Abstract
We report the detailed transcriptomic profiles of human innate myeloid cells using RNA sequencing. Monocytes migrate from blood into infected or wounded tissue to differentiate into macrophages, and control inflammation via phagocytosis or cytokine secretion. We differentiated culture primary monocytes with either GM- or M-CSF to obtain pro- or anti-inflammatory macrophages, and respectively activated them with either LPS/IFNγ or anti-inflammatory cytokines. We also treated the THP-1 monocytic cell line with PMA and similar cytokines to mimic differentiation and activation. We detected thousands of expression and alternative-splicing changes during monocyte-to-macrophage differentiation and activation, and a net increase in exon inclusion. MBNL1 knockdown phenocopies several alternative-splicing changes and strongly impairs PMA differentiation, suggesting functional defects in monocytes from Myotonic Dystrophy patients. This study provides general insights into alternative splicing in the monocyte–macrophage lineage, whose future characterization will elucidate their contribution to immune functions, which are altered in immunodeficiencies, autoimmunity, atherosclerosis and cancer.
INTRODUCTION
The human immune system exhibits a remarkable diversity and versatility in ontogeny, morphology and function, thus offering great opportunities for regulatory network analyses. Monocytes are myeloid cells of the mononuclear phagocytic system which are critical for cellular innate immunity, as they contribute to homeostasis by clearing cellular debris and toxic particles, and also fight pathogens (1,2). Upon infection or wound, blood monocytes migrate into tissues and differentiate into macrophages, which directly combat infection through a pro-inflammatory response, or subsequently control it by anti-inflammatory (wound healing) response (3).
Cultured primary monocytes are differentiated by Granulocyte-Macrophage Colony Stimulating Factor or GM-CSF (CSF-2) or by Macrophage Colony Stimulating Factor or M-CSF (CSF-1), respectively adopting mostly pro- or anti-inflammatory properties, and these cells are often referred to as M1 or M2 macrophages (Figure 1A) (3). The THP-1 cell line, which recapitulates some monocyte functions, can be differentiated into macrophage-like cells by phorbol myristic acid (PMA) (4,5). Most tissue-resident macrophages are deposited at early developmental stages, are maintained by self-renewal, and may be generated without a monocytic stage like brain microglia (2,6,7), yet monocyte-derived macrophages are important for defence against infection. Recent efforts are cataloguing the heterogeneity of macrophages in both humans and rodents (8).
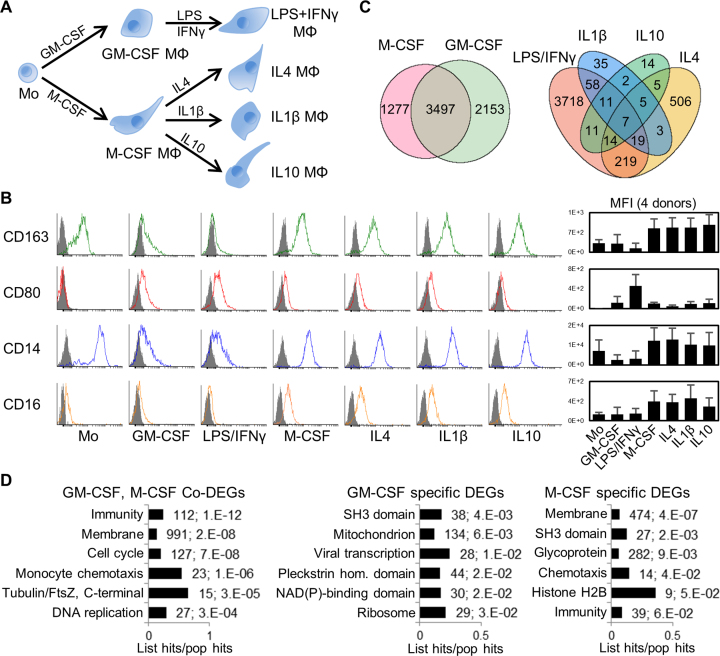
Figure 1.
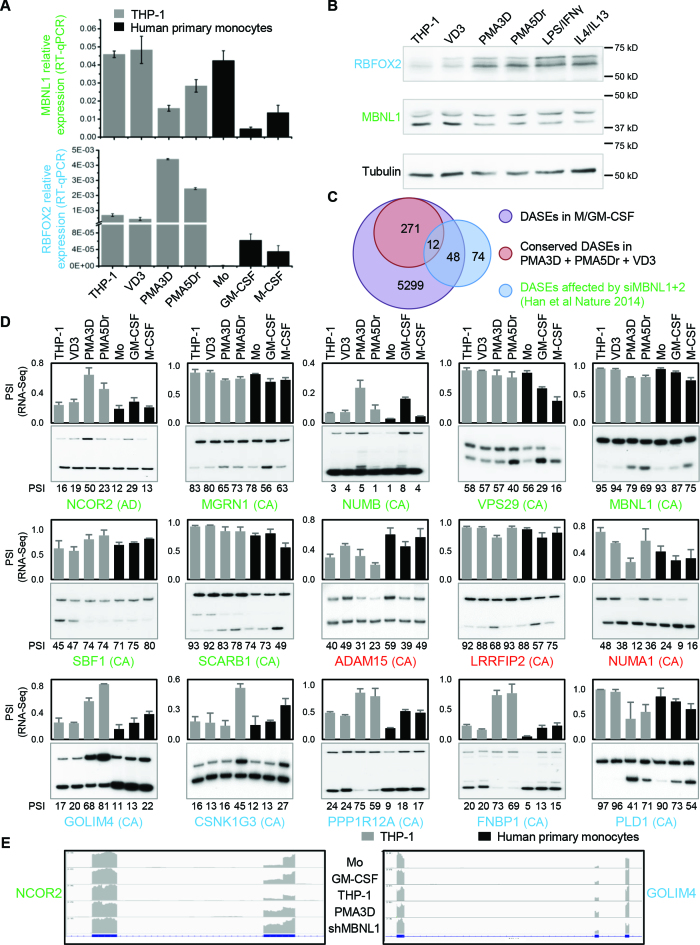
Human primary monocyte differentiation protocol and differentially expressed genes (DEGs). (A) Diagram of differentiation and polarization of human primary monocytes (Mo) to macrophages (Mϕ) in culture. We performed RNA-seq for the seven monocyte and macrophage conditions. (B) Flow cytometry measurements of four representative surface markers on monocytes and macrophages (more in Supplementary Figure S2b). Right, median fluoresce intensity (MFI) of four donors. As expected, CD14 and the haemoglobin scavenger receptor CD163 in monocytes were downregulated only in GM-CSF macrophages. The T-cell co-stimulator CD80 and the Immunoglobulin G Fc receptor CD16 were upregulated only in GM-CSF or M-CSF macrophages, respectively, while CD80 is further increased by LPS/IFNγ. (C) Venn diagrams with numbers of differentially expressed genes (DEGs) with fold change ≥2 (q-value ≤0.05) for GM-CSF and M-CSF treated macrophages versus monocytes (left), and for the four polarizations (right). The enrichment of common M/GM-CSF DEGs is highly significant (P ∼ 0, one-tailed Fisher's Exact test). (D) Bar charts with the top six enriched clusters of DEGs upon M/GM-CSF stimulations, using DAVID bioinformatics resources. List hits, number of genes in our list that belong to each gene category; Population hits (pop hits), total number of genes that belong to each gene category in the whole DAVID database. Barcharts display gene category and percentage of our list hits in the population hits in the y- and x-axis, respectively. Each bar includes our list hits number and the benjamini q-value of the cluster.
Monocyte or macrophage receptor binding to microorganisms elicits phagocytosis or cytokine secretion via deep changes in gene expression (6). The best known stimulus pertains to the Gram-negative bacteria-derived lipopolysaccharide (LPS) binding to Toll-Like Receptor 4 (TLR4), which results in inflammatory response that is enhanced by gamma-interferon (IFNγ) (9,10). Anti-inflammatory macrophages are polarized by cytokines such as IL4 for tissue repair, allergic and anti-parasitic responses, IL1β for humoral immunity and T-helper 2 response, or IL10 for anti-inflammatory response and tissue repair (Figure 1a) (11,12). These polarization protocols exemplify the great plasticity of macrophages in culture which is even higher in the body.
Monocytes and macrophages are deeply associated with many types of human disease at multiple levels. Their defective responses result in enhanced infections, while an excessive or uncontrolled function causes allergy or autoimmunity, such as in acute inflammation or sepsis (3), in obese adipose tissue leading to insulin resistance and diabetes (3,13), and in atherosclerosis (14). Tumour associated macrophages play a dual role in cancer, as they fight solid tumors early on, and later they promote metastasis and confer poor prognosis (3,15). Finally, Myelodysplastic Syndromes (MDS), which are diverse pre-malignancies with reduced myeloid differentiation, cytopenia and susceptibility for acute myeloid leukemia, are caused by frequent and recurrent Splicing Factor (SF) mutations whose prognostic value and pathogenesis mechanisms are currently under evaluation (16,17). The frequent SF mutations in MDS illustrate that splicing plays a crucial role in the development and function in the myeloid lineage.
Immune-cell differentiation and activation involve widespread transcriptomic changes, reflecting regulation via transcription and mRNA decay (18), yet alternative splicing (AS) is becoming important (19,20). The complexity of the human transcriptome and proteome, across and within tissues, is largely expanded by AS virtually regulating all intron-containing transcripts (21,22). The AS types, such as cassette exon inclusion, intron retention or alternative splice sites, result in mRNAs which can encode protein isoforms with altered, antagonistic or unrelated functions. AS patterns are dictated by cis-acting elements, comprising exonic or intronic splicing enhancers or silencers, which activate or repress splicing via binding to trans-acting SFs, and each SF regulates up to thousands AS events (23). AS also depends on RNA secondary structure (24), transcription and chromatin (25). RNA-sequencing (RNA-seq) studies showed a prevalence of tissue-specific AS (26–28).
AS has been largely studied in lymphocytes but less so in myeloid cells (29). AS and transcription affect distinct gene subsets upon T-cell activation (19), and AS changes from cell lines and primary T cells differ in the response pathways and stimuli dependence. Recent transcriptomic studies characterized AS in Hematopoietic Stem Cells (30) as well as in terminal rodent and human erythropoiesis (31,32), terminal murine granulopoiesis (33) and megacaryopoiesis (31), revealing increased intron retention as a feature of late-stage red blood cells and mature granulocytes. Even though the RNA profiles of THP-1 and primary monocytes, resting and LPS-activated macrophages are well characterized and master transcriptional and epigenetic regulators are known, there is insufficient information on AS (4,5,9,10). LPS-activated mouse macrophages slow down splicing in certain pre-mRNAs thus favoring AS (34,35). Furthermore, splicing factor 3a in murine macrophages regulates AS events involved in TLR signalling (36). For human monocytes/macrophages, a recent study established the role of the quaking (QKI) RNA-binding protein in regulating AS of wound-healing macrophages and promotion of atherogenic plaques (37), and another AS study of LPS activation revealed the role of CELF1 (38). Just a few AS events are functionally studied in monocytes/macrophages, in genes like CD44 in inflammatory disorders such as rheumatoid arthritis (39), Tissue Factor in thrombogenesis, wound healing, angiogenesis and metastasis (40), and MyD88 in signalling (41). SRSF3 was shown to control IL-1β secretion upon E. coli exposure (42), yet many more SFs await characterization.
Here, we present a comprehensive RNA-seq analysis of THP-1 and primary cells to identify thousands of AS changes during differentiation to pro- or anti-inflammatory macrophages, and hundreds during inflammatory activation, and most of these are totally uncharted. We identified MBNL1 as an important AS regulator during differentiation. This study should help understand the regulation of these important cells and their connection to multiple pathologies, including immunodeficiencies, autoimmunity and cancer (3).
MATERIALS AND METHODS
Ethics
Informed consent to all healthy donors was obtained for all blood samples from which monocytes and macrophages were derived. Approval for the presented studies with human samples was provided by Nanyang Technological University's Institutional Review Board ethics committee (IRB-2014-10-002), and donor anonymity was ensured.
Human monocyte and macrophage culture
For primary monocyte purification, we isolated Peripheral Blood Mononuclear Cells (PBMCs) from four healthy donors by Histopaque-1077 (Sigma-Aldrich) gradient (Supplementary Figure S1A). We subsequently incubated the samples with negative selection microbeads (Pan Monocyte Isolation Kit, cat.no130-096-537, Miltenyi Biotech) to obtain purified monocytes.
We added 60 mL blood from each donor to phosphate buffered saline (PBS) with a ratio of 1:3. We gently poured 30 mL of mixture into filtered Leucosep tubes (cat. no. 227290, Greiner Bio One), followed by centrifugation at 800 × g, 15 min at room temperature and zero deceleration. Then, we carefully harvested the PBMC layer and washed it twice with MACS buffer (PBS, pH 7.2 supplemented with 0.5% BSA and 2 mM EDTA, degassed before use and stored at 4°C). We resuspended 1 × 107 PBMC cells in 30 μl MACS buffer, added by FcR blocking reagent and incubated mixtures with Biotin-Antibody Cocktail and Anti-Biotin MicroBeads. We passed mixtures through LS columns standing on magnetic separator, and we collected the flow-through with the unlabeled monocytes. After washing with MACS buffer once, we collected the monocyte sample directly for RNA isolation.
We differentiated monocytes to macrophages by culturing purified monocytes with the different cytokines for 7 days in RPMI 1640 medium (cat. no. 30-2001, ATCC) supplemented with 20% Fetal Bovine Serum (FBS; Gibco), 100 U/ml Penicillin and 100 μg/ml Streptomycin (Gibco) at 37°C and 5% CO2. We used 100 ng/ml GM-CSF to induce inflammatory macrophages, and 100 ng/ml M-CSF to induce anti-inflammatory macrophages. Every 3 days we changed the culture media and added fresh cytokines. On day 7, we polarized GM-CSF macrophages by 100 ng/ml LPS plus 50 ng/ml IFN-γ for 8 h. In turn, we separately incubated M-CSF macrophages for 8 h with 10 ng/ml of either IL-4, IL-1β or IL-10 for the three polarizations.
THP-1 culture and nucleofection
We cultured the human monocytic cell line THP-1 (cat. no. TIB-202, ATCC) in RPMI 1640 Medium (cat. no. 30-2001, ATCC) supplemented with 10% FBS, 0.05 mM 2-mercaptoethanol, 100 U/ml Penicillin and 100 μg/ml Streptomycin at 37°C and 5% CO2. We separately treated THP-1 monocyte-like cells for 3 days with 100 ng/ml phorbol ester 12-O-tetradecanoylphorbol-13-acetate (PMA, Sigma-Aldrich) or 0.5 μM 1,25-dihydroxyvitamin D3 (vitamin D3, Sigma-Aldrich) to obtain PMA3D and VD3 cells. We enhanced the differentiation of PMA3D cells by removing the PMA-containing medium at day 3, and further incubating cells in fresh medium for another 5 days to derive the PMA5Dr samples, as described (43). We stimulated the PMA3D cells with 100 ng/ml LPS plus 20 ng/ml IFN-γ for 8 h, and with 20 ng/ml IL-4 plus 20 ng/ml IL-13 for 8 h.
For microscopy, we plated primary monocytes or THP-1 cells in tissue-culture treated μ-Dish (81156, Ibidi GmbH, Germany) at a density of 1 × 105/cm2, followed by the corresponding treatments. We took images with the Zeiss Live Cell Observer.
The nucleofection of untreated and PMA3D treated THP-1 cells with dsiRNA (Integrated DNA Technologies, IDT, USA) generally followed the protocol described before (44), with dsiRNA target sequences in Supplementary Table S1. For PMA3D cells, we treated THP-1 cells with 100 ng/ml PMA for 16 h and detached cells by Accutase (A6964, Sigma-Aldrich). We mixed 1 × 106 cells with 200 pmol dsiRNA in SG nucleofector solution (Lonza, Switzerland) and electroporated cells subsequently using the FI-100 program on Amaxa 4D-Nucleofector (Lonza, Switzerland). After that, we recovered the cells in THP-1 nucleofection medium (Lonza Mouse T cell nucleofection medium supplemented with 20% human serum, 1% nonessential amino acids, 1% sodium pyruvate and 1% penicillin/streptomycin/L-glutamine) for 4 h. Then we changed the medium back to normal differentiation medium with 100 ng/ml PMA, and we cultured the cells for 2 days. For untreated THP-1 nucleofection, we collected cells directly for electroporation, and after recovery we cultured them for another 3 days before RNA or protein isolation.
Lentiviral transduction
We inserted the RBFOX2 and MBNL1 shRNAs (Supplementary Table S1) into the pLKO.1 vector for gene knockdown. We packaged the recombinant lentiviruses using a second generation system (Addgene, Lentiviral Guide: https://www.addgene.org/viral-vectors/lentivirus/lenti-guide/#second-generation). We used Lipofectamine 2000 Reagent (Life Technologies, USA) to transfect HEK293T cells with three plasmids encoding pPol/Gag, pVSVG, and pLKO.1 harboring the corresponding shRNA. We changed the medium, and harvested the packaged lentiviral particles 72 h later. We introduced the lentiviral particles into THP-1 cells by centrifugation at 800 × g, 32°C for 30 min in the presence of 10 μg/ml Hexadimethrine bromide (polybrene, cat no H9268, Sigma). Then we cultured cells for 3 days and selected transfectants with 2 μg/ml puromycin (Sigma-Aldrich).
Western blotting
We washed cells twice with PBS and incubated with the protein lysis buffer [50 mM Tris–HCl, pH8; 150 mM NaCl; 1% Triton X-100; 10% glycerol; 1 mM EDTA; 1× cOmplete EDTA-free protease inhibitor cocktail (Roche)] for 10 min before centrifugation. We determined the protein concentration by Bradford assay (Bio-Rad). We ran 30 μg of protein by SDS-PAGE and transferred them to PVDF membrane (Bio-Rad, USA). We detected the specific proteins by the following primary antibodies: anti-MBNL1 (ab108519, Abcam), anti-RBFOX2 (A300-864A, Bethyl), anti-SNRPE (sc-21057, Santa Cruz), anti-RAVER2(ab174321, Abcam), anti-GEMIN2 (sc-32806, Santa Cruz), anti-hnRNP A1 (sc-32301), anti-KSRP (A302-021A, Bethyl), anti-GAPDH (sc-25778, Santa Cruz), anti-αTubulin (sc-8035, Santa Cruz) and anti-β-Actin (A5441, Sigma-Aldrich). We quantified the protein samples from three biological replicates using the Quantity One software (Bio-Rad, Life Science Research). We derived the relative expression of each protein from its ratio over reference protein level.
RNA pull-down
We generated double-stranded DNA templates by annealing and ligation of oligodeoxyribonucleotides (IDT) with complementary overhangs and a T7 promoter. We in vitro transcribed the RNA probes from these DNA templates with T7 mMESSAGE mMACHINE Kit (cat. no. AM1344, Life Technologies) at 37°C for 4 h, followed by DNase treatment for 15 min and RNA purification and concentration (Zymo Research). Next we end-labeled the RNAs with desthiobiotin, by attaching a single biotinylated nucleotide to the 3′ terminus of the RNA probe using T4 RNA ligase in its reaction buffer (50 mM Tris•HCl, 10 M MgCl2, 10 M DTT, 1 mM ATP; pH 7.8) at 16°C overnight, followed by RNA purification and ethanol precipitation (Pierce RNA 3′ End Desthiobiotinylation Kit, Thermo Scientific). We performed the RNA pull-down assay according to the Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Scientific). We captured the biotin-labelled RNA by streptavidin magnetic beads incubated with the RNA in capture buffer (20 mM Tris of pH 7.5, 1 M NaCl, 1 mM EDTA) for 30 min at room temperature. After two 20 mM Tris buffer (pH 7.5) washes, we added nuclear extract to the RNA-Protein binding buffer (20 mM Tris of pH 7.5, 50 mM NaCl, 2 mM MgCl2, 0.1% Tween-20) and incubated them with the RNA at 4°C for 2 h. We washed the beads three times with the wash buffer (20 mM Tris of pH 7.5, 10 mM NaCl, 0.1% Tween-20) before incubating them with the default biotin elution buffer at 37°C for 30 min. We collected the eluates and analysed them with the flow-through by Western Blotting.
RNA extraction and RT-PCR
We washed monocytes/macrophages with cold PBS three times, followed by addition of 600 μl lysis buffer with 2-mercaptoethanol directly to 1 × 106 cells and then mixing until cells lysed. We extracted total RNA using PureLink RNA Mini Kit (Life Technologies, USA). We extracted total RNA strictly following the kit's manual as stated above and processed for RT-qPCR freshly, or immediately stored in –20°C for short period (less than 2 weeks) and –80°C for longer time (less than 6 months). We removed residual DNA by TURBO DNase (Ambion) treatment for 30 min, followed by heat-inactivation and RNA purification. We measured RNA concentration by spectrophotometry (NanoDrop, Thermo Scientific) and checked RNA integrity by agarose gel. We used only high quality RNA for downstream experiments. We processed cDNA directly for real-time PCR or stored it immediately at –20°C for less than 2 weeks, or at –80°C for up to 6 months. We reverse transcribed 1 μg RNA to cDNA with M-MuLV reverse transcriptase (New England Biolabs), dNTPs and oligo-dT in 10 μl of volume at 42°C for 1 h, followed by 15 min of inactivation at 65°C. We performed quantitative RT–PCR using the SYBR Green Master Mix (Thermo Fisher Scientific) on CFX96 Touch Real-Time PCR Detection System (Bio-Rad). We indicate primer sequences in Supplementary Table S2, with all amplicons shorter than 200 bp. We used hACTB (β-Actin) as qPCR reference gene. We calculated the relative expression of three biological replicates of each target gene by the equation 2(Cq(β-Actin)-Cq(target gene)). We plotted means and standard deviation.
We performed semi-quantitative RT-PCR to validate or detect alternative splicing (AS) events. We designed primer pairs to map in the flanking exons of each target AS event, with sequences in Supplementary Table S3. We radioactively labeled each forward primer with γ-32P-ATP (Perkin-Elmer, USA) using T4 PolyNucleotide Kinase (New England Biolabs, USA), followed by purification with Microspin G-25 columns (GE Healthcare). We carried out the PCRs using Go-Taq DNA polymerase (Promega, USA) for 25–33 cycles. We separated the PCR products with 8% native polyacrylamide gel electrophoresis (PAGE) at 200 V for 5 h in TBE buffer, followed by vacuum-drying and exposure to phosphorimaging screen. We used Typhoon imager (GE Healthcare) for signal scanning, and we quantified the band intensities by ImageQuant TL software (GE Healthcare).
RNA-sequencing library preparation
After removing medium, we washed macrophages twice with cold PBS to get rid of floating cells. We washed THP-1 cells twice in PBS by centrifugation. We isolated total RNA using RNeasy Mini Kit together with QIAshredder (Qiagen). We removed the residual DNA by 30-min DNase treatment (TURBO DNA-free Kit, Ambion) at 37°C. Subsequently, we purified RNA with RNA Clean and Concentrator (Zymo Research), and checked its concentration and integrity using ribogreen assay and bioanalyzer (Agilent Technologies 2100 Bioanalyzer). We only processed RNA samples with RIN above 8. We prepared RNA libraries using the TruSeq Stranded mRNA Library Prep Kit (Illumina). In brief, we purified poly-A containing mRNA molecules by poly-T oligos attached to magnetic beads. We cleaved and fragmented the mRNA into small pieces, followed by reverse transcription of the first cDNA strand using random primers. We replaced dTTP with dUTP in the second strand synthesis by using DNA Polymerase I and RNase H, so as to track the strand information in the following steps. We ligated the cDNA fragments with the adapter and purified them. We enriched the molecules by PCR to generate the final cDNA library. We used HiSeq 2500 System Rapid mode (Illumina) for RNA-sequencing, giving rise to paired-end reads of 101 bp. We show total read numbers of each sample in Supplemental Table S4.
RNA-seq data analysis
We first confirmed the original read quality of our RNA-seq data by FastQC (Simon Andrews, Babraham Bioinformatics). For gene expression analysis, we aligned the paired-end reads to human genome (UCSC-hg19) using Tophat2 (45) with Bowtie2 (46). In RNA-seq data analysis, we used the default parameters for each tool unless otherwise stated. For Tophat2, we allowed a maximum of two mismatches and set two parameters as –library-type = fr-firststrand and –read-realign-edit-dist = 0. We used Cufflinks, Cuffmerge and Cuffdiff (47) for transcripts assembly and quantification of the Tophat2 output. We masked out annotated transfer-messenger RNA, ribosomal RNA and mitochondrial transcripts in Cuffdiff analysis by using the parameter –mask-file with corresponding GTF file. We obtained 87–92% of aligned reads to the genome with Tophat2 (Supplementary Table S4, except one replica). The Pearson correlation (R2) for Log10(FPKM) (Fragments Per Kilobase of transcript per Million fragments mapped) for each individual condition across donor pairs was on the range of 0.90–0.96, highlighting the strong reproducibility of our data (Supplementary Figure S3; samples from different donors are not identical). We used the common FPKM (fragments per kilobase of transcript per million fragments mapped) as the expression-levels measure for each gene or transcript.
To detect differential AS events (DASEs), we used three softwares, rMATS (48), MISO (49) and SpliceTrap (50). To run rMATS, we adopted the alignment outputs of Tophat2. Our read type is paired-end (-t paired) and with length of 101bp (-len 101), while the analysis type was the default unpaired. In SpliceTrap running, we first mapped the original paired-end reads to SpliceTrap database hg19, with low cutoff. We performed postAnalysis subsequently, with 2 junction reads required per junction (-j 2) and low cutoff (-c L). Finally, we compared exon inclusion ratio between each two samples using SpliceChange. In MISO analysis, first we used the Tophat2 alignment results to estimate isoform expression levels within each sample, with the command miso –run. We included the –paired-end option, and we set the mean and standard deviation of insert length to 350 and 150, respectively. Then we summarized the MISO outputs for each sample, with the command summarize_miso –summarize-samples, to obtain confidence intervals for the probabilistic framework. Next, we made pairwise comparisons between samples, using compare_miso –compare-samples, to detect differentially expressed isoforms. In our MISO analysis, we covered the following AS types: skipped exons (SE), alternative 3′/5′ splice sites (A3SS, A5SS), mutually exclusive exons (MXE) and Retained introns (RI).
For MISO, we took the ‘sample_posterior_mean’ average among different replicates as the final exon inclusion level for each condition, and used Bayes_factor min and median as a criteria of statistical significance among the replicates. We obtained the final alternative spliced exon list of MISO for each comparison by applying the following cutoffs: inclusion level difference ≥0.05; Bayes_factor Min ≥1 and Bayes_factor Median ≥10. For SpliceTrap, we also used the inclusion ratio average of all replicates as the final exon inclusion level, and we calculated the P-value using paired Student's t-test. We adopted the following cutoffs to get the final AS list: inclusion level difference ≥0.05 and P-value ≤0.001. For the rMATS AS list we just applied the cut-offs of inclusion level difference ≥0.05 and P-value ≤0.05.
For the co-detected DASEs among the three tools, we retrieved the coordinate annotations of each exon from each tools output, including the information of event type (AA, AD, CA, IR, MXE), chromosome, strand, start and end position of AS exons and flanking exons on the chromosome. DASEs harboring the same information detected by two or three tools were named tri- or co-detected DASEs, as we labeled them in Supplementary Figure S5.
Flow cytometry
We harvested cells and washed them with 1X Phosphate buffered saline (PBS) before incubation with 2 μl of 1 mg/ml goat serum (Sigma-Aldrich) for 5 min for Fc receptors blocking on ice prior to antibody staining. We stained cells according to manufacturer's protocol with fluorochrome-coupled antibodies listed in Supplementary Table S5. We incubated each sample with primary antibodies or the matching isotypes for 15 min on ice. Then we washed cells twice in buffer and resuspended them in 300 μl of PBS. We took flow cytometric measurements using five lasers LSR Fortessa X-20 (Becton Dickinson). We performed data analysis by Cyflogic (v.1.2.1).
RESULTS
Major changes in gene expression upon monocyte-to-macrophage differentiation and activation
We have completed a high-depth RNA-seq profiling in human blood-derived monocyte differentiation and activation. We used Ficoll gradient and negative selection to isolate human monocytes from peripheral blood mononuclear cells of four donors, yielding >96% purity as judged by FACS analysis of CD14 (Supplementary Figure S1). Then we used different cytokines to differentiate these monocytes to macrophages, and activated them with different signalling molecules (Figure 1A). In brief, we differentiated monocytes to macrophages with either GM-CSF or M-CSF for seven days, and cells adhered to plate (Supplementary Figure S2A). We polarized the GM-CSF macrophages with LPS/IFNγ, and the M-CSF macrophages with either IL1β, IL10 or IL4, each for 8 h. We confirmed the efficiency of these protocols by cell morphology and extensive surface-marker profiling by FACS (Figure 1B and Supplementary Figure S2B, and antibodies in Supplementary Table S5).
From all four donors, we obtained ∼300 million paired-end reads per each of the seven conditions (Supplementary Figure S3, details in Supplementary Methods). Using a cutoff of ≥2-fold increase or decrease in FPKM, we identified thousands of differentially expressed genes (DEGs) in macrophages compared to monocytes (Figure 1C, left Venn diagram; Supplementary Table S6). A total of 2153 and 1277 DEGs are specific to GM-CSF and M-CSF macrophages respectively, while the 3497 common changes are highly enriched. Gene Ontology (GO) analysis with DAVID for common and M-CSF or GM-CSF-specific DEGs revealed most enriched clusters in immunology-relevant categories such as immunity, membrane and chemotaxis (Figure 1D). DEGs specific to either M- or GM-CSF macrophages show enrichment in different pathways (Figure 1D, left), highlighting that certain changes in gene expression drive some of the different properties of these macrophages, while other DEGs might be a consequence of the differentiation process.
Compared to the corresponding resting macrophages, LPS/IFNγ-activation induced many more DEGs than the three anti-inflammatory polarizations (Figure 1C, right diagram). GO enrichment analysis showed that LPS/IFNγ responses clustered in innate immunity and specific pathways such as pleckstrin-homology domain, while anti-inflammatory activations were enriched in different immunity pathways like c-type lectin-like and chemotaxis.
In parallel, we treated THP-1 cells with PMA to mimic macrophage differentiation, for three days (PMA3D) and for three days followed by five days rest with fresh medium (PMA5dr) (43). Also we incubated THP-1 cells with vitamin D3 (VD3) which activates certain immune pathways (51). We also induced PMA3D cells with either LPS plus IFNγ, or with IL4 plus IL13, to mimic the LPS/IFNγ and IL4 treatments of blood-derived macrophages. These six conditions showed the expected morphological changes (Supplementary Figure S2C) and FACS profiles for five markers (Supplementary Figure S2D). RNA-seq revealed common DEGs for THP-1 differentiation and polarization (Supplementary Figure S4A; Supplementary Table S6).
Many RNA binding protein genes change their expression upon monocyte-to-macrophage differentiation and activation
Several genes encoding RNA Binding Proteins (RBPs) change expression by ≥2-fold during differentiation and LPS/IFNγ-activation of primary cells (Figure 2A). These RBPs included several SFs such as RBM6, SRRM2, and SRSF5 for M/GM-CSF, and RBM7/11/17, TIA1, SRSF12, FUBP1, MBNL2 and RBFOX2 for LPS/IFNγ activation. Among the RBPs as DEGs in THP-1 differentiation and activation (Supplementary Figure S4A and B), we confirmed by Western Blotting the PMA-induced downregulation of SNRPE, GEMIN2 and RAVER2, while we found that hnRNPA1 or KSRP levels did not change (Supplementary Figure S4C). This experiment shows that changes in RNA levels lead to differences in protein levels which should establish the splicing patterns in each condition.
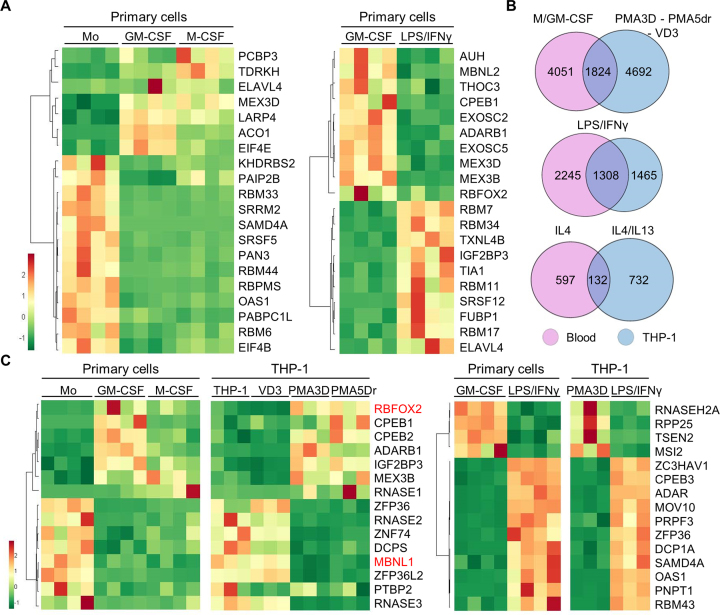
Figure 2.
DEGs common in primary and THP-1 cells, and identification of RBPs important for macrophage differentiation or polarization. (A) Heat map of mRNA expression (FPKM) for the top 20 RBPs as DEGs in primary cells, either upon M- or GM-CSF differentiation, or induced by LPS/IFNγ. The four columns under each condition represent the RNA-seq values for four donors. (B) Venn diagrams showing the proportion of DEGs with fold change ≥2 (q-value ≤0.05) between the indicated conditions in primary and THP-1 cells. We used genes instead of transcripts for this comparison. The overlap for analogous stimuli between primary and THP-1 cells is highly significant (P < 10−50 for all three tests, one-tailed Fisher's Exact test). (C) Heat map of mRNA expression (FPKM) levels for the top 15 RBPs as DEGs both in differentiation or activation of primary cells and THP-1, as labelled on top.
We found 1824 DEGs in common with M/GM-CSF in primary cells and PMA/VD3 in THP-1 cells, which is about one-third of the DEGs in each category (Figure 2B). A number of RBPs consistently change expression upon primary-cell and THP-1 differentiation (Figure 2C, left), which might account for their analogous morphological and functional features such as cell adherence and expansion, enhanced granularity, etc. GM-CSF and PMA differentiated macrophages exhibit upregulation of CPEB1 and CPEB2 translation regulators, RBFOX2, and other RBPs like IGF2BP3 and MEX3B. In turn, commonly upregulated genes by M-CSF and PMA include RNA degradation enzyme RNASE1. Importantly, M/GM-CSF and PMA macrophages strongly downregulate SFs MBNL1 and PTBP2, degradation enzymes RNASE2 and RNASE3, scavenger decapping enzyme DCPS, mRNA destabilizing factors ZFP36 (also known as TTP) and ZFP36L2, and the putative RBP ZNF74.
Among the DEGs upon LPS/IFNγ activation of both GM-CSF and PMA3D macrophages (Figure 2C, right), we detected upregulation of translation regulators CPEB2, CPEB3 and SAMD4A, splicing factor PRPF3, antiviral protein ZC3HAV1, adenosine-to-inosine editing enzyme ADAR, decapping enzyme DCP1A, exosome subunit for RNA degradation PNPT1, ARE-destabilizing factor ZFP36, microRNA biogenesis factor MOV10, and RBPs with unknown function like RBM43. In turn, we also saw downregulation of antiviral protein ZC3HAV1L, tRNA processing factors RPP25 (RNaseP) and TSEN2 (tRNA splicing endonuclease), and translation repressor MSI2. Some of these RBPs might be responsible for fine tuning the anti-inflammatory responses triggered by TLR4 stimulation.
Alternative splicing changes upon monocyte-to-macrophage differentiation and activation
To detect differential AS events (DASEs) by comparing percentage spliced in (PSI) between conditions, we used rMATS (48), MISO (49) and SpliceTrap (50) altogether with a high statistical threshold to minimize the false positives. We used three tools not to miss many true AS changes that are only detected by one of them, because just few DASEs are co-detected by two or three tools for each condition (Supplementary Figure S5A, top). We found thousands of DASEs in GM- or M-CSF induced macrophages (Figure 3A, Supplementary file AS_blood_gene_list), with a strong enrichment of shared events, with 2435 in 1549 genes. In turn 1933 and 1262 DASEs in 1327 and 903 genes are specific to GM- and M-CSF, respectively (Figure 3A, left). Interestingly, both M/GM-CSF macrophages exhibit higher levels of long isoforms, or higher inclusion of cassette exons (Table 1), suggesting that either an increase in splicing activator/s or a decrease in repressor/s is associated with macrophages as opposed to monocytes. The most common type of DASEs in differentiation and activation is cassette exon followed by mutually exclusive exons, and by intron retention and alternative 5′ or 3′ splice sites with similar levels (Supplementary Figure S5B, two left diagrams; Supplementary Table S7).
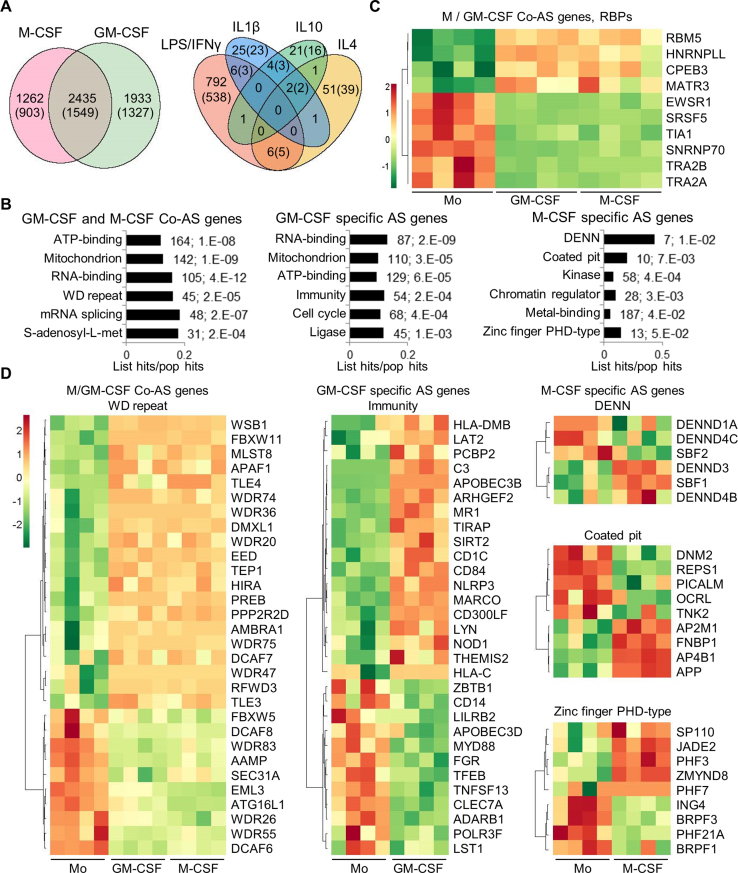
Figure 3.
Alternative splicing in primary monocyte differentiation and polarization. (A) Venn diagrams with the numbers of DASEs for differentiation and activation as indicated (PSI change ≥0.05; SpliceTrap P-value ≤0.001, MISO BFmin ≥1 & BFmedian ≥10, MATS FDR ≤0.05). In parentheses, number of genes affected by DASEs. The enrichment of common M/GM-CSF DASEs is highly significant (P∼0, one-tailed Fisher's Exact test). (B) Bar charts with the top enriched clusters of DASEs upon M- and/or GM-CSF stimulation using DAVID, as in Figure 1D. (C) Heat map showing PSI of top ten RBPs (mostly splicing regulators) as DASEs upon both M/GM-CSF treatments. Only the DASE with the highest PSI change is plotted for each RBP. (D) Heat maps for the PSI changes of DASEs in different enriched clusters of monocyte differentiation. Only DASEs with the highest PSI change (one per gene) are shown.
Table 1. Binary comparisons of differential AS events.
| Inclusion events | Skipping events | |||
|---|---|---|---|---|
| Human primary monocytes | Mo | GM-CSF | 2574 | 1794 |
| Mo | M-CSF | 2264 | 1433 | |
| GM-CSF | LPS/IFNγ | 448 | 357 | |
| M-CSF | IL4 | 34 | 27 | |
| M-CSF | IL1β | 25 | 13 | |
| M-CSF | IL10 | 17 | 12 | |
| THP-1 cell line | THP-1 | PMA3D | 305 | 267 |
| THP-1 | PMA5Dr | 349 | 211 | |
| THP-1 | VD3 | 1142 | 519 | |
| PMA3d | LPS/IFNγ | 224 | 131 | |
| PMA3d | IL4/IL13 | 133 | 75 |
Certain pathways are commonly enriched and others are specific to DASEs in M- or GM- CSF macrophages (Figure 3B), which are different from the pathways affected in the DEGs (Figure 1D), as seen in other cell types (19,27,28). DASEs in both M/GM-CSF macrophages versus monocytes frequently affect RBPs and SFs in particular, such as TRA2A/B isoforms, TIA1 or HNRNPLL (Figure 3C). Other pathways enriched in both or either M- or GM-CSF macrophage DASEs reveal known functions in ‘immunity’ or ‘coated pit’ (related to endocytosis) (Figure 3D), or cluster in sets of genes whose functions and coordinated AS changes remain to be characterized in these cells (‘WD’, ‘DENN’ and ‘zinc-finger PHD-type’ protein families). Finally, ∼800 AS changes upon LPS/IFNγ activation also demand characterization, compared to the very few detected changes by either IL4, IL1β or IL10 treatments, and with only 13 DASEs in common between the LPS/IFNγ and at least one anti-inflammatory activation.
We also measured the DASEs for THP-1 differentiation by PMA and VD3, and the activation by LPS/IFNγ or IL4/13 (Supplementary file AS_thp1_gene_list). We found hundreds of DASEs for differentiation, with many more in common between PMA3D and PMA5Dr than between these two and VD3 (Figure 4A, left, and Table 1). We also detected nearly twice as many DASEs for LPS/IFNγ compared to IL4/13 (Figure 4A, right, and Table 1). The DASE co-detection by more than one tool is very low (Supplementary Figure S5A, bottom), and the AS type distribution in THP-1 conditions shows a similar pattern as above, with the exception of higher frequency of mutually exclusive exons in LPS/IFNγ stimulation (Supplementary Figure S5B, right). By comparing these THP-1 DASEs with those of analogous primary cells, we found a small yet enriched overlap for macrophage differentiation and for LPS/IFNγ activation, but no overlap for IL4 (Figure 4B).
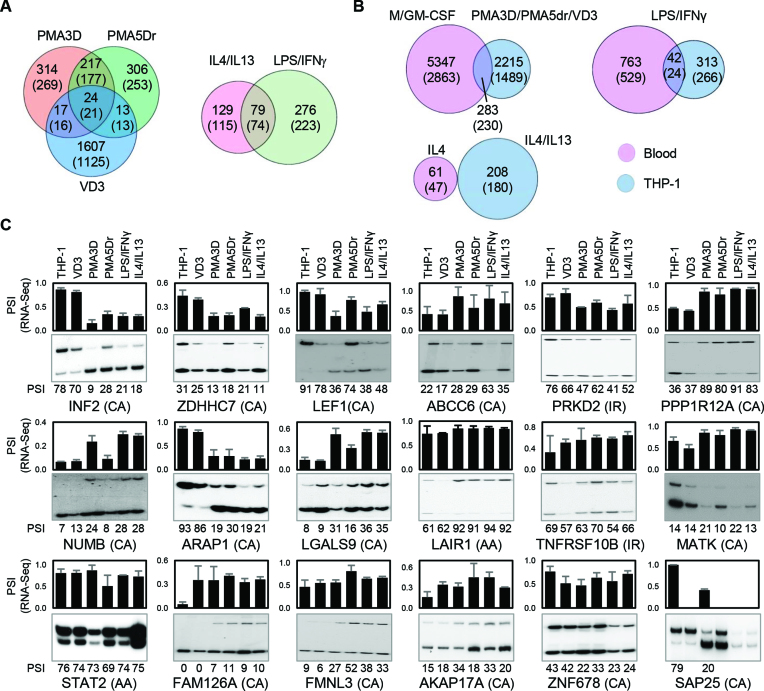
Figure 4.
Alternative splicing in THP-1 cells. (A) Venn diagrams with the numbers of DASEs (PSI change ≥0.05; SpliceTrap P-value ≤0.001, MISO BFmin ≥1 & BFmedian ≥10, MATS FDR ≤0.05) for the indicated treatments of PMA/VD3 differentiation (left) and polarization (right). In parentheses, number of genes affected by DASEs. The PMA shared events are highly enriched, while the overlap with VD3 DASEs is also higher than expected (P ∼ 0 and P < 10−5, one-tailed Fisher's Exact test). (B) Venn diagrams showing the numbers of DASEs that are common in primary and THP-1 cells upon analogous stimuli. In parentheses, number of genes affected by DASEs. The overlaps for differentiation and LPS activation are significantly enriched (both P < 10−10, Fisher's Exact test). (C) Radioactive RT-PCR validation of DASEs identified in THP-1 differentiation. Bar chart on top shows the RNA-seq-derived PSI of each DASE with its type in parenthesis, with cassette exons (CA), intron retention (IR) and alternative 3′ splice sites (AA). The radioactive PAGE shows the RT-PCR results with their PSI quantification at the bottom of each lane. The lack of some graph bars in SAP25 is due to the lack of detection in certain conditions, yet it shows a strong reduction in cassette exon inclusion by PMA. All DASEs shown here are validated except MATK and ZNF678 (Supplementary Figure S7A).
We tested as many as 45 DASEs in THP-1 (Figure 4C for 17 DASEs not counting SAP25, and 28 more in Supplementary Figure S6), plus 14 more in both THP-1 and primary cells (Figure 5D, not counting NUMB as it is repeated), for a total of 59. For all the PSI data points, we used the correlation coefficient between RNA-seq and RT-PCR, so as to monitor the trend in PSI changes across conditions rather than precise PSI values which are biased by amplification cycles (Supplementary Figure S7A–C). We considered a total of 47 (80%) as ‘validated’ as their correlation coefficient exceeds the cutoff of 0.4, while 33 (56%) are highly reliable with a coefficient >0.8 (Supplementary Figure S7D). A good example of validated DASE is INF2 cassette exon which, in both RNA-seq and RT-PCR, switches from predominant inclusion in untreated or VD3-treated THP-1 cells to strong skipping in both PMA treatments and in LPS/IFNγ or IL4/13 polarization. The 80% validation rate shows an overall high concordance between RNA-seq and RT-PCR.
Figure 5.
Regulation of MBNL1 and RBFOX2 expression upon primary monocyte-to-macrophage differentiation and their potential AS targets. ( A) Quantitative real-time RT-PCR confirms the changes in mRNA levels of MBNL1 (top) and RBFOX2 (bottom) during THP-1 and primary cell differentiation. Y axis indicates CT values of target relative to β-Actin control. (B) Western blotting shows that MBNL1 and RBFOX2 proteins are respectively down- and upregulated upon differentiation of primary monocytes and THP-1 cells, with tubulin as loading control. (C) Venn diagram with the numbers of DASEs for primary monocyte and THP-1 differentiation versus previously identified DASEs affected by siMBNL1+2 in HeLa and HEK293T (52). The overlap is significantly enriched (P < 10−25, one-tailed Fisher's Exact test). (D) Radioactive RT-PCR validation of potential MBNL1 and/or RBFOX2 DASEs conserved in THP-1 and primary cell differentiation. AS type in parenthesis, including cassette exons (CA) and alternative 5′ splice sites (AD). Green genes indicate DASEs affected by siMBNL1+2, blue genes have one or more RBFOX2 binding sites within the target exon and/or 300nt in flanking introns, and red genes have both. Top bar chart shows the RNA-seq-derived PSI of each DASE, while PAGE shows the RT-PCR results with PSI quantification of three replicates at the bottom of each lane. All these DASEs are validated (Supplementary Figure S7B). (E) RNA-seq screenshot profiles for two DASEs, with the read density for each sequence in gray aligned to the exon(box)-intron(line) structure below.
Overall, we identified a large number of DASEs during monocyte-to-macrophage differentiation and activation, which are regulated by one or likely more SFs. The best candidate AS regulators are the SFs with strongest expression changes during each process. Below we characterize two of these SFs.
Changes in RBFOX2 and MBNL1 expression are associated with differential AS between monocytes and macrophages
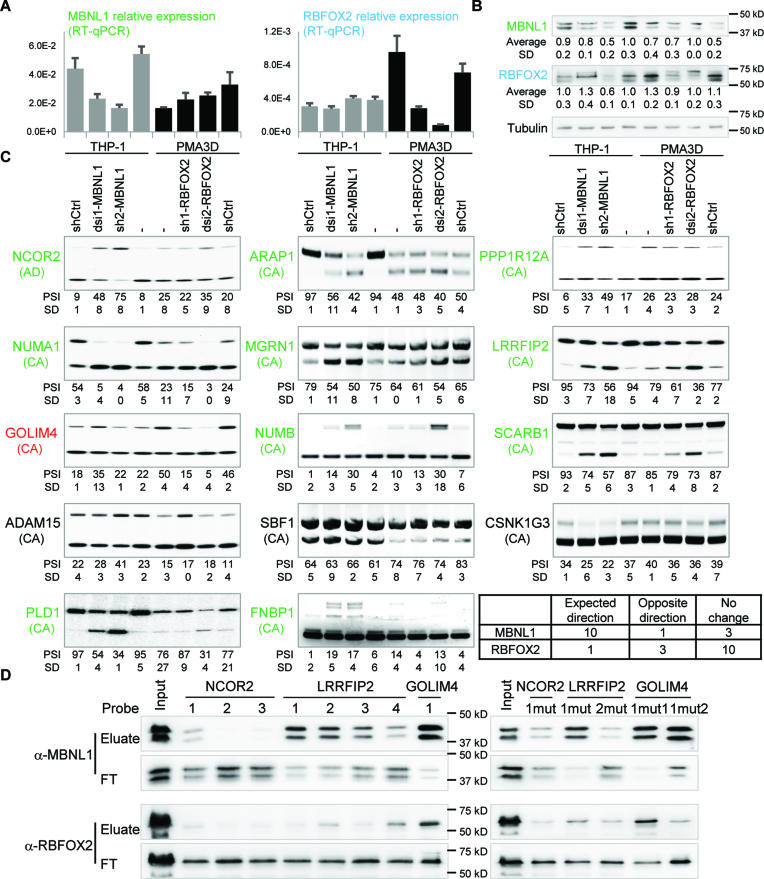
Next we sought out to establish the role of RBFOX2 and MBNL1 in regulating AS during monocyte-to-macrophage differentiation. Our RNA-seq data revealed an increase in RBFOX2 expression upon GM-CSF or PMA treatment of primary or THP-1 cells, respectively, and a decrease in MBNL1 in M/GM-CSF and PMA treated cells (Figure 2C). While MBNL2 did not change expression during monocyte-to-macrophage differentiation, MBNL3 was also downregulated but did not pass the statistical cutoff, so we focused on MBNL1 only. We confirmed the MBNL1 and RBFOX2 findings by real-time RT-PCR (Figure 5A). Furthermore, these mRNA changes led to the corresponding changes of RBFOX2 and MBNL1 protein levels upon PMA treatment of THP-1 cells as seen by western blotting (Figure 5B), suggesting that these SFs might regulate a fraction of the DASEs during monocyte differentiation.
Consistent with a role of MBNL1 in monocyte AS, we found a large overlap of our DASEs during monocyte differentiation and the DASEs for a previous study of MBNL1/2 knockdown in HEK293 and HeLa cell lines (52) (Figure 5C). Specifically, among the 134 DASEs upon shMBNL1/2 in this previous study, we identified 60 in our primary monocyte differentiation under GM/M-CSF treatment, with 12 also detected in THP-1 differentiation. In addition, we found hundreds of DASEs upon primary monocyte differentiation with RBFOX2 binding sites (UGCAUG) in the regulated exons or 300 nt of the flanking introns, with more than 80 DASEs in common with THP-1 differentiation. We validated 15 DASEs by radioactive RT-PCR (Figure 5D). Among these, ten were affected by the above-mentioned knockdown of MBNL1/2 in the previous study (green gene names), three harbour one or more RBFOX2 binding sites (red gene names), and five additional ones were not previously documented as MBNL1 targets but had RBFOX2 binding sites (blue gene names). Locations of RBFOX2 binding sites are indicated in Supplementary Figure S8. All these DASEs showed a consistent change in AS in both primary monocytes treated with M- and/or GM-CSF, and in THP-1 cells incubated with PMA. From these, inclusion of the cassette exon increased upon differentiation in seven DASEs (NUMB, SBF1, ADAM15, GOLIM4, GSNK1G3, PPP1R12A and FNBP1), decreased in seven DASEs (MGRN1, VPS29, MBNL1, SCARB1, LRRFIP2, NUMA1 and PLD1), and switched 5′ss usage in NCOR2. This AS profiles suggest that the two SFs might regulate these DASEs in monocyte-to-macrophage differentiation.
These DASEs might be biologically relevant in the context of monocyte-to-macrophage differentiation, such as in GOLIM4 (golgi integral membrane protein 4) involved in trafficking between the cis Golgi and endosomes (53), or NCOR2 (Nuclear Receptor Corepressor 2) as transcriptional repressor via chromatin remodelling (Figure 5e) (54).
MBNL1 regulates AS during monocyte-to-macrophage differentiation
To test the role of MBNL1 and RBFOX2 in AS during monocyte differentiation, we managed to knock these two SFs down in THP-1 cells by two different methods: by stably expressing a short-hairpin RNA (shRNA) using lentiviral transduction, and by transient nucleofection of dicer-substrate RNA (dsiRNA), targeting two different exons (Supplementary Table S1). As MBNL1 and RBFOX2 are respectively down- and upregulated upon monocyte-to-macrophage differentiation, we knocked these SFs down when they are highly expressed, i.e. in untreated THP-1 for MBNL1 and in PMA3D cells for RBFOX2. By real-time RT-PCR and Western Blotting, we observed that these knockdowns reduced the target RNAs and proteins to different extents (Figure 6A and B). In the western blot, each band in the MBNL1 doublet probably contains more than one MBNL1 isoform, but based on the AS of exon 7 and 8 in our RNA-Seq/RT-PCR, the lower band most likely contains isoform(s) with exon 7, while the upper band might contain isoforms with exon 6 spliced directly to 8 (55).
Figure 6.
MBNL1 protein regulates 11 out of 14 DASEs during monocyte-to-macrophage differentiation. (A) Quantitative real-time RT-PCR shows effective knockdown of MBNL1 mRNA by dsi1-MBNL1 nucleofection and sh2-MBNL1 transduction (left), and knockdown of RBFOX2 mRNA by dsi2-RBFOX2 nucleofection and sh1-RBFOx2 transduction (right), all in THP-1. Knockdown of MBNL1 or RBFOX2 has no major effect on one another's mRNA. (B) Western blotting shows MBNL1 protein downregulation by sh2-MBNL1 and slightly by dsi1-MBNL1, as well as RBFOX2 downregulation by both sh1-RBFOX2 and dsi2-RBFOX2. Each knockdown did not affect the other protein with the exception of sh2-MBNL1 cells showing some RBFOX2 down-regulation, which was not seen in dsi1-MBNL1 and shRNA control. Lane labels at the bottom. (C) Radioactive RT-PCR showing that MBNL1 knockdown in THP-1 cells changed 10 DASEs (green labels) in the same direction as PMA3D treatment, while both MBNL1 and RBFOX2 knockdowns in PMA3D cells changed the DASE of GOLIM4 (red) in the expected direction. AS type indicated in parenthesis, including cassette exons (CA) and alternative 5′ splice sites (AD). Bottom right corner, mini-table summary of the 14 DASE changes upon SF knockdown. (D) Western blotting analysis of RNA pulldowns with 139-nt probes for NCOR2 (probes 1–3), LIRRFIP2 (probes 1–4), and GOLIM4 (probe 1), numbered from 5′ to 3′ (Supplementary Figure S8). Wild-type probes on the left and mutants on the right. FT, flow-through.
We expected that the MBNL1 targets change AS upon knockdown in THP-1 in the same direction than upon PMA differentiation. Among the eight predicted MBNL1 targets we tested, six showed the same change in AS upon MBNL1 knockdown (especially in sh2-MBNL1) and PMA, which included NCOR2, MGRN1, NUMB, SCARB1, LRRFIP2, NUMA1, but not SBF1 and ADAM15 (Figure 6C). Interestingly, five out of the other six DASEs also clearly responded to MBNL1 knockdown in the expected direction, including GOLIM4, CSNK1G3, PPP1R12A, FNBP1, PLD1, ARAP1 but not CSNK1G3. Even though sh2-MBNL1 slightly reduces RBFOX2 protein, all confirmed DASEs show consistent effects with dsi1-MBNL1, which does not have such cross-effect. Our finding that eleven of the fourteen tested DASEs are regulated by MBNL1 in monocytes suggest that this SF is an important AS regulator in these cells.
In turn, we expected that the RBFOX2 targets change AS upon knockdown in the reverse direction as in PMA treatment of THP-1 cells, but only GOLIM4 among the nine predicted targets showed this pattern (Figure 6C). Knockdown of RBFOX2 had no consistent effect on the AS of ADAM15, CSNK1G3, FNBP1, and PPP1R12A. In addition, dsi2-RBFOX2 changed AS of LRRFIP2, NUMA1, PLD1 and ARAP1 in the opposite direction as the expected one, but these effects are less reliable because they are not shared with sh1-RBFOX2, and because this knockdown also slightly upregulates MBNL1 protein. Thus, most of the RBFOX2 AS targets in macrophages remain to be identified.
Next, we performed RNA pulldowns to investigate whether MBNL1 and RBFOX2 regulate DASEs by direct binding to the target RNAs. Based on predictions of MBNL1 and RBFOX2 motifs by SpliceAid2 (56), we designed probes for exon 47 of NCOR2 and exon 19 of LRRFIP2 because both are clearly regulated by MBNL1 (Figure 6C), as well as exon 7 of GOLIM4, regulated by both SFs (Supplementary Figure S8). RNA pulldowns show that RBFOX2 binds to probes with its cognate motif (Figure 6D, left), with a stronger signal for GOLIM4 probe 1 than for LRRFIP2 probe 4. This binding is specific because it is dramatically reduced by mutating the two motifs in GOLIM4 (Figure 6D, GOLIM4-1mut2). Consistent with the presence of MBNL1 binding sites in all probes, MBNL1 is pulled down by all except NCOR2 probes 2 and 3 (Figure 6D, left), at different efficiencies. MBNL1 binding was abrogated in the mutant LRRFIP2 probe 2 (Figure 6D, right) but not in other mutant probes, suggesting that MBNL1 might bind these RNAs at multiple sites. These RNA pulldowns are consistent with MBNL1-mediated regulation of DASEs during monocyte-to-macrophage differentiation via direct binding to these targets.
MBNL1 but not RBFOX2 is essential for monocyte-to-macrophage differentiation
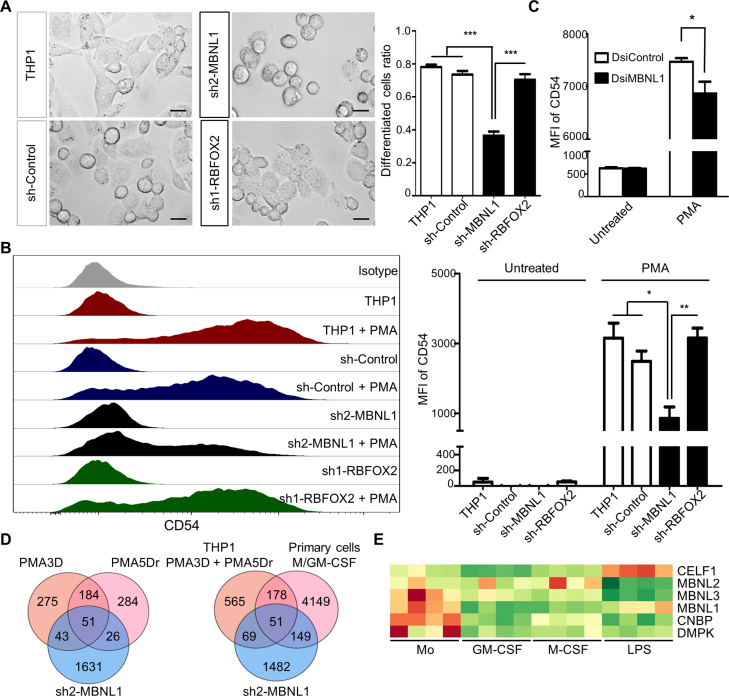
Finally, we addressed the functional significance of these two SFs in THP-1 differentiation by PMA. Our abovementioned results suggest that MBNL1 knockdown in THP-1 cells might recapitulate some of the phenotypic changes induced by PMA. To test this hypothesis, we induced PMA differentiation in the stable MBNL1 and RBFOX2 knockdown cells, in comparison to differentiation in the original and control shRNA THP-1 cells. Surprisingly, we found a very strong impairment of PMA differentiation upon MBNL1 knockdown, as most PMA-treated THP-1 cells remained round-shaped (Figure 7A) like monocytes, and the macrophage-specific cell adhesion marker CD54 was far less induced in these cells compared to controls (Figure 7B). Consistently, MBNL1 knockdown with dsi1 showed a smaller yet significant reduction of PMA-triggered CD54 compared to control (Figure 7C). The fact that dsi1 and sh2 target different MBNL1 sequences strongly argue for the specificity of these results, and the degree of knockdown corresponds to the magnitude of the phenotypic change. In turn, we found no visible phenotype upon RBFOX2 downregulation (Figure 7A and B). We also performed RNA-seq on the THP-1 cells with MBNL1 knockdown, and found an enriched overlap with the DASEs induced by PMA, and in either with PMA and/or M/GM-CSF macrophages, which should account for the impaired differentiation phenotype (Figure 7D; Supplementary files AS_shMBNL1_gene_list and DEG_gene_list). In summary, these results establish the key role of MBNL1 in PMA differentiation of THP-1 cells, which suggests that this factor is also crucial for primary monocyte differentiation.
Figure 7.
Knockdown of MBNL1 causes a strong deficiency in PMA differentiation of THP-1 cells. (A) Left, phase-contrast microscope images showing THP-1 differentiation upon PMA3D treatment, with control and shRNA-expressing cells. Right, quantification of PMA-differentiated cells. We counted the extended irregular shaped cells as differentiated, while small round cells as un-differentiated. Differentiated cells ratio = number of differentiated cells divided by total. Bar chart derived from three biological replicates with ∼100 counted cells per replicate. (B) Left, FACS profiles of CD54 expression in control, MBNL1 or RBFOX2 shRNA knockdown in untreated THP-1 and PMA3D. Right, median fluoresce intensity (MFI) of three biological replicates. (C) MFI of THP-1 and PMA3D cells treated with indicated dsiRNAs. (D) Venn diagrams of RNA-seq-detected DASEs (only with rMATS) upon MBNL1 knockdown of THP-1 cells in common with those in PMA differentiation (left) and in M/GM-CSF differentiation of primary cells (right). The overlaps between MBNL1 DASEs and those for PMA conditions or both PMA and M/GM-CSF are significantly enriched (both P < 10−30, one-tailed Fisher's Exact test). (E) Heatmap of RNA levels of MBNL1/3, CELF1, CNBP and DMPK in primary monocyte-to-macrophage differentiation and LPS/IFNγ activation, whose expression patterns suggest a possible monocyte dysfunction in myotonic dystrophies.
DISCUSSION
We unveiled thousands of DASEs during human monocyte-to-macrophage differentiation and activation in culture primary cells and THP-1 cell line. Importantly, from the many RBPs and SFs with strong expression changes, MBNL1 appears to be a major regulator of AS in monocytes and is essential for PMA differentiation of THP-1 cells. This study should be a major stepping stone for the detailed characterization of AS regulation in these important myeloid cells and their connection to disease and therapy.
AS is a major regulatory process in human monocytes and macrophages
We found that AS plays a prominent role in both M- and GM-CSF differentiation of primary monocytes, with many DASEs in common and many that are specific to these stimuli. The differential pathways that are regulated by DASEs upon these treatments strongly suggest that AS contributes to the distinct properties of these cells. As GM- and M-CSF macrophages have mostly pro- and anti-inflammatory properties and are associated with different disease states, their differential regulation is crucial to understand and manipulate these cells (3). For instance, repressing tumor associated macrophages with a wound healing phenotype (like M-CSF macrophages) might have therapeutic benefit (3,15). QKI was shown to regulate AS in M-CSF macrophages (37), consistent with our RNAseq data showing a ∼30% increase in its RNA levels specific to these cells. We only detected a reduction in expression of SF3A1-3 and SRSF3 in macrophages, despite the reported role of the SF3A subcomplex in AS during TLR4 stimulation of mouse macrophages (36), and the role of SRSF3 in IL1β production (42). Our study adds MBNL1 as an important AS regulator in monocytes.
We also revealed hundreds of DASEs in polarization of GM-CSF macrophages by LPS/IFNγ, but very few in the IL4/IL1β/IL10 polarizations of M-CSF macrophages, with almost none in common. This finding might simply reflect that AS does not play a prominent role in anti-inflammatory responses of macrophages, as opposed to differentiation and LPS/IFNγ activation. Alternatively, even though we used well-established protocols, the lack of AS changes in anti-inflammatory polarizations could be due to the different doses (100 ng/ml LPS versus 10 ng/ml IL4/IL1β/IL10), to treatment times (8 h in all), or to M-CSF macrophages being already fully activated. In THP-1, a lower PMA dose of 10 ng/mL results in stronger IL4/IL13 activation (57), suggesting that an adjustment of cytokine treatments might reveal more DASEs. Finally, we confirmed the upregulation of CELF1 by LPS/IFNγ (38) (Figure 7E).
Known and new RBPs as regulators of monocytes and macrophages
Our study revealed a large number of RBPs that strongly change expression (≥2-fold up or down) in the different primary monocytes and macrophages. RBPs that also change in analogous THP-1 conditions may contribute to the common function of these cells. Encouragingly, we found many RBPs previously implicated in monocyte/macrophage functions. For differentiation, the previously known DEGs include (i) CPEB1 depletion in macrophages, which enhances IL6 production upon LPS treatment, by regulating translation of other mRNAs (58); (ii) macrophage-enhanced downregulation of RNASE2 and RNASE3, which encode mostly eosinophil-derived secretory ribonucleases that fight infection (59); (iii) ZFP36 downregulation, which in turn mediate the AU-Rich Element-dependent degradation of TNF-α mRNA in liver macrophages, among other targets (60); ZFP36 and ZFP36L2 are also respectively up and downregulated by LPS treatment of mouse macrophages (61). The remaining conserved RBPs are uncharacterized in monocytes/macrophages, and might also regulate their functions.
For LPS/IFNγ activation, the RBPs as conserved DEGs in primary and THP-1 cells that have been previously characterized include: (i) the p150 ADAR isoform upregulated by the interferon-inducible promoter (62); (ii) ZC3HAV1 upregulated in macrophages along other antiviral genes (63); (iii) increased DCP1A expression which regulates IL6 secretion in THP-1 (64); (iv) PNPT1 upregulation which induces pro-inflammatory cytokines IL6 and IL8 (65); (v) MSI2 downregulation, which controls progression of Chronic Myeloid Leukemia via NUMB (66). The biological significance of the LPS/IFNγ-induced expression changes of the other RBPs remains to be elucidated.
Several transcription factors that control monocytes and/or macrophages are acquiring diagnostic, prognostic or therapeutic value, like Nuclear Factor kappa B (NF-κB) associated with cancer inflammation and tumor progression (67), or Peroxisome Proliferator-activated Receptor gamma (PPARγ) and Kruppel-like factor 4 (KLF4) associated with protection from obesity-induced insulin resistance (68,69). Furthermore, microRNAs play a role in endotoxin tolerance (70). Certain RBPs/SFs, other than QKI in atherogenesis (37), should be soon linked to monocyte/macrophage disease.
Role of RBFOX2 and MBNL1 during monocyte-to-macrophage differentiation
Several findings initially supported a role of RBFOX2 and MBNL1 as AS regulators in human monocyte differentiation, including their respective up- and downregulation during differentiation of both primary monocytes and THP-1 cells, and the overlap between our differentiation DASEs and known MBNL1 targets in other cells (52). However, our analysis of the role of these SFs upon knockdown in THP-1 only clarified the picture for MBNL1. For RBFOX2, only one DASE (GOLIM4) changed AS in the expected direction upon knockdown, indicating that the RBFOX2 targets in macrophages remain to be identified. Nevertheless, RBFOX2 knockdown in PMA-treated THP-1 cells had no visible phenotype, as these cells remained differentiated and attached, strongly suggesting that this factor is dispensable for the establishment or maintenance of the PMA differentiated state. It could also be that the slight knockdown is not sufficient or that other paralogs compensate for the RBFOX2 reduction. Thus, further studies are required to establish the role of RBFOX2 in macrophages.
Remarkably, many DASEs changed AS upon MBNL1 knockdown in the same direction as during monocyte differentiation (Figures 6C and 7C), arguing that the overall MBNL1 reduction, rather than the changes in the relative expression of MBNL1 splice isoforms, largely caused the differential AS patterns between monocytes and macrophages. Our dataset includes known and new MBNL1 targets, and for the two DASEs we checked by RNA pulldowns the regulation may be via direct binding (Figure 6D), to be confirmed by other approaches to monitor binding within cells such as Cross-Linking and ImmunoPrecipitation (CLIP). Most importantly, MBNL1 knockdown in THP-1 very strongly impaired PMA differentiation, indicating that the MBNL1-mediated regulation of AS or other processes such as mRNA localization (71) is essential for the differentiation of this cell line and probably of primary monocytes.
MBNL1 in human monocyte/macrophage physiology and disease
MBNL1 increases expression and regulates three developmental transitions, which are embryonic stem cell differentiation (52), the transition between fetal and adult tissues, majorly including muscle, heart and brain (72–75), and terminal erythroid differentiation (76). In contrast, our human primary monocyte-to-macrophage differentiation model shows >2-fold downregulation of total MBNL1 RNA and at least one protein isoform (Figure 6A and B). Albeit monocytes and macrophages are both mature cells, monocytes proliferate much more than macrophages, hence in these myeloid cells low MBNL1 is associated with lower proliferation, as opposed to stem cells. Furthermore, the loss of PMA-induced THP-1 differentiation upon MBNL1 knockdown could be caused by a putative MBNL1 function in ‘priming’ these monocytes for the differentiation process. An alternative explanation based on MBNL1’s role in developmental transitions is that its knockdown might turn THP-1 cells into a more de-differentiated state which renders these cells unresponsive to PMA. Consistent with this notion, the DASEs of PMA-treated THP-1 cells and upon MBNL1 knockdown do not show a perfect overlap, highlighting the role of other SFs in PMA differentiation, and that MBNL1 knockdown might have turned these cells into a new state, perhaps reminiscent of precursors in the myeloid lineage. Future experiments should extend this finding to primary cells and fully characterize the identity of these MBNL1 knockdown cells. Furthermore, such follow-up should address whether MBNL1 overexpression in macrophages disrupts their identity and/or function.
The important AS regulation by MBNL1 in human monocytes and its role during differentiation revealed a putative unexpected connection to Myotonic Dystrophies DM1 and DM2. DM1/2 are caused by repeat expansions in RNAs which sequester MBNL1, thus resulting in a loss of function of this SF and turning the AS in adult tissues to that of undifferentiated cells, mainly resulting in muscular dystrophy, myotonia as well as cardiac and neurological problems (73,77). The causative genes with the expansions, DMPK for the severe DM1 and CNBP (ZNF9) for the milder DM2, need to be expressed in the cells together with MBNL1 for the loss of function to occur. Indeed, both DMPK and CNBP are highly expressed in human monocytes compared to macrophages (Figure 7E), suggesting that human monocytes in DM patients suffer from a loss of function of MBNL1, and that monocyte-derived macrophages would be defective in these patients. Furthermore, CELF1, whose gain of function upon phosphorylation-induced stabilization contributes to DM1 (78), is strongly induced by LPS (38), suggesting that activated inflammatory macrophages might also exhibit phenotypic defects in DM1 patients. DM patients show abnormally high blood insulin together with its low binding to INSR (insulin receptor) in monocytes (79,80). Even though MBNL1 supports strong binding to insulin by inducing inclusion of INSR exon 11 (81), this cassette exon showed very low inclusion level in all our samples from primary and THP-1 cells, as seen before (82), suggesting that the high insulin levels in DM patients are caused by cells other than monocytes. DM monocytes also exhibit abnormal expression of IgG-Fc receptors resulting in hypercatabolism of IgG (83), which could be caused by aberrant AS (or other processes) due to MBNL1 loss of function in monocytes and perhaps other leukocytes. Indeed, we did detect a few DASEs in FCGR1B/2A/2B/3A genes in primary monocytes and macrophages to be characterized in future studies. Beyond these disruptions, other monocyte functions and probably their ability to differentiate to macrophages could be compromised in DM patients leading to immunosuppression, underlying the characteristic muscle, cardiac and neurological disorders. This immunosuppression should be mild if any, otherwise it would have been clinically described, or perhaps the monocytic defects are compensated by the two paralogs MBNL2/3 (84) or by other factors. Indeed, a recent study showed differential alterations of AS patterns upon different degrees of MBNL1 knockdown (85), which highlights the high sensitivity of deregulated AS events to free MBNL1 levels which could also be tissue specific. Further studies should clarify the functional properties of monocytes and macrophages in DM patients.
In brief, here we report an extensive and detailed characterization of AS in human monocytic differentiation and activation, and established an important yet intriguing role of MBNL1 in AS of monocytes.
DATA AVAILABILITY
We deposited the raw fastq files in the Sequence Read Archives (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SRP139891 of Bioproject PRJNA449980.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Roca lab for useful advice. We are grateful to Daniela Moses for RNA library preparation and for running the Illumina Hi-Seq machine. We also thank Christiane Ruedl and Sin Tiong Ong for advice.
Author contributions: H.L. performed most of the experiments and in silico analyses, with critical contributions from P.A.L. F.Z. was instrumental in setting the bioinformatics pipeline with supervision from J.Z. M.S.M.W. performed many of the A.S. validations, and S.X. did some of the last experiments with MBNL1. X.R. conceived the study and wrote the manuscript with help from coauthors, mainly from H.L.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Academic Research Fund Tier 2 [MOE2013-T2-1-101 ARC 45/13, MOE2016-T2-2-104(S)], both from Singapore's Ministry of Education (https://www.olga.moe.gov.sg/T2/default.aspx). The funders played no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. Funding for open access charge: Academic Research Fund Tier 2 [MOE2016-T2-2-104(S)].
Conflict of interest statement. None declared.
REFERENCES
- 1. Auffray C., Sieweke M.H., Geissmann F.. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009; 27:669–692. [DOI] [PubMed] [Google Scholar]
- 2. Ginhoux F., Jung S.. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014; 14:392–404. [DOI] [PubMed] [Google Scholar]
- 3. Biswas S.K., Chittezhath M., Shalova I.N., Lim J.Y.. Macrophage polarization and plasticity in health and disease. Immunol. Res. 2012; 53:11–24. [DOI] [PubMed] [Google Scholar]
- 4. Kohro T., Tanaka T., Murakami T., Wada Y., Aburatani H., Hamakubo T., Kodama T.. A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage. J. Atheroscler. Thromb. 2004; 11:88–97. [DOI] [PubMed] [Google Scholar]
- 5. Qin Z. The use of THP-1 cells as a model for mimicking the function and regulation of monocytes and macrophages in the vasculature. Atherosclerosis. 2012; 221:2–11. [DOI] [PubMed] [Google Scholar]
- 6. Lavin Y., Mortha A., Rahman A., Merad M.. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015; 15:731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Varol C., Mildner A., Jung S.. Macrophages: development and tissue specialization. Annu. Rev. Immunol. 2015; 33:643–675. [DOI] [PubMed] [Google Scholar]
- 8. Ginhoux F., Schultze J.L., Murray P.J., Ochando J., Biswas S.K.. New insights into the multidimensional concept of macrophage ontogeny, activation and function. Nat. Immunol. 2016; 17:34–40. [DOI] [PubMed] [Google Scholar]
- 9. Alasoo K., Martinez F.O., Hale C., Gordon S., Powrie F., Dougan G., Mukhopadhyay S., Gaffney D.J.. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci. Rep. 2015; 5:12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sharif O., Bolshakov V.N., Raines S., Newham P., Perkins N.D.. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC Immunol. 2007; 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gordon S., Martinez F.O.. Alternative activation of macrophages: mechanism and functions. Immunity. 2010; 32:593–604. [DOI] [PubMed] [Google Scholar]
- 12. Mantovani A., Sozzani S., Locati M., Allavena P., Sica A.. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002; 23:549–555. [DOI] [PubMed] [Google Scholar]
- 13. Lumeng C.N., Bodzin J.L., Saltiel A.R.. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007; 117:175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Moore K.J., Tabas I.. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011; 145:341–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qian B.Z., Pollard J.W.. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010; 141:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Inoue D., Bradley R.K., Abdel-Wahab O.. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev. 2016; 30:989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M. et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011; 478:64–69. [DOI] [PubMed] [Google Scholar]
- 18. Stumpo D.J., Lai W.S., Blackshear P.J.. Inflammation: cytokines and RNA-based regulation. Wiley Interdiscip. Rev. RNA. 2010; 1:60–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martinez N.M., Pan Q., Cole B.S., Yarosh C.A., Babcock G.A., Heyd F., Zhu W., Ajith S., Blencowe B.J., Lynch K.W.. Alternative splicing networks regulated by signaling in human T cells. RNA. 2012; 18:1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O’Connell R.M., Rao D.S., Baltimore D.. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012; 30:295–312. [DOI] [PubMed] [Google Scholar]
- 21. Lee Y., Rio D.C.. Mechanisms and regulation of alternative pre-mRNA splicing. Annu. Rev. Biochem. 2015; 84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Busch A., Hertel K.J.. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip. Rev. RNA. 2012; 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Warf M.B., Berglund J.A.. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010; 35:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Naftelberg S., Schor I.E., Ast G., Kornblihtt A.R.. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem. 2015; 84:165–198. [DOI] [PubMed] [Google Scholar]
- 26. Licatalosi D.D., Darnell R.B.. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010; 11:75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R. et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012; 338:1587–1593. [DOI] [PubMed] [Google Scholar]
- 28. Merkin J., Russell C., Chen P., Burge C.B.. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012; 338:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez N.M., Lynch K.W.. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol. Rev. 2013; 253:216–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen L., Kostadima M., Martens J.H., Canu G., Garcia S.P., Turro E., Downes K., Macaulay I.C., Bielczyk-Maczynska E., Coe S. et al. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014; 345:1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edwards C.R., Ritchie W., Wong J.J., Schmitz U., Middleton R., An X., Mohandas N., Rasko J.E., Blobel G.A.. A dynamic intron retention program in the mammalian megakaryocyte and erythrocyte lineages. Blood. 2016; 127:e24–e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pimentel H., Parra M., Gee S.L., Mohandas N., Pachter L., Conboy J.G.. A dynamic intron retention program enriched in RNA processing genes regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2016; 44:838–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wong J.J., Ritchie W., Ebner O.A., Selbach M., Wong J.W., Huang Y., Gao D., Pinello N., Gonzalez M., Baidya K. et al. Orchestrated intron retention regulates normal granulocyte differentiation. Cell. 2013; 154:583–595. [DOI] [PubMed] [Google Scholar]
- 34. Bhatt D.M., Pandya-Jones A., Tong A.J., Barozzi I., Lissner M.M., Natoli G., Black D.L., Smale S.T.. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012; 150:279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pandya-Jones A., Bhatt D.M., Lin C.H., Tong A.J., Smale S.T., Black D.L.. Splicing kinetics and transcript release from the chromatin compartment limit the rate of Lipid A-induced gene expression. RNA. 2013; 19:811–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O’Connor B.P., Danhorn T., De Arras L., Flatley B.R., Marcus R.A., Farias-Hesson E., Leach S.M., Alper S.. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015; 11:e1004932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Bruin R.G., Shiue L., Prins J., de Boer H.C., Singh A., Fagg W.S., van Gils J.M., Duijs J.M., Katzman S., Kraaijeveld A.O. et al. Quaking promotes monocyte differentiation into pro-atherogenic macrophages by controlling pre-mRNA splicing and gene expression. Nat. Commun. 2016; 7:10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin J., Hu Y., Nunez S., Foulkes A.S., Cieply B., Xue C., Gerelus M., Li W., Zhang H., Rader D.J. et al. Transcriptome-Wide analysis reveals modulation of human macrophage inflammatory phenotype through alternative splicing. Arterioscler. Thromb. Vasc. Biol. 2016; 36:1434–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grisar J., Munk M., Steiner C.W., Amoyo-Minar L., Tohidast-Akrad M., Zenz P., Steiner G., Smolen J.S.. Expression patterns of CD44 and CD44 splice variants in patients with rheumatoid arthritis. Clin. Exp. Rheumatol. 2012; 30:64–72. [PubMed] [Google Scholar]
- 40. Srinivasan R., Bogdanov V.Y.. Splice variants of Tissue Factor and integrin-mediated signaling. Thromb. Res. 2012; 129(Suppl. 2):S34–S37. [DOI] [PubMed] [Google Scholar]
- 41. Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J.. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 2003; 197:263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moura-Alves P., Neves-Costa A., Raquel H., Pacheco T.R., D’Almeida B., Rodrigues R., Cadima-Couto I., Chora A., Oliveira M., Gama-Carvalho M. et al. An shRNA-based screen of splicing regulators identifies SFRS3 as a negative regulator of IL-1beta secretion. PLoS One. 2011; 6:e19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daigneault M., Preston J.A., Marriott H.M., Whyte M.K., Dockrell D.H.. The identification of markers of macrophage differentiation in PMA-stimulated THP-1 cells and monocyte-derived macrophages. PLoS One. 2010; 5:e8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maess M.B., Wittig B., Lorkowski S.. Highly efficient transfection of human THP-1 macrophages by nucleofection. J. Vis. Exp. 2014; e51960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L.. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L.. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012; 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Katz Y., Wang E.T., Airoldi E.M., Burge C.B.. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat. Methods. 2010; 7:1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu J., Akerman M., Sun S., McCombie W.R., Krainer A.R., Zhang M.Q.. SpliceTrap: a method to quantify alternative splicing under single cellular conditions. Bioinformatics. 2011; 27:3010–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Neme A., Nurminen V., Seuter S., Carlberg C.. The vitamin D-dependent transcriptome of human monocytes. J. Steroid Biochem. Mol. Biol. 2016; 164:180–187. [DOI] [PubMed] [Google Scholar]
- 52. Han H., Irimia M., Ross P.J., Sung H.K., Alipanahi B., David L., Golipour A., Gabut M., Michael I.P., Nachman E.N. et al. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013; 498:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Linstedt A.D., Mehta A., Suhan J., Reggio H., Hauri H.P.. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell. 1997; 8:1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen J.D., Evans R.M.. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995; 377:454–457. [DOI] [PubMed] [Google Scholar]
- 55. Tran H., Gourrier N., Lemercier-Neuillet C., Dhaenens C.M., Vautrin A., Fernandez-Gomez F.J., Arandel L., Carpentier C., Obriot H., Eddarkaoui S. et al. Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of Muscleblind-like-1 isoforms. J. Biol. Chem. 2011; 286:16435–16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Piva F., Giulietti M., Burini A.B., Principato G.. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum. Mutat. 2012; 33:81–85. [DOI] [PubMed] [Google Scholar]
- 57. Jin C., Wu L., Li J., Fang M., Cheng L., Wu N.. Multiple signaling pathways are involved in the interleukine-4 regulated expression of DC-SIGN in THP-1 cell line. J. Biomed. Biotechnol. 2012; 2012:357060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ivshina M., Alexandrov I.M., Vertii A., Doxsey S., Richter J.D.. CPEB regulation of TAK1 synthesis mediates cytokine production and the inflammatory immune response. Mol. Cell. Biol. 2015; 35:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Acharya K.R., Ackerman S.J.. Eosinophil granule proteins: form and function. J. Biol. Chem. 2014; 289:17406–17415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Carballo E., Lai W.S., Blackshear P.J.. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998; 281:1001–1005. [DOI] [PubMed] [Google Scholar]
- 61. Wang K.T., Wang H.H., Wu Y.Y., Su Y.L., Chiang P.Y., Lin N.Y., Wang S.C., Chang G.D., Chang C.J.. Functional regulation of Zfp36l1 and Zfp36l2 in response to lipopolysaccharide in mouse RAW264.7 macrophages. J. Inflamm. (Lond.). 2015; 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blango M.G., Bass B.L.. Identification of the long, edited dsRNAome of LPS-stimulated immune cells. Genome Res. 2016; 26:852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oshiumi H., Mifsud E.J., Daito T.. Links between recognition and degradation of cytoplasmic viral RNA in innate immune response. Rev. Med. Virol. 2016; 26:90–101. [DOI] [PubMed] [Google Scholar]
- 64. Seto E., Yoshida-Sugitani R., Kobayashi T., Toyama-Sorimachi N.. The assembly of EDC4 and Dcp1a into processing bodies is critical for the translational regulation of IL-6. PLoS One. 2015; 10:e0123223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sarkar D., Fisher P.B.. Molecular mechanisms of aging-associated inflammation. Cancer Lett. 2006; 236:13–23. [DOI] [PubMed] [Google Scholar]
- 66. Ito T., Kwon H.Y., Zimdahl B., Congdon K.L., Blum J., Lento W.E., Zhao C., Lagoo A., Gerrard G., Foroni L. et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010; 466:765–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Biswas S.K., Sica A., Lewis C.E.. Plasticity of macrophage function during tumor progression: regulation by distinct molecular mechanisms. J. Immunol. 2008; 180:2011–2017. [DOI] [PubMed] [Google Scholar]
- 68. Liao X., Sharma N., Kapadia F., Zhou G., Lu Y., Hong H., Paruchuri K., Mahabeleshwar G.H., Dalmas E., Venteclef N. et al. Kruppel-like factor 4 regulates macrophage polarization. J. Clin. Invest. 2011; 121:2736–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Odegaard J.I., Ricardo-Gonzalez R.R., Goforth M.H., Morel C.R., Subramanian V., Mukundan L., Red Eagle A., Vats D., Brombacher F., Ferrante A.W. et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature. 2007; 447:1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nahid M.A., Satoh M., Chan E.K.. MicroRNA in TLR signaling and endotoxin tolerance. Cell. Mol. Immunol. 2011; 8:388–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang E.T., Cody N.A., Jog S., Biancolella M., Wang T.T., Treacy D.J., Luo S., Schroth G.P., Housman D.E., Reddy S. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012; 150:710–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A.. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Konieczny P., Stepniak-Konieczna E., Sobczak K.. MBNL proteins and their target RNAs, interaction and splicing regulation. Nucleic Acids Res. 2014; 42:10873–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lin X., Miller J.W., Mankodi A., Kanadia R.N., Yuan Y., Moxley R.T., Swanson M.S., Thornton C.A.. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum. Mol. Genet. 2006; 15:2087–2097. [DOI] [PubMed] [Google Scholar]
- 75. Miller J.W., Urbinati C.R., Teng-Umnuay P., Stenberg M.G., Byrne B.J., Thornton C.A., Swanson M.S.. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000; 19:4439–4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cheng A.W., Shi J., Wong P., Luo K.L., Trepman P., Wang E.T., Choi H., Burge C.B., Lodish H.F.. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood. 2014; 124:598–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Thornton C.A., Wang E., Carrell E.M.. Myotonic dystrophy: approach to therapy. Curr. Opin. Genet. Dev. 2017; 44:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kuyumcu-Martinez N.M., Wang G.S., Cooper T.A.. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol. Cell. 2007; 28:68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mishra S.K., Chetty S., Kataoka M.. Abnormal insulin receptor binding in cultured monocytes in myotonic muscular dystrophy. Biochem. Med. Metab. Biol. 1992; 47:161–167. [DOI] [PubMed] [Google Scholar]
- 80. Moxley R.T. 3rd, Livingston J.N., Lockwood D.H., Griggs R.C., Hill R.L.. Abnormal regulation of monocyte insulin-binding affinity after glucose ingestion in patients with myotonic dystrophy. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:2567–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Echeverria G.V., Cooper T.A.. Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron. Nucleic Acids Res. 2014; 42:1893–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Benecke H., Flier J.S., Moller D.E.. Alternatively spliced variants of the insulin receptor protein. Expression in normal and diabetic human tissues. J. Clin. Invest. 1992; 89:2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Banerjee D., McClintock J., Silver M.M., Hudson A.J.. Monocyte IgG-Fc receptors in myotonic dystrophy. Clin. Exp. Immunol. 1982; 50:572–578. [PMC free article] [PubMed] [Google Scholar]
- 84. Fardaei M., Rogers M.T., Thorpe H.M., Larkin K., Hamshere M.G., Harper P.S., Brook J.D.. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002; 11:805–814. [DOI] [PubMed] [Google Scholar]
- 85. Wagner S.D., Struck A.J., Gupta R., Farnsworth D.R., Mahady A.E., Eichinger K., Thornton C.A., Wang E.T., Berglund J.A.. Dose-dependent regulation of alternative splicing by MBNL proteins reveals biomarkers for myotonic dystrophy. PLoS Genet. 2016; 12:e1006316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We deposited the raw fastq files in the Sequence Read Archives (SRA) of the National Center for Biotechnology Information (NCBI) under accession number SRP139891 of Bioproject PRJNA449980.