Abstract
Background
Cyanobacteria, oxygenic photoautotrophic prokaryotes, can be engineered to produce various valuable chemicals from solar energy and CO2 in direct processes. The concept of photosynthetic production of isobutanol, a promising chemical and drop-in biofuel, has so far been demonstrated for Synechocystis PCC 6803 and Synechococcus elongatus PCC 7942. In Synechocystis PCC 6803, a heterologous expression of α-ketoisovalerate decarboxylase (Kivd) from Lactococcus lactis resulted in an isobutanol and 3-methyl-1-butanol producing strain. Kivd was identified as a bottleneck in the metabolic pathway and its activity was further improved by reducing the size of its substrate-binding pocket with a single replacement of serine-286 to threonine (KivdS286T). However, isobutanol production still remained low.
Results
In the present study, we report on how cultivation conditions significantly affect the isobutanol production in Synechocystis PCC 6803. A HCl-titrated culture grown under medium light (50 μmol photons m−2 s−1) showed the highest isobutanol production with an in-flask titer of 194 mg l−1 after 10 days and 435 mg l−1 at day 40. This corresponds to a cumulative isobutanol production of 911 mg l−1, with a maximal production rate of 43.6 mg l−1 day−1 observed between days 4 and 6. Additional metabolic bottlenecks in the isobutanol biosynthesis pathway were further addressed. The expression level of KivdS286T was significantly affected when co-expressed with another gene downstream in a single operon and in a convergent oriented operon. Moreover, the expression of the ADH encoded by codon-optimized slr1192 and co-expression of IlvC and IlvD were identified as potential approaches to further enhance isobutanol production in Synechocystis PCC 6803.
Conclusion
The present study demonstrates the importance of a suitable cultivation condition to enhance isobutanol production in Synechocystis PCC 6803. Chemostat should be used to further increase both the total titer as well as the rate of production. Furthermore, identified bottleneck, Kivd, should be expressed at the highest level to further enhance isobutanol production.
Electronic supplementary material
The online version of this article (10.1186/s13068-018-1268-8) contains supplementary material, which is available to authorized users.
Keywords: Synechocystis, Cumulative titer, Cultivation condition, Metabolic bottleneck, Co-expression
Background
Isobutanol is a branched-chain, four-carbon alcohol used in various manufacturing industries. It is recognized as a promising gasoline additive candidate due to its higher energy content, lower vapor pressure, and lower water solubility compared to ethanol, the most commonly used gasoline supplement in the market today [1]. Isobutanol can be used as a drop-in fuel in the petroleum infrastructure without causing engine and pipeline corrosion [2]. All these advantages make isobutanol an attractive chemical. In addition, isobutanol can be produced via biological routes.
The 2-keto acid pathway is a branched-chain amino acids synthesis pathway, and the 2-ketoisovalerate produced via this pathway can be decarboxylated by an α-ketoisovalerate decarboxylase into isobutyraldehyde and then further converted to isobutanol by an alcohol dehydrogenase. This isobutanol biosynthesis pathway has been widely engineered into various microorganisms, such as Escherichia coli (E. coli) [3–8], Saccharomyces cerevisiae (S. cerevisiae) [9–11], Bacillus subtilis (B. subtilis) [12], Corynebacterium glutamicum (C. glutamicum) [13, 14], and cyanobacteria [15–18]. Specifically, in the unicellular cyanobacterium Synechocystis PCC 6803 (referred hereafter as Synechocystis), it was demonstrated that a heterologous expression of an α-ketoisovalerate decarboxylase (Kivd) from Lactococcus lactis (L. lactis) results in an isobutanol and 3-methyl-1-butanol (3M1B)-producing strain [17]. In addition, the heterologously expressed Kivd was identified as a bottleneck in this isobutanol synthetic pathway, e.g., isobutanol production level correlated positively with Kivd expression levels. Moreover, an endogenous alcohol dehydrogenase in Synechocystis showed a significant high activity towards isobutanol production [17]. In a following study, the substrate-binding pocket of Kivd was engineered to gain higher activity towards isobutanol rather than 3M1B production. ST (KivdS286T), with a replacement of serine-286 to threonine, showed both the highest increase in total catalytic activity and a preferential shift towards isobutanol production [18].
However, the isobutanol titer observed in all the engineered Synechocystis strains still remained low and it was unclear whether other metabolic bottlenecks (besides Kivd), or the culture condition employed, were limiting the isobutanol synthesis. Herein, in the present study, we investigate how to adjust the cultivation conditions to further enhance isobutanol production in Synechocystis. In addition, we address the existence of the other potential metabolic bottlenecks than Kivd in the isobutanol synthesis pathway when introduced into the Synechocystis cells.
Methods
Strains and cultivation conditions
Escherichia coli strain DH5αZ1 (Invitrogen) was used for cloning and conjugation work. The cells were grown at 37 °C in LB medium (agar or liquid) supplemented with 50 μg ml−1 kanamycin (Sigma-Aldrich). The glucose-tolerant Synechocystis strain was used in this study. Synechocystis seed cultures were grown under 35 μmol photons m−2 s−1 at 30 °C in BG11 with addition of 50 μg ml−1 kanamycin. Experimental cultures were inoculated as OD750 = 0.1 with a total volume of 25 ml in BioLite 25 cm2 plug-sealed tissue culture flasks (Thermo Fisher Scientific). The medium used was BG11 medium with addition of 50 μg ml−1 kanamycin and 50 mM NaHCO3 (Sigma-Aldrich). For examining different light intensities, the flasks were shaken horizontally at 120 rpm, at 30 °C, under 15, 50, and 100 μmol photons m−2 s−1, respectively. Light was measured using LI-190SB Quantum sensor. For the examining of different pH adjustment methods, 50 μmol photon m−2 s−1 light-intensity growth condition was employed. For HEPES-buffered cultures, 50 mM HEPES (Sigma-Aldrich) was added into the flasks every second day with the supplement media. For the HCl-titrated cultures, 37% HCl (Sigma-Aldrich), maximal 40 μl, was added every day to adjust the pH of the culture between 7 and 8. pH was measured using MColorpHast™ pH-indicator strips (pH 6.5–10) (Merck). 2 ml of culture was sampled from each flask every second day for OD750 and products measurements, and replaces with 2 ml fresh BG11 medium with addition of 50 μg ml−1 kanamycin and 500 mM NaHCO3. For the tolerance test, mid-log-phase pEEK2-ST cells were washed with fresh BG11 media and resuspended in BG11 media with 50 mM NaHCO3 to OD750 = 0.5 in plug-sealed tissue culture flasks (Thermo Fisher Scientific). Three isobutanol concentrations (321 mg l−1, 642 mg l−1, and 962 mg l−1) and two isobutyraldehyde concentrations (632 mg l−1 and 948 mg l−1) were examined. OD750 of the cultures were measured after 24 h.
Plasmid construction
The self-replicating plasmid pEEK2-ST from our previous study [18], containing a strong constitutive promoter Ptrccore and the engineered Kivd named ST, was used as template to make further constructs in this study.
Transformation of Synechocystis via conjugation
Escherichia coli cargo cells and E. coli HB101 helper cells with the plasmid pRL443-AmpR were grown at 37 °C. The overnight cultures were centrifuged at 3000 rpm for 5 min and resuspended in fresh liquid LB medium without antibiotics. Wild-type Synechocystis cells (200 μl) were mixed with 1 ml cargo cells and 1 ml helper cells, and the mixture was incubated under 100 μmol photons m−2 s−1 at 30 °C for 1.5 h before being spread on a filter on BG11 agar plate for another 48 h incubation.
For colony selection and maintenance, filters were changed onto new BG11 agar plates with 50 μg ml−1 kanamycin. Gene-specific primers were used to analyze the colonies and the correct colonies were inoculated into 6-well plates for further use.
Crude protein extraction and SDS-PAGE/western immunoblot
Proteins were extracted from day 2 cultures; cells were harvested by centrifugation at 5000 rpm, 4 °C, for 10 min. The pellet was washed in 2 ml PBS and recollected by centrifugation, 5000 rpm, 4 °C, for 10 min, and resuspended in 200 µl PBS with Protease Arrest (GBioscience). This mixture went through freeze–thaw process and was disrupted by acid-washed 425–600 μm-diameter glass beads (Sigma-Aldrich) using a Precellys-24 Beadbeater (Bertin Instruments), program 3 × 30 s. Centrifugation was performed twice at 12,000 rpm, 4 °C, 1 min each, to get a clean supernatant-containing soluble proteins. Protein concentration was determined by the DC protein assay (Bio-Rad). 5 μg soluble proteins from each strain were separated by SDS-PAGE, using Mini-PROTEAN TGX TM gels (Bio-Rad), and transferred to PVDF membrane (Bio-Rad). Anti-Strep-tag II (abcam) antibody was used to detect Strep-tagged Kivd through the standard techniques, while anti-ATPase western immunoblot was done on the same samples as loading control. This western immunoblot was repeated three times for determining the relative Kivd expression level in all strains.
Optical density measurement and product quantification
Optical density was measured every day in 96-well plates using a micro-plate reader (HIDEX, Plate Chameleon). Every second day, 2 ml culture was sampled from each flask and centrifuged at 5000 rpm, for 10 min. 1305 μl of the supernatant was transferred into a 15 ml screw cap tube, and mixed with 45 μl 3000 mg l−1 internal standard (1-pentanol) and 450 μl extraction solvent dichloromethane (DCM). The mixture was shaken on Multi-Tube Vortexer VX-2500 (VWR) at maximum speed for 5 min and then centrifuged at 5000 rpm, 4 °C, for 10 min. The DCM phase (bottom) was transferred into 1.5 ml clear glass gas chromatography (GC) vials (VWR). The extracted samples were analyzed on a PerkinElmer GC 580 system equipped with a flame ionization detector and an Elite-WAX Polyethylene Glycol Series Capillary column, 30 m × 0.25 mm × 0.25 μm (PerkinElmer). Nitrogen was the carrier gas, with 10 ml min−1 flow rate. The temperatures of injector and detector were 220 °C and 240 °C, respectively. The initial oven temperature was 50 °C and then raised to 100 °C with a rate of 10 °C min−1 followed by a rise to 180 °C with a rate of 20 °C min−1. The GC results were analyzed using TotalChrom Navigator version 6.3.2.
Results and discussion
Modification on the cultivation condition
In the present study, we attempted to investigate how to improve cultivation conditions for enhanced isobutanol production in Synechocystis. A strain, pEEK2-ST, solely expressing KivdS286T on a self-replicating vector pEEK2, generated for enhanced isobutanol production in our previous study [18] was used for the cultivation experiments. Initially, we examined growth and isobutanol production under different light intensities (Fig. 1a). All experimental cultures were grown in plug-sealed tissue culture flasks to avoid product evaporation. We inoculated the cultures as OD750 = 0.1 from seed culture grown in cotton-cap E-flasks under 35 μmol photons m−2 s−1.
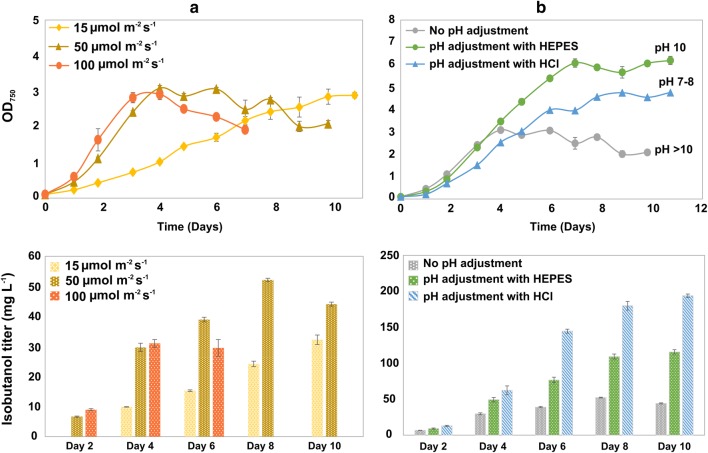
Fig. 1.
Growth and isobutanol titer produced from engineered Synechocystis PCC 6803 strain pEEK2-ST in different cultivation conditions. a Growth and isobutanol production titer of the cells grew under 15 μmol photons m−2 s−1, 50 μmol photons m−2 s−1, and 100 μmol photons m−2 s−1 light intensities without any pH adjustment. b Growth and isobutanol production titer of the cells grew under 50 μmol photons m−2 s−1 with different pH adjustments. The target pH range for the pH adjustment, using either HEPES or HCl, was between pH 7 and 8. The pH labeled on the growth curve was the final pH measured in the end of the cultivation. Results represent the mean of three biological replicates; error bars represent standard deviation
In the first 3 days, cells under 100 μmol photons m−2 s−1 grew significantly faster than under the other two light intensities examined. However, from day 4, cells under 100 μmol photons m−2 s−1 became yellowish and the OD750 started to drop quickly with a completely bleached phenotype at day 7. Moreover, the isobutanol titer observed in cultures under 100 μmol photons m−2 s−1 did not show any significant superiority compared to the cultures under lower light intensities. In addition, the empty vector control strain showed similar growth trends under these three different light conditions (Additional file 1: Figure S1). Moreover, in an isobutanol and isobutyraldehyde tolerance test, we did not observe a significant effect on cells growth in media with maximal 962 mg l−1 isobutanol (Additional file 1: Figure S2). This indicates that the phenotype of the cells under different light intensities does not correlate with the isobutanol production levels. The unhealthy phenotype of the cells under 100 μmol photons m−2 s−1 after day 4 was most likely due to the potential photoinhibition from the relative high light [19–22].
We observed the highest OD750 and isobutanol production from cells grown under 50 μmol photons m−2 s−1. These cells also maintained a longer steady state than the cells under 100 μmol photons m−2 s−1 and the isobutanol in-flask titer reached 52 mg l−1 at day 8. Furthermore, the cells under 15 μmol photons m−2 s−1 showed the slowest growth rate with an isobutanol in-flask titer of 32 mg l−1 at day 10 when the cultures went into steady stage. Based on these observations, we selected 50 μmol photons m−2 s−1 as light intensity for further investigations.
Since we grew the cells in sealed flasks, the supplementary of NaHCO3 every second day became the carbon supply for photosynthesis. In addition, Synechocystis cells in the early stationary phase naturally alkalize their living environment to pH around 10. As a consequence, the alkaline condition generated by both NaHCO3 and cells may cause changes in carbon equilibrium in the growth media or cell metabolism. Therefore, we compared two widely used pH adjustment approaches, HEPES buffer and HCl titration, where, in the control group, cells were grown without any pH adjustment.
The control group showed the least growth, fastest cell death, and the lowest isobutanol production compared to the cultures with pH adjustment. HEPES-buffered cultures showed the highest OD750, while the HCl-titrated cultures showed the highest isobutanol production (Fig. 1b). The dramatic differences between the cultures with and without pH adjustment may indicate that, in the sealed cultivation system applied, pH plays an important role to affect carbon supply. When the pH was above 10, most of the carbon existed in the form of CO32− in the media, which cannot be up taken by the cells. When the pH in the culture was adjusted, the carbon equilibrium shifted forward to generate relatively more HCO3− and CO2. At this stage, the cells can assimilate more carbon, resulting in enhanced growth and isobutanol production. However, the HEPES-buffered cultures grew to a higher OD750 than the HCl-titrated culture, though the pH in the later cultures was closer to the theoretical optimal pH for Synechocystis [23] (Fig. 1b). This may be due to HEPES being much milder than HCl and better for maintaining enzyme structure and cell physiological characters [24, 25].
To our surprise, the HCl-titrated cultures stayed in stationary phase(s) for long time, and we continuously observed isobutanol production from these cultures until day 46. The cultivation was terminated when the isobutanol in-flask titer had dropped for three continues measurements. A maximal in-flask isobutanol titer of 435 mg l−1 was observed at day 40 when the OD750 was 4.7 (Fig. 2). This titer is 1.5 times higher than the reported net isobutanol concentration observed in another isobutanol producing Synechocystis strain under mixotrophic cultivation condition at an OD730 above 12 [26]. After day 40, the isobutanol in-flask titer started to drop, the isobutanol produced between two measurements was not enough to overcome the dilution caused by the sampling of 2 ml culture and the supplement of 2 ml fresh media every second day. In the end of 46 days culturing, the in-flask isobutanol titer was 397 mg l−1 and the cumulative titer was as high as 911 mg l−1. Moreover, we also calculated the in-flask and cumulative titer of the by-product 3-methyl-1-butanol (3M1B). A maximal in-flask titer of 105 mg l−1 was observed at day 40 with a final cumulative titer of 225 mg l−1 at day 46. In addition, we divided the 46 days cultivation period into growth and steady-state phases, the isobutanol production rate in the growth phase was significantly higher than that in any of the steady-state phases (Table 1). The highest isobutanol production rate of 43.6 mg l−1 day−1 was observed between days 4 and 6, coinciding with maximal growth (Table 1 and Fig. 2). This demonstrates that maintaining the culture in the growth phase may be an optimal approach to further enhance isobutanol production. Therefore, a chemostat cultivation should be used in the future.
Fig. 2.

Growth, isobutanol/3M1B in-flask and cumulative titer observed in a long-term cultivation experiment. The OD750 was measured every day and product titer was measured every second day. S1, S2, and S3 represent different periods in the steady-state phase. Results represent the mean of three biological replicates and error bars represent standard deviation
Table 1.
Isobutanol production rates in the different phases of the long-term cultivation of engineered Synechocystis PCC 6803
| Time | In-flask (mg l−1 day−1) | Cumulative (mg l−1 day−1) | |
|---|---|---|---|
| Growth phase | Days 2–10 | 22.6 ± 0.4 | 26.6 ± 0.3 |
| Steady-state phase (total) | Days 10–46 | 5.7 ± 0.2 | 19.0 ± 0.3 |
| Steady-state phase (S1) | Days 10–22 | 11.2 ± 0.5 | 21.9 ± 0.4 |
| Steady-state phase (S2) | Days 23–35 | 10.3 ± 1.5 | 25.9 ± 1.8 |
| Steady-state phase (S3) | Days 36–46 | − 4.2 ± 0.7 | 12.9 ± 0.8 |
| Maximal rate | Days 4–6 | 41.1 ± 3.5 | 43.6 ± 3.3 |
Further investigation on the isobutanol synthesis pathway
Despite modifying the cultivation condition, we also aimed to further investigate the isobutanol synthesis pathway to identify other potential metabolic bottleneck enzyme(s) than Kivd [18]. In Synechococcus, an additional heterologous expression of acetolactate synthase (AlsS) from Bacillus subtilis (B. subtilis), acetohydroxy acid isomeroreductase (IlvC) and dihydroxy acid dehydratase (IlvD) from E. coli (Fig. 3a) enhanced isobutanol production 2.5 times [16]. Therefore, in the present study, we examined the heterogenous AlsS, IlvC, and IlvD discussed above, and the corresponding endogenous enzymes in Synechocystis. Moreover, we also attempted to investigate the effect on isobutanol production when expressing an additional copy of the endogenous alcohol dehydrogenase (ADH) encoded by codon-optimized slr1192, since the native ADHs may not be sufficient to convert the enhanced level of isobutyraldehyde produced by KivdS286T.
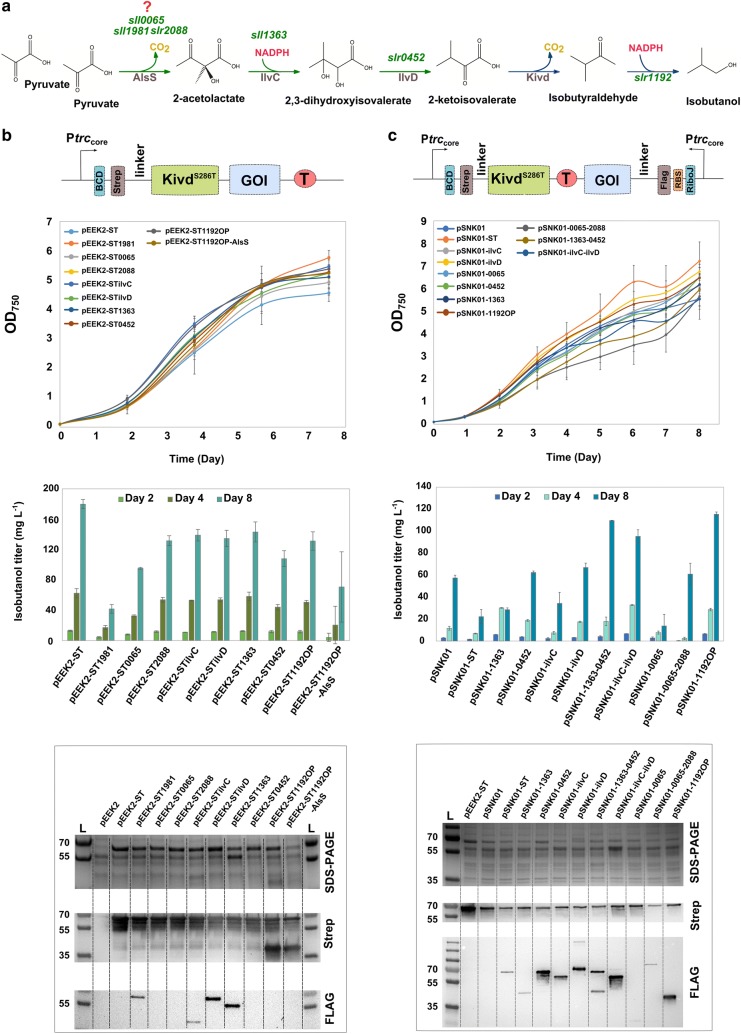
Fig. 3.
Schematic overview of the isobutanol synthesis pathway, the genetic constructs for identifying respective bottleneck, and the results from the engineered Synechocystis PCC 6803 strains containing different constructs. a Isobutanol synthesis pathway. The heterogenous enzymes are colored in brown: acetolactate synthase (AlsS) from Bacillus subtilis (B. subtilis), acetohydroxy acid isomeroreductase (IlvC), dihydroxy acid dehydratase (IlvD) from E. coli, and α-ketoisovalerate decarboxylase (Kivd) from L. lactis. The endogenous genes are colored in green. The gene-encoding alcohol dehydrogenase used here is the codon-optimized slr1192 (slr1192OP). b Genetic concepts, growth curve, isobutanol titer, SDS-PAGE, and western immunoblot from the engineered strains containing single operon. GOI Gene of interest. KivdS286T and slr1192OP-encoded alcohol dehydrogenase were strep-tagged and the other genes were flag-tagged. BCD is the bicistronic design for providing more reliable expression. c Growth curve, isobutanol titer, SDS-PAGE, and Western blot from the engineered strains containing convergent-oriented operons. KivdS286T was strep-tagged and the other genes were Flag-tagged. RiboJ is a self-cleaving ribozyme for providing more reliable expression. Size of the proteins are: AlsS: 62 kDa, IlvC: 54 kDa, IlvD: 65 kDa, KivdS286T: 61 kDa, Sll1981: 60 kDa, Sll1363: 40 kDa, Slr0452: 59 kDa, Slr1192: 36 kDa, Sll0065: 21 kDa, Slr2088: 68 kDa
It was unclear whether Synechocystis has a native AlsS or only an acetohydroxy acid synthase (AHAS), which is a bi-functional enzyme catalyzing the reaction from pyruvate to acetolactate and also the reaction from pyruvate and α-ketoglutarate to α-acetohydroxybutyrate [27]. Thus, we attempted to overexpress the endogenous gene-encoding potential AlsS and AHAS in Synechocystis (Fig. 3a) to examine their effects on isobutanol production. We initially generated eight engineered Synechocystis strains (Additional file 1: Figure S3A) where each strain contained an operon-expressing KivdS286Tand one of the genes to be examined. Unfortunately, we could not get transformants for the strain containing KivdS286T and AlsS (B. subtilis). Similar result has been reported in Clostridium cellulolyticum (C. cellulolyticum); no transformants could be obtained when expressing this AlsS with high transcription level. This may be due to the high activity of AlsS [28]. Therefore, we constructed another strain-containing AlsS in the end of the operon-expressing KivdS286T and ADH (Additional file 1: Figure S3B).
The growth and isobutanol titer from the obtained engineered strains were compared to the control strain pEEK2-ST. The variations in isobutanol titer from the three biological replicates of strain pEEK2-ST1192OP-AslS were very high and no AlsS expression could be detected using western immunoblots from any of the replicates (data not shown). Besides, isobutanol production from strain pEEK2-ST1981 was five times lower than from the control strain pEEK2-ST (Fig. 3b). This could be interpreted as an evidence for the α-KGD function but not an AlsS function of the enzyme encoded by sll1981, since the overexpression of sll1981 might direct more carbon flux towards the TCA cycle instead of isobutanol synthesis [29–32]. Surprisingly, strain pEEK2-ST still showed the highest isobutanol in-flask titer, which corresponded to its highest expression level of KivdS286T among all the examined strains (Fig. 3b). In addition, the expression of sll0065, slr2088, and slr0452 could not be detected using anti-Flag tag western immunoblot. Thus, two hypotheses could be made: (i) there is no other bottleneck(s) than Kivd in the isobutanol synthesis pathway. Thus, the strain with the highest KivdS286T expression level showed the highest isobutanol production. (ii) There is other bottleneck(s), but could not be evaluated due to the low protein expression levels.
Therefore, to increase protein expression levels, we expressed KivdS286T solely in a PtrccoreBCD driven operon and the other gene(s) to be examined in a convergent-oriented operon driven by PtrccoreRiboJ (Additional file 1: Figure S3C). A double BioBrick terminator B0015 was applied between these two operons to avoid read through. The new control strain pSNK01 contains the operon-expressing KivdS286T and a convergent operon-expressing ccdB. The addition of the convergent-oriented operon successfully increased the expression levels of all the enzymes to be examined. Strains pSNK01-1363-0452 and pSNK01-ilvc-ilvD showed significantly higher isobutanol production than the control strain. However, strains pSNK01-1363 and pSNK01-ilvC showed only half of isobutanol production compared to the control, and strains pSNK01-0452 and pSNK01-ilvD did not show any improvement in isobutanol production (Fig. 3c). These results indicate that acetohydroxy acid isomeroreductase (IlvC) may be a bottleneck enzyme in the isobutanol synthesis pathway, but the intermediate metabolite that produced may have negative effects on the metabolism in the Synechocystis cells. Therefore, dihydroxy acid dehydratase needs to be co-expressed with acetohydroxy acid isomeroreductase for enhanced metabolic flux towards isobutanol production. Furthermore, compared to the control strain, strain pSNK01-1192OP showed more than five times higher isobutanol titer, which means that the additional copy of ADH increased the conversion of isobutyraldehyde to isobutanol. To increase the level of KivdS286T, the strain pSNK01-ST containing two copies of KivdS286T was made. Unfortunately, this strain showed less than half of the isobutanol titer compared to the control strain (Fig. 3c). This may be due to the lower KivdS286T expression level in both operons compared to that in the control strain or some mutations might occur when the construct was conjugated into Synechocystis and the activity of KivdS286T was interrupted.
Strain pSNK01-0065 showed significantly lower isobutanol titer compared to all the other engineered strains, this may indicate that the protein encoded by sll0065 is similar to the small subunit of the AHAS from S. cerevisiae which functions as the regulatory subunit that inhibits the catalytic subunit of AHAS in response to a feedback signal from excess levels of valine [33]. Interestingly, strain pSNK01-0065-2088 showed similar isobutanol titer but much lower OD750 compared to the control strain, meaning that the isobutanol production per cell was higher in strain pSNK01-0065-2088. This may confirm the function of slr2088 as the catalytic subunit of AHAS in Synechocystis. Unfortunately, we were unable to obtain any transformants for strain pSNK01-2088, which may be due to the similar problem occurred when expressing AlsS from B. subtilis.
Notably, all the strains containing two convergent operons showed much lower KivdS286T expression levels and thereby much less isobutanol production than strain pEEK2-ST (Fig. 3c). This may be due to the transcriptional interference between two operons caused by the leakage of terminator, though the convergent-oriented operons were claimed as the best composition to have high expression level for the genes in both operons [34, 35]. Therefore, the results indicate that keeping a high expression level of KivdS286T is an essential requirement for high isobutanol production in engineered strains of Synechocystis. Expressing KivdS286T alone on a high-copy number self-replicating vector is so far the best option to obtain the highest expression in Synechocystis, whereas all the other genetic modifications may be done in the genome. Furthermore, genome-scale metabolic models should be applied in the future to further identify potential metabolic bottlenecks and enhance the carbon flux towards isobutanol production.
Conclusions
In summary, the present study demonstrates the importance of applying suitable cultivation condition for enhancing isobutanol production in Synechocystis. The isobutanol titer was dramatically increased in cells cultivated under 50 μmol photons m−2 s−1 and regularly adjusted to a pH between 7 and 8. A cumulative isobutanol titer of 911 mg l−1 was obtained after 46 days with a maximal isobutanol production rate of 43.6 mg l−1 day−1 observed between days 4 and 6. More frequently, maybe even continuously, removal of the produced isobutanol will further increase both the total titer as well as the rate of production. We also observed that the expression level of KivdS286T should be kept as high as possible in Synechocystis to enhance isobutanol production. Finally, the present study suggested that IlvC and IlvD (heterogenous or endogenous) may be expressed together for further enhancement of isobutanol production. In conclusion, this study provides more detailed metabolic engineering possibilities for direct photosynthetic isobutanol production in Synechocystis.
Additional file
Additional file 1: Figure S1. Growth of the Synechocystis PCC 6803 empty vector control strain under different light intensities and with different pH adjustments. Results represent the mean of three biological replicates, error bars represent standard deviation. Figure S2. Tolerance test for isobutanol and isobutyraldehyde. Three isobutanol concentrations and two isobutyraldehyde concentrations were tested in plug-sealed tissue flasks. The chemicals were added into pEEK2-ST cultures with OD750 = 0.5. The change on OD750 in each culture after 24 h cultivation was analyzed here. Results represent the mean of three biological replicates, error bars represent standard deviation. Figure S3. Schematic overview of the constructs for identifying potential bottlenecks. All the constructs are generated on the self-replicating vector pEEK2. A: Schematic overviews of the constructs with single operon expressing KivdS286T and one of the respective genes to be examined. B: The construct generated to reduce the expression level of AlsS. C: Schematic overview of the constructs with two convergent orientated operons with a double terminator BBa_B0015 in between.
Authors’ contributions
RM designed all the experiments, generated all the genetic constructs, and analyzed the data. HX performed the cultivation experiments and analyzed the data. RM wrote the manuscript. HX commented on the manuscript. PL supervised the work and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Siri Norlander from Microbial Chemistry, Department of Chemistry—Ångström (Uppsala University) is thanked for constructing the pSNK01 plasmid.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by the European Union Horizon 2020 Framework Programme under the grant agreement number 640720 (Photofuel), and the NordForsk NCoE program “NordAqua” (Project Number 82845).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rui Miao, Email: rui.miao@kemi.uu.se.
Hao Xie, Email: haoxie1993@gmail.com.
Peter Lindblad, Email: peter.lindblad@kemi.uu.se.
References
- 1.Lu J, Brigham CJ, Gai CS, Sinskey AJ. Studies on the production of branched-chain alcohols in engineered Ralstonia eutropha. Appl Microbiol Biotechnol. 2012;96:283–297. doi: 10.1007/s00253-012-4320-9. [DOI] [PubMed] [Google Scholar]
- 2.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488:320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi S, Wu T-Y, Eckl E-M, Hawkins SD, Buelter T, Liao JC. Engineering the isobutanol biosynthetic pathway in Escherichia coli by comparison of three aldehyde reductase/alcohol dehydrogenase genes. Appl Microbiol Biotechnol. 2009;85:651–657. doi: 10.1007/s00253-009-2085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baez A, Cho K-M, Liao JC. High-flux isobutanol production using engineered Escherichia coli: a bioreactor study with in situ product removal. Appl Microbiol Biotechnol. 2011;90:1681–1690. doi: 10.1007/s00253-011-3173-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith KM, Liao JC. An evolutionary strategy for isobutanol production strain development in Escherichia coli. Metab Eng. 2011;13:674–681. doi: 10.1016/j.ymben.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Liu Z, Liu P, Xiao D, Zhang X. Improving isobutanol production in metabolically engineered Escherichia coli by co-producing ethanol and modulation of pentose phosphate pathway. J Ind Microbiol Biotechnol. 2016;43:851–860. doi: 10.1007/s10295-016-1751-9. [DOI] [PubMed] [Google Scholar]
- 7.Desai SH, Rabinovitch-Deere CA, Fan Z, Atsumi S. Isobutanol production from cellobionic acid in Escherichia coli. Microb Cell Fact. 2015;14:52. doi: 10.1186/s12934-015-0232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trinh CT, Li J, Blanch HW, Clark DS. Redesigning Escherichia coli metabolism for anaerobic production of isobutanol. Appl Environ Microbiol. 2011;77:4894–4904. doi: 10.1128/AEM.00382-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen X, Nielsen KF, Borodina I, Kielland-Brandt MC, Karhumaa K. Increased isobutanol production in Saccharomyces cerevisiae by overexpression of genes in valine metabolism. Biotechnol Biofuels. 2011;4:21. doi: 10.1186/1754-6834-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo T, Tezuka H, Ishii J, Matsuda F, Ogino C, Kondo A. Genetic engineering to enhance the Ehrlich pathway and alter carbon flux for increased isobutanol production from glucose by Saccharomyces cerevisiae. J Biotechnol. 2012;159:32–37. doi: 10.1016/j.jbiotec.2012.01.022. [DOI] [PubMed] [Google Scholar]
- 11.Yuan J, Chen X, Mishra P, Ching C-B. Metabolically engineered Saccharomyces cerevisiae for enhanced isoamyl alcohol production. Appl Microbiol Biotechnol. 2017;101:465–474. doi: 10.1007/s00253-016-7970-1. [DOI] [PubMed] [Google Scholar]
- 12.Li S, Wen J, Jia X. Engineering Bacillus subtilis for isobutanol production by heterologous Ehrlich pathway construction and the biosynthetic 2-ketoisovalerate precursor pathway overexpression. Appl Microbiol Biotechnol. 2011;91:577–589. doi: 10.1007/s00253-011-3280-9. [DOI] [PubMed] [Google Scholar]
- 13.Smith KM, Cho K-M, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blombach B, Riester T, Wieschalka S, Ziert C, Youn J-W, Wendisch VF, et al. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Atsumi S, Higashide W, Liao JC. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat Biotechnol. 2009;27:1177–1180. doi: 10.1038/nbt.1586. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Shen CR, Liao JC. Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosynth Res. 2014;120:301–310. doi: 10.1007/s11120-014-9987-6. [DOI] [PubMed] [Google Scholar]
- 17.Miao R, Liu X, Englund E, Lindberg P, Lindblad P. Isobutanol production in Synechocystis PCC 6803 using heterologous and endogenous alcohol dehydrogenases. Metab Eng Commun. 2017;5:45–53. doi: 10.1016/j.meteno.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miao R, Xie H, Ho FM, Lindblad P. Protein engineering of α-ketoisovalerate decarboxylase for improved isobutanol production in Synechocystis PCC 6803. Metab Eng. 2018;47:42–48. doi: 10.1016/j.ymben.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Scheller HV, Haldrup A. Photoinhibition of photosystem I. Planta. 2005;221:5–8. doi: 10.1007/s00425-005-1507-7. [DOI] [PubMed] [Google Scholar]
- 20.Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI. Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta BBA Bioenerg. 2007;1767:414–421. doi: 10.1016/j.bbabio.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 21.Nishiyama Y, Yamamoto H, Allakhverdiev SI, Inaba M, Yokota A, Murata N. Oxidative stress inhibits the repair of photodamage to the photosynthetic machinery. EMBO J. 2001;20:5587–5594. doi: 10.1093/emboj/20.20.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Mustila H, Magnuson A, Stensjö K. Homologous overexpression of NpDps2 and NpDps5 increases the tolerance for oxidative stress in the multicellular cyanobacterium Nostoc punctiforme. FEMS Microbiol Lett. 2018;365(18):fny198. doi: 10.1093/femsle/fny198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang JJ, Kolodny NH, Redfearn JT, Allen MM. The acid stress response of the cyanobacterium Synechocystis sp. strain PCC 6308. Arch Microbiol. 2002;177:486–493. doi: 10.1007/s00203-002-0419-1. [DOI] [PubMed] [Google Scholar]
- 24.Eley JH. The use of HEPES as a buffer for the growth of the cyanobacterium Anacystis nidulans. Appl Microbiol Biotechnol. 1988;28:297–300. doi: 10.1007/BF00250459. [DOI] [Google Scholar]
- 25.Wehr JD, Brown LM, Vanderelst IE. Hydrogen ion buffering of culture media for algae from moderately acidic, oligotrophic waters. J Phycol. 1986;22:88–94. doi: 10.1111/j.1529-8817.1986.tb02520.x. [DOI] [Google Scholar]
- 26.Varman AM, Xiao Y, Pakrasi HB, Tang YJ. Metabolic engineering of Synechocystis sp. strain PCC 6803 for isobutanol production. Appl Environ Microbiol. 2013;79:908–914. doi: 10.1128/AEM.02827-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouhen OM-E, Joset F. Biosynthesis of the branched-chain amino acids in the cyanobacterium Synechocystis PCC 6803: existence of compensatory pathways. Curr Microbiol. 2002;45:94–98. doi: 10.1007/s00284-001-0097-2. [DOI] [PubMed] [Google Scholar]
- 28.Higashide W, Li Y, Yang Y, Liao JC. Metabolic engineering of Clostridium cellulolyticum for production of isobutanol from cellulose. Appl Environ Microbiol. 2011;77:2727–2733. doi: 10.1128/AEM.02454-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badger MR, Price GD. CO2 concentrating mechanisms in cyanobacteria: molecular components, their diversity and evolution. J Exp Bot. 2003;54:609–622. doi: 10.1093/jxb/erg076. [DOI] [PubMed] [Google Scholar]
- 30.Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, et al. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria function and phylogenetic analysis. J Biol Chem. 2002;277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 31.Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008;59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 32.Chatterjee A, Dastidar KG, Maitra S, Das-Chatterjee A, Dihazi H, Eschrich K, et al. sll 1981, an acetolactate synthase homologue of Synechocystis sp. PCC6803, functions as l-myo-inositol 1-phosphate synthase. Planta. 2006;224:367–379. doi: 10.1007/s00425-006-0221-4. [DOI] [PubMed] [Google Scholar]
- 33.Takpho N, Watanabe D, Takagi H. High-level production of valine by expression of the feedback inhibition-insensitive acetohydroxy acid synthase in Saccharomyces cerevisiae. Metab Eng. 2018;46:60–67. doi: 10.1016/j.ymben.2018.02.011. [DOI] [PubMed] [Google Scholar]
- 34.Zhu T, Xie X, Li Z, Tan X, Lu X. Enhancing photosynthetic production of ethylene in genetically engineered Synechocystis sp. PCC 6803. Green Chem. 2015;17:421–434. doi: 10.1039/C4GC01730G. [DOI] [Google Scholar]
- 35.Wang X, Lei G, Wu X, Wang F, Lai C, Li Z. Expression, purification and characterization of sll1981 protein from cyanobacterium Synechocystis sp. PCC6803. Protein Expr Purif. 2017;139:21–28. doi: 10.1016/j.pep.2017.07.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Growth of the Synechocystis PCC 6803 empty vector control strain under different light intensities and with different pH adjustments. Results represent the mean of three biological replicates, error bars represent standard deviation. Figure S2. Tolerance test for isobutanol and isobutyraldehyde. Three isobutanol concentrations and two isobutyraldehyde concentrations were tested in plug-sealed tissue flasks. The chemicals were added into pEEK2-ST cultures with OD750 = 0.5. The change on OD750 in each culture after 24 h cultivation was analyzed here. Results represent the mean of three biological replicates, error bars represent standard deviation. Figure S3. Schematic overview of the constructs for identifying potential bottlenecks. All the constructs are generated on the self-replicating vector pEEK2. A: Schematic overviews of the constructs with single operon expressing KivdS286T and one of the respective genes to be examined. B: The construct generated to reduce the expression level of AlsS. C: Schematic overview of the constructs with two convergent orientated operons with a double terminator BBa_B0015 in between.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.