Abstract
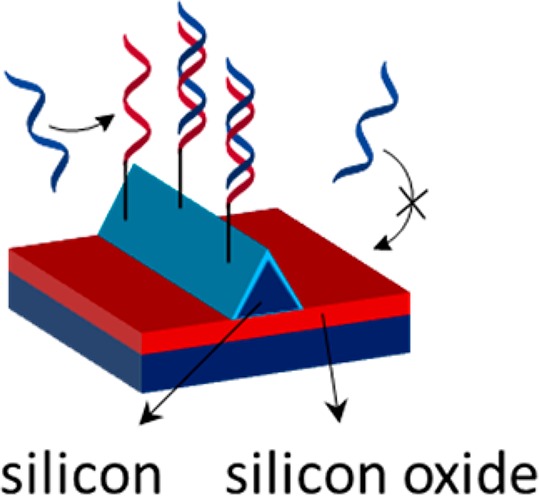
Silicon nanowire chips can function as sensors for cancer DNA detection, whereby selective functionalization of the Si sensing areas over the surrounding silicon oxide would prevent loss of analyte and thus increase the sensitivity. The thermal hydrosilylation of unsaturated carbon–carbon bonds onto H-terminated Si has been studied here to selectively functionalize the Si nanowires with a monolayer of 1,8-nonadiyne. The silicon oxide areas, however, appeared to be functionalized as well. The selectivity toward the Si–H regions was increased by introducing an extra HF treatment after the 1,8-nonadiyne monolayer formation. This step (partly) removed the monolayer from the silicon oxide regions, whereas the Si–C bonds at the Si areas remained intact. The alkyne headgroups of immobilized 1,8-nonadiyne were functionalized with PNA probes by coupling azido-PNA and thiol-PNA by click chemistry and thiol–yne chemistry, respectively. Although both functionalization routes were successful, hybridization could only be detected on the samples with thiol-PNA. No fluorescence was observed when introducing dye-labeled noncomplementary DNA, which indicates specific DNA hybridization. These results open up the possibilities for creating Si nanowire-based DNA sensors with improved selectivity and sensitivity.
Introduction
Early diagnostics of diseases, in particular cancer, is receiving increasing attention as early detection promises higher curing rates and/or prolonged survival.1 Detecting tumor DNA is preferably noninvasive, for example based on blood2 or urine samples.3 As a sensor, for screening or disease monitoring, lab-on-a-chip configurations are attractive since analysis outside the hospital is possible, for example by the general practitioner or even at home as a do-it-yourself test.4,5 Ideally, the DNA detection is highly specific, that is, for the targeted biomarker DNA only, in particular, for a well-recognized marker sequence, and highly sensitive, that is, able to detect the biomarker DNA at low concentrations, even in the presence of a large amount of background DNA.
Surface chemistry can be used to specifically capture tumor DNA.6,7 For this purpose, a specific DNA or peptide nucleic acid (PNA) oligo can be attached as a probe sequence to the surface, which consists of the complementary strand for the disease-specific DNA sequence. PNA is an artificially synthesized polymer that resembles DNA but contains a neutral peptide-like backbone instead of a negatively charged deoxyribose phosphate backbone.8 The spacing between the nucleotides is equal for DNA and PNA, which makes PNA–DNA hybridization possible. For sensing purposes, PNA is preferred as a probe since PNA–DNA interactions are stronger than DNA–DNA interactions due to the lack of electrostatic repulsion, and PNA–DNA recognition often also shows a better selectivity.9
Sensors to detect PNA–DNA hybridization rely on signal transduction based on, for example, surface plasmon resonance10 or electronic measurements,11−14 and the latter are frequently based on silicon (Si) nanowires on a chip. Hydrosilylation chemistry, in which unsaturated carbon–carbon bonds are coupled onto oxide-free, H-terminated Si surfaces, is commonly used when fabricating Si sensors for DNA detection.11−15 Hydrosilylation is advantageous because the resulting Si–C bonds are stable in aqueous environment, and the absence of an insulating silicon oxide (SiOx) layer improves the electrical contact with the underlying substrate and thus the sensitivity of the sensor as well.14 Nonetheless, any adsorption of analyte DNA outside the sensor area, either specific or nonspecific, would result in a loss of sensitivity. Therefore, the sensitivity of a DNA sensor device can be significantly improved when the probe is only bound at the sensing area.16,17 Here, we focus on chips with Si nanowires surrounded by SiOx,18 of which the Si regions should be functionalized selectively with the DNA or PNA probe. Hydrosilylation could potentially be used to selectively functionalize the Si nanowires over the SiOx surroundings, as has been suggested in the literature.15,19,20 This material-selective functionalization has, however, not yet been studied in detail.
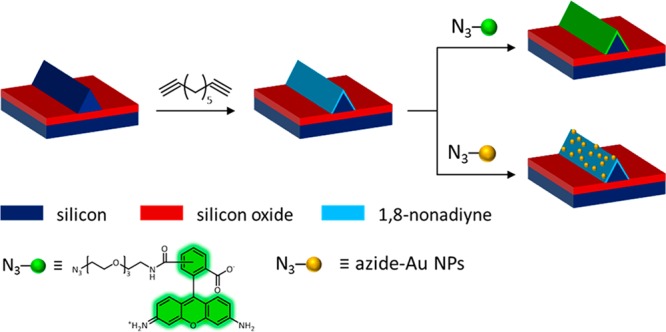
Here, we study the selective functionalization of the sensing area of a chip, that is, its Si nanowires, whereas the surrounding SiOx should remain unfunctionalized. Specifically, the dialkyne 1,8-nonadiyne is coupled to H-terminated Si in order to achieve material-selective functionalization. Whereas most examples from literature are based on photochemical hydrosilylation, that is, coupling under illumination with light, we have used thermal hydrosilylation since this technique generally yields monolayers with a higher coverage.21 The freestanding alkyne group of the 1,8-nonadiyne monolayer can be functionalized subsequently with azide or thiol moieties by copper-catalyzed click chemistry22,23 and thiol–yne chemistry,24,25 respectively. First, the specific functionalization of Si nanowires on a chip is tested by click chemistry with dummy molecules, that is, with an azide-functionalized dye and with azide-functionalized nanoparticles (NPs) to enable characterization by fluorescence microscopy and high-resolution scanning electron microscopy (HR-SEM), respectively. Thereafter, azido-PNA and thiol-PNA are coupled onto the 1,8-nonadiyne monolayer as a proof of concept for a biosensor. Although we do not aim for developing a complete sensing device here, PNA–DNA hybridization with complementary DNA (cDNA) is investigated as a proof of principle using fluorescence microscopy and quartz crystal microbalance (QCM) measurements.
Results and Discussion
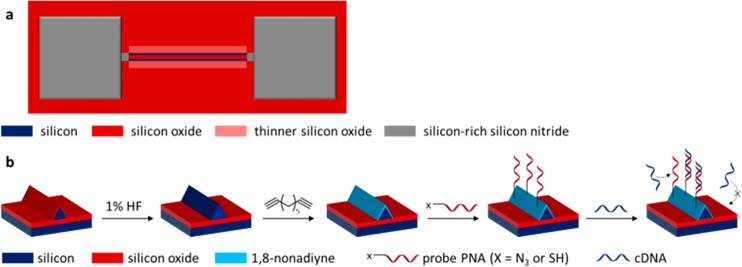
The chip studied for selective functionalization is based on Si nanowires as sensing areas surrounded by inactive SiOx, as reported before (Scheme 1a).18 A sensor consists of two Si nanowires with a triangular cross section, bridging two contact pads. Each substrate contained several sensors with the same design but different nanowire lengths. For proof of principle tests for selective chemical functionalization, chips with silicon-rich silicon nitride contact pads instead of metal contacts were used,18 which did not allow for electrical characterization. Instead, fluorescence microscopy, HR-SEM, and QCM were used to verify the selective functionalization routes. In the nanowire fabrication process, two rectangular areas of SiOx, next to the Si nanowires, are slightly etched (Scheme 1a). The composition of these areas is similar to the surrounding SiOx, but the difference in thickness can be seen as a contrast in the microscopy images.
Scheme 1. (a) Schematic Illustration of the Chips with Si Nanowires and (b) Schematic Illustration of the Material-Selective Monolayer Formation and Subsequent Probe PNA Modification onto H-Terminated Si Nanowires Surrounded by SiOx.
The process to functionalize the Si nanowires specifically with probe PNA, while not functionalizing the SiOx areas, is shown in Scheme 1b. First, a 4 min immersion in an aqueous solution of 1% hydrofluoric acid (HF) leads to removal of the thin oxide layer from the Si nanowires. The surroundings, including the rectangular areas around the nanowires, consist of a thick (>120 nm) SiOx layer, which is only marginally removed by the HF dip. Nonetheless, the treatment needs to be controlled well to avoid under-etching and, thereby, potential removal of the Si nanowires. Subsequent monolayer formation with 1,8-nonadiyne targets the H-terminated Si nanowires. Functionalized PNA can be coupled thereafter onto the alkyne headgroup by click chemistry with azido-PNA26,27 or thiol–yne chemistry with thiol-PNA.28,29 Introducing cDNA onto this chip should result in specific and spatioselective binding onto the probe-functionalized nanowires.
Selective monolayer formation at the Si nanowires was tested using click chemistry with dummy compounds (Scheme 2). As stated above, monolayer formation of 1,8-nonadiyne was first performed to functionalize the H-terminated Si nanowires. Click chemistry with an azide-functionalized dye or azide-functionalized gold (Au) NPs was used to allow characterization by fluorescence microscopy and HR-SEM, respectively, in order to probe the success and the selectivity of the preceding monolayer formation step.
Scheme 2. Schematic Illustration of the Click Chemistry Routes Tested at Si Nanowires Functionalized with a 1,8-Nonadiyne Monolayer.
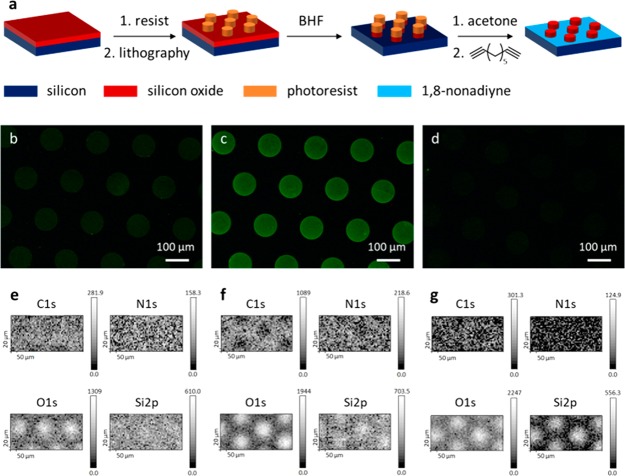
To properly discriminate between monolayer formation at the Si and SiOx regions, first tests were performed on patterns larger than the nanowires on chip (150 nm diameter). Using photolithography, patterns of SiO2 dots were created with a diameter of 100 μm and a thickness of 160 nm, surrounded by Si–H due to SiO2 removal on these resist-free areas (Figure 1a). Immediately afterward, a monolayer of 1,8-nonadiyne was formed on the patterned substrate by thermal hydrosilylation (160 °C) of the pure 1,8-nonadiyne.23,30 After click chemistry with an azide-functionalized dye, fluorescence imaging was expected to show nonfluorescent SiO2 dots surrounded by fluorescent Si. Figure 1b showed, however, the inverted pattern with a higher intensity at the dots compared to the surrounding Si. This observation does not necessarily mean that the coverage of the dye is higher at the dots, as Si is known to quench fluorescence.31
Figure 1.
(a) Schematic illustration of the formation of SiO2 dots surrounded by H-terminated Si and subsequent material-selective monolayer formation (BHF = buffered hydrogen fluoride), (b–d) Fluorescence microscopy images (exposure time 1 s) of SiO2/Si patterns functionalized with (b) 1,8-nonadiyne and azide-functionalized dye, (c) the same sequence as (b) with an extra BHF dip (10 s) after the 1,8-nonadiyne monolayer formation, and (d) a control sample without 1,8-nonadiyne, and (e–g) elemental mapping of the C 1s, N 1s, O 1s, and Si 2p regions on the SiO2/Si patterned substrates of (e) panels b, (f) c, and (g) d.
X-ray photoelectron spectroscopy (XPS) elemental mapping showed more O atoms at the dots, as expected due to the SiO2 composition (Figure 1e). The expected contrast in C and N was, however, hardly distinguishable. This cannot be due to physisorption of the dye, as a control sample without a 1,8-nonadiyne monolayer did not show any fluorescence (Figure 1d) nor N atoms (Figure 1g) by fluorescence microscopy and XPS, respectively. We therefore attribute these observations to, here undesired, 1,8-nonadiyne monolayer formation at the SiO2 dots, occurring simultaneous to the desired functionalization of the Si areas outside the dots. This was supported by the deconvoluted N 1s XPS spectrum (data not shown), where the formation of a triazole moiety at the dots was confirmed by the formation of two bands at 399 and 402 eV in the N 1s region. This means that the azide-functionalized dye is covalently bound at the SiO2 dots. Furthermore, on a planar SiO2 substrate the contact angle changed from hydrophilic (<20°) after a 1% HF dip to 77.8° ± 1.2 after the 1,8-nonadiyne reaction, which is comparable to a 1,8-nonadiyne monolayer on Si–H (vide infra). This nonselective functionalization of oxidized and unoxidized Si by hydrosilylation has been observed before.32 There, a 2 min 2% HF dip was sufficient to remove the monolayer from the oxidized regions.32 Here, a buffered hydrogen fluoride (BHF) dip for 10 s lowered the contact angle to <20°, which indicates removal of any undesired monolayer at the SiO2 parts. At the SiO2 parts, the monolayer is bound through SiO–C bonds, which are chemically sensitive to BHF.32 In contrast, the Si–C bound monolayer should withstand the BHF treatment, as was verified on a planar Si substrate (data not shown).
When an extra BHF dip was performed between the 1,8-nonadiyne monolayer formation and the click chemistry step on a patterned sample, an even higher fluorescence intensity was observed at the dots (Figure 1c). Nonetheless, the XPS elemental mapping showed more C and Si at the areas outside the dots (Figure 1f), as expected from the selective presence of a 1,8-nonadiyne monolayer. The difference in composition was also reflected by the Si 2p element spectra, in which oxidized Si was observed at the dots (Figure S1a) and mainly unoxidized Si outside the dots (Figure S1b). For the N 1s signal, however, hardly a difference could be detected between the Si and SiO2 regions in the mappings (Figure 1f). The element spectra recorded at (Figure S1c) and outside (Figure S1d) the dots showed the presence of N atoms at both areas. Nevertheless, the highest intensity was observed at the Si areas, as expected. The deconvoluted spectra showed two bands at 399 and 402 eV in the N 1s region for both areas (Figure S1c,d), which are characteristic for the formation of a triazole moiety. Any physisorbed azide-containing compound would have appeared at 405 eV,23 which was not observed in these spectra. All these observations denote undesired 1,8-nonadiyne monolayer formation at the SiO2 dots, albeit to a lesser extent than the desired monolayer formation at the Si areas.
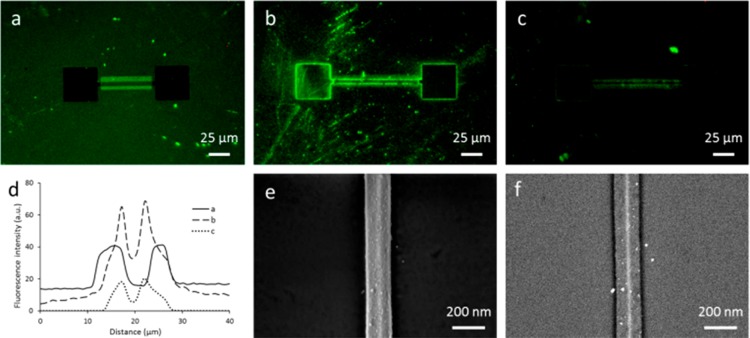
The monolayer formation process was transferred onto chips with Si nanowires surrounded by SiOx. After 1,8-nonadiyne monolayer formation, click chemistry was performed with an azide-functionalized dye (azide-fluor 488) or 10 nm azide-functionalized Au NPs. Both fluorescence microscopy (Figure 2a, d) and HR-SEM images (Figure 2e, f) showed successful functionalization of the Si nanowires, both with the dye and the Au NPs, as indicated by a bright fluorescence and dots with a bright contrast in the HR-SEM images, respectively. A control sample without 1,8-nonadiyne was only slightly fluorescent upon treatment with the azide-functionalized dye under click chemistry conditions (Figure 2c,d), although no fluorescent signal was expected at all. For the HR-SEM image, an energy-selective backscattering (ESB) detector was used to display compositional variations on the sample based on atomic number (Figure 2f). This shows the selective presence of Au NPs on the Si nanowire only. Whereas the HR-SEM images indicate specific functionalization, the background fluorescence observed at the oxidized areas in Figure 2a could denote nonspecific physisorption and undesired 1,8-nonadiyne monolayer formation, as observed above for the SiO2 dots pattern. The higher background fluorescence in the rectangular areas around the Si nanowires is expected to be due to a higher surface roughness, which could lead to a higher monolayer coverage. An extra BHF dip between the 1,8-nonadiyne monolayer formation and the click chemistry step resulted in a more defined presence of the dye at the nanowires only (Figure 2b,d). Furthermore, the fluorescence intensity of the SiOx background generally decreased, thus, indicating less undesired presence of the dye. The fluorescent patterns in the background are attributed to roughening of the SiOx areas by BHF etching. Thus, material-selective functionalization at the nanowires seems to be possible, although removal of 1,8-nonadiyne from the SiOx areas is a necessary step.
Figure 2.
Selective functionalization of Si nanowires on chips with a 1,8-nonadiyne monolayer characterized by (a–d) fluorescence microscopy after click chemistry with an azide-functionalized dye (azide-fluor 488, exposure time 2 s), and (e, f) HR-SEM imaging after click chemistry with azide-functionalized Au NPs. The fluorescence microscopy images include (a) a chip treated with 1,8-nonadiyne and azide-functionalized dye, (b) a chip treated additionally with a 10 s BHF dip after the 1,8-nonadiyne monolayer formation, (c) a control sample without 1,8-nonadiyne, and (d) the corresponding fluorescence intensity profiles averaged over the entire length of the nanowires. The HR-SEM images include (e) an InLens zoom-in image of a Si nanowire and (f) the corresponding ESB image to show a contrast in elements.
In order to allow future use of the nanowire chips for DNA detection, surface chemistry should allow specific DNA hybridization. Tests on PNA-DNA hybridization were first performed on planar Si substrates. A monolayer of 1,8-nonadiyne was formed on H-terminated Si (Scheme S1). Subsequently, two functionalization routes were used to couple PNA probes onto the freestanding alkyne moiety, that is, click chemistry with azido-PNA (Scheme S2a) and thiol–yne chemistry with thiol-PNA (Scheme S2b). In the latter reaction, potentially two thiol groups may bind to one alkyne headgroup.25
Click chemistry was performed as described above and resulted in a change of the contact angle from 78.3° ± 2.2 for a 1,8-nonadiyne monolayer to 50.1° ± 1.4 after azido-PNA coupling. This lowering in contact angle indicates azido-PNA coupling to the surface, as the increased hydrophilicity is expected from the polar structural groups.33 Thiol–yne chemistry was performed by exposing the 1,8-nonadiyne monolayer to a solution of thiol-PNA in phosphate-buffered saline (PBS) under illumination with a 365 nm light source. The thiol-PNA-functionalized surface changed the contact angle from 87.6° ± 1.1 after 1,8-nonadiyne to 46.5° ± 3.2 after thiol-PNA, again indicating a hydrophilic surface and thus proper functionalization. XPS measurements confirmed the coupling of PNA for both routes by the atomic percentages of N and S, which elements are absent in the 1,8-nonadiyne monolayer but increase to 16% N after click chemistry (each azido-PNA molecule contains 94 N atoms) and 0.26% S after thiol–yne chemistry (each thiol-PNA molecule contains 1 S atom). As a very rough estimation, the N/C and S/C ratios were used to calculate the degrees of functionalization, without taking into account the signal penetration depth. This resulted in a surface coupling of about 10% and 65% for the azido-PNA and thiol-PNA (assuming a maximum of 1 PNA molecule per alkyne headgroup), respectively. Thus, azido-PNA and thiol-PNA have been successfully coupled onto 1,8-nonadiyne monolayers.
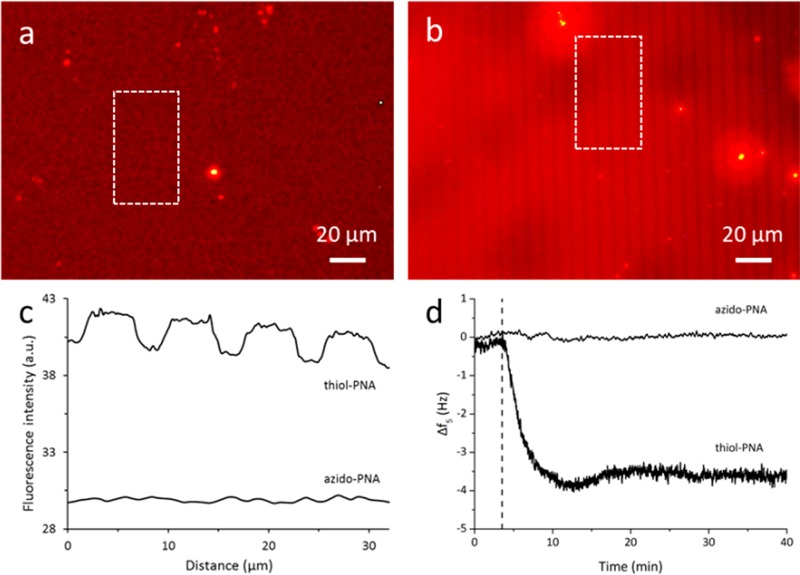
Hybridization tests were performed at micrometer-sized lines of azido-PNA and thiol-PNA to be able to visualize hybridization with dye-labeled cDNA by a contrast in the fluorescence signal. On a fully formed 1,8-nonadiyne monolayer, lines of PNA were created by microcontact printing (μCP). Azido-PNA was microcontact printed using Cu(I)(CH3CN)4PF6 and TBTA as stabilizing ligand,34 as opposed to the use of a Cu(II) salt with ascorbic acid for the click reaction described above. Seen the different procedure, XPS was used again to verify whether the azido-PNA coupling was successful. On a separate sample, an atomic percentage of 12% N was observed after μCP, which indicates a successful coupling. The yield of the click reaction is comparable to the Cu(II) reaction described above (16% N) when taking into account the maximum coverage of 2/3 due to the spacing of the μCP stamp (10 μm diameter, 5 μm spacing) and the use of a different azido-PNA sequence. For μCP of the thiol–yne reaction,24 the stamp with lines (5 μm diameter, 3 μm spacing) was inked with a thiol-PNA solution in PBS, equal to the samples that were fully immersed. As a difference, the substrate was illuminated through the stamp. After μCP, hybridization with a fluorescently labeled cDNA (dye-cDNA, rhodamine) did not result in the expected fluorescent pattern for the substrate with azido-PNA (Figure 3a,c). For the samples functionalized with thiol-PNA, however, fluorescent lines were observed after hybridization (Figure 3b,c).
Figure 3.
(a, b) Fluorescence microscopy images (exposure time 20 s) after hybridization with dye-cDNA on Si substrates with a 1,8-nonadiyne monolayer functionalized by μCP of (a) azido-PNA and (b) thiol-PNA, and (c) the corresponding fluorescence intensity profiles of the original images, as averaged over the dashed rectangles shown in panels (a) and (b); (d) QCM-D measurements on Si sensors with azido-PNA or thiol-PNA attached to a 1,8-nonadiyne monolayer, showing the fifth resonance frequency overtone (Δf5) when adding a 3 μM cDNA (azido-PNA) or 2 μM cDNA (thiol-PNA) solution in buffer; the vertical dashed line indicates the time at which the flow of cDNA was started.
The hybridization step was quantified further using QCM with dissipation monitoring (QCM-D), where a decrease in resonance frequency reflects an increase in mass at the surface. The frequency was monitored while flowing cDNA over PNA-functionalized QCM sensors (Figure 3d). These measurements supported the observations of the fluorescence microscopy images. No hybridization was observed for the azido-PNA surface, whereas the thiol-PNA-functionalized QCM sensor showed a decrease of the resonance frequency upon addition of a 2 μM cDNA solution. This reflects successful PNA-DNA hybridization for the thiol-PNA substrates. As a rough estimation, the Sauerbrey equation was used to convert the observed frequency change (3.4 Hz) into a mass change, giving an adsorbed mass of about 12 ng/cm2. In the best case, that is, assuming no water adsorption, this mass change corresponds to a cDNA coverage of about 10–12 mol/cm2, which is comparable to values reported before in the literature for PNA/DNA hybridization at surfaces.10,35 Considering the azido-PNA substrates, the reason for the absence of hybridization is unknown, as the presence of azido-PNA was confirmed by XPS. The low degree of azido-PNA coupling (estimated to be 10% as mentioned before) might partly explain the absence of (detectable) hybridization, although QCM-D should have been sensitive enough to detect even small amounts of hybridization. Two different azido-PNA sequences were tested, including a sequence similar to the thiol-PNA oligonucleotide, which was expected to be successful seen the positive μCP and QCM-D results. Backfilling of the 1,8-nonadiyne monolayer with azide-functionalized tetra(ethylene glycol) as antifouling layer did not improve the results either.
The successful hybridization on thiol-PNA samples described above was transferred onto Si nanowire chips as a proof of concept. After applying the same functionalization route to couple thiol-PNA onto a 1,8-nonadiyne monolayer, hybridization with dye-functionalized DNA was characterized using fluorescence microscopy. Immersion in a dye-cDNA solution resulted in a clear fluorescence signal (Figure 4a,d), the intensity profile of which is comparable to the signal observed in Figure 2a. When adding a dye-functionalized noncomplementary DNA (dye-ncDNA) onto the PNA monolayer, no fluorescence could be detected (Figure 4b,d), which indicates that the PNA-DNA interactions are specific. A control sample without 1,8-nonadiyne monolayer did not show fluorescence either after immersion in dye-cDNA (Figure 4c,d), which indicates that there is no physisorption of dye-cDNA in the absence of PNA. Consequently, the fluorescence observed in Figure 4a, in particular, in the SiOx areas, is likely due to the, here undesired, presence of a 1,8-nonadiyne monolayer with PNA at the surrounding SiOx. As described above, implementation of a BHF step may remove the coupling to the SiOx areas fully or partially (but was not further attempted here).
Figure 4.
Fluorescence microscopy images of Si nanowires on chips functionalized with a 1,8-nonadiyne monolayer and thiol-PNA, after adding (a) dye-cDNA, (b) dye-ncDNA, and (c) a control sample without 1,8-nonadiyne, immersed in dye-cDNA, and (d) the corresponding fluorescence intensity profiles of the main images, where the profiles of (b) and (c) are located at zero intensity. The exposure time is 50 ms for the main images and 2 s for the insets.
Summary and Conclusions
In summary, selective functionalization of Si nanowires on SiOx substrates appeared impossible in a direct way. Hydrosilylation of 1,8-nonadiyne led to a covalently bound monolayer at both the Si–H and the SiO2 regions, as shown by fluorescence microscopy and XPS after click chemistry with an azide-functionalized dye. An extra BHF dip after 1,8-nonadiyne monolayer formation was used to partly remove the monolayer from the oxidized regions. This seemed to result in successful local functionalization at the Si nanowires only, although the BHF treatment only resulted in a minor contrast between the Si and SiO2 regions for surfaces patterned at a larger scale. The reason for this apparent difference between the substrates is still unknown. Thus, selective functionalization of Si over SiOx seems to be possible when using an extra (B)HF treatment, but this step requires more optimization to increase the selectivity.
Monolayers of 1,8-nonadiyne functionalized with probe PNA were used to test the hybridization with cDNA at the surface. Azido-PNA and thiol-PNA were successfully coupled onto the 1,8-nonadiyne monolayer, as confirmed by contact angle and XPS measurements. For unknown reasons, no hybridization could be detected on the samples with azido-PNA. Nonetheless, successful hybridization of cDNA onto the substrates with thiol-PNA was confirmed by fluorescence microscopy and QCM-D measurements. On nanowire chips, hybridization was only observed when using cDNA and not for the noncomplementary sequence, which indicates specificity toward a disease-specific DNA sequence.
To increase the selectivity of the 1,8-nonadiyne monolayer formation on Si, the thermal hydrosilylation route could be replaced by another type of hydrosilylation. For example, the selectivity for functionalization of Si–H versus oxidized Si has shown to be higher for the photochemical version.32 Alternatively, the reaction could be performed in the dark, since the oxidized areas then keep a low contact angle (33°), whereas 1-alkynes could still react onto Si–H with relatively high yield.36 Furthermore, a one-step reaction could be performed with a mixture of silane-based and alkyne-based molecules, which preferably graft onto the oxidized and unoxidized regions, respectively.19
All in all, a proof of principle was shown for PNA/DNA hybridization after thiol-PNA coupling, which is required to further develop the Si nanowire sensor. Further research is needed to validate whether the tumor DNA can be detected at concentrations low enough for early diagnostics and in physiological solutions, that is, in the presence of a lot of other background DNA.
Experimental Section
Materials
Boron-doped p-type Si wafers (⟨100⟩-oriented, 100 mm diameter, single side polished, resistivity 5–10 Ω·cm, thickness 525 μm) were obtained from Okmetic (Finland). Chips with Si nanowires were fabricated as reported before18 and consisted of two Si nanowires with a triangular cross section, bridging two silicon-rich silicon nitride contact pads surrounded by SiOx. Chips without metal contacts were used, which did not allow for electrical characterization. Si-coated QCM sensors QSX-Si, consisting of Au electrodes with 200 nm sputtered, polycrystalline Si (resonance frequency of 5 MHz), were obtained from LOT-QuantumDesign GmbH.
Acetone (pure, VWR), acetonitrile (ACS grade, CH3CN, Merck), l-ascorbic acid (>99%, Sigma-Aldrich), azide-fluor 488 (>90%, Sigma-Aldrich), buffered hydrogen fluoride (VLSI, BHF, 7:1, Technic France), copper(II) sulfate pentahydrate (99.995% metals basis, CuSO4·5H2O, Sigma-Aldrich), dimethyl sulfoxide (anhydrous, >99.9%, DMSO, Sigma-Aldrich), ethanol (absolute, VWR), ethylenediaminetetraacetic acid disodium salt dihydrate (>99%, EDTA, Sigma-Aldrich), hydrofluoric acid 1% (aqueous, VLSI, Technic France), hydrogen peroxide (33%, H2O2, VWR), O-(2-azidoethyl)-O′-methyl-triethylene glycol (azido-TEG, >90%, Sigma-Aldrich), phosphate-buffered saline powder (pH 7.4, results in 10 mM PBS with 0.138 M NaCl, Sigma-Aldrich), photoresist OiR 906–12 or OiR 907–17 (Fujifilm), resist developer OPD 4262 (Fujifilm), sodium chloride (>99.5%, NaCl, Sigma-Aldrich), sodium citrate monobasic (>99%, Sigma-Aldrich), sodium dodecyl sulfate (SDS, >99%, Sigma-Aldrich), sulfuric acid (95%, H2SO4, VWR), tetrakis(acetonitrile)copper(I) hexafluorophosphate (Cu(I)(CH3CN)4PF6, Sigma-Aldrich), tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Sigma-Aldrich), and Tween-20 (Aldrich) were used as received. SSC buffer 20× consisted of 3 M sodium chloride and 0.3 M sodium citrate at pH 7.0 in water. Tris(benzyltriazolylmethyl)amine (TBTA) was synthesized according to a procedure from the literature.37 Hexane was obtained from a solvent purification system (MB SPS-800). Milli-Q water with a resistivity >18 MΩ·cm was obtained from a Milli-Q Integral water purification system (Merck Millipore). Glassware used for the hydrosilylation reaction was dried overnight at 120 °C. The dialkyne 1,8-nonadiyne (98%, Sigma-Aldrich) was dried over molecular sieves (0.3 nm). Dichloromethane (99.7%, Actu-All) was dried over anhydrous magnesium sulfate (Merck). Azide-functionalized Au NPs of 10 nm diameter were obtained from NanoCS, with a particle concentration of 0.5 mg/mL in water (based on Au salt, 2.8 × 1013 particles/mL), a size distribution <15%, and a poly(ethylene glycol) linker between the NPs and the azide groups.
The used (n)cDNA sequences were obtained from Eurofins Genomics and included 5′-GCG TGC CAA CGC GCT GCG CAT-3′ (100 μM in water) as cDNA for azido-PNA1 and 5′-AGC TGG TGG CGT AG-3′ (100 μM in water) as cDNA for azido-PNA2 and thiol-PNA. The latter cDNA was obtained both with and without fluorescent rhodamine at its 5′ end. As dye-ncDNA, the sequence 5′-CTA CGC CAC CAG CT-3′ was obtained with a rhodamine dye at the 5′ end.
PNA Synthesis
PNA commercial monomers, 2-[2-(Fmoc-amino)ethoxy]ethoxyacetic acid (Fmoc-AEEA or Fmoc-O) and 3-{2-[2-(2-{2-[3-(pyridin-2-yldisulfanyl)-propionylamino]-ethoxy}-ethoxy)-ethoxy]-ethoxy}-propionic acid (SPDP-PEG4) spacers were purchased from Link Technologies. All other chemicals and solvents were obtained from Sigma-Aldrich, Alfa Aesar, or Scharlab, and used without any further purification. Dimethylformamide (DMF) was dried over 0.4 nm molecular sieves and purged with nitrogen to avoid the presence of dimethylamine.
The PNA sequences were synthesized by solid phase methodologies based on Fmoc strategy, as reported earlier,38,39 by adding a coupling step with either 2-azidoacetic acid or SPDP-PEG4 (using HBTU/DIPEA coupling) as the final step before cleavage. The synthesis of the PNAs was performed manually in polypropylene reactors for Solid Phase Synthesis using a Chemmatrix Rink Amide resin preloaded with Fmoc-Glycine in 5 μmol scale, on a Syro I parallel peptide synthesizer. The protocol used for Fmoc-based chemistry contains the following modules: (a) deprotection with 20% piperidine in DMF (twice 8 min), (b) coupling with PNA monomer (5 equiv at 0.05 M), HBTU (5 equiv at 0.05 M)/DIPEA (10 equiv, 0.1 M) in dry DMF (2 min activation followed by 40 min each), and (c) capping with acetic anhydride/DIPEA in dry DMF, ratio 5:6:95 (twice, for 2 min).
Fmoc-AEEA spacers and azido acetic acid linker were introduced using HBTU/DIPEA coupling with the same conditions described above (5 equiv). The SPDP-PEG4 spacer was introduced under the same conditions using HBTU/DIPEA overnight coupling.
After the automatic synthesis, PNAs were cleaved from the resin using TFA/m-Cresol/TFMSA/thioanisole 6:2:1:1 solution and precipitated in ethyl ether. After removal of the ether layer, PNAs were dissolved in water and purified using reversed phase HPLC with a semipreparative column XTerra Prep RP18 (7.8 × 300 mm, 10 μm) with a gradient elution. Gradient: 100% A for 5 min, then from 0% to 50% B in 30 min at 4 mL/min flow (A: water + 0.1% trifluoroacetic acid; B: acetonitrile + 0.1% trifluoroacetic acid).
PNAs identity and purity were confirmed using UPLC-ESI system (Waters Acquity ultra performance LC HO6UPS-823M, with Waters SQ detector equipped with Waters UPLC BEH 300, 50 × 2.1 mm, 1.7 μm, C18) at 35 °C. A flow rate of 0.25 mL/min was used with the following solvent systems: (A) 0.2% FA in H2O and (B) 0.2% FA in MeCN (FA = formic acid). The column was flushed for 0.9 min with solvent A, then a gradient from 0 to 50% B in 6.6 min was used.
PNAs have been quantified using a UV–vis spectrophotometer (Lamba BIO 20 PERKIN ELMER) using as ε (260 nm) of the nucleobases the followings: adenine 13700, cytosine 6600, guanine 11700, and thymine 8600.
Azido-PNA1
X-O-O-GCA-GCG-CGT-TGG-CAC-Gly-NH2 (X = azidoacetyl, O = [2-(2-aminoethoxy)ethoxy]acetyl, 297 μM in water, probe for bladder cancer with an azide group at the N terminus (5′)): yield, 11%; Rt, 3.21 min (Figure S2). Calculated ε (260 nm): 147800 M–1 cm–1. ESI-MS (Figure S2): Calcd MW 4534.38; m/z Calcd (found): 1134.60 (1134.60) [MH4]4+, 907.88 (907.93) [MH5]5+, 756.73 (756.79) [MH6]6+, 648.77 (648.65) [MH7]7+.
Azido-PNA2
X-O-O-CTA CGC CAC CAG CT-Gly-NH2 (X = 2-azidoacetyl, O = [2-(2-aminoethoxy)ethoxy]acetyl, 272 μM in water, wild type probe for KRas colon cancer biomarker with an azide group at the N terminus (5′)): yield, 10%; Rt, 2.92 min (Figure S3). Calculated ε (260 nm): 127900 M–1 cm–1. ESI-MS (Figure S3): Calcd MW 4147.07; m/z Calcd (found): 1383.36 (1383.29) [MH3]3+, 1037.77 (1037.54) [MH4]4+, 830.41 (830.37) [MH5]5+, 692.18 (692.06) [MH6]6+, 593.44 (593.30) [MH7]7+.
(Protected) Thiol-PNA
SPDP-dPEG4-CTA CGC CAC CAG CT-Gly-NH2 (SPDP = 3-(2-pyridyldithio)propionyl, PEG = poly(ethylene glycol), 369 μM in water, wild type probe for KRas colon cancer biomarker with a thiol group at the N terminus (5′)): yield, 21%; Rt, 3.33 min (Figure S4). Calculated ε (260 nm): 127900 M–1 cm–1. ESI-MS (Figure S4): Calcd MW 4218.28; m/z Calcd (found): 1055.57 (1055.42) [MH4]4+, 855.66 (844.61) [MH5]5+, 704.05 (703.94) [MH6]6+, 603.61 (603.56) [MH7]7+, 528.29 (528.23) [MH8]8+, 469.70 (469.71) [MH9]9+. The thiol-PNA was deprotected from the PDP group by adding 1 mM TCEP in PBS.
Silicon Oxide Patterning
To make a pattern of SiO2 dots, first a 160 nm thick SiO2 layer was grown by wet oxidation on a cleaned Si p(100) wafer. A photoresist layer was spin coated on the front side (OiR 906–12, 6000 rpm, 30 s), baked at 95 °C for 90 s, patterned using standard photolithography (3 s UV exposure), immersed in resist developer (OPD 4262, 45 s), and baked at 120 °C for 10 min. This resulted in a hexagonal array of resist dots with both a diameter and spacing of 100 μm, which was used as a mask to etch away the surrounding SiO2 layer by 135 s immersion in an aqueous BHF solution. After resist removal by acetone rinsing, the resulting substrate contained SiO2 dots surrounded by H-terminated Si. Without extra 1% HF dip, a 1,8-nonadiyne monolayer was formed following the procedure described below.
Monolayer Formation of 1,8-Nonadiyne
To form a 1,8-nonadiyne monolayer on Si substrates by hydrosilylation (Scheme S1), the pure 1,8-nonadiyne solution was first degassed by four freeze–pump–thaw cycles. The Si substrates, that is, planar Si pieces or Si nanowires on chip, were cleaned by 5 min ultrasonication in acetone and for the chips an additional 25 min piranha cleaning (95% H2SO4 and 33% H2O2 mixed at 3:1 v/v). A hydrogen-terminated surface was created by 2 and 4 min exposure to an aqueous 1% HF solution to remove the native oxide, respectively. After rinsing in Milli-Q water and drying in a nitrogen stream, the substrates were immersed in the degassed 1,8-nonadiyne solution inside a nitrogen glovebox. A round-bottom reaction flask was equipped with a capillary as a nitrogen inlet and a reflux condenser. The hydrosilylation reaction was performed overnight under a low continuous nitrogen flow at 160 °C. Afterward the samples were cleaned by immersion in hexane, rinsing with dichloromethane, rinsing with ethanol, 5 min ultrasonication in dichloromethane to remove any physisorbed material, and subsequently dried in a stream of nitrogen.
Click Chemistry with Azide-Functionalized Dye, Au NPs, PNA, or TEG
Copper-catalyzed azide–alkyne cycloaddition (click chemistry, Schemes S1 and S2a) was used to couple the fluorescent dye azide-fluor 488, azide-functionalized Au NPs, azide-functionalized PNA1, or azide-functionalized TEG onto a 1,8-nonadiyne monolayer. The substrate was overnight incubated with 25 μL of the azide solution (2 mM azide-fluor 488 in water, azide-functionalized Au NPs as received, 297 μM azido-PNA1 in water, 2 mM azide-TEG in water) and 25 μL of the catalyst solution (2 mM Cu(II)SO4·5H2O, 80 mM l-ascorbic acid in water (for the azide-dye, azido-PNA1, and azide-TEG click chemistry) or in DMSO (for the azide-NPs)) in a silicone isolator (Electron Microscopy Sciences). A glass slide on top was used to avoid solvent evaporation. Afterward, the samples were sequentially rinsed with water, ethanol, immersed in acetone to remove the glue of the isolator, and sonicated in PBS with 0.05% v/v Tween-20 for 2 min (azido-PNA1) or 5 min (azide-dye, azide-Au NPs, and azide-TEG). After rinsing with a 0.05% w/v EDTA solution in water to remove any copper traces, the substrate was dried under nitrogen.
Thiol–yne Chemistry with Thiol-PNA
Thiol–yne chemistry (Scheme S2b) was used to couple thiol-PNA onto a 1,8-nonadiyne monolayer. The substrate was covered with a 10 μM solution of thiol-PNA in PBS. The reaction was performed for 1 h under illumination by a 365 nm light source (4 W) at a 0.5 cm distance. Subsequently, the sample was sonicated in PBS for 1 min, rinsed with water, and dried under nitrogen.
Microcontact Printing of Azido-PNA or Thiol-PNA
Poly(dimethylsiloxane) (PDMS) stamps were prepared by casting the precursor poly(dimethylsiloxane) and curing agent (Sylgard 184, Dow Corning) at 10:1 volume ratio onto a Si master. Air bubbles were removed by vacuum for 30 min, and the stamps were cured overnight at 60 °C. Before μCP, the cut stamps (10 μm lines and 5 μm spacing for azido-PNA2, and 5 μm lines and 3 μm spacing for thiol-PNA) were oxidized by oxygen plasma (power tuned at 40 mA) for 30 s. The stamp for click chemistry was inked with 75 μL of azido-PNA2 (272 μM in water) and 25 μL of catalyst solution (2 mM Cu(I)(CH3CN)4PF6 and 2 mM TBTA in CH3CN/ethanol, ratio 2:1 v/v) for 4 min. After drying in a stream of nitrogen, the stamp was brought into conformal contact with the substrate for 2 h. Subsequently, the printed substrate was rinsed with ethanol and water, and dried under nitrogen. For the thiol–yne reaction, the stamp was inked with 40 μL of thiol-PNA (25 μM in PBS) for 4 min. After drying the stamp under nitrogen, the stamp was brought into conformal contact with the substrate for 75 min under UV illumination (365 nm (4 W) at a 0.5 cm distance). Afterward, the substrate was rinsed with PBS and water, and dried in a stream of nitrogen.
PNA-DNA Hybridization
Hybridization with dye-(n)cDNA was performed by covering the PNA-monolayer-containing sample with a 2 μM solution of dye-(n)cDNA in buffer (5× SSC, optionally with 0.2% w/v SDS). The reaction was performed for 2 h at room temperature under aluminum foil. Afterward, the samples were sonicated in PBS with 0.05% v/v Tween-20 for 2 min, rinsed with water, and dried in a stream of nitrogen.
Contact Angle Measurements
Static contact angles were measured with Milli-Q water on a Krüss G10 Contact Angle Measuring Instrument equipped with a CCD camera. Contact angle values were determined automatically by a drop shape analysis software. Contact angles were measured directly after the reaction and averaged over at least three drops.
Fluorescence Microscopy
Fluorescence microscopy images were recorded in air on an Olympus inverted research microscope IX71 equipped with a mercury burner U-RFL-T as light source and a digital Olympus DP70 camera. To image the fluorescence of the azide-fluor 488 dye, blue excitation (490 ≤ λex ≤ 510 nm) and green emission (520 ≤ λem ≤ 550 nm) were filtered using a Chroma filter cube. For the rhodamine-labeled DNA sequences, green excitation (510 ≤ λex ≤ 550 nm) and red emission (λem ≥ 590 nm) were filtered using an Olympus filter cube. Intensity profiles were obtained by a rectangular average over a part of the surface.
X-ray Photoelectron Spectroscopy
XPS measurements were performed on a Quantera SXM setup from Physical Electronics equipped with an Al Kα X-ray source (1486.6 eV). A takeoff angle of 45° was used, and collected spectra were calibrated on the C 1s peak at 284.8 eV.
High-Resolution Scanning Electron Microscopy
HR-SEM images of nanowires on a chip were obtained with a Zeiss Merlin HR-SEM system with an InLens or ESB detector, operated at a typical acceleration voltages of 1.4 kV.
Quartz Crystal Microbalance with Dissipation Monitoring
QCM-D sensograms were recorded using a Q-Sense E4 module (Biolin Scientific) with two peristaltic pumps. Si-coated QCM sensors were cleaned by 5 min immersion in a piranha solution (95% H2SO4 and 33% H2O2 mixed at 3:1 v/v) and 5 min ultrasonication in acetone. To expose only the active sensor area to 1% HF, the remaining areas of the Si QCM sensors were first protected by photoresist. The active area at the top side of the sensor was covered with a small suction cup, after which photoresist OiR 907–17 was spin coated three times (1000 rpm, 30 s). After baking for 10 min at 120 °C, the entire back side was covered with photoresist using the same spin coating parameters. After a 3 min 1% HF dip, the resist was removed by acetone rinsing, and the substrates were immediately modified with a monolayer of 1,8-nonadiyne and azido-PNA1 or thiol-PNA, as described above. Afterward, QCM-D measurements were started by sequentially recording a baseline in Milli-Q water and buffer (PBS for azido-PNA1 and 5× SSC with 0.2% w/v SDS for thiol-PNA) until stable. Hybridization was tested with 3 μM cDNA (azido-PNA1) or 2 μM cDNA (thiol-PNA) solutions in the same buffer. The flow rate was set at 100 μL/min, and the temperature was kept at 22 °C. The sensograms were treated with a linear baseline correction to correct for a drift in the signal.
Acknowledgments
Songyue Chen and Jan van Nieuwkasteele are acknowledged for their help on the Si nanowire chips. Roberto Ricciardi is kindly thanked for his help with the QCM-D experiments and sharing his experience on the PNA/DNA hybridization. Carlo Nicosia is thanked for the synthesis of TBTA. J.V., W.V., and J.H. acknowledge The Netherlands Organization for Scientific Research (NWO) for financial support (MESA+ School for Nanotechnology, Grant 022.003.001 and FOM Project 13CO12-2). S.K., R.C., and J.H. acknowledge financial support from the European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 633937, Project ULTRAPLACAD.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.langmuir.8b02401.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Lee S.; Huang H.; Zelen M. Early detection of disease and scheduling of screening examinations. Stat. Methods Med. Res. 2004, 13 (6), 443–456. 10.1191/0962280204sm377ra. [DOI] [PubMed] [Google Scholar]
- Best M. G.; Sol N.; Kooi I.; Tannous J.; Westerman B. A.; Rustenburg F.; Schellen P.; Verschueren H.; Post E.; Koster J.; Ylstra B.; Ameziane N.; Dorsman J.; Smit E. F.; Verheul H. M.; Noske D. P.; Reijneveld J. C.; Nilsson R. J. A.; Tannous B. A.; Wesseling P.; Wurdinger T. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28 (5), 666–676. 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel J. H.; Ren H.; Sin M. L. Y.; Liao J. C.; Chae J. Rapid bladder cancer cell detection from clinical urine samples using an ultra-thin silicone membrane. Analyst 2016, 141 (2), 652–660. 10.1039/C5AN01616A. [DOI] [PubMed] [Google Scholar]
- Mir M.; Homs A.; Samitier J. Integrated electrochemical DNA biosensors for lab-on-a-chip devices. Electrophoresis 2009, 30 (19), 3386–3397. 10.1002/elps.200900319. [DOI] [PubMed] [Google Scholar]
- Gardeniers J. G. E.; van den Berg A. Lab-on-a-chip systems for biomedical and environmental monitoring. Anal. Bioanal. Chem. 2004, 378 (7), 1700–1703. 10.1007/s00216-003-2435-7. [DOI] [PubMed] [Google Scholar]
- Scheres L.; ter Maat J.; Giesbers M.; Zuilhof H. Microcontact Printing onto Oxide-Free Silicon via Highly Reactive Acid Fluoride-Functionalized Monolayers. Small 2010, 6 (5), 642–650. 10.1002/smll.200901650. [DOI] [PubMed] [Google Scholar]
- Calabretta A.; Wasserberg D.; Posthuma-Trumpie G. A.; Subramaniam V.; van Amerongen A.; Corradini R.; Tedeschi T.; Sforza S.; Reinhoudt D. N.; Marchelli R.; Huskens J.; Jonkheijm P. Patterning of Peptide Nucleic Acids Using Reactive Microcontact Printing. Langmuir 2011, 27 (4), 1536–1542. 10.1021/la102756k. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E.; Egholm M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999, 1 (1–2), 89–104. [PubMed] [Google Scholar]
- Schwarz F. P.; Robinson S.; Butler J. M. Thermodynamic comparison of PNA/DNA and DNA/DNA hybridization reactions at ambient temperature. Nucleic Acids Res. 1999, 27 (24), 4792–4800. 10.1093/nar/27.24.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.; Germini A.; Sforza S.; Corradini R.; Marchelli R.; Knoll W. Effect of ionic strength on PNA-DNA hybridization on surfaces and in solution. Biointerphases 2007, 2 (2), 80–88. 10.1116/1.2746871. [DOI] [PubMed] [Google Scholar]
- Cai W.; Peck J. R.; van der Weide D. W.; Hamers R. J. Direct electrical detection of hybridization at DNA-modified silicon surfaces. Biosens. Bioelectron. 2004, 19 (9), 1013–1019. 10.1016/j.bios.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Wei F.; Sun B.; Guo Y.; Zhao X. S. Monitoring DNA hybridization on alkyl modified silicon surface through capacitance measurement. Biosens. Bioelectron. 2003, 18 (9), 1157–1163. 10.1016/S0956-5663(03)00002-2. [DOI] [PubMed] [Google Scholar]
- Michaels P.; Alam M. T.; Ciampi S.; Rouesnel W.; Parker S. G.; Choudhury M. H.; Gooding J. J. A robust DNA interface on a silicon electrode. Chem. Commun. 2014, 50 (58), 7878–7880. 10.1039/C4CC03418J. [DOI] [PubMed] [Google Scholar]
- Bunimovich Y. L.; Shin Y. S.; Yeo W. S.; Amori M.; Kwong G.; Heath J. R. Quantitative real-time measurements of DNA hybridization with alkylated nonoxidized silicon nanowires in electrolyte solution. J. Am. Chem. Soc. 2006, 128 (50), 16323–16331. 10.1021/ja065923u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. J.; Chua J. H.; Chee R. E.; Agarwal A.; Wong S. M.; Buddharaju K. D.; Balasubramanian N. Highly sensitive measurements of PNA-DNA hybridization using oxide-etched silicon nanowire biosensors. Biosens. Bioelectron. 2008, 23 (11), 1701–1707. 10.1016/j.bios.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Zhang M.; Huang J.; Cui W.; Pang W.; Zhang H.; Zhang D.; Duan X. Kinetic studies of microfabricated biosensors using local adsorption strategy. Biosens. Bioelectron. 2015, 74, 8–15. 10.1016/j.bios.2015.05.061. [DOI] [PubMed] [Google Scholar]
- Lifson M. A.; Basu Roy D.; Miller B. L. Enhancing the Detection Limit of Nanoscale Biosensors via Topographically Selective Functionalization. Anal. Chem. 2014, 86 (2), 1016–1022. 10.1021/ac401523e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. Y.; Bomer J. G.; van der Wiel W. G.; Carlen E. T.; van den Berg A. Top-Down Fabrication of Sub-30 nm Monocrystalline Silicon Nanowires Using Conventional Microfabrication. ACS Nano 2009, 3 (11), 3485–3492. 10.1021/nn901220g. [DOI] [PubMed] [Google Scholar]
- Seitz O.; Fernandes P. G.; Mahmud G. A.; Wen H. C.; Stiegler H. J.; Chapman R. A.; Vogel E. M.; Chabal Y. J. One-Step Selective Chemistry for Silicon-on-Insulator Sensor Geometries. Langmuir 2011, 27 (12), 7337–7340. 10.1021/la200471b. [DOI] [PubMed] [Google Scholar]
- Masood M. N.; Chen S.; Carlen E. T.; van den Berg A. All-(111) Surface Silicon Nanowires: Selective Functionalization for Biosensing Applications. ACS Appl. Mater. Interfaces 2010, 2 (12), 3422–3428. 10.1021/am100922e. [DOI] [PubMed] [Google Scholar]
- Sun Q. Y.; de Smet L. C. P. M.; van Lagen B.; Giesbers M.; Thune P. C.; van Engelenburg J.; de Wolf F. A.; Zuilhof H.; Sudholter E. J. R. Covalently attached monolayers on crystalline hydrogen-terminated silicon: Extremely mild attachment by visible light. J. Am. Chem. Soc. 2005, 127 (8), 2514–2523. 10.1021/ja045359s. [DOI] [PubMed] [Google Scholar]
- Li Y.; Cai C. Z. Click Chemistry-Based Functionalization on Non-Oxidized Silicon Substrates. Chem. - Asian J. 2011, 6 (10), 2592–2605. 10.1002/asia.201100294. [DOI] [PubMed] [Google Scholar]
- Ciampi S.; Bocking T.; Kilian K. A.; James M.; Harper J. B.; Gooding J. J. Functionalization of acetylene-terminated monolayers on Si(100) surfaces: A click chemistry approach. Langmuir 2007, 23 (18), 9320–9329. 10.1021/la701035g. [DOI] [PubMed] [Google Scholar]
- Wendeln C.; Rinnen S.; Schulz C.; Arlinghaus H. F.; Ravoo B. J. Photochemical Microcontact Printing by Thiol-Ene and Thiol-Yne Click Chemistry. Langmuir 2010, 26 (20), 15966–15971. 10.1021/la102966j. [DOI] [PubMed] [Google Scholar]
- Bhairamadgi N. S.; Gangarapu S.; Caipa Campos M. A.; Paulusse J. M. J.; van Rijn C. J. M.; Zuilhof H. Efficient Functionalization of Oxide-Free Silicon(111) Surfaces: Thiol-yne versus Thiol-ene Click Chemistry. Langmuir 2013, 29 (14), 4535–4542. 10.1021/la400007y. [DOI] [PubMed] [Google Scholar]
- Devaraj N. K.; Miller G. P.; Ebina W.; Kakaradov B.; Collman J. P.; Kool E. T.; Chidsey C. E. D. Chemoselective covalent coupling of oligonucleotide probes to self-assembled monolayers. J. Am. Chem. Soc. 2005, 127 (24), 8600–8601. 10.1021/ja051462l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S. Y.; Chung W.-y.; Lee H. K.; Park M. S.; Park H. G. Direct and nondestructive verification of PNA immobilization using click chemistry. Biochem. Biophys. Res. Commun. 2008, 376 (4), 633–636. 10.1016/j.bbrc.2008.08.147. [DOI] [PubMed] [Google Scholar]
- Escorihuela J.; Banuls M. J.; Puchades R.; Maquieira A. Site-specific immobilization of DNA on silicon surfaces by using the thiol-yne reaction. J. Mater. Chem. B 2014, 2 (48), 8510–8517. 10.1039/C4TB01108B. [DOI] [PubMed] [Google Scholar]
- Meziane D.; Barras A.; Kromka A.; Houdkova J.; Boukherroub R.; Szunerits S. Thiol-yne Reaction on Boron-Doped Diamond Electrodes: Application for the Electrochemical Detection of DNA-DNA Hybridization Events. Anal. Chem. 2012, 84 (1), 194–200. 10.1021/ac202350c. [DOI] [PubMed] [Google Scholar]
- Veerbeek J.; Firet N. J.; Vijselaar W.; Elbersen R.; Gardeniers H.; Huskens J. Molecular Monolayers for Electrical Passivation and Functionalization of Silicon-Based Solar Energy Devices. ACS Appl. Mater. Interfaces 2017, 9 (1), 413–421. 10.1021/acsami.6b12997. [DOI] [PubMed] [Google Scholar]
- Danos L.; Greef R.; Markvart T. Efficient fluorescence quenching near crystalline silicon from Langmuir-Blodgett dye films. Thin Solid Films 2008, 516 (20), 7251–7255. 10.1016/j.tsf.2007.12.103. [DOI] [Google Scholar]
- Mischki T. K.; Donkers R. L.; Eves B. J.; Lopinski G. P.; Wayner D. D. M. Reaction of Alkenes with Hydrogen-Terminated and Photooxidized Silicon Surfaces. A Comparison of Thermal and Photochemical Processes. Langmuir 2006, 22 (20), 8359–8365. 10.1021/la060797t. [DOI] [PubMed] [Google Scholar]
- Cattani-Scholz A.; Pedone D.; Dubey M.; Neppl S.; Nickel B.; Feulner P.; Schwartz J.; Abstreiter G.; Tornow M. Organophosphonate-based PNA-functionalization of silicon nanowires for label-free DNA detection. ACS Nano 2008, 2 (8), 1653–1660. 10.1021/nn800136e. [DOI] [PubMed] [Google Scholar]
- Nicosia C.; Cabanas-Danés J.; Jonkheijm P.; Huskens J. A Fluorogenic Reactive Monolayer Platform for the Signaled Immobilization of Thiols. ChemBioChem 2012, 13 (6), 778–782. 10.1002/cbic.201200062. [DOI] [PubMed] [Google Scholar]
- Hvastkovs E. G.; Buttry D. A. Characterization of Mismatched DNA Hybridization via a Redox-Active Diviologen Bound in the PNA-DNA Minor Groove. Langmuir 2009, 25 (6), 3839–3844. 10.1021/la803284r. [DOI] [PubMed] [Google Scholar]
- Scheres L.; Arafat A.; Zuilhof H. Self-assembly of high-quality covalently bound organic monolayers onto silicon. Langmuir 2007, 23 (16), 8343–8346. 10.1021/la701359k. [DOI] [PubMed] [Google Scholar]
- Chan T. R.; Hilgraf R.; Sharpless K. B.; Fokin V. V. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org. Lett. 2004, 6 (17), 2853–2855. 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- Bertucci A.; Manicardi A.; Candiani A.; Giannetti S.; Cucinotta A.; Spoto G.; Konstantaki M.; Pissadakis S.; Selleri S.; Corradini R. Detection of unamplified genomic DNA by a PNA-based microstructured optical fiber (MOF) Bragg-grating optofluidic system. Biosens. Bioelectron. 2015, 63, 248–254. 10.1016/j.bios.2014.07.047. [DOI] [PubMed] [Google Scholar]
- Manicardi A.; Bertucci A.; Rozzi A.; Corradini R. A Bifunctional Monomer for On-Resin Synthesis of Polyfunctional PNAs and Tailored Induced-Fit Switching Probes. Org. Lett. 2016, 18 (21), 5452–5455. 10.1021/acs.orglett.6b02363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.