Abstract
During carcinogenesis, cells are exposed to increased replication stress due to replication fork arrest at sites of DNA lesions and difficult to replicate genomic regions. Efficient fork restart and DNA repair are important for cancer cell proliferation. We previously showed that the ADP-ribosyltransferase PARP10 interacts with the replication protein proliferating cell nuclear antigen and promotes lesion bypass by recruiting specialized, non-replicative DNA polymerases. Here, we show that PARP10 is overexpressed in a large proportion of human tumors. To understand the role of PARP10 in cellular transformation, we inactivated PARP10 in HeLa cancer cells by CRISPR/Cas9-mediated gene knockout, and overexpressed it in non-transformed RPE-1 cells. We found that PARP10 promotes cellular proliferation, and its overexpression alleviates cellular sensitivity to replication stress and fosters the restart of stalled replication forks. Importantly, mouse xenograft studies showed that loss of PARP10 reduces the tumorigenesis activity of HeLa cells, while its overexpression results in tumor formation by non-transformed RPE-1 cells. Our findings indicate that PARP10 promotes cellular transformation, potentially by alleviating replication stress and suggest that targeting PARP10 may represent a novel therapeutic approach.
INTRODUCTION
Adenosine diphosphate (ADP) ribosylation is a post-translational modification that has recently emerged as an important regulatory factor in both DNA and cancer biology. The poly-ADP-ribose polymerase (PARP) family of ADP-ribosyltransferases encompasses 17 enzymes with a PARP catalytic domain in their C-termini (1,2). PARP1, the founding member of the family, and the closely related PARP2 and PARP3, catalyze formation of poly-ADP-ribose chains on themselves and a number of other substrates. PARP1 plays major roles in regulating DNA transcription, repair and replication. Depletion or inhibition of PARP1 results in spontaneous death of cells with homologous recombination (HR) DNA repair deficiency, and thus PARP1 inhibitors are used in clinical treatment of breast and ovarian tumors with BRCA1 or BRCA2 mutations (3–6).
In contrast to PARP1, which catalyzes poly-ADP-ribose chain formation, PARP10 (also known as ARTD10) and other members of the PARP family catalyze the transfer of a single ADP-ribose molecule (process known as mono-ADP-ribosylation, or MARylation) (7). In line with this, the functions of PARP10 are distinct from those of PARP1. PARP10 was originally identified as a Myc-interacting protein (8). Subsequently, it has been proposed to be important for the G1/S cell-cycle transition (9) as well as for caspase-dependent apoptosis (10). More recently, it was shown that PARP10 can suppress cytokine-induced activation of the NFκB pathway (11), and plays roles in mitochondrial oxidation (12) and cell migration (13).
We have previously uncovered an unexpected involvement of PARP10 in DNA repair (14,15). We showed that PARP10 interacts with the replication protein proliferating cell nuclear antigen (PCNA), an essential polymerase co-factor (14,16) which recruits PARP10 to replication forks. We found that the interaction with PCNA is mediated by the PIP-box (PCNA-interacting peptide motif) sequence QEVVRAFY at position 834–841 in PARP10. One of the well-described roles of PCNA is promoting the stability and progress of replication machineries during stress conditions. Unrepaired DNA lesions, secondary DNA structures, repetitive elements and other non-canonical DNA structures can arrest the progression of replicative DNA polymerases (17,18). Unless efficiently restarted, stalled replication forks can disassemble, resulting in DNA strand breaks and genomic instability. One mechanism that restarts stalled replication forks is translesion DNA synthesis (TLS), which employs specialized polymerases able to accommodate modified DNA bases in their active sites, to bypass fork arresting structures (16,17). Upon replication fork arrest, mono-ubiquitination of PCNA at Lys164 promotes recruitment of TLS polymerases, which possess PIP and ubiquitin-interacting motifs, to restart the stalled fork (19,20). We previously showed that PARP10 downregulation results in reduced levels of PCNA ubiquitination, impaired recruitment of the TLS polymerase Rev1 to sites of DNA damage and sensitivity to replication arresting drugs such as hydroxyurea (HU) (14). In line with this, by employing a plasmid-based reporter of TLS activity, we showed that PARP10 is required for efficient TLS. This activity requires PCNA interaction, as TLS levels could be restored by re-expression of wild-type PARP10 but not of a PARP10 variant harboring a mutation of the 8-residue PIP-box sequence (14).
During cellular transformation, increased proliferation is associated with replication stress and frequent replication fork arrest (18). Replication stress is a major barrier to oncogene-induced proliferation as it activates the DNA damage and replication stress checkpoints leading to cell-cycle arrest and/or senescence (21,22). Suppression of this mechanism is an essential step in carcinogenesis. By restarting stalled replication forks, TLS suppresses DNA damage accumulation and allows completion of DNA replication, thereby enabling cellular proliferation and potentially promoting transformation (17). Because of the role of PARP10 in TLS that we previously described, we decided to investigate how PARP10 affects transformation and cancer proliferation. Here, we show that PARP10 expression promotes in vitro cellular proliferation and in vivo tumor growth, potentially by promoting replication fork stability and suppressing replication stress.
MATERIALS AND METHODS
Cell culture and protein techniques
Human HeLa and RPE-1 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. For PARP10 gene knockout, the commercially available PARP10 CRISPR/Cas9 KO plasmid was used (Santa Cruz Biotechnology sc-406703). Single cells were FACS-sorted into 96-well plates using a BD FACSAria II instrument. Following clonal expansion, resulting mono-clonal cultures were screened by western blot for PARP10 protein levels. For exogenous PARP10 expression, pLV-puro-TRE lentiviral constructs encoding wild-type or the ΔPARP variant spanning amino acids 1–834 (lacking the PIP motif and PARP catalytic domain), with the Myc-epitope tag EQKLISEEDL at the N-terminus, were obtained from Cyagen. Infected cells, stably expressing the tetracycline transactivator element, were selected by puromycin. For induction of expression, cells were grown in the presence of 2 μg/ml doxycycline. Cell extracts, chromatin fractionation and western blot experiments were performed as previously described (14,23,24). Antibodies used for western blot were: PARP10 (Novus NB100–2157), GAPDH (Santa Cruz Biotechnology sc-47724), Polη (Cell Signaling Technology 13848), PCNA (Cell Signaling Technology 2586), Ubiquityl-PCNA Lys164 (Cell Signaling Technology 13439).
Functional assays
Apoptosis was quantified using the FITC Annexin V kit (Biolegend 640906). For cell-cycle profiles, cells were fixed in 4% paraformaldehyde and stained with FxCycle PI reagent (Invitrogen F10797). EdU incorporation was assayed using the Click-iT Plus kit (Invitrogen C10633). For clonogenic experiments, 250 cells were seeded in 6-well plates and, 2 weeks later, stained with Crystal violet or trypsinized and counted using an automated cell counter for quantification of cellular proliferation. When HU sensitivity was analyzed, cells were incubated with 0.2 mM HU immediately after seeding; 72 h later, media was replaced and plates were stained with Crystal violet 2 weeks later. For time-course proliferation experiments, 500 cells were seeded in wells of 96-well plates and cellular viability was scored at indicated days using the CellTiterGlo reagent (Promega G7572).
DNA fiber assay
Cells were incubated with 100 μM CldU for 30 min, washed with phosphate-buffered saline (PBS) and incubated with 100 μM IdU for another 45 min (for HeLa cells) or 90 min (for RPE-1 cells), either by itself, or in the presence of 0.2 mM HU as indicated. Next, cells were harvested and DNA fibers were obtained using the FiberPrep kit (Genomic Vision). DNA fibers were stretched on glass slides using the FiberComb Molecular Combing instrument (Genomic Vision). Slides were incubated with primary antibodies (Abcam 6326 for detecting CIdU; BD 347580 for detecting IdU; Millipore Sigma MAB3034 for detecting DNA), washed with PBS and incubated with Cy3, Cy5 or BV480—coupled secondary antibodies (Abcam 6946, Abcam 6565 and BD Biosciences 564879). Following mounting, slides were imaged using a Leica SP5 confocal microscope. At least 100 tracts were quantified for each sample.
RNA sequencing and mutation burden analyses
Total RNA was extracted using Trizol (Invitrogen) and cDNA libraries were prepared using the NEXTflex Illumina Rapid Directional RNA-Seq Library Prep Kit (BioO Scientific). Libraries were run on Illumina NovaSeq 6000 for 100 cycles (pair end). For data analyses, Broad Institute’s GATK Best Practice workflow for polymorphism calling from RNA-seq data (25) was followed. Ensembl reference genome (GRCh38) and known variants from the Broad Institute’s resource bundle were used as reference. Recalibrated reads were subjected to MuTect2 analyses for somatic polymorphism calling. Additional filters including germline read depth >5, and germline variant fraction <0.03 were applied to remove germline variants.
Mouse xenograft studies
Cells (either 5 million or 10 million, as indicated in the ‘Results’ section) were suspended in PBS and mixed 1:1 with Matrigel Matrix (Corning 354234). Cells were then injected into both flanks of 4–6 weeks old athymic nude female mice (Charles River Laboratories stock #490). For the experiments involving cells with exogenous PARP10 expression, mice were also administered 2 mg/ml doxycycline in their drinking water (supplemented with 5 mg/ml sucrose) starting the day of injection.
Statistical analyses
With the exception of the DNA fiber combing and the xenograft data, the statistical analysis performed was the TTEST (two-tailed, equal variance), using PRISM software. For the DNA fiber combing and the xenograft data, the Mann–Whitney test was performed. Statistical significance is indicated for each graph (ns = not significant, for P > 0.05; * for P < 0.05; ** for P < 0.01; *** for P < 0.001; **** for P < 0.0001).
RESULTS
Loss of PARP10 inhibits proliferation of HeLa cells and increases sensitivity to replication stress
To gain insights into a possible involvement of PARP10 in carcinogenesis, we mined the publicly available cBioPortal database (26) for PARP10 gene expression and mutation in cancer samples. Strikingly, through a variety of cancer types and datasets, PARP10 was almost exclusively amplified with almost no PARP10 gene mutations or deletions found (Supplementary Figure S1A). This pattern is very similar to that of known oncogenes (such as MYC), and contrasts that of known tumor suppressors, such as BRCA2—for which gene mutations or deletions are predominant, while few amplifications are found (Supplementary Figure S1A). This pattern is specific to PARP10, as the related mono-ADP-ribosyltransferase PARP14 shows a seemingly random pattern of gene amplification and deletion/mutations (Supplementary Figure S1A). Moreover, survey of cancer-specific TCGA datasets, which also include mRNA quantification, showed that up to 19% of all breast tumors and 32% of all ovarian tumors have increased PARP10 expression (gene amplification and/or mRNA upregulation) (Supplementary Figure S1B). These findings suggest that PARP10 may function as an oncogene and promote transformation.
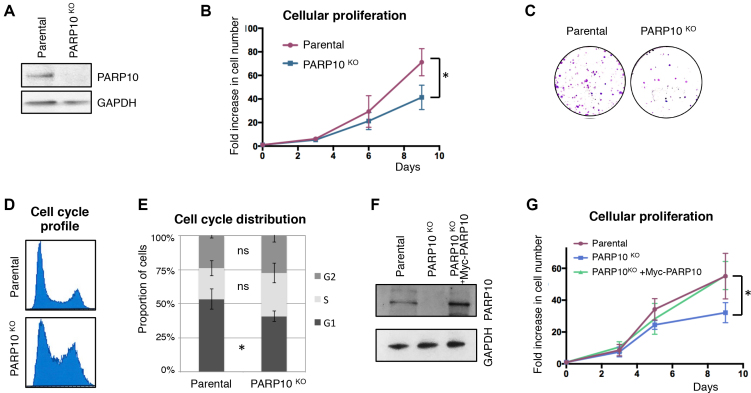
To evaluate its impact on proliferation of cancer cells, we knocked-out PARP10 in HeLa cells using CRISPR/Cas9 technology (Figure 1A). PARP10-deleted HeLa cells showed reduced proliferation (Figure 1B and C) and an altered cell-cycle profile, with increased accumulation of cells in S and G2 phases, (Figure 1D and E) but without any significant difference in EdU incorporation (not shown). Importantly, stable re-expression of wild-type Myc-tagged PARP10 from a lentiviral construct (Figure 1F) could rescue the proliferation defect of the PARP10-knockout cells (Figure 1G). These findings indicate that loss of PARP10 reduces cancer cell proliferation.
Figure 1.
Loss of PARP10 impairs proliferation of HeLa cells. (A) Western blot showing loss of PARP10 expression in HeLa cells with CRISPR/Cas9-mediated PARP10 knockout. (B) PARP10-knockout HeLa cells show reduced proliferation rates. The average of three experiments with error bars representing standard deviations is shown. The asterisk indicates statistical significance (using the two-tailed equal variance TTEST). (C) Representative clonogenic assay showing reduced proliferation of PARP10-knockout HeLa cells. (D) Representative PI flow cytometry profile showing an altered cell-cycle distribution in PARP10-knockout HeLa cells. (E) Quantification of cell-cycle distribution in control and PARP10-knockout HeLa cells. The average of four experiments, with error bars as standard deviations, is shown. Statistical significance was calculated using the two-tailed equal variance TTEST. (F) Western blot showing the re-expression of PARP10, with a Myc-tag, in PARP10-knockout HeLa cells. (G) Exogenous PARP10 expression rescues the proliferation defect of PARP10-deleted HeLa cells. The average of four experiments with error bars representing standard deviations is shown. The asterisk indicates statistical difference between the PARP10KO and PARP10KO + Myc-PARP10 samples.
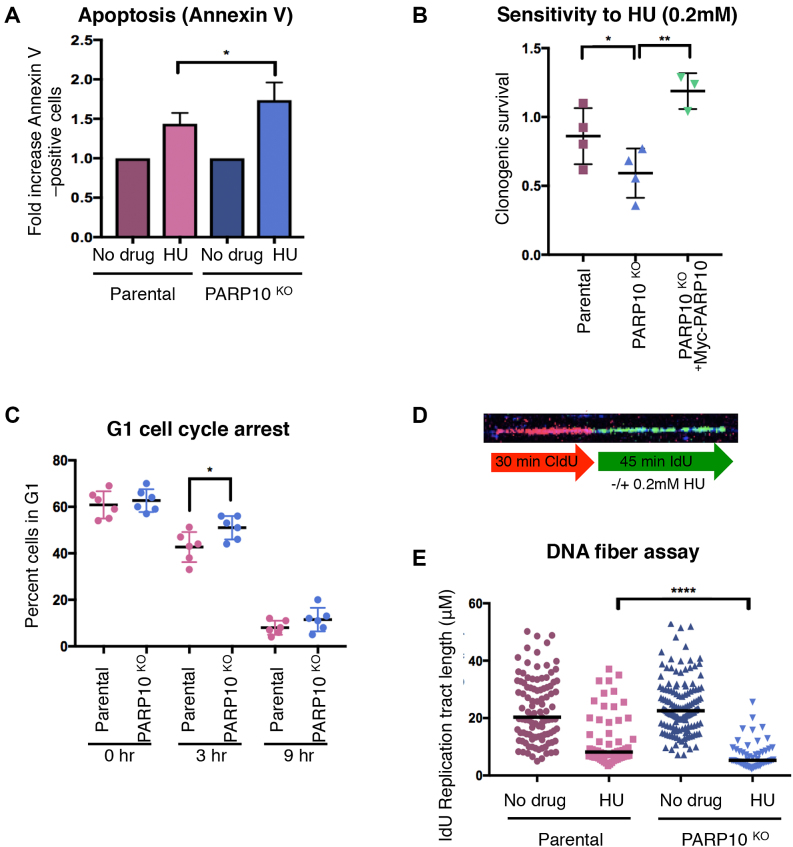
We next tested the ability of PARP10-deleted HeLa cells to handle replication stress. Exposure to HU increased apoptosis and reduced clonogenic survival, which was restored by Myc-PARP10 re-expression (Figure 2A and B). Acute HU treatment induced G1/S cell-cycle arrest to a similar extent in both control and PARP10-knockout cells. However, upon removal of drug and re-plating in fresh media, PARP10-knockout cells showed a slight delay in re-starting replication (Figure 2C and Supplementary Figure S2), suggesting reduced ability to recover from replication stress. Altogether, these results argue that PARP10 is important for alleviating replication stress.
Figure 2.
Loss of PARP10 results in sensitivity to replication stress. (A) Annexin V apoptosis experiment showing increased apoptosis in PARP10-knockout cells following HU treatment. Cells were treated with 1 mM HU for 24 h. Data are shown as normalized to the control (no drug treatment) condition for each cell line. The mean of six experiments with error bars as standard deviations is shown. (B) Clonogenic assay showing that PARP10-knockout cells are sensitive to HU, and re-expression of PARP10 corrects this sensitivity. For each cell line, the ratios of HU-treated to non-treated are presented. The mean and standard deviation are shown. (C) Quantification of the proportion of cells in G1 at the indicated time points after release from HU (1 mM for 24 h). The mean and standard deviation are shown. Representative flow cytometry histograms are shown in Supplementary Figure S2. (D) Schematic representation of the DNA fiber combing assay condition, including a representative micrograph. (E) DNA fiber combing assay showing reduced replication fork progression in PARP10-knockout cells upon HU exposure. Shown is the quantification of the IdU tract length, with the median values marked. Representative micrographs for each condition are presented in Supplementary Figure S3.
Next, we investigated the progression and stability of individual replication factories at the molecular level, using the DNA fiber combing assay. Cells were grown in the presence of thymidine analogs CldU and IdU consecutively, with 0.2 mM HU being added during the IdU incubation (Figure 2D). Quantification of the IdU tract length showed a significant reduction in replication tract length in PARP10-knockout cells (Figure 2E and Supplementary Figure S3), indicating that PARP10 plays a role in fork elongation under replication stress conditions.
PARP10 overexpression promotes cellular proliferation and replication fork progression
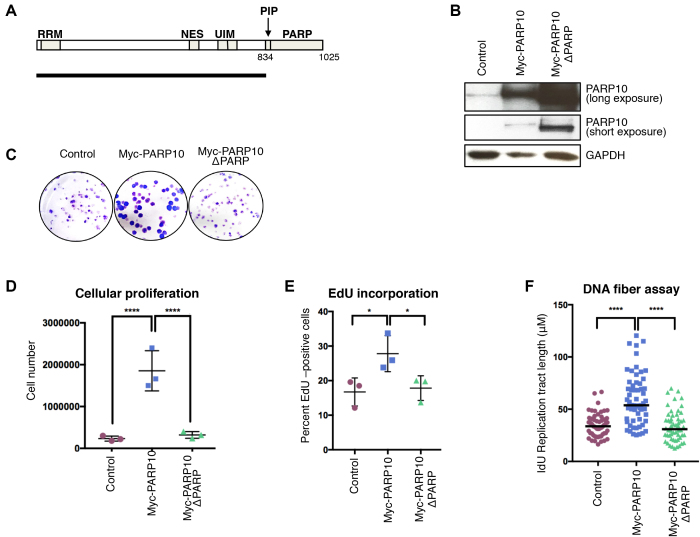
PARP10 overexpression was identified in a large proportion of tumors (Supplementary Figure S1), suggesting that PARP10 may act as an oncogene to promote transformation. To address this, we overexpressed PARP10 in the non-transformed, hTERT-immortalized human epithelial cell line RPE-1 using a stable, lentiviral-mediated, doxycycline-inducible system (Figure 3A and B). As a control, we also overexpressed a PARP10 variant lacking the PCNA-interacting PIP-box and the catalytic PARP domain (PARP10-ΔPARP, spanning residues 1–834). PARP10 overexpression resulted in increased growth rates of RPE-1 cells, while PARP10-ΔPARP overexpression did not alter proliferation (Figure 3C and D).
Figure 3.
Overexpression of PARP10 promotes proliferation of non-transformed RPE-1 cells. (A) Schematic representation of PARP10 domain organization. The black line underneath shows the length of the PARP10-ΔPARP variant (spanning residues 1–834) which lacks the PCNA-interacting PIP motif and the catalytic PARP domain. RRM: RNA recognition motif; NES: nuclear localization signal; UIM: ubiquitin interacting motifs; PIP: PCNA-interacting motif; PARP: catalytic ADP-ribosyltransferase domain. (B) Western blot showing the overexpression of Myc-tagged PARP10 wild-type and ΔPARP in RPE-1 cells. (C–E) Overexpression of wild-type, but not PARP-deleted PARP10, promotes proliferation of RPE-1 cells. (C) Representative clonogenic assay. (D) Quantification of cell number from clonogenic assays using CellTiterGlo reagent. The mean and standard deviation are shown. (E) Quantification of EdU-incorporating cells. EdU was added to the media for 45 min prior to harvesting. The mean with standard deviation is shown. Representative flow cytometry plots are shown in Supplementary Figure S4. (F) DNA fiber combing assay showing increased replication tracts in PARP10-overexpressing RPE-1 cells under normal (no drug treatment) conditions. Shown is the quantification of the IdU tract length, with the median values marked.
To examine the mechanistic basis for their increased proliferation, we measured DNA synthesis rates in PARP10-overexpressing RPE-1 cells. We grew cells in the presence of thymidine analog EdU for 45 min and quantified EdU incorporation using Click chemistry. Compared to control, a higher proportion of PARP10-overexpressing cells showed EdU incorporation, while PARP10-ΔPARP overexpression did not affect EdU incorporation rates (Figure 3E and Supplementary Figure S4). Moreover, DNA fiber combing indicated that, under normal growth conditions (no drug treatment), PARP10 overexpression results in longer DNA tracts (Figure 3F), which was not the case for PARP10-ΔPARP overexpression. These findings indicate that PARP10 promotes replication fork progression in non-transformed cells.
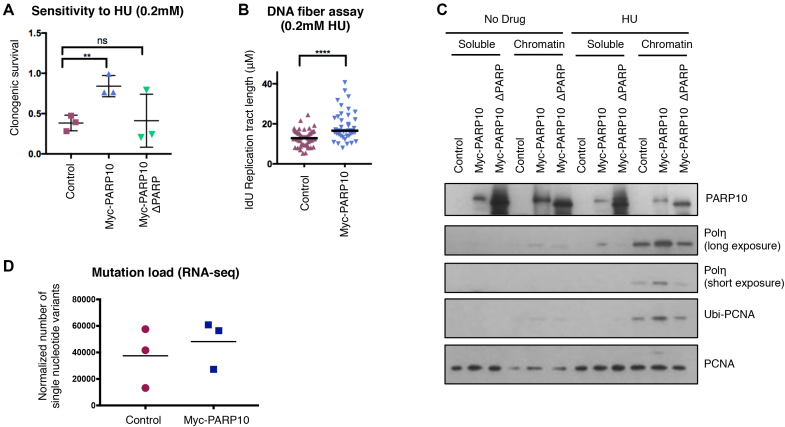
We next investigated if replication fork elongation upon PARP10 overexpression is coupled with increased resistance to replication stress. Clonogenic assays indicated that PARP10 overexpressing cells, but not PARP10-ΔPARP-overexpressing cells, were resistant to HU (Figure 4A). Moreover, PARP10-overexpressing cells showed longer replication tracts in the presence of HU in the DNA fiber combing assay (Figure 4B), suggesting that PARP10 overexpression promotes the ability of the replication machinery to restart stalled forks.
Figure 4.
Overexpression of PARP10 in RPE-1 suppresses replication stress and promotes engagement of mutagenic TLS polymerase Polη. (A) Clonogenic assay showing that PARP10-overexpressing RPE-1 cells are resistant to HU. For each cell line, the ratios of HU-treated to non-treated are presented. The mean and standard deviation are shown. (B) DNA fiber combing assay showing increased replication tracts in PARP10-overexpressing RPE-1 cells under HU treatment. Shown is the quantification of the IdU tract length, with the median values marked. (C) Chromatin fractionation experiments showing increased chromatin recruitment of TLS polymerase Polη in PARP10-overexpressing RPE-1 cells following HU treatment (2 mM for 24 h). A repeat experiment, as well as quantification of band intensities are provided in Supplementary Figure S5. (D) Mutation load in control and PARP10-overexpressing RPE-1 cells calculated from RNA-seq data. Three independent clones each were grown for 30 generations in the presence of doxycycline to induce PARP10 expression. The number of point mutations from loci with at least 10 reads was calculated, and normalized against genome coverage. The mean values are shown (P = 0.55).
Next, we attempted to decipher how PARP10 promotes fork stability under replication stress. We previously showed that PARP10 downregulation reduces the levels of ubiquitinated PCNA (14). In line with this, overexpression of PARP10 increased PCNA ubiquitination in RPE-1 cells (Figure 4C). The TLS polymerase Polη was previously shown to be recruited to chromatin and promote DNA synthesis upon HU treatment (27). As PCNA ubiquitination targets Polη to stalled forks (20), we reasoned that PARP10 overexpression may result in increased Polη engagement to promote replication under HU conditions. Indeed, chromatin fractionation experiments showed that RPE-1 cells overexpressing wild-type, but not the ΔPARP variant, have increased chromatin loading of Polη upon HU exposure (Figure 4C and Supplementary Figure S5). These results suggest that PARP10 promotes PCNA ubiquitination and subsequent TLS polymerase Polη engagement to enhance the restart of stalled replication forks and allow DNA synthesis under replication stress conditions.
As TLS polymerases are inherently error-prone, we hypothesized that their engagement would result in increased mutagenesis. To test for this, we grew three independent clones of control and PARP10-overexpressing RPE-1 cells for 25 generations (in the presence of doxycycline to induce PARP10 overexpression) and subjected them to RNA sequencing. We then calculated the number of single nucleotide variants identified in each sample (limiting this analysis to loci with at least 10 reads, to support the call). When normalized to genome coverage, we found a trend toward increased mutation burden in PARP10-overexpressing cells (Figure 4D). While not statistically significant, this trend is suggestive of an impact of PARP10 overexpression on mutation rates. Overall, these results support the model that PARP10-mediated replication stress suppression may involve, at least in part, mutagenic fork restart by the TLS polymerase Polη.
PARP10 regulates tumor growth in vivo
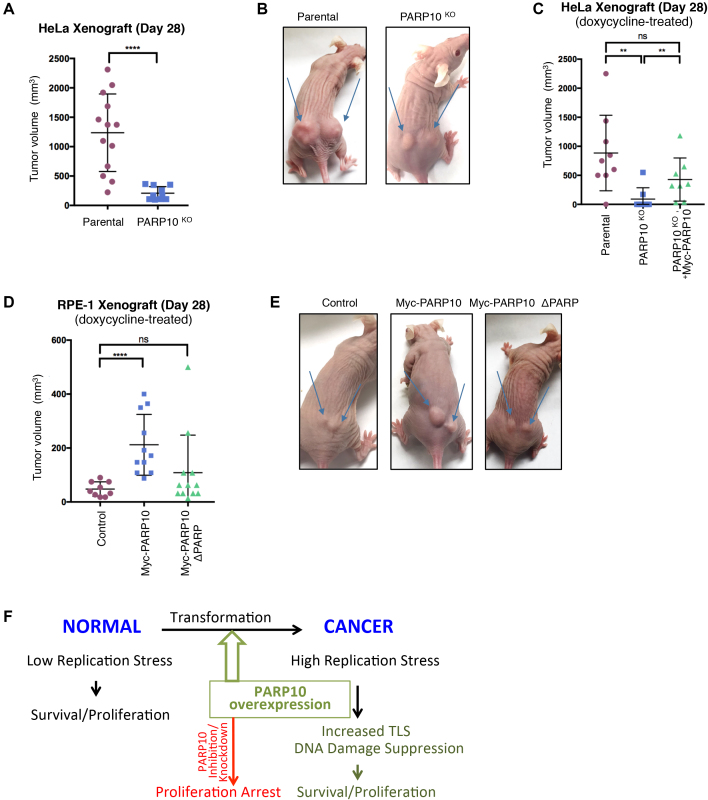
Because of the strong effect of PARP10 on in vitro cellular proliferation that we uncovered here, we decided to investigate the impact of PARP10 on in vivo tumor growth using xenograft models. First, we tested tumor formation by HeLa PARP10-knockout cells. We subcutaneously injected 5 million matrigel-embedded cells into each flank of athymic nude mice, and monitored tumor growth. As expected, wild-type HeLa cells generated robust tumors within 4 weeks from the time of injection. In contrast, PARP10-knockout cells showed severely impaired tumor formation capacity (Figure 5A and B) indicating that PARP10 is necessary for tumor growth in vivo. To rule out possible off-target effects generated by the CRISPR/Cas9 genome editing system in the PARP10-knockout cells, we repeated the experiment and included HeLa PARP10-knockout cells corrected by doxycycline-induced expression of Myc-tagged PARP10. For this experiment, 10 million matrigel-embedded cells were injected, and mice were administered doxycycline in their drinking water starting at the day of injection. Cells re-expressing Myc-PARP10 could form tumors similar to parental cells (Figure 5C), thus firmly establishing that PARP10 is specifically required for tumor growth in vivo.
Figure 5.
PARP10 promotes tumor growth in vivo. (A and B) PARP10 deletion reduces tumor formation by HeLa cells. (A) Quantification of tumor size, 28 days after subcutaneous injection. A total of eight mice were used for each condition. The mean and standard deviation are shown. (B) Representative images of tumor formation by HeLa control and PARP10-knocokout cells at day 28. (C) Exogenous re-expression of Myc-tagged PARP10 restores tumor formation ability. Quantification of tumor size, 28 days after subcutaneous injection is shown. For each condition, five mice were used, which were administered doxycycline in their drinking water to induce exogenous Myc-PARP10 expression. The mean and standard deviation are shown. (D and E) PARP10 overexpression promotes tumor formation by non-transformed RPE-1 cells. (D) Quantification of tumor size, 28 days after subcutaneous injection. For each condition, seven mice were used, which were administered doxycycline in their drinking water to induce exogenous Myc-PARP10 expression. The mean and standard deviation are shown. (E) Representative images of tumor formation by RPE-1 cells at day 28. (F) Model showing the involvement of PARP10 in carcinogenesis. PARP10 overexpression during transformation confers protection against replication stress by increasing TLS. Targeting PARP10 may reduce proliferation of cancer cells.
As PARP10 is overexpressed in a significant proportion of human cancers, suggestive of an oncogenic role (Supplementary Figure S1), we next tested tumor formation by PARP10-overexpresing non-transformed RPE-1 cells. To this end, we injected 10 million matrigel-embedded RPE-1 cells (control, PARP10-overexpressing or PARP10-ΔPARP overexpressing) in each flank of athymic nude mice, which were also administered doxycycline. Consistent with the non-transformed status of RPE-1 cells, previous studies have shown that these cells do not induce tumor formation in immunocompromised mice (28). In line with this, in our study, control RPE-1 cells induced negligible growth at the site of injection. In contrast, PARP10-overexpressing RPE-1 cells generated noticeable tumors (Figure 5D and E), albeit much smaller than those generated by HeLa cells within the same time frame. Importantly, PARP10-ΔPARP-overexpressing RPE-1 cells lacked tumor formation ability (Figure 5D and E). Altogether, these results suggest that PARP10 has oncogene-like properties.
DISCUSSION
We show here that PARP10 is upregulated in large number of human tumors, and its overexpression promotes cellular proliferation in vitro and tumor formation in vivo (Figures 3–5 and Supplementary Figure S1). We propose that PARP10 upregulation contributes to alleviating replication stress during cellular transformation, through increasing TLS and thus suppressing DNA damage accumulation (Figure 5F). In line with this, removal of PARP10 from cancer cells severely impairs replication stress resistance and tumor formation in vivo (Figures 1, 2 and 5).
While significantly smaller than tumors generated by HeLa cells within the same time frame, tumor formation by PARP10-overexpressing RPE-1 cells (Figure 5) is nevertheless a significant and unexpected finding. Xenograft tumor formation by RPE-1 cells was previously established as a model for investigating and validating oncogenic mechanisms. Known oncogenes, such as Ras, have been shown to promote tumor formation in this system, which was subsequently used to confirm oncogenesis by recently discovered oncogenes such as PGBD5 (28,29). Our work thus suggests that PARP10 functions as an oncogene to promote tumor growth. In line with the concept of oncogene addiction (30), loss of PARP10 reduces proliferation of cancer cells (Figure 1). Thus, PARP10 may represent a novel target in cancer therapy.
Other roles of PARP10 have been recently described, which may mediate its impact on proliferation. One study showed that PARP10 impacts the mitochondrial oxidation process (12). While we cannot exclude an impact of this process on the tumor-promoting activity of PARP10, our data indicate that PARP10 enhances replication fork elongation at the molecular level, thus arguing that replication stress suppression through TLS engagement is at least one of the components of the oncogenic activity of PARP10. Another recent paper described a role for PARP10 in cell motility through regulating Aurora A activity (13). Interestingly, the authors also created PARP10-knockout HeLa cells and found increased cell motility in vitro, and increased metastasis in vivo (using an experimental approach in which they injected the HeLa cells in the tail vein of Balb/C mice and measured lung metastases). As the experimental setup is different from our study, we believe that these are likely to be two separate functions of PARP10. Indeed, preliminary RNA-sequencing studies (not shown) indicate that the top two cellular processes upregulated in PARP10-overexpressing cells are DNA replication and cell cycle (in line with their increased proliferation). At the same time, a number of processes linked to cell adhesion (extracellular matrix, fibronectin) are found within the top five downregulated pathways—indicating a separate, inhibitory activity of PARP10 against cell adhesion. In any case, our results indicate that PARP10 knockout results in reduced proliferation in vitro and tumor formation in vivo, and both phenotypes can be rescued by exogenous re-expression of PARP10. Moreover, we show that these proliferation defects correlate with replication stress hypersensitivity, and lower rates of fork progression under replication stress conditions. Thus, regardless of any effects of PARP10 on cell motility, our data support a role for PARP10 in alleviating replication stress and promoting proliferation.
Our work suggests that PARP10 overexpression enhances engagement of TLS polymerases such as Polη to promote DNA synthesis under endogenous and exogenous replication stress. Indeed, PARP10 overexpressing cells have longer replication tracts under both control (no drug) and HU treatment (Figures 3 and 4). While our DNA fiber combing assays cannot differentiate between increased fork speed versus more efficient restart of stalled forks, our results showing engagement of Polη suggest that PARP10 overexpression acts through fork restart. Thus, our studies provide additional support for the model that, by alleviating replication stress, TLS promotes cellular transformation (17). Our findings that PARP10-overexpressing cells may potentially have increased mutation load, coupled with our previous study showing that PARP10 downregulation reduces mutation rates (14), suggest that PARP10 expression may be associated with increased mutagenesis. This induction of genomic instability by PARP10 may contribute to its oncogenic activity, but it is likely that other PARP10 functions play roles in this activity.
At this time, it is still unclear exactly how PARP10 may promote TLS, and if PCNA interaction is required for this. Its effect of promoting PCNA ubiquitination (see our previous work (14) and Figure 4), suggests that PARP10 activity may directly enhance the enzymatic process of PCNA ubiquitination, either by making PCNA a better substrate or by increasing the activity of ubiquitin ligases such as RAD18 toward PCNA (16). Alternatively, PARP10 may act downstream of this modification by stabilizing it against de-ubiquitination by USP1. Indeed, we previously showed that PARP10 preferentially interacts with ubiquitinated PCNA, through its ubiquitin-interacting motifs (14). Perhaps this results in shielding of PCNA ubiquitination against USP1. This model is in line with the findings reported here, that PARP10 acts in a dose-dependent manner to enhance PCNA ubiquitination. On the other hand, our results using the PARP10-ΔPARP mutant may indicate that the catalytic activity of PARP10 is required for this activity. In line with this, in preliminary studies (not shown) we observed that PARP10 is able to ADP-ribosylate PCNA in vitro. We speculate that this modification is in turn allowing PCNA ubiquitination levels to build up, thus increasing TLS polymerase recruitment to promote DNA synthesis under replication stress conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dipanjan Chowdhury and Katherine Choe for the RPE-1 cells, and the Penn State College of Medicine Flow Cytometry and Imaging cores. This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco CURE Funds. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society [ACS-IRG-13–043-01]; Pennsylvania Department of Health (to C.M.N.); National Institutes of Health [ES026184 to G.L.M.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gibson B.A., Kraus W.L.. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat. Rev. Mol. Cell Biol. 2012; 13:411–424. [DOI] [PubMed] [Google Scholar]
- 2. Kalisch T., Ame J.C., Dantzer F., Schreiber V.. New readers and interpretations of poly(ADP-ribosyl)ation. Trends Biochem. Sci. 2012; 37:381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T.. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005; 434:913–917. [DOI] [PubMed] [Google Scholar]
- 4. Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C. et al. . Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005; 434:917–921. [DOI] [PubMed] [Google Scholar]
- 5. Fong P.C., Boss D.S., Yap T.A., Tutt A., Wu P., Mergui-Roelvink M., Mortimer P., Swaisland H., Lau A., O’Connor M.J. et al. . Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N. Engl. J. Med. 2009; 361:123–134. [DOI] [PubMed] [Google Scholar]
- 6. Murai J. Targeting DNA repair and replication stress in the treatment of ovarian cancer. Int. J. Clin. Oncol. 2017; 22:619–628. [DOI] [PubMed] [Google Scholar]
- 7. Kleine H., Poreba E., Lesniewicz K., Hassa P.O., Hottiger M.O., Litchfield D.W., Shilton B.H., Luscher B.. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol. Cell. 2008; 32:57–69. [DOI] [PubMed] [Google Scholar]
- 8. Yu M., Schreek S., Cerni C., Schamberger C., Lesniewicz K., Poreba E., Vervoorts J., Walsemann G., Grotzinger J., Kremmer E. et al. . PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene. 2005; 24:1982–1993. [DOI] [PubMed] [Google Scholar]
- 9. Chou H.Y., Chou H.T., Lee S.C.. CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J. Biol. Chem. 2006; 281:15201–15207. [DOI] [PubMed] [Google Scholar]
- 10. Herzog N., Hartkamp J.D., Verheugd P., Treude F., Forst A.H., Feijs K.L., Lippok B.E., Kremmer E., Kleine H., Luscher B.. Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J. 2013; 280:1330–1343. [DOI] [PubMed] [Google Scholar]
- 11. Verheugd P., Forst A.H., Milke L., Herzog N., Feijs K.L., Kremmer E., Kleine H., Luscher B.. Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nat. Commun. 2013; 4:1683. [DOI] [PubMed] [Google Scholar]
- 12. Marton J., Fodor T., Nagy L., Vida A., Kis G., Brunyanszki A., Antal M., Luscher B., Bai P.. PARP10 (ARTD10) modulates mitochondrial function. PLoS One. 2018; 13:e0187789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao Y., Hu X., Wei L., Song D., Wang J., You L., Saiyin H., Li Z., Yu W., Yu L. et al. . PARP10 suppresses tumor metastasis through regulation of Aurora A activity. Oncogene. 2018; 37:2921–2935. [DOI] [PubMed] [Google Scholar]
- 14. Nicolae C.M., Aho E.R., Vlahos A.H., Choe K.N., De S., Karras G.I., Moldovan G.L.. The ADP-ribosyltransferase PARP10/ARTD10 interacts with proliferating cell nuclear antigen (PCNA) and is required for DNA damage tolerance. J. Biol. Chem. 2014; 289:13627–13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shahrour M.A., Nicolae C.M., Edvardson S., Ashhab M., Galvan A.M., Constantin D., Abu-Libdeh B., Moldovan G.L., Elpeleg O.. PARP10 deficiency manifests by severe developmental delay and DNA repair defect. Neurogenetics. 2016; 17:227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choe K.N., Moldovan G.L.. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol. Cell. 2017; 65:380–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zafar M.K., Eoff R.L.. Translesion DNA synthesis in cancer: molecular mechanisms and therapeutic opportunities. Chem. Res. Toxicol. 2017; 30:1942–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S.. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002; 419:135–141. [DOI] [PubMed] [Google Scholar]
- 20. Kannouche P.L., Wing J., Lehmann A.R.. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004; 14:491–500. [DOI] [PubMed] [Google Scholar]
- 21. Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L.V., Kolettas E., Niforou K., Zoumpourlis V.C. et al. . Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006; 444:633–637. [DOI] [PubMed] [Google Scholar]
- 22. Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A. Jr, Kastrinakis N.G., Levy B. et al. . Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005; 434:907–913. [DOI] [PubMed] [Google Scholar]
- 23. Choe K.N., Nicolae C.M., Constantin D., Imamura Kawasawa Y., Delgado-Diaz M.R., De S., Freire R., Smits V.A., Moldovan G.L.. HUWE1 interacts with PCNA to alleviate replication stress. EMBO Rep. 2016; 17:874–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicolae C.M., Aho E.R., Choe K.N., Constantin D., Hu H.J., Lee D., Myung K., Moldovan G.L.. A novel role for the mono-ADP-ribosyltransferase PARP14/ARTD8 in promoting homologous recombination and protecting against replication stress. Nucleic Acids Res. 2015; 43:3143–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. . The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E. et al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013; 6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Feraudy S., Limoli C.L., Giedzinski E., Karentz D., Marti T.M., Feeney L., Cleaver J.E.. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene. 2007; 26:5713–5721. [DOI] [PubMed] [Google Scholar]
- 28. Henssen A.G., Koche R., Zhuang J., Jiang E., Reed C., Eisenberg A., Still E., MacArthur I.C., Rodriguez-Fos E., Gonzalez S. et al. . PGBD5 promotes site-specific oncogenic mutations in human tumors. Nat. Genet. 2017; 49:1005–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., Weinberg R.A.. Creation of human tumour cells with defined genetic elements. Nature. 1999; 400:464–468. [DOI] [PubMed] [Google Scholar]
- 30. Weinstein I.B., Joe A.K.. Mechanisms of disease: oncogene addiction–a rationale for molecular targeting in cancer therapy. Nat. Clin. Pract. Oncol. 2006; 3:448–457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.