Abstract
Fanconi Anemia (FA) is characterized by bone marrow failure, congenital abnormalities, and cancer. Of over 20 FA-linked genes, FANCJ uniquely encodes a DNA helicase and mutations are also associated with breast and ovarian cancer. fancj−/− cells are sensitive to DNA interstrand cross-linking (ICL) and replication fork stalling drugs. We delineated the molecular defects of two FA patient-derived FANCJ helicase domain mutations. FANCJ-R707C was compromised in dimerization and helicase processivity, whereas DNA unwinding by FANCJ-H396D was barely detectable. DNA binding and ATP hydrolysis was defective for both FANCJ-R707C and FANCJ-H396D, the latter showing greater reduction. Expression of FANCJ-R707C or FANCJ-H396D in fancj−/− cells failed to rescue cisplatin or mitomycin sensitivity. Live-cell imaging demonstrated a significantly compromised recruitment of FANCJ-R707C to laser-induced DNA damage. However, FANCJ-R707C expressed in fancj-/- cells conferred resistance to the DNA polymerase inhibitor aphidicolin, G-quadruplex ligand telomestatin, or DNA strand-breaker bleomycin, whereas FANCJ-H396D failed. Thus, a minimal threshold of FANCJ catalytic activity is required to overcome replication stress induced by aphidicolin or telomestatin, or to repair bleomycin-induced DNA breakage. These findings have implications for therapeutic strategies relying on DNA cross-link sensitivity or heightened replication stress characteristic of cancer cells.
INTRODUCTION
Understanding the pathological basis of disease-causing mutations is a potentially insightful approach to development of treatments and cures for enigmatic conditions. We have been particularly engaged in characterizing missense mutant alleles of helicase genes that underlie chromosomal instability disorders punctuated by accelerated aging and a predisposition to cancer. Fanconi Anemia (FA) Complementation Group J (FANCJ) is an excellent example of such a genetic disorder. Two percent of FA patients are attributed to pathogenic variants in the FANCJ gene (1). FA is complex with an every-growing list of genes (currently over 20) that encode proteins implicated in a specialized DNA repair pathway tailored for the correction of DNA interstrand cross-links (ICLs). ICLs represent a very lethal form of DNA damage because by their very nature, they strongly interfere with DNA replication as well as virtually all aspects of DNA metabolism (2). Among the essential genes in the FA pathway is a single DNA helicase, designated FANCJ that is implicated in homologous recombination (HR) repair of double-strand breaks (DSB) that arise from processing of an ICL or may occur directly from endogenous replication stress or exposure to agents that cause closely clustered breaks in each strand or simultaneous breaks in both strands (3). In addition to its FA linkage, FANCJ has been found in multiple studies to be associated with breast and ovarian cancer (4). Therefore, its disease relevance is very strong, yet FANCJ remains one of the most poorly understood of the FA gene products in terms of its pathway function(s). Moreover, FANCJ appears to operate outside the classic FA pathway of ICL repair because its deficiency results in a more generalized poor response to agents that impose replication stress other than ICLs (5–7).
To develop a greater understanding of FANCJ’s molecular and cellular roles, we have characterized two missense mutations residing in the helicase gene coding sequence that are linked to FA. The two FANCJ mutations are quite distinct in terms of their effects on molecular functions. Our analysis has elucidated a novel catalytic threshold whereby FANCJ can fulfill its function in response to a DNA polymerase inhibitor or G-quadruplex binding ligand that induces replication stress or a chemical that introduces DNA strand breaks; however, even a partially functional helicase that displays compromised recruitment to DNA damage is unable to restore cross-link resistance. We propose a model in which FANCJ remodeling of stalled replication forks requires only modest helicase activity on short duplexes and is independent of the classic FA pathway, whereas its involvement in ICL repair requires rapid recruitment to the damaged DNA and more processive DNA unwinding to suppress the persistence of single-stranded DNA and Rad51, indicative of improperly or incompletely processed recombinant DNA intermediates.
MATERIALS AND METHODS
Plasmid DNA constructions
FANCJ-R707C and FANCJ-H396D mutations were generated by the Quik Change site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions using respective primers (Supplementary Table S1) in the vector pEGFP-C2 (Clontech). Mutations were confirmed by DNA sequencing.
Recombinant proteins
Expression and purification of wild-type and mutant FANCJ proteins were performed using baculovirus encoding FANCJ-wild type (WT), FANCJ-R707C and FANCJ-H396D with a C-terminal FLAG tag. The baculovirus constructs were infected in High Five insect cells, and the recombinant FANCJ proteins were purified with modifications to a protocol previously described (8). Briefly, cell pellets were resuspended in buffer A (10 mM Tris–HCl (pH 7.5), 130 mM NaCl, 1% Triton X-100, 10 mM NaF, 10 mM NaPi, 10 mM NaPPi). Cells were lysed in the presence of protease inhibitors (Roche Applied Science) for 45 min at 4°C with mild agitation and centrifuged at 21 000 × g for 10 min at 4°C. The supernatant was incubated with FLAG antibody resin (Sigma) for 2 h at 4°C. The resin was washed twice with buffer B (50 mM Tris–HCl (pH 7.4), 1 mM EDTA, and 0.5% Nonidet P-40) supplemented with 500 mM NaCl followed by buffer B supplemented with 150 mM NaCl. FANCJ was eluted twice with 4 μg/ml 3× FLAG peptide (Sigma) in buffer D (25 mM Tris–HCl (pH 7.4), 100mM NaCl, 10% glycerol, 0.1% Tween 20, 5 mM Tris (2-carboxyethyl) phosphine hydrochloride; Sigma) for 1 h. FLAG-tagged FANCJ proteins were dialyzed for 2 h against buffer D using a dialysis tube with a 50-kDa molecular mass cut-off (Tube-O-DIALYZER™), and aliquots were frozen in liquid nitrogen and stored at -80°C. RPA was purified as described before (9). Protein concentration was determined by the Bradford assay using BSA as a standard.
DNA substrates
Polyacrylamide gel electrophoresis purified oligonucleotides used for the preparation of DNA substrates were purchased from Lofstrand Labs or IDT (Integrated DNA Technologies) and are listed in Supplementary Table S2. The forked duplex DNA substrates, as well as the unimolecular G4 PolyZic1 and tetra-molecular TPG4 DNA substrates were prepared from the indicated oligonucleotides as previously described (8,10–11).
Radiometric helicase assays
Helicase assay reaction mixtures (20 μl) contained 40 mM Tris–HCl (pH 7.4), 25 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 2% glycerol, 100 ng/μl bovine serum albumin, 2 mM ATP and forked duplex (19 bp) DNA substrate (10 fmol) or G4 DNA substrate (5 fmol), with the indicated concentrations of FANCJ and/or RPA. Helicase reactions were initiated by the addition of FANCJ and incubated at 30°C for 15 min unless otherwise indicated. Helicase reactions were terminated by addition of stop buffer (74% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue, 10 mM KCl, 20 mM EDTA). Reaction products were resolved on non-denaturing 12% and 10% (19:1 acrylamide:bisacrylamide) polyacrylamide gels for the forked duplex and TPG4 DNA substrates, respectively. The resolved radiolabeled DNA species from products of helicase reaction mixtures were visualized with Typhoon 9400 phosphor-imaging and analyzed with ImageQuant 5.2 software (GE Healthcare).
For the unimolecular Poly(A) Zic1-G4 DNA substrate (5 fmol), helicase reactions (20 μl) were performed in reaction buffer containing a 20-fold excess of peptide–nucleic acid complementary oligonucleotide (10,12). G4 helicase reactions were terminated by the addition of 20 μl of stop buffer, and reaction products were resolved on 15% nondenaturing polyacrylamide gels. Radiolabeled DNA products were visualized and analyzed quantitatively as described above.
Protein trap kinetic helicase assays
Protein trap kinetic assays were performed as described before (13). Briefly, FANCJ-WT (0.6 nM), FANCJ-R707C (1.9 nM) and FANCJ-H396D (4.8 nM) were preincubated for 3 min at 24°C with 5 nM of the radiolabeled forked duplex DNA substrate. After 3 min, ATP (2 mM) and 500 nM oligo dT200 (to serve as protein trap) was added simultaneously to the reaction mixture and incubated at 30°C. Aliquots (20 μl) of the reaction mixture were quenched at 10-s intervals with stop Buffer containing a 10-fold excess of unlabeled oligonucleotide with the same sequence as the labeled strand. Products of helicase reaction mixtures were then resolved on 12% polyacrylamide gels and visualized as described under ‘Helicase assays’.
ATP hydrolysis assays
ATP hydrolysis was measured using [γ−32P] ATP (PerkinElmer Life Sciences) and analyzed by thin layer chromatography (TLC) on polyethyleneimine-cellulose plates (Mallinckrodt Baker). The standard reaction mixture (20 μl of total volume) contained 40 mM Tris–HCl (pH 7.4), 25 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 2% glycerol, 100 ng/μl bovine serum albumin, 250 μM [γ−32P] ATP, 2.1 nM M13mp18 single-stranded DNA circle, and 60 nM FANCJ protein and was incubated at 30°C. For Km determinations, final ATP concentrations ranged from 30 to 8000 μM. For kcat determinations, ATP concentration was 4.4 mM. The product was spotted onto a PEI-cellulose TLC plate and resolved by using 0.5 M LiCl and 1 M formic acid as the carrier solvent. The TLC plate was exposed to a phosphorimaging screen for 1 h, and the radiolabeled species were visualized and analyzed as described above.
Electrophoretic mobility shift assay
EMSA’s were as described previously (8). Briefly, protein/DNA binding mixtures (20 μl) contained the indicated concentrations of FANCJ and 0.5 nM concentrations of the specified 32P-end labeled DNA substrate in the same reaction buffer as used for helicase assays (see above) except ATP was excluded. The binding mixtures were incubated at 24°C for 30 min after the addition of FANCJ. After incubation, 3 μl of loading dye (74% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue) were added to each mixture, and samples were loaded onto native 5% (19:1 acrylamide:bisacrylamide) polyacrylamide gels and electrophoresed at 200 V for 2 h at 4°C using 1× Tris borate-EDTA as the running buffer. The resolved radiolabeled species were visualized and analyzed as described above.
Streptavidin displacement assays
Streptavidin displacement assay were performed as described before (14). Briefly, reaction mixtures contained 40 mM Tris–HCl (pH 7.6), 25 mM KCl, 2 mM MgCl2, 2% glycerol, 100 ng/μl bovine serum albumin, 2 mM ATP, 0.5 nM streptavidin-bound biotinylated oligonucleotide (X12-1-52-BIOT13), and the indicated concentrations of FANCJ helicase. For the assays, 0.5 nM of biotinylated oligonucleotide was preincubated with 100 nM streptavidin (Sigma) for 10 min at 37°C. Reactions were initiated by adding FANCJ prior to the addition of biotin (1 μM) and incubated at 30°C for 15 min, followed by a quench with the addition of 10 μl of stop buffer (50 mM EDTA, 40% glycerol, 0.9% SDS, 0.05% bromophenol blue, and 0.05% xylene cyanol). Products were resolved on nondenaturing 12% polyacrylamide gel (19:1 acrylamide/bisacrylamide). The resolved radiolabeled species were visualized and analyzed as described above.
Single-molecule fluorescence-based helicase assays
The DNA hairpin substrate preparation and experimental and analysis procedures for single-molecule measurements of DNA unwinding activity have been described in detail (15,16). Briefly, a 537 bp DNA hairpin with a 25 nt single-stranded DNA gap that facilitates FANCJ binding was tethered to a surface and a 2.7 μM magnetic bead via digoxigenin/anti-digoxigenin and biotin-streptavidin linkages, respectively. The unwinding assays were performed at 30°C, in the presence of 0.5–2 nM FANCJ in the modified helicase buffer containing 40 mM Tris–HCl (pH 7.4), 25 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 300 ng/ml bovine serum albumin, 0.01% Tween-20, and 2 mM ATP. The unwinding of the DNA hairpin duplex by FANCJ was measured in real-time by tracking the tethered magnetic bead at 200 Hz from images acquired with a CCD camera using Labview-based image analysis. The unwinding activity was further analyzed using a custom T-test based analysis program (Igor) to identify unwinding events and obtain measures of the unwinding rate and run-length (15,17).
Size exclusion chromatography
Purified recombinant FANCJ protein was applied to a Superdex-200 size exclusion column (GE Healthcare) using an AKTA FPLC (GE Healthcare) as described previously (8). Proteins were detected by a UV detector. The column was calibrated using standard molecular mass markers containing blue dextran (2000 kDa), thyroglobulin (670 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa), and aprotinin (6.5 kDa) (Sigma).
Cell lines and survival assays
Chicken DT40 cells were cultured in RPMI-1640 medium supplemented with 9% (v/v) fetal calf serum, 1% (v/v) chicken serum, 2 mM l-glutamine, 50 μM 2-mercaptoethanol, 1% (v/v) penicillin-streptomycin in a 5% CO2 incubator at 39°C. The fancj−/−, fancc−/− and fancj−/− fancc−/− knockout cell lines were previously described (18). fancj−/− blm−/− cells were constructed by transfecting FANCJ knockout constructs described by (14) in blm−/− cells described by (19). The DT40 cell lines were transfected with plasmids encoding GFP, GFP-FANCJ-WT and GFP-FANCJ-R707C and GFP-FANCJ-H396D and selected by G418 resistance using a procedure previously described (20).
Survival assays were carried out as described previously by colony formation assays containing 1.4% (w/v) methylcellulose (21) using increasing concentrations of cisplatin (CisPt), aphidicolin (APH), telomestatin (TMS), mitomycin C (MMC), bleomycin (Bleo) and Mirin as indicated in the figures.
Chk1 phosphorylation
fancj−/− DT40 cells transfected with GFP empty vector or vector expressing GFP-FANCJ recombinant proteins were either not treated (DMSO) or treated with various dosages of APH (0–40 μM) for 60 min. Cells were harvested and 2 million cells were lysed in 2× SDS buffer, followed by sonication and heated at 95°C for 5 min. Cell lysates were spun at 11 000 × g for 10 min, and proteins from supernatant were resolved on 8–12% polyacrylamide gels (nuPAGE (Invitrogen)). After immune-blotting, Chk1 phosphorylation was detected by a phosphor Chk1-S245 antibody (Cell Signaling). Cellular actin (Sigma) was used as a loading control.
G4 antibody staining
The isogenic fancj−/− transfected cell lines were treated with DMSO or 5 μM TMS for 6 h. Cells were fixed with freshly prepared 4% formaldehyde and stained for immunoreactive G4 using a mouse anti-G4 1H6 monoclonal antibody (1:1000, Millipore) previously described (22). Immunofluorescence analyses were performed with a Zeiss LSM 510 META inverted Axiovert 200M laser scan microscope with a Plan-Apochromat 63× 1.4-numerical- aperature oil immersion differential interference contrast objective lens. Images were captured with a CCD camera and analyzed using LSM Browser software (Zeiss).
Chromosome aberrations
Chromosome aberrations were restricted to macrochromosome variation using a method described previously (23). Briefly cells were treated with CisPt (2 μM, 14 h) followed by 1% colcemid (Sigma) for 2 h. Cells were harvested and hypotonically swollen with 0.9% sodium citrate for 15 min at room temperature, and fixed with freshly prepared 5 ml of methanol:acetic acid (3:1). Fixed cells were dropped onto chilled glass slides, immediately flame, dried, and stained with DAPI. Images were captured in a Zeiss LSM 510 META inverted Axiovert 200M laser scan microscope with a Plan-Apochromat 100× 1.4-numerical- aperature oil immersion differential interference contrast objective lens. Images were captured with a CCD camera and analyzed using LSM Browser software (Zeiss).
Immunofluorescence cellular localization
For BrdU detection to mark single-stranded DNA, fancj−/− cells transfected with the indicated FANCJ plasmid were grown in the presence of 20 μM BrdU (Sigma) for 24 h. Cells were then treated with CisPt (2 μM) or APH (200 nM) for 14 h, harvested, and processed as described below for immunofluorescent detection of BrdU foci using a mouse anti-BrdU antibody (1:100, BD Pharmingen™) and goat anti-mouse Alexa flour 488 secondary antibody (1:1000 Invitrogen). For immunofluorescent cellular detection of γ-H2AX or Rad51 foci, cells were fixed with freshly prepared 4% formaldehyde at room temperature for 15 min. Fixed cells were washed 4 times with PBS and treated with 0.5% Triton X-100 solution (Sigma) at room temperature for 10 min. After washing with PBS (4 times), cells were blocked with 10% goat serum (Sigma) overnight at 4°C. Indirect immunostaining was performed by first incubating cells with mouse anti-γ-H2AX monoclonal antibody (1:500, Millipore) or rabbit anti-Rad51 antibody (1:250, Calbiochem) for 4 h at room temperature or overnight at 4°C. After four washes in PBS with 0.1% Tween 20, cells were incubated with Alexa Fluor 488 goat anti-mouse IgG (1:500, Invitrogen) or Alexa Fluor 633 goat anti-mouse IgG (1:500, Invitrogen) for 1 h at room temperature. Cells were washed 4 times with PBS containing 0.1% Tween 20 and coated with Prolong Gold anti-fade reagent containing DAPI (Invitrogen). Coverslips were placed on chamber slides, and cells were cured at room temperature in the dark for 24 h. Immunofluorescence analyses were performed with a Zeiss LSM 510 META inverted Axiovert 200M laser scan microscope with a Plan-Apochromat 63× 1.4-numerical- aperature oil immersion differential interference contrast objective lens. Images were captured with a CCD camera and analyzed using LSM Browser software (Zeiss).
Laser irradiation and confocal microscopy
Immunofluorescence localization of GFP-tagged FANCJ proteins to laser-activated psoralen-ICLs (Pso-ICLs) or laser-induced DSBs in vivo was performed as previously described (24).
DNA fiber studies
DNA fiber spreads in DT40 cells were prepared as described before (25) with slight modifications. Briefly, cells were first pulse-chased with CldU at 25 μM for 20 min and then with IdU at 250 μM for the indicated times. Cells were treated with 200 nM of APH at the time of IdU labeling. Cells were harvested and washed in ice-cold PBS and resuspended in ice-cold PBS at a concentration of 0.5 × 106 cells/ml. A sample of cells (volume: 2 μl) was mixed with lysis buffer (10 μl; 200 mM Tris–HCl, pH 7.4, 50 mM EDTA, 0.5% SDS) on a glass slide. Slides were tilted ∼ 15° to allow cell extract to move down the slide. The DNA spreads were air-dried and fixed in a mixture of methanol/acetic acid (3:1, v:v). The slides were incubated in a solution of HCl (2.5 M) for 60 min, neutralized in a buffer (400 mM Tris–HCl, pH 7.4) for 10 min, washed in PBS, and immuno-stained, as described before (25). Staining of the slides with antibodies specific to CldU and IdU was done sequentially. The antibodies and dilutions used for staining were: rat anti-BrdU (CldU), 1:200; Daylight 647 goat anti-rat, 1:100; mouse anti-BrdU (IdU), 1:40, and Daylight 488 goat anti-mouse, 1:100. Imaging of the slides was carried out using a Zeiss Axiovert 200 M microscope with the Axio Vision software packages (Zeiss).
RESULTS
Patient-derived R707C and H396D mutations reside in distinct regions of the FANCJ helicase core domain
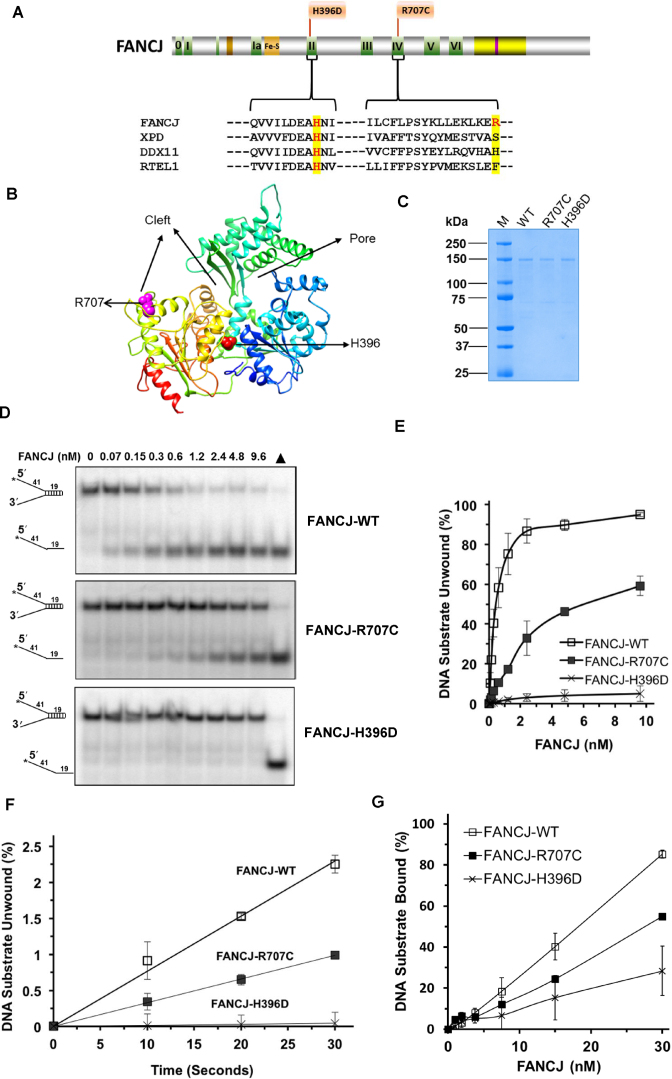
In this work, we characterized two FA patient-derived missense mutations corresponding to single amino acid substitutions: Histidine (H) 396 mutated to Aspartic Acid (D) (26); and Arginine (R) 707 mutated to Cysteine (C) (27) (Figure 1A). Histidine (H)-396 resides in motif II, also known as the Walker B motif, and is highly conserved among Fe-S containing superfamily (SF2) family of DNA helicases which includes FANCJ, XPD, DDX11 and RTEL-1. Arginine (R)-707 resides in motif IV and is variable in the human Fe–S helicases with serine (S) in XPD, Histidine (H) in DDX11 and Phenylalanine (F) in RTEL-1 (Figure 1A). Using the sequence-related Fe–S cluster helicase Thermoplasma acidophilum (Ta) XPD (28) for homology modeling of FANCJ, the predicted location of the R707 and H396 amino acids are shown in pink and red, respectively (Figure 1B). Visual inspection of the homology model suggests that R707 would be located near the DNA binding cleft and H396 resides at the interface between the two conserved RecA domains. The corresponding recombinant FANCJ wild-type (WT) and mutant proteins were expressed in insect cells and purified for biochemical characterization (Figure 1C).
Figure 1.
FA-linked FANCJ-R707C and FANCJ-H396D mutations and their biochemical characterization. (A) Human FANCJ depicting helicase core domain with canonical motifs indicated by colored boxes and FA-linked FANCJ mutations R707C and H396D are shown. The conservation in motif II and IV is shown with the sequence alignment of human FANCJ, XPD, DDX11 and RTEL1. BLM binding region (yellow) and BRCA1 binding site (purple) are shown. (B) Predicted structure of FANCJ based on TaXPD (28) as a template using Phyre2 (61). The locations of R707 and H396 are shown in pink and red, respectively. Based on TaXPD structure, the cleft and pore in the modeled structure of FANCJ is highlighted. (C) A Coomassie-blue stained SDS-polyacrylamide gel showing 2 μg of purified homogenous recombinant proteins FANCJ-WT, FANCJ-R707C, and FANCJ-H396D. kD (kilodalton) represents molecular mass protein standards. (D) FANCJ mutations H396D and R707C differentially affected DNA helicase activity. Representative native polyacrylamide gel images showing helicase reaction products from reaction mixtures containing the indicated FANCJ proteins incubated with 0.5 nM forked duplex (19 bp) DNA substrate at 30°C for 15 min as described in Materials and Methods. Filled triangle represents heat-denatured DNA substrate control. (E) Quantitative analysis of helicase activity by FANCJ proteins is shown with standard deviation (S.D.) indicated by error bars. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D. (F) Unwinding kinetics by FANCJ helicase proteins as determined by protein trap kinetics assays. Reactions with the indicated FANCJ protein (FANCJ-WT (0.6 nM), FANCJ-R707C (1.9 nM), or FANCJ-H396D (4.8 nM)) in the presence of 5 nM forked duplex (19 bp) DNA substrate were initiated by simultaneous addition of 100-fold excess of dT200 oligo (serve as protein trap) and ATP at 30°C and aliquots quenched at 5 s intervals. Quantitative analyses of DNA unwinding by FANCJ-WT (open square), FANCJ-R707C (filled square), and FANCJ-H396D (open cross) under conditions simulating single-turnover are shown. (G) DNA binding by FANCJ wild-type and mutant proteins. The indicted concentrations of the specified FANCJ protein were incubated with the forked duplex (19 bp) DNA substrate (0.5 nM) at 24°C for 30 min. Radiolabeled DNA and protein-DNA complexes were resolved by EMSA (Supplementary Figure S1), as described in Materials and Methods. Quantitative analysis of % DNA substrate bound by FANCJ is shown with S.D. indicated by error bars. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D.
FANCJ missense mutations differentially affect DNA helicase activity
The effect of FANCJ mutations H396D and R707C on FANCJ helicase activity were initially assessed using a forked 19 base-pair (bp) duplex DNA substrate flanked by noncomplementary 5′ and 3′ single-stranded tails. FANCJ-WT was able to efficiently unwind the DNA substrate during the 15 min incubation period (Figure 1D), consistent with previous reports (8,14). FANCJ-R707C retained partial helicase activity (Figure 1D), whereas unwinding by FANCJ-H396D was barely detectable (Figure 1D). At 2.4 nM enzyme concentration, FANCJ-WT unwound 86% of the substrate compared to 32% of the substrate unwound by FANCJ-R707C (Figure 1E), indicating that the R707C mutation partially impaired FANCJ-catalyzed DNA unwinding of the forked 19 bp DNA substrate.
Single turn-over kinetic analysis of FANCJ helicase activity
To further characterize the partially reduced DNA unwinding by FANCJ-R707C, we performed kinetic assays with a protein trap to simulate single-turnover conditions as described (13) (Figure 1F). For this, we used a concentration of FANCJ-R707C that yielded a similar level of helicase activity compared to FANCJ-WT under the multi-turnover 15 min reaction. FANCJ-WT (0.6 nM), FANCJ-R707C (1.9 nM) and FANCJ-H396D (4.8 nM) were first allowed to bind 5 nM of forked duplex (19 bp) substrate. Subsequently, reactions were initiated by the addition of ATP and 500 nM of dT200 oligonucleotide which served as a protein trap to capture excess FANCJ not bound to substrate at the end of the pre-incubation or FANCJ which dissociated during the reaction incubation period when ATP was present. The initial rates of DNA unwinding by the FANCJ proteins were determined from time-points take in 10 s intervals (Figure 1F). FANCJ-WT unwound the fork DNA substrate at a rate of 0.15 bp s−1 FANCJ monomer−1. FANCJ-R707C unwound at a rate of 0.017 bp s−1 FANCJ monomer−1. FANCJ-H396D displayed barely detectable unwinding, consistent with observations in multi-turnover conditions. Based on these results, we conclude that the FANCJ-R707C mutant displays a significantly reduced but still detectable rate of unwinding compared to FANCJ-WT under conditions simulating single-turnover.
Assessment of DNA binding by FANCJ mutant and wild-type proteins
The differential effects of the R707C and H396D mutations on FANCJ helicase activity prompted us to test the recombinant proteins for their ability to bind DNA substrate by electrophoretic mobility shift assays (EMSA). As shown quantitatively in Figure 1G and by gel image in Supplementary Figure S1, FANCJ-WT bound the forked duplex substrate to a greater extent than either mutant; however, FANCJ-R707C bound the forked duplex better than FANCJ-H396D. Both FANCJ-R707C and FANCJ-H396D bound a 45-mer single-stranded DNA oligonucleotide much less efficiently than FANCJ-WT (Supplementary Figure S1B and C). FANCJ-WT, as well as the two FANCJ mutant proteins, bound poorly if at all to a blunt duplex DNA substrate or a synthetic replication fork structure with duplex leading and lagging strand arms (Supplementary Figure S1D and E), indicating that single-stranded DNA is an important component of the nucleic acid substrate in order for the helicase to bind. Based on the results, we conclude that the R707C and H396D mutations significantly compromise DNA binding by FANCJ.
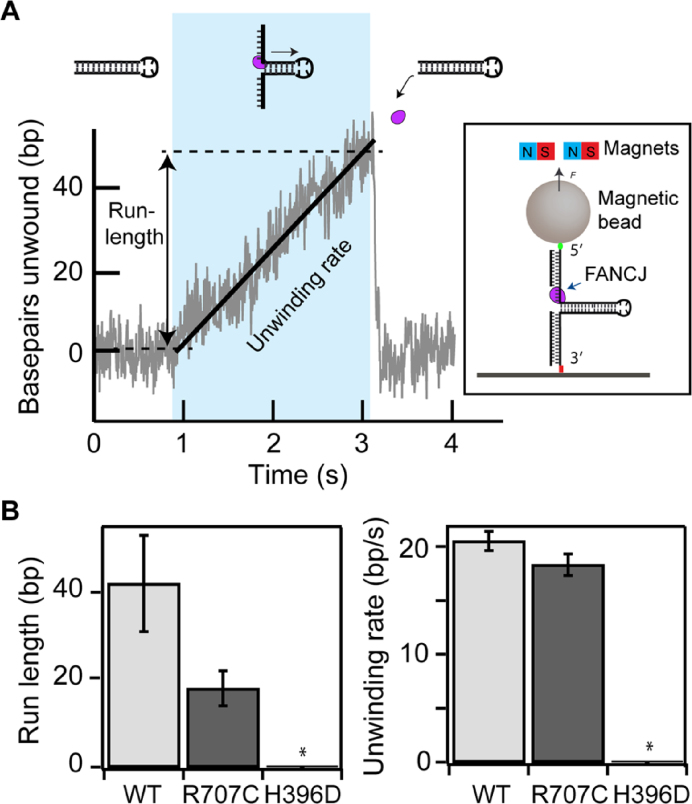
Single-molecule measurements of FANCJ unwinding rate and processivity
The differences in binding affinity and unwinding kinetics among the three FANCJ constructs are consistent with multiple mechanistic interpretations of the effects of the R707C and H396D mutations on FANCJ helicase activity. To resolve the mechanistic effects of the mutations, we employed a magnetic-tweezers based single-molecule DNA unwinding assay (Figure 2A and Materials and Methods). In this assay, the DNA hairpin unwinding activity of individual FANCJ helicases was probed by directly measuring the increase in extension accompanying the opening of the 537 bp hairpin (Figure 2B). Consistent with the ensemble helicase assays above, unwinding activities were observed for FANCJ-WT and FANCJ-R707C, but no activity was observed for FANCJ-H396D. The unwinding rates of FANCJ-R707C and FANCJ-WT were comparable, whereas the processivity, reflected in the average run-length (maximum number of bp unwound before the hairpin rezips) of the R707C mutant was less than half of the WT enzyme (Figure 2B, Supplementary Figures S2 and S3, and Supplementary Table S3). These results indicate that the R707C mutation affects DNA binding, consistent with the binding studies above, but that it has no effect on the catalytic activity per se. Rather, the decrease in DNA binding stability reduced the processivity more than two-fold relative to the WT enzyme.
Figure 2.
Single-molecule DNA hairpin unwinding assay and unwinding characteristics of wild-type and mutant FANCJ enzymes. (A) DNA hairpin unwinding as a function of time showing a DNA hairpin unwinding event (blue shaded region). Individual unwinding events are characterized by their run-length (maximum number of bp unwound before the hairpin rezips) and the unwinding rate (slope of a line fit to the bp unwound as a function of time). Cartoon of a DNA hairpin (not to scale) is shown in which each end is tethered, respectively, to a surface and a magnetic bead that is held under constant tension by applying a magnetic field via external magnets (inset). The number of bp unwound is determined from the height of the magnetic bead. (B) The average run-lengths and unwinding rates of FANCJ-R707C and FANCJ-WT. The unwinding activity of FANCJ-H396D was not detected (*).
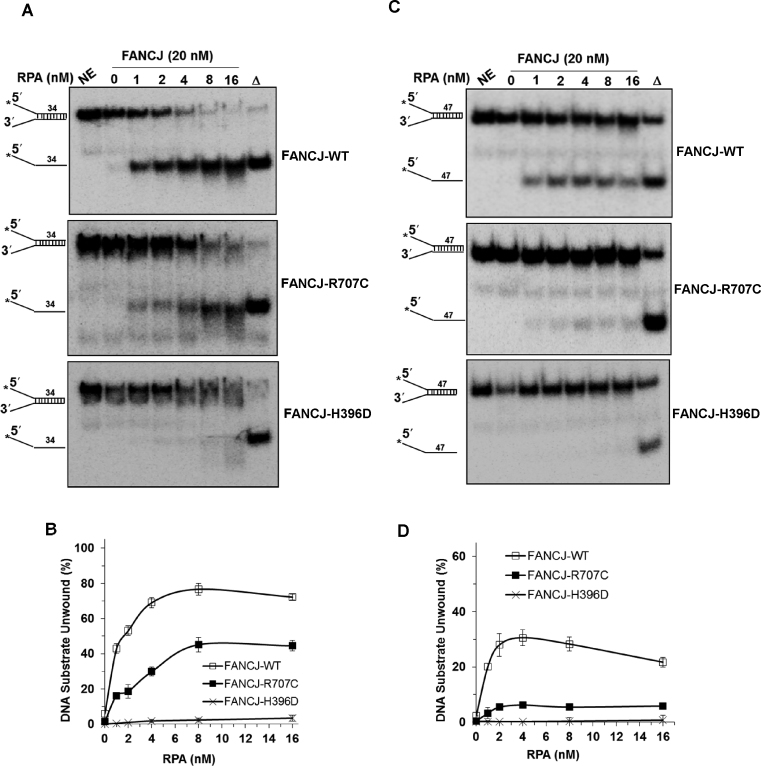
FANCJ-R707C poorly unwinds longer duplex DNA substrates even in the presence of its interacting partner RPA
FANCJ physically and functionally interacts with RPA such that RPA stimulates FANCJ’s DNA unwinding in a specific manner (29). RPA’s ability to interact with FANCJ and stimulate its helicase activity is likely to be important to remodel stalled forks and/or generate single-stranded DNA during a key DNA repair event (e.g. strand resection, strand invasion, or processing of the recombinant D-loop intermediate). The ability of FANCJ-R707C to rescue APH or TMS sensitivity, but not CisPt sensitivity, prompted us to examine the ability of RPA to stimulate its DNA unwinding on substrates with longer DNA duplex regions that the helicase is likely to encounter and act upon in a cellular context.
FANCJ-WT (20 nM) activity alone displayed poor activity on a 34-bp forked duplex DNA substrate under conditions that the helicase unwound a 19-bp forked duplex to near completion at a concentration of 2.4 nM (Figure 3A and B), consistent with its previously observed poor helicase activity on a DNA substrate with a 47 bp duplex length (29). However, in the presence of 1 nM RPA, FANCJ-WT unwound over 40% of the 34 bp DNA substrate; moreover, increasing percentages of the substrate were unwound by FANCJ in an RPA concentration-dependent manner, achieving approximately 80% of the substrate unwound in the presence of 8 nM RPA. Although detectable unwinding of the 34 bp substrate by FANCJ-R707C was observed in the presence of increasing RPA concentrations, the stimulation by RPA was more modest compared to what was observed for FANCJ-WT. Nonetheless, at 8 nM RPA, FANCJ-R707C unwound ∼45% of the 34 bp substrate, indicating that the FANCJ-RPA functional interaction was partially preserved. In contrast, FANCJ-H396D failed to unwind the 34 bp forked duplex substrate throughout the RPA titration.
Figure 3.
FANCJ-R707C displays poor unwinding of longer DNA duplexes even in the presence of its interacting partner RPA. Representative native polyacrylamide gel images showing helicase reaction products from the reactions mixtures containing indicated FANCJ proteins (20 nM) in the presence or absence of the specified concentrations of the single-stranded DNA binding protein RPA with 0.5 nM forked duplex DNA substrate (34 bp (A) or 47 bp (C)) incubated at 30°C for 15 min. (B, D) Quantitative analyses of fork duplex unwinding by FANCJ-proteins is shown for 34 bp (B) or 47 bp (D) substrates with S.D. indicated by error bars. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D.
We also tested a 47 bp forked duplex DNA substrate and observed that FANCJ-R707C displayed very low but detectable helicase activity in the presence of RPA (Figure 3C and D). The maximal amount of 47 bp substrate unwound was ∼5%. FANCJ-WT unwound significantly greater percentages of the 47 bp substrate in the presence of RPA, achieving a maximal 30% substrate unwound. FANCJ-H396D, as expected, failed to measurably unwind the 47 bp substrate at all RPA concentrations tested. Based on these results, we conclude that RPA stimulated FANCJ-R707C, but not FANCJ-H396D, on DNA substrates of 34 bp and 47 bp; however, the strength of RPA stimulation on FANCJ-R707C helicase was highly dependent on the length of duplex in the substrate. Moreover, FANCJ-R707C helicase activity on the 47 bp forked duplex substrate was strongly reduced, even in the presence of RPA, consistent with its reduced processivity evidenced by the single-molecule studies.
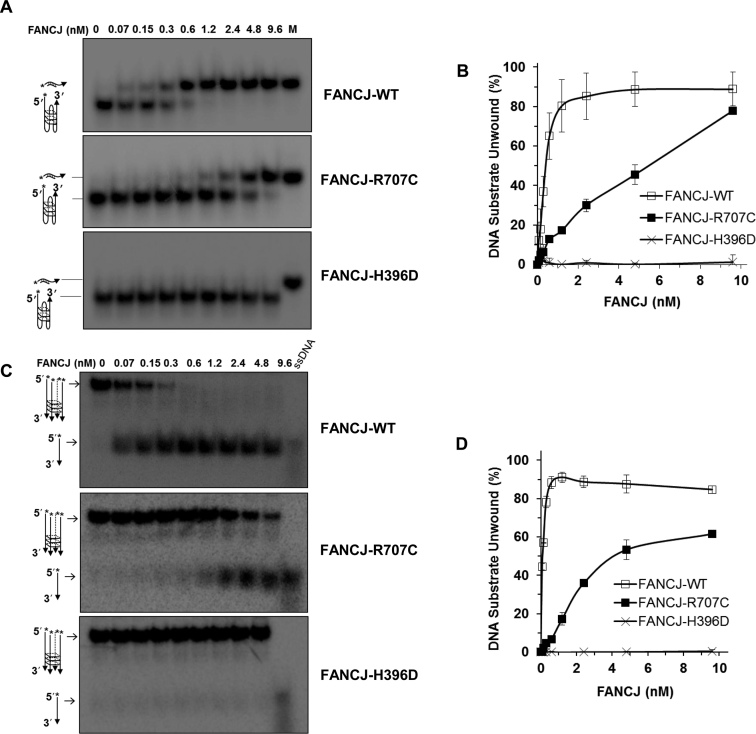
Effect of FANCJ mutations on G-quadruplex resolvase activity
FANCJ is specialized among Fe-S cluster helicases in its ability to unwind intramolecular and intermolecular G quadruplex substrates (7,10). This led us to test the effect of R707C and H396D mutations on FANCJ’s ability to unwind such G4 substrates in vitro. FANCJ-R707C retained partial unwinding activity on a unimolecular G4 DNA substrate compared to FANCJ-WT, whereas FANCJ-H396D was barely detectable (Figure 4A and B). Similar results were obtained when the FANCJ proteins were tested on a tetra-stranded G4 DNA substrate (Figure 4C and D).
Figure 4.
FANCJ G-quadruplex DNA unwinding is abolished by the H396D mutation but only partially compromised by the R707C mutation. (A, B) Helicase reactions were performed with the indicated FANCJ proteins in the presence of 0.5 nM of Poly (A) Zic1 intramolecular G4 DNA substrate and 5-fold molar excess of complementary PNA oligonucleotide at 30°C for 15 min. Representative gel images of FANCJ reaction mixture products analyzed by native polyacrylamide gel electrophoresis are shown with the conversion of unwound G4 substrate into duplex DNA in Panel A. M represents a positive control marker of duplex DNA generated after annealing of single strand G4 DNA and complementary oligonucleotide. Quantitative analysis of intramolecular G4 unwinding by FANCJ-proteins is shown with S.D, indicated by error bars in Panel B. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D. (C, D) Helicase reactions were performed with the indicated FANCJ proteins in the presence of 0.5 nM of tetra-stranded G4 DNA (TPG4) substrate at 30°C for 15 min. Representative gel images of FANCJ reaction mixture products analyzed by native polyacrylamide gel electrophoresis are shown with the conversion of unwound G4 substrate into single-stranded DNA in Panel C. 5′radiolabeled ssDNA as a marker is shown. Quantitative analysis of fork duplex unwinding of FANCJ-proteins is shown with S.D, indicated by error bars in Panel D. Open square, FANCJ-WT; filled square, FANCJ-R707C; open cross, FANCJ-H396D.
Effect of FANCJ mutations on ATPase activity
To further characterize the effects of the FANCJ helicase core mutations on its catalytic activity, we assessed ATP hydrolysis by determining the Michaelis–Menton parameter Km and the turn-over rate constant, kcat (Table 1). Km values for FANCJ-R707C and FANCJ-H396D were determined to be 150 and 92 μM, respectively, lower than that of FANCJ-WT (505 μM). ATP hydrolysis by the FANCJ proteins was analyzed by calculating kcat values from experiments conducted in the presence of 4.4 mM ATP, which is ∼8-fold greater than Km value for FANCJ-WT. FANCJ-R707C and FANCJ-H396D displayed kcat values of 921 and 339 min−1, respectively, which is decreased 3-fold and 8-fold compared to the kcat value of 2730 min−1 for FANCJ-WT (Table 1). The decreased kcat values for ATP hydrolysis by the FANCJ mutant proteins correlated with the effects of the respective mutations on DNA binding and helicase activity. The reduced Km values for ATP hydrolysis by either FANCJ mutant protein may reflect a compromised ability of FANCJ-R707C or FANCJ-H396D to release the bound ATP that is less efficiently hydrolyzed. Calculation of the specificity constant (kcat/Km) suggests that FANCJ-H396D is significantly compromised in its efficiency to hydrolyze ATP, more so than FANCJ-R707C.
Table 1. ATPase kinetic rate constants of FANCJ wild-type and mutant proteins.
| Protein | K m | k cat | k cat/Km |
|---|---|---|---|
| (μM)a,b,c | (min-1)a,b,d | (min-1μM-1) | |
| FANCJ-WT | 505 ± 90 | 2730 ± 90 | 5.4 |
| FANCJ-R707C | 150 ± 50 | 921 ± 30 | 2.3 |
| FANCJ-H396D | 92 ± 30 | 339 ± 15 | 10 |
aFANCJ concentrations were 60 nM.
bM13mp18 ssDNA concentration was 2.1 nM.
cATP concentration ranged from 30 μM to 8000 μM.
dATP was 4.4 mM.
Ability of FANCJ-R707C and FANCJ-H396D to disrupt protein-DNA complexes
FANCJ has the ability to disrupt protein-DNA interactions through its motor function (14). To address the effects of the R707C and H396D mutations on FANCJ catalytic activity in this capacity, we assessed the ability of the mutant proteins to displace streptavidin bound to a biotinylated oligonucleotide, which is estimated to be of a very strong affinity (apparent Kd ∼ 10−15 M (30)). Compared to FANCJ-WT, both FANCJ-R707C and FANCJ-H396D were significantly impaired in streptavidin displacement activity with the greater effect on FANCJ-H396D (Supplementary Figure S4).
R707C mutation negatively affects FANCJ’s ability to dimerize
Previously we had reported that FANCJ dimerizes and that the dimeric form of FANCJ displays greater DNA binding ability as well as increased specific activity for ATP hydrolysis and DNA unwinding (8). These findings led us to test the effects of the H396D and R707C mutations on FANCJ’s assembly state. By size exclusion chromatography, it was determined that FANCJ-WT and FANCJ-H396D eluted from a Superdex200 HR column in two major peaks, one corresponding to a dimer and the other corresponding to a monomer (Supplementary Figure S5). In contrast, FANCJ-R707C was primarily found in the fraction corresponding to a monomer, indicating that the R707C mutation impaired the ability of FANCJ to oligomerize.
Genetic characterization of FANCJ mutant alleles
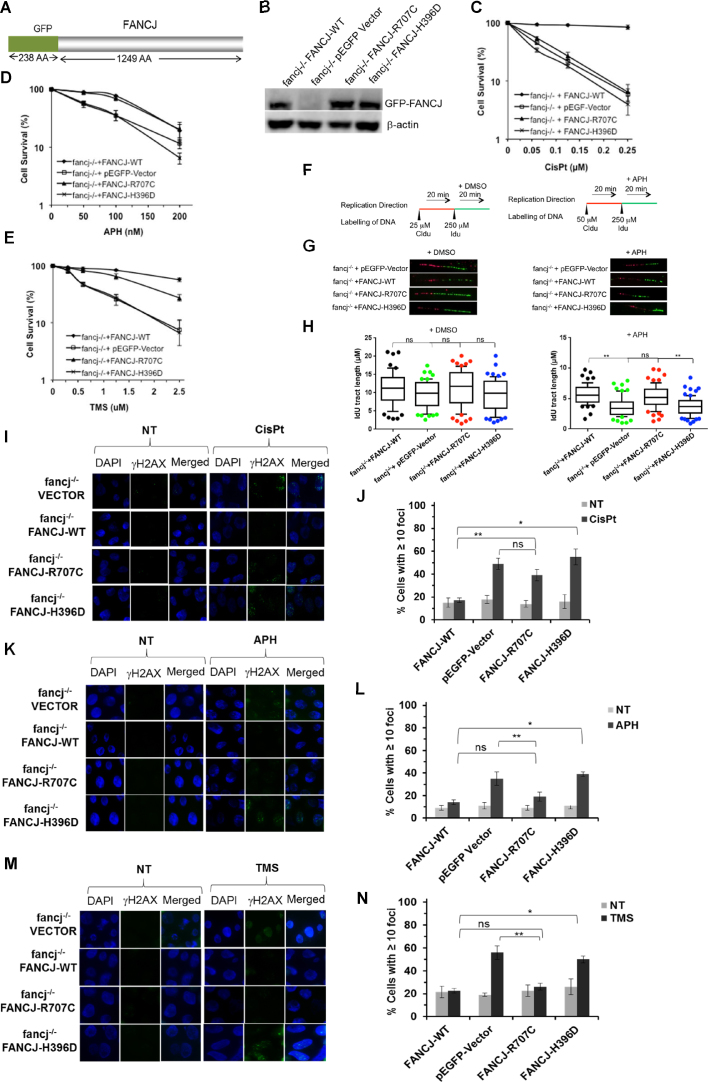
To characterize the functional impact of the two patient-derived FANCJ missense mutations, we performed a series of genetic complementation assays. Because chicken and human FANCJ are conserved (14,31), we exploited DT40 cells harboring a deletion mutation in the fancj gene to assess the functionality of human GFP-tagged wild-type or mutant FANCJ proteins (Figure 5A) that are stably expressed in the indicated cell lines after a process of transfection, selection for antibiotic resistance, colony isolation, and verification of exogenous GFP-tagged FANCJ protein expression. Depending on the isolate FANCJ-H396D and FANCJ-R707C were expressed at a 2- to 3-fold greater level that exogenously expressed FANCJ-WT in the fancj-/- cells, as judged by western blot analysis of total cell lysates (Figure 5B). At least three sub-clones for each stably transfected DT40 cell line were analyzed to verify the reproducibility of genetic complementation assays in the indicated cell lines for sensitivity to the DNA cross-linker CisPt. In addition, APH that inhibits DNA polymerase α (32) or TMS that specifically binds to G-quadruplex DNA (33,34) were tested on the cell lines. FANCJ-deficient cells were previously shown to be sensitive to all of these agents (8,14,35).
Figure 5.
FANCJ-R707C is a separation-of-function mutant that partially rescues sensitivity to the replication stress inducing agents aphidicolin or telomestatin but fails to restore cisplatin resistance. (A) Schematic diagram of green fluorescence protein (GFP)-FANCJ recombinant protein expressed in fancj−/− cells for genetic complementation assays. (B) Western blot analysis of whole cell lysate protein (40 μg) from fancj−/− DT40 cells transfected with plasmid encoding (pEGFP-Vector), pEGFP-FANCJ-WT (FANCJ-WT), pEGFP-FANCJ-R707C (FANCJ-R707C), or pEGFP-FANCJ-H396D (FANCJ-H396D). Protein was detected with antibody against FANCJ or actin (as a loading control, 10% loaded). (C-E) Cell survival assays, as measured by methyl cellulose colony formation, for the indicated fancj−/− cells expressing mutant or wild-type FANCJ proteins. Cells were exposed to the indicated concentrations of CisPt (C), APH (D) or TMS (E). Filled square, fancj−/− cells transfected with FANCJ-WT; open square, fancj−/− cells transfected with pEGFP-Vector; filled triangle, fancj−/− cells transfected with FANCJ-R707C; open cross- fancj−/− cells transfected with FANCJ-H396D. (F, G) FANCJ-R707C restores normal replication restart after APH exposure. Schematic representation of the protocol used to track DNA replication fibers is shown in Panel F. Cells were pulse-chased with CldU (red label), and then labeled with IdU (green label) for the indicated times. (G) Representative images of fluorescently-labeled DNA fibers from the indicated cell lines treated with DMSO or APH. (H) Box and whiskers graphs indicating the 10–90 percentile of the IdU tract length (μm) for ongoing forks. The data, presented as mean ± standard error of the mean (s.e.m.), are based on the measurement of at least 100 DNA fibers from two independent experiments. (I–N), DNA damage, as marked by immunofluorescent detection of γ-H2AX foci, in the indicated fancj−/− transfected cell lines. fancj−/− cells transfected with pEGFP-Vector, pEGFP-FANCJ-WT (FANCJ-WT), pEGFP-FANCJ-R707C (FANCJ-R707C) or pEGFP-FANCJ-H396D (FANCJ-H396D) were exposed for 14 h to: CisPt (1 μM) (I, J); APH (200 nM) (K, L); TMS (5 μM) (M, N). Immunofluorescence detection of γ-H2AX foci by Alexa fluor 488 is shown along with DAPI, or merged (DAPI and Alexa fluor 488). Quantitative analyses of γ-H2AX foci are shown with S.D. (ns-not significant; **P < 0.05, *P < 0.01 (Student's t-test).
Initially, colony formation assays were performed to assess the effect of the DNA cross-linker CisPt on cell survival (8,14). fancj−/− cells expressing FANCJ-R707C or FANCJ-H396D were highly sensitive to CisPt (Figure 5C). fancj−/− cells complemented with FANCJ-WT exhibited only a 12% reduction in colonies at a CisPt concentration of 0.25 μM, whereas fancj−/− cells expressing FANCJ-R707C or FANCJ-H396D displayed only a 10% colony survival, comparable to that observed for cells transfected with empty vector. The CisPt sensitivity of fancj−/− cells expressing FANCJ-H396D or FANCJ-R707C prompted us to assess for chromosome aberrations. Metaphase spreads revealed that chromosome aberrations were significantly elevated in the cell lines expressing the FANCJ mutant proteins at a level comparable to the fancj−/− vector cells (Supplementary Figure S6).
Next, we assessed colony formation for the respective fancj−/− transfected cell lines after exposure to APH, which yielded some unexpected results (Figure 5D). fancj−/− cells expressing FANCJ-R707C retained a level of APH resistance comparable to those cells exogenously expressing FANCJ-WT, a finding that was very different from what was determined for CisPt. In contrast, fancj−/− cells expressing FANCJ-H396D displayed cell survival after APH exposure very much the same as fancj−/− cells transfected with empty vector. The pathway for resumption of daughter strand synthesis after polymerase inhibition may be affected by FANCJ helicase status, given its importance in the replication stress response. To address this, DNA synthesis rates were estimated by tract lengths acquired from sequential pulse experiments with the various fancj−/− transfected cell lines. Replication track analysis was performed after labeling the cells with CldU to track ongoing fibers and then with IdU to measure their progression in the presence or absence of APH (Figure 5F). In the absence of APH, measurement of IdU indicated that fancj−/− vector cells did not show any significant reduction in fiber length (Figure 5G and H). However, in the presence of APH (200 nM, 20 min), fancj−/− vector cells showed ∼35% reduction in IdU tract lengths compared to fancj−/− cells exogenously expressing FANCJ-WT. fancj−/− cells expressing FANCJ-R707C did not show any significant reduction in IdU tract length, whereas fancj−/− cells expressing FANCJ-H396D showed ∼35% reduction in IdU length, similar to what was observed for fancj−/− vector cells. Based on these results, we conclude that FANCJ-R707C, but not FANCJ-H3986D, enables cells to respond to replication stress induced by APH in a manner similar to fancj-/- cells exogenously expressing FANCJ-WT.
We next examined the sensitivity of the fancj−/− transfected cell lines to the G4 stabilizing ligand TMS (Figure 5E). Here, we observed that cells expressing FANCJ-R707C were mildly sensitive to TMS, whereas cells expressing FANCJ-H396D were highly sensitive to the compound. Because TMS specifically binds to G4 structures (7,34), we wanted to determine if TMS-treated cells expressing FANCJ-R707C which was partially active on G4 DNA substrates would show a lower level of immune-reactive G4. Consistent with our previous observation (22), fancj−/− vector cells exposed to TMS displayed a 2.5-fold increase in G4 staining foci compared to fancj−/− cells expressing FANCJ-WT (Supplementary Figure S7). TMS exposure to fancj−/− cells expressing FANCJ-R707C displayed G4 staining foci similar to those cells expressing FANCJ-WT, whereas fancj−/− cells expressing FANCJ-H396D displayed elevated G4 staining comparable to fancj−/− vector cells. These results, taken together with the TMS survival data, suggest that G4 accumulation in cells expressing helicase-dead FANCJ-H396D or completely lacking FANCJ, underlies the observed sensitivity to the G4 ligand. In contrast, cells expressing the partially helicase-active FANCJ-R707C were largely resistant to TMS-induced G4 staining and only mildly sensitive to the G4 ligand.
The ability of FANCJ-R707C expressed in fancj−/− cells to rescue APH or TMS sensitivity but not CisPt sensitivity as measured by survival assays raised the question if the separation of function mutant FANCJ-R707C would rescue sensitivity to agents that induce ICL lesions or DSBs that could be repaired by HR in cells undergoing replication. To address this, the transfected fancj-/- cell lines were exposed to the cross-linking agent MMC or the drug Bleo which directly introduces DSBs. As shown in Supplementary Figure S8, FANCJ-R707C failed to restore MMC resistance, but partially rescued Bleo sensitivity. These results suggest that the partially helicase active FANCJ-R707C mutant protein can function to some extent in repair of Bleo-induced DSBs, but not in CisPt- or MMC-induced cross-links.
Stalled or regressed replication forks are known to undergo degradation by Mre11 exonuclease, which is proposed to contribute to chromosomal instability (36). Previously, it was reported that FA proteins play a role in fork protection from degradation during conditions of replication stress (37). Therefore, we tested if the Mre11 exonuclease inhibitor Mirin affects survival of fancj-/- cells transfected with empty vector or expressing helicase-inactive FANCJ-H396D that are exposed to the DNA polymerase inhibitor APH. Consistent with previously published results for Chinese hamster ovary cells (38), Mirin alone caused reduced cell survival as measured by colony formation; however, cells expressing FANCJ-WT or FANCJ-R707C showed greater resistance than cells expressing FANCJ-H396D or no FANCJ (Supplementary Figure S9). Moreover, Mirin failed to confer resistance to APH or CisPt irrespective of FANCJ status (Supplementary Figure S9).
FANCJ-H396D, but not FANCJ-R707C, localizes efficiently to laser-induced DNA damage
The inability of FANCJ-R707C or FANCJ-H396D expressed in fancj−/− cells to rescue their sensitivity to the DNA cross-linking agents CisPt or MMC raised the question if these mutant proteins were able to localize to sites of DNA damage. To address this, we examined their recruitment kinetics to laser-induced DSBs or Pso-ICLs introduced at localized areas within the nuclei of asynchronized U2OS cells, as visualized by live-cell imaging. Representative cells showing the recruitment pattern of wild-type and mutant GFP-FANCJ proteins to DSBs or ICLs are displayed in Supplementary Figure S10A and C, and quantification found in Supplementary Figure S10B and D, respectively. Consistent with previously published results for FANCJ-WT (24), FANCJ-H396D recruited to both DSBs and ICLs rapidly. However, FANCJ-R707C recruited inefficiently or hardly at all to laser-induced DSBs and failed to detectably recruit to laser-activated Pso-ICLs. Based on these results, we conclude that FANCJ-H396D was proficient in its recruitment to sites of laser-induced DNA damage, whereas FANCJ-R707C was seriously compromised in its recruitment to DSBs and defective altogether in its recruitment to laser-activated Pso-ICLs.
DNA damage induction in cells expressing FANCJ mutants
Given the evidence that FANCJ operates in HR repair to correct lesions arising directly from endogenous or exogenous stress, or during the process of ICL repair (5), we wanted to assess markers of HR repair in dividing cells expressing the FANCJ mutant proteins. First, we examined γ-H2AX foci, a well-known maker of DNA damage, particularly DSBs (39). Exposure of fancj−/− cells to CisPt (2 μM, 14 h) resulted in a 3-fold increase in cells with ≥10 γ-H2AX foci compared to the same cells exogenously expressing FANCJ-WT (Figure 5I and J), consistent with our previous results (8). CisPt-treated fancj−/− cells expressing either FANCJ-R707C or FANCJ-H396D displayed a 2.5-fold or 4-fold increase in γ-H2AX foci, respectively, as compared to fancj−/− cells. Therefore, neither FANCJ mutant effectively suppressed DNA damage in cells exposed to a DNA cross-linking agent.
Next, we monitored γ-H2AX foci levels in the same cell lines exposed to APH (200 nM, 14 h). In this case, APH exposure to fancj−/− vector cells resulted in a 3.5-fold increase in cells with ≥10 γ-H2AX (Figure 5K and L). However, fancj−/− cells expressing FANCJ-R707C did not show a statistically significant increase in γ-H2AX foci, whereas fancj−/− cells expressing FANCJ-H396D displayed a 4-fold increase. Based on these results, we conclude that FANCJ-H396D failed to suppress DNA damage induced by APH-stalling of replication forks. In contrast, FANCJ-R707C functioned in this capacity. We then tested the effect of cellular exposure to the G4 ligand TMS (2.5 μM, 14 h) on γ-H2AX foci and observed that the nearly 3-fold increase in γ-H2AX in fancj−/− cells was suppressed by the expression of FANCJ-R707C, but not FANCJ-H396D (Figure 5M and N). This finding was in the same line with that for APH. Furthermore, the results from the γ-H2AX induction assays were consistent with those obtained from the cell survival measurements, indicating that FANCJ-R707C, but not FANCJ-H396D, rescued the sensitivity of cells lacking FANCJ-WT to APH or TMS, agents that induce replication stress. However, FANCJ-R707C was not able to rescue the survival deficiency or DNA damage induction caused by exposure to the cross-linking agent CisPt.
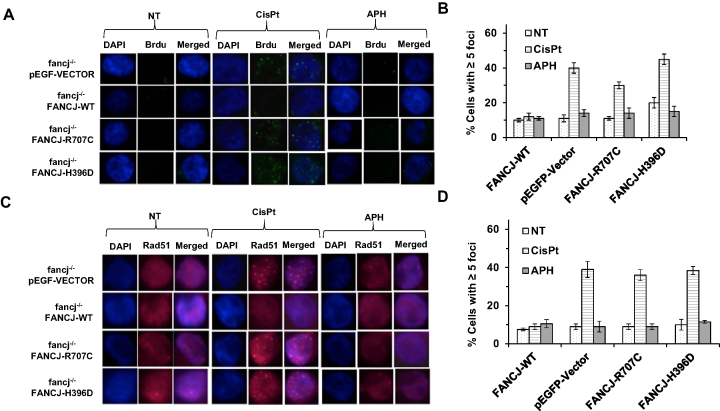
FANCJ-R707C fails to suppress single-stranded DNA or Rad51 persistence in cells exposed to CisPt
The inability of either FANCJ-R707C or FANCJ-H396D to suppress the DSBs induced by CisPt suggested that the HR step to correct the processed ICL might be defective. Strand resection initiated by the Mre11-Rad50-NBS1 (MRN) complex along with CtIP and extended by BLM interactions with the 5′ processing nucleases DNA2 or EXO-1 results in long 3′ single-stranded DNA tails coated by RPA which are subsequently replaced with Rad51, the major strand exchange protein that mediates invasion into sister chromatid duplex to form a displacement loop (D-loop) involving the nucleoprotein filament (40). Using its motor ATPase function, FANCJ was previously shown to disrupt fixed D-loop DNA substrates (41), destabilize Rad51 nucleoprotein filaments (41), and inhibit Rad51 DNA strand exchange activity (41) in vitro. In addition, FANCJ physically and functionally interacts with the Mre11 (24) and BLM (42) proteins, suggesting that FANCJ modulates single-stranded DNA resection and/or Rad51 strand exchange. Therefore, we sought to determine if fancj−/− cells expressing FANCJ-R707C or FANCJ-H396D show an abnormal level of CisPt-induced single-stranded DNA or Rad51 foci in vivo. For detection of single-stranded DNA, we pre-labeled cells with 5-bromodeoxyuridine (BrdU), and subsequently stained the cells that had been exposed to CisPt (2 μM, 14 h) with a BrdU antibody that specifically recognizes the modified nucleotide when it is in the single-stranded state (43); therefore, by avoiding DNA denaturation, the single-stranded DNA arising from post-ICL DNA processing could be visualized. BrdU staining of the CisPt-treated fancj−/− cells expressing FANCJ-H396D or FANCJ-R707C behaved like fancj−/− cells transfected with empty vector, showing a three- to four-fold increase in percentage of cells with ≥5 BrdU foci (Figure 6A and B). In contrast, CisPt-treated fancj−/− cells exogenously expressing FANCJ-WT did not show any difference in BrdU staining compared to untreated cells, suggesting that any single-stranded DNA generated during ICL repair was promptly repaired via the HR pathway. Similar experiments performed with cells that had been prelabeled with BrdU and subsequently exposed to APH (200 nM, 14 h) revealed no differences in BrdU foci compared to untreated cells irrespective of FANCJ status (Figure 6A and B), suggesting that persistent single-stranded DNA is not generated at APH-stalled replication forks under the experimental conditions.
Figure 6.
FANCJ-R707C fails to suppress single-stranded DNA or Rad51 persistence in cells exposed to CisPt. fancj−/− cells transfected with pEGFP-Vector, pEGFP-FANCJ-WT (FANCJ-WT), pEGFP-FANCJ-R707C (FANCJ-R707C) or pEGFP-FANCJ-H396D (FANCJ-H396D) were exposed for 14 h to CisPt (1 μM) (A, B) or APH (200 nM) (C, D). Note that for BrdU staining, cells were pre-labeled with BrdU prior to drug exposure as described in Materials and Methods. (A, C) Immunofluorescent BrdU foci (A) or Rad51 foci (C) were detected by Alexa fluor 488 and are shown along with DAPI, or merged (DAPI and Alexa fluor 488). (B, D) Quantitative analyses of BrdU foci (B) or Rad51 foci (D) are shown with S.D. indicated by error bars.
Next, we performed similar experiments to immuno-stain for Rad51 in the various fancj−/− cell lines exposed to either CisPt or APH (Figure 6C and D). As was observed for BrdU foci, CisPt-treated fancj−/− cells expressing either FANCJ-H396D or FANCJ-R707C behaved like fancj−/− cells transfected with empty vector, showing an approximately four-fold increase in percentage of cells with ≥5 Rad51 foci. In contrast, CisPt-treated fancj−/− cells exogenously expressing FANCJ-WT did not show any difference in Rad51 staining compared to untreated cells, suggesting that Rad51 filaments do not persist after ICL-induced DNA damage when FANCJ-WT is present. Similar experiments performed with cells exposed to APH did not show any difference in Rad51 foci compared to untreated cells irrespective of FANCJ status (Figure 6C and D), suggesting that Rad51 filament formation does not occur or persist at APH-stalled replication forks.
FANCJ deficiency does not perturb Chk1 activation induced by aphidicolin
The poor replication fork recovery of FANCJ-deficient cells from APH exposure may be attributed to a role of FANCJ in remodeling stalled forks, activation of the Chk1/ATR-dependent S-phase checkpoint, or a combination of the two. To assess if the replication checkpoint plays a role in the ability of FANCJ-R707C to rescue APH sensitivity of expression of fancj−/− cells, we examined Chk1-Ser 345 phosphorylation, a well-known marker of the ATR checkpoint (44). We observed that a 1 h exposure of APH concentrations as low as 200 nM induced robust Chk1-Ser 345 phosphorylation in DT40 cells. fancj−/− vector cells, or fancj−/− cells expressing FANCJ-H396D, FANCJ-R707C, or FANCJ-WT displayed Chk1 phosphorylation in a comparable manner (Supplementary Figure S11). Based on these results, we suggest that the partially active FANCJ-R707C helicase plays a direct role in restarting APH-stalled replication forks that does not involve modulation of the ATR checkpoint.
FANCJ-R707C partially rescues aphidicolin or telomestatin sensitivity independent of the FA pathway
The FA pathway of ICL repair is generally believed to be a linear one involving sequential steps: (i) ICL recognition by the FA core complex; (ii) FANCD2/FANCI mono-ubiquitination and chromatin association; (iii) ICL unhooking by structure-specific nucleases; (iv) translesion DNA synthesis past the ICL remnant; (v) HR repair of the DSB (3,45). Although FANCJ is believed to operate in the HR step of ICL repair, its precise mechanism is not well understood and the dependence of FANCJ on other FA proteins and associated factors in the DNA damage or replication stress response has attracted interest. Recently it was shown that FANCD2, FANCJ and BRCA2 cooperate in promoting replication fork recovery following stalling by APH in a process that is independent to their involvement in the FA pathway (6). This led us to ask if genetic rescue of fancj−/− cells to APH or TMS by FANCJ-R707C in our experimental model was dependent on an upstream player of the classic FA ICL repair pathway, namely FANCC, a member of the FA core complex. Firstly, as expected, FANCJ-R707C or FANCJ-WT, failed to rescue sensitivity to the DNA cross-linker CisPt in fancj−/− cells that were also deficient in FANCC (Supplementary Figure S12). However, for APH or TMS, expression of FANCJ-R707C partially rescued the sensitivity of fancj−/− fancc−/− cells (Supplementary Figure S12), indicating that the ability of FANCJ-R707C to help cells cope with replication stress by a DNA polymerase inhibitor or G4 ligand was not dependent on FANCC.
Given evidence that BLM helicase is important for the replication stress response and interacts physically and functionally with FANCJ (42), we wanted to assess if genetic rescue by FANCJ-R707C of sensitivity to CisPt, APH or TMS was dependent on BLM. Unfortunately, fancj−/− blm−/− cells grew very poorly (Supplementary Figure S13A) and cell cycle analyses revealed that they accumulate in sub-G1 (Supplementary Figure S13B). Although the growth defects of these cells precluded further analysis with FANCJ variants, the results indicate that BLM is required even under endogenous stress conditions to compensate for lack of FANCJ function.
DISCUSSION
The FANCJ-R707C amino acid substitution in motif IV of the helicase core domain represents a partial loss-of-function mutant in a biochemical sense. The reduced DNA binding, ATPase, and helicase activity rendered by the FANCJ-R707C mutation can be rationalized by homology modeling with the TaXPD crystal structure in which the R707 position is projected to reside near the single-stranded DNA binding cleft (28). The FANCJ-H396D mutation in motif II, the Walker B box implicated in ATP binding and hydrolysis, nearly completely inactivates catalytic activity of FANCJ, consistent with the TaXPD homology model in which the invariant histidine resides at the interface of the two RecA domains responsible for the motor ATPase function of the enzyme. Because the ATPase activity of FANCJ is strongly coupled to DNA binding (11,46), we surmise that the negative effect of the H396D mutation on DNA binding as well as the amino acid substitution's direct impact on ATP hydrolysis are responsible for the strong reduction in the kcat for ATP hydrolysis. Further insight to the molecular interactions of FANCJ with DNA and ATP awaits its structural characterization.
The FANCJ-R707C mutation clearly interfered with dimerization, which would significantly contribute to its reduced DNA binding and catalytic activity based on our previous observations that the dimeric form of FANCJ shows greater specific activity for DNA binding, ATP hydrolysis and helicase activity compared to the FANCJ monomer (8). Thus, the FANCJ-R707C mutation is a dimerization-defective mutant, similar to a previously described Q25A mutation in the Q motif residing just upstream of the Walker A (motif I) and Walker B (motif II) boxes of FANCJ (8). However, unlike FANCJ-R707C the recombinant FANCJ-Q25A mutant was completely inactive as a helicase on forked duplex or G4 DNA substrates. Moreover, FANCJ-Q25A, like FANCJ-R707C, failed to rescue for CisPt resistance. In contrast to FANCJ-R707C but similar to FANCJ-H396D, the helicase-dead Q25A mutant failed to confer resistance to the G4 ligand TMS. Therefore, the complete inactivation of FANCJ helicase activity exerted by the H396D or the Q25A substitution is likely responsible for the inability of the mutated FANCJ to confer resistance to ICL-induced DNA damage or TMS-stabilized G4 DNA structures.
The FANCJ-R707C mutant retained catalytic DNA unwinding activity on a short (19 bp) forked duplex or 5’ tailed G4 DNA substrate; moreover, FANCJ-R707C-calayzed DNA unwinding of the longer 34 bp forked substrate was stimulated by its interacting partner, RPA. Therefore, the R707C mutation has not drastically altered the overall global structure and functionality of FANCJ; however, FANCJ-R707C helicase activity is significantly compromised on the longer 47 bp forked duplex substrate, even in the presence of its interacting partner, RPA. In addition, the motor ATPase-driven displacement of streptavidin from a biotinylated oligonucleotide is significantly compromised for the FANCJ-R707C mutant. Therefore, it is reasonable to postulate that the apparent 3-fold reduced kcat for ATP hydrolysis is associated with the pronounced decrease in accomplishing more arduous tasks, i.e. unwinding of relatively long duplexes or disruption of high affinity protein-DNA interactions. The single-molecule results are consistent with this general scenario, and permit a refinement of the mechanistic consequences of the R707C mutation. The run-length of FANCJ-R707C was significantly reduced whereas the unwinding rate was comparable to WT, suggesting that the R707C mutation directly affects processivity by reducing DNA binding stability, in line with the measured reduction in DNA affinity for the R707C mutant. The apparent decrease in kcat for ATP hydrolysis measured in ensemble assays is likely influenced by the reduced processivity and lower binding affinity.
We propose that the partial loss of biochemical activity attributed to the R707C mutation is manifested in the FANCJ-R707C separation-of-function allelic effects assayed in vivo. FANCJ-R707C expressed at a level comparable to exogenous FANCJ-WT failed to rescue CisPt or MMC sensitivity. In contrast, FANCJ-R707C rescued sensitivity to the DNA polymerase inhibitor APH or G4 ligand TMS. The simplest interpretation of the biochemical and genetic data is that a lower catalytic threshold of FANCJ helicase activity is required for the cellular response to DNA polymerase inhibition or small molecule-induced stabilization of G-quadruplexes compared to repair of ICLs. The observation that the catalytically defective FANCJ-H396D mutant fails to rescue APH or TMS sensitivity establishes a window for detecting the requisite level of FANCJ catalytic activity required for the replication stress response to APH or TMS. Furthermore, comparison to FANCJ-R707C indicates that processivity and DNA binding are key determinants for FANCJ’s role in response to APH or TMS.
FANCJ-R707C (or FANCJ-H396D) could be co-immunoprecipitated with MLH1 (FANCJ’s protein partner in cross-link repair (47)) from HeLa whole cell extracts (Supplementary Figure S14), suggesting that the helicase core domain mutations did not drastically interfere with FANCJ’s ability to interact with the mismatch repair factor; however, the partially helicase active FANCJ-R707C was defective in dimerization and showed compromised recruitment to sites of DNA damage. The marked decrease in unwinding long and difficult DNA substrates or displacing proteins can be attributed to the same decrease in affinity that decreases processivity. For the long DNA substrate, the effect on processivity is obvious. In the other two cases, if the forward rate of the enzyme decreases, or it spends more time in a weak DNA binding state as a result of the impediments to translocation (G-quadruplex or bound protein), then the R707C mutant enzyme will have a much higher probability of detaching and failing to resolve the G4 or displace streptavidin than the WT enzyme. This would persist even in the presence of RPA.
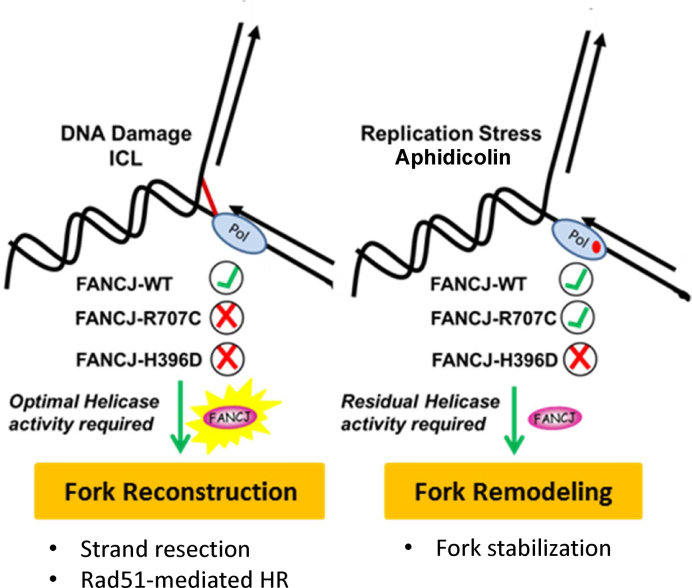
To explain the separation-of-function incurred by the R707C mutation, we surmise FANCJ-R707C retains the ability to catalyze unwinding of relatively short DNA duplexes in order to remodel stalled forks during replication or to resolve secondary DNA structure that impedes DNA synthesis (Figure 7). In addition, FANCJ-R707C confers partial resistance to Bleo, a drug that induces single-strand breaks and DSBs characterized by 5′-staggered ends or blunt ends (48) that are repaired at least in part by nonhomologous end-joining (49). Consistent with the Bleo rescue, FANCJ-R707C was able to recruit to DSBs, albeit less efficiently than FANCJ-H396D or FANCJ-WT (24). However, the catalytically compromised FANCJ-R707C that recruits poorly to ICL DNA damage is unable to perform its more extensive role(s) in HR required for fork reconstruction after the unhooking step of ICL repair (e.g. strand resection, strand invasion, or maturation of a D-loop intermediate, Rad51 dissociation). The distinctive nature of CisPt-induced DNA damage versus TMS-stabilized secondary structure or APH-stalled DNA synthesis is evident by the immunofluorescent detection of γ-H2AX foci, single-stranded DNA, or Rad51 in cells exposed to these agents under the described experimental conditions. Fork remodeling after APH stalling or G4 secondary structure resolution is less destructive to the fork and does not require the extensive nucleolytic DNA processing and subsequent Rad51-mediated strand exchange invoked for completion of ICL repair by HR with the sister chromatid duplex. Presumably, at stalled forks the limited single-stranded DNA produced by a minimal threshold of FANCJ catalytic activity (sufficiently performed by FANCJ-R707C) and MRE11-dependent DNA degradation is not of sufficient length to load RPA and hence Rad51 after fork arrest. However, after ICL-induced DNA damage, extensive single-stranded DNA is generated by strand resection machinery at the broken replication fork such that Rad51 is loaded for HR repair to ensue in a manner that requires optimal FANCJ catalytic activity for its downstream role.
Figure 7.
Model depicting the divergent roles of FANCJ in DNA repair and the replication stress response, as evidenced by the characterization of the FANCJ-R707C separation of function mutant. A lower catalytic threshold of FANCJ helicase activity is required for the cellular response to DNA polymerase inhibition or G-quadruplex accumulation compared to repair of interstrand cross-links.
Greenberg et al. reported that FANCJ-depleted HeLa cells displayed an elevated S-phase accumulation after low-dose aphidicolin treatment, suggesting a poor response to S-phase DNA damage (50). However, FANCJ depletion in HeLa cells did not affect IR-dependent phosphorylation of CHK1 at Ser 317 or CHK2 phosphorylation, suggesting that FANCJ acts independently of CHK1/CHK2 when DNA damage is encountered (50). We observed that FANCJ-deficient cells exposed to APH were not defective in CHK1 activation, suggesting that FANCJ’s ability to promote efficient restart of replication upon APH-induced DNA polymerase α stalling is not likely to involve ATR/Chk1 checkpoint activation. This finding is distinct from the earlier observation that Chk1 activation is dependent on FANCJ in U2OS cells exposed to a high (10 mM) concentration of hydroxyurea (51). Nonetheless, a threshold of FANCJ helicase activity is required for efficient response to replication stress as determined by our replication tract analysis in which fancj−/− cells expressing partially active FANCJ-R707C behaved similar to the same cells exogenously expressing FANCJ-WT, whereas helicase-dead FANCJ-H396D expressing cells behaved like fancj−/− vector cells showing shorter replication tracts.
Cross-talk between FANCJ and the BRCA pathway in the replication stress response is likely. The BRCA2/FANCD1 gene was found to protect nascently synthesized DNA strands at stalled replication forks (caused by hydroxyurea (HU)-induced nucleotide depletion) from degradation by the MRE11 nuclease; however, replication track analyses suggested that while BRCA2 status did not affect replication fork restart, BRCA2 may suppress uncoupling of leading strand and lagging strand synthesis (36). A separation-of-function BRCA2 C-terminal mutation that destabilizes its interaction with RAD51 crippled its fork protection role but not its ability to perform HR of DSBs, suggesting that BRCA2 requirements for fork protection versus HR of DSBs are distinct (36). Subsequently, it was reported that the FANCD2 and BRCA1 (FANCS) genes also protect stalled forks from MRE11 degradation in a RAD51-dependent pathway, but (like BRCA2) no defects in fork recovery were observed (37). Further studies showed that RAD51 and BRCA2 behave in epistatic fashion with FANCD2 to stabilize stalled forks and that FANCD2-deficient cells were protected from HU-induced degradation of nascent strands by either expression of an ATPase-dead version of RAD51 that is highly stable on DNA or elevated expression of wild-type RAD51 (37). Finally, assessment of BRCA2-defective cells expressing a S3291A variant that is functional in DSB repair but unable to protect stalled forks to ensure survival after exposure to the DNA cross-linking agent mitomycin c (MMC) showed only moderate sensitivity at high dose, suggesting that the functions of BRCA2 during ICL repair versus stalled fork stabilization could be delineated (37).
Our observations that fancj−/− cells (as well as fancj−/− cells exogenously expressing FANCJ-H396D) were markedly sensitive to the MRE11 exonuclease inhibitor Mirin is similar to the drug's negative effect on clonogenic survival reported for BRCA2-deficient cells (38). Interestingly, expression of FANCJ-R707C conferred greater resistance to Mirin than FANCJ-H396D, suggesting that limited processive DNA unwinding by FANCJ is required for cell survival under conditions of attenuated MRE11 nucleolytic activity. In addition, pre-exposure of fancj−/− cells (or fancj−/− cells expressing FANCJ-H396D) to Mirin did not suppress their sensitivity to aphidicolin, suggesting that fork protection from MRE11-dependent degradation does not prominently affect survival of FANCJ helicase-deficient cells experiencing replication stress under pharmacological conditions of DNA polymerase α inhibition.
Recently, the Sobeck lab determined from cell-based experiments that a non-ubiquitinated isoform of FANCD2 functioning with FANCJ and BRCA2 is sufficient to promote efficient replication fork recovery from APH exposure (6). Our results showing that the FA core complex member FANCC is not required for APH resistance are consistent with this; furthermore, the FANCJ-R707C mutant behaved like FANCJ-WT in this capacity, indicating that only partial FANCJ catalytic activity is necessary. Clearly, FANCJ operates outside the classic FA pathway of ICL repair to confer resistance to replication stress caused by DNA polymerase α inhibition ((6); current study) or G4 ligand exposure (7). Interestingly, while BLM-deficient cells are sensitive to agents that stall replication forks (52), the role of BLM appears to be in fork restart and not fork protection (52). Further studies are required to determine if FANCJ (which interacts with BLM (42)) also plays a role in fork restart or if it has any other role to help cells cope with replication stress.
How is the biochemical and genetic characterization of the disease-causing FANCJ mutations relevant from a biomedical perspective? Characterization of helicase missense mutations genetically linked to a chromosomal instability disorder like FA (that is prone to cancer) or mutations associated with non-Mendelian cancers might yield insight to improved diagnosis or intervention (53). The FANCJ-R707C allele was identified along with a FANCJ-W647C mutation in the second allele in an individual with FA (27). The FANCJ-W647C mutant was determined to be completely inactive for its helicase activity (Supplementary Figure S15) and displayed a dramatic 17-fold reduction in its kcat for ATP hydrolysis (Supplementary Table S4). Therefore, cells from the FA individual with the allelic combination of FANCJ-R707C and FANCJ-W647C would only retain the partial helicase activity catalyzed by the FANCJ-R707C protein. In another individual with FA, the FANCJ-H396D allele was described with a second allelic FANCJ-R251C mutation (26). Previously, the FANCJ-R251C mutant was characterized genetically and biochemically and found to be completely inactive for its helicase activity and fail to complement fancj−/− cells for cisplatin resistance (54). Therefore, the cells from this FA individual would be completely devoid of FANCJ helicase activity. Unfortunately, the medical data for the individuals with either the FANCJ-R707C / FANCJ-W647C or FANCJ-H396D/FANCJ-R251C allelic mutations were unavailable, and patients expressing homozygous FANCJ-H396D or FANCJ-R707C alleles have not been reported; consequently, we were unable to assess genotype-phenotype relationships in terms of clinical features. However, the biochemical and genetic characterization of these FANCJ missense mutants suggest that cellular sensitivity to a DNA cross-linking agent like CisPt is a good marker for FA, whereas cellular sensitivity to an agent like APH or TMS that induces replication stress is not a strong indicator for the molecular diagnosis of FA.
From a cancer viewpoint, there is great interest in the field of chemotherapy drug discovery to develop strategies that exploit the replicative stress of rapidly dividing cancer cells to cause catastrophic devastation of the cell proliferation machinery (55). In personalized medicine, the treatment strategy for a cancer patient may take into account the existing personal genome sequence of the individual to tailor an appropriate intervention strategy. We believe that DNA helicases, like FANCJ, play critical roles in the DNA damage response elicited by many chemotherapy drugs or radiation (56,57). In our current work, we discovered a FANCJ missense mutation that differentially affects the cellular response to a chemotherapeutic ICL-inducing agent versus a replication inhibitor APH that has undergone preclinical studies for potentiation of cisplatin cytotoxicity (58), or a representative G4 ligand that has attracted interest for pharmaceutical use to target telomeres or oncogene promoters to induce cancer cell senescence (59,60). With further research, characterization of separation of function alleles like FANCJ-R707C may lead to novel cancer treatment strategies. Moreover, virtual docking of small molecules at the R707 site in FANCJ may lead to the identification of a compound that only partially affects FANCJ catalytic activity; lead compound development may ultimately produce a compound that sensitizes cancer cells with heightened replicative sensitivity to the helicase-specific agent. With the advent of whole genome high-throughput sequencing and vast small molecule screens, it is conceivable that specialized DNA metabolic enzymes like helicases may become very useful targets to eliminate cancers and reduce the non-specific cytotoxicity incurred by many currently used chemotherapy regimens.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr Peter Lansdorp (University Medical Center Groningen) for the 1H6 G4 monoclonal antibody. We also thank Dr Jochen Kuper (University of Wuerzburg) for helpful advice on FANCJ homology model.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health, NIA, NHLBI and the Italian Association for Cancer Research AIRC grant [14171 to D.B.]. Funding for open access charge: Intramural Research Program of the National Institutes of Health, NIA.
Conflict of interest statement. None declared.
REFERENCES
- 1. Wang A.T., Smogorzewska A.. SnapShot: Fanconi anemia and associated proteins. Cell. 2015; 160:354. [DOI] [PubMed] [Google Scholar]
- 2. Mamrak N.E., Shimamura A., Howlett N.G.. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017; 31:93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kottemann M.C., Smogorzewska A.. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013; 493:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cantor S.B., Guillemette S.. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011; 7:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brosh R.M. Jr, Cantor S.B.. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014; 5:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raghunandan M., Chaudhury I., Kelich S.L., Hanenberg H., Sobeck A.. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015; 14:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu Y., Shin-ya K., Brosh R.M. Jr. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008; 28:4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu Y., Sommers J.A., Loiland J.A., Kitao H., Kuper J., Kisker C., Brosh R.M. Jr. The Q motif of Fanconi anemia group J protein (FANCJ) DNA helicase regulates its dimerization, DNA binding, and DNA repair function. J. Biol. Chem. 2012; 287:21699–21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hass C.S., Lam K., Wold M.S.. Repair-specific functions of replication protein A. J. Biol. Chem. 2012; 287:3908–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bharti S.K., Sommers J.A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M.P., Kisker C., Brosh R.M. Jr. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013; 288:28217–28229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta R., Sharma S., Sommers J.A., Jin Z., Cantor S.B., Brosh R.M. Jr. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005; 280:25450–25460. [DOI] [PubMed] [Google Scholar]
- 12. Giri B., Smaldin P.J., Thys R.G., Creacy S.D., Routh E.D., Hantgan R.R., Lattmann S., Nagamine Y., Akman S.A., Vaughn J.P.. Gr resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucl. Acid. Res. 2011; 39:7161–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khan I., Suhasini A.N., Banerjee T., Sommers J.A., Kaplan D.L., Kuper J., Kisker C., Brosh R.M. Jr. Impact of age-associated cyclopurine lesions on DNA repair helicases. PLoS One. 2014; 9:e113293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Y., Sommers J.A., Suhasini A.N., Leonard T., Deakyne J.S., Mazin A.V., Shin-Ya K., Kitao H., Brosh R.M. Jr. Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010; 116:3780–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seol Y., Strub M.P., Neuman K.C.. Single molecule measurements of DNA helicase activity with magnetic tweezers and t-test based step-finding analysis. Methods (San Diego, Calif.). 2016; 105:119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harami G.M., Seol Y., In J., Ferencziova V., Martina M., Gyimesi M., Sarlos K., Kovacs Z.J., Nagy N.T., Sun Y. et al. Shuttling along DNA and directed processing of D-loops by RecQ helicase support quality control of homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E466–E475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seol Y., Neuman K.C.. Magnetic tweezers for single-molecule manipulation. Methods Mol. Biol. (Clifton, N.J.). 2011; 783:265–293. [DOI] [PubMed] [Google Scholar]
- 18. Kitao H., Nanda I., Sugino R.P., Kinomura A., Yamazoe M., Arakawa H., Schmid M., Innan H., Hiom K., Takata M.. FancJ/Brip1 helicase protects against genomic losses and gains in vertebrate cells. Genes Cells. 2011; 16:714–727. [DOI] [PubMed] [Google Scholar]
- 19. Wang W., Seki M., Narita Y., Sonoda E., Takeda S., Yamada K., Masuko T., Katada T., Enomoto T.. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 2000; 19:3428–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamamoto K., Hirano S., Ishiai M., Morishima K., Kitao H., Namikoshi K., Kimura M., Matsushita N., Arakawa H., Buerstedde J.M. et al. Fanconi anemia protein FANCD2 promotes immunoglobulin gene conversion and DNA repair through a mechanism related to homologous recombination. Mol. Cell. Biol. 2005; 25:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishiai M., Kitao H., Smogorzewska A., Tomida J., Kinomura A., Uchida E., Saberi A., Kinoshita E., Kinoshita-Kikuta E., Koike T. et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 2008; 15:1138–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sonoda E., Sasaki M.S., Buerstedde J.M., Bezzubova O., Shinohara A., Ogawa H., Takata M., Yamaguchi-Iwai Y., Takeda S.. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998; 17:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Suhasini A.N., Sommers J.A., Muniandy P.A., Coulombe Y., Cantor S.B., Masson J.Y., Seidman M.M., Brosh R.M. Jr. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell. Biol. 2013; 33:2212–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwab R.A., Niedzwiedz W.. Visualization of DNA replication in the vertebrate model system DT40 using the DNA fiber technique. J. Visual. Exp.: JoVE. 2011; e3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chandrasekharappa S.C., Lach F.P., Kimble D.C., Kamat A., Teer J.K., Donovan F.X., Flynn E., Sen S.K., Thongthip S., Sanborn E. et al. Massively parallel sequencing, aCGH, and RNA-Seq technologies provide a comprehensive molecular diagnosis of Fanconi anemia. Blood. 2013; 121:e138–e148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Levitus M., Waisfisz Q., Godthelp B.C., de Vries Y., Hussain S., Wiegant W.W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M.A., Pals G. et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 2005; 37:934–935. [DOI] [PubMed] [Google Scholar]
- 28. Kuper J., Wolski S.C., Michels G., Kisker C.. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J. 2012; 31:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gupta R., Sharma S., Sommers J.A., Kenny M.K., Cantor S.B., Brosh R.M. Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007; 110:2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Green N.M. Avidin. Adv. Protein Chem. 1975; 29:85–133. [DOI] [PubMed] [Google Scholar]
- 31. Bridge W.L., Vandenberg C.J., Franklin R.J., Hiom K.. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 2005; 37:953–957. [DOI] [PubMed] [Google Scholar]
- 32. Huberman J.A. New views of the biochemistry of eucaryotic DNA replication revealed by aphidicolin, an unusual inhibitor of DNA polymerase alpha. Cell. 1981; 23:647–648. [DOI] [PubMed] [Google Scholar]
- 33. Kim M.Y., Vankayalapati H., Shin-Ya K., Wierzba K., Hurley L.H.. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J. Am. Chem. Soc. 2002; 124:2098–2099. [DOI] [PubMed] [Google Scholar]
- 34. De Cian A., Guittat L., Shin-ya K., Riou J.F., Mergny J.L.. Affinity and selectivity of G4 ligands measured by FRET. Nucleic Acids Symp. Ser. (Oxf.). 2005; 235–236. [DOI] [PubMed] [Google Scholar]
- 35. Schwab R.A., Nieminuszczy J., Shin-ya K., Niedzwiedz W.. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013; 201:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ying S., Hamdy F.C., Helleday T.. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012; 72:2814–2821. [DOI] [PubMed] [Google Scholar]
- 39. Kuo L.J., Yang L.X.. Gamma-H2AX - a novel biomarker for DNA double-strand breaks. In vivo (Athens, Greece). 2008; 22:305–309. [PubMed] [Google Scholar]
- 40. Kowalczykowski S.C. An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harbor Perspect. Biol. 2015; 7:a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sommers J.A., Rawtani N., Gupta R., Bugreev D.V., Mazin A.V., Cantor S.B., Brosh R.M. Jr. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 2009; 284:7505–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suhasini A.N., Rawtani N.A., Wu Y., Sommers J.A., Sharma S., Mosedale G., North P.S., Cantor S.B., Hickson I.D., Brosh R.M. Jr. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 2011; 30:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raderschall E., Golub E.I., Haaf T.. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. U.S.A. 1999; 96:1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zachos G., Rainey M.D., Gillespie D.A.. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol. Cell. Biol. 2005; 25:563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lopez-Martinez D., Liang C.C., Cohn M.A.. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell. Mol. Life Sci.: CMLS. 2016; 73:3097–3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D.M.. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng M., Litman R., Xie J., Sharma S., Brosh R.M. Jr, Cantor S.B.. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007; 26:3238–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen J., Stubbe J.. Bleomycins: towards better therapeutics. Nat. Rev. Cancer. 2005; 5:102–112. [DOI] [PubMed] [Google Scholar]
- 49. Ray S., Breuer G., DeVeaux M., Zelterman D., Bindra R., Sweasy J.B.. DNA polymerase beta participates in DNA End-joining. Nucleic Acids Res. 2018; 46:242–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Greenberg R.A., Sobhian B., Pathania S., Cantor S.B., Nakatani Y., Livingston D.M.. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006; 20:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gong Z., Kim J.E., Leung C.C., Glover J.N., Chen J.. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010; 37:438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davies S.L., North P.S., Hickson I.D.. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007; 14:677–679. [DOI] [PubMed] [Google Scholar]
- 53. Suhasini A.N., Brosh R.M. Jr. Disease-causing missense mutations in human DNA helicase disorders. Mut. Res. 2013; 752:138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo M., Vidhyasagar V., Ding H., Wu Y.. Insight into the roles of helicase motif Ia by characterizing Fanconi anemia group J protein (FANCJ) patient mutations. J. Biol. Chem. 2014; 289:10551–10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dobbelstein M., Sorensen C.S.. Exploiting replicative stress to treat cancer. Nat. Rev. Drug Discov. 2015; 14:405–423. [DOI] [PubMed] [Google Scholar]
- 56. Aggarwal M., Brosh R.M. Jr. Hitting the bull's eye: novel directed cancer therapy through helicase-targeted synthetic lethality. J. Cell. Biochem. 2009; 106:758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brosh R.M., Jr DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer. 2013; 13:542–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sargent J.M., Elgie A.W., Williamson C.J., Taylor C.G.. Aphidicolin markedly increases the platinum sensitivity of cells from primary ovarian tumours. Br. J. Cancer. 1996; 74:1730–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hale T.K., Norris G.E., Jameson G.B., Filichev V.V.. Helicases, G4-DNAs, and drug design. ChemMedChem. 2014; 9:2031–2034. [DOI] [PubMed] [Google Scholar]
- 60. Portugal J., Barcelo F.. Noncovalent Binding to DNA: Still a target in developing anticancer agents. Curr. Med. Chem. 2016; 3:4108–4134. [DOI] [PubMed] [Google Scholar]
- 61. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015; 10:845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.