Abstract
The RGG domain, defined as closely spaced Arg-Gly-Gly repeats, is a DNA and RNA-binding domain in various nucleic acid-binding proteins. Translocated in liposarcoma (TLS), which is also called FUS, is a protein with three RGG domains, RGG1, RGG2 and RGG3. TLS/FUS binding to G-quadruplex telomere DNA and telomeric repeat-containing RNA depends especially on RGG3, comprising Arg-Gly-Gly repeats with proline- and arginine-rich regions. So far, however, only non-specific DNA and RNA binding of TLS/FUS purified with buffers containing urea and KCl have been reported. Here, we demonstrate that protein purification using a buffer with high concentrations of urea and KCl decreases the G-quadruplex binding abilities of TLS/FUS and RGG3, and disrupts the β-spiral structure of RGG3. Moreover, the Arg-Gly-Gly repeat region in RGG3 by itself cannot form a stable β-spiral structure that binds to the G-quadruplex, because the proline- and arginine-rich regions induce the β-spiral structure and the G-quadruplex-binding ability of RGG3. Our findings suggest that the G-quadruplex-specific binding abilities of TLS/FUS require RGG3 with a β-spiral structure stabilized by adjacent proline- and arginine-regions.
INTRODUCTION
The Arg-Gly-Gly repeat (RGG) domain is an evolutionarily conserved sequence found in combination with other DNA- or RNA-binding and unknown functional domains in proteins that are responsible for a variety of regulatory processes, including gene expression, pre-mRNA splicing, RNA transport and DNA damage signaling (1,2). The high density of glycine in the RGG domain suggests that it is not a rigid protein structure, and the RGG domain is thought to be a sequence- and structure-non-specific RNA-binding domain (RBD) (3). Nucleolin has an RGG domain with an RNA recognition motif (RRM) and inhibits c-myc promoter activity (4). The RRM–RGG domain in nucleolin binds to the G-quadruplex in the c-myc promoter responsible for its transcriptional activity. The RGG domain alone, however, is not sufficient for binding to G-quadruplex DNA and repressing c-myc promoter activity. In contrast, spectroscopic analysis and molecular modeling of the RGG repeat suggest that the RGG repeat forms a helical β-spiral comprising several β-turn structures, and the RGG domain of human fragile X mental retardation protein (FMRP), which has a central role in fragile X syndrome, binds to the specific RNA structure (3,5–9). Solution and crystal structures of the RGG domain in FMRP with in vitro-selected G-quadruplex-containing duplex RNA reveal that the β-turn in the RGG domain mainly recognizes the groove of a duplex–quadruplex junction (8,9). To reveal the biologic roles of RGG domain-containing proteins in cells, it is important to confirm the nucleic acid-binding specificities of each RGG domain.
Translocated in liposarcoma (TLS), also called FUS, is a multifunctional protein involved in transcription, mRNA splicing and mRNA transport from the nucleus to the cytoplasm, and contains three RGG domains (RGG1, RGG2 and RGG3), RRM and zinc finger domains (10). We previously reported that full-length TLS/FUS and RGG3 specifically bind to G-quadruplex human telomere DNA (Htelo) and telomeric repeat-containing RNA (TERRA) in vitro (11–13). In vivo, this protein regulates telomere length and histone modification by associating with histone methyl transferase in the telomere region (11). RGG3 has proline-rich and arginine-rich regions with RGG repeats, and phenylalanine and tyrosine adjacent to RGG are the key amino acids for specific binding to G-quadruplex DNA and RNA, respectively (12,13). On the other hand, Cech et al. (14) and Schwartz et al. (15) showed that TLS/FUS and RGG3 purified with buffer containing high concentrations of urea and KCl bind only weakly to DNA and RNA, irrespective of the structure and sequence. To investigate the reasons for the different DNA- and RNA-binding abilities to TLS/FUS and RGG3, we compared the DNA- and RNA-binding activities and specificities of TLS/FUS and RGG3 under different purification methods. We found that high concentrations of urea and KCl in the purification step decreased G-quadruplex-binding activity and the specificity of RGG3 due to changes in the β-spiral structure of RGG3, even if the high concentrations of urea and KCl were removed from the buffer for reaction of the protein and G-quadruplex. Moreover, the proline- and arginine-rich region in RGG3 promoted the G-quadruplex-binding activities of TLS/FUS and RGG3, and the β-spiral formation of RGG3, whereas the RGG repeat sequence alone was not sufficient for G-quadruplex binding. Our findings suggest that the nucleic acid-binding activity of the RGG domain in TLS/FUS depends on a soft secondary protein structure that is stabilized by neighboring domains.
MATERIALS AND METHODS
Plasmid constructs
The TLS/FUS, RGG1, RGG2-ZnF and RGG3 cDNAs cloned into the pGEX6P-1 vector to express an N-terminal glutathione S-transferase (GST) fusion protein were described previously (16). TLS/FUSΔP and RGG3ΔP expression vectors encoding TLS/FUS amino acids 1–526 with deletion of 449–468 and 469–526, respectively, were obtained by deletion using a KOD-Plus- Mutagenesis Kit (Toyobo, Tokyo, Japan) with the RGG3 domain or TLS/FUS in the pGEX6P-1 vector used as the template and the following primers, respectively: for TLS/FUSΔP, ΔP forward 1 d(CTT ACA CTG GTT GCA TTC ATT CCT CC) and ΔP reverse 1 d(TAC GGG GAT GAT CGT CGT GGT GG); for RGG3ΔP, ΔP forward 2 d(GGA TCC CAG GGG CCC CTG GAA C) and ΔP reverse 1. TLS/FUSΔR and RGG3ΔR expression vectors contained a polymerase chain reaction-amplifiable region encoding TLS/FUS amino acids 1–506 and 449–506, respectively, cloned into pGEX6P-1 using the following sets of primers: for TLS/FUSΔR, ΔR forward 1 d(CGG GAT CCA TGG CCT CAA ACG ATT ATA CCC) and ΔR reverse 1 d(GCC TCG AGT TAA AAG CCA CCT CTG TCC CC); for RGG3ΔR, ΔR forward 2 d(CGG GAT CCG CCC CTA AAC CAG ATG GC) and ΔR reverse 1. All constructs were verified by automated DNA sequencing. All DNA oligomers were obtained from Operon Biotechnologies (Tokyo, Japan). The RNA oligomers used for the electrophoretic mobility shift assay (EMSA) were obtained from Hokkaido System Science Co., Ltd. (Hokkaido, Japan).
Expression and purification of glutathione S-transferase fusion proteins
All recombinant proteins were fused at the N-terminus to GST and overexpressed in Escherichia coli. The E. coli strain BL21 (DE3) pLysS-competent cells were transformed with the vectors, and transformants were grown at 37°C in Luria Bertani medium containing ampicillin (0.1 mg/ml). Protein expression was induced at A600 = 0.6 with 0.1 mM isopropyl β-D-1-thiogalactopyranoside. The cells were then grown for an additional 16 h at 25°C and harvested by centrifugation (6400 × g for 20 min). The E. coli pellets were resuspended in each of three buffers; 150 mM KCl buffer (50 mM Tris–HCl [pH 7.5], 150 mM KCl, 1 mM dithiothreitol [DTT] and 1000 units/gram pellet micrococcal nuclease [Takara, Japan]); 1 M KCl buffer (50 mM Tris–HCl [pH 7.5], 1 M KCl, 1 mM DTT and 1000 units/gram pellet micrococcal nuclease); and 1 M urea buffer (50 mM Tris–HCl [pH 7.5], 150 mM KCl, 1 M urea, 1 mM DTT and 1000 units/gram pellet micrococcal nuclease). The supernatants containing the expressed proteins were lysed by sonication (model UR-20P, Tomy Seiko, Japan) and centrifuged at 16 200 × g for 15 min at 4°C. The resulting supernatants were applied to a 1-ml GSTrap FF column (GE Healthcare, USA) and washed with 20 ml of 100 mM KCl buffer (50 mM Tris–HCl [pH 7.5], 100 mM KCl). GST tags were cleaved using buffer containing 8 units/ml PreScission protease (GE Healthcare) on a GSTrap FF column for 16 h at 4°C, and the protein was eluted with 100 mM KCl buffer. Purified protein was analyzed by size-exclusion chromatography with a Superdex 200 Increase 10/300 GL column (GE Healthcare), which revealed that the protein formed a monomer, not an oligomer. The protein concentrations were determined using a BCA Protein Assay Kit (Thermo Scientific, USA). All proteins were stored at 4°C and used within 12 h after purification.
Electrophoretic mobility shift assay
32P-Labeled oligonucleotide annealing and quadruplex formation were induced by heating samples to 95°C on a thermal heating block and cooling to 4°C at a rate of 2°C/min in 50 mM Tris–HCl (pH 7.5) in the presence of 100 mM KCl. Binding reactions were performed in a final volume of 20 μl using 1 nM labeled oligonucleotide and 250 nM (Figures 1 and 3) or various concentrations of each purified protein (Figure 6) in a binding buffer (50 mM Tris–HCl [pH 7.5], 0.1 mg/ml bovine serum albumin, 1 μg/ml calf thymus DNA and 100 mM KCl). After incubating the samples for 1 h at 4°C, they were loaded on a 6% polyacrylamide (acrylamide/bisacrylamide = 19:1) non-denaturing gel. Both the gel and the electrophoresis buffer contained 0.5× TBE buffer (45 mM Tris base, 45 mM boric acid and 0.5 mM EDTA) with 50 mM KCl. Electrophoresis was performed at 10 V/cm for 2 h at 4°C. The gels were exposed in a phosphorimager cassette and imaged by a Personal Molecular Imager FX (Bio-Rad Laboratories, USA). To determine the equilibrium dissociation constants (Kd), the data from four replicate experiments were plotted as ϕ (1 fraction of free DNA) versus the protein concentration, which is equal to the protein at which half of the free DNA is bound. The Kd was extracted by non-linear regression using Microsoft Excel 2011 and the following equation: ϕ = [P]/{Kd + [P]}.
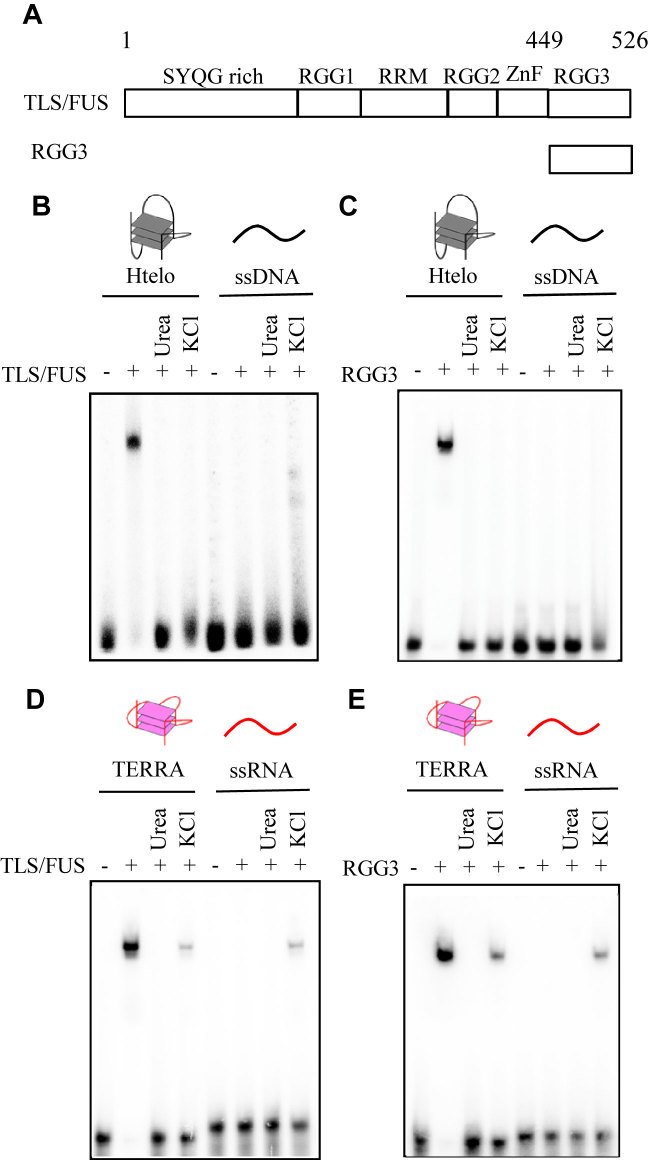
Figure 1.
Effects of urea and KCl on binding of TLS/FUS and RGG3 to the G-quadruplex. (A) Schematic illustration of TLS/FUS and RGG3. Binding ability of DNA and RNA to TLS/FUS (B and D) and RGG3 (C and E) purified under different conditions was analyzed by EMSA. TLS/FUS and RGG3 purified in the presence of 150 mM KCl (lanes 2 and 6), 1 M urea and 150 mM KCl (lanes 3 and 7) or 1 M KCl (lanes 4 and 8) were incubated with DNA or RNA. EMSA was performed with TLS/FUS or RGG3 and either 32P-labeled Htelo, ssDNA, TERRA or ssRNA. The nucleic acid–protein complexes were resolved by 6% polyacrylamide gel electrophoresis and visualized by autoradiography.
Figure 3.
Identification of the regions in TLS/FUS and RGG3 responsible for G-quadruplex binding. (A) Schematic illustration of TLS/FUS and truncated TLS/FUS (TLS/FUSΔP and TLS/FUSΔR). EMSA was performed with TLS/FUS, TLS/FUSΔP or TLS/FUSΔR, and either 32P-labeled Htelo and ssDNA (B, D and F) or TERRA and ssRNA (C, E and G). (H) Schematic illustration of RGG3 and truncated RGG3 (RGG3ΔP and RGG3ΔR). RGG sequences are underlined. Red, blue and green show proline from 449 to 448, arginine from 506 to 526 and aspartic acid from 468 to 506, respectively. EMSA was performed with RGG3, RGG3ΔP or RGG3ΔR, and either 32P-labeled Htelo and ssDNA (I, K and M) or TERRA and ssRNA (J, L and N). The nucleic acid–protein complexes were resolved by 6% polyacrylamide gel electrophoresis and visualized by autoradiography.
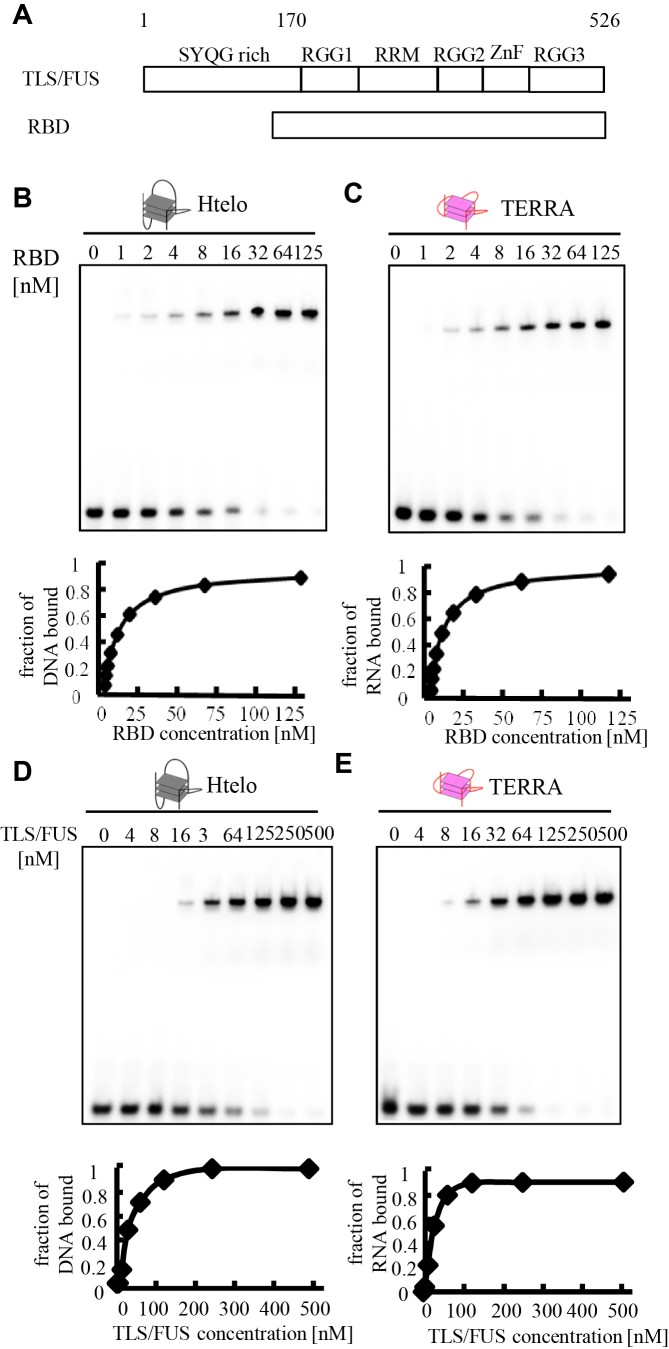
Figure 6.
Binding affinity of TLS/FUS and RBD to Htelo or TERRA. (A) Schematic illustration of TLS/FUS and RBD. The nucleic acid concentration was fixed at 1 nM while the concentration of the protein added to the binding reaction was varied, as indicated above each lane. The equilibrium-binding curve was obtained by calculating the fraction of Htelo (B and D) or TERRA (C and E) bound at each protein concentration. The binding constant (Kd) was determined by fitting to the equation. The nucleic acid–protein complexes were resolved by 6% polyacrylamide gel electrophoresis and visualized by autoradiography.
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy was performed as described previously (16). CD spectra were recorded on a model J-820 instrument (Jasco). The CD spectra of proteins in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl as specified were recorded using a 0.2-cm pathlength cell at 20°C. For each spectrum, the spectrum of the corresponding buffer was subtracted, and these data were not further processed (e.g. by smoothing).
RESULTS
Purification with high concentrations of urea and KCl decreased the binding activities of full-length TLS/FUS and RGG3 to the G-quadruplex
To investigate the effect of 1 M urea and 1 M KCl in the buffer used for protein purification on nucleic acid-binding activity, we compared the nucleic acid-binding activities of TLS/FUS and RGG3 purified with or without 1 M urea or 1 M KCl, and the buffer for protein–nucleic acid reactions was changed to 100 mM KCl buffer without urea (Figure 1). All purified proteins discussed in this paper were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Supplementary Figure S1). EMSA showed that TLS/FUS and RGG3, purified with 150 mM KCl buffer followed by 100 mM KCl buffer, bound to the G-quadruplex Htelo and TERRA but not to single-stranded DNA (ssDNA) and ssRNA (Figure 1B–E, lanes 2 and 6, Table 1). These results were consistent with our previous findings, indicating that TLS/FUS and RGG3 specifically bound to the G-quadruplex with calculated dissociation constants of 10 ± 1 nM (RGG3-telomere DNA) and 11 ± 1 nM (RGG3-TERRA) (12). TLS/FUS and RGG3 purified with 1 M urea or 1 M KCl followed by 100 mM KCl buffer, however, did not bind to the G-quadruplex or ssDNA (Kd > 250 nM; Figure 1B and C, lanes 3, 4, 7 and 8), consistent with the findings of Cech et al. (14) that RGG3 bound to G-quadruplex DNA and ssDNA with calculated dissociation constants of 730 nM (TLS/FUS-telomere DNA) and 690 nM (TLS/FUS-ssDNA). Moreover, TLS/FUS and RGG3 purified with 1 M KCl showed non-specific binding to G-quadruplex TERRA and ssRNA, and TLS/FUS and RGG3 proteins purified with 1 M urea did not bind to G-quadruplex TERRA and ssRNA (Figure 1D and E, lanes 3, 4, 7 and 8). These findings indicated that the G-quadruplex binding activities and specificities of TLS/FUS and RGG3 are decreased by high concentrations of urea and KCl—even if these reagents are removed from the reaction buffer for protein–nucleic acid binding.
Table 1. Sequence of oligonucleotides used in EMSA.
| Name | Sequence |
|---|---|
| TERRA | r(UUAGGG)4 |
| ssRNA | r(UUAGGGUUAGUGUUAGUGUUAGGG) |
| Htelo | d[AGGG(TTAGGG)3] |
| ssDNA | d(AGGGTTAGTGTTAGTGTTAGGG) |
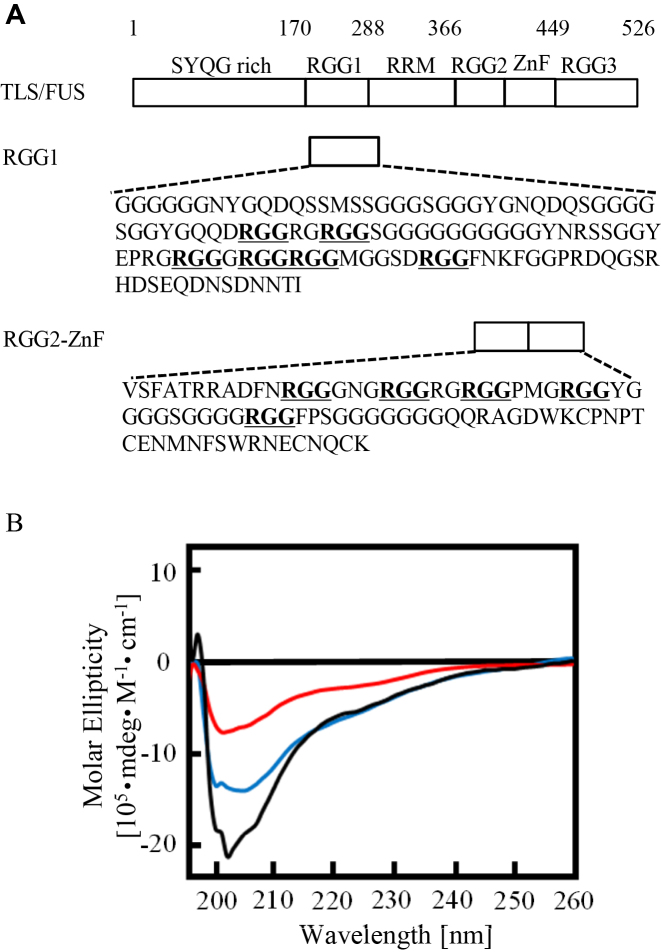
The β-spiral structure of RGG3 purified with high concentrations of urea and KCl was disordered
Urea in aqueous solution changes the hydrophobic environment around proteins and the hydrogen bonding between amino acids, thus denaturing the proteins (17). Spectroscopic analysis and molecular modeling of RGG repeats suggest that RGG repeats form a helical β-spiral comprising several β-turn structures. To investigate the effect of urea and KCl on the RGG3 structure, we performed CD spectroscopy studies of RGG3 purified with high concentrations of urea or KCl (Figure 2). The CD of RGG3 purified in the presence of 150 mM KCl without urea showed a negative band at 202 nm, which is characteristic of the β-spiral and consistent with the results of previous CD spectroscopy studies (5,7). On the other hand, the negative peak at 202 nm of RGG3 purified with a high concentration of urea or KCl in the buffer was decreased and shifted to 205 nm, especially in the case of a high concentration of KCl. The CD and nucleic acid-binding activities of RGG3 purified with 1 M urea buffer were consistent with the activities of denatured RGG3 at 50°C for 30 min (Supplementary Figure S2). These findings indicate that purification conditions that include 1 M urea or KCl induces denaturation of the β-spiral structure of RGG3, even if each buffer was exchanged with 100 mM KCl without urea after purification.
Figure 2.
CD spectra of RGG3 purified under different buffer conditions. Line colors: black, 150 mM KCl; red, 1 M urea and 150 mM KCl; and blue, 1 M KCl. The concentration of protein was 5 μM.
The proline- and arginine-rich region adjacent to the RGG repeat in RGG3 is necessary for high-affinity and specific binding to the G-quadruplex
Previously, we reported that the RGG repeat region, the proline- and arginine-rich regions of RGG3 in Ewing’s sarcoma protein, is necessary for G-quadruplex DNA binding activities and specificities (16). To investigate the effects of the proline- (amino acids 449–468) and arginine-rich regions (amino acids 506–526) of RGG3 in TLS/FUS for G-quadruplex binding activities, we performed EMSA with the G-quadruplex and deleted mutants without proline- (TLS/FUSΔP) or arginine-rich regions (TLS/FUSΔR) in TLS/FUS or proline- (RGG3ΔP) or arginine-rich regions (RGG3ΔR) in RGG3, respectively (Figure 3). TLS/FUSΔP and TLS/FUSΔR did not bind to the G-quadruplex, or to single- and double-stranded nucleic acids (Figure 3D–G and Supplementary Figure S3). These findings indicate that the proline- and arginine-rich regions are required for specific binding of TLS/FUS to the G-quadruplex. Furthermore, G-quadruplex binding activities of RGG3ΔP were not detected, whereas RGG3ΔR weakly bound to the G-quadruplex, and single-stranded and double-stranded nucleic acids without specific binding (Figure 3K–N and Supplementary Figure S4). The proline- and arginine-rich regions (RGG3ΔRGG) alone were not sufficient to bind to nucleic acids (Supplementary Figure S5B and C). In particular, the nucleic acid-binding activities of RGG3 purified with 1 M urea buffer were consistent with those of RGG3ΔP. Furthermore, the nucleic acid-binding activities of RGG3 purified with 1 M KCl buffer were consistent with those of RGG3ΔR. These findings suggest that the proline- and arginine-rich regions of RGG3 in TLS/FUS are required for specific binding to the G-quadruplex.
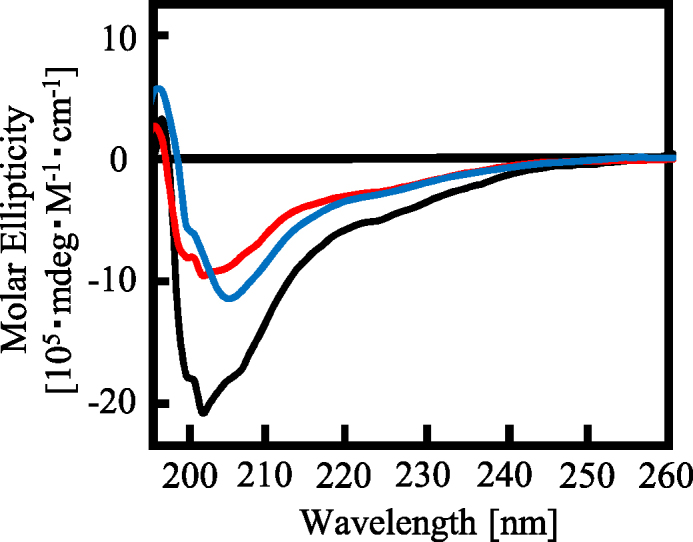
The proline- and arginine-rich regions beside the RGG repeat are necessary for RGG3 to form the β-spiral structure
The effects of the proline- and arginine-rich regions on the RGG repeat in RGG3 to form the β-spiral structure are unknown. To investigate the structure of the RGG repeat with or without the proline- and arginine-rich regions, we performed CD spectroscopy studies of RGG3ΔP and RGG3ΔR (Figure 4). The results showed that the negative peak at 202 nm of RGG3ΔP and RGG3ΔR was decreased and shifted to 205 nm, especially in the case of RGG3ΔR. The CD spectrum of RGG3 purified with 1 M KCl buffer and 1 M urea buffer was similar to that of RGG3ΔR and RGG3ΔP, respectively. The CD spectrum of RGG3ΔP did not show a shoulder signal at 200 nm, consistent with the CD signal of the poly proline II helix (18). In addition, the CD spectrum of the proline- and arginine-rich regions alone, RGG3ΔRGG, did not show the typical negative peak at 202 nm of a β-spiral structure (Supplementary Figure S5D). These findings suggest that the proline- and arginine-rich regions in RGG3 induce the formation of the β-spiral by the RGG repeat.
Figure 4.
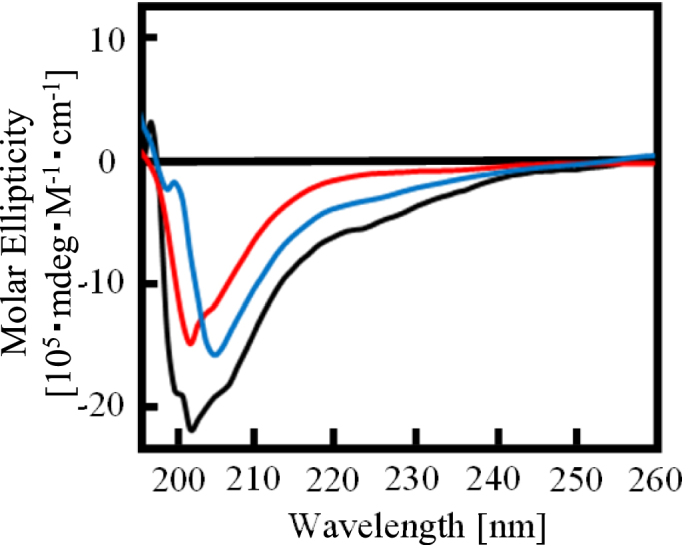
Effect of the arginine- and proline-rich region on the β-spiral formation of RGG3. CD spectra of RGG3, RGG3ΔP and RGG3ΔR. Line colors: black, RGG3; red, RGG3ΔP; and blue, RGG3ΔR. The concentration of protein was 5 μM.
TLS/FUS has three RGG domains (RGG1, RGG2 and RGG3). RGG3, which comprises an Arg-Gly-Gly repeat with proline- and arginine-rich regions, bound to G-quadruplex telomere DNA and telomeric-repeat containing RNA (11–13). On the other hand, RGG1 and RGG2, comprising an Arg-Gly-Gly repeat without proline- and arginine-rich regions, do not show specific binding to the G-quadruplex in vitro (11). To investigate the structures of RGG1 and RGG2, we analyzed the CD spectra of RGG1 and RGG2 (Figure 5). The CD spectrum of both RGG1 and RGG2 showed a decreased negative peak at 202 nm compared with that of RGG3. These findings suggest that RGG1 and RGG2 do not form a stable β-spiral structure.
Figure 5.
Comparison of the CD of RGG1, RGG2 and RGG3. (A) Amino acid sequences of RGG1 and RGG2. (B) Line colors: black RGG3; red RGG2-ZnF; blue RGG1. The concentration of protein was 5 μM.
RNA binding domain containing RGG1 and RGG2 facilitates RGG3 binding with the G-quadruplex, whereas a SYQG-rich domain inhibits the binding
Previously, we demonstrated that TLS/FUS bound to the G-quadruplex formed Htelo and TERRA (11). Especially, the RGG3 domain specifically bound to the G-quadruplex with calculated dissociation constants of 10 ± 1 nM (RGG3-telomere DNA) and 11 ± 1 nM (RGG3-TERRA) (12). On the other hand, binding specificities and constants of the TLS/FUS and RGG3 domain purified in the presence of 1 M urea and 1 M KCl were decreased, and other RGG domains in TLS/FUS were associated with G-quadruplex binding (14,15). To compare the binding affinity of the G-quadruplex and TLS/FUS or RNA binding domain (RBD) containing RGG1, RGG2 and RGG3 purified without a high concentration of urea and KCl, we examined G-quadruplex binding of TLS/FUS and RBD purified in the presence of 100 mM KCl without urea (Figure 6). The mobility shift data of RBD with Htelo or TERRA were fitted to a hyperbolic equation to give a Kd of 6.7 ± 2 and 6.2 ± 2 nM, respectively (Figure 6B and C). These results are consistent with previous results (14) suggesting that regions other than RGG3 in the RBD associate with the G-quadruplex, which slightly enhances RGG3 binding. On the other hand, the dissociation constants of TLS/FUS with telomere DNA or TERRA were 37 ± 8 and 28 ± 3 nM, indicating weaker binding with the G-quadruplex than RGG3 and RBD (Figure 6D and E). The weaker binding of TLS/FUS to the G-quadruplex was consistent with the results of a previous binding assay, in which the SYQG-rich domain bound to the C terminal domain containing RBD (19). It was reported that non-coding RNA inhibits the binding between the SYQG-rich domain and RBD by binding to RBD itself and accelerates HAT inhibition by the SYQG-rich domain (19). These results suggest that the SYQG-rich domain might affect the DNA/RNA binding of RBD.
DISCUSSION
Previously, we reported that phenylalanine and tyrosine in RGG3 are responsible for G-quadruplex DNA and RNA binding, respectively (13). Especially, tyrosine in RGG3 recognizes the 2′-OH of the riboses of the G-quadruplex loops (12). Furthermore, an NMR-based binding assay revealed that RGG3 interacts with the G-tetrad of the G-quadruplex structure formed by Htelo and TERRA (20). This paper reports that the G-quadruplex binding activities of TLS/FUS and RGG3 depend on the proline- and arginine-rich region in the C-terminal region of TLS/FUS and the purification conditions with regard to the concentrations of urea and KCl. CD spectroscopy of RGG3 revealed a β-spiral structure, but RGG3ΔP, RGG3ΔR or RGG3 purified with high concentrations of urea or KCl did not form the β-spiral structure. These findings suggest that the β-spiral structure of RGG3 is important for the specific binding to the G-quadruplex.
In particular, urea in aqueous solution changes the hydrophobic environment around the proteins and the hydrogen bonding between amino acids, thus denaturing the proteins (17). A high concentration of urea might disrupt the hydrogen bonding required to form the β-spiral structure of RGG3. Actually, the CD spectrum of denatured RGG3 under high heat conditions, which disrupts hydrogen bonding, is similar to that of RGG3 purified with urea. Moreover, RGG3 purified with urea and RGG3ΔP had a similar CD spectrum and nucleic acid-binding activities, resulting in a decrease of the negative peak at 202 nm and low binding activities to DNA and RNA. A previous report indicated that the proline-rich domain forms a hydrophobic proline helix and induces the peptide to fuse with the C-terminal to form an α-helix (21). The hydrophobic proline-rich domain in RGG3 might promote the β-spiral formation, which comprises several β-turn structures of Arg-Gly-Gly and aromatic amino acids with hydrogen bonds.
A high concentration of KCl might change the RGG3 structure by screening electrostatic interactions between the amino acids in the structure (22). Moreover, RGG3 purified with 1 M KCl and RGG3ΔR had a similar CD spectrum and nucleic acid-binding activities, resulting in a negative peak shift to 205 nm and non-specific weak binding activities to DNA and RNA. RGG3 contains arginine as a cationic-charged amino acid in the C-terminal domain and Arg-Gly-Gly repeats and aspartic acids between each Arg-Gly-Gly sequence as anionic-charged amino acids (Figure 3H). The β-spiral structure might not be induced only by hydrogen bonding, but also by electrostatic interactions in RGG3.
The RGG domain is a conserved sequence of DNA- or RBDs in several proteins. RGG1 and RGG2 in TLS/FUS cannot form a stable β-spiral structure and bind specifically to the G-quadruplex, while RGG3 can do both. Previously, we reported the importance of a proline- and arginine-rich region of the RGG domain in Ewing’s sarcoma protein for specific binding to G-quadruplex DNA (16). The proline- and arginine-rich region, however, was not able to bind G-quadruplex DNA. Therefore, it was unclear why both regions were necessary for G-quadruplex DNA binding of the RGG domain. Furthermore, the RGG domain is thought to be a disordered nucleic acid-binding region (1). Our findings suggest that each of the structures of the RGG domain is affected by its neighbor domain, thus the function of the RGG domain might depend on its secondary structure. To our knowledge, this is the first report to describe the importance of β-spiral formation for binding to the G-quadruplex by the RGG domain. These findings contribute to reveal the nucleic acid-binding activities of each RGG domain conserved in several DNA- or RNA-binding proteins. The function and localization of RGG domain-containing proteins in vivo are suggested to be regulated by arginine methylation due to protein arginine methyltransferase (1,23). The conformational and functional changes induced by arginine modifications of the RGG domains require further investigation.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
JGC-S Scholarship Foundation; Ministry of Education, Culture, Sports, Science, and Technology of Japan, Grant-in-Aid for Scientific Research (C) [17K05930 to T.O.]. Funding for open access charge: JGC-S Scholarship Foundation; Ministry of Education, Culture, Sports, Science, and Technology of Japan, Grant-in-Aid for Scientific Research (C) [17K05930 to T.O.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Thandapani P., O’Connor T.R., Bailey T.L., Richard S.. Defining the RGG/RG motif. Mol. Cell. 2013; 50:613–623. [DOI] [PubMed] [Google Scholar]
- 2. Sun X., Ali M.S.S.H., Moran M.. The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 2017; 474:2925–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burd C.G., Dreyfuss G.. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994; 265:615–621. [DOI] [PubMed] [Google Scholar]
- 4. Gonzalez V., Hurley L.H.. The C-terminus of nucleolin promotes the formation of the c-myc G-quadruplex and inhibition c-myc promoter activity. Biochemistry. 2010; 49:9706–9714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghisolfi L., Joseph G., Amalric F., Erard M.. The Glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J. Biol. Chem. 1992; 267:2955–2959. [PubMed] [Google Scholar]
- 6. Dreyfuss G., Matunis M.J., Pinol-Roma S., Burn C.G.. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 1993; 62:289–321. [DOI] [PubMed] [Google Scholar]
- 7. Renugopalakrishnan V. A 27-mer tandem repeat polypeptide in bovine amelogenin: synthesis and CD spectra. J. Pept. Sci. 2002; 8:139–143. [DOI] [PubMed] [Google Scholar]
- 8. Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Plonskai A., Chen C., Clain D. et al. . Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Vasilyev N., Polonskaia A., Darnell J.C., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA be a β-turn in the RGG motif of FMRP. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schwartz J.C., Cech T.R., Parker R.R.. Biochemistry properties and biological functions of FET proteins. Annu. Rev. Bichem. 2015; 84:355–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takahama K., Takada A., Tada S., Shimizu M., Sayama K., Kurokawa R., Oyoshi T.. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013; 20:341–350. [DOI] [PubMed] [Google Scholar]
- 12. Takahama K., Oyoshi T.. Specific binding of modified RGG domain in TLS/FUS to G-quadruplex RNA: tyrosine in RGG domain recognize 2′-OH of the riboses of loops in G-quadruplex. J. Am. Chem. Soc. 2013; 135:18016–18019. [DOI] [PubMed] [Google Scholar]
- 13. Takahama K., Miyawaki A., Shitara T., Mitsuya K., Morikawa M., Harihara M., Kino K., Yamamoto A., Oyoshi T.. G-quadruplex DNA- and RNA-specific-binding proteins engineered from the RGG domain of TLS/FUS. ACS Chem. Biol. 2015; 10:2564–2569. [DOI] [PubMed] [Google Scholar]
- 14. Wang X., Schwartz J.C., Cech T.R.. Nucleic acids-binding specificity of human FUS protein. Nucleic Acids Res. 2015; 43:7535–7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ozdilek B., Thompson V.F., Ahmed N.S., Wjote C.I., Batey R.T., Schwartz J.C.. Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 2017; 45:7984–7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Takahama K., Sugimoto C., Arai S., Kurokawa R., Oyoshi T.. Loop lengths of G-quadruplex structures affect the G-quadruplex DNA binding selectivity of the RGG motif in Ewing's Sarcoma. Biochemistry. 2011; 50:5369–5378. [DOI] [PubMed] [Google Scholar]
- 17. Zhou R., Li J., Hua L., Yang Z., Berne B.J.. Comment on ‘Urea-mediated protein denaturation: A consensus view’. J. Phys. Chem. B. 2011; 115:1323–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sakurada Y., Yonehara R., Kataoka M., Gekko K.. Secondary-structure analysis of denatured proteins by vacuum-ultraviolet circular dichroism spectroscopy. Biophys. J. 2007; 92:4088–4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R.. Induced ncRNA allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008; 454:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kondo K., Mashima T., Oyoshi T., Yagi R., Kurokawa R., Kobayashi N., Nagata T., Katahira M.. Plastic roles of phenylalanine and tyrosine residues of TLS/FUS in complex formation with the G-quadruplexes of telomeric DNA and TERRA. Sci. Rep. 2018; 8:2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Montclare J.K., Schepartz A.. Miniature homeodomains: high specificity without an N-terminal arm. J. Am. Chem. Soc. 2003; 125:3416–3417. [DOI] [PubMed] [Google Scholar]
- 22. Song B., Cho J.-H., Raleigh D.P.. Ionic-strength-dependent effects in protein folding: Analysis of rate equilibrium free-energy relationships and their interpretation. Biochemistry. 2007; 46:14206–14214. [DOI] [PubMed] [Google Scholar]
- 23. Yamaguchi A., Kitajo K.. The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS One. 2012; 7:e49267. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.