Abstract
DNA synthesis by DNA polymerases (dPols) is central to duplication and maintenance of the genome in all living organisms. dPols catalyze the formation of a phosphodiester bond between the incoming deoxynucleoside triphosphate and the terminal primer nucleotide with the release of a pyrophosphate (PPi) group. It is believed that formation of the phosphodiester bond is an endergonic reaction and PPi has to be hydrolyzed by accompanying pyrophosphatase enzymes to ensure that the free energy change of the DNA synthesis reaction is negative and it can proceed in the forward direction. The fact that DNA synthesis proceeds in vitro in the absence of pyrophosphatases represents a long-standing conundrum regarding the thermodynamics of the DNA synthesis reaction. Using time-resolved crystallography, we show that hydrolysis of PPi is an intrinsic and critical step of the DNA synthesis reaction catalyzed by dPols. The hydrolysis of PPi occurs after the formation of the phosphodiester bond and ensures that the DNA synthesis reaction is energetically favorable without the need for additional enzymes. Also, we observe that DNA synthesis is a two Mg2+ ion assisted stepwise associative SN2 reaction. Overall, this study provides deep temporal insight regarding the primary enzymatic reaction responsible for genome duplication.
INTRODUCTION
In all living organisms, deoxyribonucleic acid (DNA) is synthesized by DNA polymerases (dPols), and these enzymes play a central role in genome duplication. Also, dPols are essential for many applications in research and biotechnology such as DNA sequencing, gene cloning, and Polymerase Chain Reaction-based diagnostic kits. In both natural and artificial settings, dPols catalyze the template-directed addition of deoxynucleoside triphosphates (dNTPs) to the primer strand (1–5). Mg2+ ions play a critical role in the polymerization reaction, and dPols extends the primer in 5′ to 3′ direction. The formation of a phosphodiester bond (pB bond) between the α-phosphate of the incoming dNTP and the 3′- hydroxyl group of the terminal primer nucleotide represents the primary chemical reaction catalyzed by dPols. This reaction also involves breakage of the bond between the α-phosphate, and the bridging oxygen (αO bond) between the α- and β- phosphates resulting in the release of the pyrophosphate (PPi) moiety as a by-product of the synthesis reaction.
The reaction involving incorporation of a nucleotide into the primer through the formation of the phosphodiester bond along with the release of PPi is expected to have a low free energy change (1,6,7). To ensure that the reaction moves in the forward direction, it is believed that the PPi moiety is cleaved by an accompanying pyrophosphatase enzyme to render a large negative free energy change (∼7 kcal/mol) to the DNA synthesis reaction (1,8–10). However, DNA synthesis can proceed smoothly in vitro in the absence of any pyrophosphatase enzyme, and there is no satisfactory explanation available for this long-standing thermodynamic conundrum.
Based on structural and biochemical studies, two distinct mechanisms have been proposed for the synthesis of the pB bond by dPols. In the classical two-metal mechanism, three conserved acidic residues in the palm subdomain bind two Mg2+ ions (11). Structures of dPols in complex with DNA and dNTP showed that one Mg2+ (Metal B) coordinates two active-site acidic residues and the triphosphate moiety of the incoming nucleotide (12–17). The other Mg2+ (Metal A) coordinates all three active-site acidic residues and 3′OH of the primer terminus (13,18,19) and is thought to facilitate the formation of the attacking oxyanion by lowering the pKa of the primer terminus 3′O–. The in-line nucleophilic attack by the 3′O– oxyanion on the α-phosphate should lead to the formation of a penta-coordinated bipyramidal transition state. The metal B may stabilize the developing negative charge on the triphosphate moiety during the nucleophilic attack. According to this model, the reaction is hypothesized to be a concerted SN2-type reaction wherein there are simultaneous formation and breakage of the pB and αO bonds, respectively.
In the recent past, the appearance of an additional divalent metal ion (Metal C) which is distinct from the catalytic (Metal A) and nucleotide (Metal B) metal ions has been reported (19–21). The metal ion (Metal C) coordinates the non-bridging oxygens on the phosphates of what previously were α- and β-phosphates of the incoming nucleotide and is predicted to assist breakage of the αO bond. According to this model, the binding of metal C is the rate-limiting step of the DNA polymerase reaction (21,22). Both the classical two-metal-ion and the new three-metal-ion mechanisms propose a concerted SN2-type reaction scheme. However, based on theoretical analysis, it has been suggested earlier that DNA synthesis by dPols may follow either a concerted, stepwise associative or a stepwise dissociative reaction scheme (23,24).
To elucidate the different stages of the DNA synthesis reaction, we have employed DNA polymerase IV (PolIV) from Escherichia coli. PolIV is a member of the Y-family of dPols and capable of template-dependent dNTP incorporation (18,25,26). We have conducted time-resolved crystallography on DNA synthesis by PolIV and obtained periodic snapshots of the reaction. Our studies show that hydrolysis of the pyrophosphate moiety is an inherent part of the DNA synthesis reaction catalyzed by dPols. This step ensures that the nucleotide incorporation reaction is energetically favorable without the need for accompanying pyrophosphatases. Also, we observed that the DNA synthesis is a two Mg2+ ion assisted stepwise associative SN2 reaction wherein the formation of the pB bond and dissolution of the αO bond occur sequentially. Overall, the study provides insight regarding the mechanism utilized by dPols to synthesize DNA during DNA replication.
MATERIALS AND METHODS
Protein purification and primer extension assays
PolIV was purified as mentioned before (18,27). Primer extension assays were carried out at different pH and incubation times to unearth conditions wherein the nucleotide addition reaction is considerably slowed down. The primer extension assays were carried out with the following DNA substrate:
1. Template A
3′-GCATGAGCATCCGTAATCACACTGGTCGACAAGTCCATC CGTGCCATCCT-5′
5′-XCGTACTCGTAGGCAT-3′
The concentrations of protein, DNA and incoming nucleotide used were 100 nM, 100 nM and 25 μM, respectively and the reaction products were processed as mentioned previously (28). It was seen that at pH 5.2, the rate of reaction was reduced by >60-fold at 37°C (Supplementary Figure S1).
In crystallo DNA synthesis
An 18mer self-annealing oligonucleotide was used for crystallization and has the following sequence: 5′ TCTAGGGTCCTAGGACCC 3′. The ternary complexes were reconstituted by mixing PolIV (0.3 mM) with dsDNA (0.36 mM) followed by addition of 5 mM dTTP (GE Healthcare) or 5 mM dTMPPnP (Jena Biosciences). Crystallization was carried out in the absence of metal ion and crystals were obtained in 0.1 M acetate (pH 5.2) and 5–12% MPD. The in crystallo reaction was initiated by transferring crystals to a cryosolution containing 0.1 M acetate (pH 5.2) and 30% MPD with 5 mM MgCl2. The crystals were incubated for different time periods at 4°C (to further reduce the rate of the nucleotide incorporation reaction) and then flash frozen in liquid nitrogen to stop the reaction. Subsequently, X-ray diffraction data were collected from the frozen crystals. The protocol for preparing crystals utilized here is distinct from that employed in previous time-resolved crystallography studies carried out on DNA polymerases β and η, wherein crystals were grown in the presence of Ca2+ first and then incubated with high concentrations (∼100 mM) of Mg2+ or with Mn2+.
Structure determination and crystallographic refinement
For the crystals prepared with dTTP, X-ray diffraction data were collected at the BM14 beamline of ESRF. Data were processed using iMosflm and the SCALA program in CCP4 (29,30). Using PHASER, the structures corresponding to different time points were determined using the structure of PolIVdA:dTMPnPP (4IR1) as a search model (18,31). Before molecular replacement, the primer nucleotide, incoming nucleotide, and metal ions were removed from the coordinate file of the search model. The MR solutions were subjected to three cycles of refinement in PHENIX, and Fo – Fc maps were calculated for the resolution range of 2.15–40Å for all datasets and viewed at 3.3σ for comparison (Figure 2). The electron density maps (omit Fo – Fc) prepared at the highest resolution possible for each dataset (Supplementary Figure S5) were nearly identical to the previous ones (Figure 2) and in agreement with the all the major conclusions of the analysis. The terminal primer nucleotide and incoming dTTP were positioned at appropriate locations in the electron density maps using Coot, and the structures were refined until convergence using PHENIX (32). For the crystal prepared with dTTP and incubated with 50 mM MgCl2 for 30 min (Supplementary Figure S6), the data were collected at the BM14 beamline of ESRF, and were processed using iMosflm and Scala. Structure solution and refinement were carried out using the same protocol used for structures obtained using 5 mM MgCl2.
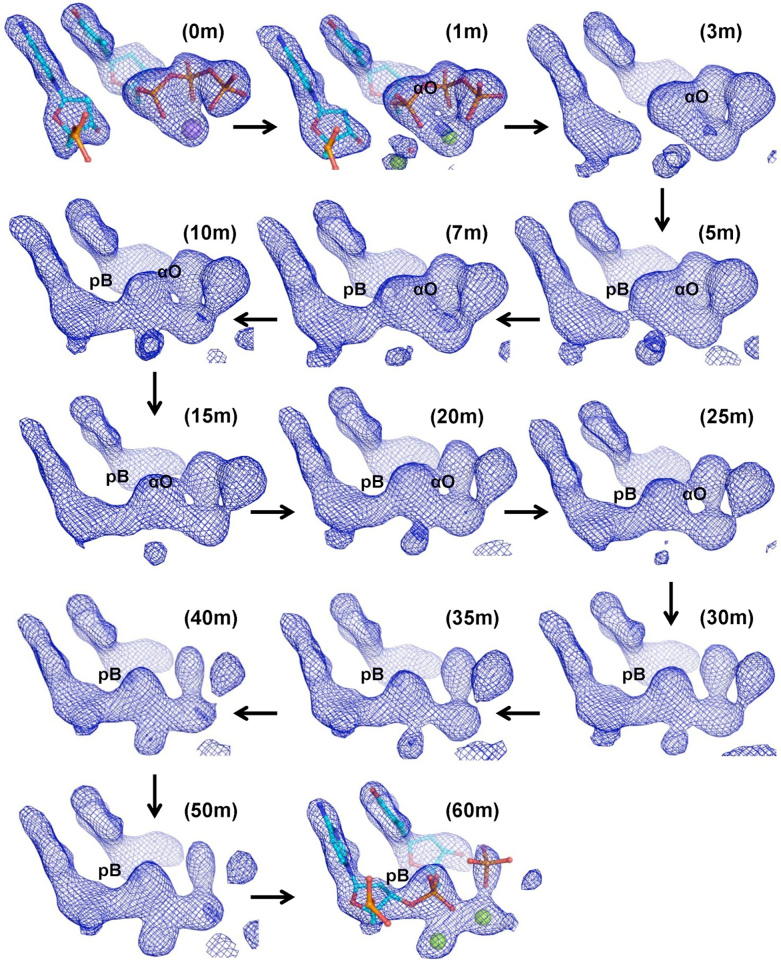
Figure 2.
Electron density maps depicting the sequence of events during phosphodiester (pB) bond formation: Omit electron density maps (Fo – Fc) calculated to a resolution of 2.15 Å are displayed at a contour of 3.3. These maps show that the electron density corresponding to formation of pB appears first (5, 7 and 10 m) followed by dissolution of αO (15, 20 and 25m) and subsequent hydrolysis of the PPi moiety to phosphate ions (30, 35, 40, 50 and 60 m). The terminal primer nucleotide and the incoming dTTP are displayed here in stick representation and colored by element. Na+ and Mg2+ ions are shown as purple and green spheres respectively.
For the crystals obtained with dTMPPnP (Figure 5), diffraction data were collected at the ID29 beamline of ESRF and data was processed using XDS, Aimless and Pointless (33–36). Structure solution and refinement were carried out using the same protocol used for the structures with dTTP.
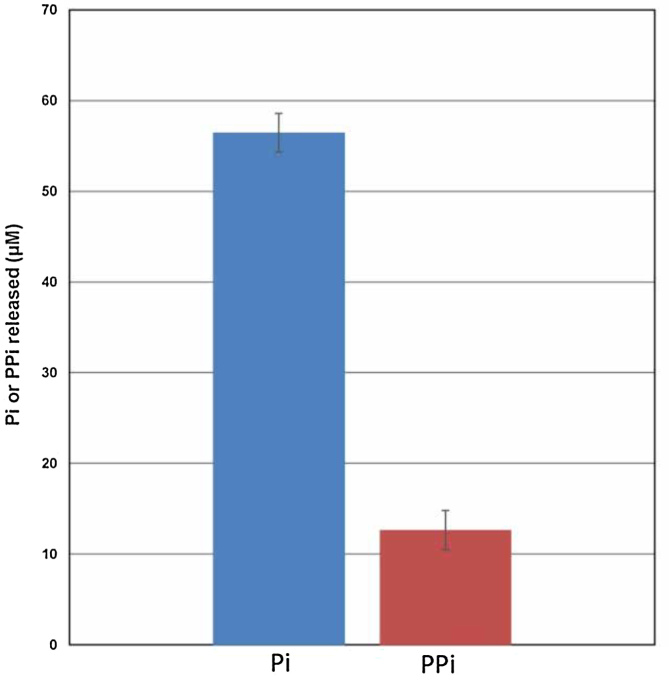
Figure 5.
Comparison of PPi and Pi formation during DNA synthesis. The displayed graph shows that, during DNA synthesis, the amount of Pi generated is >5-fold as compared to PPi.
For all the refined structures, the majority of the residues were in the favorable regions of the Ramachandran plot with only 1% of residues in the disallowed regions. All the refined structures show the presence of two complexes of PolIV:DNA:dTTP:Mg2+ which, based on the chain IDs of the components, are called ABC and FGH. The electron density maps were clearer for the FGH complex, and hence this complex was selected for analysis. The electron density maps were prepared in PHENIX and viewed using Coot (37). The structures were also analyzed using PyMOL (Schrodinger Inc.), and all the figures were generated using PyMOL.
Primer extension assay with modified nucleotides
The modified deoxynucleoside triphosphates dTMPnPP, dTMPPnP, dGMPnPP and dGMPPnP were obtained from Jena Biosciences. Primer extension assay was conducted with different dPols (PolIV, Dbh, Dpo4, MsDpo4, Taq, Klenow exo-, PfPrex, Pol II and M-MuLV RT) and the substrate DNAs utilized were as follows:
Template A
3′-GCATGAGCATCCGTAATCACACTGGTCGACAAGTCCATC CGTGCCATCCT-5′
5′-XCGTACTCGTAGGCAT-3′
Template C
3′-GCATGAGCATCCGTACTCACACTGGTCGACAAGTCCATC CGTGCCATCCT-5′
5′-XCGTACTCGTAGGCAT-3′
where X = 5′ 6FAM label.
Protein, DNA and incoming nucleotide (normal or modified) were added to a final concentration of 10 nM, 50 nM and 2 μM, respectively. The reaction mixture was incubated for 30 min at 37°C, and the reaction products were processed as mentioned previously (28).
Phosphate and pyrophosphate assay
To quantitate the level of phosphates and pyrophosphates generated during the DNA synthesis reaction, primer extension assays were carried out initially, as mentioned previously. The following DNA substrate was utilized:
Template A
3′-GCATGAGCATCCGTAATCACACTGGTCGACAAGTCCATC CGTGCCATCCT-5′
5′-GTACTCGTAGGCAT-3′
The reaction mixture included 100 nM of PolIV, 15 μM DNA duplex and 2 mM of all four dNTPs. After incubation at 37°C for 1 h, the reaction was terminated by adding EDTA solution to a final concentration of 20 mM. The amount of phosphate in the reaction mix was determined using the Phosphate Assay Kit (Sigma-Aldrich) wherein the sensor reagent is a proprietary formulation of the Malachite green dye. 100 μl of this reagent was added to 50 μl of the reaction mix followed by incubation at room temperature for 30 min, and subsequently, absorbance was recorded at 630 nm on a Spectramax M5 plate reader (Molecular Devices). The amount of pyrophosphate was determined using the Pyrophosphate assay kit (MAK168, Sigma) wherein the sensor reagent is a fluorogenic PPi sensor. 50 μl of the reaction mix was added to 50 μl of sensor reagent followed by 30 min of incubation at room temperature. After incubation, the fluorescence intensity was measured (λex = 320 nm and λem = 456 nm) on a Spectramax M5 plate reader (Molecular Devices). To enable accurate quantitation, initially standard curves were plotted for both Pi and PPi. All measurements were done in triplicate and with appropriate controls.
RESULTS
Entry of Mg2+ ions triggers conformational changes
Primer extension assays conducted under different reaction conditions shows that the rate of the polymerization reaction was slowed down considerably at a pH of 5.2 (Supplementary Figure S1). Crystals of the PolIV:DNA:dTTP complex were grown at this pH in the absence of Mg2+ ions. The crystals were soaked in Mg2+ for 0, 1, 3, 5, 7, 10, 15, 20, 25, 30, 35, 40, 50 and 60 min at 4°C and then frozen. X-ray diffraction data were collected from the frozen crystals, and the data collection and refinement statistics are displayed in Supplementary Table S1.
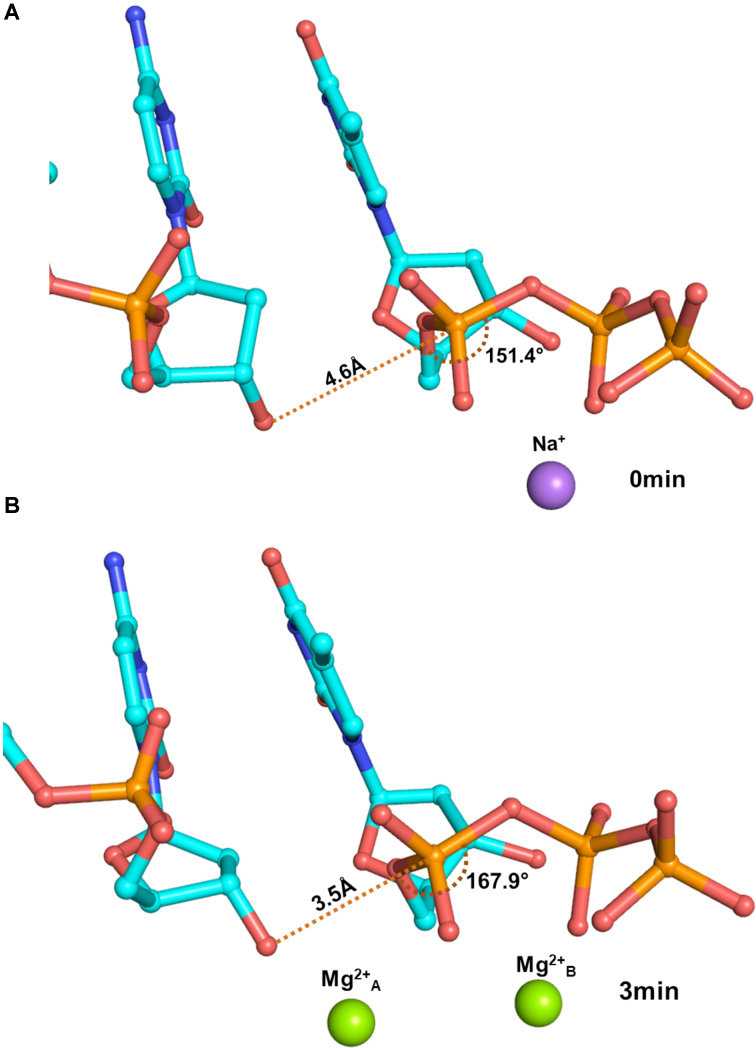
Alignment of the ground state structure (0 min), where no metal ions are present, with the complex obtained 1 min after addition of the Mg2+ ions, showed only marginal differences. The incoming nucleotide, therefore, attains the conformation compatible with productive catalysis in the absence of the co-factor ion (Supplementary Figure S2). The density for both the metal ions appeared 1min after the addition of Mg2+ ions and the activation of the 3′-hydroxyl group was observed immediately after the metal ions occupied their respective locations. The sugar pucker of the terminal nucleotide of the primer changed from C2′-endo to C3′-endo (Supplementary Figure S3). Also, the incoming nucleotide moved towards the 3′OH of the primer terminus. As a result of these changes, the distance between the 3′O– of the terminal primer nucleotide and the α-phosphate of the incoming dTTP reduced from 4.6 to 3.5 Å (Figure 1). Furthermore, the angle formed by the 3′O–, the α-phosphate and the bridging oxygen between α- and β- phosphates increased from 150° to ∼170° (Figure 1). The conformational changes triggered by binding of Mg2+ ions, therefore facilitate the in-line nucleophilic attack by the 3′O– of the terminal primer nucleotide on the α-phosphate of the incoming dTTP.
Figure 1.
Conformational changes triggered by the Mg2+ ions. The terminal primer nucleotide and the incoming dTTP are displayed here in stick representation and coloured by element. Upon entry of two Mg2+ ions, the terminal primer nucleotide and the incoming dTTP undergo conformational changes such that the distance between the 3′O– and the α-phosphate decreases from 4.6 Å (0 min) to 3.5 Å (3 min) and the angle between the 3′O–, the β-phosphate and the bridging oxygen between the α- and β- phosphates increases from 150° to 170°. The conformational changes, therefore, facilitate the in-line nucleophilic attack by the 3′O– on the α-phosphate.
Phosphodiester bond formation is a stepwise associative SN2 reaction
From 5min onwards, electron density developed gradually between the 3′-end of the primer and α-phosphate of the incoming dTTP. By 10 min, the pB bond was completely formed resulting in the formation a pentavalent intermediate. In this intermediate, the α-phosphate is simultaneously bonded to the 3′O– of the primer terminus, the bridging oxygen bonded with the β-phosphate, the bridging oxygen bonded with 5′ carbon of the deoxyribose sugar and two oxygen atoms (Supplementary Figure S4). After 10 min, we observed that there is a gradual decrease in the density of the αO bond (Figure 2, Supplementary Figure S5). The analysis of the omit Fo – Fc maps showed that the pB bond formation and αO bond dissolution happen sequentially and not concomitantly. The DNA synthesis reaction, therefore, follows the stepwise associative SN2 scheme (23). Further, the DNA synthesis reaction may also represent an example of the alternate two-step mechanistic model of SN2(P5) reactions proposed recently by Kolodiazhnyi and Kolodiazhna (38). In this alternate model, the pentavalent intermediate exists in a lower-energy state between two higher-energy transition states corresponding to, in this case, the formation of pB bond and dissolution of the αO bond.
Unlike postulated by the classical two-metal-ion mechanism, the DNA synthesis reaction does not follow a concerted SN2 reaction scheme as the pB bond formed first followed by dissolution of the αO bond (Movie S1) (11). Additionally, there was no electron density present in the appropriate location for a third Mg2+ (Metal C) ion in the maps as observed in the case of DNA polymerases β and η. Hence, our studies show that two Mg2+ ions are adequate to enable synthesis of the pB bond (Figure 2 & Supplementary Figure S5). This observation gave rise to the possibility that the third metal ion observed in earlier studies may be due to the use of Mn2+ or high concentrations of Mg2+. The electron density maps for a crystal incubated with 50 mM MgCl2 for 30 min showed the presence of density corresponding to the third Mg2+ (Metal C) ion (Supplementary Figure S6). Hence, the third metal ion may play no actual part in DNA synthesis.
Also, the time-resolved experiment clearly shows that a pentavalent intermediate is formed and decomposes gradually during the DNA synthesis reaction (Figure 2 & Supplementary Figure S5). The selection of the multivalent phosphorous element to form the backbone of genetic material may in part be due to the need to form this transient pentavalent intermediate during formation of the DNA polymer (39).
Pyrophosphate hydrolysis is critical for DNA synthesis
The products of the nucleotide incorporation reaction are the extended primer strand which has increased in length by one nucleotide, and the PPi moiety from the dNTP. Surprisingly, we observed an additional step in the reaction wherein there is hydrolysis of the PPi moiety to generate two individual phosphate ions- βPi and γPi corresponding to the β- and γ-phosphate of dTTP, respectively (Figure 2 & Movie S1). The Arg49 residue involved in stabilizing the PPi in the active site moved away after breakdown of PPi (Supplementary Figure S7). Consistent with the observed importance of the R49 residue, the R49→A mutant protein lost the ability to catalyze the DNA synthesis reaction (Supplementary Figure S8). After hydrolysis of the PPi moiety, γPi diffused out first, and the two metal ions were retained in the complex. The mechanism of PPi release by dPols has been the subject of scrutiny in the past, and our study shows that this happens through the hydrolysis of PPi into free phosphates (40,41).
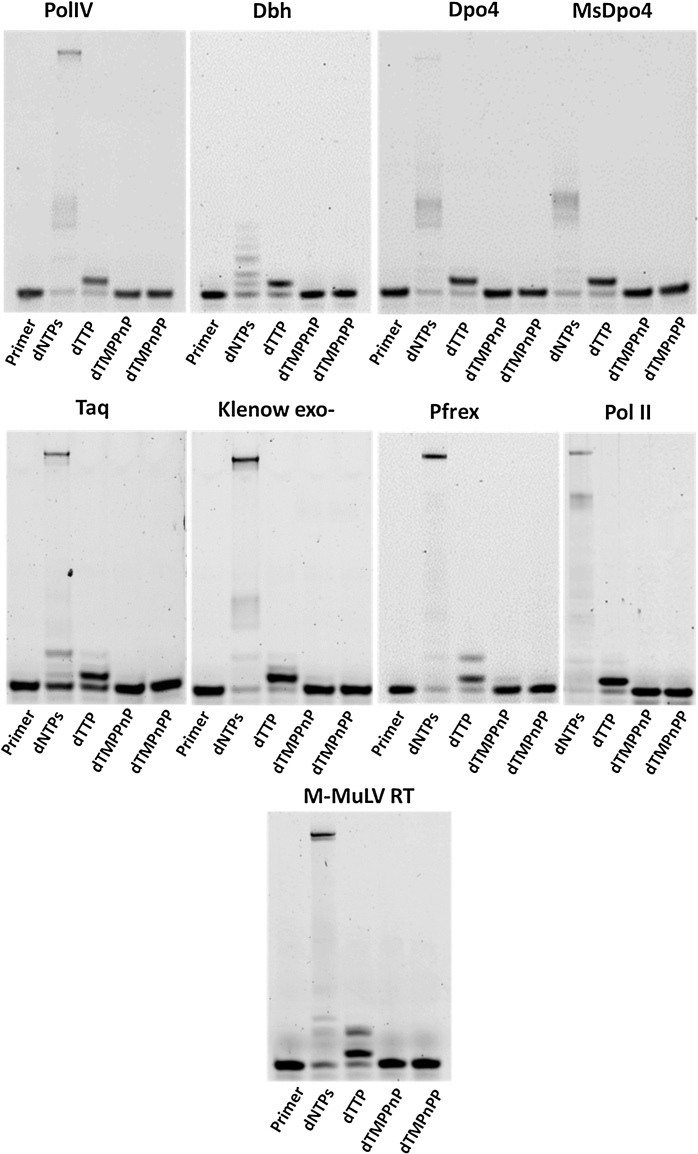
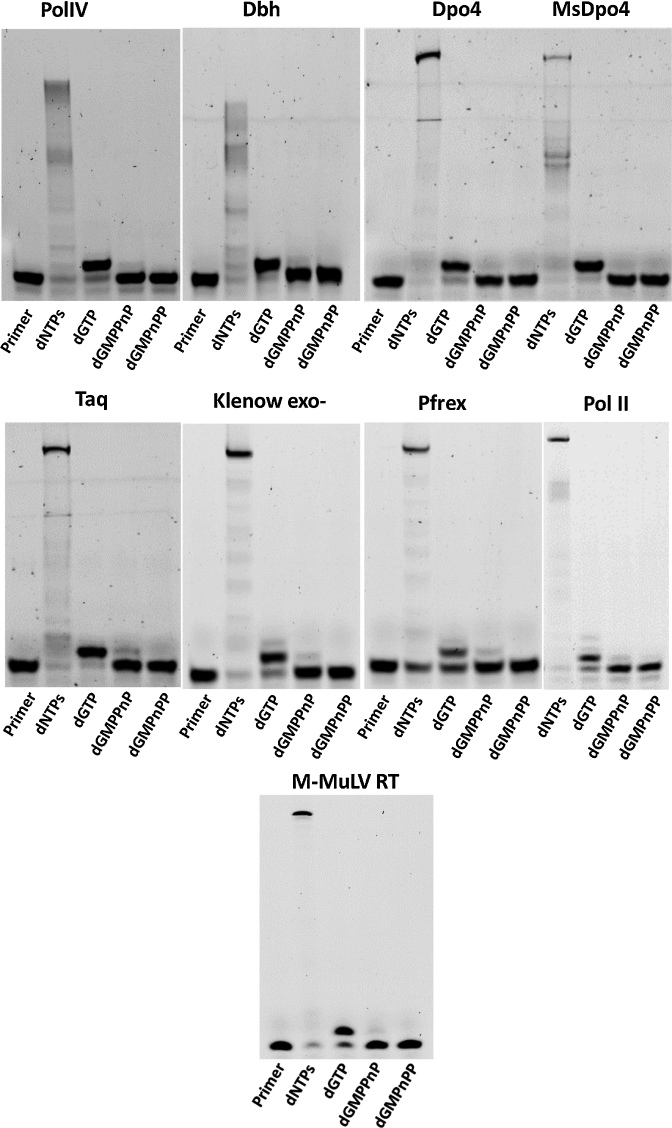
To confirm the importance of breakdown of the PPi in the completion of the polymerization reaction, we conducted primer extension assays using the modified nucleotides dTMPPnP and dGMPPnP, wherein the bond between the β- and γ-phosphates is non-hydrolyzable. If PPi is the final by-product of the reaction, the polymerases should incorporate dTMPPnP and dGMPPnP opposite template dA and dC respectively, as there is no modification in the bond between the α- and β-phosphates. The dPols tested include PolIV (E. coli), Dpo4 (Sulfolobus solfataricus), Dbh (Sulfolobus acidocaldarius), MsDpo4 (Mycobacterium smegmatis), the Klenow fragment of DNA polymerase I (E. coli), the polymerase module of the Pfprex protein (Plasmodium falciparum), DNA polymerase II (E. coli) and M-MuLV RT (Moloney Murine Leukemia Virus). PolIV, Dpo4, Dbh and MsDpo4 belong to the Y-family of dPols, the Klenow fragment and Pfprex are representatives of the A-family, DNA Polymerase II is a member of the B-family, and M-MuLV RT belongs to the Reverse Transcriptase family. All the tested dPols failed to incorporate the modified nucleotides (Figures 3 and 4). These studies show that the hydrolysis of PPi for completion of the synthesis reaction may be a conserved feature of all dPols.
Figure 3.
Primer Extension Assays with template dA and incoming dTTP, dTMPnPP and dTMPPnP. Opposite template dA, different dPols added dTTP and could not add the α-β modified dTMPnPP. The β-γ modified dTMPPnP was also not incorporated into the primer by these enzymes and this observation validates the importance of PPi hydrolysis in the DNA synthesis reaction.
Figure 4.
Primer Extension Assays with template dC and incoming dGTP, dGMPnPP and dGMPPnP. The tested dPols added dGTP and could not add the α-β modified dGMPnPP opposite template dC. The β-γ modified dGMPPnP was also not incorporated into the primer and therefore, hydrolysis of PPi is important for completion of the DNA synthesis reaction.
Biochemical assays that assess the amount of inorganic phosphate (Pi) and PPi generated during the dNTP incorporation reaction showed that the level of Pi produced is much higher than that of PPi. This observation is in line with the inference that PPi hydrolysis is critical for the completion of the DNA synthesis reaction (Figure 5).
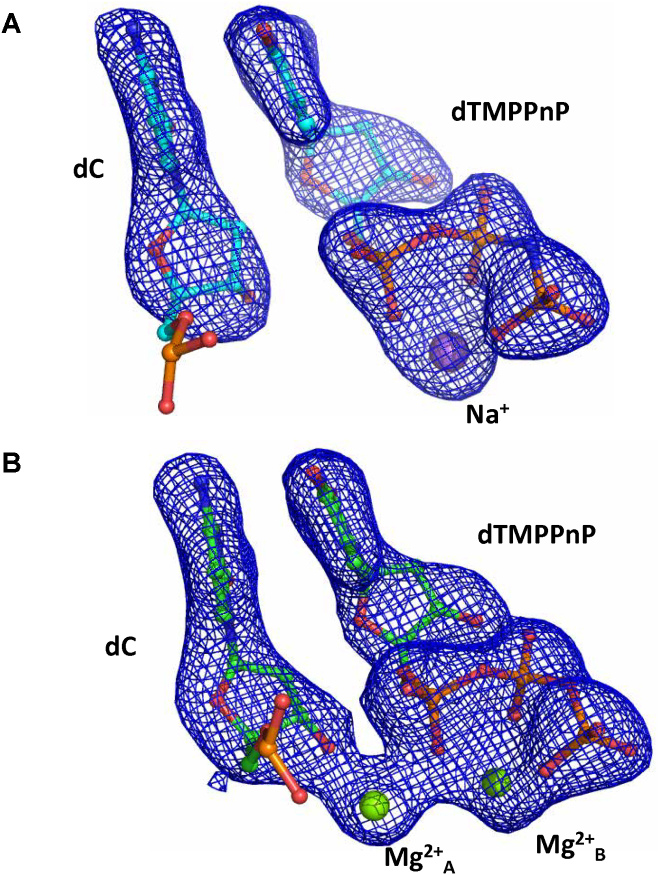
X-ray diffraction data were collected from crystals of PolIVDNA:dTMPPnP complex soaked with Mg2+ for 0min and 60min (Supplementary Table S2). The corresponding electron density maps showed that on Mg2+ ion entry, the sugar pucker of the terminal primer nucleotide changes and the incoming nucleotide moves closer to the terminal primer nucleotide. The distance between the 3′O- of the terminal primer nucleotide and the α-phosphate of dTMPPnP is 3.3 Å and the angle formed between these two atoms and the bridging oxygen between α- and β- phosphates is 171° (Supplementary Figure S9). However, the reaction does not proceed to completion; incorporation does not occur (Figure 6) and, this observation reinforces the importance of PPi hydrolysis.
Figure 6.
DNA synthesis reaction does not occur in the presence of dTMPPnP. Omit Fo – Fc maps are displayed at a contour of 3.3 for the PolIVDNA:dTMPPnP complex before (A) and 60 min after the addition of Mg2+ (B). The electron density maps show that although the 3′-OH is activated, the DNA synthesis reaction does not proceed further. These observations are in line with the inference that cleavage of the pyrophosphate moiety is critical for the irreversible completion of the dNTP incorporation reaction.
Temporal sequence of events during the dNTP incorporation reaction
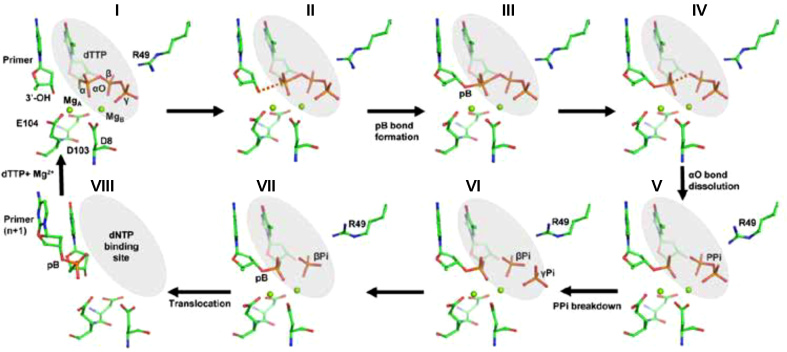
Based on our studies, the following series of events may occur during the DNA synthesis reaction catalyzed by PolIV (Figure 7 & Movie S2). Initially, binding of dNTP along with Mg2+ ions initiates the DNA synthesis reaction in the DNA polymerase active site. (I) Once all the components required for the DNA synthesis are assembled in the correct location and conformation, Mg2+ ion activates the 3′-OH of the primer nucleotide, and this results in the change in the sugar pucker from C2′-endo to C3′-endo form, (II-V) phosphodiester bond (pB) formation results in the appearance of a penta-covalent transition state which is followed by αO bond dissolution and release of PPi, (VI) PPi is further hydrolyzed to two phosphate ions followed by reorientation of the Arg49 residue. (VII) γPi diffuses out first (VIII) both metal ions and βPi are released followed by DNA translocation so that the newly incorporated nucleotide moves out of the nucleotide-binding site to make way for the next nucleotide to enter, and a new cycle of DNA synthesis can start. The identity of the incoming nucleotide will be determined by the next unpaired template nucleotide. DNA replication is hypothesized to have existed in DNA viruses even before the emergence of the Last Universal Common Ancestor (42). Hence, it is possible that the proposed scheme may have appeared early in evolution and may be common for all dPols (43,44).
Figure 7.
Mechanism of incorporation of dTTP opposite dA by DNA polymerase IV. The different steps associated with the DNA synthesis reaction catalyzed by PolIV are displayed. The terminal primer nucleotide (dC), the incoming nucleotide (dTTP) and active site residues are shown in stick representation and colored according to element. The Mg2+ ions are shown in the form of green spheres.
DISCUSSION
Our studies reveal that breakdown of the PPi moiety is critical for completion of the DNA synthesis reaction. The conversion of PPi to Pi (ΔG = −7 kcal/mol) is essential to render a large overall negative free energy change (ΔG = −6.5 kcal/mol) to the DNA synthesis reaction (45,46). It was previously believed that dPols act in tandem with pyrophosphatase enzymes that cleave the PPi, so that the coupling of the two reactions provides an overall negative free energy change (1,8). It is clear from this study that the PPi hydrolysis step is inherently part of the complete dNTP incorporation reaction catalyzed by dPols and therefore, the DNA synthesis reaction carried out by dPols is energetically favorable without the need of any other enzyme activity. The entropic penalty imposed by nucleotide incorporation is expected to be substantial since the mobility of the dNTP reduces drastically on polymerization (24,47). PPi hydrolysis may also serve to reduce the effect of reduction in entropy on the energetics of the DNA synthesis reaction. The energy released due to the breakdown of PPi may also aid in translocation of DNA on the polymerase so that the next unpaired template nucleotide reaches the active site and a new round of DNA synthesis can begin.
PPi is known to participate in pyrophosphorolysis, which is the reverse of the polymerization reaction and involves excision of the terminal nucleotide of the primer strand (48). Pyrophosphorolysis results in restoration of the dNTP and the length of the primer reduces by one nucleotide. The hydrolysis of the PPi moiety ensures that the probability of the pyrophosphorolysis reaction is minimal and DNA synthesis is irreversible.
Recently, a study on DNA polymerase β has shown that utilization of dNTPs that are non-hydrolyzable at the β-γ position leads to the promotion of the pyrophosphorolysis reaction instead of the DNA synthesis reaction (49). Also, the utilization of a PPi analog, with an imido- linkage has been shown to promote pyrophosphorolysis (50). Moreover, modification at the β- and γ- phosphates of dNTPs results in a drastic reduction or complete failure of incorporation of the incoming nucleotide by different dPols (51–53). These observations are in line with the inference that cleavage of the PPi is critical for completion of the DNA synthesis. It has been suggested previously that dissociation of metal A from the polymerase prevents pyrophosphorolysis (21). However, our studies indicate that the hydrolysis of PPi may be the primary strategy to avoid the reversal of the synthesis reaction. The breakdown of PPi will, therefore allow for smooth progression of the replication fork and ensure that the cell is not subjected to replication stress.
The cleavage of the PPi will contribute towards enforcing fidelity. The presence of a mispair in the active site will lead to distortion of the spatial alignment of the PPi group with respect to the enzyme residues and the Mg2+ ion. As a result, the rate at which PPi group is broken down will reduce considerably and thus affect completion of the synthesis reaction (52–55). Although the PPi moiety can potentially move to bind in the correct orientation after the dissolution of the αO bond, pyrophosphorolysis might happen before PPi has attained the right configuration for cleavage and thus prevent incorporation of the wrong nucleotide. It has been shown that the average ΔG° for incorporation of the correct dNTP is −5.2 kcal/mol and that for the addition of the incorrect dNTP is −0.13 kcal/mol (56). These observations suggest that the PPi moiety is cleaved only when the correct dNTP is incorporated and are in line with the inference that cleavage of PPi moiety by dPols will contribute towards ensuring fidelity of the DNA synthesis reaction.
The modification of the γ-phosphate has been shown to affect the ability of viral reverse transcriptase to incorporate nucleotides, and hence, it is possible that PPi cleavage may be an evolutionarily conserved feature of all replicative polymerases (57). Enhancement in pyrophosphorolysis activity has been implicated in the ability of mutant viral polymerases to remove chain terminating inhibitors that are used as drugs such as zidovudine (58,59). It is believed that these mutations enhance the affinity for PPi, and thus promote the reverse reaction. However, our studies raise the possibility that these mutations might prevent/reduce cleavage of PPi by the enzyme and thus promote pyrophosphorolysis.
Overall, we provide concrete experimental evidence that hydrolysis of the pyrophosphate moiety is an intrinsic and critical step of the DNA synthesis reaction. Also, DNA synthesis is a stepwise associative SN2 reaction assisted by two Mg2+ ions. The study brings to light the mechanism of the fundamental reaction responsible for genome duplication and the insight obtained from this study may aid the development of improved PCR-based diagnostic kits and novel therapeutic strategies against retroviruses.
DATA AVAILABILITY
The Structure factors and the refined co-ordinates have been deposited in the PDB with the following codes: 5YUR (0 min), 5YUS (1 min), 5YUT (3 min), 5YUU (5 min), 5YUV (7 min), 5YUW (10 min), 5YV3 (15 min), 5YUX (20 min), 5YUY (25 min), 5YV4 (30 min), 5YUZ (35 min), 5YV0 (40 min), 5YV1 (50 min), 5YV2 (60 min), 5YYD (dTMPPnP: 0 min), 5YYE (dTMPPnP: 60 min) and 5ZLV (50 mM MgCl2; dTTP: 30 min).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Minakshi Sharma and Mary K. Johnson for purifying Pfprex and MsDpo4, respectively; Dr Dinakar M. Salunke (ICGEB, New Delhi) and Prof. Jayant B. Udgaonkar (NCBS, Bangalore) for critically reading the manuscript; Prof. S. Ramaswamy (inStem, Bangalore) for discussions. We thank the X-ray diffraction facility located at the Regional Centre for Biotechnology. DTN thanks Dr. Hassan Belrhali & Dr Babu Manjashetty (BM14 beamline, ESRF) and Dr Danielle de Sanctis (ID29 beamline, ESRF) for help with X-ray diffraction data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Regional Centre for Biotechnology; Data collection at the BM14 beamline of ESRF (Grenoble, France) was supported by the BM14 project—a collaboration between DBT, EMBL and ESRF; Data collection at ID29 was facilitated by the ESRF Access Program of RCB which is supported by Department of Biotechnology, Government of India [BT/INF/22/SP22660/2017]. Funding for open access charge: Intramural Funding from Regional Centre for Biotechnology.
Conflict of interest statement. None declared.
REFERENCES
- 1. Watson J.D., Baker T.A., Bell S.P., Gann A., Levine M., Losick R.. Molecular Biology of the Gene. 2013; 7th ednPearson; (Chapter 3 and Chapter 9) [Google Scholar]
- 2. Bessman M.J., Kornberg A., Lehman I.R., Simms E.S.. Enzymic synthesis of deoxyribonucleic acid. Biochim. Biophys. Acta. 1956; 21:197–198. [DOI] [PubMed] [Google Scholar]
- 3. Berdis A.J. Mechanisms of DNA polymerases. Chem. Rev. 2009; 109:2862–2879. [DOI] [PubMed] [Google Scholar]
- 4. Johansson E., Dixon N.. Replicative DNA polymerases. Cold Spring Harb. Perspect. Biol. 2013; 5:a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aschenbrenner J., Marx A.. DNA polymerases and biotechnological applications. Curr. Opin. Biotechnol. 2017; 48:187–195. [DOI] [PubMed] [Google Scholar]
- 6. Peller L. On the free-energy changes in the synthesis and degradation of nucleic acids. Biochemistry. 1976; 15:141–146. [DOI] [PubMed] [Google Scholar]
- 7. Peller L. Thermodynamic factors in the synthesis of two-stranded nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 1966; 55:1025–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lapenta F., Monton Silva A., Brandimarti R., Lanzi M., Gratani F.L., Vellosillo Gonzalez P., Perticarari S., Hochkoeppler A.. Escherichia coli DnaE polymerase couples pyrophosphatase activity to DNA replication. PLoS One. 2016; 11:e0152915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burke C.R., Luptak A.. DNA synthesis from diphosphate substrates by DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:980–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aravind L., Koonin E.V.. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998; 26:3746–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Steitz T.A. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999; 274:17395–17398. [DOI] [PubMed] [Google Scholar]
- 12. Nair D.T., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Human DNA polymerase iota incorporates dCTP opposite template G via a G.C + Hoogsteen base pair. Structure. 2005; 13:1569–1577. [DOI] [PubMed] [Google Scholar]
- 13. Nair D.T., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005; 309:2219–2222. [DOI] [PubMed] [Google Scholar]
- 14. Swan M.K., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K.. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase delta. Nat. Struct. Mol. Biol. 2009; 16:979–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nair D.T., Johnson R.E., Prakash S., Prakash L., Aggarwal A.K.. Replication by human DNA polymerase-iota occurs by Hoogsteen base-pairing. Nature. 2004; 430:377–380. [DOI] [PubMed] [Google Scholar]
- 16. Silverstein T.D., Johnson R.E., Jain R., Prakash L., Prakash S., Aggarwal A.K.. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010; 465:1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zahn K.E., Averill A.M., Aller P., Wood R.D., Doublie S.. Human DNA polymerase theta grasps the primer terminus to mediate DNA repair. Nat. Struct. Mol. Biol. 2015; 22:304–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sharma A., Kottur J., Narayanan N., Nair D.T.. A strategically located serine residue is critical for the mutator activity of DNA polymerase IV from Escherichia coli. Nucleic Acids Res. 2013; 41:5104–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura T., Zhao Y., Yamagata Y., Hua Y.J., Yang W.. Watching DNA polymerase eta make a phosphodiester bond. Nature. 2012; 487:196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Freudenthal B.D., Beard W.A., Shock D.D., Wilson S.H.. Observing a DNA polymerase choose right from wrong. Cell. 2013; 154:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gao Y., Yang W.. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016; 352:1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yang W., Weng P.J., Gao Y.. A new paradigm of DNA synthesis: three-metal-ion catalysis. Cell Biosci. 2016; 6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kamerlin S.C., Sharma P.K., Prasad R.B., Warshel A.. Why nature really chose phosphate. Q. Rev. Biophys. 2013; 46:1–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ram Prasad B., Warshel A.. Prechemistry versus preorganization in DNA replication fidelity. Proteins. 2011; 79:2900–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kottur J., Nair D.T.. Reactive oxygen species play an important role in the bactericidal activity of quinolone antibiotics. Angew. Chem. Int. Ed. Engl. 2016; 55:2397–2400. [DOI] [PubMed] [Google Scholar]
- 26. Kottur J., Sharma A., Gore K.R., Narayanan N., Samanta B., Pradeepkumar P.I., Nair D.T.. Unique structural features in DNA polymerase IV enable efficient bypass of the N2 adduct induced by the nitrofurazone antibiotic. Structure. 2015; 23:56–67. [DOI] [PubMed] [Google Scholar]
- 27. Sharma A., Nair D.T.. Cloning, expression, purification, crystallization and preliminary crystallographic analysis of MsDpo4: a Y-family DNA polymerase from Mycobacterium smegmatis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011; 67:812–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sharma A., Nair D.T.. MsDpo4-a DinB homolog from mycobacterium smegmatis-Is an Error-Prone DNA polymerase that can promote G:T and T:G mismatches. J. Nucleic Acids. 2012; 2012:285481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leslie A.W., Powell H.. Read R, Sussman J. Evolving Methods for Macromolecular Crystallography. 2007; 245:Netherlands: Springer; 41–51. [Google Scholar]
- 30. Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G.. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L. C., Read R. J.. Phase crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Evans P.R., Murshudov G.N.. How good are my data and what is the resolution. Acta Crystallogr. D Biol. Crystallogr. 2013; 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Evans P.R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011; 67:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006; 62:72–82. [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K.. Coot: model building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Kolodiazhnyi O.I., Kolodiazhna A. O.. Nucleophilic substitution at phosphorus: stereochemistry and mechanisms. Tetrahedron: Assymetry. 2017; 28:24 [Google Scholar]
- 39. Westheimer F.H. Why nature chose phosphates. Science. 1987; 235:1173–1178. [DOI] [PubMed] [Google Scholar]
- 40. Da L.T., Wang D., Huang X.. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 2012; 134:2399–2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Genna V., Gaspari R., Dal Peraro M., De Vivo M.. Cooperative motion of a key positively charged residue and metal ions for DNA replication catalyzed by human DNA Polymerase-eta. Nucleic Acids Res. 2016; 44:2827–2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Forterre P. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 2002; 5:525–532. [DOI] [PubMed] [Google Scholar]
- 43. Cheetham G.M., Jeruzalmi D., Steitz T.A.. Transcription regulation, initiation, and “DNA scrunching” by T7 RNA polymerase. Cold Spring Harb. Symp. Quant. Biol. 1998; 63:263–267. [DOI] [PubMed] [Google Scholar]
- 44. Yao N.Y., O’Donnell M.E.. Evolution of replication machines. Crit. Rev. Biochem. Mol. Biol. 2016; 51:135–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fazakerley G.V., Sowers L.C., Eritja R., Kaplan B.E., Goodman M.F.. Structural and dynamic properties of a bromouracil-adenine base pair in DNA studied by proton NMR. J. Biomol. Struct. Dyn. 1987; 5:639–650. [DOI] [PubMed] [Google Scholar]
- 46. M.Davies J., J.Poole R., Sanders D.. The computed free energy change of hydrolysis of inorganic pyrophosphate and ATP: apparent significance. for inorganic-pyrophosphate-driven reactions of intermediary metabolism. Biochim. Biophys. Acta (BBA) - Bioenergetics. 1993; 1141:29–36. [Google Scholar]
- 47. Minetti C.A., Remeta D.P., Miller H., Gelfand C.A., Plum G.E., Grollman A.P., Breslauer K.J.. The thermodynamics of template-directed DNA synthesis: base insertion and extension enthalpies. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:14719–14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pandey M., Patel S.S., Gabriel A.. Kinetic pathway of pyrophosphorolysis by a retrotransposon reverse transcriptase. PLoS One. 2008; 3:e1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shock D.D., Freudenthal B.D., Beard W.A., Wilson S.H.. Modulating the DNA polymerase beta reaction equilibrium to dissect the reverse reaction. Nat. Chem. Biol. 2017; 13:1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rozovskaya T., Tarussova N., Minassian S., Atrazhev A., Kukhanova M., Krayevsky A., Chidgeavadze Z., Beabealashvilli R.. Pyrophosphate analogues in pyrophosphorolysis reaction catalyzed by DNA polymerases. FEBS Lett. 1989; 247:289–292. [DOI] [PubMed] [Google Scholar]
- 51. Martynov B.I., Shirokova E.A., Jasko M.V., Victorova L.S., Krayevsky A.A.. Effect of triphosphate modifications in 2′-deoxynucleoside 5′-triphosphates on their specificity towards various DNA polymerases. FEBS Lett. 1997; 410:423–427. [DOI] [PubMed] [Google Scholar]
- 52. Sucato C.A., Upton T.G., Kashemirov B.A., Batra V.K., Martinek V., Xiang Y., Beard W.A., Pedersen L.C., Wilson S.H., McKenna C.E. et al. Modifying the beta,gamma leaving-group bridging oxygen alters nucleotide incorporation efficiency, fidelity, and the catalytic mechanism of DNA polymerase beta. Biochemistry. 2007; 46:461–471. [DOI] [PubMed] [Google Scholar]
- 53. Sucato C.A., Upton T.G., Kashemirov B.A., Osuna J., Oertell K., Beard W.A., Wilson S.H., Florian J., Warshel A., McKenna C.E. et al. DNA polymerase beta fidelity: halomethylene-modified leaving groups in pre-steady-state kinetic analysis reveal differences at the chemical transition state. Biochemistry. 2008; 47:870–879. [DOI] [PubMed] [Google Scholar]
- 54. Lecomte P., Doubleday O.P., Radman M.. Evidence for an intermediate in DNA synthesis involving pyrophosphate exchange. A possible role in fidelity. J. Mol. Biol. 1986; 189:643–652. [DOI] [PubMed] [Google Scholar]
- 55. Vaisman A., Ling H., Woodgate R., Yang W.. Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 2005; 24:2957–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olson A.C., Patro J.N., Urban M., Kuchta R.D.. The energetic difference between synthesis of correct and incorrect base pairs accounts for highly accurate DNA replication. J. Am. Chem. Soc. 2013; 135:1205–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Arzumanov A.A., Semizarov D.G., Victorova L.S., Dyatkina N.B., Krayevsky A.A.. Gamma-phosphate-substituted 2′-deoxynucleoside 5′-triphosphates as substrates for DNA polymerases. J. Biol. Chem. 1996; 271:24389–24394. [DOI] [PubMed] [Google Scholar]
- 58. Arion D., Parniak M.A.. HIV resistance to zidovudine: the role of pyrophosphorolysis. Drug Resist. Updat. 1999; 2:91–95. [DOI] [PubMed] [Google Scholar]
- 59. Urban S., Fischer K.P., Tyrrell D.L.. Efficient pyrophosphorolysis by a hepatitis B virus polymerase may be a primer-unblocking mechanism. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:4984–4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Structure factors and the refined co-ordinates have been deposited in the PDB with the following codes: 5YUR (0 min), 5YUS (1 min), 5YUT (3 min), 5YUU (5 min), 5YUV (7 min), 5YUW (10 min), 5YV3 (15 min), 5YUX (20 min), 5YUY (25 min), 5YV4 (30 min), 5YUZ (35 min), 5YV0 (40 min), 5YV1 (50 min), 5YV2 (60 min), 5YYD (dTMPPnP: 0 min), 5YYE (dTMPPnP: 60 min) and 5ZLV (50 mM MgCl2; dTTP: 30 min).