Abstract
Previous studies have shown that common variants of the gene coding for FK506-binding protein 51 (FKBP5), a critical regulator of glucocorticoid sensitivity, affect vulnerability to stress-related disorders. In a previous report, FKBP5 rs1360780 was identified as a functional variant because of its effect on gene methylation. Here we report evidence for a novel functional FKBP5 allele, rs3800373. This study assessed the association between rs3800373 and post-traumatic chronic pain in 1607 women and men from two ethnically diverse human cohorts. The molecular mechanism through which rs3800373 affects adverse outcomes was established via in silico, in vivo, and in vitro analyses. The rs3800373 minor allele predicted worse adverse outcomes after trauma exposure, such that individuals with the minor (risk) allele developed more severe post-traumatic chronic musculoskeletal pain. Among these individuals, peritraumatic circulating FKBP5 expression levels increased as cortisol and glucocorticoid receptor (NR3C1) mRNA levels increased, consistent with increased glucocorticoid resistance. Bioinformatic, in vitro, and mutational analyses indicate that the rs3800373 minor allele reduces the binding of a stress- and pain-associated microRNA, miR-320a, to FKBP5 via altering the FKBP5 mRNA 3′UTR secondary structure (i.e., is a riboSNitch). This results in relatively greater FKBP5 translation, unchecked by miR-320a. Overall, these results identify an important gene–miRNA interaction influencing chronic pain risk in vulnerable individuals and suggest that exogenous methods to achieve targeted reduction in poststress FKBP5 mRNA expression may constitute useful therapeutic strategies.
SIGNIFICANCE STATEMENT FKBP5 is a critical regulator of the stress response. Previous studies have shown that dysregulation of the expression of this gene plays a role in the pathogenesis of chronic pain development as well as a number of comorbid neuropsychiatric disorders. In the current study, we identified a functional allele (rs3800373) in the 3′UTR of FKBP5 that influences vulnerability to chronic post-traumatic pain in two ethnic cohorts. Using multiple complementary experimental approaches, we show that the FKBP5 rs3800373 minor allele alters the secondary structure of FKBP5 mRNA, decreasing the binding of a stress- and pain-associated microRNA, miR-320a. This results in relatively greater FKBP5 translation, unchecked by miR-320a, increasing glucocorticoid resistance and increasing vulnerability to post-traumatic pain.
Keywords: chronic pain, FKBP5, functional genetics, microRNA, post-traumatic stress, RNA structure
Introduction
Post-traumatic chronic musculoskeletal pain (MSP) is common, morbid, and a major public health problem (Carroll et al., 2008; McLean et al., 2014; Ulirsch et al., 2014). As with other stress-related disorders, the pathogenesis of chronic poststress MSP remains poorly understood. This lack of knowledge is a barrier to the development of improved preventive/treatment interventions.
Increasing evidence suggests that individual differences in the post-traumatic expression of FK506-binding protein 51 (FKBP5) may influence poststress MSP outcomes. FKBP5 is a critical protein within the hypothalamic–pituitary–adrenal (HPA) axis that regulates glucocorticoid receptor (GR) sensitivity (Denny et al., 2000; Zhang et al., 2008). Among European Americans (EAs) experiencing two common traumatic stress exposures, motor vehicle collision (MVC) and sexual assault, FKBP5 genetic variants, including rs3800373, have been found to predict poststress MSP severity (Bortsov et al., 2013). Consistent with these human data, recent animal data indicate that FKBP5 plays an important role in pain processing (Maiarù et al., 2016).
Although studies regarding FKBP5 variants and MSP outcomes are increasing, to date a far larger set of literature has evaluated the influence of FKBP5 variants on post-traumatic psychological sequelae (Zannas et al., 2016), which are commonly comorbid with post-traumatic MSP (Asmundson et al., 2002; Bair et al., 2003; Liu and Chen, 2014). In these studies, three FKBP5 single nucleotide polymorphisms (SNPs) in high linkage disequilibrium (rs3800373, rs9296158, and rs1360780) have been identified as most likely to be functional (Binder et al., 2004; Koenen et al., 2005; Roy et al., 2010). One of these SNPs, rs1360780, has been shown to be functional and to affect FKBP5 transcription via DNA methylation (Klengel et al., 2013).
Highly influential genes such as FKBP5 can have multiple functional alleles (Hormozdiari et al., 2017). Identifying additional functional variants in this lynchpin protein, if present, would be valuable to gain further insight into pathophysiologic mechanisms and inform potential preventive/treatment interventions. In the current study, we present evidence that rs3800373, in the 3′UTR of FKBP5, is a second FKBP5 functional allele affecting individual vulnerability to post-traumatic MSP and related stress disorders. We demonstrate a stress-dependent association between the rs3800373 allele and chronic MSP in two different ethnic cohorts and show that, among distressed trauma survivors with the rs3800373 risk allele, circulating FKBP5 mRNA levels are positively correlated with cortisol and glucocorticoid receptor (NR3C1) mRNA levels. In silico, in vitro, and in vivo human data suggest that the rs3800373 risk allele reduces the binding of a stress- and pain-associated microRNA, miR-320a, to FKBP5 via altering the RNA secondary structure. Together, these data identify a specific molecular mechanism by which a common genetic variant affects individual vulnerability to chronic MSP development after traumatic stress and novel molecular targets for potential future intervention.
Materials and Methods
Study design, setting, and eligibility criteria.
Two prospective longitudinal studies enrolling male and female European or African American (AA) individuals (18–65 years of age) presenting to the emergency department (ED) within 24 h of MVC were used to study chronic post-traumatic MSP outcomes. The details of these sister studies have been described previously (Platts-Mills et al., 2011; Linnstaedt et al., 2016c). The studies were approved by the institutional review boards at the data coordinating center (The University of North Carolina at Chapel Hill) and at all participating hospitals. Each study participant provided written informed consent before enrollment.
Eligible and consenting participants provided blood samples in the ED and completed an ED interview evaluation. Interview evaluations were performed by research assistants at the time of the ED visit using a web-based survey with explicit definitions of variables. Injury characteristics and medications administered in the ED were obtained by data extraction from the ED medical record. Six weeks after the MVC, participants completed a follow-up interview by telephone, online, or via mail. Participant demographic characteristics (including age, gender, income, height, weight, and educational attainment) were obtained from the ED medical record and from participant self-report.
Assessments and outcome definitions.
MVC-related overall MSP in the past week was assessed 6 weeks after MVC using the modified Regional Pain Scale (Wolfe, 2003). MSP intensity in each region and overall pain was evaluated via a 0 (no MSP) to 10 (maximum possible MSP) numeric rating scale (Farrar et al., 2001).
MVC-related post-traumatic stress (PTS) symptoms were assessed 6 months after MVC using the Impact of Event Scale: Revised (Weiss, 2007). This 22-item questionnaire includes avoidance, intrusion, and hyperarousal subscales. Scores range from 22 to 110.
Distress in the ED was measured using the Peritraumatic Distress Inventory, a 13-item questionnaire assessing the level of distress experienced immediately after a traumatic event (Brunet et al., 2001). Each item on the questionnaire was evaluated via a numeric rating scale from 0 (no distress) to 4 (high distress). A median cutoff score of 18 and 23 was used to identify those with substantial distress in the European American and African American populations, respectfully. [Previous studies have demonstrated increased peritraumatic distress in African Americans vs European Americans (Clark et al., 1999; Roberts et al., 2011)].
Life trauma before MVC was assessed using the Life Events Checklist for DSM-5 (LEC-5; Gray et al., 2004). The LEC-5 assesses exposure to 16 events known to potentially result in distress and includes one additional item addressing any other extraordinarily stressful event. A composite score for each participant was generated by tallying the total number of reported life events occurring to the individual.
Litigants were classified based on whether they said yes to the following question at the 6 week follow-up assessment: “Have you hired a lawyer, or are you working with a lawyer, because of injuries or other health problems that were related to your motor vehicle accident?”
DNA collection and genotyping.
European American samples were genotyped as described previously (Bortsov et al., 2013). For African American samples, study personnel collected blood samples at the time of enrollment using PAXgene DNA tubes. After DNA purification (PAXgene blood DNA kit, QIAGEN), genotyping using the Infinium Multi-Ethnic Global Array (Illumina) was performed. DNA from an individual with a known genotype (NA19819, 1000 genomes) and two repeat samples were included in each genotyping batch (96 samples) to ensure genotypic accuracy and reliability. SNP rs3800373 was in the Hardy–Weinberg equilibrium (p > 0.05) and had an excellent call rate (>99%).
Experimental design and statistical analysis.
Sociodemographic characteristics of the sample were summarized using standard descriptive statistics. The relationship between rs3800373 and chronic MSP outcomes in European Americans and African Americans after MVC were assessed using general linear models. In these models, β coefficients denote the magnitude of change in MSP severity occurring with each unit increase (positive β) or decrease (negative β) in the independent variable. A dominant genetic model (homozygous for the major allele vs heterozygous and homozygous for the minor allele) was used to match analyses leading to the main outcomes from our previous study (Bortsov et al., 2013). Based on the results of previous studies, participant age, sex, and study site were included in the model as covariates, and interactions between rs3800373 and both peritraumatic distress and participant sex were assessed by including corresponding product terms in the model (McLean et al., 2011, 2014; Bortsov et al., 2013; Linnstaedt et al., 2015a, 2016a,b). Marginal means corresponding to overall MSP levels were derived from the full general linear models. Associations between FKBP5 mRNA and cortisol levels, NR3C1 mRNA, or miR-320a were evaluated using Pearson correlation coefficients. All analyses were performed using R (R Project for Statistical Computing, RRID:SCR_001905) or SPSS software (SPSS, RRID:SCR_002865), and all graphs were made for publication using GraphPad Prism (GraphPad Software, RRID:SCR_002798).
RNA collection and sequencing.
For RNA sample collection, research assistants collected blood samples in the ED at the time of enrollment using PAXgene RNA tubes. Total RNA (including miRNA) was isolated using the PAXgene blood miRNA kit (QIAGEN) and stored at −80°C until use. RNA concentration and purity were measured using NanoDrop 1000 (Nanodrop Technologies).
For small RNA sequencing, template libraries for miRNA Next Generation Sequencing were produced from 1.0 μg of total RNA using two similar protocols. An initial set of 82 samples (randomly selected from 907 participant cohort) were prepped using an adaptation of published protocols as described previously (Pfeffer et al., 2005; Linnstaedt et al., 2015b). A second set of 96 samples (also randomly selected) were prepped using TruSeq Small RNA library prep kits according to the manufacturer's specifications (Illumina). Six samples were library prepped using both methods. Twelve barcoded libraries were combined per lane and sequenced on a HiSeq 2000 (Illumina). Raw sequence reads were processed using a custom bioinformatics pipeline as described previously (Linnstaedt et al., 2015b) and were normalized using upper-quartile normalization. To normalize potential technical biases between the two methods of library preparation, the sequencing reads were adjusted using the methodology described by Johnson et al. (2007). Of the 172 samples with miRNA sequencing data, 66 samples also had total RNA sequencing data and were used for analyses in this study.
For total RNA (excluding miRNA) sequencing, template libraries were produced from 600 ng of total RNA using the Ovation Human Blood RNA-Seq Library Systems kit (NuGen) according to the manufacturer's specifications. Libraries were multiplexed in groups of six and sequenced on a HiSeq 2000 at The University of North Carolina at Chapel Hill High Throughput Sequencing Facility. Raw sequencing reads were aligned to the human hg19 genome assembly using STAR (version 2.4.2a; Dobin et al., 2013). The expression level of each transcript was estimated via quantification of gene and isoform expression from RNA-Seq data, RSEM (Li and Dewey, 2011), using the University of California Santa Cruz known gene transcript and gene definitions. Raw RSEM read counts for all samples were normalized to the overall upper quartile (Bullard et al., 2010) before comparison and visualization.
Plasma collection and measurement of cortisol levels.
For plasma collection, research assistants collected blood samples in the ED at the time of enrollment using EDTA tubes. (Blood samples collected in the early morning between 5:00 A.M. and noon were not considered for analysis to avoid potential confounding by early-morning spikes in cortisol levels.) To separate plasma, EDTA blood tubes were centrifuged within 1 h of collection at 1500 × g for 15 min, and plasma was frozen at −20°C until use.
Cortisol was measured using the MILLIPLEX MAP Steroid/Thyroid Hormone Magnetic Bead Panel-Endocrine Multiplex Assay (STTHMAG-21K, Millipore). This assay was designed to measure cortisol, estradiol, and progesterone and have <10% cross-reactivity; samples were prepped according to the manufacturer's specifications and assayed on a Bio-Plex 200 system (Bio-Rad) using version 6.0 Bio-Plex Manager software (RRID:SCR_014330). Data were exported to Excel after manual outlier removal to achieve optimal five-parameter logistic fit probabilities of the standard curves.
Bioinformatics.
The miRdSNP online database (http://mirdsnp.ccr.buffalo.edu/) was used to assess whether rs3800373 is predicted to directly affect miRNA binding and to determine miRNAs predicted to bind 300 nucleotides upstream or downstream of rs3800373. The RNAsnp RNA folding algorithm (http://rth.dk/resources/rnasnp/) was used to determine whether the presence of the major versus minor allele of rs3800373 was likely to have an effect on the RNA secondary structure within the SNP-related binding region. To follow up on the RNAsnp results, the partition and Fold programs from the RNAstructure suite (Mathews, 2006; Reuter and Mathews, 2010) were used to find the base-pairing probabilities and the minimum free energy (MFE) structures, respectively, of the 3′UTR for the region containing selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) data. SHAPE reactivities were incorporated as a pseudo-free energy term in the nearest neighbor prediction model (Deigan et al., 2009). For both prediction algorithms, the maximum base-pairing distance was set at 200 nucleotides, and otherwise standard parameters were used for prediction and the SHAPE pseudo-free energy term.
DNA constructs.
Lentiviral constructs containing either a firefly luciferase gene (pL-SV40-GL3) or a renilla luciferase gene (pL-SV40-RLUC) were used for dual luciferase assays (Gottwein and Cullen, 2010). The 3′UTR of FKBP5 was amplified from HEK293T cells using primers: ATGATGCTCGAGGTGAACTCGGCCCCTCCTCAATGG and ATGATGGAATTCGCTGACTTGGCAATCCATATGCAG. The resulting 2125 bp product was cloned downstream of the firefly luciferase gene in pL-SV40-GL3 using XhoI and EcoRI restriction enzyme sites. This newly created construct, pL-GL3-FKBP5-maj, was sequenced to confirm the major allele, T, at position rs3800373 in the FKBP5 3′UTR and was then used as a template to mutate the major allele to the minor allele, G, using the QuickChange II Site-Directed Mutagenesis kit (Promega) and primers GCTTAATTTTTAAAAACAAAATCTGTAGGGTTGACAAATAAATAGTTGCTCTTCTAC and GTAGAAGAGCAACTATTTATTTGTCAACCCTACAGATTTTGTTTTTAAAAATTAAGC. This new construct, pL-GL3-FKBP5-min, was then sequenced to confirm the presence of the minor allele and the absence of additional introduced mutations.
A shortened version of the FKBP5 3′UTR, FKBP5-truncated, was amplified from the full-length 3′UTR using primers ATCATCCTCGAGCACAGUACUUCCUCCCAGC and ATCATCGAATTCGGGTTGCCAAATGCATTCC. This shortened version is 588 nucleotides and contains only the genomic region of the 3′UTR used to predict structural changes using RNAsnp. Mutations in three nucleotide positions that were predicted to disrupt complementarity between miR-320a and the FKBP5 3′UTR seed-binding region were engineered into FKBP5-truncated using primers CCAUGCCGGCUUUUUUGUCUUUUUAAGAUUAUUGGC and CAAAAAAGCCGGCATGGATCCTAAATGAC, to create a new construct termed “miR-320a mut.”
Additionally, mutations were made to the FKBP5-truncated construct to increase or decrease nucleotide accessibility (i.e., create double-stranded regions vs single-stranded regions) in the miR-320a-binding region. We first used the online algorithms mFold (Zuker, 2003) and RNAstructure (Reuter and Mathews, 2010) to monitor structural changes in response to nucleotide mutations in the region of the miR-320a-binding site (but that did not change the sequence of the exact binding region). To the 3′UTR with the major allele, we changed 31 nucleotides between sequence coordinates 35574855 and 35574832 on 6p21.31 from CATTTAGGATCCATGCAGCTTTT to CCAACAAATTTAGGATCCATGCAGCTTTTCAACAAACUAUGUAAAUAGAUCCACT. To the 3′UTR with the minor allele, we changed 20 nucleotides between sequence coordinates 35574818 and 35574793 on 6p21.31 from ATTATTGGCTCATAAGCATATGTATA to ACAAAAAAAGCTGCATGGATCCTAAATGA. These FKBP5 3′UTR sequences were synthesized and ligated into the EcoR1/Xho1 restriction sites in pL-SV40-GL3 (catalog #U7355DC190, GenScript) to generate pL-GL3-FKBP5-BasePaired and pL-GL3-FKBP5-Accessible. For the predicted structures resulting from the above mutations, see Figure 6A.
Figure 6.
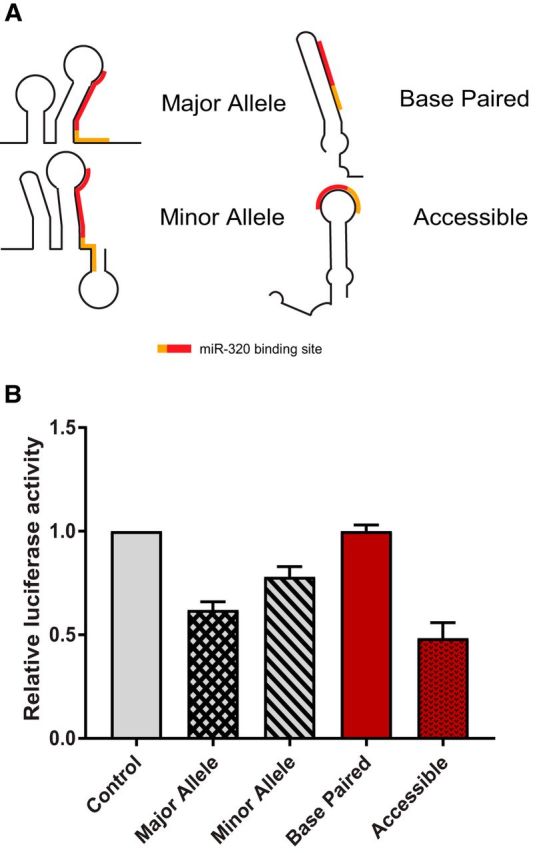
The RNA secondary structure in the FKBP5 3′UTR influences miR-320a-binding efficiency. A, Schematics of SHAPE informed RNA secondary structures for the FKBP5 3′UTR in the presence of the major allele (top left) or the minor allele (bottom left) and schematics of predicted RNA secondary structures for the FKBP5 3′UTR with nucleotide changes that either increase base pairing in the vicinity of the miR-320a-binding site (base paired, top right) or decrease nucleotide complementarity (accessible, bottom right). The miR-320a-binding site is indicated by a red line on each schematic, and the seed region of miR-320a is indicated by an orange line. B, In vitro dual luciferase assays assessing miR-320a binding to the FKBP5 3′UTR in the presence of each construct represented by images in A. Error bars (SD) are derived from three independent experiments performed in duplicate.
The genomic sequences for miR-99a, miR-122, and miR-320a were amplified from HEK293T cells using primers ATCATCGAATTCGCATCCTTAGAACTCAGC and ATCATCCTCGAGGTTGAGAATTGAAGCCTGC, ATCATCGAATTCGCTCAAGATGCTTGTACC and ATCATCCTCGAGGAGAGGCAGGGTTCAGC, and ATCATCGAATTCTCTCCGCGCCGGGC and ATCATCCTCGAGCAGCTACCCCGAAGCTCC, respectively. The resulting products, ∼400–500 bp in length containing the full pre-miRNA sequence, were cloned into a pcDNA backbone using EcoRI and XhoI restriction sites. These expression constructs increased expression of each miRNA >10-fold when transfected into HEK293T cells. The pLCE-s320a sponge construct and associated controls, pLCE and pLCE-sCXCR4, were generously donated from the Cullen laboratory at Duke University.
Cell culture.
HEK293T cells were grown at 37°C, 5% CO2, in DMEM with 10% fetal bovine serum (FBS) and 1% gentamicin. Human neuroblastoma SK-N-AS cells (catalog #CRL-2137, ATCC; RRID:CVCL_1700) were cultured in DMEM with 10% FBS, 0.1 mm nonessential amino acids, and 1% gentamicin. Other neuroblastoma cell lines used to test miR-320a and FKBP5 expression levels (BE-2C, SH-SY5Y, and IMR-32) were grown as recommended by ATCC guidelines. All four neuroblastoma cell lines expressed both RNA molecules. SK-N-AS cells were selected for this study based on similar expression levels of both miR-320a and FKBP5 mRNA.
Dual luciferase reporter assays.
The binding of miR-99a, miR-122, and miR-320a to the FKBP5 3′UTR was assessed using FLUC- and RLUC-based indicator vectors pL-SV40-GL3 and pL-SV40-RLUC. HEK293T cells (catalog #CRL-3216, ATCC; RRID:CVCL_0063) were cotransfected with 120 fmol of the miRNA expression construct, 5 fmol of pL-SV40-RLUC, and 30 fmol of either pL-GL3-FKBP5-maj or pL-GL3-FKBP5-min using Fugene 6 transfection reagent (Promega). Seventy-two hours after transfection, cells were collected and lysed. RLUC and FLUC levels were measured on a SpectraMax microplate reader (Molecular Devices) after adding substrates from dual luciferase reporter kits to the lysate (Promega); FLUC values were normalized to RLUC values, and relative luciferase activity was calculated between the compared samples. Statistically significant differences in miRNA binding between FKBP5 3′UTRs with either the major or the minor alleles was determined using Mann–Whitney U tests.
Generation of neuroblastoma cells stably knocked down for miR-320a expression.
To knock down miR-320a expression in SK-N-AS cells, we stably transduced the cells with pLCE-s320, a miR-320 sponge construct. Such “sponge” constructs have been described previously (Ebert et al., 2007). Viral transductants for miRNA sponges were produced in HEK293T cells using a third-generation lentiviral packaging system (Dull et al., 1998) consisting of pMD2-VSVG, pRSV-REV, pMDLgp, and either pLCE-s320 (s320), pLCE (control), or pLCE-sCXCR4 (control). Each sponge construct contained the GFP gene. HEK293T cells were transfected using Fugene 6 reagent (Promega). Forty-eight hours after transfection, viral medium was collected, filtered, and concentrated using Amicon Ultra centrifugal columns and added to the culture medium atop the SK-N-AS cells, and integration of the sponge sequence was monitored by GFP expression. Cells with the top 30% mean GFP fluorescence were sorted using FACS analysis (ARIA II) and grown in culture until sufficient numbers of cells were available for RNA isolation via TRIzol (Invitrogen) or for protein isolation via RIPA buffer (Pierce).
RT-qPCR and Western blotting.
For analysis of FKBP5 mRNA expression from cell-culture studies, RNA was first treated with DNaseI (New England Biolabs (NEB)) to remove genomic DNA. Random primers were used for reverse transcription (High Capacity Reverse Transcription kit, Invitrogen), and SYBR reagents were used for qPCR with transcript-specific primers (Invitrogen). Primer sequences CTCCCTAAAATTCCCTCGAATGC and CCCTCTCCTTTCCGTTTGGTT were used to detect FKBP5 mRNA, and primers GGAGCGAGATCCCTCCAAAAT and GGCTGTTGTCATACTTCTCATGG were used to detect GAPDH control RNA. Taqman reagents were used to detect miR-320a expression levels and control RNA (RNU48) in RNA isolated from human neuroblastoma cell lines (Invitrogen; assay ID: 478594_mir and assay ID: 001006).
For analysis of FKBP5 mRNA expression from human cohort RNA samples (that were not sequenced and that corresponded to those samples used for cortisol analysis), 40 ng of total RNA was reverse transcribed using random primers (High Capacity Reverse Transcription kit, Invitrogen). For qPCR, TaqMan reagents were used with intron-spanning transcript-specific primers (Invitrogen) for FKBP5 (human assay ID: Hs01561006_m1) and the TATA-box binding protein (human assay ID: Hs00427620_m1).
To compare FKBP5 protein levels, protein was isolated from SK-N-AS cells using RIPA buffer with added protease inhibitors (Thermo Fisher Scientific). The total protein concentration was measured with a BCA protein assay (Pierce), and equal amounts of protein (5 μg per lane) were loaded onto a 4–12% Bis-Tris gel (Invitrogen) and analyzed by Western blotting. Primary rabbit anti-FKBP5 (catalog #8245S; RRID:AB_10831198) and primary rabbit anti-β-actin (catalog #4970; RRID:AB_2223172) were obtained from Cell Signaling Technology. Secondary anti-rabbit IgG HRP was obtained from Cell Signaling Technology (catalog #7074; RRID:AB_2099233). Signals were developed using the ECL Western Blotting Substrate (Pierce), and signal intensities were quantified using ImageJ (RRID:SCR_003070).
SHAPE.
SHAPE data were obtained in vivo using the EBV immortalized lymphoblastoid cell line generated from Yoruban male 19098 (1000 Genomes–Coriell Institute for Medical Research), which is heterozygous for rs3800373 (Genomes Project Consortium et al., 2015). These cells were grown in RPMI 1640 with 2 mm l-glutamine and 15% FBS. Approximately 50 million cells were grown, spun down, and resuspended in 1 ml of folding buffer (100 mm Na-HEPES, pH 8.0, 100 mm NaCl, and 10 mm MgCl2) supplemented with 400 U of RNase inhibitor (NEB). Cells were kept on ice and sonicated at 10% power for three 10 s intervals (Model 500 Sonic Dismembrator, Thermo Fisher Scientific). The lysate was incubated at 37°C for 10 min. SHAPE treatment was performed over 5 min with the addition of DMSO or three separate additions of 1-methyl-7-nitroisatoic anhydride (1M7) with a final concentration of 30 mm 1M7 and 10% DMSO. The DMSO alone provides a background control. RNA was extracted from the lysate and depleted of ribosomal RNA (Thermo Fisher Scientific Trizol LS, 5 PRIME PhaseLock Heavy, Invitrogen Purelink RNA columns, and Thermo Fisher Scientific RiboMinus Eukaryote System version 2 from Invitrogen). SHAPE-MaP has been described previously (Siegfried et al., 2014; Smola et al., 2015). Briefly, reverse transcription was performed with SuperScript II (Thermo Fisher Scientific) under buffering conditions (Mn2+ buffer) to convert adducts into mutations (NEB random nonamers). Primers were designed to be specific to the 3′ region of the mRNA containing rs3800373: CCTACACGACGCTCTTCCGATCTNNNNNTCCTCCCAGCATTGCTACTG and GACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNTGCTCAATCTGTTTACCCGT, which was amplified by PCR (NEB Q5 HotStart Polymerase). Secondary PCR was performed to add TruSeq barcodes. An Illumina MiSeq Sequencing Platform was used to sequence the treated and background samples as a paired-end, 2 × 300 multiplex run.
Allele-specific sorting and SHAPE profile calculation.
Sequencing reads were aligned to the FKBP5 3′UTR using Bowtie2 version 2.2.9 (Langmead and Salzberg, 2012). The aligned reads were then split into separate FASTQ files by the allele at the rs3800373 SNP position from cell lines heterozygous at this allele. For each allele, the SHAPE profile of the amplified region was derived from the allele-specific sorted sequencing reads using the ShapeMapper pipeline version 1.2 (Siegfried et al., 2014). The SHAPE reactivity at each position corresponds to the difference between the mutation rate in the treatment condition and the mutation rate in the background control, normalizing the mutation rates by a multiplier so that the distribution of SHAPE reactivities matched that of reference mRNA.
SHAPE reactivities were used as a pseudo-free energy term in the nearest-neighbor thermodynamic model of RNA folding. This approach significantly improves the accuracy of structure prediction from positive predictive values of ∼0.5, on average, in the absence of experimental data to around 0.9 when SHAPE data are included (Deigan et al., 2009; Washietl et al., 2012; Hajdin et al., 2013; Sükösd et al., 2013; Siegfried et al., 2014). The base-pairing probabilities provide a representation of the structural ensemble of the 3′UTR and the average accessibility of each nucleotide. It is important to note that although SHAPE-directed modeling improves the prediction accuracy of MFE structures by >25%, the conformations remain models (Deigan et al., 2009).
Results
Participants
Baseline characteristics of participants in the AA MVC study (Linnstaedt et al., 2016c; n = 930) are shown in Table 1. Within the cohort, 763 participants provided DNA, were genotyped, and completed 6 week pain questionnaires. The miRNA sequencing was performed in another subset of 172 of 763 participants, total RNA sequencing was performed in a subset of 96 of 763 participants, and plasma cortisol assays were performed in a subset of 80 of 163 participants. Cortisol, miRNA, and total RNA data did not always overlap because plasma was obtained from only a subset of study participants and because RNA samples were selected for sequencing before planning these specific analyses. (RNA samples were selected for sequencing from individuals with relatively high or low levels of chronic MSP and RNA integrity scores of seven or greater.) Baseline characteristics of participants in the EA MVC study (Platts-Mills et al., 2011; n = 948) are also shown in Table 1.
Table 1.
Baseline characteristics of participants in the European American and African American MVC studies
| Characteristic | European American | African American |
|||
|---|---|---|---|---|---|
| Genotyping | Genotyping | mRNA | Micro RNA | Cortisol | |
| Enrolled, n | 948 | 930 | 96 | 172 | 80 |
| Age, years, mean (SD) | 35.6 (13.3) | 35.1 (12.7) | 30.5 (10.3) | 34.6 (11.8) | 32.3 (10.4) |
| Females, n (%) | 575 (60.7) | 578 (62.2) | 66 (68.8) | 107 (62.2) | 48 (60.0) |
| Education, n (%) | |||||
| 8–11 years | 42 (4.4) | 71 (7.6) | 2 (2.0) | 10 (5.8) | 11 (13.8) |
| High school | 184 (19.4) | 290 (31.2) | 31 (32.3) | 48 (27.9) | 29 (36.3) |
| Post-high school | 57 (6.0) | 42 (4.5) | 3 (3.1) | 5 (2.9) | 5 (6.3) |
| Some college | 312 (32.9) | 338 (36.3) | 42 (43.8) | 77 (44.8) | 25 (31.3) |
| College graduate | 238 (25.1) | 137 (14.7) | 15 (15.6) | 25 (14.5) | 7 (8.8) |
| Post-graduate studies | 113 (11.9) | 36 (3.9) | 3 (3.1) | 6 (3.5) | 3 (3.8) |
| Overall pain severity, 0–10 NRS, mean (SD) | |||||
| Month prior to MVC | 1.6 (2.6) | 2.2 (3.3) | 1.1 (2.4) | 1.6 (2.9) | 1.9 (3.2) |
| ED | 5.5 (2.4) | 7.2 (2.1) | 7.1 (2.2) | 7.2 (2.1) | 7.4 (1.8) |
| Week 6 | 3.8 (2.8) | 6.0 (2.9) | 4.8 (4.1) | 5.5 (3.4) | 6.2 (2.5) |
| Peritraumatic distress in the ED, mean (SD) | 19.2 (9.9) | 22.2 (11.6) | 24.3 (11.6) | 23.3 (11.5) | 23.5 (11.5) |
NRS, Numeric rating scale.
Relationship between FKBP5 allele rs3800373 and chronic MSP in EAs after MVC
In EA individuals, we previously identified a main effect of rs3800373 genotype [minor allele frequency (MAF), 29%] on chronic MSP severity 6 weeks after MVC (Bortsov et al., 2013). In subsequent studies, we and others have found that the influence of genetic variants on vulnerability to post-traumatic MSP are often sex and/or stress dependent (Linnstaedt et al., 2016a,b; Meloto et al., 2016). We therefore performed additional analyses of EA cohort data to assess for potential interactions between sex and/or participant peritraumatic distress and the influence of rs3800373 on 6 week post-MVC MSP severity. No interaction with sex was present (β = 0.052, p = 0.892), but the influence of rs3800373 on chronic MSP severity was stress dependent (β = 0.787, p = 0.035).
Relationship between FKBP5 allele rs3800373 and chronic MSP in AAs after MVC
To determine whether this stress-dependent effect of rs3800373 on post-MVC MSP vulnerability replicated across ethnicity, we repeated these analyses in our AA MVC cohort (MAF, 44%). Consistent with EA MVC cohort findings, no interaction with sex was present (β = 0.208, p = 0.661), but the influence of rs3800373 on chronic MSP severity was stress dependent (β = 1.36, p = 0.002). These associations persisted after adjusting for opioid administration in the ED and/or use of opioids at 6 weeks.
Influence of peritraumatic distress on MSP outcome is allele dependent
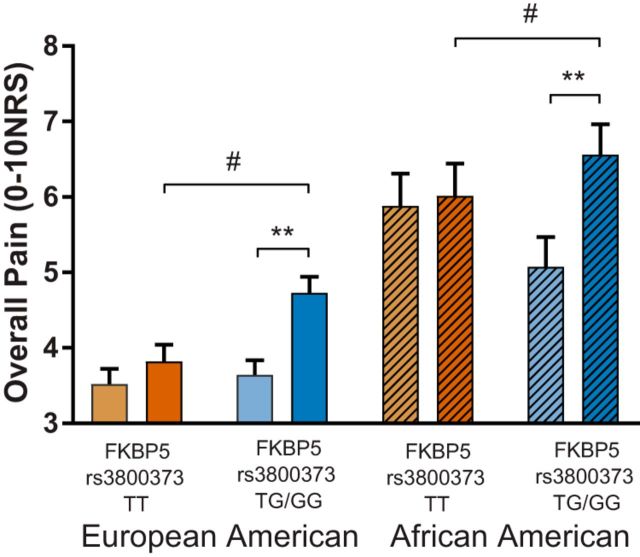
Among individuals with the rs3800373 major allele, peritraumatic distress severity had relatively little impact on chronic MSP development (Fig. 1). In contrast, in both the AA and EA cohorts, individuals with increased subjective distress in the immediate aftermath of MVC and one or more copies of the minor allele developed more severe chronic MSP 6 weeks after MVC than nondistressed individuals with one or more copies of the minor allele (EA: 4.76 vs 3.43, p < 0.001; AA: 6.70 vs 5.29, p < 0.001; Fig. 1). (The presence or absence of one or more copies of the rs3800373 minor allele did not directly influence peritraumatic distress in either cohort; data not shown.) In the EA cohort, distressed individuals with the rs3800373 minor allele also developed more severe chronic post-MVC MSP than distressed individuals with the major allele (4.76 vs 3.85, p = 0.001). This difference was also present at the trend level in the AA cohort (6.70 vs 6.11, p = 0.061; Fig. 1). In the AA cohort, increased peritraumatic distress also only predicted increased PTS symptoms at 6 weeks among individuals with the rs3800373 minor allele. Point estimates for the above findings did not significantly change when adjusting for previous life trauma (assessed in AA cohort only), opioid use, or litigation status (both cohorts).
Figure 1.
FKBP5 × stress interaction on chronic pain severity after motor vehicle collision in EA (plain bars) and AA (striped bars) individuals. Shown is the interaction between the FKBP5 SNP rs3800373 allele and ED peritraumatic distress (light bars, low levels of distress; dark bars, high levels of distress) on pain severity measured 6 weeks after MVC. Individuals with increased peritraumatic distress and at least one copy of the minor allele (TG/GG, blue shading) reported more severe pain relative to individuals with less peritraumatic distress (**p < 0.001) and relative to individuals with two copies of the major allele (TT, orange shading; #p < 0.05). Similar effects of FKBP5 × stress interactions were seen in both EAs (n = 844, left) and AAs (n = 763, right).
Allele-dependent relationship between circulating cortisol levels and FKBP5 mRNA expression
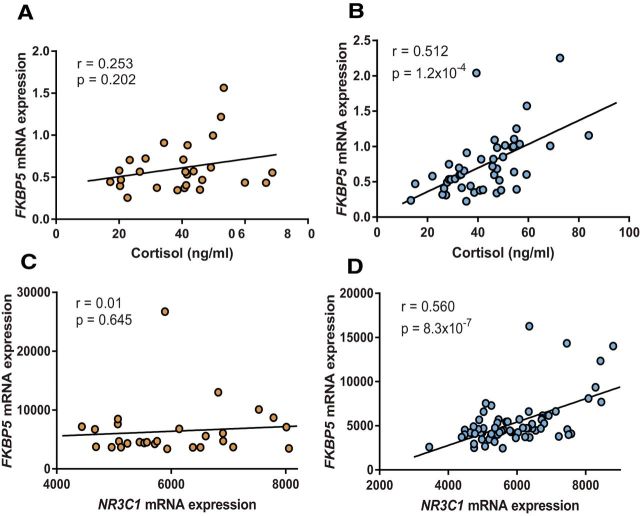
Cortisol induces the expression of FKBP5 (Vermeer et al., 2003), and FKBP5 reduces the nuclear transport of cortisol required for negative feedback inhibition, resulting in relatively increased cortisol levels (Tatro et al., 2009). Both animal and human data indicate that increased FKBP5 and cortisol levels influence the development of chronic pain (Khasar et al., 2008; Bortsov et al., 2013; Maiarù et al., 2016, 2018). To evaluate whether the FKBP5 rs1360780 allele affects negative feedback inhibition, we assessed the relationship between circulating cortisol and FKBP5 mRNA levels in the immediate aftermath of MVC among individuals with the major versus minor allele. Among AA MVC cohort participants with the major allele (n = 80), there was no correlation between circulating cortisol and FKBP5 mRNA expression (r = 0.253, p = 0.202; Fig. 2A). In contrast, among individuals with the rs3800373 minor allele, there was a strong positive correlation between cortisol levels and FKBP5 mRNA expression (r = 0.512, p = 1.2 × 10−4; Fig. 2B). (Of note, low allele-specific subgroup sizes did not permit us to further subgroup these relationships according to distress level.) Increasing levels of FKBP5 in response to increased cortisol levels among individuals with the minor allele may contribute to chronic pain development, both via direct effects (Maiarù et al., 2016, 2018) and/or via elevated cortisol levels, which can promote chronic pain via the sensitization of primary afferents (Khasar et al., 2008).
Figure 2.
Allele-dependent relationships between circulating FKBP5 mRNA expression levels and circulating cortisol levels or between FKBP5 mRNA and circulating NR3C1 mRNA expression levels in the early aftermath of MVC. A, Correlation between plasma cortisol levels and circulating FKBP5 mRNA expression levels in individuals with two copies of the rs3800373 protective/major allele (n = 27, r = 0.253, p = 0.202). B, Correlation between plasma cortisol levels and circulating FKBP5 mRNA expression levels in individuals with at least one copy of the risk/minor allele (n = 51, r = 0.512, p = 1.2 × 10−4). C, Correlation between circulating NR3C1 and FKBP5 mRNA expression levels in individuals with two copies of the rs3800373 protective allele (n = 28, r = 0.091, p = 0.645). D, Correlation between circulating NR3C1 and FKBP5 mRNA expression levels in individuals with at least one copy of the rs3800373 risk allele (n = 66, r = 0.560, p = 8.3 × 10−7). Pearson correlation coefficients (r) and p values were calculated using bivariate analyses.
Allele-dependent relationship between circulating FKBP5 mRNA and NR3C1 mRNA expression
Because translocation of cortisol into the nucleus is also known to downregulate GR expression (Silva et al., 1994), we also evaluated whether the FKBP5 rs1360780 allele affects the relationship between FKBP5 mRNA levels and GR receptor (NR3C1) mRNA levels. Among individuals with two copies of the major allele, there was no correlation between FKBP5 and NR3C1 mRNA levels (r = 0.091, p = 0.645; Fig. 2C). However, in individuals with one or more copies of the minor allele, FKBP5 and NR3C1 mRNA levels exhibited a strong positive correlation (r = 0.560, p = 8.3 × 10−7; Fig. 2D). As with the data presented above, these results are consistent with the expected molecular consequences of reduced HPA axis negative feedback inhibition.
rs3800373-dependent miR-320a-binding efficiency
The rs3800373 is located in the 3′UTR of FKBP5 (Fig. 3A). To evaluate potential molecular mechanisms through which the rs3800373 allele might influence post-MVC MSP development, we first used web-based bioinformatics algorithms, including two databases for general regulatory function, RegulomeDB and SNPinfo. In addition, we also used a database specific to miRNA binding, miRdSNP, because rs3800373 is located in the 3′UTR where microRNAs primarily bind (Xu and Taylor, 2009; Boyle et al., 2012; Bruno et al., 2012). The RegulomeDB algorithm, which identifies regions with DNase hypersensitivity, binding sites for transcription factors, and biochemically characterized promoter regions, found no regulatory elements associated with rs3800373. The SNPinfo database (using the Function Prediction module) indicated that rs3800373 did not affect RNA splicing, transcription factor-binding sites, or other queried regulatory elements. However, the miRdSNP online algorithm indicated that three miRNA, miR-320a, miR-99a, and miR-122, are predicted to bind FKBP5 mRNA within 300 nucleotides upstream or downstream of rs3800373 (Fig. 3A).
Figure 3.
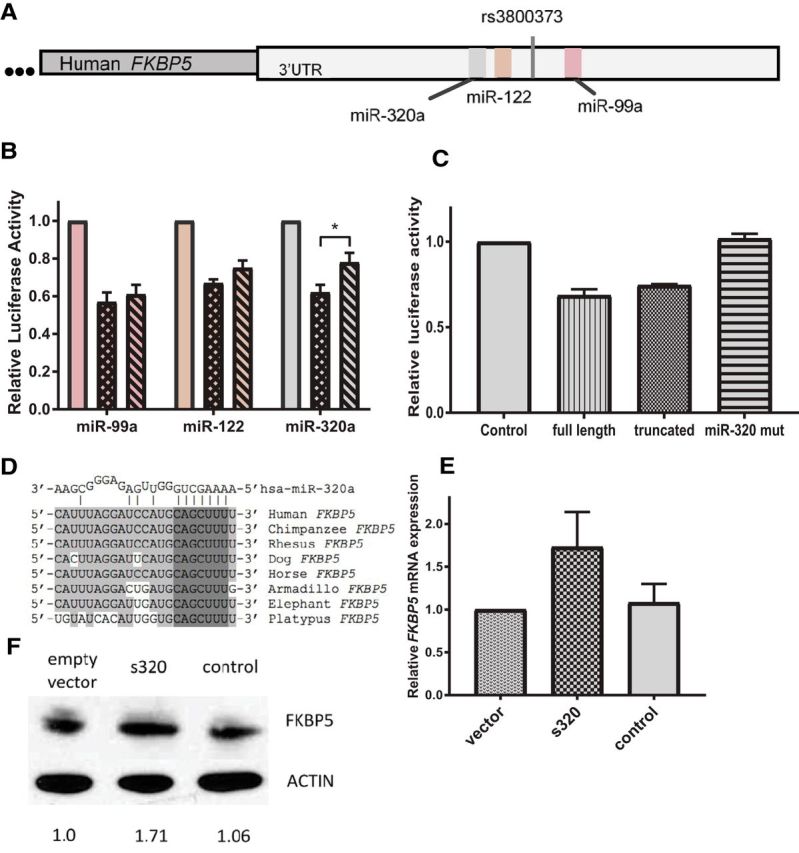
A, miR-320a binds and regulates the FKBP5 3′UTR and does so in an allele-dependent manner. The rs3800373 is located in the 3′UTR of FKBP5. The three closest predicted microRNA-binding sites are miR-320a, miR-122, and miR-99a, which are 134, 66, and 234 nucleotides from rs3800373, respectively. B, In vitro dual luciferase assays assessing miRNA binding to the FKBP5 3′UTR in the presence of the major allele (checkered bars), the minor allele (striped bars), and the control (plain bars). Error bars (SD) are derived from at least three independent experiments performed in duplicate, and the difference in binding between the major and minor allele constructs for miR-320a was determined using a Mann–Whitney U test (p = 0.025). C, miR-320a binding to the FKBP5 3′UTR in the presence of a 3′UTR construct containing 2125 nucleotides of the FKBP5 3′UTR (full length), 588 nucleotides of the FKBP5 3′UTR surrounding the risk allele (truncated), and mutation of the seed-binding region for miR-320a (miR-320 mut) that conserves the predicted RNA secondary structure of the 3′UTR. D, Nucleotide conservation among mammals at the predicted miR-320a-binding site in FKBP5. The miR-320a seed region is indicated by dark gray nucleotides, and conserved nucleotides outside of the seed region are indicated by light gray nucleotides. E, FKBP5 mRNA expression in the SK-N-AS neuroblastoma cell line knocked down for miR-320a (s320). mRNA expression levels are represented relative to two controls, a vector-only control (vector) and a nonrelated sponge sequence (control). Error bars (SD) are derived from two independently sorted cell lines for each condition measured in triplicate F, FKBP5 protein expression in the SK-N-AS neuroblastoma cell line for the two sponge controls (vector and control) and the miR-320a sponge (s320). Western blots were performed for each of the independently sorted cell lines. Protein expression quantitation was measured using ImageJ software, and the values represent the average band intensity calculated for each Western blot, adjusted for background signal intensity. *p < 0.05.
To assess miRNA binding to the 3′UTR of FKBP5, we created luciferase reporter constructs. In in vitro analyses, all three miRNA could bind to the FKBP5 3′UTR in the presence of either the major or minor allele (Fig. 3B). However, for miR-320a, there was a statistically significant decrease in binding efficiency in the presence of the minor allele (p = 0.025; Fig. 3B). Shortening the size of the 3′UTR surrounding rs3800373 had no effect on miR-320a-binding efficiency, but mutation of the predicted miR-320a seed-binding region abolished binding activity (Fig. 3C). This miR-320a seed-binding region is highly conserved among mammals (Fig. 3D). Together, these in silico and in vitro data indicate that the rs3800373 minor allele reduces the ability of miR-320a to bind FKBP5, resulting in increased FKBP5 levels. These results are consistent with previous reports indicating that increased expression of FKBP5 mRNA is associated with glucocorticoid resistance and adverse neuropsychiatric outcomes (Denny et al., 2000; Scammell et al., 2001; Binder et al., 2004; Segman et al., 2005; Klengel et al., 2013) and could at least partially account for the observed allele-dependent relationships between FKBP5 mRNA and NR3C1 mRNA and cortisol levels reported above.
Expression of miR-320a, FKBP5, and miR-320a regulation of FKBP5
To evaluate whether miR-320a regulates FKBP5 expression in an endogenous setting, we examined whether miR-320a regulates FKBP5 mRNA and protein levels in a neuroblastoma cell line. To do so, we knocked down miR-320a levels using lentiviral sponge constructs in SK-N-AS cells. Compared with cells expressing the control sponge, cells knocked down for miR-320a (s320) exhibited higher FKBP5 mRNA (Fig. 3E) and protein expression (Fig. 3F) levels.
rs3800373-dependent influence of miR-320a on chronic MSP development after MVC
In a previous pilot study using our AA MVC cohort (n = 69), we found that miR-320a expression in the hours after MVC predicts adverse MSP outcomes (Linnstaedt et al., 2016d). Using an expanded cohort (n = 159), we evaluated whether miR-320a predicts post-MVC MSP severity in a manner consistent with FKBP5 rs3800373 allele-dependent miR-320a regulation. If an allele-dependent regulation was occurring, then a statistical interaction between the FKBP5 rs3800373 allele and miR-320a would be expected. In general linear modeling, a significant interaction between the rs3800373allele and miR-320a was observed (t = −2.51, p = 0.013; Table 2). In stratified analyses, reduced circulating miR-320a expression in the hours after MVC predicted increased chronic MSP after MVC (standardized β = −0.233, p = 0.014) among individuals with one or more copies of the minor allele, but not among individuals who were homozygous for the FKBP5 rs3800373 major allele (standardized β = 0.185, p = 0.211). These results provide further support for the hypothesis that among individuals with the rs3800373 minor allele, there is less efficient miR-320a regulation of FKBP5 mRNA.
Table 2.
Linear regression model examining the relationship between candidate predictors and chronic pain severity 6 weeks after MVC in African American individuals (n = 159)
| Variablea | β | SE | t | p value |
|---|---|---|---|---|
| Age | 0.05 | 0.024 | 2.25 | 0.025 |
| Sex | −0.085 | 0.591 | −0.14 | 0.885 |
| Peritraumatic distressb | 0.06 | 0.024 | 2.58 | 0.011 |
| rs3800373c | 2.39 | 1.25 | 1.92 | 0.056 |
| miR-320a | 0.01 | 0.001 | 0.17 | 0.195 |
| rs3800373 * miR-320a | −0.01 | 0.001 | −2.51 | 0.013 |
aStudy site and education level were also included as a categorical variables.
bDistress was assessed in the emergency department using the Peritraumatic Distress Inventory (Brunet et al., 2001).
cA dominant genetic model was used for rs3800373.
Allele-dependent relationship between miR-320a and FKBP5 following MVC
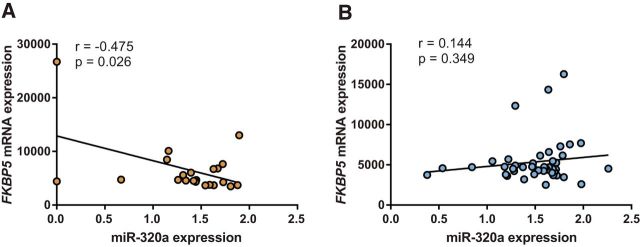
Consistent with in vitro data (Fig. 3A–E), RNA expression levels in blood obtained from AAs in the hours after MVC also support the hypothesis that miR-320a regulation of FKBP5 is rs3800373 allele dependent (Fig. 4). Among individuals with the FKBP5 rs3800373 major allele, miR-320a and FKBP5 mRNA levels were negatively correlated (r = −0.475, p = 0.026; Fig. 4A). In contrast, among those with one or more copies of the minor allele, no correlation was observed (r = 0.144, p = 0.349; Fig. 4B). These data suggest that miR-320a is only able to bind and reduce FKBP5 mRNA levels among individuals with the major allele.
Figure 4.
The relationship between FKBP5 mRNA levels and miR-320a levels in individuals after motor vehicle collision is dependent on the FKBP5 3′UTR allele, rs3800373. In the early aftermath of MVC, FKBP5 mRNA and miR-320a expression is negatively correlated in individuals with the protective allele (A; major allele, n = 22, r = −0.475, p = 0.026) but is not correlated in individuals with at least one copy of the risk allele (B; minor allele, n = 44, r = 0.144, p = 0.349). Pearson correlation coefficients (r) and p values were calculated using bivariate analyses.
Effect of rs3800373 on FKBP5 mRNA 3′UTR secondary structure
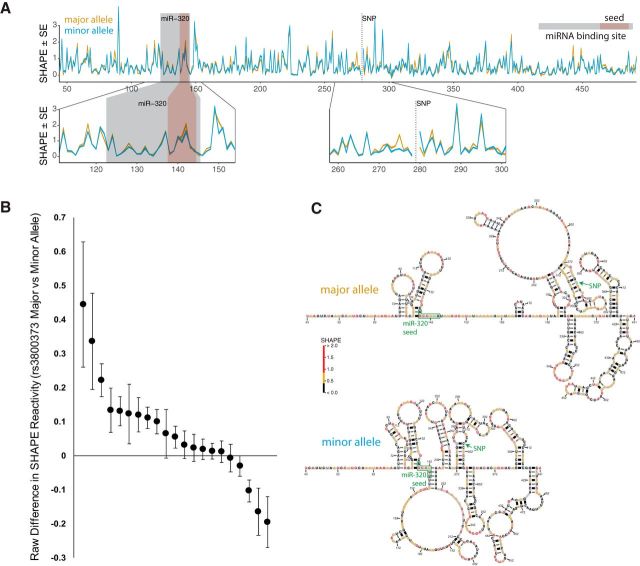
As shown in Figure 3A, rs3800373 and miR-320a are distally located in the FKBP5 3′UTR. Therefore, rs3800373 cannot directly affect miR-320a-binding efficiency. However, genetic variants in the 3′UTR can also affect miRNA binding by altering the RNA secondary structure (Arnold et al., 2012). We evaluated whether rs3800373 might affect the RNA folding/secondary structure using in silico analyses (RNAsnp). The algorithm predicted a statistically significant difference in RNA structure depending on the rs3800373 allele (p = 0.0136). We used SHAPE (Wilkinson et al., 2006; Siegfried et al., 2014) to experimentally compare RNA structural folding in RNA transcripts from each allele. SHAPE is an established method for chemically probing the RNA secondary structure in vivo, based on the selective chemical reactivity of the 2′-hydroxyl groups of flexible bases compared with bases involved in pair bonding or other interactions (Wilkinson et al., 2006; Siegfried et al., 2014). Yoruban cell line 19098 is heterozygous for rs3800373 and thus was selected for SHAPE analyses; lysis of these cells was performed using nondenaturing conditions that preserve the secondary structure of the RNA. Allele-specific preprocessing of sequencing reads was performed before SHAPE reactivity calculation to allow for controlled comparison of the allele-specific RNA structure (Siegfried et al., 2014). Results indicated allele-specific differences in RNA structure, both immediately surrounding rs3800373 and extending throughout the assayed region (Fig. 5A).
Figure 5.
The rs3800373 affects the RNA secondary structure of the FKBP5 3′UTR. A, Top, SHAPE data for a ∼500 nt region of the FKBP5 3′UTR encompassing rs3800373 and the miR-320-binding site. Gold, Major (protective) allele; blue, minor (risk) allele; gray box, miR-320a-binding site; red box, miR-320a seed site; dotted line, position of rs3800373. RNA SHAPE reactivities indicate widespread structural differences throughout the assayed FKBP5 3′UTR transcript based on the allele at rs3800373. Bottom, Magnified SHAPE reactivity plots for the miR-320a-binding site. SHAPE reactivity is slightly higher for the major allele in the miR-320a seed site (left), indicating reduced base pairing. The thickness of the lines indicates the SE on the measurements of reactivity. B, Raw difference in SHAPE reactivity data from A for the 21 individual nucleotides (black dots) in the miR-320a-binding region. These values were computed by comparing SHAPE reactivity data in the major allele structure versus SHAPE reactivity data in the minor allele structure. Higher SHAPE reactivity values indicate a greater degree of accessibility (i.e., less base pairing). Therefore, nucleotides with a more open structure in the context of the major allele (n = 16) are represented by positive values, and nucleotides with a more open structure in the context of the minor allele are represented by negative values (n = 5). Raw differences are shown along with confidence intervals based on four separate assessments of SHAPE reactivity in this genomic region. C, SHAPE-informed minimum free energy structures for the rs3800373 major (top) and minor (bottom) alleles, with SNP and miR-320a seed sites annotated. Nucleotide colors represent measured SHAPE reactivity as indicated by the color bar.
Because the mRNA structure can dramatically affect the accessibility of a 3′UTR to microRNA binding (Kertesz et al., 2007), we next evaluated whether the miR-320a-binding site in the FKBP5 3′UTR was affected by the presence of the rs3800373 allele. SHAPE reagents act by probing the relative flexibility of the RNA backbone, with higher reactivity generally indicating unpaired nucleotides (Kutchko and Laederach, 2017). Given that our SHAPE experiments relied on next-generation sequencing, we were able to accurately estimate the error on each nucleotide's reactivity as a function of sequencing depth (Smola et al., 2015). We therefore determined whether a nucleotide's reactivity was significantly different based on the allele at rs3800373. Of the 21 nucleotides in the miR-320-binding site, 16 nucleotides showed higher reactivity in the context of the major allele (10 of these nucleotides showed a statistically significant difference, p < 0.01; Fig. 5B). In comparison, only five nucleotides showed more reactivity in the context of the minor allele, with three of these showing a statistically significant difference (p < 0.01; Fig. 5B). Overall, this analysis of SHAPE reactivity data indicates that in the context of the rs3800373 major allele, the miR-320a-binding site is likely to be more accessible to miR-320a binding (i.e., less base paired and having a more open, single-stranded structure in the key binding region). This result is consistent with the in vitro experiments reported above showing less efficient binding of miR-320a to the FKBP5 3′UTR containing the minor/risk allele (Fig. 3B).
Using SHAPE-directed reactivities as a pseudo-free energy term, we next performed thermodynamic modeling to identify the MFE RNA structures of the FKBP5 3′UTR in the presence of the rs3800373 major or the minor allele (Fig. 5C). This modeling indicated that the MFE structure for the FKBP5 3′UTR with the major allele is less compact (i.e., less base paired) overall and in the region of the miR-320-binding site. In contrast, in the minor allele, MFE modeling indicated that the FKBP5 3′UTR was more compact and more sterically hindered by proximate base-paired stem loop structures (Fig. 5C). Overall, these modeling data suggest that rs3800373 alters base pairing in the miR-320a-binding site by shifting the RNA secondary structure in the context of the major allele to a more open/accessible conformation.
Together, the above results suggest that rs3800373 acts as a functional riboSNitch (Solem et al., 2015) by affecting the FKBP5 3′UTR RNA secondary structure and the resultant efficiency of miR-320a binding to the FKBP5 3′UTR. Furthermore, these results suggest that in individuals with the rs3800373 risk allele, the FKBP5 3′UTR is less accessible to repression by miR-320a, leading to increased expression of FKBP5 mRNA, elevated glucocorticoid levels, and reduced negative feedback inhibition.
Effect of mutations that alter the FKBP5 3′UTR secondary structure on miR-320a-binding efficiency
The above results indicate that rs3800373 likely affects FKBP5 mRNA expression via alterations to the 3′UTR secondary structure and, more specifically, via alterations to a miR-320a-binding site. Therefore, we next sought to better understand the effects of site accessibility (as determined by base-pairing interactions within the mRNA; Kertesz et al., 2007) on miR-320a binding in the context of the FKBP5 3′UTR. To do this, we made nucleotide changes in the FKBP5 3′UTR in the vicinity of the miR-320a-binding site that either force the RNA to base pair at the miR-320a-binding site (Fig. 6A, base paired) or force the RNA to be single stranded at the miR-320a binding-site (Fig. 6A, accessible).
Using in vitro luciferase reporter assays, we measured miR-320a binding to both RNA structural variants shown in Figure 6A. Consistent with our results showing decreased miR-320a binding in the context of the minor allele 3′UTR (that was shown via SHAPE analysis to exhibit increased base pairing in the miR-320a-binding site), we observed little to no binding when the RNA structure was fully base paired (Fig. 6B, base paired). In contrast, when the miR-320a-binding site was made more accessible through disruption of sequence complementarity, we observed increased miR-320a binding relative to either native major or minor allele structure (Fig. 6B, accessible). Together, these results indicate that miR-320a binding is sensitive to RNA structural changes in the FKBP5 3′UTR and support the hypothesis that rs3800373 reduces miR-320a binding to FKBP5 mRNA, resulting in relatively greater FKBP5 translation unchecked by miR-320a.
Discussion
The FKBP5 protein has an important influence on vulnerability to a number of post-traumatic neuropsychiatric disorders, including chronic pain (Binder, 2009; Bortsov et al., 2013; Maiarù et al., 2016, 2018; Zannas et al., 2016). In this study, we identified a novel microRNA-dependent mechanism by which a common genetic variant in the 3′UTR of FKBP5 affects chronic pain vulnerability after MVC. Building on previous work, we first identified an allele (rs3800373) in the 3′UTR of FKBP5 that influences vulnerability to chronic post-traumatic pain in two ethnic cohorts. Then, using complementary experimental approaches, including in silico, in vivo, and mutational analyses, we showed that the FKBP5 rs3800373 minor allele reduces the ability of microRNA miR-320a to bind FKBP5 and decreases its expression by altering the RNA secondary structure of the FKBP5 3′UTR. We show that, consistent with an inability of miR-320a binding to act as a counterbalancing regulatory mechanism restraining FKBP5 expression, among individuals with the minor allele, increasing cortisol or NR3C1 mRNA levels were associated with increasing FKBP5 mRNA levels.
The in silico analyses, SHAPE analyses and corresponding structural models, and mutational experiments described above support the hypothesis that the rs3800373 SNP alters the FKBP5 3′UTR secondary structure. Such a polymorphism has previously been termed a “riboSNitch” (Halvorsen et al., 2010; Ritz et al., 2012). However, this is the first study to demonstrate that a riboSNitch affects the efficiency of microRNA binding in the 3′UTR of a highly influential gene. Of the three microRNA identified to bind to FKBP5 in this study, only one microRNA, miR-320a, showed differential binding based on this riboSNitch. These data are consistent with previous evidence suggesting that miR-320a may be an important moderator of stress responses and the development of chronic post-traumatic pain. We previously found that peritraumatic miR-320a levels predict post-MVC pain outcomes in distressed individuals and that miR-320a binds multiple stress- and pain-associated transcripts. Circulating levels of miR-320a have also been associated with multiple other musculoskeletal pain conditions (Orlova et al., 2011; Bjersing et al., 2015; Devaux et al., 2015; Qureshi et al., 2015) and shown to be modulated by stress (Li et al., 2014). Importantly, previous studies have shown that miR-320a is expressed in nervous tissues that express FKBP5 and that are involved in chronic MSP pathogenesis (Landgraf et al., 2007; Madathil et al., 2011). Notably, it is possible that other miRNA known to bind FKBP5 and affect stress-related outcomes, such as miR-15 (Volk et al., 2016), might also be affected by alterations in the FKBP5 3′UTR RNA secondary structure induced by the rs3800373 minor allele.
The functional allele identified in this study, rs3800373, is now the second FKBP5 allele shown to influence vulnerability to post-traumatic sequelae. Klengel et al. (2013) previously demonstrated that rs1360780, in intron 2 of FKBP5, affects the methylation status of FKBP5 in intron 7. The rs1360780 and rs3800373 risk alleles are in high linkage disequilibrium (D′ = 0.89, r2 = 0.78), indicating that the effect of the risk alleles at each SNP generally occur in concert. The relative influence of each allele/mechanism on FKBP5 mRNA regulation and protein levels in different tissues, at different timepoints, and on different post-traumatic neuropsychiatric sequelae (e.g., pain, post-traumatic stress, depression, somatic symptoms) should be explored in future studies, as this may give greater insight into pathogenic mechanisms of post-traumatic neuropsychiatric sequelae and potential preventative interventions.
Interestingly, whereas the rs3800373 risk allele did not increase the risk of increased peritraumatic distress among the two human longitudinal cohorts assessed, a significant interaction was observed between peritraumatic distress and FKBP5 rs3800373 allele influence. Only individuals with substantial peritraumatic distress who also had the rs3800373 minor (risk) allele experienced worse post-traumatic pain outcomes. These data provide a molecular example of the common clinical observation that not all those with a risk factor (in this case, peritraumatic distress) develop the adverse outcome (chronic MSP). These data are consistent with the hypothesis that individuals with greater stress/distress experience greater stress-system activation and increased FKBP5 expression and that among such individuals, counterbalancing miR-320a downregulation of FKBP5 plays a particularly important role. This hypothesis should be evaluated in future studies with larger subsamples.
Available data suggest that FKBP5 levels may influence chronic pain outcomes via a number of mechanisms. In animal models, FKBP5 expression in the CNS has been shown to increase pain-related inflammatory mediators such as TNF-α, IL-1β, and IL-6 (Elenkov et al., 2000; Sternberg, 2006). Such mediators may promote allodynia and hyperalgesia by sensitizing peripheral and central afferents directly (Ferreira, 1993; Maier et al., 1993; Fukuoka et al., 1994) and/or by sensitizing CNS neurons via an afferent feedback mechanism (Ferreira, 1993; Maier et al., 1993; Fukuoka et al., 1994; Watkins et al., 1994, 1995a,b). In addition to influencing inflammatory mediators that elicit pain responses, increased FKBP5 levels enhance circulating glucocorticoid levels, which in concert with elevated catecholamine levels cause widespread hyperalgesia in animal models by sensitizing primary afferents (Khasar et al., 2008). FKBP5 may also be a critical factor in shifting glucocorticoid receptor activation after stress exposure from antinociceptive to pronociceptive (Maiarù et al., 2016).
Several limitations should be noted when interpreting the results of this study. First, to avoid confounding by population stratification, our studies only enrolled European American or African American individuals. The generalizability of our findings to other race/ethnic groups is unknown. However, because of population heterogeneity in allele frequencies and linkage disequilibrium patterns in the two cohorts differ (Morris, 2011; Marigorta and Navarro, 2013; Hormozdiari et al., 2014; Wen et al., 2015), the consistent association between rs3800373 and pain outcomes across both cohorts increases the likelihood that the allele is indeed functional. Second, we did not perform DNA methylation studies on samples from our two cohorts. Therefore, we were not able to assess the relative contribution of rs1360780 and rs3800373 on methylation and microRNA regulation of FKBP5 in these samples. (We hope to obtain funding to do this in the future.) Third, different cutoffs of peritraumatic distress were used in the European and African American populations since these cutoffs were based on median scores and the distribution of distress levels in the two cohorts varied substantially. However, while applying the median distress level cutoff used in European Americans to the African American cohort greatly reduced subsample sizes, it did not significantly influence effect size estimates observed in general linear modeling. Fourth, mRNA and miRNA expression levels reported in this study were measured in blood tissue (vs neuronal tissues that might be more relevant to chronic pain physiology). However, RNA expression patterns in the blood and CNS overlap significantly (Sullivan et al., 2006; Liu et al., 2010; Daskalakis et al., 2014; Hensman Moss et al., 2017), a number of previous studies have linked RNA expression in the blood (including expression of FKBP5) with neurological disease outcomes (Binder et al., 2004; Segman et al., 2005; Lai et al., 2011; Breen et al., 2018), and the stress response that appears to be involved in the pathogenesis of post-MVC MSP (McLean et al., 2005; McLean, 2011; Linnstaedt et al., 2016b) is primarily systemic and involves circulating stress and immune factors that communicate via the blood.
In conclusion, the findings of the present study indicate that, in addition to regulation via methylation levels, FKBP5 expression is regulated by miR-320a in a rs3800373 allele-dependent fashion. Identification of this additional functional SNP in FKBP5 increases understanding of genetic risk factors for post-traumatic MSP, uncovers an important gene–miRNA interaction influencing chronic pain risk, identifies the specific mechanism by which this genetic risk allele alters miRNA regulation, and suggests potential therapeutic approaches (e.g., exogenous methods to achieve targeted reduction in poststress FKBP5 mRNA expression via miRNA mimics or siRNA administration).
Footnotes
This work was supported by the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (Grant R01AR060852 to S.A.M.); The Mayday Fund (S.D.L. and S.A.M.); a Future Leaders in Pain grant from The American Pain Society (S.D.L.); and the National Human Genome Research Institute (Grant R01HG008133 to A.L.). We thank the participants for taking part in this study as well as The University of North Carolina BioSpecimen Facility for the storage, accessioning, and disbursement of biological samples.
The authors declare no competing financial interests.
References
- Arnold M, Ellwanger DC, Hartsperger ML, Pfeufer A, Stümpflen V (2012) Cis-acting polymorphisms affect complex traits through modifications of microRNA regulation pathways. PLoS One 7:e36694. 10.1371/journal.pone.0036694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmundson GJ, Coons MJ, Taylor S, Katz J (2002) PTSD and the experience of pain: research and clinical implications of shared vulnerability and mutual maintenance models. Can J Psychiatry 47:930–937. 10.1177/070674370204701004 [DOI] [PubMed] [Google Scholar]
- Bair MJ, Robinson RL, Katon W, Kroenke K (2003) Depression and pain comorbidity: a literature review. Arch Intern Med 163:2433–2445. 10.1001/archinte.163.20.2433 [DOI] [PubMed] [Google Scholar]
- Binder EB. (2009) The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 34:S186–S195. 10.1016/j.psyneuen.2009.05.021 [DOI] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Pütz B, Papiol S, Seaman S, Lucae S, Kohli MA, Nickel T, Künzel HE, Fuchs B, Majer M, Pfennig A, Kern N, Brunner J, Modell S, Baghai T, Deiml T, et al. (2004) Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36:1319–1325. 10.1038/ng1479 [DOI] [PubMed] [Google Scholar]
- Bjersing JL, Bokarewa MI, Mannerkorpi K (2015) Profile of circulating microRNAs in fibromyalgia and their relation to symptom severity: an exploratory study. Rheumat Int 35:635–642. 10.1007/s00296-014-3139-3 [DOI] [PubMed] [Google Scholar]
- Bortsov AV, Smith JE, Diatchenko L, Soward AC, Ulirsch JC, Rossi C, Swor RA, Hauda WE, Peak DA, Jones JS (2013) Polymorphisms in the glucocorticoid receptor co-chaperone FKBP5 predict persistent musculoskeletal pain after traumatic stress exposure. Pain 154:1419–1426. 10.1016/j.pain.2013.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, Cherry JM, Snyder M (2012) Annotation of functional variation in personal genomes using RegulomeDB. Genome Res 22:1790–1797. 10.1101/gr.137323.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Tylee DS, Maihofer AX, Neylan TC, Mehta D, Binder EB, Chandler SD, Hess JL, Kremen WS, Risbrough VB, Woelk CH, Baker DG, Nievergelt CM, Tsuang MT, Buxbaum JD, Glatt SJ (2018) PTSD blood transcriptome mega-analysis: shared inflammatory pathways across biological sex and modes of trauma. Neuropsychopharmacology 43:469–481. 10.1038/npp.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Weiss DS, Metzler TJ, Best SR, Neylan TC, Rogers C, Fagan J, Marmar CR (2001) The peritraumatic distress inventory: a proposed measure of PTSD criterion A2. Am J Psychiatry 158:1480–1485. 10.1176/appi.ajp.158.9.1480 [DOI] [PubMed] [Google Scholar]
- Bruno AE, Li L, Kalabus JL, Pan Y, Yu A, Hu Z (2012) miRdSNP: a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes. BMC Genomics 13:44. 10.1186/1471-2164-13-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S (2010) Evaluation of statistical methods for normalization and differential expression in mRNA-seq experiments. BMC Bioinformatics 11:94 10.1186/1471-2105-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll LJ, Holm LW, Hogg-Johnson S, Côté P, Cassidy JD, Haldeman S, Nordin M, Hurwitz EL, Carragee EJ, van der Velde G, Peloso PM, Guzman J (2008) Course and prognostic factors for neck pain in whiplash-associated disorders (WAD): results of the bone and joint decade 2000–2010 task force on neck pain and its associated disorders. Spine 33:S83–92. 10.1097/BRS.0b013e3181643eb8 [DOI] [PubMed] [Google Scholar]
- Clark R, Anderson NB, Clark VR, Williams DR (1999) Racism as a stressor for African Americans: a biopsychosocial model. Am Psychologist 54:805 10.1037/0003-066X.54.10.805 [DOI] [PubMed] [Google Scholar]
- Daskalakis NP, Cohen H, Cai G, Buxbaum JD, Yehuda R (2014) Expression profiling associates blood and brain glucocorticoid receptor signaling with trauma-related individual differences in both sexes. Proc Natl Acad Sci U S A 111:13529–13534. 10.1073/pnas.1401660111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM (2009) Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci U S A 106:97–102. 10.1073/pnas.0806929106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG (2000) Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology 141:4107–4113. 10.1210/endo.141.11.7785 [DOI] [PubMed] [Google Scholar]
- Devaux Y, Mueller M, Haaf P, Goretti E, Twerenbold R, Zangrando J, Vausort M, Reichlin T, Wildi K, Moehring B, Wagner DR, Mueller C (2015) Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med 277:260–271. 10.1111/joim.12183 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L (1998) A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA (2007) MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 4:721–726. 10.1038/nmeth1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elenkov IJ, Wilder RL, Chrousos GP, Vizi ES (2000) The sympathetic nerve–an integrative interface between two supersystems: the brain and the immune system. Pharmacol Rev 52:595–638. [PubMed] [Google Scholar]
- Farrar JT, Young JP Jr, LaMoreaux L, Werth JL, Poole RM (2001) Clinical importance of changes in chronic pain intensity measured on an 11-point numerical pain rating scale. Pain 94:149–158. 10.1016/S0304-3959(01)00349-9 [DOI] [PubMed] [Google Scholar]
- Ferreira SH. (1993) The role of interleukins and nitric oxide in the mediation of inflammatory pain and its control by peripheral analgesics. Drugs 46 [Suppl. 1]:1–9. [DOI] [PubMed] [Google Scholar]
- Fukuoka H, Kawatani M, Hisamitsu T, Takeshige C (1994) Cutaneous hyperalgesia induced by peripheral injection of interleukin-1 beta in the rat. Brain Res 657:133–140. 10.1016/0006-8993(94)90960-1 [DOI] [PubMed] [Google Scholar]
- Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR (2015) A global reference for human genetic variation. Nature 526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Cullen BR (2010) A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest. J Virol 84:5229–5237. 10.1128/JVI.00202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Litz BT, Hsu JL, Lombardo TW (2004) Psychometric properties of the life events checklist. Assessment 11:330–341. 10.1177/1073191104269954 [DOI] [PubMed] [Google Scholar]
- Hajdin CE, Bellaousov S, Huggins W, Leonard CW, Mathews DH, Weeks KM (2013) Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci U S A 110:5498–5503. 10.1073/pnas.1219988110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A (2010) Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6:e1001074. 10.1371/journal.pgen.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensman Moss DJ, Flower MD, Lo KK, Miller JR, van Ommen GB, 't Hoen PA, Stone TC, Guinee A, Langbehn DR, Jones L, Plagnol V, van Roon-Mom WM, Holmans P, Tabrizi SJ (2017) Huntington's disease blood and brain show a common gene expression pattern and share an immune signature with Alzheimer's disease. Sci Rep 7:44849. 10.1038/srep44849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E (2014) Identifying causal variants at loci with multiple signals of association. Genetics 198:497–508. 10.1534/genetics.114.167908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormozdiari F, Zhu A, Kichaev G, Ju CJ, Segrè AV, Joo JWJ, Won H, Sankararaman S, Pasaniuc B, Shifman S, Eskin E (2017) Widespread allelic heterogeneity in complex traits. Am J Hum Genet 100:789–802. 10.1016/j.ajhg.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A (2007) Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8:118–127. 10.1093/biostatistics/kxj037 [DOI] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284. 10.1038/ng2135 [DOI] [PubMed] [Google Scholar]
- Khasar SG, Burkham J, Dina OA, Brown AS, Bogen O, Alessandri-Haber N, Green PG, Reichling DB, Levine JD (2008) Stress induces a switch of intracellular signaling in sensory neurons in a model of generalized pain. J Neurosci 28:5721–5730. 10.1523/JNEUROSCI.0256-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM, Pace TW, Mercer KB, Mayberg HS, Bradley B, Nemeroff CB, Holsboer F, Heim CM, Ressler KJ, Rein T, Binder EB (2013) Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 16:33–41. 10.1038/nn.3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen KC, Saxe G, Purcell S, Smoller JW, Bartholomew D, Miller A, Hall E, Kaplow J, Bosquet M, Moulton S, Baldwin C (2005) Polymorphisms in FKBP5 are associated with peritraumatic dissociation in medically injured children. Mol Psychiatry 10:1058–1059. 10.1038/sj.mp.4001727 [DOI] [PubMed] [Google Scholar]
- Kutchko KM, Laederach A (2017) Transcending the prediction paradigm: novel applications of SHAPE to RNA function and evolution. Wiley Interdisciplinary Rev RNA 8:e1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CY, Yu SL, Hsieh MH, Chen CH, Chen HY, Wen CC, Huang YH, Hsiao PC, Hsiao CK, Liu CM (2011) MicroRNA expression aberration as potential peripheral blood biomarkers for schizophrenia. PloS One 6:e21635. 10.1371/journal.pone.0021635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, et al. (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–1414. 10.1016/j.cell.2007.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12:323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Li H, Li T, Fan J, Zhao RC, Weng X (2014) MicroRNA expression profile of dexamethasone-induced human bone marrow-derived mesenchymal stem cells during osteogenic differentiation. J Cell Biochem 115:1683–1691. 10.1002/jcb.24831 [DOI] [PubMed] [Google Scholar]
- Linnstaedt SD, Hu J, Bortsov AV, Soward AC, Swor R, Jones J, Lee D, Peak D, Domeier R, Rathlev N, Hendry P, McLean SA (2015a) Mu-opioid receptor gene A118 G variants and persistent pain symptoms among men and women experiencing motor vehicle collision. J Pain 16:637–644. 10.1016/j.jpain.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Walker MG, Parker JS, Yeh E, Sons RL, Zimny E, Lewandowski C, Hendry PL, Damiron K, Pearson C, Velilla MA, O'Neil BJ, Jones J, Swor R, Domeier R, Hammond S, McLean SA (2015b) MicroRNA circulating in the early aftermath of motor vehicle collision predict persistent pain development and suggest a role for microRNA in sex-specific pain differences. Mol Pain 11:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Bortsov AV, Soward AC, Swor R, Peak DA, Jones J, Rathlev N, Lee DC, Domeier R, Hendry PL, McLean SA (2016a) CRHBP polymorphisms predict chronic pain development following motor vehicle collision. Pain 157:273–279. 10.1097/j.pain.0000000000000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Walker MG, Riker KD, Nyland JE, Hu J, Rossi C, Swor RA, Jones JS, Diatchenko L, Bortsov AV, Peak DA, McLean SA (2016b) Genetic variant rs3750625 in the 3′UTR of ADRA2A affects stress-dependent acute pain severity after trauma and alters a microRNA-34a regulatory site. Pain 158:230–239. 10.1097/j.pain.0000000000000742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Hu J, Liu AY, Soward AC, Bollen KA, Wang HE, Hendry PL, Zimny E, Lewandowski C, Velilla MA, Damiron K, Pearson C, Domeier R, Kaushik S, Feldman J, Rosenberg M, Jones J, Swor R, Rathlev N, McLean SA (2016c) A methodology of AA CRASH: a prospective observational study evaluating the incidence and pathogenesis of adverse post-traumatic sequelae in African-Americans experiencing motor vehicle collision. BMJ Open 6:e012222. 10.1136/bmjopen-2016-012222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnstaedt SD, Riker KD, Walker MG, Nyland JE, Zimny E, Lewandowski C, Hendry PL, Damiron K, Pearson C, Velilla MA, Jones J, Swor RA, Domeier R, McLean SA (2016d) MicroRNA 320a predicts chronic axial and widespread pain development following motor vehicle collision in a stress-dependent manner. J Orthop Sports Phys Ther 46:911–919. 10.2519/jospt.2016.6944 [DOI] [PubMed] [Google Scholar]
- Liu DZ, Tian Y, Ander BP, Xu H, Stamova BS, Zhan X, Turner RJ, Jickling G, Sharp FR (2010) Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J Cereb Blood Flow Metab 30:92–101. 10.1038/jcbfm.2009.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu MG, Chen J (2014) Preclinical research on pain comorbidity with affective disorders and cognitive deficits: challenges and perspectives. Prog Neurobiol 116:13–32. 10.1016/j.pneurobio.2014.01.003 [DOI] [PubMed] [Google Scholar]
- Madathil SK, Nelson PT, Saatman KE, Wilfred BR (2011) MicroRNAs in CNS injury: potential roles and therapeutic implications. Bioessays 33:21–26. 10.1002/bies.201000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiarù M, Tochiki KK, Cox MB, Annan LV, Bell CG, Feng X, Hausch F, Géranton SM (2016) The stress regulator FKBP51 drives chronic pain by modulating spinal glucocorticoid signaling. Sci Transl Med 8:325ra319. 10.1126/scitranslmed.aab3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiarù M, Morgan OB, Mao T, Breitsamer M, Bamber H, Pöhlmann M, Schmidt MV, Winter G, Hausch F, Géranton SM (2018) The stress regulator Fkbp51: a novel and promising druggable target for the treatment of persistent pain states across sexes. Pain 159:1224–1234. 10.1097/j.pain.0000000000001204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier SF, Wiertelak EP, Martin D, Watkins LR (1993) Interleukin-1 mediates the behavioral hyperalgesia produced by lithium chloride and endotoxin. Brain Res 623:321–324. 10.1016/0006-8993(93)91446-Y [DOI] [PubMed] [Google Scholar]
- Marigorta UM, Navarro A (2013) High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genet 9:e1003566. 10.1371/journal.pgen.1003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH. (2006) RNA secondary structure analysis using RNAstructure. Curr Protoc Bioinformatics Chapter 12:Unit 12.6. 10.1002/0471250953.bi1206s13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean SA. (2011) The potential contribution of stress systems to the transition to chronic WAD. Spine 36:S226—S232. 10.1097/BRS.0b013e3182387fb4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean SA, Clauw DJ, Abelson JL, Liberzon I (2005) The development of persistent pain and psychological morbidity after motor vehicle collision: integrating the potential role of stress response systems into a biopsychosocial model. Psychosom Med 67:783–790. 10.1097/01.psy.0000181276.49204.bb [DOI] [PubMed] [Google Scholar]
- McLean SA, Diatchenko L, Lee YM, Swor RA, Domeier RM, Jones JS, Jones CW, Reed C, Harris RE, Maixner W, Clauw DJ, Liberzon I (2011) Catechol O-methyltransferase haplotype predicts immediate musculoskeletal neck pain and psychological symptoms after motor vehicle collision. J Pain 12:101–107. 10.1016/j.jpain.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean SA, Ulirsch JC, Slade GD, Soward AC, Swor RA, Peak DA, Jones JS, Rathlev NK, Lee DC, Domeier RM, Hendry PL, Bortsov AV, Bair E (2014) Incidence and predictors of neck and widespread pain after motor vehicle collision among US litigants and nonlitigants. Pain 155:309–321. 10.1016/j.pain.2013.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meloto CB, Bortsov AV, Bair E, Helgeson E, Ostrom C, Smith SB, Dubner R, Slade GD, Fillingim RB, Greenspan JD, Ohrbach R, Maixner W, McLean SA, Diatchenko L (2016) Modification of COMT-dependent pain sensitivity by psychological stress and sex. Pain 157:858–867. 10.1097/j.pain.0000000000000449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP. (2011) Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 35:809–822. 10.1002/gepi.20630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova IA, Alexander GM, Qureshi RA, Sacan A, Graziano A, Barrett JE, Schwartzman RJ, Ajit SK (2011) MicroRNA modulation in complex regional pain syndrome. J Transl Med 9:195. 10.1186/1479-5876-9-195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T (2005) Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276. 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- Platts-Mills TF, Ballina L, Bortsov AV, Soward A, Swor RA, Jones JS, Lee DC, Peak DA, Domeier RM, Rathlev NK, Hendry PL, McLean SA (2011) Using emergency department-based inception cohorts to determine genetic characteristics associated with long term patient outcomes after motor vehicle collision: methodology of the CRASH study. BMC Emerg Med 11:14. 10.1186/1471-227X-11-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi RA, Tian Y, McDonald MK, Capasso KE, Douglas SR, Gao R, Orlova IA, Barrett JE, Ajit SK, Sacan A (2015) Circulating microRNA signatures in rodent models of pain. Mol Neurobiol 53:3416–3427. 10.1007/s12035-015-9281-4 [DOI] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH (2010) RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11:129. 10.1186/1471-2105-11-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz J, Martin JS, Laederach A (2012) Evaluating our ability to predict the structural disruption of RNA by SNPs. BMC Genomics 13 [Suppl. 4]:S6. 10.1186/1471-2164-13-S4-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Gilman SE, Breslau J, Breslau N, Koenen KC (2011) Race/ethnic differences in exposure to traumatic events, development of post-traumatic stress disorder, and treatment-seeking for post-traumatic stress disorder in the united states. Psychol Med 41:71–83. 10.1017/S0033291710000401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Gorodetsky E, Yuan Q, Goldman D, Enoch MA (2010) Interaction of FKBP5, a stress-related gene, with childhood trauma increases the risk for attempting suicide. Neuropsychopharmacology 35:1674–1683. 10.1038/npp.2009.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scammell JG, Denny WB, Valentine DL, Smith DF (2001) Overexpression of the FK506-binding immunophilin FKBP51 is the common cause of glucocorticoid resistance in three new world primates. Gen Comp Endocrinol 124:152–165. 10.1006/gcen.2001.7696 [DOI] [PubMed] [Google Scholar]
- Segman RH, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev AY (2005) Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol Psychiatry 10:500–513. 10.1038/sj.mp.4001636 [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM (2014) RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11:959–965. 10.1038/nmeth.3029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva CM, Powell-Oliver FE, Jewell CM, Sar M, Allgood VE, Cidlowski JA (1994) Regulation of the human glucocorticoid receptor by long-term and chronic treatment with glucocorticoid. Steroids 59:436–442. 10.1016/0039-128X(94)90013-2 [DOI] [PubMed] [Google Scholar]
- Smola MJ, Rice GM, Busan S, Siegfried NA, Weeks KM (2015) Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10:1643–1669. 10.1038/nprot.2015.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solem AC, Halvorsen M, Ramos SB, Laederach A (2015) The potential of the riboSNitch in personalized medicine. Wiley Interdisciplinary Rev RNA 6:517–532. 10.1002/wrna.1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg EM. (2006) Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol 6:318–328. 10.1038/nri1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sükösd Z, Swenson MS, Kjems J, Heitsch CE (2013) Evaluating the accuracy of SHAPE-directed RNA secondary structure predictions. Nucleic Acids Res 41:2807–2816. 10.1093/nar/gks1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Fan C, Perou CM (2006) Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet 141:261–268. 10.1002/ajmg.b.30272 [DOI] [PubMed] [Google Scholar]
- Tatro ET, Everall IP, Kaul M, Achim CL (2009) Modulation of glucocorticoid receptor nuclear translocation in neurons by immunophilins FKBP51 and FKBP52: implications for major depressive disorder. Brain Res 1286:1–12. 10.1016/j.brainres.2009.06.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulirsch JC, Ballina LE, Soward AC, Rossi C, Hauda W, Holbrook D, Wheeler R, Foley KA, Batts J, Collette R, Goodman E, McLean SA (2014) Pain and somatic symptoms are sequelae of sexual assault: results of a prospective longitudinal study. Eur J Pain 18:559–566. 10.1002/j.1532-2149.2013.00395.x [DOI] [PubMed] [Google Scholar]
- Vermeer H, Hendriks-Stegeman BI, van der Burg B, van Buul-Offers SC, Jansen M (2003) Glucocorticoid-induced increase in lymphocytic FKBP51 messenger ribonucleic acid expression: a potential marker for glucocorticoid sensitivity, potency, and bioavailability. J Clin Endocrinol Metab 88:277–284. 10.1210/jc.2002-020354 [DOI] [PubMed] [Google Scholar]
- Volk N, Pape JC, Engel M, Zannas AS, Cattane N, Cattaneo A, Binder EB, Chen A (2016) Amygdalar microRNA-15a is essential for coping with chronic stress. Cell Rep 17:1882–1891. 10.1016/j.celrep.2016.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Stadler PF, Kellis M (2012) RNA folding with soft constraints: reconciliation of probing data and thermodynamic secondary structure prediction. Nucleic Acids Res 40:4261–4272. 10.1093/nar/gks009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LR, Wiertelak EP, Goehler LE, Smith KP, Martin D, Maier SF (1994) Characterization of cytokine-induced hyperalgesia. Brain Res 654:15–26. 10.1016/0006-8993(94)91566-0 [DOI] [PubMed] [Google Scholar]
- Watkins LR, Goehler LE, Relton J, Brewer MT, Maier SF (1995a) Mechanisms of tumor necrosis factor-alpha (TNF-alpha) hyperalgesia. Brain Res 692:244–250. 10.1016/0006-8993(95)00715-3 [DOI] [PubMed] [Google Scholar]
- Watkins LR, Maier SF, Goehler LE (1995b) Immune activation: the role of pro-inflammatory cytokines in inflammation, illness responses and pathological pain states. Pain 63:289–302. 10.1016/0304-3959(95)00186-7 [DOI] [PubMed] [Google Scholar]
- Weiss DS. (2007) The Impact of Event Scale: revised. In: Wilson JP, Tang CS (eds) Cross-Cultural Assessment of Psychological Trauma and PTSD, pp 219–238. Boston, MA: Springer. [Google Scholar]
- Wen X, Luca F, Pique-Regi R (2015) Cross-population joint analysis of eQTLs: fine mapping and functional annotation. PLoS Genet 11:e1005176. 10.1371/journal.pgen.1005176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM (2006) Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1:1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- Wolfe F. (2003) Pain extent and diagnosis: development and validation of the regional pain scale in 12,799 patients with rheumatic disease. J Rheumatol 30:369–378. [PubMed] [Google Scholar]
- Xu Z, Taylor JA (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 37:W600–W605. 10.1093/nar/gkp290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannas AS, Wiechmann T, Gassen NC, Binder EB (2016) Gene–stress–epigenetic regulation of FKBP5: clinical and translational implications. Neuropsychopharmacology 41:261–274. 10.1038/npp.2015.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clark AF, Yorio T (2008) FK506-binding protein 51 regulates nuclear transport of the glucocorticoid receptor β and glucocorticoid responsiveness. Investigative ophthalmology and visual science 49:1037–1047. 10.1167/iovs.07-1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic acids research 31:3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]