Deafferentation of motor neurons as a result of defective sensory-motor connectivity in the spinal cord is an important early event in spinal muscular atrophy. Shorrock et al. report that this process is reversible in mice and results from disruption of a novel UBA1/GARS pathway that controls sensory neuron fate.
Keywords: motor neuron disease, spinal muscular atrophy, Charcot-Marie-Tooth disease, UBA1, GARS
Abstract
Deafferentation of motor neurons as a result of defective sensory-motor connectivity is a critical early event in the pathogenesis of spinal muscular atrophy, but the underlying molecular pathways remain unknown. We show that restoration of ubiquitin-like modifier-activating enzyme 1 (UBA1) was sufficient to correct sensory-motor connectivity in the spinal cord of mice with spinal muscular atrophy. Aminoacyl-tRNA synthetases, including GARS, were identified as downstream targets of UBA1. Regulation of GARS by UBA1 occurred via a non-canonical pathway independent of ubiquitylation. Dysregulation of UBA1/GARS pathways in spinal muscular atrophy mice disrupted sensory neuron fate, phenocopying GARS-dependent defects associated with Charcot-Marie-Tooth disease. Sensory neuron fate was corrected following restoration of UBA1 expression and UBA1/GARS pathways in spinal muscular atrophy mice. We conclude that defective sensory motor connectivity in spinal muscular atrophy results from perturbations in a UBA1/GARS pathway that modulates sensory neuron fate, thereby highlighting significant molecular and phenotypic overlap between spinal muscular atrophy and Charcot-Marie-Tooth disease.
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive form of motor neuron disease with an incidence of 1 in ∼10 000 live births (Sugarman et al., 2012; Verhaart et al., 2017; Groen et al., 2018b). SMA is characterized primarily by degeneration of lower motor neurons in the anterior horn of the spinal cord. This is accompanied by proximal muscle weakness and atrophy, resulting in a progressive decline in motor function and, in severe cases, paralysis and death (Kolb and Kissel, 2011; Shorrock et al., 2018). Recently, compelling evidence has been put forward from several studies demonstrating that sensory neuron defects are a significant feature of SMA pathogenesis. For example, a reduction in numbers of myelinated dorsal root axons has been reported (Ling et al., 2010), alongside significant reductions in the number of proprioceptive synapses formed onto motor neurons in the spinal cord (Ling et al., 2010; Mentis et al., 2011; Fletcher et al., 2017). The deafferentation of motor neurons as a result of defective sensory-motor connectivity is now known to be a critical early event in the pathogenesis of SMA, where it represents a primary cause of motor neuron dysfunction (Ling et al., 2010; Mentis et al., 2011; Simon et al., 2017). Similarly, it has previously been shown that sensory nerve endings innervating embryonic footpads display defective outgrowth and altered neuron terminal structure in SMA mice (Jablonka et al., 2006). Although defects in sensory neuron outgrowth and altered β-actin protein and mRNA localization have been reported in cultured sensory neurons from SMA mice (Jablonka et al., 2006), the molecular mechanisms underlying defective sensory-motor connectivity in SMA are still to be established.
In ∼95% of cases, SMA is caused by homozygous deletion of the survival motor neuron 1 gene (SMN1), which encodes the survival motor neuron protein (SMN) (Lefebvre et al., 1995). Interestingly, however, not all forms of SMA are caused by deletions or mutations in the SMN1 gene. Mutations in the ubiquitin-like modifier activating enzyme 1 gene (UBA1) cause a rare form of SMA known as X-linked SMA (XL-SMA) (Ramser et al., 2008; Dlamini et al., 2013; Jedrzejowska et al., 2015). UBA1 encodes the primary enzyme, UBA1, responsible for activating ubiquitin as the first step in the ubiquitin-conjugation pathway, to mark proteins for degradation by the proteasome (Groen and Gillingwater, 2015). XL-SMA is clinically similar to SMA and is characterized by loss of lower motor neurons, muscle weakness, hypotonia and a lack of reflexes. In addition, congenital contractures and fractures are also commonly associated with XL-SMA (Ramser et al., 2008; Dlamini et al., 2013; Jedrzejowska et al., 2015). Mutations in UBA1 underlying XL-SMA are predicted to cause instability of the enzyme, leading to a reduction in expression levels of UBA1 (Ramser et al., 2008; Lao et al., 2012; Balak et al., 2017).
Recent findings have suggested that altered ubiquitin homeostasis is a core molecular feature of SMN1-dependent SMA, with reduced UBA1 expression central to this disruption in a range of animal models and in SMA patient-derived iPSC motor neurons (Aghamaleky Sarvestany et al., 2014; Wishart et al., 2014; Fuller et al., 2015; Powis et al., 2016). Suppression or pharmacological inhibition of UBA1 is sufficient to induce an SMA-like neuromuscular phenotype in zebrafish (Wishart et al., 2014) and, similarly, pharmacological inhibition of UBA1 in Schwann cells recapitulates the defective myelination phenotype observed in SMA (Aghamaleky Sarvestany et al., 2014; Hunter et al., 2014). Consistent with this, therapies that restore UBA1 have shown beneficial effects on neuromuscular phenotypes in several SMA models (Powis et al., 2016). SMA mice treated with adeno-associated virus serotype 9 (AAV9) expressing full-length human UBA1 cDNA (AAV9-UBA1) show increased survival compared to untreated SMA mice, along with improvements in systemic (e.g. heart and liver) pathology. SMA mice treated with AAV9-UBA1 also showed a rescue of the number of motor neuron cell bodies in the spinal cord, an increase in muscle fibre diameter, and rescue of NMJ innervation defects compared to untreated SMA mice (Powis et al., 2016).
Because of the wide ranging therapeutic benefits of treatment with AAV9-UBA1 and the relevance of UBA1-mediated degeneration for multiple types of SMA, here, we set out to determine whether UBA1-dependent pathways are responsible for mediating defective sensory-motor connectivity in SMA and to identify UBA1 dependent mediators of degeneration. We report that elevation of UBA1 levels via AAV9 gene therapy in vivo was sufficient to correct sensory-motor connectivity defects in SMA mice, acting via UBA1’s regulation of aminoacyl-tRNA synthetases, including GARS, in sensory neurons.
Materials and methods
Study approvals
All animal studies were approved by the internal ethics committee at the University of Edinburgh and were performed under the authority of relevant project and personal licenses from the UK Home Office.
Animal models and UBA1 overexpression in vivo
Taiwanese SMA mice (Smn−/−; SMN2tg/0) (Hsieh-Li et al., 2000; Riessland et al., 2010), on a congenic FVB background, were originally obtained from Jackson Laboratories (strain no. 005058) and maintained according to established breeding protocols (Riessland et al., 2010). Phenotypically-normal heterozygous (Smn+/−; SMN2tg/0) littermates were used as controls. Wild-type FVB mice were obtained from in-house breeding stocks at the University of Edinburgh. Mice were maintained under standard specific pathogen-free conditions and retrospectively genotyped using standard PCR protocols.
To overexpress UBA1 in vivo, mice were injected with AAV9-UBA1 intravenously on the day of birth (Powis et al., 2016). AAV9-UBA1 (Vigene) was administered at a concentration of 7 × 1013 viral genomes. Mice underwent general chilled anaesthesia and were then injected with 10 µl of AAV9-UBA1 into the facial vein using a Hamilton syringe fitted with a Hamilton 33-gauge RN needle. Litters of mice were randomly assigned to treatment groups. For all subsequent analyses, the observer was blinded to the disease/treatment status of the mice.
Tissue isolation and preparation
Mice were sacrificed by overdose of anaesthetic at postnatal Day 8, a late-symptomatic stage of disease progression. Spinal cord dissection was performed as previously described (Powis and Gillingwater, 2016). For quantitative fluorescent western blot analysis, isolated spinal cords were snap-frozen on dry ice and stored at −80°C. For immunohistochemistry, spinal cords were fixed in 4% paraformaldehyde (PFA) (Electron Microscopy Sciences) for 24 h and cryopreserved in 30% sucrose for 24 h at 4°C. Lumbar regions of spinal cord were embedded in O.C.T. (Cell Path), sectioned at 25 μm on a cryostat, immediately collected onto SuperFrost Plus™ microscope slides (Thermo Scientific) and stored at −20°C.
For immunohistochemistry of lumbar dorsal root ganglia (DRG), spinal columns were dissected from postnatal Day 8 mice, cut transversely at the T13 vertebrae, fixed for 6 h in 4% PFA, and cryopreserved in 30% sucrose for 24 h at 4°C. Lumbar and lower thoracic regions were embedded in O.C.T. and cryosectioned at 12 µm. Using the T13 DRGs, spinal columns were aligned so that the DRGs on each side of the column were symmetrical, sections from L1 and L2 DRGs were collected onto SuperFrost Plus™ microscope slides. For quantitative fluorescent western blot analysis lumbar dorsal root ganglia were dissected out of the spinal column using a previously described method (Sleigh et al., 2016). For each mouse, three DRGs from each half of the lumbar spinal column were dissected so that western blots were performed on six DRGs per mouse.
Cell culture and transfection
HEK293 cells (European Collection of Authenticated Cell Cultures) were grown in high glucose Dulbecco’s modified Eagle medium (Life Technologies) supplemented with 10% heat inactivated foetal bovine serum (FBS; Sigma), penicillin/streptavidin (Invitrogen) and l-glutamine (Invitrogen). HEK293 cells were transfected with RNAiMAX (Invitrogen) and 2.5 µM Silencer® Select Validated UBA1 siRNA (s601, targeted against exons 24 and 25; Life Technologies), or with Lipofectamine™ (Invitrogen) and 3.5 µg pCMV6-XL4-UBA1 plasmid (Origene) for proteomics samples. For proteomics, three separate transfections were combined per sample with three samples per condition. For co-transfection of multiple plasmids, 1.25 µg of each plasmid was used and the same amount of DNA and Lipofectamine™/RNAiMax was added to each well: pCMV6-XL4-UBA1 (Origene), pCMV6-Entry (Origene); pEGFP-GARS-N2 (gift from Dr Antonelli Antonellis), pEGFP-N1 (gift from Prof. Mike Cousin); pcDNA3.1-HA-Ubiquitin (gift from Prof. Yeh), Addgene plasmid #18712 (Kamitani et al., 1997), pcDNA3.1+ (gift from Prof. Mike Cousin); pcDNA3.1-SMN-V5-His. Negative control 2 siRNA (Life Technologies) was used as a control for UBA1 siRNA. For all co-transfections, cells were harvested 48 h after transfection.
Ubiquitylation assay
Following transfection, 10 µM of MG132 proteasome inhibitor was added to HEK293 cells for 90 min. Cells were lysed in RIPA buffer (Fisher Scientific) containing 1% protease inhibitor cocktail (Life Technologies), 1% phosphatase inhibitor (Life Technologies) and 10 mM N-ethylmaleimide (Sigma). The lysate was incubated with 0.5 µg anti-GFP primary antibody for 1 h 20 min at 4°C before incubation with protein A magnetic beads (Life Technologies) for 40 mins at 4°C. Immunoprecipitates were then washed four times in NP40 buffer. Immunoprecipitates and input controls were probed with the indicated antibodies.
Protein extraction and western blotting
Protein was extracted in RIPA buffer with 1% protease inhibitor cocktail, homogenized using a motorized disposable pestle mixer. Protein concentration was determined using a BCA assay (ThermoScientific). Proteins were separated on a 4–12% gradient gel (Novex) and blotted using PVDF membrane stacks using the iBlot® system (Life Technologies). Western blotting was performed using primary antibodies against SMN (1:1000, BD Transduction Laboratories), UBA1 (1:1000, Thermo, PA5–17274), GARS (1:2500, Abcam, ab42905), YARS (1:1000, Abcam, ab154819), HA (1:1000, CST, 3724P), GFP (1:5000, Abcam, ab290) and the appropriate IRDye® secondary antibodies (LICOR). Tissue and cell-type appropriate loading controls were: histone H3 (1:5000, Abcam, ab1791), CoxIV (1:1000, Abcam, ab14744), α-Tubulin (1:5000, Abcam, ab7291), GAPDH (1: 2500, Abcam, ab9484) and Ponceau S total protein stain. Membranes were scanned on an Odyssey® imager (LICOR; Eaton et al., 2013) and analysed using ImageStudio (LICOR).
Immunohistochemistry
Spinal cord and spinal column sections were permeabilized in 0.3% Triton™ X-100 (Sigma) in PBS for 10 min at room temperature, blocked in 4% BSA, 0.1% Triton™ X-100 in PBS for 1 h and incubated in primary antibodies in a 1:4 dilution of blocking solution at 4°C for 24 h. Slides were washed in PBS and incubated with appropriate Alexa Fluor®-labelled secondary antibodies (1:400; Life Technologies) for 1 h at room temperature and subsequently in DAPI (Life Technologies) for 10 min. For immunohistochemistry of NF200, peripherin, parvalbumin and cleaved caspase 3, spinal column sections were thawed at room temperature for 1 h and permeabilized in 0.3% Triton™ X-100 in PBS (PBST) for 3 × 10 mins at room temperature, and blocked in 10% BSA in PBST for 1 h. Sections were then incubated with primary antibodies in blocking solution, at 4°C for 24 h. Slides were washed and sections incubated with the appropriate Alexa Fluor®-labelled secondary antibodies (1:400; Life Technologies) in PBS for 1 h at room temperature and incubated in DAPI for 10 min. All slides were mounted and coverslipped in a 10% Mowiol® solution (Polyscience). The following primary antibodies were used in this study: GARS, Abcam, ab42905, 1:500; UBA1a, CST, 4890S, 1:200; ChAT, Millipore, AB144P, 1:100; VGlut1, synaptic systems, 135 302, 1:1000; SMI32, Covance, SMI-32R, 1:1000; NF200, Sigma, N0142, 1:500; peripherin, Merck, AB1530, 1:500; parvalbumin, Swant, PV27, 1:1000; βIII-tubulin, Abcam, ab41489, 1:500; cleaved caspase-3, CST, 9661, 1:500.
Imaging and analysis
Imaging was performed on a Nikon A1R FILM at the IMPACT facility, University of Edinburgh, or a Zeiss LSM710 confocal microscope. For distribution analysis, all imaging was performed at constant confocal settings. Z-stack images were taken and individual motor neurons selected for analysis based on an intact nucleus and an intact cell body, as ascertained by DAPI and ChAT, respectively; motor neurons were analysed in the plane where the nuclear outline was most clearly defined. To count the number of VGLUT1 synapses per ChAT-positive motor neuron soma, Z-stack images were taken with a slice interval of 0.2 µm and a pixel size of 0.08 µm throughout the whole section thickness to include the whole cell body (Mentis et al., 2011). Whole DRGs were imaged in a single plane and selected for imaging and subsequent analysis based on structural integrity. Only sections that included a full cross-section through the centre of the DRG were imaged. One left and one right DRG were imaged per lumbar segment from L1 and L2 for each mouse. For distribution analysis of sensory neurons, seven larger area neurons and seven smaller area neurons were analysed per DRG. All example images of DRGs are from the L2 segment. Distribution analysis was performed in Fiji using the XOR and measurement functions (Ferreira and Rasband, 2012) to measure the intensity of the protein of interest in the nucleus and cytoplasm independent of each other (Supplementary Fig. 1) (Zhang et al., 2015). Neuron area was measured by outlining the cell profile (NF200 or peripherin) in Fiji. Sensory neurons were only analysed if a clear nuclear outline was observed. The cell counter plugin in Fiji was used to count NF200- or peripherin-positive neurons (Sleigh et al., 2017), and to count the number of VGLUT1 synapses per ChAT-positive soma.
Label-free proteomics
HEK293 cell pellets were lysed in SDT lysis buffer containing 100 mM Tris-HCl (pH 7.6; Sigma), 4% (w/v) SDS (VWR). Protein concentration was determined using a BCA assay (ThermoScientific). Aliquots (100 μg) of each HEK293 lysate were processed through filter-aided sample preparation involving buffer exchange to 8 M urea and alkylation with 50 mM iodoacetamide. Double digestion with trypsin (Roche, sequencing grade) was then performed, initially for 4 h, then overnight, at 30°C. Samples were desalted by washes with 0.1% trifluoroacetic acid and then 70% acetonitrile. Each sample was separated by injection onto a nanoflow LC-MS/MS Ultimate 3000 RSLC (Thermo Scientific) system coupled to a linear ion trap Orbitrap hybrid mass spectrometer (LTQ Orbitrap Velos Pro, Thermo Scientific) via a nanoelectrospray ion source (Thermo Scientific). Peptides from each digest were separated with a linear gradient of 2–40% acetonitrile, 0.1% formic acid over 124 min with a constant flow of 300 nl/min. Full-scan mass spectrometry (MS) survey spectra were acquired in the LTQ Oribitrap with a resolution of 60 000; this was followed by IT-MS/MS scans for the 15 most intense peptide ions. Data were acquired using Xcalibur™ software. Alongside the nine samples, four quality control samples were processed, each of which consisted of a mixture of the nine samples.
Proteomic data analysis
Raw proteomic data were imported into Progenesis for analysis of relative ion abundance and peptide characterization. The MS/MS output were converted into 2D representations for each sample; these were then aligned to a quality control samples with all alignment scores >90%. The data were subsequently filtered where all ions with a charge >5 were removed, as were features detected <23 min and >137 min. The runs for the different biological replicates of each condition were combined and statistical P-values were automatically generated for the peptides in Progenesis software through a one-way ANOVA on the ArcSinh transform of the normalized data. The peptides were then filtered and those with a P-value <0.05 or a power >0.8 were removed (Supplementary Fig. 1A). The remaining data were then exported for identification of individual proteins using the IPI-Homo sapiens database via Mascot Search Engine (V2.3.2) in which 712 proteins were identified; statistical analysis was carried out automatically in Mascot. Proteins identified in Mascot were then imported into Progenesis for filtering and further analysis. Peptide conflicts were removed and proteins were filtered to eliminate those with less than two unique peptides; proteins with a P-value < 0.05 or a fold-change <1.1 in either UBA1 overexpression compared to control or knockdown compared to control were removed (Supplementary Fig. 1A). This resulting dataset contained 222 proteins, which showed the largest significant variation in expression following modulation of UBA1 expression levels (Supplementary Fig. 1B and C). UBA1 itself was removed from the dataset for all subsequent analysis.
The Database for Annotation, Visualization and Integrated Discovery (DAVID) was used to identify enriched biological themes within the filtered proteomics dataset; functional annotation clustering was performed in which enriched terms are grouped together to identify enriched functional clusters. Modified Fischer’s exact P-values for each term are automatically calculated during this analysis; annotation terms that belong to the same proteins are then grouped together and assigned an enrichment score, which is the geometric mean of the P-values for all the terms in that cluster. The enrichment score is the −log10 of the P-value so an enrichment score of 1.3 is equivalent to a P-value of 0.05. This software was also used to characterize the functions associated with the protein clusters generated in BioLayout. BioLayout (http://www.biolayout.org) was used to identify UBA1-dependent clusters of proteins based on the expression of the proteins across UBA1 overexpression, control and UBA1 knockdown. The network graphs in this study were generated using a Pearson correlation (set to 0.98) and Markov clustering algorithm (Enright et al., 2002). The expression profiles are displayed as mean normalized abundance (pareto scale) with standard error of the mean (SEM) for each condition. Ingenuity pathway analysis (IPA; www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/) was used as previously described (Wishart et al., 2007). The significance of an association between the dataset and a canonical pathway was defined by a Fischer’s exact P-value (Savli et al., 2008). A 1.1-fold change threshold filter was applied in IPA to each dataset analysed and only experimentally observed interactions were selected for each analysis.
Statistical analyses
All statistical analyses were performed in Prism 6 (GraphPad). Statistical analyses were performed using unpaired two-tailed Student’s t-test or a one-way ANOVA with a Tukey’s multiple comparison test for parametric data, and a Mann-Whitney test or a Kruskal-Wallis test with Dunn’s multiple comparisons test for non-parametric data, as appropriate. Data are reported as mean ± SEM.
Data availability
Raw proteomics data files from this study are freely available for download from http://dx.doi.org/10.7488/ds/2405.
Results
Restoration of UBA1 with AAV9-UBA1 rescues sensory-motor connectivity defects in spinal muscular atrophy mice
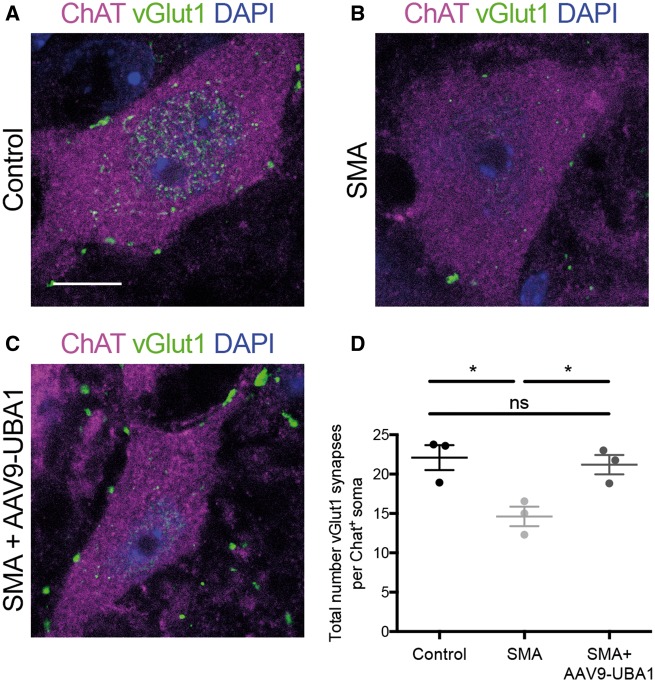
To establish whether UBA1-dependent pathways linked to SMA (Aghamaleky Sarvestany et al., 2014; Hunter et al., 2014; Wishart et al., 2014; Fuller et al., 2015; Powis et al., 2016) contribute to the development of defective sensory-motor connectivity in SMA, we initially investigated the effects of systemic restoration of UBA1 on sensory-motor connectivity in SMA mice. Synaptic inputs formed onto lower motor neurons in the spinal cord were identified using immunohistochemical labelling of VGLUT1 (Ling et al., 2010; Mentis et al., 2011; Fletcher et al., 2017; Simon et al., 2017). Quantification of proprioceptive inputs onto lower motor neurons in the lumbar region of spinal cord revealed a significant reduction by 34% in symptomatic (postnatal Day 8) ‘Taiwanese’ SMA mice compared to littermate controls (Fig. 1A, B and D), confirming previous reports from different mouse models of SMA (Ling et al., 2010; Mentis et al., 2011). Systemic restoration of UBA1 levels, via intravenous delivery of AAV9-UBA1 on the day of birth (Powis et al., 2016), completely restored the number of VGLUT1-positive synaptic inputs onto motor neurons in SMA mice (Fig. 1C and D). Thus, restoration of UBA1 was sufficient to ameliorate defective sensory-motor connectivity in SMA mice, suggesting that UBA1-dependent pathways play a significant role in the regulation of sensory neuron health in vivo and contribute directly to sensorimotor aspects of disease pathogenesis in SMA.
Figure 1.
Rescue of sensory-motor connectivity defects in the spinal cord of SMA mice following systemic restoration of UBA1. (A–C) Lumbar motor neurons from control, SMA and SMA+AAV9-UBA1 mice labelled with ChAT (magenta), VGLUT1 (green) and DAPI. Scale bar = 10 µm. (D) Quantification of the number of VGLUT1 synapses per ChAT-positive motor neuron soma. n = 3 mice per condition, n > 20 motor neurons analysed per mouse. ns = not significant, *P < 0.05.
Label-free proteomics identifies UBA1-dependent proteins
To identify downstream targets of UBA1 likely to be mediating its effects on sensory-motor connectivity in SMA, label-free proteomics was performed on HEK293 cells where UBA1 had been either overexpressed (using a UBA1 plasmid) or knocked down (using UBA1 siRNA). HEK293 cells were chosen for the proteomics screen to enable dissection of UBA1 dependent pathways relevant for degeneration in SMA, independent of the reduction in SMN expression seen in SMA. Western blot confirmed a 10.11-fold increase in levels of UBA1 protein in cells overexpressing UBA1 (Fig. 2A and B) and a 76% reduction in UBA1 protein levels in cells transfected with UBA1 siRNA (Fig. 2A and C). Following filtering of the raw proteomic data (Supplementary Fig. 2), 222 unique proteins were identified with a fold-change >10% after either overexpression or knockdown of UBA1. Of the proteins that revealed expression changes, the majority were upregulated following UBA1 overexpression and conversely downregulated following UBA1 knockdown (Supplementary Fig. 2).
Figure 2.
Label-free proteomics analysis identifies tRNA synthetases as UBA1-dependent proteins. (A) UBA1 expression in HEK293 cells used for proteomics screen showing UBA1 overexpression (OE) and knockdown (KD) compared to control. CoxIV and H3 = loading control. (B and C) Quantification of UBA1 overexpression (B) and knockdown (C). Ctrl = control; **P < 0.01, ***P < 0.001. (D) BioLayout clustering 3D representation of proteomic expression across UBA1 overexpression, control and UBA1 knockdown. Nodes represent proteins and edges represent similarity of expression between proteins and clusters of proteins with similar expression profiles are indicated by different colours. Cluster numbers are indicated. (E and F) Expression profile means ± SEM in log scale for the two UBA1-dependent clusters: cluster 1 (E) and cluster 4 (F). (G and H) IPA analysis showing the top network for UBA1 overexpression/control (G) and UBA1 knockdown/control (H), proteins highlighted in magenta are upregulated, proteins highlighted in green are downregulated. See also Supplementary Figs 2–5 and Supplementary Table 1.
Gene ontology term enrichment analysis was performed on the filtered proteomic data in DAVID, identifying aminoacyl-tRNA synthetases (also known as tRNA ligases), translation elongation factors, small molecule synthesis, and protein and RNA transport as significantly enriched gene ontology term clusters with the highest enrichment scores (Table 1). Gene ontology terms enriched with lower enrichment score included ubiquitin-like conjugation, protein complex assembly, and ATP and nucleotide binding (Table 1); functions related to the ubiquitin conjugation process. A similar analysis performed in IPA confirmed RAN signalling (a process involved in nuclear transport) and tRNA charging (a function of aminoacyl-tRNA synthetases) as significantly enriched canonical pathways. Nine of the 39 proteins involved in the tRNA charging canonical pathway were present in this dataset (Supplementary Table 1).
Table 1.
Gene ontology term enrichment of proteins changed following modulation of UBA1 expression
| GO term cluster | Enrichment score |
|---|---|
| Translation elongation | 4.56 |
| tRNA ligase activity | 4.31 |
| Nucleic acid synthesis | 4.10 |
| Cytoskeleton | 3.30 |
| Protein transport and localisation | 3.03 |
| RNA transport and localisation | 2.95 |
| Protein complex assembly | 2.85 |
| ATP and nucleotide binding | 2.55 |
| Glycolysis | 2.45 |
| Cytoskeletal binding | 2.36 |
| Ubl conjugation | 2.25 |
| Membrane bound vesicle | 2.04 |
| GTPase binding | 2.02 |
See also Supplementary Table 1.
To identify specific UBA1 target proteins, BioLayout analyses were performed whereby clusters of proteins were generated based on the similarity of protein expression profiles. This analysis revealed two main groups of proteins modified downstream of UBA1, each of which was further subdivided, generating six subclusters of proteins (Fig. 2D). Two of the clusters (clusters 1 and 4) showed UBA1-dependent expression profiles (Fig. 2E and F), while the other four clusters showed a prominent expression change in only one experimental condition (Supplementary Fig. 3). The first UBA1-dependent cluster of proteins, cluster 1, contained proteins that were upregulated following UBA1 overexpression and downregulated following UBA1 knockdown (Fig. 2E). Subsequent analysis in DAVID revealed that the proteins in this cluster function as aminoacyl-tRNA synthetases or in glycolysis, translation elongation or assembly of protein complexes. Cluster 4 showed the opposite UBA1-dependent expression profile (downregulation following UBA1 overexpression and upregulations following UBA1 knockdown; Fig. 2F) in which proteins function in transport and localization of protein, RNA and nucleic acids. Interestingly, the functions of the UBA1-dependent clusters overlap with the enriched gene ontology terms and canonical pathways (Table 1 and Supplementary Table 1); thus, indicating that these are key protein families and functions changed on modulation of UBA1 protein levels.
To investigate UBA1 dependency at the level of individual proteins and interaction networks, a network analysis was performed using IPA software. Interestingly, the top protein network generated for each condition (overexpression or knockdown of UBA1) was composed of the same proteins, in which 25 of 30 proteins present in the network behaved in a UBA1-dependent manner (Fig. 2G and H; see Supplementary Figs 4 and 5 for expanded networks), suggesting that modulation of UBA1 expression causes widespread reciprocal changes in expression in discrete networks of proteins.
Importantly, our analyses repeatedly highlighted aminoacyl-tRNA synthetases (including GARS, YARS, HARS and WARS) as key UBA1 downstream targets (Fig. 2G and H). This finding was of particular interest and relevance due to known links between this family of enzymes and neurodegenerative conditions, with mutations in several of these genes causing hereditary neuropathies (Kapur et al., 2017). More specifically, the GlyRS protein (GARS) encoded by GARS was of particular interest because of its known role in Charcot-Marie-Tooth disease type 2D (CMT2D), a neuromuscular disorder with phenotypic similarity to SMA (Antonellis et al., 2003; Sivakumar et al., 2005; James et al., 2006; Motley et al., 2010).
UBA1 regulates GARS expression independent of its role in ubiquitylation
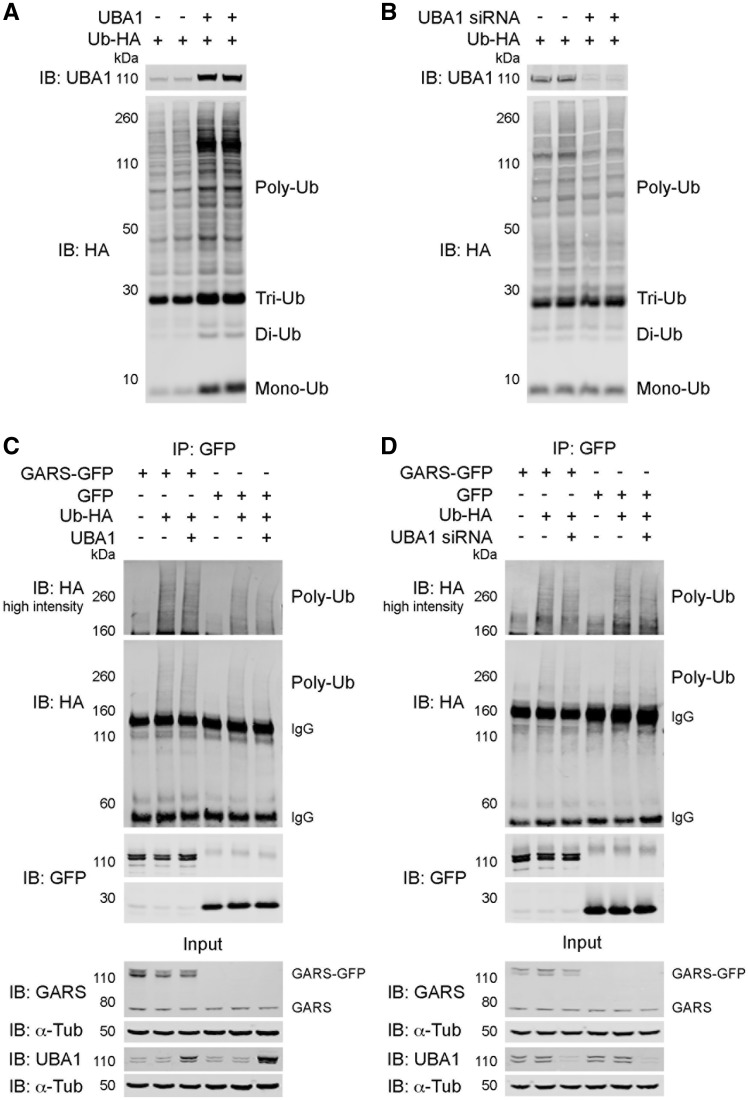
As the canonical role of UBA1 concerns priming of the ubiquitylation cascade (Groen and Gillingwater, 2015), we next sought to investigate whether the downstream protein targets of UBA1 identified in our proteomics screen were showing different levels of expression due to direct perturbations in their ubiquitylation status. To investigate the effect of UBA1 modulation on overall ubiquitylation levels, HEK293 cells were transfected with ubiquitin (Ub-HA) and a UBA1 plasmid, UBA1 siRNA or a suitable control vector. As expected, following overexpression of UBA1 overall polyubiquitylation of substrate proteins was increased (Fig. 3A). Following UBA1 knockdown, there was a concomitant, albeit less prominent, change in overall ubiquitylation status with a modest reduction in polyubiquitylation of substrate proteins (Fig. 3B). Overall, this confirms that modulation of UBA1 levels leads to changes in ubiquitylation of substrate proteins, demonstrating that UBA1 can influence the expression of downstream targets through differential ubiquitylation.
Figure 3.
UBA1 influences GARS expression through a non-canonical function. (A and B) HEK293 cells were transfected with ubiquitin-HA (Ub-HA) and UBA1 (A) or UBA1 siRNA (B); western blot of UBA1 and ubiquitin (immunoblotted for HA tag), showing polyubiquitylated substrate proteins (Poly-Ub), and free triubiquitin (Tri-Ub), diubiquitin (Di-Ub) and monoubiquitin (Mono-Ub). (C and D) HEK293 cells were transfected with GARS-GFP or GFP along with Ub-HA and UBA1 (C) or UBA1 siRNA (D). Input control samples were immunoblotted for UBA1 and GARS; immunoprecipitation (IP) with GFP; IP samples were immunoblotted for GFP and HA. IgG bands and polyubiquitylation (Poly-Ub) smears are indicated. High intensity immunoblots show polyubiquitylation smears imaged at increased laser power. See also Supplementary Fig. 6.
To investigate whether the UBA1-dependent tRNA-synthetase GARS was being differentially ubiquitylated following modulation of UBA1 expression, a protein-specific ubiquitylation assay was performed. As the ubiquitylation pattern of SMN is well characterized (Chang et al., 2004; Kwon et al., 2013; Abera et al., 2016), the ubiquitylation assay, in which the proteins of interest were co-transfected, was initially optimized using SMN, confirming the ubiquitylation pattern of SMN (Supplementary Fig. 6). Surprisingly, following overexpression of UBA1, in the presence of both GARS and ubiquitin, there was no change in the overall ubiquitylation of GARS (Fig. 3C). Similarly, following UBA1 knockdown, there was also no change in the ubiquitylation of GARS compared to control UBA1 expression (Fig. 3D). Thus, modulation of UBA1 expression did not affect the ubiquitylation of GARS, suggesting that UBA1 influences the expression of GARS through a non-canonical pathway independent of its role in the ubiquitylation cascade.
Perturbations in UBA1 lead to disruption of GARS in spinal muscular atrophy mice
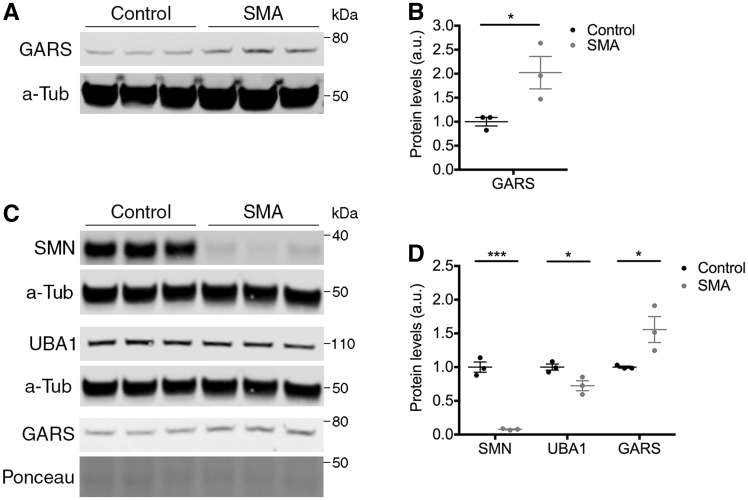
To explore the potential role and relevance of the UBA1/GARS pathway in SMA pathogenesis further, we next wanted to establish whether GARS protein was disrupted in SMA in vivo. Furthermore, to investigate the wider effects of UBA1 on tRNA synthetases, we quantified the expression of the tRNA-synthetase YARS in SMA mice; mutations in YARS cause dominant intermediate CMT type C (DI-CMTC) (Jordanova et al., 2006). Importantly, it has previously been demonstrated that both UBA1 and SMN expression are reduced in spinal cord from Taiwanese SMA mice (Powis et al., 2016; Groen et al., 2018a). GARS showed an increase of 100% in spinal cord from SMA mice at postnatal Day 8 compared to control littermates (Fig. 4A and B) whilst YARS showed a reduction of 37% in SMA spinal cord (Supplementary Fig. 7A and B); thereby validating expression changes in two different tRNA-synthetases in SMA.
Figure 4.
GARS is dysregulated in neuronal tissue from SMA mice. (A and B) Western blot (A) and quantification (B) of GARS protein levels in spinal cord from late-symptomatic SMA mice and control littermates. α-Tubulin (α-Tub) = loading control. (C and D) Western blot (C) and quantification (D) of SMN, UBA1 and GARS protein levels in DRG from late-symptomatic SMA mice and control littermates. α-Tubulin (α-Tub), total protein (Ponceau): loading controls. n = 3 mice per condition. See also Supplementary Figs 7 and 8.
Interestingly, however, there was no significant difference in GARS expression specifically within lower motor neuron bodies in the spinal cord (Supplementary Fig. 8A and B), suggesting that modulations in UBA1/GARS pathways are unlikely to underlie motor neuron pathology in SMA. In stark contrast, expression changes of GARS were present in the dorsal root ganglia (DRG), which contain sensory neuron bodies (Fig. 4C and D). As protein levels of SMN and UBA1 have not previously been reported for DRGs in control and SMA mice, their expression was quantified by western blot analysis. As expected, a significant reduction of SMN was observed in DRGs (Fig. 4C) with an overall reduction of 92% in SMA mice compared to controls (Fig. 4D). This is consistent with the reduction of SMN protein reported in SMA patients (Lefebvre et al., 1997) and the reduced levels of Smn transcript reported in DRGs from ‘delta7’ SMA mice (Mentis et al., 2011). Similarly, UBA1 was reduced by 28% in late-symptomatic SMA DRGs compared to controls (Fig. 4C and D), consistent with the magnitude of UBA1 reduction reported in spinal cord from late-symptomatic Taiwanese SMA mice (Powis et al., 2016). Consistent with the change in expression seen in spinal cord, GARS expression was upregulated by 56% in SMA DRGs compared to controls (Fig. 4C and D), while YARS expression showed no change in SMA DRGs (Supplementary Fig. 7A and B). Thus, suppression of UBA1 downstream of SMN leads to a significant increase in GARS protein levels in SMA mice in vivo, with DRGs being particularly affected.
Identification of a sensory neuron fate phenotype in spinal muscular atrophy mice
The increased levels of GARS we observed in SMA were consistent with changes in GARS protein levels reported in CMT2D, where they are notably increased during neonatal development and disease progression (Achilli et al., 2009; Motley et al., 2010, 2011). It has recently been shown that disruption to sensory neuron fate is a significant feature of disease pathogenesis in CMT2D mice, leading to an alteration in the functional subpopulations of sensory neurons in the DRG (Sleigh et al., 2017). We therefore wanted to establish whether similar GARS-dependent phenotypes were contributing to the sensory-motor connectivity defects that occur in SMA.
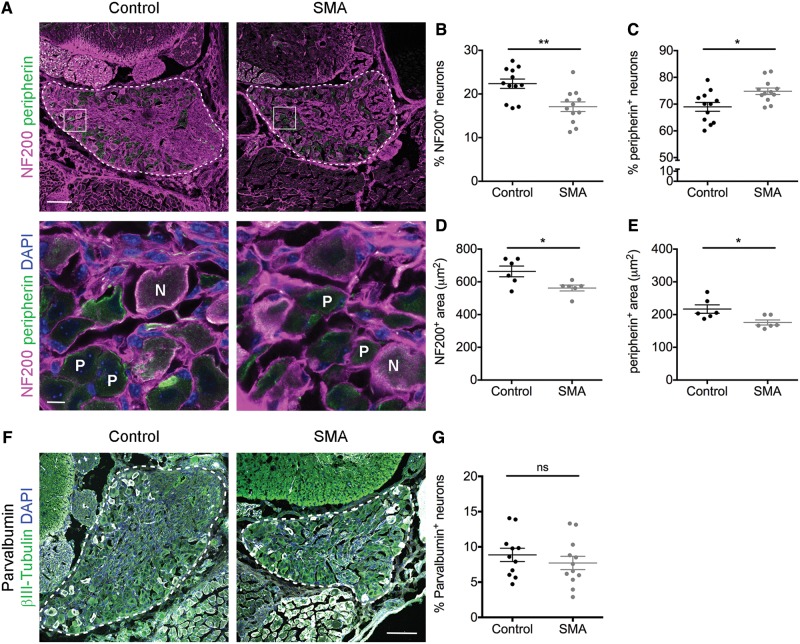
Large area sensory neurons within DRGs are predominantly either mechanoreceptors (sense touch) or proprioceptors (sense position and movement), both of which stain positive for NF200: ∼20% of NF200-positive neurons are proprioceptors and the remaining 80% are mechanoreceptors (Sleigh et al., 2017). Smaller area sensory neurons stain positive for peripherin and are mostly nociceptive. In CMT2D mice there is a severity-dependent disruption to the fate of sensory neurons, with a decrease in the percentage of NF200-positive neurons and an increase in the percentage of peripherin-positive neurons, leading to concordant behavioural deficits (Sleigh et al., 2017).
Qualitatively, DRGs from SMA mice appeared smaller with fewer NF200-positive neurons compared to control mice (Fig. 5A and B). Quantification revealed a significant 24% reduction in the percentage of NF200-positive neurons relative to control (Fig. 5B), with a concomitant increase in peripherin-positive neurons by 85% relative to control (Fig. 5C). CMT2D mice have a reduction in the area of sensory neuron cell bodies (Sleigh et al., 2017). Likewise, NF200-positive neurons were significantly smaller in SMA mice than in controls, with an average reduction of 101 µm2 per neuron (Fig. 5D). Similarly, peripherin-positive neurons were also significantly smaller in SMA mice (Fig. 5E), thereby confirming that SMA DRGs were indeed smaller than those in control animals, as has previously been reported for other organs in SMA (Powis et al., 2016; Thomson et al., 2017; Maxwell et al., 2018). Despite the overall reduced size and disruption to sensory neuron fate in SMA DRGs, these phenotypes were not due to overt cell death in the SMA DRGs (Supplementary Fig. 9), a finding consistent with previous reports in CMT2D (Sleigh et al., 2017).
Figure 5.
Sensory neuron fate is disrupted in SMA dorsal root ganglia. (A) Spinal column sections from lumbar segments 1 and 2 of late-symptomatic SMA and control mice labelled with NF200 (magenta) and peripherin (green). DRG are outlined in the top panels, boxes indicate the area in bottom panel. Nuclei of cells in bottom panels are labelled with DAPI. N = NF200-positive neurons; P = peripherin-positive neurons. Scale bars = 100 µm (top), 10 µm (bottom). (B and C) Quantification of the percentage of NF200-positive (NF200+) (B) and peripherin-positive (peripherin+) (C) sensory neurons. n = 3 mice per condition, n = 4 DRGs per mouse. (D and E) Quantification of the area of NF200+ (D) and peripherin+ (E) sensory neurons. n = 3 mice per condition, n = 2 DRGs per mouse (seven NF200+ and seven peripherin+ neurons per DRG were analysed). (F) Spinal column sections from lumbar segments 1 and 2 of late-symptomatic SMA and control mice labelled with parvalbumin (white), beta-III tubulin (green) and DAPI (blue), dorsal root ganglia (DRG) are outlined. Scale bar = 100 µm. (G) Quantification of parvalbumin-positive sensory neurons as a percentage of beta-III tubulin-positive sensory neurons. n = 3 mice per condition, n = 4 DRGs per mouse. *P < 0.05, **P < 0.01. See also Supplementary Fig. 9.
Because of the identification of a reduction in the proportions of NF200-positive sensory neurons in SMA, we next wanted to investigate whether mechanoreceptors and proprioceptors were differentially affected. Quantification of the overall percentage of sensory neurons that stain positive for parvalbumin, a proprioceptor specific marker, revealed no significant difference in the proportion of parvalbumin-positive sensory neurons in SMA DRGs compared to control (Fig. 5F and G). This is consistent with a previous report in a different mouse model of SMA (Mentis et al., 2011), and demonstrated that, as in CMT2D (Sleigh et al., 2017), both subtypes of NF200-positive sensory neurons were equally affected leading to the disruption to sensory neuron fate seen in SMA.
Taken together, these findings reveal a novel sensory neuron fate phenotype associated with increased levels of GARS in SMA mice, highlighting significant pathological overlap between SMA and CMT2D at the level of sensory neurons.
Dysregulation of UBA1/GARS-dependent pathways in sensory neurons from spinal muscular atrophy mice
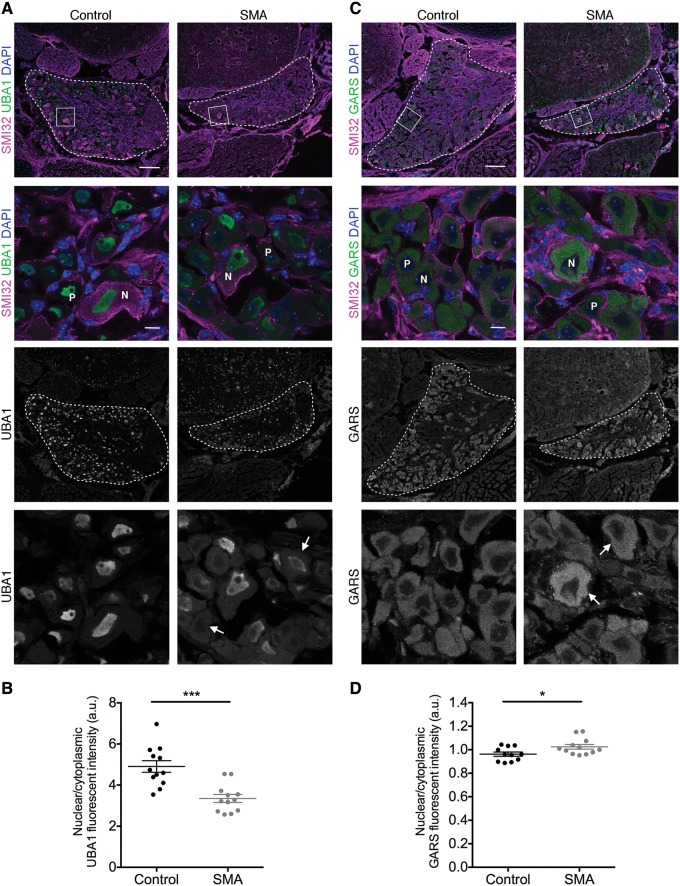
To investigate links between UBA1/GARS-dependent pathways and disrupted sensory neuron fate in SMA further, the ratio of UBA1 and GARS expression was investigated at the level of individual sensory neurons within the DRG. There was less prominent nuclear labelling and more pronounced cytoplasmic labelling of UBA1 in DRGs from SMA mice, indicating a shift in the subcellular distribution of UBA1 (Fig. 6A). Quantification revealed a significant reduction of the nuclear-cytoplasmic ratio (NCR) by 24% in SMA mice compared to controls (control = 5.34, SMA = 4.07; Fig. 6B). Sensory neurons also showed a significant increase in the NCR of GARS in SMA compared to controls (Fig. 6C and D). Notably, however, larger diameter neurons revealed a selective increase in the intensity of GARS labelling (Fig. 6C). Interestingly, here changes in GARS expression occurred in the opposite direction to changes in UBA1 expression, consistent with changes at the whole tissue level as detected by western blot analysis of whole DRGs and spinal cord (Fig. 4). Thus, sensory neuron cell fate phenotypes observed in SMA mice correlate directly with parallel expression changes in key proteins of the UBA1/GARS pathway.
Figure 6.
Dysregulation of UBA1 and GARS in dorsal root ganglia sensory neurons from SMA mice. (A and C) Spinal column sections from lumbar segments 1 and 2 of late-symptomatic SMA and control mice labelled with UBA1a (A) or GARS (C) (green), SMI32 (magenta) and DAPI. DRG are outlined in overview panels, boxes indicate the area in lower panels. Arrows in bottom panel indicate sensory neurons with altered UBA1 (A) or GARS (C) expression. N = NF200-positive neurons; P = peripherin-positive neurons. Scale bars = 100 µm (overview), 10 µm (lower panels). (B and D) Late-symptomatic SMA DRG sensory neurons show a reduction in the nuclear to cytoplasmic ratio of UBA1 (B) and an increase in the nuclear to cytoplasmic ratio of GARS (D) compared to control mice. n = 3 mice per condition, n = 4 DRGs per mouse (14 sensory neurons were analysed per DRG); *P < 0.05, ***P ≤ 0.001.
Restoration of UBA1 rescues sensory neuron fate phenotypes in spinal muscular atrophy mice
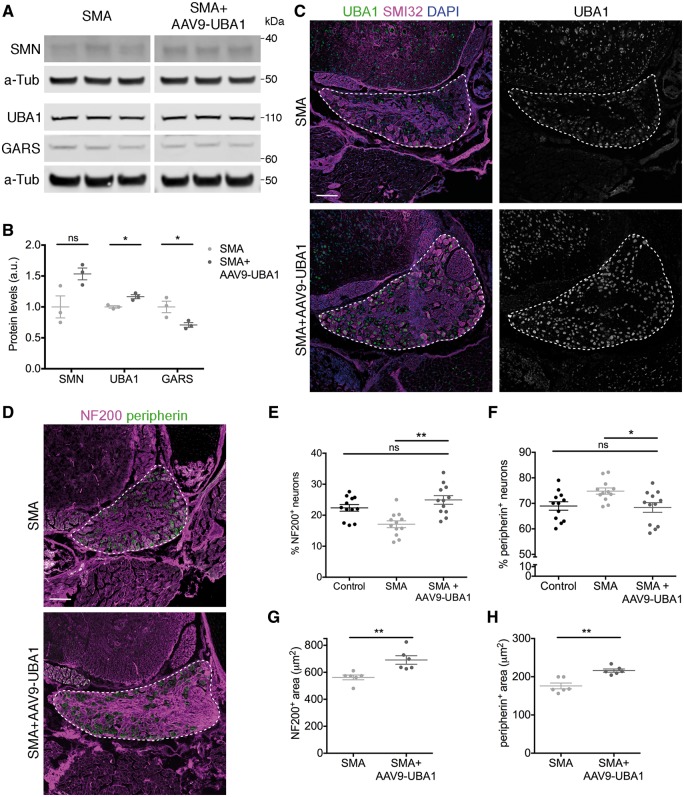
Finally, we wanted to establish whether the rescue of sensory-motor connectivity observed in the spinal cord of SMA mice treated with AAV9-UBA1 was due to correction of sensory neuron fate phenotypes in the DRG and if this disruption to sensory neuron fate in SMA mice was directly dependent on changes in the UBA1/GARS pathway. Quantitative western blot performed on lumbar DRGs from FVB mice and FVB mice injected with AAV9-UBA1 confirmed transduction of DRG sensory neurons, with an increase in UBA1 expression by 33% in mice injected with AAV9-UBA1, but no change in SMN expression (Supplementary Fig. 10A and B). Immunofluorescence confirmed transduction of sensory neurons at the cellular level (Supplementary Fig. 10C). Similarly, SMA mice and SMA mice injected with AAV9-UBA1 revealed a significant increase in UBA1 levels, by 17% compared to non-injected animals at postnatal Day 8, but no change in SMN expression (Fig. 7A and B). Upregulation of UBA1 was also evident at the cellular level within individual sensory neurons of the DRG (Fig. 7C). Importantly, upregulation of UBA1 using AAV9-UBA1 reduced GARS levels by 29% (Fig. 7A and B), back toward the levels observed in DRGs from control mice.
Figure 7.
Restoration of UBA1 in SMA mice reverses GARS dysregulation and rescues sensory neuron cell fate phenotypes. (A and B) Representative fluorescent western blot (A) and quantification (B) of SMN, UBA1 and GARS in dorsal root ganglia from late-symptomatic SMA mice and SMA mice injected with AAV9-UBA1 (SMA+AAV9-UBA1). α-Tubulin (α-Tub): loading control. n = 3 mice per condition. (C) Spinal column sections from lumbar segments 1 and 2 of late-symptomatic SMA and SMA+AAV9-UBA1 mice labelled with UBA1a (green), SMI32 (magenta) and DAPI. (D) Spinal column sections from lumbar segments 1 and 2 of late-symptomatic SMA and SMA+AAV9-UBA1 mice labelled with NF200 (magenta) and peripherin (green). (C and D) Dorsal root ganglia (DRG) are outlined. (E and F) Quantification of the percentage of NF200-positive (NF200+) (E) and peripherin-positive (peripherin+) (F) sensory neurons. Data from control mice is shown as reference. n = 3 mice per condition, n = 4 DRGs per mouse. (G and H) Quantification of the area of NF200+ (G) and peripherin+ (H) sensory neurons. n = 3 mice per condition, n = 2 DRGs per mouse (seven NF200+ and seven peripherin+ neurons per DRG were analysed); ns = not significant. *P < 0.05, **P < 0.01. See also Supplementary Figs 10 and 11.
To determine whether AAV9-UBA1-mediated correction of GARS levels rescued sensory neuron fate phenotypes, we assessed sensory neuron subtype proportions in SMA mice and SMA mice injected with AAV9-UBA1. Overexpression of UBA1 in SMA mice fully rescued the disruption to sensory neuron fate (Fig. 7D) whereby the percentage of NF200-positive neurons was increased by 46% in SMA mice injected with AAV9-UBA1 relative to SMA mice (Fig. 7E). There was also a significant reduction in the percentage of peripherin-positive neurons in SMA mice injected with AAV9-UBA1 by 9% relative to SMA mice (Fig. 7F). Compared to data obtained from control mice, there was no significant difference in the percentage of NF200- or peripherin-positive neurons in SMA mice treated with AAV9-UBA1 (Fig. 7E and F). Interestingly, following treatment with AAV9-UBA1 in SMA mice there was a significant increase in the percentage of parvalbumin-positive neurons, from 8% of the total sensory neuron population in SMA mice, to 13% in SMA mice injected with AAV9-UBA1 (Supplementary Fig. 11A and B). This indicates that AAV9-UBA1 predominantly affects proprioceptive sensory neurons.
Overexpression of UBA1 in SMA mice also rescued the area of sensory neurons with an increase in the area of NF200-positive neurons by an average of 129 μm2 per neuron (Fig. 7G) and an increase in peripherin-positive neuron area by an average of 40 μm2 in treated SMA mice (Fig. 7H). Thus, restoration of UBA1 levels using viral gene delivery in SMA mice was sufficient to correct GARS protein levels in sensory neurons and fully rescue the disruption to sensory neuron fate.
Discussion
Taken together, data from the current study demonstrate that UBA1 is a major regulator of sensory-motor connectivity phenotypes in SMA. Aminoacyl-tRNA synthetases were identified as key UBA1 downstream targets, revealing a novel UBA1/GARS dependent pathway that mediates sensory neuron cell fate through a non-canonical pathway distinct from differential ubiquitylation. Restoration of UBA1 in SMA mice using a gene therapy approach revealed that correction of UBA1 levels, and downstream UBA1/GARS pathways, was sufficient to rescue sensory neuron cell fate phenotypes, and sensory-motor connectivity defects in the spinal cord, in SMA mice (Supplementary Fig. 12). Because of the known role of mutant GARS in mediating sensory neuron pathology in CMT2D, our findings provide experimental evidence of a significant molecular and phenotypic overlap between SMA and CMT.
The finding that the newly-identified UBA1/GARS pathway was regulated independently of the ubiquitylation status of GARS adds further support to a growing body of evidence suggesting that UBA1 can influence its downstream targets and pathways through non-canonical functions. For example, functions of UBA1 outside of the canonical ubiquitylation cascade have previously been reported in Atg8-dependent autophagy (Chang et al., 2013). Our findings therefore extend our understanding of the broad range of pathways influenced by UBA1, and confirm the existence of multiple, non-canonical functions for UBA1 in vivo. Consistent with this model, recent studies have suggested that UBA6 ubiquitylates a larger proportion of the proteome than previously thought and indicated that ubiquitylation of GARS may be performed by UBA6 (Liu et al., 2017b).
Moreover, the tissue-specific nature of UBA1-dependent perturbations in GARS and YARS identified here echoes previous work in which downstream molecular targets of UBA1 were found to vary depending on the tissue and organ being examined (Wishart et al., 2014). For example, therapies treating the accumulation of β-catenin downstream of UBA1 in SMA, rescued neuromuscular but not systemic/non-neuromuscular pathologies in Taiwanese SMA mice (Wishart et al., 2014). Furthermore, it has previously shown that AAV9-UBA1 increases SMN expression in hearts from both SMA and control mice compared to untreated SMA and control mice, respectively (Powis et al., 2016). Here, we report that AAV9-UBA1 does not increase SMN expression in either SMA (Fig. 7A and B) or control DRGs (Supplementary Fig. 10A and B), indicating that modest SMN restoration may contribute to the rescue of some SMA phenotypes, but not the restoration of sensory neuron phenotypes following AAV9-UBA1 treatment. This suggests that therapies restoring UBA1, which lead to widespread rescue of SMA phenotypes (Powis et al., 2016), are likely to be modulating numerous, tissue-specific downstream pathways of UBA1, thereby revealing a complex spatiotemporal influence of ubiquitin pathways on disease pathogenesis in SMA and related conditions.
While sensory neuron pathology has previously been identified in models of SMA (Jablonka et al., 2006; Ling et al., 2010; Mentis et al., 2011; Fletcher et al., 2017), the disruption to sensory neuron fate and reduction in the size of sensory neurons within the DRG reported here provides important insights into the pathogenesis of sensory-motor connectivity defects in SMA. A recent study suggested that motor neuron hyperexcitability in SMA occurs specifically due to defects in proprioceptive sensory neurons, with the onset of motor neuron dysfunction correlating with sensory-motor connectivity defects in SMA. Importantly, blocking neurotransmission specifically in proprioceptive neurons in wild-type mice caused severe motor defects, shortened lifespan and rendered motor neurons dysfunctional (Fletcher et al., 2017). Together this suggests that restoration of proprioceptive synapses in SMA leads to increased functionality of motor neurons. Our finding that defects in sensory neuron fate specifically target NF200-positive neurons in SMA, and that restoration of UBA1 was sufficient to rescue sensory neuron fate in the DRG alongside sensory-motor connectivity in the spinal cord, suggests that UBA1/GARS-dependent regulation of this sensory neuron subtype plays a critical role in the pathogenesis of both motor and sensory phenotypes in SMA. It also highlights the possibility that UBA1/GARS-mediated restoration of sensory-motor connectivity leads to improvements in motor neuron function which, along with rescue of other motor neuron and muscle specific defects through alternative UBA1-mediated pathways, cause the phenotypic improvements in neuromuscular pathology previously reported in AAV9-UBA1 treated SMA mice (Powis et al., 2016).
Importantly, sensory pathology in SMA patients is becoming increasingly well characterized (Supplementary Table 2). Several reports of patients with SMA type 1 indicate abnormal sensory conduction (Duman et al., 2013) or absence of sensory responses (Reid et al., 2016), as well as axonal degeneration in sensory nerves (Rudnik-Schoneborn et al., 2003). In earlier studies of presumed SMA cases, axonal degeneration of sensory neurons was also identified, alongside ballooned neurons and chromatolysis within DRGs (Marshall and Duchen, 1975; Carpenter et al., 1978; Murayama et al., 1991). Furthermore, a genetically-confirmed case of XL-SMA showed nodules within the DRG, indicating a loss of sensory neurons (Supplementary Table 2) (Dlamini et al., 2013).
Our finding of a key role for GARS in regulating sensory neuron phenotypes and sensory-motor connectivity in SMA, provides experimental support for the hypothesis that there is significant molecular and phenotypic overlap between SMA and CMT2D. Furthermore, the finding of altered YARS expression in SMA spinal cord indicates a wider overlap of spinal muscular atrophies and peripheral neuropathies. Interestingly, several cases of co-segregation of SMA and CMT have been reported (Supplementary Table 2) (Jedrzejowska et al., 2008; Fernandez et al., 2016), indicating that the pathologies of these two diseases can occur simultaneously. In addition to this, several genes have now been identified where mutations can cause a range of phenotypes incorporating both CMT and SMA-like diseases. GARS is one such gene, where mutations can cause CMT2D, distal-SMA type V or more classical infantile SMA (Supplementary Table 2) (Antonellis et al., 2003; James et al., 2006; Eskuri et al., 2012). Similarly, mutations in MORC2 can cause CMT type 2Z or SMA (Schottmann et al., 2016; Sevilla et al., 2016) and mutations in IGHMBP2 have been shown to cause both CMT type 2S and SMA with respiratory distress type 1 (SMARD1; Supplementary Table 2) (Grohmann et al., 2001; Cottenie et al., 2014; Pedurupillay et al., 2016; Liu et al., 2017a). Moreover, mutations in LRSAM1, an E3 ubiquitin ligase, cause CMT type 2G (Peeters et al., 2016) thereby implicating ubiquitin pathways in the pathogenesis of CMT. Similarly, mutations in PIEZO2, which encodes a mechanosensitive ion channel responsible for mechanosensation of light touch and proprioception, can cause a neuromuscular disease characterized by muscle atrophy, mild sensory involvement, delayed motor milestones and scoliosis (Chesler et al., 2016; Delle Vedove et al., 2016); suggesting that defects within sensory neurons themselves can lead to muscle defects and delayed motor development. Together, not only does this highlight considerable overlap between the phenotypes of CMT and SMA, but it also demonstrates considerable overlap of genetic pathways and molecular causes of these diseases, hinting at the potential of shared therapeutic opportunities for these neuromuscular conditions.
Supplementary Material
Acknowledgements
The authors thank Elena Perenthaler for help with optimising the ubiquitylation assay and other members of the Gillingwater laboratory and Natalie Courtney for valuable input and discussion. The authors also thank the IMPACT facility at the University of Edinburgh.
Glossary
Abbreviations
- AAV9
adeno-associated virus serotype 9
- CMT
Charcot-Marie-Tooth disease
- DRG
dorsal root ganglion
- SMA
spinal muscular atrophy
- SMN
survival motor neuron protein
Funding
This work was supported by funding from the Euan MacDonald Centre for Motor Neurone Disease Research, SMA Europe, the SMA Trust UK Consortium (all to T.H.G.), the Wellcome Trust (to J.N.S., E.J.N.G and T.H.G), the Darwin foundation and the BBSRC (to T.M.W), the European Union’s Horizon 2020 Research and Innovation programme under grant agreement 739572, the Wellcome Trust Senior Investigator Award (107116/Z/15/Z) and a UK Dementia Research Institute Foundation award (all to G.S.).
Competing interests
T.H.G. is Chair of the Scientific and Clinical Advisory Board of the SMA Trust.
References
- Abera MB, Xiao J, Nofziger J, Titus S, Southall N, Zheng W et al. . ML372 blocks SMN ubiquitination and improves spinal muscular atrophy pathology in mice. JCI Insight 2016; 1: e88427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achilli F, Bros-Facer V, Williams HP, Banks GT, AlQatari M, Chia R et al. . An ENU-induced mutation in mouse glycyl-tRNA synthetase (GARS) causes peripheral sensory and motor phenotypes creating a model of Charcot-Marie-Tooth type 2D peripheral neuropathy. Dis Model Mech 2009; 2: 359–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghamaleky Sarvestany A, Hunter G, Tavendale A, Lamont DJ, Llavero Hurtado M, Graham LC et al. . Label-free quantitative proteomic profiling identifies disruption of ubiquitin homeostasis as a key driver of Schwann cell defects in spinal muscular atrophy. J Proteome Res 2014; 13: 4546–57. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ et al. . Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet 2003; 72: 1293–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balak CD, Hunter JM, Ahearn ME, Wiley D, D’Urso G, Baumbach-Reardon L. Functional characterizations of rare UBA1 variants in X-linked spinal muscular atrophy. F1000Res 2017; 6: 1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter S, Karpati G, Rothman S, Watters G, Andermann F. Pathological involvement of primary sensory neurons in Werdnig-Hoffmann disease. Acta Neuropathol 1978; 42: 91–7. [DOI] [PubMed] [Google Scholar]
- Chang HC, Hung WC, Chuang YJ, Jong YJ. Degradation of Survival Motor Neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int 2004; 45: 1107–12. [DOI] [PubMed] [Google Scholar]
- Chang TK, Shravage BV, Hayes SD, Powers CM, Simin RT, Wade Harper J et al. . Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol 2013; 15: 1067–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler AT, Szczot M, Bharucha-Goebel D, Ceko M, Donkervoort S, Laubacher C et al. . The role of PIEZO2 in human mechanosensation. N Engl J Med 2016; 375: 1355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottenie E, Kochanski A, Jordanova A, Bansagi B, Zimon M, Horga A et al. . Truncating and missense mutations in IGHMBP2 cause Charcot-Marie Tooth disease type 2. Am J Hum Genet 2014; 95: 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delle Vedove A, Storbeck M, Heller R, Holker I, Hebbar M, Shukla A et al. . Biallelic loss of proprioception-related PIEZO2 causes muscular atrophy with perinatal respiratory distress, arthrogryposis, and scoliosis. Am J Hum Genet 2016; 99: 1206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlamini N, Josifova DJ, Paine SM, Wraige E, Pitt M, Murphy AJ et al. . Clinical and neuropathological features of X-linked spinal muscular atrophy (SMAX2) associated with a novel mutation in the UBA1 gene. Neuromuscul Disord 2013; 23: 391–8. [DOI] [PubMed] [Google Scholar]
- Duman O, Uysal H, Skjei KL, Kizilay F, Karauzum S, Haspolat S. Sensorimotor polyneuropathy in patients with SMA type-1: electroneuromyographic findings. Muscle Nerve 2013; 48: 117–21. [DOI] [PubMed] [Google Scholar]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 2002; 30: 1575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskuri JM, Stanley CM, Moore SA, Mathews KD. Infantile onset CMT2D/dSMA V in monozygotic twins due to a mutation in the anticodon-binding domain of GARS. J Peripher Nerv Syst 2012; 17: 132–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez RM, Pecina A, Munoz-Cabello B, Antinolo G, Borrego S. Co-segregation of a homozygous SMN1 deletion and a heterozygous PMP22 duplication in a patient. Clin Case Rep 2016; 4: 879–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T, Rasband W. ImageJ user guide—IJ 1.46r. 2012. Available from:https://imagej.nih.gov/ij/docs/guide/146–30.html.
- Fletcher EV, Simon CM, Pagiazitis JG, Chalif JI, Vukojicic A, Drobac E et al. . Reduced sensory synaptic excitation impairs motor neuron function via Kv2.1 in spinal muscular atrophy. Nat Neurosci 2017; 20: 905–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller HR, Mandefro B, Shirran SL, Gross AR, Kaus AS, Botting CH et al. . Spinal muscular atrophy patient iPSC-Derived motor neurons have reduced expression of proteins important in neuronal development. Front Cell Neurosci 2015; 9: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen EJN, Gillingwater TH. UBA1: at the crossroads of ubiquitin homeostasis and neurodegeneration. Trends Mol Med 2015; 21: 622–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen EJN, Perenthaler E, Courtney NL, Jordan CY, Shorrock HK, van der Hoorn D et al. . Temporal and tissue-specific variability of SMN protein levels in mouse models of spinal muscular atrophy. Hum Mol Genet 2018a; 27: 2851–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen EJN, Talbot K, Gillingwater TH. Advances in therapy for spinal muscular atrophy: promises and challenges. Nat Rev Neurol 2018b14, 214–24. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Schuelke M, Diers A, Hoffmann K, Lucke B, Adams C et al. . Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet 2001; 29: 75–7. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH et al. . A mouse model for spinal muscular atrophy. Nat Genet 2000; 24: 66–70. [DOI] [PubMed] [Google Scholar]
- Hunter G, Aghamaleky Sarvestany A, Roche SL, Symes RC, Gillingwater TH. SMN-dependent intrinsic defects in Schwann cells in mouse models of spinal muscular atrophy. Hum Mol Genet 2014; 23: 2235–50. [DOI] [PubMed] [Google Scholar]
- Jablonka S, Karle K, Sandner B, Andreassi C, von Au K, Sendtner M. Distinct and overlapping alterations in motor and sensory neurons in a mouse model of spinal muscular atrophy. Hum Mol Genet 2006; 15: 511–18. [DOI] [PubMed] [Google Scholar]
- James PA, Cader MZ, Muntoni F, Childs AM, Crow YJ, Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology 2006; 67: 1710–12. [DOI] [PubMed] [Google Scholar]
- Jedrzejowska M, Jakubowska-Pietkiewicz E, Kostera-Pruszczyk A. X-linked spinal muscular atrophy (SMAX2) caused by de novo c.1731C>T substitution in the UBA1 gene. Neuromuscul Disord 2015; 25: 661–6. [DOI] [PubMed] [Google Scholar]
- Jedrzejowska M, Ryniewicz B, Kabzinska D, Drac H, Hausmanowa-Petrusewicz I, Kochanski A. A patient with both Charcot-Marie-Tooth disease (CMT 1A) and mild spinal muscular atrophy (SMA 3). Neuromuscul Disord 2008; 18: 339–41. [DOI] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M et al. . Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet 2006; 38: 197–202. [DOI] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 1997; 272: 28557–62. [DOI] [PubMed] [Google Scholar]
- Kapur M, Monaghan CE, Ackerman SL. Regulation of mRNA translation in neurons-A matter of life and death. Neuron 2017; 96: 616–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb SJ, Kissel JT. Spinal muscular atrophy: a timely review. Arch Neurol 2011; 68: 979–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon DY, Dimitriadi M, Terzic B, Cable C, Hart AC, Chitnis A et al. . The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell 2013; 24: 1863–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao T, Chen S, Sang N. Two mutations impair the stability and function of ubiquitin-activating enzyme (E1). J Cell Physiol 2012; 227: 1561–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L et al. . Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80: 155–65. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A et al. . Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16: 265–9. [DOI] [PubMed] [Google Scholar]
- Ling KK, Lin MY, Zingg B, Feng Z, Ko CP. Synaptic defects in the spinal and neuromuscular circuitry in a mouse model of spinal muscular atrophy. PLoS One 2010; 5: e15457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Li X, Hu Z, Mao X, Zi X, Xia K et al. . IGHMBP2-related clinical and genetic features in a cohort of Chinese Charcot-Marie-Tooth disease type 2 patients. Neuromuscul Disord 2017a; 27: 193–9. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhao B, Sun L, Bhuripanyo K, Wang Y, Bi Y et al. . Orthogonal ubiquitin transfer identifies ubiquitination substrates under differential control by the two ubiquitin activating enzymes. Nat Commun 2017b; 8: 14286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A, Duchen LW. Sensory system involvement in infantile spinal muscular atrophy. J Neurol Sci 1975; 26: 349–59. [DOI] [PubMed] [Google Scholar]
- Maxwell GK, Szunyogova E, Shorrock HK, Gillingwater TH, Parson SH. Developmental and degenerative cardiac defects in the Taiwanese mouse model of severe spinal muscular atrophy. J Anat 2018; 232: 965–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L et al. . Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron 2011; 69: 453–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A et al. . Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet 2011; 7: e1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Talbot K, Fischbeck KH. GARS axonopathy: not every neuron’s cup of tRNA. Trends Neurosci 2010; 33: 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama S, Bouldin TW, Suzuki K. Immunocytochemical and ultrastructural studies of Werdnig-Hoffmann disease. Acta Neuropathol 1991; 81: 408–17. [DOI] [PubMed] [Google Scholar]
- Pedurupillay CR, Amundsen SS, Baroy T, Rasmussen M, Blomhoff A, Stadheim BF et al. . Clinical and molecular characteristics in three families with biallelic mutations in IGHMBP2. Neuromuscul Disord 2016; 26: 570–5. [DOI] [PubMed] [Google Scholar]
- Peeters K, Palaima P, Pelayo-Negro AL, Garcia A, Gallardo E, Garcia-Barredo R et al. . Charcot-Marie-Tooth disease type 2G redefined by a novel mutation in LRSAM1. Ann Neurol 2016; 80: 823–33. [DOI] [PubMed] [Google Scholar]
- Powis RA, Gillingwater TH. Selective loss of alpha motor neurons with sparing of gamma motor neurons and spinal cord cholinergic neurons in a mouse model of spinal muscular atrophy. J Anat 2016; 228: 443–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powis RA, Karyka E, Boyd P, Come J, Jones RA, Zheng Y et al. . Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy. JCI Insight 2016; 1: e87908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramser J, Ahearn ME, Lenski C, Yariz KO, Hellebrand H, von Rhein M et al. . Rare missense and synonymous variants in UBE1 are associated with X-linked infantile spinal muscular atrophy. Am J Hum Genet 2008; 82: 188–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid D, Zinger Y, Raheja D. Sensory neuronopathy in spinal muscular atrophy: a case presentation. J Clin Neuromuscul Dis 2016; 18: 44–6. [DOI] [PubMed] [Google Scholar]
- Riessland M, Ackermann B, Forster A, Jakubik M, Hauke J, Garbes L et al. . SAHA ameliorates the SMA phenotype in two mouse models for spinal muscular atrophy. Hum Mol Genet 2010; 19: 1492–506. [DOI] [PubMed] [Google Scholar]
- Rudnik-Schoneborn S, Goebel HH, Schlote W, Molaian S, Omran H, Ketelsen U et al. . Classical infantile spinal muscular atrophy with SMN deficiency causes sensory neuronopathy. Neurology 2003; 60: 983–7. [DOI] [PubMed] [Google Scholar]
- Savli H, Szendroi A, Romics I, Nagy B. Gene network and canonical pathway analysis in prostate cancer: a microarray study. Exp Mol Med 2008; 40: 176–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottmann G, Wagner C, Seifert F, Stenzel W, Schuelke M. MORC2 mutation causes severe spinal muscular atrophy-phenotype, cerebellar atrophy, and diaphragmatic paralysis. Brain 2016; 139 (Pt 12): e70. [DOI] [PubMed] [Google Scholar]
- Sevilla T, Lupo V, Martinez-Rubio D, Sancho P, Sivera R, Chumillas MJ et al. . Mutations in the MORC2 gene cause axonal Charcot-Marie-Tooth disease. Brain 2016; 139 (Pt 1): 62–72. [DOI] [PubMed] [Google Scholar]
- Shorrock HK, Gillingwater TH, Groen EJN. Overview of current drugs and molecules in development for spinal muscular atrophy therapy. Drugs 2018; 78: 293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon CM, Dai Y, Van Alstyne M, Koutsioumpa C, Pagiazitis JG, Chalif JI et al. . Converging mechanisms of p53 activation drive motor neuron degeneration in spinal muscular atrophy. Cell Rep 2017; 21: 3767–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A et al. . Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain 2005; 128 (Pt 10): 2304–14. [DOI] [PubMed] [Google Scholar]
- Sleigh JN, Dawes JM, West SJ, Wei N, Spaulding EL, Gomez-Martin A et al. . Trk receptor signaling and sensory neuron fate are perturbed in human neuropathy caused by Gars mutations. Proc Natl Acad Sci USA 2017; 114: E3324–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleigh JN, Weir GA, Schiavo G. A simple, step-by-step dissection protocol for the rapid isolation of mouse dorsal root ganglia. BMC Res Notes 2016; 9: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM et al. . Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72 400 specimens. Eur J Hum Genet 2012; 20: 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson AK, Somers E, Powis RA, Shorrock HK, Murphy K, Swoboda KJ et al. . Survival of motor neurone protein is required for normal postnatal development of the spleen. J Anat 2017; 230: 337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaart IEC, Robertson A, Wilson IJ, Aartsma-Rus A, Cameron S, Jones CC et al. . Prevalence, incidence and carrier frequency of 5q-linked spinal muscular atrophy—a literature review. Orphanet J Rare Dis 2017; 12: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart TM, Mutsaers CA, Riessland M, Reimer MM, Hunter G, Hannam ML et al. . Dysregulation of ubiquitin homeostasis and beta-catenin signaling promote spinal muscular atrophy. J Clin Invest 2014; 124: 1821–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart TM, Paterson JM, Short DM, Meredith S, Robertson KA, Sutherland C et al. . Differential proteomics analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wlds) gene. Mol Cell Proteomics 2007; 6: 1318–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P et al. . The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 2015; 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw proteomics data files from this study are freely available for download from http://dx.doi.org/10.7488/ds/2405.