Abstract
Messenger RNA translation is a complex process that is still poorly understood in eukaryotic organelles like mitochondria. Growing evidence indicates though that mitochondrial translation differs from its bacterial counterpart in many key aspects. In this analysis, we have used ribosome profiling technology to generate a genome-wide snapshot view of mitochondrial translation in Arabidopsis. We show that, unlike in humans, most Arabidopsis mitochondrial ribosome footprints measure 27 and 28 bases. We also reveal that respiratory subunits encoding mRNAs show much higher ribosome association than other mitochondrial mRNAs, implying that they are translated at higher levels. Homogenous ribosome densities were generally detected within each respiratory complex except for complex V, where higher ribosome coverage corroborated with higher requirements for specific subunits. In complex I respiratory mutants, a reorganization of mitochondrial mRNAs ribosome association was detected involving increased ribosome densities for certain ribosomal protein encoding transcripts and a reduction in translation of a few complex V mRNAs. Taken together, our observations reveal that plant mitochondrial translation is a dynamic process and that translational control is important for gene expression in plant mitochondria. This study paves the way for future advances in the understanding translation in higher plant mitochondria.
INTRODUCTION
The functioning of eukaryotic cells relies on the integrated activity of several highly specialized subcellular organelles. Among them, mitochondria represent essential energy conversion powerhouses that produce most of the energy of cells through aerobic respiration. Mitochondria have an exogenous origin and the prevailing view is that they arose from the endosymbiotic acquisition of an ancient α-proteobacteria (1,2). Throughout evolution, the adoption of mitochondria has been associated with a substantial reduction in their genetic content. Modern mitochondrial genomes encode only a handful of mRNAs (33 in Arabidopsis thaliana) that are translated within the organelle. This requires the existence of a complete gene expression machinery whose most factors are encoded by nuclear genes. Gene expression in mitochondria therefore involves the cooperation between highly degenerated bacterial-like features inherited from the original endosymbiont and eukaryotic-derived nuclear-encoded trans-factors that appeared throughout evolution. This dual origin has resulted in complex mRNA expression processes involving a high number of post-transcriptional modifications (3,4). In recent years, significant progress has been made concerning certain aspects of gene expression in plant mitochondria (4). However, mitochondrial translation, the last level of mRNA expression that is also the least prone to easy molecular analyses, has remained largely unexplored in plants. Nevertheless, we know that the mitochondrial translation machinery differs profoundly from its bacterial counterpart in many essential aspects, and that the major translation-associated components are substantially different from those found in bacteria (3,5,6). In particular, the rRNA and protein composition of mitoribosomes differ extensively from those of Escherichia coli ribosomes (7, 8). Additionally, the constraints associated with the translation of mRNAs encoding mostly hydrophobic respiratory subunits have led to highly specialized translation mechanisms guided by membrane-associated mitochondrial ribosomes (mitoribosomes) to facilitate cotranslational insertion of mitochondria-encoded proteins into the inner mitochondrial membrane (9). The divergence with bacteria is also particularly obvious for the translation initiation step since plant mitochondrial mRNAs lack the typical ribosome anchoring motifs in their 5′ leaders that are used in prokaryotes to help position the start codon at the P-site of the ribosome (7). Thus, we currently do not know how mitochondrial ribosomes are recruited to 5′ untranslated regions (UTR), nor how the correct translation initiation codon is recognized by the small ribosomal subunit. The high degree of sequence divergence among 5′ leaders of plant mitochondrial mRNAs has suggested a ribosome recruitment mechanism involving gene-specific cis-sequences and trans-factors (10,11). Up until now only two proteins belonging to the large family of pentatricopeptide repeat (PPR) proteins have been shown to promote mitochondrial translation initiation in higher plants (12,13). The mechanisms by which these PPR proteins help to recruit the mitoribosome toward the AUG codon of their target mitochondrial mRNA have, however, not yet been determined. Moreover, the sequence of mitochondrial transcripts in plants is extensively modified through a process called RNA editing in which more than 400 precisely-selected cytidine residues are converted into uridines. In general, C-to-U RNA editing restores conserved codons of coding sequences and is thus essential for the proper function of mitochondrial proteins and subsequently the activity of the organelle. However, a large number of these sites are not edited to 100%, rendering plant mitochondrial mRNA sequences heterogeneous (14,15). How mitochondrial translation and RNA editing machineries functionally coordinate remains an open question. Contradictory results have been obtained with regard to a few mitochondrial transcripts (16–20), but a genome-wide view of the functional relationship between RNA editing and translation is currently lacking.
The recent development of ribosome profiling technology has greatly facilitated the detailed measurement of translation at the single-nucleotide level and on a genome-wide scale (21). Its general principal consists in the purification and subsequent sequencing of short mRNA segments covered by the translating ribosomes. These mRNA segments are called ribosome footprints and measure about 30-nt long in most species (22). The statistical analysis of their distribution reveals the identity and boundaries of translated RNA regions and provides measurements of their relative translational levels. This technology has recently revealed interesting functional aspects about ribosome behavior in plastid and human mitochondria, but had been never used to study plant mitochondrial translation (23–25). In order to gain a comprehensive view of the Arabidopsis mitochondrial translatome and the functioning of the plant mitochondrial machinery, we have prepared and sequenced total ribosome footprints from Arabidopsis inflorescences and mapped them to the Arabidopsis mitochondrial genome. Notably, this approach has allowed us to estimate and compare the translational activity among mitochondrial mRNAs, to see how mitochondrial translation is reorganized in respiratory mutants and whether edited mRNAs preferentially associate with mitochondrial polysomes.
MATERIALS AND METHODS
Plant material
Arabidopsis (A. thaliana) Col-0 plants were obtained from the Arabidopsis Stock Centre of Institut National de la Recherche Agronomique in Versailles (http:// dbsgap.versailles.inra.fr/portail/). Arabidopsis mtsf1 and mtl1 mutants were previously described (13,26). Plants were grown in a greenhouse in long day conditions for 10–12 weeks before use. Flowers were harvested simultaneously for all genotypes, snap-frozen in liquid nitrogen and stored at −80°C until use.
Ribosome footprints preparation
Total ribosome footprints were prepared from a mix of flower buds and open flowers following a procedure adapted from (23,27). About 2.5 g of flowers were ground in liquid nitrogen and resuspended in 15 ml of ribosome extraction buffer (0.2 M sucrose, 0.2 M KCl, 40 mM Tris-acetate, pH 8, 10 mM MgCl2, 10 mM β-mercaptoethanol, 2% polyoxyethylene 10 tridecyl ether, 1% Triton X-100, 100 μg/ml chloramphenicol and 100 μg/ml cycloheximide). The obtained solution was centrifuged twice for 10 min at 15 000 g at 4°C to remove the cell debris and the supernatant was transferred to a new tube. To estimate the RNA concentration contained in the supernatant, total RNA was extracted from a 0.5 ml aliquot using the TRI Reagent® (Sigma-Aldrich) following the manufacturer’s instructions. The supernatant was then digested with 10 U/OD260nm/ml of RNAse I (Ambion), and incubated 1 h at 25°C to degrade unprotected RNA. The solution was then layered on an equal volume of a sucrose cushion (1 M Sucrose, 0.1 M KCl, 40 mM Tris-acetate, pH 8, 15 mM MgCl2, 5 mM β-mercaptoethanol, 100 μg/ml chloramphenicol and 100 μg/ml cycloheximide) and centrifuged for 3 h at 55 000 g at 4°C in a TFT 65-13 rotor (Kontron). The obtained pellet was resuspended in 0.6 ml of RNA extraction buffer I (0.1 M EGTA pH 8, 0.1 M NaCl, 1% sodium dodecyl sulphate (SDS), 10 mM Tris–HCl pH 8 and 1 mM ethylenediaminetetraacetic acid (EDTA)), and RNAs were extracted with the TRI Reagent® (Sigma-Aldrich) following the manufacturer’s recommendations. Ribosomal footprints were further purified on 17% (v/v) acrylamide-bisacrylamide (19:1) gel made with 7 M urea and 1 × Tris-acetate-EDTA (TAE) buffer. For this, 600 μg of RNAs were electrophoresed in two separated 15 cm-wide wells. The gel was pre-run for 1 h at 50°C and then run for 15–16 h at 90 V at the same temperature. A mix comprising 100 ng of 27-mer and 33-mer RNA oligonucleotides was loaded on both gel extremities and used as size marker. After staining for 30 min in 1× SYBR Gold (Life Technologies) diluted in 1× TAE, RNA contained in the 27 and 33 nt regions were separately excised from the gel. The gels slices were stored at least 2 h at -20°C and then physically disrupted by centrifugation through a needle hole made in a 0.5 ml microcentrifuge tube nested in an outer 1.5 ml collection microtube. The tube assembly was centrifuged at maximum speed for 5 min. RNAs were eluted overnight from gel fragments by passive diffusion in 5 ml of RNA extraction buffer II (10 mM Tris–HCl pH 8, 1 mM EDTA, 0.1 M NaCl, and 0.2% SDS) at 4°C on a rotating wheel. The recovered RNAs were then isolated with the TRI Reagent® (Sigma-Aldrich) following the manufacturer's instructions, resuspended in 30 μl of RNAse-free water and stored at −80°C until use.
Ribosome footprints were depleted from ribosomal RNA with the Ribo-Zero Plant kit (Illumina) following manufacturer’s recommendations. Sequencing libraries were prepared with the TruSeq Small RNA library preparation kit (Illumina). Next generation sequencing was performed by Beckman (single end, 50 nt).
Total RNA quantification
The abundance of mitochondrial mRNAs contained in each genotype used was determined by quantitative RT-PCR as described in (13). Primers were designed to quantify the abundance of each exon of mitochondrial mRNAs of known function, except for the nad5 exon 3 that is too small to be quantified by quantitative RT-PCR (see Supplementary Table S1).
RNA-seq analysis
A total of 5 μg of RNA (derived from the initial step of ribosome footprint purifications) were depleted from ribosomal RNA with the Ribo-Zero Plant kit (Illumina) following manufacturer’s recommendations. Sequencing libraries and next generation sequencing was performed by Beckman (single end, 50 nt).
Bioinformatic analysis
From raw data to alignment files
Raw data were first trimmed to remove the 3′ adapter sequence with CutAdapt 1.9.1 (28) configured with −e 0.12, −m 24, −M 35 and -a options to select a read size in a range from 24 to 35 nt allowing 12% of error. The trimmed reads were then mapped against the rRNA sequences of A. thaliana (www.arabidopsis.org) with Bowtie 1.1.2 (29) set up with –all and –un options. The –un option was used to select all unmapped reads. These filtered reads were finally mapped against the complete Col-0 mitochondrial genome (JF729201.1) or only the coding sequences with Bowtie 1.1.2. The latter alignments were configured with two mismatches allowed (−n 2) and only uniquely mapped reads were selected (−m 1). The sam formatted files generated by the aligner were converted to sorted and indexed bam formatted files using the Samtools program (30). All the pipeline was automatized through the use of Snakemake workflow engine version 3.5.2 (31).
Differential expression
The number of reads for each gene was calculated with featureCounts 1.5.0-p2 program (32) and normalized with DESeq2 method (33) through SARTools R Package (34). Snakemake workflow engine was also used to automatize the steps of this analysis.
For the analysis of editing, we generated new alignment files by mapping the filtered reads on the non-edited Col-0 mitochondrion genome and the edited one. These alignments were done with zero mismatch allowed and selecting only uniquely mapped reads. Then, with a homemade Python3 script, we parsed the alignment files and extracted the percentage of C and U for each mitochondrial editing position listed in (14). Using this approach, editing efficiencies were calculated from both Ribo-seq and RNA-seq sequencing data that were generated in the course of the present analysis.
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE104028.
Mapping of RNA-seq reads to mitochondrial mRNA extremities
Reads alignments were performed with Bowtie 1.1.2 (29).
RESULTS
Most mitochondrial ribosome footprints in Arabidopsis measure 27 and 28 nt
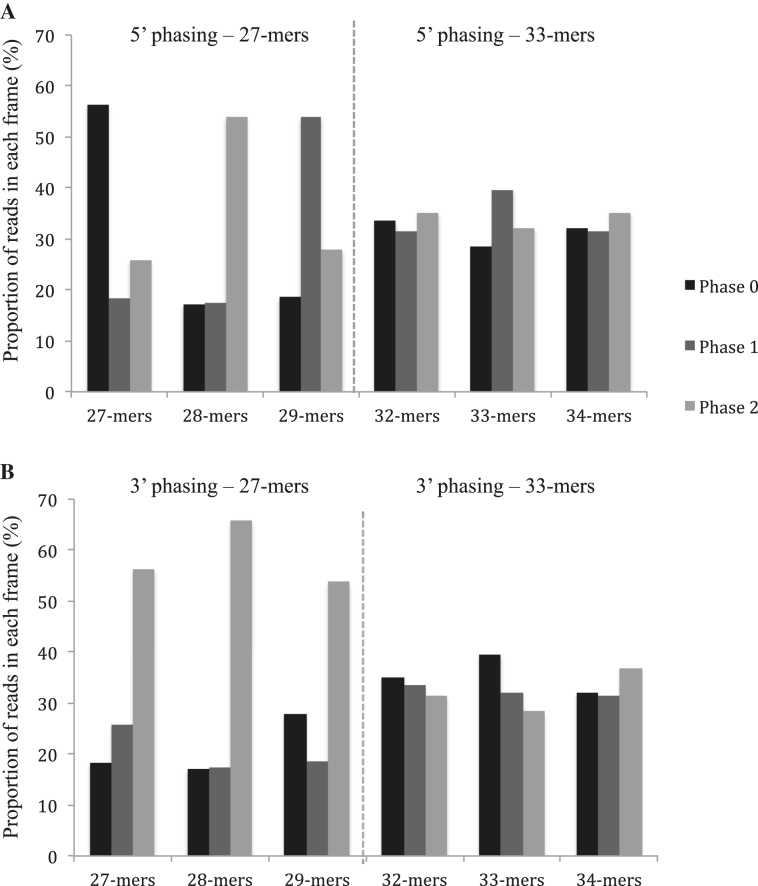
To get a comprehensive view of all ribosome-associated mRNA regions in Arabidopsis mitochondria, we generated total ribosome footprints and mapped the obtained reads to the Arabidopsis mitochondrial genome. To optimize the recovery of mitochondrial ribosome footprints from total extracts, polysomes were prepared from Arabidopsis flowers which are known to contain high amounts of active mitochondria (35). The presence of 1% Triton in the extraction buffer allowed us to extract both soluble and membrane-bound mitochondrial polysomes and thereby obtain good ribosome protected fragment (RPF) coverage along all mitochondrial transcripts. Since the size of selected RNA fragments from preparative gels can strongly influence the recovery of the desired ribosome footprints, we built upon a recent publication indicating that ribosome footprints in human mitochondria showed a bimodal distribution peaking at 27 and 33 nt (24). To determine whether Arabidopsis mitoribosome footprints measured either or both of these sizes, RNA bands at 27 and 33 bases were extracted from preparative polyacrylamide gels and sequenced. Sequence analysis of the reads mapping to the mitochondrial genome indicated that the 27-nt RNA population mostly contained fragments of 27, 28 and 29 bases, whereas the 33-nt band was essentially composed of 32, 33 and 34-base RNA fragments (Supplementary Figure S1). The strongest evidence that these fragments corresponded to ribosomal footprints was their biased repartition relative to the start codon of mitochondrial coding sequences. Indeed, ribosomes move triplet by triplet, and we were expecting to observe the same regularity for their footprints. In fact, the two RNA categories show very different behavior regarding their bias to the start codon (Figure 1). The longest fragments (32/34-mers) displayed a perfect random distribution in the three possible reading frames whereas the smallest fragments (27/29-mers) showed a clear repartition bias. We could effectively observe a bias toward the phase 2 for all the k-mers when analyzing their 3′ extremities, whereas the phase bias differed for each k-mer at their 5′ end. Mapping to mitochondrial genes further revealed that the 27 and 28-nt reads mapped primarily to coding sequences and to a much lesser extent to UTRs and introns (Supplementary Figure S2A). In contrast, the 32/34-mers mapped mostly to the 5′UTRs of genes and much less frequently to mitochondrial coding sequences (Supplementary Figure S2B). Overall, this preliminary analysis showed us that the majority of Arabidopsis mitochondrial ribosomes footprints are 27–29 nt long, and that those with 32–34 nt contribute very poorly to the mitoribosome footprint population. Consequently, all our subsequent analyses were done after recovery of the 27–29 nt RNA band from preparative gels.
Figure 1.
The 27-nt RPF population shows a better periodicity relative to mitochondrial gene start codons compared to the 33-nt RNA population. (A) 5′ phasing of the 27-mer and 33-mer populations. (B) 3′ phasing of the 27-mer and 33-mer populations.
Analysis of ribosome footprint distributions along mitochondrial genes sequences
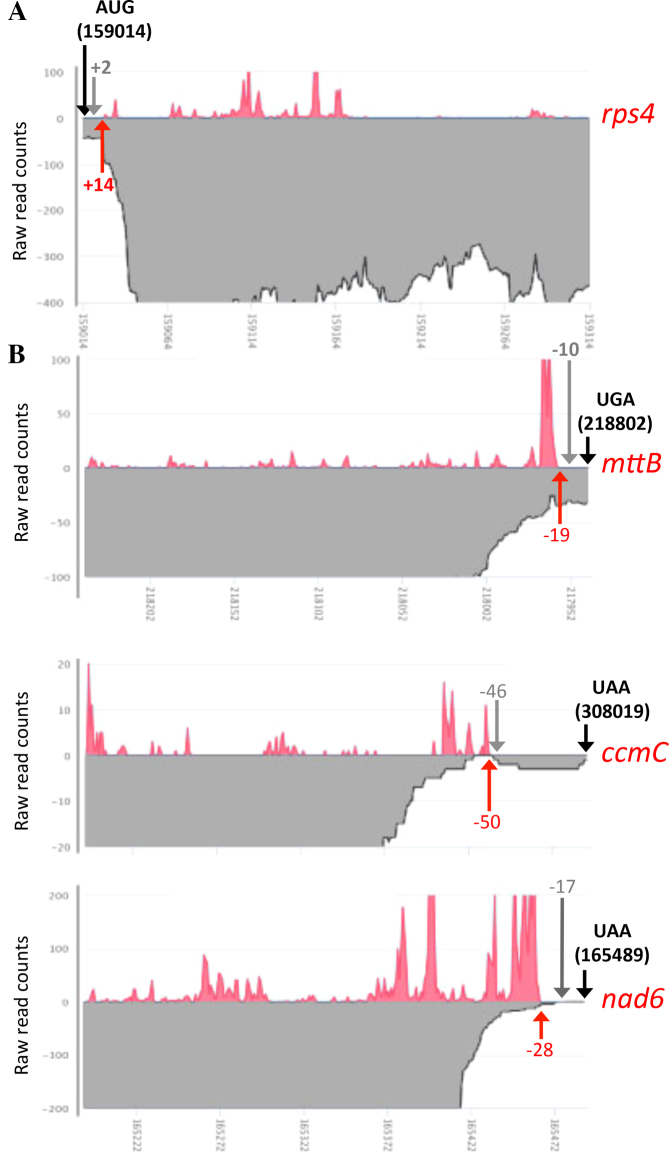
A first global analysis of the distribution of Arabidopsis mitochondrial RPF was done from a total of 900 million high quality reads obtained from wild-type plants. Less than 2% (1.9%) of these reads could be aligned to the mitochondrial genome of the Col-0 accession (JF729201.1) and 38% to the nuclear and plastid genomes, indicating that mitochondrial ribosome footprints represent a mere fraction of total ribosome footprints generated from flowers. The vast majority of reads aligned with exons corresponding to known mitochondrial genes, further supporting that they were indeed mitochondrial ribosome footprints. Despite its relative small size, the Arabidopsis mitochondrial genome contains a large number of duplicated regions, preventing the correct mapping of RPF along the entire sequence. This was particularly obvious for atp6-1 and atp6-2 genes that share identical 3′ regions. The partial overlap between the rps3 and rpl16 genes was also problematic in this respect. Despite these classical limitations, the alignments obtained allowed us firstly to examine the translation initiation and termination sites of genes of known function. For most genes, the ribosome footprint distributions were coherent with the proposed positions of start and stop codons (http://www.arabidopsis.org/). We focused on the few mitochondrial transcripts for which the translational initiation and termination codons were shown to be removed by post-transcriptional mRNA processing. This concerns the rps4 mRNA that has been shown to be processed two bases downstream of its AUG and the nad6, mttB and ccmC mRNAs that are matured 17, 10 and 46 nt upstream of their annotated stop codon, respectively (36,37). We first used the RNA-seq data we obtained from Col-0 plants to check these transcript extremities. This allowed us to unambiguously confirm that the vast majority rps4 transcripts are effectively devoid of any AUG (Figure 2A) and that nad6 and ccmC mRNAs do not carry any translational stop codon (Figure 2B). The situation is more mitigated for mttB, however most mttB mRNAs lack any translational termination codon. In agreement with these truncated mRNA ends, we observed that the ribosome footprints on rps4 initiate 14 bases downstream of the annotated AUG (Figure 2A). Similarly, the RPF distributions along nad6 and ccmC terminate a few bases upstream of their mapped 3′ extremity and do not extend until the predicted stop codon (Figure 2B). No ribosome footprints was found to cover the annotated stop codon of mttB and the presence of a high RPF peak just upstream suggest a ribosome pausing site resulting from the post-transcriptional removal of the terminal UGA. Altogether, these observations suggest that the translation initiation and termination in plants show surprising flexibility and that the plant translational machinery evolved ways to efficiently decode mRNAs devoid of conventional AUG or STOP codons.
Figure 2.
Ribosome footprint densities near the extremities of Arabidopsis mitochondrial mRNAs for which the start or the stop codons are removed by post-transcriptional processing. Ribosome footprint distributions are shown as red curves and RNA-seq read densities are depicted in gray. (A) Close-up view showing the first 300 nt of the rps4 mRNA. (B) Close-up view showing the last 300 nt of the nad6, mttB and ccmC mRNAs. Black arrows materialize the positions of the theoretical translation start or stop codons and their coordinate on the mitochondrial genome of Col-0 is specified. Red arrows and the associated numbers indicate the start (rps4) or the end (nad6, mttB and ccmC) of observed Ribo-seq signals, respectively to either the translational start or stop codon of each transcript. Gray arrows and the associated numbers indicate the position of the 5′ (rps4) or 3′ (nad6, mttB and ccmC) mRNA extremities mapped by circular RT-PCR in (36,37).
To potentially identify new coding regions within the mitochondrial genome, we also searched for RPF signals located outside of annotated regions. We looked for regions of at least 100 bases long, comprising a minimum of 10 ribosome footprints overall and in which 70% of the nucleotides are covered by at least 1 RPF. The 5′ and 3′ extremities of these regions were defined when a minimum of 40 nt with no RPF was encountered. Using these criteria, no signal outside of known genes could be identified, therefore, our approach did not reveal any new translated protein-coding ORFs in Arabidopsis mitochondria.
Arabidopsis mitochondrial mRNAs show highly different ribosome association levels and ribosome entry mechanisms
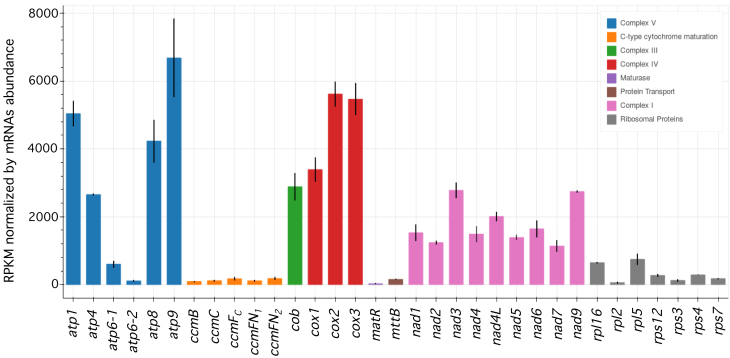
To estimate and compare the translational efficiencies of Arabidopsis mitochondrial mRNAs, we determined the relative density of ribosome footprints per gene. To allow comparison between genes, the calculated densities were normalized to both transcript length and abundance. This approach revealed clear differences in ribosome density between mRNAs (Figure 3). Globally, mRNAs encoding respiratory chain subunits showed much higher ribosome loading levels compared to other mitochondrial transcripts, implying a greater translational activity for these genes. Within each respiratory complex, small differences in translational efficiencies were observed except for the ATP synthase complex (V). The atp1, atp4, atp8 and mostly atp9 showed significantly higher ribosome densities compared to atp6-1 and atp6-2. Interestingly, the two copies of the atp6 gene also appear to be translated at different levels, and the atp6-1 gene showed much higher ribosome density compared to atp6-2 (Figure 3 and Supplementary Figure S3). All other mitochondrial genes, notably the ones coding for c-type cytochrome maturation and ribosomal proteins, had particularly low ribosome loading densities along their transcripts suggesting that they are translated at low levels. Low levels of ribosome footprints were also detected for the matR and mttb coding sequences, supporting the proposed functionality of these two genes (38,39).
Figure 3.
Protein-coding mitochondrial mRNAs show large differences in ribosome footprint densities in Arabidopsis. RPF densities normalized to both mRNA length and abundance are shown. Mitochondrial mRNAs are indicated and color-coded according to the respiratory complex or the functional category to which they belong. Means ± SE (n = 2).
The relative distributions of ribosome footprints along the five di-cistronic mitochondrial transcripts (37) showed different patterns, implying different mechanisms to guide the ribosomes toward the downstream cistrons. Regarding the nad4L-atp4 (3300 reads per kilobase million (RPKM)) for nad4L versus 4500 for atp4) and rpl5-cob (1150 versus 4600 RPKM) transcripts, the ribosome density in the downstream cistron appeared to be higher compared to the upstream open reading frame (Supplementary Figure S4). An internal entry of mitoribosomes just upstream of the translation start codon of the downstream cistron in these two cases may explain this increase. The translation of rps3-rpl16 also implies independent entry of mitoribosomes, as the two orfs overlap and are encoded on different reading frames. Conversely, the ribosome density along the downstream open reading frame was much lower in the case of the nad3-rps12 mRNA (4900 versus 450 RPKM), suggesting a less efficient translation of rps12 (Supplementary Figure S4). This could be explained either by an inefficient internal entry of ribosomes upstream of rps12 or by a weak re-initiation downstream of nad3.
The translation of mitochondrial mRNAs is not dependent on their editing status
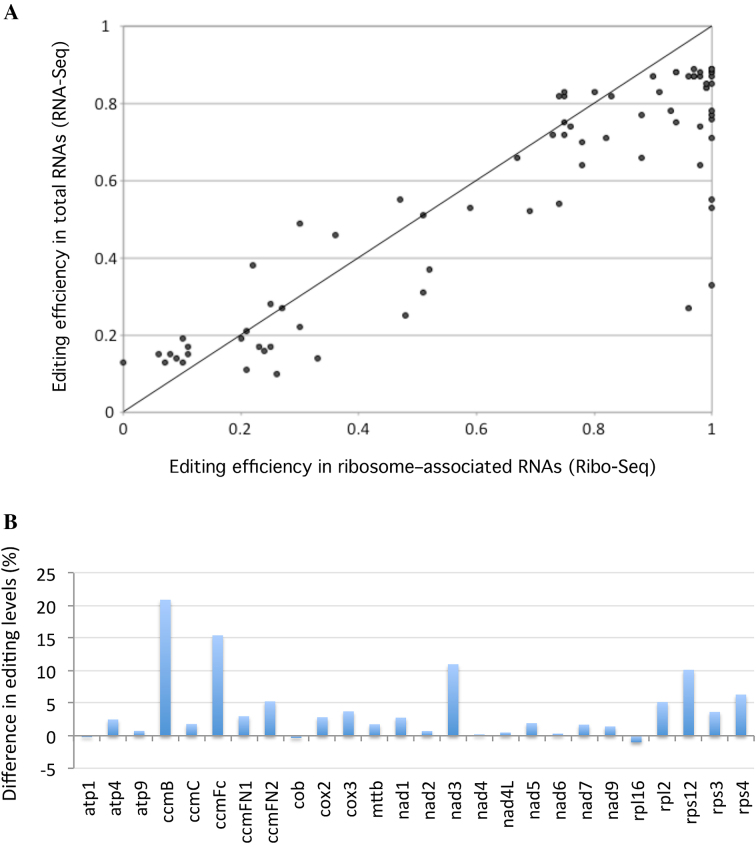
The C-to-U RNA editing process corrects more than 400 positions in plant mitochondrial transcripts (14,15). Most editing events reestablish a correct identity to codons so that edited mRNAs encode functionally optimal mitochondrial proteins (15). Nevertheless, many mitochondrial sites are not edited to 100% and a large proportion of plant mitochondrial mRNAs can encode potentially sub-optimal, if not inactive, proteins. To see whether the plant mitochondrial translation machinery preferentially associates with fully edited transcripts or not, we evaluated the editing status of transcripts associated with Arabidopsis mitochondrial ribosomes. We calculated the editing levels of all known mitochondrial RNA editing sites from both total and ribosome-associated RNAs (Supplementary Table S2). Of the 434 previously identified editing sites (14), 81 appeared to be covered by less than 10 ribosome footprints and were therefore excluded from further analysis because of insufficient coverage. Among the 353 remaining sites, most partially edited sites remained partially edited in polysome-associated mRNAs, revealing at the level of the entire mitochondrial transcriptome that complete editing of mitochondrial mRNAs is not an absolute prerequisite for their translation. A slight increase in C-to-U transition efficiencies could, however, be detected at many positions in polysomal mRNAs. In effect, we observed that 70% of sites (58 out of 80) whose editing levels range between 10 and 90% showed increased editing levels when contained in mitoribosome-associated mRNAs compared to total mRNAs (Figure 4A). The editing increase for these more-edited sites is around 16% overall and most of them are distributed in different mitochondrial transcripts (Figure 4B). Nevertheless, quite a few are contained in ccmB, ccmFc, nad3 and rps12 RNAs, whose global editing levels are much higher in ribosome-protected RNAs compared to total RNAs (Figure 4B). Most of the sites whose editing levels reach almost 100% when associated with ribosomes are contained in these four mitochondrial transcripts.
Figure 4.
Mitochondria-encoded mRNAs are more edited when they associate with mitoribosomes. (A) C-to-U editing levels calculated from total (RNA-seq) and ribosome-associated (Ribo-seq) RNA populations are presented. To identify potential variations in editing efficiencies, only sites that are partially edited and whose editing efficiencies is comprised between 10 and 90% in total RNAs were considered in the analysis. (B) Average editing levels of mitochondrial mRNAs were determined from both ribosome footprint and total RNA populations. The differences of editing levels calculated from both RNA samples (polysomal RNA—total RNA) are shown.
Mitochondrial translation is reorchestrated in respiratory complex I mutants
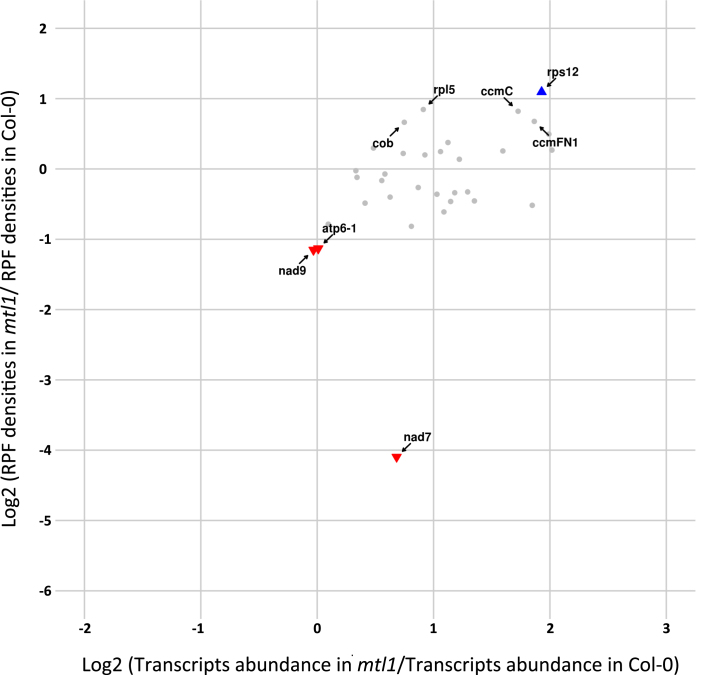
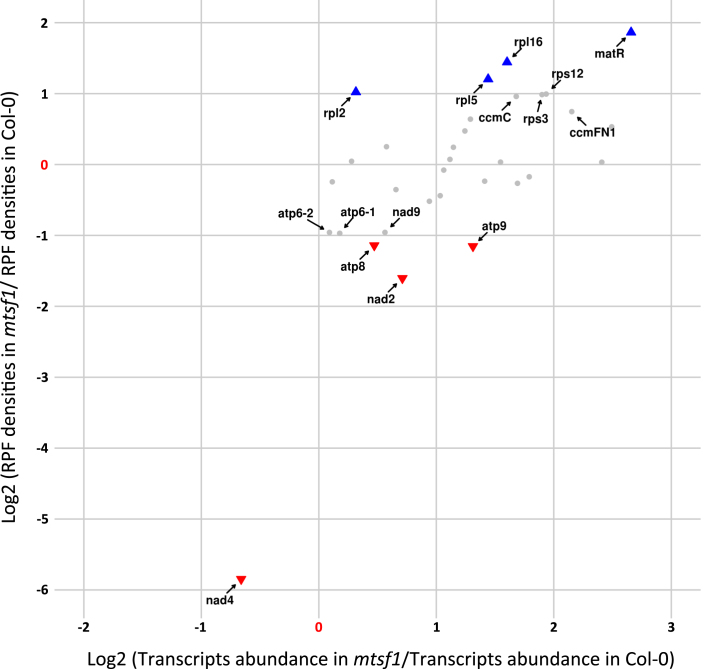
To better understand the plasticity of mitochondrial translation and the extent to which it participates in regulating gene expression in the organelle, we determined mitochondrial translational changes in two complex I respiratory mutants. For this approach, we chose the mtl1 and mtsf1 mutants, which are two ppr mutants affected in the translation of nad7 mRNA and in the stability of nad4 transcript respectively (13,26). These two mutants show different growth retardation phenotypes, with mtsf1 plants being much more impaired in their development than the mtl1 plants. In both mutants, the ribosome densities and the steady state levels of mitochondrial mRNAs were quantified. In response to the lack of respiratory complex I, we observed a global over-accumulation of most mitochondrial transcripts in both mutants (Supplementary Figure S5). The increase in transcript abundance varied among genes and was comprised between 1.5- and 4.5-folds. Interestingly, this change did not correlate with an equivalent increase in ribosome density for most transcripts. Effectively, many mRNA species showing higher abundance contained either similar or even lower ribosome loading compared to the wild-type, suggesting that mitochondrial translation is a tightly regulated process that does not simply follow the changes in transcript abundance (Figures 5 and 6; Supplementary Figures S6 and 7). Otherwise, the major transcript expression defects that are known to occur in mtl1 and mtsf1 mutants were confirmed by our analysis. Regarding mtl1, our ribosome profiling data confirmed the strong and specific reduction of nad7 mRNA translation despite a 1.7-time over-accumulation of its transcript (Figure 5 and Supplementary Figures S6). A strong reduction in the association of ribosomes with nad4 mRNA was detected in mtsf1 (Figure 6 and Supplementary Figures S7), which correlated with the strong destabilization of the nad4 transcript, excluding a role of MTSF1 in nad4 translation. For other transcripts, the comparison of mtsf1 and mtl1 translation profiles identified differences, but interesting overlaps were also observed. Among mRNAs whose translation varied most significantly in both mutants (P-value < 0.05 and RP log2(FC) ≥ or ≤ 1) several corresponded to ribosomal protein genes. Of the seven genes showing increased ribosome occupancy in mtsf1 compared to the wild-type, five corresponded to ribosomal protein mRNAs, namely rps3, rps12, rpl2, rpl5 and rpl16 (Figure 6). In the mtl1 mutant, rps12 and rpl5 transcripts also showed significant increases in ribosome loading in comparison to the wild-type (Figure 5). The translation of the nad9 transcript also appeared to vary in a similar manner in both mutants. An ∼2-fold decrease in nad9 translation was measured in both mutants, whereas the abundance of nad9 transcripts remained the same in mtl1 and was slightly increased in mtsf1 compared to the wild-type (Figures 5 and 6; Supplementary Figure S5). Interestingly, moderate but significant reductions in ribosome loading on several F1FO-ATP synthase transcripts were also detected in both mutants. This concerned the atp8, atp9, atp6-1 and atp6-2 transcripts in mtsf1 plants and the atp6-1 transcript in mtl1 plants (Figures 5 and 6).
Figure 5.
Differential transcriptional and translational profiles of mitochondria-encoded mRNAs in mtl1 and wild-type plants. For each mitochondrial mRNA, relative RPF densities (Y-axis) and transcript abundance (X-axis) are reported. Genes whose names are indicated show significant changes in ribosome occupancies compared to the wild-type (P-value < 0.05). Colored triangles show genes whose changes in ribosome loads are larger than 2-fold compared to wild-type plants.
Figure 6.
Differential transcriptional and translational profiles of mitochondria-encoded mRNAs in mtsf1 and wild-type plants. For each mitochondrial mRNA, relative RPF densities (Y-axis) and transcript abundance (X-axis) are reported. Genes whose names are indicated show significant changes in ribosome occupancies compared to the wild-type (P-value < 0.05). Colored triangles show genes whose changes in ribosome loads are larger than 2-fold compared to wild-type plants.
DISCUSSION
Unlike in human and yeast mitochondria, Arabidopsis mitoribosome footprints measure 27–29 nt long
Messenger RNA translation is a complex multi-step process that is still poorly understood at the molecular level in genome-containing organelles like mitochondria. Growing evidence indicates that although these organelle are of prokaryotic origin, mitochondrial translation has significantly diverged from its bacterial counterpart. Additionally, mitochondrial translation displays considerable species-specific specialization that evolved to adapt translation requirements to mitochondrial genome organization (40). These adaptations are exemplified by the variable composition and structure of mitoribosomes that differ not only between species, but also from modern bacterial ribosomes (41–43). In this analysis, we have used ribosome profiling technology to generate a genome-wide snapshot of mitochondrial translation in Arabidopsis, and obtained the first global analysis of a plant mitochondrial translatome. This approach revealed that Arabidopsis mitochondrial ribosome footprints measure mainly 27 and 28 nt in length and thereby do not seem to follow a bimodal distribution peaking at 27 and 33 nt as previously found for human mitochondria (24). In effect, the 27 and 28 nt RPFs presented a frame-specific distribution whereas those of 33 bases did not show any periodicity with respect to the start codons of mitochondrial genes (Figure 1). Additionally, the 27 and 28 nt RPFs aligned to translationally active mitochondrial coding sequences (Supplementary Figure S2A), as shown by the lack of coverage of nad7 mRNA in the mtl1 mutant that is specifically deficient in the translation of this transcript (Supplementary Figure S8; (13)). In contrast, the 33-nt RNA population aligned poorly with Arabidopsis mitochondrial coding sequences (Supplementary Figure S2B) and was accordingly found to represent a very minor fraction of mitoribosome-protected fragments in maize (25). Thus, these fragments make only a modest contribution to the mitoribosome footprint population in Arabidopsis. Interestingly, a recent study indicated that mitoribosome footprints in yeast measure around 38 nt (44). Fragments of this size are almost inexistent in the maize mitoribosome RPF population (25) and do not appear, therefore, to be major representatives of mitoribosome footprints in plants. The discrepancy between human, yeast and Arabidopsis regarding the size distribution of mitochondrial RPFs is currently unclear, but may result from differences in sample preparation or in mitoribosome structures between plants, yeast and animals (24).
A global alignment to the mitochondrial genome showed that mitochondrial RPFs are distributed over all the open reading frames of most genes of known function. For mRNAs like rps4, ccmC, nad6 or mttB for which the start or the stop codons are removed post-transcriptionally the RPF distributions coincide with their mapped 5′ or the 3′ extremities (Figure 2), confirming that despite the truncations in their coding sequence these transcripts are still competent for translation. Indeed, our RNA-seq data revealed that ccmC, nad6 and mttB mRNAs bear highly heterogeneous 3′ extremities and that most transcripts contain more or less extensive deletions at the end of the coding sequence (Figure 2B). The RPF distributions suggest that production of full-length CcmC and Nad6 proteins relies on the translation of the longest transcript forms that, nonetheless, lack any stop codon (37). Together, these observations raise questions about the mechanisms guiding mitoribosomes toward translation initiation sites in plants and also how they are released from 3′-truncated transcripts lacking stop codons such as ccmC or nad6. As suggested in mammalian mitochondria, specialized translational release factors or peptidyl hydrolases could intervene to release plant mitoribosomes stalled at the 3′ extremity of mRNAs lacking stop codons (45). Several lines of evidence also support that translation initiation in plant mitochondria is a relaxed process with mitoribosomes accommodating different kinds of non-AUG start codons or initiating translation internally in di-cistronic mRNAs as suggested for nad4L-atp4, rpl5-cob and rps3-rpl16 (Supplementary Figure S4). Accordingly, we find that mitochondrial orfs like ccmFN2, mttB and matR that do not start with a classical AUG codon (37) are actively translated by mitoribosomes (Figure 3). Almost nothing is known about translation initiation and termination in plant mitochondria and major genetic and biochemical analyses will be required to shed light on the behavior of plant mitoribosomes.
Arabidopsis mitochondrial mRNAs are translated to highly different levels
The number of RPFs per gene, normalized by both transcript size and steady-state abundance, was used as a proxy to estimate the translational level of each mitochondria-encoded Arabidopsis mRNA. Assuming that the average of translation elongation rates are similar for all mitochondrial genes, this approach revealed large differences in ribosome coverage between mitochondrial protein-coding transcripts. We effectively observed that respiratory subunit mRNAs show much higher ribosome association than other types of mRNAs, notably those coding for ribosomal and c-type cytochrome maturation proteins, which appear to be translated at very low levels from our analysis (Figure 3). These results are coherent with recent quantification of plant mitochondrial proteomes which indicated that steady-state levels of respiratory chain proteins are abundant and well represented in mitochondrial proteome lists, whereas proteins of the genetic apparatus are much less abundant (46,47). The c-type cytochrome maturation proteins, the MttB transporter or the MatR maturase are present at such low levels in mitochondria that they are generally not detected at all in plant mitochondrial proteomes. The high sensitivity of the ribosome profiling technology allowed us to detect the translational activity of these very poorly translated mitochondrial mRNAs, strongly supporting their functionality. Among respiratory complexes subunits, we observed very similar ribosome densities for genes encoding proteins belonging to the same complex, except for the F1FO-ATP synthase. Large differences were detected between complex V mRNAs, particularly for the atp1, atp4, atp8 and atp9 transcripts, which showed higher RPF coverage than other atp mRNAs (Figure 3). The atp1 and atp9 mRNAs encode the c and α subunits of the F1FO-ATP synthase, respectively, and are present at 10 and 3 copies in complex V. The higher ribosome densities of these two transcripts, therefore, correlates with their relative stoichiometry in complex V, corroborating the proportional translation of the different ATP synthase subunits previously described in E. coli (48). Conversely, the higher translational activity of atp8 compared to other single-copy complex V subunits (atp4 and atp6) cannot be explained by its higher stoichiometry in the F1FO-ATP synthase, but suggest higher requirements for this specific subunit for reasons that need to be clarified. Interestingly, we also observed a large difference in mitoribosome coverage between the two copies of atp6 encoded in the Arabidopsis mitochondrial genomes, with atp6-1 being translated significantly more than atp6-2 (Supplementary Figure S3). The two genes show small sequence variations in their 5′ UTRs, which likely account for their differential association with mitoribosomes. Together, the large differences in ribosome densities between the various mitochondrial mRNAs reveal that translational control is an important component of gene expression in plant mitochondria. How this control operates at the molecular level is currently unclear, as we still do not know how ribosomes are recruited to mitochondrial mRNAs in the absence of Shine and Dalgarno (SD) sequences in their 5′ UTR (7). This control can operate at the level of translation initiation, which is often proposed as being the most rate-limiting step of translation in most systems (49,50). The combinatory action of cis-acting elements within the 5′UTRs and gene-specific trans-factors may be responsible for the differential recruitment of ribosomes on mitochondrial mRNAs. Nonetheless, to date very few translational activators have been found to operate in plant mitochondria (12,13,51–53) and the way by which they control translation of their cognate mRNA target remains to be determined. In bacteria, SD-like sequences within coding sequences were shown to impact translation elongation and to have a major effect on mRNA translation rates (54,55). The lack of anti-SD sequence at the 3′ end of the mitochondrial SSU rRNA strongly suggests that this mechanism cannot account for the differential translation of Arabidopsis mitochondrial mRNAs. Much remains to be discovered, therefore, before we understand how mitochondrial translation is regulated in plant mitochondria.
Mitoribosome-associated mRNAs are more edited than in total RNAs
Mitochondrial mRNAs from land plants undergo hundreds of specific C-to-U changes by RNA editing (15). In the present analysis, of the 353 Arabidopsis editing sites for which we obtained more than 10 reads per sites in both RNAseq and Riboseq data, 80 showed partial editing with C-to-U changes comprised between 10 and 90%. The accessibility of non-edited or partially edited mitochondrial mRNAs by the mitochondrial translation machinery has long remained an intriguing question. The translation of non-edited mitochondrial mRNA was previously suggested in a few specific cases (16–20), but no genome-wide view was available to date. By comparing C-to-U transition efficiencies in total and ribosome-associated mRNAs we observed that most partially edited sites remained incompletely edited in polysomal mRNAs confirming at a genome-wide level that complete editing of mitochondrial mRNAs is not an absolute pre-requisite for their translation (Figure 4A). Nonetheless, around 70% of these sites showed, on average, a 16% increase in their editing levels when loaded on mitoribosomes (Figure 4A and B). The origin of this increase is currently unclear, as no obvious inter-functionality between the mitochondrial translational and editing machineries has yet been revealed. As indicated, large differences in C-to-U transition efficiencies are consistently observed between editing sites in plant mitochondria (14). These differences likely result from variable functional constraints between sites, making some of them less efficiently recognized and/or processed by the editing machinery than others. Maybe, the accessibility of RNA editing sites to the editosome complex is increased when mRNAs are being translated, generating the rather global raise in editing efficiency that we observed in our riboseq data. In particular, the strong RNA helicase activity of translating ribosomes may be responsible for rendering editing sites more accessible to the editing machinery. Even with this increase, our results indicate that partially edited mRNAs are decoded by mitoribosomes, confirming at the level of the entire mitochondrial transcriptome that transcript recruitment by mitochondrial polysomes does not a priori discriminate between unedited and edited RNAs. A proportion of mitochondrial polypeptides derives thus from the translation of unedited mRNAs, making ‘unedited’ proteins a non-negligible part of plant mitochondrial proteomes. Post-translational processes must therefore be at play to limit any potential negative effects of these likely sub-optimal ‘unedited’ polypeptides.
Mitochondrial translation is reorganized in Arabidopsis complex I mutants
The regulation of mitochondrial gene expression is fundamental not only to ensure proportional synthesis of both mitochondria and nuclear-encoded subunits belonging to the same complexes but also to adapt mitochondrial activity to the changing requirements of plants throughout their life cycle or in response to environmental stimuli. The basis of this regulation is not well understood, however several lines of evidence support that most of the control is exerted at the post-transcriptional and/or at the post-translational levels (5,56–58). To better understand to what extent mitochondrial translation participates in these regulatory circuits and gain hints about its plasticity, we measured mitochondrial translational and transcriptional changes in two contrasted complex I Arabidopsis mutants that accumulate either no complex I (mtl1) or a truncated version of it (mtsf1). Although both mutants have strong delays in their development compared to wild-type, the growth defects exhibited by the mtsf1 mutant were more severe than those of mtl1 (13,26). First, in response to complex I deficiency, we observed a 2- to 5-fold over-accumulation of most mitochondrial transcripts in mutants compared to wild-type (Supplementary Figure S5). Nonetheless, this increase in transcript abundance was not associated with an equivalent rise in ribosome coverage along mitochondrial mRNAs (Figures 5 and 6). Our analysis thus revealed that mitochondrial translation is a precisely regulated process that does not simply follow the changes in transcript abundance and that a transcript-specific translational control clearly operates in plant mitochondria. Significant changes in ribosome loading for several transcripts were, however, detected in the mutants. The most important translational change concerned the strong decrease in nad7 ribosome occupancy in mtl1, confirming previously published data for this mutant (13). The impact on other mitochondrial genes was less pronounced, although some interesting tendencies were uncovered. The first one concerned the 2-fold decrease in nad9 translation in both mutants, suggesting that the lack of functional complex I may induce a negative feedback control on the translation of this specific mRNA. As we did not detect any significant decrease in the ribosome loading of other complex I mRNAs, this means that complex I deficiency does not induce a global down-regulation of all nad gene translation. Interestingly, and despite a significant rise in transcript abundance, the translation of several mRNAs encoding ATP synthase subunits was lower in both complex I mutants. This negative translational control impacted a few more atp genes in mtsf1 than in mtl1 (Figures 5 and 6) and may, therefore, be correlated with the gravity of developmental alterations associated with complex I deficiency. It has recently been demonstrated that complex I-lacking Arabidopsis mutants compensate for the deficit in ATP production derived from the respiratory chain by inducing higher metabolic fluxes through glycolysis and the tricarboxylic acid cycle (59). Our analysis thus reveals that these increased metabolic fluxes correlate with a slight reduction in the translation of mitochondria-encoded ATP synthase subunits. Whether or not this reduction results from a negative feedback loop induced by the reduced activity of the respiratory chain in complex I mutants remains to be clarified. Interestingly, we also observed higher ribosome association with several ribosomal protein encoding mRNAs in both mtsf1 and mtl1 mutants. The ribosome densities of four ribosomal protein mRNA transcripts in mtsf1 (rps12, rpl2, rpl5, rpl16) and two in mtl1 (rps12, rpl5) appeared to be increased compared to the wild-type. These observations corroborate nicely recent analyses showing that reducing the production of a mitochondria-targeted ribosomal protein resulted in an altered translation pattern in Arabidopsis mitochondria (60). In this mutant context, ribosomal proteins were translated more efficiently at the expense of respiratory chain encoding mRNAs. Our results suggest that an analogous reorganization of mitochondrial translational occurs in respiratory mutants, further demonstrating that plant mitoribosomes do not translate mRNAs non-selectively, but that a certain level of translational control likely operates in plant mitochondria.
Concluding remarks
Using ribosome profiling technology, we have generated a genome-wide snapshot of mitochondrial translation in Arabidopsis and monitored certain aspects of its dynamics. We show that mitochondria-encoded mRNAs are differentially translated and that proportional translation levels correlate with the stoichiometry of respiratory chain subunits, in particular for complex V. Moreover, a slight reorganization of mitochondrial translation could be detected in complex I respiratory mutants involving a reduction in ribosome densities for F1FO-ATP synthase mRNAs and an increase in the translation of several ribosomal protein encoding transcripts. We thus reveal that mitochondrial translation is controlled at the level of individual mRNAs and that it participates in regulating the activity of plant mitochondria. Our analysis also reveals that translation of mitochondria-encoded transcripts can be satisfactorily measured from total ribosome footprint extracts and does not necessarily require the purification of mitochondria.
DATA AVAILABILITY
The data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE104028.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institut National de la Recherche Agronomique (INRA) [UMR 1318]; French Ministère de l’Enseignement et de la Recherche and Fondation pour la Recherche Médicale [FDT20160736470 to N.P.]; LabEx Saclay Plant Sciences-SPS [ANR-10-LABX-0040-SPS]; INCa [RiboTEM 2014-092]; ANR [Rescue_Ribosome ANR-17-CE12-0024-01 to O.N.]. Funding for open access charge: IJPB; INRA [UMR 1318].
Conflict of interest statement. None declared.
REFERENCES
- 1. Gray M.W. Mitochondrial evolution. Science. 1999; 283:1476–1481. [DOI] [PubMed] [Google Scholar]
- 2. Gray M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012; 4:a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lightowlers R.N., Rozanska A., Chrzanowska-Lightowlers Z.M.. Mitochondrial protein synthesis: figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014; 588:2496–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hammani K., Giegé P.. RNA metabolism in plant mitochondria. Trends Plant Sci. 2014; 19:380–389. [DOI] [PubMed] [Google Scholar]
- 5. Janska H., Kwasniak M.. Mitoribosomal regulation of OXPHOS biogenesis in plants. Front. Plant Sci. 2014; 5:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ott M., Amunts A., Brown A.. Organization and regulation of mitochondrial protein synthesis. Annu. Rev. Biochem. 2015; 85:77–101. [DOI] [PubMed] [Google Scholar]
- 7. Bonen L. Daniell H, Chase CD. Translational Machinery in Plant Organelles. Molecular Biology and Biotechnology of Plant Organelles. 2004; Dordrecht: Springer; 323–345. [Google Scholar]
- 8. Greber B.J., Ban N.. Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 2016; 85:103–132. [DOI] [PubMed] [Google Scholar]
- 9. Pfeffer S., Woellhaf M.W., Herrmann J.M., Forster F.. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2015; 6:1–8. [DOI] [PubMed] [Google Scholar]
- 10. Hazle T., Bonen L.. Comparative analysis of sequences preceding protein-coding mitochondrial genes in flowering plants. Mol. Biol. Evol. 2007; 24:1101–1112. [DOI] [PubMed] [Google Scholar]
- 11. Choi B., Acero M.M., Bonen L.. Mapping of wheat mitochondrial mRNA termini and comparison with breakpoints in DNA homology among plants. Plant Mol. Biol. 2012; 80:539–552. [DOI] [PubMed] [Google Scholar]
- 12. Manavski N., Guyon V., Meurer J., Wienand U., Brettschneider R.. An essential pentatricopeptide repeat protein facilitates 5′ maturation and translation initiation of rps3 mRNA in maize mitochondria. Plant Cell. 2012; 24:3087–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haïli N., Planchard N., Arnal N., Quadrado M., Vrielynck N., Dahan J., Francs-Small des C.C., Mireau H.. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrial NADH DEHYDROGENASE SUBUNIT7 mRNA in Arabidopsis. Plant Physiol. 2016; 170:354–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bentolila S., Oh J., HANSON M.R., Bukowski R.. Comprehensive high-resolution analysis of the role of an arabidopsis gene family in RNA editing. PLoS Genet. 2013; 9:e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takenaka M., Zehrmann A., Verbitskiy D., Härtel B., Brennicke A.. RNA Editing in plants and its evolution. Annu. Rev. Genet. 2013; 47:335–352. [DOI] [PubMed] [Google Scholar]
- 16. Lu B., Hanson M.R.. Fully edited and partially edited nad9 transcripts differ in size and both are associated with polysomes in potato mitochondria. Nucleic Acids Res. 1996; 24:1369–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu B., Wilson R.K., Phreaner C.G., Mulligan R.M., Hanson M.R.. Protein polymorphism generated by differential RNA editing of a plant mitochondrial rps12 gene. Mol. Cell. Biol. 1996; 16:1543–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lu B., Hanson M.R.. A single homogeneous form of ATP6 protein accumulates in petunia mitochondria despite the presence of differentially edited atp6 transcripts. Plant Cell. 1994; 6:1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phreaner C.G., Williams M.A., Mulligan R.M.. Incomplete editing of rps12 transcripts results in the synthesis of polymorphic polypeptides in plant mitochondria. Plant Cell. 1996; 8:107–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams M.A., Tallakson W.A., Phreaner C.G., Mulligan R.M.. Editing and translation of ribosomal protein S13 transcripts: unedited translation products are not detectable in maize mitochondria. Curr. Genet. 1998; 34:221–226. [DOI] [PubMed] [Google Scholar]
- 21. Ingolia N.T., Ghaemmaghami S., Newman J.R.S., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brar G.A., Weissman J.S.. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat. Rev. Mol. Cell Biol. 2015; 16:651–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zoschke R., Watkins K.P., Barkan A.. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell. 2013; 25:2265–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rooijers K., Loayza-Puch F., Nijtmans L.G., Agami R.. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013; 4:2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chotewutmontri P., Barkan Alice. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 2016; 12:e1006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haïli N., Arnal N., Quadrado M., Amiar S., Tcherkez G., Dahan J., Briozzo P., Colas des Francs-Small C., Vrielynck N., Mireau H.. The pentatricopeptide repeat MTSF1 protein stabilizes the nad4 mRNA in Arabidopsis mitochondria. Nucleic Acids Res. 2013; 41:6650–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Baudin-Baillieu A., Hatin I., Legendre R., Namy O.. Translation analysis at the genome scale by ribosome profiling. Methods Mol. Biol. 2016; 1361:105–124. [DOI] [PubMed] [Google Scholar]
- 28. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2015; 17:10–12. [Google Scholar]
- 29. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Köster J., Rahmann S.. Snakemake–a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 32. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 33. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Varet H., Brillet-Guéguen L., Coppée J.-Y., Dillies M.-A.. SARTools: a DESeq2- and edgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS One. 2016; 11:e0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang J., Struck F., Matzinger D.F., Levings C.S.. Flower-enhanced expression of a nuclear-encoded mitochondrial respiratory protein is associated with changes in mitochondrion number. Plant Cell. 1994; 6:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Raczynska K.D., Le Ret M., Rurek M., Bonnard G., Augustyniak H., Gualberto J.M.. Plant mitochondrial genes can be expressed from mRNAs lacking stop codons. FEBS Lett. 2006; 580:5641–5646. [DOI] [PubMed] [Google Scholar]
- 37. Forner J., Weber B., Thuss S., Wildum S., Binder S.. Mapping of mitochondrial mRNA termini in Arabidopsis thaliana: t-elements contribute to 5′ and 3′ end formation. Nucleic Acids Res. 2007; 35:3676–3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sultan L.D., Mileshina D., Grewe F., Rolle K., Abudraham S., Głodowicz P., Niazi A.K., Keren I., Shevtsov S., Klipcan L. et al. The reverse transcriptase/RNA maturase protein MatR is required for the splicing of various group II introns in brassicaceae mitochondria. Plant Cell. 2016; 28:2805–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carrie C., Wei enberger S., Soll J.R.. Plant mitochondria contain the protein translocase subunits TatB and TatC. J. Cell Sci. 2016; 129:3935–3947. [DOI] [PubMed] [Google Scholar]
- 40. Burger G., Gray M.W., Lang B.F.. Mitochondrial genomes: anything goes. Trends Genet. 2003; 19:709–716. [DOI] [PubMed] [Google Scholar]
- 41. Amunts A., Brown A., Toots J., Scheres S.H.W., Ramakrishnan V.. The structure of the human mitochondrial ribosome. Science. 2015; 348:95–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Greber B.J., Bieri P., Leibundgut M., Leitner A., Aebersold R., Boehringer D., Ban N.. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015; 348:303–308. [DOI] [PubMed] [Google Scholar]
- 43. Desai N., Brown A., Amunts A., Ramakrishnan V.. The structure of the yeast mitochondrial ribosome. Science. 2017; 355:528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Couvillion M.T., Soto I.C., Shipkovenska G., Churchman L.S.. Synchronized mitochondrial andcytosolic translation programs. Nature. 2016; 533:499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mai N., Chrzanowska-Lightowlers Z.M.A., Lightowlers R.N.. The process of mammalian mitochondrial protein synthesis. Cell Tissue Res. 2017; 367:5–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salvato F., Havelund J.F., Chen M., Rao R.S.P., Rogowska-Wrzesinska A., Jensen O.N., Gang D.R., Thelen J.J., Møller I.M.. The potato tuber mitochondrial proteome. Plant Physiol. 2014; 164:637–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rao R.S.P., Salvato F., Thal B., Eubel H., Thelen J.J., Møller I.M.. The proteome of higher plant mitochondria. Mitochondrion. 2017; 33:22–37. [DOI] [PubMed] [Google Scholar]
- 48. Li G.-W., Burkhardt D., Gross C., Weissman J.S.. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014; 157:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sonenberg N., Hinnebusch A.G.. Regulation of translation initiation in eukaryotes: mechanismsand biological targets. Cell. 2009; 136:731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shah P., Ding Y., Niemczyk M., Kudla G., Plotkin J.B.. Rate-limiting steps in yeast protein translation. Cell. 2013; 153:1589–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kazama T., Nakamura T., Watanabe M., Sugita M., Toriyama K.. Suppression mechanism of mitochondrial ORF79 accumulation by Rf1 protein in BT-type cytoplasmic male sterile rice. Plant J. 2008; 55:619–628. [DOI] [PubMed] [Google Scholar]
- 52. Uyttewaal M., Arnal N., Quadrado M., Martin-Canadell A., Vrielynck N., Hiard S., Gherbi H., Bendahmane A., Budar F., Mireau H.. Characterization of raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for ogura cytoplasmic male sterility. Plant Cell. 2008; 20:3331–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Uyttewaal M., Mireau H., Rurek M., Hammani K., Arnal N., Quadrado M., Giegé P.. PPR336 is associated with polysomes in plant mitochondria. J. Mol. Biol. 2008; 375:626–636. [DOI] [PubMed] [Google Scholar]
- 54. Li G.-W., Oh E., Weissman J.S.. The anti-Shine–Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2013; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. O’Connor P.B.F., Li G.W., Weissman J.S., Atkins J.F., Baranov P.V.. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 2013; 29:1488–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Giegé P., Hoffmann M., Binder S., Brennicke A.. RNA degradation buffers asymmetries of transcription in Arabidopsis mitochondria. EMBO Rep. 2000; 1:164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Giegé P., Sweetlove L.J., Cognat V., Leaver C.J.. Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell. 2005; 17:1497–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Holec S., Lange H., Kühn K., Alioua M., Börner T., Gagliardi D.. Relaxed transcription in Arabidopsis mitochondria is counterbalanced by RNA stability control mediated by polyadenylation and polynucleotide phosphorylase. Mol. Cell. Biol. 2006; 26:2869–2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kühn K., Obata T., Feher K., Bock R., Fernie A.R., Meyer E.H.. Complete mitochondrial complex I deficiency induces an Up-Regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 2015; 168:1537–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwasniak M., Majewski P., Skibior R., Adamowicz A., Czarna M., Sliwinska E., Janska H.. Silencing of the nuclear RPS10 gene encoding mitochondrial ribosomal protein alters translation in arabidopsis mitochondria. Plant Cell. 2013; 25:1855–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE104028.