Abstract
Several studies have described functional peptides encoded in RNA that are considered to be noncoding. Telomerase RNA together with telomerase reverse transcriptase and regulatory proteins make up the telomerase complex, the major component of the telomere length-maintaining machinery. In contrast to protein subunits, telomerase RNA is expressed constitutively in most somatic cells where telomerase reverse transcriptase is absent. We show here that the transcript of human telomerase RNA codes a 121 amino acid protein (hTERP). The existence of hTERP was shown by immunoblotting, immunofluorescence microscopy and mass spectroscopy. Gain-of-function and loss-of-function experiments showed that hTERP protects cells from drug-induced apoptosis and participates in the processing of autophagosome. We suggest that hTERP regulates crosstalk between autophagy and apoptosis and is involved in cellular adaptation under stress conditions.
INTRODUCTION
Noncoding RNAs are defined as regulatory RNAs that do not possess a coding region for protein synthesis (1). However, more precise analyses have revealed that long noncoding RNAs often contain small open reading frames (ORFs) (2,3), and many long noncoding RNAs have been found in polysome-containing fractions via ribosome profiling approaches (4,5). The regulatory functions of several peptides encoded in noncoding RNAs were recently determined (6–8). Such peptides may be involved in the regulation of different aspects of the development of organisms, the maintenance of cellular homeostasis, cell differentiation, and the regulation of biological and physiological activity.
Telomerase RNA (TERC) (9) is considered to be noncoding RNA that, together with telomerase reverse transcriptase (TERT) (10) and regulatory proteins (9), make up the telomerase complex, the major component of the telomere length-maintaining machinery (11). The mature form of hTERC contains 451 nt, although longer transcripts have been determined by reverse transcription followed by polymerase chain reaction (RT-PCR) (9) and rapid amplification of cDNA 3′-end (3′-RACE) (12) approaches (Figure 1A). Telomerase is active in germ cells, stem cells, in cells from tissue characterized by active proliferation capacity and in most cancer cells. The expression of TERT is switched off during differentiation of stem cells (13). However, the expression of hTERC has been observed in the majority of somatic cells regardless of the stage of differentiation (14). An alternative function of telomerase RNA in preventing apoptosis has also been suggested (15,16).
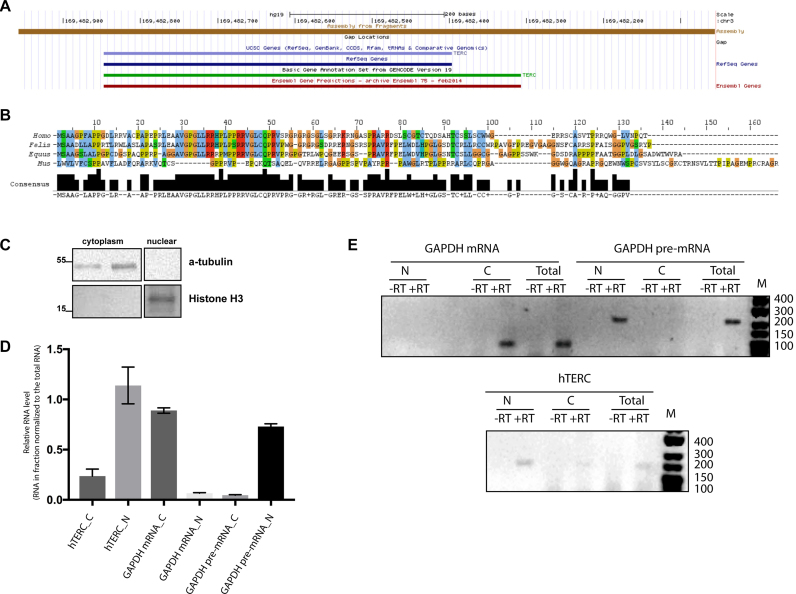
Figure 1.
Coding capacity of the hTERC primary transcript. (A) Structure of genome locus around the hTERC revealed by UCSC Genome Browser. (B) Alignment of TERPs from human, cat, horse and mouse using JalView. Amino acid residues are colored in ClustalX colors. (C) Western blot analysis of nuclear and cytoplasmic extracts of HEK293T cells for α-tubulin and histone H3. (D) RNA prepared from nuclear and cytoplasmic fractions of HEK293T cells was subjected to the RT-qPCR for hTERC and GAPDH pre- and mRNA. Mean fold change for RNA in nuclear and cytoplasmic fractions relative to the same RNA in total cellular extract. Mean ± SD was calculated for three biological replicates. Statistical significance was calculated by GraphPad Software. (E) RNA prepared from nuclear and cytoplasmic fractions of HEK293T cells was subjected to the RT-PCR for hTERC and GAPDH unspliced (pre-) and spliced (mRNA) transcript. Products of RT-PCR from nuclear cytoplasmic and total fractions were resolved in agarose gel. –RT lanes represent the products obtained in PCR reactions by omitting the RT enzyme in a mock reaction to rule out the DNA contamination.
The primary transcript of hTERC is synthesized by RNA polymerase II (17). It was shown that the yeast telomerase RNA contains a poly(A)-tail (18) that is necessary for messenger RNA (mRNA) stabilization (19). m7G-cap modification of the 5′-end of hTERC occurs co-transcriptionally, and further methylation resulting in the appearance of the trimethyl-guanosine cap inherent to small nuclear RNAs (snRNAs) occurs in Cajal bodies (20). Cytoplasmic localization of hTERC was demonstrated under the conditions of affected processing mechanism and in normal cells (21–23). Surprisingly, ribosome profiling analysis revealed that polysomes contained more hTERC than hTERT mRNA (4,5). These properties of the nascent hTERC transcript suggest a protein coding capacity of this RNA.
MATERIALS AND METHODS
Cell culture
Human HEK293T, VA13 and HT1080 cells were grown in DMEM/F12-Ham medium, supplemented with l-glutamine, 10% fetal bovine serum (FBS), 100 units/ml penicillin and 100 μg/ml streptomycin. Human Jurkat cell line cells were maintained in Roswell Park Memorial Institute (RPMI) medium supplemented with 10% FBS, 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were cultured at 37°C and 5% CO2. Cultures were frequently examined under an inverted microscope for confluency and viability. Human HEK293T cells were transiently co-transfected with plasmids expressing vector, wild-type hTERC (hTERCwt), hTERCstU or hTERCstop and lentiviral pMDLg/pRRE, pRSV-Rev and phCMV-VSV-G plasmids using the calcium phosphate method. Lentiviral particles were harvested and used for HEK293T cell infections, as described (24). Cells were analyzed using a Nikon Ti2000 microscope, and GFP-positive cells were sorted using a FACSAriaIII cell sorter (BD Biosciences). The data were analyzed using FACSDiva software. Cells were tested for mycoplasma contamination and found negative.
To genome editing, HEK293T cells were transfected with the help of Lipofectamine 3000 followed by single-cell sorting of GFP-positive cells.
Plasmids
hTERC sequences were obtained from the BAC-clone RP11–816J6 from Roswell Park Cancer Institute by PCR with primers hTERC Full Fw 5′-CGGACGCATCCCACTGAGC-3′ and hTERC Full Rv 5′-CATTAAAGGAACACAATTTCCAATG-3′. The hTERP coding sequence was obtained by PCR with primers hTERP ORF Fw 5′-GATATCATCAGCTGCTGGCC-3′ and hTERP ORF Rv 5′-CTCGAGGGTTTGGGGGTTCAC-3′. The PCR product and pET32b(+) vector were restricted by EcoRV and XhoI and ligated. The TrxA domain was deleted from pET32b(+)hTERP by NdeI restriction and religation.
hTERCwt (875 nt) was amplified by PCR with primers hTERC Full Fw 5′-CGGACGCATCCCACTGAGC-3′ and hTERC Rv 5′-GGGCCCATTAAAGGAACACAATTTCCAATG-3′. The obtained PCR product was digested by EcoRI and Bsp120I and ligated with the LeGo-iG2 vector that was restricted by EcoRI and NotI. hTERCstU and hTERCstop were obtained by site-directed mutagenesis hTERCwt plasmid using a Quick Change II Site-Directed mutagenesis Kit (Agilent Technologies) and primers hTERCstU Fw 5′-GTTCATTCTAGAGCAAACAAAAAATTTCAGCTGCTGGCCCGTTCGCCCC-3′ and hTERCstU Rv 5′-GGGGCGAACGGGCCAGCAGCTGAAATTTTTTGTTTGCTCTAGAATGAAC-3′ for the hTERCstU plasmid and hTERCstop Fw 5′-GCGGCGGGTCGCCTGACCAGCCCCCGAAC-3′ and hTERCstop Rv 5′-GTTCGGGGGCTGGTCAGGCGACCCGCCGC-3′ for the hTERCstop plasmid.
Constructs for gene inactivation and genome editing were created on a basis of pX458 plasmid (25) using primers sghTERC Fw 5′-CACCGAAACAAAAAATGTCAGCTGC-3′ and sghTERC Rv 5′-AAACGCAGCTGACATTTTTTGTTTC-3′.
All constructs were verified by sequencing.
Nuclear and cytoplasmic RNA quantification
Fractions for cytoplasmic and nuclear RNAs were prepared according to Wang et al. (26). Approximately 5 × 106 cells were used for purification of total RNA and for fractionation on nuclear and cytoplasmic fractions. Cells were washed with phosphate buffered saline (PBS) three times and scrapped with a cell scrapper. Obtained cells were centrifuged, washed with reticulocyte standard buffer (RSB) (Tris–HCl (pH 7.4) 10 mM, MgCl2 3 mM and NaCl 10 mM). Cytoplasmic fraction was extracted by RSBG40 (Tris–HCl (pH 7.4) 10 mM, MgCl2 3 mM, NaCl 10 mM, glycerol 10%, Nonidet-P40, dithiothreitol (DTT) 0.5 mM) buffer. Nuclei was washed with RSBG40 and 1/10 volume of detergent (3/3% w/w sodium deoxycholate and 6.6% v/v Tween 20). Pellet was used as a nuclear fraction. RNA and proteins were extracted by TRIZOL (Invitrogen), according to the manufacturer’s instructions. Western blot analysis for histone H3 (Abcam (ab21054), 1:5000) and α-tubulin (Abcam (ab18251), 1:2000) proved the separation of nuclear and cytoplasmic fractions. Total, cytoplasmic and nuclear RNA was treated with DNAse I followed by reverse transcription with random primer and obtained complementary DNA (cDNA) was used for PCR. Quantification of the hTERC in cytoplasmic and nuclear fractions was performed with primers specific to the hTERC, GAPDH mRNA (GAPDH PCR Fw 5′-TGCACCACCAACTGCTTAGC-3′ and GAPDH PCR Rv 5′-GGCATGGACTGTGGTCATGAG-3′) and GAPDH pre-mRNA (GAPDH pre Fw 5′-CCACCAACTGCTTAGCACC-3′ and GAPDH pre Rv 5′-CTCCCCACCTTGAAAGGAAAT-3′). To confirm the specificity of primers, products obtained by reverse transcription followed by quantitative PCR (RT-qPCR) were sequenced.
RT-qPCR
RNA was isolated using a PureLink RNA Mini Kit (Ambion) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using a Maxima First Strand Kit cDNA Synthesis for RT-qPCR (Thermo Scientific) according to the manufacturer’s protocol. Quantitative PCR of cDNA was performed in triplicate using the primers hTERC PCR Fw 5′-GTGGTGGCCATTTTTTGTCTAAC-3′ and hTERC PCR Rv 5′-TGCTCTAGAATGAACGGTGGAA-3′ and a reaction mixture with SYBR Green nucleic acid staining (Invitrogen) on a CFX96 Real-Time system (Bio-Rad). The PCR cycle parameters were as follows: 95°C for 10 min and 35 cycles with denaturation at 95°C for 30 s, annealing at 58°C for 30 s and extension at 72°C for 40 s. The relative quantification in gene expression was determined using the 2−ΔΔCt method (27). The relative expression level of RNA was calibrated by the geometric mean of GAPDH mRNA (GAPDH PCR Fw 5′-TGCACCACCAACTGCTTAGC-3′ and GAPDH PCR Rv 5′-GGCATGGACTGTGGTCATGAG-3′) and U2 RNA (U2 PCR Fw 5′-GCTTCTCGGCCTTTTGGC-3′ and U2 PCR Rv 5′-GTGCACCGTTCCTGGAGGT-3′) levels to minimize experimental variation. Minus–RT controls by omitting the RT enzyme in a mock reaction to rule out DNA contamination were performed in every RT-qPCR analysis.
Recombinant protein expression
Expression of hTERP fused with S-tag and 6-His epitopes in Escherichia coli was induced by 0.1 mM isopropyl b-D-1-thiogalactopyranoside (IPTG) at 18°C for 16 h. Recombinant protein was purified by Ni-NTA-Sepharose under denaturing conditions. The obtained protein was dialyzed against buffer (100 мМ NaH2PO4, 10 мМ Tris–HCl) containing 2 M urea and used for rabbit immunization.
Antibodies
Polyclonal antibodies against full-length hTERP fused with 6-His and expressed in E. coli were produced in rabbits after immunization with recombinant protein. Serum was purified using HiTrap Protein G (GE Healthcare Life Sciences) followed by affinity fast protein liquid chromatography using an AKTA purifier system. Recombinant protein from the same purification that was used for rabbit immunization was conjugated in a HiTrap NHS HP column (GE Healthcare Life Sciences). The obtained column was used for affinity purification of the IgG antibody fraction eluted from a HiTrap Protein G column.
Immunofluorescence
For immunofluorescence, cells were grown on coverslips to subconfluence. The cells were rinsed by 0.1% Tween-20 in PBS and fixed in 4% formaldehyde in PBS for 10 min. For nonadherent Jurkat cells, fixation was followed by a washing step and smearing the cell suspension to a gelatin-coated slide. Cells were permeabilized in 1% Triton X-100 in PBS for 15 min at room temperature, washed twice in PBS, incubated for 1 h in blocking solution (PBS, 1% bovine serum albumin (BSA), 0.1% Tween 20) and for 1 h at room temperature or overnight at 4°C with primary antibodies in blocking solution. The primary antibodies used were anti-hTERP (1:15 in 0.1% Tween 20 in PBS; 0.014 mg/ml stock), anti-MAP LC3 α/β (Santa Cruz Biotech (sc-398822)) and anti-SQSTM1/p62 (Abcam (ab56416)). Cells were washed three times with 0.1% Triton/PBS, followed by incubation with fluorescein isothiocyanate-conjugated goat anti-rabbit IgG antibodies (Invitrogen, 1:2400 in 0.1% Tween 20 in PBS) or Alexa555-labeled goat anti-rabbit IgG antibodies (Invitrogen, 1:2000 in 0.1% Tween 20 in PBS) or AlexaFluor488 anti-mouse IgG antibodies (Thermo Fisher Scientific (A-11029)) for 1 h at room temperature. The cells were washed again three times as stated above, incubated with 300 nM 4′,6-diamidine-2-phenylindole (DAPI) for 1 min at room temperature and mounted in Mowioil onto glass slides. Confocal fluorescence images were obtained using a Nikon C2 confocal system or a Nikon Ti-Eclipse fluorescent system. Images were taken with a 40× NA 0.95 objective or a 100× NA 1.40 oil objective and acquired using standard NIS-Elements software. Possible crosstalk between fluorochromes was avoided by careful selection of imaging conditions.
Immunoblotting
Fractions of soluble cytoplasmic and nuclear proteins were prepared according to Rosner et al. (28). Approximately 1–2 × 107 cells were used for this procedure. To yield ready-to-use extraction buffers, protease inhibitors were supplemented by adding a protease inhibitors cocktail (Roche). RIPA-buffer extracts were used for analysis of total cellular proteins. The extracts were either immediately used or stored at −80°C until further use. Protein concentration was determined by measuring the OD 280 using a NanoDrop 2000 (Thermo Fisher). Equal amounts of protein extracts (30 μg of nuclear, 60 μg of cytoplasmic and 20 μg of total extracts) were boiled for 5 min in HU buffer (200 mM Tris–HCl (pH 6.8), 8 M urea, 5% w/v sodium dodecyl sulphate (SDS), 1 mM ethylenediaminetetraacetic acid, 100 mM DTT, Bromophenol Blue), and proteins were separated on 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) gels under denaturing conditions. Immunoblotting was performed according to standard methods using anti-hTERP polyclonal antibodies (1:250 in 1.5% BSA in tris-buffered saline with 0,05% Tween-20 (TBST); 0.014 mg/ml stock), anti-MAP LC3 α/β (Santa Cruz Biotech (sc-398822), 1:1000), anti-SQSTM1/p62 (Abcam (ab56416)), anti-H3_HRP (Abcam (ab21054), 1:5000) or anti-GAPDH (Thermo Fisher Scientific (39–8600), 1:5000) primary antibodies and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (G21234, Invitrogen, 1:2500) or HRP-conjugated anti-mouse IgG (Thermo Fisher Scientific (G21–040), 1:2000) secondary antibodies for 1 h at room temperature. Detection was performed using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences) according to the manufacturer’s protocols.
Trypsinolysis
PAGE gel bands were fixed in 20% CH3OH/10% CH3COOH/70% H2O for 30 min and washed twice with H2O. Gels were cut in 1 × 1 mm pieces, transferred into tubes and treated with 10 mM DTT and 100 mM NH4HCO3 for 30 min at 56°С. Proteins were alkylated with 55 mM iodoacetamide in 100 mM NH4HCO3 for 20 min in the dark. The gel pieces were dehydrated by the addition of 100% acetonitrile and treated with 150 μl trypsin solution (40 mM NH4HCO3, 10% acetonitrile, 20 ng/μl Trypsin Gold, mass spectrometry grade, Promega). Samples were incubated for 60 min at 40°С and for 16–18 h at 37°С. Peptides were extracted once with 5% formic acid in H2O and twice by 50% acetonitrile with 5% formic acid. Extracts were joined and vacuum-dried at 45°С. The precipitate was diluted in 15 μl of 5% acetonitrile with 0.1% formic acid.
LC-MS analysis
Analysis was performed using a TripleTOF 5600+ mass-spectrometer with a NanoSpray III ion source (ABSciex, Canada) coupled with a NanoLC Ultra 2D+ nano-HPLC system (Eksigent). The high pressure liquid chromatography (HPLC) system was configured in trap-elute mode. For the sample loading buffer and buffer A, a mixture of 98.9% water, 1% methanol and 0.1% formic acid (v/v) was used. Buffer B was composed of 99.9% acetonitrile and 0.1% formic acid (v/v). Samples were loaded on a trap column (Chrom XP C18 3 μm 120 Å 350 μm × 0.5 mm (Eksigent, Dublin, CA)) at a flow rate of 3 μl/min for 10 min and eluted through a separation column (3C18-CL-120 (3 μm 120 Å) 75 μm × 150 mm (Eksigent, Dublin, CA)) at a flow rate of 300 nl/min. The gradient was from 5 to 40% of buffer B in 60 min. The column and the pre-column were regenerated between runs by a blank-injection gradient (5 × 10 min 5–95% B short gradients) followed by equilibration with 5% of buffer B for 25 min. Peptide identification was performed in information-dependent acquisition (IDA) mass-spectrometer mode. Each IDA experiment included one survey MS1 scan followed by 50 dependent MS2 scans. The MS1 acquisition parameters were as follows: the mass range for analysis and subsequent ion selection for MS2 analysis was 300–1250 m/z, and the signal accumulation time was 250 ms. Ions for MS2 analysis were selected based on intensity with a threshold of 400 cps and a charge state from 2 to 5. The MS2 acquisition parameters were as follows: resolution of the quadrupole was set to UNIT (0.7 Da), the measurement mass range was 200–1800 m/z, the ion beam focus was optimized to obtain the maximal sensitivity and the signal accumulation time was 50 ms for each parent ion. Collision-activated dissociation was performed with nitrogen gas with collision energy ramping from 25 to 55 V within 50 ms of signal accumulation time. Analyzed parent ions were sent to a dynamic exclusion list for 15 s in order to get an MS2 spectra at the chromatographic peak apex (the minimum peak width throughout the gradient was ∼30 s).
Identification analysis of mass spectrometry data
Raw data were analyzed with ProteinPilot 4.5 revision 1656 (ABSciex) using the search algorithm Paragon 4.5.0.0 revision 1654 (ABSciex) and standard settings to search the database containing SwissProt DB and the target protein sequence. The following parameters were used for the search: alkylation of cysteine by iodoacetamide, trypsin digestion, TripleTOF 5600 equipment, and including a deep search with additional statistical analysis of results reliability. The spectra were grouped with default settings by the ProGroup algorithm within ProteinPilot. Statistical analysis of the results reliability (and identification of the threshold value of unused score) was performed using the ProteomicS Performance Evaluation Pipeline Software (PSPEP) algorithm within ProteinPilot.
Directed LC-MS/MS analysis
Directed peptide search by LC-MS/MS was performed using the acquisition cycle containing the MS1 survey scan (for further peptide peak RT alignment between different runs check) and the ProductOf scan for the parent ion with m/z corresponding to the peptide of interest, LEAAVGPGLLR (m/z 548.3). The parameters were as follows: CE, 45; accumulation time, 100 ms; mass range, 100–2000 amu. Spectra were processed with PeakView 2.1 Software (ABSciex, Canada).
Cell treatment and viability measurements
Apoptosis was induced by etoposide and doxorubicin treatment. Approximately 10 000 cells per well were seeded the day before drug treatment in 96-well plates. Cells were treated with drug concentrations from 0.1 to 12 μM. Dimethyl sulfoxide (DMSO) stocks of drugs were diluted in medium. Cells treated with DMSO diluted in medium were used as a control. After 48-h incubation with drugs, 100 μg/well 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent was added to the cells, and cells were incubated for 3 h. The medium was aspirated, and 100 μl DMSO was added to each well to dissolve the purple precipitate. After 3 h of incubation under gentle swirling in the dark, the absorbance was measured at 555 nm using a plate reader (Victor X5, Perkin Elmer). All pipetting was performed in a Janus Extended liquid handling station (Perkin Elmer). Triplicate wells were analyzed for each drug concentration. The results were reproduced in three independent experiments.
Autophagy inhibition
Cells were seeded at 3.0 × 103/cm2 in wells of a 6-well plate and treated with chloroquine (C66288, Sigma-Aldrich) (10 μM) for 24 or 6 h.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7.0 software (La Jolla, CA, USA). Statistical significance was determined using the one- and two-way ANOVA, and differences between the analyzed samples were determined using the Sidak’s or Dunett’s multiple comparison tests. Each experiment was replicated three to five times.
RESULTS
Vertebrate telomerase RNAs code for protein
Bioinformatics analysis of the hTERC gene revealed that the ORF started from the AUG codon at position 176 nt followed by 363 nt, which corresponds to the premature form of telomerase RNA coding 121 amino acid residues (Supplementary Figure S1a). This protein was termed hTERP—human TElomerase RNA Protein. The molecular weight of full-length hTERP is 13 kDa (pI of 11.7). It is enriched by Arg, Gly, Pro and Ser (these four amino acids residues make up to 50% of the total residue count (Supplementary Table S1)). TERP may have at least two isoforms: a full-length isoform from the primary transcript (13.3 kDa) and a shorter isoform from mature hTERC (9.9 kDa). Truncated mRNA may be translated by ribosomes that stalled at the ragged position and released by the No-Go mechanism (29).
Putative proteins encoded by telomerase RNA genes of Fellus catus, Equus cabballus and Mus musculus homologous to hTERP were predicted and aligned (Figure 1B; Supplementary Figure S1b and Supplementary Table S2). The aligned sequences showed 40% or more identity (Supplementary Table S2), similar length (113 amino acid residues for F. catus, 106 amino acid residues for E. cabballus and 139 amino acid residues for M. musculus) and amino acids composition (Supplementary Table S1). All analyzed proteins are enriched in Arg, Gly and Pro.
We performed an analysis of the PRIDE Archive (30), a proteomics data repository, to search for hTERP peptides. The peptide search was initiated by virtually cutting the protein of interest, hTERP, using Peptide Cutter software (31). The obtained peptides were used as input data for PRIDE analysis. Eleven unique peptides that originated from hTERP were found in MS/MS data of proteins among the interactomes with transcription factors (32), tyrosine phosphatases (33) and CDK (34) in HEK293T cells (Supplementary Table S3).
hTERC localizes in the nucleus and in the cytoplasm of HEK293T cells
To be translated, RNA should be localized in the cytoplasm. It was shown previously that hTERC forms cytoplasmic bodies (so-called cyTER) under conditions of hTERC processing defects (22,23). However, the small portion of the similar structures may be observed under normal conditions (21,23). To prove the localization of hTERC in the cytoplasm, we purified nuclear and cytoplasmic RNA and proteins. To confirm the quality of purification obtained nuclear and cytoplasmic fractions were analyzed by western blot (Figure 1C) and by RT-PCR (Figure 1D). We detected α-tubulin in the cytoplasm fractions and histone H3 in the nuclear fraction by western blot (Figure 1C). RT-PCR analysis of the distribution of the GAPDH mRNA demonstrated that unspliced pre-mRNA localized in the nuclear (Figure 1D and E) and only spliced mRNA was detected in the cytoplasm fraction (Figure 1D and E). We observed the efficient separation of the cellular lysates. To quantify the distribution of hTERC and spliced and unspliced forms of GAPDH mRNA in cytoplasm and nuclear fractions, we performed RT-qPCR analysis (Figure 1D). To compare the quantity of RNA in the cytoplasm and nucleus, we normalized the level of RNA in cytoplasmic or nuclear fractions to the amount of the same RNA in the total cellular RNA. The quantification revealed that spliced form of GAPDH mRNA is localized in the cytoplasm and the unspliced form of GAPDH pre-mRNA in the nucleus. We observed 20% of hTERC in the cytoplasmic fraction (Figure 1D and E). The presence of hTERC in the cytoplasm is consistent with the translational capacity of this RNA.
hTERP protein is present in the cell as proved by three independent approach
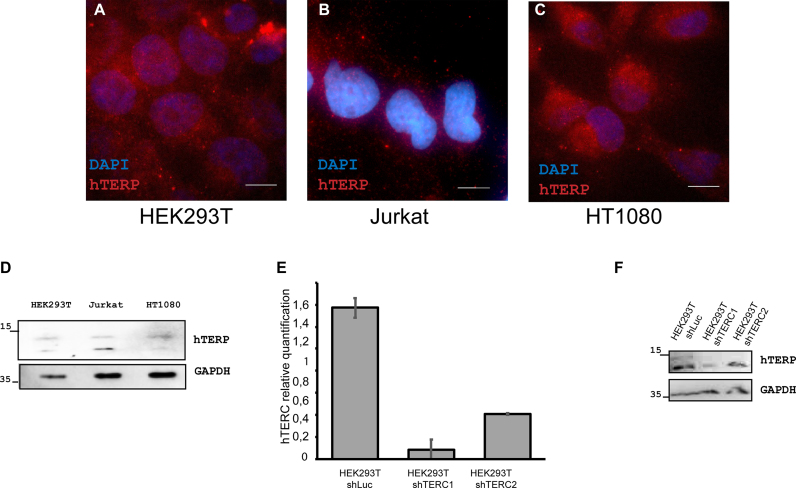
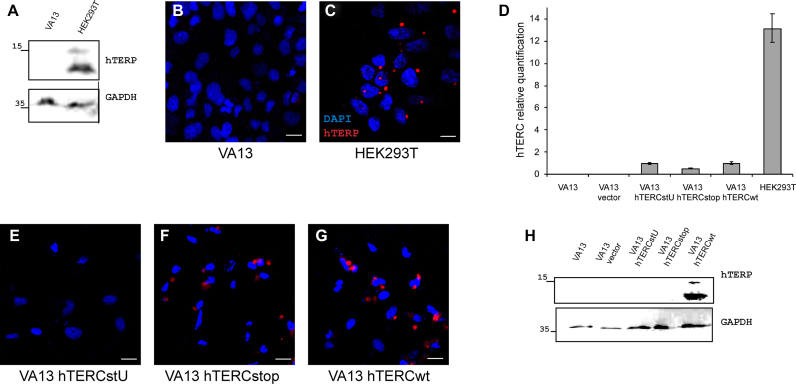
To show the presence of hTERP in cells, antibodies were generated in rabbits by immunization with recombinant 6His-tagged TERP protein purified from E. coli (Supplementary Figure S2a–c). Antibodies were purified, characterized (Supplementary Figures S2d and e and S3) and used for immunofluorescence microscopy and western blot analysis of telomerase-positive (HEK293T, HT1080 and Jurkat) cell lines and a telomerase-negative (VA13) cell line. The expression levels of telomerase RNA in cells were measured by RT-qPCR analysis, which confirmed the absence of hTERC in VA13 cells and showed variations in the amount of hTERC in telomerase-positive cells (HEK293T, HT1080 and Jurkat) (Supplementary Figure S4a). Positive signals due to antibody staining were detected for all telomerase-positive cells (HEK293T, HT1080 and Jurkat) (Figure 2A–D) but were absent in telomerase-negative cells (VA13) (Figure 3B). The knockdown of the hTERC in HEK293T cells performed with two different short hairpin RNAs (shRNAs) (35,36) (Figure 2E) decreased the level of hTERP as was shown by western blot (Figure 2F). We did not observe the hTERP staining in VA13 cells either by western blot (Figure 3A) or by immunofluorescence (Figure 3B) under conditions of HEK293T cells positive staining (Figure 3A and C). We expressed wild-type and mutant forms of hTERC in VA13 cells and confirmed their expression by RT-qPCR (Figure 3D). The expression of mutant hTERC in which the AUG codon (start codon for hTERP) was mutated to AUU (hTERCstU) (Supplementary Figure S1C) did not result in the appearance of immunofluorescence signals in VA13 cells (Figure 3E) and in the positive staining in western blot (Figure 3H). However, the expression of truncated form of hTERP (17aa) obtained by insertion of the preliminary stop-codon (Supplementary Figure S1c) resulted in the appearance of immunofluorescence signal in VA13 cells (Figure 3F) similar to the signal observed in HEK293T cells under the same magnification (Figure 3C). Since the polyclonal antibodies were used for staining, it is not surprising that we were able to detect the truncated form of the protein by immunofluorescence but not by western blot (Figure 3H). Transient expression of the exogenous telomerase RNA gene (875 nt) that encodes full-length hTERP protein induced immunofluorescent signals in VA13 cells (Figure 3G) similar to the signal observed in HEK293T cells under the same magnification (Figure 3C) and positive staining in western blot (Figure 3H).
Figure 2.
hTERP is expressed in telomerase-positive cells. (A–C) Immunolocalization of hTERP in HEK293T (A), Jurkat (B) and HT1080 (C) cells; blue, DAPI-stained nuclei; red, hTERP; scale bars, 5 μm. (D) Western blot analysis of HEK293T, Jurkat and HT1080 cells for the presence of hTERP. GAPDH staining was used as a loading control. (E) Levels of hTERC in HEK293T shLuc (expressed shRNA targeted luciferase mRNA (negative control)), HEK293T shTERC1 and HEK293T shTERC2 (expressed shRNAs targeted hTERC) normalized to the hTERC level in HEK293T cells. Mean ± SD was calculated for three biological replicates. (F) Western blot analysis of HEK293T shLuc (expressed shRNA targeted luciferase mRNA (negative control)), HEK293T shTERC1 and HEK293T shTERC2 (expressed shRNAs targeted hTERC) for the presence of hTERP. GAPDH staining was used as a loading control.
Figure 3.
hTERC encodes an ORF. (A) Western blot analysis of HEK293T and VA13 cells for the presence of hTERP. GAPDH staining was used as a loading control. (B and C) Immunolocalization of hTERP in VA13 (B) and HEK293T (C); blue, DAPI-stained nuclei; red, hTERP; scale bars, 10 μm. (D) Levels of hTERC in VA13, VA13 vector, VA13 hTERCstU, VA13 hTERCstop, VA13 hTERCwt (VA13 expressed empty vector or wild-type or mutant forms of hTERC) and HEK293T. Mean ± SD was calculated for three biological replicates. (E–G) Immunofluorescence analysis of VA13 hTERCstU (E), VA13 hTERCstop (F) and VA13 hTERCwt (G) (VA13 cells expressed wild-type or mutant forms of hTERC); blue, DAPI-stained nuclei; red hTERP; scale bars, 10 μm. (H) Western blot analysis of VA13, VA13 vector, VA13 hTERCstU, VA13 hTERCstop and VA13 hTERCwt cells for the presence of hTERP. GAPDH staining was used as a loading control.
Additional analysis revealed the presence of hTERP in both the nuclear and cytoplasmic fractions of HEK293T cells and the absence of hTERP in VA13 cell extracts (Supplementary Figure S2e), confirming the data obtained by immunofluorescence analysis. Both the full-length form (13.3 kDa) corresponding to premature hTERC and the shorter form (9.9 kDa) corresponding to mature hTERC of hTERP can be seen in Figure 2D and F (Supplementary Figure S2e). We observed multiple bands of hTERP oligomers by SDS-PAGE separation of recombinant protein and in western blots of cellular extracts (Supplementary Figure S2b–f). Oligomerization may be induced by the N-terminal domain of hTERP, which is similar to the oligomerization domain of p62 protein.
We used LC-MS/MS peptide identification to directly confirm the existence of hTERP. Purified recombinant protein was used as a reference. Five peptides corresponding to hTERP with confidence >99% were identified (Supplementary Table S4). The total protein unused score was 7.02, and the sequence coverage was 27.27% (for peptides with confidence >95%) (Supplementary Table S4). One unique for hTERP tryptic peptide (LEAAVGPGLLR) was identified with high confidence (Supplementary Figure S5a and b and Supplementary Table S4). The uniqueness of the selected peptide was also verified by the BlastP algorithm (http://blast.ncbi.nlm.nih.gov/) against a nonredundant protein database, and only one target corresponding to the peptide from human TERP with 100% identity and an E-value of 0.42 was found.
LC-MS/MS analysis was used to detect hTERP in HEK293T cells. A targeted MS/MS approach with the m/z of the selected unique hTERP peptide (LEEAVGPGLLR) determined in the control experiment with the recombinant protein was used for hTERP identification (Supplementary Figure S5c). This unique hTERP tryptic peptide was found in analyzed samples from HEK293T cells extracted from PAGE (Supplementary Figures S2g and h and S5c) as well as from total HEK293T proteins. To identify more peptides, hTERP was concentrated by immunoprecipitation. We used the recombinant protein purified from E. coli as a reference. Recombinant protein and the elution fraction after hTERP immunoprecipitation from HEK293T extracts were trypsinized from PAGE. LC-MS/MS analysis detected three peptides that corresponded to hTERP (Supplementary Figure S6).
Increased level of hTERP protects cell from drug-induced apoptosis
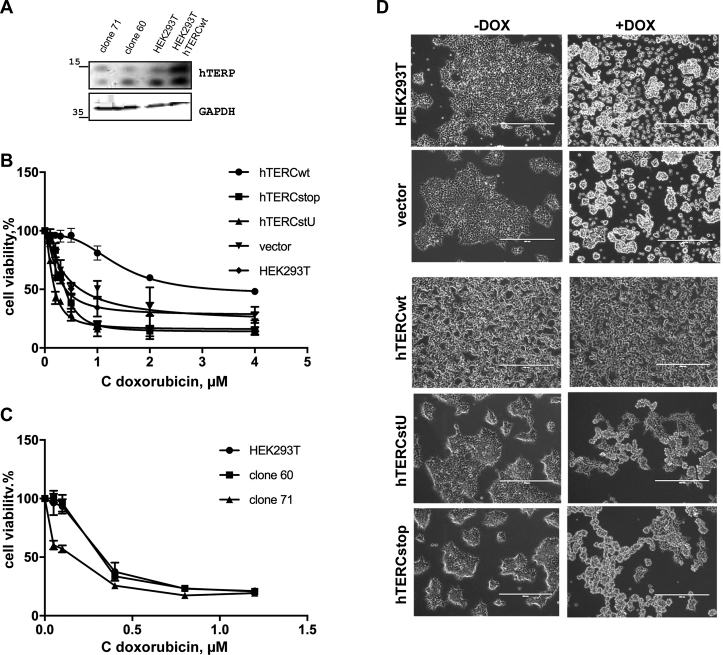
Loss-of-function and gain-of-function approaches were utilized to investigate the functional role of hTERP in cells. HEK293T cells with mutated ORF were obtained by the CRISPR/Cas9 technique. After numerous attempts, we were able to detect 10 clones with a mutation in hTERC ORF. None of the survived clones contained a mutation that led to complete protein disappearance. Only four clones contained double allele mutations. In all cases, we observed at least one allele that contained various deletions, but these deletions retained the translational frame in hTERP ORF. We expected that 66% of analyzed alleles would have an altered ORF after CRISPR/Cas9 treatment (mutations at 2 of 3 positions would induce an ORF shift). The analysis revealed that only 16% (in contrast to the expected 66%) of alleles possessed the frame-shift (Supplementary Table S5). Two clones were chosen for further analysis. Clone 60 characterized by double allele deletion of 15 nt was predicted to delete five amino acid residues from the N-terminus (Supplementary Figure S1d). Clone 71 contained one allele with a 7-nt deletion with disrupted ORF and another allele with a 12-nt deletion but conserved ORF; thus, TERP lost four amino acids in the N-terminal region (Supplementary Figure S1d). TERC level increased by 3-fold in clone 71 and decreased by 2-fold in clone 60, as revealed by RT-qPCR (Supplementary Figure S4b). Telomerase activity did not change significantly in clone 71 and was undetectable in clone 60 (Supplementary Figure S4c), as measured by RQ-TRAP (37). Deletion of 15 nt obtained in clone 60 affects the pseudoknot structural element of the hTERC (Supplementary Figure S4d) that is crucial for the telomerase activity (38). However, the clone 71 had different deletions in two alleles. The pseudoknot element was impaired in one allele but rescued in the second one in which the mutations changed the template boundary element in a way that allows telomerase to function (39) (Supplementary Figure S4d). The level of hTERP protein decreased slightly in clones 60 and 71 (Figure 4A).
Figure 4.
hTERP protects cells from drug-induced apoptosis. (A) hTERP level in HEK293T expressing wild-type hTERC and in clones 60 and 71 in comparison with the hTERP level in wild-type HEK293T cells. (B) The viability of wild-type HEK293T cells and HEK293T cells with exogenous expression of hTERCwt or expressing a mutant form of hTERC was analyzed by MTT assays after doxorubicin treatment. Titration curves showing the percentage of living cells after 48 h of treatment with apoptosis-inducing compounds. (C) The viability of wild-type HEK293T cells, clones 60 and 71 was analyzed by MTT assays after doxorubicin treatment. Titration curves showing the percentage of living cells after 48 h of treatment with apoptosis-inducing compounds. (D) Morphology of HEK293T cells and cells exogenously expressing hTERCwt or a mutant form of hTERC after 25 h treatment with 0.2 μM doxorubicin. Left panels, untreated cells; right panels, treated cells.
Stable HEK293T cell lines overexpressing wild-type (hTERCwt) and two mutant forms (Supplementary Figure S1c) were produced by lentiviral transduction. The hTERCstU mutant contained a mutation at the start AUG codon that impaired ORF, and the hTERCstop cell line contained a substitution of C 229 for A that resulted in the appearance of a preliminary stop codon and the predicted synthesis of a short N-terminal peptide (17aa) instead of hTERP protein (Supplementary Figure S1c). The level of hTERC increased for 1.5–3-fold than in HEK293T cells as was revealed by RT-qPCR (Supplementary Figure S4b) and telomerase activity increased for 2.5–3 times in comparison to HEK293T cells (Supplementary Figure S4c) for all obtained cell lines. The level of hTERP increased in HEK293T cells expressing wild-type hTERC (Figure 4A) and decreased slightly in clones 60 and 71.
It was previously shown that increased levels of hTERC provide anti-apoptotic defense in human immune cells (16). This effect was independent of telomerase activity and was suggested to be an alternative function of hTERC. It should be mentioned that the region of hTERC involved in the anti-apoptotic functions of hTERC corresponds to the ORF encoded hTERP, thus suggesting that this function may belong to hTERP. To address the role of hTERP in protecting cells from apoptosis, all cell lines were treated with doxorubicin or etoposide, which are known to induce apoptosis (40,41). Standard MTT assays (42) were used to measure the level of cell survival upon treatment with increasing amounts of drugs. Significant anti-apoptotic protection was observed for the cell line that additionally expressed hTERCwt, corresponding up to 80% of survived cells even after 48 h of treatment with high concentrations of the drugs (Figure 4B;Supplementary Figure S7 and Supplementary Table S6). The cells that additionally expressed mutant hTERC (hTERCstU, hTERCstop) died in a manner similar to untransfected HEK293T cells and HEK293T cells transduced with an empty vector (vector) (Figure 4B, Supplementary Figure S7). Decreased cell survival was observed for clone 71 (Figure 4C, Supplementary Table S6), which expressed mutant hTERP that lacked four amino acids in the N-terminal domain (Supplementary Figure S1d). Significant morphological changes in cells of all lines were observed after 25 h treatment with 0.2 μg/ml doxorubicin, except for the line expressing hTERCwt, which produced additional amounts of wild-type hTERP (Figure 4D). These data indicate that it is not the level of hTERC and not the telomerase activity (Supplementary Figure S4b and c), but the production of hTERP (Figure 4A, Supplementary Figure S7) determines the level of survived cells upon drug-induced apoptosis. The additional expression of hTERP protects cells from the drug-induced apoptosis and the decreasing of the hTERP level reduces the cell survival rate.
hTERP mutations affect the autophagosome formation
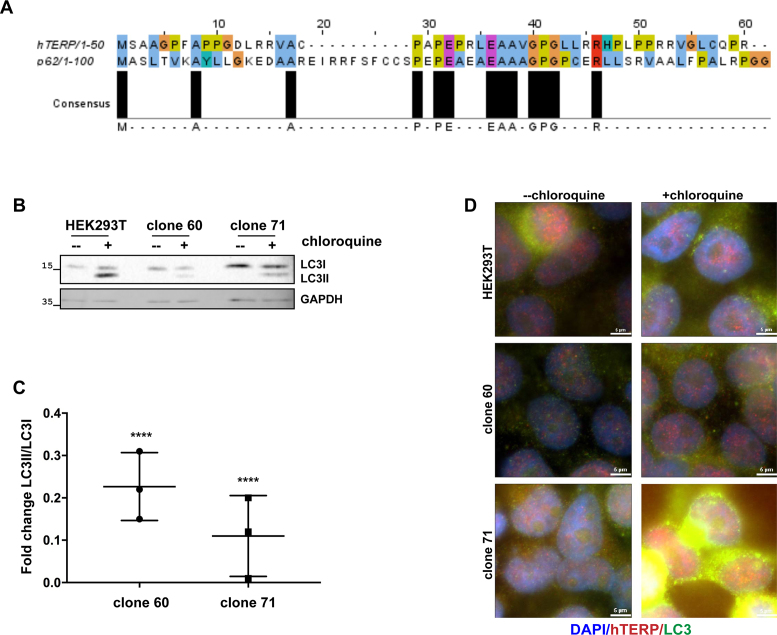
Under various circumstances, autophagy is employed as a mechanism of cellular protection under apoptosis-inducing conditions. The N-terminal 1–37 sequence of hTERP exhibited 30% identity and 60% similarity to the 1–50 N-terminal amino acids of p62 (43) protein of humans (Figure 5A). p62 or SQSTM1 (sequestosome) is a protein involved in ubiquitinated protein recycling through autophagosome degradation (43,44). It is known that p62 binds ubiquitinated proteins and targets them to autophagosomes for degradation. The N-terminal motif of hTERP is similar to the p62 motif, which corresponds to the oligomerization of p62 (45,46) and its binding with leukocyte-specific kinase (47). p62 regulates the processing of LC3 protein into the form involved in autophagosome formation. We suggested that hTERP may play a role in autophagy–apoptosis crosstalk due to the homology with p62. We used LC3 protein as a marker of autophagy function. LC3I is a soluble cytoplasmic protein that conjugates with phosphatidylethanolamine to form LC3II. The processed form of LC3 (LC3II) recruits autophagophores (48) and stimulates the curving of the membrane, leading to autophagosome formation. Newly formed autophagosomes fuse with lysosomes, and the components that had been recruited to the autophagosome and proteins that participated in autophagosome maturation are then digested by lysosome enzymes. LC3II is degraded during autophagy progression after autophagosome and lysosome fusion (48). Treating cells with chloroquine prevents autophagosome fusion with lysosomes, causing the processed form of LC3 (LC3II) to accumulate in cells (48). We observed decreased levels of LC3 in clone 60 and increased levels of LC3 in clone 71 compared to HEK293T cells (Figure 5B), suggesting that hTERP may influence the amount of LC3 in cells via its N-terminal domain. The conversion of LC3I to LC3II was impaired both in clone 60 and clone 71 compared to in HEK293T cells when autophagosome fusion with lysosomes was blocked by chloroquine treatment (Figure 5B–D). Immunofluorescence analysis of LC3 and hTERP revealed their partial co-localization in HEK293T cells (Supplementary Figure S8). p62 levels were unchanged in clones 60 and 71 compared with HEK293T cells (Supplementary Figure S9a), and no differences in intracellular localization of p62 were observed with and without chloroquine treatment (Supplementary Figure S9b).
Figure 5.
hTERP is involved in autophagy regulation. (A) Alignment of the N-terminus regions of hTERP and p62 was performed by JalView. Amino acids are colored in ClustalX colors. (B) Immunoblotting of lysates from HEK293T cells and clone 71 and clone 60 for LC3 in normal conditions and after treatment with 10 μM chloroquine for autophagy inhibition. (C) Quantification of the conversion rate of LC3I to LC3II. The conversion rate of LC3I to LC3II was determined from three independent western blots (mean ± SEM(standard error of the mean)) and quantified by GraphPad Software. The ‘****’ indicates P < 0.0001 by Dunett’s multiple comparison test. (D) Representative immunofluorescence images of HEK293T cells and of clone 60 and clone 71 with altered hTERP ORF in normal conditions and after treatment with 10 μM chloroquine for autophagy inhibition stained for hTERP and LC3; blue, DAPI-stained nuclei; red, hTERP and green, LC3; scale bars, 5 μm.
DISCUSSION
Telomerase RNA was discovered as noncoding RNA that is essential for telomerase complex formation and action (9). Telomerase RNA provides the structural scaffold for telomerase complex formation (49), contains template region for synthesis of telomeric repeats (50) and participates in processive telomere repeat synthesis (51). Number of alternative functions for TERT were described: participating in different kinase pathways regulation, localization in mitochondria during stress and other (52–54). Alternative functions of telomerase RNA are less evident. However, it was shown that hTERC is involved in cellular response to the DNA damage conditions due to regulation of activity of ATM/ATR kinases (15). The level of p53 increased under conditions when hTERC expression was reduced (15). Increased level of hTERC or mutations that prevent the association of hTERC with hTERT protected cells from drug-induced apotosis (16). Knockout of TERC in mice promoted the defects in tissue renewal, wound healing, dysfunctions of mitochondria that may be caused by general metabolism impairments. Mouse TERC (mTERC) deficiency resulted in glucose uptake and insulin secretion impairment (55,56) that may correlate with increased activity of the mammalian target of rapamycin (mTOR) in mTERC-deficient mice (57).
TERC in vertebrates is synthesized by RNA–polymerase II as a precursor (17), contains cap-structure at 5′-end (20) and may be polyadenylated (18). A small degree of cytoplasmic localization (22) and association with polysome fraction (4) for hTERC were shown. hTERC primary structure analysis revealed that it codes for protein. The alignment of TERP sequences from different vertebrates has shown some similarity with the majority of analyzed organisms (Figure 1B and Supplementary Figure S1b). The more significant differences were observed when we compared mice TERP, which is started from Leu (CUG-start codon), with the other vertebrates TERPs. The unconserved primary structure of TERP may indicate that the function of TERP and its peptide may not be conserved in rodents. We have proved that protein named hTERP is indeed expressed in the HEK293T cells by combination of several alternative approaches described above.
We proved the anti-apoptotic defense under drug treatment conditions by hTERP and observed that ectopic expression of wild-type, but not mutated at AUG-codon (start codon for hTERP) hTERC contributes to protection of cells. Thus, protein hTERP encoded in TERC but not TERC itself may provide the alternative functions of TERC described earlier (15). Previously reported data about defects in autophagy progression in telomerase-deficient mice promoted our interest to the hTERP involvement in autophagy regulation. Autophagy is the mechanism that activates during nutrient deficiency and has anti-apoptotic protective effect (58). We observed the autophagy progression defects as well as increased sensitivity to the apoptosis of cells expressing mutated hTERC obtained after CRISPR/Cas9 treatment.
Our data allow us to propose that hTERP plays a role in autophagy regulation. We observed the delayed LC3 processing—one of the initial steps of autophagosome formation. The fact that mutations in the N-terminal domain of hTERP affect LC3I conversion to LC3II and the influence of these mutations on LC3 concentration in cells provides the evidence that hTERP may be involved in the correct processing of LC3 protein and autophagosome maturation that may explain the influence of hTERP on cell survival under drug-induced apoptosis conditions.
Our findings are in agreement with previously obtained data of nontelomeric functions of telomerase RNA (16) and provide further evidence that the novel protein hTERP encoded in telomerase RNA plays a role in cell survival. hTERP is involved in the protection of cells from stress and helps cells survive and adapt under unfavorable conditions. The coding capacity of hTERC may explain the constitutive expression of this RNA in the majority of somatic cells. Our data provide new evidence that RNAs that have well-established function in the cell as noncoding can have noncanonical coding capacity, and the resulting peptide discussed herein is involved in the regulation of essential processes in living cells. The detailed molecular mechanism of hTERP involvement in the regulation of autophagy and apoptosis remains to be understood.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Foundation for Basic Research [17–04-00948 A]; Russian Science Foundation [16–14-10047]; Lomonosov Moscow State University Development Program [PNR 5.13]. Funding for open access charge: Russian Science Foundation [16-14-10047].
Conflict of interest statement. None declared.
REFERENCES
- 1. Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C. et al. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. [DOI] [PubMed] [Google Scholar]
- 2. Kim M.-S., Pinto S.M., Getnet D., Nirujogi R.S., Manda S.S., Chaerkady R., Madugundu A.K., Kelkar D.S., Isserlin R., Jain S. et al. A draft map of the human proteome. Nature. 2014; 509:575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Slavoff S.A., Mitchell A.J., Schwaid A.G., Cabili M.N., Ma J., Levin J.Z., Karger A.D., Budnik B.A., Rinn J.L., Saghatelian A.. Peptidomic discovery of short open reading frame-encoded peptides in human cells. Nat. Chem. Biol. 2013; 9:59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Heesch S., van Iterson M., Jacobi J., Boymans S., Essers P.B., de Bruijn E., Hao W., MacInnes A.W., Cuppen E., Simonis M.. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014; 15:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guttman M., Russell P., Ingolia N.T., Weissman J.S., Lander E.S.. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell. 2013; 154:240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Anderson D.M., Anderson K.M., Chang C.-L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R. et al. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015; 160:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T. et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016; 351:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Matsumoto A., Pasut A., Matsumoto M., Yamashita R., Fung J., Monteleone E., Saghatelian A., Nakayama K.I., Clohessy J.G., Pandolfi P.P.. mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature. 2017; 541:228–232. [DOI] [PubMed] [Google Scholar]
- 9. Feng J., Funk W.D., Wang S.S., Weinrich S.L., Avilion A.A., Chiu C.P., Adams R.R., Chang E., Allsopp R.C., Yu J.. The RNA component of human telomerase. Science. 1995; 269:1236–1241. [DOI] [PubMed] [Google Scholar]
- 10. Nakamura T.M., Morin G.B., Chapman K.B., Weinrich S.L., Andrews W.H., Lingner J., Harley C.B., Cech T.R.. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997; 277:955–959. [DOI] [PubMed] [Google Scholar]
- 11. Bodnar A.G., Ouellette M., Frolkis M., Holt S.E., Chiu C.P., Morin G.B., Harley C.B., Shay J.W., Lichtsteiner S., Wright W.E.. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998; 279:349–352. [DOI] [PubMed] [Google Scholar]
- 12. Tseng C.-K., Wang H.-F., Burns A.M., Schroeder M.R., Gaspari M., Baumann P.. Human telomerase RNA processing and quality control. Cell Rep. 2015; 13:2232–2243. [DOI] [PubMed] [Google Scholar]
- 13. Ulaner G.A., Hu J.-F., Vu T.H., Giudice L.C., Hoffman A.R.. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998; 58:4168–4172. [PubMed] [Google Scholar]
- 14. Yashima K., Maitra A., Rogers B.B., Timmons C.F., Rathi A., Pinar H., Wright W.E., Shay J.W., Gazdar A.F.. Expression of the RNA component of telomerase during human development and differentiation. Cell Growth Differ. 1998; 9:805–813. [PubMed] [Google Scholar]
- 15. Kedde M., le Sage C., Duursma A., Zlotorynski E., van Leeuwen B., Nijkamp W., Beijersbergen R., Agami R.. Telomerase-independent regulation of ATR by human telomerase RNA. J. Biol. Chem. 2006; 281:40503–40514. [DOI] [PubMed] [Google Scholar]
- 16. Gazzaniga F.S., Blackburn E.H.. An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity. Blood. 2014; 124:3675–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hinkley C.S., Blasco M.A., Funk W.D., Feng J., Villeponteau B., Greider C.W., Herr W.. The mouse telomerase RNA 5"-end lies just upstream of the telomerase template sequence. Nucleic Acids Res. 1998; 26:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapon C., Cech T.R., Zaug A.J.. Polyadenylation of telomerase RNA in budding yeast. RNA. 1997; 3:1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 19. Teixeira M.T., Förstemann K., Gasser S.M., Lingner J.. Intracellular trafficking of yeast telomerase components. EMBO Rep. 2002; 3:652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jády B.E., Bertrand E., Kiss T.. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body–specific localization signal. J. Cell Biol. 2004; 164:647–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cabili M.N., Dunagin M.C., McClanahan P.D., Biaesch A., Padovan-Merhar O., Regev A., Rinn J.L., Raj A.. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015; 16:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shukla S., Schmidt J.C., Goldfarb K.C., Cech T.R., Parker R.. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nat. Struct. Mol. Biol. 2016; 23:286–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen D., Grenier St-Sauveur V., Bergeron D., Dupuis-Sandoval F., Scott M.S., Bachand F.. A Polyadenylation-Dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 2015; 13:2244–2257. [DOI] [PubMed] [Google Scholar]
- 24. Weber K., Bartsch U., Stocking C., Fehse B.. A multicolor panel of novel lentiviral ‘gene ontology’ (LeGO) vectors for functional gene analysis. Mol. Ther. J. Am. Soc. Gene Ther. 2008; 16:698–706. [DOI] [PubMed] [Google Scholar]
- 25. Zhang J., Nuebel E., Wisidagama D.R.R., Setoguchi K., Hong J.S., Van Horn C.M., Imam S.S., Vergnes L., Malone C.S., Koehler C.M. et al. Measuring energy metabolism in cultured cells, including human pluripotent stem cells and differentiated cells. Nat. Protoc. 2012; 7:1068–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y., Zhu W., Levy D.E.. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006; 39:356–362. [DOI] [PubMed] [Google Scholar]
- 27. Schmittgen T.D., Livak K.J.. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008; 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 28. Rosner M., Schipany K., Hengstschläger M.. Merging high-quality biochemical fractionation with a refined flow cytometry approach to monitor nucleocytoplasmic protein expression throughout the unperturbed mammalian cell cycle. Nat. Protoc. 2013; 8:602–626. [DOI] [PubMed] [Google Scholar]
- 29. Ito-Harashima S., Kuroha K., Tatematsu T., Inada T.. Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007; 21:519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vizcaíno J.A., Csordas A., del-Toro N., Dianes J.A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T. et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016; 44:D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M., Appel R., Bairoch A.. Walker J. Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook. 2005; NY: Humana Press; 571–607. [Google Scholar]
- 32. Li X., Wang W., Wang J., Malovannaya A., Xi Y., Li W., Guerra R., Hawke D.H., Qin J., Chen J.. Proteomic analyses reveal distinct chromatin-associated and soluble transcription factor complexes. Mol. Syst. Biol. 2015; 11:775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li X., Tran K.M., Aziz K.E., Sorokin A.V., Chen J., Wang W.. Defining the protein-protein interaction network of the human protein tyrosine phosphatase family. Mol. Cell Proteomics. 2016; 15:3030–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xu S., Li X., Gong Z., Wang W., Li Y., Nair B.C., Piao H., Yang K., Wu G., Chen J.. Proteomic analysis of the human cyclin-dependent kinase family reveals a novel CDK5 complex involved in cell growth and migration. Mol. Cell Proteomics. 2014; 13:2986–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vasilkova D.V., Azhibek D.M., Zatsepin T.S., Naraikina Y.V., Prassolov V.S., Prokofjeva M.M., Zvereva M.I., Rubtsova M.P.. Dynamics of human telomerase RNA structure revealed by antisense oligonucleotide technique. Biochimie. 2013; 95:2423–2428. [DOI] [PubMed] [Google Scholar]
- 36. Azhibek D., Zvereva M., Zatsepin T., Rubtsova M., Dontsova O.. Chimeric bifunctional oligonucleotides as a novel tool to invade telomerase assembly. Nucleic Acids Res. 2014; 42:9531–9542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim N.W., Wu F.. Advances in quantification and characterization of telomerase activity by the telomeric repeat amplification protocol (TRAP). Nucleic Acids Res. 1997; 25:2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marrone A., Sokhal P., Walne A., Beswick R., Kirwan M., Killick S., Williams M., Marsh J., Vulliamy T., Dokal I.. Functional characterization of novel telomerase RNA (TERC) mutations in patients with diverse clinical and pathological presentations. Haematologica. 2007; 92:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xin Z.-T., Beauchamp A.D., Calado R.T., Bradford J.W., Regal J.A., Shenoy A., Liang Y., Lansdorp P.M., Young N.S., Ly H.. Functional characterization of natural telomerase mutations found in patients with hematologic disorders. Blood. 2007; 109:524–532. [DOI] [PubMed] [Google Scholar]
- 40. Wang S., Konorev E.A., Kotamraju S., Joseph J., Kalivendi S., Kalyanaraman B.. Doxorubicin induces apoptosis in normal and tumor cells via distinctly different mechanisms intermediacy of H2O2- and p53-dependent pathways. J. Biol. Chem. 2004; 279:25535–25543. [DOI] [PubMed] [Google Scholar]
- 41. Hahm S.-H., Chung J.H., Agustina L., Han S.-H., Yoon I.-S., Park J.-H., Kang L.-W., Park J.W., Na J.J., Han Y.S.. Human MutY homolog induces apoptosis in etoposide-treated HEK293 cells. Oncol. Lett. 2012; 4:1203–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983; 65:55–63. [DOI] [PubMed] [Google Scholar]
- 43. Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.-A., Outzen H., Øvervatn A., Bjørkøy G., Johansen T.. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007; 282:24131–24145. [DOI] [PubMed] [Google Scholar]
- 44. Johansen T., Lamark T.. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011; 7:279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wilson M.I., Gill D.J., Perisic O., Quinn M.T., Williams R.L.. PB1 domain-mediated heterodimerization in NADPH oxidase and signaling complexes of atypical protein kinase C with Par6 and p62. Mol. Cell. 2003; 12:39–50. [DOI] [PubMed] [Google Scholar]
- 46. Lamark T., Perander M., Outzen H., Kristiansen K., Øvervatn A., Michaelsen E., Bjørkøy G., Johansen T.. Interaction codes within the family of mammalian phox and Bem1p domain-containing proteins. J. Biol. Chem. 2003; 278:34568–34581. [DOI] [PubMed] [Google Scholar]
- 47. Joung I., Strominger J.L., Shin J.. Molecular cloning of a phosphotyrosine-independent ligand of the p56lck SH2 domain. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:5991–5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T.. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000; 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zappulla D.C., Cech T.R.. Yeast telomerase RNA: a flexible scaffold for protein subunits. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shippen-Lentz D., Blackburn E.H.. Functional evidence for an RNA template in telomerase. Science. 1990; 247:546–552. [DOI] [PubMed] [Google Scholar]
- 51. Blackburn E.H., Collins K.. Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb. Perspect. Biol. 2011; 3:a003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Singhapol C., Pal D., Czapiewski R., Porika M., Nelson G., Saretzki G.C.. Mitochondrial telomerase protects cancer cells from nuclear DNA damage and apoptosis. PLoS One. 2013; 8:e52989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miwa S., Saretzki G.. Telomerase and mTOR in the brain: the mitochondria connection. Neural Regen. Res. 2017; 12:358–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chiodi I., Mondello C.. Telomere-independent functions of telomerase in nuclei, cytoplasm, and mitochondria. Front. Oncol. 2012; 2:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kuhlow D., Florian S., von Figura G., Weimer S., Schulz N., Petzke K.J., Zarse K., Pfeiffer A.F.H., Rudolph K.L., Ristow M.. Telomerase deficiency impairs glucose metabolism and insulin secretion. Aging. 2010; 2:650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Missios P., Zhou Y., Guachalla L.M., von Figura G., Wegner A., Chakkarappan S.R., Binz T., Gompf A., Hartleben G., Burkhalter M.D. et al. Glucose substitution prolongs maintenance of energy homeostasis and lifespan of telomere dysfunctional mice. Nat. Commun. 2014; 5:4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheng H., Fan X., Lawson W.E., Paueksakon P., Harris R.C.. Telomerase deficiency delays renal recovery in mice after ischemia-reperfusion injury by impairing autophagy. Kidney Int. 2015; 88:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mariño G., Niso-Santano M., Baehrecke E.H., Kroemer G.. Self-consumption: the interplay of autophagy and apoptosis. Nat. Rev. Mol. Cell Biol. 2014; 15:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.