Abstract
Aberrant chromatin transformation dysregulates gene expression and may be an important driver of tumorigenesis. However, the functional role of chromosomal dynamics in tumorigenesis remains to be elucidated. Here, using in vitro and in vivo experiments, we reveal a novel long noncoding (lncing) driver at chr12p13.3, in which a novel lncRNA GALNT8 Antisense Upstream 1 (GAU1) is initially activated by an open chromatin status, triggering recruitment of the transcription elongation factor TCEA1 at the oncogene GALNT8 promoter and cis-activates the expression of GALNT8. Analysis of The Cancer Genome Atlas (TCGA) clinical database revealed that the GAU1/GALNT8 driver serves as an important indicative biomarker, and targeted silencing of GAU1 via the HKP-encapsulated method exhibited therapeutic efficacy in orthotopic xenografts. Our study presents a novel oncogenetic mechanism in which aberrant tuning of the chromatin state at specific chromosomal loci exposes factor-binding sites, leading to recruitment of trans-factor and activation of oncogenetic driver, thereby provide a novel alternative concept of chromatin dynamics in tumorigenesis.
INTRODUCTION
The dynamic structure of chromatin exists in two conformational states: heterochromatin (closed chromatin) and euchromatin (open chromatin); changes in chromatin structure alter the accessibility of the DNA to regulatory factors during transcription, replication, recombination and DNA damage repair (1–3). Open chromatin exposes binding surfaces for many free transcription factors in the nucleus, whereas closed chromatin exhibits increased density and obscures protein-binding sites (4). Disruption of chromatin dynamics leads to aberrant gene expression, chromosomal translocation, nonexistent DNA repair, oncogenesis and even necrosis (5,6). For example, the histone chaperone subunit SPT16 can accelerate the dynamic exchange of histone proteins and reinitiate RNA synthesis upon damage removal (7). Open chromatin facilitates protein-DNA binding in the TCF7L2 risk allele, inducing a higher transcription level relative to the non-risk allele and resulting in type 2 diabetes (8). Therefore, chromatin dynamics have attracted increasing attention in the pathogenesis of human disease.
As chromatin dynamics play a key role in the maintenance of homeostasis, aberrant transformation of dynamic chromatin dysregulates protein-coding gene expression and may be an important inducer of tumorigenesis. An increasing number of accessible chromatin regions have been identified in the genome of prostate cancer, and such prostate cancer-specific chromatin dynamics have formed a signature by which prostate cancer patients can be identified (9). In addition, dysregulated chromatin structure at the MYC region can directly result in overexpression of the oncogene MYC, thereby inducing many kinds of cancers (10,11). Moreover, chromosomal changes can also simultaneously affect multiple neighboring genes. For example, repressed chromatin in the INK4b–ARF–INK4a protein-coding gene cluster inhibits the expression of this cluster and accelerates tumor formation (12,13). Thus, exploration of the novel chromosomal drivers of tumorigenesis is potentially interesting.
More intriguingly, in addition to protein-coding gene clusters, chromatin dynamics can also manipulate long non-coding RNA (lncRNA) expression to influence tumor formation. Chromosomal condensation activates the lncRNA H19 promoter and triggers tumor formation (14). In addition, chromatin dynamics can simultaneously induce two lncRNAs at chr13q14.3 that target NF-κB in leukemia (15). However, whether dynamic changes in chromosome structure can simultaneously regulate the transcription of long non-coding RNA and protein-coding genes, especially in tumorigenesis, remains uncertain.
At present, the role of chromosomal dynamics in the co-regulation of lncRNAs and proteins in tumorigenesis remain to be elucidated. We thus aimed to identify chromatin dynamics that simultaneously regulate long non-coding RNA and protein-coding genes and reveal their potential mechanism in tumorigenesis. In our study, we identified for the first time a novel dynamic chromosomal driver that accelerates tumor progression by co-activating a gene cluster in chr12p13.32 that includes a novel lncRNA GAU1 and a protein-coding gene GALNT8, thereby presenting a novel dynamic mechanism of chromatin tuning.
MATERIALS AND METHODS
RNA extraction, library construction and Illumina sequencing (RNA-Seq)
Total RNA was extracted from retinoblastoma and para-tumor samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). We confirmed RNA integrity using the 2100 Bioanalyzer (Agilent Technologies, USA) and measured RNA concentration in a Qubit 2.0 fluorometer using the Qubit RNA Assay Kit (Life Technologies, Carlsbad, CA, USA). We prepared libraries from 100 ng of total RNA using an Illumina TruSeq RNA Sample Prep Kit (San Diego, CA, USA) following the manufacturer's protocol. In total, 12 libraries were sequenced using the Illumina Hiseq platform (San Diego, CA, USA). The mRNA levels of the unigenes were identified using TopHat v2.0.9 and Cufflinks and normalized by the Fragments Per Kilobase of exon model per Million mapped reads (FPKM). We used the criteria of false discovery rate (FDR) <0.01 and fold changes <0.5 or >2.0 (<−1 or >1 log2 ratio value, P value < 0.05) to identify differentially expressed genes.
Cell culture
The human SK-N-AS and 293T cell lines were cultured in DMEM (GIBCO) supplemented with 10% certified heat-inactivated fetal bovine serum (FBS; GIBCO), penicillin (100 U/ml), and streptomycin (100 mg/ml) at 37°C in a humidified 5% CO2 atmosphere. RPE1 and ARPE19 cells were cultured in DMEM/F12 medium (GIBCO), and Y79 and WERI-RB1 cells were cultured in RPMI 1640 medium (GIBCO). HSC cells were obtained as follows: sterile peripheral nerve tissue from a 28-year-old man was cut into pieces and then digested in 0.1% collagenase A (GIBCO) for 18 h at 37°C; the pieces were then pipetted from the turbid liquid to PBS without Ca2+ and Mg2+. After passing the cells through a filter with a 100-mm pore size, the cell-containing solutions were centrifuged for 5 min; the supernatant was subsequently discarded, and the pellet was resuspended in 10% FBS DMEM to yield a single-cell suspension. The cells were seeded at a density of 1 × 105/cm2 and cultured as described above.
Formaldehyde-assisted isolation of regulatory elements (FAIRE)
FAIRE was performed as previously described (16). Briefly, 37% formaldehyde was added to a final concentration of 1% followed by the addition of 2.5 M glycine to quench the formaldehyde. The samples were then centrifuged at 300–500 g for 5 min at 4°C to collect the fixed cells. The pellets were washed with 10 ml of 1× PBS for a total of three washes. The fixed cells were then resuspended in 1 ml of cold lysis buffer and sonicated to achieve an average DNA fragment size of ∼200–400 bp. The supernatant was transferred to a fresh 1.5-ml tube and incubated with 1 μl of DNase-free RNaseA for 30 min at 37°C. Subsequently, 300 μl of phenol/chloroform/isoamyl alcohol (Sangon, Shanghai, China) was added, and the sample was vortexed for 10 s followed by centrifugation at 12 000 g for 5 min. The aqueous (top) layer was then transferred, and the extraction process was repeated three times, pooling all the aqueous solutions. Subsequently, 1/10 volume of 3 M sodium acetate, 2 volumes of 95% ethanol and 1 μl of 20 mg/ml glycogen were added, and the solution was incubated at −80°C for at least 30 min. The DNA was then purified using a DNA Clean-up Kit (AxyPrep).
TCGA dataset
To validate the potential role of GAU1 and GALNT8 in retinoblastoma and neuroblastoma, we searched The Cancer Genome Atlas (TCGA; http://www.cbioportal.org) and the R2 (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) Dorsman and Versteeg database, which provided gene expression data and follow-up information on 88 neuroblastoma cases and gene expression data in 76 retinoblastoma cases.
RNA-FISH
The RNA-FISH assay was performed as previously described (17). Briefly, cells were fixed with 4% formaldehyde/10% acetic acid and stored overnight in 70% ethanol. The fluorescence-labeled single-strand probes were synthesized and then hybridized. U2 and GAPDH oligos were purchased from Sangon. To increase the stability of RNA foci, RNA signals were detected with a tyramide-Alexa Fluor 546 signal amplification kit (Invitrogen). After labeling, fluorescence signals were detected using a microscope (BX41; Olympus). Optical sections of 0.5-mm thickness were collected with SlideBook 5.0 (Intelligent Imaging Innovations).
Immunohistochemical staining
For immunohistochemical staining, tissues sections were incubated at 4°C overnight with a rat anti-human antibody (Proteintech, USA) at a dilution of 1:100. Sections were then rinsed in PBS-T (PBS containing 0.05% Triton X-100), and biotinylated anti-rat secondary antibody was added at a 1:500 dilution and incubated at room temperature for 1 h. After two washes with PBS-T, the slides were incubated with streptavidin-horseradish peroxidase (BD Biosciences, USA) and diaminobenzidine substrate for colorimetric development.
Nuclear fractionation
The cellular fraction was isolated as previously described. In brief, 107 cells were harvested; resuspended in 1 ml of ice-cold DEPC-PBS, 1 ml of buffer C1 (1.28 M sucrose, 40 mM Tris–HCl [pH 7.5], 20 mM MgCl2, 4% Triton X-100), and 3 ml of RNase-free water; and incubated for 15 min on ice. Then, cells were centrifuged for 15 min at 3000 rpm; the supernatant containing the cytoplasmic component and the pellet containing the nuclear fraction were kept for RNA extraction.
Luciferase reporter system
HEK-293T cells cultured in a six-well plate were co-transfected with 2 μg of the firefly luciferase reporter vector containing the forward or reverse fragment of the first exon or full length of GAU1 or GALNT8 or the gap between the genes. Transfections were performed in triplicate and were repeated in three independent experiments. Forty-eight hours after transfection, luciferase activity was analyzed using a Dual-Luciferase Reporter Assay System (Promega, USA) per the manufacturer's protocol. Firefly luciferase values were normalized to renilla luciferase values to control for transfection efficiency.
Plasmid construction
The dCas9 KRAB vector (WEF1A-dCas9-SV40NLS-KRAB-P2A-Hygromycin-WPRE) and SgRNA vector (hU6-sgRNA-EF1A-Puro-WPRE) were used in our system. The two shRNA sequences targeting GALNT8 were generated by PCR and then cloned into the pGIPZ lentivirus vector (System Biosciences, USA). The GAU1 overexpression cassette was generated by PCR and cloned into the CMV-MCS-EF1α-ZsGreen1-PGK-Puro vector (Addgene).
Lentivirus packaging and generation of stable cell lines
The Lipofectamine 3000 reagent (Invitrogen) was incubated with Opti-MEM I Reduced Serum Medium (GIBCO) and used to transfect 239T cells with 3 mg of the PCMV-GAU1, pGIPZ-shGALNT8-1, pGIPZ-shGALNT8-2 or pMD2.D plasmids or 6.0 mg of the PsPax plasmid. Six hours after transfection, the medium was replaced with 10 ml of fresh medium. The supernatant containing the viruses was collected at 48 and 72 h. The virus-containing solution was filtered and concentrated. Viruses carrying the dCas9-KRAB and sgRNA were premixed 1:1, and 50 μl of the virus was added into 1 ml of serum. Twenty-four hours prior to transfection, tumor cells were seeded at 2.0 × 105 cells per well in a six-well plate. The medium was replaced with virus-containing supernatant supplemented with 10 ng/ml polybrene (Sigma-Aldrich). After 48 h, the medium was replaced with fresh medium. Cells were selected by incubation with 4 mg/ml puromycin (InvivoGen) for 2 weeks. Cells transfected with the dCas9 virus were subsequently selected by incubating with 6 mg/ml hygromycin (InvivoGen) for 2 weeks. Colonies were selected and expanded for further analyses.
Cell cycle analysis
A total of 106 cells were centrifuged at 485 g for 5 min, washed once with PBS, resuspended in 1 ml of PBS and fixed overnight in 75% ethanol at 220°C. Fixed cells were washed three times with 10 ml of PBS, resuspended in 200–400 ml of PBS containing 10 ml of RNase (Qiagen) and incubated at 37°C for 30 min in the dark. Cells were subjected to FACS analysis by the Flow Cytometry Facility.
Colony formation assay
A 1-ml volume of 0.6% agar complete medium was spread in each well of a six-well plate to form the bottom layer, and 5000 cells were resuspended in 1.0 ml of 0.3% agar complete medium and seeded as the upper layer. The cells were cultured with 300 ml of complete medium for 3–4 weeks. The colonies in soft agar were stained with 0.005% crystal violet and then photographed.
Small interfering RNA
siRNA oligos targeting GAU1 (siRNA-1:5′-CAGACAUGCACUUCCAUGU-3′; siRNA-2: 5′-GACACAGAAAAGUCACACA-3′; siRNA-3: 5′-CAGCAUUGAAAU UGAUCAA-3′) and negative control siRNA were purchased from GenePharma. siRNA transfections were performed with 50 nM siRNA and Lipofectamine RNAiMAX (Life Technologies) as previously described.
Western blot
Cells were harvested at the indicated times and rinsed twice with PBS. Cell extracts were prepared with lysis buffer and centrifuged at 13 000 g for 30 min at 4°C. Protein samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 7.5% (w/v) polyacrylamide gels and transferred to polyvinylidene fluoride membranes. After blocking with 5% BSA for 1 h at room temperature, the membrane was incubated with 2.5 μg/ml antibody in 5% BSA overnight at 4°C. The membrane was then incubated with a secondary antibody conjugated to a fluorescent tag (Invitrogen). The band signals were visualized and quantified using the Odyssey Infrared Imagining System (LI-COR, USA). The following antibodies were used in this study: Anti-GALNT8 (Abcam, USA), Anti-TCEA1(Abcam, USA) and GAPDH (Sigma-Aldrich).
Intravitreal injection
The animal experiments were approved by the Shanghai Jiao Tong University Animal Care and Use Committee and conducted in accordance with the animal policies of the Shanghai JiaoTong University and the guidelines established by the National Health and Family Planning Commission of China. The cells were harvested by trypsinization and washed twice with PBS (GIBCO). BALB/c nude mice (male, 6 weeks old) were used for the study. Each animal was first anesthetized with the topical anesthetic Benoxil. Methocel eye drops were used to avoid drying of the eyes. Injections were performed using a surgical microscope. A 2-μl volume of sterile phosphate buffered saline containing 2 × 104 Y79 human retinoblastoma cells were injected into the vitreous of each eye through the sclera using a Hamilton syringe. After the injection, the eyes were treated with antibiotic eye drops. All mice were sacrificed by cervical dislocation 40 days after implantation for the tumor formation analysis or 72 days after intravitreal injection for the survival analysis.
Delivery of siRNA using poly-HKP
The branched histidine (H) and lysine (K) polymers used in this study were R-KR-KR-KR, where R = [HHHKHHHKHHHKHHH]2KH4NH4 or KHHHKHHH KHHHKHHHK and H3K4b, a branched polymer with the same core and structure as previously described. The HKP was dissolved in aqueous solution for a concentration of 10 mM and then mixed with a siRNA aqueous solution at a ratio of 4:1 by mass, forming nanoparticles with an average size of 150–200 nm in diameter.
ChIRP
A total of 2 × 107 cells were resuspended in precooled PBS buffer and crosslinked with 3% formaldehyde at room temperature on an end-to-end shaker for 30 min. The reaction was quenched by adding 125 mM glycine for 5 min, and the solution was centrifuged at 1000 RCF for 3 min. Pre-binding probes (5 for TT, 1 for NC and PC, 100 pmol per 2 × 107 cells) were added to streptavidin beads for 30 min; then, the unbound probes were removed by washing. The beads were mixed with the cell lysate and hybridized at 37°C overnight on an end-to-end shaker. Subsequently, the beads were washed five times with 1 ml of prewarmed wash buffer for 5 min per wash. At the last wash step, 1/20 of the beads were reserved for qPCR analysis. Then, 100 μl of elution buffer containing 20 U benzonase was added, and the protein was eluted at 37°C for 1 h. The supernatant was transferred to a new low-binding tube, and the beads were eluted again with 100 μl of elution buffer. The two supernatants were combined, and the crosslink was reversed at 95°C for 30 min. Subsequently, the protein was precipitated with 0.1% SDC and 10% TCA at 4°C for 2 h followed by centrifugation at top speed. The pellets were then washed with prechilled 80% acetone three times.
RNA-chromatin immunoprecipitation (RNA-ChIP)
RNA-ChIP was performed using the EZ-Magna RIP kit (Millipore) per the manufacturer's instructions as previously described. In brief, 107 cells were lysed with RIP lysis buffer with one freeze-thaw cycle. Cell extracts were coimmunoprecipitated with anti-TCEA1, anti-RBMX, anti-MCM2 or anti-CBX3 (Abcam), and the retrieved RNA was subjected to real-time qPCR analysis. Normal rabbit IgG was used as a negative control.
Chromatin immunoprecipitation (ChIP)
ChIP was conducted using an EZ-Magna ChIP A/G kit (Millipore) according to the manufacturer's instructions and a previously reported protocol. The anti-TCEA1, anti-RBMX, anti-CBX3 and anti-MCM2 antibodies used for RNA-ChIP were also applied for ChIP. Anti-normal mouse IgG was used as a negative control, and anti-RNA polymerase-II (pol-II; Millipore) was used as a positive control. Primers for ChIP-qPCR are listed in Supplementary Table S1.
RESULTS
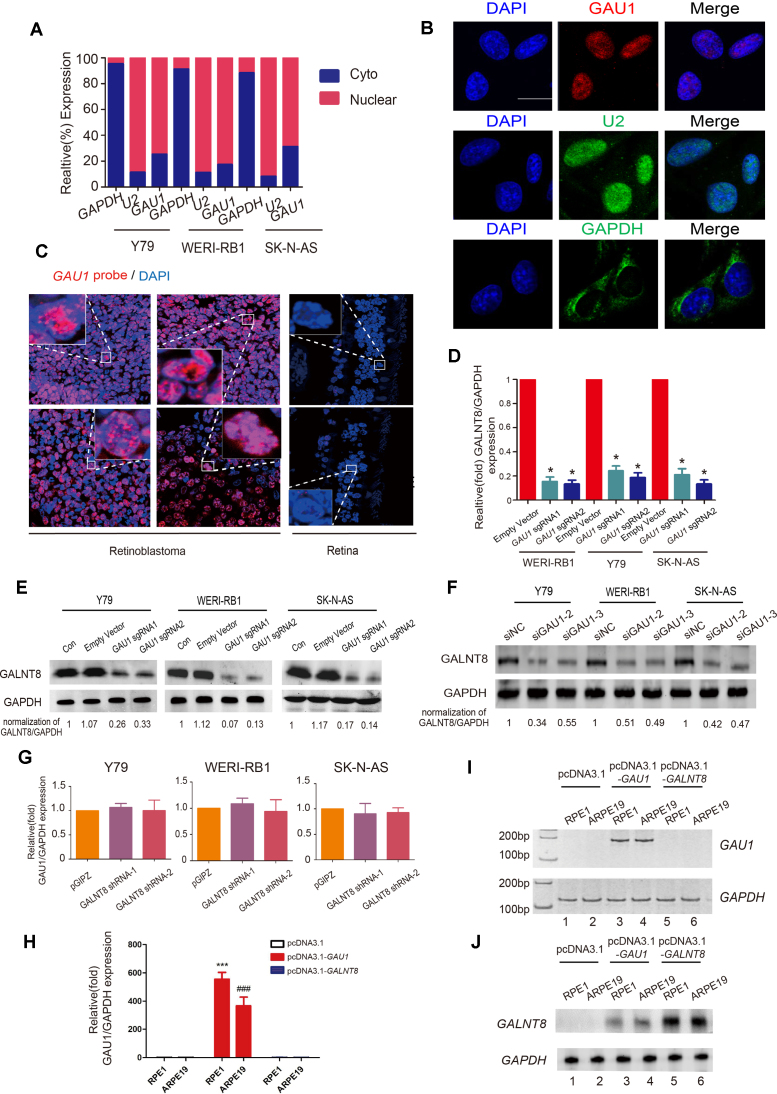
Open chromatin status leads to co-activation of GAU1 lncRNA and GALNT8 in retinoblastoma and neuroblastoma
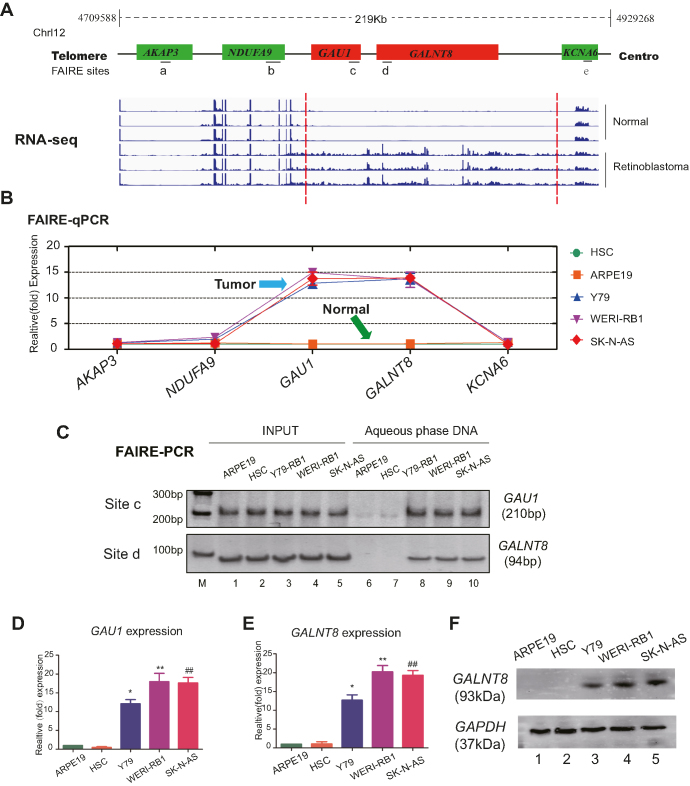
To investigate the status of dynamic chromosomal structure in tumors, genome-wide RNA-Seq (NODE, http://www.biosino.org/node/project/detail/NODEP0037193, GEO, https://www.ncbi.nlm.nih.gov/geo/, GEO Accession number: GSE111168) was first carried out to map the potential gene variations, and formaldehyde-assisted isolation of regulatory elements (FAIRE) was subsequently performed to determine the status of the chromosomes (Figure 1A, red chart). As expected, by aligning the two maps, we discovered that the chromosomal region chr12p13.32 had an open status in retinoblastoma and neuroblastoma cells; this region encodes the uncharacterized lncRNA LOC101929549 and the protein-coding gene GALNT8. In contrast, the neighboring genes NAUFA9, AKAP3 and KCNA6 remained in a closed chromosomal state in tumor and normal cells (Figure 1B and C). Gene expression analysis showed that both GALNT8 and LOC101929549 were significantly overexpressed in neuroblastoma and retinoblastoma cells (Figure 1D–F; Supplementary Figure S1A, lanes 10–12). Of note, the lncRNA LOC101929549 presented weak expression in other tumors (uveal melanoma and conjunctival melanoma) (Supplementary Figure S1A, lanes 4–9). Since histone modification is an important indicator in dynamic chromatin tuning, we then detected the changes of histone acetylation and methylation across LOC101929549/GALNT8 locus (Supplementary Figure S2A). As expected, the histone signals of open chromatin (H3K4me, H3K4me3, H3K9ac and H3K27ac) were significantly increased at LOC101929549/GALNT8 locus in retinoblastoma and neuroblastoma cell as compared with normal cells (Supplementary Figure S2B and C), while no significant histone modification was observed in negative site (Supplementary Figure S2D). These data show that the open chromatin status co-drives the expression of LOC101929549 lncRNA and GALNT8 in tumors.
Figure 1.
The chromatin status and transcription of 12p13.32 in tumors. (A) RNA-sequence analysis was performed to evaluate the transcriptome in three retinoblastoma samples and normal para-tumor samples. Red Chart: highly expressed locus in Chr12p13.32. Green Chart: lowly expressed locus in Chr12p13.32. (B and C) Quantification of the FAIRE at Chr12p13.32 indicates an open chromatin state in tumor cells (retinoblastoma: Y79 and WERI-Rb1; neuroblastoma: SK-N-AS). The normal cells were HSC and ARPE19 cells. Input DNA was used as a positive control. (D and E) Real-time PCR showed that GAU1/GALNT8 cluster mRNA expression is upregulated in tumor cells. M: marker; *P< 0.05 and **P< 0.01 compared with ARPE19; and ##P< 0.01 compared with HSC. (F) Western blot showed that the GALNT8 protein is overexpressed in tumor cells (Y79, WERI-Rb1 and SK-N-AS) compared to normal cells (HSC and ARPE19).
Next, we determined whether LOC101929549 encoded a new transcript in neuroblastoma and retinoblastoma cells. The previously reported LOC101929549 lncRNA was 1294 bp in length with four exons according to the National Center for Biotechnology Information (NCBI) database. However, after rapid amplification of cDNA ends (RACE) detection, we identified a novel 1339-bp transcript (Supplementary Figure S1B and C). More precisely, exons 2 and 3 were consistent with the predicted exons, but exon 1 had an additional 57-bp fragment at the 5′ terminus, and exon 4 was extended by 4 bp. This novel transcript also had an additional poly-A tail at the 3′ terminus compared with the predict sequence (Supplementary Figure S1D).
Using the GENCODE annotation of the human genome, we then started to search the non-coding evidence for this novel transcript (Supplementary Figure S3A). We failed to detect the important Kozak sequence for translational initiation in GAU1 lncRNA, the coding potential calculator (CPC) also showed that the full-length transcript had no protein-coding potential (Supplementary Figure S3B). In addition, we cloned the predicted two GAU1 ORFs into pcDNA3.1 vector with an enhanced green fluorescent protein (EGFP) tag (Supplementary Figure S3C). We then transfected the EGFP-tagged expressing vectors into SK-N-AS cells, similar to HOTAIR (Supplementary Figure S3D, lane 4) or No-DNA negative control (Supplementary Figure S3D, lane 5), the immunofluorescence analysis showed that EGFP did not observed in GAU1 group (Supplementary Figure S3D, lanes 2 and 3), while GALNT8 positive control showed strong fluorescence (Supplementary Figure S3D, lane 1). Similarly, western-blot analysis also showed that GAU1 could not code for protein (Supplementary Figure S3E, lanes 2 and 3). To further investigate the coding potential of GAU1 lncRNA, by following the principle (18), a series of 5′UTR-ORF-GFPmut constructs were generated (Supplementary Figure S3F). As expected, substantial expression of the GFP fusion protein was observed in known peptide-coding lncRNA HOXB-AS3 (18) and wild type GFP transfected cells (Supplementary Figure S3G lanes 1 and 3; Supplementary Figure S3H, lanes 1 and 3). However, we did not detect the expression of the ORFs of GAU1 or HOTAIR (Supplementary Figure S3G, lanes 4–6; Supplementary Figure S3H, lanes 4–6). Collectively, these data show that the isoform of the LOC101929549 lncRNA that was identified near GALNT8 is a novel non-coding transcript in tumors; we therefore named it GALNT8 Antisense Upstream 1 (GAU1, GenBank: MF576259).
In addition, we cloned the predicted two GAU1 ORFs into pcDNA3.1 vector with an enhanced green fluorescent protein (EGFP) tag (Supplementary Figure S3C). We then transfected the EGFP-tagged expressing vectors into SK-N-AS cells, similar to HOTAIR (Supplementary Figure S3D, lane 4) or No-DNA negative control (Supplementary Figure S3D, lane 5), the immunofluorescence analysis showed that EGFP did not observed in GAU1 group (Supplementary Figure S3D, lanes 2-3), while GALNT8 positive control showed strong fluorescence (Supplementary Figure S3D, lane 1). Similarly, western-blot analysis also showed that GAU1 could not code for protein (Supplementary Figure S3E, lanes 2 and 3).
Suppression of the GAU1/GALNT8 cluster inhibits tumor progression in vitro and in vivo
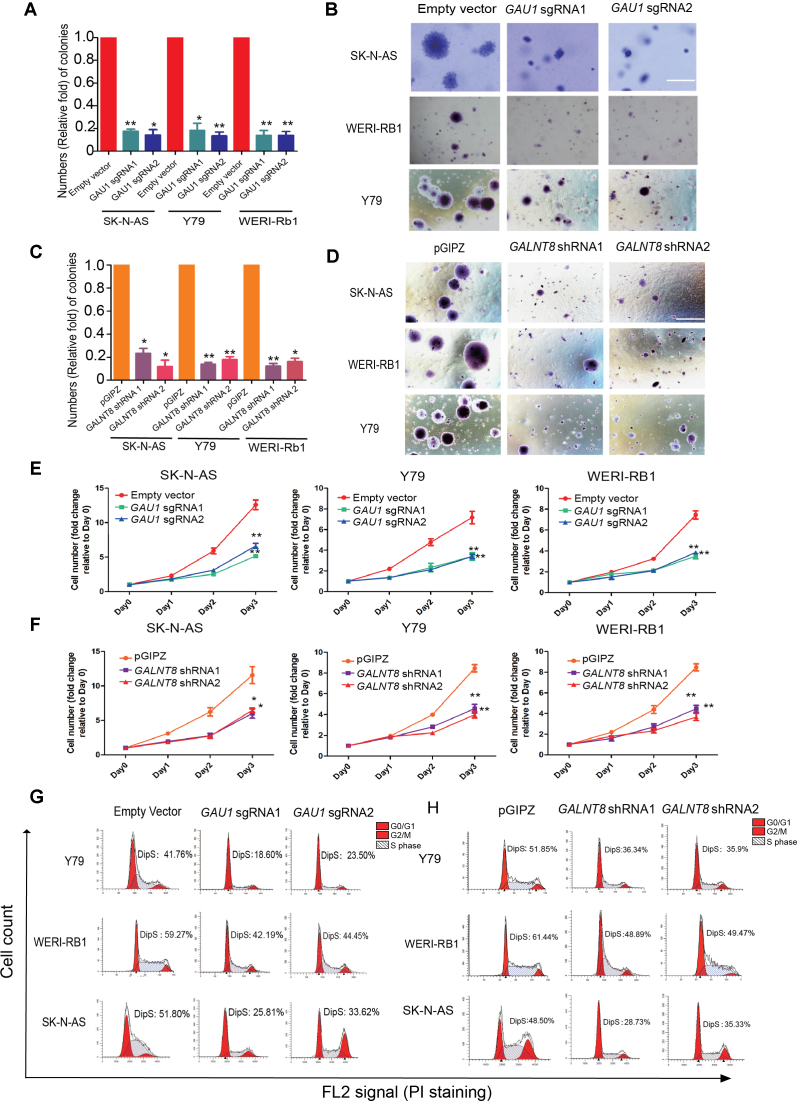
To determine whether this novel GAU1/GALNT8 cluster could alter tumor behavior, it was necessary to identify the promoters of GAU1 and GALNT8. GALNT8 and GAU1 have opposite transcriptional directions in the genome, and an additional 428-bp fragment was identified between GAU1 and GALNT8. Using a gene promoter reporter assay, we identified the 303-bp core promoter of GAU1 (Supplementary Figure S4A, g fragment) and the 294-bp promoter of GALNT8 (Supplementary Figure S4A, d fragment) (Supplementary Figure S4B). Next, we knocked down GAU1 lncRNA expression using the dCas9-KRAB system without splicing the genomic DNA. Two sgRNAs were then designed to repress the expression of GAU1 using the dCas9-KRAB vector (Supplementary Figure S4C). As expected, GAU1 expression levels were successfully knocked down to ∼20% of its original expression levels in tumor cells (Supplementary Figure S4D). In addition, GALNT8 expression was significantly knocked down by two different shGALNT8s (Supplementary Figure S4E). Next, we examined cell colony formation ability in vitro using a classical soft agar assay. Compared with the empty vector group, tumor cell colony formation was significantly decreased after GAU1 silencing (Figure 2A). We also observed that the GAU1 knockdown cells formed very small colonies (Figure 2B). Additionally, the data demonstrated that colony formation by the three GALNT8-silenced tumor cells was significantly reduced (Figure 2C). Moreover, after GALNT8 knockdown, we observed that cell colonies were remarkably smaller than those formed by wild-type tumor cells (Figure 2D). In addition, the CCK8 assays showed that the cell growth was significantly inhibited in GAU1-silenced (Figure 2E) and GALNT8-suppressed tumor cells (Figure 2F). Moreover, significant decreases in GAU1-silenced (Figure 2G) and GALNT8-suppressed tumor cells (Figure 2H) in S phase were observed.
Figure 2.
The oncogenetic role of the GAU1/GALNT8 cluster in tumors in vitro. (A) Quantification of visible colonies. The colony number in the empty vector group was set as 100%. All the experiments were performed in triplicate, and the relative colony formation rates are shown as the mean ± SEM. *P<0.05 and **P<0.01; scale bar: 5 mm. (B) A soft agar tumor formation assay was performed to determine the colony formation ability of GAU1-silenced tumor cells (Y79, WERI-Rb1 and SK-N-AS). (C) Quantification of visible colonies. The colony number in the empty vector group was set as 100%. All the experiments were performed in triplicate, and the relative colony formation rates are shown as the mean ± SEM. *P< 0.05; scale bar: 5 mm. (D) A soft agar assay was performed to determine the colony formation ability of GALNT8-suppessed tumor cells (Y79, WERI-Rb1 and SK-N-AS). (E and F) CCK8 assay was performed to measure 3-day cell growth rate after GAU1 knock down. (H) CCK8 assay after GALNT8 silencing in tumor cells. (G) Flow cytometry analysis was performed to determine the percentage of cells in the different cell cycle phases. The percentage of cells in S phase decreased after GAU1 was knocked down in Y79, WERI-Rb1 and SK-N-AS cells. (H) The percentage of cells in S phase decreased after GALNT8 was knocked down in Y79, WERI-Rb1 and SK-N-AS cells. The X axial represents FL2 tunnel captured PI staining signals and the Y axial represents cell counts. Fcs data was automatically analyzed by Flowjo.
Next, we overexpressed GAU1 or GALNT8 in normal cells (RPE1 and ARPE19) (Supplementary Figures S5A and B). We observed the cell colonies were significantly increased in GAU1-overexpressed and GALNT8-overexpressed normal cells (Supplementary Figures S5C and D). Moreover, the cell proliferation was also promoted, either in GAU1-overexpressed or in GALNT8-overexpressed normal cells (Supplementary Figures S5E and F). In GAU1-overexpressed or GALNT8-overexpressed tumor cells (Supplementary Figures S6A and B), however, we did not observe the significant changes, either in cell growth (Supplementary Figure S6C) or in soft agar colony formation (Supplementary Figures S6D and E).
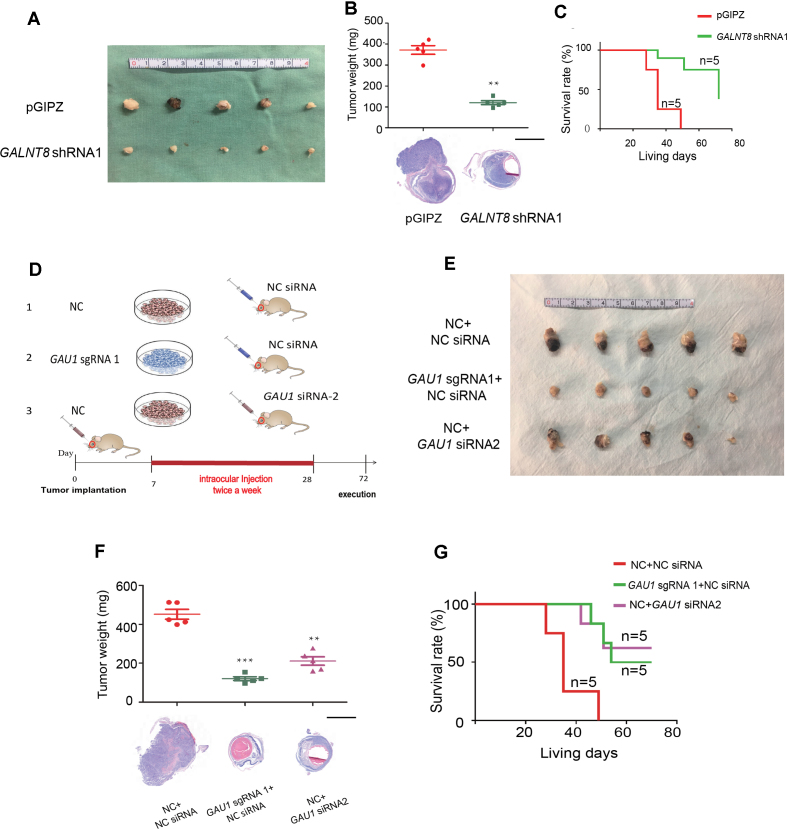
To determine the role of the GAU1/GALNT8 cluster in tumor characteristics in vivo, we performed orthotopic transplantation of Y79 retinoblastoma cells in the eyes of mice. Since we did not detect the significant changes of tumor characteristics in vitro in GAU1-overexpressed or GALNT8-overexpressed tumor cells, we thus focused on the oncogenetic role of the GAU1/GALNT8 by silencing of GAU1 or suppression of GALNT8 in tumor cells. In the GAU1-silenced group (Figure 3A, Group a), we found that both tumor volume (Figure 3B, second panel) and weight (Figure 3C, lane 2) were significantly reduced compared with the empty vector group (Figure 3A, Group b). Since siRNA has potential applications in the clinic, we designed three HKP-scrambled siGAU1s, and the most efficient No. 2 siGAU1 (Supplementary Figure S7A) was injected into the eye to form orthotopic xenografts. As expected, the siGAU1 group (Figure 3A, Group c) also exhibited a remarkable reduction in tumor formation both in terms of size (Figure 3B, third panel) and weight (Figure 3C, lane 3). Most importantly, after knockdown of GAU1 lncRNA, the survival rate of mice was significantly extended in both the sgGAU1 and siGAU1 groups (Figure 3D). Since in vitro experiment showed that GALNT8 shRNA1 could obtain more efficient efficacy of tumor-inhibition, we then chose GALNT8 shRNA1 for next in vivo experiment. As expected, tumor volume (Figure 3E) and weight (Figure 3F) were notably reduced in the GALNT8 knockdown group. In addition, we observed an increased survival rate after silencing GALNT8 via the classical shRNA method (Figure 3G). Taken together, these data indicate that GAU1 lncRNA and GALNT8 play an oncogenetic regulatory role and that the GAU1/GALNT8 cluster may serve as a potential cancer-driving chromosomal region.
Figure 3.
Suppression of the GAU1/GALNT8 cluster elicits a therapeutic effect in vivo. (A) Schematic of the animal experiment group. Group A: control cells were injected into the vitreous, and scrambled siRNA was subsequently injected into the vitreous with an HKP vector twice per week (from the 7th day to 28th day after implantation); Group B: GAU1 knockdown cells were injected and scrambled siRNA was administered as described above; and Group C: negative control cells were injected and GAU1 siRNA was administered as described above. (B): Representative image of the orthotopic xenograft formed by Y79 cells injected into the vitreous with or without GAU1 knockdown by sgRNA or siRNA at 40 days after implantation; n = 5. (C) Top: bar graph shows tumor volumes formed by the indicated Y79 cells in mouse vitreous. Bottom: representative images of H&E staining for the evaluation of tumor formation. n = 5; scale bar: 2 mm. (D) Survival analysis of mice following vitreous implantation with Y79 cells with or without GAU1 knockdown by sgRNA or siRNA; n = 5. (E) General photograph of orthotopic xenograft at 40 days after implantation by the injection of Y79 cells into the vitreous with or without GALNT8 knockdown; n = 5. (F) Top: bar graph shows tumor volumes formed by the indicated Y79 cells in mouse vitreous. Bottom: representative images of H&E staining for the evaluation of tumor formation. n = 5; scale bar: 2 mm. (G) Survival analysis of mice following vitreous implantation with Y79 cells with or without GALNT8 knockdown; n = 5.
The GAU1/GALNT8 cluster serves as a potential indicative biomarker in tumors
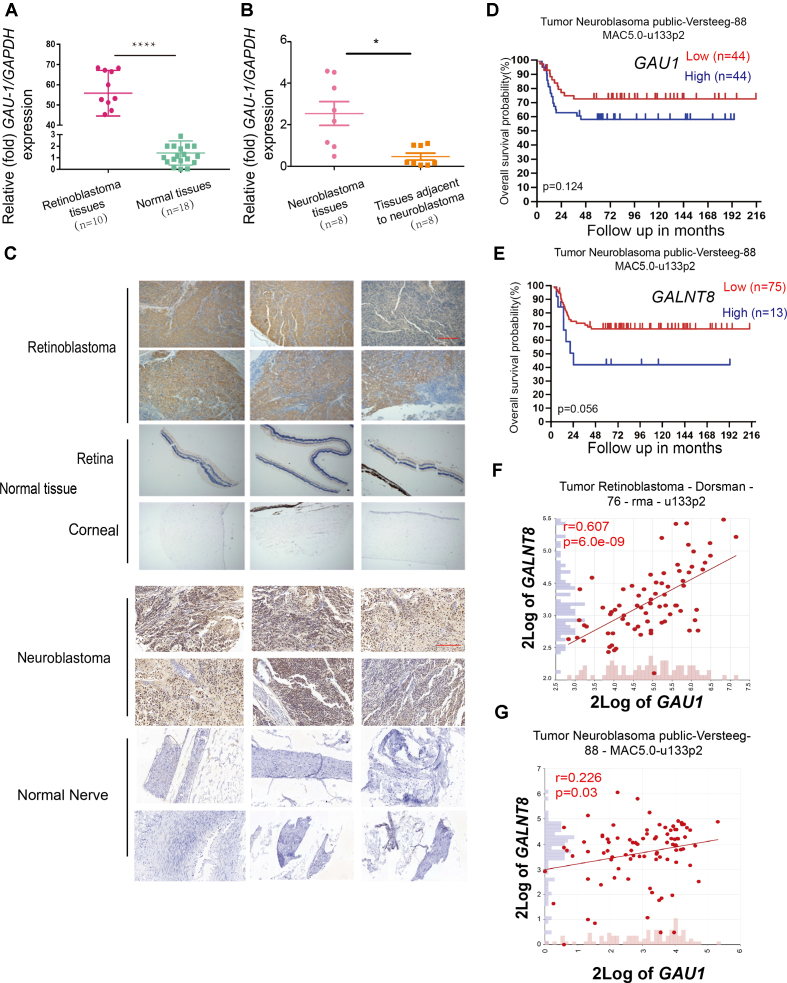
We were interested in exploring the potential diagnostic role of the GAU1/GALNT8 cluster in tumors, as real-time PCR showed that GAU1 was significantly elevated in retinoblastoma (P0.001) and neuroblastoma (P< 0.05) tissues compared with normal tissues (Figure 4A and B; Supplementary Tables S2 and S3). Using immunohistochemical staining, we also demonstrated GALNT8 expression in tumor tissues, including retinoblastoma and neuroblastoma, while normal control tissues exhibited weak GALNT8 staining (Figure 4C). To explore the prognostic role of this cluster in tumors, we investigated the overall survival probability. As expected, The Cancer Genome Atlas (TCGA; Versteeg database; http://www.cbioportal.org) showed that higher GAU1 or GALNT8 levels correlated with worse prognosis (Figure 4D and E). Moreover, the R2 (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) Dorsman database showed that GAU1 and GALNT8 expression was highly relevant (r = 0.607, P = 6.0e-09) in retinoblastoma clinical tumor cases (Figure 4F). In addition, R2 Versteeg database presented GAU1 and GALNT8 was relevant (r = 0.226, P = 0.03) in neuroblastoma clinical tumor cases (Figure 4G). These clinical data showed that the GAU1/GALNT8 cluster serves as an important indicative biomarker in tumors and that targeting the GAU1/GALNT8 cluster may be an effective tumor therapy.
Figure 4.
Clinical significance of the GAU1/GALNT8 cluster in tumors. (A and B) GAU1 expression levels in retinoblastoma tissues (A) and neuroblastoma tissues (B). GAU1 was highly expressed in retinoblastoma and neuroblastoma tissues compared with normal tissues; ****P< 0.0001. *P< 0.05. (C) Immunohistochemical staining of GALNT8 in tumor and normal tissues. GALNT8 expression was higher in tumor sections from six retinoblastoma patients and six neuroblastoma patients than in normal tissues (original magnification, 200×); scale bar: 30 μm. (D and E) Correlation between GAU1 mRNA expression and neuroblastoma patient survival in the TCGA dataset. Overall patient survival in groups with high and low GAU1 expression was analyzed using a Kaplan-Meier survival curve. (F and G) GAU1 expression strongly correlated with GALNT8 expression in clinical tumor samples from the Dorsman and Versteeg database.
GAU1 is a nuclear lncRNA and regulates the expression of GLANT8
Next, we examined the cellular location of the mature GAU1 transcript. By isolating nuclear and cytoplasmic RNA, we showed that GAU1 was mainly present in the nucleus in retinoblastoma (Y79 and WERI-Rb1 cell lines) and neuroblastoma (SK-N-AS cell line) (Figure 5A; Supplementary Figure S8A). RNA fluorescence in situ hybridization (RNA-FISH) further confirmed that GAU1 was mainly distributed in the nucleus (Figure 5B). In addition, RNA-FISH demonstrated that GAU1 RNA was abundant in the cell nucleus of retinoblastoma tissues, whereas very weak signals were observed in normal retina tissues (Figure 5C).
Figure 5.
GAU1 is distributed in the nucleus and cis-promotes GALNT8 expression. (A) Localization of mature GAU1 transcripts. GAU1 was mainly present in the nucleus. GAPDH and U2 RNA served as a positive control for the cytoplasmic and nuclear fractions, respectively. (B) Representative RNA-FISH images showed that the GAU1 signal overlapped with DAPI staining. The scale bars represent 10 μm. (C) RNA-FISH was performed with GAU1 oligos on clinical retinoblastoma samples and normal control samples. Scale bar: 20 μm. (D) Real-time PCR showed that GAU1 upregulated GALNT8 mRNA expression. Empty vector: negative control group transfected with the empty dCas9-KRAB vector. sgRNA1 and sgRNA2: the first and second sequences used with the dCas9-KRAB system to knock out GAU1 expression; **P< 0.05. (E and F) Western blot confirmed that protein levels of GALNT8 was decreased after GAU1 silencing by sgRNAs and siRNAs (G and H) RT-PCR and realtime-PCR detected the expression of GAU1 after transfecting with pcDNA3.1-GAU1 or pcDNA3.1-GALNT8. (I) Western blot showed that GALNT8 upregulated after GAU1 overexpression. pcDNA3.1-GALNT8 was conducted as positive control. (J) Real-time PCR showed that GAU1 expressional levels remained unchanged after knocking down GALNT8. pGIPZ: negative control group transfected with empty pGIPZ vector.
Since nuclear lncRNAs usually coordinate the transcriptional regulation of nearby protein-coding genes, we investigated whether GLANT8 expression was determined by GAU1 lncRNA. After silencing GAU1 with sgRNAs (Supplementary Figure S8B), GALNT8 expression was significantly reduced in tumor cells at both the mRNA (Figure 5D) and protein levels (Figure 5E). Similarly, we knocked down the GAU1 expression using two additional siRNAs, the data showed that the mRNA and protein of GALNT8 was significantly reduced (Supplementary Figure S8C, Figure 5F). However, GAU1 expression levels were unchanged after GALNT8 suppression (Figure 5G). To further confirm the regulatory relationship between the GAU1 and GALNT8, after constructing GAU1 expressing vector with a strong PCMV promoter, we then overexpressed GAU1 in GAU1-deficient RPE1 and ARPE19 normal cells (Figure 5I, lanes 3–4; H), we observed a significant increase in protein and mRNA level of GALNT8 in normal cells (Figure 5J, lanes 5–6, Supplementary Figure S8D). However, the expression of GAU1 did not change (Figure 5I, lanes 5–6) after GALNT8 overexpression in normal cells (Figure 5J, lanes 5–6). In addition, we also overexpressed GAU1 in tumor cells (Supplementary Figure S9A, lanes 4–6; S9B), the mRNA and protein levels of GALNT8 was also significantly increased (Supplementary Figure S9C lanes 4–6, Supplementary Figure S9D). Similarly, we did not observe the obvious changes in expression of GAU1 (Supplementary Figure S9A, lanes 7–9) after GALNT8 overexpression in tumor cells (Supplementary Figure S9C, lanes 7–9). These data show that the nuclear lncRNA GAU1 cis-regulates the expression of its downstream target gene, GALNT8.
GAU1 interacts with the GALNT8 promoter and the transcription factor TCEA1
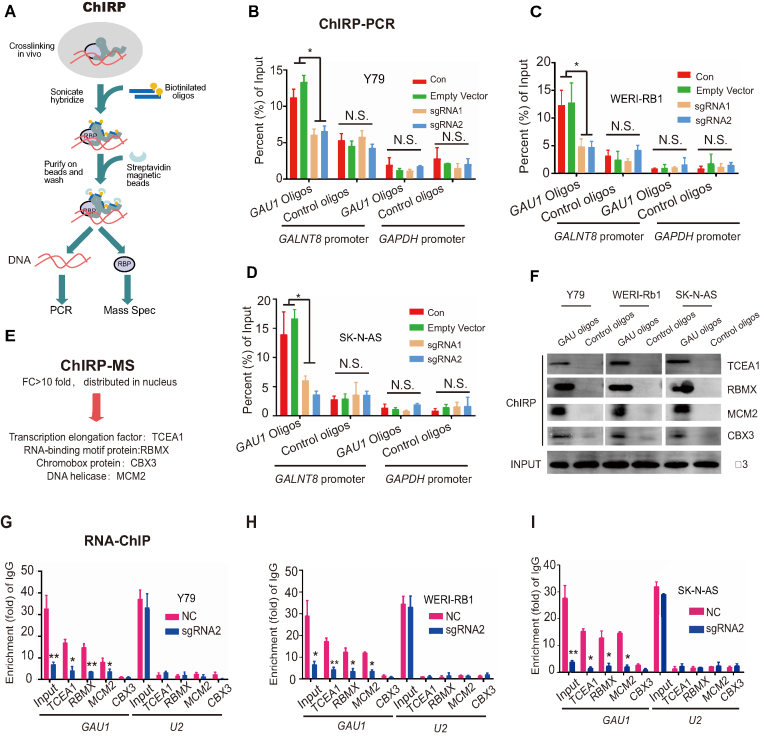
To determine the precise mechanism underlying GAU1-guided GALNT8 regulation, chromatin isolation by RNA purification (ChIRP) was performed with biotin-labeled oligos (Figure 6A). Because GAU1 could regulate GALNT8 expression, the GALNT8 promoter was chosen as the detection site, while the GAPDH promoter served as the negative control. ChIRP-PCR showed that GAU1 was enriched in the GALNT8 promoter region in three tumor cell lines. However, we did not observe the enrichment of GAU1 at the GALNT8 promoter after suppression of GAU1 (Figure 6B–D), indicating that GAU1 could directly bind to the GALNT8 promoter.
Figure 6.
GAU1 interacts with the GALNT8 promoter and TCEA1. (A) Introduction of the ChIRP method. (B–D) GAU1 oligo indicates the biotinylated antisense oligonucleotides against the GAU1 lncRNA. Negative oligo indicates the scrambled oligonucleotides. The GAPDH promoter region was selected as the negative control. The value obtained for the untreated cells was set as 1. ImageJ was used to quantify the binding of GAU1 to the GALNT8 promoter in the ChIRP assay. All the experiments were performed in triplicate and are presented as the mean ± SEM; *P<0.05. Con: untreated cells; and empty vector: cells transfected with the empty dCas9-KRAB vector. (E) ChIRP-MS revealed that four proteins bind to GAU1. (F) Western blot was used to verify the ChIRP-MS results. (G–I) Real-time PCR analysis of the binding of GAU1 to MCM2, TCEA1, RBMX and CBX3 using samples from the RNA-ChIP assay. IgG antibody and U2 RNA were used as negative controls. ImageJ was used to quantify the binding of GAU1 to TCEA1, RBMX, MCM2 and CBX3 using samples from the RNA-ChIP assay. All the experiments were performed in triplicate and are presented as the mean ± SEM; *P< 0.05 and **P< 0.01.
Next, to identify GAU1-interacting proteins, ChIRP-MS, in which the ChIRP-purified proteins were identified through mass spectrometry, was performed (Figure 6E). By screening and analyzing these peptide signals, four proteins (TCEA1, RBMX, MCM2 and CBX3) were found to be enriched in the ChIRP lysate (Supplementary Figure S10A, Supplementary Table S4). After analysis by western blot assay, we confirmed that these four proteins interacted with GAU1 (Figure 6F). Moreover, an RNA-ChIP experiment showed that GAU1 interacted with the four proteins (TCEA1, RBMX, MCM2 and CBX3) in tumor cells (Figure 6G–I). No obvious enrichment was observed for the non-specific control U2 (Supplementary Figure S10B). We thus focused on these four factors to identify the key protein involved in the regulation of GALNT8.
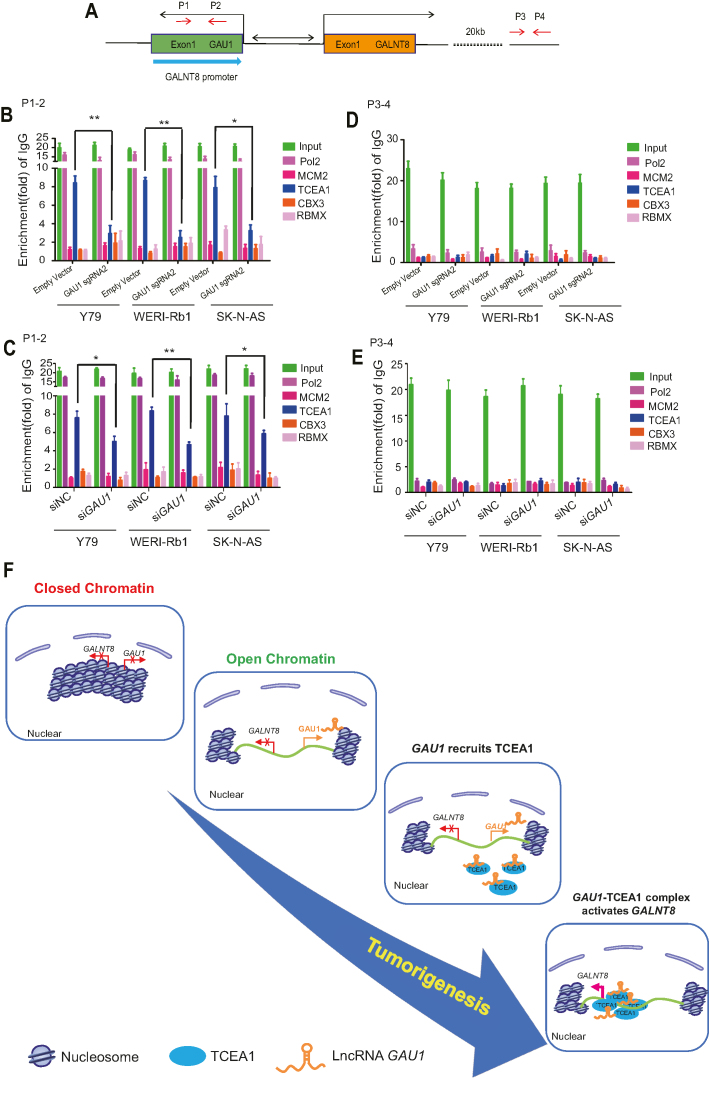
GAU1 recruits TCEA1 to activate GALNT8 expression
After confirming that GAU1 could bind to TCEA1, RBMX and MCM2 by ChIRP and RNA-ChIP, it was necessary to determine whether these GAU1-binding proteins could directly interact with the GALNT8 promoter (Figure 7A). A ChIP experiment demonstrated that only TCEA1 interacted with the GALNT8 promoter (P1–P2 site) in tumor cells (Figure 7B, line 4), while the other three proteins did not bind to the GALNT8 promoter (Figure 7B, Supplementary Figure S11B, lines 3, 5 and 6). Furthermore, TCEA1 could not interact with the GALNT8 promoter after GAU1 silencing, either by sgRNA (Supplementary Figure S11B, line 11) or siRNA (Supplementary Figure S11C, line 11). We did not observe notable enrichment of all four candidate proteins at the negative site (P3-P4) (Figure 7D–E). We also confirmed the interaction between TCEA1 and the GALNT8 promoter by real-time PCR (Figure 7D). In addition, we found that TCEA1 could bind to GAU1 promoter in tumor cells (Supplementary Figure S12B, lanes 1–3; S12C). while we failed to detect the TCEA1 binding in the normal cell (Supplementary Figure S12B, lanes 4-5; S12C). Since many studies have shown that efficient gene transcription requires recruitment of the transcription elongation factor TCEA1, also called TFIIS, to the target gene promoter (19), we detected whether TCEA1 expression was regulated by GAU1. Western blot showed that the global level of TCEA1 was similar in tumor and normal cells (Supplementary Figure S13A). In addition, we showed that TCEA1 expression levels did not change after GAU1 suppression (Supplementary Figure S13B), demonstrating that GAU1 influences the chromosome binding ability of TCEA1 but not its global expression. Taken together, these data show that GAU1 recruits TCEA1 to the GALNT8 promoter and activates the expression of GLANT8.
Figure 7.
TCEA1 is recruited to the GALNT8 promoter. (A) Schematic of sites in the GALNT8 promoter as detected using the ChIP assay. (B–E) RT-PCR examination of the binding of TCEA1 to the GALNT 8 promoter using samples from the ChIP assay. The value of the IgG group was set as 1. All the experiments were performed in triplicate and are presented as the mean ± SEM; *P< 0.05. The value obtained for the negative control cells was set as 1. (F) Schematic model of the GAU1/GALNT8 cluster in tumorigenesis. Normally, the chromatin at the GAU1/GALNT8 cluster is tightly closed. However, transition of the closed chromatin to the open status exposes the protein-binding sites, leading to recruitment of the trans-factor TCEA1 and resulting in the alteration of GAU1/GALNT8 gene transcription. Blue circle: TCEA1 protein; and green line: GAU1/GLANT8 cluster.
DISCUSSION
Chromatin dynamics refer to a dynamic change in the open or close status of chromatin, which usually influences the expression of several neighboring genes simultaneously (20). Studies have shown that upregulation of the INK4b–ARF–INK4a gene cluster by removing the repressive H2A.Z significantly inhibits tumor growth (12,13). In addition, chromatin dynamics induce the overexpression of two lncRNAs at 13q14.3 that are also associated with leukemia progression (15). Here, we revealed for the first time a novel chromosomal driver at chr12p13.32 in which a novel lncRNA GAU1 and a protein-coding gene GALNT8 is activated by a dynamic change of chromatin status, thereby presenting a novel dynamic style of chromatin tuning that accelerates tumorigenesis (Figure 7E).
The previously identified GAU1 lncRNA (LOC101929549) is 1,294 bp in length according to the University of California, Santa Cruz (UCSC) and the NCBI databases. In our study, however, we report a novel 1339-bp isoform of LOC101929549 named lncRNA GALNT8-antisense-upstream1 (GAU1), a novel transcript that differs from the previously determined genomic boundaries by 57 bp at the 5′ terminus and by 4 bp and a poly-A tail at the 3′ terminus. These data show that this isoform of the LOC101929549 lncRNA identified in retinoblastoma and neuroblastoma is a novel non-coding transcript; we therefore named it GALNT8-antisense upstream 1 (GALNT8-AU1 or GAU1; GenBank: MF576259). To date, the role of LOC101929549 or GAU1 remains unclear. Here, we show for the first time that GAU1 is a novel oncoRNA that promotes tumorigenesis in neuroblastoma and retinoblastoma.
It should be noted that the GALNT8 gene (polypeptide N-acetylgalactosaminyl transferase 8) was originally identified as an enzyme that initiates mucin-like O-glycosylation of specific proteins (21). The GALNT8 gene encodes a 637-amino acid type-II membrane protein and has been shown to contribute to the response to interferon therapy against chronic hepatitis C (22). In addition, GALNT8 is expressed in the cephalic mesoderm and hatching gland of zebrafish during early developmental stages (23). Thus far, the regulatory role of the GALNT8 gene in tumors is unknown. In this report, however, we clearly demonstrate that GALNT8 contributes to tumorigenesis and is a novel oncogene in tumor development.
Since the lncRNA GAU1 cis-regulates GALNT8 expression, we investigated the precise mechanism underlying GAU1-mediated GALNT8 transcription. Recently, dynamic changes at specific chromosomal loci have been shown to expose protein-binding sites, thereby allowing the recruitment of trans-factors and resulting in the alteration of gene transcription (24). Therefore, we focused on a type of transcription elongation factor that increases the transcriptional activity of their target genes (19,25). Here, we first demonstrate that dynamic changes in chromosomal structure lead to availability of the binding surface for an important transcription elongation factor, TCEA1 (also called TFIIS), thereby promoting GALNT8 expression. Most importantly, after the lncRNA GAU1 is transcribed, GAU1 itself guides TCEA1 protein to the GLANT8 promoter following the classic lncRNA-recruitment model, thereby substantially activating the expression of GLANT8. It also should be explained that we for the first time found the interaction of TCEA1 and GAU1 lncRNA, therefore, there was no available known lncRNA serving as a positive binding control of TCEA1 in our RIP analysis. Comparable situation was also occurred in the interaction of another novel lncRNA lnc-DC and STAT3 (26). Although we cannot eliminate other genetic or epigenetic factors regulating GALNT8 expression, this was the first study to suggest that GLANT8 transcription is highly dependent on the transcription elongation factor TCEA1. The dynamic tuning of chromatin by switching between the closed/off and open/on statuses may provide a novel alternative theory on the activation of oncogenes in neuroblastoma and retinoblastoma.
It should be emphasized that HKP is an effective carrier composed of histidine and lysine for the delivery of nucleic acid into cells (27); thus, histidine-lysine-rich polymer (HKP)-encapsulated siRNA has been applied in clinical trials to inhibit target genes (28). HKP-encapsulated siRNA has been shown to accumulate in established tumors in MDA-MB-435 xenografts (29). In addition, studies have shown that HKP-encapsulated Raf-1 and RHBDF1 siRNA could significantly inhibit tumorigenesis (27). To our knowledge, in this study, we showed for the first time that HKP-encapsulated GAU1 siRNA could be delivered through intraocular injection and significantly inhibit retinoblastoma tumor formation and increase survival time. Efficient delivery of siRNA by HKPs indiciates that it may be a feasible therapeutic approach to treating retinoblastoma, a fatal intraocular tumor in children.
It should be noted that retinoblastoma is the most common malignant intraocular tumor of childhood and is caused by inactivation of both alleles of the RB gene, which results in the defective formation of pRB protein (tumor suppressor), thereby affecting the cell cycle and resulting in uncontrolled cell proliferation (RB tumorigenesis) (30). Following mutation of the RB1 gene, abnormal changes in genetic or epigenetic processes, such as MYCN amplification, DNA methylation, histone modification, non-coding RNAs, were also observed (31–33). Studies have shown that H3K4me3 and H3K9/14ac of the spleen tyrosine kinase (SYK) promoter causes the hyperexpression of SYK, which results in RB progression (31). In our study, however, we demonstrated for the first time that activation of GAU1/GALNT8 was required for RB progression, thus furthering our understanding of RB tumorigenesis. Similarly, neuroblastoma, another pediatric malignant tumor originating from precursor neural crest cells (34), is caused by a critical oncogene, MYCN (35). Aberrant MYCN amplification leads to deceased apoptosis and increased angiogenesis in neuroblastoma (35–37). Here, we suggest that activation of the GAU1/GALNT8 cluster is strongly associated with poor prognosis among patients with neuroblastoma. Most intriguingly, although neuroblastoma and retinoblastoma are two distinct tumors (38), activation of the GAU1/GALNT8 cluster exhibited therapeutic efficacy in both cases. Therefore, further exploration of the GAU1/GALNT8 cluster as a potential biomarker and therapeutic target for neuroblastoma and retinoblastoma is of great interest.
Dynamic chromatin tuning modulates gene expression by altering the accessibility of DNA to transcription factors, thus influencing either coding gene clusters or non-coding clusters in tumor progression (39). A study has shown that chromatin condensation suppresses HOXA protein-coding gene cluster expression and induces breast cancer (40). Additionally, open chromatin upregulates a lncRNA cluster at 13q14.3, thus contributing to leukemia progression (15). However, evidence that shows chromatin dynamics can simultaneously regulate a lncRNA and a protein-coding gene is not available. To our knowledge, we identified for the first time that dynamic tuning of chromatin co-drives the activation of a gene cluster that includes the lncRNA GAU1 and the protein-coding gene GALNT8, thereby inducing tumorigenesis. Our study presents a novel alternative concept of chromatin dynamics in tumorigenesis. Chromatin transformation occurs in a variety of diseases, and it is possible that the chromatin state plays regulatory roles by modulating both lncRNA and protein-coding genes rather than regulating protein-coding genes clusters or lncRNA clusters alone. Therefore, our findings form the basis for the exploration of chromatin dynamics biology and provide potential targets for the diagnosis and treatment of disease.
DATA AVAILABILITY
GALNT8-AU1 or GAU1; GenBank: MF576259;RNA-Seq of retinoblastoma samples (NODE, http://www.biosino.org/node/project/detail/NODEP0037193, GEO, https://www.ncbi.nlm.nih.gov/geo/, GEO Accession number: GSE111168).
Supplementary Material
ACKNOWLEDGEMENTS
Special thanks to Prof. Pan Qiuhui, Dr Tian Ruicheng and Dr Wang Jing in the Shanghai Children's Medical Center (SCMC) for providing precious neuroblastoma samples. Thanks to Prof. Wang Zefeng and Dr Cao Ruifang for assisting in the uploading RNA-sequencing data to the NODE database during revision. Sincere thanks to all retinoblastoma and neuroblastoma patients enrolled in our study and wish them a good health.
Authors Contributions: In this report, P.W.C. and R.B.J. designed and performed the experiments and drafted the manuscript; R.B.J., P.H. were responsible for sample collection and data analysis; H.Y.N., H.X.W., X.Y.H., Y.Y.S., C.D.Z. assisted revision; S.Y.W. uploaded the sequencing data to the GEO database. H.Z. and X.Q.F. discussed, revised and approved the manuscript. All authors approved this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31470757, 81772875]; ShuGuang Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation [17SG19]; Outstanding Yong Medical Scholar of Shanghai Municipal Commission of Health and Family Planning [2017YQ067]; Program for Professor of Special Appointment (Eastern Scholar) at the Shanghai Institutions of Higher Learning [1410000159]; SMC-ChenXing Yong Scholar Program [2014, Class B]; Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant [20161317]; Outstanding Yong Scholar Grant of Shanghai JiaoTong University School of Medicine [16XJ11002]. Funding for open access charge: National Natural Science Foundation of China [31470757, 81772875]; ShuGuang Project of Shanghai Municipal Education Commission and Shanghai Education Development Foundation [17SG19]; Outstanding Yong Medical Scholar of Shanghai Municipal Commission of Health and Family Planning [2017YQ067]; Program for Professor of Special Appointment (Eastern Scholar) at the Shanghai Institutions of Higher Learning [1410000159]; SMC-ChenXing Yong Scholar Program [2014, Class B]; Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant [20161317]; Outstanding Yong Scholar Grant of Shanghai JiaoTong University School of Medicine [16XJ11002].
Conflict of interest statement. None declared.
REFERENCES
- 1. Lara-Astiaso D., Weiner A., Lorenzo-Vivas E., Zaretsky I., Jaitin D.A., David E., Keren-Shaul H., Mildner A., Winter D., Jung S. et al. . Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014; 345:943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dabin J., Fortuny A., Polo S.E.. Epigenome maintenance in response to DNA damage. Mol. Cell. 2016; 62:712–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Price B.D., D’Andrea A.D.. Chromatin remodeling at DNA double-strand breaks. Cell. 2013; 152:1344–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ball A.R. Jr., Yokomori K.. Damage site chromatin: open or closed. Curr. Opin. Cell Biol. 2011; 23:277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gondor A. Dynamic chromatin loops bridge health and disease in the nuclear landscape. Semin. Cancer Biol. 2013; 23:90–98. [DOI] [PubMed] [Google Scholar]
- 6. Qu K., Zaba L.C., Satpathy A.T., Giresi P.G., Li R., Jin Y., Armstrong R., Jin C., Schmitt N., Rahbar Z. et al. . Chromatin accessibility landscape of cutaneous T cell lymphoma and dynamic response to HDAC inhibitors. Cancer Cell. 2017; 32:27–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dinant C., Ampatziadis-Michailidis G., Lans H., Tresini M., Lagarou A., Grosbart M., Theil A.F., van Cappellen W.A., Kimura H., Bartek J. et al. . Enhanced chromatin dynamics by FACT promotes transcriptional restart after UV-induced DNA damage. Mol. Cell. 2013; 51:469–479. [DOI] [PubMed] [Google Scholar]
- 8. Groop L. Open chromatin and diabetes risk. Nat. Genet. 2010; 42:190–192. [DOI] [PubMed] [Google Scholar]
- 9. Nevedomskaya E., Stelloo S., van der Poel H.G., de Jong J., Wessels L.F., Bergman A.M., Zwart W.. Androgen receptor DNA binding and chromatin accessibility profiling in prostate cancer. Genom Data. 2016; 7:124–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uslu V.V., Petretich M., Ruf S., Langenfeld K., Fonseca N.A., Marioni J.C., Spitz F.. Long-range enhancers regulating Myc expression are required for normal facial morphogenesis. Nat. Genet. 2014; 46:753–758. [DOI] [PubMed] [Google Scholar]
- 11. Shi J., Whyte W.A., Zepeda-Mendoza C.J., Milazzo J.P., Shen C., Roe J.S., Minder J.L., Mercan F., Wang E., Eckersley-Maslin M.A. et al. . Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev. 2013; 27:2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu S., Wang H., Pan H., Shi Y., Li T., Ge S., Jia R., Zhang H., Fan X.. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4-hub in melanoma. Cancer Lett. 2016; 381:41–48. [DOI] [PubMed] [Google Scholar]
- 13. Lazorthes S., Vallot C., Briois S., Aguirrebengoa M., Thuret J.Y., St Laurent G., Rougeulle C., Kapranov P., Mann C., Trouche D. et al. . A vlincRNA participates in senescence maintenance by relieving H2AZ-mediated repression at the INK4 locus. Nat. Commun. 2015; 6:5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ay F., Vu T.H., Zeitz M.J., Varoquaux N., Carette J.E., Vert J.P., Hoffman A.R., Noble W.S.. Identifying multi-locus chromatin contacts in human cells using tethered multiple 3C. BMC Genomics. 2015; 16:121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garding A., Bhattacharya N., Claus R., Ruppel M., Tschuch C., Filarsky K., Idler I., Zucknick M., Caudron-Herger M., Oakes C. et al. . Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the In Cis downregulation of a gene cluster that targets NF-kB. PLoS Genet. 2013; 9:e1003373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D. et al. . A map of open chromatin in human pancreatic islets. Nat. Genet. 2010; 42:255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xing Y., Wen X., Ding X., Fan J., Chai P., Jia R., Ge S., Qian G., Zhang H., Fan X.. CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Mol. Ther. 2017; 25:1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang J.Z., Chen M., Chen, Gao X.C., Zhu S., Huang H., Hu M., Zhu H., Yan G.R.. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol. Cell. 2017; 68:171–184. [DOI] [PubMed] [Google Scholar]
- 19. Shema E., Kim J., Roeder R.G., Oren M.. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Mol. Cell. 2011; 42:477–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Voss T.C., Schiltz R.L., Sung M.H., Yen P.M., Stamatoyannopoulos J.A., Biddie S.C., Johnson T.A., Miranda T.B., John S., Hager G.L.. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011; 146:544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. White K.E., Lorenz B., Evans W.E., Meitinger T., Strom T.M., Econs M.J.. Molecular cloning of a novel human UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase, GalNAc-T8, and analysis as a candidate autosomal dominant hypophosphatemic rickets (ADHR) gene. Gene. 2000; 246:347–356. [DOI] [PubMed] [Google Scholar]
- 22. Nakano R., Maekawa T., Abe H., Hayashida Y., Ochi H., Tsunoda T., Kumada H., Kamatani N., Nakamura Y., Chayama K.. Single-nucleotide polymorphisms in GALNT8 are associated with the response to interferon therapy for chronic hepatitis C. J. Gen. Virol. 2013; 94:81–89. [DOI] [PubMed] [Google Scholar]
- 23. Nakayama Y., Nakamura N., Kawai T., Kaneda E., Takahashi Y., Miyake A., Itoh N., Kurosaka A.. Identification and expression analysis of zebrafish polypeptide alpha-N-acetylgalactosaminyltransferase Y-subfamily genes during embryonic development. Gene Expr. Patterns. 2014; 16:1–7. [DOI] [PubMed] [Google Scholar]
- 24. Bohmdorfer G., Wierzbicki A.T.. Control of chromatin structure by long noncoding RNA. Trends Cell Biol. 2015; 25:623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xu Y., Bernecky C., Lee C.T., Maier K.C., Schwalb B., Tegunov D., Plitzko J.M., Urlaub H., Cramer P.. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nat. Commun. 2017; 8:15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X.. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014; 344:310–313. [DOI] [PubMed] [Google Scholar]
- 27. Yan Z., Zou H., Tian F., Grandis J.R., Mixson A.J., Lu P.Y., Li L.Y.. Human rhomboid family-1 gene silencing causes apoptosis or autophagy to epithelial cancer cells and inhibits xenograft tumor growth. Mol. Cancer Ther. 2008; 7:1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Q.R., Zhang L., Luther P.W., Mixson A.J.. Optimal transfection with the HK polymer depends on its degree of branching and the pH of endocytic vesicles. Nucleic Acids Res. 2002; 30:1338–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leng Q., Scaria P., Lu P., Woodle M.C., Mixson A.J.. Systemic delivery of HK Raf-1 siRNA polyplexes inhibits MDA-MB-435 xenografts. Cancer Gene Ther. 2008; 15:485–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goodrich D.W., Wang N.P., Qian Y.W., Lee E.Y., Lee W.H.. The retinoblastoma gene product regulates progression through the G1 phase of the cell cycle. Cell. 1991; 67:293–302. [DOI] [PubMed] [Google Scholar]
- 31. Zhang J., Benavente C.A., McEvoy J., Flores-Otero J., Ding L., Chen X., Ulyanov A., Wu G., Wilson M., Wang J. et al. . A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012; 481:329–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnston A.J., Kirioukhova O., Barrell P.J., Rutten T., Moore J.M., Baskar R., Grossniklaus U., Gruissem W.. Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet. 2010; 6:e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. De La Rosa-Velazquez I.A., Rincon-Arano H., Benitez-Bribiesca L., Recillas-Targa F.. Epigenetic regulation of the human retinoblastoma tumor suppressor gene promoter by CTCF. Cancer Res. 2007; 67:2577–2585. [DOI] [PubMed] [Google Scholar]
- 34. Dimaras H., Kimani K., Dimba E.A., Gronsdahl P., White A., Chan H.S., Gallie B.L.. Retinoblastoma. Lancet. 2012; 379:1436–1446. [DOI] [PubMed] [Google Scholar]
- 35. Felsher D.W. Role of MYCN in retinoblastoma. Lancet Oncol. 2013; 14:270–271. [DOI] [PubMed] [Google Scholar]
- 36. Cheung N.K., Zhang J., Lu C., Parker M., Bahrami A., Tickoo S.K., Heguy A., Pappo A.S., Federico S., Dalton J. et al. . Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012; 307:1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chipumuro E., Marco E., Christensen C.L., Kwiatkowski N., Zhang T., Hatheway C.M., Abraham B.J., Sharma B., Yeung C., Altabef A. et al. . CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014; 159:1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kamihara J., Bourdeaut F., Foulkes W.D., Molenaar J.J., Mosse Y.P., Nakagawara A., Parareda A., Scollon S.R., Schneider K.W., Skalet A.H. et al. . Retinoblastoma and neuroblastoma predisposition and surveillance. Clin. Cancer Res. 2017; 23:e98–e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Adam R.C., Fuchs E.. The yin and yang of chromatin dynamics in stem cell fate selection. Trends Genet. 2016; 32:89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Novak P., Jensen T., Oshiro M.M., Wozniak R.J., Nouzova M., Watts G.S., Klimecki W.T., Kim C., Futscher B.W.. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006; 66:10664–10670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GALNT8-AU1 or GAU1; GenBank: MF576259;RNA-Seq of retinoblastoma samples (NODE, http://www.biosino.org/node/project/detail/NODEP0037193, GEO, https://www.ncbi.nlm.nih.gov/geo/, GEO Accession number: GSE111168).