Abstract
DNA interstrand crosslinks (ICLs) covalently join opposing strands, blocking both replication and transcription, therefore making ICL-inducing compounds highly toxic and ideal anti-cancer agents. While incisions surrounding the ICL are required to remove damaged DNA, it is currently unclear which endonucleases are needed for this key event. SNM1A has been shown to play an important function in human ICL repair, however its suggested role has been limited to exonuclease activity and not strand incision. Here we show that SNM1A has endonuclease activity, having the ability to cleave DNA structures that arise during the initiation of ICL repair. In particular, this endonuclease activity cleaves single-stranded DNA. Given that unpaired DNA regions occur 5′ to an ICL, these findings suggest SNM1A may act as either an endonuclease and/or exonuclease during ICL repair. This finding is significant as it expands the potential role of SNM1A in ICL repair.
INTRODUCTION
DNA damage by interstrand crosslinks (ICLs) inhibit strand separation required for transcription and replication. Failure to repair ICL damage leads to replication fork collapse, double-strand breaks, chromosomal aberrations, and the induction of apoptosis (1). As a result, ICL-inducing agents are effective, and often first-line, treatments for many cancers (1).
In eukaryotic cells, ICL lesions are largely recognized and repaired during S-phase when replication forks converge at an ICL lesion (2,3). The converged replication fork is recognized by the Fanconi anemia (FA) core complex which monoubiquitinates heterodimeric FANCI-FANCD2 (4). Ubiquitinated FANCI-FANCD2 recruits structure-specific endonucleases directly and indirectly though the scaffolding protein SLX4, which interacts with FANCD2 (Ub) (5). While incision on one strand commits the cell to repair, a second cut is required to fully unhook an ICL from duplex DNA. Lesion tolerant polymerases subsequently access the DNA and bypass the unhooked ICL using translesion synthesis (TLS) (6). Homologous recombination, using an undamaged sister chromatid, restarts the replication fork to complete repair (7). ICL repair that occurs outside of S-phase is FA-independent and relies on several factors involved in nucleotide excision repair (NER) (8). In this case, it is thought that ICLs are recognized during transcription or directly within distorted ICL-containing duplex DNA (9,10).
ICLs damage both DNA strands and therefore neither strand is readily available to act as template prior to ICL unhooking. It is thought that the endonuclease XPF-ERCC1 is recruited to the ICL and makes an incision during the unhooking of ICL damage (5,11). Which endonuclease makes the other incision to fully unhook the ICL is not clear (11). Prime candidates for this role include MUS81, FAN1 and SLX1; however, depletion and/or knockout of these endonucleases do not show the same hypersensitivity as XPF depletion (5,12–16). The only nuclease shown to be epistatic with XPF-ERCC1 is SNM1A, as knockdown of SNM1A and/or ERCC1 show similar hypersensitivity to ICL-inducing agents (17). Although this evidence suggests that SNM1A may act as an endonuclease in ICL unhooking, no endonuclease activity has been reported thus far (18,19).
SNM1A is a member of the β-CASP family of nucleases, defined by their metallo β-lactamase (MBL) and CASP homology domains (20,21). SNM1A is the functional homolog of yeast Pso2, which participates exclusively in the repair of ICLs in Saccharomyces cerevisiae (22,23). Pso2, like several other β-CASP nucleases, demonstrates both endonuclease and 5′-3′ phosphate-dependent exonuclease activity (24–29). Surprisingly, SNM1A has only been characterized as an exonuclease in vitro (18,19,22). Given the conserved dual nuclease activities of the β-CASP family, it is unclear why SNM1A would not function as an endonuclease as well.
Here, we report a single-strand specific endonuclease activity for human SNM1A. SNM1A cleaves DNA structures containing a region of single-stranded DNA in vitro and in yeast. We further show that SNM1A is able to process an ICL-containing substrate, initially as an endonuclease and then as an exonuclease. These findings expand the possible role of SNM1A beyond solely functioning as an exonuclease in repair.
MATERIALS AND METHODS
Cloning and transformation
Full length (1–1040) and truncated (698–1040) SNM1A were amplified by PCR and cloned into pDONR201 via Gateway cloning (Invitrogen). For protein expression in Escherichia coli, SNM1A was sub-cloned into pDEST-544 containing a N-terminal His6-tag, NusA fusion protein, and TEV protease cleavage site. pDest-544 was a gift from Dominic Esposito (Addgene plasmid # 11519). For protein complementation in yeast, SNM1A was sub-cloned into yeast expression vector pAG-423. pAG423GAL-ccdB was a gift from Susan Lindquist (Addgene plasmid # 14149). Escherichia coli strain TOP 10 (Invitrogen) was used for plasmid amplification. All plasmids and constructs generated in this study were verified by DNA sequencing (MOBIX Lab).
Endonuclease-dependent survival (transposon) assay
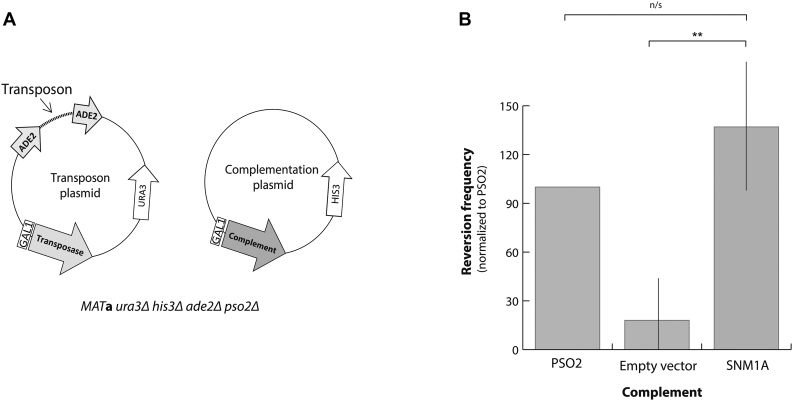
Complementation vectors were transformed into pso2Δ S. cerevisiae. Cultures were grown in histidine-lacking minimal media with 2% raffinose for 24 h at 30°C. Cells were induced by addition of 2% galactose then grown for another 24 h at 30°C. Cells were serially diluted in phosphate buffered saline and plated on histidine-lacking agar ± adenine. Plates were incubated at 30°C for three days and colonies on adenine-containing agar were enumerated to determine total number of cells. Colonies on adenine-lacking plates were incubated for 3 weeks before enumeration to determine reversion frequency. Relative reversion frequency was calculated by dividing colony forming units (CFUs) from the adenine-lacking plates by the CFUs from the adenine-containing plates, normalized to PSO2.
Protein expression and purification
SNM1ANΔ698, SNM1A NΔ698(D736A/H737A) and SNM1ANΔ608 were expressed in Star pRARE pLysS E. coli (Invitrogen), and induced at 0.700 OD600 with 1 mM IPTG at 25°C overnight. Cells were resuspended in Nickel A buffer (20 mM HEPES pH 7.5, 1 M NaCl, 30 mM imidazole, 0.5 mM TCEP, 10% glycerol) with protease inhibitors and lysed with four passes through a French press at 10 000 psi. The lysate was clarified by centrifugation and filtration. The sample was loaded onto a HisTrap Fast Flow nickel-chelating column (GE Healthcare) and eluted with 300 mM imidazole. The sample was diluted to 250 mM NaCl using Ion Exchange Buffer A (20 mM HEPES pH 7.5, 0.5 mM TCEP, 10% glycerol) then loaded onto a Q-sepharose HP column (GE Healthcare). SNM1A was eluted with a linear gradient from 250 to 650 mM NaCl. After pooling SNM1A-containing fractions, the sample was diluted to 250 mM NaCl with Ion Exchange Buffer A and treated with TEV protease overnight to remove the His6-NusA fusion protein. The sample was then run through an SP-HP sepharose column (GE Healthcare) and eluted with a linear gradient from 250 to 750 mM NaCl. Purified Pso2 was prepared as previously described in (25).
Preparation of structure-specific oligonucleotides
Oligonucleotides (Supplementary 1) were constructed with a 6-FAM or Cy3 fluorescent label (BioBasic) and purified using 20% denaturing PAGE. Substrates were annealed in 10 mM magnesium chloride, 10 mM Tris pH 7.5 and 100 mM NaCl. DNA requiring intramolecular interactions were annealed by heating to 90°C for 10 min and flash cooling on ice for 2–3 min. The remaining oligonucleotides were annealed using 1.5 molar equivalents of non-labeled complementary DNA and heated to 90°C for 10 min before slowly cooling to room temperature. Annealed DNA was native PAGE purified and re-suspended in water to 1 μM.
Preparation of site-specific interstrand crosslinked oligonucleotides
DNA (10 μM) was annealed as above. To crosslink DNA, dsDNA was incubated in 25 mM triethanolamine, 1 mM Na2EDTA, pH 7.2 with 40× excess of methanol-dissolved SJG-136 at 37°C overnight. Crosslinked DNA was ethanol precipitated and resuspended into formamide buffer for denaturing PAGE purification. Crosslinked DNA was detected at 526 nm using the GelDoc-EZ (BioRad) and bands were excised to separate crosslinked and uncrosslinked DNA. Crosslinked DNA was eluted at room temperature, ethanol precipitated and resuspended to 1 μM.
Nuclease assays
SNM1A nuclease assays were performed in 50 mM Tris acetate pH 7.2, 75 mM potassium acetate, 10 mM magnesium chloride, 1 mM dithiothreitol, 100 μg/μl BSA with ∼0.1 μM of fluorescently-labeled DNA. Pso2 reactions were performed with 50 mM sodium chloride, 10 mM Tris-hydrochloride pH 7.9, 10 mM magnesium chloride, 1 mM DTT. Reactions were initiated by the addition of protein, incubated at 37°C, then stopped with formamide loading buffer (95% formamide, 10 mM EDTA). Products of were separated using 20% denaturing PAGE and detected at 526 nm using the Typhoon Imager (GE Healthcare) or ChemiDoc-XRS (BioRad).
RESULTS
SNM1A complements Pso2 endonuclease activity in yeast
Pso2 has been shown to have endonuclease activity in vitro and in vivo (18,25). Since SNM1A has been shown to be the functional homolog of Pso2 (22), we reasoned that SNM1A might also possess endonuclease activity. To test this possibility, we used a previously described yeast assay that monitors cell survival dependent on the activity of an endonuclease (25,30). Briefly, ade2Δ yeast were transformed with a vector containing an inducible transposase and an ADE2 gene interrupted by insertion of a transposable element (Figure 1A). Induction of transposition results in incorporation of residual hairpin-capped ends within the ADE2 gene that require endonuclease processing prior to restoration. Failure to repair the ADE2 gene results in sensitivity to adenine depletion. In the absence of PSO2, ADE2 reversion frequency is significantly reduced (Figure 1B), however, complementation with PSO2 or SNM1A restores ADE2 reversion. The small deviation between PSO2 and SNM1A reversion frequency likely reflects differences in gene expression and/or regulation. Nevertheless, this result clearly suggests that SNM1A protein possesses hairpin-opening endonuclease activity similar to Pso2.
Figure 1.
SNM1A has hairpin-opening endonuclease activity. (A) Vectors for hairpin-opening assay in yeast. (B) Analysis of SNM1A endonuclease activity in pso2Δ yeast. Reversion frequency was normalized to PSO2 complemented yeast. Assays were performed in quadruplicate where error bars show the standard deviation for all assays. Pso2 expression was below detection by monoclonal anti-polyHis antibody (Sigma: H1029). ** denotes a P-value <0.01 compared to empty vector using one-tail t-test.
Characterization of SNM1A endonuclease activity
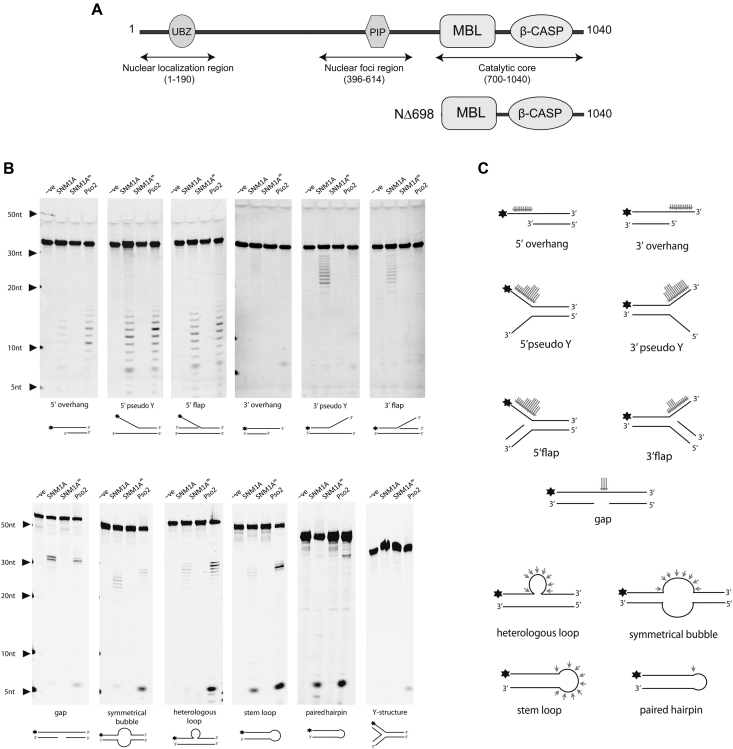
Since SNM1A complemented Pso2 endonuclease activity in yeast, we further characterized the specificity of SNM1A endonuclease activity in vitro. The catalytic domain of SNM1A (Figure 2A; residues 698-1040) was expressed and purified (Supplementary 2). As previously reported (18,19,22,31), SNM1A demonstrated phosphate-dependent 5′-3′ exonuclease activity (Supplementary 3A). In order to specifically isolate any potential endonuclease activity of SNM1A, modifications to DNA oligonucleotide substrates were required. 5′-Fluorophore-labeled (5′F) substrates were constructed to ensure 5′ exonuclease digestion of the initial substrate was blocked and any products resulting from endonuclease activity would accumulate. Using this strategy, a panel of substrates was designed to encompass a variety of intermediates that possibly arise during replication fork collapse and ICL repair, including overhangs, flaps, gaps, bubbles and hairpins. Wild-type SNM1A, but not SNM1AD736A/H737A (SNM1AM) digested these substrates (Figure 2B; Supplementary 3B), indicating that SNM1A possess endonuclease activity. Cleavage analysis on all substrates (Figure 2C) further demonstrated SNM1A exclusively cuts regions of unpaired DNA. Pso2 was tested in parallel for comparative analysis. Overall, cleavage patterns for SNM1A and Pso2 were very similar as one would expect for functional homologs (Figure 2B).
Figure 2.
SNM1A has structure-specific endonuclease activity. (A) Domain boundaries of SNM1A construct. (B) In vitro analysis of SNM1A endonuclease activity. SNM1A (0.18 μM), SNM1AM with catalytic mutations D736A/H737A (0.18 μM) and Pso2 (0.45 μM) were incubated with 5′ labeled DNA (0.1 μM). Reactions were stopped after 120 minutes. Products were resolved using 20% denaturing PAGE and imaged with Chemi-Doc (BioRad) or Typhoon Imager (GE Healthcare) at 526 nm. (C) Summary of SNM1A endonuclease cleavage events. Cleavage efficiency from at least three independent experiments were quantified using ImageLab or ImageJ. Long, medium, and short arrows indicate the relative efficiency of cleavage at that position.
To ensure that observed nuclease activities were SNM1A-dependent, a catalytic mutant (SNM1AM) was purified and assayed in parallel with wildtype. SNM1AM failed to generate any nuclease products on all substrates tested (Figure 2B; Supplementary 3B). Importantly, all endonuclease and 5′ phosphate-dependent exonuclease activities were co-purified only in fractions containing SNM1A (Supplementary 2), once again highlighting the SNM1A-dependence of these activities.
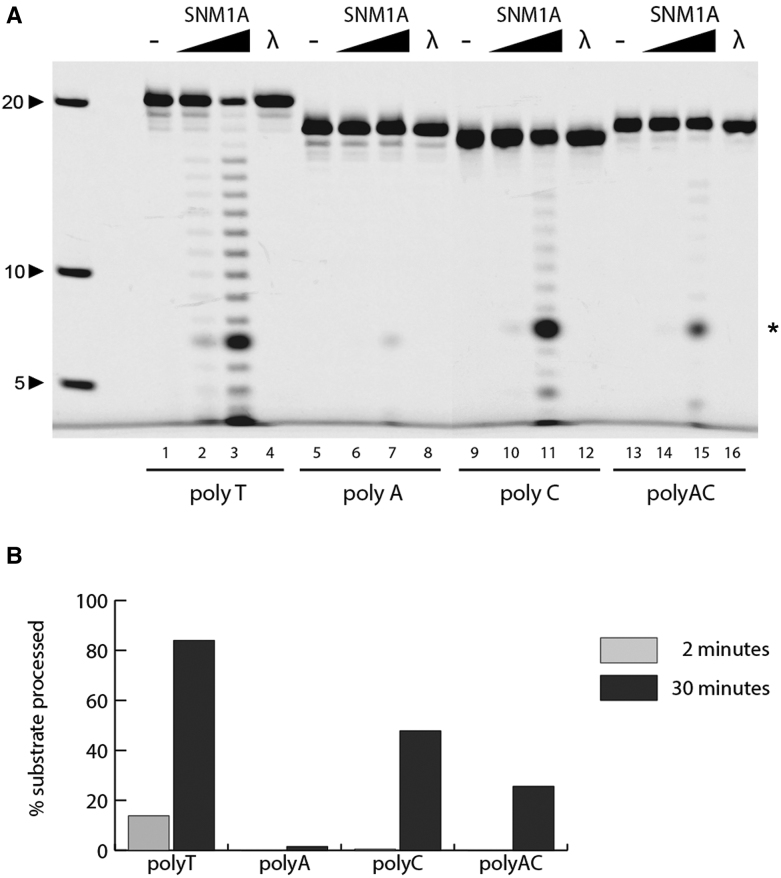
To further define the minimal substrate requirements for SNM1A endonuclease activity, 5′-fluorophore-labeled ssDNA homopolymers were generated and tested as before (Figure 3). Lambda exonuclease was included as a negative control, demonstrating the 5′ fluorophore was sufficient to block 5′-exonuclease activity. SNM1A cleaved polyT and polyC substrates with higher efficiency compared to a polyA substrate (Figure 3B), suggesting an overall preference for endonuclease cleavage at pyrimidine bases. This observation is consistent with the endonuclease base preference reported for another DNA β-CASP nuclease, Artemis (32).
Figure 3.
SNM1A has single-strand specific endonuclease activity. (A) In vitro time course analysis of SNM1A nuclease activity on 5′ fluorescently-labeled ssDNA. SNM1A (0.2 μM) or Lambda exonuclease (5 units) were incubated with indicated ssDNA (0.1 μM). Reactions were stopped after 0, 2 or 30 min. Products were resolved using 20% denaturing PAGE and imaged with the ChemiDoc XRS (BioRad) at 526 nm. (B) Quantification of substrate processed at indicated time points. Substrates were quantified using ImageLab. * denotes an aberrant fluorophore-dependent product (Supplementary 4).
SNM1A processes ICL lesions using both endonuclease and exonuclease activity
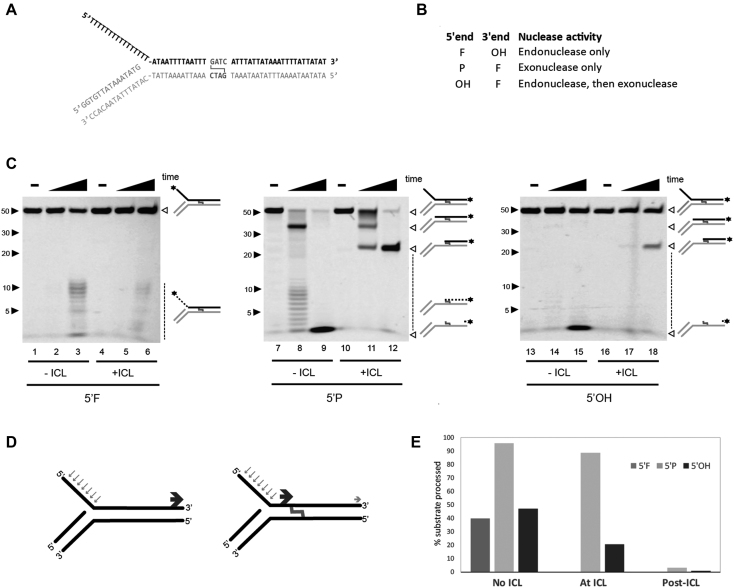
Currently, the role of SNM1A in ICL repair has been limited to acting as an exonuclease based on activity observed in vitro (12,17,33). Given the finding here that SNM1A has both endonuclease and exonuclease activity, we tested the possibility that both activities could contribute to the processing of an ICL lesion. We created a substrate to emulate ICL-dependent replication fork stalling with a 5′ flap and a downstream ICL incorporated in the duplex region (Figure 4A). The single-stranded 5′ flap and 3′ duplex DNA of the fork represents the lagging and leading strands, respectively. SJG-136 was used to generate a crosslink since this ICL is heat reversible and facilitated analysis of products. Analysis of 5′ or 3′ labeled substrates permitted comparison of distinct SNM1A functions: endonuclease, exonuclease, and translesional (Figure 4B). The purity of the crosslinked substrates is demonstrated in Supplementary 5. As shown in Figure 4C, uncrosslinked and crosslinked substrates containing a 5′ fluorophore (Figure 4C, lanes 1–3 and lanes 4–6, respectively) generated a similar series of endonuclease products within the unpaired region. Some preference for cleavage was apparent close to the junction between single- and double-strand DNA. When the same uncrosslinked substrate contained a 5′ phosphate (5′P) and was labeled on the 3′ end, fully digested products indicative of 5′ exonuclease activity were observed (Figure 4C, lanes 8-9). Incorporating an ICL in the 5′ phosphate substrate resulted in a strong block of exonuclease progression at the crosslink (Figure 4C, lanes 11 and 12). The fact that products were not readily observed past the ICL suggests that SNM1A failed to use its exonuclease activity to bypass the ICL under these conditions. Finally, using a flap substrate containing a 5′hydroxyl (5′OH) and 3′ label, SNM1A initiated processing first as an endonuclease and subsequently acted as an exonuclease, trimming up to the ICL crosslink. Taken together, these findings demonstrate that both endonuclease and exonuclease activities of SNM1A can be used to process an ICL-containing substrate.
Figure 4.
SNM1A can generate a nick to initiate trimming to the crosslink. (A) DNA substrate design mimicking stalled replication fork. (B) Substrate end modifications and corresponding nuclease activities monitored. F, fluorophore; OH, hydroxyl; P, phosphate. (C) In vitro analysis of SNM1A nuclease activities on 5′flap crosslinked DNA. SNM1A (0.4 μM) was incubated with substrate (0.1μM) as shown in (A) with or without an SJG-136 crosslink. Substrates are labeled as indicated in (B). Reactions were stopped after 0, 2 and 30 min. Products were resolved using 20% denaturing PAGE and imaged with the ChemiDoc XRS (BioRad) at 526 nm. (D) Summary of nuclease events on 5′ flap substrate with or without an SJG crosslink. Blue arrows indicate initial endonuclease cleavage. Dark gray arrows indicate exonuclease product. Light grey arrow indicates translesional nuclease product. (E) Quantification of product formed in the absence of the ICL, at the ICL, or past the ICL. Products were quantified using ImageLab.
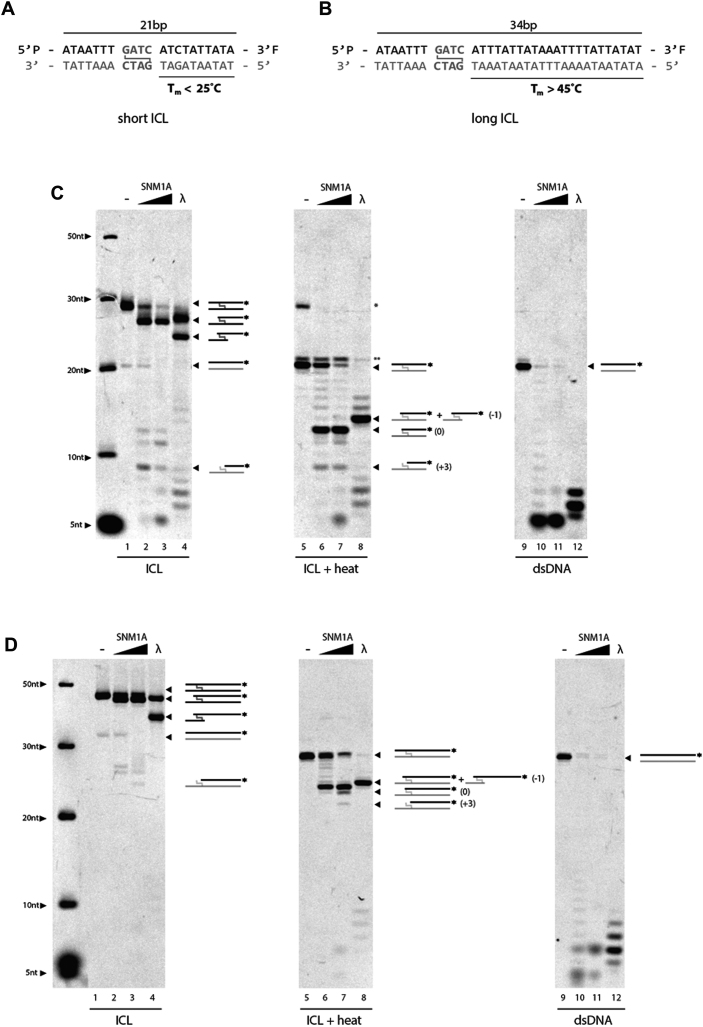
Given that SNM1A had been previously shown to bypass ICL adducts using exonuclease activity, we wondered why SNM1A failed to have the same activity under experimental conditions used here. We first considered possible differences in substrate design. As shown in Figure 5, we observed that the integrity of an SJG-136 crosslink was dependent on substrate length. When the crosslink was placed in the context of shorter 21 bp duplex (with lower thermal stability) (Figure 5A), the crosslink failed to fully block exonuclease activity of SNM1A, as shown with both intact (Figure 5C, lanes 2 and 3), as well as heat-reversed crosslinked products (Figure 5C, lanes 6 and 7) past the ICL. However, when the same crosslink was placed into longer 34 bp DNA (Figure 5B), the crosslink more efficiently blocked SNM1A exonuclease activity (Figure 5D, lanes 2, 3, 6 and 7). Consistent with this trend, the substrate with 41 bp duplex used in Figure 4 fully blocked translesion exonuclease activity. We also considered the possibility that the substrate length influenced crosslink stability. Since Lambda exonuclease lacks the ability to bypass DNA lesions (34), substrates were tested for activity by Lambda exonuclease. Strikingly, Lambda exonuclease generated products beyond the ICL in Figure 5C (lanes 4 and 8) and 5D (lanes 4 and 8), and the extent of digestion was dependent on substrate length, suggesting these lower molecular weight products are not indicative of translesion processing. The fact that both Lambda and SNM1A behaved the same in regard to their ability to bypass the covalent bond by SJG-136 further suggests that the block observed in SNM1A translesion activity (Figure 4C) was substrate-, and not protein-, dependent. Given this finding, it is likely that the efficiency of SNM1A translesion exonuclease activity will be dependent on the chemical nature and overall structure of the crosslink encountered.
Figure 5.
Stability of SJG-136 crosslinks are DNA length-dependent. Short (A) and long (B) ICL substrates with 3′ fluorescent label. In vitro analysis of SNM1A translesional nuclease activity on short ICL (C) or long ICL (D). SNM1A (0.2 μM) or Lambda exonuclease (5 units) were incubated with duplex DNA substrate (0.2 μM) with (left and middle; lanes 1–8) or without (right; lanes 9–12) an SJG-136 crosslink. Reactions were stopped after 0, 5 and 60 min. Crosslinked products were heat denatured (shown in middle panels) to disrupt the crosslink and allow visualization of the top strand alone. A schematic of products is shown with the dark strand represents the labelled DNA visible on the gel. Products were resolved using 20% denaturing PAGE and imaged with the ChemiDoc XRS (BioRad) at 526 nm. * denotes residual crosslinked DNA following heat treatment. ** denotes unphosphorylated substrate.
DISCUSSION
Repair of ICL damage remains a poorly understood process despite its importance for preventing Fanconi anemia and cancer. Although many proteins participate in ICL repair, perhaps the most important are those that initiate repair by forming incisions on either side of the ICL lesion. Several nucleases have been implicated in lesion processing; of these, only XPF-ERCC1 endonuclease has been demonstrated to generate strand incisions required for lesion unhooking (11,35). Although SNM1A has been shown to be epistatic with XPF-ERCC1 in response to ICL damage (17), its role in repair remains unclear. Results presented here demonstrate SNM1A possesses endonuclease activity that is sufficient for partial unhooking of an ICL lesion.
Endonuclease activity of SNM1A
The importance of SNM1A in human ICL repair has only recently been appreciated (17), despite its yeast homologue (Pso2) having been shown to play an essential role in ICL repair (23,36,37). Prior studies demonstrating that SNM1A is able to rescue ICL repair defects in pso2-deficient cells suggest that SNM1A and Pso2 share common activities required for ICL processing (22). Although both SNM1A and Pso2 exhibit 5′ phosphate-dependent exonuclease activity (18,24) only Pso2 has been reported to possess endonuclease function (25).
With this in mind, we tested the possibility that SNM1A may also function as an endonuclease. SNM1A has been shown to complement Pso2 in response to ICL damage (22); however, whether this was due to exonuclease or endonuclease activity was not tested. To more directly investigate if SNM1A could function as an endonuclease in yeast, we used an assay that monitors the ability to open DNA hairpins. In this assay, repair of DNA hairpins is Pso2-dependent (30). The ability of SNM1A to complement a pso2 deletion (Figure 1) suggested that SNM1A may function as an endonuclease. To further investigate this possibility, the catalytic domain of SNM1A (residues 698–1040) was purified and directly tested for endonuclease activity. SNM1A was found to nick DNA substrates containing unpaired regions, confirming its ability to act as an endonuclease. The finding that SNM1A contains endonuclease activity similar to Pso2 (Figure 2) is significant, because it is the first demonstration that these proteins, which are able to complement for repair of ICL lesions in yeast, share both exonuclease and endonuclease activity. This finding opens the possibility that either exonuclease and/or endonuclease activity may be required for ICL repair. Further studies using SNM1A separation-of-function mutants will be required to address the biological contribution of each nuclease activity.
The finding that SNM1A displays endonuclease activity was somewhat surprising considering previous reports had suggested otherwise (18,19). Failure to detect endonuclease activity in prior studies likely resulted from differences in substrate design. Since SNM1A exonuclease activity is significantly more robust than its endonuclease activity (Supplementary 6), substrates labelled at the 3′ end fail to capture this activity. Endonuclease products can only be directly observed using 5′ fluorescently-labeled substrates since these products are not turned over by 5′-3′ exonuclease activity (Figures 2 and 3). In addition, differences in reported nuclease activity may reflect variation in protein construct design between studies. Prior studies analyzed full length (18) or truncated SNM1A (residues 608–1040) (19). We found that addition of residues N-terminal (residues 608–698) to the catalytic domain (698–1040) reduced both exonuclease and endonuclease activity (Supplementary 7).
Implications for SNM1A endonuclease function in ICL repair
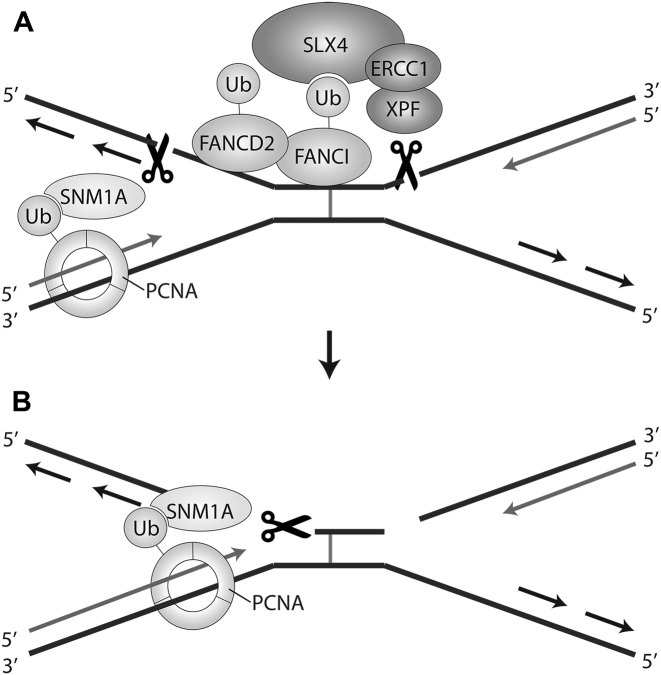
A longstanding question in the field of ICL repair is which endonucleases are required for generating nicks on both sides of an ICL during unhooking. Work presented here suggests that the endonuclease activity of SNM1A may participate in ICL unhooking. SNM1A was sufficient to initiate processing of an ICL substrate mimicking a stalled replication fork (Figure 4). Although several endonucleases could potentially function as an endonuclease in ICL unhooking, only SNM1A has been shown to work in the same pathway as XPF-ERCC1, which is thought to make a nick during unhooking (17). In agreement with the observed epistasis, it has been suggested that XPF-ERCC1 may create an initial nick that serves as substrate for SNM1A 5′ exonuclease action (12,17,33). This mechanism is not consistent, however, with the preferred 3′ flap endonuclease activity of XPF (38). If XPF-ERCC1 were to act on the 3′ flap of the leading strand created by convergent replication forks, the nick would be 3′ to the ICL, resulting in SNM1A exonuclease trimming away from the ICL. During replication-dependent ICL repair, regions of unpaired DNA occur on the lagging strand 5′ to the ICL, providing a potential substrate for SNM1A endonuclease activity. A nick generated by SNM1A endonuclease activity in the lagging strand could generate the necessary 5′ phosphate for SNM1A exonuclease activity to further trim the strand up to and beyond the ICL, as illustrated in Figure 6.
Figure 6.
Model for SNM1A nuclease activities in replication-dependent ICL repair. Monoubiquitination of FANCD2-FANCI recruits SLX4, which in turn recruits 3′ flap endonuclease XPF-ERCC1 to create the first ICL unhooking incision. SNM1A is recruited to the site of repair, possibly through PCNA interaction, cutting unpaired DNA on the lagging strand 5′ to the ICL. SNM1A could then trim the DNA using 5′-3′ exonuclease activity. The ICL could then be further bypassed by SNM1A translesion exonuclease activity (33) or TLS polymerases (6).
Replication protein A (RPA) is known to coat ssDNA on the lagging strand, potentially limiting access by SNM1A. While it is not known if SNM1A directly binds RPA, SNM1A does interact with 53BP1, which mediates RPA hyper-phosphorylation in response to DNA damage (39,40). Signalling among these proteins may result in RPA displacement, allowing SNM1A access to ssDNA for strand cleavage. Since RPA has been shown to activate cleavage by XPF (33,38), we speculate that nicking by XPF could provide a signal for further incision, similar to what has been demonstrated for the coupled incisions by XPF and XPG, mediated through RPA positioning during NER (41).
Given the central role of unhooking in ICL repair, it is not surprising that functional redundancy would exist among endonucleases. Other nucleases that act on a 5′ flap could substitute for SNM1A endonuclease cleavage. The best candidate for this endonuclease is FAN1 (42). SNM1A and FAN1 share both endonuclease and 5′ phosphate-dependent exonuclease activity (13,43), and appear to be recruited to ICL damage by interaction with PCNA (44,45). Redundancy between SNM1A and FAN1 may help ensure 5′ cleavage and further ICL trimming occur efficiently in a variety of contexts. In support of this relationship, a double knockout of pso2 and fan1 results in extreme hypersensitivity in S. pombe, while single knockouts result in milder phenotypes (46). Recent studies using mouse embryonic fibroblasts have also demonstrated functional redundancy of FAN1 and SNM1A in response to ICL damage (47). Further studies will be required to fully understand how SNM1A and FAN1 activities are coordinated during ICL repair.
The finding here that SNM1A has both exonuclease and endonuclease activity opens new possibilities for how SNM1A may function in repair of ICL damage. Further investigation, using separation-of-function mutants, will be required to more clearly define the contribution of each nuclease activity in a more biological context.
Supplementary Material
ACKNOWLEDGEMENTS
Thank you to Tracy Tiefenbach and Wendy Chen for work on Pso2, and Michelle Dowling for additional work on ICL substrates. We also thank Alexa Mordherst, Maryan Graiss, Sarah Zhou and Justin Ching-Johnson for contributions on this SNM1A project. We also thank Dr Sara Andres for comments on the initial manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institute of Health Research [MOP-89903]; Canadian Cancer Society [702157]. Funding for open access charge: Canadian Institute of Health Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Deans A.J., West S.C.. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011; 11:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang J., Dewar J.M., Budzowska M., Motnenko A., Cohn M.A., Walter J.C.. DNA interstrand cross-link repair requires replication-fork convergence. Nat. Struct. Mol. Biol. 2015; 22:242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Knipscheer P., Räschle M., Schärer O.D., Walter J.C.. Replication-Coupled DNA interstrand Cross-Link repair in xenopus egg extracts. Methods Mol. Biol. (Clifton, N.J.). 2012; 920:221–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim H., D’Andrea A.D.. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev. 2012; 26:1393–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang J., Walter J.C.. Mechanism and regulation of incisions during DNA interstrand cross-link repair. DNA Repair (Amst). 2014; 19:135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ho T.V., Schärer O.D.. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 2010; 51:552–566. [DOI] [PubMed] [Google Scholar]
- 7. Hinz J.M. Role of homologous recombination in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 2010; 51:582–603. [DOI] [PubMed] [Google Scholar]
- 8. Wood R.D. Mammalian nucleotide excision repair proteins and interstrand crosslink repair. Environ. Mol. Mutagen. 2010; 51:520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kato N., Kawasoe Y., Williams H., Gottesman M.E., Takahashi T.S., Correspondence J.G., Coates E., Roy U., Shi Y., Beese L.S. et al. Sensing and processing of DNA interstrand crosslinks by the mismatch repair pathway. Cell Rep. 2017; 21:1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Enoiu M., Jiricny J., Schärer O.D.. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic Acids Res. 2012; 40:8953–8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klein Douwel D., Boonen R.A.C.M., Long D.T., Szypowska A.A., Räschle M., Walter J.C., Knipscheer P.. XPF-ERCC1 Acts in unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol. Cell. 2014; 54:460–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sengerová B., Wang A.T., McHugh P.J.. Orchestrating the nucleases involved in DNA interstrand cross-link (ICL) repair. Cell Cycle. 2011; 10:3999–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. MacKay C., Déclais A.-C., Lundin C., Agostinho A., Deans A.J., MacArtney T.J., Hofmann K., Gartner A., West S.C., Helleday T. et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010; 142:65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hanada K., Budzowska M., Modesti M., Maas A., Wyman C., Essers J., Kanaar R.. The structure-specific endonuclease Mus81–Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006; 25:4921–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhou W., Otto E.A., Cluckey A., Airik R., Hurd T.W., Chaki M., Diaz K., Lach F.P., Bennett G.R., Gee H.Y. et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat. Genet. 2012; 44:910–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castor D., Nair N., Déclais A.-C., Lachaud C., Toth R., Macartney T.J., Lilley D.M.J., Arthur J.S.C., Rouse J.. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol. Cell. 2013; 52:221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang A.T., Sengerová B., Cattell E., Inagawa T., Hartley J.M., Kiakos K., Burgess-Brown N.A., Swift L.P., Enzlin J.H., Schofield C.J. et al. Human SNM1a and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes Dev. 2011; 25:1859–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hejna J., Philip S., Ott J., Faulkner C., Moses R.. The hSNM1 protein is a DNA 5′-exonuclease. Nucleic Acids Res. 2007; 35:6115–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sengerová B., Allerston C.K., Abu M., Lee S.Y., Hartley J., Kiakos K., Schofield C.J., Hartley J.A., Gileadi O., McHugh P.J.. Characterization of the human SNM1A and SNM1B/Apollo DNA repair exonucleases. J. Biol. Chem. 2012; 287:26254–26267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Callebaut I., Moshous D., Mornon J.-P., de Villartay J.-P.. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 2002; 30:3592–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cattell E., Sengerová B., McHugh P.J.. The SNM1/Pso2 family of ICL repair nucleases: from yeast to man. Environ. Mol. Mutagen. 2010; 51:635–645. [DOI] [PubMed] [Google Scholar]
- 22. Hazrati A., Ramis-Castelltort M., Sarkar S., Barber L.J., Schofield C.J., Hartley J.A., McHugh P.J.. Human SNM1A suppresses the DNA repair defects of yeast pso2 mutants. DNA Repair (Amst). 2008; 7:230–238. [DOI] [PubMed] [Google Scholar]
- 23. Henriques J.A.P., Moustacchi E.. Isolation and characterization of pso mutants sensitive to photo-addition of psoralen derivatives in Saccharomyces cerevisiae. Genetics. 1980; 95:273–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li X., Hejna J., Moses R.E.. The yeast Snm1 protein is a DNA 5 -exonuclease. DNA Repair (Amst). 2005; 4:163–170. [DOI] [PubMed] [Google Scholar]
- 25. Tiefenbach T., Junop M.. Pso2 (SNM1) is a DNA structure-specific endonuclease. Nucleic Acids Res. 2012; 40:2131–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li S., Chang H.H., Niewolik D., Hedrick M.P., Pinkerton A.B., Hassig C.A., Schwarz K., Lieber M.R.. Evidence that the DNA endonuclease ARTEMIS also has intrinsic 5′-exonuclease activity. J. Biol. Chem. 2014; 289:7825–7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de la Sierra-Gallay I.L., Zig L., Jamalli A., Putzer H.. Structural insights into the dual activity of RNase J. Nat. Struct. Mol. Biol. 2008; 15:206–212. [DOI] [PubMed] [Google Scholar]
- 28. Mandel C.R., Kaneko S., Zhang H., Gebauer D., Vethantham V., Manley J.L., Tong L.. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006; 444:953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ma Y., Pannicke U., Lu H., Niewolik D., Schwarz K., Lieber M.R.. The DNA-dependent protein kinase catalytic subunit phosphorylation sites in human Artemis. J. Biol. Chem. 2005; 280:33839–33846. [DOI] [PubMed] [Google Scholar]
- 30. Yu J., Marshall K., Yamaguchi M., Haber J.E., Weil C.F.. Microhomology-dependent end joining and repair of transposon-induced DNA hairpins by host factors in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004; 24:1351–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Allerston C.K., Lee S.Y., Newman J.A., Schofield C.J., McHugh P.J., Gileadi O.. The structures of the SNM1A and SNM1B/Apollo nuclease domains reveal a potential basis for their distinct DNA processing activities. Nucleic Acids Res. 2015; 43:11047–11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu J., Li S., Zhang X., Wang L.-C., Niewolik D., Schwarz K., Legerski R.J., Zandi E., Lieber M.R.. DNA-PKcs regulates a single-stranded DNA endonuclease activity of Artemis. DNA Repair (Amst). 2010; 9:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdullah U.B., Mcgouran J.F., Brolih S., Ptchelkine D., El-Sagheer A.H., Brown T., Mchugh P.J.. RPA activates the XPF-ERCC1 endonuclease to initiate processing of DNA interstrand crosslinks. EMBO J. 2017; 36:2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattes W.B. Lesion selectivity in blockage of lambda exonuclease by DNA damage. Nucleic Acids Res. 1990; 18:3723–3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhagwat N., Olsen A.L., Wang A.T., Hanada K., Stuckert P., Kanaar R., D’Andrea A., Niedernhofer L.J., McHugh P.J.. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol. Cell. Biol. 2009; 29:6427–6437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonatto D., Revers L.F., Brendel M., Henriques J.A.P.. The eukaryotic Pso2/Snm1/Artemis proteins and their function as genomic and cellular caretakers. Brazilian J. Med. Biol. Res. = Rev. Bras. Pesqui. medicas e Biol. 2005; 38:321–334. [DOI] [PubMed] [Google Scholar]
- 37. Siede W., Brendel M.. Isolation and characterization of yeast mutants with thermoconditional sensitivity to the bifunctional alkylating agent nitrogen mustard. Curr. Genet. 1981; 4:145–149. [DOI] [PubMed] [Google Scholar]
- 38. de Laat W.L., Appeldoorn E., Jaspers N.G., Hoeijmakers J.H.. DNA structural elements required for ERCC1-XPF endonuclease activity. J. Biol. Chem. 1998; 273:7835–7842. [DOI] [PubMed] [Google Scholar]
- 39. Yoo E., Kim B.U., Lee S.Y., Cho C.H., Chung J.H., Lee C.-H.. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene. 2005; 24:5423–5430. [DOI] [PubMed] [Google Scholar]
- 40. Richie C.T., Peterson C., Lu T., Hittelman W.N., Carpenter P.B., Legerski R.J.. hSnm1 colocalizes and physically associates with 53BP1 before and after DNA damage. Mol. Cell. Biol. 2002; 22:8635–8647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsunaga T., Park C.H., Bessho T., Mu D., Sancar A.. Replication protein A confers structure-specific endonuclease activities to the XPF-ERCC1 and XPG subunits of human DNA repair excision nuclease. J. Biol. Chem. 1996; 271:11047–11050. [DOI] [PubMed] [Google Scholar]
- 42. Zhao Q., Xue X., Longerich S., Sung P., Xiong Y.. Structural insights into 5′ flap DNA unwinding and incision by the human FAN1 dimer. Nat. Commun. 2014; 5:5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pizzolato J., Mukherjee S., Schärer O.D., Jiricny J.. FANCD2-associated nuclease 1, but not exonuclease 1 or flap endonuclease 1, is able to unhook DNA interstrand Cross-links in Vitro. J. Biol. Chem. 2015; 290:22602–22611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang K., Moldovan G.-L., D’Andrea A.D.. RAD18-dependent recruitment of SNM1A to DNA repair complexes by a ubiquitin-binding zinc finger. J. Biol. Chem. 2010; 285:19085–19091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Porro A., Berti M., Pizzolato J., Bologna S., Kaden S., Saxer A., Ma Y., Nagasawa K., Sartori A.A., Jiricny J.. FAN1 interaction with ubiquitylated PCNA alleviates replication stress and preserves genomic integrity independently of BRCA2. Nat. Commun. 2017; 8:1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fontebasso Y., Etheridge T.J., Oliver A.W., Murray J.M., Carr A.M.. The conserved Fanconi anemia nuclease Fan1 and the SUMO E3 ligase Pli1 act in two novel Pso2-independent pathways of DNA interstrand crosslink repair in yeast. DNA Repair (Amst). 2013; 12:1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thongthip S., Bellani M., Gregg S.Q., Sridhar S., Conti B.A., Chen Y., Seidman M.M., Smogorzewska A.. Fan1 deficiency results in DNA interstrand cross-link repair defects, enhanced tissue karyomegaly, and organ dysfunction. Genes Dev. 2016; 30:645–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.