Abstract
Toxoplasma gondii virulence depends on the expression of factors packed into specific organelles such as rhoptry and microneme. Although virulence factor expression is tightly regulated, the molecular mechanisms controlling their regulation remain poorly understood. ApiAP2 are a family of conserved transcription factors (TFs) that play an important role in regulating gene expression in apicomplexan parasites. TgAP2XI-5 is able to bind to transcriptionally active promoters of genes expressed during the S/M phase of the cell cycle, such as virulence genes (rhoptries and micronemes genes). We identified proteins interacting with TgAP2XI-5 including a cell cycle-regulated ApiAP2 TF, TgAP2X-5. Using an inducible knock-down strategy and RNA-seq, we demonstrated that the level of expression of number of virulence factors transcripts is affected by the disruption of TgAP2X-5 expression. While TgAP2X-5 disruption has mild effect on parasite invasion, it leads to the strain avirulence in mice. To better understand the molecular mechanisms at stake, we investigated the binding of TgAP2XI-5 at promoters in the TgAP2X-5 mutant strain in a genome-wide assay. We show that disruption of TgAP2X-5 expression leads to defects in TgAP2XI-5 binding to multiple rhoptry gene promoters. Taken together, these data suggest a cooperative contribution of two ApiAP2 TF in the regulation of virulence genes in T. gondii.
INTRODUCTION
Toxoplasma gondii is a unicellular eukaryotic pathogen. It belongs to the Apicomplexa phylum, which encompasses some of the deadliest pathogens of medical and veterinary importance, including Plasmodium (the causative agent of malaria), Cryptosporidium (responsible for cryptosporidiosis) and Eimeria (coccidiosis). Toxoplasma gondii is an obligate intracellular parasite that leads to the development of focal central nervous system infections in patients with HIV/AIDS. In addition, Toxoplasma is also a clinically important opportunistic pathogen that can cause birth defects in the offspring of newly infected mothers. Toxoplasma gondii has a complex life cycle, which is characterized by multiple differentiation steps that are essential for its survival in both the human and definitive feline host. The common source of infection for humans is by ingesting either oocysts shed by infected cats or cyst-contaminated meats. Sporozoites and bradyzoites are contained in oocyst and cyst, respectively. These two developmental stages can differentiate into rapidly growing tachyzoites, which is the causative parasitic form responsible for the clinical manifestations in humans. Bradyzoites have the ability to evade the immune system and resist common drug treatments, therefore causing a chronic infection. However, they are also capable of reverting back to the more virulent tachyzoite stage in immunocompromised individuals.
As other apicomplexan parasites, T. gondii possesses specific organelles, namely micronemes and rhoptries, which contain proteins of critical importance for invasion, the establishment of the infection and the control of host-cell expression. These virulence factors are of significance for many aspects of virulence, for example, certain rhoptries proteins are major determinants of virulence in mice (1). Tachyzoites are able to divide rapidly inside the host cell through a specific binary replication pattern called endodyogeny (2). In 6 h, T. gondii tachyzoites of the most virulent Type I strain perform mitosis, assembly of the cytoskeletal and membrane elements that form the pellicle, loading of the growing bud with organelles and finally, emergence of new, fully formed, invasive daughters from the mother cell (3). Notably, the parasites cell cycle is divided in three phases (G1, S and M) while the G2 is apparently absent (4). The T. gondii cell cycle is a highly coordinated process where nuclear division must be synchronized with the formation of new organelles and their packaging into a newly formed parasite (4). This process implies a tight regulation of the expression of the virulence factors that must be transcribed and translated during a short window of time when organelles are formed de novo to be trafficked and packed into them. This was nicely illustrated by the transcriptome of cell cycle-synchronized tachyzoites showing that most rhoptries and micronemes transcripts have highly dynamic profiles with a peak of expression during S and M phases of the cell cycle when the corresponding organelles are formed de novo (5). As suggested for Plasmodium (6), Toxoplasma may have adapted a ‘just in time’ mode of expression whereby transcripts and proteins are produced right when their function is needed (5).
However, how gene expression is controlled in these parasites remains poorly understood. A number of sequence-specific DNA motifs where shown to be important for promoter activity (7). Mechanisms such as epigenetics were also shown to be involved in T. gondii gene regulation (8, 9). An analysis of apicomplexan genomes uncovered a family of putative transcription factors (TF) that are characterized by the possession of one or more AP2 DNA-binding domains (10). Intriguingly, more than 60 putative AP2 TFs are currently annotated in the T. gondii genome (www.toxodb.org) and among them, 24 transcripts appear to be cell cycle dependent (5). Plasmodium and Cryptosporidium AP2 proteins, which are mostly conserved in T. gondii, were shown to bind specific DNA motifs using protein-binding microarrays and recombinant proteins encompassing their respective AP2 DNA-binding domains (11). The ability of Plasmodium ApiAP2 TFs to regulate specific subsets of genes was underlined in several studies (12–16). Moreover, a comprehensive survey of the biological function of P. berghei ApiAP2 TFs has shown that a large number of genes were co-regulated by different ApiAP2 TFs, indicating that they may act in complex to regulate gene expression (17). Recently, multiple T. gondii ApiAP2 TFs were shown to control cyst formation, indicating that ApiAP2 TFs may play an important role in T. gondii gene regulation (18–21). However, the role of T. gondii ApiAP2s in regulating the expression during the tachyzoite stage remains poorly investigated. To date, only one T. gondii ApiAP2 TF was shown to bind to promoters by ChIP–chip assay (22). TgAP2XI-5 binds to transcriptionally active promoters of genes bearing a tachyzoite cell cycle-regulated expression profile peaking during the S/M phase, among which there are a number of virulence genes (22). Surprisingly, TgAP2XI-5 transcript and protein expression profile is unchanged during the tachyzoite cell cycle, indicating that other mechanisms could control its cell cycle-regulated biological function. To address this question, we identified the proteins that were complexed to TgAP2XI-5 and uncovered its association with another ApiAP2 TF, TgAP2X-5. We described that TgAP2X-5, a cell cycle-regulated ApiAP2 TF, is involved in the regulation of the expression of a number of virulence factors through its interaction with TgAP2XI-5. These data are indicative of the presence of cooperative regulation of virulence genes by an ApiAP2 TF complex in the parasite T. gondii.
MATERIALS AND METHODS
Parasite tissue culture and manipulation
Tachyzoite of T. gondii type I RHΔhxgprtΔku80 and RHΔhxgprtΔku80 TATi strain (containing an anhydrotetracycline (ATc)-inducible system and high homologous recombination; (23)), were propagated in vitro in human foreskin fibroblasts (HFF) using Dulbeccos’s modified Eagles medium supplemented with 10% fetal calf serum (FCS), 2 mM glutamine and 1% penicillin-streptomycin. Toxoplasma gondii tachyzoites were grown in ventilated tissue culture flasks at 37°C and 5% CO2. Prior to RNA, protein purification and for ChIP, intracellular parasites were purified by sequential syringe passage with 17-gauge and 26-gauge needles and filtration through a 3-μm polycarbonate membrane filter.
Generation of transgenic T. gondii strains
All primers used for polymerase chain reaction (PCR) are listed in Supplementary Table S1. Transgenic TgAP2XI-5-myc, TgAP2X-5-HA and TgHP-HA parasites were generated with a knock-in strategy. Genomic DNA of type I RHΔhxgprtΔku80 strain was isolated with the Wizard Genomic DNA Purification kit (Promega). DNA fragment of 2-kb upstream of the stop codon of TgAP2XI-5 (TGGT1_216220), TgAP2X-5 (TGGT1_237090) and a hypothetical protein (HP) TgHP (TGGT1_297460) genes were amplified using genomic DNA and cloned into either pLIC.myc.DHFR or pLIC.3xHA.HXGPRT vectors (generously provided by Dr Carruthers, U. Michigan) (24). Transgenes were introduced by electroporation into 10 × 106 freshly egressed tachyzoites of T. gondii strains. RHΔhxgprtΔku80 strain was transfected with 15 μg of the plasmid pLIC HP-HA (linearized NcoI), pLIC AP2X-5-HA (linearized ApaI) and/or pLIC AP2XI-5-myc (linearized ApaI). Stable transformants were selected in the presence of 2 μM pyrimethamine (for the DHFR selection) and 25 μg/ml mycophenolic acid and 50 μg/ml xanthine (for the HXGPRT selection). Clonal lines were obtained by limiting dilution. The TgAP2X-5 inducible knock-down (iKD) line was generated using a plasmid with the DHFR cassette containing genomic fragments encompassing 2-kb upstream the gene and 2 kb from the predicted ATG. RHΔhxgprtΔku80 TATi parasites were transfected with 30 μg of the iKD digested plasmid (NotI and ApaI) followed by 2 μM pyrimethamine selection. For complementation, 50 μg of a synthetized pUPRT plasmid containing 3-kb upstream the predicted ATG and the full length c-myc tagged TgAP2X-5 gene (GeneCust) was co-transfected with the pSAG1::Cas9-U6::sgUPRT plasmid (25) in the iKD TgAP2X-5 strain to facilitate the insertion into the uracil phophoribosyltransferase locus. Stable transgenic and cloned parasites were selected with 5 μM 5-fluoro-2′-deoxyuridine (FUDR). Transgenic TgAP2XI-5-TAP was generated with a knock-in strategy as described above by transfecting 15 μg of the plasmid pLIC AP2XI-5-TAP (linearized ApaI) in 10 × 106 TgAP2X-5 iKD and iKD TgAP2X-5 complemented tachyzoites followed by chloramphenicol selection (34 mg/ml).
Parasite growth assay
For parasite growth assay, RHΔhxgprtΔku80 TATi, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains were pre-treated 48 h with ATc. Then, 105 parasites per well in a 24-well plate were inoculated on HFF monolayer grown on coverslips for 4 h in normal media or media with ATc and let grow 29 h, before 4% paraformaldehyde (PFA) fixation. The number of parasites per vacuole was counted for 100 vacuoles for each condition and performed twice after TgEno2 staining. Three independent experiments were performed.
Invasion assay
Two-color invasion assay was adapted from (26). Briefly, 2 × 106 freshly egressed tachyzoites were first sedimented on ice for 30 min on HFF monolayer grown on coverslips in 24-well plates. Invasion was allowed during 90 min at 37°C, extensively washed to eliminate unbound parasites and fixed in 2% PFA in Hank’s Balanced Salt Solution. Attached parasites were stained with mouse anti-TgSag1 and Alexa Fluor-594. After subsequent permeabilization with 0.01% saponin for 15 min, a second staining was performed using rabbit anti-TgGAP45 and Alexa Fluor-488. The number of extracellular and intracellular parasites was counted from nine fields (three times per coverslips) from three independent biological experiments.
In vivo experiments
A group of 4 female BALB/C mice (7-week-old) were intraperitoneally injected with 2 × 106 tachyzoites of parental (RHΔhxgprtΔku80 TATi), iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains. The drinking water was supplemented with 0.2 mg/ml of ATc and 5% of sucrose to suppress TgAP2X-5 HA expression. Survival was monitored over 15 days. All animal experiments were performed following the Pasteur Institute of Lille guidelines on animal study board, which conforms to the Amsterdam Protocol on animal protection and welfare. The animal work also complied with the French law (no. 87–848 dated 19 October 1987) and the European Communities Amendment of Cruelty to Animals Act 1976. All animals were fed with regular diet and all procedures were in accordance with national regulations on animal experimentation and welfare authorized by the French Ministry of Agriculture and Veterinary committee (permit number: 2016-0049).
Antibodies
The anti-TgEno2 rabbit (27), anti-TgSag1 T41E5 mouse (28), anti-TgChromo1 mouse (29) and anti-TgGAP45 rabbit (kindly provided by Dr Soldati) antibodies were used in IFA at 1:1000, 1:1000, 1:200 and 1:10 000, respectively. Anti-HA rabbit (Eurogentech) and rat (Invitrogen) antibodies were used at 1:500 in IFA and in western blots. Anti-myc mouse (ThermoFisher) was used at 1:200 in IFA and 1:500 in western blot. Anti-TgROP2 mouse T52D1 (kindly provided by Dr Lebrun) was used at 1:500 in western blot. Anti-TgActin mouse was used at 1:1000 in western blot.
Immunofluorescence assay and confocal microscopy
Intracellular parasites were fixed with 4% PFA for 15 min, followed by three phosphate-buffered saline (PBS) washes. Parasites were permeabilized with 0.1% Triton X-100 in PBS containing 0.1% glycine for 20 min at room temperature. Samples were blocked with FCS in the same buffer and the primary antibodies were added on parasites also in the same buffer for 1 h at room temperature. Secondary antibody coupled to Alexa Fluor-488 or to Alexa Fluor-594 (Molecular probes) diluted at 1:1000 was added in addition to DAPI for nucleus staining. Confocal imaging was performed with a ZEISS LSM880 Confocal Microscope or Apotome Microscope at 63 magnification. All images were processed using Carl Zeiss ZEN software.
Co-immunoprecipitation
Intracellular parasites (5 × 108 tachyzoites) of the TgAP2XI-5-myc, TgAP2XI-5-myc/TgAP2X-5-HA and TgAP2XI-5-myc/TgHP-HA RHΔhxgprtΔku80 strains were purified on a 3-μm filter and washed three times with PBS. The parasite pellet was resuspended in 1 ml of NEB1 buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM 1,4-Dithiothreitol (DTT), 0.1 mM ethylenediaminetetraacetic acid (EDTA), 0.65% NP40 and 0.5 mM Phenylmethanesulfonyl fluoride (PMSF)), incubated on ice for 10 min and centrifuged at 1500 × g for 10 min at 4°C. The pellet was then resuspended with 100 μl of buffer NEB2 (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 0.5 mM DTT, 25% glycerol and 0.5 mM PMSF) for 10 min on ice and centrifuged at 12 000 × g for 10 min at 4°C. The supernatant was kept as the nuclear extract. Then, 30 μl of beads Pierce™ Anti-c-Myc-Agarose and Pierce™ HA Epitope Tag Antibody, Agarose Conjugate (Thermo Scientific) were washed twice with Tris buffer saline (TBS) 1× solution (50 mM Tris–HCl, 150 mM NaCl and 0.5 mM PMSF) and centrifugated at 2500 × g for 1 min between each wash. Beads were saturated by using 5 μl of bovine serum albumin (1 mg/ml) in 500 μl of TBS 1× under stirring for 15 min at 4°C, and washed twice as previously mentioned. The nuclear extract was added to 4 ml of TBS 1× containing the anti-c-Myc or HA Epitope beads and incubated overnight at 4°C under stirring. The next day, beads were washed five times with 3 ml of 1× TBS-T (1× TBS, 1% Tween and 0.5 mM PMSF) and one time with 3 ml of final wash buffer (62.5 mM Tris pH 6.8 and 0.5 mM PMSF), by centrifugation at 2500 × g for 1 min between each wash. Immunoprecipitated proteins were eluted twice with 75 μl of elution buffer (62.5 mM Tris pH 6.8, 2% sodium dodecyl sulphate (SDS), 0.1M DTT), then boiled at 95°C for 5 min and finally centrifuged at 14 000 × g for 1 min. Both elution were pooled and analyzed by western blotting before proteomics analysis.
Mass-spectrometry proteomic analysis
After denaturation at 100°C in 5% SDS, 5% β-mercaptoethanol, 1 mM EDTA, 10% glycerol and 10 mM Tris pH 8 buffer for 3 min, protein samples were fractionated on a 10% acrylamide SDS-polyacrylamide gel electrophoresis gel. The electrophoretic migration was stopped as soon as the protein sample entered 1 cm into the separating gel. The gel was briefly labeled with Coomassie Blue, and five bands, containing the whole sample, were cut. In gel, digestion of gel slices was performed as previously described (30).
An UltiMate 3000 RSLCnano System (Thermo Fisher Scientific) was used for separation of the protein digests. Peptides were automatically fractionated onto a commercial C18 reversed phase column (75 μm × 150 mm, 2 μm particle, PepMap100 RSLC column, Thermo Fisher Scientific, temperature 35°C). Trapping was performed during 4 min at 5 μl/min, with solvent A (98% H2O, 2% ACN and 0.1% FA). Elution was performed using two solvents A (0,1% FA in water) and B (0.1% FA in ACN) at a flow rate of 300 nl/min. Gradient separation was 3 min at 5% B, 37 min from 5% B to 30% B, 5min to 80% B and maintained for 5 min. The column was equilibrated for 10 min with 5% buffer B prior to the next sample analysis.
The eluted peptides from the C18 column were analyzed by Q-Exactive instruments (Thermo Fisher Scientific). The electrospray voltage was 1.9kV, and the capillary temperature was 275°C. Full MS scans were acquired in the Orbitrap mass analyzer over m/z 300–1200 range with resolution 35 000 (m/z 200). The target value was 5.00E + 05. Ten most intense peaks with charge state between 2 and 4 were fragmented in the Higher-energy collisional dissociation (HCD) collision cell with normalized collision energy of 27%, and tandem mass spectrum was acquired in the Orbitrap mass analyzer with resolution 17 500 at m/z 200. The target value was 1.00E+05. The ion selection threshold was 5.0E+04 counts, and the maximum allowed ion accumulation times were 250 ms for full MS scans and 100 ms for tandem mass spectrum. Dynamic exclusion was set to 30 s.
Proteomic data analysis
Raw data collected during nanoLC-MS/MS analyses were processed and converted into *.mgf peak list format with Proteome Discoverer 1.4 (Thermo Fisher Scientific). MS/MS data were interpreted using search engine Mascot (version 2.4.0, Matrix Science, London, UK) installed on a local server. Searches were performed with a tolerance on mass measurement of 0.2 Da for precursor and 0.2 Da for fragment ions, against a composite target decoy database (50 620 total entries) built with three strains of T. gondiiToxoDB.org database (strains ME49, GT1 and VEG, release 12.0, September 2014, 25 264 entries) fused with the sequences of recombinant trypsin and a list of classical contaminants (46 entries). Cysteine carbamidomethylation, methionine oxidation, protein N-terminal acetylation and cysteine propionamidation were searched as variable modifications. Up to one trypsin, missed cleavage was allowed. For each sample, peptides were filtered out according to the cut-off set for proteins hits with peptides taller than eight residues, ion score > 35, identity score > 10 and a false positive rate of 0.7%. SAINT analysis was performed as previously described (31, 32), except that three biological replicates were used for the bait (TgAP2XI-5-myc) and negative control (parental strain).
Library preparation and RNA-seq
RNA was extracted as previously described, followed by genomic DNA removal and cleaning using an RNase-free DNase I Amplification Grade kit (Sigma). An Agilent 2100 Bioanalyzer was used to assess the integrity of the RNA samples. Only RNA samples having an RNA integrity score between 8 and 10 were used. Library preparation was performed using the TruSeq Stranded mRNA Sample Preparation kit (Illumina) according to the manufacturer’s instructions. Libraries were validated using a Fragment Analyzer and quantified by quantitative PCR (qPCR) (ROCHE LightCycler 480). Clusters were generated on a flow cell within a cBot using the Cluster Generation Kit (Illumina), and libraries were sequenced as 50-bp reads on a HiSeq 2000 using a sequence by synthesis technique (Illumina).
Image analysis and base calling were performed using the HiSeq control software and real-time analysis component. Demultiplexing was performed using Illumina’s conversion software (bcl2fastq 2.17). The quality of the data was assessed using FastQC from the Babraham Institute and the Illumina software Sequence Analysis Viewer. Potential contaminants were investigated with the FastQ Screen software from the Babraham Institute.
RNA-seq reads were aligned to the T. gondii genome (ToxoDB-25_TgondiiGT1_Genome.fasta from the ToxoDB Toxoplasma Genomics Resource, downloaded on 31 August 2015) with a set of gene model annotations (ToxoDB-25_TgondiiGT1.gff from the ToxoDB Toxoplasma Genomics Resource, downloaded on 31 August 2015) using the splice junction mapper TopHat 2.0.13 (33). Final read alignments having more than three mismatches were discarded. Gene counting was performed using HTSeq-count 0.6.1p1 (union mode) (34). Because the data come from a strand-specific assay, the read must be mapped to the opposite strand of the gene. Before statistical analysis, genes with <15 reads (combining all the analyzed samples) were filtered out. Differentially expressed genes were identified using the Bioconductor package edgeR 3.6.7 (35). The data were normalized using the relative log expression (RLE) normalization factors. Genes with adjusted P-values < 5% (according to the False discovery rate (FDR) method from Benjamini–Hochberg) were declared differentially expressed. The full dataset is deposited at the GEO database under the accession number GSE106864.
Chromatin immunoprecipitation
ChIP was performed using a protocol previously described (22) on TgAP2XI-5-TAP, iKD TgAP2X-5/TgAP2XI-5-TAP and iKD TgAP2X-5 complemented/TgAP2XI-5-TAP strains. Briefly, intracellular parasites were grown 40 h with ATc and fixed for 10 min at room temperature using 1% formaldehyde. DNA was processed by sonication using the Bioruptor device for 10 min at 4°C with a 30 s on/off cycle. Protein–DNA complexes were then diluted in IP Dilution Buffer (16.7 mM Tris–HCl pH8, 167 mM NaCl, 1.2 mM EDTA, 0.01% SDS, 1.1% Triton and 0.5 mM PMSF) and incubated ON with IgG Agarose beads (Amersham Biosciences). The next day, beads were washed five times with ChIP wash buffer (50 mM Tris–HCl pH8, 250 mM NaCl, 1% NP-40, 1% desoxycholic acid (Fischer Scientific MW 414.5 Cat.# BP349-100) and 0.5 mM PMSF) and eluted twice in 75 μl of ChIP elution buffer (50 mM NaHCO3 and 1% SDS). ChIP DNA purification was carried out as previously described in (8). Amplification of immunoprecipitated DNA and 10 ng input DNA was performed using the GenomePlex® Complete Whole Amplification (WGA) kit (Sigma). A tilling microarray was designed based on version 6 of the T. gondii ME49 genome (version 7.3) and printed by Agilent Technologies (22). The microarray encompasses more than 983 000 features representing the entire genome with an average coverage of one oligonucleotide every 63 bp. Purified ChIP material was processed according to the Agilent Mammalian ChIP-on-chip protocol version 10.11, and labeled DNA was hybridized to an Agilent T. gondii tiling array for 40 h at 67°C (G4481-90010; Agilent Technologies). Microarrays were washed and scanned according to the manufacturer’s protocol, and the results were processed with the Genomic Workbench Standard edition. The mPeak software (36) was used with default values to identify peaks among the TgAP2XI-5 ChIP on chip experiments. The full dataset is deposited at the GEO database under the accession number GSE109086.
Real-time quantitative PCR (RT-qPCR)
All primers were designed using Primer3Plus (https://primer3plus.com/cgi-bin/dev/primer3plus.cgi) and are listed in Supplementary Table S1. Tachyzoite RNA was isolated using Trizol Reagent (cat #15596018, Invitrogen) according to manufacturer’s protocol, and purified after an RNase-free DNase I Amplification grade treatment (Sigma). An Agilent 2100 Bioanalyzer (RNA 6000 Nano kit) was used to assess the integrity of the RNA samples that were then quantified with a NanoVue Plus Spectrophotometer (GE Healthcare). Total cDNA was generated using the Maxima Reverse Transcriptase kit (Thermo Scientific) with 5 μg of total RNA and oligo (dT) in a final volume of 20 μl as recommended by the manufacturer. Real-time qPCR (RT-qPCR) was performed using the SYBR® Selected Master Mix (Life technologies) without ROX according to manufacturer’s instructions, using 3 μM of each primer, ∼10 ng of cDNA in nuclease-free water to a final volume of 20 μl. The Mx3000P system (Agilent Technologies) was used with the following cycling conditions: 1 cycle of 50°C for 2 min, 1 cycle of 95°C for 10 min, 40 cycles of 95°C for 30 s, 60°C for 30 s before fluorescence detection followed by a dissociation curve determined with 1 cycle of 95°C for 1 min, 55°C for 30 s and 95°C for 30 s. Quantification cycles (Cq) were determined using the MxPro qPCR software (Agilent Technologies). Change in expression of individual genes is represented by relative fold change in the Cq-values as ΔΔCq (TgEno2 served as reference gene). All experiments were performed on separate biological experiments in duplicate (ChIP) or triplicate (RNA-seq).
Bioinformatic tools
The Phyre2 web portal (http://www.sbg.bio.ic.ac.uk/phyre2) (37) was used to produce homology-based model of the TgAP2X-5 AP2 domain to predict its three-dimensional structure. The MEME Suite sequence analysis tool (GOMo application; (38)) and the Regulatory Sequence Analysis Tools (39) were used to highlight overrepresented motif on promoters of genes downregulated in iKD TgAP2X-5 strain. Analysis was performed on the 2000 bp downstream the transcription start as the 5′ untranslated region is not known for all genes identified.
Statistical analysis
All data were analyzed with Graph Pad Prism software (San Diego, CA, USA). Differences in the means were assessed by two-way ANOVA and Student’s t-test. In both cases, P < 0.05 was considered as significant.
RESULTS
TgAP2XI-5 interacts with TgAP2X-5
To identify proteins that may regulate the activity of TgAP2XI-5, we performed an immunoprecipitation of this protein using an epitope tagged version of TgAP2XI-5. The strain expressing TgAP2XI-5-myc was produced using a knock-in strategy that allowed the tagging of the gene at its endogenous locus and its expression under its own promoter. TgAP2XI-5-myc was immunoprecipitated and the proteins that co-immunoprecipitated were identified by mass spectrometry analysis (Supplementary Table S2). As a negative control, we performed the same procedure using protein extracts from the parental strain. In three experiments, we identified peptides from the TgAP2XI-5 protein in the IP fraction from the TgAP2XI-5-myc strain but not from the parental strain (only one peptide was identified in one of the control experiments). The protein that was identified as co-immunoprecipitated with TgAP2XI-5-myc in the three experiments with more than one unique peptide is presented in Table 1.
Table 1. Identified proteins after TgAP2XI-5-myc protein immunoprecipitation.
| Total number of peptide | ||||||||
|---|---|---|---|---|---|---|---|---|
| Protein name | Protein accession numbers | Protein molecular weight (Da) | Sample 01 control - | Sample 01 Myc | Sample 02 control - | Sample 02 Myc | Sample 03 control - | Sample 03 Myc |
| AP2 domain TF AP2XI-5 | TGGT1_216220 | 89 485 | 25 | 19 | 1 | 20 | ||
| AP2 domain TF AP2X-5 | TGGT1_237090 | 211 315 | 3 | 8 | 4 | |||
The table includes the protein name, accession number, predicted molecular weight and the number of unique peptide identified for each experiment. Three controls (Control1–3; Parental line immunoprecipitated with the myc antibodies) and immunoprecipitation (AP2XI-5 elution 1–3) experiments were performed. Blank cells represent 0 peptides identified for the corresponding protein. We present here the protein that was identified as co-immunoprecipitated with TgAP2XI-5-myc in the three experiments with more than one unique peptide.
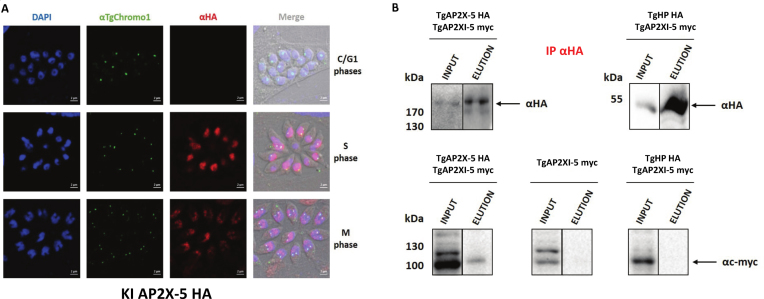
We identified peptides originating from another ApiAP2 TF, TgAP2X-5, with the protein extracts from the TgAP2XI-5-myc strain but not from the parental strain. After TgAP2XI-5, TgAP2X-5 presents the highest number of peptides identified present in the IP fraction and no peptides in the controls. We also ran the SAINT software (31) to identify the proteins with the highest probability of interacting with TgAP2XI-5. This analysis yielded two proteins with high probability scores (>0.9, see Supplementary Table S2). TGGT1_260810 was not considered for further analysis since it contains a putative transmembrane domain. TgAP2X-5 was further characterized. TgAP2X-5 is a cell cycle-regulated gene as identified by transcriptomics (5) and using an epitope tagged strain using the knock-in strategy (Figure 1A). Using IFA and a marker of the cell cycle (TgChromo1, a marker of pericentromeric chromatin), we showed that TgAP2X-5 is a nuclear protein predominately expressed during the S phase (two centromere dots) and M phase (two centromere dots and bell-shaped nucleus) of the cell cycle. TgAP2X-5 expression was barely detectable during the G1 phase (one TgChromo1 dot) of the cell cycle (Figure 1A). The AP2 domain amino-acid sequence of TgAP2X-5 is not well conserved in other apicomplexan parasites in contrast to that of TgAP2XI-5. However, bioinformatic prediction indicates that it presents the canonical secondary structure features common to AP2 DNA binding domain with three β-strands (contained in one beta-sheet) and an α-helix (Supplementary Figure S1A).
Figure 1.
TgAP2X-5, an ApiAP2 protein is cell cycle-regulated and interacts with TgAP2XI-5. (A) Expression of the TgAP2X-5 protein (anti-HA antibody) during the tachyzoite cell cycle by confocal imaging. Anti-TgChromo1 antibody was used as marker of the cell cycle. DAPI was used to stain the nucleus. The cell cycle stage is indicated on the right side of the figure. Scale bar is indicated on the lower right side of each image. (B) TgAP2X-5 interacts with TgAP2XI-5. Nuclear extract of 3 × 108 parasites for the TgAP2X-5-myc alone (negative control) or the TgAP2X-5-HA/TgAP2X-5-myc strain or TgHP-HA/TgAP2X-5-myc strain were immunoprecipitated using anti-HA antibodies. The immunoblots were probed with anti-HA antibody (upper panel) to detect both HA-tagged proteins in the input and eluates. These immunoblots were reprobed with anti-myc antibody (lower panel) to detect the TgAP2XI-5-myc protein in the input and eluates.
To verify the interaction between TgAP2X-5 and TgAP2XI-5, we performed a reverse IP using a strain where both genes are endogenously tagged (TgAP2X-5-HA and TgAP2XI-5-myc). As a negative control, we also performed an IP on a strain only expressing TgAP2XI-5-myc or on a strain expressing TgAP2XI-5-myc and TgHP-HA, a protein constitutively expressed and not found in the TgAP2XI-5 co-IPed proteins (Figure 1B). After performing the IP with an anti-HA antibody, we were able to detect both TgAP2X-5-HA and TgHP-HA proteins (Figure 1B, upper panel). However, when we probed the same blots using an anti-myc antibody, only the eluates from the TgAP2X-5-HA/TgAP2XI-5-myc strain were positive for the presence of TgAP2XI-5-myc indicating that TgAP2X-5 is indeed able to co-immunoprecipitate TgAP2XI-5 (Figure 1B, lower left panel). We failed to detect TgAP2XI-5 in the eluates from the strain only expressing TgAP2XI-5-myc or expressing TgAP2XI-5-myc and TgHP-HA confirming the specificity of the TgAP2X-5/TgAP2XI-5 interaction. We also confirmed this interaction using an IP with an anti-myc antibody (Supplementary Figure S1B) with the same negative controls.
TgAP2X-5 depletion cause a downregulation of multiple virulence genes
To better characterize the biological role of TgAP2X-5, we used a promoter replacement strategy to produce an iKD mutant strain. In this strain, the expression of the tgap2x-5 transcript is under the control of ATc (Supplementary Figure S2A). When ATc is added to the culture media, the inducible promoter is repressed and the tgap2x-5 transcript is no longer produced. We simultaneously added an HA-tag to the N-terminus of the gene in order to follow the protein expression (Supplementary Figure S2A). The correct insertion of the construction at the TgAP2X-5 locus was validated by genomic PCR (Supplementary Figure S2B). Western blots of total protein extracts from this transgenic parasite line revealed the expression of a tagged polypeptide at the predicted size (210 kDa) in the absence of ATc (Supplementary Figure S2C). In the presence of ATc, we observed a drastic reduction in the signal from the tagged protein, with an undetectable level after 48 h of treatment (Supplementary Figure S2C) that was confirmed by RT-qPCR (Supplementary Figure S2D). We also produced an iKD TgAP2X-5 complemented strain expressing the TgAP2X-5-myc-tagged protein from a dispensable locus (uprt) under the control of its own promoter (Supplementary Figure S3A). In presence of ATc, this strain expressed similar level of tgap2x-5 transcript than the parental strain as assayed by RT-qPCR (Supplementary Figure S3B). Moreover, the cell-cycle expression of TgAP2X-5-myc is conserved with a peak of expression during the S phase of the cell cycle as assessed by IFA using TgChromo1 as a cell-cycle marker (Supplementary Figure S3C).
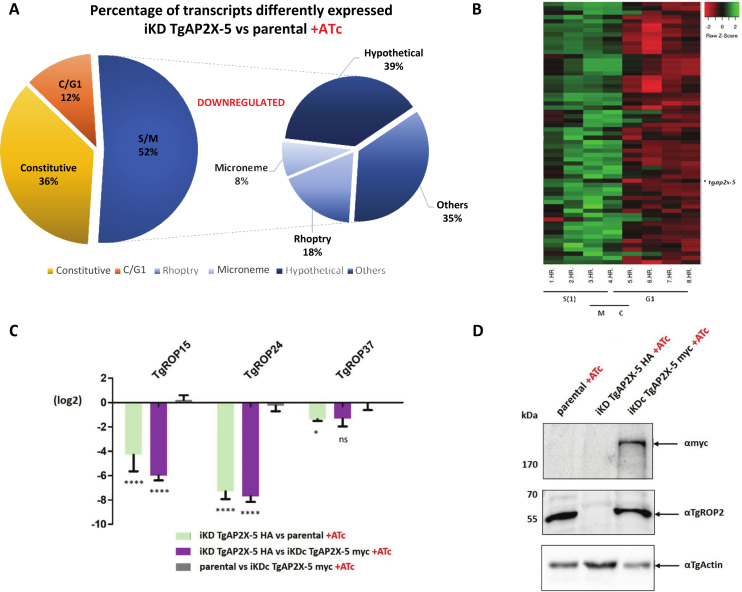
Since TgAP2X-5 is a potential TF, we examined the differential expression of genes when this protein is depleted (in presence of ATc) by RNA-sequencing (RNA-seq). Total RNA was purified from tachyzoites of the iKD TgAP2X-5 strain grown under ATc treatment for 48 h (in triplicate). RNA-seq reads were aligned to the T. gondii genome with a set of gene model annotations using the splice junction mapper TopHat 2.0.13. The data were normalized using the RLE normalization factors. Genes with adjusted P-values < 5% (according to the FDR method from Benjamini–Hochberg) were considered differentially expressed. By using edgeR (35), data analysis revealed significant changes in the transcription profile in the ATc treated iKD TgAP2X-5 parasites when compared to the parental, with 153 genes downregulated and 70 upregulated (Supplementary Table S3). The predominance of downregulated genes suggests a potential role for TgAP2X-5 in activation of gene expression. The majority of the genes with a differential expression were cell cycle-regulated with a peak of expression during the S/M phases of the cell cycle as TgAP2X-5 (Figure 2A and B). Among them were found transcripts coding for virulence factors such as rhoptries and micronemes proteins. We use RT-qPCR to verify the results obtained by RNA-seq and showed that the selected virulence genes had their expression downregulated when TgAP2X-5 is depleted while these gene had WT levels (Figure 2C) in the iKD TgAP2X-5 complemented strain. We also verified that this transcript downregulation was translating into a decreased protein levels. For that, we performed a western-blot using anti-TgROP2 antibody and showed a drastic reduction of TgROP2 expression in the iKD TgAP2X-5 strain in presence of ATc that returned to the WT level in the iKD TgAP2X-5 complemented strain (Figure 2D). Of note, only one annotated rhoptry protein (ROP20) is found in the overexpressed gene list (Supplementary Table S3).
Figure 2.
TgAP2X-5 depletion alters the expression of multiple virulence genes. (A) Pie chart representing the cell-cycle expression of transcripts downregulated in the iKD TgAP2X-5 strain compared to parental line under ATc treatment. Peak cell-cycle expression was identified based on available data (5) and the EdgeR test was used for RNA-seq data processing. The right panel represent the proportion of genes that were annotated as micronemes, rhoptries or hypotheticals proteins. Others represents gene that were annotated but were not micronemes, rhoptries or hypotheticals proteins. (B) Heat map of all individual transcript downregulated in the iKD TgAP2X-5 strain presenting an S/M transcription profile according to the cell-cycle expression profiles (5). (C) Total RNA purified from parental, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains under ATc treatment were analyzed by RT-qPCR for ROPs genes. Values are represented as the log2 ratio. Comparison of the relative transcript level was made between the iKD TgAP2X-5 strain and the parental strain in presence of ATc (green bars), the iKD TgAP2X-5 strain and the iKD TgAP2X-5 complemented strain in presence of ATc (purple bars) and the parental strain and the iKD TgAP2X-5 complemented strain in presence of ATc (gray bars). RT-qPCR and RNA-seq data are reported in this graph. A two way ANOVA-test was performed, ****P < 0.0001; **P < 0.01; *P < 0.05; ns P > 0.05 mean ± s.d. (n = 3). (D) Western blot of total protein extract from parental, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains in the presence of ATc was probed with anti-myc to detect the presence of TgAP2X-5 protein expressed from the uprt locus (upper panel). Western were also probed with an anti-TgROP2 antibodies, to detect TgROP2 which transcript is highly downregulated in the iKD TgAP2X-5 strain (middle panel). TgActin was probed as a loading control (lower panel).
TgAP2X-5 depletion has a drastic effect on in vivo virulence
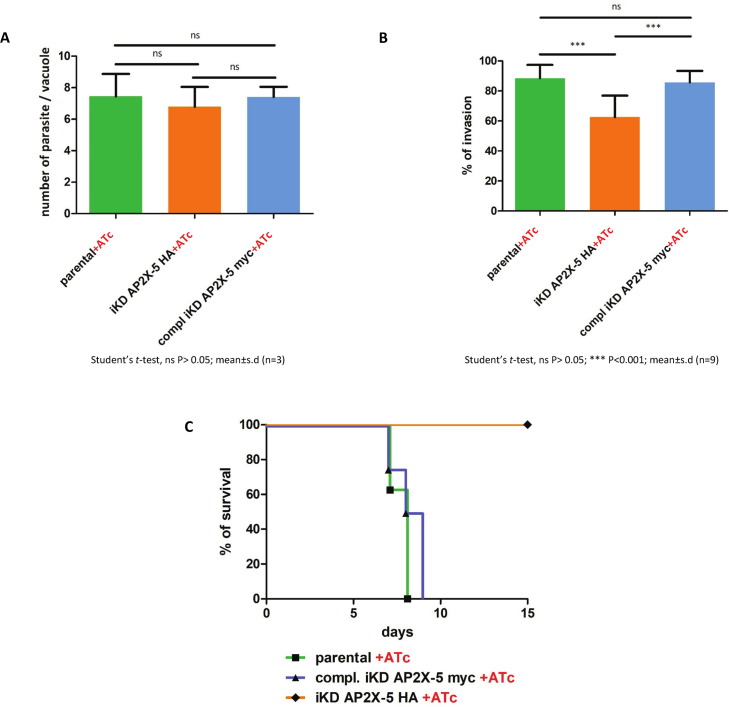
To further characterize the defects caused by TgAP2X-5 depletion, we performed standard growth and invasion assays (Figure 3). We tested the ability of the parasite to grow under ATc treatment and recorded the number of parasites per vacuole at a given time (Figure 3A). We observed that the parental strain showed similar growth to that of the iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains. This indicates that the growth of the iKD TgAP2X-5 parasite is not significantly impaired by the absence of TgAP2X-5 (Figure 3A). We also tested the ability of the parasites to invade host cells (Figure 3B). In this assay, the protocol allows to distinguish extracellular parasites that are attached to the host cell from intracellular parasites after invasion. While the parental strain with ATc showed similar number of invaded parasites to that of the iKD TgAP2X-5 complemented strain, we noticed that the iKD TgAP2X-5 strain invaded significantly less the host cells (Figure 3B). Altogether, these data indicate that the parasites depleted for TgAP2X-5 are affected in terms of their invasion ability. We then tested the iKD TgAP2X-5 strain for virulence in vivo (Figure 3C). As a control, we also infected mice with the parental strain and the iKD TgAP2X-5 complemented strain in presence of ATc. Mice infected with the iKD TgAP2X-5 strain survived the infection while the parental and iKD TgAP2X-5 complemented strains were able to kill mice within 10 days, indicating a drastic effect of the depletion of the TgAP2X-5 protein on in vivo virulence (Figure 3C). Mice that survived the iKD TgAP2X-5 strain infection were indeed infected since their sera responded to a whole parasite extract in western blots (Supplementary Figure S4).
Figure 3.
TgAP2X-5 depletion has a drastic effect on in vivo virulence. (A) Parasite growth was assayed for parental, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains under ATc treatment for 24 h. The number of parasite per vacuole was scored for a minimum of 100 vacuoles and the average number of parasite is represented in this graph. A Student’s t-test was performed, ns P > 0.05; mean ± s.d. (n = 3). (B) Host cell invasion assay. Parental, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains pretreated with ATc were allowed to invade host cell for 90 min and then fixed and processed for a two-color invasion assay. The experiment was performed three-times in triplicate yielding nine measurements. A Student’s t-test was performed, ns P > 0.05; ***P < 0.001; mean ± s.d. (C) Survival of mice infected with parental, iKD TgAP2X-5 and iKD TgAP2X-5 complemented strains was determined for 15 days under ATc treatment. Mice were infected with 2.106 parasites intraperitoneally.
TgAP2X-5 depletion prevents TgAP2XI-5 binding to a number of promoters
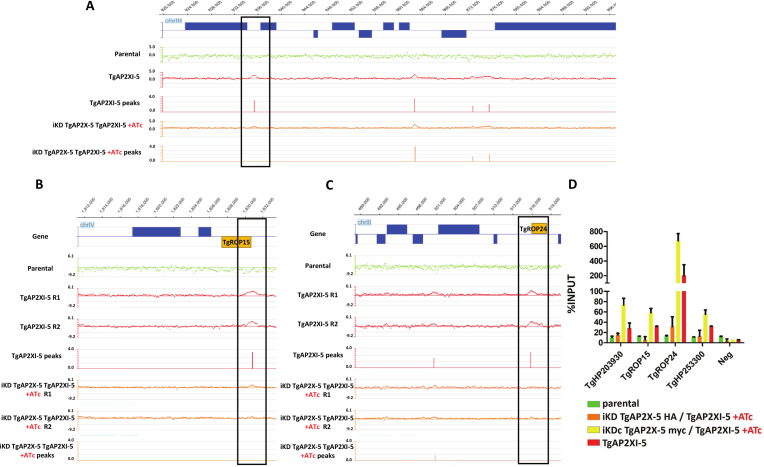
By RNA-seq, we showed that TgAP2X-5 regulates directly or indirectly the expression of number of virulence genes. We attempted several time to perform chromatin immunoprecipitation using tagged version of the gene but we repeatedly failed to show any enrichment at promoters (Supplementary Figure S5A). Moreover, we were unable to find any enrichment of a potential DNA motif in the promoter of genes that were influenced by TgAP2X-5 depletion (Supplementary Figure S5B). Since TgAP2X-5 and TgAP2XI-5 interact (Figure 1), we measured the ability of TgAP2XI-5 to bind to target promoters in absence of TgAP2X-5 using a genome-wide assay. We produced ChIP data using a tagged version of TgAP2XI-5 for the parental strain as well as for the iKD TgAP2X-5 strain in presence of ATc. As a negative control, we used the parental strain with an untagged version of TgAP2XI-5 (Supplementary Figure S6). Using the mPeak software, we found that more than 98% of the ChIP peaks found in a previously published study (22) overlapped with the present ChIP data using a tagged version of TgAP2XI-5. When comparing the TgAP2XI-5 ChIP peaks identified by the mPeak software in presence and in absence of TgAP2X-5, we found that 92 promoters showed a decrease of TgAP2XI-5 binding in the iKD TgAP2X-5 strain in presence of ATc (Supplementary Table S4) while most of the other TgAP2XI-5 binding sites remained unaffected (Figure 4A and Supplementary Figure S6). Among the 153 genes that were downregulated in absence of TgAP2X-5, only 63 corresponding promoters are bound by TgAP2XI-5 in normal conditions. Among these, 13% are downregulated in the TgAP2X-5 mutant and showed a decreased level of TgAP2XI-5 binding (Supplementary Figure S7 and Table S4). For example, TgROP24 and TgROP15 were downregulated in the TgAP2X-5 mutant and also impacted in terms of binding of TgAP2XI-5 in the iKD TgAP2X-5 strain in presence of ATc (Figure 4B and C). We confirmed by qPCR the results obtained in the genome-wide assay for several loci (Figure 4D). We also confirmed that in the iKD TgAP2X-5 complemented strain, the level of TgAP2XI-5 binding was similar to the parental strain (Figure 4D). These results indicate that for a number of promoters, the binding of TgAP2XI-5 is dependent on the presence of TgAP2X-5.
Figure 4.
TgAP2X-5 depletion prevents TgAP2XI-5 binding to a number of promoters. (A) ChIP-on-chip data are represented as the log2 ratio of the hybridization signal given by DNA immunoprecipitated over the signal given by the non-enriched input DNA. The log2 ratio of each oligonucleotide present on the tiled microarray has been plotted at the respective genomic position. Genes above the horizontal axis are encoded the forward strand whereas those below are encoded the reverse strand. The scale of the Y-axis was kept identical for all representation to allow the comparison of signals. TgAP2XI-5 ChIP-chip assay is represented for the TgAP2XI-5 protein in the parental strain (RHΔhxgprtΔku80, green, negative control) or TgAP2XI-5 tagged strain (red, positive control) or TgAP2XI-5 tagged in the iKD TgAP2X-5 strain in presence of ATc (orange). The associated peaks identified by the mPeak algorithm are also provided. A portion of the Toxoplasma gondii chromosome III is represented. The boxed promoter presents a lower enrichment of TgAP2XI-5 in the iKD TgAP2X-5 strain in presence of ATc. (B) Representation of the ChIP-chip experiments for a portion of chromosome IV which encompasses the TgROP15 gene (yellow). Two repeats (R1 and R2) are presented for each ChIP experiment along with the mPeak identified peaks. (C) Representation of the ChIP-chip experiments for a portion of chromosome III which encompasses the TgROP24 gene (yellow). Two repeats (R1 and R2) are presented for each ChIP experiment along with the mPeak identified peaks. (D) Quantitative PCR was performed on the TgAP2XI-5 ChIP assays performed in the iKD TgAP2X-5/TgAP2XI-5 strain (orange), iKD TgAP2X-5 complemented/TgAP2XI-5 strain (yellow), TgAP2XI-5-tagged strain (red) and parental strain (negative control, green). Amplification was carried out on regions within the promoter of each of the six genes listed. The enrichment for corresponding ChIP DNA samples is presented as a percentage of INPUT for each target.
DISCUSSION
The complex life cycle of most apicomplexan parasites suggests a tight regulation of gene expression to meet the requirements of changing environments. Transcriptome and the characterization of promoters studies have shown that at least, a part of these regulation is based on the transcriptional control of gene expression (7, 40). However, the characterization of the TFs responsible for the control of the promoter’s activities remained elusive until the discovery of ApiAP2 TFs and their function in regulating gene expression. In T. gondii, we described the promoter binding activity of TgAP2XI-5 which is able to bind to hundreds of promoters of genes transcriptionally active and mostly regulated during the S/M phase of the cell cycle (22). Interestingly, TgAP2XI-5 is one of the most conserved ApiAP2 TF among apicomplexan parasites, not only in the AP2 DNA binding domain (22), but also in another region which was identified as a potent trans-activator domain (41). TgAP2XI-5 is most likely an activator of transcription of S/M specific genes in T. gondii, as suggested by its association with active promoters (22). However, its constitutive protein expression profile (22) suggested that its activity may be regulated by other proteins. We identified a cell cycle-regulated ApiAP2 TF, TgAP2X-5, as a potential partner of TgAP2XI-5. This interaction is specific and can be direct or indirect. As most of ApiAP2 proteins in T. gondii, TgAP2X-5 is a large protein with no other recognizable motif than the AP2 DNA binding domain. Therefore, no potential interaction domain could be identified in TgAP2X-5. However, when examining the protein regions that are conserved among coccidian parasites, a region adjacent to the AP2 domain is well conserved while most of the protein is not (with the exception of the AP2 DNA binding domain). Interestingly, crystallography of a Plasmodium falciparum ApiAP2 DNA binding domain suggested that dimerization of two AP2 domains could occur (42). Therefore, heterodimerization of two AP2 domains might occur in the TgAP2XI-5/TgAP2X-5 complex. Further studies could aim at decipher the function of this potential interaction domain.
Cooperativity of TF is a very common feature on eukaryotic promoters to fine tune the level of transcription of each gene (43). However, it has been poorly explored in apicomplexan parasites. In P. falciparum, an ApiAP2 protein recruits a bromo-domain containing protein indicating that cooperativity of transcriptional regulators takes place in this parasite (44). In T. gondii, ApiAP2 TFs were found in complex with chromatin modifiers (45, 46), but interaction of two ApiAP2 was never reported to our knowledge. Interestingly, identification of the protein associated with TgAP2XI-5 did not reproducibly yield chromatin modifiers or other gene regulation related proteins with the exception of TgAP2X-5 (although the absence of detection does not mean the absence of the protein). In Plasmodium, the characterization of ApiAP2 TFs showed that several of these proteins are able to co-regulate similar set of genes indicating that cooperativity of ApiAP2 TFs indeed exists in this parasite (17). Together, these data suggest that cooperativity may be a widespread mechanism to regulate the activity of apicomplexan promoters.
TgAP2X-5 depletion alters the ability of TgAP2XI-5 to bind to number of its target promoters. However, we were not able to identify genomic regions of significant enrichment of TgAP2X-5 using ChIP-chip. This could be explained by technical difficulties that hampered our ability to appropriately perform ChIP with this protein. Alternatively, TgAP2X-5 may not be able to contact DNA with enough affinity to be detected by this technique. Nevertheless, we cannot exclude that TgAP2X-5 is able to directly bind DNA. Therefore, TgAP2X-5 may interact with TgAP2XI-5 but its potential DNA binding activity remain to be proven. The dependence of TgAP2XI-5 for its DNA binding to a subset of promoters may therefore depend on the presence of TgAP2X-5 in the complex through direct or indirect protein interactions. TgAP2X-5 may also facilitate the accessibility of TgAP2XI-5 to DNA in a specific chromatin environment. Many factors can influence the DNA binding abilities of a TF on a promoter containing the TF target sequence, among which DNA unwinding, bending and chromatin interactions (47). Alternatively, TgAP2X-5 may induce a change of conformation of TgAP2XI-5 allowing the binding to specific promoters. Homodimerization of the PF14_0633 protein AP2 domain upon DNA binding induces conformational changes in these domains lead by a domain-swapping mechanism (42). Assuming that AP2 domains of TgAP2XI-5 and TgAP2X-5 interact, this mechanism could lead to increased affinity to selected promoter. Multi-component TFs are well known in other eukaryotes among which NF-κB that is composed of two subunits (48).
RNA-seq of TgAP2X-5-depleted parasite revealed that a subset of virulence factors are downregulated. Among these, annotated rhoptry (ROP) proteins are over-represented with fewer microneme (MIC) proteins present in the list of genes downregulated. This is not surprising since the timing of expression of these transcripts and proteins is shifted during the S/M phase of the cell cycle (5), with ROP transcripts being expressed earlier (during S phase) than the MIC transcripts. This correlates well with the TgAP2X-5 protein expression pattern. Similarly, RON proteins, which represent an intermediate expression pattern are not affected by TgAP2X-5 depletion. Number of cell cycle-regulated transcripts were identified as downregulated in the iKD TgAP2X-5 strain, are annotated as hypothetical and may represent potential candidates for new ROP proteins. As shown for the PfAP2-I protein (44), TgAP2X-5 regulates only a subset of virulence factors indicating that other ApiAP2 may work in concert to regulate the expression of the full set of ROP and MIC transcripts. The particular subset of ROPs that are regulated by TgAP2X-5 may be linked by some functional particularities. For example, they might be exported together to the newly form organelles as it is the case for MIC proteins (49).
Nevertheless, the genes regulated by TgAP2X-5 are involved in virulence in vivo since their downregulation is enough to attenuate the virulence of the mutant parasites. The lack of apparent phenotype in vitro when testing growth may be linked to the function of some of the ROP proteins in manipulating the host cell. Indeed, knock-out for TgROP5, TgROP16 or TgROP18 did not yield in vitro growth defects but instead drastic virulence phenotypes (50–52). It is likely that the combination of the downregulation of multiple genes is the cause of this phenotype as single gene knock-outs were not enough to produce an avirulent strain. It is the case for TgROP15, TgROP46 and TgROP47 (53) and for TgROP13 (54).
The promoter of the transcripts impacted by TgAP2X-5 are not all bound by the TgAP2XI-5 protein (only 42% are bound by TgAP2XI-5). This indicates that TgAP2X-5 may interact with other proteins to regulate those genes. Alternatively, it may directly bind to the promoter of these genes. Taken together, these data indicate that the regulation of gene expression in apicomplexan parasites is probably dependent on the crucial involvement of numbers of TF regulating the same promoters. This implies that transcriptional regulation of gene expression in these parasites is a multifactorial process which probably involves TFs, chromatin modification and remodeling enzymes and regulation of Pol II activity on the promoters.
We showed here that two ApiAP2 TFs play a crucial role in regulating virulence factors expression in the apicomplexan parasite T. gondii. The presence of TgAP2X-5 at the right timing and in the right amount allows the efficient binding of TgAP2XI-5 to certain promoters. This is the first evidence of cooperation of ApiAP2 TF for the regulation of gene expression.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank the BioImaging Center Lille for access to instruments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National de la Recherche Scientifique; Institut National de la Santé et de la Recherche Médicale; Agence Nationale de la Recherche (ANR) [ANR-13-JSV3-0006-01 to M.G.]. Funding for open access charge: Agence Nationale de la Recherche [ANR-13-JSV3-0006-01].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shwab E.K., Jiang T., Pena H.F.J., Gennari S.M., Dubey J.P., Su C.. The ROP18 and ROP5 gene allele types are highly predictive of virulence in mice across globally distributed strains of Toxoplasma gondii. Int. J. Parasitol. 2016; 46:141–146. [DOI] [PubMed] [Google Scholar]
- 2. Francia M.E., Striepen B.. Cell division in apicomplexan parasites. Nat. Rev. Microbiol. 2014; 12:125–136. [DOI] [PubMed] [Google Scholar]
- 3. Striepen B., Jordan C.N., Reiff S., van Dooren G.G.. Building the perfect parasite: cell division in apicomplexa. PLoS Pathog. 2007; 3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gubbels M.-J., White M., Szatanek T.. The cell cycle and Toxoplasma gondii cell division: tightly knit or loosely stitched. Int. J. Parasitol. 2008; 38:1343–1358. [DOI] [PubMed] [Google Scholar]
- 5. Behnke M.S., Wootton J.C., Lehmann M.M., Radke J.B., Lucas O., Nawas J., Sibley L.D., White M.W.. Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One. 2010; 5:e12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bozdech Z., Llinas M., Pulliam B.L., Wong E.D., Zhu J., DeRisi J.L.. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003; 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gissot M., Kim K., Schaap D., Ajioka J.W.. New eukaryotic systematics: a phylogenetic perspective of developmental gene expression in the Apicomplexa. Int. J. Parasitol. 2009; 39:145–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gissot M., Kelly K.A., Ajioka J.W., Greally J.M., Kim K.. Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog. 2007; 3:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hakimi M.-A., Deitsch K.W.. Epigenetics in Apicomplexa: control of gene expression during cell cycle progression, differentiation and antigenic variation. Curr. Opin. Microbiol. 2007; 10:357–362. [DOI] [PubMed] [Google Scholar]
- 10. Balaji S., Babu M.M., Iyer L.M., Aravind L.. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005; 33:3994–4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Campbell T.L., De Silva E.K., Olszewski K.L., Elemento O., Llinás M.. Identification and genome-wide prediction of DNA binding specificities for the ApiAP2 family of regulators from the malaria parasite. PLoS Pathog. 2010; 6:e1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuda M., Iwanaga S., Shigenobu S., Mair G.R., Janse C.J., Waters A.P., Kato T., Kaneko I.. Identification of a transcription factor in the mosquito-invasive stage of malaria parasites. Mol. Microbiol. 2009; 71:1402–1414. [DOI] [PubMed] [Google Scholar]
- 13. Yuda M., Iwanaga S., Shigenobu S., Kato T., Kaneko I.. Transcription factor AP2-Sp and its target genes in malarial sporozoites. Mol. Microbiol. 2010; 75:854–863. [DOI] [PubMed] [Google Scholar]
- 14. Iwanaga S., Kaneko I., Kato T., Yuda M.. Identification of an AP2-family protein that is critical for malaria liver stage development. PLoS One. 2012; 7:e47557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuda M., Iwanaga S., Kaneko I., Kato T.. Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:12824–12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sinha A., Hughes K.R., Modrzynska K.K., Otto T.D., Pfander C., Dickens N.J., Religa A.A., Bushell E., Graham A.L., Cameron R. et al. A cascade of DNA-binding proteins for sexual commitment and development in Plasmodium. Nature. 2014; 507:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Modrzynska K., Pfander C., Chappell L., Yu L., Suarez C., Dundas K., Gomes A.R., Goulding D., Rayner J.C., Choudhary J. et al. A knockout screen of ApiAP2 genes reveals networks of interacting transcriptional regulators controlling the plasmodium life cycle. Cell Host Microbe. 2017; 21:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walker R., Gissot M., Croken M.M., Huot L., Hot D., Kim K., Tomavo S.. The Toxoplasma nuclear factor TgAP2XI-4 controls bradyzoite gene expression and cyst formation. Mol. Microbiol. 2012; 87:641–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hong D.-P., Radke J.B., White M.W.. Opposing transcriptional mechanisms regulate toxoplasma development. mSphere. 2017; 2:e00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang S., Holmes M.J., Radke J.B., Hong D.-P., Liu T.-K., White M.W., Sullivan W.J.. Toxoplasma gondii AP2IX-4 regulates gene expression during bradyzoite development. mSphere. 2017; 2:e00054-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Radke J.B., Lucas O., De Silva E.K., Ma Y., Sullivan W.J. Jr, Weiss L.M., Llinas M., White M.W.. ApiAP2 transcription factor restricts development of the Toxoplasma tissue cyst. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:6871–6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walker R., Gissot M., Huot L., Alayi T.D., Hot D., Marot G., Schaeffer-Reiss C., Van Dorsselaer A., Kim K., Tomavo S.. Toxoplasma transcription factor TgAP2XI-5 regulates the expression of genes involved in parasite virulence and host invasion. J. Biol. Chem. 2013; 288:31127–31138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sheiner L., Demerly J.L., Poulsen N., Beatty W.L., Lucas O., Behnke M.S., White M.W., Striepen B.. A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog. 2011; 7:e1002392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huynh M.-H., Carruthers V.B.. Tagging of endogenous genes in a Toxoplasma gondii strain lacking Ku80. Eukaryot. Cell. 2009; 8:530–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen B., Brown K.M., Lee T.D., Sibley L.D.. Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. mBio. 2014; 5:e01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lentini G., Kong-Hap M., El Hajj H., Francia M., Claudet C., Striepen B., Dubremetz J.-F., Lebrun M.. Identification and characterization of Toxoplasma SIP, a conserved apicomplexan cytoskeleton protein involved in maintaining the shape, motility and virulence of the parasite. Cell. Microbiol. 2015; 17:62–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dzierszinski F., Mortuaire M., Dendouga N., Popescu O., Tomavo S.. Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J. Mol. Biol. 2001; 309:1017–1027. [DOI] [PubMed] [Google Scholar]
- 28. Couvreur G., Sadak A., Fortier B., Dubremetz J.F.. Surface antigens of Toxoplasma gondii. Parasitology. 1988; 97:1–10. [DOI] [PubMed] [Google Scholar]
- 29. Gissot M., Walker R., Delhaye S., Huot L., Hot D., Tomavo S.. Toxoplasma gondii chromodomain protein 1 binds to heterochromatin and colocalises with centromeres and telomeres at the nuclear periphery. PLoS One. 2012; 7:e32671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Miguet L., Béchade G., Fornecker L., Zink E., Felden C., Gervais C., Herbrecht R., Van Dorsselaer A., van Dorsselaer A., Mauvieux L. et al. Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J. Proteome Res. 2009; 8:3346–3354. [DOI] [PubMed] [Google Scholar]
- 31. Choi H., Larsen B., Lin Z.-Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z.S., Tyers M., Gingras A.-C., Nesvizhskii A.I.. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods. 2011; 8:70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Skarra D.V., Goudreault M., Choi H., Mullin M., Nesvizhskii A.I., Gingras A.-C., Honkanen R.E.. Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5. Proteomics. 2011; 11:1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anders S., Pyl P.T., Huber W.. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zheng M., Barrera L.O., Ren B., Wu Y.N.. ChIP-chip: data, model, and analysis. Biometrics. 2007; 63:787–796. [DOI] [PubMed] [Google Scholar]
- 37. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Buske F.A., Bodén M., Bauer D.C., Bailey T.L.. Assigning roles to DNA regulatory motifs using comparative genomics. Bioinformatics. 2010; 26:860–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C. et al. RSAT 2015: Regulatory sequence analysis tools. Nucleic Acids Res. 2015; 43:W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sullivan W.J., Hakimi M.A.. Histone mediated gene activation in Toxoplasma gondii. Mol. Biochem. Parasitol. 2006; 148:109–116. [DOI] [PubMed] [Google Scholar]
- 41. Pino P., Sebastian S., Kim E.A., Bush E., Brochet M., Volkmann K., Kozlowski E., Llinás M., Billker O., Soldati-Favre D.. A tetracycline-repressible transactivator system to study essential genes in malaria parasites. Cell Host Microbe. 2012; 12:824–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lindner S.E., De Silva E.K., Keck J.L., Llinás M.. Structural determinants of DNA binding by a P. falciparum ApiAP2 transcriptional regulator. J. Mol. Biol. 2010; 395:558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ogata K., Sato K., Tahirov T.H., Tahirov T.. Eukaryotic transcriptional regulatory complexes: cooperativity from near and afar. Curr. Opin. Struct. Biol. 2003; 13:40–48. [DOI] [PubMed] [Google Scholar]
- 44. Santos J.M., Josling G., Ross P., Joshi P., Orchard L., Campbell T., Schieler A., Cristea I.M., Llinás M.. Red blood cell invasion by the malaria parasite is coordinated by the PfAP2-I transcription factor. Cell Host Microbe. 2017; 21:731–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Saksouk N., Bhatti M.M., Kieffer S., Smith A.T., Musset K., Garin J., Sullivan W.J., Cesbron-Delauw M.F., Hakimi M.A.. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 2005; 25:10301–10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang J., Dixon S.E., Ting L.-M., Liu T.-K., Jeffers V., Croken M.M., Calloway M., Cannella D., Hakimi M.A., Kim K. et al. Lysine acetyltransferase GCN5b interacts with AP2 factors and is required for Toxoplasma gondii proliferation. PLoS Pathog. 2014; 10:e1003830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Slattery M., Zhou T., Yang L., Dantas Machado A.C., Gordân R., Rohs R.. Absence of a simple code: how transcription factors read the genome. Trends Biochem. Sci. 2014; 39:381–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pereira S.G., Oakley F.. Nuclear factor-κB1: Regulation and function. Int. J. Biochem. Cell Biol. 2008; 40:1425–1430. [DOI] [PubMed] [Google Scholar]
- 49. Kremer K., Kamin D., Rittweger E., Wilkes J., Flammer H., Mahler S., Heng J., Tonkin C.J., Langsley G., Hell S.W. et al. An overexpression screen of Toxoplasma gondii Rab-GTPases reveals distinct transport routes to the micronemes. PLoS Pathog. 2013; 9:e1003213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Reese M.L., Zeiner G.M., Saeij J.P.J., Boothroyd J.C., Boyle J.P.. Polymorphic family of injected pseudokinases is paramount in Toxoplasma virulence. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:9625–9630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saeij J.P.J., Boyle J.P., Coller S., Taylor S., Sibley L.D., Brooke-Powell E.T., Ajioka J.W., Boothroyd J.C.. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 2006; 314:1780–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor S., Barragan A., Su C., Fux B., Fentress S.J., Tang K., Beatty W.L., Hajj H.E., Jerome M., Behnke M.S. et al. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 2006; 314:1776–1780. [DOI] [PubMed] [Google Scholar]
- 53. Wang J.-L., Li T.-T., Elsheikha H.M., Chen K., Zhu W.-N., Yue D.-M., Zhu X.-Q., Huang S.-Y.. Functional characterization of rhoptry kinome in the virulent toxoplasma gondii RH Strain. Front. Microbiol. 2017; 8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Turetzky J.M., Chu D.K., Hajagos B.E., Bradley P.J.. Processing and secretion of ROP13: a unique Toxoplasma effector protein. Int. J. Parasitol. 2010; 40:1037–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.