Abstract
Sense codon reassignment to unnatural amino acids (uAAs) represents a powerful approach for introducing novel properties into polypeptides. The main obstacle to this approach is competition between the native isoacceptor tRNA(s) and orthogonal tRNA(s) for the reassigned codon. While several chromatographic and enzymatic procedures for selective deactivation of tRNA isoacceptors in cell-free translation systems exist, they are complex and not scalable. We designed a set of tRNA antisense oligonucleotides composed of either deoxy-, ribo- or 2′-O-methyl ribonucleotides and tested their ability to efficiently complex tRNAs of choice. Methylated oligonucleotides targeting sequence between the anticodon and variable loop of tRNASerGCU displayed subnanomolar binding affinity with slow dissociation kinetics. Such oligonucleotides efficiently and selectively sequestered native tRNASerGCU directly in translation-competent Escherichia coli S30 lysate, thereby, abrogating its translational activity and liberating the AGU/AGC codons. Expression of eGFP protein from the template harboring a single reassignable AGU codon in tRNASerGCU-depleted E. coli lysate allowed its homogeneous modification with n-propargyl-l-lysine or p-azido-l-phenylalanine. The strategy developed here is generic, as demonstrated by sequestration of tRNAArgCCU isoacceptor in E. coli translation system. Furthermore, this method is likely to be species-independent and was successfully applied to the eukaryotic Leishmania tarentolae in vitro translation system. This approach represents a new direction in genetic code reassignment with numerous practical applications.
INTRODUCTION
Genetic encoding of unnatural amino acids (uAAs) is a powerful approach for production of polypeptides with novel chemistries and properties (1–3). Polypeptides carrying uAAs are used as molecular probes, biosensors, drug lead candidates and functionalized biologics. While the majority of earlier efforts for uAA incorporation focused on the reassignment of nonsense codons, the idea of exploiting the degeneracy of the genetic code has recently gained popularity (2,4–6). This approach is attractive due to the large number of redundant sense codons and the lack of competition from the release factors that decrease the efficiency of the nonsense codon reassignment. Yet, the main obstacle for the efficient sense codon reassignment is the competition of native aminoacylated tRNA(s) with the orthogonal tRNA carrying uAA for the chosen sense codon. Hence, effective elimination of the competing endogenous tRNA(s) is critical for the success of this approach.
The concept of sense codon reassignment was initially validated on the example of the rare AGG (Arg) codon in Escherichia coli that was successfully reassigned to various uAAs in vivo (4,7,8). These methods required elimination of the competing endogenous Arg-tRNA, achieved through multiple rounds of genetic knockouts and gene complementation. Generally, sense codon reassignment is difficult to perform in vivo due to abundance of the redundant codons in the endogenous reading frames and the lack of methods for selective elimination of native tRNAs. A radical solution to these problems is construction of organisms with synthetic genomes where both the redundant codons and their cognate native tRNAs are eliminated. Recent publications reported progress toward the goal of complete elimination of redundant codons in E. coli (9). However, deletion of the redundant tRNAs may have broad and unexpected effects on the organism as the former have other functions in addition to genetic decoding (10). Furthermore, it is hard to predict to what extent the freed codons will be miscoded by the remaining native tRNAs. Finally, due to the immense cost of synthetic genome construction, this approach is likely to be limited to bacteria for the foreseeable future. Another avenue for expansion of genome coding capacity is the use of non-canonical codons such as quadruplet codons or codons with unnatural nucleotide structure. While impressive progress has been made in both directions, their broad applicability remains to be established (11–13).
An alternative approach that addresses the above mentioned problems is the use of in vitro translation systems, where the levels and identities of the translation reaction components can be easily controlled. Selective depletion of tRNA isoacceptors for amino acids encoded either by mixed codon families or by the families with high wobble restrictions would free the respective codons for decoding with orthogonal tRNAs. Suga's group demonstrated the feasibility of this approach for production of peptides containing different uAAs in a fully reconstituted E. coli in vitro translation system (6). Favorable competition of orthogonal tRNAs (o-tRNAs) for the reassigned codons is achieved by maintaining ultra-high concentration of their uAA pre-charged species. If applied to protein translation such high tRNA excess may affect the yield and fidelity of the protein synthesis. In contrast, sense codon reassignment using E. coli S30 lysate (14) would reduce the technical and economic barrier for technology adoption and provide a platform for rapid generation of protein probes for EPR, NMR, FRET analysis as well as production of proteins with other desired chemistries and functionalities such as post-translational modification and bioorthogonal reactive groups (15,16). Given that S30 lysate has been adopted to large scale protein manufacturing, the availability of compatible sense codon reassignment methodology would enable industrial production of therapeutic proteins functionalized with novel chemistries (17,18).
Previously, several elegant approaches for removal of the entire tRNA pool from the E. coli lysate either by using RNase A-coated beads or ethanolamine-functionalized matrix have been devised (19–23). Such tRNA-depleted lysates could then be supplemented with either native or synthetic tRNAs, thus restoring translation. One of the main limitations of these approaches is the tradeoff between the tRNA depletion efficiency and the remaining translational activity of the lysate (22). The observation that the fully recombinant PURE system contains co-purified protein-complexed tRNA fractions large enough to support efficient translation leads us to conclude that complete and selective tRNA removal is difficult to achieve (22).
Building on the idea of in vitro translation system reconstitution, several groups exploited the ability of oligonucleotides to inactivate a particular tRNA in the total tRNA fraction via hetero-duplex formation. Heating and annealing of the total tRNA fraction with tRNA-specific oligonucleotides was employed to accelerate the RNA-DNA hybridization (24). Subsequent treatment with RNaseH was shown to deactivate both tRNAAsp and tRNAPhe. These could then be replaced with uAA charged tRNAs. Combination of such tRNA complements with RNase A-treated lysate allowed partial reassignment of Asp- and Phe-codons to uAAs (24). The partial efficacy of this method is likely due to the incomplete depletion of tRNAAsp and tRNAPhe despite the repeated RNaseH treatment. Furthermore, both chosen codons were from the split codon boxes, thus their reassignment would result in a loss of corresponding native amino acids as protein building blocks. The sense codon reassignment approach is likely to be most useful when applied to codons of four- and six-fold degenerate amino acids as originally proposed by Kanda for tRNASerGCU (19,24).
As an efficient method for the selective tRNA inactivation is still outstanding, we addressed this bottleneck by designing the antisense oligonucleotides that selectively sequester chosen tRNA isoacceptors directly in the translation-competent cell lysate. By using the lysate in combination with the orthogonal tRNA charged with an unnatural amino acid, we demonstrate site-selective incorporation of the uAA into the newly synthesized polypeptides. Both reactions can be performed in batch and are fully scalable. Importantly, we demonstrate that this approach is not restricted to E. coli and can be applied to eukaryotic cell-free translation systems.
MATERIALS AND METHODS
Materials
The antisense oligonucleotides were synthesized by Integrated DNA Technologies. The sequences of all oligonucleotides used in this study are summarized in Supplementary Table S1. The p-azido-l-phenylalanine (AzF) and n-propargyl-L-lysine (PrK) were purchased from SynChem and Sirius Fine Chemicals, respectively. Ethanolamine-sepharose used for tRNA purification was prepared by coupling ethanolamine to epoxy-activated Sepharose 6B (GE Healthcare) as described (5).
Microscale thermophoresis (MST) analysis of tRNA interaction with oligonucleotides
The equilibrium dissociation constants between antisense oligonucleotides and synthetic tRNASerGCU were measured using Monolith NT.115 from NanoTemper according to the manufacturer's protocol. Briefly, the unlabeled tRNA molecules were prepared at 8 μM concentration and serially diluted fourfold to the lowest concentration of 0.03 nM in buffer A (20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 100 mM NaCl, 0.05 mg/ml BSA, 0.05% Tween 20, 2% PEG 8000). BSA and Tween 20 were added to reduce the unspecific binding while PEG 8000 was expected to act as a molecular crowding agent. The equal volumes of Cy3-labeled antisense oligonucleotides dissolved in buffer A were added to the serially diluted tRNA samples to the final concentration of 10 nM. After incubation at room temperature for 3 h to approach equilibrium the mixtures were transferred into the NT.115 standard treated capillaries (NanoTemper Technologies). The real-time measurements were performed at 80% LED (blue channel) and 80% MST power (IR laser). The results were analyzed and the Kd values were calculated using nonlinear fit to law of mass action equation as implemented in Monolith software. The same procedure was used to measure the equilibrium dissociation constant between antisense oligonucleotide M5-1 and Cy3-labeled tRNA with the concentration of the former varied from 0.08 to 125 nM while the concentration of the latter kept at 5 nM.
Gel shift assay
Modified oligonucleotides (M1-M8) at the final concentration of 0.5 μM were incubated with 2.5 μM of indicated tRNA in a total reaction volume of 10 μl using TMN buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 100 mM NaCl) (25) at 37°C for 15 min. The reaction was then chilled on ice and diluted with the equal volume of cold RNA loading dye containing 95% formamide and 0.025% bromphenol blue. Aliquots of 5 μl were loaded onto 8% denaturing PAGE gel made in TBE buffer with 6 M urea and 2.5 mM MgCl2. The gel electrophoresis was performed in TBE buffer supplemented with 2.5 mM MgCl2 at 70 V for 10 min followed by the voltage increase to 120 V until the dye front reached the end of the gel. After electrophoresis, the gel was stained with SYBR Green II dye for 15 min and scanned for fluorescence using Gel Doc XR Imaging System (Bio-Rad).
tRNA labeling
tRNA labeling was performed by oxidation of its 3′ terminus with sodium periodate (Sigma Aldrich) and subsequent reaction with hydrazide-Cy3 (Lumiprobe Corp.). Briefly, 15 μM tRNA and 100 mM NaOAc (pH 5) in a total volume of 1 ml were mixed and placed on ice for 5 min. After adding NaIO4 to 3 mM concentration, the oxidation reaction was performed in the dark for 30 min. The tRNA was then precipitated with ethanol and washed once with 70% ethanol. After re-suspending the pellet to 0.5 mM final concentration in the solution of 0.1 M NaOAc (pH5.0), the tRNA was supplemented with 5 mM Cy3-hydrazide which was dissolved in DMSO and incubated with agitation in the dark at room temperature for 1 h. After diluting the sample 10 times with 0.1 M NaOAc, tRNA was ethanol precipitated. Labeled tRNA was then purified on the POROS® R1 10 μm column (Applied Biosystems) using buffers B and C containing 0.1 M triethylamine acetate (pH 5.2) and either 1% (Buffer B) or 90% (Buffer C) of acetonitrile using 1–20% linear gradient in 9 min run. After HPLC purification, the labeled tRNA fraction was precipitated by ethanol twice, re-suspended in water and stored at −20°C.
Determination of the dissociation rate of oligonucleotide: tRNA complex
The koff of M5-1: tRNASerGCU-Cy3 complex was measured in competition experiments. Briefly, the complex was formed by incubating 5 nM of tRNASerGCU-Cy3 and 15 nM of M5-1 in buffer A at room temperature for 1 h. Subsequently unlabeled tRNASerGCU was added to the final concentration of 1500 nM and the fluorescence decrease at λex548/λem561 was monitored over time using Fluoromax spectrometer (Horiba). The koff value was calculated by fitting the resulting data to the one phase exponential decay equation using Prism software.
E. coli in vitro protein translation assay
The E. coli S30 cell lysate was prepared from BL21-Gold(DE3) strain as described previously (14). For inactivation of selected tRNAs, the E. coli S30 cell lysate was incubated with antisense oligonucleotides at indicated concentration at 37°C for 5 min and then chilled on ice. The antisense oligonucleotides were added from 400 μM stock solution to less than 1/10th volume of that of the lysate. The oligonucleotide-treated lysate can be used directly for assembly of the in vitro translation or stored frozen at −80°C.
The DNA templates coding for eGFP ORFs with biased codon usage were designed and constructed as described in Supplementary methods and their sequences are provided in the Supplementary Table S2. Cell-free translation reactions with eGFP-coding templates were performed following the previously developed protocol (14) with minor modifications. Briefly, 10 μl reaction contained 3.57 μl S30 lysate (parental or treated with antisense oligonucleotide), 10 mM Mg(OAc)2, 2 mM DTT, 2% PEG 8000, 87 mM HEPES–KOH pH 7.4, 1× protease inhibitor cocktail (Complete™ EDTA-free, Roche), 0.1 mg/ml folic acid, 1 mM rNTP mix, 15 mM acetyl phosphate, 25 mM creatine phosphate, 1.6 mM of amino acids R, C, W, M, D and E, 0.6 mM of the other 14 canonical amino acids, 0.05 mg/ml T7 RNA polymerase, 45 U/ml creatine phosphokinase and 50–100 ng/ml DNA template, in the presence or absence of suppressor tRNA indicated in each experiment. The real-time eGFP fluorescence was monitored using Synergy™ HTX multi-mode microplate reader (BioTek) at 30°C for 3–5 h using 485 nm excitation and 528 nm emission wavelengths. The yield of eGFP protein was estimated using serial dilutions of purified eGFP in PBS buffer as a reference. The calibration curve was generated using linear regression by plotting their fluorescence measured by Synergy microplate reader versus total protein concentration measured using Nanodrop (Thermo Scientific).
Production of synthetic tRNAs and orthogonal aminoacyl-tRNA synthetases
The sequences of synthetic tRNAs used in this work are provided in Supplementary Table S3. These synthetic tRNAs were prepared and purified as described before (22). Briefly, the DNA templates with 5′-T7 promoter and tRNA coding sequences were synthesized by overlap PCR. The resulting products were purified by ethanol precipitation and then dissolved in water. The T7 RNA polymerase run-off transcription reactions were performed in 40 mM HEPES–KOH (pH 7.9), 18 mM Mg(OAc)2, 2 mM spermidine, 40 mM DTT, 5 mM of each rNTP, 0.25 U/ml yeast inorganic pyrophosphatase, 0.1 mg/ml T7 RNA polymerase and 0.25 μM DNA template at 32°C for 3 h. The tRNA transcripts were purified by affinity chromatography using ethanolamine–sepharose matrix (5).
The orthogonal aminoacyl-tRNA synthetases PylRSAF and AzFRS were expressed in E. coli and purified by Ni2+ affinity chromatography and gel filtration as described previously (5).
Mass spectrometry
In vitro expressed eGFP proteins were incubated with anti-GFP VHH-Sepharose matrix (26) at room temperature for 30 min. The matrix was washed 3 times with PBS buffer containing 0.1% Triton and once with PBS buffer only. The protein was eluted with SDS PAGE loading buffer after pre-incubation at 98°C for 5 min and then separated on NuPAGE 4–12% Bis–Tris Gel (Thermo Fisher Scientific). The corresponding band was cut out and digested with sequencing grade trypsin following the in-gel tryptic digestion protocol from Thermo Scientific. The peptides containing supernatant were lyophilized for at least 3 h. The samples were dissolved in 20 μl of 0.1% formic acid and subjected to LC–MS/MS analysis using a Shimadzu Nexera uHPLC interfaced with a 5600 TripleTOF with a duo electrospray ion source (AB SCIEX). The 10 μl sample was injected onto a Zorbax C18 1.8 μm column (Agilent) using buffers D and E both containing 0.1% formic acid and either none (Buffer D) or 90% (Buffer E) of acetonitrile. The peptides were separated using 1–40% linear gradient over 50 min. The ion spray voltage was set to 5500 V. The mass spectrometer acquired 200 ms full scan of TOF MS spectra followed by up to 10, 200 ms of full scan of product ion spectra (MS/MS) in an information-dependent acquisition (IDA) mode. The criteria for selection of ions, from the initial TOF MS survey spectrum, for product ion MS/MS analysis was based upon the ions having a charge state of +2 to +5, exceeding 100 counts intensity and being one of the 10 most intense ions. Full scan TOF MS spectra were acquired over the mass range of 300–2000 m/z and MS/MS spectra (for product ion) between 100 and 1800 m/z. The data was processed and analyzed using Analyst TF 1.6 software (AB SCIEX) to assess the identity of the amino acid incorporated at the desired position.
Preparation of tRNA-dependent LTE translation system
The L. tarentolae extract (LTE) was prepared as described before (27) and stored at −80°C in buffer F (45 mM HEPES–KOH pH 7.6, 10 mM KOAc, 10.5 mM Mg(OAc)2 and 0.5 mM rGTP) for tRNA depletion. Typically 2.5 ml of LTE was re-buffered using NAP-25 column (GE healthcare) equilibrated with buffer G (25 mM KCl, 10 mM NaCl, 1.1 mM Mg(OAc)2, 0.1 mM EDTA, 10 mM HEPES–KOH pH7.5 and 100 mM KOAc). After re-buffering the lysate was incubated with 0.8 ml of settled ethanolamine–sepharose matrix at 4°C for 30 min with orbital shaking followed by flow-through collection. The flow-through from matrix washing with 1 ml of buffer G containing 180 mM KOAc was also collected and combined with the previous one, snap frozen and stored at −80°C.
Total tRNA mixture was prepared from L. tarentolae cell culture by phenol extraction following the modified Zubay's method (28). In brief, the L. tarentolae culture was inoculated into 1 L of TB medium (tryptone 12 g/l, yeast extract 24 g/l, glycerol 8 ml/l, glucose 1 g/l, KH2PO4 2.31 g/l, K2HPO4 2.54 g/l) and grew at 26°C until OD600 reached 3–3.5 (1.0–1.3 × 108 cells/ml). The cells were pelleted by centrifugation at 2500 g and washed with PBS. The nucleic acids were then extracted twice from the pelleted cells with 88% redistilled phenol, and precipitated by addition of 0.05 volume of 4 M KOAc and double volume of absolute ethanol. The tRNA precipitate was extracted with 1 M NaCl twice. The supernatants from the two NaCl extraction were combined and precipitated by addition of two volumes of ethanol. After two repeats, the crude tRNA fraction was dissolved in the nuclease-free water.
The specific tRNA species from the total L. tarentolae tRNA mixture were inactivated using antisense oligonucleotides (Supplementary Table S1). The isolated total tRNA at 3 μg/μl final concentration was mixed with antisense oligonucleotides at 60 μM final concentration in water and incubated at 37°C for 30 min. The translation reaction was set up using modified standard conditions (27). The reaction (10 μl) contained 50% tRNA-depleted LTE (5 μl), 1.67 μl of 3 μg/μl of antisense-oligonucleotide treated total tRNA, 4.45 mM Mg(OAc)2, 0.24 mM spermidine, 2 mM DTT, 40 mM creatine phosphate, 20 mM HEPES–KOH pH 7.6, 1% (v/v) PEG 3000, 1× protease inhibitor (Complete™ EDTA-free, Roche), 0.14 mM of each amino acid, 1.5 mM rNTP mix (ATP, GTP, UTP and CTP), 0.01 mM anti-splice leader DNA oligonucleotide, 0.1 mg/ml T7 RNA polymerase, 40 U/ml creatine phosphokinase, 50–100 ng/μl DNA template, with or without 20 μM AGC-suppressor tRNA (L. tarentolae tRNAGlyGCU) as indicated.
For inactivation of selected tRNAs directly in LTE, the cell extract was incubated with antisense oligonucleotides at 30 μM concentration at 37°C for 5 min and then cooled down on ice prior to assembly of the in vitro translation reaction. The 10 μl reactions contained 50% antisense oligonucleotide-treated LTE (5 μl), Mg(OAc)2 at 3.5 mM and other components as described above for the reconstituted system. The progress of eGFP translation was monitored by fluorescence increase on the Synergy microplate reader at 27°C for 3–5 h using λex485/λem528.
RESULTS
Antisense oligonucleotides can selectively and efficiently sequester their tRNA targets
Inactivation of a specific tRNA in the context of in vitro translation system is a challenging task as the size, shape and physical stability of tRNAs are similar. Most of the unique primary sequence determinants are embedded in the secondary and tertiary structures and have low solvent accessibility. The unpaired single-stranded segments of the anticodon loop and the 3′-CCA end are short and share significant similarity among tRNAs. Thus, we set out to devise a generally applicable approach for selective tRNA inactivation by exploiting the isoacceptor-specific differences in the primary structure.
Antisense oligonucleotides have been extensively used in vivo and in vitro to target eukaryotic or bacterial RNAs by promoting their cleavage or impairing their interactions with the cellular machinery (29,30). With introduction of novel oligonucleotide chemistries the antisense technology moved from the simple linear oligonucleotides to highly modified polymers with improved membrane penetration, nuclease resistance and stronger base pairing (30–32). The successful design of antisense oligonucleotides depends largely on the ability to identify a primary hybridization site in RNA that is not obstructed by the high order structures (33). This is particularly true for tRNAs that are highly structured since in addition to secondary structure the final L-shaped conformation is held together by tertiary interactions between D- and T-arms and additionally strengthened by Mg2+ ions and base-modifications (34).
Based on these considerations we set out to design oligonucleotides targeting isoacceptors from six-fold degenerate codon families that are most suited for reassignment. As the first candidate we chose the tRNASerGCU isoacceptor as its depletion would allow reassignment of AGC/AGU codons to uAA while leaving UCN codon box for serine encoding. The antisense oligonucleotides were designed to target the regions spanning from D-stem down to anticodon-loop or from the anticodon-loop to the variable loop (Figure 1A). The oligonucleotides include DNA- (M1, M6), RNA- (M2, M7), 2′-O-methylated (2′OMe) oligonucleotides (M5, M8) as well as mixed DNA/2′OMe (M3) and RNA/2′OMe oligonucleotides (M4). A Cy3 fluorophore was attached to their 5′-termini in order to enable microscale thermophoresis (MST)-based interaction analysis (35) of these polymers with the synthetic tRNASerGCU (Figure 1B).
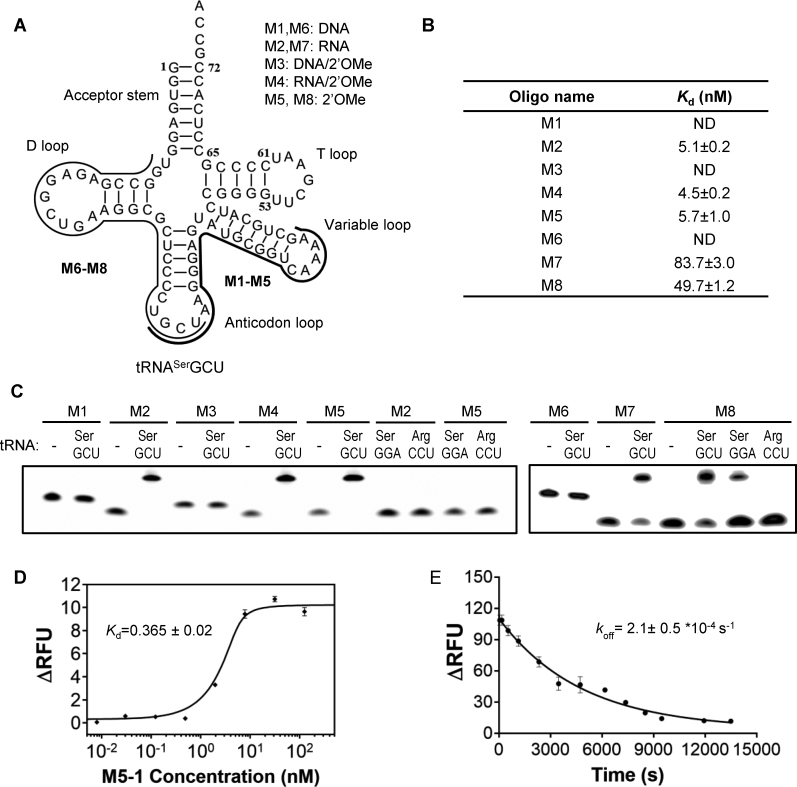
Figure 1.
Design and analysis of oligonucleotides targeting E. coli tRNASerGCU. (A) Schematic representation of the antisense oligonucleotides targeting anticodon/variable loop (M1-M5) or D-arm/anticodon regions (M6-M8) of tRNASerGCU. (B) Table of the equilibrium dissociation constant (Kd) of the antisense oligonucleotides for tRNASerGCU measured by Microscale Thermophoresis (MST). The measurements were performed on the mixtures of 10 nM solution of fluorescently labeled antisense oligonucleotide supplemented with increasing concentrations of tRNASerGCU. The Kd values were calculated using a nonlinear fit equation of the law of mass action and represented averages of three experiments (Supplementary Figure S1). ND—no detectable binding at the test condition. (C) Denaturing PAGE analysis of 5 pmol of Cy3-labled antisense oligonucleotide mixed with 25 pmole of the indicated tRNAs. The positions of the oligonucleotides were visualized by the fluorescent scanning of the gel. (D) Interaction analysis between oligonucleotide M5-1 and Cy3-labeled tRNASerGCU. The M5-1 oligonucleotide has the same sequence as M5, but is lacking 5′-Cy3 fluorophore. The affinity was measured as described in (B) but using a 5 nM solution of Cy3-labeled tRNA titrated with the increasing concentrations of M5-1. The error bars represent standard deviations of three independent experiments. (E) Determination of the dissociation rate of M5-1: tRNASerGCU–Cy3 complex. In the experiment mixture of 15 nM M5-1 with 5 nM tRNASerGCU–Cy3 was pre-incubated at room temperature for 1 h and supplemented with unlabeled tRNASerGCU to the final concentration of 1500 nM. The decrease of fluorescence signal was fitted as a single exponential decay leading to koff value of 2.1 × 10−4 s−1. The mean values and standard deviations were calculated from two independent experiments.
The M2 (RNA), M4 (partially methylated RNA) and M5 (fully methylated RNA) oligonucleotides targeting the anticodon/variable loop region of tRNASerGCU displayed similar Kd values of ∼5 nM while the Kd values for M7 (RNA) and M8 (fully methylated RNA) targeting the D-arm/anticodon region of tRNA were ∼80 and 50 nM, respectively. The lower efficiency of M7 and M8 is expected as they target an entire D-stem and most likely adopt either hairpin or dimer structure themselves, thus increasing the activation barrier for complex formation with the target. Unmodified (M1 and M6) and partially modified (M3) DNA oligonucleotides showed no detectable binding to tRNASerGCU. These observations stress the importance of chemical composition of antisense oligonucleotides and are consistent with the hybrid thermal stability and binding affinity increasing in the order: DNA/RNA < RNA/RNA ≤ 2′OMe/RNA (36–38). Although 2′OMe/RNA and RNA/RNA duplex stabilities are comparable (38,39), 2’OMe modifications conferring an enhanced entropic stabilization to the hybrid base pairs (40) would be expected to grant better RNA strand displacement abilities to the antisense oligonucleotide for the faster tRNA structure invasion.
In order to confirm the observed interactions using a more direct method we employed a gel-shift assay. As can be seen in Figure 1C the RNA- (M2, 4, 5, 7, 8) but not DNA-based oligonucleotides (M1, 3, 6) formed stable complexes with tRNA. In agreement with the determined affinities, oligonucleotides targeting the anticodon/variable loop region (M2, 4, 5) were more efficient binders, with complex formation saturating after 15 min incubation at 37°C. The oligonucleotides targeting D-arm/anticodon region (M7, 8) displayed weaker interactions and could not be fully complexed even when using 5-fold excess of tRNASerGCU.
To confirm the specificity of the observed interactions, we tested the ability of M2 and M5 to recognize the tRNASerGGA isoacceptor which is the closest homologue of tRNASerGCU showing 46% identity within the targeted anticodon/variable loop region. We observed no complex formation using the gel shift assay (Figure 1C). In contrast, M8 oligonucleotide, targeting the D-arm/anticodon region, displayed some affinity toward tRNASerGGA that shares ∼77% of homology with the tRNASerGCU within this region. Neither of these oligonucleotides formed complexes with tRNAArgCCU. These results show that appropriately designed oligonucleotides targeting the less conserved region could selectively form complexes with the target tRNA molecules.
As the fluorescent dye at the 5′-end of oligonucleotide could potentially influence its 5′-terminal base pairing affecting the measured affinity (41,42), we repeated the MST experiments after relocating Cy3 fluorophore to the 3′-terminus of tRNASerGCU. We monitored the fluorescence change of labeled tRNA while adding increasing concentrations of the unlabeled version of M5 oligonucleotide termed M5-1. The fit of the data resulted in a Kd value of 0.4 nM, which is ∼15 times lower than that measured between Cy3-labeled oligonucleotide (M5) and unlabeled tRNA (Figure 1B and D) indicating the negative impact of the label on complex formation.
As the designed oligonucleotides are intended for application in the in vitro translation system it is critical to have the kinetic parameters of their interaction with tRNA as these are expected to affect their ability to sequester tRNA from the translation machinery (43). To obtain this information we measured the koff by monitoring the fluorescence signal of tRNASerGCU-Cy3: M5-1 complex upon addition of more than 100-fold molar excess of unlabeled tRNA. The fit of the fluorescent decay to a first order reaction resulted in koff value of 2.1 × 10−4 s−1 (Figure 1E). Using the obtained Kd and koff values we then calculated the kon value of 5.8 × 105 M−1 s−1. This association rate suggests that addition of micromolar concentration of oligonucleotides to the in vitro translation system would rapidly complex the targeted tRNA. As the residence time of M5-1: tRNA complex of ∼80 min is more than half of the typical lifetime of the in vitro translation reaction, we expect that slow dissociation of the complex should prevent tRNA recruitment by the translational machinery. This is particularly important for the depletion of the protein-bound fraction of tRNA that is responsible for the pervasive background activity (22). Furthermore, the long residence time is expected to kinetically enhance the selectivity of the oligonucleotides toward its target over the other isoacceptors in the system (44).
Design of the fluorescent reporter system for monitoring of oligonucleotide-mediated tRNA depletion in E. coli cell-free expression system
In the next step we wanted to evaluate the ability of the developed antisense oligonucleotides to inactivate native tRNASerGCU in the E. coli in vitro translation system. To this end we designed DNA templates coding for eGFP with biased selection for serine codons. We introduced two AGC-codons decoded by tRNASerGCU at consecutive positions 21 and 22 while encoding remaining serines with one AGT and ten TCC codons (Figure 2A and Supplementary Table S2). We expected that translational pausing at the consecutive AGC would lead to the decrease in eGFP expression and serve as a measure of functional tRNASerGCU concentration in the system (22). To confirm that fluorescence reduction is a direct consequence of tRNASerGCU sequestration, an efficient AGC-codon suppressor based on tRNAGlyUCC was constructed by replacing its anticodon with GCU (unpublished observation). Due to its sequence divergence from the tRNASerGCU this tRNA would not form a sequence-specific complex with the oligonucleotide and was expected to rescue the eGFP expression if its reduction was caused by ribosome pausing at AGC codons.
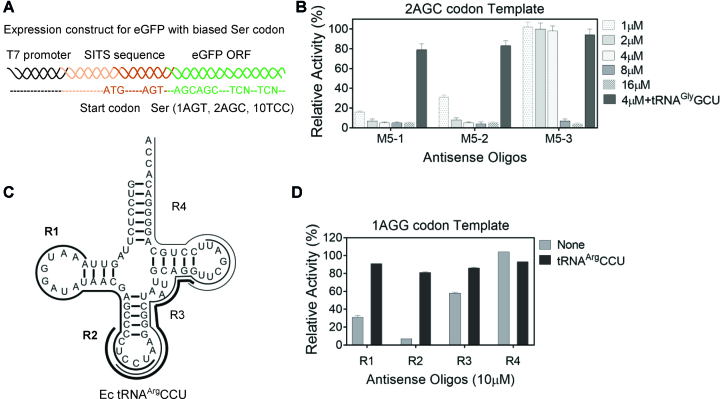
Figure 2.
Testing the ability of the antisense oligonucleotides to selectively inactivate tRNA isoacceptors in E. coli cell-free translation system. (A) The schematic representation of the eGFP fluorescent reporter construct for evaluation of tRNASerGCU levels in cell-free system. The wt eGFP-coding ORF designed with biased Ser-codons is prefaced by Species Independent Translation Initiation Sequence (SITS) (27) that enables translation in both pro- and eukaryotic cell-free systems. (B) Translational activity of the 2AGC-codon template in E. coli lysate pre-treated with oligonucleotides in the presence or absence of synthetic tRNAGlyGCU. The M5-sequence analogs M5-1,-2,-3 contain various percentages of modified residues (100%, 75% and 46%, respectively). The concentration of the antisense oligonucleotides for lysate treatment is indicated while tRNAGlyGCU is added to final concentration of 20 μM in the translation reaction. The relative translation activity was calculated as the percentage of activity of the parental lysate. (C) The schematic representation of the methylated RNA oligonucleotides targeting four regions of tRNAArgCCU (R1, 2, 3, 4). Each oligonucleotide is represented by a black line with thickness proportionate to their inhibition efficiency as shown in Figure 2D. (D) Translational activity of eGFP-coding template harboring single AGG (Arg) codon (Supplementary Table S2) in E. coli lysate treated with 10 μM oligonucleotides R1–R4. The expression activity is represented as described in (B). Data in B and D represent the mean and standard deviations of at least two independent experiments.
To measure the extent of tRNA depletion using the developed reporter we incubated the lysate with the oligonucleotide at 37°C for 5 min after which the translation reaction was assembled and fluorescence was monitored for 3 h (Figure 2B and Supplementary Figure S2). We found that the oligonucleotide-mediated fluorescence reduction could be reversed by the addition of synthetic tRNAGlyGCU and the extent of recovery was roughly proportionate to the affinities between oligonucleotides and tRNASerGCU (Figure 1B and Supplementary Figure S2).
Given the ability of methylation for endowing oligonucleotides with high affinity for tRNA and resistance to nuclease degradation (45), we decided to test the effect of the modification level on the efficiency of translation inhibition and recovery in the in vitro translation reaction. We designed two partially modified variants of M5-1 based on the alignment between tRNASerGCU and the rest of Ser isoacceptors. In these variants termed M5-2 and M5-3 the methylated nucleotides remained at the tRNASerGCU-specific positions while the respective 6 (25%) or 13 (>50%) unmodified residues were inserted at the least degenerate positions in an attempt to reduce potential cross-reactivity toward the other tRNA homologs (Supplementary Table S1). As can be seen in the Figure 2B we observed a gradual increase in IC90 with the increase in the number of unmodified residues being below 2 μM for M5-1 and M5-2 and ≥8 μM for M5-3. The recovery of the translational activity with synthetic tRNAGlyGCU suppressor for the first two oligonucleotides reached more than 80% of the untreated reaction (Figure 2B). The site-selective modifications did not increase the inhibition effect and the fully methylated M5-1 oligonucleotide was so far the most effective tRNA binder designed.
In order to establish whether the developed approach is generally applicable and not confined to tRNASerGCU, we selected tRNAArgCCU which decodes the AGG codon from the mixed codon family of arginine. Although the AGG-codon is also decoded at lower efficiency by the native tRNAArgUCU we conjectured that its low abundance in E. coli would still allow the detection of tRNAArgCCU inactivation in the translation reaction. Following the considerations described above we designed four 2′OMe antisense oligonucleotides R1-R4 targeting different regions of tRNAArgCCU from 5′ to 3′ (Figure 2C). R1 and R3 target two loops of tRNAs while R4 targets the 3′-end. R2 is slightly shifted to the 5′ of the tRNA compared to R3. As shown in the Figure 2D, R2 reduced the translation of eGFP template containing a single AGG codon to less than 10% of the control translation level. The translation inhibition could be reverted by the addition of the excess of synthetic tRNAArgCCU resulting in eGFP production reaching 81% of the untreated lysate level. These experiments demonstrate that the methylated oligonucleotide-mediated inactivation of tRNAs is generally applicable.
Identifying the functionally essential elements in the sequence of M5-1 oligonucleotide
As the M5-1 antisense oligonucleotide performed very well in the in vitro translation reactions we set out to reveal its mechanism of action and identify the key structural elements responsible for its activity. M5-1 spans two structurally separate entities of the tRNA including the anticodon and variable stem-loops. There is additional complexity introduced by the N6-threonylcarbamoyl modification of native tRNASerGCU on the A37 (t6A37) of the anticodon loop which is expected to destabilize the hybrid in this region (46).
To dissect the structure/function relationship of M5-1 we produced a range of its truncated variants (Figure 3A). In M5T1 and M5T2 the respective 5′-dinucleotide or most of the variable loop spanning region were deleted. The M5T3 was lacking most of its 3′ anticodon loop complementary region, while M5T4 was entirely devoid of the anticodon arm spanning part. By monitoring the eGFP fluorescence in the translation reaction assay we found that the removal of most of the variable loop complementary fragment (M5T2) or the fragment spanning the anticodon arm (M5T4) led to a dramatic reduction in tRNA targeting efficiency (Figure 3B). Truncation of only two bases from the 5′ of M5-1 (M5T1) had a moderate effect with IC90 increasing from 2 to >8 μM (compare Figures 2B and 3B) while the removal of 3′-terminal region matching the anticodon loop (M5T3) did not cause any significant change suggesting its functional redundancy.
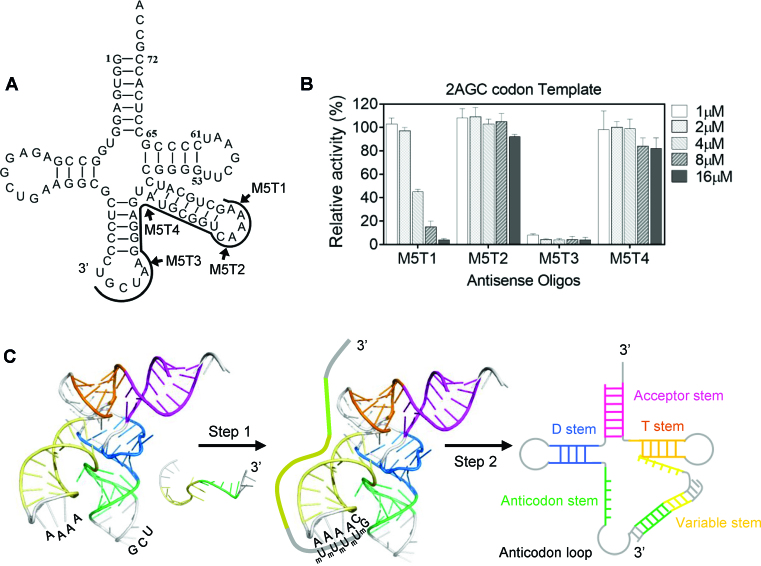
Figure 3.
Truncation analysis of M5-1 oligonucleotides. (A) Schematic representation of the M5-1 truncations. The black line shows the region of tRNASerGCU targeted by M5-1 oligonucleotide. M5T1 and M5T2 are truncated from 5′-end while M5T3 and M5T4 are truncated from 3′-end (indicated in the figure by the arrows). (B) Translational activity of 2AGC-codon eGFP-coding template in antisense oligo-treated E. coli lysate. The used concentrations of the oligonucleotides are coded by increasing shading of the graphs. The error bars represent standard deviations of at least two independent experiments. (C) Proposed mechanism of antisense oligonucleotide: tRNA interaction. The crystal structure of a tRNASec (PDB: 3W3S) was used to represent the E. coli tRNASerGCU. The loop regions are shown in grey while the stems are in colour. The nucleotides in the variable loop and the anticodon loop that are expected to be solution-exposed are marked. The antisense oligonucleotide is shown as an unstructured single-stranded sequence. The initial binding (Step 1) of the methylated antisense oligonucleotide to tRNASerGCU is proposed to initiate at the four consecutive adenosines of the variable loop facilitating duplex propagation (Step 2).
In order to construct a physically meaningful tRNA: oligonucleotide interaction model we examined the crystal structure of tRNASerGGA from Thermus thermophilus (PDB: 1Ser) that was the closest homologue of E. coli tRNASerGCU (47). In this structure the part of variable loop was invisible indicating its dynamic nature while in the related structure of tRNASec (PDB: 3W3S) (48) the variable tetraloop adopted a classical U-turn with three adenosine bases stacked and exposed to the solvent (Supplementary Figure S3). Based on the comparative analysis of these related structures, we conjectured that the respective adenosines in the variable loop of E. coli tRNASerGCU would be also stacked providing a pre-formed anchoring point for docking of the antisense oligonucleotide.
We hypothesized that the complex formation between the antisense oligonucleotide and tRNA under physiological conditions was hindered by the high activation barrier. The successful transition through the major free-energy maxima would depend on the likelihood of elementary rearrangements at initial oligonucleotide anchoring step and following duplex propagation through the tRNA stem and/or tertiary structure (Figure 3C). Our data stresses the importance of the initial anchoring event showing that deletion of only two nucleotides from the 5′-terminus of the antisense oligonucleotide affected the formation of inhibitory complex while further truncation of the successive uridines completely abrogated its binding activity. Therefore, four consecutive adenosine residues in the variable loop of tRNA are likely to serve as a platform for initial anchoring of the oligonucleotide on tRNA similar to that observed in group I introns (49). In turn, 2′OMe residues in the antisense oligonucleotide are expected to accelerate heteroduplex propagation into the tRNA stem by shifting the elementary equilibriums toward the hybrid at each successive base-pairing step. The truncated oligonucleotide limited to targeting the variable arm was unable to abrogate tRNA function and additional complementarity to the 3′-part of the anticodon stem was necessary to gain the activity. This may be a consequence of insufficient destabilization of the overall tRNA tertiary structure that is required for efficient heteroduplex propagation.
In summary, these results demonstrate that modified antisense oligonucleotides can potently and selectively deactivate the native E. coli tRNAs of choice directly in the in vitro translation system. While the oligonucleotide design algorithm and choice of the optimal nucleotide chemistry call for further development, the approach is sufficiently simple and expected to suit many applications.
Using oligonucleotide-mediated tRNA inactivation for sense codon reassignment in vitro
The ability of the designed antisense oligonucleotides to selectively inactivate the endogenous tRNAs in the E. coli lysate prompted us to test the suitability of this approach for site-selective incorporation of uAA into polypeptides. Of the two liberated serine codons the AGU codon was chosen for reassignment since its natural decoding by tRNASerGCU relies on wobble-pairing reducing its productive selection on ribosome (50). This was exploited to minimize the background interference from the residual trace amounts of unsequestered tRNASerGCU in the oligonucleotide-treated lysate (data not shown) by utilizing o-tRNA with non-native ACU anticodon perfectly matching AGU for incorporation of uAA. To test this, we redesigned the eGFP ORF to include a unique AGU codon at position 21 (Supplementary Table S2) and attempted its reassignment to n-propargyl-l-lysine and p-azido-l-phenylalanine by modifying the anticodon of orthogonal tRNA in the o-tRNACUA/aminoacyl-tRNA synthetase (aaRS) pairs previously established for these uAAs (Figure 4A) (5). To avoid possible recognition of split arginine codons AGG/A by ACU-harbouring o-tRNA we replaced all instances of these rare codons in the template with synonymous CGN. In exceptional cases, when AGG/A were present in the ORF we resorted to the use of o-tRNA carrying GCU anticodon which would specifically decode C/U-ending but not A/G-ending codons thereby ensuring the translation fidelity.
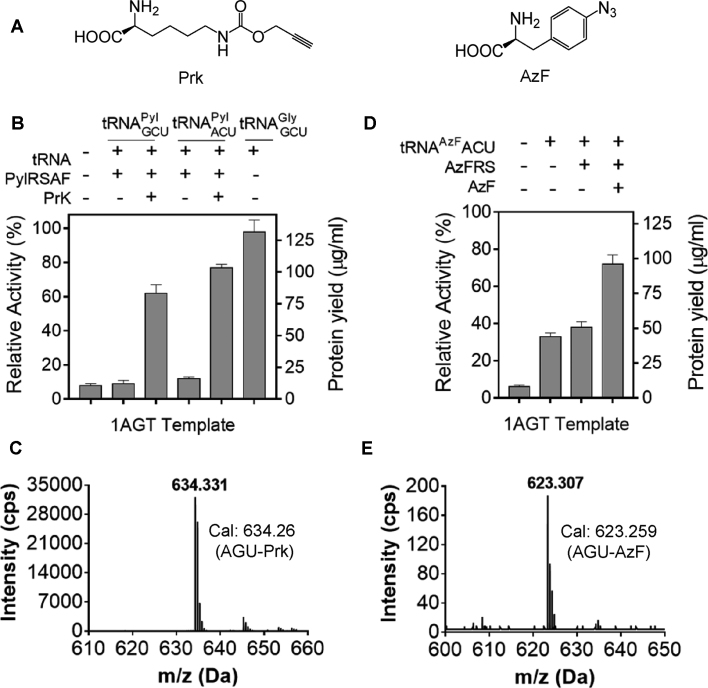
Figure 4.
Reassignment of AGU codon (Ser) in the open reading frame of eGFP to n-propargyl-l-lysine and p-azido-L-phenylalanine. (A) Structures of n-propargyl-l-lysine (Prk) and p-azido-l-phenylalanine (AzF). (B) Reassignment of AGU codon to Prk using eGFP-coding template with single AGT. The bar chart shows relative fluorescence of the M5-1 oligonucleotide treated lysate in the presence or absence of 10 μM of tRNAPyl, 30 μM of PylRSAF as well as 1 mM PrK. To confirm AGU codon suppression specificity tRNAGlyGCU was used as a positive control. The eGFP fluorescence produced in parental lysate was used to calculate the relative translation activity. The protein yield was quantified using dilutions of the purified eGFP as a standard. (C) LC–MS/MS analysis of Prk incorporation via AGU codon in eGFP using tRNAPylACU, PylRSAF and PrK. The mass of the detected peptide of 634.33 Da matches the calculated mass for PrK-modified peptide (634.26 Da). (D) Reassignment of AGU codon to AzF using eGFP template from (B). The translational activity of the M5-1 oligonucleotide treated lysate were measured in the presence or absence of 10 μM of tRNAAzFACU, AzFRS and 1 mM AzF. The error bars in B and D represent standard deviations of three independent experiments. (E) LC–MS/MS analysis of AzF incorporation at AGU codon of the eGFP ORF using tRNAAzFACU, AzFRS and AzF. The detected mass of 623.31 Da matches the predicted mass of 623.26 Da of AzF-modified peptide.
Reassignment of AGU codon (Ser) to n-propargyl-L-lysine (PrK)
To reassign AGU codon to PrK we used the pyrrolysine (Pyl) system (5) consisting of tRNAPyl (Supplementary Table S2), pyrrolysyl-tRNA synthetase variant (PylRSAF) and Prk. As seen in Figure 4B, M5-1-treated lysate displayed 8% of residual translation efficiency when programmed with the DNA template harboring a unique AGT codon thereby indicating an efficient inactivation of native tRNASerGCU isoacceptor. Supplementation of the reaction with only PylRSAF and tRNAPyl did not result in the synthesis of the eGFP reporter indicating their orthogonality to E. coli translation system while PrK addition into the reaction containing tRNAPylACU/PylRS could restore the translation to 80% indicating that ∼90% of protein was modified with uAA. The expression yield of the modified protein reached around ∼100 μg/ml. The resulting eGFP protein was purified, trypsin digested and analyzed by LC–MS/MS confirming that Prk efficiently replaced serine at AGU codon (Figure 4C) as no Ser-containing peptides were detected.
Reassignment of AGU codon (Ser) to p-azido-l-phenylalanine (AzF)
Next we attempted to reassign AGU codon to AzF using tRNATyr derivative (5) with grafted ACU anticodon (tRNAAzFACU) (Supplementary Table S2) and AzF-tRNA synthetase (AzFRS) derived from tyrosyl-tRNA synthetase of Methanocaldococcus jannaschii (Mj) (51) (Figure 4D). When M5-1-treated lysate was supplemented with only tRNAAzFACU ∼35% of eGFP translation was recovered indicating its low orthogonality to E. coli system. Further addition of AzF and AzFRS increased the eGFP expression level to ∼70% indicating occurrence of sense codon suppression via incorporation of AzF at AGU codon. This was confirmed by the subsequent LC-MS/MS analysis of purified eGFP (Figure 4E).
We conclude that the developed approach can be used to incorporate Prk and AzF into peptides and proteins providing bioorthogonal handles for further chemical and biochemical manipulation with the resulting polypeptides.
Applying oligonucleotide-mediated tRNA inactivation to eukaryotic cell-free translation system
The diversity among eukaryotic tRNA isodecoders (having the same anticodon but different sequences elsewhere in the body) varies significantly (52). For instance, more than 400 tRNA genes in human and 275 in yeast genome produce 274 and 51 different tRNA species, respectively. The genome of Leishmania, a unicellular eukaryote, comprises 82 genes coding for 48 tRNA species (53). Dependent on sequence similarity within the given subset of isodecoders, it may be necessary to design more than one antisense oligonucleotides targeting multiple isodecoders in order to free the chosen codon for reassignment.
Physical targeting of tRNAs with antisense oligonucleotides in the in vitro translation reaction is expected to be broadly applicable owing to conservative nature of tRNA structural arrangement. To test this experimentally we made use of Leishmania tarentolae extract (LTE) -based cell-free translation system developed in our laboratory (27). This system was shown to perform on par with human HeLa system in its ability to produce multi-domain human proteins in folded states (54). The ability to incorporate uAAs into such proteins would be very advantageous for their analysis or endowment with enhanced or novel activities. As in the above described case we chose to target tRNASerGCU which is represented by two isodecoders in Leishmania differing only at position 6. The high similarity makes depletion of these isodecoders straightforward by using unique antisense oligonucleotide targeting common sequence elements.
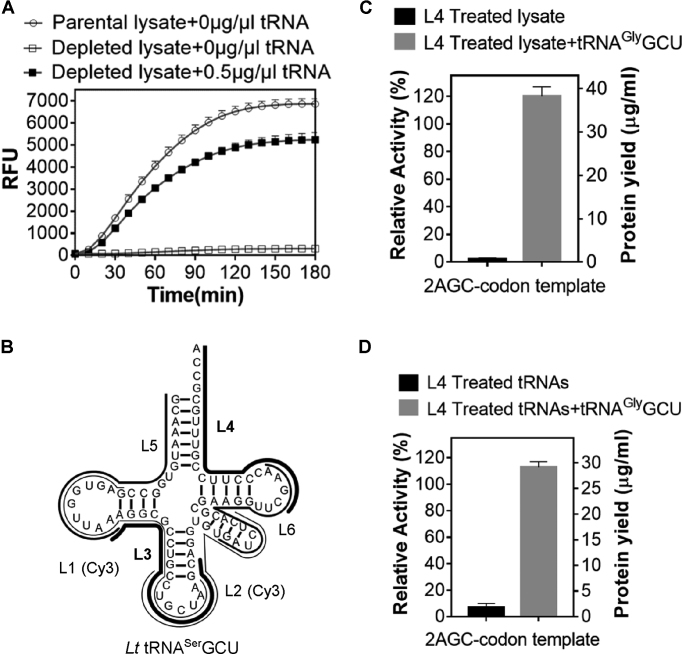
As LTE is much less characterized and more prone to inactivation at prolonged incubation than the E. coli cell-free system we initially sought to devise a ‘hybrid procedure’ that would allow manipulating tRNAs composition and structure without affecting the rest of the translational machinery. Given this, we prepared a tRNA-depleted LTE by chromatography on ethanolamine matrix which unspecifically binds total tRNA fraction at certain ionic strength (22). As expected, translational activity of such lysate was tRNA-dependent (Figure 5A). We then manually designed L1 and L2 2′OMe-oligonucleotides that target D-/anticodon loop and anticodon/variable loop regions of L. tarentolae tRNASerGCU. L3 oligonucleotide targeted the 3′-part of the D-loop containing several consecutive adenosines while L4 was complementary to the 3′-part of the tRNA (Figure 5B). Oligonucleotides L5 and L6 were designed using an online RNA design tool, OligoWalk, which utilized hybridization thermodynamics to predict the best interfering or antisense binders against the target RNA of unkown structure (55) (Supplementary Figure S4). All oligonucleotides were synthesized in fully-methylated form with L1 and L2 also carrying Cy3 fluorophore at their 5′-termini (Figure 5B). The oligonucleotides were tested for their ability to inactivate L. tarentolae tRNASerGCU in the context of the unfractionated native tRNAs. Following incubation at 37°C for 30 min the oligonucleotide/total tRNA mixtures and all tRNA-depleted LTE were used to reconstitute the in vitro translation reaction programmed with eGFP template carrying two consecutive AGC codons. L4 was found to be most effective at sequestering tRNASerGCU as judged by the reduction in the fluorescence yield to less than 10% of control level. The fluorescence could be efficiently recovered upon addition of the chimeric L. tarentolae tRNAGlyCCC with grafted GCU anticodon (tRNAGlyGCU) (Figure 5C).
Figure 5.
2′OMe antisense oligonucleotide mediated inactivation of tRNASerGCU in the eukaryotic cell-free translation system based on L. tarentolae extract (LTE). (A) Monitoring of eGFP production in the total tRNA-depleted LTE translation system upon its supplementation with the indicated amount of the total L. tarentolae tRNA. (B) The schematic representation of the 2′OMe antisense oligos to L. tarentolae tRNASerGCU (L1–L6). (C) Inactivation of tRNASerGCU by antisense oligonucleotides in the context of total L. tarentolae tRNA. The resulting tRNA mixture and all tRNA-depleted LTE was used for reconstitution of the L. tarentolae in vitro translation system programmed by 2AGC-codon eGFP-coding template. The final concentration of the antisense oligonucleotide in the translation reaction was 10 μM. (D) 2′OMe antisense oligonucleotide mediated inactivation of L. tarentolae tRNASerGCU in LTE lysate. The LTE lysate was incubated with L4 oligo at 37°C for 5 min and then used for translation of 2AGC-codon eGFP-coding template. The final concentration of the antisense oligonucleotide in the translation reaction was 15 μM. In all experiments tRNAGlyGCU-suppressor was added to 20 μM final concentration. The eGFP fluorescence produced by translation reactions supplemented with untreated tRNA mixture or a parental lysate shown in C and D, respectively, were used to calculate the relative translation activity. The protein yield was quantified using purified recombinant eGFP. The error bars in A, C and D represent standard deviations of at least two independent experiments.
We subsequently added the L4 oligonucleotide directly to the LTE lysate and observed significant translational inhibition (Figure 5D) which could also be rescued by L. tarentolae tRNAGlyGCU (Figure 5D) confirming the selective inactivation of the native tRNASerGCU. These experiments demonstrate that once hybridized the developed oligonucleotides can efficiently block translation at the chosen codon validating the general applicability of the 2′OMe-oligonucleotide-mediated tRNA sequestration approach.
DISCUSSION
In this study, we present a new approach for sense codon reassignment in the in vitro translation systems. The approach is based on the idea that individual tRNA isoacceptors can be selectively inactivated by the antisense oligonucleotides in the context of in vitro translation system and replaced with their synthetic orthogonal analogs carrying unnatural amino acids (uAA).
We demonstrate that the combination of rational design and nucleotide modifications allows us to create oligonucleotide M5-1 that binds E. coli tRNASerGCU with subnanomolar affinity while showing no detectable binding with its closest homologue tRNASerGGA. The equilibrium and kinetic analysis of the M5-1 interaction with its target tRNA revealed subnanomolar affinity with a very slow off-rate. These parameters enable selective tRNA inactivation in the translation system by promoting a long lived tRNA: oligonucleotide complex which prevents recruitment of the tRNA by translational machinery. The relatively low association rate of M5-1 and tRNASerGCU at 5.8 × 105 M−1 s−1 could be compensated by the use of high oligonucleotide concentrations that would accelerate complex formation. This enables oligonucleotide deployment directly in the E. coli S30 lysate with no observable off-target effects. We confirm the general applicability of this approach by creating inhibitory oligonucleotides against E. coli tRNAArgCCU. The best oligonucleotide design targets the sequence from the anticodon stem to the beginning of T-loop in tRNAArgCCU.
The truncation analysis allowed us to propose a mechanism for oligonucleotide-mediated tRNASerGCU sequestration. According to our model the single-stranded oligonucleotide initially anchors to the solvent-facing stack of nucleotides in the tRNA loop. Further propagation of heteroduplex from the loop into the double stranded and higher order structure-constrained parts leads to tRNA unfolding. The heteroduplex stabilized by modifications in the oligonucleotide traps the tRNA impeding its return to the native conformation. Importantly, the appropriately designed oligonucleotides were able to suppress over 90% of protein production that could be restored to nearly original levels with synthetic tRNA suppressors.
In order to achieve robust uAA incorporation, the heterogeneous tRNA with corresponding anticodon should be orthogonal to the host translation machinery and efficiently charged with uAA by orthogonal aaRS. We showed that using AGU codon instead of AGC for reassignment helped to minimize the background competition from the trace amount of native tRNASerGCU which evaded sequestration. We demonstrated that tRNAPylACU based on previously established tRNAPylCUA amber suppressor fully retained the orthogonality and read AGU codon more efficiently than its variant with GCU anticodon (Figure 4B). The n-propargyl-L-lysine (PrK) charged tRNAPylACU could support AGU codon suppression in the oligonucleotide-treated lysate reaching ∼80% suppression efficiency of untreated lysate. The resulting protein was homogeneously modified with Prk, as confirmed by the LC-MS/MS analysis, indicating high efficiency of sense codon reassignment. Further experiments demonstrated that ACU anticodon transplantation onto the orthogonal amber suppressor tRNA from M. jannaschii (o-tRNAAzFACU) led to its unspecific recognition by the endogenous E. coli aaRS. However, supplementation of p-azido-l-phenylalanine (AzF) and p-azido-L-phenylalanine-tRNA synthetase (AzFRS) to the reaction containing o-tRNAAzFACU enabled efficient AzF incorporation at AGU codon indicating that high concentration of AzF and AzFRS was able to shift the balance toward uAA charging by outcompeting the o-tRNA aminoacylation by the endogenous aaRS (56,57). In these experiments, we replaced all split arginine codons AGG/AGA in the template with synonymous CGN to avoid possible recognition of the former codons by A34-harbouring o-tRNA.
The yield of PrK-containing protein in the presented cell-free translation system reached 100 μg/ml. This amount is suitable for many analytical applications. For instance, Prk and AzF could be further conjugated to fluorophores to create probes for spectroscopic analysis (15), such as monitoring protein conformational dynamics by single-molecule FRET (5). It could also be used to generate proteins with site-specific post-translational modifications (phosphorylation, glycosylation, ubiquitination, lipidation etc.) (3) or produce cyclized peptides by co-incorporating AzF or chloroacetyl group (58). This codon reassignment platform can be used in conjunction with mRNA display or other display systems enabling identification of peptides and proteins with novel binding properties (59). The use of continuous exchange expression format is expected to scale-up the yields to milligrams per ml (14) which would allow the preparative production of protein probes for methods relying on large sample quantities (such as NMR spectroscopy). Scalability of cell-free translation opens up another avenue for presented technology to be used for production of antibody-drug conjugates and modified virus-like particles (18).
We subsequently applied the developed approach for inactivation of L. tarentolae tRNASerGCU in the context of total tRNA mixture and directly in L. tarentolae cell extract. The designed antisense oligonucleotides were effective in both contexts thereby liberating AGC/AGU codons. This demonstrates that the developed approach is not restricted to a particular cell-free system and is likely generally applicable. Yet, further work on optimization of orthogonal tRNA/aaRS pairs in LTE system might be required to reassign the liberated codons to various uAAs.
One of the important considerations for antisense oligonucleotide design is the tRNA modifications, which can indirectly affect heteroduplex formation by increasing the overall stability of tRNA (60–62) or directly perturb the Watson-Crick base-pairing with the oligonucleotide if present within the oligonucleotide targeted region of tRNA (63,64). For example, M5-1 oligonucleotide and its truncated variant (M5T3) lacking 3′-extension complementary to A37-G34 part of the anticodon loop of E. coli tRNASerGCU both showed similar activities consistently with the presence of duplex-destabilizing N6-threonylcarbamoyl modification at A37 (63) (Figure 3). Surprisingly, 3′-extension of L4 oligonucleotide (Figure 5B) to further complement the two consecutive modified uridines (5′-methyluridine and pseudouridine at positions 54 and 55, respectively) in the T-loop of Leishmania tRNASerGCU decreased its ability to form a complex with the tRNA (data not shown) despite these modifications demonstrating a stabilizing effect on the respective base-pairs (65,66). Therefore, the effect of tRNA modifications on stability and kinetics of complex formation with antisense oligonucleotide can not be always predicted while optimization of tRNA: oligonucleotide hybridization conditions, such as Mg2+ concentration, incubation time and temperature, would be required to promote heteroduplex formation especially in the case of heavily modified eukaryotic tRNAs. For instance, 5 min incubation of L4 oligonucleotide with Leishmania cell extract at 27°C was not enough to effectively sequester tRNASerGCU (data not shown) and increasing the incubation temperature to 37°C was required to achieve significant translation inhibition (Figure 5D).
The best antisense oligonucleotides identified for E. coli tRNASerGCU, E. coli tRNAArgCCU, L. tarentolae tRNASerGCU do not complement the same sequence motifs in their tRNA targets. The anticodon and variable loops as well as the CCA tail are likely to present the most accessible single-stranded regions in these tRNAs for the oligonucleotides to anchor and further merge into the tRNA structure. We assume that 2′-O-methylated residues in the antisense oligonucleotide accelerate heteroduplex propagation finally locking the denatured tRNA in a stable complex. Rational design of oligonucleotides needs to be combined with the functional screening to identify the best performing variants. It is worth noting that the algorithm-based oligonucleotide design implemented in the OligoWalk software failed to produce efficient tRNA binders. Future systematic analysis of 12–30 nts RNA oligonucleotide arrays spanning the entire tRNAs appear to be a promising strategy for identifying the most effective oligonucleotides and establishing their design criteria. Besides 2′-O-methylation, exploration of other oligonucleotide chemistries such as Locked Nucleic Acid (LNA), phosphorodiamidate morpholino oligomers (PMOs) as well as phosphorothioate derivatives (32,67,68) with a goal of increasing the heteroduplex stability and preventing their nucleolytic degradation represents the future strategy for designing of even more efficient antisense oligonucleotides to other tRNA species.
In conclusion, the approach reported here stands out in its simplicity as it only requires a short incubation of the cell lysate with oligonucleotides before assembly of the translation reaction. The approach provides a new avenue for sense codon reassignment and uAA incorporation into recombinant polypeptides. Unlike previously reported approaches, this method is chromatography-independent and fully scalable, potentially allowing its industrial applications. This opens up a route for creation of peptides and proteins with novel properties, such as drug-antibody conjugates, bioactive peptides with improved bioavailability, synthetic vaccines as well as enzymes with enhanced or novel catalytic activities.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jason Whitfield for critical reading of the manuscript. We thank Alun Jones for his help on performing the LC–MS/MS experiment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Australian Research Council [LP150100689 to K.A.]; National Health and Medical Research Council [APP1037320 to K.A.]. Funding for open access charge: Australian Research Council (to K.A.).
Conflict of interest statement. The authors declare the following competing financial interest(s): K.A. holds equity in Molecular Warehouse ltd which commercially exploits sense codon reassignment technology.
REFERENCES
- 1. Liu C.C., Schultz P.G.. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010; 79:413–444. [DOI] [PubMed] [Google Scholar]
- 2. O’Donoghue P., Ling J., Wang Y.S., Soll D.. Upgrading protein synthesis for synthetic biology. Nat. Chem. Biol. 2013; 9:594–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dumas A., Lercher L., Spicer C.D., Davis B.G.. Designing logical codon reassignment – expanding the chemistry in biology. Chem. Sci. 2015; 6:50–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mukai T., Yamaguchi A., Ohtake K., Takahashi M., Hayashi A., Iraha F., Kira S., Yanagisawa T., Yokoyama S., Hoshi H. et al. Reassignment of a rare sense codon to a non-canonical amino acid in Escherichia coli. Nucleic Acids Res. 2015; 43:8111–8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cui Z., Mureev S., Polinkovsky M.E., Tnimov Z., Guo Z., Durek T., Jones A., Alexandrov K.. Combining sense and nonsense codon reassignment for Site-Selective protein modification with unnatural amino acids. ACS Synth. Biol. 2017; 6:535–544. [DOI] [PubMed] [Google Scholar]
- 6. Iwane Y., Hitomi A., Murakami H., Katoh T., Goto Y., Suga H.. Expanding the amino acid repertoire of ribosomal polypeptide synthesis via the artificial division of codon boxes. Nat. Chem. 2016; 8:317–325. [DOI] [PubMed] [Google Scholar]
- 7. Zeng Y., Wang W., Liu W.R.. Towards reassigning the rare AGG codon in Escherichia coli. Chembiochem. 2014; 15:1750–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee B.S., Shin S., Jeon J.Y., Jang K.S., Lee B.Y., Choi S., Yoo T.H.. Incorporation of unnatural amino acids in response to the AGG codon. ACS Chem. Biol. 2015; 10:1648–1653. [DOI] [PubMed] [Google Scholar]
- 9. Ostrov N., Landon M., Guell M., Kuznetsov G., Teramoto J., Cervantes N., Zhou M., Singh K., Napolitano M.G., Moosburner M. et al. Design, synthesis, and testing toward a 57-codon genome. Science. 2016; 353:819–822. [DOI] [PubMed] [Google Scholar]
- 10. Phizicky E.M., Hopper A.K.. tRNA biology charges to the front. Gene Dev. 2010; 24:1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J.W.. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010; 464:441–444. [DOI] [PubMed] [Google Scholar]
- 12. Malyshev D.A., Dhami K., Lavergne T., Chen T., Dai N., Foster J.M., Correa I.R. Jr, Romesberg F.E.. A semi-synthetic organism with an expanded genetic alphabet. Nature. 2014; 509:385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malyshev D.A., Romesberg F.E.. The expanded genetic alphabet. Angew. Chem. Int. Ed. Engl. 2015; 54:11930–11944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schwarz D., Junge F., Durst F., Frolich N., Schneider B., Reckel S., Sobhanifar S., Dotsch V., Bernhard F.. Preparative scale expression of membrane proteins in Escherichia coli-based continuous exchange cell-free systems. Nat. Protoc. 2007; 2:2945–2957. [DOI] [PubMed] [Google Scholar]
- 15. Quast R.B., Mrusek D., Hoffmeister C., Sonnabend A., Kubick S.. Cotranslational incorporation of non-standard amino acids using cell-free protein synthesis. FEBS Lett. 2015; 589:1703–1712. [DOI] [PubMed] [Google Scholar]
- 16. Loscha K.V., Herlt A.J., Qi R., Huber T., Ozawa K., Otting G.. Multiple-site labeling of proteins with unnatural amino acids. Angew. Chem. Int. Ed. Engl. 2012; 51:2243–2246. [DOI] [PubMed] [Google Scholar]
- 17. Carlson E.D., Gan R., Hodgman C.E., Jewett M.C.. Cell-free protein synthesis: applications come of age. Biotechnol. Adv. 2012; 30:1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hong S.H., Kwon Y.C., Jewett M.C.. Non-standard amino acid incorporation into proteins using Escherichia coli cell-free protein synthesis. Front. Chem. 2014; 2:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanda T., Takai K., Yokoyama S., Takaku H.. An easy cell-free protein synthesis system dependent on the addition of crude Escherichia coli tRNA. J. Biochem. 2000; 127:37–41. [DOI] [PubMed] [Google Scholar]
- 20. Salehi A.S.M., Smith M.T., Schinn S.M., Hunt J.M., Muhlestein C., Diray-Arce J., Nielsen B.L., Bundy B.C.. Efficient tRNA degradation and quantification in Escherichia coli cell extract using RNase-coated magnetic beads: A key step toward codon emancipation. Biotechnol. Prog. 2017; 33:1401–1407. [DOI] [PubMed] [Google Scholar]
- 21. Frankel A., Roberts R.W.. In vitro selection for sense codon suppression. RNA. 2003; 9:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui Z., Stein V., Tnimov Z., Mureev S., Alexandrov K.. Semisynthetic tRNA complement mediates in vitro protein synthesis. J. Am. Chem. Soc. 2015; 137:4404–4413. [DOI] [PubMed] [Google Scholar]
- 23. Ahn J.H., Hwang M.Y., Oh I.S., Park K.M., Hahn G.H., Cho C.Y., Kim D.M.. Preparation method for Escherichia coliS30 extracts completely dependent upon tRNA addition to catalyze cell-free protein synthesis. Biotechnol. Bioproc. E. 2006; 11:420–424. [Google Scholar]
- 24. Kanda T., Takai K., Hohsaka T., Sisido M., Takaku H.. Sense codon-dependent introduction of unnatural amino acids into multiple sites of a protein. Biochem. Biophys. Res. Commun. 2000; 270:1136–1139. [DOI] [PubMed] [Google Scholar]
- 25. Thisted T., Sorensen N.S., Wagner E.G., Gerdes K.. Mechanism of post-segregational killing: Sok antisense RNA interacts with Hok mRNA via its 5′-end single-stranded leader and competes with the 3′-end of Hok mRNA for binding to the mok translational initiation region. EMBO J. 1994; 13:1960–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kubala M.H., Kovtun O., Alexandrov K., Collins B.M.. Structural and thermodynamic analysis of the GFP:GFP-nanobody complex. Protein Sci. 2010; 19:2389–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mureev S., Kovtun O., Nguyen U.T., Alexandrov K.. Species-independent translational leaders facilitate cell-free expression. Nat. Biotechnol. 2009; 27:747–752. [DOI] [PubMed] [Google Scholar]
- 28. Zubay G. Isolation and fractionation of soluble ribonucleic acid. J. Mol. Biol. 1962; 4:347–356. [Google Scholar]
- 29. Bennett C.F., Swayze E.E.. RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform. Annu. Rev. Pharmacol. Toxicol. 2010; 50:259–293. [DOI] [PubMed] [Google Scholar]
- 30. Rasmussen L.C., Sperling-Petersen H.U., Mortensen K.K.. Hitting bacteria at the heart of the central dogma: sequence-specific inhibition. Microb. Cell Fact. 2007; 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Khorkova O., Wahlestedt C.. Oligonucleotide therapies for disorders of the nervous system. Nat. Biotechnol. 2017; 35:249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lundin K.E., Gissberg O., Smith C.I.. Oligonucleotide therapies: the past and the present. Hum. Gene Ther. 2015; 26:475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sczakiel G., Far R.K.. The role of target accessibility for antisense inhibition. Curr. Opin. Mol. Ther. 2002; 4:149–153. [PubMed] [Google Scholar]
- 34. Giege R., Juhling F., Putz J., Stadler P., Sauter C., Florentz C.. Structure of transfer RNAs: similarity and variability. Wiley Interdiscip. Rev. RNA. 2012; 3:37–61. [DOI] [PubMed] [Google Scholar]
- 35. Jerabek-Willemsen M., Andre T., Wanner R., Roth H.M., Duhr S., Baaske P., Breitsprecher D.. MicroScale Thermophoresis: Interaction analysis and beyond. J. Mol. Struct. 2014; 1077:101–113. [Google Scholar]
- 36. Lesnik E.A., Freier S.M.. Relative thermodynamic stability of DNA, RNA, and DNA:RNA hybrid duplexes: relationship with base composition and structure. Biochemistry. 1995; 34:10807–10815. [DOI] [PubMed] [Google Scholar]
- 37. Majlessi M., Nelson N.C., Becker M.M.. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 1998; 26:2224–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ecker D.J., Vickers T.A., Bruice T.W., Freier S.M., Jenison R.D., Manoharan M., Zounes M.. Pseudo–half-knot formation with RNA. Science. 1992; 257:958–961. [DOI] [PubMed] [Google Scholar]
- 39. Inoue H., Hayase Y., Imura A., Iwai S., Miura K., Ohtsuka E.. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987; 15:6131–6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yildirim I., Kierzek E., Kierzek R., Schatz G.C.. Interplay of LNA and 2′-O-methyl RNA in the structure and thermodynamics of RNA hybrid systems: a molecular dynamics study using the revised AMBER force field and comparison with experimental results. J. Phys. Chem. B. 2014; 118:14177–14187. [DOI] [PubMed] [Google Scholar]
- 41. Spiriti J., Binder J.K., Levitus M., van der Vaart A.. Cy3-DNA stacking interactions strongly depend on the identity of the terminal basepair. Biophys. J. 2011; 100:1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milas P., Gamari B.D., Parrot L., Krueger B.P., Rahmanseresht S., Moore J., Goldner L.S.. Indocyanine dyes approach free rotation at the 3′ terminus of A-RNA: a comparison with the 5′ terminus and consequences for fluorescence resonance energy transfer. J. Phys. Chem. B. 2013; 117:8649–8658. [DOI] [PubMed] [Google Scholar]
- 43. Pan A.C., Borhani D.W., Dror R.O., Shaw D.E.. Molecular determinants of drug-receptor binding kinetics. Drug Discov. Today. 2013; 18:667–673. [DOI] [PubMed] [Google Scholar]
- 44. Copeland R.A., Pompliano D.L., Meek T.D.. Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 2006; 5:730–739. [DOI] [PubMed] [Google Scholar]
- 45. Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S.M., McGee D., Guinosso C.J., Cook P.D.. Characterization of fully 2′-modified oligoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995; 23:2019–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murphy F.V.T., Ramakrishnan V., Malkiewicz A., Agris P.F.. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004; 11:1186–1191. [DOI] [PubMed] [Google Scholar]
- 47. Biou V., Yaremchuk A., Tukalo M., Cusack S.. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994; 263:1404–1410. [DOI] [PubMed] [Google Scholar]
- 48. Itoh Y., Sekine S., Suetsugu S., Yokoyama S.. Tertiary structure of bacterial selenocysteine tRNA. Nucleic Acids Res. 2013; 41:6729–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gate J.H., Gooding A.R., Podell E., Zhou K.H., Golden B.L., Szewczak A.A., Kundrot C.E., Cech T.R., Doudna J.A.. RNA tertiary structure mediation by adenosine platforms. Science. 1996; 273:1696–1699. [DOI] [PubMed] [Google Scholar]
- 50. Ledoux S., Olejniczak M., Uhlenbeck O.C.. A sequence element that tunes Escherichia coli tRNA(Ala)(GGC)to ensure accurate decoding. Nat Struct Mol Biol. 2009; 16:359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chin J.W., Santoro S.W., Martin A.B., King D.S., Wang L., Schultz P.G.. Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002; 124:9026–9027. [DOI] [PubMed] [Google Scholar]
- 52. Goodenbour J.M., Pan T.. Diversity of tRNA genes in eukaryotes. Nucleic Acids Res. 2006; 34:6137–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chan P.P., Lowe T.M.. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2016; 44:D184–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gagoski D., Polinkovsky M.E., Mureev S., Kunert A., Johnston W., Gambin Y., Alexandrov K.. Performance benchmarking of four cell-free protein expression systems. Biotechnol. Bioeng. 2016; 113:292–300. [DOI] [PubMed] [Google Scholar]
- 55. Lu Z.J., Mathews D.H.. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008; 36:W104–W108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sherman J.M., Rogers M.J., Soll D.. Competition of aminoacyl-tRNA synthetases for tRNA ensures the accuracy of aminoacylation. Nucleic Acids Res. 1992; 20:2847–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wohlgemuth I., Pohl C., Mittelstaet J., Konevega A.L., Rodnina M.V.. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos. Trans. R. Soc. B. 2011; 366:2979–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Iwasaki K., Goto Y., Katoh T., Suga H.. Selective thioether macrocyclization of peptides having theN-terminal 2-chloroacetyl group and competing two or three cysteine residues intranslation. Org Biomol Chem. 2012; 10:5783–5786. [DOI] [PubMed] [Google Scholar]
- 59. Passioura T., Suga H.. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chem. Commun. (Camb.). 2017; 53:1931–1940. [DOI] [PubMed] [Google Scholar]
- 60. Lorenz C., Lunse C.E., Morl M.. tRNA modifications: Impact on structure and thermal adaptation. Biomolecules. 2017; 7:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Phizicky E.M., Alfonzo J.D.. Do all modifications benefit all tRNAs. FEBS Lett. 2010; 584:265–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 63. Motorin Y., Muller S., Behm-Ansmant I., Branlant C.. Identification of modified residues in RNAs by reverse transcription-based methods. Methods Enzymol. 2007; 425:21–53. [DOI] [PubMed] [Google Scholar]
- 64. Sipa K., Sochacka E., Kazmierczak-Baranska J., Maszewska M., Janicka M., Nowak G., Nawrot B.. Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA. 2007; 13:1301–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kierzek E., Malgowska M., Lisowiec J., Turner D.H., Gdaniec Z., Kierzek R. et al. The contribution of pseudouridine to stabilities andstructure of RNAs. Nucleic Acids Res. 2014; 42:3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Motorin Y, Helm M et al. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011; 2:611–631. [DOI] [PubMed] [Google Scholar]
- 67. Vester B., Wengel J.. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004; 43:13233–13241. [DOI] [PubMed] [Google Scholar]
- 68. Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014; 24:374–387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.