Abstract
One of the first events to occur upon DNA damage is the local opening of the compact chromatin architecture, facilitating access of repair proteins to DNA lesions. This early relaxation is triggered by poly(ADP-ribosyl)ation by PARP1 in addition to ATP-dependent chromatin remodeling. CHD4 recruits to DNA breaks in a PAR-dependent manner, although it lacks any recognizable PAR-binding domain, and has the ability to relax chromatin structure. However, its role in chromatin relaxation at the site of DNA damage has not been explored. Using a live cell fluorescence three-hybrid assay, we demonstrate that the recruitment of CHD4 to DNA damage, while being poly(ADP-ribosyl)ation-dependent, is not through binding poly(ADP-ribose). Additionally, we show that CHD3 is recruited to DNA breaks in the same manner as CHD4 and that both CHD3 and CHD4 play active roles in chromatin remodeling at DNA breaks. Together, our findings reveal a two-step mechanism for DNA damage induced chromatin relaxation in which PARP1 and the PAR-binding remodeler activities of Alc1/CHD1L induce an initial chromatin relaxation phase that promotes the subsequent recruitment of CHD3 and CHD4 via binding to DNA for further chromatin remodeling at DNA breaks.
INTRODUCTION
One of the earliest steps in the DNA damage response (DDR) is rapid relaxation of chromatin in the vicinity of the DNA lesions. Chromatin relaxation is crucial for efficient DNA repair (1), promoting the loading of repair proteins at DNA breaks (2). This early step in the DNA damage response is heavily reliant on poly(ADP-ribose) (PAR) polymerase 1 (PARP1), which acts as a sensor of DNA damage, binding to DNA breaks and promoting auto-activation (3). Activated PARP1 PARylates target proteins including itself, core histones as well as the histone linker H1 (4) and more recently, has been shown to initiate chromatin relaxation to sites of DNA damage (5–7). The formation of PAR at DNA breaks has two main consequences. Firstly, PAR itself induces chromatin relaxation through the addition of a large number of negative-charges onto chromatin (8). Secondly, PAR promotes recruitment of PAR-binding effectors such as the macrodomain-containing chromatin remodeler, Alc1/CHD1L (9,10). We recently demonstrated the involvement of Alc1 in the rapid relaxation of chromatin at break sites, however, it was clear that DNA damage induced chromatin remodeling does not solely rely on the activity of Alc1 but requires additional ATP-dependent mechanisms (5,11). In addition to Alc1, several remodelers, such as SMARCA5, CHD2 and CHD4, have been previously described to recruit to DNA damage in a PARylation-dependent manner (6,12–16), however, their role in DNA damage-induced chromatin relaxation has not been investigated.
In this study, we focused on the CHD4 chromatin remodeler, and its close homologue CHD3 (17,18), integral members of two distinct histone deacetylase containing NuRD complexes (19–22). In addition to a repressive role in transcription regulation (23,24), CHD4 was shown to efficiently decompact the LacO array when recruited through RNF8 or directly tethered to it and has been shown to play a role in DNA damage repair and BRCA1 assembly (15,16,25,26). Knock-down of CHD4 impairs the repair of double strand breaks by homologous recombination (HR) but not via non-homologous end joining (NHEJ) (25) implicating a role for CHD4 in regulating the balance between the HR and NHEJ repair pathways. What is currently unknown, however, is whether CHD4 directly contributes to chromatin remodeling during the DDR. Furthermore, despite both the human CHD4 and the Drosophila homologue dMi2 demonstrating PAR binding in vitro (27,28), the exact mechanism driving the recruitment of CHD4 to DNA lesions remains unclear as it does not bear a canonical PAR-binding domain. Recently, it has been shown that while CHD4 and CHD3 form distinct NuRD complexes, they share some functionality and that CHD3 also recruits to UV-induced DNA lesions, similar to CHD4 (22). In the present work, we deciphered the molecular mechanism responsible for the accumulation of these remodelers at DNA breaks and assessed their specific contribution during chromatin remodeling in response to DNA damage induction.
MATERIALS AND METHODS
Plasmids and siRNA
PARP1-mCherry (9), YFP-macroH2A1.1 macrodomain (29), EGFP-Alc1 and PATagRFP-H2B (5) were described previously. H2B-PAGFP was a kind gift from J. Ellenberg (30). GFP-CHD4 was a kind gift from S. Polo (13). GFP-CHD3 was a kind gift from H. Hoffmeister (22). piRFP670-N1 (31), wild-type and ATPase-dead mutant Alc1 (5) were used to generate iRFP-Alc1 WT and iRFP-Alc1 E175Q via Gibson Assembly. mCherry was amplified from pmCherry-C1 (Clontech) (Forward 5′- ATTAGGCCGCGGACCATGGACGTGAGCAAG-3′ and Reverse 5′- ATATCCCGGGAGGACTTGTACAGCTCGTC-3′) and used to exchange GFP from GFP-CHD4 using SacII and XmaI to create mCherry-CHD4. The GFP-nanobody previously described (32) was amplified (Forward 5′-AAAACTTAGGTCTAGATCCGGTGG-3′ and Reverse 5′-AAAACATATGGTGATGGTGATGG-3′) and LacI was amplified from pmCherry-LacI-H3 (33) (Forward 5′-AAAACATATGGTGAAACCAGTAACG-3′ and Reverse 5′-AAAAGCGGCCGCTTATCTAGATCCGGTGG-3′). The GFP-nanobody insert was digested with NheI and NdeI and the LacI insert was digested with NdeI and NotI and pcDNA3.1 was digested with NheI and NotI before being used together in a single ligation reaction to produce the GFP nanobody-LacI fusion. Silencer Select siRNA (Thermo Scientific) were used for siRNA-mediated knockdown of target proteins (siCHD4-1 #s2984, siCHD4-2 #s2985, siCHD3-1 #s2980, siCHD3-2 #s2981, siScr #4390843).
Cell culture
U2OS, U2OS-2B2 (34), U2OS-Alc1KO and U2OS-PARP1KO (5) were cultured in DMEM (4.5 g/L glucose) supplemented with 10% fetal bovine serum (Life Technologies), 2 mM glutamine (Sigma), 100 μg/mL penicillin and 100 U/mL streptomycin (Sigma) in a humidified incubator at 37°C with 5% CO2.
U2OS cells were transfected with H2B-PAGFP using XtremeGENE HP (Roche) and selected with Geneticin containing media to produce U2OS cells stably expressing H2B-PAGFP. After initial selection, cells were maintained in growth media supplemented with 500 μg/ml Geneticin (Life Technologies).
For transient expression of plasmids, cells were transfected 12–24 h after seeding into 8-well Lab-Tek II chambered coverglass (Thermo Scientific) with XtremeGENE HP or Xfect (Clontech) according to the manufactures instructions and incubated for 48 h prior to imaging.
For siRNA-mediated knockdown, cells were transfected with 2 pmol of siRNA per well of an 8-well Lab-Tek using Lipofectamine RNAiMAX (Invitrogen) according to the manufactures instructions and incubated for 72 h prior to imaging.
For live-cell imaging, cells were seeded on LabTek II chambered coverglass (Thermo Scientific). For Hoechst presensitization, growth medium was aspirated from the Lab-Tek and replaced with fresh medium containing 0.3 μg/ml Hoechst 33342 for 1 h at 37°C. Immediately prior to imaging, growth medium was replaced with CO2-independent imaging medium (Phenol Red-free Leibovitz's L-15 medium (Life Technologies) supplemented with 20% fetal bovine serum, 2 mM glutamine, 100 μg/ml penicillin and 100 U/ml streptomycin).
Olaparib (3 μM) was used for PARP inhibition for at least 30 min before experiments. For hypotonic treatments the cells were bathed with normal imaging medium diluted with distilled water (35:65, v/v) as previously described (35).
Fluorescence imaging and laser microirradiation
Except for the images shown in Figure 2D, Supplementary Figures S3C and E the fluorescence imaging was performed on two different confocal systems composed of an inverted microscope body (Zeiss AxioObserver Z1 or Nikon Ti-E) and a spinning-disk scan head CSU-X1 from Yokogawa at a rotation speed of 5000 rpm. We used either a C-Apo 63× water immersion objective lens (N.A. 1.2) or a Plan APO 63× oil immersion objective lens (N.A. 1.4). Images were acquired on an AxioCam HRm CCD camera (Zeiss) or a sCMOS ORCA Flash 4.0 camera (Hamamatsu). For laser microirradiation at 405 nm, we used dedicated single-point scanning head (UGA-42 firefly from Rapp OptoElectronic or iLas2 from Roper Scientific) coupled to the epifluorescence backboard of the microscope (Supplementary Figure S1A). For DNA damage induction and photoactivation of H2B tagged proteins, the power density of the 405 nm laser was set to 1 μJ/μm2 at the sample level. The laser power was measured and adjusted at the beginning of each experiment to ensure reproducibility. Cells were maintained at 37°C in the absence of CO2.
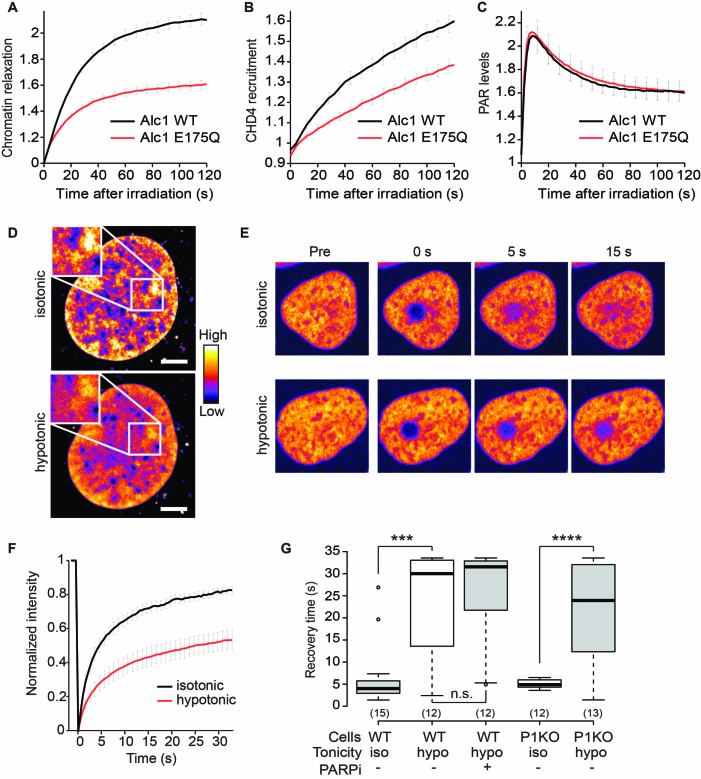
Figure 2.
Chromatin relaxation is sufficient to promote the binding of CHD4 to chromatin. (A–C) Chromatin relaxation (A), CHD4 recruitment (B) and PAR levels (C) in cells over-expressing wild-type Alc1 (Alc1 WT, black) or the ATPase-dead (Alc1 E175Q, red) mutant after UV-microirradiation. (D) Pseudocolored confocal images of the same Hoechst-stained nucleus in isotonic and hypotonic media. Scale bar is 5 μm. (E) U2OS cells expressing GFP-CHD4 in isotonic or hypotonic solutions before (Pre) and after photobleaching (F, G) Normalized FRAP curves (F) and recovery times (G) of GFP-CHD4 in isotonic (iso) or hypotonic (hypo) solutions in wild-type (WT) or PARP1 knockout (P1KO) cells. PARP inhibition is denoted by PARPi. The look-up-table shown on panel (D) is valid for all pseudocolored images.
The images shown in Figure 2D, Supplementary Figures S3C and E were acquired on a Leica SP8 confocal microscope with a Plan APO 63× oil immersion objective lens (N.A. 1.4) and a pinhole set at one Airy unit and a pixel size of 60 nm. Hoechst staining was excited with a 405 nm laser and the emission band was chosen to optimize fluorescence collection. Pixel-to-pixel contrast was calculated using a custom made ImageJ macro.
Fluorescence correlation spectroscopy (FCS)
FCS experiments were performed on a Leica SP8 confocal microscope equipped with a Plan APO 63×/1.2 N.A. water immersion objective. GFP fluorescence was excited with a 488 nm laser and emission selected by a bandpass filter at 500–550 nm. Laser power used for FCS measurements was adjusted to minimize photobleaching and avoid the induction of photodamage in Hoechst-sensitized cells. Single photons were detected and counted using a τ-Single Photon Avalanche Photodiode and a PicoHarp module from PicoQuant. Each FCS acquisition lasted 45 s to reduce the noise on the autocorrelation curves. For each nucleus, FCS acquisitions were performed before and 1–2 min after laser microirradiation at 405 nm. Cells were maintained at 37°C using a heating chamber. To estimate the residence time of GFP-tagged CHD4 in the focal volume, autocorrelation curves were fitted with a one-species model assuming pure diffusion and neglecting the contribution of the photo-physics of GFP using the Fluctuation Analyzer 4G software (36).
Fluorescence recovery after photobleaching
FRAP experiments were performed on an inverted confocal spinning-disk (Nikon Ti-E body and Yokogawa CSU-X1 scan head) using a 63x oil immersion objective lens (N.A. 1.4) and a sCMOS ORCA Flash 4.0 camera (Hamamatsu) for image acquisition. Local bleaching within a 3 μm diameter circular area in the cell nucleus was performed using a dedicated single-point scanning head (iLas2 from Roper Scientific) coupled to the epifluorescence backboard of the microscope. To estimate fluorescence recovery kinetics, the mean fluorescence intensity inside the bleached area was measured by automatic segmentation using a custom-made Matlab (MathWorks) routine. This routine also allowed background subtraction from the intensity measurements and correction for photobleaching due to imaging by dividing the intensity in the bleached area with the one measured for the whole nucleus. The recovery time was the time required to recover half of the fluorescence signal lost upon initial photobleaching.
For simultaneous DNA damage induction and FRAP used in Supplementary Figure S1D, Hoechst sensitized nuclei expressing H2B-PATagRFP and GFP-CHD4 were irradiated along a 16 μm line crossing the nucleus for 350 ms. The 405 nm light was set to 1 μJ/μm2 at the sample level and we adjusted the power of 488 nm line to allow photobleaching without inducing DNA damage. To control the latter, we verified that no recruitment of PARP1 was observed when micro-irradiating Hoechst sensitized nuclei only at 488 nm (not shown). Images were streamed for the first 60 s to collect images for FRAP and every 4 s for the remaining 4 min for recruitment analysis. To estimate fluorescence recovery kinetics, the mean fluorescence intensity inside the bleached area, as defined by photoactivated H2B, was measured, corrected for bleaching and background subtraction and normalized to pre-bleach intensity.
Quantification of chromatin relaxation and protein recruitment at DNA breaks
The chromatin relaxation assay has been previously described in detail (5,37). Briefly, photoactivable H2B (PTagRFP-H2B or PAGFP-H2B) was irradiated with a 405 nm laser for photoactivation highlighting the irradiated chromatin region and, in cells treated with Hoechst, to simultaneously induce DNA damage (Supplementary Figure S1B). The width of the photoactivated chromatin line, which is used as a readout to assess chromatin relaxation, was then estimated at different time-points after irradiation by automatic segmentation of the images using a custom-made Matlab routine, which is available upon request. The width of the segmented DNA damaged line was normalized to the first image post irradiation (Supplementary Figure S1C).
A related assay was used to quantitatively analyze recruitment kinetics at DNA breaks for a given protein of interest (37). Cells were expressing the protein of interest tagged with GFP or YFP together with H2B-PATagRFP. Using similar 405 nm irradiation conditions used for the chromatin relaxation assay, the outline of the segmented area defined by the photoactivated chromatin line was used as a mask to measure the mean fluorescence intensity of the protein of interest in the damaged area. After background subtraction, photobleaching due to imaging was corrected by dividing the intensity in the DNA damage area with the one measured for the whole nucleus. The intensities at the damaged area were normalized to the intensity prior to irradiation measured by applying the irradiation mask obtained for the first image post-irradiation to the last image pre-irradiation. These different image analysis steps were performed using a custom-made Matlab routine (Supplementary Figure S1B and C).
Statistical significance
P-values were calculated using a Welch Two Sample t-test. On the boxplots, * refers to P < 0.05, ** to P < 0.01, *** to P < 0.001, **** to P < 0.0001 and n.s. to non significant.
Western blotting
Whole cell extracts from siRNA treated samples was collected and resolved by SDS-PAGE. Samples were transferred to a PVDF membrane at 150 mA for 4 h, blocked with 5% milk in TBST (10 mM Tris–HCl, pH 7.6, 150 mM NaCl, 0.2% Tween-20) for 1 h and incubated in primary antibody—1:1000 anti-CHD4 #39289 (Active Motif), 1:10 000 anti-tubulin T9026 (Sigma), 1:500 anti-CHD3 (ab109195), 1:10 000 anti-PARP1 (rabbit polyclonal, (5)), 1:1000 anti-Alc1 (affinity purified rabbit polyclonal, (5))—in 1% milk in TBST overnight. Primary antibodies were detected using anti-mouse or anti-rabbit HRP-conjugated IgG (BioRad) diluted 1:10 000 in 1% milk in TBST. The blots were developed using WesternBright ECL-spray (Advansta).
Immunofluorescence
Cells were washed once with PBS and fixed for 15 min in 4% paraformaldehyde. After two washes with PBS cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min. Cells were washed twice for 10 min in PBS before being blocked in blocking buffer (5% BSA and 0.2% Tween-20 in PBS) for 1 h. Blocking buffer was removed and cells were incubated in primary antibody (1:1000 anti-CHD4 #39289 (Active Motif), 1:1000 anti-gammaH2AX (NB100-78356 Novus)) in blocking buffer overnight at 4°C. Cells were washed three times with 0.1% Triton X-100 in PBS before incubation with anti-mouse Alexa Fluor 488 conjugated IgG or anti-rabbit Alexa Fluor 555 conjugated IgG (Life Technologies) diluted 1:500 in blocking buffer at room temperature for 1 hour in the dark. Cells were washed twice with 0.1% Triton X-100 in PBS and counterstained with Hoechst (1 μg/ml in PBS) for 10 min. Images were obtained on Zeiss AxioObserver Z1 and Yokogawa CSU-X1 scan head using a Plan Apo 20× air objective (N.A 0.8) and an AxioCam HRm CCD camera.
RESULTS
PAR-dependent recruitment of CHD4 at DNA breaks is not mediated through PAR-binding
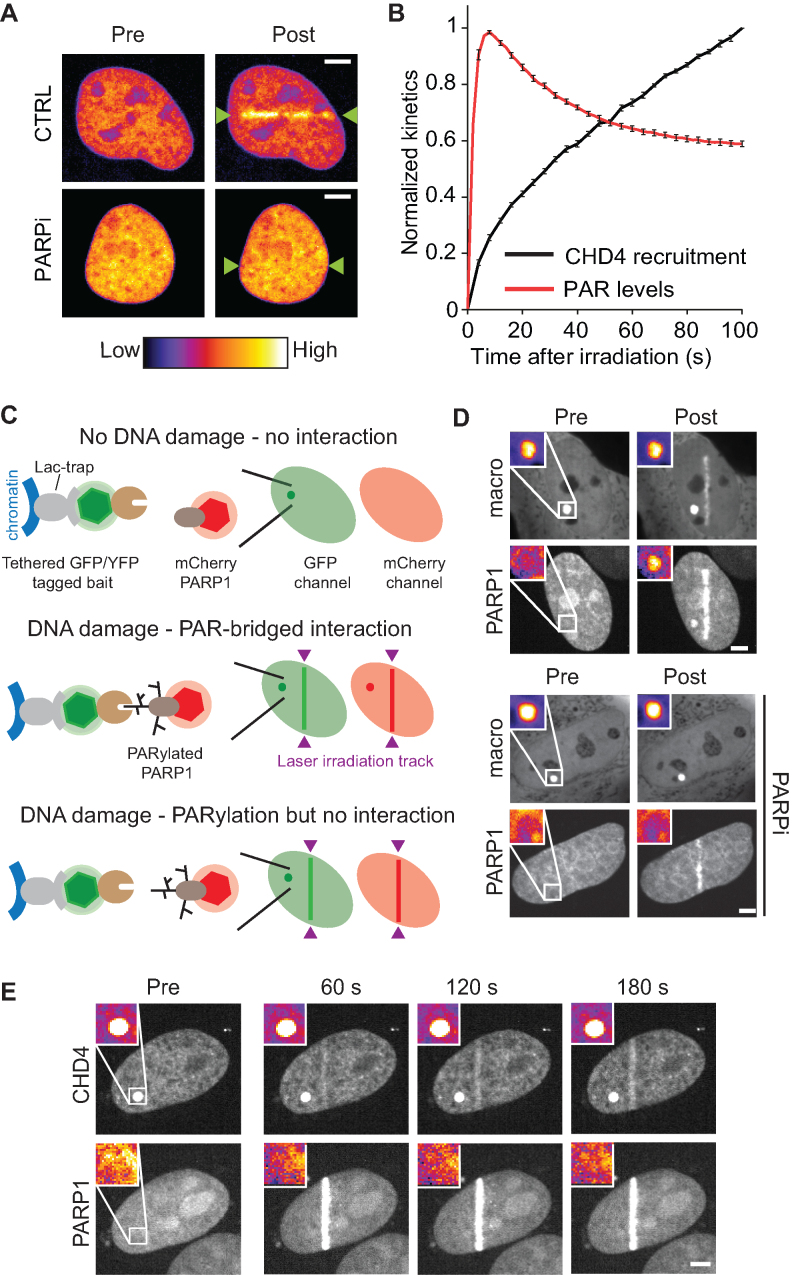
CHD4 has been reported to recruit to sites of DNA damage in a PARP1-dependent manner (13–16) and more recently, the HMG-box-like domain of CHD4 was shown to bind PAR in vitro (27) suggesting that CHD4 is recruited at DNA breaks through direct binding to PAR chains generated by the PARP1 activity. While our observations also confirm that the recruitment of CHD4 to DNA lesions is suppressed by PARP inhibition (Figure 1A), removing the HMG-box-like domain of CHD4 does not affect the recruitment of this protein to DNA breaks (27). Furthermore, when comparing the kinetics of PAR and CHD4 accumulation at DNA breaks, we found that while PAR levels quickly increase at DNA breaks to peak ∼10s post-irradiation (Figure 1B, red), closely matching the recruitment kinetics observed for Alc1 (5), the accumulation of CHD4 in the damaged area is much slower (Figure 1B, black). Performing laser irradiation at both 405 and 488 nm to simultaneously induce DNA breaks and bleach GFP-tagged CHD4 showed that fluorescence recovery at DNA breaks occurs much faster than the recruitment itself (Supplementary Figure S1D), indicating that the slow accumulation of CHD4 at DNA breaks cannot be simply attributed to major diffusion hindrance attributed to the large size of the NuRD complex to which CHD4 belongs. The discrepancy between the kinetics of PAR and CHD4 accumulation at DNA breaks thus suggests that the mechanism responsible for Alc1 accumulation at DNA lesions, which involves the direct binding of Alc1 to PAR via its macrodomain (9), may not hold true for CHD4.
Figure 1.
The PAR-dependent recruitment of CHD4 to sites of DNA damage is not triggered by PAR binding. (A) Recruitment of GFP-CHD4 to sites of microirradiation 120 seconds after DNA damage in the presence (PARPi) or absence (CTRL) of PARP inhibitor. Microirradiation is highlighted with green arrows. (B) Normalized kinetics of PAR levels as measured by mH2A1.1 macrodomain recruitment (red) and CHD4 recruitment (black) after microirradiation. Kinetics are normalized at a maximum of 1. (C) Schematic of PAR binding assay. The LacI-GFP trap is used to tether the GFP or YFP bait-protein to the LacO array and appears as a bright spot within the nucleus. In a non-damaged situation, there is no interaction of mCherry-PARP1 with the bait protein. Upon UV-irradiation induced DNA damage, both mCherry-PARP1 and the GFP/YFP-bait protein will recruit to the site of damage. If the bait protein is capable of binding PAR, PARylated mCherry-PARP1 that is released from the site of damage will interact with the bait protein at the LacO array, resulting in an increase in mCherry signal at the LacO array. If the bait protein cannot bind PAR, there will be no enrichment of mCherry-PARP1 at the LacO array. (D) YFP-macrodomain of mH2A1.1 recruits PARylated PARP1, which is abolished by PARP inhibition (PARPi). Inset shows the magnified LacO array. Post-irradiation images are shown at 120 seconds. (E) GFP-CHD4 does not recruit PARylated PARP1 after microirradiation. Post-irradiation images are shown at the indicated time points post-irradiation. Scale bars are 5 μm. The look-up-table shown on panel (A) is valid for all pseudocolored images.
In order to directly assess the ability of CHD4 to bind PAR in the cellular environment, we developed a specific fluorescence three-hybrid assay (Figure 1C) that relies on measuring the accumulation of PARylated PARP1 released from the site of DNA damage on a lacO-anchored ‘ADP-ribose-binding’ protein. The assay allows us to simultaneously observe the accumulation of the protein of interest at the site of microirradiation-induced DNA damage and its ability to interact with PARylated PARP1 at the LacO array. To validate the system, we tethered the PAR-binding YFP-macroH2A1.1 macrodomain to the LacO array using a LacI-GFP-trap in cells co-expressing mCherry-PARP1 (29,32). Upon DNA damage, YFP-mH2A1.1 macrodomain and mCherry-PARP1 readily recruit to the site of microirradiation-induced damage, while mCherry-PARP1 also accumulates at the LacO-tethered mH2A1.1 macrodomain (Figure 1D). The accumulation of PARP1 at the LacO tethered macrodomain was lost upon PARP inhibition while PARP1s recruitment to the site of damage was not affected (38), further validating that the tethered macrodomain interacts with PARylated PARP1 released from the sites of DNA damage (Figure 1D). Using this assay, we did not observe recruitment of PARylated PARP1 to tethered GFP-CHD4 (Figure 1E) indicating that this remodeler is not a bona fide PAR-binding protein. Based on these findings it is unlikely that the recruitment of CHD4 at DNA breaks is driven by direct binding to PAR chains.
CHD4 is recruited to sites of DNA damage as a consequence of chromatin relaxation
An alternative hypothesis to explain the accumulation of CHD4 at DNA breaks is that the PARylation-induced chromatin relaxation facilitates the accessibility of DNA for DNA-binding proteins, a mechanism proposed for transcription factors recruited at DNA breaks (39). To test this possibility, we first analyzed the effect of enhancing chromatin relaxation on the accumulation of CHD4 at DNA breaks through the overexpression of the ATP-dependent remodeler Alc1 (5). Overexpression of wild-type Alc1 induces a robust increase in DNA damage-induced chromatin relaxation as compared to cells expressing the ATPase-dead Alc1 (Figure 2A and Supplementary Figure S2A). The enhanced chromatin relaxation correlates with increased CHD4 recruitment at DNA breaks (Figure 2B and Supplementary Figure S2B) while PAR levels remain unaffected (Figure 2C) suggesting that chromatin relaxation per se promotes the recruitment of CHD4 at DNA breaks. We found that the accumulation of CHD4 at DNA breaks is not mediated through direct binding to Alc1. Using the F3H assay, no interaction was observed between LacO tethered Alc1-GFP and mCherry-CHD4 in either the absence of DNA damage or after laser irradiation, while PARP1 enriched at the LacO tethered Alc1 upon DNA damage (Supplementary Figure S2C). Furthermore, in Alc1 knock-out cells we observed reduced, but not fully suppressed, recruitment of CHD4 (Supplementary Figure S2D) Since chromatin relaxation at DNA breaks is only partially impaired in Alc1 knocked-out cells (5), the reduction of CHD4 accumulation at DNA breaks in these cells supports the hypothesis that CHD4 accumulation is controlled by chromatin relaxation and not direct binding to Alc1.
The recruitment of CHD4 to sites of DNA damage is also associated with increased binding as shown by an increase in residency time determined through FCS measurements (Supplementary Figure S2E and F). If enhanced binding of CHD4 to relaxed chromatin is sufficient to trigger the recruitment of this remodeler to sites of DNA damage, one would expect that CHD4 shows tighter binding to more relaxed chromatin independently of the DDR. We therefore used tonic shifts to tune the chromatin compaction state (35) before examining CHD4 dynamics. Switching from isotonic to hypotonic medium lead to a rapid and uniform relaxation of the chromatin structure (Figure 2D). Using Fluorescence Recovery After Photobleaching (FRAP), we found that chromatin relaxation by hypotonic treatment was associated with reduced mobility of GFP-CHD4 indicative of its enhanced binding to relaxed chromatin (Figure 2E–G). This reduced mobility upon hypotonic treatment was also observed upon PARP inhibition or in PARP1 knockout cells, demonstrating that this effect is independent of PARP1 signaling. These findings suggest that the initial PAR-dependent chromatin relaxation directly drives the increased binding of CHD4 to sites of DNA damage we observed, which ultimately leads to the recruitment of this remodeler in the damaged area.
CHD4 knock-down reduces chromatin relaxation at sites of DNA damage and leads to increased DNA damage signaling
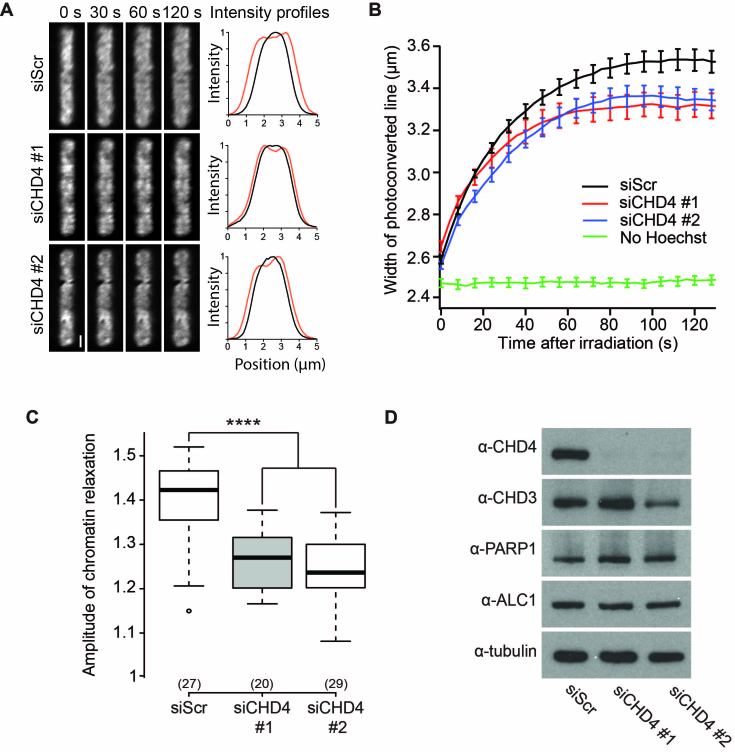
To investigate the potential role of CHD4 during chromatin remodeling at DNA damage sites, we used an in vivo chromatin relaxation assay where Hoechst-presensitized nuclei expressing the core histone H2B fused to photoactivable GFP are microirradiated with 405 nm light to simultaneously induce DNA damage and photoactivate H2B-PAGFP in a line crossing the nucleus (Figure 3A, Supplementary Figure S1B and C). The thickness of the photoactivated line is measured over time allowing assessment of the change in the chromatin compaction state occurring upon DNA damage induction (37). Using this assay, we were able to show that the knockdown of CHD4 resulted in a strong reduction of chromatin relaxation at sites of DNA damage (Figure 3B and C). Additionally, to assess the effect of CHD4 depletion on DNA repair signaling, we examined γ-H2AX levels upon X-ray irradiation with CHD4 knock-down using two independent siRNAs, observing increased and prolonged signaling (Supplementary Figure S3A and B).
Figure 3.
CHD4 contributes to chromatin relaxation at sites of DNA damage. (A) Confocal image sequence of the photoconverted chromatin region upon micro-irradiation at 405 nm in U2OS nuclei expressing H2B-PAGFP presensitized by Hoechst labeling and that have been treated with scrambled or CHD4 siRNA. The enlargement of the photoconverted line at 120 s post-irradiation is used to assess chromatin relaxation at sites of DNA damage. Intensity profiles show the thickness of the line (μm) at 0 s (Black) and 120 s (Red). Scale bars are 2 μm. (B) Chromatin relaxation curves showing the kinetics of chromatin relaxation as measured by the width of the photoconverted line of scrambled (siScr, Black) or CHD4 knock-down (siCHD4 #1, Red, siCHD4 #2, Blue) cells. A ‘no damage’ control, nuclei without Hoechst sensitization, is shown in green. (C) Effect of two independent CHD4 (siCHD4) knockdowns on DNA damage-induced chromatin relaxation as compared to scrambled siRNA treatment (siScr). Boxplots show chromatin relaxation 120 seconds after microirradiation. (D) Western blot showing knockdown efficiency of CHD4 and expression levels of CHD3, PARP1 and Alc1. Tubulin is used as a loading control.
We have previously reported that the preexisting chromatin structure can influence the level of DNA damage induced chromatin relaxation (5). As CHD4 is part of the histone deacetylase containing NuRD complex (19–21), knock-down of CHD4 could alter the overall acetylation status of chromatin, shifting to an increased acetylation state and an overall increase in chromatin relaxation. We measured chromatin texture in Hoechst stained nuclei to determine the changes to chromatin structure using hypotonic treatment, which promotes chromatin relaxation, as a control (35). We observed a decrease in pixel-to-pixel contrast in hypotonic treated nuclei, consistent with a more relaxed chromatin landscape as compared to isotonic treatment (Supplementary Figure S3C). This was accompanied by impaired chromatin relaxation upon DNA damage (Supplementary Figure S3D). Importantly, the knock-down of CHD4 did not alter the chromatin architecture prior to DNA damage induction (Supplementary Figure S3E) indicating the reduced chromatin relaxation upon DNA damage is due the loss of the remodeling function of CHD4.
Since CHD4 plays a central role in transcriptional regulation (40), we also ensured CHD4 knock-down did not alter the expression of PARP1 and Alc1, the two early players in chromatin remodeling at DNA breaks (Figure 3D). We also found that knocking-down CHD4 does not impair the expression of the closely related remodeler CHD3. Altogether, these findings demonstrate CHD4 plays a direct role in chromatin relaxation at sites of DNA damage.
CHD3, a close homologue to CHD4, displays similar functions to CHD4 at sites of DNA damage
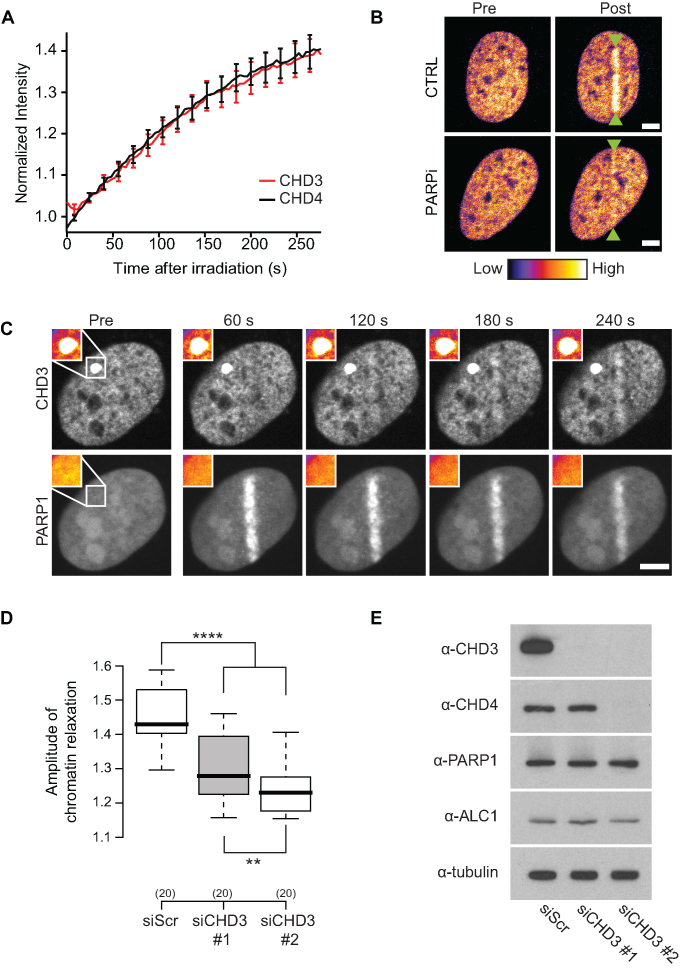
CHD3 is a chromatin remodeler homologous to CHD4, with the two remodelers sharing close to 72% homology (17,18,22). These two remodelers belong to distinct NuRD complexes and display similar, yet not fully overlapping, functional roles in the context of transcriptional regulation (22). We wondered if the shared functionalities of the two remodelers also hold true during the DNA damage response. CHD3 is recruited to sites of DNA damage with very similar kinetics to CHD4 (Figure 4A) and the recruitment of CHD3 to sites of DNA damage was also strongly impaired upon PARylation inhibition (Figure 4B). Using the F3H assay, we tethered CHD3 to the LacO array and found no evidence of binding between CHD3 and PARyted PARP1 (Figure 4C), suggesting that CHD3, similar to CHD4, is recruited to sites of DNA damage due to increased binding to chromatin relaxed by the PARP1/Alc1 remodeling activities. Finally, we used two siRNAs to mediate the knockdown of CHD3. While both siRNAs lead to a reduction of the chromatin relaxation process (Figure 4D), siCHD3 #2 showed a significantly stronger reduction in the decondensation. When levels of CHD3 and CHD4 were compared (Figure 4E), siCHD3 #2 also very strongly depleted CHD4. These findings reveal that both CHD3 and CHD4 play redundant roles during the chromatin relaxation process at DNA breaks.
Figure 4.
CHD3 is recruited to sites of DNA damage and is required for DNA damage induced chromatin relaxation. (A) Recruitment kinetics of GFP-CHD3 (red) and GFP-CHD4 (black) to sites of DNA damage. (B) Recruitment of GFP-CHD3 to sites of microirradiation 120 seconds after DNA damage in the presence (PARPi) or absence (CTRL) of PARP inhibitor. Microirradiation is highlighted with green arrows. Scale bars are 5 μm. (C) GFP-CHD3 is tethered to the LacO array and co-expressed with mCherry-PARP1. Upon DNA damage, PARP1 recruits to microirradiation induced DNA breaks but does not accumulate at the tethered GFP-CHD3, indicating that CHD3 does not bind PAR. Scale bars are 5 μm. Inset shows the magnified LacO array. (D) Effect of two independent CHD3 (siCHD3) knockdowns on DNA damage-induced chromatin relaxation as compared to scrambled siRNA treatment (siScr). Boxplots show chromatin relaxation 120 seconds after microirradiation (E) Western blot showing knockdown efficiency of CHD3 and expression levels of CHD4, PARP1 and Alc1. Tubulin is used as a loading control.
DISCUSSION
The complex chromatin remodeling mechanisms that occur during the early stages of the DDR aim to arrange chromatin into a repair competent conformation allowing repair proteins easy access to DNA breaks while keeping the broken ends in close proximity, avoiding deleterious recombination (41). Poly(ADP-ribosy)lation plays a key role in the initiation of DNA damage-induced chromatin relaxation. The presence of negatively charged PAR-chains along the chromatin fiber may initiate chromatin loosening (8) but additional ATP-dependent remodeling also occurs through the recruitment of PAR-dependent chromatin remodelers such as Alc1 (5,11). While Alc1 contributes significantly to relaxation, additional ATP-dependent processes are required to achieve a maximal relaxed state. The fact that few chromatin remodelers display known PAR-binding motifs suggest a second mode of protein recruitment is responsible for bringing these factors to the site of damage. In this study, we show that it is the initial PAR-dependent chromatin relaxation that facilitates the subsequent recruitment of CHD3 and CHD4 to the site of damage via increased binding to relaxed chromatin. Indeed, this initial PAR-dependent relaxation may facilitate the accumulation of DNA-binding proteins at the DNA breaks without the need for the activation of additional signaling pathways (39). The recruitment of the remodeler CHD2, which was also reported to be PAR-dependent, shows similar recruitment kinetics to the one we measured for CHD4, i.e. much slower than the increase of PAR levels upon irradiation (12). It is then likely that CHD2, which does not bear a canonical PAR-binding domain, recruits to DNA breaks through enhanced binding to chromatin relaxed by early PAR-dependent remodeling processes. It is worth noting that such chromatin relaxation-driven recruitment may occur upon cellular stresses other than DNA damage that lead to the activation of PAR-signaling. In Drosophila, the heat-shock-induced puffing of the Hsp70 locus in the larval salivary gland polytene chromosomes is PAR dependent (42). Murawska et al. later showed that the Drosophila homologue of CHD4, dMi2, is recruited to heat-shock responsive genes in a PARP-dependent manner and identified the tandem chromodomain of dMi2 as crucial for this recruitment (28). This tandem chromodomain was found to bind PAR chains but is also a well-known DNA-binding domain (28). Given our observations with CHD3 and CHD4, the recruitment of dMi2 at heat-shock genes could be also facilitated by PAR-dependent puffing of the polytene chromosomes.
Two alternative hypotheses could explain the enhanced binding of CHD3 and CHD4 to relaxed chromatin at sites of DNA damage. The first is that the presence of PAR chains along the DNA fiber, in combination with the nucleosome repositioning activity of the PAR-dependent chromatin remodeler Alc1 (5,43,44), shapes chromatin into a specific conformation in which DNA is more exposed, promoting the binding of diffusible DNA-binding proteins such as CHD3 and CHD4. The second hypothesis is that the increased binding of CHD3 or CHD4 to relaxed chromatin area is promoted by the reduced macromolecular crowding encountered in this area. The nucleus is a highly crowded environment with recent reports estimating that the volume fraction occupied by chromatin is, on average, equal to 30% (45). It is then possible that the lowering of the crowding level induced by chromatin relaxation at sites of DNA damage decreases volume exclusion, thus allowing more CHD3 and CHD4 proteins to fit in the relaxed area, similar to what was observed for GFP arrays in centromeric heterochromatin foci relaxed by osmotic shift (35). The macromolecular crowding theory also predicts that diffusion-limited binding reactions, which describes the dynamics of many DNA-binding proteins (30), will display increased association rates upon reduced crowding while dissociation rates remain unaffected (46). While it is challenging to experimentally differentiate between these scenarios or a mixture of the two, both will have the same outcome: increased binding of CHD3 and CHD4 at sites of DNA damage.
Overall, our findings suggest a model in which the chromatin relaxation at sites of DNA damage occurs in two consecutive phases (Figure 5). First, PARylation initiates the relaxation of chromatin directly through the recruitment and activation of the PAR-binding remodeler Alc1. This initial chromatin relaxation then allows the recruitment of CHD3 and CHD4 and subsequent chromatin remodeling through enhanced binding to relaxed chromatin but independently of interactions with PAR chains. Interestingly, CHD2, similar to CHD3 and CHD4, contributes to chromatin relaxation but, while CHD4 seems to be involved in DNA repair via HR, CHD2 rather promotes NHEJ (12). This raises the interesting possibility that, after an initial relaxation phase generic to all repair mechanisms, remodelers specific to HR or NHEJ may be required at later stages of the chromatin remodeling required at sites of DNA damage.
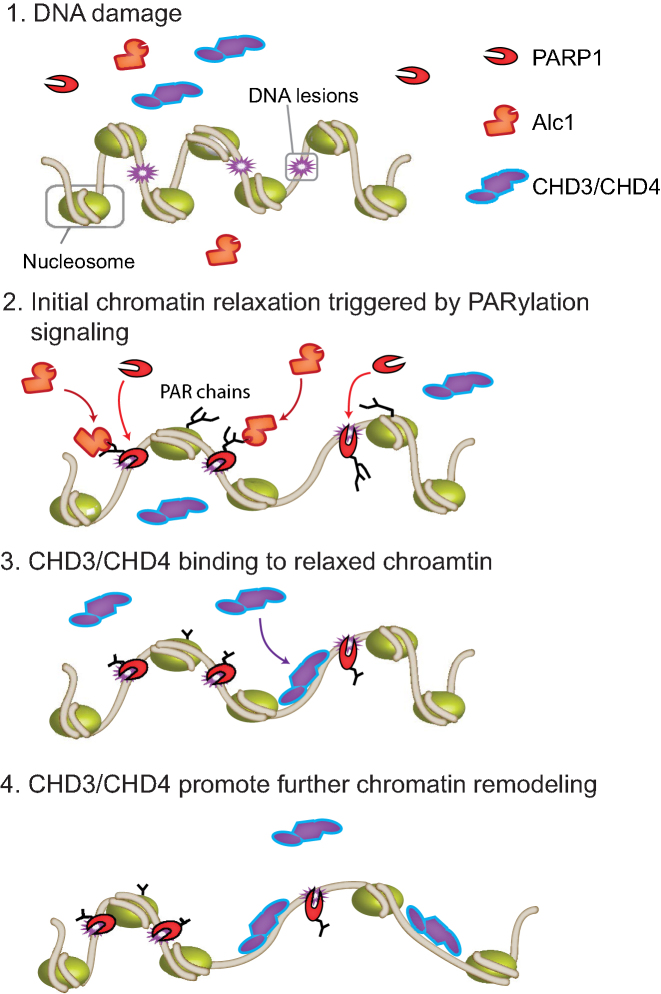
Figure 5.
Model of two-phase chromatin remodeling at sites of DNA damage. DNA damage promotes the recruitment of PARP1 where it is activated. Activated PARP1 will PARylate proteins at the site of damage, including itself, and promote the recruitment of the chromatin remodeler, Alc1. Alc1 remodeling activity leads to chromatin relaxation in the vicinity of the DNA break. CHD3 and CHD4 bind to the relaxed chromatin and further remodel chromatin.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Microscopy Rennes Imaging Center (BIOSIT, Université Rennes 1) for technical assistance.
Author Contributions: R.S., S.H. and G.T. designed the experiments. R.S. and S.H. completed the experiments within the manuscript. H.S. completed preliminary chromatin relaxation assays. C.C. generated cell lines and DNA constructs. S.H. and G.T. conceived and supervised this study. R.S., S.H. and G.T. wrote the manuscript. All authors read and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Worldwide Cancer Research [14-1315]; Hungarian Academy of Sciences [LP2017-11/2017 to G.T.]; European Union [FP7-PEOPLE-2011-CIG, ChromaTranscript project]; Ligue contre le Cancer du Grand-Ouest (committees 35 and 72); Fondation ARC pour la recherche sur le cancer [20161204883 to S.H.]; Bayerisch-Französische Hochschulzentrum (BFHZ) [FK_02_16 to G.T. and S.H.]; People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (FP7/2007-2013) under REA grant agreement [PCOFUND-GA-2013-609102], through the PRESTIGE program coordinated by Campus France [PRESTIGE-2017-2-0042 to R.S.]. Funding for open access charge: Magyar Tudományos Akadémia.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cleaver J.E. Nucleosome structure controls rates of excision repair in dna of human cells. Nature. 1977; 270:451–453. [DOI] [PubMed] [Google Scholar]
- 2. Murr R., Loizou J.I., Yang Y.-G., Cuenin C., Li H., Wang Z.-Q., Herceg Z.. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006; 8:91–99. [DOI] [PubMed] [Google Scholar]
- 3. Ali A.A.E., Timinszky G., Arribas-Bosacoma R., Kozlowski M., Hassa P.O., Hassler M., Ladurner A.G., Pearl L.H., Oliver A.W.. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012; 19:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D’Amours D., Desnoyers S., D'Silva I., Poirier G.G.. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999; 342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 5. Sellou H., Lebeaupin T., Chapuis C., Smith R., Hegele A., Singh H.R., Kozlowski M., Bultmann S., Ladurner A.G., Timinszky G. et al. The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage. Mol. Biol. Cell. 2016; 27:3791–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smeenk G., Wiegant W.W., Marteijn J.A., Luijsterburg M.S., Sroczynski N., Costelloe T., Romeijn R.J., Pastink A., Mailand N., Vermeulen W. et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 2013; 126:889–903. [DOI] [PubMed] [Google Scholar]
- 7. Strickfaden H., McDonald D., Kruhlak M.J., Haince J.F., Th’ng J.P.H., Rouleau M., Ishibashi T., Corry G.N., Ausio J., Underhill D.A. et al. Poly(ADP-ribosyl)ation-dependent transient chromatin decondensation and histone displacement following laser microirradiation. J. Biol. Chem. 2016; 291:1789–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poirier G.G., de Murcia G., Jongstra-Bilen J., Niedergang C., Mandel P.. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc. Natl. Acad. Sci. U.S.A. 1982; 79:3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottschalk A.J., Timinszky G., Kong S.E., Jin J., Cai Y., Swanson S.K., Washburn M.P., Florens L., Ladurner A.G., Conaway J.W. et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:13770–13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahel D., Horejsi Z., Wiechens N., Polo S.E., Garcia-Wilson E., Ahel I., Flynn H., Skehel M., West S.C., Jackson S.P. et al. Poly(ADP-ribose)-Dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009; 325:1240–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kruhlak M.J., Celeste A., Dellaire G., Fernandez-Capetillo O., Muller W.G., McNally J.G., Bazett-Jones D.P., Nussenzweig A.. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 2006; 172:823–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Luijsterburg M.S., de Krijger I., Wiegant W.W., Shah R.G., Smeenk G., de Groot A.J., Pines A., Vertegaal A.C., Jacobs J.J., Shah G.M. et al. PARP1 links CHD2 mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell. 2016; 61:547–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polo S.E., Kaidi A., Baskcomb L., Galanty Y., Jackson S.P.. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010; 29:3130–3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chou D.M., Adamson B., Dephoure N.E., Tan X., Nottke A.C., Hurov K.E., Gygi S.P., Colaiacovo M.P., Elledge S.J.. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18475–18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Luijsterburg M.S., Acs K., Ackermann L., Wiegant W.W., Bekker-Jensen S., Larsen D.H., Khanna K.K., van Attikum H., Mailand N., Dantuma N.P.. A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure. EMBO J. 2012; 31:2511–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Smeenk G., Wiegant W.W., Vrolijk H., Solari A.P., Pastink A., van Attikum H.. The NuRD chromatin–remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010; 190:741–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seelig H.P., Renz M., Targoff I.N., Ge Q., Frank M.B.. Two forms of the major antigenic protein of the dermatomyositis-specific Mi-2 autoantigen. Arthritis Rheumatism. 1996; 39:1769–1771. [DOI] [PubMed] [Google Scholar]
- 18. Woodage T., Basrai M.A., Baxevanis A.D., Hieter P., Collins F.S.. Characterization of the CHD family of proteins. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:11472–11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., LeRoy G., Seelig H.P., Lane W.S., Reinberg D.. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998; 95:279–289. [DOI] [PubMed] [Google Scholar]
- 20. Xue Y., Wong J., Moreno G.T., Young M.K., Cote J., Wang W.. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell. 1998; 2:851–861. [DOI] [PubMed] [Google Scholar]
- 21. Tong J.K., Hassig C.A., Schnitzler G.R., Kingston R.E., Schreiber S.L.. Chromatin deacetylation by an ATP-dependent nucleosome remodelling complex. Nature. 1998; 395:917–921. [DOI] [PubMed] [Google Scholar]
- 22. Hoffmeister H., Fuchs A., Erdel F., Pinz S., Grobner-Ferreira R., Bruckmann A., Deutzmann R., Schwartz U., Maldonado R., Huber C. et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017; 45:10534–10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cai Y., Geutjes E.J., de Lint K., Roepman P., Bruurs L., Yu L.R., Wang W., van Blijswijk J., Mohammad H., de Rink I. et al. The NuRD complex cooperates with DNMTs to maintain silencing of key colorectal tumor suppressor genes. Oncogene. 2014; 33:2157–2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Denslow S.A., Wade P.A.. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007; 26:5433–5438. [DOI] [PubMed] [Google Scholar]
- 25. Qi W.J., Chen H.Y., Xiao T., Wang R.X., Li T., Han L.P., Zeng X.L.. Acetyltransferase p300 collaborates with chromodomain helicase DNA-binding protein 4 (CHD4) to facilitate DNA double-strand break repair. Mutagenesis. 2016; 31:193–203. [DOI] [PubMed] [Google Scholar]
- 26. Pan M.R., Hsieh H.J., Dai H., Hung W.C., Li K.Y., Peng G., Lin S.Y.. Chromodomain Helicase DNA-binding Protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to Poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J. Biol. Chem. 2012; 287:6764–6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Silva A.P.G., Ryan D.P., Galanty Y., Low J.K.K., Vandevenne M., Jackson S.P., Mackay J.P.. The N-terminal region of chromodomain helicase DNA-binding Protein 4 (CHD4) is essential for activity and contains a high mobility group (HMG) Box-like-domain that can bind Poly(ADP-ribose). J. Biol. Chem. 2016; 291:924–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murawska M., Hassler M., Renkawitz-Pohl R., Ladurner A., Brehm A.. Stress-induced PARP activation mediates recruitment of Drosophila Mi-2 to promote heat shock gene expression. PLoS Genet. 2011; 7:e1002206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Timinszky G., Till S., Hassa P.O., Hothorn M., Kustatscher G., Nijmeijer B., Colombelli J., Altmeyer M., Stelzer E.H., Scheffzek K. et al. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat. Struct. Mol. Biol. 2009; 16:923–929. [DOI] [PubMed] [Google Scholar]
- 30. Beaudouin J., Mora-Bermúdez F., Klee T., Daigle N., Ellenberg J.. Dissecting the contribution of diffusion and interactions to the mobility of nuclear proteins. Biophys. J. 2006; 90:1878–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shcherbakova D.M., Verkhusha V.V.. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013; 10:751–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kubala M.H., Kovtun O., Alexandrov K., Collins B.M.. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010; 19:2389–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bowman A., Lercher L., Singh H.R., Zinne D., Timinszky G., Carlomagno T., Ladurner A.G.. The histone chaperone sNASP binds a conserved peptide motif within the globular core of histone H3 through its TPR repeats. Nucleic Acids Res. 2016; 44:3105–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Czarna A., Berndt A., Singh H.R., Grudziecki A., Ladurner A.G., Timinszky G., Kramer A., Wolf E.. Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function. Cell. 2013; 153:1394–1405. [DOI] [PubMed] [Google Scholar]
- 35. Walter A., Chapuis C., Huet S., Ellenberg J.. Crowded chromatin is not sufficient for heterochromatin formation and not required for its maintenance. J. Struct. Biol. 2013; 184:445–453. [DOI] [PubMed] [Google Scholar]
- 36. Wachsmuth M., Conrad C., Bulkescher J., Koch B., Mahen R., Isokane M., Pepperkok R., Ellenberg J.. High-throughput fluorescence correlation spectroscopy enables analysis of proteome dynamics in living cells. Nat. Biotechnol. 2015; 33:384–389. [DOI] [PubMed] [Google Scholar]
- 37. Lebeaupin T., Smith R., Huet S., Timinszky G.. Poly(ADP-Ribose)-dependent chromatin remodeling in DNA repair. Methods Mol. Biol. (Clifton, N.J.). 2017; 1608:165–183. [DOI] [PubMed] [Google Scholar]
- 38. Murai J., Huang S.Y., Das B.B., Renaud A., Zhang Y., Doroshow J.H., Ji J., Takeda S., Pommier Y.. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012; 72:5588–5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Izhar L., Adamson B., Ciccia A., Lewis J., Pontano-Vaites L., Leng Y., Liang A.C., Westbrook T.F., Harper J.W., Elledge S.J.. A Systematic analysis of factors localized to damaged chromatin reveals PARP-Dependent recruitment of transcription factors. Cell Rep. 2015; 11:1486–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang Z., Brown K.E., Carroll T., Taylor B., Vidal I.F., Hendrich B., Rueda D., Fisher A.G., Merkenschlager M.. A high-resolution map of transcriptional repression. eLife. 2017; 6:e22767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lebeaupin T., Sellou H., Timinszky G., Huet S.. Chromatin dynamics at DNA breaks: what, how and why. Aims Biophys. 2015; 2:458–475. [Google Scholar]
- 42. Tulin A., Spradling A.. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science. 2003; 299:560–562. [DOI] [PubMed] [Google Scholar]
- 43. Singh H.R., Nardozza A.P., Moller I.R., Knobloch G., Kistemaker H.A.V., Hassler M., Harrer N., Blessing C., Eustermann S., Kotthoff C. et al. A Poly-ADP-Ribose trigger releases the Auto-Inhibition of a chromatin remodeling oncogene. Mol. Cell. 2017; 68:860–871. [DOI] [PubMed] [Google Scholar]
- 44. Lehmann L.C., Hewitt G., Aibara S., Leitner A., Marklund E., Maslen S.L., Maturi V., Chen Y., van der Spoel D., Skehel J.M. et al. Mechanistic Insights into autoinhibition of the oncogenic chromatin remodeler ALC1. Mol. Cell. 2017; 68:847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ou H.D., Phan S., Deerinck T.J., Thor A., Ellisman M.H., O'Shea C.C.. ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017; 357:eaag0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou H.X., Rivas G., Minton A.P.. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 2008; 37:375–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.