Abstract
The tumor suppressor, breast cancer susceptibility gene 1 (BRCA1), plays an integral role in the maintenance of genome stability and, in particular, the cellular response to DNA damage. Here, the emerging role of BRCA1 in nonhomologous end-joining-mediated DNA repair following DNA damage will be reviewed, as well as the activation of apoptotic pathways. The control of these functions via DNA damage-induced BRCA1 shuttling will also be discussed, in particular BRCA1 shuttling induced by erlotinib and irradiation. Finally, the potential targeting of BRCA1 shuttling as a novel strategy to sensitize cells to DNA damage will be entertained.
Keywords: BRCA1, DNA damage, DNA repair, homologous recombination, nonhomologous end-joining
Introduction
Cells are constantly subjected to a variety of insults that endanger the integrity and fidelity of the genome. However, several processes are in place to prevent or resolve the potential damage incurred, including DNA damage response pathways to initiate cell cycle checkpoints, execute repair of DNA damage, and activate programmed cell death [1]. The breast cancer susceptibility gene 1 (BRCA1) plays a central role in this manner.
BRCA1 functions in a number of cellular processes, including chromatin remodeling, protein ubiquitination, DNA replication, DNA repair, regulation of transcription, cell cycle checkpoint control and apoptosis [2–7]. Disruption of any or all of these processes may contribute to the increased risk for carcinogenesis, as seen in carriers of germline BRCA1 mutations [7]. Regulation of BRCA1 function occurs through a variety of mechanisms, including transcriptional control, protein– protein interactions and post-translational modification [2–9]. BRCA1 is a nuclear–cytoplasmic shuttling protein and its functions may be controlled via active shuttling between cellular compartments [8–12].
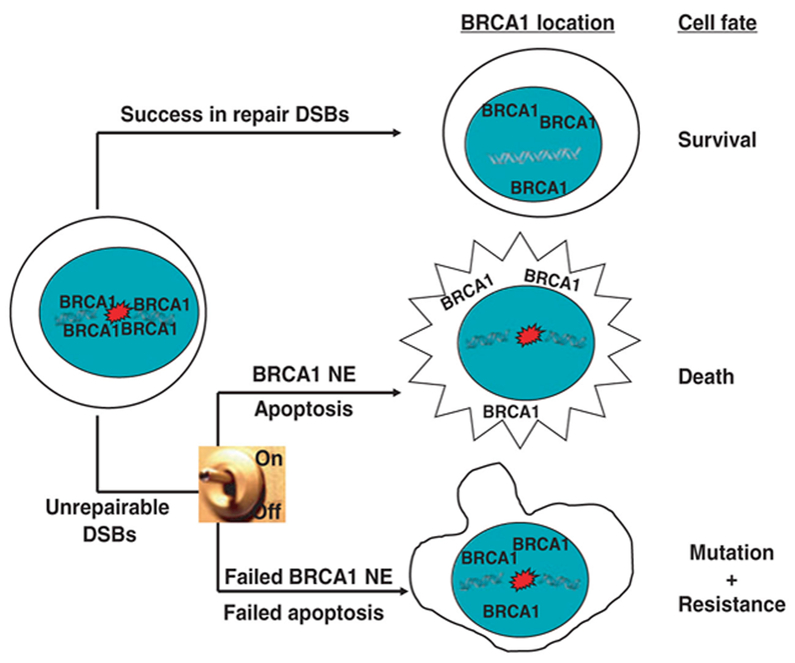
This review will focus specifically on the emerging role of BRCA1 in the repair of DNA double strand breaks (DSBs) through regulating nonhomologous end-joining (NHEJ), one of the two major repair pathways. Additionally, DNA damage-induced regulation of BRCA1 shuttling to various cellular compartments to control its functions will be discussed. We propose a model by which BRCA1 protects the genome integrity through facilitating the repair of damaged DNA and, if unsuccessful, is shuttled to other cellular compartments to activate cell death pathways to eliminate cells with persistent DNA lesions (Fig. 1).
Fig. 1.
Model depicting how BRCA1 protects the genome. BRCA1 protects the genome through facilitating the repair of damaged DNA and, if unsuccessful, is shuttled to other cellular compartments to activate cell death pathways to eliminate cells with persistent DNA lesions. In contrast, survival of cells carrying damaged DNA will lead to genomic instability and resistance to DNA damage-based cancer therapy.
BRCA1 and DNA repair
In response to DNA damage, the initial signaling cascade involves the ataxia telangiectasia mutated ⁄ ataxia telangiectasia and Rad3-related-dependent phosphorylation of the histone variant H2AX and its subsequent localization at sites of DNA damage [13,14]. This, in turn, recruits other signaling and repair proteins to DNA breaks to resolve the damaged DNA [15]. Discussion of these specific factors has been extensively reviewed [16,17] and is beyond the scope of this article.
Two major processes exist in cells and compete for DSB sites to repair these DNA lesions: homologous recombination (HR) and NHEJ [18,19]. NHEJ is an efficient and the predominant mechanism of DSB repair throughout all phases of the cell cycle. In contrast, HR is less efficient and repairs DSBs mostly during the late S and G2 phases of the cell cycle, but results in high fidelity repair [20,21]. BRCA1 is integral in the DNA damage response and serves to maintain genomic fidelity by playing a central role in controlling these pathways. The role of BRCA1 in HR is extensively reviewed elsewhere [17,22] and therefore will not be discussed here. However, emerging evidence suggests the importance of BRCA1 in NHEJ, in particular precise end-joining, and will be subsequently reviewed.
BRCA1 and NHEJ
Alternative to HR, cells can rejoin DSBs via NHEJ without extensive sequence homologies. Two major subpathways exist for NHEJ: the canonical (or conservative) NHEJ (C-NHEJ) pathway and the alternative NHEJ (A-NHEJ) pathway [23–31]. The C-NHEJ pathway, which is dependent on the DNA-PK ⁄ Ku70 ⁄ 80 and XRCC4 ⁄ ligase IV protein complex, can precisely repair the DSB when the physical structures at the ends are compatible. In contrast, the A-NHEJ pathway, which is independent of Ku80 and depends on Mre11 [32–35], repairs the DSBs by searching and using flanking microhomologies. This results in deletions at the junction and is highly mutagenic.
The exact role of BRCA1 in NHEJ, however, has not been well defined. Studies have yielded conflicting results, from enhanced NHEJ to suppressed NHEJ to no effect [26,36–40]. For example, BRCA1-deficient mouse embryonic stem cells were found to exhibit increased nonhomologous random integration [36,37], In contrast, cell extracts derived from BRCA1-deficient mouse embryonic fibroblasts had reduced end-joining activity [39]. Additionally, Chk2-mediated phosphorylation of the serine 988 residue of BRCA1 has been shown to promote precise religation while suppressing error-prone repair processes [26,38,40]. This discrepancy may be due to the differing involvement of BRCA1 in the various NHEJ subpathways. However, given BRCA1’s role in maintaining genome integrity, it has been hypothesized that BRCA1 enhances precise C-NHEJ while suppressing the error-prone A-NHEJ. In support of this notion, precise NHEJ was shown to require BRCA1 [25,41].
Interestingly, Chk2-mediated phosphorylation of BRCA1 at serine 988 was reported to promote error-free HR and precise NHEJ while preventing error-prone A-NHEJ [25,26,38]. Furthermore, ataxia telangiectasia mutated-mediated phosphorylation of BRCA1 at serine 1423 and serine 1524 was found to be important in precise end-joining activity [25]. BRCA1 was also found to rapidly accumulate at sites of laser irradiation-induced DSBs [42]. This recruitment was dependent on interaction with Ku80, which is intimately involved in precise NHEJ [42]. Our recent data further showed that BRCA1 directly interacts with Ku80 and stabilizes binding of Ku80 to the ends of DSBs (unpublished). These results indicate that BRCA1 may directly regulate NHEJ through physical interaction to control the activity of the DNA-PK ⁄ Ku80 protein complex. Another possible mechanism by which BRCA1 promotes high fidelity repair is by protecting DNA ends from resection by exonucleases, such as Mre11[43].
BRCA1 functions as a central regulator of genome maintenance. One such role is to regulate the repair of damaged DNA. As multiple competing pathways exist in a cell to resolve the DNA lesion, BRCA1 serves to promote high fidelity repair processes, including both HR and C-NHEJ, while suppressing mutagenic and error-prone pathways.
DNA damage-induced BRCA1 shuttling
BRCA1 serves a multitude of functions in the DNA damage response, one of which is to promote high fidelity repair of damaged DNA. Regulation of these functions is complex and involves a variety of mechanisms, one of which includes nuclear–cytoplasmic shuttling.
BRCA1 is a shuttling protein [9–12]. When nuclear, BRCA1 controls high fidelity repair of damaged DNA. In contrast, BRCA1 has been shown to enhance p53-independent apoptosis when cytoplasmic [10,11]. Two nuclear localization signals reside within BRCA1, which target it to the nucleus in an importin alpha / beta manner [12,44]. Two nuclear export sequences (NESs) exist at the N-terminus of BRCA1, which transports BRCA1 to the cytoplasm through the chromosome region maintenance 1 (CRM1) / exportin pathway [45,46]. Specific details regarding the BRCA1 nuclear localization signals and NESs are discussed elsewhere [12].
BRCA1 shuttling can also be regulated via protein– protein interaction. The BRCA1-associated RING domain protein (BARD1) has been shown to bind and mask the BRCA1 NES located at the N-terminal RING domain, thereby preventing nuclear export of BRCA1 through CRM1 [10,11]. At the C-terminus of BRCA1, the BRCA1 C-terminus (BRCA) domain has been shown to play a crucial role in the nuclear import of BRCA1 through association with numerous other proteins, including p53, CtIP and BACH, in response to DNA damage [47–49]. Mutations that target the BRCT region of BRCA1 have been shown to exclude BRCA1 from the nucleus by blocking nuclear import[45]. Reciprocally, human breast cancer cells with deficiency in p53 function have been shown to exhibit aberrant BRCA1 shuttling (Jiang et al., manuscript submitted). The critical region of this regulation appears to reside in the BRCT domain of BRCA1. The BRCT domain also acts in conjunction with the RING domain to facilitate the formation of nuclear foci following DNA damage [50,51]. Thus, the control of BRCA1 subcellular localization is potentially an important mechanism by which BRCA1-mediated repair of DNA damage can be regulated.
Irradiation-induced BRCA1 shuttling
The subcellular redistribution of BRCA1 is an important regulatory mechanism in the cellular response to DNA damage [8,9]. It has been previously reported that following irradiation, BRCA1 is exported out of the nucleus [8,9]. This effect occurs as early as 4 h after irradiation and persists 50 h after irradiation. This redistribution of BRCA1 follows a dose-dependent manner and utilizes a CRM1-dependent mechanism. Additionally, as irradiation-induced DNA damage can trigger checkpoints, BRCA1 nuclear export following irradiation could be a function of cell cycle [8,9]. Although there is variation in BRCA1’s localization between the G1, S, and G2 / M phases, DNA damage-induced BRCA1 nuclear export occurs in all phases of the cell cycle. In particular, cells subjected to irradiation- or UV-induced DNA damage were found to redistribute BRCA1 phosphorylated at serine 988 to perinuclear regions [8]. Interestingly, DNA damage-induced BRCA1 nuclear export was abrogated when p53 was rendered dysfunctional [9]. As p53 is intimately involved in the activation of DNA damage-induced checkpoints as well as apoptosis, this interplay between p53 and the regulation of BRCA1 shuttling may be an interesting mechanism by which communication between DNA repair and cell death pathways ensures elimination of cells that retain persistent DNA damage.
Erlotinib-induced BRCA1 shuttling
The epidermal growth factor receptor (EGFR) family functions in modulating proliferation, differentiation and survival, and has become the target of novel cancer therapeutic strategies [52]. Aberrant expression and dysregulation of any EGFR can be found in several cancers, including lung, pancreas, head and neck, brain and breast. Interestingly, the EGFR family has been shown to interact with the DNA damage pathways [53,54]. Blockade of EGFR signaling results in alteration of the DNA damage response [53–56].
In particular, erlotinib has been shown to decrease irradiation-induced expression of Rad51 and to enhance radiation-induced apoptosis, suggesting a potential role of erlotinib in influencing the DNA damage response [54,55]. Accordingly, erlotinib treatment of breast cancer cells suppresses HR capacity independent of cell cycle effects [54]. This correlates with accumulation of persistent γ-H2AX nuclear foci, which is a well-characterized in situ marker of chromosomal DSBs[18]. Erlotinib treatment results in a significant shift of BRCA1 to the cytoplasm [54]. As nuclear BRCA1 plays a central role in the DNA damage response, and in particular repair, these results again provide a link between BRCA1 localization and the DNA damage response.
BRCA1 and apoptosis
In addition to the repair of damaged DNA, BRCA1 plays a role in apoptosis. Overexpression of BRCA1 induces apoptosis [6]. This process has been linked to the DNA damage response and the c-Jun N-terminal kinase pathway [57,58] and depends on its nuclear export [10,11]. Conversely, BARD1, which binds and masks the BRCA1 NES to prevent BRCA1 nuclear export, inhibits BRCA1-mediated apoptosis [11]. The apoptotic pathway stimulated by BRCA1 is independent of p53.
BRCA1 also stimulates apoptosis in chemotherapyand UV-treated cells [59,60]. Exogenous expression of BRCA1 also enhanced this cytoxic response. The mechanism of BRCA1-mediated apoptosis involves caspase 3-mediated cleavage of BRCA1 to a 90 kDa fragment (BRCA1-p90) [59,60]. This fragment comprises the C-terminal region of BRCA1 and is mainly localized to the cytoplasm. Expression of BRCA1-p90 was sufficient to promote cell death and to increase cytotoxicity to cisplatin chemotherapy [59].
Other mechanisms of BRCA1-mediated apoptosis include activation of caspase 3 in response to DNA damage-induced phosphorylation of BRCA1 [61]. This in turn disrupts the interaction between X-linked inhibitor of apoptosis protein and caspase 9. Caspase 9 subsequently cleaves caspase 3 and hence activates the apoptotic cascade. Additionally, BRCA1 apoptotic activity may be linked to its mitochondrial localization [62]. Taken together, these findings demonstrate that the cytoplasmic subcellular localization of BRCA1 plays an important role in regulating BRCA1-mediated apoptosis.
Targeting BRCA1 localization
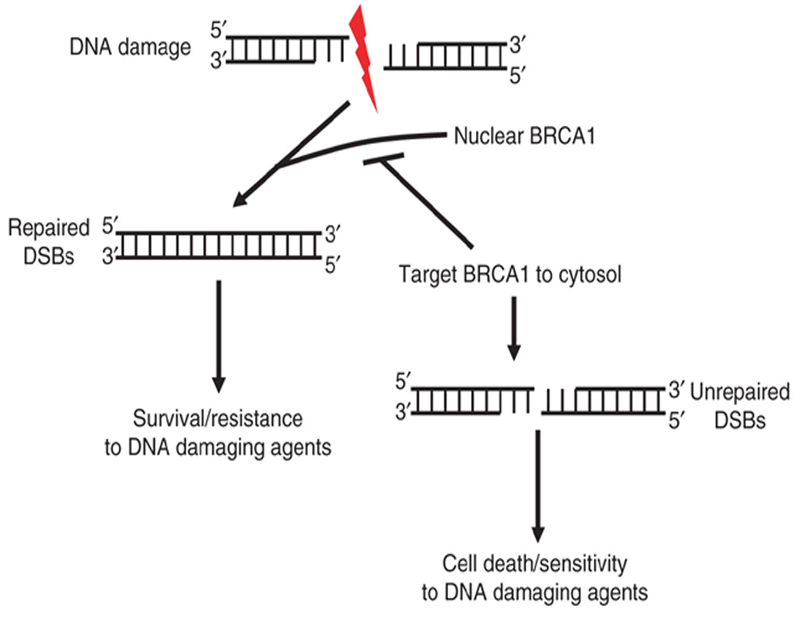
Given the multiple roles that BRCA1 plays in the DNA damage response, including repair and activation of apoptosis, it is intriguing to hypothesize that following DNA damage, BRCA1 facilitates the repair of DNA in the nucleus and, if not successful, is exported out of the nucleus to initiate apoptotic pathways in the cytoplasm. Furthermore, the targeting of BRCA1 sub-cellular localization (i.e. deplete nuclear BRCA1) may be a potential avenue by which tumor cells can be sensitized to DNA-damaging agents (Fig. 2). In this subsequent section, a potential clinical application whereby altering BRCA1 localization will enhance the therapeutic response will be addressed.
Fig. 2.
Model depicting potential targeting of BRCA1 to the cytoplasm to inhibit repair of DSBs and to sensitize cells to DNA-damaging agents. Following DNA damage, BRCA1 facilitates the repair of DNA in the nucleus. By targeting BRCA1 subcellular localization (i.e. depletion of nuclear BRCA1 or translocation of BRCA1 DNA-damaging agents.
One strategy by which BRCA1 localization can be targeted is by altering the interaction between BRCA1 and BARD1, which binds BRCA1 at the N-terminal RING domain and masks the BRCA1 NES to prevent BRCA1 nuclear export [10,11]. Previous reports have shown that ectopic expression of the N-terminal RING domain fragment peptide tr-BRCA1, which also contains the BRCA1 NES and BARD1 binding site, can effectively shift BRCA1 to the cytosol [10,11,54,63]. Importantly, this action is as effective as irradiation-induced BRCA1 nuclear export and does not require p53. Given these results, tr-BRCA1 could be a potential tool to target BRCA1 localization to enhance the cytotoxic response to DNA-damaging agents.
In support of this notion, tr-BRCA1-mediated trans-location of BRCA1 to the cytosol has been shown to sensitize breast cancer cells to erlotinib [54]. Additionally, tumor cells with aberrant p53, which do not exhibit DNA damage-induced BRCA1 nuclear export, were found to be more resistant to the DNA-damaging agents cisplatin and irradiation. Sensitivity to these agents was rescued upon restoration of BRCA1 shuttling by tr-BRCA1 (Jiang et al., manuscript submitted). Thus, these findings substantiate the targeting of BRCA1 shuttling as a novel strategy to enhance the cytotoxic response to DNA-damaging agents.
Conclusion
BRCA1 is essential in maintaining genomic stability and controlling the cellular response to genotoxic stress. Precise regulation of these BRCA1 functions is of obvious importance from an oncological and cell survival perspective. One emerging target is BRCA1 localization and shuttling, as sequestration of BRCA1 away from the nucleus may switch BRCA1 function from repair in the nucleus to activation of cell death signals in the cytoplasm. The potential targeting of BRCA1 shuttling may be a novel avenue by which manipulation of BRCA1 localization can control cellular function and sensitivity to therapy. Furthermore, BRCA1 shuttling / localization itself may be a functional biomarker to predict a tumor response to therapy.
Abbreviations
- BARD1
BRCA1-associated RING domain protein
- BRCA1
breast cancer susceptibility gene 1
- BRCT
BRCA1 C-terminus
- CRM1
chromosome region maintenance 1
- DSB
double strand breaks
- EGFR
epidermal growth factor receptor
- HR
homologous recombination
- NES
nuclear export sequence
- NHEJ
nonhomologous end-joining
References
- 1.Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell Growth Differ 108, 171–182. [DOI] [PubMed] [Google Scholar]
- 2.Kerr P & Ashworth A (2001) New complexities for BRCA1 and BRCA2. Curr Biol 11, R668–R676. [DOI] [PubMed] [Google Scholar]
- 3.Pierce AJ, Stark JM, Araujo FD, Moynahan ME, Berwick M & Jasin M (2001) Double-strand breaks and tumorigenesis. Trends Cell Biol 11, S52–S59. [DOI] [PubMed] [Google Scholar]
- 4.Scully R, Puget N & Vlasakova K (2000) DNA polymerase stalling, sister chromatid recombination and the BRCA genes. Oncogene 19, 6176–6183. [DOI] [PubMed] [Google Scholar]
- 5.Venkitaraman AR (2002) Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 108, 171–182. [DOI] [PubMed] [Google Scholar]
- 6.Shao N, Chai YL, Shyam E, Reddy P & Rao VN (1996) Induction of apoptosis by the tumor suppressor protein BRCA1. Oncogene 13, 1–7. [PubMed] [Google Scholar]
- 7.Linger RJ & Kruk PA (2010) BRCA1 16 years later: risk-associated BRCA1 mutations and their functional implications. FEBS J 277, 3086–3096. [DOI] [PubMed] [Google Scholar]
- 8.Okada S & Ouchi T (2003) Cell cycle differences in DNA damage-induced BRCA1 phosphorylation affect its subcellular localization. J Biol Chem 278, 2015–2020. [DOI] [PubMed] [Google Scholar]
- 9.Feng Z, Kachnic L, Zhang J, Powell SN & Xia F (2004) DNA damage induces p53-dependent BRCA1 nuclear export. J Biol Chem 279, 28574–28584. Epub 22004 Apr 28515. [DOI] [PubMed] [Google Scholar]
- 10.Fabbro M, Rodriguez JA, Baer R & Henderson BR (2002) BARD1 induces BRCA1 intranuclear foci formation by increasing RING-dependent BRCA1 nuclear import and inhibiting BRCA1 nuclear export. J Biol Chem 277, 21315–21324. [DOI] [PubMed] [Google Scholar]
- 11.Fabbro M, Schuechner S, Au WWY & Henderson BR (2004) BARD1 regulates BRCA1 apoptotic function by a mechanism involving nuclear retention. Exp Cell Res 298, 661–673. [DOI] [PubMed] [Google Scholar]
- 12.Thompson ME (2010) BRCA1 16 years later: nuclear import and export processes. FEBS J 277, 3072–3078. [DOI] [PubMed] [Google Scholar]
- 13.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ et al. (2002) Genomic instability in mice lacking histone H2AX. Science 296, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogakou EP, Pilch DR, Orr AH, Ivanova VS & Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273, 5858–5868. [DOI] [PubMed] [Google Scholar]
- 15.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M & Bonner WM (2000) A critical role for his-tone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10, 886–895. [DOI] [PubMed] [Google Scholar]
- 16.Jackson SP & Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huen MS, Sy SM & Chen J (2009) BRCA1 and its toolbox for the maintenance of genome integrity. Nat Rev Mol Cell Biol 23, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khanna K & Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 19.van Gent DC, Hoeijmakers JH & Kanaar R (2001) Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2, 196–206. [DOI] [PubMed] [Google Scholar]
- 20.Lim DS, Vogel H, Willerford DM, Sands AT, Platt KA & Hasty P (2000) Analysis of ku80-mutant mice and cells with deficient levels of p53. Mol Cell Biol 20, 3772–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RD & Jasin M (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19, 3398–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J & Powell SN (2005) The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res 3, 531–539. [DOI] [PubMed] [Google Scholar]
- 23.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP et al. (2007) IgH class switching and translocations use a robust non-classical end-joining pathway. Nature 449, 478–482. [DOI] [PubMed] [Google Scholar]
- 24.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA et al. (2007) Rag mutations reveal robust alternative end joining. Nature 449, 483–486. [DOI] [PubMed] [Google Scholar]
- 25.Wang H-C, Chou W-C, Shieh S-Y & Shen C-Y (2006) Ataxia telangiectasia mutated and checkpoint kinase 2 regulate BRCA1 to promote the fidelity of DNA end-joining. Cancer Res 66, 1391–1400. [DOI] [PubMed] [Google Scholar]
- 26.Zhuang J, Zhang J, Willers H, Wang H, Chung JH, van Gent DC, Hallahan DE, Powell SN & Xia F (2006) Checkpoint kinase 2-mediated phosphorylation of BRCA1 regulates the fidelity of nonhomologous end-joining. 10.1158/0008–5472.CAN-05–3278 Cancer Res 66, 1401–1408. [DOI] [PubMed] [Google Scholar]
- 27.Bennardo N, Cheng A, Huang N & Stark JM (2008) Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guirouilh-Barbat J, Rass E, Plo I, Bertrand P & Lopez BS (2007) Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc Natl Acad Sci USA 104, 20902–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L & Lopez BS (2004) Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell 14, 611–623. [DOI] [PubMed] [Google Scholar]
- 30.Liang F, Romanienko PJ, Weaver DT, Jeggo PA & Jasin M (1996) Chromosomal double-strand break repair in Ku80-deficient cells. Proc Natl Acad Sci USA 93, 8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P & de Villartay JP (2007) V(D)J and immunoglobulin class switch recombinations: a paradigm to study the regulation of DNA end-joining. Oncogene 26, 7780–7791. [DOI] [PubMed] [Google Scholar]
- 32.Xie A, Kwok A & Scully R (2009) Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol 16, 814–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rass E, Grabarz A, Plo I, Gautier J, Bertrand P & Lopez BS (2009) Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol 16, 819–824. [DOI] [PubMed] [Google Scholar]
- 34.Dinkelmann M, Spehalski E, Stoneham T, Buis J,Wu Y, Sekiguchi JM & Ferguson DO (2009) Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol 16, 808–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhuang J, Jiang G, Willers H & Xia F (2009) The exonuclease function of human Mre11 promotes deletional non-homologous end-joining. J Biol Chem 9, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moynahan ME, Chiu JW, Koller BH & Jasin M (1999) Brca1 controls homology-directed DNA repair. Mol Cell 4, 511–518. [DOI] [PubMed] [Google Scholar]
- 37.Snouwaert JN, Gowen LC, Latour AM, Mohn AR, Xiao A, DiBiase L & Koller BH (1999) BRCA1 deficient embryonic stem cells display a decreased homologous recombination frequency and an increased frequency of non-homologous recombination that is corrected by expression of a Brca1 transgene. Oncogene, 18), 7900–7907. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, Chung JH, Powell SN & Xia F (2004) Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol 24, 708–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong Q, Boyer TG, Chen P-L & Lee W-H (2002) Deficient nonhomologous end-joining activity in cell-free extracts from Brca1-null fibroblasts. Cancer Res 62, 3966–3970. [PubMed] [Google Scholar]
- 40.Zhong Q, Chen C-F, Chen P-L & Lee W-H (2002) BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J Biol Chem 277, 28641–28647. [DOI] [PubMed] [Google Scholar]
- 41.Bau D-T, Fu Y-P, Chen S-T, Cheng T-C, Yu J-C, Wu P-E & Shen C-Y (2004) Breast cancer risk and the DNA double-strand break end-joining capacity of nonhomologous end-joining genes are affected by BRCA1. Cancer Res 64, 5013–5019. [DOI] [PubMed] [Google Scholar]
- 42.Wei L, Lan L, Hong Z, Yasui A, Ishioka C & Chiba N (2008) Rapid recruitment of BRCA1 to DNA double-strand breaks is dependent on its association with Ku80. Mol Cell Biol 28, 7380–7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paull TT, Cortez D, Bowers B, Elledge SJ & Gellert M (2001) From the cover: direct DNA binding by Brca1. Proc Natl Acad Sci USA 98, 6086–6091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CF, Li S, Chen Y, Chen PL, Sharp ZD & Lee WH (1996) The nuclear localization sequences of the BRCA1 protein interact with the importin-alpha subunit of the nuclear transport signal receptor. J Biol Chem 271, 32863–32868. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez JA & Henderson BR (2000) Identification of a functional nuclear export sequence in BRCA1. J Biol Chem 15, 15. [DOI] [PubMed] [Google Scholar]
- 46.Thompson ME, Robinson-Benion CL & Holt JT (2005) An amino-terminal motif functions as a second nuclear export sequence in BRCA1. J Biol Chem 280, 21854–21857. [DOI] [PubMed] [Google Scholar]
- 47.Chai YL, Cui J, Shao N, Shyam E, Reddy P & Rao VN (1999) The second BRCT domain of BRCA1 proteins interacts with p53 and stimulates transcription from the p21WAF1 / CIP1 promoter. Oncogene 18, 263–268. [DOI] [PubMed] [Google Scholar]
- 48.Li S, Chen PL, Subramanian T, Chinnadurai G, Tomlinson G, Osborne CK, Sharp ZD & Lee WH (1999) Binding of CtIP to the BRCT repeats of BRCA1 involved in the transcription regulation of p21 is disrupted upon DNA damage. J Biol Chem 274, 11334–11338. [DOI] [PubMed] [Google Scholar]
- 49.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA et al. (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105, 149–160. [DOI] [PubMed] [Google Scholar]
- 50.Au WW & Henderson BR (2005) The BRCA1 RING and BRCT domains cooperate in targeting BRCA1 to ionizing radiation-induced nuclear foci. J Biol Chem 280, 6993–7001. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez JA, Au WW & Henderson BR (2004) Cytoplasmic mislocalization of BRCA1 caused by cancer-associated mutations in the BRCT domain. Exp Cell Res 293, 14–21. [DOI] [PubMed] [Google Scholar]
- 52.Scaltriti M & Baselga J (2006) The epidermal growth factor receptor pathway: a model for targeted therapy. Clin Cancer Res 12, 5268–5272. [DOI] [PubMed] [Google Scholar]
- 53.Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R & Rodemann HP (2005) Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem 280, 31182–31189. [DOI] [PubMed] [Google Scholar]
- 54.Li L, Wang H, Yang ES, Arteaga CL & Xia F (2008) Erlotinib attenuates homologous recombinational repair of chromosomal breaks in human breast cancer cells. Cancer Res 68, 9141–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chinnaiyan P, Huang S, Vallabhaneni G, Armstrong E, Varambally S, Tomlins SA, Chinnaiyan AM & Harari PM (2005) Mechanisms of enhanced radiation response following epidermal growth factor receptor signaling inhibition by erlotinib (Tarceva). Cancer Res 65, 3328–3335. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka T, Munshi A, Brooks C, Liu J, Hobbs ML & Meyn RE (2008) Gefitinib radiosensitizes non-small cell lung cancer cells by suppressing cellular DNA repair capacity. Clin Cancer Res 14, 1266–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD et al. (1999) Induction of GADD45 and JNK / SAPK-dependent apoptosis following inducible expression of BRCA1. Cell 97, 575–586. [DOI] [PubMed] [Google Scholar]
- 58.Thangaraju M, Kaufmann SH & Couch FJ (2000) BRCA1 facilitates stress-induced apoptosis in breast and ovarian cancer cell lines. J Biol Chem 275, 33487–33496. [DOI] [PubMed] [Google Scholar]
- 59.Dizin E, Ray H, Suau F, Voeltzel T & Dalla Venezia N (2008) Caspase-dependent BRCA1 cleavage facilitates chemotherapy-induced apoptosis. Apoptosis 13, 237–246. [DOI] [PubMed] [Google Scholar]
- 60.Zhan Q, Jin S, Ng B, Plisket J, Shangary S, Rathi A, Brown KD & Baskaran R (2002) Caspase-3 mediated cleavage of BRCA1 during UV-induced apoptosis. Oncogene 21, 5335–5345. [DOI] [PubMed] [Google Scholar]
- 61.Martin SA & Ouchi T (2005) BRCA1 phosphorylation regulates caspase-3 activation in UV-induced apoptosis. Cancer Res 65, 10657–10662. [DOI] [PubMed] [Google Scholar]
- 62.Maniccia AW, Lewis C, Begum N, Xu J, Cui J, Chipitsyna G, Aysola K, Reddy V, Bhat G, Fujimura Y et al. (2009) Mitochondrial localization, ELK-1 transcriptional regulation and growth inhibitory functions of BRCA1, BRCA1a, and BRCA1b proteins. J Cell Physiol 219, 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown MA, Nicolai H, Howe K, Katagiri T, Lalani E, Simpson KJ, Manning NW, Deans A, Chen P, Khanna KK et al. (2002) Expression of a truncated Brca1 protein delays lactational mammary development in transgenic mice. Transgenic Res 11, 467–478. [DOI] [PubMed] [Google Scholar]