Abstract
The use of paired Cas9 nickases instead of Cas9 nuclease drastically reduces off-target effects. Because both nickases must function for a nickase pair to make a double-strand break, the efficiency of paired nickases can intuitively be expected to be lower than that of either corresponding nuclease alone. Here, we carefully compared the gene-disrupting efficiency of Cas9 paired nickases with that of nucleases. Interestingly, the T7E1 assay and deep sequencing showed that on-target efficiency of paired D10A Cas9 nickases was frequently comparable, but sometimes higher than that of either corresponding nucleases in mammalian cells. As the underlying mechanism, we found that the HNH domain, which is preserved in the D10A Cas9 nickase, has higher activity than the RuvC domain in mammalian cells. In this study, we showed: (i) the in vivo cleavage efficiency of the HNH domain of Cas9 in mammalian cells is higher than that of the RuvC domain, (ii) paired Cas9 nickases are sometimes more efficient than individual nucleases for gene disruption. We envision that our findings which were overlooked in previous reports will serve as a new potential guideline for tool selection for CRISPR-Cas9-mediated gene disruption, facilitating efficient and precise genome editing.
INTRODUCTION
Programmable nucleases and nickases are versatile tools for targeted gene editing in cells and organisms (1). For such enzymes to be useful, high on-target efficiency and a low frequency of off-target effects are required. RNA-guided Cas9 nucleases, which generate double-strand breaks (DSBs) at targeted sites in the genome, have significant off-target effects (2–4). A mutant version of Cas9, Cas9 nickase (Cas9n), generates single-strand nicks in DNA. Paired nickases, which effectively create DSBs by generating two single-strand breaks a short distance apart at a targeted site, have been introduced to reduce such off-target activity (5–8). Several independent studies have clearly shown that the frequencies of off-target effects generated by paired nickases are orders of magnitude lower than those of nucleases targeting either of the two nickase sites (5,6,9). However, the efficiencies of nucleases and paired nickases have not been extensively compared, although it has been proposed that the on-target efficiency of paired nickases would be frequently lower or at least comparable to that of either nuclease (5,6,10). Given that the efficiency of Cas9 nuclease is significantly affected by the amount of plasmid DNA encoding the single guide RNA (sgRNA) and Cas9 (3) that is transfected into cells, an exact comparison may require both the transfection of the same amount of plasmid DNA and the use of a control plasmid in the nuclease group to allow equal expression of sgRNA and Cas9 or Cas9n. Three previous studies that compared the on-target efficiencies of nucleases and paired nickases did not monitor the transfection efficiency or use such a control plasmid (5,6,11).
For a nickase pair to make a DSB, both nickases must be functional. Thus, the efficiency of paired nickases can usually be expected to be comparable or slightly lower than that of either nuclease alone. However, to our surprise, a careful comparison of the efficiencies of 28 D10A Cas9 nickase pairs with their corresponding 56 Cas9 nucleases showed that the paired nickases frequently exhibited comparable or sometimes significantly higher on-target efficiency than the individual nucleases. Infrequently, we found that the paired nickases showed significantly lower on-target efficiency than the corresponding nucleases. Interestingly, paired nickases sometimes lead to significant generation of insertions or deletions (indels) despite insignificant mutation frequencies associated with either of the corresponding nucleases. We also elucidated that the underlying mechanism is the superior activity of the HNH domain, which is preserved in the D10A Cas9 nickase, compared to that of the RuvC domain. We envision that our findings will serve as a potential new guideline for efficient genome editing tool selection and facilitate the use of Cas9 nickases and nucleases for efficient and specific genome engineering strategies.
MATERIALS AND METHODS
Nuclease and nickase constructs
The hSpCas9n nickase (Cas9n; D10A)-expressing pX335 plasmid (#42335) and hSpCas9 nuclease-expressing pX330 plasmid (#42230) (12) were purchased from Addgene (Cambridge, MA, USA). Using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA), we modified the sgRNA cloning sites of pX335 and pX330 by adding two guanine nucleotides at the 5′ end of the sgRNA (5′-GGX20 sgRNA) (6) or one guanine nucleotide at the 5′ end of the sgRNA (5′-GX20/GX19 sgRNA) and by changing the BbsI restriction enzyme recognition site (GAAGAC) to that of BsaI (GGTCTC). These nuclease and nickase constructs with no tagged fluorescent proteins were used for surrogate reporter assay. To generate U6-sgRNA-Cas9n-IRES-eGFP and U6-sgRNA-Cas9n-IRES-mCherry plasmids, IRES-eGFP and IRES-mCherry, respectively, were PCR-amplified using MSCV-IRES-eGFP and IRES3-mCherry-CL plasmids (kindly provided by Prof. Chung Hee Yong and Prof. Chang Hwan Park, respectively, Hanyang University, South Korea) as templates and then inserted between the Cas9n sequence and the 3′ NLS of pX335. Similarly, we inserted IRES-eGFP and IRES-mCherry between the Cas9 sequence and the 3′ NLS of pX330 to generate U6-sgRNA-Cas9-IRES-eGFP and U6-sgRNA-Cas9-IRES-mCherry. To generate RuvC-mutant Cas9 (D10A), HNH-mutant Cas9 (H840A or N863A), site directed mutagenesis was performed using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The full sequences and maps of these vectors are shown in Supplementary Notes 1–4 and Supplementary Figure S1, respectively.
The sgRNA target sequences were cloned into the vectors as previously described (13). Briefly, oligonucleotides containing each target sequence were synthesized (Bioneer, Seoul, South Korea) and annealed in vitro using a thermocycler. The vector was digested with BsaI restriction enzyme and ligated with the annealed oligonucleotides. Oligonucleotide sequences are listed in Supplementary Tables S1–6.
Reporters
Flow cytometric reporter plasmids containing a Cas9 target sequence were constructed as previously described (13,14). Briefly, oligonucleotides including target sequences (Supplementary Table S7) were synthesized (Bioneer, Daejon, South Korea) and annealed in vitro using a thermocycler (95°C for 5 min and then ramped down to 25°C at 5°C/min). The annealed oligonucleotides were ligated into the reporter vectors digested with EcoRI and BamHI.
Cell culture
Human embryonic kidney 293T (HEK293T) cells, human cervical cancer (HeLa) cells, human chronic myelogenous leukemia (K562) cells, mouse embryonic fibroblast cell line (NIH3T3) and mouse neuroblastoma (Neuro-2a) cells were purchased from American Type Culture Collection (Manassas, VA). HEK293T, HeLa, NIH3T3 and Neuro-2a cells were maintained in Dulbecco's Modified Eagle Medium (DMEM; Invitrogen, Carlsbad, CA, USA), K562 cells were maintained in Roswell Park Memorial Institute medium (RPMI-1640; Invitrogen, Carlsbad, CA, USA) supplemented with 100 units ml−1 penicillin, 100 μg ml–1 streptomycin, and 10% fetal bovine serum.
Transfection
Cells were transfected with mixtures of plasmids encoding Cas9n (or Cas9)-IRES-eGFP and Cas9n (or Cas9)-IRES-mCherry) at a 1:1 weight ratio using polyethyleneimine (HEK293T and Neuro-2a cells; linear, MW∼25,000; Polysciences, Warrington, PA) or Neon (HeLa and NIH3T3 cells; Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The Neon transfection conditions used for HeLa were 1005 pulse voltage, 35 pulse width and 2 pulse number, and those for NIH3T3 cells were 1400 pulse voltage, 20 pulse width and 2 pulse number. Cells were analyzed three days after transfection.
Flow cytometry
Cells were transfected with plasmids encoding Cas9n (or Cas9)-IRES-eGFP and Cas9n (or Cas9)-IRES-mCherry). Three days after transfection, cells were trypsinized and resuspended in 2% FBS in PBS. The transfection efficiency was monitored by the expression of fluorescent proteins such as eGFP and mCherry using a flow cytometer (FACSAria II; BD Biosciences). Untransfected cells and cells transfected with either Cas9n-IRES-eGFP or Cas9n-IRES-mCherry alone were used as controls.
T7E1 assay
The T7E1 assay was performed as previously described (15,16). Briefly, genomic DNA was isolated using the Wizard Genomic DNA purification Kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. The region including the paired nickase or nuclease target site was nested PCR-amplified using appropriate primers (Supplementary Table S8). PCR was performed in two different reactions. The first PCR reaction condition was: Template: 300 ng genomic DNA; denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C (extension time was 1 min/kb); and 25 PCR cycles. The second PCR reaction condition was: Template: First PCR amplicon sample: distilled water, diluted to 1:200 ratio; denaturation at 95°C for 30 s, annealing at 60°C for 30 s, extension at 72°C; and 35 PCR cycles. The amplicons were denatured by heating and annealed to allow formation of heteroduplex DNA, which was treated with 5 units of T7 endonuclease 1 (New England Biolabs) for 20 min at 37°C followed by analysis using 2% agarose gel electrophoresis. Mutation frequencies were calculated as previously described based on the band intensities using ImageJ software and the following equation (17): mutation frequency (%) = 100 × (1 − (1 − fraction cleaved)1/2), where the fraction cleaved is the total relative density of the cleavage bands divided by the sum of the relative density of the cleavage bands and uncut bands.
Sequencing analysis
Sequencing of the genomic region including the target sequence was performed as previously described (13,14). Briefly, PCR amplicons that included nuclease or paired nickase target sites were cloned into the T-Blunt vector (Promega, Madison, WI). Cloned plasmids were sequenced using the primers used for PCR amplification.
Targeted deep sequencing
The on-target regions about 300 bp within the genomic DNA were amplified using Pfu DNA polymerase (Promega, Madison, WI). Equal amounts of the PCR amplicons were subjected to paired-end read sequencing using Illumina MiSeq at Bio Medical Laboratories. The region including the paired nickase or nuclease target sites were nested PCR-amplified using appropriate primers (Supplementary Table S9). Indels mapped around the Cas9 nuclease or nickase cleavage site (3 bp upstream of the PAM) were considered to be the result of NHEJ-mediated mutagenesis induced by the nuclease or paired nickases. The deep sequencing data have been deposited to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra).
Surrogate reporter assay
The episomal reporter assay was performed as previously described (13). HEK293T cells were transfected with Cas9 (Addgene #42230)- or Cas9n (D10A and H840A)-encoding plasmid (Addgene #51130 and #51129), sgRNA-encoding plasmid and reporter plasmid at 1:1:1 weight ratio. Three days after transfection, the cells were analyzed using a flow cytometer (FACSAria II; BD Biosciences). Cells transfected with reporters alone were used as an analysis control (13).
Statistical analysis
All data were expressed as means ± S.E.M. and statistical analysis was conducted using GraphPad Prism 5. The paired t-test was used to compare the mutation frequencies of paired nickases and either corresponding nuclease. A P value < 0.05 was considered statistically significant.
RESULTS
Gene disruption efficiencies of paired Cas9 nickases vs. Cas9 nucleases in human and mouse cells
To monitor transfection efficiency, we inserted genes encoding fluorescent proteins (eGFP or mCherry) into the plasmid vectors that encode sgRNA (5′GGX20) (6) and Cas9 or Cas9n (D10A) (Supplementary Figures S1 and S2). Furthermore, in both nuclease groups, we used a control plasmid expressing a control sgRNA so that an equal amount of plasmid DNA encoding Cas9 or Cas9n and sgRNA would be transfected in the three groups, i.e. a nickase pair, one nuclease and the other nuclease.
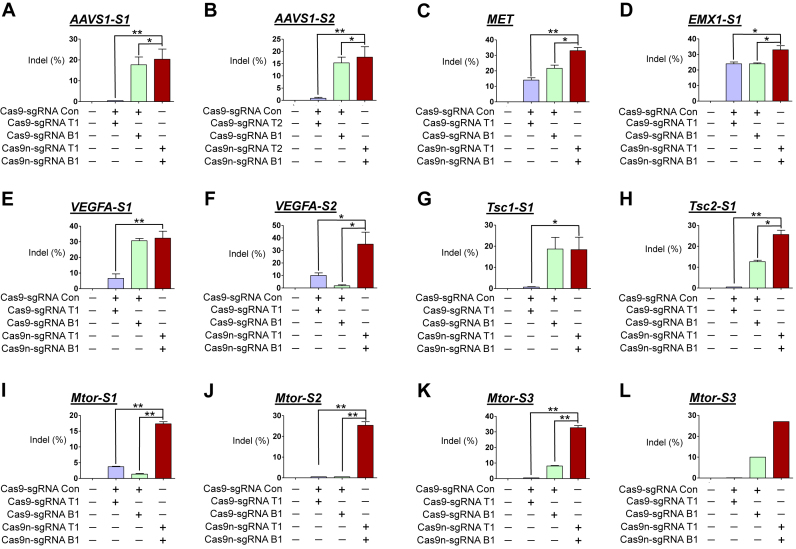
HEK293T cells were transfected with plasmids encoding either a nuclease or a nickase pair targeting six loci in human HEK293T cells. Flow cytometry showed that the fraction of eGFP- and mCherry-positive cells in these three cell populations was comparable (Supplementary Figures S3 and S4), indicating similar transfection or delivery of plasmids in each group. However, to our surprise, for all the six analyzed loci, the T7E1 assay showed that the indel frequencies in cells treated with paired nickases were higher than those in at least one of the two cell populations treated with the corresponding nucleases coupled with sgRNA complementary to the top or bottom strands (hereafter referred to as top [T] and bottom [B] sgRNAs; Supplementary Figure S2C) and were at least comparable to those in cells treated with nucleases with higher activities (Figure 1A–F). Out of six analyzed loci, cells transfected with plasmids encoding either a nuclease or a nickase pair targeting two genes (EMX1-S1 and MET) were subjected to capillary sequencing. Sequencing results showed that the mutation frequency in cells treated with paired nickases was 47% versus 25% and 27% in cells treated with nuclease targeting of the EMX1-S1 gene, while paired nickases was 44% versus 20% and 22% in cells treated with nuclease targeting of the MET gene (Supplementary Figure S5), corroborating the higher efficiency of paired nickases as compared with those of nucleases. Notably, at two out of the six analyzed loci, although the activities of one of the two nucleases were below the detection limit of the T7E1 assay, the corresponding paired nickases showed significant indel-generating activities (Figure 1A and B).
Figure 1.
Gene disruption efficiencies of paired D10A Cas9 nickases vs. Cas9 nucleases using sgRNA (5′GGX20) and control sgRNA in human and mouse cells. HEK293T were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, sgRNAs targeting AAVS1-S1 (T1 and B1; A), AAVS1-S2 (T2 and B1; B), MET (T1 and B1; C), EMX1-S1 (T1 and B1; D), VEGFA-S1 (T1 and B1; E), VEGFA-S2 (T1 and B1; F). NIH3T3 were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, sgRNAs targeting Tsc1-S1 (T1 and B1; G), Tsc2-S1 (T1 and B1; H), Mtor-S1 (T1 and B1; I), Mtor-S2 (T1 and B1; J), Mtor-S3 (T1 and B1; K) and fluorescent proteins (i.e., eGFP or mCherry). A sgRNA that does not target the respective genes was used as a control and is designated as Con. The frequency of Cas9 nuclease- or paired nickases-driven mutations as determined by the T7E1 assay are shown using bar graph. Error bars were derived from three independent experiments (n = 3). For brevity, statistical significance was shown only for comparison between a group-of-interest and the paired nickase group, except for the negative control group; *P< 0.05, **P< 0.01 by paired t-test. (L) Cas9 nuclease- or paired nickase- driven indels determined by the deep sequencing.
To determine whether paired nickases result in a higher mutation frequency than either corresponding nuclease in other species as well, we next compared the mutation frequencies of paired nickases and nucleases targeting five loci in mouse NIH3T3 cells. Flow cytometry showed that the fraction of eGFP- and mCherry-positive cells in these three cell populations was comparable (Supplementary Figure S6), suggesting that similar plasmid transfection occurred in each group. Similar to findings observed in human cells, the T7E1 assay showed that the indel frequencies in cells treated with paired nickases were higher than those in at least one of the two cell populations treated with the corresponding nucleases and were at least comparable to those in cells treated with nucleases with higher activities at all five loci (Figure 1G–K). Out of the five analyzed loci, the activities of one of the two nucleases at two loci (Mtor-S3, Tsc2-S1) and those of both nucleases at one locus (Mtor-S2) were below the detection limit of the T7E1 assay. However, the corresponding paired nickases showed significant indel-generating activities at all five analyzed loci, which is compatible with the results observed in human cells. We also performed deep sequencing of a representative locus, Mtor-S3; the indel frequencies in the cells treated with nucleases were 0.067% and 10.4%, whereas that of cells treated with the corresponding paired nickases was 27% (Figure 1L, Supplementary Table S10), suggesting that paired nickases are frequently more efficient than the corresponding nucleases.
From a practical point of view, two important factors need to be considered to make a valid comparison between paired nickases vs nucleases. First, the addition of two guanine nucleotides at the 5′ end of sgRNA (5′GGX20) exhibited greater specificity, but in some cases the nuclease activity was significantly reduced (6). This could lead to a misleading comparison between nuclease and paired nickases. Second, the total amount of transfected DNA is sometimes a limiting factor. Although the use of nontargeting sgRNA is appropriate for comparing the efficiencies of nucleases and paired nickases, one might choose to increase the amount of transfected plasmid encoding a nuclease (i.e. Cas9 and sgRNA) for enhanced gene disruption instead of using paired nickases. Thus, we replaced 5′GGX20 sgRNA with 5′GX20 or 5′GX19 sgRNA and doubled the amount of transfected Cas9 nuclease plasmids for further comparison analysis.
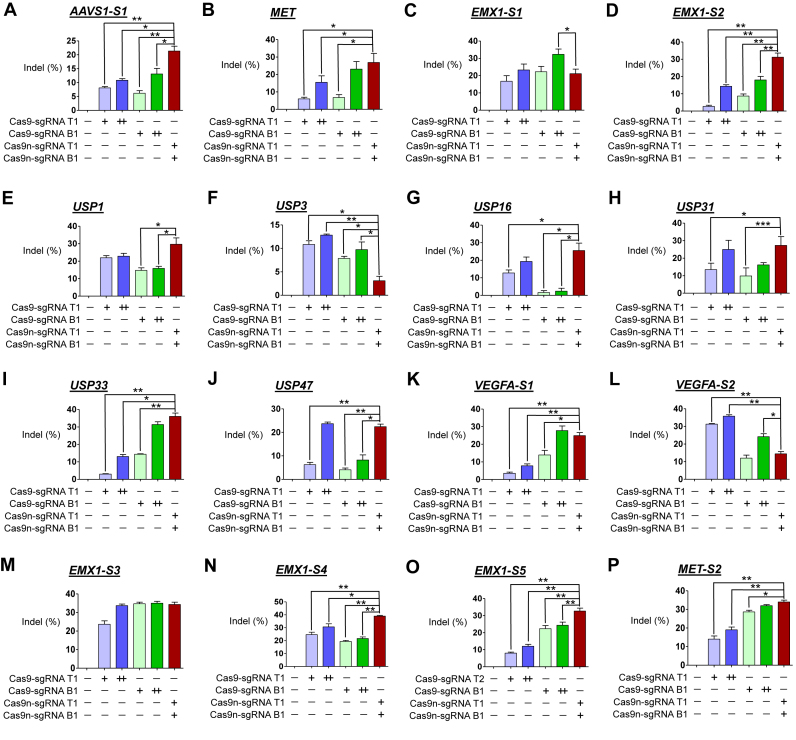
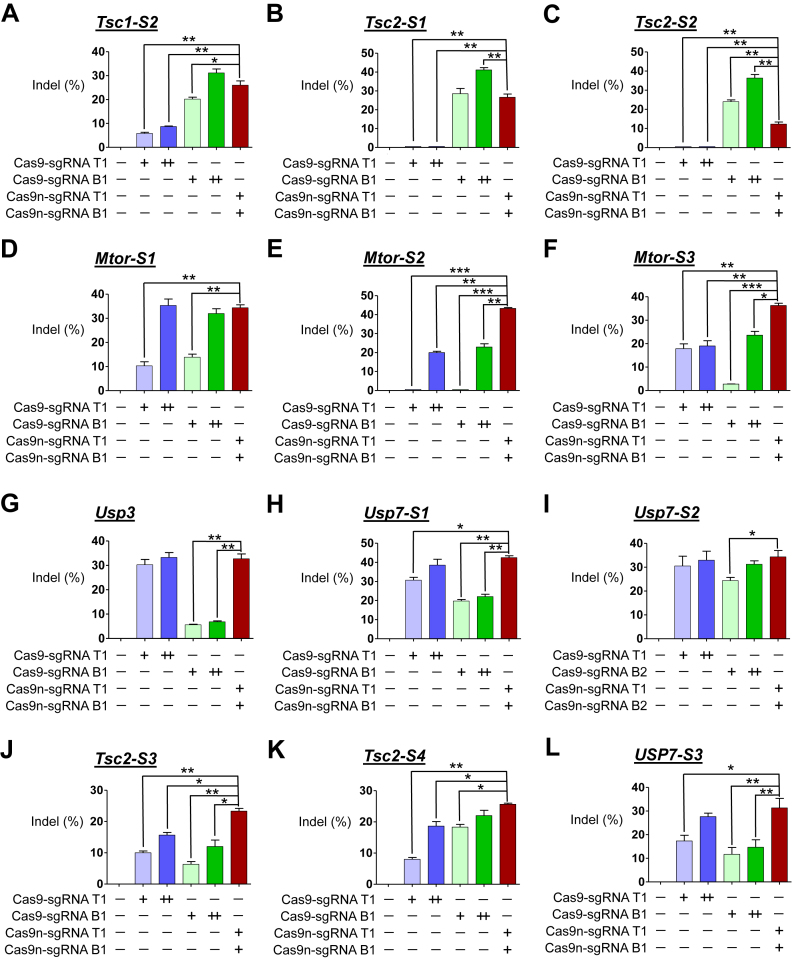
Next, we compared the indel frequencies of cells transfected with plasmids encoding paired nickases and those transfected with a doubled amount of nuclease-encoding plasmid targeting 16 loci in human HEK293T cells without control sgRNA. The T7E1 assay showed that increasing the amount of transfected plasmids encoding Cas9 nuclease and sgRNA from 1 μg to 2 μg led to an increase in indel frequencies at all 16 analyzed loci (Figure 2) which is compatible with a previous report (3). Similar comparison analysis was performed on 12 loci in mouse Neuro-2a cells without control sgRNA. The T7E1 assay showed that increasing the amount of transfected plasmids encoding Cas9 nuclease and sgRNA from 1 to 2 μg led to an increase in indel frequencies at all 10 analyzed loci (Figure 3), except for two loci (Tsc2-S1 and Tsc2-S2; Figure 3B and C), no changes in indel frequencies were observed. Importantly, the mutation frequencies in cells treated with paired nickases were sometimes higher than those in at least one of two cell populations treated with the doubled amount of corresponding nucleases and were frequently comparable to those in cells treated with the doubled amount of nucleases with higher activities (Figures 2 and 3). Furthermore, in some cases, the mutation frequencies in the paired nickases group were significantly lower than those in either nuclease group, suggesting that paired nickases are not always superior to nucleases regarding efficiency. Taken together, these results show that both of the methods, i.e. replacing 5′GGX20 sgRNA with 5′GX20/5′GX19 sgRNA and transfecting plasmids with double the amount of corresponding nucleases, did not significantly affect the results, showing overall comparable or higher efficiency of paired nickases over individual nuclease.
Figure 2.
Gene disruption efficiencies of paired Cas9 nickases vs. double the amount of Cas9 nucleases using sgRNA (5′GX20 or 5′GX19) without control sgRNA in human cells. (A–P) Cas9 paired nickase- or nuclease-driven mutations in the human genes detected by the T7E1 assay. HEK293T were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, 5′GX20 sgRNAs targeting AAVS1-S1 (T1 and B1; A), MET (T1 and B1; B), EMX1-S1 (T1 and B1; C), EMX1-S2 (T1 and B1; D), USP1 (T1 and B1; E), USP3 (T1 and B1; F), USP16 (T1 and B1; G), USP31 (T1 and B1; H), USP33 (T1 and B1; I), USP47 (T1 and B1; J), VEGFA-S1 (T1 and B1; K), and VEGFA-S2 (T1 and B1; L). HEK293T were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, 5′GX19 sgRNAs targeting EMX1-S3 (T1 and B1; M), EMX1-S4 (T1 and B1; N), EMX1-S5 (T2 and B1; O) and MET-S2 (T1 and B1; P). The frequency of Cas9 nuclease- or paired nickases-driven mutations as determined by the T7E1 assay are shown using bar graph. Error bars were derived from three independent experiments (n = 3). For brevity, statistical significance was shown only for comparison between a group-of-interest and the paired nickase group, except for the negative control group; *P< 0.05, **P< 0.01, ***P< 0.001 by paired t-test. ‘+’ and ‘++’ denote 1 and 2 μg concentrations of Cas9 nucleases or paired Cas9 nickases using top or bottom sgRNAs, respectively.
Figure 3.
Gene disruption efficiencies of paired Cas9 nickases versus double the amount of Cas9 nucleases using sgRNA (5′GX20 or 5′GX19) without control sgRNA in mouse cells. (A–L) paired Cas9 nickase- or nuclease-driven mutations in the mouse genes detected by the T7E1 assay. Neuro-2a were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, 5′GX20 sgRNAs targeting Tsc1-S2 (T1 and B1; A), Tsc2-S1 (T1 and B1; B), Tsc2-S2 (T1 and B1; C), Mtor-S1 (T1 and B1; D), Mtor-S2 (T1 and B1; E), Mtor-S3 (T1 and B1; F), Usp3 (T1 and B1; G), Usp7-S1 (T1 and B1; H) and Usp7-S2 (T1 and B2; I). Neuro-2a were analyzed 3 days after transfection with plasmids encoding paired Cas9 nickases or Cas9 nuclease, 5′GX19 sgRNAs targeting Tsc2-S3 (T1 and B1; J), Tsc2-S4 (T1 and B1; K) and mUSP7-S3 (T1 and B1; L). The frequency of Cas9 nuclease- or paired nickases-driven mutations as determined by the T7E1 assay are shown using bar graph. Error bars were derived from three independent experiments (n = 3). For brevity, statistical significance was shown only for comparison between a group-of-interest and the paired nickase group, except for the negative control group; *P< 0.05, **P< 0.01, ***P< 0.001 by paired t-test. ‘+’ and ‘++’ denote 1 and 2 μg concentrations of Cas9 nucleases or paired Cas9 nickases using top or bottom sgRNAs, respectively.
Next, we checked mutation indel frequencies for three human genes (MET, USP33 and VEGFA-S1) in other human cell lines such as K562 (Supplementary Figure S7A–C), and HeLa (Supplementary Figure S7D-F), and two mouse genes (Tsc2-S2 and Mtor-S2) in a mouse cell line NIH3T3 (Supplementary Figure S8). The cells were treated with paired nickases and the corresponding nuclease; paired nickases exhibited similar higher mutation frequency or higher than those in at least one of the two cell populations treated with the corresponding nucleases, suggesting that the comparable or higher efficiency of paired nickases over the individual nuclease are not cell specific (Supplementary Figures S7 and S8).
Comparison of the efficiencies of nucleases and paired nickases
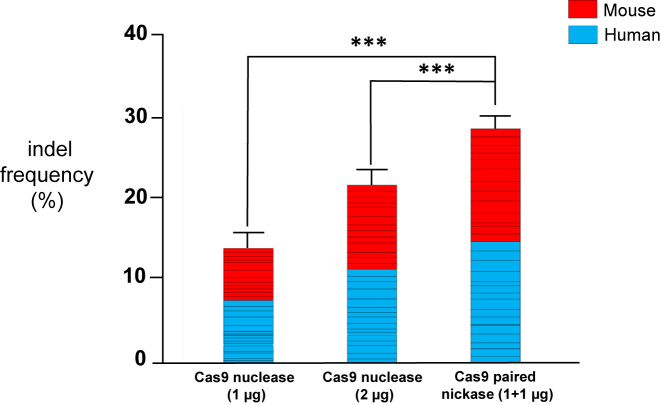
We next summarized the T7E1 assay results obtained in both human and mouse cells to compare the efficiency of nucleases and paired nickases in general. The average mutation frequency of cells treated with paired nickases was 28.0 ± 1.7%, significantly higher than the frequency in cells treated with nucleases at a 1 μg concentration using top or bottom sgRNAs (the average ratio of nickase pair/nuclease = 5.3 ± 2.9, P < 0.001; the average mutation frequency of nucleases = 13.8 ± 1.3%; please see Figure 4, Supplementary Table S11). Similarly, the average mutation frequency of cells treated with paired nickases was 28.0 ± 1.7%, significantly higher than the frequency in cells treated with nucleases at twice the concentration (2 μg) using top or bottom sgRNAs (the average ratio of nickase pair/nuclease = 1.4 ± 0.1, P < 0.001, and the average mutation frequency of nucleases = 21.5 ± 1.2%; please see Figure 4, Supplementary Table S12).
Figure 4.
Comparison of gene disruption efficiencies of paired Cas9 nickases versus Cas9 nucleases in human and mouse cells. Comparison of indel frequencies associated with nickase pairs and corresponding nucleases or double the amount of nucleases as measured using the T7E1 assay in human and mouse cells. Sixteen pairs of sgRNAs targeting human genes and twelve pairs of sgRNAs targeting mouse genes were used in HEK293T cells and Neuro-2a cells, respectively. The average values from three independent experiments are shown. Representative bar graphs from each experiment are shown in detail in Figures 2 and 3. Error bars represent s.e.m. ***P< 0.001 by paired t-test.
When the mutation frequency associated with each nickase pair was individually compared with that of either nuclease, in 77% (43/56), 7% (4/56), and 9% (5/56) of comparisons, the mutation frequency associated with the nickase pair was significantly higher (P < 0.05), trending higher (0.05 < P < 0.1) without statistical significance, and comparable (P > 0.1), respectively (Supplementary Table S11). In contrast, only 3.5% (2/56) showed that the mutation frequency associated with each nickases pair was significantly lower than the cell populations treated with the nucleases using top or bottom sgRNAs, the remaining 3.5% (2/56) showed that the mutation frequency associated with each nickases pair was higher than those in at least one of two cell populations treated with the corresponding nucleases (Supplementary Table S11), corroborating that the activity of nickase pairs is overall superior to that of nucleases with infrequent exceptions.
However, when the mutation frequency associated with each nickase pair was individually compared with that of either nuclease at twice the concentration, in 50% (28/56), 9% (5/56), and 29% (16/56) of comparisons, the mutation frequency associated with the nickase pair was significantly higher (P < 0.05), trending higher (0.05 < P < 0.1) without statistical significance, and comparable (P > 0.1), respectively (Supplementary Table S12). In contrast, only 7% (4/56) showed that the mutation frequency associated with each nickases pair was significantly lower than the cell populations treated with the double the amount of nucleases using top or bottom sgRNAs. However, 5% (3/56) showed that the mutation frequencies in cells treated with paired nickases were higher than those in at least one of two cell populations treated with the doubled amount of corresponding nucleases and were at least comparable to those in cells treated with the doubled amount of nucleases with higher activities (Supplementary Table S12), suggesting that the efficiency of paired nickases for gene disruption is sometimes higher or comparable to that of a nuclease at twice the concentration.
The Cas9 HNH domain has higher cleavage activity than the RuvC domain
To elucidate the mechanism underlying the higher indel generating activity of paired D10A nickases as compared with nucleases, we compared the activities of the two nuclease domains of Cas9: the HNH domain and the RuvC domain, which are preserved in D10A and H840A Cas9 nickases (18), respectively. Although an H840A mutation was reported to convert Cas9 into a nicking enzyme, this mutant has low levels of activity in mammalian cells compared with another mutant N863A which has been found to inactivate the HNH domain (19). Thus, we first compared the indel generating efficiencies of D10A and H840A or N863A nickases at the same target sequence. Using the same sgRNA pairs and the same target sequences that were used in the studies described above, we compared the indel frequencies of HEK293T cells transfected with plasmids encoding D10A Cas9 or H840A or N863A Cas9. The T7E1 assay showed that transfection of plasmids encoding D10A Cas9 and a sgRNA pair led to significant gene disruption at frequencies that ranged from 15% to 31% at all five analyzed loci, whereas the indel frequency was below the detection limit (<0.5%) in all cell populations transfected with plasmids encoding H840A Cas9 or N863A Cas9 and the same sgRNA pair (Supplementary Figure S9). Although we did not observe detectable indels using H840A Cas9 or N863A Cas9, N863A Cas9 can induce significant indels in some target sequences (20).
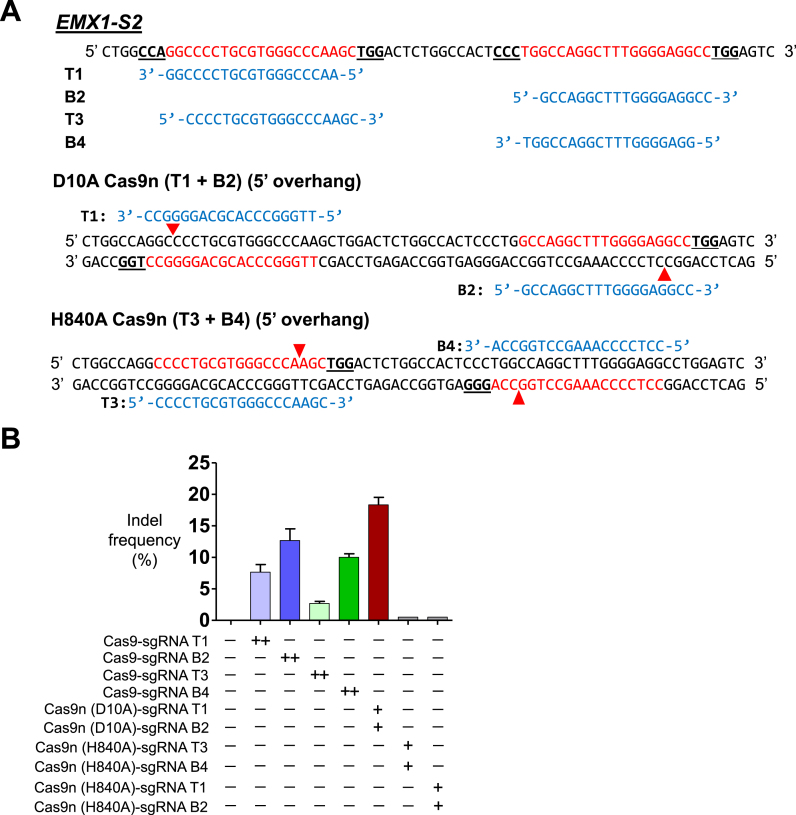
One can speculate that the lack of indel generating efficiency by H840A or N863A Cas9 could be due to the 3′ overhang generated by H840A or N863A Cas9 (6,7,20). To rule out the effects of the 3′ overhang, we attempted to design sgRNAs so that the cleavages by D10A and H840A paired nickases would lead to a DSB with a 5′ overhang. We found an endogenous locus in the human genome where a D10A sgRNA pair and a H840A sgRNA pair could be designed in a reverse-identical orientation so that the same sequences could be targeted using the two sgRNA pairs (Figure 5A). The T7E1 assay showed that the average mutation frequencies in cells transfected with plasmids encoding the Cas9 nuclease and T1, B2, T3 and B4 sgRNAs at twice the concentration were 7.6%, 12%, 2.6% and 10%, respectively (Figure 5B). The mutation frequency in cells transfected with plasmids encoding D10A Cas9 and T1 and B2 sgRNAs was 18%, whereas the frequency in cells transfected with plasmids encoding H840A Cas9 and T3 and B4 sgRNAs was below the detection limit, suggesting that the HNH domain may be more efficient than the RuvC domain.
Figure 5.
Comparison of indel generation efficiencies of paired Cas9 D10A nickases versus paired Cas9 H840A nickases using reverse-identical 5′GX19 sgRNAs without control sgRNA. (A) Sequence of the human EMX1-S2 locus with designed sgRNAs (T1, B2, T3 and B4). Reverse-identical sgRNAs (T1 and T3; B2 and B4) are shown in blue letters. sgRNA target sites (T1, B2, T3 and B4) are indicated by red letters and PAM sequences are marked by bold underlined letters. The cleavage by D10A Cas9 nickase and the sgRNA pair of T1 and B2 leads to a double-strand break with a 5′ overhang. Similarly, the cleavage by H840A Cas9 nickase and the sgRNA pair of T3 and B4 also leads to a double strand break with a 5′ overhang. Red arrowheads indicate the cleavage site. (B) The frequency of double the amount of Cas9 nuclease- or paired nickase-driven mutations as determined by the T7E1 assay are shown using bar graph. Error bars were derived from three independent experiments (n = 3). ‘+’ and ‘++’ denote 1 and 2 μg concentrations of Cas9 nucleases or paired Cas9 nickases using top or bottom sgRNAs, respectively.
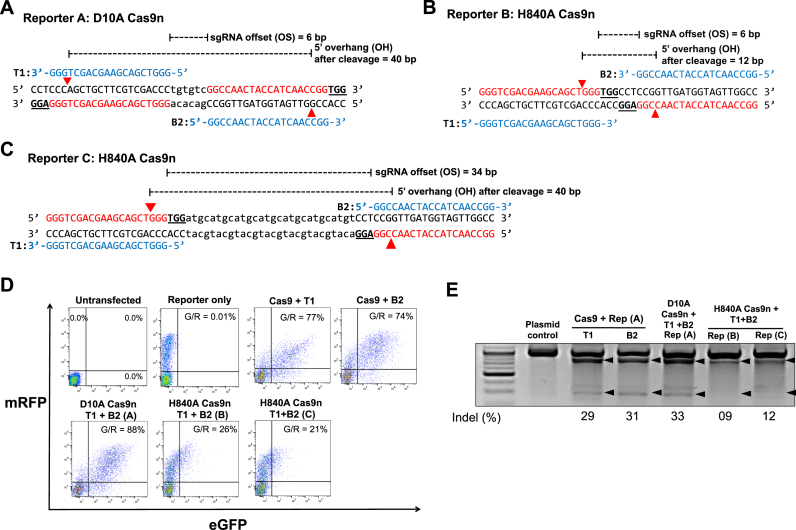
When two sgRNAs are identical in a reverse orientation, we cannot rule out the possibility that the sgRNA activities are different as observed in Figure 5, a potentially compounding factor when comparing D10A and H840A nickase activity. Thus, we next used an identical sgRNA pair to compare D10A and H840A nickases. For this purpose, we prepared a palindromic sgRNA pair (T1 and B2) and three target DNAs (Figure 6A–C). Target DNA A can be cleaved by the sgRNA pair and D10A nickase with a 6 bp offset (OS), leading to a DSB with a 40 bp 5′ overhang (OH). Target DNA B can be cleaved by the sgRNA pair and H840A nickase with the same offset as target DNA A cleaved by D10A nickase, leading to a DSB with a 12 bp 5′ overhang. Target DNA C can be cleaved by the sgRNA pair and H840A nickase with a 34 bp offset, leading to a DSB with the same length 5′ overhang as target DNA A cleaved by D10A.
Figure 6.
Comparison of indel generation efficiencies of paired Cas9 D10A nickases vs. paired Cas9 H840A nickases using identical 5′GX20 sgRNAs without control sgRNA. (A–C) Three surrogate reporters containing sequences that can be targeted with a fixed sgRNA pair (T1 and B2; shown in blue letters) were constructed. sgRNA target sites (T1 and B2) are indicated by red letters and PAM sequences are marked by bold underlined letters. (A) A sequence that can be targeted by D10A Cas9n and the sgRNA pair with a +6 bp offset. The cleavage is expected to lead to a 40 bp 5′ overhang. (B) A sequence that can be targeted by H840A Cas9n and the sgRNA pair with a +6 bp offset. The cleavage is expected to lead to a 12 bp 5′ overhang. (C) A sequence that can be targeted by H840A Cas9n and the sgRNA pair with a +28 bp offset. The cleavage is expected to lead to a 40 bp 5′ overhang. (D–E) HEK293T cells were analyzed 3 days after transfection with a plasmid encoding Cas9 nickase (D10A or H840A) or double the amount of Cas9 nuclease, plasmids encoding sgRNAs (T1 and B2), and a reporter plasmid containing the target sequence shown above (A, B or C). (D) Representative flow cytometry. The percentages of GFP+ cells in the total RFP+ cell population (G/R) are shown (e.g. G/R = 77%). (E) The mutation frequencies in the target sequence of the transfected reporter plasmids detected by the T7E1 assay.
Non-homologous end joining (NHEJ) surrogate reporters, previously used to determine the activity of programmable nucleases (13), were modified to contain target DNAs A, B or C to compare the activity of D10A and H840A nickase pairs. When transfected into mammalian cells, this reporter constitutively expresses RFP and conditionally expresses GFP in the presence of programmable nuclease activity that generates indels in the target sequence (Supplementary Figure S10). When co-transfected with plasmids encoding programmable nucleases, the percentage of GFP+ cells among the total RFP+ cells (i.e. transfected cells) reflects the nuclease activity. Here, flow cytometry showed that the frequency of GFP+ cells relative to RFP+ transfected cells (hereafter, G/R) was 0.01% in the reporter only-transfected cell population and 77% and 74% in cell populations transfected with double the amount of plasmids encoding Cas9 nuclease, and T1 and B2 sgRNAs, respectively, and reporter plasmids containing target DNA A (Figure 6D), suggesting that both T1 and B2 sgRNA have significant activity in the presence of Cas9 and that this reporter assay has minimal background signal. The G/R was 88% in the cells cotransfected with the reporter plasmid containing target DNA A and plasmids encoding D10A Cas9 nickase and T1 and B2 sgRNAs, versus 26% and 21% in the cells cotransfected with reporter plasmids containing target DNA B and C, respectively, and plasmids encoding H840A Cas9 nickase and T1 and B2 sgRNAs (Figure 6D), suggesting that D10A nickase has higher cleavage activity than H840A nickase. We next isolated the reporter plasmid and performed the T7E1 assay to evaluate the indel frequency in the target DNA. The mutation frequency in target DNA A was 33%, versus 9% and 12% in target DNA B and C, respectively (Figure 6E), corroborating that the cleavage activity of D10A nickase is higher than that of H840A nickase. We conducted three more similar experiments using different transfection methods and different amounts of plasmids and observed a similar tendency (Supplementary Figure S11). The higher cleavage activity of D10A nickase versus H840A nickase indicates that the Cas9 HNH domain has a higher cleavage activity than the RuvC domain.
DISCUSSION
In this study, we report that paired Cas9 nickases are sometimes more efficient for gene disruption than nucleases. Paired Cas9 nickases have been introduced to reduce off-target effects associated with Cas9 nucleases. Given that both members of a nickase pair must function to induce a DSB, the on-target efficiency of a nickase pair may usually be expected to be comparable or slightly lower than that of a nuclease targeting either of the two nickase sites. This intuitive assumption that the activity of a Cas9 nickase pair would be at best comparable to that of a Cas9 nuclease might be one of the reasons for the limited use of paired nickases compared to nucleases, which is partly supported by the number of publications; >3000 articles report on Cas9 nuclease-mediated genome editing, whereas fewer than 80 describe Cas9 nickase-mediated editing. Given that knowledge about the general efficiency of programmable nucleases can facilitate genome editing (1,21), our study will be of great value to the users of Cas9 nucleases and nickases. In addition, our findings are compatible with the recent study from the Cotta-Ramusino group which demonstrated that a high rate of gene conversion and DNA lesions were found in RuvC-mutant Cas9 (D10A)-induced 5′ overhangs compared with HNH-mutant Cas9 (N863A)-induced 3′ overhangs (20). In particular, different ends generated by the Cas9 variants govern different repair pathways. For instance, D10A Cas9-induced 5′ overhangs yield the highest gene correction by HDR compared to N863A-induced 3′ overhangs, whereas N863A-induced 3′ overhangs are primarily engaged in insertions featuring microhomologies (20).
Two studies proposed that the mutation efficiencies of paired nickases are comparable to those of the corresponding nucleases (5,6). Moreover, the mutation frequencies of paired nickases are higher than those of the nucleases using the top sgRNA if the offsets of the sgRNA pairs are in the optimum range of –4 to 20 bp (5), which is compatible with our conclusion (Supplementary Tables S11 and S12). However, when the sgRNA pairs are offset by ←4 or >20 bp, the mutation frequencies associated with paired nickases are significantly lower than those associated with either nuclease, which could be a confounding factor leading to the previous interpretation that the two mutation frequencies were comparable.
In this study, we carefully compared the efficiencies between nucleases and paired nickases in both human and mouse cell lines with different guide RNA structure, i.e. sgRNA (5′GGX20), which exhibited greater specificity (6), and sgRNA (5′GX20 or 5′GX19). Our results showed that the mutation frequencies in cells treated with paired nickases were comparable or higher than those in at least one of two cell populations treated with corresponding nucleases (Figures 1–3). We noticed, however, that there were several differences between our comparison studies on paired nickases vs nucleases and those conducted by others. First, we used a proper control plasmid which was cotransfected in the nuclease group so that the same amount of sgRNA and Cas9 or Cas9n would be expressed. Second, only the transfected cell batches showing the fraction of eGFP- and mCherry-positive cells from three groups (i.e., a nickase pair, one nuclease and the other nuclease) were comparable in expression and were subjected for further mutation frequency analysis. Third, a careful comparison was conducted by subjecting equal amounts of genomic DNA obtained from the tested groups for PCR amplification and further DNA cleavage analysis. Fourth, we analyzed guide pairs at least three to four times more than the other groups, which led us to confidently draw a conclusion on the overall comparable or sometimes higher efficiencies of paired nickases over the corresponding nuclease. Thus, considering all these technical factors, a deterministic approach to designing appropriately positioned paired sites will lead to improved editing efficiency and suggests that the use of paired Cas9n over Cas9 nuclease can be beneficial and can be a good choice for future experimental strategies.
Paired Cas9 nickases have benefits relative to Cas9 nucleases. Paired nickases are more efficient and specific than Cas9 nucleases. In addition, the overhangs generated by paired nickases can be used for targeted gene insertion via NHEJ (5,10,22). One drawback of paired Cas9 nickases, however, is the requirement for two sgRNAs instead of one. Based on a consideration of these advantages and disadvantages, researchers can choose a nickase pair or nuclease that best fits their experimental conditions and purposes. In addition, in a minor fraction of cases, the activities of paired nickases are comparable or slightly lower than that of one of the nucleases.
Despite barely detectable off-target effects, paired nickases have not been predominantly used in genome editing studies, partly because it has been thought that their on-target efficiency would be lower than that of nucleases. However, we showed here that the efficiency of paired nickases is mostly comparable, but occasionally superior to that of either corresponding nuclease. In addition, our paired nickase-encoding vectors allow the monitoring of transfection and the flow cytometric enrichment of mutant cells. We envision that our findings will suggest a new guideline for genome editing tool selection and greatly facilitate the use of paired Cas9 nickases, which may be useful replacements for Cas9 nucleases when efficient and specific genome editing is required.
DATA AVAILABILITY
National Center for Biotechnology Information (NCBI) Sequence Read Acrchive (SRA): The deep sequencing data from this study are available under accession code SRP135732. Source data for Figure 1.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arun Pandian Chandrasekaran, Soumydip Das and Myungjae Song for the technical assistance.
Author contributions: S.R. and H.K. designed the experiments and wrote the manuscript. R.G., B.S. and S.R. performed the experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea [HI16C1009]; National Research Foundation of Korea [2017M3A9C6061361]. Funding for open access charge: Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health and Welfare, Republic of Korea [HI16C1009]; National Research Foundation of Korea [2017M3A9C6061361].
Conflict of interest statement. None declared.
REFERENCES
- 1. Kim H., Kim J.S.. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014; 15:321–334. [DOI] [PubMed] [Google Scholar]
- 2. Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D.. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013; 31:822–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013; 31:827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R.. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013; 31:839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cho S.W., Kim S., Kim Y., Kweon J., Kim H.S., Bae S., Kim J.S.. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014; 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M.. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013; 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shen B., Zhang W., Zhang J., Zhou J., Wang J., Chen L., Wang L., Hodgkins A., Iyer V., Huang X. et al. Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods. 2014; 11:399–402. [DOI] [PubMed] [Google Scholar]
- 9. Guilinger J.P., Thompson D.B., Liu D.R.. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014; 32:577–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kocher T., Peking P., Klausegger A., Murauer E.M., Hofbauer J.P., Wally V., Lettner T., Hainzl S., Ablinger M., Bauer J.W. et al. Cut and paste: efficient homology-directed repair of a dominant negative KRT14 Mutation via CRISPR/Cas9 nickases. Mol. Ther. 2017; 25:2585–2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramakrishna S., Cho S.W., Kim S., Song M., Gopalappa R., Kim J.S., Kim H.. Surrogate reporter-based enrichment of cells containing RNA-guided Cas9 nuclease-induced mutations. Nat. Commun. 2014; 5:3378. [DOI] [PubMed] [Google Scholar]
- 14. Kim H., Um E., Cho S.R., Jung C., Kim H., Kim J.S.. Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat. Methods. 2011; 8:941–943. [DOI] [PubMed] [Google Scholar]
- 15. Kim Y.H., Ramakrishna S., Kim H., Kim J.S.. Enrichment of cells with TALEN-induced mutations using surrogate reporters. Methods. 2014; 69:108–117. [DOI] [PubMed] [Google Scholar]
- 16. Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.S.. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009; 19:1279–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guschin D.Y., Waite A.J., Katibah G.E., Miller J.C., Holmes M.C., Rebar E.J.. A rapid and general assay for monitoring endogenous gene modification. Methods Mol. Biol. 2010; 649:247–256. [DOI] [PubMed] [Google Scholar]
- 18. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bothmer A., Phadke T., Barrera L.A., Margulies C.M., Lee C.S., Buquicchio F., Moss S., Abdulkerim H.S., Selleck W., Jayaram H. et al. Characterization of the interplay between DNA repair and CRISPR/Cas9-induced DNA lesions at an endogenous locus. Nat. Commun. 2017; 8:13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ding Q., Regan S.N., Xia Y., Oostrom L.A., Cowan C.A., Musunuru K.. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell. 2013; 12:393–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maresca M., Lin V.G., Guo N., Yang Y.. Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res. 2013; 23:539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
National Center for Biotechnology Information (NCBI) Sequence Read Acrchive (SRA): The deep sequencing data from this study are available under accession code SRP135732. Source data for Figure 1.