Abstract
Cold shock proteins are multifunctional RNA/DNA binding proteins, characterized by the presence of one or more cold shock domains. In humans, the best characterized members of this family are denoted Y-box binding proteins, such as Y-box binding protein-1 (YB-1). Biological activities range from the regulation of transcription, splicing and translation, to the orchestration of exosomal RNA content. Indeed, the secretion of YB-1 from cells via exosomes has opened the door to further potent activities. Evidence links a skewed cold shock protein expression pattern with cancer and inflammatory diseases. In this review the evidence for a causative involvement of cold shock proteins in disease development and progression is summarized. Furthermore, the potential application of cold shock proteins for diagnostics and as targets for therapy is elucidated.
Background
Imagine proteins that are conserved in both structure and function, that can be found in almost all organisms from bacteria to humans (except yeast), and have been detected in almost every cellular compartment. Add to this the ability to regulate not only their own expression, but the expression of a number of disease-associated genes, and to orchestrate multiple cellular processes, including proliferation and differentiation. Who are these jack-of-all-trades? Enter our protagonists, the cold shock proteins.
Members of the cold shock protein family
Cold shock proteins are among the most evolutionarily conserved proteins [1–3]. Their distinguishing characteristic is the presence of one or more cold shock domains (CSD), which possess nucleic acid binding properties (see Fig. 1 and Table 1). This endows these proteins with pleiotropic functions, such as the regulation of transcription, translation, and splicing [4, 5].
Fig. 1.
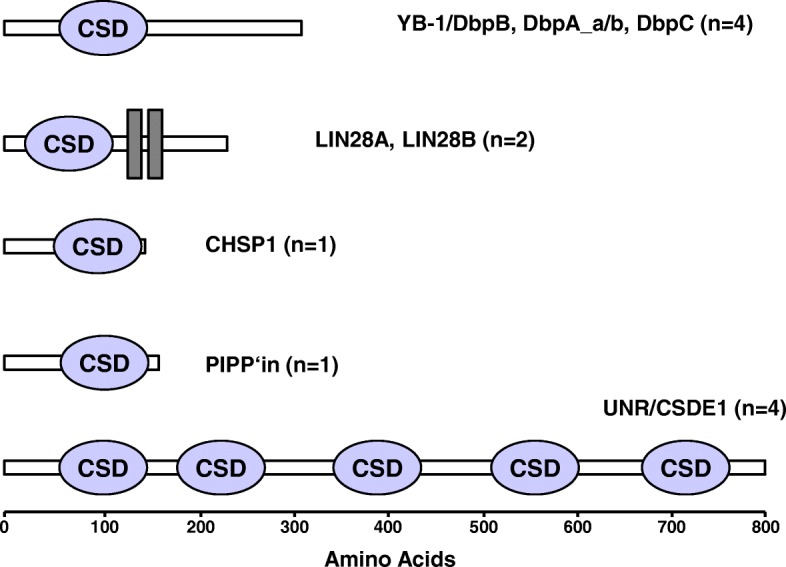
The human cold shock domain proteins. The five groups of human cold shock proteins are presented. The number of proteins in each group is indicated within the brackets. The cold shock domain (CSD) is presented in blue. Lin28 contains two additional zinc finger domains (grey bars). The numbers below indicate the approximate number of amino acids. Structure predictions were performed using the SMART software [215]
Table 1.
Nomenclature of the human cold shock domain proteins.
| Gene | Gene synonym | Protein | Alternative names |
|---|---|---|---|
| YBX1 | MSY1 | YB-1 | CSDB, DbpB, NSEP1, EF1A |
| YBX2 | MSY2 | DbpC | Contrin |
| YBX3 | MSY3/MSY4 | DbpA* | CSDA, ZONAB, oxyR, NF-GMB, YB-2 |
| CARHSP1 | CARHSP1 | CSDC1, CRHSP-24, CHSP1 | |
| CSDC2 | PIPPin | ||
| CSDE1 | UNR* | ||
| LIN28A | LIN28A | CSDD1 | |
| LIN28B | LIN28B | CSDD2 |
The gene names (italics), common names (bold), as well as commonly used alternative names are presented for each protein. Abbreviations are as follows: Y-box binding protein 1, 2, 3 (YBX1, YBX2, YBX3), mouse Y-box protein 1, 2, 3, 4 (MSY1, MSY2, MSY3, MSY4), cold shock domain A, B, C1, C2, D1, D2, E1 (CSDA-CSDE1), calcium-regulated heat stable protein 1 (CARHSP1, CHSP1), calcium regulated heat stable protein 24 kDa (CRHSP-24), abnormal cell lineage protein 28 homolog A, B (LIN28A), DNA binding protein A, B, C (DbpA, DbpB, DbpC), Y-box binding protein 1, 2 (YB-1, YB-2), upstream of N-Ras (UNR), nuclease sensitive element binding protein 1 (NSEP1), enhancer factor I subunit A (EF1A, rat), ZO-1-associated nucleic acid-binding protein (ZONAB), oxidative stress regulatory protein (oxyR), nuclear factor that binds the GM-CSF promoter b (NF-GMB). *Alternatively spliced protein: DbpA has two isoforms, which differ by a single domain of ~ 70 amino acids, whereas the UNR isoforms differ by 31 amino acids
Cold shock proteins were initially identified in bacteria, where a sudden drop in temperature (from 37 °C to 10 °C) induced a 200-fold increase in cold shock protein A (CspA) expression within minutes, which was independent of transcriptional activity [3, 6]. This rapid inducibility is conserved amongst species [7]. A recent study revisited the original observation using genome-wide methods to analyze the global changes occurring in bacteria during the cold shock response [8]. The authors identified RNase R and CspA to be the major players. RNase R appears to be responsible for degrading misfolded RNAs, while CspA melts double-stranded RNAs to enable translation.
In humans, the predominant group of cold shock domain proteins is denoted the Y-box protein family. The prototypic member is Y-box binding protein-1 (YB-1), also known as DNA binding protein B (DbpB), encoded by the gene YBX1. Two additional family members exist, DNA binding protein A (DbpA) and C (DbpC), which are encoded by the genes YBX3 and YBX2, respectively.
Whereas Ybx2 expression is restricted to germ cells [9], Ybx1 and Ybx3 are ubiquitously expressed during development. However, following birth the expression of Ybx3 (DbpA) is down-regulated in most tissues, the exceptions being heart, skeletal muscle, blood vessels, and testis [10, 11]. In humans, two isoforms of DbpA are reported (DbpA_a and DbpA_b), which differ by an alternatively spliced exon that encodes the 69 amino acid long unique domain located adjacent to the CSD [12, 13].
The Ybx1 knockout mouse is embryonic lethal indicating an important role during development [14]. The Ybx3 knockout is viable, however the Ybx1/Ybx3 double knockout shows a more severe developmental phenotype indicating overlapping activities during development [15].
Another developmentally important cold shock protein expressed in humans is Lin28, which was first characterized as a developmental factor in C. elegans [16]. However, it was its potential for cellular reprogramming that brought it into the spotlight, as together with Oct3, Sox2, and Nanog, Lin28 is able to revert differentiated cells into their pluripotent state [17]. In addition to the cold shock domain, Lin28A/B are unique in that they also possess two CCHC type zinc fingers, which form a knuckle domain that also participates in nucleic acid binding [18]. Of particular note is the ability of Lin28 to repress let-7 miRNAs, e.g. thereby regulating glucose metabolism [18, 19]. let-7 also targets Lin28 creating a double-negative feedback loop [20]. In addition to miRNAs, Lin28 also binds to mRNAs, participating in a number of ribonucleoprotein complexes, such as P-bodies and stress granules, to regulate translation [21].
A further member of the human cold shock protein family is the calcium-regulated heat-stable protein 1 (CARHSP1); a 24 kDa protein also known as CRHSP-24. Originally identified as a substrate of the calcium/calmodulin-regulated protein phosphatase calcineurin [22], CARHSP1 is a paralog of the brain-specific cold shock protein PIPPin [23]. CARHSP1 binds to and stabilizes tumor necrosis factor (TNF) mRNA within P-bodies and exosomes [24].
PIPPin expression is restricted to brain, where it binds mRNA to regulate translation [25–29]. PIPPin is found with ribonucleoprotein complexes, where it interacts with other RNA binding proteins, e.g. hnRNP A1, hnRNP K, and YB-1 [30].
The final member of this family is denoted upstream of N-RAS (UNR) [31, 32]. This gene was initially identified as a regulator of N-Ras expression [33–36]. Later it was discovered that UNR encodes a protein possessing 5 cold shock domains, which undergoes alternative splicing (see Fig. 1) [37–39]; the gene was then renamed cold shock domain containing E1 (CSDE1). Like the other cold shock proteins, UNR/CSDE1 binds single-stranded DNA or RNA [37, 40, 41]. UNR works together with the polypyrimidine-tract-binding protein (PTB) to regulate translation and mRNA stability [42, 43]. The generation of Unr knockout mice demonstrated that, like Ybx1, it is essential for mouse development. Further characterization demonstrated that Unr maintains the pluripotent state of embryonic stem cells [44, 45].
As mentioned above, cold shock proteins are components of ribonucleoprotein complexes. Two recent studies using proximity biotinylation to map components of the stress granules identified YB-1, DbpA, CSDE1, and Lin28B [46, 47]. Additionally, CHSP1 (a paralog of PIPPin) was shown to colocalize with G3BP1, an initiator of stress granule formation in human cells [24, 48, 49].
Cold shock proteins: Thinking in regulatory feedforward and feedback loops
Cells undergo stress in many ways, e.g. via interferon release in response to viral infection, the presence of lipopolysaccharide produced by bacteria, or profibrotic factors released by immune cells during inflammation. The binding of these factors to their cell surface receptors activates kinases, which phosphorylate the cold shock proteins; here we use YB-1 as an example (see Fig. 2). Upon activation, these RNA/DNA chaperones release specific mRNA, thereby enabling a rapid translational response and translocate to the nucleus to regulate gene expression. In many ways this is similar to the unfolded protein response (UPR) observed for heat shock proteins [50]. The uptake of YB-1 by cells, which is secreted as an RNA:protein complex [51, 52], uniquely positions this cold shock protein to participate in cellular reprogramming by modulating the expression of numerous target genes. Many of these target genes are themselves known to regulate various aspects of disease both intra- and extracellularly (see Table 2) and can induce cold shock protein expression, e.g. PDGF-B and TGF-β. This is envisioned to result in a feedforward amplification loop that prolongs inflammation, promotes cell proliferation and immune cell infiltration, as well as drives fibrosis, analogous to an avalanche [5, 53]. Indeed this scenario has recently been documented, supporting our goal for targeted intervention. How this circuit is terminated is unclear, however the development of cold shock protein targeting “neutralizing” antibodies presents one possibility [54]. Other potential mechanisms include the inducible proteolytic degradation of YB-1 protein, microRNA-mediated inhibition of YB-1 expression, and the induction of protein tyrosine phosphatase activity to counteract the kinase-mediated phosphorylation/activation that induces nuclear protein translocation [55–58].
Fig. 2.
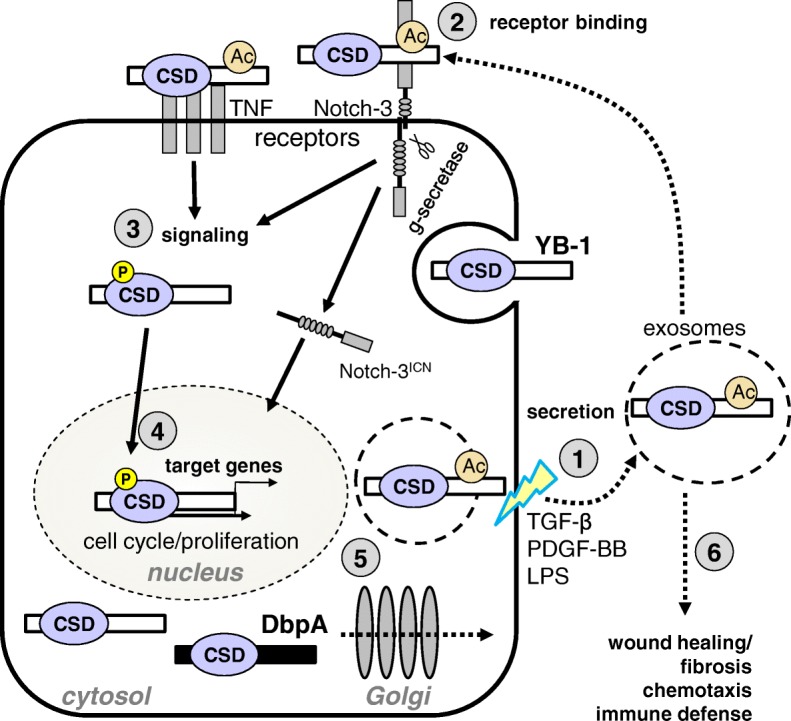
Potential amplification loop for YB-1 in inflammation. (1) Extracellular stimuli (e.g. TGF-β, PDGF-B, LPS) activate cells and induce YB-1 secretion. (2) YB-1 binds to specific membrane associated receptors on the cell surface inducing intracellular signaling cascades that result in kinase activation. YB-1 can also be endocytosed. (3) Activated kinases (e.g. Akt/PKB, ERK, JAK2, RSK) phosphorylate cytoplasmic YB-1 (indicated by the yellow circle), inducing its nuclear translocation. (4) In the nucleus, YB-1 activates the transcription of target genes, as well as induces its own expression and that of DbpA. Cold shock proteins are rapidly induced in response to cell stress, due in part to the existence of preformed complexes of cold shock proteins with their cognate mRNA. (5) Activated cells may also secrete YB-1, which may then act in either an autocrine or paracrine manner. Activated cells may also secrete DbpA via the Golgi. (6) Extracellular YB-1 has mitogenic activity that promotes wound healing/fibrosis. YB-1 also contributes to the recruitment of immune cells to the site of inflammation; directly via its chemoattractant activity or indirectly via the products of its target genes, e.g. CCL5/RANTES. Extracellular activities for DbpA await elucidation. Abbreviations: acetylation (Ac); cold shock domain (CSD); DNA binding protein A (DbpA); lipopolysaccharide (LPS); phosphorylation (P); platelet-derived growth factor B homodimer (PDGF-BB); transforming growth factor beta (TGF-β); tumor necrosis factor (TNF); Y-box binding protein 1 (YB-1)
Table 2.
Genes regulated by cold shock proteins in disease
| Protein | Disease | Target Cell | Mode of Action | Target Gene | Ref. |
|---|---|---|---|---|---|
| YB-1 | Sepsis | neutrophils, macrophages | N.D. | Toll-like receptor 4 (TLR4) CXCL-1 | [123] |
| T-cell activation Autoimmunity Inflammation |
T-helper cells | binding and stabilization of mRNA | Interleukin 2 (IL-2) | [129, 204] | |
| Allergic asthma | activated eosinophils | stabilization and up-regulation of mRNA transcripts | GM-CSF | [140] | |
| embryonic lung fibroblasts | suppression of gene transcription | GM-CSF | [205] | ||
| Chronic liver disease | activated hepatic stellate cells | induction of expression; antagonizes TGFβ signaling | Smad7 | [153] | |
| Chronic liver disease | rat hepatoma cells (FAO) | suppression of gene transcription | Mrp2 | [206] | |
| Kidney transplant rejection | primary monocytes | activation of gene transcription | RANTES/CCL5 | [126] | |
| Kidney transplant rejection | differentiated macrophages | suppression of gene transcription | RANTES/CCL5 | [126] | |
| Neointimal hyperplasia Atherosclerosis | vascular smooth muscle cells | activation of gene transcription | RANTES/CCL5 | [127] | |
| Endometriosis | peritoneal macrophages | activation of gene transcription and recruitment of inflammatory cells | RANTES/CCL5* | [207, 208] | |
| Chronic kidney disease Interstial kidney disease | proximal tubular cells | control of translation | TGFβ | [132, 209] | |
| Mesangioproliferative glomerulonephritis | endothelial cells | gene transcription | PDGF-B | [111] | |
| Mesangioproliferative glomerulonephritis | renal cells | gene transcription, secretion | PDGF-B | [138] | |
| Tubulointerstial nephritis | renal cells, macrophages | gene transcription, secretion, differentiation, phagocytosis | RANTES/CCL5 MCP-1/CCL2 IL-10 | [124, 203] | |
| Dysregulated angiogenesis | repression of VEGF promotor | VEGF | [210] | ||
| Calcineurin inhibitor mediated kidney fibrosis | mesangial cells | binding and stabilization of mRNA | Collagen | [136] | |
| Anti-Thy1.1 nephritis | mesangial cells | gene transcription, secretion | Notch-3 | [54] | |
| Type II diabetes | skeletal muscle | gene transcription, signal pathways | PTP1B | [55] | |
| T-ALL | T cell | Cell cycle | Cdk6 | [181] | |
| CHSP1 | Inflammation Sepsis | macrophages | enhancement of mRNA stability | TNF | [24] |
| DbpA | Dysregulated angiogenesis | fibroblasts | repression of VEGF promoter | VEGF | [130, 210] |
| Hepatocellular carcinoma | hepatocytes | [211–214] | |||
| Mesangioproliferative glomerulonephritis | renal cells | gene transcription, secretion | DbpA | [13] |
For the studied cold shock domain proteins, the disease, target cell, mode of action, and target genes are listed, together with the relevant citation. In sepsis, the mode of action has not been determined (N.D.). Modified from Lindquist et al. [4].
Cold shock proteins function in the cellular response to stress
Components of stress granules and P-bodies have been implicated in the cellular stress response [59, 60]. Under ‘normal’ conditions, stress granules form when translation initiation is stalled. The RNA binding proteins G3BP1 or TIA-1 are key components of stress granule formation, as they possess the ability for self-association. Over-expression of either protein has been shown to induce stress granule formation even in the absence of stress [49, 61, 62]. Using mRNA as a scaffold, these proteins form homo- or hetero-oligomeric ribonucleoprotein complexes; self-assembly is mediated by intrinsically disordered regions (IDRs) within the RNA binding protein(s); also referred to as low complexity regions [63–68]. Several genetic mutations associated with neurodegenerative diseases have been identified that influence the self-assembly of RNA binding proteins (e.g. transactive response DNA-binding protein (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS)). Both are known to form prion-like protein aggregates; an activity attributed to their low complexity regions [67, 68]. The more we learn about the molecular mechanisms underlying protein aggragation diseases, the greater the number of RNA binding proteins identified [69–71]. The mutations identified within these diseaseassociated proteins typically favor cytoplasmic localization, facilitate protein aggregation, or prevent granulophagy; the clearance of stress granules by autophagosomes [49, 66, 70, 72]. Recently, the expansion of intronic GGGGCC repeats within C9ORF72 was identified as a common cause of ALS/FTD [73]. C9ORF72 interacts with endosomes and is required for normal vesicle trafficking, therefore the loss of C9ORF72 observed with G4-repeat expansion may affect granulophagy. Alternatively, the G4-repeats of C9ORF72 have been proposed to inhibit the neuroprotective effects mediated by tiRNAs binding to the cold shock domain of YB-1 [74].
As a known component of stress granules, YB-1 also possesses the ability for self-assembly [75]. YB-1 has been shown to form amyloid-like fibrils, an activity attributed to its C-terminal domain, which is composed of alternating regions of positive or negatively charged amino acids that form a zipper-like structure as well as contributes to its RNA binding activity [76–81]. Interestingly, the oligomerization of YB-1 is induced by a select set of RNAs [79]. In the context of neurodegeneration, YB-1 and G3BP1 have been shown to compete with TDP-43 and FUS for mRNA binding and thereby induce the release of prion-like protein aggregates that have formed [82]. To complicate matters further, in human sarcoma YB-1 activates G3BP1 mRNA thereby controlling both the expression levels of G3BP1 and the subsequent nucleation of stress granule formation [83]. Indeed cold shock is one trigger of stress granule assembly in mammals [84]. Stress granules have been implicated in the pathophysiology for a number of neurodegenerative diseases, including Alzheimer’s, amyotrophic lateral sclerosis (ALS), fronto-temporal dementia (FTD), spinocerebellar ataxia (SCA), and Huntington’s disease [49, 71, 85]. Here we propose possible mechanisms where cold shock proteins may play a critical role in the pathophysiology of these diseases. When granulophagy is defective either due to an inability to degrade protein aggregates or to system overload, i.e. when the rate of production exceeds degradation, stress granules that would normally undergo autophagy become lysosomes [64]. The autophagic pathway intersects with both the classical and the unconventional pathways of protein secretion [86, 87]. YB-1 is secreted via a non-classical pathway involving ATP-binding cassette transporters and microvesicles, as well as post-translational modification of two C-terminal lysine residues (K301/K304) [88, 89]. Non-canonical K27-linked ubiquitination of YB-1 was shown to be required for its interaction with tumor susceptibility gene 101 (TSG101), a component of multivesicular bodies (MVBs) [90]. Fusion of MVB with the plasma membrane is required for the release of exosomes [91]. Since YB-1 is a component of exosomes, required for the sorting of mRNAs [51, 52, 92–94], it remains to be determined whether stress granule clearance coincides with the pathway of exosome formation and YB-1 secretion. If these pathways are indeed one in the same, does this apply to both cytoplasmic and nuclear stress granules? Should this hypothesis hold true for YB-1, it will be of interest to see whether it also applies to other cold shock proteins, such as DbpA or CSDE1, that have already been identified as components of both stress granules and exosomes [95, 96].
Cold shock proteins in disease
The decisive data for a causal relationship between cold shock proteins and disease comes from cancer. The role of YB-1 as an oncoprotein was secured when it was demonstrated that 100% of YB-1 transgenic mice over-expressing the protein developed invasive tumors [97]. YB-1 and DbpA expression is upregulated in cancer and nuclear localization indicates a poor prognosis [57, 98]. In the nucleus, cold shock proteins bind to single- and double-stranded DNA and serve as transcriptional regulators. In tumors, nuclear YB-1 correlates with enhanced expression of the multidrug resistance protein 1 (MDR1) [99–105]. Cells in which YB-1 expression has been ablated using small inhibitory RNA fail to proliferate and were recently shown to prevent tumor growth by disrupting angiogenesis [106].
YB-1 can also be secreted [88]. Acetylation and ubiquitination of YB-1 have both been shown to play roles in regulating secretion as well as intracellular stability [58, 90, 107, 108]. YB-1 can be proteolytically cleaved and extracellular YB-1, and/or fragments thereof, is found in the serum of patients, binds to cell surface receptors, and exerts extracellular activities, e.g. enhancing proliferation and induces migration of immune cells [56, 89, 109–114].
Serum YB-1 levels are increased in cancer patients and the occurrence of extracellular YB-1 or its fragments may serve as a useful marker for cancer, as ~ 80% of patients tested positive for the YB-1/p18 fragment, whereas inflammatory diseases did not correlate with positive results [98, 112–115].
Lin28 reactivation is also found in a number of cancers, where Lin28 appears to contribute to the formation of cancer stem cells [18]. The role of Lin28 in cancer has been extensively reviewed elsewhere [116]. Similar to Lin28, Unr also regulates the differentiation state of cells [44]. Due to its ability to regulate the expression of several proto-oncogenes, UNR has also been investigated in cancer [117–119]. In prostate cancer, a novel regulatory activity of HEPSIN on UNR was identified [120, 121]. UNR expression levels have also been demonstrated as a prognostic biomarker for survival in pancreatic ductal adenocarcinoma [122].
For inflammatory and fibrotic diseases, the data for the role of cold shock proteins appears more associative. The initial data came from animal studies on Ybx1 heterozygous mice, which express only half the amount of YB-1 compared to wild type. The induction of disease in experimental models such as sterile sepsis or unilateral ureter obstruction identified non-redundant roles for YB-1 in the development of inflammation and fibrosis [123, 124]. These activities are mediated in part by YB-1-dependent gene regulation of pro-inflammatory factors (PDGF-B, VEGF, IL-2, GM-CSF, EGF, TGF-β, CCL2, CCL5, and CXCR4) [111, 125–134] as well as fibrosis-related genes (MMP2, Col1a1, and Col2a1) (see Table 2) [135–137]. In mesangioproliferative glomerulonephritis, cold shock protein expression is clearly induced; an effect mediated by PDGF-B, and regulates mesangial cell proliferation [13, 138]. In atherosclerosis, YB-1 contributes to neointima formation by modulating CCL5 expression [126, 127, 139]. In asthma, YB-1 promotes eosinophil survival by stabilizing granulocyte macrophage-colony-stimulating factor mRNA [140, 141]. Successful approaches to ameliorate diseases by targeting YB-1 activities have been demonstrated [124, 142–146].
From molecules to intervention strategies: Rationale for cold shock protein targeting
We propose that cold shock proteins represent verifiable targets for therapeutic intervention and envision strategies aimed at targeting cold shock proteins directly or targeting cold shock protein-dependent mechanisms. This goal is supported by the following observations that link the prototypic cold shock protein YB-1 with other key molecule activities. For the latter, intervention strategies have already proven to be successful.
YB-1 regulates NF-κB activation. In the absence of YB-1, NF-κB activation is defective [147, 148].
YB-1 regulates IL-2 production. CD28 co-stimulation is required for T cell activation and the induction of autocrine IL-2 production. CD28 signals stabilize IL-2 mRNA. YB-1 is one of the essential RNA binding proteins that mediate this activity [129].
YB-1 interacts with p53. Nuclear YB-1 regulates p53 function by inhibiting its ability to induce apoptosis, however it does not influence p53’s regulation of cell cycle [149–151].
YB-1 and TGF-β counter-regulate one another. It was recently demonstrated that TGF-β induces miR-216a, which suppresses YB-1 expression. YB-1 suppresses Tsc22, which serves as an enhancer for Col1a2 expression [152]. Additionally, we have shown that YB-1 mediates the anti-fibrotic effect of interferon-gamma, directly competes for Smad3 binding to p300/CBP [153].
Molecular pathways are not per se pathological, but rather part of regulatory networks. A prolonged or permanent dysregulation results in diseases, especially those of an inflammatory or malignant nature. Developing targeted therapies requires insight into the molecular pathways of underlying diseases, as pivotal cell decisions are dependent on the “activation” of key molecules. Examples of such molecules are provided in the following.
NF-κB; diseases: Cancer, inflammatory, and autoimmune
Nuclear factor binding near the kappa-light-chain gene in B cells (NF-κB) are a family of inducible transcription factors that control inflammatory gene expression [154–157]. In many cancers, NF-κB is constitutively active and localized to the nucleus. Therefore many anti-tumor therapies seek to block NF-κB activity as a means to inhibit tumor growth or to sensitize tumor cells to conventional therapies, such as chemotherapy. The extensive involvement of NF-κB in inflammation and disease has also established it as a therapeutic target. Indeed, many common synthetic (e.g., aspirin, ibuprofen, glucocorticoids) and traditional medicines (e.g., green tea, curcumin) target the NF-κB pathway. To date, over 800 compounds have been shown to inhibit NF-κB signaling (such as anatabine, disulfiram, dithiocarbamates, olmesartan). Many natural products (including anti-oxidants) that have been promoted to have anti-cancer and anti-inflammatory activity have also been shown to inhibit NF-κB.
IL-2; diseases: Autoimmune and organ transplantation; cancer, viral infection, and vaccination
Interleukin-2 (IL-2) is essential for lymphocyte survival, differentiation, and proliferation [158–161]. Therefore, many immunosuppressive drugs (such as corticosteroids, cyclosporine A, and tacrolimus) used to treat autoimmune diseases or suppress graft rejection work by inhibiting the production of IL-2 by antigen-activated T cells. Sirolimus blocks intracellular IL-2R signaling, thereby preventing the clonal expansion of activated T cells. The extracellular effects of IL-2 are abrogated by monoclonal antibody application. The use of antibody induction after kidney transplantation has increased to 60% in the past decade and roughly one half of the induction agent used is anti-interleukin-2 receptor alpha antibody (IL-2RA, i.e. basiliximab or daclizumab). In combination with calcineurin inhibitors, IL-2RAs have been shown to reduce the incidence of acute rejection without increasing risks of infections or malignancies in kidney transplantation.
Recombinant IL-2 has been approved for the treatment of cancers (malignant melanoma, renal cell cancer) and has been tested in clinical trials for the treatment of chronic viral infections, and as an adjuvant for vaccines.
p53; disease: Cancer
Tumor protein p53 (p53) is a tumor suppressor and the most frequently mutated gene in human cancers [162–165]. People who possess only one functional copy of the p53 gene have a higher incidence of tumor development. The p53 gene can also be damaged by chemical mutagenesis or radiation, as well as p53 protein inactivated by viruses (e.g. human papillomavirus). p53 itself does not bind to DNA, but rather exerts its influence via its complex interactions with transcription factors and regulators. p53 mutants are associated with changes in chromatin structure, leading to genetic instability and alterations in cell cycle regulation as well as cellular metabolism. Mutant p53 has been shown to act downstream of the TNF receptor to prolong and enhance NF-κB activation thereby driving tumor-promoting inflammation and enhancing chemokine secretion. The p53 pathway inhibitors nutlin and PRIMA-1 reactivate p53 function, enhancing its anti-proliferative activity and thereby sensitizing cancer cells to apoptosis [166].
TGF-β; diseases: Organ fibrosis, cancer, immune suppression
Transforming growth factor-β (TGF-β) promotes fibroblast proliferation, differentiation, and survival. In addition to inducing cytokine secretion, TGF-β upregulates the synthesis of collagens and extracellular matrix, making it a therapeutic target in fibrotic diseases [167]. TGF-β also induces the epithelial-mesenchymal xtransition (EMT); an important step in tumor progression, thus making it a target for anti-cancer therapy [168]. Strategies to target TGF-β include neutralizing monoclonal antibodies targeting TGF-β, monoclonal antibodies targeting the integrin αvβ6 are aimed at preventing the activation of latent TGF-β, and small molecules targeting TGF-β receptor activity. Additionally, some commonly used drugs, e.g. the kinase inhibitor imatinib mesylate, appear to also block TGF-β activities and abrogate fibrotic responses [169, 170]. However, inhibiting TGF-β can also have unwanted effects, such as enhanced immune cell activation (due to the loss of TGF-β-mediated inhibition), hindering implantation during pregnancy, and impaired wound healing (within a normal response to injury).
Outlook
Diagnostics and therapy with interventions targeting cold shock proteins
Cold shock protein expression is a suitable biomarker for diverse disease activities [112–114]. The presence of extracellular cold shock proteins, and/or fragments thereof, may serve diagnostic purposes. Beyond their diagnostic potential, we envision that therapeutic interventions targeting cold shock proteins may reduce disease burden, as YB-1 is expected to target pathways distinct from those targeted by current therapies. Therefore, we anticipate at least in some cases synergistic activity with existing therapies.
At present cold shock protein research is on the verge of entering clinical trials in different fields, especially for advanced cancer disease (ongoing trials adopt a vaccination strategy against YB-1 epitope in HER2-negative stage III-IV breast cancer or an oncolytic virotherapy in bladder cancer). In experimental disease models intervention strategies targeting YB-1 reduced inflammation and organ fibrosis [124, 142, 143, 171]. HSc025 was identified in a natural products screen for compounds that suppressed collagen gene expression, i.e. fibrosis [172]. HSc025 promotes nuclear translocation of YB-1, which acts as a suppressor of the gene COL1A2 (collagen type I alpha 2) thereby reducing fibrosis [137, 142, 153, 171, 173–175].
Another compound is the natural product fisetin (3,7,3′,4′-tetrahydroxyflavone); a polyphenolic compound found in plants, also called a flavonoid, that demonstrated anti-cancer as well as anti-inflamatory activity [176, 177]. Fisetin blocks the Akt-mediated phosphorylation of Ser102 within the CSD [144, 178]. However, an inhibition of p70S6K, a member of the ribosomal S6 kinase (RSK) family, has also been reported [179]. Molecular modeling proposed that fisetin binds to the CSD of YB-1; if such binding prevents YB-1 from being phosphorylated then this proposal would unify these reports, as both kinases phosphorylate Ser102 [144, 180, 181]. Regardless of the mechanism of action, fisetin prevents the nuclear translocation of YB-1 by preventing phosphorylation of the CSD.
Developing topics in the cold shock protein field
Pro-inflammatory factors, like TNF, activate NF-κB, which induces miR-155 expression. Increased miR-155 suppresses CARHSP1, which stabilizes TNF mRNA; thus, this negative feedback loop relieves chronic inflammation and was shown to play a protective role during atherosclerosis [182].
The modulation of tumor necrosis factor receptor signaling by extracellular cold shock proteins is relevant to a number of diseases, including preeclampsia, diabetic nephropathy, systemic lupus erythematosus, liver fibrosis, and infectious diseases where TNF plays a central role in disease pathology [183]. Additionally, TNF promotes expansion of JAK2V617F positive cells in myeloproliferative neoplasms [184]. Extracellular cold shock proteins are also topics of interest, as is their potential role in fetal-maternal communication during implantation.
Receptor Notch-3 is a developmental receptor that plays an important role in stem cell maintenance as well as in cell differentiation. Known roles include the development thymocytes as well as hepatocellular carcinoma. Strong expression is also found in the placenta and uterus suggesting an important role in pregnancy. Extracellular YB-1 serves as a noncanonical ligand for receptor Notch-3 and therefore its ability to modulate receptor Notch-3 signaling is of relevance [88, 89]. Progranulin has recently been demonstrated as a Notch ligand [185] and therefore YB-1/progranulin may also modulate Notch signaling, which may be of relevance in a number of diseases, e.g. diabetic nephropathy, systemic lupus erythematosus, liver fibrosis, and infectious disease.
The participation of extracellular cold shock proteins in inter-organ communication is another important emerging idea (i.e. endocrine activity). Liver-kidney interactions have recently been described for nonalcoholic fatty liver disease (NAFLD) [186]. Here, the liver is an important source of pro-inflammatory cytokines, which modulate inflammation and renal injury [187, 188]. Chronic kidney disease induces intestinal dysbiosis, which contributes to systemic inflammation (via the production of uremic toxins) thereby promoting NAFLD. Inflammation also drives renal fibrosis, which further reduces kidney function, in so doing enhances the levels of uremic toxins within the blood, creating a self-perpetuating multiorgan disease [189]. Several pro-inflammatory cytokines as well as bacterial toxins, e.g. lipopolysaccharide, induce cold shock protein secretion, which binds to TNF receptors and receptor Notch-3 [89]. Therefore we believe that extracellular cold shock proteins are intimately involved in this cycle.
Finally, evidence is emerging that cold shock proteins may regulate the formation of protein aggregates in neurodegenerative diseases [82]. The role of exosomes in the spreading of neurodegenerative and prion diseases is well documented [190–192]. However, it remains to be determined whether stress granules do indeed serve as precursors for exosomes and if so, to what extent they contribute to the spread of neurodegenerative diseases versus the detoxification of cells by removing protein aggregates or perhaps both. Additionally, it remains to be shown whether the targeting of cold shock proteins in this context might be of therapeutic benefit.
Since many components of stress granules and P-bodies are also targets of autoantibodies, the question remains as to whether this pathway contributes to the generation of autoantibodies against YB-1 [193–196]. Certainly the RNA:protein complexes described as “beads on a string” possess the essential elements (i.e. multiple repeating epitopes) required for the successful activation of B-cells [80, 197].
Post-translational modifications of cold shock proteins
The number of post-translational modifications identified within cold shock proteins is continually growing [198]. A recent paper described O-GlcNAcylation of YB-1; a post-translational modification linking nutrient and stress sensing to transcriptional and translational regulation [199, 200]. This novel modification was shown to contribute to the oncogenic potential of YB-1 in hepatocellular carcinoma (HCC) and appears to exert its activity within the nucleus, since it also requires phosphorylation of Ser102 within the CSD. O-GlcNAcylation is mediated by the enzyme O-GlcNAc transferase (OGT), which is known to promote liver cancer as well as a number of diseases, such as diabetes and neurodegeneration [200, 201]. Since O-GlcNAcylation of NF-κB potentiates its acetylation [202], it will be interesting to see whether a similar effect is also found for the acetylation of YB-1. As you see from this example, there is still much work to be done in linking a particular post-translational modification to specific protein activities. To extrapolate this idea further, it remains to be seen whether there are cell-specific modifications or activities of the cold shock proteins and whether these apply to particular compartments within the cell (e.g. nucleus, mitochondria, exosomes, etc.). Here, it is anticipated that CRISPR/Cas technology will help in creating and characterizing cell lines with specific point mutations targeting a particular modified amino acid. However, there is still much work to be done in identifying and characterizing cell-specific activities of the cold shock proteins. Our recent study demonstrating cell-specific activities of YB-1 in monocytes and macrophages is likely merely the tip of the iceberg [203]. There are still numerous organs, cell types, and cell subsets (e.g. Th1 versus Th2 cells) awaiting characterization. Therefore strategies aimed at deleting Ybx1 in specific tissues and/or cell types must consider possible developmental effects when characterizing the phenotypes of such cells. Add to this the presence of cold shock proteins within exosomes and thus their extracellular activities and we have a long road ahead to fully understand the complex behavior and activities of these fascinating proteins in both health and disease. Here, the application of high-throughput omics technologies will be essential to keep track of the changes going on within such cells on both the transcriptional as well as translational levels.
Acknowledgements
The authors would like to thank Dr. Sabine Brandt for helpful discussion.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG): SFB 854, project A01, grants ME-1365/7–2 and ME-1365/9–1 to PRM, and LI-1031/4–1 to JAL.
Abbreviations
- Ac
Acetylation
- CARHSP1
Calcium-regulated heat-stable protein 1
- CBP
CREB-binding protein
- CCL2
Chemokine (C-C motif) ligand 2
- CCL5
Chemokine (C-C motif) ligand 5
- CRHSP-24
Calcium-regulated heat-stable protein of 24 kDa
- CSD
Cold shock domain
- CspA
Cold shock protein A
- CXCR4
C-X-C motif chemokine receptor 4
- DbpA
DNA binding protein A
- DbpB
DNA binding protein B
- DbpC
DNA binding protein C
- EGF
Epidermal growth factor
- EMT
Epithelial-mesenchymal transition
- FUS/TLS
Fused in sarcoma/translocated in liposarcoma
- GM-CSF
Granulocyte-macrophage colony-stimulating factor
- HER2
Human epidermal growth factor receptor 2
- hnRNP
Heterogeneous nuclear ribonucleoprotein
- IL-2
Interleukin-2
- IL-2RA
Interleukin-2 receptor alpha
- JAK
Janus kinase
- LPS
Lipopolysaccharide
- MDR1
Multidrug resistance protein 1
- mRNA
Messenger RNA
- NAFLD
Nonalcoholic fatty liver disease
- NF-κB
Nuclear factor binding near the kappa-light-chain gene in B cells
- PDGF-B
Platelet-derived growth factor subunit B
- TDP-43
transactive response DNA-binding protein
- TGF-β
Transforming growth factor beta
- TNF
Tumor necrosis factor
- UNR
Upstream of N-Ras
- UPR
Unfolded protein response
- VEGF
Vascular endothelial growth factor
- YB-1
Y-box binding protein-1
Authors’ contributions
JAL and PRM wrote and edited the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wolffe AP, Tafuri S, Ranjan M, Familari M. The Y-box factors: a family of nucleic acid binding proteins conserved from Escherichia coli to man. New Biol. 1992;4(4):290–298. [PubMed] [Google Scholar]
- 2.Wolffe AP. Structural and functional properties of the evolutionarily ancient Y-box family of nucleic acid binding proteins. BioEssays. 1994;16(4):245–251. doi: 10.1002/bies.950160407. [DOI] [PubMed] [Google Scholar]
- 3.Jones PG, Inouye M. The cold-shock response--a hot topic. Mol Microbiol. 1994;11(5):811–818. doi: 10.1111/j.1365-2958.1994.tb00359.x. [DOI] [PubMed] [Google Scholar]
- 4.Lindquist JA, Brandt S, Bernhardt A, Zhu C, Mertens PR. The role of cold shock domain proteins in inflammatory diseases. J Mol Med (Berl). 2014;92(3):207–216. doi: 10.1007/s00109-014-1136-3. [DOI] [PubMed] [Google Scholar]
- 5.Brandt S, Raffetseder U, Djudjaj S, Schreiter A, Kadereit B, Michele M, Pabst M, Zhu C, Mertens PR. Cold shock Y-box protein-1 participates in signaling circuits with auto-regulatory activities. Eur J Cell Biol. 2012;91(6–7):464–471. doi: 10.1016/j.ejcb.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Gottesman S. Chilled in translation: adapting to bacterial climate change. Mol Cell. 2018;70(2):193–194. doi: 10.1016/j.molcel.2018.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Graumann PL, Marahiel MA. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23(8):286–290. doi: 10.1016/S0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, Burkhardt DH, Rouskin S, Li GW, Weissman JS, Gross CA. A stress response that monitors and regulates mRNA structure is central to cold shock adaptation. Mol Cell. 2018;70(2):274–286. doi: 10.1016/j.molcel.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder E, Soundararajan R, Sharma M, Dearth A, Smith B, Braun RE. Compound heterozygosity for Y box proteins causes sterility due to loss of translational repression. PLoS Genet. 2015;11(12):e1005690. doi: 10.1371/journal.pgen.1005690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berghella L, De Angelis L, De Buysscher T, Mortazavi A, Biressi S, Forcales SV, Sirabella D, Cossu G, Wold BJ. A highly conserved molecular switch binds MSY-3 to regulate myogenin repression in postnatal muscle. Genes Dev. 2008;22(15):2125–2138. doi: 10.1101/gad.468508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima WR, Parreira KS, Devuyst O, Caplanusi A, N'Kuli F, Marien B, Van Der Smissen P, Alves PM, Verroust P, Christensen EI, Terzi F, Matter K, Balda MS, Pierreux CE, Courtoy PJ. ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells. J Am Soc Nephrol. 2010;21(3):478–488. doi: 10.1681/ASN.2009070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasegawa SL, Doetsch PW, Hamilton KK, Martin AM, Okenquist SA, Lenz J, Boss JM. DNA binding properties of YB-1 and dbpA: binding to double-stranded, single-stranded, and abasic site containing DNAs. Nucleic Acids Res. 1991;19(18):4915–4920. doi: 10.1093/nar/19.18.4915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu C, Sauter E, Schreiter A, van Roeyen CR, Ostendorf T, Floege J, Gembardt F, Hugo CP, Isermann B, Lindquist JA, Mertens PR. Cold shock proteins mediate GN with Mesangioproliferation. J Am Soc Nephrol. 2016;27(12):3678–3689. doi: 10.1681/ASN.2015121367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fan L, Jones SN, Padden C, Shen Q, Newburger PE. Nuclease sensitive element binding protein 1 gene disruption results in early embryonic lethality. J Cell Biochem. 2006;99(1):140–145. doi: 10.1002/jcb.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu ZH, Books JT, Ley TJ. Cold shock domain family members YB-1 and MSY4 share essential functions during murine embryogenesis. Mol Cell Biol. 2006;26(22):8410–8417. doi: 10.1128/MCB.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57(1):49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- 17.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 18.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22(9):474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, Shyh-Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI, Consortium D, Investigators M, et al. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 21.Mayr F, Heinemann U. Mechanisms of Lin28-mediated miRNA and mRNA regulation--a structural and functional perspective. Int J Mol Sci. 2013;14(8):16532–16553. doi: 10.3390/ijms140816532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groblewski GE, Yoshida M, Bragado MJ, Ernst SA, Leykam J, Williams JA. Purification and characterization of a novel physiological substrate for calcineurin in mammalian cells. J Biol Chem. 1998;273(35):22738–22744. doi: 10.1074/jbc.273.35.22738. [DOI] [PubMed] [Google Scholar]
- 23.Schafer C, Steffen H, Krzykowski KJ, Goke B, Groblewski GE. CRHSP-24 phosphorylation is regulated by multiple signaling pathways in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2003;285(4):G726–G734. doi: 10.1152/ajpgi.00111.2003. [DOI] [PubMed] [Google Scholar]
- 24.Pfeiffer JR, McAvoy BL, Fecteau RE, Deleault KM, Brooks SA. CARHSP1 is required for effective tumor necrosis factor alpha mRNA stabilization and localizes to processing bodies and exosomes. Mol Cell Biol. 2011;31(2):277–286. doi: 10.1128/MCB.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castiglia D, Scaturro M, Nastasi T, Cestelli A, Di Liegro I. PIPPin, a putative RNA-binding protein specifically expressed in the rat brain. Biochem Biophys Res Commun. 1996;218(1):390–394. doi: 10.1006/bbrc.1996.0068. [DOI] [PubMed] [Google Scholar]
- 26.Nastasi T, Scaturro M, Bellafiore M, Raimondi L, Beccari S, Cestelli A, di Liegro I. PIPPin is a brain-specific protein that contains a cold-shock domain and binds specifically to H1 degrees and H3.3 mRNAs. J Biol Chem. 1999;274(34):24087–24093. doi: 10.1074/jbc.274.34.24087. [DOI] [PubMed] [Google Scholar]
- 27.Nastasi T, Muzi P, Beccari S, Bellafiore M, Dolo V, Bologna M, Cestelli A, Di Liegro I. Specific neurons of brain cortex and cerebellum are PIPPin positive. Neuroreport. 2000;11(10):2233–2236. doi: 10.1097/00001756-200007140-00034. [DOI] [PubMed] [Google Scholar]
- 28.Raimondi L, D'Asaro M, Proia P, Nastasi T, Di Liegro I. RNA-binding ability of PIPPin requires the entire protein. J Cell Mol Med. 2003;7(1):35–42. doi: 10.1111/j.1582-4934.2003.tb00200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bono E, Compagno V, Proia P, Raimondi L, Schiera G, Favaloro V, Campo V, Donatelli M, Di Liegro I. Thyroid hormones induce sumoylation of the cold shock domain-containing protein PIPPin in developing rat brain and in cultured neurons. Endocrinology. 2007;148(1):252–257. doi: 10.1210/en.2006-0660. [DOI] [PubMed] [Google Scholar]
- 30.Di Liegro CM, Schiera G, Proia P, Saladino P, Di Liegro I. Identification in the rat brain of a set of nuclear proteins interacting with H1 degrees mRNA. Neuroscience. 2013;229:71–76. doi: 10.1016/j.neuroscience.2012.10.072. [DOI] [PubMed] [Google Scholar]
- 31.Anderson EC, Catnaigh PO. Regulation of the expression and activity of Unr in mammalian cells. Biochem Soc Trans. 2015;43(6):1241–1246. doi: 10.1042/BST20150165. [DOI] [PubMed] [Google Scholar]
- 32.Ray S, Catnaigh PO, Anderson EC. Post-transcriptional regulation of gene expression by Unr. Biochem Soc Trans. 2015;43(3):323–327. doi: 10.1042/BST20140271. [DOI] [PubMed] [Google Scholar]
- 33.Jeffers M, Paciucci R, Pellicer A. Characterization of unr; a gene closely linked to N-ras. Nucleic Acids Res. 1990;18(16):4891–4899. [PMC free article] [PubMed] [Google Scholar]
- 34.Jeffers M, Pellicer A. Multiple intragenic elements regulate the expression of the murine N-ras gene. Oncogene. 1992;7(11):2115–2123. [PubMed] [Google Scholar]
- 35.Jacquemin-Sablon H, Dautry F. Organization of the unr/N-ras locus: characterization of the promoter region of the human unr gene. Nucleic Acids Res. 1992;20(23):6355–6361. doi: 10.1093/nar/20.23.6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boussadia O, Amiot F, Cases S, Triqueneaux G, Jacquemin-Sablon H, Dautry F. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 1997;420(1):20–24. doi: 10.1016/S0014-5793(97)01479-8. [DOI] [PubMed] [Google Scholar]
- 37.Jacquemin-Sablon H, Triqueneaux G, Deschamps S, le Maire M, Doniger J, Dautry F. Nucleic acid binding and intracellular localization of unr, a protein with five cold shock domains. Nucleic Acids Res. 1994;22(13):2643–2650. doi: 10.1093/nar/22.13.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doniger J, Landsman D, Gonda MA, Wistow G. The product of unr, the highly conserved gene upstream of N-ras, contains multiple repeats similar to the cold-shock domain (CSD), a putative DNA-binding motif. New Biol. 1992;4(4):389–395. [PubMed] [Google Scholar]
- 39.Boussadia O, Jacquemin-Sablon H, Dautry F. Exon skipping in the expression of the gene immediately upstream of N-ras (unr/NRU) Biochim Biophys Acta. 1993;1172(1–2):64–72. doi: 10.1016/0167-4781(93)90270-N. [DOI] [PubMed] [Google Scholar]
- 40.Ferrer N, Garcia-Espana A, Jeffers M, Pellicer A. The unr gene: evolutionary considerations and nucleic acid-binding properties of its long isoform product. DNA Cell Biol. 1999;18(3):209–218. doi: 10.1089/104454999315420. [DOI] [PubMed] [Google Scholar]
- 41.Triqueneaux G, Velten M, Franzon P, Dautry F, Jacquemin-Sablon H. RNA binding specificity of Unr, a protein with five cold shock domains. Nucleic Acids Res. 1999;27(8):1926–1934. doi: 10.1093/nar/27.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mitchell SA, Brown EC, Coldwell MJ, Jackson RJ, Willis AE. Protein factor requirements of the Apaf-1 internal ribosome entry segment: roles of polypyrimidine tract binding protein and upstream of N-ras. Mol Cell Biol. 2001;21(10):3364–3374. doi: 10.1128/MCB.21.10.3364-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sawicka K, Bushell M, Spriggs KA, Willis AE. Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36(Pt 4):641–647. doi: 10.1042/BST0360641. [DOI] [PubMed] [Google Scholar]
- 44.Elatmani H, Dormoy-Raclet V, Dubus P, Dautry F, Chazaud C, Jacquemin-Sablon H. The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage. Stem Cells. 2011;29(10):1504–1516. doi: 10.1002/stem.712. [DOI] [PubMed] [Google Scholar]
- 45.Ju Lee H, Bartsch D, Xiao C, Guerrero S, Ahuja G, Schindler C, Moresco JJ, Yates JR, 3rd, Gebauer F, Bazzi H, Dieterich C, Kurian L, Vilchez D. A post-transcriptional program coordinated by CSDE1 prevents intrinsic neural differentiation of human embryonic stem cells. Nat Commun. 2017;8(1):1456. doi: 10.1038/s41467-017-01744-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Markmiller S, Soltanieh S, Server KL, Mak R, Jin W, Fang MY, Luo EC, Krach F, Yang D, Sen A, Fulzele A, Wozniak JM, Gonzalez DJ, Kankel MW, Gao FB, Bennett EJ, et al. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172(3):590–604. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Youn JY, Dunham WH, Hong SJ, Knight JDR, Bashkurov M, Chen GI, Bagci H, Rathod B, MacLeod G, Eng SWM, Angers S, Morris Q, Fabian M, Cote JF, Gingras AC. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol Cell. 2018;69(3):517–532. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- 48.Hou H, Wang F, Zhang W, Wang D, Li X, Bartlam M, Yao X, Rao Z. Structure-functional analyses of CRHSP-24 plasticity and dynamics in oxidative stress response. J Biol Chem. 2011;286(11):9623–9635. doi: 10.1074/jbc.M110.177436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahboubi H, Stochaj U. Cytoplasmic stress granules: dynamic modulators of cell signaling and disease. Biochim Biophys Acta. 2017;1863(4):884–895. doi: 10.1016/j.bbadis.2016.12.022. [DOI] [PubMed] [Google Scholar]
- 50.Gulow K, Bienert D, Haas IG. BiP is feed-back regulated by control of protein translation efficiency. J Cell Sci. 2002;115(Pt 11):2443–2452. doi: 10.1242/jcs.115.11.2443. [DOI] [PubMed] [Google Scholar]
- 51.Kang S, Lee TA, Ra EA, Lee E, Choi H, Lee S, Park B. Differential control of interleukin-6 mRNA levels by cellular distribution of YB-1. PLoS One. 2014;9(11):e112754. doi: 10.1371/journal.pone.0112754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kossinova OA, Gopanenko AV, Tamkovich SN, Krasheninina OA, Tupikin AE, Kiseleva E, Yanshina DD, Malygin AA, Ven'yaminova AG, Kabilov MR, Karpova GG. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim Biophys Acta. 2017;1865(6):664–673. doi: 10.1016/j.bbapap.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Castellana B, Aasen T, Moreno-Bueno G, Dunn SE, Ramon y Cajal S. Interplay between YB-1 and IL-6 promotes the metastatic phenotype in breast cancer cells. Oncotarget. 2015;6(35):38239–38256. doi: 10.18632/oncotarget.5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raffetseder U, Rauen T, Boor P, Ostendorf T, Hanssen L, Floege J, En-Nia A, Djudjaj S, Frye BC, Mertens PR. Extracellular YB-1 blockade in experimental nephritis upregulates Notch-3 receptor expression and signaling. Nephron Exp Nephrol. 2011;118(4):e100–e108. doi: 10.1159/000324209. [DOI] [PubMed] [Google Scholar]
- 55.Fukada T, Tonks NK. Identification of YB-1 as a regulator of PTP1B expression: implications for regulation of insulin and cytokine signaling. EMBO J. 2003;22(3):479–493. doi: 10.1093/emboj/cdg067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sorokin AV, Selyutina AA, Skabkin MA, Guryanov SG, Nazimov IV, Richard C, Th'ng J, Yau J, Sorensen PH, Ovchinnikov LP, Evdokimova V. Proteasome-mediated cleavage of the Y-box-binding protein 1 is linked to DNA-damage stress response. EMBO J. 2005;24(20):3602–3612. doi: 10.1038/sj.emboj.7600830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blenkiron C, Hurley DG, Fitzgerald S, Print CG, Lasham A. Links between the oncoprotein YB-1 and small non-coding RNAs in breast cancer. PLoS One. 2013;8(11):e80171. doi: 10.1371/journal.pone.0080171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dong W, Wang H, Shahzad K, Bock F, Al-Dabet MM, Ranjan S, Wolter J, Kohli S, Hoffmann J, Dhople VM, Zhu C, Lindquist JA, Esmon CT, Grone E, Grone HJ, Madhusudhan T, et al. Activated protein C ameliorates renal ischemia-reperfusion injury by restricting Y-box binding Protein-1 ubiquitination. J Am Soc Nephrol. 2015;26(11):2789–2799. doi: 10.1681/ASN.2014080846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11(4):371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J Cell Biol. 2003;160(6):823–831. doi: 10.1083/jcb.200212128. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochim Biophys Acta. 2015;1849(7):861–870. doi: 10.1016/j.bbagrm.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Treeck B, Protter DSW, Matheny T, Khong A, Link CD, Parker R. RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome. Proc Natl Acad Sci U S A. 2018;115(11):2734–2739. doi: 10.1073/pnas.1800038115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Protter DS, Parker R. Principles and properties of stress granules. Trends Cell Biol. 2016;26(9):668–679. doi: 10.1016/j.tcb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bounedjah O, Desforges B, Wu TD, Pioche-Durieu C, Marco S, Hamon L, Curmi PA, Guerquin-Kern JL, Pietrement O, Pastre D. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 2014;42(13):8678–8691. doi: 10.1093/nar/gku582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Maharana S, Wang J, Papadopoulos DK, Richter D, Pozniakovsky A, Poser I, Bickle M, Rizk S, Guillen-Boixet J, Franzmann TM, Jahnel M, Marrone L, Chang YT, Sterneckert J, Tomancak P, Hyman AA, et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360(6391):918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, Grishin NV, Frantz DE, Schneider JW, Chen S, Li L, Sawaya MR, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149(4):753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Han TW, Kato M, Xie S, Wu LC, Mirzaei H, Pei J, Chen M, Xie Y, Allen J, Xiao G, McKnight SL. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 2012;149(4):768–779. doi: 10.1016/j.cell.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 69.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19(R1):R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ito Daisuke, Hatano Mami, Suzuki Norihiro. RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Science Translational Medicine. 2017;9(415):eaah5436. doi: 10.1126/scitranslmed.aah5436. [DOI] [PubMed] [Google Scholar]
- 71.Maziuk B, Ballance HI, Wolozin B. Dysregulation of RNA binding protein aggregation in neurodegenerative disorders. Front Mol Neurosci. 2017;10:89. doi: 10.3389/fnmol.2017.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153(7):1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi Y, Lin S, Staats KA, Li Y, Chang WH, Hung ST, Hendricks E, Linares GR, Wang Y, Son EY, Wen X, Kisler K, Wilkinson B, Menendez L, Sugawara T, Woolwine P, et al. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat Med. 2018;24(3):313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ivanov P, O'Day E, Emara MM, Wagner G, Lieberman J, Anderson P. G-quadruplex structures contribute to the neuroprotective effects of angiogenin-induced tRNA fragments. Proc Natl Acad Sci U S A. 2014;111(51):18201–18206. doi: 10.1073/pnas.1407361111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 76.Guryanov SG, Filimonov VV, Timchenko AA, Melnik BS, Kihara H, Kutyshenko VP, Ovchinnikov LP, Semisotnov GV. The major mRNP protein YB-1: structural and association properties in solution. Biochim Biophys Acta. 2013;1834(2):559–567. doi: 10.1016/j.bbapap.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 77.Selivanova OM, Guryanov SG, Enin GA, Skabkin MA, Ovchinnikov LP, Serdyuk IN. YB-1 is capable of forming extended nanofibrils. Biochemistry (Mosc) 2010;75(1):115–120. doi: 10.1134/S0006297910010153. [DOI] [PubMed] [Google Scholar]
- 78.Guryanov SG, Selivanova OM, Nikulin AD, Enin GA, Melnik BS, Kretov DA, Serdyuk IN, Ovchinnikov LP. Formation of amyloid-like fibrils by Y-box binding protein 1 (YB-1) is mediated by its cold shock domain and modulated by disordered terminal domains. PLoS One. 2012;7(5):e36969. doi: 10.1371/journal.pone.0036969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kretov DA, Curmi PA, Hamon L, Abrakhi S, Desforges B, Ovchinnikov LP, Pastre D. mRNA and DNA selection via protein multimerization: YB-1 as a case study. Nucleic Acids Res. 2015;43(19):9457–9473. doi: 10.1093/nar/gkv822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skabkin MA, Kiselyova OI, Chernov KG, Sorokin AV, Dubrovin EV, Yaminsky IV, Vasiliev VD, Ovchinnikov LP. Structural organization of mRNA complexes with major core mRNP protein YB-1. Nucleic Acids Res. 2004;32(18):5621–5635. doi: 10.1093/nar/gkh889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kloks CP, Spronk CA, Lasonder E, Hoffmann A, Vuister GW, Grzesiek S, Hilbers CW. The solution structure and DNA-binding properties of the cold-shock domain of the human Y-box protein YB-1. J Mol Biol. 2002;316(2):317–326. doi: 10.1006/jmbi.2001.5334. [DOI] [PubMed] [Google Scholar]
- 82.Abrakhi S, Kretov DA, Desforges B, Dobra I, Bouhss A, Pastre D, Hamon L. Nanoscale analysis reveals the maturation of neurodegeneration-associated protein aggregates: grown in mRNA granules then released by stress granule proteins. ACS Nano. 2017;11(7):7189–7200. doi: 10.1021/acsnano.7b03071. [DOI] [PubMed] [Google Scholar]
- 83.Somasekharan SP, El-Naggar A, Leprivier G, Cheng H, Hajee S, Grunewald TG, Zhang F, Ng T, Delattre O, Evdokimova V, Wang Y, Gleave M, Sorensen PH. YB-1 regulates stress granule formation and tumor progression by translationally activating G3BP1. J Cell Biol. 2015;208(7):913–929. doi: 10.1083/jcb.201411047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hofmann S, Cherkasova V, Bankhead P, Bukau B, Stoecklin G. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol Biol Cell. 2012;23(19):3786–3800. doi: 10.1091/mbc.e12-04-0296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shukla S, Parker R. Hypo- and hyper-assembly diseases of RNA-protein complexes. Trends Mol Med. 2016;22(7):615–628. doi: 10.1016/j.molmed.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Deretic V, Jiang S, Dupont N. Autophagy intersections with conventional and unconventional secretion in tissue development, remodeling and inflammation. Trends Cell Biol. 2012;22(8):397–406. doi: 10.1016/j.tcb.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–240. doi: 10.1016/j.tcb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 88.Frye BC, Halfter S, Djudjaj S, Muehlenberg P, Weber S, Raffetseder U, En-Nia A, Knott H, Baron JM, Dooley S, Bernhagen J, Mertens PR. Y-box protein-1 is actively secreted through a non-classical pathway and acts as an extracellular mitogen. EMBO Rep. 2009;10(7):783–789. doi: 10.1038/embor.2009.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rauen T, Raffetseder U, Frye BC, Djudjaj S, Muhlenberg PJ, Eitner F, Lendahl U, Bernhagen J, Dooley S, Mertens PR. YB-1 acts as a ligand for Notch-3 receptors and modulates receptor activation. J Biol Chem. 2009;284(39):26928–26940. doi: 10.1074/jbc.M109.046599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palicharla VR, Maddika S. HACE1 mediated K27 ubiquitin linkage leads to YB-1 protein secretion. Cell Signal. 2015;27(12):2355–2362. doi: 10.1016/j.cellsig.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 91.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126(Pt 24):5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 92.Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. elife. 2016;5. 10.7554/eLife.19276. [DOI] [PMC free article] [PubMed]
- 93.Suresh PS, Tsutsumi R, Venkatesh T. YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling. Eur J Cell Biol. 2018;97(3):163–167. doi: 10.1016/j.ejcb.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 94.Yanshina DD, Kossinova OA, Gopanenko AV, Krasheninina OA, Malygin AA, Venyaminova AG, Karpova GG. Structural features of the interaction of the 3′-untranslated region of mRNA containing exosomal RNA-specific motifs with YB-1, a potential mediator of mRNA sorting. Biochimie. 2018;144:134–143. doi: 10.1016/j.biochi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 95.Keerthikumar S, Chisanga D, Ariyaratne D, Al Saffar H, Anand S, Zhao K, Samuel M, Pathan M, Jois M, Chilamkurti N, Gangoda L, Mathivanan S. ExoCarta: a web-based compendium of Exosomal cargo. J Mol Biol. 2016;428(4):688–692. doi: 10.1016/j.jmb.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Simpson Richard J., Kalra Hina, Mathivanan Suresh. ExoCarta as a resource for exosomal research. Journal of Extracellular Vesicles. 2012;1(1):18374. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bergmann S, Royer-Pokora B, Fietze E, Jurchott K, Hildebrandt B, Trost D, Leenders F, Claude JC, Theuring F, Bargou R, Dietel M, Royer HD. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer Res. 2005;65(10):4078–4087. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 98.Kosnopfel C, Sinnberg T, Schittek B. Y-box binding protein 1--a prognostic marker and target in tumour therapy. Eur J Cell Biol. 2014;93(1–2):61–70. doi: 10.1016/j.ejcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 99.Asakuno K, Kohno K, Uchiumi T, Kubo T, Sato S, Isono M, Kuwano M. Involvement of a DNA binding protein, MDR-NF1/YB-1, in human MDR1 gene expression by actinomycin D. Biochem Biophys Res Commun. 1994;199(3):1428–1435. doi: 10.1006/bbrc.1994.1390. [DOI] [PubMed] [Google Scholar]
- 100.Bargou RC, Jurchott K, Wagener C, Bergmann S, Metzner S, Bommert K, Mapara MY, Winzer KJ, Dietel M, Dorken B, Royer HD. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3(4):447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 101.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28(23):7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kuwano M, Oda Y, Izumi H, Yang SJ, Uchiumi T, Iwamoto Y, Toi M, Fujii T, Yamana H, Kinoshita H, Kamura T, Tsuneyoshi M, Yasumoto K, Kohno K. The role of nuclear Y-box binding protein 1 as a global marker in drug resistance. Mol Cancer Ther. 2004;3(11):1485–1492. [PubMed] [Google Scholar]
- 103.Janz M, Harbeck N, Dettmar P, Berger U, Schmidt A, Jurchott K, Schmitt M, Royer HD. Y-box factor YB-1 predicts drug resistance and patient outcome in breast cancer independent of clinically relevant tumor biologic factors HER2, uPA and PAI-1. Int J Cancer. 2002;97(3):278–282. doi: 10.1002/ijc.1610. [DOI] [PubMed] [Google Scholar]
- 104.Oda Y, Ohishi Y, Saito T, Hinoshita E, Uchiumi T, Kinukawa N, Iwamoto Y, Kohno K, Kuwano M, Tsuneyoshi M. Nuclear expression of Y-box-binding protein-1 correlates with P-glycoprotein and topoisomerase II alpha expression, and with poor prognosis in synovial sarcoma. J Pathol. 2003;199(2):251–258. doi: 10.1002/path.1282. [DOI] [PubMed] [Google Scholar]
- 105.Shen H, Xu W, Luo W, Zhou L, Yong W, Chen F, Wu C, Chen Q, Han X. Upregulation of mdr1 gene is related to activation of the MAPK/ERK signal transduction pathway and YB-1 nuclear translocation in B-cell lymphoma. Exp Hematol. 2011;39(5):558–569. doi: 10.1016/j.exphem.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 106.Setoguchi K, Cui L, Hachisuka N, Obchoei S, Shinkai K, Hyodo F, Kato K, Wada F, Yamamoto T, Harada-Shiba M, Obika S, Nakano K. Antisense oligonucleotides targeting Y-box binding Protein-1 inhibit tumor angiogenesis by downregulating Bcl-xL-VEGFR2/-tie axes. Mol Ther Nucleic Acids. 2017;9:170–181. doi: 10.1016/j.omtn.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.di Martino O, Troiano A, Guarino AM, Pollice A, Vivo M, La Mantia G, Calabro V. DeltaNp63alpha controls YB-1 protein stability: evidence on YB-1 as a new player in keratinocyte differentiation. Genes Cells. 2016;21(6):648–660. doi: 10.1111/gtc.12373. [DOI] [PubMed] [Google Scholar]
- 108.Chibi M, Meyer M, Skepu A, DJ GR, Moolman-Smook JC, Pugh DJ. RBBP6 interacts with multifunctional protein YB-1 through its RING finger domain, leading to ubiquitination and proteosomal degradation of YB-1. J Mol Biol. 2008;384(4):908–916. doi: 10.1016/j.jmb.2008.09.060. [DOI] [PubMed] [Google Scholar]
- 109.van Roeyen CR, Scurt FG, Brandt S, Kuhl VA, Martinkus S, Djudjaj S, Raffetseder U, Royer HD, Stefanidis I, Dunn SE, Dooley S, Weng H, Fischer T, Lindquist JA, Mertens PR. Cold shock Y-box protein-1 proteolysis autoregulates its transcriptional activities. Cell Commun Signal. 2013;11:63. doi: 10.1186/1478-811X-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kim ER, Selyutina AA, Buldakov IA, Evdokimova V, Ovchinnikov LP, Sorokin AV. The proteolytic YB-1 fragment interacts with DNA repair machinery and enhances survival during DNA damaging stress. Cell Cycle. 2013;12(24):3791–3803. doi: 10.4161/cc.26670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stenina OI, Poptic EJ, Di Corleto PE. Thrombin activates a Y box-binding protein (DNA-binding protein B) in endothelial cells. J Clin Invest. 2000;106(4):579–587. doi: 10.1172/JCI9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tacke F, Kanig N, En-Nia A, Kaehne T, Eberhardt CS, Shpacovitch V, Trautwein C, Mertens PR. Y-box protein-1/p18 fragment identifies malignancies in patients with chronic liver disease. BMC Cancer. 2011;11:185. doi: 10.1186/1471-2407-11-185. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 113.Tacke F, Galm O, Kanig N, Yagmur E, Brandt S, Lindquist JA, Eberhardt CS, Raffetseder U, Mertens PR. High prevalence of Y-box protein-1/p18 fragment in plasma of patients with malignancies of different origin. BMC Cancer. 2014;14:33. doi: 10.1186/1471-2407-14-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rohr I, Braicu EI, En-Nia A, Heinrich M, Richter R, Chekerov R, Dechend R, Heidecke H, Dragun D, Schafer R, Gorny X, Lindquist JA, Brandt S, Sehouli J, Mertens PR. Y-box protein-1/p18 as novel serum marker for ovarian cancer diagnosis: a study by the tumor Bank ovarian Cancer (TOC) Cytokine. 2016;85:157–164. doi: 10.1016/j.cyto.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 115.Ferreira AR, Bettencourt M, Alho I, Costa AL, Sousa AR, Mansinho A, Abreu C, Pulido C, Macedo D, Vendrell I, Pacheco TR, Costa L, Casimiro S. Serum YB-1 (Y-box binding protein 1) as a biomarker of bone disease progression in patients with breast cancer and bone metastases. J Bone Oncol. 2017;6:16–21. doi: 10.1016/j.jbo.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang S, Baltimore D. RNA-binding protein Lin28 in cancer and immunity. Cancer Lett. 2016;375(1):108–113. doi: 10.1016/j.canlet.2016.02.050. [DOI] [PubMed] [Google Scholar]
- 117.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22(39):8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- 118.Grosset C, Chen CY, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu AB. A mechanism for translationally coupled mRNA turnover: interaction between the poly(a) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103(1):29–40. doi: 10.1016/S0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 119.Wurth L, Papasaikas P, Olmeda D, Bley N, Calvo GT, Guerrero S, Cerezo-Wallis D, Martinez-Useros J, Garcia-Fernandez M, Huttelmaier S, Soengas MS, Gebauer F. UNR/CSDE1 drives a post-transcriptional program to promote melanoma invasion and metastasis. Cancer Cell. 2016;30(5):694–707. doi: 10.1016/j.ccell.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 120.Wu Q, Parry G. Hepsin and prostate cancer. Front Biosci. 2007;12:5052–5059. doi: 10.2741/2447. [DOI] [PubMed] [Google Scholar]
- 121.Zhang C, Zhang M, Wu Q, Peng J, Ruan Y, Gu J. Hepsin inhibits CDK11p58 IRES activity by suppressing unr expression and eIF-2alpha phosphorylation in prostate cancer. Cell Signal. 2015;27(4):789–797. doi: 10.1016/j.cellsig.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 122.Martinez-Useros J, Georgiev-Hristov T, Fernandez-Acenero MJ, Borrero-Palacios A, Indacochea A, Guerrero S, Li W, Cebrian A, Gomez Del Pulgar T, Puime-Otin A, Del Puerto-Nevado L, Rodriguez-Remirez M, Perez N, Celdran A, Gebauer F, Garcia-Foncillas J. UNR/CDSE1 expression as prognosis biomarker in resectable pancreatic ductal adenocarcinoma patients: a proof-of-concept. PLoS One. 2017;12(8):e0182044. doi: 10.1371/journal.pone.0182044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hanssen L, Alidousty C, Djudjaj S, Frye BC, Rauen T, Boor P, Mertens PR, van Roeyen CR, Tacke F, Heymann F, Tittel AP, Koch A, Floege J, Ostendorf T, Raffetseder U. YB-1 is an early and central mediator of bacterial and sterile inflammation in vivo. J Immunol. 2013;191(5):2604–2613. doi: 10.4049/jimmunol.1300416. [DOI] [PubMed] [Google Scholar]
- 124.Wang J, Gibbert L, Djudjaj S, Alidousty C, Rauen T, Kunter U, Rembiak A, Enders D, Jankowski V, Braun GS, Floege J, Ostendorf T, Raffetseder U. Therapeutic nuclear shuttling of YB-1 reduces renal damage and fibrosis. Kidney Int. 2016;90(6):1226–1237. doi: 10.1016/j.kint.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 125.Alidousty C, Rauen T, Hanssen L, Wang Q, Alampour-Rajabi S, Mertens PR, Bernhagen J, Floege J, Ostendorf T, Raffetseder U. Calcineurin-mediated YB-1 dephosphorylation regulates CCL5 expression during monocyte differentiation. J Biol Chem. 2014;289(31):21401–21412. doi: 10.1074/jbc.M114.562991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raffetseder U, Rauen T, Djudjaj S, Kretzler M, En-Nia A, Tacke F, Zimmermann HW, Nelson PJ, Frye BC, Floege J, Stefanidis I, Weber C, Mertens PR. Differential regulation of chemokine CCL5 expression in monocytes/macrophages and renal cells by Y-box protein-1. Kidney Int. 2009;75(2):185–196. doi: 10.1038/ki.2008.457. [DOI] [PubMed] [Google Scholar]
- 127.Krohn R, Raffetseder U, Bot I, Zernecke A, Shagdarsuren E, Liehn EA, van Santbrink PJ, Nelson PJ, Biessen EA, Mertens PR, Weber C. Y-box binding protein-1 controls CC chemokine ligand-5 (CCL5) expression in smooth muscle cells and contributes to neointima formation in atherosclerosis-prone mice. Circulation. 2007;116(16):1812–1820. doi: 10.1161/CIRCULATIONAHA.107.708016. [DOI] [PubMed] [Google Scholar]
- 128.Dhawan L, Liu B, Pytlak A, Kulshrestha S, Blaxall BC, Taubman MB. Y-box binding protein 1 and RNase UK114 mediate monocyte chemoattractant protein 1 mRNA stability in vascular smooth muscle cells. Mol Cell Biol. 2012;32(18):3768–3775. doi: 10.1128/MCB.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen CY, Gherzi R, Andersen JS, Gaietta G, Jurchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14(10):1236–1248. [PMC free article] [PubMed] [Google Scholar]
- 130.Coles LS, Diamond P, Lambrusco L, Hunter J, Burrows J, Vadas MA, Goodall GJ. A novel mechanism of repression of the vascular endothelial growth factor promoter, by single strand DNA binding cold shock domain (Y-box) proteins in normoxic fibroblasts. Nucleic Acids Res. 2002;30(22):4845–4854. doi: 10.1093/nar/gkf615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Diamond P, Shannon MF, Vadas MA, Coles LS. Cold shock domain factors activate the granulocyte-macrophage colony-stimulating factor promoter in stimulated Jurkat T cells. J Biol Chem. 2001;276(11):7943–7951. doi: 10.1074/jbc.M009836200. [DOI] [PubMed] [Google Scholar]
- 132.Fraser DJ, Phillips AO, Zhang X, van Roeyen CR, Muehlenberg P, En-Nia A, Mertens PR. Y-box protein-1 controls transforming growth factor-beta1 translation in proximal tubular cells. Kidney Int. 2008;73(6):724–732. doi: 10.1038/sj.ki.5002719. [DOI] [PubMed] [Google Scholar]
- 133.Berquin IM, Pang B, Dziubinski ML, Scott LM, Chen YQ, Nolan GP, Ethier SP. Y-box-binding protein 1 confers EGF independence to human mammary epithelial cells. Oncogene. 2005;24(19):3177–3186. doi: 10.1038/sj.onc.1208504. [DOI] [PubMed] [Google Scholar]
- 134.Basaki Y, Hosoi F, Oda Y, Fotovati A, Maruyama Y, Oie S, Ono M, Izumi H, Kohno K, Sakai K, Shimoyama T, Nishio K, Kuwano M. Akt-dependent nuclear localization of Y-box-binding protein 1 in acquisition of malignant characteristics by human ovarian cancer cells. Oncogene. 2007;26(19):2736–2746. doi: 10.1038/sj.onc.1210084. [DOI] [PubMed] [Google Scholar]
- 135.Mertens PR, Harendza S, Pollock AS, Lovett DH. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272(36):22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- 136.Hanssen L, Frye BC, Ostendorf T, Alidousty C, Djudjaj S, Boor P, Rauen T, Floege J, Mertens PR, Raffetseder U. Y-box binding protein-1 mediates profibrotic effects of calcineurin inhibitors in the kidney. J Immunol. 2011;187(1):298–308. doi: 10.4049/jimmunol.1100382. [DOI] [PubMed] [Google Scholar]
- 137.Higashi K, Inagaki Y, Suzuki N, Mitsui S, Mauviel A, Kaneko H, Nakatsuka I. Y-box-binding protein YB-1 mediates transcriptional repression of human alpha 2(I) collagen gene expression by interferon-gamma. J Biol Chem. 2003;278(7):5156–5162. doi: 10.1074/jbc.M208724200. [DOI] [PubMed] [Google Scholar]
- 138.van Roeyen CR, Eitner F, Martinkus S, Thieltges SR, Ostendorf T, Bokemeyer D, Luscher B, Luscher-Firzlaff JM, Floege J, Mertens PR. Y-box protein 1 mediates PDGF-B effects in mesangioproliferative glomerular disease. J Am Soc Nephrol. 2005;16(10):2985–2996. doi: 10.1681/ASN.2004111009. [DOI] [PubMed] [Google Scholar]
- 139.Raffetseder U, Liehn EA, Weber C, Mertens PR. Role of cold shock Y-box protein-1 in inflammation, atherosclerosis and organ transplant rejection. Eur J Cell Biol. 2012;91(6–7):567–575. doi: 10.1016/j.ejcb.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 140.Capowski EE, Esnault S, Bhattacharya S, Malter JS. Y box-binding factor promotes eosinophil survival by stabilizing granulocyte-macrophage colony-stimulating factor mRNA. J Immunol. 2001;167(10):5970–5976. doi: 10.4049/jimmunol.167.10.5970. [DOI] [PubMed] [Google Scholar]
- 141.Esnault S, Malter JS. Hyaluronic acid or TNF-alpha plus fibronectin triggers granulocyte macrophage-colony-stimulating factor mRNA stabilization in eosinophils yet engages differential intracellular pathways and mRNA binding proteins. J Immunol. 2003;171(12):6780–6787. doi: 10.4049/jimmunol.171.12.6780. [DOI] [PubMed] [Google Scholar]
- 142.Higashi K, Tomigahara Y, Shiraki H, Miyata K, Mikami T, Kimura T, Moro T, Inagaki Y, Kaneko H. A novel small compound that promotes nuclear translocation of YB-1 ameliorates experimental hepatic fibrosis in mice. J Biol Chem. 2011;286(6):4485–4492. doi: 10.1074/jbc.M110.151936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Imai J, Hozumi K, Sumiyoshi H, Yazawa M, Hirano K, Abe J, Higashi K, Inagaki Y, Mine T. Anti-fibrotic effects of a novel small compound on the regulation of cytokine production in a mouse model of colorectal fibrosis. Biochem Biophys Res Commun. 2015;468(4):554–560. doi: 10.1016/j.bbrc.2015.10.123. [DOI] [PubMed] [Google Scholar]
- 144.Khan MI, Adhami VM, Lall RK, Sechi M, Joshi DC, Haidar OM, Syed DN, Siddiqui IA, Chiu SY, Mukhtar H. YB-1 expression promotes epithelial-to-mesenchymal transition in prostate cancer that is inhibited by a small molecule fisetin. Oncotarget. 2014;5(9):2462–2474. doi: 10.18632/oncotarget.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Gunasekaran Vinoth Prasanna, Nishi Kumari, Sivakumar Dakshinamurthy, Sivaraman Thirunavukkarasu, Mathan Ganeshan. Identification of 2,4-dihydroxy-5-pyrimidinyl imidothiocarbomate as a novel inhibitor to Y box binding protein-1 (YB-1) and its therapeutic actions against breast cancer. European Journal of Pharmaceutical Sciences. 2018;116:2–14. doi: 10.1016/j.ejps.2017.09.019. [DOI] [PubMed] [Google Scholar]
- 146.Ciani F, Tafuri S, Troiano A, Cimmino A, Fioretto BS, Guarino AM, Pollice A, Vivo M, Evidente A, Carotenuto D, Calabro V. Anti-proliferative and pro-apoptotic effects of Uncaria tomentosa aqueous extract in squamous carcinoma cells. J Ethnopharmacol. 2018;211:285–294. doi: 10.1016/j.jep.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 147.Prabhu L, Mundade R, Wang B, Wei H, Hartley AV, Martin M, McElyea K, Temm CJ, Sandusky G, Liu Y, Lu T. Critical role of phosphorylation of serine 165 of YBX1 on the activation of NF-kappaB in colon cancer. Oncotarget. 2015;6(30):29396–29412. doi: 10.18632/oncotarget.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martin M, Hua L, Wang B, Wei H, Prabhu L, Hartley AV, Jiang G, Liu Y, Lu T. Novel serine 176 phosphorylation of YBX1 activates NF-kappaB in colon cancer. J Biol Chem. 2017;292(8):3433–444. [DOI] [PMC free article] [PubMed]
- 149.Inoue K, Fry EA, Frazier DP. Transcription factors that interact with p53 and Mdm2. Int J Cancer. 2016;138(7):1577–1585. doi: 10.1002/ijc.29663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Homer C, Knight DA, Hananeia L, Sheard P, Risk J, Lasham A, Royds JA, Braithwaite AW. Y-box factor YB1 controls p53 apoptotic function. Oncogene. 2005;24(56):8314–8325. doi: 10.1038/sj.onc.1208998. [DOI] [PubMed] [Google Scholar]
- 151.Okamoto T, Izumi H, Imamura T, Takano H, Ise T, Uchiumi T, Kuwano M, Kohno K. Direct interaction of p53 with the Y-box binding protein, YB-1: a mechanism for regulation of human gene expression. Oncogene. 2000;19(54):6194–6202. doi: 10.1038/sj.onc.1204029. [DOI] [PubMed] [Google Scholar]
- 152.Kato M, Wang L, Putta S, Wang M, Yuan H, Sun G, Lanting L, Todorov I, Rossi JJ, Natarajan R. Post-transcriptional up-regulation of Tsc-22 by Ybx1, a target of miR-216a, mediates TGF-{beta}-induced collagen expression in kidney cells. J Biol Chem. 2010;285(44):34004–34015. doi: 10.1074/jbc.M110.165027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dooley S, Said HM, Gressner AM, Floege J, En-Nia A, Mertens PR. Y-box protein-1 is the crucial mediator of antifibrotic interferon-gamma effects. J Biol Chem. 2006;281(3):1784–1795. doi: 10.1074/jbc.M510215200. [DOI] [PubMed] [Google Scholar]
- 154.Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell. 2017;168(1–2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Basak S, Behar M, Hoffmann A. Lessons from mathematically modeling the NF-kappaB pathway. Immunol Rev. 2012;246(1):221–238. doi: 10.1111/j.1600-065X.2011.01092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hayden MS, Ghosh S. Regulation of NF-kappaB by TNF family cytokines. Semin Immunol. 2014;26(3):253–266. doi: 10.1016/j.smim.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Maubach G, Schmadicke AC, Naumann M. NEMO links nuclear factor-kappaB to human diseases. Trends Mol Med. 2017;23(12):1138–1155. doi: 10.1016/j.molmed.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 158.Abbas Abul K., Trotta Eleonora, R. Simeonov Dimitre, Marson Alexander, Bluestone Jeffrey A. Revisiting IL-2: Biology and therapeutic prospects. Science Immunology. 2018;3(25):eaat1482. doi: 10.1126/sciimmunol.aat1482. [DOI] [PubMed] [Google Scholar]
- 159.Malek TR, Yu A, Zhu L, Matsutani T, Adeegbe D, Bayer AL. IL-2 family of cytokines in T regulatory cell development and homeostasis. J Clin Immunol. 2008;28(6):635–639. doi: 10.1007/s10875-008-9235-y. [DOI] [PubMed] [Google Scholar]
- 160.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12(3):180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 161.Malek TR. The biology of interleukin-2. Annu Rev Immunol. 2008;26:453–479. doi: 10.1146/annurev.immunol.26.021607.090357. [DOI] [PubMed] [Google Scholar]
- 162.Braithwaite AW, Del Sal G, Lu X. Some p53-binding proteins that can function as arbiters of life and death. Cell Death Differ. 2006;13(6):984–993. doi: 10.1038/sj.cdd.4401924. [DOI] [PubMed] [Google Scholar]
- 163.Meek DW. Regulation of the p53 response and its relationship to cancer. Biochem J. 2015;469(3):325–346. doi: 10.1042/BJ20150517. [DOI] [PubMed] [Google Scholar]
- 164.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 165.Charni M, Aloni-Grinstein R, Molchadsky A, Rotter V. p53 on the crossroad between regeneration and cancer. Cell Death Differ. 2017;24(1):8–14. doi: 10.1038/cdd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Duffy MJ, Synnott NC, McGowan PM, Crown J, O'Connor D, Gallagher WM. p53 as a target for the treatment of cancer. Cancer Treat Rev. 2014;40(10):1153–1160. doi: 10.1016/j.ctrv.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 167.Loeffler I, Wolf G. Transforming growth factor-beta and the progression of renal disease. Nephrol Dial Transplant. 2014;29(Suppl 1):i37–i45. doi: 10.1093/ndt/gft267. [DOI] [PubMed] [Google Scholar]
- 168.Yingling JM, Blanchard KL, Sawyer JS. Development of TGF-beta signalling inhibitors for cancer therapy. Nat Rev Drug Discov. 2004;3(12):1011–1022. doi: 10.1038/nrd1580. [DOI] [PubMed] [Google Scholar]
- 169.Wallace E, Gewin L. Imatinib: novel treatment of immune-mediated kidney injury. J Am Soc Nephrol. 2013;24(5):694–701. doi: 10.1681/ASN.2012080818. [DOI] [PubMed] [Google Scholar]
- 170.Gordon J, Spiera R. Imatinib and the treatment of fibrosis: recent trials and tribulations. Curr Rheumatol Rep. 2011;13(1):51–58. doi: 10.1007/s11926-010-0146-6. [DOI] [PubMed] [Google Scholar]
- 171.Brandt S, Mertens PR. A remedy for kidney disease successfully alters the cold shock protein response during inflammation. Kidney Int. 2016;90(6):1148–1150. doi: 10.1016/j.kint.2016.07.041. [DOI] [PubMed] [Google Scholar]
- 172.Hasegawa M, Matsushita Y, Horikawa M, Higashi K, Tomigahara Y, Kaneko H, Shirasaki F, Fujimoto M, Takehara K, Sato S. A novel inhibitor of Smad-dependent transcriptional activation suppresses tissue fibrosis in mouse models of systemic sclerosis. Arthritis Rheum. 2009;60(11):3465–3475. doi: 10.1002/art.24934. [DOI] [PubMed] [Google Scholar]
- 173.Inagaki Y, Kushida M, Higashi K, Itoh J, Higashiyama R, Hong YY, Kawada N, Namikawa K, Kiyama H, Bou-Gharios G, Watanabe T, Okazaki I, Ikeda K. Cell type-specific intervention of transforming growth factor beta/Smad signaling suppresses collagen gene expression and hepatic fibrosis in mice. Gastroenterology. 2005;129(1):259–268. doi: 10.1053/j.gastro.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 174.Higashi K, Inagaki Y, Fujimori K, Nakao A, Kaneko H, Nakatsuka I. Interferon-gamma interferes with transforming growth factor-beta signaling through direct interaction of YB-1 with Smad3. J Biol Chem. 2003;278(44):43470–43479. doi: 10.1074/jbc.M302339200. [DOI] [PubMed] [Google Scholar]
- 175.Norman JT, Lindahl GE, Shakib K, En-Nia A, Yilmaz E, Mertens PR. The Y-box binding protein YB-1 suppresses collagen alpha 1(I) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J Biol Chem. 2001;276(32):29880–29890. doi: 10.1074/jbc.M103145200. [DOI] [PubMed] [Google Scholar]
- 176.Syed DN, Adhami VM, Khan MI, Mukhtar H. Inhibition of Akt/mTOR signaling by the dietary flavonoid fisetin. Anti Cancer Agents Med Chem. 2013;13(7):995–1001. doi: 10.2174/18715206113139990129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gupta SC, Tyagi AK, Deshmukh-Taskar P, Hinojosa M, Prasad S, Aggarwal BB. Downregulation of tumor necrosis factor and other proinflammatory biomarkers by polyphenols. Arch Biochem Biophys. 2014;559:91–99. doi: 10.1016/j.abb.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 178.Adhami VM, Syed DN, Khan N, Mukhtar H. Dietary flavonoid fisetin: a novel dual inhibitor of PI3K/Akt and mTOR for prostate cancer management. Biochem Pharmacol. 2012;84(10):1277–1281. doi: 10.1016/j.bcp.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Syed DN, Chamcheu JC, Khan MI, Sechi M, Lall RK, Adhami VM, Mukhtar H. Fisetin inhibits human melanoma cell growth through direct binding to p70S6K and mTOR: findings from 3-D melanoma skin equivalents and computational modeling. Biochem Pharmacol. 2014;89(3):349–360. doi: 10.1016/j.bcp.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sutherland BW, Kucab J, Wu J, Lee C, Cheang MC, Yorida E, Turbin D, Dedhar S, Nelson C, Pollak M, Leighton Grimes H, Miller K, Badve S, Huntsman D, Blake-Gilks C, Chen M, et al. Akt phosphorylates the Y-box binding protein 1 at Ser102 located in the cold shock domain and affects the anchorage-independent growth of breast cancer cells. Oncogene. 2005;24(26):4281–4292. doi: 10.1038/sj.onc.1208590. [DOI] [PubMed] [Google Scholar]
- 181.Gieseler-Halbach S, Meltendorf S, Pierau M, Weinert S, Heidel FH, Fischer T, Handschuh J, Braun-Dullaeus RC, Schrappe M, Lindquist JA, Mertens PR, Thomas U, Brunner-Weinzierl MC. RSK-mediated nuclear accumulation of the cold-shock Y-box protein-1 controls proliferation of T cells and T-ALL blasts. Cell Death Differ. 2017;24(2):371–383. doi: 10.1038/cdd.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li X, Kong D, Chen H, Liu S, Hu H, Wu T, Wang J, Chen W, Ning Y, Li Y, Lu Z. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Sci Rep. 2016;6:21789. doi: 10.1038/srep21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Kalliolias GD, Ivashkiv LB. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat Rev Rheumatol. 2016;12(1):49–62. doi: 10.1038/nrrheum.2015.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sollazzo D, Forte D, Polverelli N, Romano M, Perricone M, Rossi L, Ottaviani E, Luatti S, Martinelli G, Vianelli N, Cavo M, Palandri F, Catani L. Crucial factors of the inflammatory microenvironment (IL-1beta/TNF-alpha/TIMP-1) promote the maintenance of the malignant hemopoietic clone of myelofibrosis: an in vitro study. Oncotarget. 2016;7(28):43974–43988. doi: 10.18632/oncotarget.9949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Altmann C, Vasic V, Hardt S, Heidler J, Haussler A, Wittig I, Schmidt MH, Tegeder I. Progranulin promotes peripheral nerve regeneration and reinnervation: role of notch signaling. Mol Neurodegener. 2016;11(1):69. doi: 10.1186/s13024-016-0132-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Musso G, Cassader M, Cohney S, Pinach S, Saba F, Gambino R. Emerging liver-kidney interactions in nonalcoholic fatty liver disease. Trends Mol Med. 2015;21(10):645–662. doi: 10.1016/j.molmed.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 187.Sun YY, Li XF, Meng XM, Huang C, Zhang L, Li J. Macrophage phenotype in liver injury and repair. Scand J Immunol. 2017;85(3):166–174. doi: 10.1111/sji.12468. [DOI] [PubMed] [Google Scholar]
- 188.Zhou Z, Xu MJ, Gao B. Hepatocytes: a key cell type for innate immunity. Cell Mol Immunol. 2016;13(3):301–315. doi: 10.1038/cmi.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lindquist JA, Schneider A, Mertens PR. Regulation of endogenous brakes to kidney fibrosis: turning the view upside down. J Mol Med (Berl) 2017;95(6):571–573. doi: 10.1007/s00109-017-1540-6. [DOI] [PubMed] [Google Scholar]
- 190.Quek C, Hill AF. The role of extracellular vesicles in neurodegenerative diseases. Biochem Biophys Res Commun. 2017;483(4):1178–1186. doi: 10.1016/j.bbrc.2016.09.090. [DOI] [PubMed] [Google Scholar]
- 191.Howitt J, Hill AF. Exosomes in the pathology of neurodegenerative diseases. J Biol Chem. 2016;291(52):26589–26597. doi: 10.1074/jbc.R116.757955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Vilette D, Courte J, Peyrin JM, Coudert L, Schaeffer L, Andreoletti O, Leblanc P. Cellular mechanisms responsible for cell-to-cell spreading of prions. Cell Mol Life Sci. 2018;75(14):2575. [DOI] [PMC free article] [PubMed]
- 193.Bloch DB, Nobre RA, Yang WH. GW/P-bodies and autoimmune disease. Adv Exp Med Biol. 2013;768:61–70. doi: 10.1007/978-1-4614-5107-5_5. [DOI] [PubMed] [Google Scholar]
- 194.Yang WH, Bloch DB. Probing the mRNA processing body using protein macroarrays and “autoantigenomics”. RNA. 2007;13(5):704–712. doi: 10.1261/rna.411907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Jeoung DI, Bong Lee E, Lee S, Lim Y, Lee DY, Kim J, Kim HY, Wook Song Y. Autoantibody to DNA binding protein B as a novel serologic marker in systemic sclerosis. Biochem Biophys Res Commun. 2002;299(4):549–554. doi: 10.1016/S0006-291X(02)02685-2. [DOI] [PubMed] [Google Scholar]
- 196.Braunschweig D, Krakowiak P, Duncanson P, Boyce R, Hansen RL, Ashwood P, Hertz-Picciotto I, Pessah IN, Van de Water J. Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry. 2013;3:e277. doi: 10.1038/tp.2013.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chernov KG, Curmi PA, Hamon L, Mechulam A, Ovchinnikov LP, Pastre D. Atomic force microscopy reveals binding of mRNA to microtubules mediated by two major mRNP proteins YB-1 and PABP. FEBS Lett. 2008;582(19):2875–2881. doi: 10.1016/j.febslet.2008.07.019. [DOI] [PubMed] [Google Scholar]
- 198.Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015;43(Database issue):D512–D520. doi: 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Liu Q, Tao T, Liu F, Ni R, Lu C, Shen A. Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp Cell Res. 2016;349(2):230–238. doi: 10.1016/j.yexcr.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 200.Yang X, Qian K. Protein O-GlcNAcylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol. 2017;18(7):452–465. doi: 10.1038/nrm.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Xu W, Zhang X, Wu JL, Fu L, Liu K, Liu D, Chen GG, Lai PB, Wong N, Yu J. O-GlcNAc transferase promotes fatty liver-associated liver cancer through inducing palmitic acid and activating endoplasmic reticulum stress. J Hepatol. 2017;67(2):310–320. doi: 10.1016/j.jhep.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 202.Allison DF, Wamsley JJ, Kumar M, Li D, Gray LG, Hart GW, Jones DR, Mayo MW. Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-kappaB acetylation and transcription. Proc Natl Acad Sci U S A. 2012;109(42):16888–16893. doi: 10.1073/pnas.1208468109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bernhardt A, Fehr A, Brandt S, Jerchel S, Ballhause TM, Philipsen L, Stolze S, Geffers R, Weng H, Fischer KD, Isermann B, Brunner-Weinzierl MC, Batra A, Siegmund B, Zhu C, Lindquist JA, et al. Inflammatory cell infiltration and resolution of kidney inflammation is orchestrated by the cold-shock protein Y-box binding protein-1. Kidney Int. 2017;92(5):1157–1177. doi: 10.1016/j.kint.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 204.Seko Y, Cole S, Kasprzak W, Shapiro BA, Ragheb JA. The role of cytokine mRNA stability in the pathogenesis of autoimmune disease. Autoimmun Rev. 2006;5(5):299–305. doi: 10.1016/j.autrev.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 205.Coles LS, Diamond P, Occhiodoro F, Vadas MA, Shannon MF. Cold shock domain proteins repress transcription from the GM-CSF promoter. Nucleic Acids Res. 1996;24(12):2311–2317. doi: 10.1093/nar/24.12.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mertens PR, Martin IV, Frye BC, Rauen T, Strauch S, Pabst M, Geier A. Rat Mrp2 gene expression is regulated by an interleukin-1beta-stimulated biphasic response with enhanced transcription and subcellular shuttling of YB-1. Eur J Cell Biol. 2012;91(6–7):533–541. doi: 10.1016/j.ejcb.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 207.Silveira CG, Krampe J, Ruhland B, Diedrich K, Hornung D, Agic A. Cold-shock domain family member YB-1 expression in endometrium and endometriosis. Hum Reprod. 2012;27(1):173–182. doi: 10.1093/humrep/der368. [DOI] [PubMed] [Google Scholar]
- 208.Hornung D, Bentzien F, Wallwiener D, Kiesel L, Taylor RN. Chemokine bioactivity of RANTES in endometriotic and normal endometrial stromal cells and peritoneal fluid. Mol Hum Reprod. 2001;7(2):163–168. doi: 10.1093/molehr/7.2.163. [DOI] [PubMed] [Google Scholar]
- 209.Jenkins RH, Bennagi R, Martin J, Phillips AO, Redman JE, Fraser DJ. A conserved stem loop motif in the 5’untranslated region regulates transforming growth factor-beta(1) translation. PLoS One. 2010;5(8):e12283. doi: 10.1371/journal.pone.0012283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Coles LS, Lambrusco L, Burrows J, Hunter J, Diamond P, Bert AG, Vadas MA, Goodall GJ. Phosphorylation of cold shock domain/Y-box proteins by ERK2 and GSK3beta and repression of the human VEGF promoter. FEBS Lett. 2005;579(24):5372–5378. doi: 10.1016/j.febslet.2005.08.075. [DOI] [PubMed] [Google Scholar]
- 211.Hayashi J, Kajino K, Umeda T, Takano S, Arakawa Y, Kudo M, Hino O. Somatic mutation and SNP in the promoter of dbpA and human hepatocarcinogenesis. Int J Oncol. 2002;21(4):847–850. [PubMed] [Google Scholar]
- 212.Yasen M, Kajino K, Kano S, Tobita H, Yamamoto J, Uchiumi T, Kon S, Maeda M, Obulhasim G, Arii S, Hino O. The up-regulation of Y-box binding proteins (DNA binding protein a and Y-box binding protein-1) as prognostic markers of hepatocellular carcinoma. Clin Cancer Res. 2005;11(20):7354–7361. doi: 10.1158/1078-0432.CCR-05-1027. [DOI] [PubMed] [Google Scholar]
- 213.Yasen M, Obulhasim G, Kajino K, Mogushi K, Mizushima H, Tanaka S, Tanaka H, Hino O, Arii S. DNA binding protein a expression and methylation status in hepatocellular carcinoma and the adjacent tissue. Int J Oncol. 2012;40(3):789–797. doi: 10.3892/ijo.2011.1282. [DOI] [PubMed] [Google Scholar]
- 214.Obulhasim G, Yasen M, Kajino K, Mogushi K, Tanaka S, Mizushima H, Tanaka H, Arii S, Hino O. Up-regulation of dbpA mRNA in hepatocellular carcinoma associated with metabolic syndrome. Hepatol Int. 2013;7(1):215–225. doi: 10.1007/s12072-012-9357-4. [DOI] [PubMed] [Google Scholar]
- 215.Letunic I, Bork P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018;46(D1):D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]


