Abstract
Nucleases play important roles in nucleic acid metabolism. Some archaea encode a conserved protein known as Hef-associated nuclease (HAN). In addition to its C-terminal DHH nuclease domain, HAN also has three N-terminal domains, including a DnaJ-Zinc-finger, ribosomal protein S1-like, and oligonucleotide/oligosaccharide-binding fold. To further understand HAN’s function, we biochemically characterized the enzymatic properties of HAN from Pyrococcus furiosus (PfuHAN), solved the crystal structure of its DHH nuclease domain, and examined its role in DNA repair. Our results show that PfuHAN is a Mn2+-dependent 3′-exonuclease specific to ssDNA and ssRNA with no activity on blunt and 3′-recessive double-stranded DNA. Domain truncation confirmed that the intrinsic nuclease activity is dependent on the C-terminal DHH nuclease domain. The crystal structure of the DHH nuclease domain adopts a trimeric topology, with each subunit adopting a classical DHH phosphoesterase fold. Yeast two hybrid assay confirmed that the DHH domain interacts with the IDR peptide of Hef nuclease. Knockout of the han gene or its C-terminal DHH nuclease domain in Haloferax volcanii resulted in increased sensitivity to the DNA damage reagent MMS. Our results imply that HAN nuclease might be involved in repairing stalled replication forks in archaea.
INTRODUCTION
DNA replication and repair are vital for all living cells to stabilize their genetic information. The molecular mechanism of DNA metabolism has been studied in many kinds of organisms from the three domains of life: bacteria, eukarya and archaea (1). The bacterial and archaeal/eukaryotic replication systems evolved from two independent branches (2). Because archaea use a simplified eukaryotic DNA replication and repair system (3,4), it is an ideal model for gaining clues to the molecular mechanisms of eukaryotic nucleic acid metabolism.
Studies on replication restart in bacteria strongly suggest that many replication forks will stop when they encounter a roadblock or DNA damage. Replication restart pathways function both in bacterial and eukaryotic cells, and key roles of related proteins in these processes are conserved. RecBCD, DnaC and XerCD are the core repair factors for repairing stalled replication forks in bacteria (5). In eukaryotes, MUS81, XPF and FANCM are involved in replication restart, DNA repair, and meiotic recombination, respectively (6). However, compared with bacteria, DNA replication and repair are poorly understood in archaea. Archaeal Hef nuclease is essential for cell viability, and when the Holliday junction resolvase Hjc is absent, it provides an alternative pathway to restart stalled DNA replication forks (7). It was reported that a new nuclease, which associates with Hef, contributes to restarting the stopped replication forks during DNA replication. This Hef-associated nuclease (HAN) from Thermococcus kodakarensis (T. kodakarensis) is a 3′-exonuclease and has a classical nuclease domain from the DHH phosphoesterase superfamily (8).
The DHH phosphoesterase superfamily possesses a diagnostic motif consisting of three conserved DHH residues and commonly exists in eukaryotic, bacterial and archaeal genomes. DHH phosphoesterases have many functions in cells, such as participating in DNA repair, hydrolysing cyclic nucleoside messengers, and recycling RNA (9,10). The DHH phosphoesterase superfamily is divided into the DHHA1 and DHHA2 subfamily based on differences in the C-terminal conserved motif VII (9). The DHHA1 subsuperfamily includes bacterial RecJ nuclease, archaeal GINS-associated nuclease (GAN), nanoRNase (Nrn), HAN and YybT families. The DHHA2 subsuperfamily includes inorganic pyrophosphatase (PPase) II, yeast exopolyphosphatase, Drosophila prune protein, and pApase families. To date, additional novel DHH nucleases have been identified and characterized. Some studies have been performed to elucidate their biochemical properties and physiological roles. Each family within the DHH superfamily is characterized by different activities, substrate profiles, and in vivo functions. Bacterial RecJ has intrinsic 5′-exonuclease activity and functions primarily in DNA recombination repair (11), mismatch repair (12), and base excision repair pathways (13). Archaeal GAN is the homologue of bacterial RecJ (14) and has 5′- and 3′-exonuclease activities on ssDNA and ssRNA, respectively (15,16). Archaeal GAN forms a complex with GINS, and its nuclease activity is stimulated by GINS (15,16). It is hypothesized that archaeal GAN functions as a nuclease to remove RNA primers or to proofread mismatched RNA primers (16,17). Other possible functions of archaeal GAN include DNA repair and unwinding chromosomal DNA during DNA replication (18). Archaeal GAN and eukaryotic Cdc45 are considered to be counterparts in their respective functions for replicative helicases (19,20). The Nrn family specifically degrades short nanoRNA (2–5 nt) and pAp, as well as c-di-AMP (21). NrnA (Rv2837c) from Mycobacterium tuberculosis is a DHH-DHHA1 phosphoesterase that hydrolyses c-di-AMP, which regulate many cellular processes such as sporulation, cell wall homeostasis, virulence, etc. (22). The YybT family mainly exhibits phosphodiesterase (PDE) activity on c-di-AMP and downregulates the levels of signaling molecules (23). Similar to HAN, YybT also has an N-terminal non-catalytic domain that has ATPase activity. PPase II, which has a DHHA2 motif, can hydrolyse inorganic pyrophosphate (PPi) into phosphate (24).
During the coordinated repair of interstrand cross-linked DNA damage, both Hef and HAN potentially function as nucleases by interacting with the sliding clamp PCNA in T. kodakarensis. Detailed analysis revealed that the Hef internal disordered region (IDR) interacts with HAN, though it does not bind to PCNA and HAN simultaneously (8). During the repair process, T. kodakarensis HAN (TkoHAN) functions as an exonuclease and probably acts after strand incision at the stalled fork to start homologous recombination or to remove lesions during the repair process, similar to nucleotide excision repair (25).
Although HAN is a candidate nuclease for repairing interstrand cross-linked DNA damage (8), its role in DNA repair and/or oligo(ribo)nucleotides degradation remains unclear. To elucidate its structure and function in detail, HAN from the hyperthermophilic archaea Pyrococcus furiosus was fully biochemically characterized, and the crystal structure of its C-terminal DHH nuclease domain was solved. Subsequently, Haloferax volcanii was used as a model strain to examine its possible role(s) in DNA repair by deleting the han gene and its respective coding sequences for N-terminal non-nuclease domain and C-terminal nuclease domain and by assaying the growth curve in the presence of DNA damage reagents. Our results show that P. furiosus HAN (PfuHAN) exhibits Mn2+-dependent ssDNA/ssRNA-specific 3′-exonuclease activity. PfuHAN prefers 12–25 nt ssDNAs and ssRNAs. Meanwhile, double-stranded DNA (dsDNA) with a 3′-overhang/fork could be digested from the 3′-ssDNA termini, and completely digested products were generated. The nuclease activity of PfuHAN is derived from its C-terminal DHH domain, not its N-terminal domain. Topology of the DHH nuclease domain (DND) is similar to those of DHH phosphoesterases but exists as a trimer. Disruption of the trimer results in decreased nuclease activity. Based on results from the yeast two-hybrid assay, the DHH domain of DND interacts with the IDR domain of Hef nuclease. Interaction between ssDNA and DND of PfuHAN were stimulated, and the effects on nuclease activity were confirmed. Knockout of the han gene or its C-terminal dnd fragment resulted in increased sensitivity to the DNA damage reagent MMS in H. volcanii. Collectively, our results allow us to propose a model to interpret the function of HAN as a nuclease in repairing DNA damage generated by MMS. Our results provide new clues on the roles of HAN in genome stability in archaea.
MATERIALS AND METHODS
Materials
The oligonucleotides used in this study were purchased from Takara (Dalian, China). Escherichia coli strain DH5α was used for gene cloning, and the Rosetta2(DE3)pLysS strain was used for recombinant protein expression. The expression vectors pET28a and pDEST17 were used to express recombinant proteins. The H. volcanii strain and plasmids for gene knockout were provided by Dr Allers (University of Nottingham, UK). Pyrococcus furiosus genomic DNA was purchased from the American Type Culture Collection (ATCC). KOD-plus DNA polymerase was purchased from Toyobo. Nickel–nitrilotriacetic acid resin was purchased from Bio-Rad. The crosslinking reagent BS3 and the DNA damage reagents 4-nitroquinoline-1-oxide (4NQO), mitomycin C (MMC), and methyl methanesulfonate (MMS), methyl methanesulfonate (MMS), N3-methyladenine and adenine were purchased from Sigma–Aldrich (St Louis, MO). All other chemicals and reagents were of analytical grade.
Preparation of recombinant proteins
Pyrococcus furiosus han (ORF PF0399) and H. volcanii han (ORF Hvo1018) genes were amplified from respective genomic DNA by PCR using specific primers (Supplementary Table S1) and then inserted into pDEST17 and pET28a vectors, respectively (26). Subsequently, the plasmids for expressing the N-terminal-truncated PfuHANs, including D1 (PfuHAN305–740), D2 (PfuHAN227–740), D3 (PfuHAN201–740), D4 (PfuHAN112–740) and D5 (PfuHAN50–740), and the N-terminal single and combined DnaJ-Zf, RpS1-like, and OB-fold domains, were also constructed. Amino acid substitutions were introduced into PfuHAN or its truncated version with a QuikChange® Site-Directed Mutagenesis Kit using KOD-plus DNA polymerase and appropriate primers (Supplementary Table S1). Nucleotide sequences were confirmed by DNA sequencing.
Recombinant plasmids were introduced into the Rosetta2(DE3)pLysS strain to express recombinant proteins. Isopropylthio-β-galactoside (0.5 mM final concentration) was added to a bacterial culture (OD600 = 0.6) to induce recombinant protein expression. Recombinant proteins were purified via immobilized Ni2+ affinity chromatography as follows: the bacterial pellet was resuspended in lysis buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 5 mM β-mercaptoethanol, 5 mM imidazole, 1 mM phenylmethylsulfonyl fluoride and 10% glycerol) and then disrupted by sonication. After incubation for 15 min at 75°C, cell extracts were clarified by centrifugation at 16 000 g for 30 min. After loading the supernatants onto columns pre-equilibrated with lysis buffer, resins were washed with 100 column volumes of lysis buffer containing 20 mM imidazole. Finally, bound proteins were eluted from the column using elution buffer (20 mM Tris–HCl (pH 8.0), 300 mM NaCl, 5 mM β-mercaptoethanol, 200 mM imidazole and 10% glycerol). After verifying the purity of the eluent using 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE), preparations were dialyzed using a storage buffer (20 mM Tris–HCl (pH 8.0), 100 mM NaCl and 50% glycerol) and then stored in small aliquots at −20°C.
HvoHAN was expressed and purified according to the above protocol for PfuHAN. However, the 300 mM NaCl was replaced with 1 M KCl in lysis, washing and elution buffers. Finally, purified HvoHAN was dissolved in a storage buffer consisting of 20 mM Tris–HCl (pH 8.0), 500 mM KCl and 50% glycerol and stored at −20°C.
Biochemical characterization of exonuclease activity
PfuHAN nuclease activity was first characterized in basic buffer consisting of 20 mM Tris–HCl (pH 7.5), 50 mM NaCl, 1 mM dithiothreitol (DTT), 2.0 mM MnCl2 and 100 μg/ml BSA. After optimization of reaction buffer, all reactions (20 μl) were performed in buffer containing 20 mM Tris–HCl (pH 8.0), 40 mM NaCl, 1.0 mM DTT, 1.0 mM MnCl2 and 100 μg/ml BSA. One pmole oligonucleotides or dsDNAs with various termini (Supplementary Table S2) were incubated with specified concentrations of PfuHAN or its mutants at 55°C for indicated times. After incubation, an equal volume of stopping buffer (90% formamide, 100 mM EDTA and 0.2% SDS) was added to the reaction. Subsequently, the reactions were subjected to 8 M urea-denatured 15% polyacrylamide gel electrophoresis. After electrophoresis, the gels were imaged and quantified using an FL9500 fluorescent scanner (GE Healthcare).
Crystallization
Full-length PfuHAN and its C-terminal DHH nuclease domain (PfuHAN_DND, peptide from 305 to 739) were used for crystallization after a three-step purification including Ni2+-affinity, ion-exchange and gel-filtration chromatography. Affinity-purified proteins were fully dialyzed using buffer A (25 mM HEPES (pH 8.0), and 25 mM NaCl) and loaded onto a MonoQ cation exchange column (GE Healthcare) pre-equilibrated with buffer A. PfuHAN and PfuHAN_DND were eluted with 250–500 mM NaCl. Fractions containing target proteins were first pooled and concentrated using a 30-kDa Amicon Ultra-15 centrifugal filter (Millipore) and then further purified using a 120 ml Hiload Superdex 200 column (GE Healthcare) with a buffer containing 25 mM HEPES (pH 8.0), and 150 mM NaCl. The fractions containing PfuHAN and PfuHAN_DND were pooled and concentrated to 20 mg/ml for crystallization. Selenomethionine-labeled (SeMet) PfuHAN_DND was expressed using the E. coli strain BL21 (DE3) in a defined medium and purified similar to the native protein.
After failing to obtain a crystal for PfuHAN after multiple attempts, we tried and succeeded in crystalizing PfuHAN_DND. PfuHAN_DND crystals were grown at 4°C using the hanging drop vapor-diffusion method by mixing equal volumes of protein and reservoir solution consisting of 4% PEG 400, 100 mM sodium acetate/acetic acid (pH 5.5), 1.4 M Li2SO4, and 100 mM MgSO4. After 1 week, crystals were stabilized and cryoprotected by socking into a reservoir solution containing 20% glycerol and followed by flash cooling in liquid nitrogen. The SeMet protein was crystallized using the same method.
Data collection and structure determination
X-ray diffraction data were collected at 100 K on the BL19U1 beamline at the National Centre for Protein Sciences Shanghai (NCPSS) at the Shanghai Synchrotron Radiation Facility (SSRF). Diffraction data were indexed, integrated and scaled using HKL2000 (27,28).
The Se-derivative structure of PfuHAN_DND was determined using the AutoSol programme from the PHENIX software suite (29). The initial model was refined using REFMAC5 (30) from the CCP4 programme suite and interactively rebuilt using σA-weighted electron density maps with coefficients 2mFo-DFc and mFo-DFc in the COOT programme (31). Refinement converged to an R-factor of 18.66% and R-free of 20.18%. The stereochemical quality of the final model was assessed using MolProbity (32). All structural figures were prepared with PyMOL (DeLano Scientific). Data collection and refinement statistics for the PfuHAN_DND structure are shown in Supplementary Table S3.
Identification of the PfuHAN-bound metal ions
The metal ions bound in PfuHAN were determined as follows. First, the freeze-dried protein was digested using 2 ml HNO3 in a microwave digestion system (Anton Paar, Multiwave 3000, Austria). Then, the mixture was cooled to room temperature and adjusted to a final volume of 10 ml with Milli-Q ultrapure water. Finally, the metal ion content was measured via inductively coupled plasma mass spectrometry (ICP-MS, Thermo Fisher, iCAPQ, USA). Standard solutions for Ni2+, Mn2+ and Mg2+ ranging in concentration from 20 to 100 ppb were prepared from analytically pure reagents using Milli-Q ultrapure water. Buffer and Milli-Q ultrapure water were used as controls in the assays. No measurable quantities of metal ions were detected in the controls.
Characterization of the PfuHAN_DND trimer complex
Protein crosslinking with BS3 was performed according to the manufacturer's instructions (Thermo Scientific). The crosslinking reagent BS3, with a final concentration of 5 mM, was mixed with the protein samples (1.0–2.0 mg/ml) and dissolved in buffer consisting of 20 mM HEPES (pH 7.5), 100 mM sodium phosphate, 150 mM NaCl, and 100 mM carbonate. To crosslink the potential protein complex, mixtures were incubated at room temperature (RT) for 30 min. The reaction was terminated by adding a final concentration of 40 mM Tris–HCl (pH 7.5) and 40 mM glycine and incubated at RT for 15 min. After stopping the crosslinking reaction, samples were subjected to 15% SDS-PAGE to validate generation of the cross-linked complex.
Gene knockout and phenotype assay
The full-length han gene (ORF Hvo1018) and its 5′ and 3′ region, which respectively encode the N-terminal non-nuclease fragment and C-terminal DND, were deleted in H. volcanii using the Pop-in/Pop-out method according to the protocol reported by Allers (33). Genotypes of gene-knockout strains were first analysed by PCR using 5′ and 3′ flanking primers, and the resulting amplified fragments were sequenced to confirm gene disruption.
WT and mutant H. volcanii strains (Δhan, Δntd, Δdnd) were grown aerobically at 45°C overnight in Hv-YPC rich medium. In growth curve assays, overnight cultures were diluted to an OD600 of 0.05, and their growth was monitored for 32 h by taking OD600 readings every 4 h. The DNA damage reagents H2O2, 4NQO, MMC, and MMS were added to the liquid medium at their respective concentrations. In drop dilution assays, cultures (2.3 × 106 cells/ml) were serially diluted 10-fold with 18% ASW. Diluted cultures were spotted on YPC plates with or without DNA damage reagents and incubated at 45°C for 5–7 days. In survival percentage assays, overnight cultures were treated with 0, 2, 4 or 6 mM MMS in 18% ASW for 0.5 h and spread on YPC plates. Survival percentages were calculated by dividing the clone number of each treatment condition (2–6 mM MMS) by that of 0 mM MMS.
UPLC-Mass Spectrometry (ACQUITYTM UPLC & Q-TOF MS Premier) was used to confirm the accumulation of DNA damage in cells as described in previous methods with some modifications (34). Genomic DNAs were extracted from 50 ml H. volcanii cells as described (33) and dissolved in 90% formic acid at a final concentration of 0.5 mg/ml. The DNA solution was incubated at 70°C for 12 h to release bases, and then 10 volumes of ddH2O was added. Finally, the supernatant was collected by centrifuging at 6000 g for 10 min and vacuum dried at −50°C. The lyophilizate was dissolved in 50 μl 0.1% formic acid for analysing the content of N3-methyladenine by UPLC-MS.
Yeast two-hybrid assays for identifying interaction between HAN and hef
Yeast two-hybrid assays were performed to identify the interaction between Hef and HAN nucleases using the Matchmaker™ Gold Yeast Two-hybrid System (Clontech) according to the manufacturer's protocol. The IDR fragment of P. furiosus Hef (PfuHef) nuclease was inserted into the pGADT7 plasmid, encoding the activation domains, and the fragments corresponding to each domain of PfuHAN were inserted into the pGBKT7 plasmid, encoding the GAL4 DNA binding region. Next, constructed pGADT7 and pGBKT7 series plasmids were co-transformed into yeast strain Gold, and transformants were selected on S.D. medium lacking Leu and Trp at 30°C for 3 days. Finally, selected transformants were cultured in liquid medium and spotted onto SD plates lacking Leu, Trp, and His. Yeast cells were grown at 30°C for 4 days for analyzing potential interactions.
Homologous modeling and molecular dynamics stimulation
A ssDNA of 5′-GATGTAC-3′ was first built in PYMOL-1.8 (35), and subsequently, an optimized conformer of the ssDNA was docked into the crystal structure of PfuHAN_DND in AUTODOCK (36). The AMBER14 suit package (37) was used to construct solvent-protein systems and perform molecular dynamic simulation. For the simulation, appropriate ions were added to the system as counterions lead to neutrality. Next, the protein-DNA complex and ions were solvate in a truncated octahedron box of TIP3P waters with a water thickness of 10 Å. Long-range electrostatic interactions were calculated using the Particle Mesh Ewald (PME) algorithm (38). Bonds involving hydrogen atoms were constrained using the SHAKE algorithm (39). All systems were minimized to 40 000-step with steepest descent method, then with heating for 20 ps and equilibrating for 20 ps in the NVT ensemble at room temperature. To refine the conformers of this PfuHAN_DND-ssDNA complex, a single trajectory for the system was simulated at 300K with ff14SB force fields. Fifty nanosecond simulations were adequate for the system to converge. Furthermore, routine analysis of trajectory sampling was carried out using the CPPTRAJ in Ambertools14 (40). Calculation of the average conformer and its receptor-ligand interaction were performed using in-house programs (41,42).
RESULTS
HAN belongs to the DHH phosphoesterase superfamily
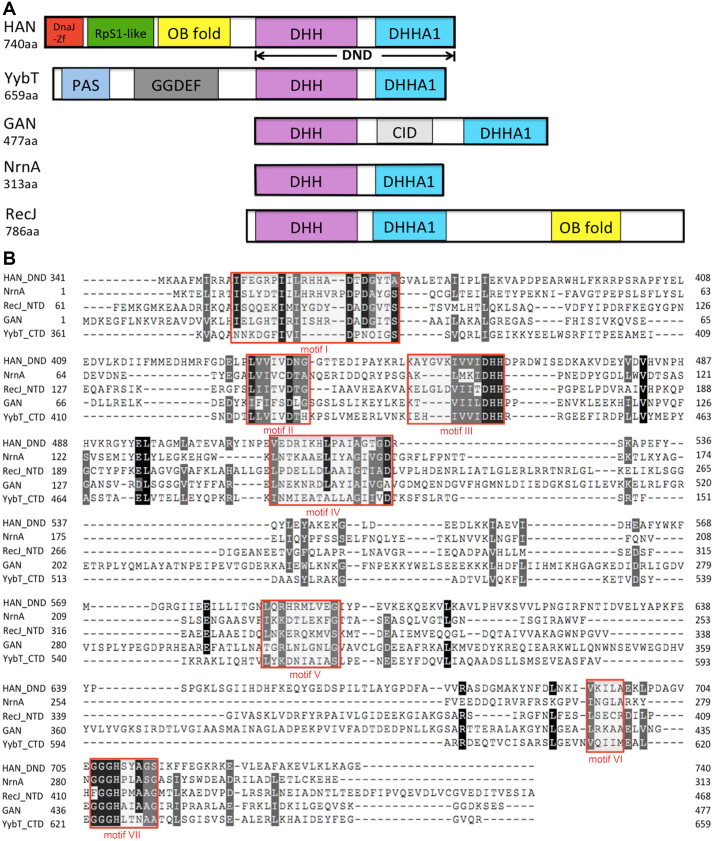
Although TkoHAN has been reported as an intrinsic 3′-exonuclease (8), little is known about its biological function(s) in vivo and in vitro. Sequence multi-alignment shows that the HAN C-terminal peptide has some sequence similarity to members of the DHH phosphoesterase superfamily, with a similar domain combination to DHH nucleases (Figure 1A). Its C-terminal peptide, ranging from aa 350 to 740, has typical motifs I-VII that are conserved in DHH phosphoesterase superfamily members (Figure 1B). HAN belongs to the DHHA1 subsuperfamily with a diagnostic motif of ‘GGGH’ (motif VII). In addition to the C-terminal DHH nuclease domain, HAN and YybT (23) also have other N-terminal domains (Figure 1A). HAN has three N-terminal domains, including a DnaJ_Zn-finger (DnaJ-Zf), a ribosomal protein S1-like (RpS1-like) domain, and an oligonucleotide/oligosaccharide-binding (OB) fold. Phylogenetic tree analysis shows that the HAN family is a specific branch separated from other DHH phosphoesterase families, including Nrn, RecJ, GAN and YybT (Supplementary Figure S1). Interestingly, HAN is only commonly present in Euryarchaeota and is not found in other phyla in archaea or bacteria (Supplementary Figure S2 and Tables S4 and S5). Conservation in motifs I-VII imply that each member of the DHH superfamily evolved from a common ancestor.
Figure 1.
Schematic representation and multi-alignment of HAN-related DHH phosphoesterase superfamily members. (A) Schematic representation of HAN, NrnA, GAN (also called archaeal RecJ), YybT and RecJ. DHH and DHHA1 indicate that the DHH domain consists of motifs I-V, and the DHHA domain consists of motifs VI and VII (motif VII is classified into DHHA1 or DHHA2 motif, based on the sequence conservation). DnaJ-Zf, RpS1-like, OB-fold, CID, PAS and GGDEF denote DnaJ type Zn-finger, ribosomal protein S1-like, and oligonucleotide/oligosaccharide-binding fold, CMG interaction domain, and Per-Arnt-Sim that are required for heme-binding and GGDEF, which originates from the conserved amino acid motif GGDEF in diguanylate cyclase, domains, respectively. (B) Sequence multi-alignment of PfuHAN and other family members of the DHHA1 phosphodiesterase subsuperfamily. One typical protein was selected from the HAN, Nrn, GAN, YybT and RecJ families and used for multi-alignment with the ClustalW programme. These proteins are PfuHAN (AAL80523), T. kodakarensis GAN (TkoGAN, BAD85441), Bacillus subtilis (B. subtilis) NrnA (BsuNrnA, NP_390803), B. subtilis RecJ (NP_390640), and B. subtilis YybT (NP_391931.1). For clarity, only DHH and DHHA1 domains from PfuHAN (PfuHAN_DND, aa341–740), N-terminal domain of BsuRecJ (BsuRecJ_NTD, aa61–468, i.e., the catalytic core of bacterial RecJ), and C-terminal domain of BsuYybT (BsuYybT_CTD, aa361–659) are used for multi-sequence alignment. Completely and partially conserved residues between the sequences are shaded in dark and light, respectively. Motifs I-VII are marked with red rectangles.
PfuHAN possesses intrinsic 3′-exonuclease activity
To understand the enzymatic properties of HAN in detail, we selected PfuHAN to characterize its nuclease activity. As a multi-domain protein, PfuHAN and its mutants showed some degradation during recombinant expression and affinity purification (Figure 2A). PfuHAN is shown in purple at a higher concentration (Supplementary Figure S3A). The purple colour is derived from the N-terminal domains (residues 1–350). After scanning the absorption values at wavelengths ranging from 220 to 750 nm, the protein showed a specific absorption peak at 350 nm (Supplementary Figure S3B).
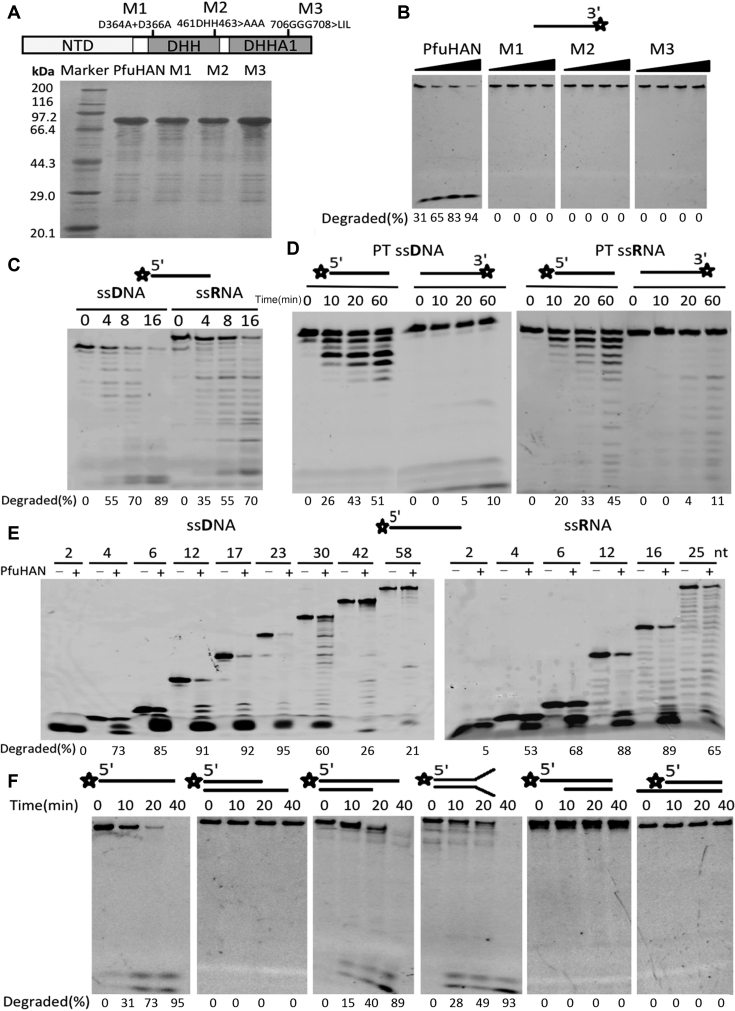
Figure 2.
Substrate preference of PfuHAN nuclease. (A) 15% SDS-PAGE of PfuHAN and its mutants. (B) Hydrolysis of ssDNA by PfuHAN and its mutants. Increasing concentrations of enzymes (3, 10, 30, and 100 nM) were incubated with a 3′-FAM-labeled 42-nt ssDNA (50 nM) in basic reaction buffer at 55°C for 20 min. (C) Preference of PfuHAN for (deoxy)ribose. A 5′-FAM-labeled 23-nt ssDNA or ssRNA (50 nM) with the same base sequences were incubated with 10 nM PfuHAN at 55°C for 0, 4, 8, and 16 min in the optimized buffer. (D) Digestion of fully phosphorothioated (PT) substrates. Fully PT ssDNAs and ssRNAs (50 nM) were incubated with 50 nM PfuHAN at 55°C for 0, 10, 20, and 60 min. (E) Length preference of PfuHAN for ssDNA and ssRNA. Different lengths of 5′-FAM-labeled ssDNAs and ssRNAs were incubated with 10 nM PfuHAN at 55°C for 20 min. (F) Degradation of DNAs with various secondary structures. DNA substrates (50 nM) were incubated with 50 nM PfuHAN at 55°C for 0, 10, 20 and 40 min. Asterisks denote the terminal fluorescent group FAM.
PfuHAN displayed strong 3′-exonuclease activities on ssDNA and released the 3′-single nucleotide product (Figure 2B). To exclude the possibility that the activity was from contaminating E. coli nucleases, site-directed mutations of conserved residues, including D364A+D366A (M1), 461DHH463>AAA (M2), and 706GGG708>LIL (M3), were used to detect the nuclease activity. All three mutants inactivated the nuclease (Figure 2B), verifying that the hydrolysis of ssDNA by PfuHAN was not caused by contaminants. Ten nM of PfuHAN is sufficient to degrade the 3′-FAM-labeled ssDNA, indicating that the FAM group does not block hydrolysis of the first phosphodiester bond.
After confirming PfuHAN nuclease activity, hydrolysis reactions were optimized using the 3′-FAM-labeled ssDNA substrate. The optimized parameters include the pH value, ion strength, divalent metal ions and Mn2+ concentration. PfuHAN showed the highest activity at pH 8.0 (Supplementary Figure S4A). Divalent ion Mn2+ is the most effective metal cofactor (Supplementary Figure S4B), with an optimal concentration of 2.0 mM (Supplementary Figure S4C). PfuHAN displayed higher activity at lower NaCl concentrations (Supplementary Figure S4D).
In addition to ssDNA, PfuHAN also showed intrinsic 3′-exonuclease activity on ssRNA (Figure 2C). However, PfuHAN showed a slight preference for ssDNA. To further confirm the hydrolysis direction of PfuHAN, fully phosphorothioated (PT) 23-nt ssDNA and ssRNA were used to characterize the degradation reactions. Results show that PfuHAN can degrade 5′-FAM-labeled fully PT ssDNA and ssRNA into ladders (Figure 2D). Only a small amount of 3′-nucleotide product was generated from 3′-FAM-labeled fully PT ssDNA, indicating that the 3′-FAM fluorescent group also inhibits degradation of the first 3′-nucleotide (Figure 2D). These results confirm that PfuHAN is a genuine 3′-exonuclease. The 3′-phosphate group strongly inhibits the removal of the first 3′-nucleotide in ssDNA and ssRNA (Supplementary Figure S5), suggesting that the free 3′-hydroxyl group is requisite for the efficient hydrolysis of the first phosphodiester bond.
Single-stranded oligonucleotides of different lengths were used to characterize the length preference of PfuHAN. Based on the degradation degree of various substrates (Figure 2E), we conclude that PfuHAN prefers 12–25 nt ssDNAs or ssRNAs. SsDNAs longer than 23 nt or shorter than 12 nt and ssRNAs shorter than 12 nt were clearly degraded at lower efficiencies. Specifically, PfuHAN could not hydrolyse 2-nt ssDNA. Similar to ssDNA, 2-nt ssRNA were not degraded by PfuHAN.
To determine whether PfuHAN possesses 3′-exonuclease activity on dsDNA, different dsDNAs with various secondary structures were used as substrates in the exonuclease assay. As shown in Figure 2F, no product was detected for dsDNAs with 3′-blunt or 3′-recessive structures. However, PfuHAN could degrade dsDNAs with a 14-nt forked or 3′-overhang termini, generating fully digested product (5′-FAM-labeled nucleotide) after long time incubation (two middle panels in Figure 2F). Although the 3′-overhang or fork of dsDNAs is longer than the 12-nt ssDNA, its cleavage activity is clearly decreased against the 12-nt ssDNA (Figure 2E and F). A possible interpretation is that the junction of single-stranded and double-stranded structures exerts some inhibition on the binding and hydrolysis of single-stranded regions.
The C-terminal DHH phosphoesterase domain provides the exonuclease activity
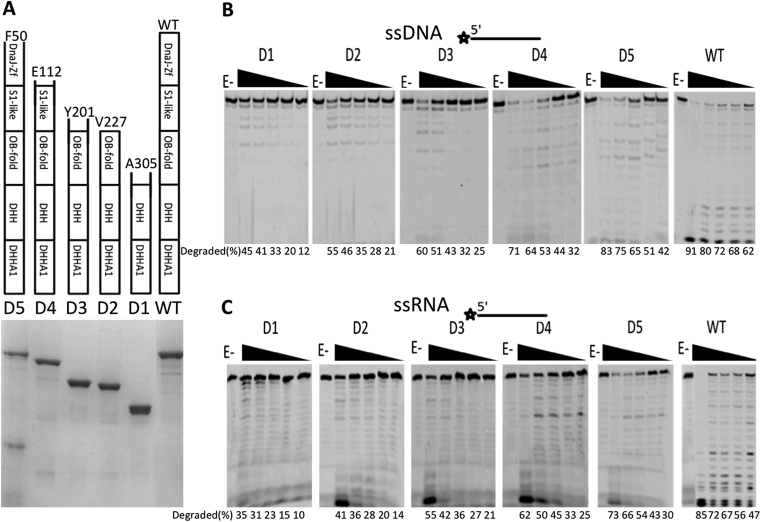
As a multi-domain protein, the function of each domain of PfuHAN needs to be determined, especially to identify the core domain(s) responsible for nuclease activity. We first confirmed that the N-terminal domains had no detectable nuclease activity (Supplementary Figure S6). To decipher the detailed function(s) of the N-terminal DNA-binding domains in hydrolysing phosphodiester bonds, five truncated PfuHAN proteins were designed (Figure 3A). Each truncated protein is 95% pure, except for truncation D5. Among them, PfuHAN-D1, which has an intact DND and a largely disrupted OB-fold domain, displayed the least efficient hydrolysis of ssDNA and ssRNA (Figure 3B and C). In contrast, PfuHAN-D2, which has a DND and intact OB-fold domain, yielded more product than did HAN-D1. The other three truncated PfuHANs with partial N-terminal domain(s) showed comparable activity to full-length PfuHAN (Figure 3B and C). In summary, the degradation activities for ssDNA and ssRNA are proportional to the length of the truncated PfuHAN protein, indicating that the N-terminal non-nuclease domains in PfuHAN might promote DND nuclease activity by assisting with substrate binding.
Figure 3.
PfuHAN nuclease activity is dependent upon the C-terminal DHH nuclease domain. (A) 15% SDS-PAGE of full-length and truncated PfuHANs. Schematic representation of the domain combinations of each protein are listed on the top of each lane in the gel. The number of starting residues for each protein is indicated. The 3′-exonuclease activities of full-length PfuHAN and the five truncated proteins were characterized by incubating enzymes with (B) ssDNA or (C) ssRNA (50 nM) at 55°C for 20 min. The concentration of PfuHAN and its truncated mutants are 0, 10, 20, 50, 100 and 200 nM.
Degradation of dsDNAs with a 7- or 14-nt 3′-overhang or fork were further characterized using the five truncated PfuHANs. Digestion of both the dsDNA duplex and 3′-ssDNA region is proportional to the length of the remaining N-terminal domain of PfuHAN (Supplementary Figure S7A and B). Native gel analysis of the conformation of dsDNA substrates with 3′-overhang or fork, used in Supplementary Figure S7A and B, showed that the dsDNAs are in duplex conformation (Supplementary Figure S7C and D). These results confirm that PfuHAN could digest a dsDNA duplex after loading onto and hydrolysing the ssDNA region of 3′-overhang or fork, which is similar to the digestion of dsDNA with 5′-overhang by bacterial RecJ nuclease (43). Similar to 3′-protruding dsDNA, digestion of different length ssDNAs is also proportional to the length of the remaining N-terminal domain of PfuHAN (Supplementary Figure S7E).
Kinetic parameters of full-length and truncated PfuHANs
To quantitatively compare the activity differences between various truncated PfuHAN proteins, the kinetics parameters of PfuHAN, D1, D2, D3, D4 and D5 were calculated by double-reciprocal plotting of the initial rate and substrate concentration (Table 1). Removal of the N-terminal domains resulted in >10-fold increase of Km values, but affected kcat values less, suggesting that these non-nuclease domains participate in ssDNA or ssRNA substrate binding during phosphodiester bond hydrolysis.
Table 1.
Comparison of Km and kcat among PfuHAN and its truncated proteins
| ssDNA | ssRNA | |||||
|---|---|---|---|---|---|---|
| K m (μM) | k cat (min−1) | k cat/Km (min−1·μM−1) | K m (μM) | k cat (min−1) | k cat/Km (min−1·μM−1) | |
| HAN | 3.6±0.2 | 0.096±0.01 | 0.027±0.003 | 2.7±0.2 | 0.064±0.005 | 0.024±0.002 |
| D1 | 58.4±0.6 | 0.056±0.004 | 0.001±0.0001 | 46.4±0.4 | 0.034±0.004 | 0.0007±0.00005 |
| D2 | 43.3±0.5 | 0.065±0.004 | 0.0015±0.0001 | 29.9±0.3 | 0.048±0.003 | 0.010±0.0004 |
| D3 | 41.4±0.3 | 0.070±0.006 | 0.0017±0.0001 | 25.2±0.3 | 0.052±0.004 | 0.0016±0.0002 |
| D4 | 14.1±0.3 | 0.084±0.007 | 0.0060±0.0001 | 10.4±0.3 | 0.058±0.004 | 0.0056±0.0005 |
| D5 | 8.8±0.3 | 0.086±0.008 | 0.010±0.001 | 6.1±0.2 | 0.062±0.005 | 0.010±0.001 |
K m and kcat were calculated by double reciprocal plotting using the initial reaction rates at various substrate concentrations (0.05, 0.1, 0.25, 0.5, 1.0 and 2.0 μM). The kinetic parameters were determined in the presence of 15 nM PfuHAN and each truncated protein. The substrates are 3′-FAM-labeled ssDNA or ssRNA, and the released 3′-FAM mononucleotide products were quantified for calculating the initial rates. All data are the means of three independent experiments.
We next attempted to examine the binding capabilities of full-length and truncated PfuHANs. Unfortunately, MST assays showed no clear substrate binding activity for PfuHAN (Supplementary Figure S8A). Next, the binding capability of full-length PfuHAN was determined under various conditions by electrophoretic mobility shift assay. PfuHAN showed very weak binding activity for ssDNA and dsDNA with 3′-overhang or forks, even in the presence of metal Mn2+ ions, and almost no bound DNAs were observed (Supplementary Figure S8B). These results suggest that it is impossible to directly observe the effect of the N-terminal domains on substrate binding. In fact, we did not observe the binding of ssDNA by five truncated PfuHAN and full-length protein (Supplementary Figure S8C).
PfuHAN_DND adopts typical topology of the DHH superfamily
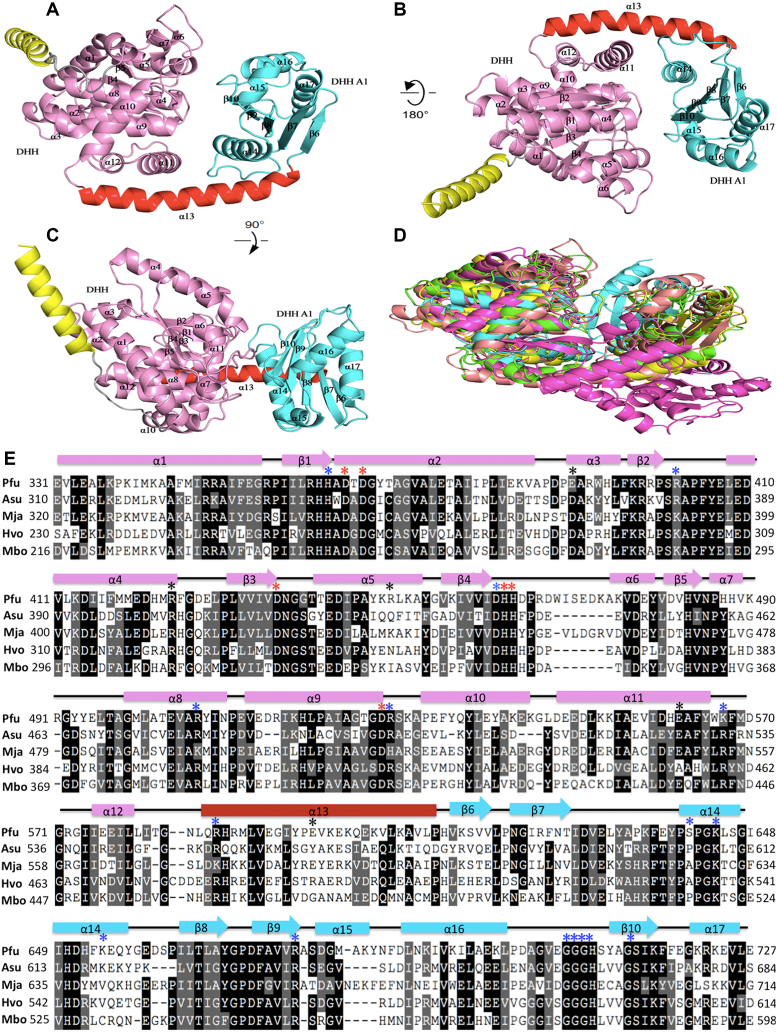
To understand the catalytic mechanism, we tried to solve the structure of PfuHAN. Despite intensive efforts, we were unable to obtain a crystal structure of full-length PfuHAN. Because the enzymatic activity is located at the C-terminal DHH nuclease domain, PfuHAN_DND (corresponding to aa 305 to 739) was used to grow a crystal instead. The crystal structure of PfuHAN_DND was determined at a 2.2-Å resolution (Supplementary Table S3). PfuHAN_DND adopts a trimer complex topology (three molecules in an asymmetric unit), and each subunit consists of an N-terminal DHH and a C-terminal DHHA1 domain connected by a long helix linker α13 (corresponding to residues 585–613) (Figure 4A–C). The N-terminal DHH domain adopts a mixed α/β fold featuring a 5-stranded parallel β-sheet at its core. The residues from motifs I-IV come together to form the active site for coordinating the divalent metal cofactor and the ordered water molecule, which is activated for nucleophilic attack on the phosphodiester bond in the backbone (44–45). The C-terminal DHHA1 domain also adopts an α/β fold with a central mixed β-sheet and helices. The DHHA1 motif (motif VII) located at the loop between helix α16 and β10 is responsible for recognizing specific substrates.
Figure 4.
Crystal structures of PfuHAN_DND and other DHH phosphoesterases. (A) The overall structure of PfuHAN_DND is shown by rotation of (B) 180° and (C) 90°. The DHH and DHHA1 domains are shown in pink and cyan, respectively. The long alpha helix connecting DHH and DHHA1 domains is shown in red. The first alpha helix belongs to the OB-fold domain and is shown in yellow. (D) Comparison of the structures of DHH superfamily members. The structures of PfuHAN_DND (pink), catalytic core (DHH and DHHA1 domain) of Thermus thermophiles RecJ (green, PDB 1IR6), TkoGAN (purple, PDB 5GHT), Streptococcus gordonii PPase (cyan, PDB 1K20), and BsuNrnA (yellow, PDB 5J21) are superposed. (E) Multiple sequence alignment of the HAN DHH-DHHA1 nuclease domains. The alignment was performed using multiple sequence alignment programmes ClustalW and was manually modified to take into account the secondary structural prediction. Completely and partially conserved residues are shaded in dark and light, respectively. Secondary structural elements in PfuHAN_DND are represented at the top of the sequences. The horizontal cylinders indicate α-helices, and the arrows indicate β-strands. The mutated conserved residues for coordinating metal ions (D364, D366, D436, H462, H463 and D528), binding substrates (H362, R402, D461, R507, R529, K567, R587, S641, K644, K654, R676, G706, G707, G708, H709 and S714), and forming trimeric complexes (E390, R449, E598, R424, and E562) are indicated with red, blue, and black asterisks, respectively, above the PfuHAN sequence. The HANs are from Pyrococcus furiosus (Pfu, PF0399), Methanocaldococcus jannaschii (Mja, MJ1198), Haloferax volcanii (Hvo, Hvo1018), Archaeoglobus sulfaticallidus (Asu, Asulf01445) and Methanoregula boonei (Mbo, Mboo1586).
Although the amino acid similarity among DHH superfamily members is less than 20%, a Dali search revealed that their three-dimensional structures are notably similar. The topology of PfuHAN_DND is similar to the structures of several DHH superfamily members (Figure 4D, and Supplementary Figure S9), including bacterial RecJs (44–46), GAN (47), NrnA (48), and PPase (49). Each of them has an N-terminal DHH and a C-terminal DHHA1/2 domain, suggesting they might have evolved from a common ancestor. However, the long alpha helix connecting the DHH and DHHA domains are slightly different. Family II PPase and BsuNrnA incorporate a split helix (Supplementary Figure S9C and E), while the other proteins adopt a long intact helix (Supplementary Figure S9A, B, D). However, BsuNrnA, RecJ, GAN, and PfuHAN_DND have conserved residues for coordinating metal ions and binding substrates (Figure 4E).
Conserved residues for binding metal ion and substrates
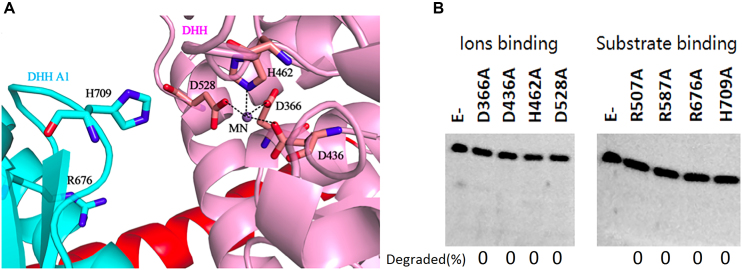
In several DHH superfamily members, such as bacterial RecJ, motifs I-IV are involved in coordinating a divalent metal cofactor that is necessary for hydrolysing the phosphodiester bond (44–46). In PfuHAN_DND, residues D366, D436, H462 and D528 (Figure 5A) are responsible for binding Mn2+. Mutation of these conserved key residues resulted in inactivation of the nuclease activity (Figure 5B, left panel), suggesting that these conserved residues are involved in coordinating Mn2+, which is the PfuHAN exonuclease cofactor. The quantitative results on bound Mn2+ by wt and mutant PfuHANs confirmed that the mutant enzymes lost the capacity to bind metal ions (Supplementary Figure S10A).
Figure 5.
Functions of conserved PfuHAN motifs. (A) Metal ion binding site. The metal ion was proven to be a Mn2+ ion by inductively coupled plasma mass spectrometry and is represented by a magenta sphere. Residues for coordinating Mn2+ are shown in stick, and the coordination bonds between the Mn2+ ion and the side chains of the residues are shown by black dashed lines. The two conserved residues R676 and H709 in the DHHA1 domain for binding substrates are also shown in stick. (B) Activity comparison between wt PfuHAN_DND and its site-directed mutants. Eight conserved residue mutants, including metal ion binding and substrate binding residues, were prepared and used to determine the effect of the conserved residues on nuclease activity. Mutant PfuHAN_DND proteins (20 nM) were incubated with ssDNA (50 nM) at 55°C for 20 min.
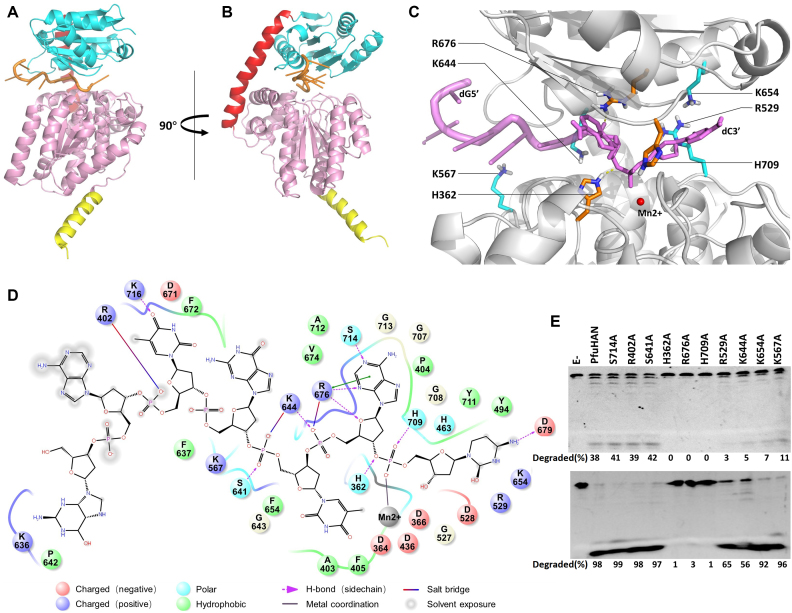
The complex structure of Deinococcus radiodurans RecJ (DraRecJ) with ssDNA showed that several conserved residues are responsible for substrate binding (45). In PfuHAN_DND, residues R507, R587, R676 and H709 (Figures 4E, 5A) correspond to the residues required for DraRecJ to bind ssDNA and are supposed to be involved in binding longer oligonucleotides. Similar to bacterial RecJ and archaeal GAN nucleases, mutation of these conserved residues in PfuHAN inactivates the nuclease (Figure 5B, right panel), indicating that these motifs are required for PfuHAN to bind substrates. However, it is impossible to verify the effect of mutated residues on substrate binding due to the low affinity of wt and mutant PfuHANs to DNA (Supplementary Figure S8B and S10B).
Because of the absence of co-crystal structures for PfuHAN_DND and ssDNA, we studied their interaction by molecular dynamics stimulation. The ssDNA is bound in the cleft between DHH and DHHA1 domains (Figure 6A–C). The interaction strength between PfuHAN_DND and ssDNA, including salt bridge, hydrogen bond and hydrophobic interaction, were analysed (Figure 6D), and the main interactional residues are shown in Figure 6C and D. Based on the computational results for the interaction, a series of residues that have strong interactions with phosphate groups, deoxyriboses, and bases were chosen to verify their roles in hydrolysing phosphodiester bonds (Figure 6E). Results show that H362, R676 and H709, which bind the two phosphate groups at the 3′-terminus via salt bridge or hydrogen bond (Figure 6D), are crucial to the enzymatic activity. However, residues R529, K567, K644 and K654, which are involved in binding the phosphate groups located far from the 3′-termimus (the third and fourth ones from the 3′-end), play a smaller role in degrading ssDNA. The two residues for binding bases R402 and S714 have no detectable effect on degradation, which is consistent with that the fact PfuHAN has no specific sequence selectivity on DNA. In summary, the residues interacting with the phosphate groups adjacent to the 3′-terminus are key for removing the 3′-nucletide.
Figure 6.
Interactions between ssDNA and PfuHAN_DND. A model of PfuHAN_DND binding a 7-nt ssDNA (5′-GATGTAC-3′) is shown in two vertical directions. (A and B) Mn2+ is represented by a purple sphere. (C) The enlarged ssDNA-binding patch of PfuHAN_DND, where the key residues for binding ssDNA are marked, and the Mn2+ is represented by a red sphere. (D) Simulated interactions between ssDNA and PfuHAN_DND. The main interaction strengths, including salt bridge, hydrogen bond, electrostatic interaction, and hydrophobic interaction, are shown. The residues ∼4 Å of ssDNA are shown, and the Mn2+ is shown as a grey sphere. (E) Degradation of ssDNA by wt and mutated PfuHAN. A 5′-FAM-labeled 23-nt ssDNA (50 nM) was incubated with 2 nM (upper panel) or 20 nM (lower panel) PfuHANs at 55°C for 20 min. The detailed protocol for homologous modeling and molecular dynamics stimulation are described in Materials and Methods. Figures were created using the PyMOL programmes (DeLano Scientific).
Interaction and function of the PfuHAN trimeric complex
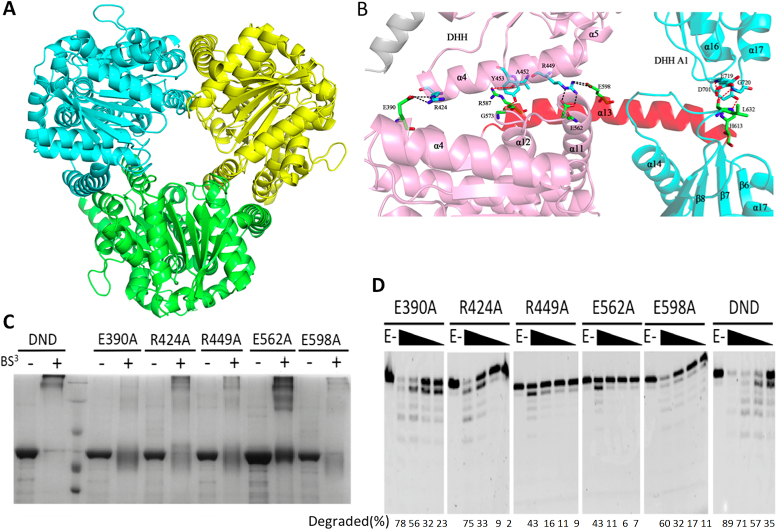
Unlike BsuNrnA, which forms a dimer via interactions between two DHHA1 domains (48), PfuHAN forms a symmetric trimer through both DHH and DHHA1 domains, as well as the helix linker α13 (Figure 7A and B). The interaction strength of each subunit is primarily dictated by hydrogen bonds and salt bridges. The residues E390, R424, R449, E562 and E598 form three salt bridges, one of which exists between E390 and R424, while the other two are between R449 and E562/E598 (Figure 7B). We disrupted the salt bridge by introducing site-directed mutations into PfuHAN_DND and analysed mutant quaternary structures (Figure 7C). The chemical reagent BS3, which can crosslink the protein complex into a polymer, was used to confirm whether the trimeric form of each mutant was disrupted. Among the five mutants, which were expected to disrupt the interactions between each subunit in the complex, the E390A, R449A and E598A mutants generated very few cross-linked polymers (Figure 7C), indicating that the three residues participate in trimer formation. However, the R424A and E562A mutants could still form polymers comparable to the WT enzyme (Figure 7C), indicating that these residues do not participate in trimer formation. Gel-filtration experiments also confirmed the trimeric form of PfuHAN_DND and the E562A mutant, as well as the single subunit form of the E390A mutant (Supplementary Figure S11). In summary, the contribution of the residues in forming the trimeric complex is in the following order: E390 > E598 > R449 > R424 > E562.
Figure 7.
The PfuHAN_DND trimer-forming mechanism. (A) Topology of the PfuHAN_DND trimer complex. Each subunit is depicted in a different colour. (B) Interaction strengths among each subunit in the complex. Key residues involved in the interaction are shown in stick. Hydrogen bonds and salt bridges are represented by red and black dashed lines, respectively. (C) Analysis of the PfuHAN_DND and mutant complexes. 15% SDS-PAGE was used to analyse the cross-linked state of PfuHAN_DND and the five mutants after reaction with the crosslinker BS3. M denotes the protein molecular weight standard consisting of 200, 116, 66, 44 and 29 kDa bands. (D) Exonuclease activity of PfuHAN_DND and its mutants. Increasing concentrations of PfuHAN_DND and the five mutants (10, 20, 50, 100 and 200 nM) were incubated with ssDNA (50 nM) at 55°C for 15 min.
As a homologous trimer, PfuHAN_DND might promote its activity by forming the trimer complex. After confirming the quaternary structures, effects of the interacting residues on nuclease activity were characterized, and all five mutants exhibited reduced activity (Figure 7D). The decreased activity of E390A, R449A, and E598A might have resulted from trimer disruption. However, because the R424A and E562A mutants, specifically the E562A, exist as partial trimers, it is possible that R424 and E562 impairs catalytic steps of key residues.
Physiological roles of HAN in resisting DNA damage
The Haloarchaea H. volcanii was used to investigate the function of HAN in vivo. Before constructing the H. volcanii Δhan mutant, we first analysed the nuclease activity of H. volcanii HAN (HvoHAN, ORF Hvo_1018). HvoHAN has the domains similar to PfuHAN and TkoHAN, while lacking the N-terminal first domain DnaJ-Zf (Supplementary Figure S12A). Three HANs show high sequence similarity and have the same conserved residues and motifs I-VII (Supplementary Figure S12B). The recombinant HvoHAN (Supplementary Figure S12C) has the same 3′-exouclease activity as PfuHAN (Supplementary Figure S12D). These results indicate that H. volcanii is suitable for studying the physiological roles of han gene in vivo.
The Δhan mutant strain was constructed in H. volcanii by deleting the entire ORF Hvo_1018 (Supplementary Figure S13A and B). The growth of the Δhan mutant was first compared with that of the wt strain under standard growth conditions in YPC medium. However, no differences in growth were noted between the wt and Δhan strains (Supplementary Figure S13C). To investigate the possible roles of han in DNA repair, its tolerance to DNA damage reagents, including H2O2, 4NQO, MMC, and MMS, was investigated in H. volcanii wt and Δhan mutant strains. These chemical reagents can promote various forms of DNA damage, such as crosslinking, single stranded breaks, etc. (50). Compared with the wt strain, deletion of han led to retarded growth in the presence of 0.5 mM H2O2, 4.0 μM 4NQO, 0.3 μM MMC or 4.0 mM MMS (Supplementary Figure S14A–D). However, no sensitivities to H2O2, 4NQO, or MMC were observed for Δhan mutants in the drop dilution plating assay (Supplementary Figure S14 E, G and H), where even higher concentrations of DNA damage reagents were used. Deletion of the han gene does not result in sensitivity to MMC in Thermococcus kodakarensis (51). Consistent with the retarded growth curve, Δhan mutants showed sensitivity to 6.0 mM MMS in the drop dilution plating assay (Supplementary Figure S14F).
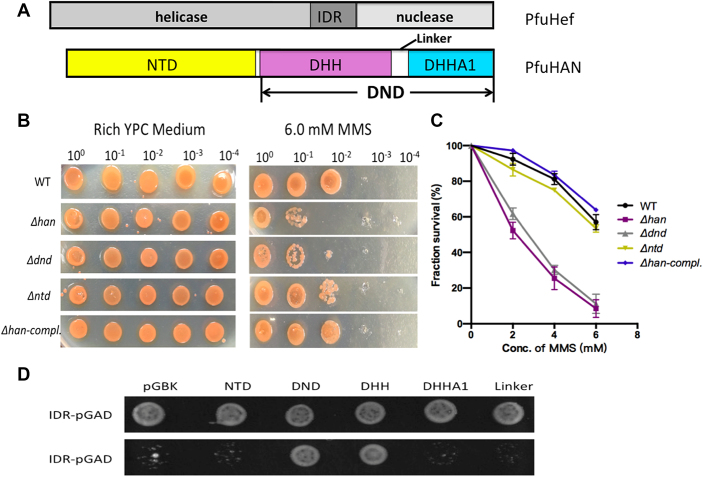
After confirming that the han gene is involved in the repair of DNA damage in response to MMS, its gene fragments encoding the N-terminal non-nuclease and C-terminal DHH nuclease peptide (Figure 8A) were each deleted (Supplementary Figure S13A and B) to determine their respective roles in resisting MMS. Our results show that only the C-terminal nuclease is required to resist MMS (Figure 8A–C). A complementary plasmid carrying the wt han gene restored the resistance of the Δhan mutant to MMS. Finally, the accumulation of DNA damage derived from MMS treatment, such as N3-methyadenine, was analysed in the mutant and wt strains (Supplementary file2). The N3-methyadenine in Δhan mutant is clearly higher than that in wt cell (Supplementary Figure S15), indicating that HAN plays a direct role in DNA repair.
Figure 8.
Interaction model of HAN and Hef and functions of han and its domains in response to MMS resistance. (A) A schematic diagram for domain combinations of PfuHAN and PfuHef. The three domains of PfuHef, the N-terminal domain and DND consisting of DHH, DHHA1 and linker region of PfuHAN are shown. (B) Sensitivities of different H. volcanii strains to MMS. H. volcanii cells (2.3 × 106 cells/ml), including wt, Δhan, Δntd, Δdnd and Δhan complemented with han plasmid, were serially diluted (10 0, 10−1, 10−2, 10−3 and 10−4) with 18% ASW and spotted on the plates with/without 6.0 mM MMS. Plates were then incubated at 45°C for 5–7 days. (C) Survival percentage curves (treated with 2.0–6.0 mM MMS) of different strains. Each strain was examined in at least three replicates, and error bars represent SEM. (D) Interaction model between PfuHAN and IDR of PfuHef. Yeast two-hybrid was used to identify the interactional domain of PfuHAN with IDR of PfuHef. The DNA fragment for encoding IDR was inserted into the pGADT7 vector, and the DNA fragments encoding N-terminal domains, DND and its DHH, DHHA1 and linker regions were inserted into the pGBKT7 vector. Then, yeast cells harbouring different combinations of these constructed pGADT7 and pGBKT7 series plasmids were selected on SD plate lacking Leu, Trp and His. The nonselective plate with addition of His was used as a control to verify the growth of all yeast cells.
To understand the interaction between HAN and the IDR of Hef nuclease (8,51) and its potential function during coordinately treating DNA damage (51), we identified the interactional domain of PfuHAN with the IDR of PfuHef via yeast two-hybrid assays. The DHH domain of C-terminal DND interacts with the IDR of PfuHef, but other domains, including the N-terminal domain, DHHA1 domain and the linker between DHH and DHHA1, do not interact with IDR (Figure 8A and D). Considering that PfuHAN interacts with the IDR of Hef nuclease via its DHH domain of DND, it might be that the DND takes part in resisting MMS via its nuclease activity and/or forming repair complex with Hef nuclease, which is responsible for repair of the halted DNA replication fork generated by the DNA damage reagents MMS, aphidicolin, and hydroxyurea (8,51–53).
DISCUSSION
Substrate specificity and hydrolysis direction
Each DHH family nuclease has a unique substrate preference, meaning that the mode of substrate binding might be different and responsible for the hydrolysis polarity. PfuHAN prefers long strand DNAs, while YybT is a hydrolytic enzyme for cyclic dinucleotides c-di-AMP. The reduced efficiency of PfuHAN to hydrolyse oligonucleotides shorter than 6 nt reflects that the length of binding patch is approximately 6 nt and cannot accommodate shorter substrates perfectly. Our simulation results on PfuHAN_DND binding ssDNA also confirmed this possibility (Figure 6A–D). Meanwhile, the binding patch is not wide enough to accommodate double-stranded substrates. Both ssDNA and ssRNA can be degraded, suggesting that HAN has no specific recognition residues or discrimination of the ribose 2′-OH. In comparison, bacterial RecJ can only hydrolyse ssDNA, not ssRNA. Although the structure of bacterial DraRecJ in complex with ssDNA has been solved and the conserved residues for binding ssDNA have been identified (46), the mechanism of excluding ssRNA, i.e. the steric gate for ribose, has not been elucidated. In contrast to bacterial RecJ, bacterial NrnA can hydrolyse short ssRNA (nanoRNA), pAp, and c-di-GMP, but not ssDNA (48,54). In the structures of NrnA, the split alpha helix connecting the DHH and DHHA1 domains further separates the two domains and forms a wide and short binding patch that supports the hydrolysis of short nanoRNA, c-di-GMP and pAp (48,54). Therefore, it might be plausible to identify the residues in bacterial RecJ that sterically repulse the ribose by comparing differences in the substrate binding patches of HAN, NrnA, and RecJ.
In addition to substrate diversity, the hydrolysis polarity of oligonucleotide is also diversified in DHH family nucleases. Bacterial RecJ is a 5′-exonuclease (55), and archaeal HAN is a 3′-exonuclease (8,51). In contrast, NrnA is a bidirectional exonuclease (48), while archaeal GAN has a more complicated hydrolytic polarity: 5′-exonuclease specific for ssDNA and 3′-exonuclease for ssRNA (15,16,56,57). The 5′-phosphate-binding pocket, which consists of the polar residues R109, R280, S371, and R373, is present above the DraRecJ active site and determines the 5′-3′ polarity of RecJ for ssDNA (46). The structure of PfuHAN_DND does not have this specific pocket for binding the 3′-nucleotide; thus, it might determine the hydrolysis polarity based on the full binding patch for single-stranded substrates (Figure 6D). The conserved residues R262 and R264 in BsuNrnA are key residues for determining the 3′-5′ digestion of nanoRNA (48). NrnA also hydrolyses the cyclic dinucleotides c-di-AMP and c-di-GMP and does not require a specific pocket for determining hydrolysis polarity (48,54). In the future, solving the co-crystal structure of HAN and ssDNA or ssRNA will be important to elucidate its catalytic mechanism, including its hydrolysis polarity.
The alpha helix connecting the DHH and DHHA domains is another structural feature for classifying the DHH family. Bacterial RecJ and PfuHAN_DND have a long intact alpha helix, while NrnA and PPase have a split one, resulting in a widened cleft for binding cyclic dinucleotides (48,49,54). Considering that NrnA and PPase can hydrolyse the phosphoester bond of non-oligonucleotide substrates, cyclic dinucleotides (48,54) and inorganic pyrophosphate (49,57), it is possible that the split alpha helix is an element that determines the substrate specificity and the kind of phosphoester bond to be hydrolyzed.
Function of N-terminal domains
Aside from the DHH nuclease domain, HAN also possesses three additional N-terminal domains. Our results demonstrated that the N-terminal DnaJ-Zf, RpS1-like, and OB-fold domains all increase the hydrolytic activity of the C-terminal DND, possibly by promoting its substrate binding. The nucleic acid binding activity of these three domains has been confirmed in other proteins. The Zn-finger domain has been reported to recognize and bind DNA sequences (58). The OB-fold and RpS1-like domains are considered to play important roles in ssRNA binding (59). In this scenario, as shown in the modeled structure of full-length PfuHAN (Supplementary Figure S16A), the N-terminal domains might increase the interactions between the substrate and catalytic domain. Our kinetic parameters, specifically the increased Km value of various N-terminal-truncated PfuHANs (Table 1), also support the key role of the N-terminal domain in substrate binding. Deletion of the C-terminal domain resulted in a slightly increased sensitivity to MMS (Figure 8B and C), suggesting that the N-terminal domain might play a little role in repairing DNA damage in response to MMS. Meanwhile, it is important to solve the N-terminal non-nuclease domain to determine its function in substrate binding.
The HAN trimer complex functions as a higher activity nuclease
To date, the structures of several DHH phosphoesterase superfamily members have been solved, and all of them adopt a similar overall topology consisting of a typical DHH domain and DHHA domain. However, their quaternary structures are diverse, including monomeric RecJ (44–46), GAN (47) and family II PPase (49,60), dimeric NrnA (48,61), and our solved trimeric PfuHAN. Exonuclease I from P. furiosus (PfuExoI), which does not belong to the DHH superfamily, shows high processive degradation and adopts a trimeric topology with an internal pore for ssDNA/ssRNA entry (62). PfuExoI displays high affinity for ssDNA and ssRNA. Unlike PfuExoI, PfuHAN exhibits very weak affinity to substrates. PfuExoI prefers ssRNA, while PfuHAN exhibits a slight preference for ssDNA. PfuExoI can degrade long substrates into 2–4 nt oligonucleotides via the coordinated hydrolysis of trimeric complex. PfuExoI and PfuHAN might coordinate to degrade DNA and RNA in vivo. Disruption of the PfuExoI complex, via D189R mutation, changes the processive cleavage into a distributive one. The modeled complex structure of PfuHAN_DND and ssDNA shows that the three substrate entries are related and form a common central passageway (Supplementary Figure S16B), suggesting PfuHAN takes a cleavage mode similar to PfuExoI trimeric complex. Disruption of the PfuHAN_DND trimer indeed changed the cleavage mode, resulting in a slow and distributive degradation of ssDNA (Figure 7D). The efficient degradation of dsDNA with 3′-overhang or fork into single nucleotide (Figure 2F, Supplementary Figure S7A and B), even for ssDNA digestion (Figure 3C and D), might require the coordinated function of three subunits in the trimeric complex.
HAN participates in DNA repair as a nuclease
Each DHH superfamily member has specific physiological roles in vivo. To date, the physiological role of HAN in DNA repair has not been reported. TkoHAN interacts with the structure-specific nuclease Hef (8), which is required for the processing of a stalled replication fork and repair-associated recombination (6,51,52). Therefore, HAN is considered to play important roles in these two processes in archaea by coordinating with the helicase and endonuclease activities of Hef (51,52). Our results confirmed that PfuHAN can digest 3′-overhanged or forked dsDNAs, which are possible immediate products during the repair of DNA damage, such as the intermediate product in a stalled replication fork by MMS (53). Meanwhile, deletion of the han gene and its DND-encoding region resulted in increased sensitivity to the DNA damage reagent MMS (Figure 8B and C), suggesting that HAN is directly involved in repairing stopped replication forks created by DNA damage reagents. The accumulation of N3-methyadenine generated by MMS in Δhan cells also implies that HAN participates in removal of DNA damages. Although N3-methyadenine was also detected in wt cell even in the absence of MMS, it might be the endogenous N6-methyladenine (m6A), which has the same molecular weight as N3-methyladenine and might be identified as the latter, in H. volcanii cells (63).
Based on our biochemical and genetic results and the interactions between DHH domain of PfuHAN and the IDR of Hef nuclease, we propose a model to interpret the possible function of HAN in DNA repair (Supplementary Figure S17). The HAN trimer interacts with three Hef nucleases via its DHH domain and the IDR of Hef. By coordinating with their interactional partner PCNA, the HAN-Hef hexamer digests a stagnantly extending strand from the 3′-end in a stopped replication fork, and reverses back the damaged base, whereby the damage will be repaired by other pathways, such as recombination repair. During regression of the stagnantly extending strand, the helicase activity of Hef might promote the degradation reaction by unwinding the dsDNA (51). Although the DnaJ-Zf domain is naturally absent in Hvo_1018, it might play a minor function during DNA repair because of the nonessentiality of whole N-terminal domain of HAN in resisting MMS. The detailed mechanism for HAN in repairing DNA damage by MMS will be further interpreted in the future.
DATA AVAILABILITY
The atomic coordinates and structural factors have been deposited into the Protein Data Bank (http://www.rcsb.org/pdb) with PDB ID code 5YH1 (Apo PfuHAN_DND).
Supplementary Material
ACKNOWLEDGEMENTS
We are extremely grateful to Professor Thorsten Allers for providing the H. volcanii strain and related plasmids. We thank the staffs from BL17B/BL18U/BL19U1/ BL19U2/BL01B beamline of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility, for assistance during data collection.
Author Contributions: X.P.L. conceived and supervised the research. X.P.L. and L.F. designed the experiments. L.F., C.C.C., D.S., C.J., Y.S. and Y.J.Z. performed the experiments. All authors analysed the data. X.P.L., W.D., X.X., H.F.C. and F.P.W. contributed reagents/materials/analysis tools. L.F., Y.S., D.S. and X.P.L. contributed to the figures. X.P.L. and L.F. wrote the paper.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31371260 and 41530967]; China Ocean Mineral Resources R&D Association [DY125-22-04]. Funding for open access charge: [31371260].
Conflict of interest statement. None declared.
REFERENCES
- 1. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M.. Eukaryotic chromosome DNA replication: where, when, and how. Annu. Rev. Biochem. 2010; 79:89–130. [DOI] [PubMed] [Google Scholar]
- 2. Leipe D.D., Aravind L., Koonin E.V.. Did DNA replication evolve twice independently. Nucleic Acids Res. 1999; 27:3389–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002; 5:525–532. [DOI] [PubMed] [Google Scholar]
- 4. Barry E.R., Bell S.D.. DNA replication in the archaea. Microbiol. Mol. Biol. Rev. 2006; 70:876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox M.M., Goodman M.F., Kreuzer K.N., Sherratt D.J., Sandler S.J., Marians K.J.. The importance of repairing stalled replication forks. Nature. 2000; 404:37–41. [DOI] [PubMed] [Google Scholar]
- 6. Ciccia A., McDonald N., West S.C.. Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 2008; 77:259–287. [DOI] [PubMed] [Google Scholar]
- 7. Lestini R., Duan Z., Allers T.. The archaeal Xpf/Mus81/FANCM homolog Hef and the Holliday junction resolvase Hjc define alternative pathways that are essential for cell viability in Haloferax volcanii. DNA Repair (Amst). 2010; 9:994–1002. [DOI] [PubMed] [Google Scholar]
- 8. Ishino S., Yamagami T., Kitamura M., Kodera N., Mori T., Sugiyama S., Ando T., Goda N., Tenno T., Hiroaki H. et al. Multiple interactions of the intrinsically disordered region between the helicase and nuclease domains of the archaeal Hef protein. J. Biol. Chem. 2014; 289:21627–21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova K.S., Koonin E.V., Kelman Z.. The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes. Biol. Direct. 2012; 7:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aravind L., Koonin E.V.. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 1998; 23:17–19. [DOI] [PubMed] [Google Scholar]
- 11. Thoms B., Borchers I., Wackernagel W.. Effects of single-strand DNases ExoI, RecJ, ExoVII, and SbcCD on homologous recombination of recBCD+ strains of Escherichia coli and roles of SbcB15 and XonA2 ExoI mutant enzymes. J. Bacteriol. 2008; 190:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Burdett V., Baitinger C., Viswanathan M., Lovett S.T., Modrich P.. In vivo requirement for RecJ, ExoVII, ExoI, and ExoX in methyl-directed mismatch repair. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dianov G., Lindahl T.. Reconstitution of the DNA base excision-repair pathway. Curr. Biol. 1994; 4:1069–1076. [DOI] [PubMed] [Google Scholar]
- 14. Rajman L.A., Lovett S.T.. A thermostable single-strand DNase from Methanococcus jannaschii related to the RecJ recombination and repair exonuclease from Escherichia coli. J. Bacteriol. 2000; 182:607–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z., Pan M., Santangelo T.J., Chemnitz W., Yuan W., Edwards J.L., Hurwitz J., Reeve J.N., Kelman Z.. A novel DNA nuclease is stimulated by association with the GINS complex. Nucleic Acids Res. 2011; 39:6114–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yuan H., Liu X.P., Han Z., Allers T., Hou J.L., Liu J.H.. RecJ-like protein from Pyrococcus furiosus has 3′-5′ exonuclease activity on RNA: implications for proofreading of 3′-mismatched RNA primers in DNA replication. Nucleic Acids Res. 2013; 41:5817–5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burkhart B.W., Cubonova L., Heider M.R., Kelman Z., Reeve J.N., Santangelo T.J.. The GAN exonuclease, or the flap endonuclease Fen1 and RNase HII are necessary for viability of. Thermococcus kodakarensis. J. Bacteriol. 2017; 199:e00141-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nagata M., Ishino S., Yamagami T., Ogino H., Simons J. R., Kanai T., Atomi H., Ishino Y.. The Cdc45/RecJ-like protein forms a complex with GINS and MCM, and is important for DNA replication in Thermococcus kodakarensis. Nucleic Acids Res. 2017; 45:10693–10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krastanova I., Sannino V., Amenitsch H., Gileadi O., Pisani F.M., Onesti S.. Structural and functional insights into the DNA replication factor Cdc45 reveal an evolutionary relationship to the DHH family of phosphoesterases. J. Biol. Chem. 2012; 287:4121–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sanchez-Pulido L., Ponting C.P.. Cdc45: the missing RecJ ortholog in eukaryotes. Bioinformatics. 2011; 27:1885–1888. [DOI] [PubMed] [Google Scholar]
- 21. Mechold U., Fang G., Ngo S., Ogryzko V., Danchin A.. YtqI from Bacillus subtilis has both oligoribonuclease and pAp-phosphatase activity. Nucleic Acids Res. 2007; 35:4552–4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang J., Bai Y., Zhang Y., Gabrielle V.D., Jin L., Bai G.. Deletion of the cyclic di-AMP phosphodiesterase gene (cnpB) in Mycobacterium tuberculosis leads to reduced virulence in a mouse model of infection. Mol. Microbiol. 2014; 93:65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rao F., See R.Y., Zhang D., Toh D.C., Ji Q., Liang Z.X.. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 2010; 285:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Young T.W., Kuhn N. J., Wadeson A., Ward S., Burges D., Cooke G.D.. Bacillus subtilis ORF yybQ encodes a manganese-dependent inorganic pyrophosphatase with distinctive properties: the first of a new class of soluble pyrophosphatase. Microbiology. 1998; 144:2563–2571. [DOI] [PubMed] [Google Scholar]
- 25. Ishino Y., Narumi I.. DNA repair in hyperthermophilic and hyperradioresistant microorganisms. Curr. Opin. Microbiol. 2015; 25:103–112. [DOI] [PubMed] [Google Scholar]
- 26. Liu X.P., Liu J.H.. The terminal 5′ phosphate and proximate phosphorothioate promote ligation-independent cloning. Protein Sci. 2010; 19:967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen V.B., Arendall W.B., Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C.. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Allers T., Ngo H.P., Mevarech M., Lloyd R.G.. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004; 70:943–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hu C.W., Lin B.H., Chao M.R.. Quantitative determination of urinary N3-methyladenine by isotope-dilution LC–MS/MS with automated solid-phase extraction. Int. J. Mass Spectrom. 2011; 300:99–107.21499524 [Google Scholar]
- 35. Schrodinger, L.LC. 2015; The PyMOL Molecular Graphics System, Version 1.8. [Google Scholar]
- 36. Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J.. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009; 30:2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Case D.A., Götz A.W., Darden T., Kolossváry I., Cheatham T.E. III, Paesani F., Simmerling C., Wu X., Roitberg A., Steinbrecher T. et al. Amber 14. Mech. Eng. 2014; 126:14. [Google Scholar]
- 38. Darden T., York D., Pedersen L.. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993; 98:10089–10092. [Google Scholar]
- 39. Ryckaert J.P., Ciccotti G., Berendsen H.J.C.. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977; 23:327–341. [Google Scholar]
- 40. Roe D.R., Cheatham T.E.. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013; 9:3084–3095. [DOI] [PubMed] [Google Scholar]
- 41. Song D., Luo R., Chen H.F.. The IDP-specific force field ff14IDPSFF improves the conformer sampling of intrinsically disordered proteins. J. Chem. Inf. Model. 2017; 57:1166–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen H., Luo R.. Binding induced folding in p53-MDM2 complex. J. Am. Chem. Soc. 2007; 129:2930–2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morimatsu K., Kowalczykowski S.C.. RecQ helicase and RecJ nuclease provide complementary functions to resect DNA for homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5133–E5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wakamatsu T., Kitamura Y., Kotera Y., Nakagawa N., Kuramitsu S., Masui R.. Structure of RecJ exonuclease defines its specificity for single-stranded DNA. J. Biol. Chem. 2010; 285:9762–9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamagata A., Kakuta Y., Masui R., Fukuyama K.. The crystal structure of exonuclease RecJ bound to Mn2+ ion suggests how its characteristic motifs are involved in exonuclease activity. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:5908–5912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cheng K., Xu H., Chen X., Wang L., Tian B., Zhao Y., Hua Y.. Structural basis for DNA 5′-end resection by RecJ. Elife. 2016; 5:e14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oyama T., Ishino S., Shirai T., Yamagami T., Nagata M., Ogino H., Kusunoki M., Ishino Y.. Atomic structure of an archaeal GAN suggests its dual roles as an exonuclease in DNA repair and a CMG component in DNA replication. Nucleic Acids Res. 2016; 44:9505–9517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmier B.J., Nelersa C.M., Malhotra A.. Structural basis for the bidirectional activity of Bacillus nanoRNase NrnA. Sci. Rep. 2017; 7:11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fabrichniy I.P., Lehtiö L., Tammenkoski M., Zyryanov A.B., Oksanen E., Baykov A.A., Lahti R., GolMBDan A.. A trimetal site and substrate distortion in a family II inorganic pyrophosphatase. J. Biol. Chem. 2007; 282:1422–1431. [DOI] [PubMed] [Google Scholar]
- 50. Fujikane R., Ishino S., Ishino Y., Forterre P.. Genetic analysis of DNA repair in the hyperthermophilic archaeon, Thermococcus kodakaraensis. Genes Genet. Syst. 2010; 85:243–257. [DOI] [PubMed] [Google Scholar]
- 51. Nagata M., Ishino S., Yamagami T., Simons J.R., Kanai T., Atomi H., Ishino Y.. Possible function of the second RecJ-like protein in stalled replication fork repair by interacting with Hef. Sci. Rep. 2017; 7:16949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lestini R., Laptenok S.P., Kühn J., Hink M.A., Schanne-Klein M.C., Liebl U., Myllykallio H.. Intracellular dynamics of archaeal FANCM homologue Hef in response to halted DNA replication. Nucleic Acids Res. 2013; 41:10358–10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A.S., Helleday T.. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005; 33:3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He Q., Wang F., Liu S., Zhu D., Cong H., Gao F., Li B., Wang H., Lin Z., Liao J. et al. Structural and biochemical insight into the mechanism of Rv2837c from Mycobacterium tuberculosis as a c-di-NMP phosphodiesterase. J. Biol. Chem. 2016; 291:14386–14387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yamagata A., Masui R., Kakuta Y., Kuramitsu S., Fukuyama K.. Overexpression, purification and characterization of RecJ protein from Thermus thermophilus HB8 and its core domain. Nucleic Acids Res. 2001; 29:4617–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ogino H., Ishino S., Kohda D., Ishino Y.. The RecJ2 protein in the thermophilic archaeon Thermoplasma acidophilum is a 3′-5′ exonuclease that associates with a DNA replication complex. J. Biol. Chem. 2017; 292:7921–7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yi G.S., Song Y., Wang W.W., Chen J.N., Deng W., Cao W., Wang F.P., Xiao X., Liu X.P.. Two archaeal RecJ nucleases from Methanocaldococcus jannaschii show reverse hydrolysis polarity: implication to their unique function in archaea. Genes. 2017; 8:E211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tsai R.Y., Reed R.R.. Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol. 1998; 18:6447–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Amblar M., Barbas A., Gomez-Puertas P., Arraiano C.M.. The role of the S1 domain in exoribonucleolytic activity: substrate specificity and multimerization. RNA. 2007; 13:317–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fabrichniy I.P., Lehtiö L., Salminen A., Zyryanov A.B., Baykov A.A., Lahti R., GolMBDan A.. Structural studies of metal ions in family II pyrophosphatases: the requirement for a Janus ion. Biochemistry. 2004; 43:14403–14411. [DOI] [PubMed] [Google Scholar]
- 61. Srivastav R., Kumar D., Grover A., Singh A., Manjasetty B.A., Sharma R., Taneja B.. Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases. Nucleic Acids Res. 2014; 42:7894–7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Miyazono K.I., Ishino S., Tsutsumi K., Ito T., Ishino Y., Tanokura M.. Structural basis for substrate recognition and processive cleavage mechanisms of the trimeric exonuclease PhoExo I. Nucleic Acids Res. 2015; 43:7122–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ouellette M., Jackson L., Chimileski S., Papke R.T.. Genome-wide DNA methylation analysis of Haloferax volcanii H26 and identification of DNA methyltransferase related PD-(D/E)XK nuclease family protein HVO_A0006. Front. Microbiol. 2015; 6:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structural factors have been deposited into the Protein Data Bank (http://www.rcsb.org/pdb) with PDB ID code 5YH1 (Apo PfuHAN_DND).