Abstract
RNA editing is one of the most common RNA level modifications that potentially generate amino acid changes similar to those resulting from genomic nonsynonymous mutations. However, unlike DNA level allele-specific modifications such as DNA methylation, it is currently unknown whether RNA editing displays allele-specificity across tissues and species. Here, we analyzed allele-specific RNA editing in human tissues and from brain tissues of heterozygous mice generated by crosses between divergent mouse strains and found a high proportion of overlap of allele-specific RNA editing sites between different samples. We identified three allele-specific RNA editing sites cause amino acid changes in coding regions of human and mouse genes, whereas their associated SNPs yielded synonymous differences. In vitro cellular experiments confirmed that sequences differing at a synonymous SNP can have differences in a linked allele-specific RNA editing site with nonsynonymous implications. Further, we demonstrate that allele-specific RNA editing is influenced by differences in local RNA secondary structure generated by SNPs. Our study provides new insights towards a better comprehension of the molecular mechanism that link SNPs with human diseases and traits.
INTRODUCTION
The regulation of biallellic gene expression is important for development in diploid organisms, and is associated with several human diseases (1). A large class of genes have been demonstrated to express only one allele through processes including random monoallelic expression (2,3), parental-specific (imprinted) expression (4–7), and allele sequence-specific expression (7). Analyses of single cells from mouse preimplantation embryos revealed that from 12 to 24% of autosomal genes appear to have random or dynamic monoallelic expression (3). In addition, hundreds of autosomal genes have been demonstrated to show imprinted and allele sequence-specific expression (6,7). Allele-specific DNA methylation is one of the factors that control allele-specific expression (8), and has been extensively explored in previous studies (9–11). These studies mainly focused on modifications that influence allele-specific regulation at the DNA level, whereas at the RNA level, important modification, such as RNA editing, which can generate RNA changes similar to genomic mutations has not been studied. Adenosine to inosine (A to I (G)) editing, which is catalyzed by the double-stranded RNA dependent ADAR1 and ADAR2 proteins, is the most common RNA editing event (12,13). The majority of RNA editing sites are located in non-coding regions of RNA transcripts, especially in inverted pairs of Alu repeats, where there is a higher propensity for the formation of double-stranded RNA structures (14,15). Other less prevalent types of RNA editing have been reported, for instance the C to U type, which is meditated by Apobec-1 using single-stranded RNA as substrate (16,17). Additionally, mismatches, bulges and loops affect the structure of double or single-stranded RNA and thus contribute to the specificity of editing (18–20). The level of editing, on the other hand, has been shown to be associated with genetic variants in natural populations of Drosophila melanogaster (21). RNA editing QTL and allele-specific RNA editing have been examined in 445 lymphoblastoid cell lines (22). However, whether modifications occurring at the RNA level, such as RNA editing, show allele-specificity across tissues and species is currently unknown.
Here, we identified RNA editing sites from three sources: (a) diverse human tissues (23), (b) brain tissues from mice generated by reciprocal crosses between three inbreed strains representing different subspecies (7) and (c) human U87MG cell line. Single nucleotide polymorphisms (SNPs) facilitated the identification of allele-specific RNA editing sites in each individual using human and mouse transcriptome data. Our findings reveal that some allele-specific RNA editing sites lead to amino acid changes in the coding regions of genes, while their associated SNPs only cause synonymous changes. Taking advantage of these allele-specific RNA editing sites, we discovered that SNPs surrounding RNA editing sites can affect secondary structure of RNA, which leads to changes in the allele-specificity of RNA editing.
MATERIALS AND METHODS
Human and mouse data
We downloaded human body epigenome map datasets (Gene Expression Omnibus (GEO) accession number GSE16256) that included 37 transcriptomes from human tissues and their matched genomic sequences from four individuals (23). The transcriptome data consisted of 6.1G reads that were sequenced as 100 bp paired-end reads. These transcriptomes and the respective genomic sequences were combined for the identification of allele-specific RNA editing sites.
A total of 67 RNA-seq datasets (Sequence Read Archive (SRA) accession code SRP056236) generated from brain tissues of reciprocal F1 hybrids generated from three wild mice strains CAST/EiJ, PWK/PhJ, and WSB/EiJ, had 2.6G paired-end reads with a length of 100 bp (7). SNPs from these three strains were identified from the downloaded Genome Sequences of Laboratory Mice Project (24). The RNA-seq dataset and SNPs were used to detect allele-specific RNA editing sites in the mouse brain.
Cell culture, DNA/RNA purification and sequencing from human U87MG cells
Human U87MG cells were obtained from ATCC and grown in DMEM with 10% fetal bovine serum (FBS) and 1% l-glutamine. Genomic DNA and total RNA were extracted together using the DNA/RNA Isolation Kit (TianGen) according to the manufacturer's protocol. The standard Illumina protocol was used to construct libraries for DNA-seq and RNA-seq on the Illumina HiSeq X ten platform. DNA-seq and RNA-seq yielded 104M reads and 106M pair-end reads, respectively, with lengths of about 150 bp.
RNA editing sites
RNA editing sites for human and mouse genomes were downloaded from the RADAR RNA editing database (v2) (25). Genomic position of the RNA editing sites were converted from hg19 or mm9 coordinates to hg38 or mm10 using the liftover tool (http://genome.ucsc.edu/cgi-bin/hgLiftOver).
Detecting allele-specific RNA editing sites
To obtain high-confidence allele-specific RNA editing sites, we first trimmed low-quality bases from the RNA-seq and DNA-seq reads using Trimmomatic 0.33 with default parameters (26), and aligned the passed reads to the reference genome using BWA mem 0.7.12 with default parameters (27). Duplicate reads were removed by Picard (https://broadinstitute.github.io/picard/).
For the human DNA-seq data, we first used Genome Analysis Toolkit (GATK) to call heterozygous SNPs for each human sample (28,29). SNPs were then filtered using the GATK variant filter module with a hard filter setting. Variants that passed the filters (QD < 2.0 || FS > 60.0 || MQ < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0) were used for the following analysis. As for the human and mouse RNA-seq data, we used WASP software to re-map reads to prevent mapping bias caused by polymorphisms (30). Mapped RNA-seq data were used for the subsequent analyses.
RNA-seq reads overlapping RNA editing sites and heterozygous SNPs were assigned to alleles with or without the edited base. Further, we only considered RNA editing sites that were covered by at least five reads for each allele for statistical testing. Differential RNA editing efficiencies of the alleles was assessed using the Fisher's exact test and chi-square test for each sample. Allele-specific RNA editing sites were identified with a P value cutoff of 0.05 and FDR (Benjamini-Hochberg multiple testing correction) less than 0.1. RNA editing sites and SNPs were annotated to genes or transcripts using ANNOVAR (31).
PCR amplification of RNA editing sites
To confirm the reliability of the RNA editing sites showing allele-specificity, we randomly selected five RNA editing sites from each of the U87MG cell line and mouse brain datasets. For the U87MG cell line data, the five RNA editing sites were distributed across three PCR amplicons. For the mouse brain data, four PCR amplicons contained the five RNA editing sites. Genomic DNA and RNA samples used for the PCR experiments were the same as those used for the DNA-seq and RNA-seq experiments described above. SuperScript II reverse transcriptase (Invitrogen) was used to convert total RNA into cDNA with oligo d(T) according to the manufacturer's instructions. For each RNA editing site, we amplified a fragment that included the SNP and the RNA editing sites from both genomic DNA and cDNA (primer information is listed in Supplementary Table S1). PCR products from genomic DNA were subjected to Sanger sequencing (ABI 3730 DNA sequencer). PCR products from cDNA were used to construct TA clones. Clones were randomly selected for Sanger sequencing on the same ABI 3730 DNA sequencer to confirm the RNA editing sties.
In vitro cellular experiments for allele-specific RNA editing
Full-length coding sequences (CDS) for both alleles of the mouse Dact3 gene with the C759T substitution (Dact3-T and Dact3-C) were separately created by chemical synthesis (Sangon). A tag sequence (5′-3′, GAT TAC AAG GAT GAC GAC GAT AAG) was included at the 3′-end of each allele. The two sequences were directionally cloned into the HindIII and XbaI sites of pcDNA3.1 (Invitrogen) and the sequences of each construct was confirmed by Sanger sequencing.
Constructs, at four different quantities (30ng, 300ng, 600ng and 3,000ng), were separately transfected into 106 N2a cells using ViaFectTm (Promega) according to the manufacturer's instructions. Transfected cells were incubated for 48h and total RNA isolated using RNeasy® Mini Kit (Qiagen). cDNA was obtained from total RNA with SuperScript II reverse transcriptase (Invitrogen). PCR was then performed using primers targeting the tag sequence in the constructs to prevent amplification of endogenous transcripts of the Dact3 gene and the products were subjected to TA cloning (Supplementary Table S1). Clones were randomly selected for Sanger sequencing on an ABI 3730 DNA sequencer to evaluate the RNA editing site (A766G) and determine the RNA editing level for each allele (C759T). The RNA editing level of each allele was calculated as the number of clones with the edited base divided by the total number of allele clones sequenced.
RNA secondary structure analysis using SHAPE method
RNAs for both alleles of the human CTSB gene with the U3181G substitution were in vitro transcribed using T7 Megascript Kit (Ambion) following the manufacturer's instructions. As described by the SHAPE protocol, RNAs were mixed with 0.5 × TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH 8.0) and heated at 95°C for 2 min. The RNAs were cooled on ice for 2 min and then 3.3 × RNA folding mix (333 mM HEPES–KOH, 20 mM MgCl2, 333 mM NaCl) was added and allowed to equilibrate at 37°C for 20 min. 1 ul of 10 × NMIA, including DMSO, was then added to the mixture and incubated at 37°C for 45 min. Modified RNAs were precipitated using ethanol. Diethylpyrocarbonte (DEPC) treated water was used in all steps.
32P labeled DNA primers were annealed to the modified RNAs at 65°C for 5 min, followed by incubation at 35°C for 5 min and then placed on ice for 1 min. SHAPE enzyme mix (5 × FS buffer (Invitrogen), 10 mM dNTP, 100 mM DTT) was added and the reaction was heated to 52°C for 1 min. Superscript III was then added, and the extension reaction was incubated at 52°C for 30 min. NaOH was then added and the mixture was heated at 95°C for 5 min. HCl was added to stop the reaction. After degradation of modified RNAs, cDNA products were loaded on an 8% urea-PAGE (8 M urea) gel. Bands were visualized using PhosphorImager, isolated and their sequences were compared to the unmodified input sequences. RNA secondary structure of transcripts containing the allele-specific RNA editing sites were predicted using the RNAStructure package with default parameters (32).
RESULTS
Allele-specific RNA editing in human tissues
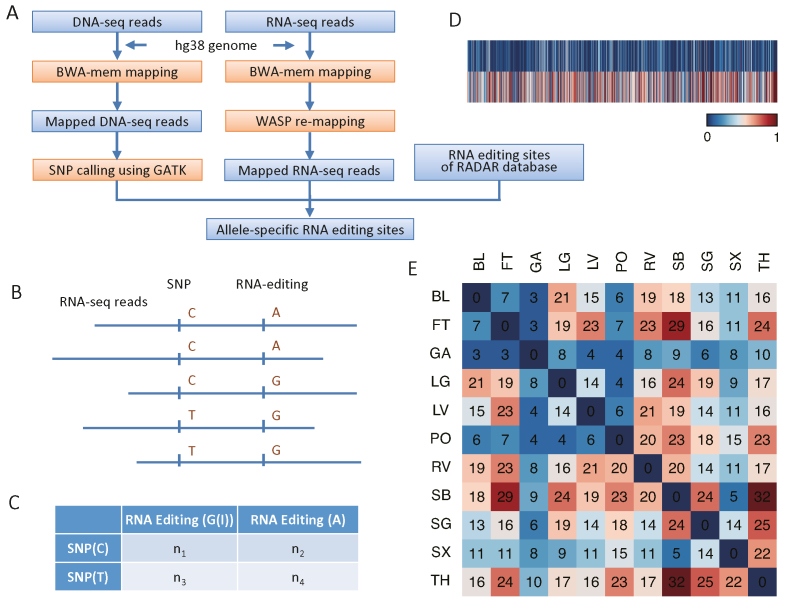
To investigate allele-specific RNA editing in human tissues, we used transcriptome data from 18 human tissue types obtained from 4 individuals and RNA editing sites from the RADAR RNA editing database (v2) (25). We identified allele-specific RNA editing sites using genes that were heterozygotes in each individual (Figure 1A). Editing sites were identified in 273 587 to 11 920 962 RNA-seq paired-end reads in the different human transcriptome samples, where reads displayed evidence of both a heterozygous SNP and an RNA-editing site (Figure 1B). Genome wide, 1 615 697 (62.7%) of the 2 576 292 RNA editing sites were in reads that contained heterozygous SNPs. To retain only high confidence allele-specific RNA editing sites, we focused on RNA editing sites that had at least 5× coverage of each SNP allele for every sample. The significance of the allele-specific RNA editing sites was assessed using Fisher's exact test and chi-square test for the reads that had both the SNPs and the RNA-editing site in each sample (Figure 1C). With a cutoff (Fisher's exact test and chi-square test P value < 0.05 and FDR < 10%), we identified 309 allele-specific RNA editing sites in 18 tissues (Figure 1D and Supplementary Table S2). Significant RNA-editing sites had a high overlap between tissues from the same individual (Figure 1E, Supplementary Figures S1 and S2), whereas the overlap in allele-specific RNA editing between tissues from different individuals was minimal (Supplementary Figure S3). This was possibly due to the small number of individuals sampled. The different results observed between individuals may be one of the molecular mechanisms contributing to diversity of humans.
Figure 1.
Identification and characterization of allele-specific RNA editing sites in human tissues. (A) Overview of the approach for identifying allele-specific RNA editing sites. The pipeline uses raw DNA-seq and RNA-seq reads as source data and was compared to RNA editing sites from the RADAR database to assess allele-specific RNA editing. (B) Reads that contain both heterozygous SNPs and RNA-editing sites. These reads were used to identify allele-specific RNA editing sties. (C) Fisher's exact test and chi-square test evaluation of allele-specific RNA editing. (D) Heatmap showing the RNA editing efficiency for each allele associated with an allele-specific RNA editing site. RNA editing efficiency was defined as the ratio of G reads number to the sum of A and G reads number. Upper row represent low efficiency and the lower row high efficiency. (E) Heatmap of overlapping numbers of allele-specific RNA editing sites for tissues from one individual. Individual 1 is used as an example to indicate a high proportion of overlap between tissues. Tissues sampled are: bladder (BL), fat (FT), gastric (GA), lung (LG), ventricle (LV), psoas (PO), right ventricle (RV), small bowel (SB), Sigmoid colon (SG), spleen (SX) and thymus (TH).
Allele-specific RNA editing in the mouse brain
At the DNA level, previous studies have often used reciprocal crosses between two distantly related strains to identify differences in allele-specific expression and allele-specific DNA methylation patterns (7,9). Those studies were primarily designed to identify imprinted–genes that show paternal or maternal origin allele-specific expression patterns. As RNA editing is critical for brain development (13,33,34), we conducted a comprehensive analysis of allele-specific RNA editing in 67 brain tissues from F1 mice generated by reciprocal crosses between three distant breeds, CAST/EiJ, PWK/PhJ and WSB/EiJ, using RNA editing sites downloaded from the RADAR database (25) to assess whether RNA editing displays an allele-specific pattern.
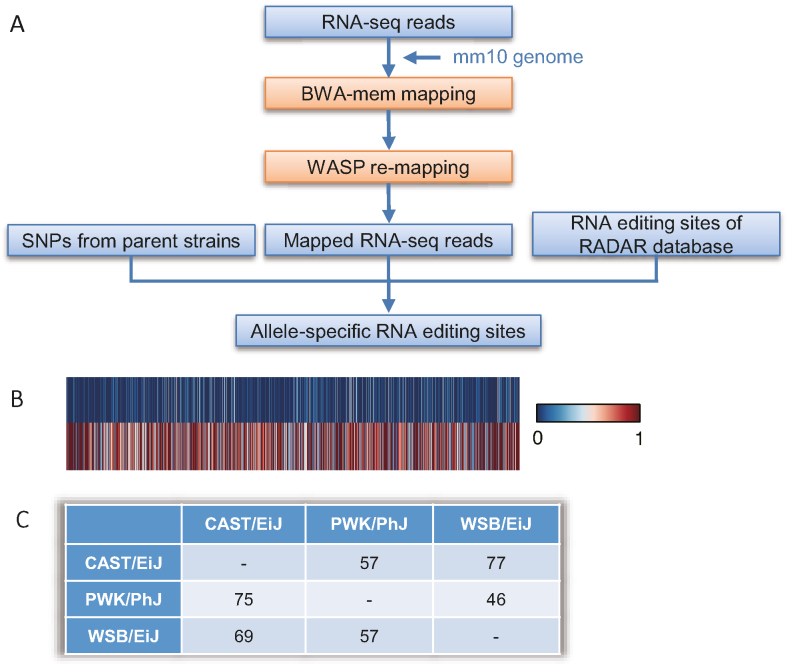
To discover allele-specific RNA sites in transcriptomes from the mouse brain, we used the same strategy used for human tissues described above (Figure 2A). A total of 184 allele-specific editing sites were identified in the 67 samples (Figure 2B and Supplementary Table S3), including 88 from the reciprocal cross between CAST/EiJ and PWK/PhJ, 90 in cross between CAST/EiJ and WSB/EiJ, and 73 in from PWK/PhJ and WSB/EiJ (Figure 2C). The editing sites in the coding region of 3 genes that displayed allele-specificity were validated by Sanger sequencing (Supplementary Table S4). A common RNA editing site (in the 3′-UTR of the Cds2 gene) was found in 44 of the 67 samples, indicating a high reliability of these detected allele-specific RNA editing sites (Table 1). We also found a high proportion of overlap in the allele-specific RNA editing sites found in the initial and reciprocal cross (Supplementary Table S5). The majority of the allele-specific editing sites discovered in the human and mouse transcripts were in the 3′-UTR of genes (Supplementary Tables S2, S3 and Supplementary Figure S4).
Figure 2.
Pipeline for allele-specific RNA editing analysis from transcriptome data from mouse brains from individuals generated by reciprocal crosses between three distinct mouse strains. (A) Procedure for identifying allele-specific RNA editing sites in the mouse. Only RNA-seq reads were used for the analysis of allele-specific RNA editing. Known mouse RNA editing sites were downloaded from the RADAR database. (B) Heatmap displaying the editing efficiency for heterozygous SNPs associated with allele-specific RNA editing sites. The editing efficiency was calculated as the ratio of G reads number to the sum of A and G reads number. Upper rows represents low efficiency. Lower row is high efficiency. (C) Number of allele-specific RNA editing sites from the different reciprocal crosses.
Table 1.
Allele-specific RNA editing sites in brain tissues from individuals generated by reciprocal crosses among three distinct breeds of mice
| Chr | Position | RNA editing type | Gene | Location of sites | Number of samples |
|---|---|---|---|---|---|
| 2 | 132309880 | A→G | Cds2 | 3′-UTR | 44 |
| 7 | 126971183 | A→G | \ | downstream | 26 |
| X | 150985529 | A→G | Gnl3l | 3′-UTR | 19 |
| 14 | 32081694 | A→G | Dph3 | 3′-UTR | 16 |
| 12 | 100207186 | A→G | Calm1 | 3′-UTR | 14 |
| 5 | 142669235 | A→G | Wipi2 | 3′-UTR | 13 |
| 2 | 132309656 | A→G | Cds2 | 3′-UTR | 10 |
To test whether the RNA editing sites display parental specificity, we used SNPs from the parental strains as markers to analyze paternal and maternal allele-specific RNA editing. We found no evidence for paternal or maternal allele-specific RNA editing sites in the mouse brain tissue transcriptomes. These results illustrate that sequence variation may be one of molecular mechanism causing allele-specific RNA editing.
Differing RNA editing efficiencies observed between tissues and a cell line
To further validate our method, we obtained paired-end DNA-seq and RNA-seq data from human U87MG cells. From DNA-seq, we extracted a total of 3 096 947 heterozygous SNPs in the U87MG genome. We used the same strategy used in human tissues and mouse brains to identify allele-specific RNA editing sites in the U87MG cells. Seven allele-specific RNA editing sites were detected in these cells (Supplementary Table S2).
To confirm RNA editing at sites linked to SNPs, we randomly selected five allele-specific RNA editing sites for experimental validation using PCR amplification, TA cloning and Sanger sequencing. Overall, all RNA editing was confirmed at the five sites linked to the SNPs (Supplementary Table S6). Our experimental results showed that one allele experienced 100% editing, while the other allele for all seven sites showed no editing (0% editing level) (Supplementary Table S2). This phenomena of absolute difference in editing was, however, not found in our examination of human tissues or mouse brain samples. This result might indicate that different regulatory mechanisms are used in vivo compared to this cell line.
Allele-specific RNA editing is associated nonsynonymous modification
Recent studies have demonstrated that many RNA editing sites have the potential to generate amino acid changes (35,36). It has further been evidenced that allele-specificity is one of the methods that directly contribute to cis-regulatory variation (37). Although almost all of the allele-specific RNA editing sites were found in the UTR, we analyzed the allele-specific RNA editing sites for their potential to cause amino acids substitutions in coding sequences. In mice, all of the SNPs identified in the coding regions of genes were synonymous. We found four of these SNPs are associated with three allele-specific RNA editing sites that lead to amino acid changes (Supplementary Table S3). In contrast, none of the SNPs located in the UTRs of protein-coding genes were associated with allele-specific RNA editing sites leading to nonsynonymous changes. In the transcriptome data from human tissues, three allele-specific RNA editing sites that cause amino acids alteration and are linked with synonymous SNPs were found.
Several studies have reported many nonsynonymous mutations are linked with particular phenotypes and diseases (38–40). Similarly, many mutations located in the untranslated regions or are synonymous have also been associated with disease (41–44). Allele-specific RNA editing potentially explain why some of the synonymous mutations cause phenotypic variation and disease. Interestingly, a SNP (chr7: 16885340(C→T)) in the Dact3 gene is associated with a nearby allele-specific RNA editing site (A to I (G), chr7:16885347) that leads to an amino acid change (arginine (R) to Glycine (G)) (Figure 3). This RNA editing site is conserved in mammals, which suggests that it has an important role in a fundamental biological process (45).
Figure 3.
Allele-specific RNA editing causes an amino acid change in the product of the Dact3 gene in the mouse. Upper section shows the two alleles with a synonymous difference. The middle part indicates the editing site, which results in a nonsynonymous substitution, and the neighboring synonymous SNP. The lower section shows the editing efficiency for the C and T alleles in samples showing allele-specific RNA editing of the editing site. The efficiency in each sample was defined as G reads number divided by A and G reads number.
SNP led to allele-specific RNA editing
In the above section, we identified a SNP associated with allele-specific RNA editing that could either be a causal mutation or is linked with a causal variant. Thus, we used the Dact3 gene in the mouse as an example and sub-cloned CDS sequences for the two alleles generated by this SNP (chr7: 16885340(C→T)). The sub-cloned sequences, and tag sequence used to distinguish them from the endogenous gene, were cloned into an expression vector. The two Dact3 allele sequences were transiently transfected into mouse neuroblastoma N2a cells and the resulting transcripts were examined for their efficiency for RNA editing (A to I (G), chr7:16885347) by Sanger sequencing (Figure 4).
Figure 4.
In vitro validation that a SNP leads to allele-specific RNA editing. Full-length coding sequences (CDS) of the two alleles of the C759T SNP of the mouse Dact3 gene were cloned into plasmids. Constructs, at four different quantities, were transfected into N2a cells. After 48 h, total RNA was isolated from the transfected cells and reverse transcribed into cDNA. Sanger sequencing of cloned RT-PCR products was performed for the analysis to determine the RNA editing levels for each allele. RNA editing levels for each allele was defined as the number of clones with the edited sites divided by the total number of the allele clones.
From this transfection experiment, we found that the C allele had a higher level of RNA editing than the T allele, which was consistent across the four concentrations of the transfected plasmids used (Figure 4 and Supplementary Table S7). Although, the results from the transfection experiment differ from the results from the allele-specific RNA editing analysis using RNA-seq data from the mouse brain, we can conclude that the synonymous SNP (chr7: 16885340(C→T)) affects allele-specific nonsynonymous RNA editing (chr7: 16885347(C→T), Arginine (R) to Glycine (G)). The difference in the allele-specific effect on RNA editing between the RNA-seq and transfection experiment might suggest that additional variants also regulate the nonsynonymous RNA editing site (chr7: 16885347(C→T)). The difference could also be attributed to different regulatory mechanism used in the cell line compared to mouse brains.
Association of allele-specific RNA editing with RNA secondary structure
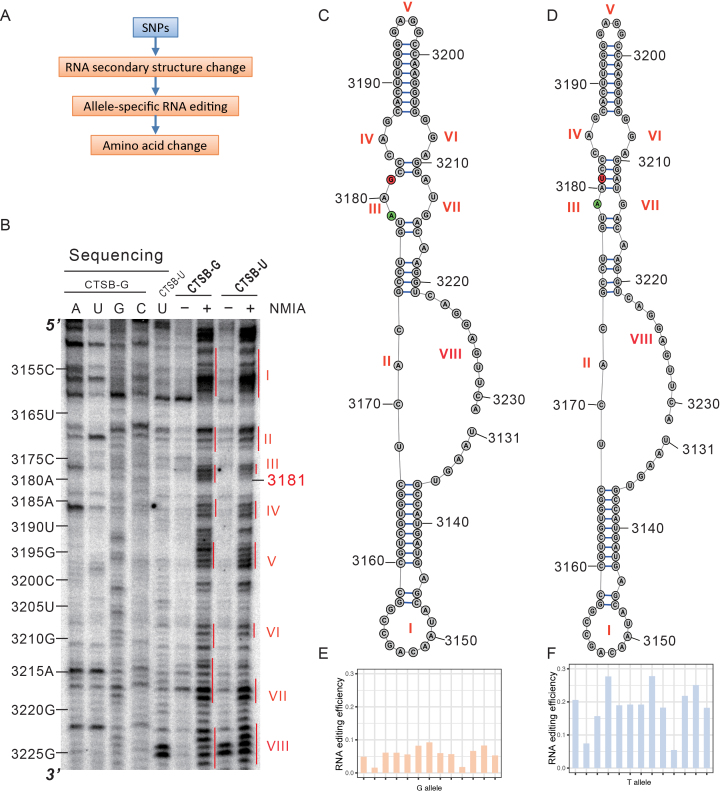
RNA editing can occur in either double-stranded RNA (dsRNA) or in single-stranded RNA (ssRNA), and many SNPs have the potential to alter local RNA secondary structure (46). Thus, a possible mechanism for allele-specific RNA editing is through changes in local RNA secondary structure due to the proximal SNPs (Figure 5A). A to I editing events are mediated by ADAR proteins acting on double-stranded RNAs (18,19). To validate a connection between allele-specific RNA editing and RNA secondary structure, we used a SHAPE analysis to obtain RNA secondary structures for the transcripts of two alleles of the human CTSB gene generated by a SNP that contains an allele-specific RNA editing site(Figure 5B). For CTSB (Figure 5C and D), the SNP associated with (causing or linked with the causal mutation) allele-specific RNA editing altered the local RNA secondary structure of the mRNA. We found that the transcript bearing double-stranded RNA structures (due to the SNP) had a higher efficiency of editing (i.e. preferential editing for one allele) (Figure 5C,D,E and F). From the above, we infer that increased levels of RNA editing seen for specific alleles (i.e. allele-specific editing) is mechanistical explained by changes in the secondary structure of the RNA caused by the closely associated SNPs (Figure 5A).
Figure 5.
RNA secondary structure and allele-specific RNA editing. (A) The effect pipeline of allele-specific RNA editing sites. SNPs influence RNA secondary structure leading to allele-specific RNA editing. Amino acid changes are one of the results of allele-specific RNA editing. (B) Denaturing gel electrophoresis of the SHAPE analysis for the G (CTSB-G) and T (CTSB-U) alleles of the human CTSB gene associated with allele-specific RNA editing. A, U, G and C indicate reverse transcription with dideoxyadenine triphosphate, dideoxyuracil triphosphate, dideoxyguanine triphosphate and dideoxycytosine triphosphate, respectively. Nucleotide numbers on the left correspond to the (–) and (+) NMIA lanes. (–) and (+) NMIA lanes only represent presence or absence of reagent. Red vertical lines represents loop structures, and are numbered in Roman numerals. (C, D) RNA secondary structures for the G (c) and T (d) alleles of the human CTSB gene predicated by the RNAStructure package, which is consistent with the result of the SHAPE analysis. Roman numerals identify the loops in (C, D) that are analyzed in (B). Loops III and VII differ in size between the two alleles. (E, F) Histograms showing editing efficiency of the G (E) and T (F) alleles displaying allele-specific RNA editing in human tissues. Editing efficiency in each sample was calculated as the ratio of edited reads to the sum of edited reads and non-edited reads.
DISCUSSION
This study represents the first assessment of allele-specific modification at the RNA level across tissues and species. We document 315 allele-specific RNA editing sites, a large proportion of which overlap between transcriptomes from different human tissues. To further characterize this phenomena of allele-specific RNA editing, we identified 184 allele-specific RNA editing sites from mouse brain tissue transcriptomes from individuals generated by reciprocal crosses between three distantly related mouse strains (7), and observed a considerable overlap in the identification of sites between crosses. From the results obtained in these two species, we can reliably conclude that allele-specific RNA editing exists in mammalian species. Similar conclusions have been reported at the DNA level, however, many of these modifications, including RNA expression and DNA methylation (4,7,9), show parental allele-specificity, likely due to inherited epigenetic modifications from the parents. Recent studies have indicated that many types of RNAs, such as piRNAs (47), tsRNAs (48) and mRNA (49), are maternally or paternally inherited. Thus, we analyzed whether the RNA editing that we observed was maternally or paternally inherited. However, we found no evidence of parental allele-specific RNA editing sites in the F1 reciprocal crosses among the three strains of mice examined, indicating that the modifications at the RNA level likely have a different genetic mechanism from the modifications observed at the DNA level. In this study, we only focused on RNA editing of mRNA and therefore the editing of other of RNAs and other RNA modification need to be investigated in future studies.
SNPs associated with allele-specific RNA editing are mainly located in the 3′-UTR or result in synonymous substitutions in the coding region. Many SNPs in UTRs as well as those that cause synonymous changes in coding regions are associated with human diseases and phenotypes (41–44). SNPs in the UTRs are thought to influence the stability of mRNAs or inhibit translation (50–53). SNPs causing synonymous changes are believed to act by altering translation efficiency and thus can influence traits (42,54). In our study, we suggest that allele-specific RNA editing that leads to amino acid changes is a potential mechanism for explaining how SNPs in UTRs or result in synonymous changes in coding regions yield diseases and phenotypes. These results provide new clues for genetic studies to explain the role of the SNPs that do not result in amino acid differences, whether in UTRs or coding regions, in the development of important traits.
By introducing point mutations, we validated that a synonymous SNP could lead to nonsynonymous allele-specific RNA editing in the mouse Dact3 gene. However, there were some differences in the efficiency of editing between out in vitro cell line model and the results observed from mouse brain tissue. We suggest three possible reasons to explain these differing results: (i) Different regulatory mechanisms are used by RNA editing in the cell line compared to tissues, (ii) we only used the coding sequence for the transfection experiment, thus other portions of the mRNA (i.e. UTR) might alter the secondary structures recognized by ADAR proteins and (iii) additional variants outside the coding sequence might also regulate allele-specific RNA editing. A new high throughput method will be needed to detect causal mutations for allele-specific RNA editing or RNA editing QTLs, which should help us identify causal variants from GWAS studies. These results also indicate that sequencing experiments at the RNA level are necessary for protein expression analyses using plasmids, as RNA editing could change protein sequences.
We also investigated possible molecular mechanisms causing the allele-specific RNA editing in the transcriptomes. Previous studies have suggested that the secondary structure of RNAs is an important factor in determining the efficiency of RNA editing (21). SNPs are known to affect the secondary structures of RNAs (46). Our findings support the conclusion that SNPs alter RNA secondary structures, and thus influence the efficiency of editing, thereby offering a potential molecular mechanism for allele-specific RNA editing (Figure 5A). To illustrate the effect of alleles at SNPs on RNA secondary structure we selected the human CTSB gene, which has both an RNA editing site and an associated SNP. Not only are there differences in the RNA secondary structures for the transcripts of these two alleles but also differences in RNA editing efficiency. In this study, we did not observe an effect of mutations in the UTRs on allele-specific editing causing amino acid substitution, thus they may have less of effect on secondary structure of RNA. However, this might also be due to a limitation of using short reads from RNA-seq data. In future, both RNA-seq and RNA secondary structure sequencing of the same sample would be needed to better elucidate allele-specific RNA editing and its association with RNA secondary structure, and further analyze the influence of SNPs in UTR regions on amino acid changes.
In summary, we analyzed allele-specific RNA editing sites in human and mouse transcriptomes and provide findings and reasons for allele-specific RNA editing, which offer new ideas on the molecular consequences of synonymous or 3′UTR SNPs associated with diseases and phenotypes.
DATA AVAILABILITY
DNA-seq and RNA-seq datasets from U87MG cells were submitted to the Genome Sequence Archive (GSA) with ID CRA000575.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dong-Dong Wu and Ling-Qun Ye for reviewing the manuscript, and Hongjie Zhang (Core Facility for Protein Sciences, Institute of Biophysics, Chinese Academy of Sciences) for technical support with autoradiography.
Author Contributions: Z.Y.Z. and Y.P.Z. designed the research. Z.Y.Z., Y.H., A.M.L. and Y.J.L. performed data collection and data analysis. Z.Y.Z., Y.J.L., H.Z., S.Q.W., D.Z. J.H.W. and Y.Q. performed experiment. Z.Y.Z., Y.H., A.M.L., N.O.O., D.M.I. and Y.P.Z. wrote and revised the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Agriculture of China [2016ZX08009003-006]; National Natural Science Foundation of China [31321002]; Chinese Academy of Sciences [QYZDB-SSW-SMC028]; Strategic Priority Research Programs (Category A) of the Chinese Academy of Sciences [XDA12010313]; State Key Laboratory of Genetic Resources and Evolution [1101090303]; The National Key R&D Program of China [2018YFA0106901]; Natural Science Basic Research Plan in Shaanxi Province of China [2017JM6024]; Natural Science Foundation of Shaanxi Provincial Department of Education [17JK0572]; Natural Science Foundation of Shandong province of China [ZR2016HB54]; N.O.O. thanks the CAS-TWAS President's Fellowship Program for Doctoral Candidates for support. Funding for open access charge: Ministry of Agriculture of China [2016ZX08009003-006].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lalande M. Parental imprinting and human disease. Annu. Rev. Genet. 1996; 30:173–195. [DOI] [PubMed] [Google Scholar]
- 2. Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A.. Widespread monoallelic expression on human autosomes. Science. 2007; 318:1136–1140. [DOI] [PubMed] [Google Scholar]
- 3. Deng Q., Ramsköld D., Reinius B., Sandberg R.. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. 2014; 343:193–196. [DOI] [PubMed] [Google Scholar]
- 4. Gregg C., Zhang J., Weissbourd B., Luo S., Schroth G.P., Haig D., Dulac C.. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010; 329:643–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gregg C., Zhang J., Butler J.E., Haig D., Dulac C.. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010; 329:682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeVeale B., Van Der Kooy D., Babak T.. Critical evaluation of imprinted gene expression by RNA–Seq: a new perspective. PLos Genet. 2012; 8:e1002600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crowley J.J., Zhabotynsky V., Sun W., Huang S., Pakatci I.K., Kim Y., Wang J.R., Morgan A.P., Calaway J.D., Aylor D.L.. Analyses of allele-specific gene expression in highly divergent mouse crosses identifies pervasive allelic imbalance. Nat. Genet. 2015; 47:353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reik W., Walter J.. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001; 2:21–32. [DOI] [PubMed] [Google Scholar]
- 9. Xie W., Barr C.L., Kim A., Yue F., Lee A.Y., Eubanks J., Dempster E.L., Ren B.. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell. 2012; 148:816–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schilling E., El Chartouni C., Rehli M.. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009; 19:2028–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shoemaker R., Deng J., Wang W., Zhang K.. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010; 20:883–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010; 79:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J.B., Church G.M.. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat. Neurosci. 2013; 16:1518–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Athanasiadis A., Rich A., Maas S.. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004; 2:e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim D.D., Kim T.T., Walsh T., Kobayashi Y., Matise T.C., Buyske S., Gabriel A.. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004; 14:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smith H.C., Bennett R.P., Kizilyer A., McDougall W.M., Prohaska K.M.. Seminars in Cell & Developmental Biology. 2012; 23:Elsevier; 258–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rosenberg B.R., Hamilton C.E., Mwangi M.M., Dewell S., Papavasiliou F.N.. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3′ UTRs. Nat. Struct. Mol. Biol. 2011; 18:230–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eggington J.M., Greene T., Bass B.L.. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011; 2:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lehmann K.A., Bass B.L.. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999; 291:1–13. [DOI] [PubMed] [Google Scholar]
- 20. Rieder L.E., Staber C.J., Hoopengardner B., Reenan R.A.. Tertiary structural elements determine the extent and specificity of messenger RNA editing. Nat. Commun. 2013; 4:2232. [DOI] [PubMed] [Google Scholar]
- 21. Ramaswami G., Deng P., Zhang R., Carbone M.A., Mackay T.F., Li J.B.. Genetic mapping uncovers cis-regulatory landscape of RNA editing. Nat. Commun. 2015; 6:8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Park E., Guo J., Shen S., Demirdjian L., Ying N.W., Lan L., Yi X.. Population and allelic variation of A-to-I RNA editing in human transcriptomes. Genome Biol. 2017; 18:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schultz M.D., He Y., Whitaker J.W., Hariharan M., Mukamel E.A., Leung D., Rajagopal N., Nery J.R., Urich M.A., Chen H.. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015; 523:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Keane T.M., Goodstadt L., Danecek P., White M.A., Wong K., Yalcin B., Heger A., Agam A., Slater G., Goodson M.. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011; 477:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ramaswami G., Jin B.L.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li H., Durbin R.. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009; 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M.. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Van De Geijn B., McVicker G., Gilad Y., Pritchard J.K.. WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 2015; 12:1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K., Li M., Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010; 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuter J.S., Mathews D.H.. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinformatics. 2010; 11:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wahlstedt H., Daniel C., Ensterö M., Öhman M.. Large-scale mRNA sequencing determines global regulation of RNA editing during brain development. Genome Res. 2009; 19:978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mehler M.F., Mattick J.S.. Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol. Rev. 2007; 87:799–823. [DOI] [PubMed] [Google Scholar]
- 35. Li J.B., Levanon E.Y., Yoon J.-K., Aach J., Xie B., LeProust E., Zhang K., Gao Y., Church G.M.. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009; 324:1210–1213. [DOI] [PubMed] [Google Scholar]
- 36. Han L., Diao L., Yu S., Xu X., Li J., Zhang R., Yang Y., Werner H.M., Eterovic A.K., Yuan Y.. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015; 28:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat. Rev. Genet. 2010; 11:533–538. [DOI] [PubMed] [Google Scholar]
- 38. Hampe J., Franke A., Rosenstiel P., Till A., Teuber M., Huse K., Albrecht M., Mayr G., De La Vega F.M., Briggs J.. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat. Genet. 2007; 39:207–211. [DOI] [PubMed] [Google Scholar]
- 39. Sookoian S., Castaño G.O., Burgueño A.L., Gianotti T.F., Rosselli M.S., Pirola C.J.. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 2009; 50:2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Smyth D.J., Cooper J.D., Bailey R., Field S., Burren O., Smink L.J., Guja C., Ionescu-Tirgoviste C., Widmer B., Dunger D.B.. A genome-wide association study of nonsynonymous SNPs identifies a type 1 diabetes locus in the interferon-induced helicase (IFIH1) region. Nat. Genet. 2006; 38:617–619. [DOI] [PubMed] [Google Scholar]
- 41. Brest P., Lapaquette P., Souidi M., Lebrigand K., Cesaro A., Vouret-Craviari V., Mari B., Barbry P., Mosnier J.-F., Hébuterne X.. A synonymous variant in IRGM alters a binding site for miR-196 and causes deregulation of IRGM-dependent xenophagy in Crohn's disease. Nat. Genet. 2011; 43:242–245. [DOI] [PubMed] [Google Scholar]
- 42. Sauna Z.E., Kimchi-Sarfaty C.. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011; 12:683–691. [DOI] [PubMed] [Google Scholar]
- 43. Conne B., Stutz A., Vassalli J.-D.. The 3′ untranslated region of messenger RNA: a molecular ‘hotspot’for pathology. Nat. Med. 2000; 6:637–641. [DOI] [PubMed] [Google Scholar]
- 44. Miyamoto Y., Mabuchi A., Shi D., Kubo T., Takatori Y., Saito S., Fujioka M., Sudo A., Uchida A., Yamamoto S.. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat. Genet. 2007; 39:529–533. [DOI] [PubMed] [Google Scholar]
- 45. Pinto Y., Cohen H.Y., Levanon E.Y.. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol. 2014; 15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wan Y., Qu K., Zhang Q.C., Flynn R.A., Manor O., Ouyang Z., Zhang J., Spitale R.C., Snyder M.P., Segal E.. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014; 505:706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brennecke J., Malone C.D., Aravin A.A., Sachidanandam R., Stark A., Hannon G.J.. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008; 322:1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.-H., Peng H., Zhang X., Zhang Y.. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016; 351:397–400. [DOI] [PubMed] [Google Scholar]
- 49. Soto J.G., Nelson B.H., Weisblat D.A.. A leech homolog of twist: evidence for its inheritance as a maternal mRNA. Gene. 1997; 199:31–37. [DOI] [PubMed] [Google Scholar]
- 50. Wang J., Pitarque M., Ingelman-Sundberg M.. 3′-UTR polymorphism in the human CYP2A6 gene affects mRNA stability and enzyme expression. Biochem. Biophys. Res. Commun. 2006; 340:491–497. [DOI] [PubMed] [Google Scholar]
- 51. Kamiyama M., Kobayashi M., Araki S.-I., Iida A., Tsunoda T., Kawai K., Imanishi M., Nomura M., Babazono T., Iwamoto Y.. Polymorphisms in the 3′ UTR in the neurocalcin δ gene affect mRNA stability, and confer susceptibility to diabetic nephropathy. Hum. Genet. 2007; 122:397–407. [DOI] [PubMed] [Google Scholar]
- 52. Laguette M.-J., Abrahams Y., Prince S., Collins M.. Sequence variants within the 3′-UTR of the COL5A1 gene alters mRNA stability: implications for musculoskeletal soft tissue injuries. Matrix Biol. 2011; 30:338–345. [DOI] [PubMed] [Google Scholar]
- 53. Clop A., Marcq F., Takeda H., Pirottin D., Tordoir X., Bibé B., Bouix J., Caiment F., Elsen J.-M., Eychenne F.. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006; 38:813–818. [DOI] [PubMed] [Google Scholar]
- 54. Kimchi-Sarfaty C., Oh J.M., Kim I.-W., Sauna Z.E., Calcagno A.M., Ambudkar S.V., Gottesman M.M.. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007; 315:525–528. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA-seq and RNA-seq datasets from U87MG cells were submitted to the Genome Sequence Archive (GSA) with ID CRA000575.