhantao Yan, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Qianqian Yin, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Kai Cao, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Yu Wei, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Jaime Rodriguez-Canales, Laboratory of Pathology, MedImmune, LLC, Gaithersburg, Maryland.
Dongshen Ma, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Yongping Wu, Department of Pathology; Laboratory of Clinical and Experimental Pathology; Xuzhou Medical University, Xuzhou, Jiangsu, P.R. China.
Literature Cited
- 1. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–70. [DOI] [PubMed] [Google Scholar]
- 2. Chuang LS, Ito Y. RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene. 2010;29(18):2605–15. [DOI] [PubMed] [Google Scholar]
- 3. Fujii M, Fujimoto N, Hiraki A, Gemba K, Aoe K, Umemura S, et al. Aberrant DNA methylation profile in pleural fluid for differential diagnosis of malignant pleural mesothelioma. Cancer sci. 2012;103(3):510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24(2):132–8. [DOI] [PubMed] [Google Scholar]
- 5. Parrella P, Poeta ML, Gallo AP, Prencipe M, Scintu M, Apicella A, et al. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10(16):5349–54. [DOI] [PubMed] [Google Scholar]
- 6. Bae YK, Brown A, Garrett E, Bornman D, Fackler MJ, Sukumar S, et al. Hypermethylation in histologically distinct classes of breast cancer. Clin Cancer Res. 2004;10(18 Pt 1):5998–6005. [DOI] [PubMed] [Google Scholar]
- 7. Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, et al. Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell. 2002;109(1):113–24. [DOI] [PubMed] [Google Scholar]
- 8. Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68(8):2551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Miranda TB, Jones PA. DNA methylation: the nuts and bolts of repression. J Cell Physiol. 2007;213(2):384–90. [DOI] [PubMed] [Google Scholar]
- 10. Song XY, Li BY, Zhou EX, Wu FX. The clinicopathological significance of RUNX3 hypermethylation and mRNA expression in human breast cancer, a meta-analysis. Onco Targets Ther. 2016;9:5339–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki M, Shigematsu H, Shames DS, Sunaga N, Takahashi T, Shivapurkar N, et al. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93(9):1029–37. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, et al. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65(17):7743–50. [DOI] [PubMed] [Google Scholar]
- 13. Subramaniam MM, Chan JY, Soong R, Ito K, Ito Y, Yeoh KG, et al. RUNX3 inactivation by frequent promoter hypermethylation and protein mislocalization constitute an early event in breast cancer progression. Breast Cancer Res Treat. 2009;113(1):113–21. [DOI] [PubMed] [Google Scholar]
- 14. Jiang Y, Tong D, Lou G, Zhang Y, Geng J. Expression of RUNX3 gene, methylation status and clinicopathological significance in breast cancer and breast cancer cell lines. Pathobiology. 2008;75(4):244–51. [DOI] [PubMed] [Google Scholar]
- 15. Huang B, Qu Z, Ong CW, Tsang YH, Xiao G, Shapiro D, et al. RUNX3 acts as a tumor suppressor in breast cancer by targeting estrogen receptor alpha. Oncogene. 2012;31(4):527–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hwang KT, Han W, Bae JY, Hwang SE, Shin HJ, Lee JE, et al. Downregulation of the RUNX3 gene by promoter hypermethylation and hemizygous deletion in breast cancer. J Korean Med Sci. 2007;22(Suppl):S24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lau QC, Raja E, Salto-Tellez M, Liu Q, Ito K, Inoue M, et al. RUNX3 is frequently inactivated by dual mechanisms of protein mislocalization and promoter hypermethylation in breast cancer. Cancer Res. 2006;66(13):6512–20. [DOI] [PubMed] [Google Scholar]
- 18. Chen W, Salto-Tellez M, Palanisamy N, Ganesan K, Hou Q, Tan LK, et al. Targets of genome copy number reduction in primary breast cancers identified by integrative genomics. Genes Chromosomes Cancer. 2007;46(3):288–301. [DOI] [PubMed] [Google Scholar]
- 19. Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW, Bang YJ, et al. Methylation of RUNX3 in various types of human cancers and premalignant stages of gastric carcinoma. Lab Invest. 2004;84(4):479–84. [DOI] [PubMed] [Google Scholar]
- 20. Tycko B. Cancer epigenetics and targeted therapies. Oncology (Williston Park, NY). 2011;25(3):228, 231. [PubMed] [Google Scholar]
- 21. Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2’-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66(10):5495–503. [DOI] [PubMed] [Google Scholar]
- 22. Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O’Brien S, Cortes J, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109(1):52–7. [DOI] [PubMed] [Google Scholar]