Abstract
We examined the assembly of DNA G-quadruplexes (G4s) into higher-order structures using atomic force microscopy, optical and electrophoretic methods, NMR spectroscopy and molecular modeling. Our results suggest that parallel blunt-ended G4s with single-nucleotide or modified loops may form different types of multimers, ranging from stacks of intramolecular structures and/or interlocked dimers and trimers to wires. Decreasing the annealing rate and increasing salt or oligonucleotide concentrations shifted the equilibrium from intramolecular G4s to higher-order structures. Control antiparallel and hybrid G4s demonstrated no polymorphism or aggregation in our experiments. The modification that mimics abasic sites (1′,2′-dideoxyribose residues) in loops enhanced the oligomerization/multimerization of both the 2-tetrad and 3-tetrad G4 motifs. Our results shed light on the rules that govern G4 rearrangements. Gaining control over G4 folding enables the harnessing of the full potential of such structures for guided assembly of supramolecular DNA structures for nanotechnology.
INTRODUCTION
G-quadruplex (G4) is a prominent example of the non-canonical four-stranded structures formed in the G-rich regions of DNA (1). The building blocks of G4s are G-tetrads—planar arrangements of four guanine residues held together by Hoogsteen hydrogen bonds. Several G-tetrads are stacked on top of each other and are stabilized by monovalent cations to form inter- or intramolecular quadruplexes. Genomic G4s are believed to play important roles in several biological processes, such as the regulation of telomere maintenance, transcription, replication and translation (2,3). Their relevance for genomic instability (non-homologous recombination and other rearrangements) has also been widely discussed (4), and elucidation of the interaction patterns of G4-prone DNA fragments may provide insight into instability-related processes.
G4s have attracted much interest in the field of nanotechnology as DNA switches (5–7), conducting elements for molecular electronics (8), nanoscaffolds (9), etc. All of the above applications require strict control over G4 folding. In this regard, much effort is being made towards the programming of G4 assembly and rearrangement. In particular, research has been focused on the identification of the determinants of the multimerization of G4s into higher-order structures. It should be noted, however, that biological roles of many higher-order structures, such as G-wires, are not well established.
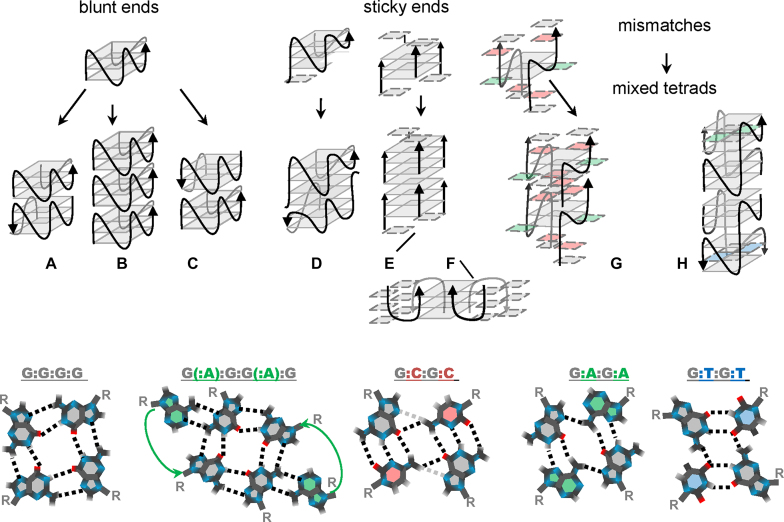
The classification of higher-order G4 structures has been outlined in a recent work by Oliviero et al. (10). According to this classification, the two ‘marginal’ types of multimers are interlocked G-wires, which are composed of mutually slipped G-rich strands, and stacks of G4s, which are held together by interquadruplex pi–pi stacking. Many previously reported structures are arguably referred to as intermediate variants. Hereafter, we will follow the basic classification but approach it from a slightly different perspective, emphasizing the roles of specific structural features (blunt/sticky ends and mixed tetrads) as drivers of multimerization (Figure 1).
Figure 1.
Schematic representations of the major types of previously reported G4 oligomers and fragments of multimers. (A) Head-to-head stacks; (B) head-to-tail stacks; (C) tail-to-tail stacks; (D) head-to-head interlocked G4s; (E) fragments of classical G-wires; (F) fragments of ‘lego’ wires; (G) head-to-head interlocked G4s with mixed tetrads and hexads; (H) stacks of interlocked G4s with mixed tetrads. The bottom panel contains top views of the G-tetrad, mixed tetrads and the hexad, colored accordingly in the top panel.
Interquadruplex stacking is typically observed for parallel G4s with ‘blunt ends’, i.e. no overhangs at the stacking interface. Stacking of two and three G4 monomers has been shown previously by electrospray mass spectrometry, and potassium ions reportedly enhance the association efficiency (11). Overhangs (3′-terminal overhangs in particular) tend to compromise interactions due to steric hindrance (12). It would be reasonable to assume that similar effects exist for other structural elements that partially shield external tetrads, e.g. lateral or diagonal loops of antiparallel G4s. In contrast, expanded pi systems, such as G(:C):G(:C):G(:C):G(:C) octads at the stacking interface, facilitate association (13).
Possible G4 stacking modes include 5′-5′ (head-to-head) (14–16), 5′-3′ (head-to-tail) (17) and 3′-3′ (tail-to-tail) (18) variants (schemes (a), (b) and (c) in Figure 1, respectively). Head-to-head interactions appear to be the most common (12). Assuming that such interactions are thermodynamically more favourable than head-to-tail interactions, for blunt-end G4s, one can expect the formation of primarily dimers rather than lengthy multimers. This also suggests that the interquadruplex stacking potential can be reprogrammed (e.g. switched from dimerization to further multimerization) by strand-polarity inversion sites, as has been demonstrated recently for G4s with external G(:C):G(:C):G(:C):G(:C) octads (10).
Another known route for programming G4 association exploits interlocking via ‘sticky ends’, such as G/GC overhangs and vacancies. Guided head-to-head dimerization (scheme (d) in Figure 1) was first described for the so-called ‘3+1’ intramolecular structures with single-G overhangs and single vacancies in 5′-terminal tetrads (19). Programming head-to-tail interlocking, which is analogous to strand arrangement in ‘classical’ G-wires (20,21), seems to be more challenging than head-to-head dimerization. We use the term ‘classical’ here to distinguish such well-known vertically interlocked structures (scheme (e) in Figure 1) from ‘lego’ G4s (22,23) which are characterized by side-by-side interlocking (scheme (f) in Figure 1).
Head-to-tail interlocking has been considered for bimolecular parallel G4s with GC sticky ends. Pairing of the sticky ends suggests formation of mixed (G:C:G:C) tetrads, as has been demonstrated for GCGGAGGCG oligomers (24) (scheme (g) in Figure 1). In the case of GCGGTG4TGGCG oligomers (25), only the 5′-terminal GC participated in pairing, ensuring head-to-head dimerization of the G4 units. Interestingly, elimination of the nonfunctional 3′-terminal sticky ends restored the propensity for further G4 multimerization due to blunt-end tail-to-tail stacking of the interlocked units. The different behaviours of the GCGGTG4TGGCG (25) and GCGGAGGCG (24) dimers suggest that internal elements of the G4 units (e.g., loops or G(:A):G:G(:A):G hexads) may influence the interlocking/stacking interfaces and, therefore, affect the G4 multimerization potential.
Additional examples of internal G4 features as drivers of multimerization have been reported in a recent work by S. Kolesnikova et al (26). First, dimerization can be programmed by introducing mismatches (non-G nucleotide residues, N) into G-runs of intramolecular G4 motifs. A single middle-run mismatch predetermines the formation of mixed G:N:G:N internal tetrads in dimers (analogously, mismatches in different G-runs can be used to engineer dimers with non-G tetrads, e.g., A:T:A:T). Surprisingly, mixed internal tetrads close to the 5′ termini were shown to hinder blunt-end stacking of the dimeric G4 units, while 3′-proximal mixed tetrads were tolerated in head-to-head stacks of dimers (scheme (h) in Figure 1).
In summary, G4 multimers are highly diverse, and there remains much to be understood regarding the rules of multimerization. In this study, we aimed to elucidate the condition-dependent polymorphism of G4 structures composed of intramolecular quadruplex motifs without mismatches, sticky ends or other known drivers of programmed multimerization. Our major goal was to identify the basic features of the G4 motif that predetermine association, and we focused on the roles of loops and general topologies (parallel, antiparallel or hybrid). We used motifs that are ubiquitous in the human genome, because the association of such fragments might occur in vivo and could account for the packaging of G-rich microsatellites (18,27–29) and G4-driven juxtaposition or for the specific alignment of the nucleic acid strands (16,30). The general regularities of G4 multimerization discussed in this study can also be useful for guided assembly of supramolecular DNA structures for nanotechnological applications.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
Oligonucleotides (ONs) were synthesized using a Biosset ASM-800 DNA synthesizer (Biosset Ltd., Russia) and standard reagents (Glen Research, USA) following standard phosphoramidite protocols. Modified ONs were synthesized using standard reagents and 5′-O-dimethoxytrityl-1′,2′-dideoxyribose-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]-phosphoramidite (GlenResearch, USA). ONs were purified by a preparative-scale reverse-phase high-performance liquid chromatography (HPLC) on a 250 mm × 4.0 mm Hypersil C18 column (Thermo Fisher Scientific, USA) with detection at λ = 260 nm and a 2.5–15% gradient of acetonitrile in 0.1 M ammonium acetate buffer at 55°C. Dimethoxytrityl protection groups were removed via treatment with 80% acetic acid for 30 min, and the detritylated ONs were further purified in a 2.5–15% gradient of acetonitrile in 0.1 M ammonium acetate buffer. The purity of all ONs was approximately 95% by HPLC. The ON concentrations were calculated from the absorbance measured above 90°C and extinction coefficients, which were approximated using the nearest-neighbor model.
Mass spectrometry
Matrix-assisted laser desorption ionization time-of-flight (MALDI TOF) mass spectrometry was used to verify the compliance of theoretical and experimental masses of ONs. MALDI TOF spectra were acquired on a Microflex mass spectrometer (Bruker, Billerica, MA, USA) in the linear mode for positive ions (typical acceleration voltage, +20 kV; pulsed ion extraction delay, 130 ns). Each spectrum was accumulated using 400 laser shots (nitrogen gas laser; 20 Hz; λ = 337 nm). A mixture of 35 g/ml 3-hydroxypicolinic acid in deionized water and 0.2 M dibasic ammonium citrate (20:1 v/v) was used as a matrix. The observed difference between the theoretical and experimental masses of ONs was less than 3 Da (Table 1).
Table 1.
G4 sequences, MS data, topologies and melting/annealing temperatures
| T m/Ta, ±1°C* | ||||||
|---|---|---|---|---|---|---|
| Code | Sequence | m/z [M+H]+, found (calculated) | G4 type | Hysteresis | 10 mM KCl, rapid preannealing | 200 mM KCl, slow preannealing |
| 2G_L0 | GGxGGxGGxGG | 3113 (3113) | Parallel | + | 67/ 45 | ≥85/NA |
| Alu-G4 | GGCGGAGGCGG | 3464 (3463) | Parallel | + | ∼55/NA | ≥85/55 |
| 2G_L1 | GGTGGTGGTGG | 3488 (3485) | Parallel | + | 49/NA | 62/55 |
| 2G_L3 | GGTTAGGTTAGGTTAGG | 5337 (5338) | Antiparallel | - | 36/NA | 48/48 |
| 3G_L0 | GGGxGGGxGGGxGGG | 4430 (4430) | Parallel | NA | ≥85 | ≥90 |
| 3G_L1 | GGGTGGGTGGGTGGG | 4800 (4802) | Parallel | NA | ≥85 | ≥90 |
| 3G_L3 | GGGTTAGGGTTAGGGTTAGGG | 6655 (6653) | Mixed, predominantly antiparallel | - | 56/NA | 74/74 |
Atomic force microscopy (AFM)
Atomic force microscopy (AFM) experiments were performed on freshly cleaved graphite surfaces rendered hydrophilic with an amphiphilic modificator (CH2)n(NCH2CO)m-NH2 (31). For rapid annealing at a low salt concentration, ONs were diluted to a final concentration of 50 nM with 5 mM Tris–HCl buffer (pH 7.6) containing 10 mM KCl. Then, the ONs were denatured at 95°C for 5 min and snap cooled on ice. Typically, 5 μl of the annealed sample was applied on the substrate surface, kept there for 5–15 s, and then removed with a nitrogen stream, making the surface dry for imaging in air. For slow annealing at a high salt concentration, ONs were diluted to a final concentration of 2 μM with a 200 mM KCl solution in water. Then, the ONs were denatured at 95°C for 5 min and cooled in a thermos overnight. Immediately before sample preparation, the annealed ONs were diluted 20–40 times with 5 mM Tris–HCl buffer (pH 7.6) containing 10 mM KCl and were applied to the substrate as described above. Low salt concentration of the dilution buffer allowed us to eliminate from the sample preparation procedure a rinsing step, which could otherwise alter the folding of G4s. All experiments were performed at least in triplicate.
AFM imaging was performed using a multimode AFM instrument with an NTEGRA Prima controller (NT-MDT, Russia) in the tapping mode with a typical scan rate of 1 Hz and a typical free amplitude of several nm. All measurements were performed in air using home-made super-sharp cantilevers: carbon spikes were grown on the tips of commercially available standard silicon cantilevers using chemical vapor deposition process, the diameter of the spikes was approximately 1 nm (31). FemtoScan Online software (ATC, Russia; http://www.femtoscanonline.com) was used to filter and present the AFM data. Standard algorithms for AFM image flattening were used (subtraction of the quadric surface and averaging by lines), and no algorithms for resolution improvement were used, i.e. the raw AFM images are presented in this article. SPM Image Magic software (https://sites.google.com/site/spmimagemagic, Alex Kryzhanovsky) was used to analyse the ON heights semi-automatically. The analysis consisted of two steps: first, individual particles were identified automatically on the images by the local maxima and their heights were calculated in respect to the local background surrounding the particles; next, the results of the automatic analysis were filtered manually when necessary.
Control ONs used in AFM experiments included A25, T30, TTAACTTCTTCACATTC and GGGG.
Circular dichroism spectroscopy and melting curve analysis
ONs were dissolved to a concentration of 5 μM in 20 mM Tris–HCl buffer (pH 7.6) containing 10 or 200 mM KCl. Two preannealing procedures were employed: for rapid annealing, the samples were denatured at 95°C for 5 min and then snap cooled on ice, and for slow annealing, the samples were denatured at 95°C for 5 min and cooled in a thermos overnight. The absorption spectra, circular dichroism (CD) spectra and melting/annealing curves of the ONs were recorded using a Chirascan spectrophotometer (Applied Photophysics, UK) equipped with a thermostatic cuvette holder. To obtain melting/annealing curves of G4s, CD was recorded during heating or cooling across the 20–90°C range (heating/cooling rate = 0.5°C/min), and the melting temperature was estimated from the maxima in the first derivative of the melting curve.
Electrophoretic analysis
Denaturing (7 M urea) and non-denaturing (no urea) 20% polyacrylamide gels were prepared in a standard Tris–borate–EDTA (TBE) buffer (pH 8). The oligonucleotide samples (100 μM) were annealed as described in the ‘Circular dichroism spectroscopy and melting curve analysis’ subsection. A mixture of 10–100-nt ssDNA fragments (low molecular weight marker, Affymetrix) was used as a control. The gels were run for 2 h at 200 V at room temperature with a standard 1× TBE running buffer (for denaturing gels) or 1× TBE with 10 mM KCl (for non-denaturing gels), stained with SYBR Gold (Thermo Fisher Scientific, USA) and analysed using a Gel Doc scanner (Bio-Rad, USA).
Nuclear magnetic resonance (NMR) spectroscopy
For nuclear magnetic resonance (NMR) spectroscopy, ONs were dissolved in 20 mM Tris–HCl buffer (pH 7.6) containing 0, 10 or 20 mM KCl. D2O was added to each sample to a final concentration of 10% for lock signal stabilization. The final ON concentration was 100 μM. All the spectra were obtained using a Bruker Avance III 500 MHz NMR spectrometer (Bruker, Billerica, MA, USA) equipped with a Prodigy TCI cryogenic triple channel inverse probe. The 1H chemical shifts were measured relative to an external standard, namely, tetramethylsilane. The 2D DOSY (diffusion) and common 1D proton spectra with water suppression were measured (the stimulated echo pulse sequence with bipolar gradient pulses was used) (32,33). The water signal in both the 1D and 2D DOSY spectra was suppressed by the WATERGATE pulse sequence with five pairs of symmetric gradients in all the acquired spectra (125 ms delay for binomial water suppression for 1D spectra, 20 ms delay for binomial water suppression for 2D DOSY spectra, and 200 ms delay for gradient recovery) (34). 2D DOSY spectra were acquired with 100 ms big delta. NMR data processing and analysis were performed using a Bruker TopSpin v.3.2 and Mnova 10 software.
Molecular modeling
3D models of the G4 dimers were created using a previously reported strategy (35) and available PDB data for a related 3-tetrad parallel G4 structure (PDB 2MB2). Terminal T residues and loops were removed; the G4 cores were positioned manually in head-to-tail (models 1, 2 and 4), head-to-head (model 3) or side-by-side (models 5 and 6) orientations. In models 3 and 4, the G-tracks were shifted to allow for single-nucleotide interlocking, and then, the connecting loops were added. After each step, the structure was subjected to molecular-mechanical optimization to eliminate possible van der Waals overlapping. The optimizations were performed using the Sybyl-X program [https://support.certara.com/software/molecular-modelling-and-simulation/sybyl-x/] and Powell's method (36) with the following settings: a non-bonded cut-off distance of 12 Å, 1000 iterations, simplex method for initial optimization, energy gradient convergence criterion of 0.05 kcal.mol−1.Å−1. The effect of the medium was taken into account by using a dielectric constant of 4. The final optimization of each structure was performed via 1.5 ns of molecular dynamics (also using Sybyl-X and the AMBER7FF99 force field) at T = 300 K.
RESULTS AND DISCUSSION
Design of G4 oligonucleotides
In this study, we aimed to clarify whether the basic internal features of quadruplex units predetermine multimerization in a condition-dependent manner and analysed blunt-ended G4 motifs, each comprising four G3/G2 tracks interrupted by short or relatively short non-G tracks (loops). Such motifs, particularly those with single-nucleotide loops, are abundant in the human genome (37,38) and are often colocalized with chromosomal fragile sites (4).
Single-nucleotide loops typically ensure parallel G4 topologies (39), and pyrimidines, especially thymine, in single-nucleotide loops are preferred over adenine in terms of overall G4 thermodynamic stability (40,41). Therefore, the genomic sequences 3G_L1 and 2G_L1, with single thymine residues between the G3/G2 tracks, were selected as representative motifs of parallel 3-tetrad and 2-tetrad G4s, respectively. The frequencies and distributions of these motifs in the human genome are presented in Supplementary Figure S1. The sequence 3G_L1 has also been used as a model 3-tetrad G4 motif in our previous studies (42).
An additional genomic sequence from our set of G4 motifs with single-nucleotide loops is Alu-G4—a ubiquitous Alu-repeat fragment (Supplementary Figure S1) referred to as PQS4.1 in previous works (43,44). Alu-G4 is a homologue of the wire-forming fragment GCGGAGGCG (24), which was discussed in the Introduction (scheme (g) in Figure 1). The thermal stability of Alu-G4 (Tm > 80°C in the presence of 100 mM KCl (44)) is remarkably high for an intramolecular 2-tetrad G4, which prompted us to investigate the secondary structure of this ON in detail and consider the possibility of oligomerization/multimerization. Importantly, Alu-G4 has no apparent drivers for interlocking (unlike its truncated version GCGGAGGCG, which has GC sticky ends (24)), although interquadruplex stacking or complex multimerization pathways involving mixed tetrad formation cannot be excluded.
The sequences 3G_L3 and 2G_L3 represent intramolecular G4 motifs with three-nucleotide (TTA) loops between the G3/G2 tracks. Three-nucleotide loops are generally long enough to be diagonal or lateral, allowing opposite-strand orientations in a G4 core. The sequence 3G_L3 is actually a human telomeric sequence that is reportedly prone to condition- and flanking sequence-dependent conformational polymorphism (45). Here, we used the blunt-ended sequence variant. As shown below, under the selected conditions, this ON adopts a hybrid topology, while its 2-tetrad analogue 2G_L3 is a typical antiparallel G4. Diagonal and/or lateral loops of hybrid and antiparallel structures partially shield the external tetrads. For such G4s, we expected no multimerization (at least via interquadruplex stacking).
We also complemented our set of natural G4s with the modified analogues 3G_L0 and 2G_L0, containing 1′,2′-dideoxyribose residues between the G3/G2-tracks. This modification has been reported to enhance the thermal stabilities of parallel G4s, including fluorescently labelled 3G_L0 analogues with 5′ and 3′ T flanks (40), but the effects of this modification on G4 multimer formation have not been considered to date. Sequences of the native and modified ONs are given in Table 1.
AFM studies
For initial analysis of the G4 structures, we used AFM (46)—a powerful technique that allows high spatial resolution of single biological molecules (47–51), including nucleic acids (31,52,53). Few previously reported AFM studies of G4 structures have demonstrated the high potential of this method. Short G4s (containing four G-tetrads) introduced into DNA origami frames have been visualized using AFM (54). Relatively long (several hundred nanometres) G-wires have been visualized with AFM as rod-like structures with diameters of ∼2 nm (55–57). Recently, periodic features of G-wires have been revealed by high-resolution solution AFM (58). However, to our knowledge, AFM has not yet been successfully applied for the systematic characterization of single, isolated, short G4s. To address this challenge, we exploited high-resolution AFM with ultra-sharp tips (59).
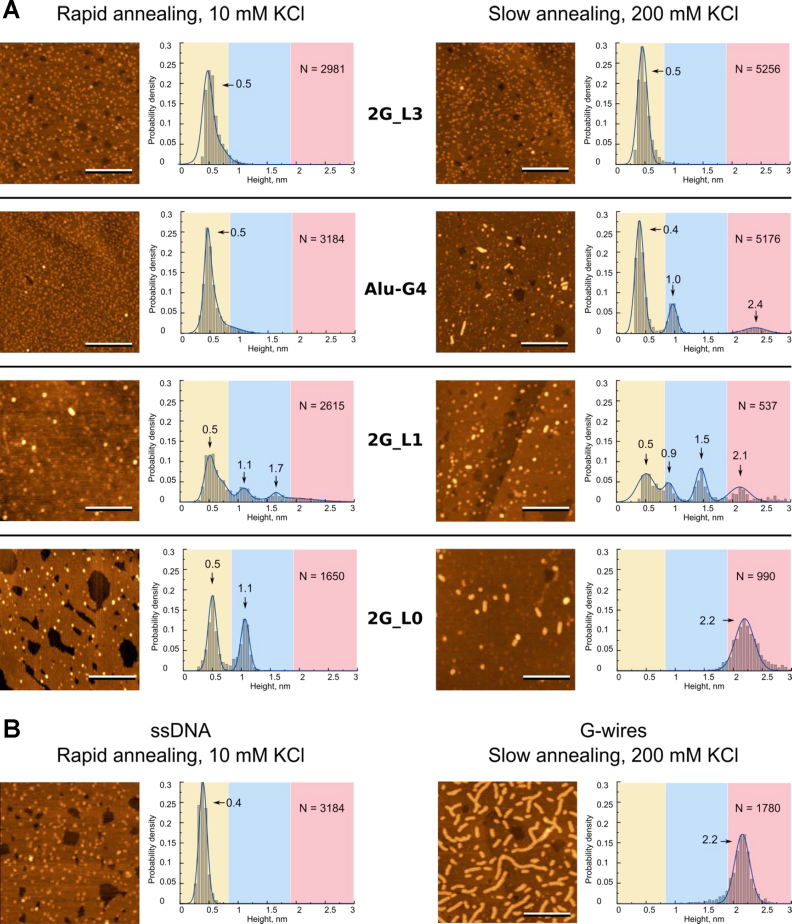
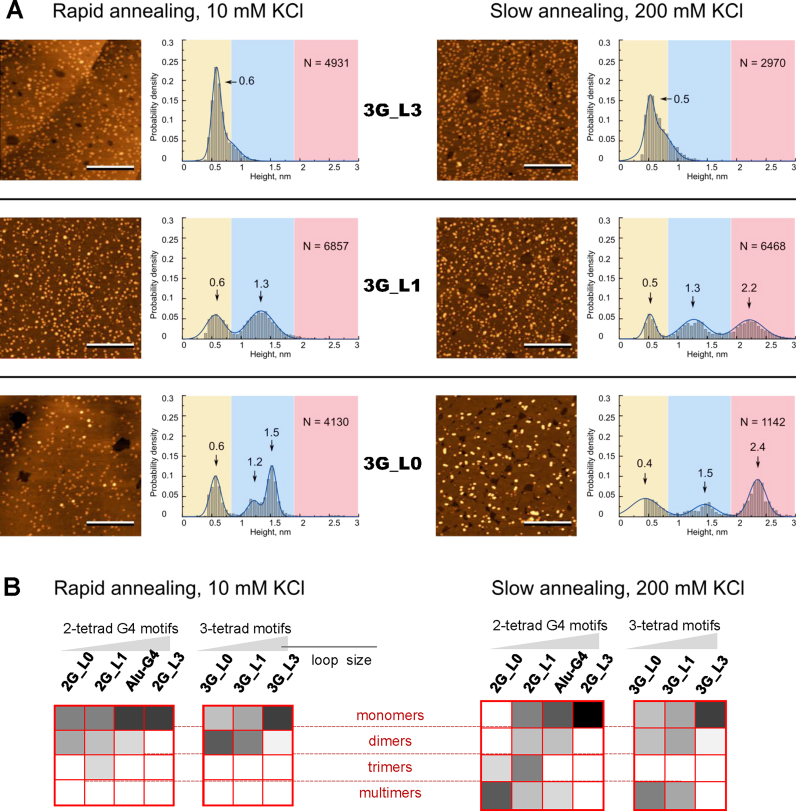
We prepared G4 samples using rapid or slow annealing at low (10 mM) or high (200 mM) KCl concentrations, respectively, as described in the Materials and Methods section. The rapid annealing procedure was meant to facilitate the formation of structures with relatively low activation energies, such as intramolecular G4s and/or meta-stable intermediates (e.g. triplexes), which were possibly prone to interlocking. The slow annealing procedure was meant to shift the equilibrium towards structures with the highest thermodynamic stability, i.e., multimers, should the G4s be prone to multimerization, and excess KCl was used to further enhance multimerization (11). The representative AFM scans of the G4 samples subjected to rapid and slow annealing and the corresponding height distribution histograms are shown in Figures 2A and 3A.
Figure 2.
Morphology of 2-tetrad G4 motifs (A) and control ONs (B) revealed by AFM. AFM height images and corresponding height histograms were obtained after rapid sample annealing at 10 mM KCl concentration (left) and slow annealing at 200 mM KCl concentration (right). The length of the scale bars is 100 nm. Control ssDNA is TTAACTTCTTCACATTC; control G-wires were obtained from GGGG. N is sample size.
Figure 3.
Morphology of 3-tetrad G4 motifs (A) and summary of relative contributions of different structures (B) revealed by AFM. AFM height images and corresponding height histograms were obtained after rapid sample annealing at 10 mM KCl concentration (left) and slow annealing at 200 mM KCl concentration (right). The length of the scale bars is 100 nm. N is sample size. Color gradient in (B) indicates the contribution of a particular structure: from 0 (white) to 100% (black).
G4s with three-nucleotide loops, namely, 2G_L3 and 3G_L3 (top panels in Figures 2A and 3A), were visualized as globular structures with heights of 0.5 ± 0.2 nm irrespective of the annealing conditions. Unmodified G4s with single-nucleotide loops, namely, Alu-G4, 2G_L1 and 3G_L1 (left middle panels in Figures 2A and 3A), were visualized as globular structures with heights of ∼0.5–1.7 nm after rapid annealing. Small globules were seen to be predominant in the Alu-G4 and 2G_L1 images (Figure 2A), and large globules were seen to be predominant in the 3G_L1 images (Figure 3A). After slow annealing, rods with heights of ∼2.1–2.2 nm (G-wires) appeared among the observed structures in all three samples (right middle panels in Figures 2A and 3A). The modified G4s 2G_L0 and 3G_L0 (bottom panels Figures 2A and 3A) exhibited generally analogous polymorphisms with enhanced yields of rods.
We used three short ONs that were not supposed to form any secondary structures (A25, T30, and TTAACTTCTTCACATTC) as ssDNA controls. Irrespective of the sample preparation procedure, all three ONs were visualized as mixtures of short threads and small globules, exhibiting a single maximum in the height histograms. In the case of TTAACTTCTTCACATTC, the height was 0.4 ± 0.2 nm (Figure 2B); the other distributions were similar (data not shown). We expected apparent heights of all the 2- and 3-tetrad G4 monomers to be approximately equal to the ssDNA height (the yellow zones of the histograms in Figures 2A and 3A).
To obtain control supramolecular G4 structures, we used the GGGG ON and the slow annealing procedure. The GGGG supramolecular structures were visualized as long rods that were 2.2 ± 0.3 nm in height (Figure 2B), which was consistent with published data on G-wires (55). We expected the apparent heights of all ‘classical’ (vertically interlocked) wires (scheme (e) in Figure 1) to be close to those of the GGGG multimers (the red zones of the histograms in Figures 2 and 3A), while the ‘lego’ wires (scheme (f) in Figure 1) were expected to resemble rods of substantially reduced heights, approximately equal in height to the G4 monomers. Dimers of the 2- and 3-tetrad G4s as well as trimers of the 2-tetrad G4s were expected to have apparent heights that were between those of the monomers and ‘classical’ wires (the blue zones of the histograms in Figures 2A and 3A).
The calculations validating our assumptions regarding the G4 monomer/dimer/trimer/multimer heights are summarized in Supplementary Figure S2. We relied on the previously reported basic parameters of G4 geometry (60,61) and assumed sorption modes that ensured maximal G4-surface interaction areas (tetrads are coplanar to the surface in all the cases considered except ‘classical’ wires). The hypothetical dimers and trimers are shown as head-to-tail stacks in Supplementary Figure S2, but this is merely an example, and head-to-head/tail-to-tail stacks or interlocked structures cannot be ruled out. Comparison of the observed heights of G4 ONs (Figures 2A and 3A) with those of control ONs (Figure 2B) and the calculated heights of monomers, dimers, trimers and wires (Supplementary Figure S2) led us to interpret the AFM data as follows:
the 0.5 ± 0.1-nm globules are folded and/or unfolded monomers (present in all G4 samples except 2G_L0 after slow annealing);
the 1.0 ± 0.1-nm globules are dimers of 2-tetrad G4s (Alu-G4, 2G_L1 and 2G_L0);
the 1.3 ± 0.2-nm globules are dimers of 3-tetrad G4s (3G_L1 and 3G_L0);
the 1.6 ± 0.1-nm globules are trimers of 2-tetrad G4s (2G_L1 only);
the 2.2 ± 0.1 nm rods are ‘classical’ G-wires (Alu-G4, 2G_L1, 2G_L0, 3G_L1 and 3G_L0 after slow annealing).
The relative contributions of the presumed structures are summarized in Figure 3B. The figure illustrates the general trends revealed by the AFM experiments:
G4s with 3-nucleotide loops exhibit no apparent polymorphism;
G4s with single-nucleotide or modified loops exhibit condition-dependent polymorphism;
slow annealing of polymorphic G4s at a high salt concentration enhances oligomerization and/or multimerization;
the yield of oligomers/multimers is particularly high in the case of modified loops.
To verify and complement our AFM results, we further characterized the G4s using optical methods, electrophoretic analysis and NMR spectroscopy.
Characterization by optical methods
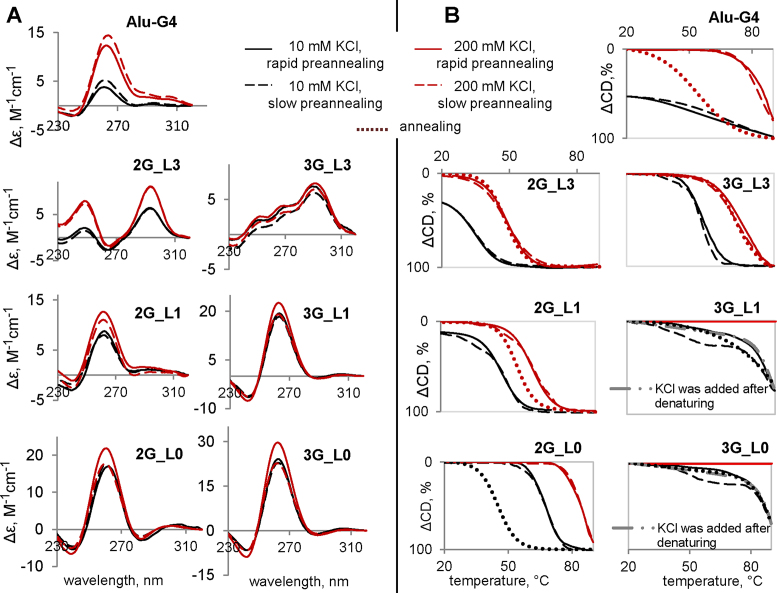
As mentioned above, compact intramolecular G4s cannot be distinguished from short ssDNAs in AFM experiments. Thus, we needed to clarify whether G4s are mostly folded or unfolded under the selected conditions. We used CD spectroscopy to confirm G4 formation and characterize the general topologies. We also performed thermal denaturation experiments to assess the stabilities of the structures. The CD spectra and melting curves recorded upon CD monitoring at G4-specific maxima are shown in Figure 4. Absorbance monitoring at 295 nm provided similar curves (data not shown).
Figure 4.
CD spectra (A) and melting/annealing curves (B) of the G4s. CD spectra were recorded at 5°C. Ellipticity is given per mole of nucleotide. The ONs (5 μM) were preannealed rapidly (solid lines) or slowly (dashed lines) in the presence of 20 mM Tris–HCl (pH 7.6) and 10 mM KCl (black lines) or 200 mM KCl (red lines).
The melting temperature (Tm) values obtained after marginal multimerization-favouring/multimerization-disfavouring preannealing procedures (rapid with 10 mМ KCl and slow with 200 mM KCl) are given in Table 1. In addition to the marginal procedures used for the AFM experiments and discussed in the previous subsection, we considered intermediate (mixed) variants (rapid with 200 mМ KCl and slow with 10 mM KCl) to separately assess the effects of salt and preannealing rates. To evaluate hysteresis and thus distinguish intramolecular structures (which are typically characterized by insignificant hysteresis) from intermolecular structures (which typically exhibit pronounced hysteresis), we also monitored CD changes upon annealing. Hysteresis was analysed at 200 mM KCl for moderately stable G4s and 10 mM KCl for highly stable G4s.
The CD spectra (Figure 4A) of G4s with single-nucleotide and modified loops (Alu-G4, 2G_L0, 2G_L1, 3G_L0 and 3G_L1) contained specific signatures of parallel G4s: a positive band at 260 nm and a negative band at 245 nm (62). The ON 2G_L3 adopted an antiparallel G4 topology, as evidenced by the positive CD band at 295 nm and the negative band at 265 nm (62), and 3G_L3 G4 adopted a mixed topology (the spectrum was similar to that reported previously for the hybrid structure (63)). The parallel G4s were predictably superior to the hybrid and antiparallel G4s in terms of thermal stabilities (39,64), and the Tm values increased with salt concentration (Table 1).
In contrast to the effect of salt, the preannealing rate had insignificant effects on Tm values but affected the CD amplitudes of highly stable G4s at 200 mM KCl. The slightly increased CD amplitudes of 3G_L1, 3G_L0 and 2G_L1 after slow preannealing (Figure 4A) may indicate additional stacking contacts (probably due to interquadruplex association). The preannealing rate also appears to influence the cooperativity of the melting process to some extent (Figure 4B): the parallel G4s had slightly diffused or broadened melting curves after slow preannealing. In the cases of 3G_L1 and 3G_L0 (10 mM KCl), the curves actually appeared to be biphasic/multiphasic, which could be due to the coexistence of different structures, probably monomers and stacks or interlocked G4s.
The parallel G4s Alu-G4, 2G-L1 and 2G-L0 exhibited pronounced hysteresis, which indicates the oligomerization/multimerization of these G4s. The hysteresis of 3G_L1 and 3G_L0 could not be analysed accurately due to the remarkably high stabilities of these G4s: the CD amplitudes decreased significantly at temperatures above 80°C, but the lower plateau was not reached at 90°C, which suggested that the Tm was ≥85°C in the presence of 10 mM KCl, and at high salt concentrations, no melting was observed. We repeated the experiments with a modified preannealing procedure (KCl was added after heating, and then the sample was cooled) and obtained similar results irrespective of the annealing rate. The hybrid/antiparallel G4s exhibited no hysteresis: the melting and annealing curves roughly coincided, which indicates intramolecular folding.
The major results can be summarized as follows:
G4 motifs with three-nucleotide loops form intramolecular antiparallel and hybrid structures;
G4 motifs with single-nucleotide and modified loops form parallel, likely intermolecular, structures;
KCl enhances the thermal stabilities of all the structures, while slow preannealing seems to slightly enhance the structural diversity;
all the G4s are folded at room temperature under the selected conditions.
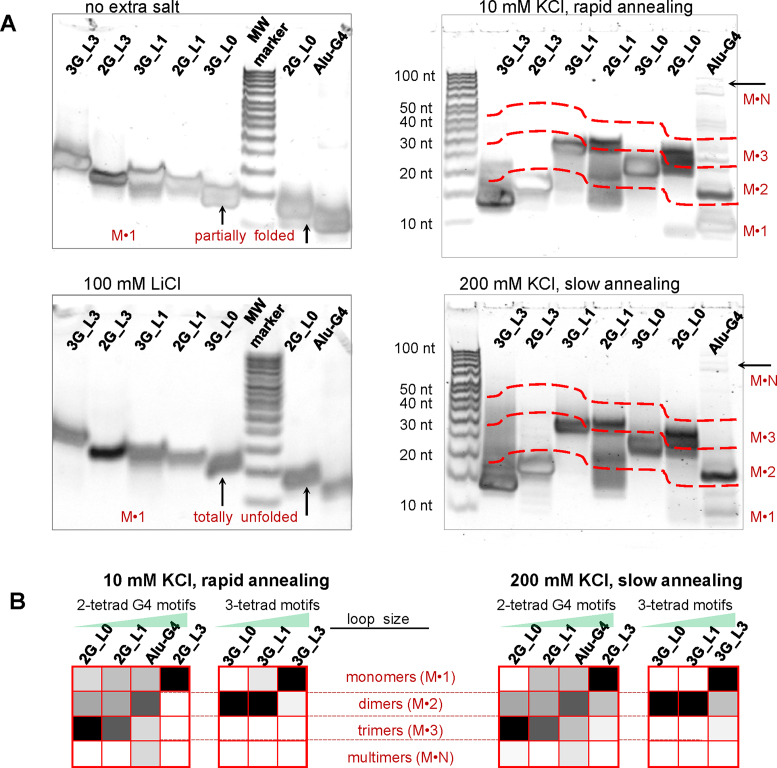
Electrophoretic studies
To additionally confirm the intermolecular folding of parallel G4s and verify the formation of dimers, trimers and/or multimers, as revealed by AFM experiments, we performed polyacrilamide gel electrophoresis (PAGE). The results are shown in Figure 5A. The experiments were first performed under denaturing conditions (left panel in Figure 5A) to estimate the mobilities of the monomers for reference. The remarkably stable parallel G4s appeared to be partially folded, even in the denaturing gel; therefore, we repeated the experiment with G4 preannealing in the presence of 100 mM LiCl to obtain fully unfolded monomers. The mobilities of the unfolded monomers generally correlated with the number of nucleotide residues; however, Alu-G4 was closer to 2G_L0 than to 2G_L1.
Figure 5.
Electrophoretic analysis of the G4s. (A) electrophorograms. Conditions: 100 μM ON, 20% denaturing (left) or non-denaturing (right) PAAG; standard TBE buffer (left) or TBE buffer with 10 mM KCl (right). Sample preannealing conditions are specified in the figure. The red dashed lines mark boundaries between presumed monomer, dimer, trimer and multimer zones. (B) Summary of the relative contributions of different structures revealed by PAGE. Color gradient indicates the contribution of a particular structure: from 0 (white) to 100% (black).
Next, we performed PAGE under non-denaturing conditions with rapid or slow sample preannealing in the presence of 10 or 200 mM KCl, respectively (right panel in Figure 5A). The folded monomers (observed for all ONs except 3G_L0 and 3G_L1) had slightly higher mobilities than the unfolded monomers (which was apparent for the longest ON, namely, 3G_L3, and less pronounced for the others). Dimers were observed for all the parallel G4s and were predominant in the cases of Alu-G4, 3G_L1 and 3G_L0. The hybrid G4 3G_L3 had a diffuse minor band in the dimer zone in addition to a major band in the monomer zone after slow annealing at a high salt concentration, which indicates minor conformational polymorphism under multimerization-favouring conditions. Trimers were observed for 2G_L1 and 2G_L0, and multimers were observed for Alu-G4 exclusively. For details on the assignment of particular bands to monomers, dimers or trimers, see Supplementary Figure S3.
The relative contributions of the presumed structures are summarized in Figure 5B. The following trends can be outlined:
antiparallel and hybrid G4s with 3-nucleotide loops exhibit no apparent polymorphism;
parallel G4s with single-T and modified loops are prone to oligomerization;
the predominant oligomers are dimers of 3-tetrad motifs and trimers of 2-tetrad motifs;
the yield of the oligomers is particularly high in the case of modified loops at high salt concentrations.
NMR studies
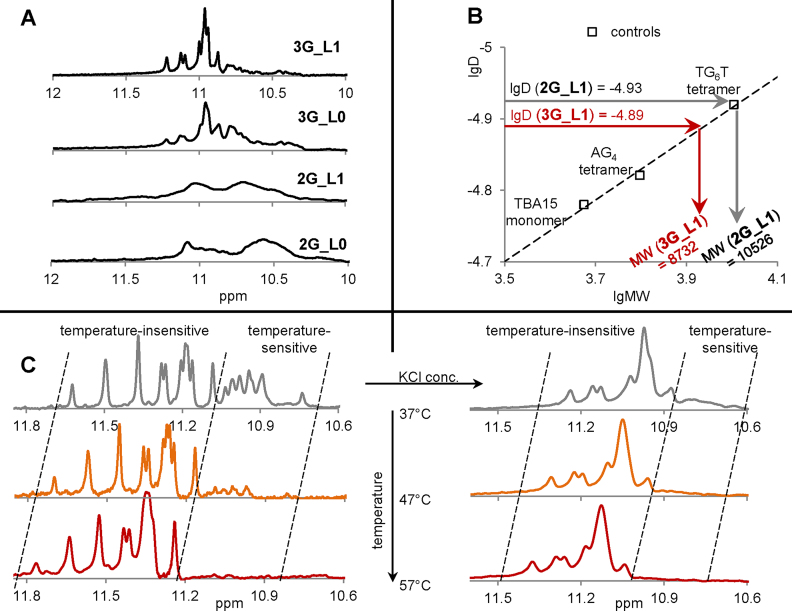
Oligomerization of G4s with single-T loops was further verified by 2D DOSY NMR spectroscopy. The modified G4s 2G_L0 and 3G_L0 were not tested, but the structures of these G4s are likely analogous to those of 2G_L1 and 3G_L1, respectively, considering the similarities in the 1H NMR spectra (the characteristic imino-region fragments are shown in Figure 6A). The 2D DOSY NMR spectra of 2G_L1, 3G_L1 and the control G4s are shown in Supplementary Figure S4, and the interpretation of the spectra is summarized in Figure 6B. The control G4s, adopting well-characterized intramolecular (TBA15 = GGTTGGTGTGGTTGG (65)) and intermolecular (AG4 and TG6T (66)) structures, were used to obtain the 2D DOSY calibration curve (D vs. MW, where D is the diffusion coefficient and MW is the molecular weight).
Figure 6.
NMR analysis of the G4s. (A) fragments of 1H-NMR spectra (imino regions). (B) DOSY-NMR results. (C) Effects of salt and temperature on 3G_L1 imino signals. Conditions: 100 μM ON, 20 mM Tris–HCl (pH 7.6); rapid preannealing of 2G_L1 and 3G_L1. The control monomer TBA15 was preannealed rapidly to facilitate intramolecular folding, and the control tetramers (AG4 and TG6T) were preannealed slowly to facilitate intermolecular folding. KCl concentrations: 10 mM (A and B), 0 (left panel in C) or 20 mM (right panel in C).
The calibration curve (linear in double logarithmic coordinates, see the dashed line in Figure 6B) was used to recalculate the experimental D values of 2G_L1 and 3G_L1 into experimental MW values. The latter values were compared with the theoretical MWs of the 2G_L1 and 3G_L1 monomers (Table 1). The experimental:theoretical MW ratio was close to 3:1 for 2G_L1 and 2:1 for 3G_L1 (10526:3484 and 8732: 4801, respectively), which is consistent with the formation of 2G_L1 trimers and 3G_L1 dimers in the presence of 10 mM KCl. Thus, the 2D DOSY NMR data are qualitatively consistent with the AFM (Figures 2 and 3) and PAGE (Figure 5) data. PAGE and AFM may have underestimated the oligomer yields.
We next exploited 1H NMR spectroscopy to further analyse the salt dependence of the 3G_L1 G4 (Figure 6C). In the case of 2G_L1, detailed analysis was impossible because of signal broadening and overlapping (Figure 6A). Salt had a substantial negative effect on the 1H NMR signal resolution (as seen by comparing the 3G_L1 spectra obtained at 0 and 20 mM KCl; left and right panels in Figure 6C, respectively); therefore, high ionic strength was avoided. Remarkably, the 3G_L1 G4 signature (imino proton signals at ∼10.6–11.8 ppm) was present even in the absence of KCl (Figure 6C, left panel). We attribute this to the stabilizing role of residual ammonium cations, which are inevitably present in ON samples after purification under standard HPLC conditions.
Studies of temperature dependency revealed that the G4 signature of 3G_L1 contains two groups of signals: a temperature-sensitive group (amplitudes decrease upon heating) and a temperature-insensitive group (no substantial decrease in amplitude upon heating). At 37°C, in the absence of KCl (left panel in Figure 6C), the chemical shifts of the partially overlapping temperature-sensitive signals were within the 10.7–11.1-ppm region. Using a well-defined singlet at 10.75 ppm (the last signal on the right) as a reference, we determined the integral of the temperature-sensitive group to be equal to 12, which is consistent with a 3-tetrad intramolecular structure. The chemical shifts of the partially overlapping temperature-insensitive signals were within the 11.1–11.7-ppm region. Using a well-defined singlet at 11.65 ppm (the last signal on the left) as a reference, we determined the integral of the temperature-insensitive group to be equal to 23–24, which suggested the presence of six G-tetrads and could indicate a G4 dimer.
The contribution of the temperature-sensitive signal group was substantial in the absence of KCl (left panel in Figure 6C) but decreased greatly with increasing salt concentration (right panel in Figure 6C), which is probably because KCl enhances oligomerization (shifting the equilibrium to 3G_L1 dimers). Importantly, all G4s identified by NMR spectroscopy, with and without the addition of KCl, are parallel, as confirmed by CD spectroscopy under the NMR spectroscopy conditions (Supplementary Figure S5).
In summary, the major results of our NMR experiments were as follows:
condition-dependent polymorphism of 3G_L1 was demonstrated;
2G_L1 was confirmed to form trimers at a moderate KCl concentration;
3G_L1 was confirmed to form dimers at a moderate KCl concentration.
Comparison of AFM, PAGE, CD and NMR spectroscopy data
Analysis of the G4s by optical methods and NMR spectroscopy provided mostly qualitative structural information, while AFM and PAGE allowed us to characterize the G4 polymorphism semi-quantitatively (Figures 3B and 5B, respectively). Both AFM and PAGE experiments revealed the same general trend: the extent of oligomerization/multimerization increased with decreasing loop size for both 2-tetrad and 3-tetrad G4 motifs. The principal differences between the AFM and PAGE results concern the following two aspects:
the extent of condition dependency (substantial dependency according to AFM data and minor dependency according to PAGE data);
oligomerization vs. multimerization of parallel G4s with single-T or modified loops (substantial yields of multimers according to AFM data and negligible yields according to PAGE data).
With regard to condition dependency, it should be noted that the methods have limitations in terms of working/running buffers and ON concentrations (concentrated G4 samples in PAGE and diluted samples in AFM). The G4 preannealing procedures were similar in the two assays, but multifold sample dilution prior to application to the AFM substrate surface likely accounts for enhanced sensitivity to initial salt concentrations.
With regard to the yields of multimers of G4s with single-T or modified loops, different oligomerization/multimerization routes should be considered. Stacking of intramolecular G4s (11) cannot be ruled out. However, the apparent differences between the AFM and PAGE results can be explained if we assume more complex multimerization routes, e.g. blunt-end stacking of interlocked dimers/trimers. Interquadruplex stacking may be disrupted under PAGE conditions, but survive AFM due to rapid fixation on the substrate surface. In contrast, completely interlocked wires would likely survive both AFM and PAGE. We hypothesize that Alu-G4 wires are completely interlocked (they are seen in native PAGE images, though Alu-G4 dimers dominate). The fact that Alu-G4 behaves differently from other 2-tetrad G4 motifs could be attributed to G-track-interrupting A and C residues, which allow for G:С pairing (possibly with G:С:G:C tetrad formation) and/or G(:A):G:G(:A):G hexad formation.
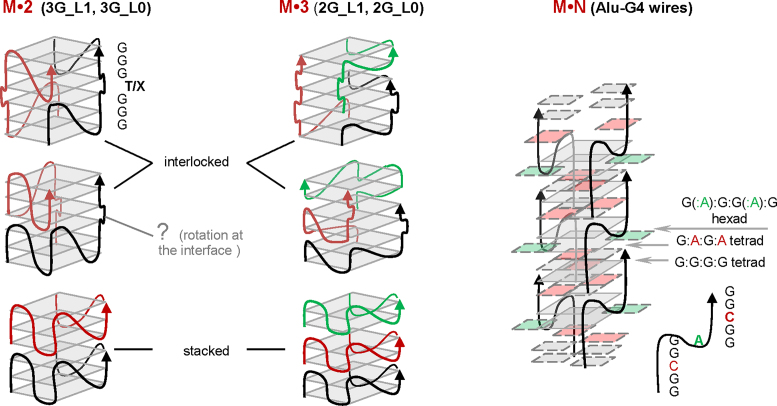
The general schemes of the presumed structures are shown in Figure 7. The structures include the following:
dimers of 3-tetrad G4s with single-T or modified loops, as revealed by AFM (Figure 2) and confirmed by PAGE (Figure 5) and NMR spectroscopy (Figure 6);
trimers of 2-tetrad G4s with single-T or modified loops, as revealed by AFM (Figure 3) and confirmed by PAGE (Figure 5) and NMR spectroscopy (Figure 6);
multimers of Alu-G4, as revealed by AFM (Figure 2) and confirmed by PAGE (Figure 5).
Figure 7.
Possible structures of G4 dimers, trimers and multimers.
All of these structures were parallel, as confirmed by CD spectroscopy (Figure 4A).
The scheme for Alu-G4 multimers (M*N in Figure 7) was drawn to be analogous to scheme (g) in Figure 1 (the structure, proposed previously for multimers of GCGGAGGCG, which is a truncated version of Alu-G4 (24)). The schemes for the interlocked dimers and trimers (M*2 and M*3 in Figure 7) were drawn to be analogous to scheme (h) in Figure 1 (26). The schemes for the stacked dimers and trimers were drawn to be analogous to scheme (b) in Figure 1 (17). We only show head-to-tail stacks because our CD data argue against other stacking modes.
Head-to-head or tail-to-tail stacking suggests opposite orientations of the tetrads at the interface. This is geometrically similar to opposite tetrad polarities and would theoretically look like an antiparallel ‘admixture’ in the CD spectrum — a minor positive band would be present at 295 nm. The assumption is grounded on reported spectra simulations and was corroborated by the analysis of modified G4s with 3′-3′ and 5′-5′ polarity inversion sites (67,68). No positive band at 295 nm can be seen in our spectra of parallel G4s (Figure 4A and Supplementary Figure S5), suggesting no stacked tetrads in opposite orientation, However, this matter is somewhat controversial. The minor antiparallel G4 signature might be absent due to relatively free rotation at the interface, and several examples of head-to-head dimers with no signs of antiparallel G4 ‘admixtures’ in the CD spectra have been reported previously (14,15,69).
Molecular modeling
We chose 3G_L1 for a more detailed analysis of possible interlocking patterns. Molecular modeling was used to clarify the unclear aspects of the interlocked M*2 schemes (Figure 7) and investigate possible alternatives (Supplementary Figure S6). The unclear aspects are associated with interfacial arrangement. For clarity, we show the interfacial single-T (or modified) loops as bulges in Figure 7, but this is merely a schematic representation. Rotation along the G4 axis would convert the bulges to loops (intermediate between lateral and propeller-type loops). Theoretically, both right-handed and left-handed interfacial helical twists are possible. Left-handed twists have been observed in G-wires with TT loops between the G4 blocks (58).
In our optimized models of vertically interlocked structures with single-T loops (models 1–4 in Supplementary Figure S6), the interfacial helical twist was right-handed (close to 28–29°), which ensured homogeneity of the G4 core. Models 1 and 2 (based on the interlocked M*2 schemes in Figure 7; the interfacial twist was taken into account) differ by the number of G-tracks that participate in interlocking (single-track- and double-track-interlocked dimers, respectively). Models 3 and 4 are loosely based on previously reported ‘3+1’ interlocked structures (scheme (d) in Figure 1, vertical interlocking via G residues rather than G-tracks). Such interlocking of blunt-ended G4s could result from G-register exchange (70) (sliding of the strands along the G4 axis) in head-to-tail or head-to-head stacks.
The 3G_L1 side-by-side-interlocked structures (models 5 and 6 in Supplementary Figure S6) are loosely based on previously reported ‘lego’ G4s (71) (scheme (f) in Figure 1) and, similarly to models 1 and 2, differ by the number of G-tracks that participate in interlocking (single-track- and double-track-interlocked dimers, respectively). The principal differences between the ‘lego’ G4s (71) and 3G_L1 side-by-side dimers (models 5 and 6) are associated with loop arrangement: pseudo-lateral interquadruplex loops were observed in the ‘lego’ structures and pseudo-diagonal loops were observed in our models. We believe that our variant of loop arrangement is reasonable because this arrangement enabled stacking of the loop nucleobases after geometry optimization (apparent for model 5 in Supplementary Figure S6), which ensured overall compactness of the structures and must be thermodynamically favourable.
Comparative analysis of the 3G_L1 models 1–6 led us to the following conclusions:
Models 1 and 2, inspired by scheme (h) in Figure 1, are the most thermodynamically favourable (the number of H-bonds and stacking contacts is maximized). Importantly, dimers 1 and 2 are theoretically prone to multimerization via blunt-end stacking, which can explain why the 3G_L1 wires were visualized as rods of an apparent height of 2.2 nm in AFM experiments.
In the alternative models 3 and 4, inspired by scheme (d) in Figure 1, the number of H-bonds is reduced: at least two tetrads are partially compromised due to the register shift. However, these models illustrate the possible conversion of blunt-ended G4s to sticky-ended G4s, which allows for an alternative multimerization route.
In models 5 and 6, inspired by scheme (f) in Figure 1, all H-bonds are maintained, but the number of stacking contacts is reduced. Importantly, dimers 5 and 6 have no potential for further multimerization, and the heights of these dimers are similar to those of monomers. This result could explain the underestimation of 3G_L1 dimers in the AFM experiments (as compared with the PAGE and NMR data).
CONCLUSIONS
We have shown that blunt-ended parallel G4s with short loops are polymorphic and prone to condition-dependent association. Increasing ON and KCl concentrations, decreasing annealing rates (thermodynamic control conditions) and decreasing loop sizes enhance G4 oligomerization and/or multimerization. Association pathways may involve both stacking and interlocking and appear to be different for G4s with single-T loops and G4s with A/C loops. G4s with modified (1′,2′-dideoxyribose) loops behave analogously to G4s with single-T loops, but the equilibrium is shifted towards higher-order structures.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Russian Science Foundation [14-25-00013 to G.E.P]. Funding for open access charge: Russian Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1. Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S.. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006; 34:5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 4. Zybailov B.L., Sherpa M.D., Glazko G.V., Raney K.D., Glazko V.I.. [G4-quadruplexes and genome instability]. Mol. Biol. (Mosk). 2013; 47:224–231. [DOI] [PubMed] [Google Scholar]
- 5. Wang F., Liu X., Willner I.. DNA switches: from principles to applications. Angew. Chem. Int. Ed. Engl. 2015; 54:1098–1129. [DOI] [PubMed] [Google Scholar]
- 6. Galer P., Wang B., Sket P., Plavec J.. Reversible pH switch of Two-Quartet G-Quadruplexes formed by human telomere. Angew. Chem. Int. Ed. Engl. 2016; 55:1993–1997. [DOI] [PubMed] [Google Scholar]
- 7. Hasuike E., Akimoto A.M., Kuroda R., Matsukawa K., Hiruta Y., Kanazawa H., Yoshida R.. Reversible conformational changes in the parallel type G-quadruplex structure inside a thermoresponsive hydrogel. Chem. Commun. (Camb.). 2017; 53:3142–3144. [DOI] [PubMed] [Google Scholar]
- 8. Livshits G.I., Stern A., Rotem D., Borovok N., Eidelshtein G., Migliore A., Penzo E., Wind S.J., Di Felice R., Skourtis S.S. et al. Long-range charge transport in single G-quadruplex DNA molecules. Nat. Nanotechnol. 2014; 9:1040–1046. [DOI] [PubMed] [Google Scholar]
- 9. Lech C.J., Phan A.T.. Ball with hair: modular functionalization of highly stable G-quadruplex DNA nano-scaffolds through N2-guanine modification. Nucleic Acids Res. 2017; 45:6265–6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Oliviero G., D’Errico S., Pinto B., Nici F., Dardano P., Rea I., De Stefano L., Mayol L., Piccialli G., Borbone N.. Self-Assembly of G-Rich oligonucleotides incorporating a 3 ‘-3 ’ inversion of polarity site: a new route towards G-Wire DNA nanostructures. ChemistryOpen. 2017; 6:599–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smargiasso N., Rosu F., Hsia W., Colson P., Baker E.S., Bowers M.T., De Pauw E., Gabelica V.. G-quadruplex DNA assemblies: loop length, cation identity, and multimer formation. J. Am. Chem. Soc. 2008; 130:10208–10216. [DOI] [PubMed] [Google Scholar]
- 12. Do N.Q., Phan A.T.. Monomer-dimer equilibrium for the 5 ‘-5’ stacking of propeller-type parallel-stranded G-quadruplexes: NMR structural study. Chem.-Eur. J. 2012; 18:14752–14759. [DOI] [PubMed] [Google Scholar]
- 13. Borbone N., Amato J., Oliviero G., D’Atri V., Gabelica V., De Pauw E., Piccialli G., Mayol L.. d(CGGTGGT) forms an octameric parallel G-quadruplex via stacking of unusual G(:C):G(:C):G(:C):G(:C) octads. Nucleic Acids Res. 2011; 39:7848–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Do N.Q., Lim K.W., Teo M.H., Heddi B., Phan A.T.. Stacking of G-quadruplexes: NMR structure of a G-rich oligonucleotide with potential anti-HIV and anticancer activity. Nucleic Acids Res. 2011; 39:9448–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuryavyi V., Cahoon L.A., Seifert H.S., Patel D.J.. RecA-binding pilE G4 sequence essential for pilin antigenic variation forms monomeric and 5′ end-stacked dimeric parallel G-quadruplexes. Structure. 2012; 20:2090–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Martadinata H., Phan A.T.. Formation of a stacked dimeric G-Quadruplex containing bulges by the 5′-Terminal region of human telomerase RNA (hTERC). Biochemistry. 2014; 53:1595–1600. [DOI] [PubMed] [Google Scholar]
- 17. Shi Y., Luo H.Q., Li N.B.. A highly sensitive resonance Rayleigh scattering method to discriminate a parallel-stranded G-quadruplex from DNA with other topologies and structures. Chem. Commun. 2013; 49:6209–6211. [DOI] [PubMed] [Google Scholar]
- 18. Kato Y., Ohyama T., Mita H., Yamamoto Y.. Dynamics and thermodynamics of dimerization of parallel G-quadruplexed DNA formed from d(TTAGn) (n = 3-5). J. Am. Chem. Soc. 2005; 127:9980–9981. [DOI] [PubMed] [Google Scholar]
- 19. Phan A.T., Do N.Q.. Engineering of interlocked DNA G-quadruplexes as a robust scaffold. Nucleic Acids Res. 2013; 41:2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsh T.C., Henderson E.. G-Wires - self-assembly of a telomeric oligonucleotide, D(Ggggttgggg), into large superstructures. Biochemistry. 1994; 33:10718–10724. [DOI] [PubMed] [Google Scholar]
- 21. Vesenka J., Henderson E., Marsh T.. Construction and examination of “G-wire” DNA. AIP Conf. Proc. 2002; 640:99–109. [Google Scholar]
- 22. Biyani M., Nishigaki K.. Structural characterization of ultra-stable higher-ordered aggregates generated by novel guanine-rich DNA sequences. Gene. 2005; 364:130–138. [DOI] [PubMed] [Google Scholar]
- 23. Usui K., Okada A., Sakashita S., Shimooka M., Tsuruoka T., Nakano S., Miyoshi D., Mashima T., Katahira M., Hamada Y.. DNA G-Wire formation using an artificial peptide is controlled by protease activity. Molecules. 2017; 22:E1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hessari N.M., Spindler L., Troha T., Lam W.C., Drevensek I., da Silva M.W.. Programmed self-assembly of a quadruplex DNA nanowire. Chem-Eur J. 2014; 20:3626–3630. [DOI] [PubMed] [Google Scholar]
- 25. Ilc T., Sket P., Plavec J., da Silva M.W., Drevensek-Olenik I., Spindler L.. Formation of G-Wires: the role of G:C-base pairing and G-quartet stacking. J. Phys. Chem. C. 2013; 117:23208–23215. [Google Scholar]
- 26. Kolesnikova S., Hubalek M., Bednarova L., Cvacka J., Curtis E.A.. Multimerization rules for G-quadruplexes. Nucleic Acids Res. 2017; 45:8684–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phan A.T. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010; 277:1107–1117. [DOI] [PubMed] [Google Scholar]
- 28. Petraccone L., Spink C., Trent J.O., Garbett N.C., Mekmaysy C.S., Giancola C., Chaires J.B.. Structure and stability of higher-order human telomeric quadruplexes. J. Am. Chem. Soc. 2011; 133:20951–20961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parkinson G.N., Lee M.P., Neidle S.. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002; 417:876–880. [DOI] [PubMed] [Google Scholar]
- 30. Kshirsagar R., Khan K., Joshi M.V., Hosur R.V., Muniyappa K.. Probing the Potential Role of Non-B DNA Structures at Yeast Meiosis-Specific DNA Double-Strand Breaks. Biophys. J. 2017; 112:2056–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Klinov D., Dwir B., Kapon E., Borovok N., Molotsky T., Kotlyar A.. High-resolution atomic force microscopy of duplex and triplex DNA molecules. Nanotechnology. 2007; 18:225102. [Google Scholar]
- 32. Johnson C.S., Jr Diffusion ordered nuclear magnetic resonance spectroscopy: principles and applications. Prog. Nucl. Magn. Reson. Spectrosc. 1999; 34:203–256. [Google Scholar]
- 33. Chou J.J., Baber J.L., Bax A.. Characterization of phospholipid mixed micelles by translational diffusion. J. Biomol. NMR. 2004; 29:299–308. [DOI] [PubMed] [Google Scholar]
- 34. Liu M., Mao X.-A., Ye C., Huang H., Nicholson J.K., Lindon J.C.. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J. Magn. Reson. 1998; 132:125–129. [Google Scholar]
- 35. Protopopova A.D., Tsvetkov V.B., Varizhuk A.M., Barinov N.A., Podgorsky V.V., Klinov D.V., Pozmogova G.E.. The structural diversity of C-rich DNA aggregates: unusual self-assembly of beetle-like nanostructures. PCCP. 2018; 20:3543–3553. [DOI] [PubMed] [Google Scholar]
- 36. Powell M.J.D. Restart procedures for the conjugate gradient method. Math. Program. 1977; 12:241–254. [Google Scholar]
- 37. Todd A.K., Johnston M., Neidle S.. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005; 33:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Huppert J.L., Balasubramanian S.. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005; 33:2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hazel P., Huppert J., Balasubramanian S., Neidle S.. Loop-length-dependent folding of G-quadruplexes. J. Am. Chem. Soc. 2004; 126:16405–16415. [DOI] [PubMed] [Google Scholar]
- 40. Rachwal P.A., Brown T., Fox K.R.. Sequence effects of single base loops in intramolecular quadruplex DNA. FEBS Lett. 2007; 581:1657–1660. [DOI] [PubMed] [Google Scholar]
- 41. Guedin A., De Cian A., Gros J., Lacroix L., Mergny J.L.. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008; 90:686–696. [DOI] [PubMed] [Google Scholar]
- 42. Varizhuk A., Ischenko D., Tsvetkov V., Novikov R., Kulemin N., Kaluzhny D., Vlasenok M., Naumov V., Smirnov I., Pozmogova G.. The expanding repertoire of G4 DNA structures. Biochimie. 2017; 135:54–62. [DOI] [PubMed] [Google Scholar]
- 43. Sekridova A.V., Varizhuk A.M., Tatarinova O.N., Severov V.V., Barinov N.A., Smirnov I.P., Lazarev V.N., Klinov D.V., Pozmogova G.E.. Conformational polymorphysm of G-rich fragments of DNA ALU-repeats. I. Potential noncanonical structures. Biomed. Khim. 2016; 62:535–543. [DOI] [PubMed] [Google Scholar]
- 44. Varizhuk A.M., Sekridova A.V., Tankevich M.V., Podgorsky V.S., Smirnov I.P., Pozmogova G.E.. Conformational polymorphysm of G-rich fragments of DNA Alu-repeats. II. the putative role of G-quadruplex structures in genomic rearrangements. Biomed. Khim. 2016; 62:630–637. [DOI] [PubMed] [Google Scholar]
- 45. Boncina M., Vesnaver G., Chaires J.B., Lah J.. Unraveling the thermodynamics of the folding and interconversion of human telomere G-Quadruplexes. Angew. Chem. Int. Ed. Engl. 2016; 55:10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Binnig G., Quate C.F., Gerber C.. Atomic force microscope. Phys. Rev. Lett. 1986; 56:930–933. [DOI] [PubMed] [Google Scholar]
- 47. Dufrene Y.F., Martinez-Martin D., Medalsy I., Alsteens D., Muller D.J.. Multiparametric imaging of biological systems by force-distance curve-based AFM. Nat. Methods. 2013; 10:847–854. [DOI] [PubMed] [Google Scholar]
- 48. Barinov N.A., Prokhorov V.V., Dubrovin E.V., Klinov D.V.. AFM visualization at a single-molecule level of denaturated states of proteins on graphite. Colloids Surf. B Biointerfaces. 2016; 146:777–784. [DOI] [PubMed] [Google Scholar]
- 49. Sheng S., Gao Y., Khromov A.S., Somlyo A.V., Somlyo A.P., Shao Z.. Cryo-atomic force microscopy of unphosphorylated and thiophosphorylated single smooth muscle myosin molecules. J. Biol. Chem. 2003; 278:39892–39896. [DOI] [PubMed] [Google Scholar]
- 50. Kodera N., Yamamoto D., Ishikawa R., Ando T.. Video imaging of walking myosin V by high-speed atomic force microscopy. Nature. 2010; 468:72–76. [DOI] [PubMed] [Google Scholar]
- 51. Protopopova A.D., Barinov N.A., Zavyalova E.G., Kopylov A.M., Sergienko V.I., Klinov D.V.. Visualization of fibrinogen αC regions and their arrangement during fibrin network formation by high-resolution AFM. J. Thromb. Haemost. 2015; 13:570–579. [DOI] [PubMed] [Google Scholar]
- 52. Raigoza A.F., Dugger J.W., Webb L.J.. Review: recent advances and current challenges in scanning probe microscopy of biomolecular surfaces and interfaces. ACS Appl. Mater. Interfaces. 2013; 5:9249–9261. [DOI] [PubMed] [Google Scholar]
- 53. Rivetti C., Guthold M., Bustamante C.. Scanning force microscopy of DNA deposited onto mica: equilibration versus kinetic trapping studied by statistical polymer chain analysis. J. Mol. Biol. 1996; 264:919–932. [DOI] [PubMed] [Google Scholar]
- 54. Endo M., Xing X., Zhou X., Emura T., Hidaka K., Tuesuwan B., Sugiyama H.. Single-Molecule manipulation of the duplex formation and dissociation at the G-Quadruplex/i-Motif site in the DNA nanostructure. ACS Nano. 2015; 9:9922–9929. [DOI] [PubMed] [Google Scholar]
- 55. Marsh T.C., Vesenka J., Henderson E.. A new DNA nanostructure, the G-wire, imaged by scanning probe microscopy. Nucleic Acids Res. 1995; 23:696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kotlyar A.B., Borovok N., Molotsky T., Cohen H., Shapir E., Porath D.. Long, monomolecular guanine-based nanowires. Adv. Mater. 2005; 17:1901–1905. [Google Scholar]
- 57. Chiorcea-Paquim A.M., Santos P.V., Eritja R., Oliveira-Brett A.M.. Self-assembled G-quadruplex nanostructures: AFM and voltammetric characterization. Phys. Chem. Chem. Phys. 2013; 15:9117–9124. [DOI] [PubMed] [Google Scholar]
- 58. Bose K., Lech C.J., Heddi B., Phan A.T.. High-resolution AFM structure of DNA G-wires in aqueous solution. Nat. Commun. 2018; 9:1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Klinov D., Magonov S.. True molecular resolution in tapping-mode atomic force microscopy with high-resolution probes. Appl. Phys. Lett. 2004; 84:2697–2699. [Google Scholar]
- 60. Lech C.J., Heddi B., Phan A.T.. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013; 41:2034–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tsvetkov V., Pozmogova G., Varizhuk A.. The systematic approach to describing conformational rearrangements in G-quadruplexes. J. Biomol. Struct. Dyn. 2016; 34:705–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Del Villar-Guerra R., Trent J.O., Chaires J.B.. G-quadruplex secondary structure from circular dichroism spectroscopy. Angew.Chem.(Int.Ed.Engl.). 2017; 57:7171–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bugaut A., Alberti P.. Understanding the stability of DNA G-quadruplex units in long human telomeric strands. Biochimie. 2015; 113:125–133. [DOI] [PubMed] [Google Scholar]
- 64. Bugaut A., Balasubramanian S.. A sequence-independent study of the influence of short loop lengths on the stability and topology of intramolecular DNA G-quadruplexes. Biochemistry. 2008; 47:689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Avino A., Fabrega C., Tintore M., Eritja R.. Thrombin binding aptamer, more than a simple aptamer: chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012; 18:2036–2047. [DOI] [PubMed] [Google Scholar]
- 66. Mergny J.L., De Cian A., Ghelab A., Sacca B., Lacroix L.. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005; 33:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gray D.M., Wen J.D., Gray C.W., Repges R., Repges C., Raabe G., Fleischhauer J.. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polarities. Chirality. 2008; 20:431–440. [DOI] [PubMed] [Google Scholar]
- 68. Randazzo A., Spada G.P., da Silva M.W.. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013; 330:67–86. [DOI] [PubMed] [Google Scholar]
- 69. Mukundan V.T., Do N.Q., Phan A.T.. HIV-1 integrase inhibitor T30177 forms a stacked dimeric G-quadruplex structure containing bulges. Nucleic Acids Res. 2011; 39:8984–8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Harkness R.W.T., Mittermaier A.K.. G-register exchange dynamics in guanine quadruplexes. Nucleic Acids Res. 2016; 44:3481–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Biyani M., Nishigaki K.. Structural characterization of ultra-stable higher-ordered aggregates generated by novel guanine-rich DNA sequences. Gene. 2005; 364:130–138. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.