Abstract
Acquisition of foreign DNA by natural transformation is an important mechanism of adaptation and evolution in diverse microbial species. Here, we characterize the mechanism of ComM, a broadly conserved AAA+ protein previously implicated in homologous recombination of transforming DNA (tDNA) in naturally competent Gram-negative bacterial species. In vivo, we found that ComM was required for efficient comigration of linked genetic markers in Vibrio cholerae and Acinetobacter baylyi, which is consistent with a role in branch migration. Also, ComM was particularly important for integration of tDNA with increased sequence heterology, suggesting that its activity promotes the acquisition of novel DNA sequences. In vitro, we showed that purified ComM binds ssDNA, oligomerizes into a hexameric ring, and has bidirectional helicase and branch migration activity. Based on these data, we propose a model for tDNA integration during natural transformation. This study provides mechanistic insight into the enigmatic steps involved in tDNA integration and uncovers the function of a protein required for this conserved mechanism of horizontal gene transfer.
INTRODUCTION
Natural competence is a physiological state in which some bacterial species can take up free DNA from the environment. Some competent species regulate the genes required for this process and, depending on the organism, competence can be induced in response to the availability of certain nutrients, quorum sensing pathways, or by DNA damage/stress (1). The Gram-negative bacterium Vibrio cholerae is activated for competence during growth on chitin, a polymer of β1,4-linked N-acetyl glucosamine (2). Chitin is the primary constituent of crustacean exoskeletons and is commonly found in the aquatic environment where this facultative pathogen resides. Soluble chitin oligosaccharides indirectly induce expression of TfoX, the master regulator of competence (3,4), which regulates expression of competence-related genes in concert with HapR, the master regulator of quorum sensing (5,6). Acinetobacter baylyi ADP1, on the other hand, is a naturally competent Gram-negative microbe that is constitutively active for competence throughout exponential growth (7).
During competence, dsDNA is bound extracellularly, however, only a single strand of this DNA is transported into the cytoplasm. Competent bacterial species may use this ingested DNA as a source of nutrients, however, if this DNA has sufficient homology to the host chromosome, the incoming DNA can also be integrated into the bacterial genome by homologous recombination (8). This process of DNA uptake and integration is referred to as natural transformation. As a result, natural transformation is an important mechanism of horizontal gene transfer and can lead to the repair of damaged DNA or facilitate acquisition of novel genetic information. Homologous recombination of single-stranded transforming DNA (tDNA) with the host chromosome requires the function of RecA, which facilitates homology searching and initiates strand invasion of tDNA through the formation of a displacement loop (D-loop). Following RecA mediated strand invasion, DNA junctions of this D-loop can then be moved in a process known as branch migration to increase or decrease the amount of tDNA integrated. Then, by a presently unresolved mechanism, this intermediate is resolved to stably integrate tDNA into the host chromosome. The molecular details involved in the integration of tDNA downstream of RecA strand invasion, however, are poorly understood.
One previously studied gene from the competent species Haemophilus influenzae, comM, is not required for DNA uptake but is required for the integration of tDNA into the host chromosome, a phenotype consistent with ComM playing a role in homologous recombination during natural transformation (9). The function of ComM, however, has remained unclear. Here, through both in vivo and in vitro characterization of ComM in V. cholerae and A. baylyi, we uncover that this protein functions as a hexameric helicase to aid in branch migration during this conserved mechanism of horizontal gene transfer.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The parent V. cholerae strain used throughout this study is E7946 (10), while the A. baylyi strain used is ADP1 (also known as BD413) (11). A description of all strains used in this study are listed in Supplementary Table S1. Strains were routinely grown in LB broth and plated on LB agar. When required, media was supplemented with 200 μg/ml spectinomycin, 50 μg/ml kanamycin, 100 μg/ml carbenicillin, 10 μg/ml trimethoprim or 40 μg/ml X-Gal.
Construction of mutants and transforming DNA
Linear PCR product was constructed using splicing-by-overlap extension PCR exactly as previously described (12). All primers used to construct and detect mutant alleles are described in Supplementary Table S2. The pBAD18 Kan plasmid used as tDNA was purified from TG1, a recA+ Escherichia coli host. Mutants were made by cotransformation exactly as previously described (13).
Transformation assays
Transformation assays of V. cholerae on chitin were performed exactly as previously described (3). Briefly, ∼108 CFU of mid-log V. cholerae were incubated statically in instant ocean (IO) medium (7g/L; aquarium systems) containing chitin from shrimp shells (Sigma) for 16–24 h at 30°C. Then, tDNA was added (500 ng for linear products containing an antibiotic resistance cassette inserted at the non-essential locus VC1807 and 1500 ng for plasmid DNA), and reactions were incubated at 30°C for 5–16 h. Reactions were outgrown with the addition of LB medium to each reaction by shaking at 37°C for 1–3 h and then plated for quantitative culture onto medium to select for the tDNA (transformants) or onto plain LB (total viable counts). Transformation efficiency is shown as the number of transformants / total viable counts. In cases where no colonies were observed, efficiencies were denoted as below the limit of detection (LOD) for the assay.
Chitin-independent transformation assays were performed exactly as previously described using strains that contain an IPTG inducible Ptac promoter upstream of the native TfoX gene (14). Briefly, strains were grown overnight with 100 μM IPTG. Then, ∼108 cells were diluted into instant ocean medium, and tDNA was added. Reactions were incubated statically for 5 h and then outgrown by adding LB and shaking at 37°C for 1–3 h. Reactions were plated for quantitative culture as described above.
For transformation of A. baylyi ADP1, strains were grown overnight in LB media. Then, ∼108 cells were diluted into fresh LB medium, and tDNA was added (∼100 ng). Reactions were incubated at 30°C with agitation for 5 h and then plated for quantitative culture as described above.
Protein expression and purification
ComM and Pif1 were cloned into StrepII expression vectors, expressed in Rosetta 2(DE3) pLysS cells using autoinduction medium (15), and purified using Strep-Tactin Sepharose (IBA). ComM protein preparations lacked detectable nuclease activity. For details please see Supplementary Methods.
Electrophoretic mobility shift assay
The ssDNA probe (BBC742) and dsDNA probe (annealed BBC742 and BBC743) were labeled using Cy5 dCTP (GE Healthcare) and Terminal deoxynucleotidyl Transferase (TdT; Promega). Reactions were composed of 10 mM Tris–HCl pH 7.5, 20 mM KCl, 1 mM DTT, 10% Glycerol, 100 μg/ml BSA, 9 nM Cy5 labeled probe, purified ComM protein at the indicated concentration, and 0.1 mM ATP where indicated. Reactions were incubated at RT for 30 min and run on 8% Tris-borate acrylamide gel at 150 V for 45 min. Probes were detected using a Chemidoc MP imaging system (Biorad).
Blue native PAGE
Blue native PAGE was performed essentially as previously described (16). Purified ComM (2.5 μM) was incubated for 30 min at room temperature in reaction buffer [10 mM Tris–HCl pH 7.5, 20 mM KCl, 1 mM DTT, 10% Glycerol] with 5 mM ATP and/or 5 μM ssDNA (oligo ABD363) where indicated. For additional details see Supplementary Methods.
Negative stain electron microscopy
For negative stain electron microscopy, sample was prepared by applying 4 μl of ComM-ATP-ssDNA solution onto a glow-discharged continuous carbon film coated copper grid (EMS) and stained with 0.75% (w/v) uranyl formate. EM micrographs were acquired using a 300 kV JEM-3200FS electron microscopy with 20-eV energy slit under low dose conditions (≤20 e−/Å2) on a Gatan UltraScan 4000 4k × 4k CCD camera. Additional details for EM image collection and analysis are provided in the Supplementary Methods. The EM electron density map has been deposited to EMDataBank.org with the accession number EMD-8575.
Phylogenetic trees
ComM homologs were identified using a protein BLAST search of diverse bacterial genomes followed by phylogenetic analysis. Starting with a broadly representative set of genomes adapted from Wu and Eisen (17), an initial ComM candidate pool was generated from a comprehensive protein BLAST search of all predicted CDS translations against the V. cholerae ComM allele with a 0.001 e-value cutoff. Subsequent sequence alignment with MUSCLE (18) and maximum likelihood phylogenetic reconstruction with FastTree (19) identified a single clade of alleles with high sequence similarity (generally well above 50%) to V. cholerae ComM (data not shown), with the remaining alleles excluded from further analysis. After manually pruning divergent alleles with alignments covering <70% of ComM, the retained sequences comprised the set of true ComM orthologs used in the final sequence alignment and ComM phylogeny reconstruction. For whole genome phylogenetic reconstruction, Phylosift (20) identified and aligned a default set of 36 highly conserved marker genes. FastTree was used for initial reconstruction, whereas RAxML (21) subsequently estimated the maximum likelihood phylogeny for a reduced set of representative genomes under the LG (22) substitution model with gamma-distributed rate variation.
Helicase and branch migration assays
Helicase assay substrates were 5′ end-labeled with T4 polynucleotide kinase (T4 PNK; NEB) and γ[32P]-ATP. The two-strand forked helicase substrates was annealed by combining equimolar amounts of a labeled oligonucleotide with the unlabeled complement and incubating at 37°C overnight.
The short three-strand branch migration substrates were constructed in a two-step annealing process. First, we incubated equimolar amounts of a labeled strand and unlabeled partial complement at 95°C for 5 min. and then slow cooled to 25°C to create a forked substrate. An equimolar amount of a third strand was then added to the resulting forked product and incubated overnight at 37°C. Following annealing, the three-stranded products were PAGE purified. Primers used for all probes and additional details can be found in the Supplementary Methods. Helicase and branch migration activity was then assessed by incubating the indicated concentrations of ComM, Pif1 or Hrq1 with 5 mM ATP and 0.1 nM DNA substrate in resuspension buffer (25 mM Na-HEPES (pH 7.5), 5% (v/v) glycerol, 300 mM NaOAc, 5 mM MgOAc and 0.05% Tween-20). Reactions were incubated at 37°C for 30 min and stopped with the addition of 1× Stop-Load dye (5% glycerol, 20 mM EDTA, 0.05% SDS, and 0.25% bromophenol blue) supplemented with 400 μg/ml SDS-Proteinase K followed by a 10-min incubation at 37°C. Reactions were then separated on 8% 19:1 acrylamide:bis-acrylamide gels in TBE buffer at 10 V/cm. Gels were then dried between layers of Whatman filter paper under vacuum at 55°C for 20 min and then exposed to a phosphor imaging screen for 24–48 h prior to scanning on a Typhoon 9210 Variable Mode Imager to image radiolabeled DNA probes. DNA unwinding and branch migration were quantified using ImageQuant 5.2 software.
The long three-strand recombination intermediates were generated by RecA-mediated strand exchange between circular single-stranded φX174 virion DNA (NEB) and PstI-linearized double-stranded φX174 DNA (NEB) essentially as previously described (23). Briefly, recombination reactions were conducted in strand exchange buffer (25 mM Tris acetate pH 8.0, 1 mM DTT, 1% glycerol, 10 mM magnesium acetate, 3 mM potassium glutamate, 10 U/ml creatine phosphokinase, 12.5 mM phosphocreatine and 50 μg/ml acetylated BSA). First, 7.1 μM RecA (NEB) was incubated with 44 μM (in nucleotides) circular single-stranded φX174 virion DNA (CSS) at 37°C for 10 min. Then, 2 mM ATP and ∼0.84 μM SSB (Promega) were added to the reaction and incubated at 37°C for 8 min. Finally, the strand exchange reactions were started by the addition of ∼16.7 μM (in nucleotides) of PstI-linearized φX174 and incubated at 37°C for an additional 20 min to allow for the generation of recombination intermediates. Reactions were then deproteinated and cleaned up using a PCR purification kit (Qiagen). To assess branch migration of recombination intermediates, the deproteinated DNA was then incubated with ComM in strand exchange buffer containing 5 mM ATP at 37°C for 10 min. Reactions were then stopped using 1X stop load dye and separated on a 0.8% agarose gel in TAE. Gels were then stained with GelGreen (Biotium), and scanned on a Typhoon 9210 Variable Mode Imager.
DNA damage assay
For DNA damage assays, ∼108 cells of a midlog V. cholerae culture in instant ocean medium were treated with the indicated concentration of MMS or MMC for 1 h at 30°C. To determine viable counts, reactions were plated for quantitative culture on LB agar.
GFP-ComM western blots
The indicated strains of V. cholerae were grown with or without 100 μM IPTG. Cell lysates were run on 10% SDS PAGE gels and electrophoretically transferred to PVDF. This membrane was then blotted with primary rabbit anti-GFP (Invitrogen) or mouse anti-RpoA (Biolegend) antibodies and a goat anti-rabbit or anti-mouse IRDye 800CW (LI-COR) secondary as appropriate. Bands were detected using a LI-COR Odyssey classic infrared imaging system.
RESULTS
ComM is required for integration of tDNA during natural transformation
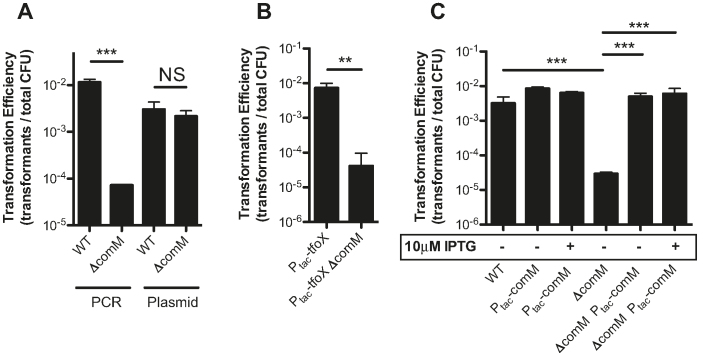
Previously, we performed an unbiased transposon-sequencing screen (Tn-seq) in V. cholerae to identify genes involved in natural transformation (3). One gene identified in that screen was VC0032, which encodes a homolog of comM from H. influenzae. ComM was previously implicated in the integration of tDNA during natural transformation in H. influenzae (9). To determine if this was also the case in V. cholerae, we performed chitin-dependent natural transformation assays using two distinct sources of tDNA. One was a linear PCR product that inserts an antibiotic resistance cassette at a non-essential locus, while the other was a replicating plasmid that lacks any homology to the host genome. We hypothesized that transformation with linear product requires both DNA uptake and chromosomal integration, while transformation with the plasmid only requires DNA uptake. To test this, we transformed a recombination deficient ΔrecA strain of V. cholerae, and found that, as expected, this strain could not be transformed with linear product but could be transformed with a replicating plasmid, consistent with plasmid transformation being recombination-independent (Supplementary Figure S1). Additionally, plasmid transformation in this assay is dependent on natural competence, as mutants in genes required for uptake (ΔpilA) (5) and cytoplasmic protection (ΔdprA) of tDNA are not transformed (5,24,25) (Supplementary Figure S1). Using this assay, we find that a comM mutant (ΔVC0032) in V. cholerae displays a ∼100-fold reduction for transformation with linear product, while rates of plasmid transformation were equal to the WT (Figure 1A). This is consistent with ComM playing a role downstream of DNA uptake, and potentially during recombination. These assays were performed on chitin to induce the natural competence of this organism. To determine if ComM is playing a role specifically downstream of competence induction, we performed a chitin-independent transformation assay by overexpressing the competence regulator TfoX. Under these conditions, a comM mutant still had reduced rates of transformation when transformed with a linear PCR product (Figure 1B). Cumulatively, these results are consistent with ComM playing a role in the late steps of transformation downstream of DNA uptake.
Figure 1.
ComM is required for integration of DNA during natural transformation. (A) Chitin-dependent natural transformation assays in the indicated strains using a linear PCR product or a replicating plasmid as tDNA. (B) Chitin-independent transformation assays of the indicated strains with linear PCR product as tDNA. (C) Complementation of comM in trans tested in chitin-dependent transformation assays using a linear PCR product as tDNA. All data are shown as the mean ± SD and are the result of at least three independent biological replicates. **P < 0.01, ***P < 0.001, NS = not significant.
To confirm that the phenotypes observed are due to mutation of comM, we complemented strains by integrating an IPTG inducible Ptac-comM construct at a heterologous site on the chromosome (at the lacZ locus). Our previous work has indicated that this expression construct is leaky (14) (Supplementary Figure S2), and consistent with this, we observe complementation even in the absence of inducer (Figure 1C).
ComM promotes branch migration through heterologous sequences in vivo
Vibrio cholerae ComM is a predicted AAA+ ATPase, and members of this family have diverse functions (26). To determine if ComM had structural similarity to any AAA+ ATPase of known function, we submitted the primary sequence of this protein to the Phyre2 server (27). Despite a lack of significant homology by BLAST, this analysis revealed structural similarity to MCM2–7, the replicative helicase of eukaryotes (28,29). As a result, we explored whether ComM functions as a helicase to promote branch migration during natural transformation.
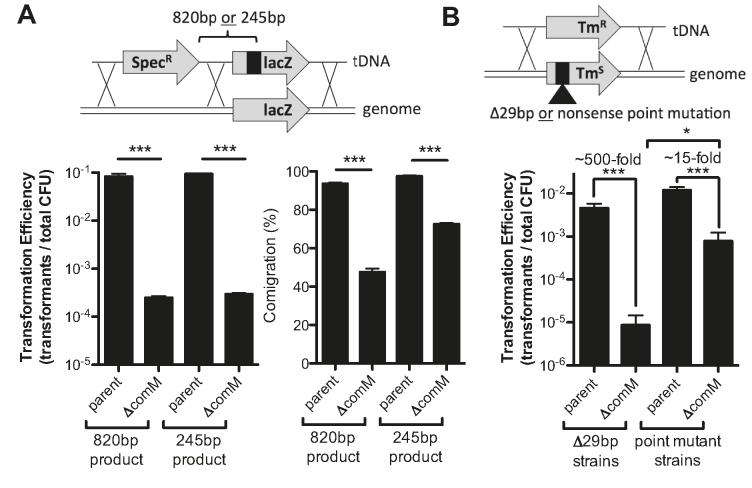
To test this in vivo, we assessed comigration of linked genetic markers on a linear tDNA product (Figure 2A). If branch migration during transformation is efficient, we hypothesized that both markers would be integrated into the host chromosome. However, if branch migration is inefficient, we hypothesized that we may observe integration of one marker but not the other. For this assay, we generated two tDNA constructs that contained a SpecR marker upstream of lacZ as well as a genetically linked point mutation in the lacZ gene that was either 820 or 245 bp downstream of the SpecR marker (Figure 2A). We selected for the SpecR marker and screened for integration of the linked lacZ mutation as an indirect measure of branch migration. To prevent post-recombination repair of the lacZ allele by the mismatch repair (MMR) system, these experiments were performed in mutS mutant backgrounds. As with previous experiments, a comM mutant is severely reduced for transformation efficiency for the SpecR marker compared to the parent (Figure 2A, left and 1A). Of those cells that integrated the SpecR marker, the comigration efficiency of the lacZ mutation was higher than 90% for both products (820 and 245 bp) in the parent strain background (Figure 2A, right), which is consistent with highly efficient branch migration in this background. When comM is deleted, however, the comigration efficiency for the lacZ mutation drops significantly for both products compared to the parent strain, and the reduction is more severe for the product where the lacZ mutation is farther away (Figure 2A, right). Cumulatively, these data suggest that comM may play a role in branch migration during natural transformation to increase the amount of tDNA integrated into the host chromosome.
Figure 2.
ComM promotes branch migration through heterologous sequences in vivo. (A) Chitin-dependent transformation assay performed using tDNA that contained linked genetic markers separated by 820 or 245 bp. (B) Chitin-dependent transformation assay performed in Tms strains (TmR marker inactivated by a nonsense point mutation or 29-bp deletion) using tDNA that would revert the integrated marker to TmR. All strains in A and B contained a mutation in mutS to prevent MMR activity. In schematics above bar graphs, X’s denote possible crossover points for homologous recombination. All data are shown as the mean ± SD and are the result of at least three independent biological replicates. ***P < 0.001.
To promote horizontal gene transfer, tDNA integrated during natural transformation must be heterologous to the host chromosome. So next, we decided to test whether ComM promotes integration of heterologous tDNA. To that end, we created strains that contain an inactivated TmR marker integrated in the chromosome. The marker was inactivated with either a 29 bp deletion or a nonsense point mutation. We then transformed these strains with tDNA that would restore the TmR marker. Again, to eliminate any confounding effects of MMR, we performed these experiments in mutS mutant backgrounds. First, we find that integration of a point mutation is similar to a 29 bp insertion in the parent strain background, indicating that in the presence of ComM, tDNA is efficiently integrated regardless of sequence heterology. In the comM mutant, however, we find that a point mutation is significantly easier to integrate compared to the 29 bp insertion (Figure 2B). This finding is consistent with ComM promoting branch migration through heterologous sequences during natural transformation; however, in its absence, the integration of heterologous sequences is unfavored.
ComM hexamerizes in the presence of ATP and ssDNA
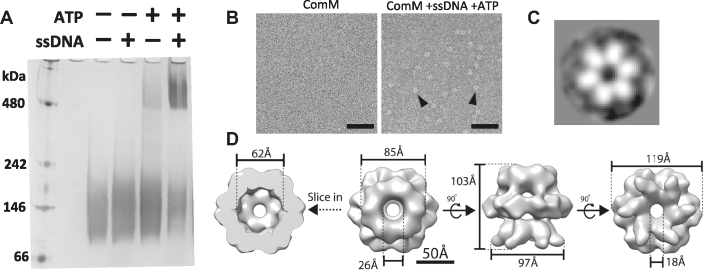
Because our in vivo data suggested that ComM acts as a branch migration factor, we next decided to test the biochemical activity of this protein in vitro. First, we determined that N-terminally tagged ComM (GFP-ComM) was functional in vivo while a C-terminally tagged fusion (ComM-GFP) was not (Supplementary Figure S2). Furthermore, recent studies indicate that comM is part of the competence regulon (30). Using a strain where comM was N-terminally tagged with GFP at the native locus, we found that comM protein levels are increased under competence inducing conditions (TfoX overexpression), indicating that native regulation of tagged comM is maintained (Supplementary Figure S2).
To characterize ComM in vitro, we expressed StrepII-ComM (N-terminal tag) in E. coli and purified it to homogeneity. The peak of recombinant ComM eluting from preparative gel filtration chromatography had a calculated molecular weight of 57 kDa (Supplementary Figure S3). As the predicted mass of ComM is 61 kDa, this suggests that ComM exists as a monomer in solution. Many helicases oligomerize in their active state, including all known MCM family helicases (31). So, we next tested whether purified ComM oligomerizes in vitro. Because ComM is a predicted ATPase and may interact with DNA, we hypothesized that these factors may be required for its oligomerization and activity. To assess oligomerization, we performed blue-native PAGE (16). In this assay, ComM appears to oligomerize robustly in the presence of ATP and ssDNA, with some oligomerization also observed in the presence of ATP alone (Figure 3A). This latter observation, however, may be due to a small amount of contaminating ssDNA that remains bound to ComM during purification. Defining the number of subunits in this oligomer was unreliable by blue-native PAGE due to lack of resolving power by the gel. However, the higher molecular weight species generated in the presence of ATP and ssDNA was likely larger than a dimer. Therefore, we attempted to observe ComM oligomers by negative stain transmission electron microscopy (TEM). In the absence of ligands, ComM particles were small and uniform, consistent with our gel filtration results and demonstrated the purity of our protein preparations. In the presence of ATP and ssDNA, we observed ring-like densities for ComM that upon 2D averaging revealed that this protein forms a hexameric ring (Figure 3B and C). Furthermore, we generated a 3D reconstruction from the negative stain TEM images of ComM in the presence of ATP and ssDNA (∼13.8 Å resolution, see Supplementary Methods), which revealed that ComM forms a three-tiered barrel-like structure with a large opening on both ends (Figure 3D). The pore on the bottom of this barrel is ∼18 Å, which can accommodate ssDNA but not dsDNA, while the 26 Å pore is able to accommodate dsDNA. The size of these pores, however, may be an underestimate due to the stain used during EM and/or ssDNA that may be bound and averaged into the 3D construction. Regardless, these data suggest that ComM forms a hexameric ring, consistent with structures adopted by many AAA+ helicases (26). ComM also oligomerized in the presence of ADP and the non-hydrolysable ATP analog AMP-PNP, indicating that ATP is not a strict requirement for hexamer formation (Supplementary Figure S4).
Figure 3.
ComM hexamerizes in the presence of ATP and ssDNA. (A) Blue-native PAGE assay of purified ComM in the indicated conditions. (B) Negative stain EM of purified ComM under the indicated conditions. Representative ring-like densities observed in the presence of 5 mM ATP and 5 μM ssDNA are indicated by black arrows. Scale bar = 50 nm. (C) A representative 2D class average of the ring-like densities observed by negative stain EM reveals a hexameric complex. (D) 3D reconstruction (∼13.8 Å resolution) of the ring complex imposing C6 symmetry.
ComM binds ssDNA and dsDNA in the presence of ATP
Because ATP was required for oligomerization in the presence of ssDNA, we hypothesized that it would also be required for DNA binding. To test this, we mutated the conserved lysine in the Walker A motif of ComM, which in other AAA+ ATPases, promotes ATP binding (26). In vivo, we find that this mutation abrogates ComM function (Supplementary Figure S5A), while protein stability is maintained (Supplementary Figure S2). To determine if ATP binding was required for ComM to bind ssDNA, we performed electrophoretic mobility shift assays (EMSAs) using purified ComM and ComMK224Ain vitro. We find that WT ComM binds both ssDNA and dsDNA in an ATP-dependent manner (Supplementary Figure S5B–D). Consistent with this, ComMK224A displays greatly reduced ssDNA binding even in the presence of ATP (Supplementary Figure S5B). Also, using non-labeled competitor DNA of differing lengths in EMSAs, we found that ComM preferentially bound ssDNA >60bp in length. Taken together, these results indicate that ATP is important for ComM to bind DNA and function during natural transformation.
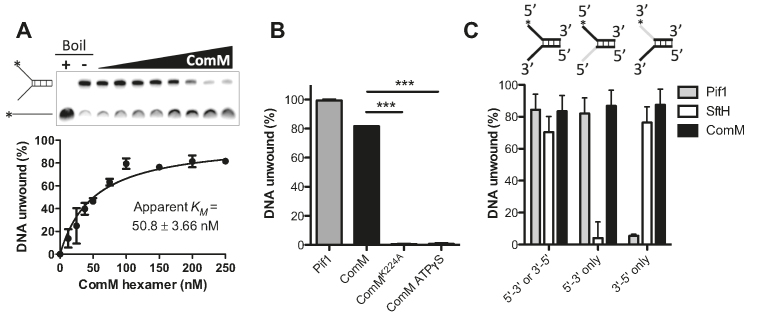
ComM has helicase activity in vitro
Thus far, our data suggest that ComM may play a role in branch migration in vivo. Some branch migration factors (e.g. RuvAB and RecG (32,33)) display helicase activity. So next, we tested the helicase activity of purified ComM in vitro. We observed enzymatic unwinding of a forked DNA substrate with increasing concentrations of ComM. Assuming that ComM hexamers are the active oligomeric form, the helicase activity had an apparent KM of 50.8 nM (Figure 4A). As expected, the purified ATP binding mutant ComMK224A did not display helicase activity in this assay compared to WT ComM and Pif1, a previously characterized helicase (34) that served as a positive control in these assays (Figure 4B). Furthermore, the non-hydrolysable ATP analog ATPγS inhibited helicase activity, which is consistent with ATP hydrolysis being required for ComM function (Figure 4B).
Figure 4.
ComM exhibits helicase activity in vitro. (A) A representative forked substrate helicase assay with increasing concentrations of purified ComM. This forked substrate has 20 bp of annealed sequence and 25 bp tails. Concentrations of ComM used (in hexamer) were 0, 10, 25, 37.5, 50, 75, 100, 150, 200 and 250 nM. Images were quantified and plotted as indicated. (B) Helicase assay using forked DNA substrate with the indicated purified protein (100 nM Pif1 and 250 nM ComM/ComMK224A hexamer) in the presence of 5 mM ATP (Columns 1, 2 and 3) or ATPγS (Column 4). (C) Helicase assays using forked DNA substrates that accommodate enzymes of either directionality or that can only be unwound by 5′ to 3′ or 3′ to 5′ activity. Directional substrates contained one ssDNA tail that is inverted relative to the remainder of the strand (inverted portion indicated in gray on the schematic above bars), thus, preventing helicase activity in one direction. Substrates were incubated with 100 nM purified ComM (hexamer), 10 nM Pif1, or 50 nM SftH. All data are shown as the mean ± SD and are the result of at least three independent replicates. ***P < 0.001.
Most helicases exhibit a preferred directionality (either 5′ to 3′ or 3′ to 5′). A forked DNA substrate, however, does not distinguish between these activities. To determine whether ComM had a preferred directionality, we tested helicase activity on forked substrates where one of the two tails is inverted (by a 3′-3′ linkage) relative to the remainder of the oligo (35). Thus, directional substrates either have two 5′ ends (to assess 5′ to 3′ directionality) or two 3′ ends (to assess 3′ to 5′ directionality). As controls in these assays, we used the unidirectional helicases Pif1 (a 5′ to 3′ helicase (34)) and SftH (a 3′ to 5′ helicase (36)). While Pif1 and SftH exhibited unidirectional helicase activity as expected, ComM unwound all of the substrates tested (Figure 4C). Taken together, these data suggest that ComM is an ATP-dependent bidirectional helicase. Bidirectional activity of a single motor protein like ComM, while uncommon, is not unprecedented (37–39).
ComM exhibits branch migration in vitro
While our data above clearly indicate that ComM is a hexameric helicase, not all helicases can promote branch migration. To more formally test whether ComM can promote branch migration, we performed in vitro branch migration assays using short 3-stranded substrates that more closely resemble the junctions of the D-loop that form during natural transformation (Supplementary Figure S6A and S6B) (40). These substrates contained a small region of heterology (indicated by the grey box), which prevents spontaneous branch migration as previously described (40). Using these substrates, we observed resolution of both the 5′ to 3′ and 3′ to 5′ substrates (Supplementary Figure S6C and D), which is consistent with the bidirectional helicase activity of ComM. This activity was inhibited when ComM was incubated with AMP-PNP, a nonhydrolyzable analog of ATP (Supplementary Figure S6C and D). And no activity was observed with ComMK224A even if incubated with ATP (Supplementary Figure S6C and D). Thus, the branch migration activity observed is ATPase-dependent. It is formally possible that resolution of these short three-stranded products was the result of only helicase activity (via the sequential removal of the unlabeled strand followed by the labeled strand or by unwinding of just the labeled strand). To address this, we also performed helicase assays using forked substrates that are derived from the oligos used to make the three-stranded branch migration substrates. This analysis indicated that ComM helicase activity on the forked substrates was markedly less efficient than its activity on the three-stranded substrates (compare Supplementary Figure S6C–D to S6E–F), which suggests that the activity observed on the three-stranded substrates is the result of bona fide branch migration and not simply helicase activity. The forked substrates used in this assay have 60 bp of annealed sequence (Supplementary Figure S6E-F), while the substrates previously used to test helicase activity only had 20 bp of annealed sequence (Figure 4A). Thus, reduced activity on the forked DNA substrates in this assay is likely attributed to poor processivity for ComM helicase activity.
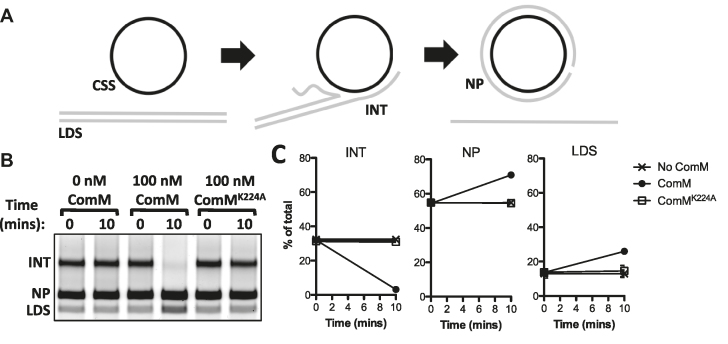
While the short three-stranded substrates used above suggest that ComM possesses branch migration activity, we further tested three-stranded branch migration using long DNA substrates. Long three-stranded substrates were generated by RecA-mediated strand exchange between circular single-stranded (CSS) and linear double-stranded (LDS) φX174 DNA (23,41). Strand exchange reactions were allowed to proceed until the majority of the LDS was reacted with the CSS to form intermediates (INT = joint molecules that have not completed strand exchange) or nicked product (NP = the product formed upon complete strand exchange). Reactions were then deproteinated, and the resulting DNA was used to assess resolution of the long three-stranded INT via branch migration (23,41). We found that ComM could efficiently drive the reaction in both directions, resolving the INT structures into both NP and LDS (Figure 5), while reactions where ComMK224A was added showed no activity. These results are consistent with ComM promoting branch migration of these long DNA substrates. Together, these results indicate that ComM exhibits branch migration on three-stranded junctions in vitro on substrates that are likely similar to the structures formed during natural transformation in vivo.
Figure 5.
ComM exhibits 3-stranded branch migration activity on long DNA substrates in vitro. (A) Schematic for RecA-mediated strand exchange between linear double stranded φX174 (LDS) and circular single-stranded φX174 (CSS), which results in the formation of intermediates (INT) that can be resolved to nicked product (NP) if strand exchange commences to completion. Strand exchange reactions were deproteinated prior to complete strand exchange, and the resulting DNA was used to assess branch migration-dependent resolution of intermediate structures (INT). (B) Representative gel where deproteinated intermediates were incubated with the proteins indicated. (C) Three independent replicates of the assay described in B were quantified, and the relative abundance of the INT, NP, and LDS are shown as the mean ± SD.
ComM is broadly conserved
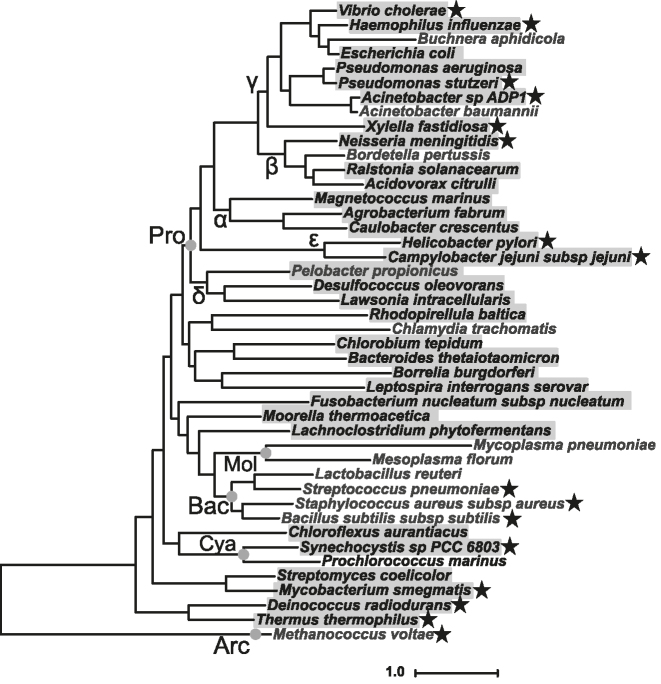
Next, we assessed how broadly conserved ComM was among bacterial species. Homologs of ComM are found almost ubiquitously among Gram-negative species, including all known Gram-negative naturally competent microbes (Figure 6 and Supplementary Figure S7). Among this group, only select species lacked a ComM homolog, suggesting that loss of ComM occurred relatively recently (Figure 6 and Supplementary Figure S7). By contrast, we see a pervasive lack of ComM homologs among species within the Bacilli and Mollicutes, (Figure 6 and Supplementary Figure S7), suggesting that ComM may have been lost in a common ancestor for these two Classes. Interestingly, all known naturally competent Gram-positive species fall within the Bacilli group, indicating that branch migration during natural transformation must occur via a ComM-independent mechanism in these microbes.
Figure 6.
ComM is broadly conserved. Estimated maximum likelihood phylogeny of diverse species based on a concatenated alignment of 36 conserved proteins identified from whole genome sequences. Species with an identified ComM homolog are highlighted in gray. Competent species are designated by a star next to the species name. Major taxa are labeled along their nodes. Pro: Proteobacteria (Greek letters indicate subdivisions); Bac: Bacilli; Mol: Mollicutes; Cya: Cyanobacteria; Arc: Archaea. Scale bar indicates distance.
ComM promotes branch migration in Acinetobacter baylyi
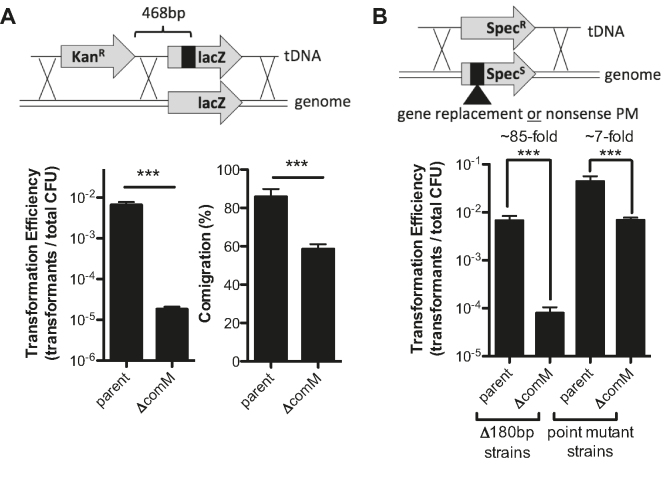
Because ComM is broadly conserved among Gram-negative naturally competent microbes, next, we tested its role during natural transformation in the model competent species A. baylyi ADP1. As in V. cholerae, a comM mutant of A. baylyi displayed greatly reduced rates of natural transformation when using linear tDNA (Figure 7A, left). Also, this mutant displayed reduced comigration of linked genetic markers, consistent with comM playing a role in branch migration (Figure 7A, right). Additionally, we observed that ComM is required for integration of tDNA containing larger regions of heterologous sequence (Figure 7B). These data are consistent with what was observed in V. cholerae (Figure 2) and suggests that ComM is a conserved branch migration factor important for natural transformation in diverse Gram-negative bacterial species.
Figure 7.
ComM promotes branch migration in Acinetobacter baylyi. (A) Transformation assay of A. baylyi using a linear tDNA product with linked genetic markers. (B) Transformation assay performed in SpecS strains with tDNA that would revert the strain to SpecR. Integration of the marker would either repair a point mutation or delete 180 bp of genomic sequence. All strains in A and B contain mutS mutations to prevent MMR activity. In schematics above bar graphs, X’s denote possible crossover points for homologous recombination. All data are shown as the mean ± SD and are the result of at least three independent biological replicates. ***P < 0.001.
RadA is not required for natural transformation in V. cholerae
While ComM is broadly conserved, it is absent in all of the known Gram-positive naturally competent species (Figure 6). In the Gram-positive Streptococcus pneumoniae, mutants of radA (also known as sms) display reduced rates of natural transformation but are not affected at the level of tDNA uptake, similar to what is observed for V. cholerae comM mutants in this study (42) and in H. influenzae (9). Similar results are also seen in radA mutants in Bacillus subtilis (43). E. coli RadA has recently been shown to promote branch migration during RecA-mediated strand exchange (23). Also, it was very recently demonstrated that RadA is a hexameric helicase that promotes bidirectional D-loop extension during natural transformation in S. pneumoniae (38). Interestingly, radA is broadly conserved and both V. cholerae and A. baylyi contain radA homologs (VC2343 and ACIAD2664, respectively). RadA, however, is not required for natural transformation in V. cholerae (Supplementary Figure S8). Also, preliminary Tn-seq data from our lab indicates that RadA is not important for natural transformation in A. baylyi. Thus, it is tempting to speculate that in Gram-positive competent species, RadA carries out the same function that ComM plays during natural transformation in Gram-negative species.
ComM is not required for DNA repair
ComM homologs are also found among diverse non-competent species (Figure 6). Moreover, many bacterial helicases are implicated in promoting branch migration during other types of homologous recombination, including during DNA repair. So next, we wanted to determine if ComM also plays a role in DNA repair independent of its role during natural transformation. ComM is poorly expressed in the absence of competence induction in V. cholerae (Supplementary Figure S2). Thus, to test the role of ComM in DNA damage, we tested survival of Ptac-tfoX and Ptac-tfoX ΔcomM strains under competence inducing conditions (i.e. ectopic expression of TfoX). A WT strain and a recA mutant were also included in these assays as controls. DNA damage was tested using methyl methanesulfonate (MMS – methylates DNA / stalls replication forks) and mitomycin C (MMC – alkylates DNA/generates interstrand DNA crosslinks) (44,45). As expected, the recA mutant was more sensitive to these treatments compared to the WT, consistent with a critical role for homologous recombination in DNA repair (Supplementary Figure S9) (46,47). The Ptac-tfoX ΔcomM mutant, however, was as resistant to these DNA damaging agents as the Ptac-tfoX strain, indicating that this branch migration factor either does not play a role during DNA repair or that the activity of this protein is redundant with other branch migration factors in the context of repair (Supplementary Figure S9). Furthermore, induction of competence (via ectopic expression of TfoX) showed little to no difference in DNA damage repair compared to the WT, indicating that natural competence plays a limited role in DNA repair in V. cholerae under the condition tested (Supplementary Figure S9).
DISCUSSION
Natural transformation is an important mechanism of horizontal gene transfer in bacterial species. It is dependent upon activation of bacterial competence or the ability to bind and take up exogenous DNA. Altogether, our in vivo and in vitro data elucidate a role for ComM as a helicase/branch migration factor that promotes the integration of tDNA during natural transformation. In our model, integration is initiated by RecA-mediated strand invasion and formation of a D-loop that generates a three-stranded intermediate structure (Figure 8). Through its bidirectional helicase and/or branch migration activity, ComM then likely promotes expansion of the D-loop, which enhances the integration of tDNA into the genome (Figure 8). Following branch migration, the junctions are resolved to mediate stable integration of tDNA by an unresolved mechanism.
Figure 8.
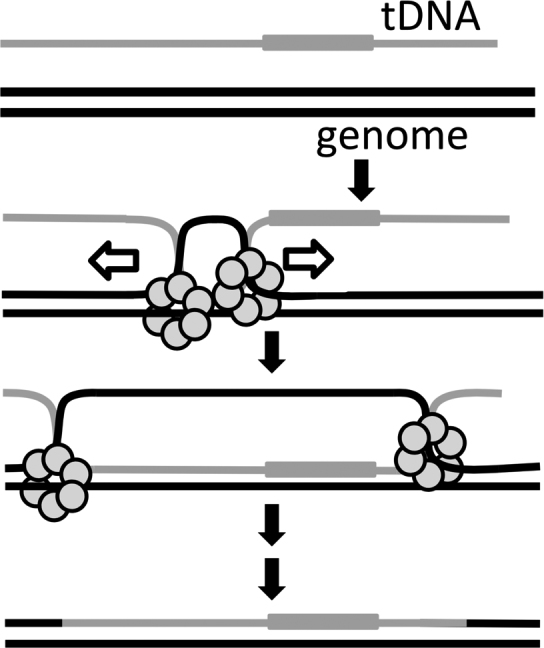
Proposed model for the role of ComM during natural transformation. ComM is shown as a hexameric ring that promotes integration of tDNA via its bidirectional helicase and/or branch migration activity. This can support integration of tDNA with a heterologous region, which is indicated by a gray box.
Our data suggest that ComM is important for the incorporation of heterologous sequences. The main drivers for evolution and maintenance of natural transformation in bacterial species are heavily debated. One model suggests that this process is largely for enhancing genetic diversity, while another hypothesis is that natural transformation evolved as a mechanism for acquisition of DNA as a nutrient (48,49). These processes are not mutually exclusive. Because ComM affects only the integration of heterologous tDNA (and not its uptake), however, the activity of this protein supports a role for natural transformation in adaptation and evolution through the acquisition of novel genetic material. Other competence genes that are involved specifically in homologous recombination (e.g. dprA) also support this hypothesis.
Our in vitro data suggest that ComM forms a hexameric ring structure in its active state similar to that of eukaryotic MCM2-7 and bacterial DnaB (31,50). Our 3D reconstruction reveals a three-tiered barrel-like complex with a ∼26-Å pore and ∼18-Å pore on the top and bottom, respectively. We propose that ComM either forms around, or is loaded onto the displaced single strand of the native genomic DNA and acts as a wedge to dissociate these strands. Alternatively, dsDNA may enter the 26-Å pore, be unwound in the central channel of the complex with single strands bring extruded through the side channels that are evident between the tiers of the barrel structure. Such side channels are not uncommon among hexameric AAA+ helicases (e.g. SV40 T-antigen (51,52) and the archaeal MCM (28,53)) and have been hypothesized to be exit channels for extruded ssDNA. The pore sizes observed in our reconstruction appear to be consistent with those found in other ring helicases, however, it has been shown that the size of the opening and channel can change depending on the nucleotide bound (ATP versus ADP) (51,54). Future work will focus on characterizing the ComM structure bound to ADP and AMP-PNP, which may help inform the structural changes associated with the catalytic cycle of this hexameric helicase.
Our phylogenetic analysis indicates that ComM is broadly conserved in bacterial species, and is largely, only excluded from the Bacilli and Mollicutes. There is also, however, evidence for isolated examples for ComM loss in species that fall outside of these two groups, which suggests that these have occurred relatively recently. Some of these represent obligate intracellular pathogens or endosymbiots, which commonly contain highly reduced genomes (e.g. Buchnera and Chlaymdia) (55,56). Another example of recent ComM loss is among Prochlorococcus species, which are related to other naturally competent cyanobacteria (e.g. Synechococcus and Synechocystis spp.). Interestingly, we found that Prochlorococcus lacked many of the genes required for natural transformation (e.g. dprA, comEA, comEC, etc.) (57), indicating that competence may have been lost in this lineage of cyanobacteria. Another notable example of ComM loss is in Acinetobacter baumanii, which is an opportunistic pathogen that is closely related to A. baylyi. Many strains of A. baumanii, including the one analyzed here (ATCC 17978), contain an AbaR-type genomic island integrated into comM (58,59). Strains that contain this horizontally transferred genomic island display low rates of natural transformation while those that lack it display higher rates of natural transformation (60), indicating that AbaR island-dependent inactivation of ComM may inhibit this mechanism of horizontal gene transfer in A. baumanii. This represents another example in a growing list of horizontally acquired genomic elements that inhibit natural transformation (61–63). Our phylogenetic analysis also indicates that ComM is highly conserved among non-competent bacterial species. This suggests that ComM may have a function outside of natural transformation. Alternatively, it is possible that many of these species are capable of natural transformation, however, the conditions required for competence induction have not yet been identified. Indeed, the inducing cue for natural transformation in V. cholerae was only discovered in 2006 (2).
Our data suggest that branch migration in Gram-positive and Gram-negative species has diverged in their dependence on distinct factors (RadA and ComM, respectively). The tract length of DNA recombined into S. pneumoniae during natural transformation is ∼2.5 kb on average (64). In H. influenzae, the mean recombination tract length is ∼14 kb (65). Thus, compared to RadA-dependent branch migration in Gram-positive species, ComM may facilitate the integration of more tDNA in Gram-negative species. Other proteins that impact DNA integration during natural transformation, however, may confound this overly simplified comparison.
ComM is not essential for natural transformation because transformants are still observed in a comM mutant. This suggests that other proteins may be involved in promoting the integration of tDNA in the absence of this branch migration factor. Also, our data suggest that these alternative branch migration factors are less efficient at incorporating tDNA with sequence heterology compared to ComM in both V. cholerae and A. baylyi. This role could be carried out by another helicase. Candidates include RuvAB, RecG, and PriA, which have all previously been implicated in branch migration (32,33,66). In fact, the primasome helicase PriA is essential for natural transformation in Neisseria gonorrhoeae. This may be due to a role in branch migration, however, PriA is also essential for restarting stalled replication forks and priA mutants have a severe growth defect (67). Also, it has been proposed that PriA helicase activity may facilitate the uptake of tDNA through the inner membrane (68). A recG homologue in Streptococcus pneumoniae (mmsA) was shown to have a mild effect on transformation when deleted (69). RecG reverses stalled replication forks and promotes branch migration opposite to the direction of RecA-mediated strand exchange in E. coli (70,71). Also, RecG acts on three-strand intermediates in vitro, which could give credence to involvement in natural transformation where such a structure is formed (41). Because RecG works counter to the direction of RecA-mediated strand exchange, however, it is currently unclear how RecG may play a role in natural transformation in the presence of ComM. Another possibility is that RecA, which has inherent ATP-dependent unidirectional branch migration activity (72,73), could promote branch migration independent of other canonical branch migration factors. Future work will focus on identifying genetically interacting partners of ComM to uncover the role of additional factors required for efficient integration of tDNA during natural transformation.
In addition to a AAA+ ATPase domain, ComM also contains two magnesium (Mg) chelatase domains. Mg chelatase domain-containing proteins have only previously been implicated in inserting Mg into protoporphyrin rings in photosynthetic organisms (74). While it is currently unclear how these domains participate in ComM function, to our knowledge, this study is the first to indicate a function for a Mg-chelatase domain-containing protein in a non-photosynthetic organism.
In conclusion, the results from this study strongly support that ComM enhances natural transformation by promoting ATP-dependent bidirectional helicase activity and/or branch migration activity, which allows for the efficient integration of heterologous tDNA. Furthermore, our data in V. cholerae and A. baylyi as well as previous work from H. influenzae indicate that this branch migration factor is a broadly conserved mechanism for integration of tDNA in diverse naturally transformable Gram-negative species.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
US National Institutes of Health Grant [AI118863 to A.B.D.]; Indiana University Collaborative Research Grant (to M.L.B.); American Cancer Society Research Scholar [RSG-16-180-01-DMC to M.L.B.]; Indiana University College of Arts and Sciences (to A.B.D.); D.T.K was supported by NIH grants [R35 GM122556 to Yves Brun and R01 GM113121 to Stephen Jacobson]. Funding for open access charge: Startup funds from the Indiana University College of Arts and Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Seitz P., Blokesch M.. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 2013; 37:336–363. [DOI] [PubMed] [Google Scholar]
- 2. Meibom K.L., Blokesch M., Dolganov N.A., Wu C.Y., Schoolnik G.K.. Chitin induces natural competence in Vibrio cholerae. Science. 2005; 310:1824–1827. [DOI] [PubMed] [Google Scholar]
- 3. Dalia A.B., Lazinski D.W., Camilli A.. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. MBio. 2014; 5:e01028–e01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yamamoto S., Mitobe J., Ishikawa T., Wai S.N., Ohnishi M., Watanabe H., Izumiya H.. Regulation of natural competence by the orphan two-component system sensor kinase ChiS involves a non-canonical transmembrane regulator in Vibrio cholerae. Mol. Microbiol. 2014; 91:326–347. [DOI] [PubMed] [Google Scholar]
- 5. Lo Scrudato M., Blokesch M.. The regulatory network of natural competence and transformation of Vibrio cholerae. PLoS Genet. 2012; 8:e1002778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lo Scrudato M., Blokesch M.. A transcriptional regulator linking quorum sensing and chitin induction to render Vibrio cholerae naturally transformable. Nucleic Acids Res. 2013; 41:3644–3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Metzgar D., Bacher J.M., Pezo V., Reader J., Doring V., Schimmel P., Marliere P., de Crecy-Lagard V.. Acinetobacter sp. ADP1: an ideal model organism for genetic analysis and genome engineering. Nucleic Acids Res. 2004; 32:5780–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lorenz M.G., Wackernagel W.. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994; 58:563–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gwinn M.L., Ramanathan R., Smith H.O., Tomb J.F.. A new transformation-deficient mutant of Haemophilus influenzae Rd with normal DNA uptake. J. Bacteriol. 1998; 180:746–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miller V.L., DiRita V.J., Mekalanos J.J.. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J. Bacteriol. 1989; 171:1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Juni E., Janik A.. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum). J. Bacteriol. 1969; 98:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dalia A.B., Lazinski D.W., Camilli A.. Characterization of undermethylated sites in Vibrio cholerae. J. Bacteriol. 2013; 195:2389–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalia A.B., McDonough E., Camilli A.. Multiplex genome editing by natural transformation. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:8937–8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dalia A.B. RpoS is required for natural transformation of Vibrio cholerae through regulation of chitinases. Environ. Microbiol. 2016; 18:3758–3767. [DOI] [PubMed] [Google Scholar]
- 15. Studier F.W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005; 41:207–234. [DOI] [PubMed] [Google Scholar]
- 16. Wittig I., Braun H.P., Schagger H.. Blue native PAGE. Nat. Protoc. 2006; 1:418–428. [DOI] [PubMed] [Google Scholar]
- 17. Wu M., Eisen J.A.. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008; 9:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Price M.N., Dehal P.S., Arkin A.P.. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010; 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Darling A.E., Jospin G., Lowe E., Matsen F.A.t., Bik H.M., Eisen J.A.. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014; 2:e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014; 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Le S.Q., Gascuel O.. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008; 25:1307–1320. [DOI] [PubMed] [Google Scholar]
- 23. Cooper D.L., Lovett S.T.. Recombinational branch migration by the RadA/Sms paralog of RecA in Escherichia coli. eLife. 2016; 5:e10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mortier-Barriere I., Velten M., Dupaigne P., Mirouze N., Pietrement O., McGovern S., Fichant G., Martin B., Noirot P., Le Cam E. et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007; 130:824–836. [DOI] [PubMed] [Google Scholar]
- 25. Suckow G., Seitz P., Blokesch M.. Quorum sensing contributes to natural transformation of Vibrio cholerae in a species-specific manner. J. Bacteriol. 2011; 193:4914–4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hanson P.I., Whiteheart S.W.. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell. Biol. 2005; 6:519–529. [DOI] [PubMed] [Google Scholar]
- 27. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brewster A.S., Wang G., Yu X., Greenleaf W.B., Carazo J.M., Tjajadi M., Klein M.G., Chen X.S.. Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaplan D.L., Davey M.J., O’Donnell M.. Mcm4,6,7 uses a “pump in ring” mechanism to unwind DNA by steric exclusion and actively translocate along a duplex. J. Biol. Chem. 2003; 278:49171–49182. [DOI] [PubMed] [Google Scholar]
- 30. Borgeaud S., Metzger L.C., Scrignari T., Blokesch M.. The type VI secretion system of Vibrio cholerae fosters horizontal gene transfer. Science. 2015; 347:63–67. [DOI] [PubMed] [Google Scholar]
- 31. Bochman M.L., Schwacha A.. The Mcm complex: unwinding the mechanism of a replicative helicase. Microbiol Mol. Biol. Rev. 2009; 73:652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lloyd R.G., Sharples G.J.. Processing of recombination intermediates by the RecG and RuvAB proteins of Escherichia coli. Nucleic Acids Res. 1993; 21:1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. West S.C. Processing of recombination intermediates by the RuvABC proteins. Annu. Rev. Genet. 1997; 31:213–244. [DOI] [PubMed] [Google Scholar]
- 34. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A.. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kaplan D.L., O’Donnell M.. DnaB drives DNA branch migration and dislodges proteins while encircling two DNA strands. Mol. Cell. 2002; 10:647–657. [DOI] [PubMed] [Google Scholar]
- 36. Yakovleva L., Shuman S.. Mycobacterium smegmatis SftH exemplifies a distinctive clade of superfamily II DNA-dependent ATPases with 3′ to 5′ translocase and helicase activities. Nucleic Acids Res. 2012; 40:7465–7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fallesen T., Roostalu J., Duellberg C., Pruessner G., Surrey T.. Ensembles of bidirectional kinesin Cin8 produce additive forces in both directions of movement. Biophys. J. 2017; 113:2055–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Marie L., Rapisarda C., Morales V., Berge M., Perry T., Soulet A.L., Gruget C., Remaut H., Fronzes R., Polard P.. Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension. Nat. Commun. 2017; 8:15638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N.. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990; 10:1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bugreev D.V., Brosh R.M. Jr, Mazin A.V.. RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 2008; 283:20231–20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whitby M.C., Lloyd R.G.. Branch migration of three-strand recombination intermediates by RecG, a possible pathway for securing exchanges initiated by 3′-tailed duplex DNA. EMBO J. 1995; 14:3302–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Burghout P., Bootsma H.J., Kloosterman T.G., Bijlsma J.J., de Jongh C.E., Kuipers O.P., Hermans P.W.. Search for genes essential for pneumococcal transformation: the RADA DNA repair protein plays a role in genomic recombination of donor DNA. J. Bacteriol. 2007; 189:6540–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Carrasco B., Fernandez S., Asai K., Ogasawara N., Alonso J.C.. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics. 2002; 266:899–906. [DOI] [PubMed] [Google Scholar]
- 44. Weng M.W., Zheng Y., Jasti V.P., Champeil E., Tomasz M., Wang Y., Basu A.K., Tang M.S.. Repair of mitomycin C mono- and interstrand cross-linked DNA adducts by UvrABC: a new model. Nucleic Acids Res. 2010; 38:6976–6984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A.S., Helleday T.. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005; 33:3799–3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goranov A.I., Kuester-Schoeck E., Wang J.D., Grossman A.D.. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J. Bacteriol. 2006; 188:5595–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Better M., Helinski D.R.. Isolation and characterization of the recA gene of Rhizobium meliloti. J. Bacteriol. 1983; 155:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Michod R.E., Wojciechowski M.F., Hoelzer M.A.. DNA repair and the evolution of transformation in the bacterium Bacillus subtilis. Genetics. 1988; 118:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Redfield R.J. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J. Hered. 1993; 84:400–404. [DOI] [PubMed] [Google Scholar]
- 50. San Martin M.C., Stamford N.P., Dammerova N., Dixon N.E., Carazo J.M.. A structural model for the Escherichia coli DnaB helicase based on electron microscopy data. J. Struct. Biol. 1995; 114:167–176. [DOI] [PubMed] [Google Scholar]
- 51. Gai D., Zhao R., Li D., Finkielstein C.V., Chen X.S.. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004; 119:47–60. [DOI] [PubMed] [Google Scholar]
- 52. Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J.A., Fanning E., Jochimiak A., Szakonyi G., Chen X.S.. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature. 2003; 423:512–518. [DOI] [PubMed] [Google Scholar]
- 53. Costa A., Pape T., van Heel M., Brick P., Patwardhan A., Onesti S.. Structural basis of the Methanothermobacter thermautotrophicus MCM helicase activity. Nucleic Acids Res. 2006; 34:5829–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Strycharska M.S., Arias-Palomo E., Lyubimov A.Y., Erzberger J.P., O'Shea V.L., Bustamante C.J., Berger J.M.. Nucleotide and partner-protein control of bacterial replicative helicase structure and function. Mol. Cell. 2013; 52:844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shigenobu S., Watanabe H., Hattori M., Sakaki Y., Ishikawa H.. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000; 407:81–86. [DOI] [PubMed] [Google Scholar]
- 56. Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R.L., Zhao Q. et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998; 282:754–759. [DOI] [PubMed] [Google Scholar]
- 57. Rocap G., Larimer F.W., Lamerdin J., Malfatti S., Chain P., Ahlgren N.A., Arellano A., Coleman M., Hauser L., Hess W.R. et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003; 424:1042–1047. [DOI] [PubMed] [Google Scholar]
- 58. Smith M.G., Gianoulis T.A., Pukatzki S., Mekalanos J.J., Ornston L.N., Gerstein M., Snyder M.. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007; 21:601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ramirez M.S., Vilacoba E., Stietz M.S., Merkier A.K., Jeric P., Limansky A.S., Marquez C., Bello H., Catalano M., Centron D.. Spreading of AbaR-type genomic islands in multidrug resistance Acinetobacter baumannii strains belonging to different clonal complexes. Curr. Microbiol. 2013; 67:9–14. [DOI] [PubMed] [Google Scholar]
- 60. Traglia G.M., Chua K., Centron D., Tolmasky M.E., Ramirez M.S.. Whole-genome sequence analysis of the naturally competent Acinetobacter baumannii clinical isolate A118. Genome Biol. Evol. 2014; 6:2235–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Dalia A.B., Seed K.D., Calderwood S.B., Camilli A.. A globally distributed mobile genetic element inhibits natural transformation of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:10485–10490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Konkol M.A., Blair K.M., Kearns D.B.. Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 2013; 195:4085–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rabinovich L., Sigal N., Borovok I., Nir-Paz R., Herskovits A.A.. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell. 2012; 150:792–802. [DOI] [PubMed] [Google Scholar]
- 64. Croucher N.J., Harris S.R., Barquist L., Parkhill J., Bentley S.D.. A high-resolution view of genome-wide pneumococcal transformation. PLoS Pathog. 2012; 8:e1002745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mell J.C., Shumilina S., Hall I.M., Redfield R.J.. Transformation of natural genetic variation into Haemophilus influenzae genomes. PLoS Pathog. 2011; 7:e1002151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Al-Deib A.A., Mahdi A.A., Lloyd R.G.. Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 1996; 178:6782–6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kline K.A., Seifert H.S.. Mutation of the priA gene of Neisseria gonorrhoeae affects DNA transformation and DNA repair. J. Bacteriol. 2005; 187:5347–5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kruger N.J., Stingl K.. Two steps away from novelty–principles of bacterial DNA uptake. Mol. Microbiol. 2011; 80:860–867. [DOI] [PubMed] [Google Scholar]
- 69. Martin B., Sharples G.J., Humbert O., Lloyd R.G., Claverys J.P.. The mmsA locus of Streptococcus pneumoniae encodes a RecG-like protein involved in DNA repair and in three-strand recombination. Mol. Microbiol. 1996; 19:1035–1045. [DOI] [PubMed] [Google Scholar]
- 70. Whitby M.C., Ryder L., Lloyd R.G.. Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell. 1993; 75:341–350. [DOI] [PubMed] [Google Scholar]
- 71. Azeroglu B., Mawer J.S., Cockram C.A., White M.A., Hasan A.M., Filatenkova M., Leach D.R.. RecG directs DNA synthesis during double-strand break repair. PLoS Genet. 2016; 12:e1005799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Cox M.M., Morrical S.W., Neuendorf S.K.. Unidirectional branch migration promoted by nucleoprotein filaments of RecA protein and DNA. Cold Spring Harb. Symp. Quant. Biol. 1984; 49:525–533. [DOI] [PubMed] [Google Scholar]
- 73. Cox M.M., Lehman I.R.. Directionality and polarity in recA protein-promoted branch migration. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:6018–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Papenbrock J., Mock H.P., Tanaka R., Kruse E., Grimm B.. Role of magnesium chelatase activity in the early steps of the tetrapyrrole biosynthetic pathway. Plant Physiol. 2000; 122:1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.