Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal degenerative motor neuron disorder of which the progression is influenced by several disease-modifying factors. Here, we investigated ELP3, a subunit of the elongator complex that modifies tRNA wobble uridines, as one of such ALS disease modifiers. ELP3 attenuated the axonopathy of a mutant SOD1, as well as of a mutant C9orf72 ALS zebrafish model. Furthermore, the expression of ELP3 in the SOD1G93A mouse extended the survival and attenuated the denervation in this model. Depletion of ELP3 in vitro reduced the modified tRNA wobble uridine mcm5s2U and increased abundance of insoluble mutant SOD1, which was reverted by exogenous ELP3 expression. Interestingly, the expression of ELP3 in the motor cortex of ALS patients was reduced and correlated with mcm5s2U levels. Our results demonstrate that ELP3 is a modifier of ALS and suggest a link between tRNA modification and neurodegeneration.
Introduction
Amyotrophic lateral sclerosis (ALS) is an adult motor neuron disorder characterized by the degeneration of motor neurons in the spinal cord, brainstem and motor cortex, resulting in muscle weakness, atrophy and spasticity. It is a progressive disease and usually fatal within 5 years after the diagnosis. Mutations in a heterogeneous set of genes, such as C9ORF72, SOD1, TARDBP and FUS have been identified to cause the familial form that accounts for 10% of all ALS. Most patients (90%) have now known family history and are classified as ‘sporadic’. Currently, there is no cure for ALS (1). The phenotypic heterogeneity of familial and sporadic ALS (2) suggests the existence of modifiers that determine disease characteristics such as site and age of onset, progression rate and duration of disease. It is important to identify these modifiers as they may be targets for therapeutic intervention, even in the absence of a known cause for ALS. One such factor is subunit 3 of the elongator complex (ELP3). An association between genetic variation in ELP3 and sporadic ALS was revealed in a genome-wide association study (3), and thereafter ELP3 was identified as a modifier of survival in patients carrying the C9orf72 repeat expansion (4).
ELP3 is the enzymatic core of the elongator complex, comprised of six subunits (ELP1-ELP6). Elongator complex promotes the side chain modification of the wobble uridine base of tRNA (position U34), a process greatly increasing fidelity and efficiency of translation (5–11). Although modification of U34 is chemically complex and still incompletely understood (12), it is known that elongator catalyzes the initial step in the formation of 5-methoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5), two modifications of the tRNA wobble uridine (5,13). In yeast, Elp3 is essential for the efficient translation of stress response transcription factors such as Atf1 and Pcr1. The absence of Elp3 results in poor expression of these factors and is lethal after stress (9). Interestingly, overexpression of two tRNAs (lysine and glutamine) rescues this stress-dependent phenotype of Elp3-deficient yeast (6,9). In higher order organisms, the absence of ELP3 seems to mainly affect the nervous system. In Caenorhabditis elegans, elpc-3 deletion abolishes tRNA uridine modification and affects translation, resulting in lower levels of neuropeptide and reduction of acetylcholine in the synaptic cleft (8). In the mouse, ELP3 regulates neuronal migration and differentiation in the developing cerebral cortex (14,15). The role of ELP3 in humans is far from understood, but recessive mutations in subunit 1 (ELP1) cause familial dysautonomia (FD), a severe hereditary sensory and autonomic neuropathy (16). ELP1 is the scaffold subunit of the complex and whose loss-of-function mutations causing FD severely affect the assembly of the complex. Accordingly, the level of modified wobble uridines was found to be reduced in the brain of FD patients (17).
ELP3 has an N-terminal S-adenosyl-methionine (SAM) domain and a C-terminal histone acetyltransferease (HAT) domain. The SAM domain contains an iron-sulfur (Fe-S) cluster that can cleave SAM to form 5′-deoxyadenosine radical and methionine. It has been shown that the SAM domain, but not the HAT domain, is required to maintain the integrity of the yeast elongator complex (18). Deletion of ELP3 is associated with hypoacetylation of histones H3 and H4 in yeast (19), through which it is thought to regulate transcription elongation, although this has been questioned (6,20). In Drosophila, Elp3 acetylates the synaptic protein Bruchpilot, resulting in dysregulated glutamatergic vesicle release (21).
Several findings suggest that ELP3 affects the nervous system through a loss-of-function mechanism. In mice, it controls the production and migration of cortical neurons (14,15,22). In ALS, the genotype increasing the risk to develop the disease was associated with lower brain expression levels of ELP3 (23). In Drosophila, loss-of-function mutations induced aberrant axonal outgrowth and synaptic defects and in zebrafish decrease of Elp3 expression induced motor axonal abnormalities similar to those induced by mutant SOD1 and mutant TDP43 (3).
We hypothesized that low ELP3 expression renders motor neurons vulnerable to neurodegeneration while overexpression is neuroprotective. In this study, we investigated this hypothesis in humans and in different ALS models (zebrafish and mouse) and found that overexpression of human ELP3 was indeed protective in zebrafish models for ALS, an effect for which the SAM domain is necessary. Furthermore, ELP3 affected muscle innervation, disease onset and survival in the mutant SOD1 mouse model for ALS. Lowering ELP3 decreased the levels of the modified tRNA nucleoside, 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U), both in the mouse and in NSC34 cells, while increasing ELP3 expression restored the levels of this modified uridine and attenuated mutant SOD1 aggregation in NSC34 cells. Finally, ELP3 levels were reduced in the motor cortex of ALS patients, which correlated with mcm5s2U levels. Altogether, our results suggest that ELP3 expression modifies motor neuron degeneration and may do so by altering tRNA modification in the central nervous system.
Results
Expression of ELP3 prevents axonopathy in two zebrafish ALS models
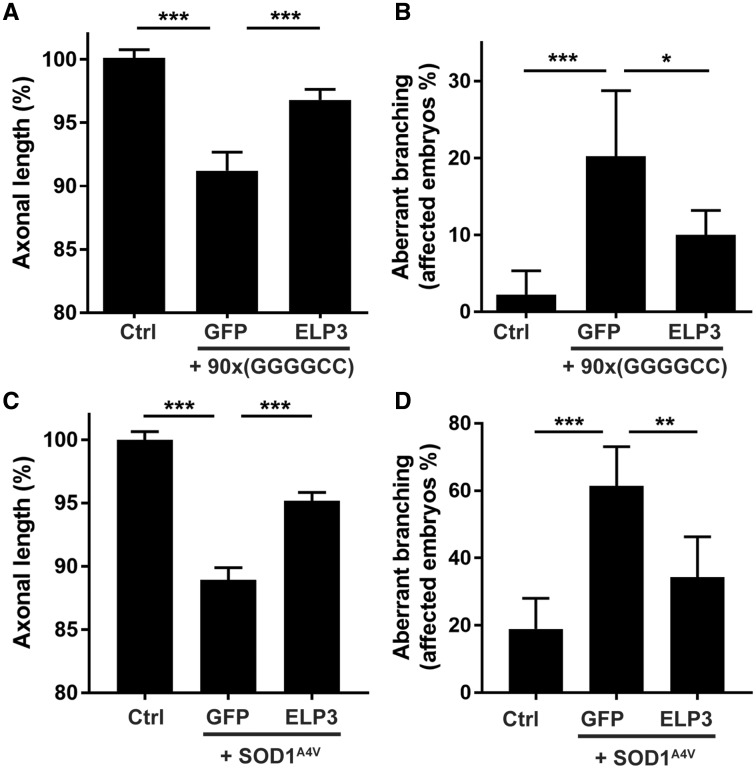
To elucidate the role of ELP3 in ALS, we first studied its effect in two zebrafish models for ALS: one for mutant C9orf72-induced ALS (C9ALS) and one for mutant SOD1-associated ALS (SOD1ALS). Injection of 90x(GGGGCC) repeat RNA, but not of 3x(GGGGCC) repeat RNA, as well as injection of mutant SOD1 RNA but not of wild-type RNA in zebrafish embryos induced a motor axonopathy, characterized by reduced axonal outgrowth and aberrant branching, as described before (23,24). Co-expressing human ELP3 RNA significantly prevented the toxic effect of the 90x(GGGGCC) repeat expansion, as it reduced the outgrowth failure by 63.6% and the number of affected embryos by 56.7% (Fig. 1A and B). Likewise, co-expression of ELP3 greatly reduced the toxicity of mutant SOD1 (SOD1A4V), reducing the outgrowth deficit by 56.8% and the number of embryos with aberrant branching with 63.5% (Fig. 1C and D). These results in two different zebrafish models suggest a protective role for ELP3 in ALS.
Figure 1.
ELP3 is neuroprotective in two zebrafish ALS models. Effect of human ELP3 in the axonopathy induced by C9orf72-associated repeat RNA (A and B) or SOD1A4V RNA (C and D), assessed by quantification of motor axon length (A and C) and affected embryos/axonal branching quantification (B and D). Co-injection of wild-type human ELP3 (ELP3) partially prevents the toxicity induced by either 90x(GGGGCC) RNA or SOD1A4V RNA. 3x(GGGGCC) RNA and SOD1WT were used as controls. Data represent mean ± S.E.M., one-way ANOVA (A and C), or mean ± 95% CI, logistic regression (B and D). n = 90. ***P < 0.001, **P < 0.01. *P < 0.05.
Ubiquitous, but not neuronal, overexpression of ELP3 attenuates the ALS-like phenotype in the SOD1G93A mouse
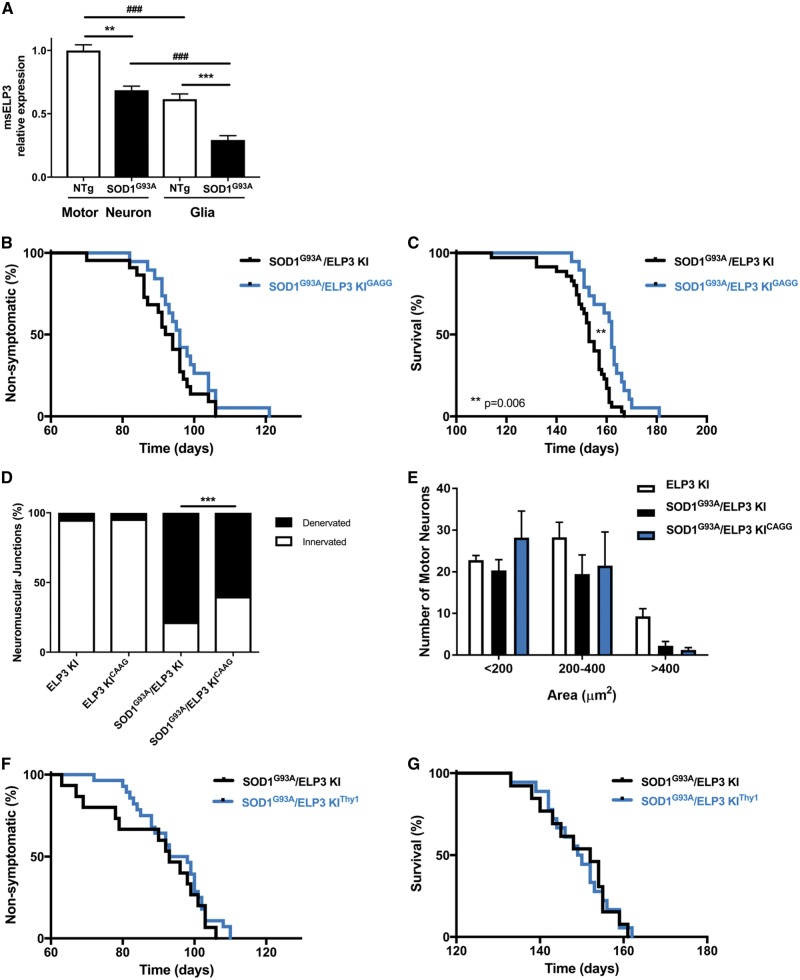
We next investigated whether a similar protection was seen in a rodent model for ALS, the SOD1G93A mouse. At symptomatic stage (post-natal day 140), spinal cord and brain ELP3 expression is reduced in this model by 30.2% and 20.0%, respectively (Supplementary Material, Fig. S1A and B). We next determined ELP3 levels in neurons versus glial cells in the SOD1G93A mouse. To do so, we laser-capture microdissected spinal motor neurons and the glia surrounding them in SOD1G93A mice (Supplementary Material, Fig. S1C and D). We found ELP3 levels to be reduced both in spinal motor neurons and in glia, by 31.4% and 52.4%, respectively, compared to non-transgenic mice. Moreover, ELP3 was less abundant in glia than in neurons both in non-transgenic mice, 38.5%, and in SOD1G93A mice, 57.5% (Fig. 2A). We then aimed to increase ELP3 levels in the SOD1G93A mouse by overexpressing human ELP3. To this end, we crossed the ELP3 KI mouse with the CAGG-CreER mouse that ubiquitously expresses the Cre recombinase (generating the ELP3 KI/CAGG-CreER mouse, herein designated ELP3 KIGAGG). The ELP3 KIGAGG mouse was then crossed with the SOD1G93A mouse, generating a triple transgenic mouse. Induction of human ELP3 expression was successfully achieved by treatment with tamoxifen at post-natal day 60 (Supplementary Material, Fig. S1E, G and K). The total ELP3 levels were increased by 1.27-fold to 1.50-fold in the spinal cord and brain of the ELP3 KIGAGG mouse, respectively (Supplementary Material, Fig. S1F and J). The levels of endogenous ELP3 and human SOD1 were not modified by the induction of human ELP3 expression (Supplementary Material, Fig. S1H, I, L and M).
Figure 2.
Overexpression of human ELP3 attenuates the ALS-like phenotype in the SOD1G93A mouse. (A) ELP3 mRNA relative expression in neurons and glia from the lumbar spinal cord of symptomatic (post-natal day 140) SOD1G93A mice. Levels were reduced by 31% and 52% in neurons and glia, respectively, compared to non-transgenic mice. Moreover, ELP3 was less abundant in glia than in neurons: 37.5% in non-transgenic mice and 57.5% in SOD1G93A mice. Data represent mean ± S.E.M., one-way ANOVA, n = 4. ***P < 0.001, **P < 0.01. (B–E) Effect of ubiquitous tamoxifen-induced expression of human ELP3 in SOD1G93A mice. Tamoxifen was administered at post-natal day 60. (B) Mean disease onset was not affected by ELP3 expression: SOD1G93A/ELP3 KI 92.3 ± 1.8 days, n = 22 versus SOD1G93A/ELP3 KICAGG 97.1 ± 2.0 days, n = 19. Data are mean ± S.E.M., log-rank test, P = 0.1671. (C) Mean survival was significantly prolonged by ELP3 expression: SOD1G93A/ELP3 KI 152.0 ± 1.8 days, n = 35 versus SOD1G93A/ELP3 KICAGG 160.7 ± 1.9 days, n = 19. Data are mean ± S.E.M., log-rank test, P = 0.006. (D) Relative quantification of the innervation of NMJs of the gastrocnemius muscle in symptomatic mice (post-natal day 150). Denervated NMJs were decreased by ELP3 expression: SOD1G93A/ELP3 KI 78.4 ± 1.0%, n = 3 versus SOD1G93A/ELP3 KICAGG 59.9 ± 2.7%, n = 3. Data represent mean ± S.E.M., one-way ANOVA, P = 0.001. (E) Quantification of ventral horn lumbar motor neurons, in function of the cell body area, in symptomatic mice (post-natal day 150). No differences were found between genotypes. Data represent mean ± S.E.M., two-way ANOVA, n = 3, P > 0.5. (F, G) Effect of neuronal-specific tamoxifen-induced expression of ELP3 in SOD1G93A mice. Tamoxifen was administered at post-natal day 60. (F) Mean disease onset was not affected by ELP3 expression: SOD1G93A/ELP3 KI 89.1 ± 3.7 days, n = 15 versus SOD1G93A/ELP3 KIThy1 94.2 ± 1.9 days, n = 28. Data are mean ± S.E.M., log-rank test, P = 0.383. (G) Mean survival was not affected by ELP3 expression: SOD1G93A/ELP3 KI 149.0 ± 2.4 days, n = 13 versus SOD1G93A/ELP3 KIThy1 149.1 ± 1.8 days, n = 18. Data are mean ± S.E.M., log-rank test, P = 0.982.
We then evaluated the effect of ubiquitous ELP3 expression in the SOD1G93A mouse. ELP3 did not affect disease onset (SOD1G93A/ELP3 KI mean 92.3 ± 1.8 days; SOD1G93A/ELP3 KIGAGG mean 97.1 ± 2.0 days; P = 0.1671), but prolonged survival (SOD1G93A/ELP3 KI mean 152.0 ± 1.8 days; SOD1G93A/ELP3 KIGAGG mean 160.7 ± 1.9 days; P = 0.006) (Fig. 2B and C). This survival benefit was associated with a protection of neuromuscular innervation of the gastrocnemius muscle. Expression of ELP3 reduced the number of denervated neuromuscular junctions (NMJs) of the gastrocnemius muscle by 23.6% (SOD1G93A/ELP3 KI 78.4%; SOD1G93A/ELP3 KIGAGG 59.9%; P < 0.0001) (Fig. 2D). Nonetheless, the number of surviving spinal motor neurons at post-natal day 150 was not affected (Fig. 2E).
In addition, we studied the effect of virally expressed human ELP3 in the mutant SOD1 mouse. AAV9 viral vector expressing either ELP3 or GFP was delivered intrathecally in SOD1G93A neonates. We found the AAV9 viral vector to be transduced in the ventral spinal cord in a non-cell specific manner, targeting both motor neurons and astrocytes (Supplementary Material, Fig. S2A). Human ELP3 expression was detected in different tissues, such as brain, spinal cord and muscle, with variable transduction efficiency (Supplementary Material, Fig. S2B and C). Increasing the expression of ELP3 delayed disease onset by 12 days (SOD1G93A:: AAV9-GFP mean 98.4 ± 2.6 days; SOD1G93A:: AAV9-ELP3 mean 110.6 ± 3.0 days, P = 0.0029) and survival by 12.5 days (SOD1G93A:: AAV9-GFP mean 147.1 ± 1.6 days; SOD1G93A:: AAV9-ELP3 mean 154.9 ± 2.1 days, P = 0.0077) (Supplementary Material, Fig. S2D and E). Viral transduction with AAV-ELP3 decreased the number of denervated NMJs of the gastrocnemius muscle (SOD1G93A:: AAV9-GFP 69.1%; SOD1G93A:: AAV9-ELP3 53.0%, P < 0.0001). Similar to the results obtained in the SOD1G93A/ELP3 KI mice, the number of surviving spinal motor neurons was not affected (Supplementary Material, Fig. S2F and G).
Because the mechanism of ALS is known to be non-motor neuron autonomous, at least in SOD1-related ALS, and because of our finding that ELP3 levels are affected both in motor neurons and in glia, we wondered whether increasing the ELP3 levels only in motor neurons was sufficient to observe the protective effect found after ubiquitous expression. We therefore expressed ELP3 in neurons by the generation of a triple transgenic mouse, in which tamoxifen treatment resulted in the expression of ELP3 in Thy1-mediated CreER expressing neurons such as motor neurons (herein designated SOD1G93A/ELP3 KIThy1) (Supplementary Material, Fig. S1N–U). Interestingly, this selective induction of ELP3 expression did not affect disease onset or survival in the SOD1G93A mouse (Fig. 2F and G), likely because either the expression of ELP3 in motor neurons alone is not sufficient (or necessary) to prolong the survival of SOD1G93A mice, or the motor neuron death process affects the expression of ELP3 during its induction.
ELP3 deletion is detrimental
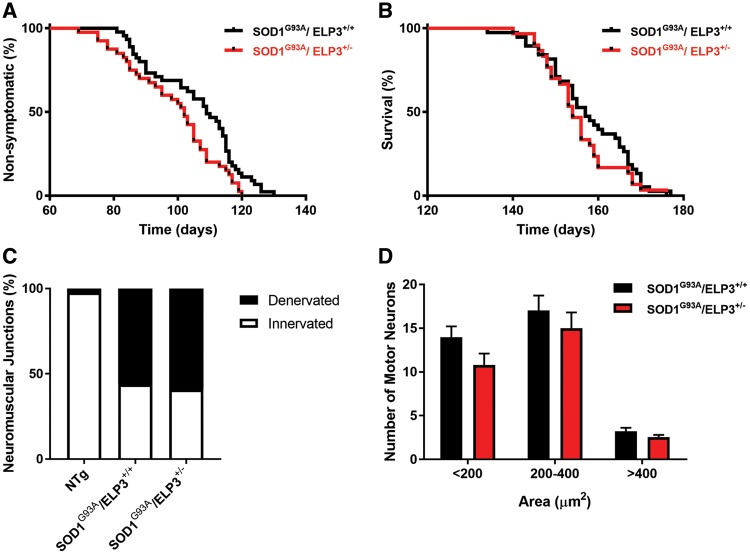
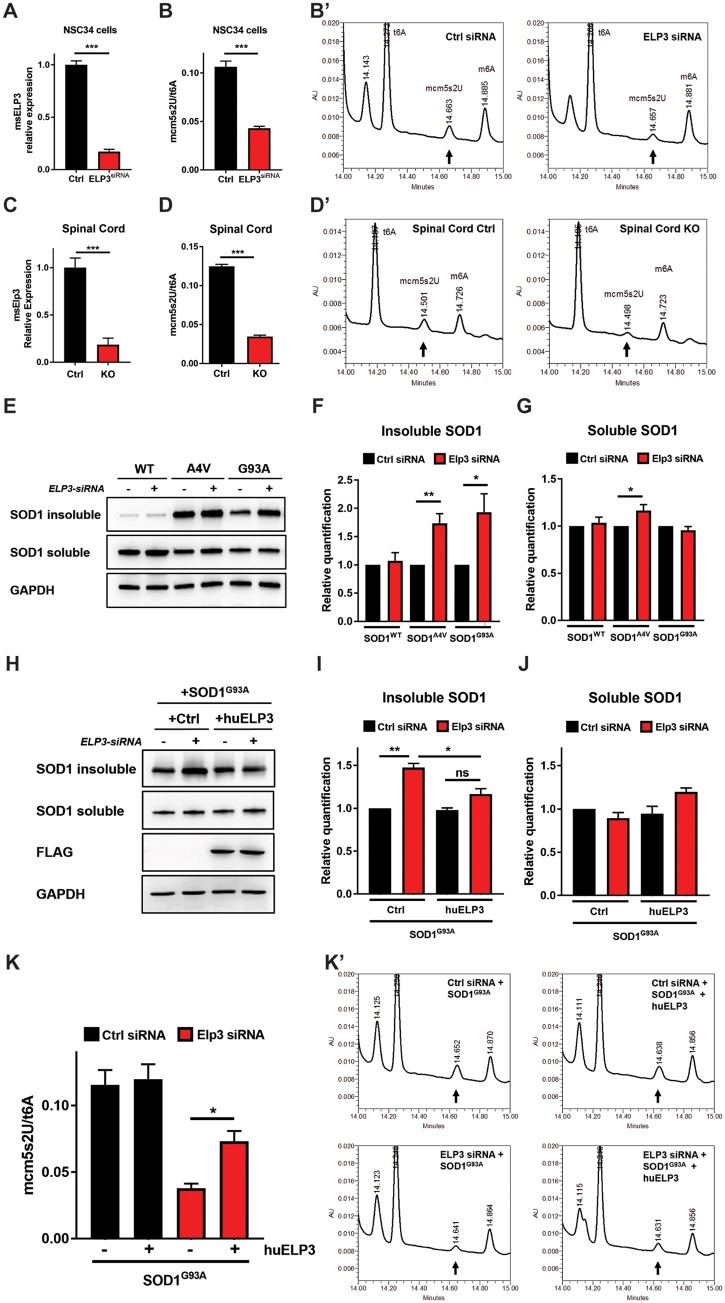
We next assessed the effect of reducing ELP3 expression in the SOD1G93A mouse. To do so, we generated an ELP3 knockout mouse, using the gene trapping technique (25). We found that absence of ELP3 is lethal around embryonic day 8, as we did not find gene-trap homozygous embryos after this stage (Supplementary Material, Fig. S3, Table S1), confirming that ELP3 is essential for mammalian embryogenesis, as reported (26). Reduction of ELP3 expression by 50% (Supplementary Material, Fig. S3A–C) did not affect embryogenesis or result in gross abnormalities in adulthood. Constitutive heterozygous deletion of ELP3 accelerated disease onset (Fig. 3A) (109 days in SOD1G93A and 102 days in SOD1G93A/ELP3+/− mice; P = 0.0230), but did not affect survival (Fig. 3B) (157 days in SOD1G93A and 154 days in SOD1G93A/ELP3+/−; P = 0.27) in the SOD1G93A mouse. Motor neuron count and neuromuscular innervation (Fig. 3C and D) did not differ between the SOD1G93A and SOD1G93A/ELP3+/− mice, most likely a reflection of the limited effect size of reducing ELP3 by only 50%. Unfortunately, we were unable to evaluate the effect of ELP3 reduction by more than 50% in the adult SOD1G93A mouse, as both ubiquitous and neuron-specific conditional knockout of Elp3 in adult mice was rapidly fatal, a phenotype not affected by the presence of mutant SOD1 (Supplementary Material, Fig. S4). In summary, the hastening of disease onset due to partial loss of ELP3 in the SOD1G93A mouse hints for a deleterious effect of ELP3 reduction in this ALS model. Moreover, these results show that ELP3 is as essential in the adult organism as it is during development.
Figure 3.
Effect of heterozygous deletion of the ELP3 gene in the SOD1G93A mouse. (A) Mean disease onset was reduced by ELP3 heterozygous deletion: SOD1G93A/ELP3+/+ 105.8 ± 2.1 days, n = 45 versus SOD1G93A/ELP3+/− 98.7 ± 2.2 days, n = 40. Data are mean ± S.E.M., log-rank test, P = 0.0141. (B) Mean survival was not affected: SOD1G93A/ELP3+/+ 157.5 ± 1.6 days, n = 38 versus SOD1G93A/ELP3+/− 155.2 ± 1.5 days, n = 30. Data are mean ± S.E.M., log-rank test, P = 0.2307. (C) Relative quantification of the innervation of NMJs of the gastrocnemius muscle in symptomatic mice (post-natal day 104). Denervated NMJs were not affected: SOD1G93A/ELP3+/+ 43.7 ±3.5%, n = 6 versus SOD1G93A/ELP3+/− 40.7 ± 10.4%, n = 3. Data represent mean ± S.E.M., two-way ANOVA, P = 0.9173. (D) Quantification of ventral horn lumbar motor neurons, in function of the cell body area, in symptomatic mice (post-natal day 104). No differences were found between genotypes. Data represent mean ± S.E.M., two-way ANOVA, n = 3, P > 0.5.
The SAM domain mediates the protective effect of ELP3 in the zebrafish
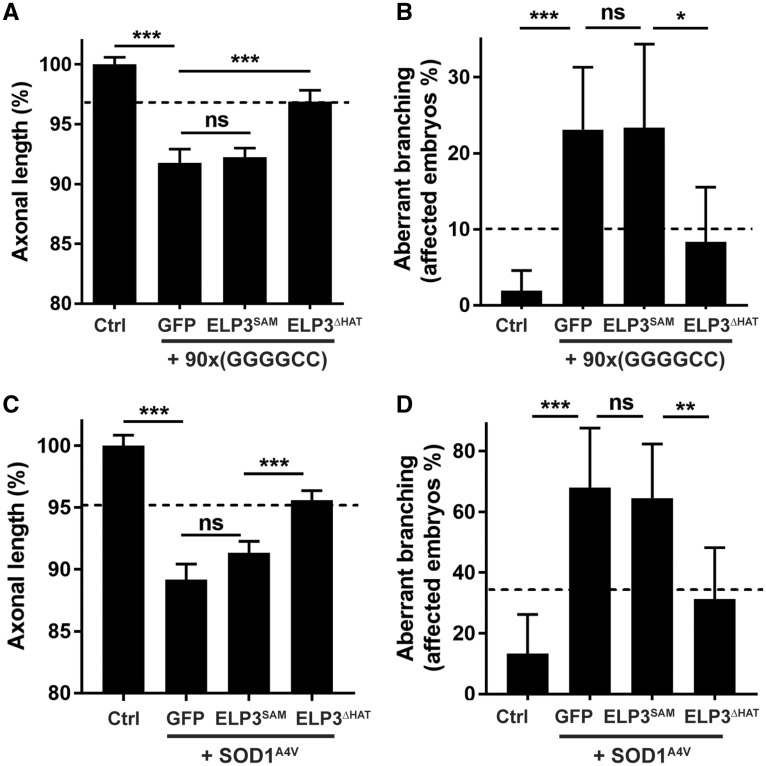
To gain insight into the molecular mechanism of ELP3, we evaluated which catalytic domain of ELP3 was mediating its protective effect. To do so, we mutated the SAM (C109/112S) and the HAT (Y529A) domains and also deleted the entire HAT domain (Supplementary Material, Fig. S5A and B), to generate proteins with reduced or no enzymatic activity (27–29). We tested the protective effect of these constructs in the C9ALS and the SOD1A4V zebrafish models. ELP3 lacking the HAT domain (ELP3ΔHAT) prevented the motor axonopathy to the same extent as the full-length ELP3 (Fig. 4), as well as the ELP3Y529A mutant (Supplementary Material, Fig. S5D). In contrast, ELP3 in which the SAM domain was inactivated (ELP3SAM) lost its protective activity in both models (Fig. 4).
Figure 4.
SAM domain, not HAT domain, mediates ELP3 protective effect in the zebrafish. Effect of ELP3 mutants in the axonopathy induced by C9orf72-associated repeat RNA (A and B) or SOD1A4V RNA (C and D), assessed by quantification of motor axon length (A and C) and affected embryos/axonal branching quantification (B and D). Deletion of the HAT domain (ELP3ΔHAT) does not affect the protective effect of ELP3 (dashed lines), whereas mutation of the SAM domain (ELP3SAM) abolishes the protective effect of ELP3. 3x(GGGGCC) RNA and SOD1WT were used as controls. Data represent mean ± S.E.M., one-way ANOVA (A and C) or mean ± 95% CI, logistic regression (B and D). n ≥ 45. ***P < 0.001, **P < 0.01, *P < 0.05.
As expected, the axonopathy induced by knockdown of ELP3 in zebrafish (3) could also be prevented by ELP3 overexpression (Supplementary Material, Fig. S5C). Of note, the SAM domain was necessary for this effect as well, as it was lost for ELP3 lacking an active SAM domain, but not for ELP3 lacking an active HAT domain (Supplementary Material, Fig. S5C). These results demonstrate that in this zebrafish model, the SAM domain, but not the HAT domain, is necessary for the protective effect of ELP3.
ELP3 regulates wobble mcm5s2U tRNA modification and affects mutant SOD1 aggregation in vitro
The modification of the wobble uridine in tRNA is the main function of ELP3. In FD, a disease caused by loss-of-function mutations in ELP1 resulting in loss of activity of the elongator complex in the nervous system, the mcm5s2U modification of tRNA is reduced (17). Interestingly, ablation of ELP3 in yeast and mice impaired proteome integrity and increases protein aggregation, by affecting codon translation rates due to defective wobble mcm5s2U tRNA modification (15,30). To gain insight into the molecular mechanism underlying the protective effect of ELP3 in ALS, we investigated the relation between ELP3 expression, wobble uridine modification and protein aggregation.
We first examined the effect of ELP3 depletion on mcm5s2U levels. In NSC34 cells, reduction of ELP3 expression by 83% dramatically decreased the levels of mcm5s2U by 60% (Fig. 5A and B). In the mouse, the lethal reduction of ELP3 levels by 90% [tamoxifen-treated ELP3lox/lox/CAGGCreER (KO) mice] reduced mcm5s2U levels in the spinal cord and brain by 72% and 75%, respectively, compared to control mice (Fig. 5C and D;Supplementary Material, Fig. S6A and B). These results confirm that ELP3 regulates the mcm5s2U wobble modification of tRNA in vitro and in vivo. In the wild-type mouse, the pool of tRNAs that should have this modification appears to be already fully modified, as conditional ubiquitous expression of human ELP3 did not increase the levels of mcm5s2U any further (Supplementary Material, Fig. S6C and D).
Figure 5.
Effect of ELP3 expression levels on wobble mcm5s2U tRNA modification and protein aggregation. (A) ELP3 mRNA relative expression in NSC34 cells, 72 h post-transfection. Levels of ELP3 were reduced by 85.0 ± 2.2% in cells transfected with ELP3-siRNA, compared to control cells. Data represent mean ± S.E.M., unpaired t-test, n = 5. ***P < 0.001. (B) mcm5s2U levels in NSC34 cells were reduced by 60.1 ± 2.1% in cells transfected with ELP3-siRNA, compared to control cells. t6A was used as internal control. Data represent mean ± S.E.M., unpaired ttest, n = 4. ***P < 0.001. (B′) Representative mcm5s2U chromatograms of the conditions analyzed in B. (C) ELP3 mRNA relative expression in spinal cord after ubiquitous tamoxifen-induced deletion of endogenous ELP3 (ELP3lox/lox/CAGGCreER mouse, designated ELP3 KO). Levels were reduced by 91.5 ± 3.9%. Data represent mean ± S.E.M., unpaired t-test, n = 3. ***P < 0.001. (D) mcm5s2U levels in spinal cord after ubiquitous tamoxifen-induced deletion of endogenous ELP3. Levels were reduced by 72.2 ± 1.7%. t6A was used as internal control. Data represent mean ± S.E.M., unpaired t-test, n = 3. ***P < 0.001. (D′) Representative mcm5s2U chromatograms of the conditions analyzed in (D). (E–J) Western blot analyses and quantification of SOD1 protein in detergent-insoluble and detergent-soluble fractions of NSC34 cells. (E–G) Aggregates of mutant SOD1 (G93A and A4V) accumulate upon siRNA-mediated depletion of ELP3, compared to control (control-siRNA, –). GAPDH was used as loading control. Data represent mean ± S.E.M., one-way ANOVA, n ≥ 4. **P < 0.01, *P < 0.05. (H–J) ELP3 overexpression reduces the accumulation of SOD1G93A aggregates induced by ELP3 depletion. FLAG was used to control ELP3 overexpression, GAPDH was used as loading control. Data represent mean ± S.E.M., one-way ANOVA, n = 3. ***P < 0.001, **P < 0.01, *P < 0.05. (K) Effect of ELP3 overexpression on the levels of mcm5s2U in NSC34 cells expressing SOD1G93A and either ELP3-siRNA or control-siRNA. Data represent mean ± S.E.M., one-way ANOVA, n = 4. *P < 0.05. (K′) Representative mcm5s2U chromatograms of the conditions analyzed in (K). ***P < 0.001, **P < 0.01, *P < 0.05.
We next evaluated the effect of ELP3 depletion on protein aggregation. Silencing ELP3 in NSC34 cells increased the total amount of aggregating proteins by 28% (Supplementary Material, Fig. S7A and B). Moreover, it induced an increase of insoluble mutant human SOD1 (SOD1A4VA, 73.7%; SOD1G93A, 93.2%), but not of wild-type human SOD1 (Fig. 5E–G). In these conditions, exogenous expression of human ELP3 significantly reduced the amount of insoluble SOD1G93A to levels similar to control (Fig. 5H–J) and, concomitantly, it restored the levels of mcm5s2U by 45.5% (Fig. 5K). These results suggest that the protective effect of ELP3 in ALS may at least in part be mediated by affecting mutant SOD1 solubility through an effect on wobble mcm5s2U tRNA modification.
ELP3 levels correlate with tRNA mcm5s2U levels in the motor cortex of sporadic ALS patients
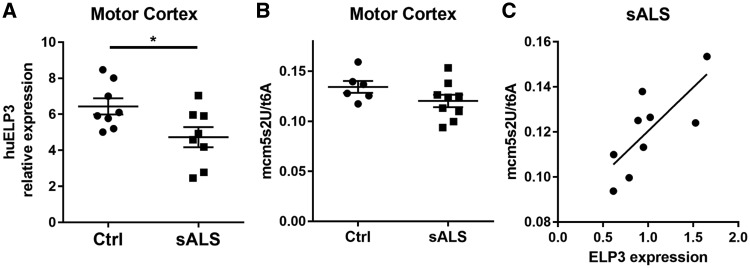
To investigate whether the levels of ELP3 and mcm5s2U tRNA are affected in ALS patients, we quantified the ELP3 mRNA in the motor cortex and the occipital cortex of sporadic ALS patients and compared it to controls. ELP3 expression was significantly reduced in the motor cortex, but not in the occipital cortex (Fig. 6A, Supplementary Material, Fig. S7C), a reduction that correlated with the levels of mcm5s2U, although mcm5s2U levels in sporadic ALS patients were not altered compared to controls (Fig. 6B and C). Together, these results suggest that ELP3 may act as a modifier of ALS through influencing tRNA modifications.
Figure 6.
ELP3 expression correlates with wobble mcm5s2U tRNA modification in sALS patients. (A) ELP3 mRNA relative expression in the motor cortex from sALS patients. Data represent mean ± S.E.M., unpaired t-test, n = 8. *P < 0.05. (B) mcm5s2U levels in the motor cortex from sALS patients. Data represent mean ± S.E.M., unpaired t-test, n ≥6. P > 0.05. (C) Levels of ELP3 mRNA correlate with mcm5s2U levels. Linear regression, n = 9, P = 0.0142.
Discussion
Loss of ELP3/elongator complex function is associated with detrimental phenotypes in yeast, fly, zebrafish, mice and humans (3,12,15). We therefore hypothesized that restoring ELP3 levels may be protective in neurodegeneration. We found ELP3 to be protective in two zebrafish ALS models, as well as in the SOD1G93A mouse. In the zebrafish, expression of human ELP3 prevented the toxicity induced by both SOD1A4V expression and 90x(GGGGCC) RNA, a novel zebrafish model for C9orf72 ALS. This result prompted us to investigate the role of ELP3 in the SOD1G93A mouse, in which we found the expression of ELP3 to be reduced in the brain and spinal cord. We expressed human ELP3 ubiquitously in pre-symptomatic (post-natal day 60) mice and found the survival to be moderately increased. The fact that the total expression of ELP3 after ubiquitous induction of human ELP3 was also moderately increased (not more than 1.5-fold of the endogenous ELP3 expression) underscores the biological relevance of these results.
A similar effect on survival was observed when human ELP3 was expressed in SOD1G93A neonates after intrathecal delivery of AAV9 viral vectors, although this effect was largely due to its effect on disease onset. The explanation for this effect on onset versus disease duration is unclear, although the difference between transgenic expression at post-natal day 60 versus viral expression at post-natal day 1 is likely to contribute to it. Nevertheless, these two approaches suggest that ELP3 expression in the SOD1G93A mouse is protective both in the pre-symptomatic stage and in the symptomatic stage. The expression of ELP3 in the SOD1G93A mouse was also associated with less denervated NMJs of the gastrocnemius muscle, compared to control mice. Together with the fact that the number of spinal motor neurons was unaffected, it suggests that ELP3 exerts its protective effect through maintaining axon integrity, more so than through maintaining motor neuron survival, and highlight the role of the NMJs as the main factor of progression and severity in ALS (31).These results are consistent with the findings from Gould et al., which dissociate motor dysfunction from motor neuron death (32). Ablation of Bax, a pro-apoptotic gene, in the SOD1G93A mouse, remarkably preserved almost completely the spinal motor neurons, but had limited effect in the reduction of the denervated NMJs and in increasing survival (32).
Moreover, we found that increasing ELP3 expression specifically in neurons in pre-symptomatic SOD1G93A mice, per se, was not sufficient to prolong their survival, suggesting a role for glial cells in the protective effect of ELP3. In agreement, the expression of ELP3 was reduced in SOD1G93A mice not only in spinal motor neurons, but, and to a larger extent, also in the glia surrounding them in which expression was reduced by more than 50%. We therefore hypothesize that increasing ELP3 expression in one or more cell types present in the glia would be protective in the SOD1G93A mouse, realizing that it is possible that the selective expression of ELP3 in either astrocytes, microglia or oligodendrocytes may not be sufficient to see the protective effect, but that a combined expression of ELP3 in different cell types is required. This is concurrent with the protective effect observed after AAV9-mediated ELP3 expression in the SOD1G93A mouse, as AAV9 viral vectors transduced nonspecifically motor neurons and glia in the spinal cord.
To confirm the role of ELP3 in neurodegeneration, we also investigated the effect of ELP3 depletion. We hypothesized that if low levels of ELP3 were associated with neurodegeneration in the fish, fly and humans (3), a reduction of ELP3 levels would exacerbate the phenotype of the SOD1G93A mouse. Heterozygous deletion of ELP3 did worsen SOD1G93A mouse phenotype, although to a limited extent. Homozygous deletion of ELP3 was embryonically lethal, as also observed when other members of the elongator complex are genetically ablated (16,26). In an attempt to bypass this lethality, we generated a conditional knockout mouse to delete ELP3 in adult mice. Interestingly, homozygous ubiquitous ELP3 deletion was very rapidly lethal in adult mice. Moreover, selective neuronal ELP3 depletion in the adult mouse was equally rapidly fatal and was accompanied by a severe neurological phenotype (Supplementary Material, Fig. S4). The Thy1CreER mouse used in this study, the SLICK-H mouse, displays strong and widespread Cre recombination in the cortex, cerebellum, hippocampus and spinal motor neurons (about 90%, data not shown) (33). Of note, several reports confirm the depletion of an elongator complex gene from a tissue/organ to be deleterious, with different outcomes (14,15,22,34,35). This rapidly progressive phenotype made the study of the effect of ELP3 reduction by more than 50% in the SOD1G93A mouse not feasible. Overall, our data confirm that ELP3 is an essential gene, both in mouse embryos and adult mice.
The molecular mechanism of the protective effect of ELP3 in ALS models was further investigated. ELP3 is the catalytic subunit of the elongator complex. In a recent study, it has been shown that defective wobble uridine tRNA modification results in slower codon translation rates which impair the proteasome, thus increasing protein aggregation (30). It is known that ELP3 plays a major role in this modification process (12). In agreement, we found that the levels of the mcm5s2U tRNA modification were largely reduced after depletion of ELP3 not only in NSC34 cells, but also in mice. Moreover, we found protein aggregation to be increased upon ELP3 ablation in NSC34 cells, similarly to yeast (30). Importantly, ELP3 depletion from NSC34 cells increased the amount of insoluble mutant SOD1, but not of wild-type SOD1. Remarkably, the effects of ELP3 depletion on mcm5s2U levels and protein aggregation in NSC34 cells were reverted by overexpressing human ELP3. Although the levels were not fully restored to control, likely due to imbalance of the elongator complex, they clearly implicate ELP3 in protein aggregation possibly through regulation of mcm5s2U tRNA modifications.
There are two catalytic domains in ELP3, the SAM and the HAT domain. Loss-of-function mutations in the SAM domain not only directly impair the catalytic activity of ELP3, but also compromise the integrity of the yeast elongator complex (18). We found that the SAM domain was necessary for the protective effect of ELP3 in the fish, and that this effect was completely independent of the HAT domain. This was unexpected as, in yeast, loss-of-function mutations in both domains of ELP3 equally affect the formation of modified wobble tRNA uredines (29). Our data suggest that at least in fish, the HAT domain is not necessary for the observed neuroprotective function of ELP3/elongator complex.
We next checked the levels of ELP3 and modified tRNA uridines in sporadic ALS patients. We found ELP3 to be reduced in the motor cortex of sALS patients, but not in the occipital cortex. Interestingly, the mcm5s2U levels correlated with ELP3 levels, indicating that mcm5s2U tRNA modifications may contribute to the pathogenesis of ALS.
Proteasome impairment is considered as one of the pathogenic factors at play in ALS. It can be triggered by different mechanisms, ER stress and a heat-shock response amongst others, resulting in the aggregation of susceptible proteins, such as SOD1, TDP43 and FUS (36). It is known that defective wobble mcm5s2U modification reduces the speed of codon translation rates (15,30), which in turn triggers proteome impairment (30). Therefore, we propose that ELP3 modifies the pathogenesis of ALS, by affecting the aggregation of susceptible proteins, through control of mcm5s2U wobble tRNA modification. Our data suggest an unexpected link between tRNA modification and motor neuron degeneration.
Materials and Methods
Plasmids, morpholinos and in vitro RNA transcription
The plasmid encoding human ELP3-FLAG under the control of a T7 promoter was purchased from Origene (Rockville, MD). The plasmids encoding ELP3 SAM (C109/112S) and ELP3 HAT (Y529A) were generated from the human ELP3-FLAG plasmid with QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). To generate the ELP3 ΔHAT vector, the human ELP3-FLAG plasmid was digested with XhoI (Thermo Fisher Scientific, Waltham, MA) and the resulting fragment subcloned into the XhoI-digested pCMV6-Entry vector, under control of a T7 promoter (Origene). To generate the AAV: ELP3-FLAG transfer plasmid, the human ELP3-FLAG plasmid was PCR-amplified with the primers 5′ CGTGCTCTAGAATGAGGCAGAAGCGGAAAGGAG and 5′ GGTACGTGACTAGTGCCGGCCGTTTAAACCTTATCG and the resulting product ligated into XbaI/SpeI (Thermo) digested AAV transfer plasmid (pZac 2.1 eGFP3 SEED). Human cDNA of SOD1WT, SOD1A4V and SOD1G93A in the pCLneo vector were kind gifts from R.H. Brown Jr (Harvard Medical School, Harvard, MA). SOD1 was cloned in the pBCM vector under control of a T3 promoter. The p90HNR construct to generate the ∼90 GGGGCC repeat RNA fragment was obtained as described (24). To synthesize RNA, all plasmids were linearized by restriction digestion, transcribed with the mMESSAGE mMACHINE®T7 or T3 kit (Ambion, Huntingdon, UK) and the resulting RNA purified with the MEGAclear™ Kit (Ambion). RNA concentration was determined by spectrophotometry (Nanodrop, Thermo). The antisense morpholino (Mo) against the start codon (ELP3-Mo; 5′-TGGCTTTCCCATCTTAGACACAATC-3′) of zebrafish ELP3 and the non-targeting control morpholino (Ctrl-Mo; 5′-CCTCTTACCTCAGTTACAATTTATA-3′) were designed and obtained from Gene Tools (Philomath, OR).
rAAV production
Recombinant adeno-associated virus (rAAV) production was performed by the Neurobiology and Gene Therapy laboratory (Katholieke Universiteit Leuven, Belgium) as described (37). The plasmids used were the construct for the AAV2/9 serotype, the pAdvDeltaF6 adenoviral helper plasmid and the AAV transfer plasmid, encoding either human ELP3 (AAV: ELP3-FLAG) or eGFP (AAV: eGFP) under control of the cytomegalovirus (CMV) promoter. Genomic copy number (GC/ml) was determined by qPCR.
Animals
Adult zebrafish (AB strain) and embryos were maintained and staged under standard laboratory conditions (38). For the intrathecal delivery of rAAV vectors, mice neonates (P0.5) were cryoanesthetized and immobilized on a styrofoam board. A total of 1 × 1010 GC of rAAV vectors were injected in the spinal canal at the lumbar level, as reported previously (39,40), with a 32G Hamilton syringe (Hamilton, Reno, NV). The maximum volume injected was 7 µl. The dye lissamine green (Sigma, ST. Louis, MO) was added to the rAAV vectors at a final concentration of 0.05% (w/v) in PBS to assess delivery accuracy. The procedure was considered successful when the rAAV vectors diffused along the spinal tract into the cerebellum/brain, and only neonates successfully injected were considered for this study. The human ELP3 conditional overexpressing mouse was generated by Ozgene (Perth, Australia). In brief, ELP3-FLAG cDNA preceded by a loxP site flanked stop cassette was inserted in the ROSA26 locus by homologous recombination (designated ELP3 KI mouse). The Elp3 conditional knockout mouse (designated ELP3lox/lox mouse) was generated by insertion of loxP sequences flanking the exon 2 of Elp3 (15). The SIGTR embryonic stem cell line AQ0461 (Mutant Mouse Recourse and Research Center, USA), containing a gene-trap cassette at intron 1 of the Elp3 gene, was used to generate the constitutive ELP3 knockout mouse. Genotyping of the constitutive ELP3 knockout mice generated by heterozygous pairings was performed using the following primers: TRAP cassette, 5′ TTA TCG ATG AGC GTG GTG GTT ATG C and 5′ GCG CGT ACATCG GGC AAA TAA TATC; Elp3 exon 1–3, 5′ TGAGGCAAAAGAGGAAAGGGGAC; 5′ CGGAGTCAGGTCCTCCGGGGC. Heterozygous mice were backcrossed for eight generations into a C57BL/6J background. The human mutant SOD1 overexpressing mouse [B6.Cg-Tg(SOD1*G93A)1Gur/J stock number 004435], the human wild-type SOD1 over-expressing mouse [B6.Cg-Tg(SOD1)2Gur/J stock number 002298], the CAGG-CreER mouse [B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J stock number 004682] and the Thy1-CreER-EYFP mouse (SLICK-H) [Tg(Thy1-cre/ERT2,-EYFP)HGfng/PyngJ stock number 012708] were purchased from the Jackson Laboratory (Ben Harbor, ME). To induce Cre-mediated recombination, mice were treated with tamoxifen (200 mg/kg/day) by oral gavage for four consecutive days. The PaGE (Paw Grip Endurance) test was used to determine disease onset by assessing the ability of the mice to hold their own weight for 60 s. In brief, the mice are placed on a wire grid, 50 cm above the platform, and turned over while hanging upside–down. When a mouse fails (drops from the grid before 60 s) and in consecutive trials cannot hold its own weight for 60 s, it is defined as symptomatic. End stage, used as a measurement of survival, was defined as the age at which mice could no longer right themselves within 30 s when placed on their back. Mice phenotypic analyses were performed in a blinded fashion. Mice were euthanized at end stage, as it corresponds to the human end-point. All mice were on a C57BL/6 background. Mice were housed in standard conditions in the specific-pathogen-free housing facility of the University of Leuven with food and water ad libitum. Experiments were performed according to the guidelines of the University of Leuven and have received ethical committee approval (project codes 230/2013, 237/2013, 125/2014 and 126/2014).
Zebrafish staining
Zebrafish work was performed as described previously (23). Briefly, 1–2 cell stage zebrafish embryos were injected with 1.76 nl of morpholino or in vitro transcribed RNA: 3x(GGGGCC), 318 pg; 90x(GGGGCC), 318 pg; SOD1WT, 1.76 ng; SOD1A4V, 1.76 ng; ELP3WT, 176 pg; ELP3ΔHAT, 176 pg; ELP3SAM, 176 pg; ELP3Y529A, 176 pg; ELP3-Mo, 6 ng; Ctrl-Mo, 6 ng. A GFP-encoding RNA was used as control when morpholinos were not injected. For whole mount staining, morphologically normal fish were dechorionated, deyolked and fixed at 30 h post fertilization (hpf) in 4% formaldehyde in phosphate-buffered saline (PBS) overnight at 4˚C. Fish were permeabilized with acetone for 1 h at −20˚C, blocked with 1%BSA/1%DMSO/PBS for 1 h at room temperature and immunostained with mouse anti-SV2 (1: 200; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA) and secondary Alexa Fluor 555 anti-mouse antibody (1: 500; Molecular Probes, Eugene, OR). Axonal length and axonal branching of motor neurons were quantified as described previously (23) using Lucia software (version 4.60, Laboratory Imaging, Prague, Czech Republic). A minimum of 15 fish per condition was analyzed per experiment.
Histological analyses
To visualize NMJs, mice were deeply anesthetized with Nembutal 10%, the gastrocnemius muscle dissected and snap frozen in liquid nitrogen-cooled isopentane. Longitudinal cryosections of 20 µm (Slee cryostat, Mainz, Germany) were fixed with 4% formaldehyde (FA) in PBS for 10 min at room temperature, blocked with 10% normal donkey serum (Sigma) in PBS-0.1% Triton X-100 (PBS-T) for 1 h at room temperature and immunostained with rabbit anti-NF-200 (1:200, # N4142, Sigma) for 16 h at 4˚C, followed by 1 h incubation at room temperature with Alexa 555 conjugated α-bungarotoxin (1: 500, # B35451 Invitrogen) and anti-rabbit Alexa 488. NMJs were counted from five sections for each muscle, using a Zeiss Imager M1 microscope and an AxioCam Mrc5 camera (Carl Zeiss, Oberkochen, Germany).
To visualize spinal cord neurons, mice were anesthetized with Nembutal 10%, transcardially perfused with PBS and subsequently with 4% FA in PBS. The lumbar spinal cord was dissected, fixed with 4% FA for 16 h at 4˚C, dehydrated in 30% sucrose and snap frozen in liquid nitrogen-cooled isopentane. Two methods were followed. To measure spinal cord neurons, cross cryosections of 20 µm were stained with cresyl violet (Sigma), followed by immersion in a 70% ethanol/10% acetic acid solution and dehydration in an increased ethanol concentration series. Sections were then mounted with PerTex® (Histolab AB, Goteborg, Sweden) and neurons visualized with a Zeiss Imager M1 microscope. Images were obtained with an AxioCam Mrc5 camera (Carl Zeiss) and the soma area of normal appearing neurons in the ventral horn of the spinal cord was measured with the AxioVision 4.8 software (Carl Zeiss). Every 10th section for a total of 10 sections were analyzed per animal. To immunolabel spinal cord neurons, cross cryosections of 20 µm were blocked with 10% normal donkey serum in PBS-T for 1 h at room temperature and immunostained with rabbit anti-Choline acetyltransferase (ChaT, 1: 500) (#AB144P, Millipore) for 16 h at 4˚C, followed by 1 h incubation at room temperature with anti-rabbit Alexa 488. Slides were mounted using ProLong Gold antifade reagent with DAPI (Thermo). Images were obtained using a Zeiss Imager M1 microscope and an AxioCam Mrc5 camera. For pathological analysis, mice were euthanized by CO2 asphyxiation followed by complete pathological examination as described previously (41). Briefly, samples were immersion fixed in 10% neutral buffered formalin (Sigma), routinely processed for paraffin embedding, sectioned at 5 μm and stained with Hematoxylin (Diapath, Martinengo, Italy) and Eosin (Diapath). Head and sternum were decalcified in a 14% solution of tetrasodium EDTA for 15 days before processing and paraffin embedding.
Laser capture microdissection
To microdissect spinal cord neurons, five non-transgenic and five SOD1G93A mice were anesthetized with Nembutal 10% and the lumbar spinal cord was dissected and embedded in optimal cutting temperature (OCT) compound. Cross cryosections of 20 µm, collected on 1.0 PEN slides (Zeiss), were stained with cresyl violet as described above. Per spinal cord, a total of 1500 motor neurons from the ventral horn, and the surrounding glia, were microdissected (PALM RoboSoftware, Zeiss) and collected in Adhesive Cap 500 opaque tubes (Zeiss).
Gene expression analyses
Total RNA was extracted with Trizol (Thermo) and precipitated with isopropyl alcohol according to the manufacturer’s instructions. cDNA was synthesized from 1 μg of total RNA with SuperScript III Reverse transcriptase (Thermo) and random hexamer primers, according to the manufacturer’s instructions. Quantitative PCR was performed by the StepOnePlus™ (Life Technologies) with TaqMan® Fast Universal PCR Master Mix (Life Technologies). Relative gene expression was determined by the 2−ΔΔct method and normalized to the average of the control group. Graphs represent the relative gene expression as calculated by the Polr2a expression. Determining relative expression by G6pdx or GAPDH confirmed the differences between the genotypes as detected with Polr2a. Digital droplet PCR was performed following Bio-Rad guidelines (Bio-Rad, Hercules, CA), using ddPCR Supermix (without dUTPs) for probes. Droplets were generated in a QX200 Droplet Generator and scanned with a QX200 Droplet Reader (Bio-Rad). Results were analyzed with QuantaSoft software from Bio-Rad. Gene expression assays were purchased from Life Technologies: Human ELP3, Hs00216429_m1 and Hs00986853_m1; mouse ELP3, Mm00804538_m1; Human SOD1, Hs00533490_m1; mouse Chat, Mm01221880_m1; mouse Slc1a2, Mm01275814_m1; Human PolR2a, Hs00172187_m1; mouse PolR2a, Mm00839502_m1, Human GAPDH, Hs99999905_m1 and mouse G6pdx, Mm00656735_g1. The Polr2a-HEX labeled Taqman assay Mm.PT.58.13811327 for the digital droplet PCR was from integrated DNA Technologies (Leuven, Belgium).
HPLC and UPLC quantification of tRNA modified uridines
Total RNA was extracted from tissue with Trizol and precipitated with isopropyl alcohol according to the manufacturer’s instructions. Total tRNA was isolated using Nucleobond RNA/DNA 80 columns (Macherey-Nagel, Düren, Germany) and precipitated with isopropyl alcohol. Total tRNA was digested to nucleosides using nuclease P1 (Sigma) and bacterial alkaline phosphatase (Sigma) and analyzed as described earlier (42). t6A was used as internal control to normalize mcm5s2U quantification.
Cell culture and transfections
NSC34 cells (Mouse Motor Neuron-Like Hybrid Cell Line, Cedarlane Laboratories, Ontario, Canada) were cultured in high glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (Greiner), 4 mm Glutamax (Invitrogen), penicillin (100 U/mL), streptomycin (100 μg/mL) and non-essential amino acids (1%). Cells were grown at 37˚C in a humidified atmosphere with 5% CO2. Cells were transiently transfected with siRNA (mouse ELP3 or mouse negative control, Ambion) and/or plasmids (indicated where appropriate) using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions, and collected 48 or 72 h after.
Protein analyses
Mouse tissue was homogenized in T-PER buffer (Thermo Fisher Scientific) supplemented with protease inhibitors (Complete, Roche) by mechanical disruption for 30 s (MagNA Lyser, Roche). NSC34 cells were lysed in the same lysis buffer. To isolate protein aggregates, lysates were sonicated (five times, duty cycle 50%, level 4) (Branson, Danbury, CT) and analyzed for protein concentration using the BSA protein assay (ThermoScientific). Samples with equal amount of protein were centrifuged at 20 000g for 30 min at 4°C. The supernatant was collected as the soluble fraction (containing ‘soluble SOD1’) and the pellet was washed once with 1 ml of ice-cold lysis buffer, centrifuged again at 20 000g for 30 min at 4°C. The pellet (detergent-resistant or insoluble fraction, containing ‘insoluble SOD1’) was then resuspended in 5% of the original lysis volume with 2× Laemmli buffer (Bio-Rad) and sonicated (three times, duty cycle 50%, level 3). When isolation of protein aggregates was not performed, lysates were centrifuged at 11 000g for 15 min at 4°C and the supernatants analyzed for protein concentration. For the immunoprecipitation assay, lysate supernatants were incubated with anti-FLAG M2 agarose beads (Sigma) overnight at 4°C. The beads were washed five times with ice-cold PBS and the proteins were eluted using the 3× FLAG peptide, according to the manufacturer’s instructions.
Samples were resolved on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and either silver stained (Pierce Silver Stain Kit, Thermo) or transferred to a polyvinyldifluoride membrane using a semi-dry blotting apparatus (TE70XP, Hoefer, San Francisco, CA). After blocking with 5% (w/v) low-fat milk in Tris-buffered saline (10 mmol/l Tris, 150 mmol/L NaCl, pH 7.6) for 1 h, the membranes were incubated overnight at 4°C with the primary antibody (mouse anti-FLAG antibody, #F3165, Sigma; rabbit anti-SOD1, #ADI-SOD-100, Enzo Life Sciences, Farmingdale, NY; mouse anti-GAPDH, #AM4300, Ambion), washed in Tris-buffered saline 1% Tween-20 and incubated with HRP-conjugated secondary antibody (Dako). After incubation, membranes were washed, developed with enhanced chemiluminescence (ECL western blotting substrate, Thermo Scientific) and scanned using a LAS4000 Biomolecular imager (GE Healthcare, Uppsala, Sweden). Images were analyzed with ImageQuant TL software (GE Healthcare).
Nerve conduction studies
Mice were anesthetized with 1% isoflurane/O2 gas inhalation and placed on a heating pad to maintain body temperature. Nerve conduction was measured using subdermal needle electrodes (Technomed Europe) and a Medelec EMG monitor (Medelec Vickers, Sidcup, UK) compatible with Synergy software (version 20.1.0.100). Compound muscle action potentials (CMAPs) were measured by supramaximal stimulation (1 pulse/s, 0.1 ms stimulus duration), the stimulating electrode was placed at the sciatic notch and the recording electrode at the level of the gastrocnemius muscle. Sensory nerve action potentials (SNAPs) were measured by supramaximal stimulation (6 pulse/s, 0.1 ms stimulus duration) at the distal tip of the tail and were measured 4 cm proximal in the tail. For SNAP recordings, multiple traces were averaged (43).
Human samples
Samples were collected after obtaining informed consent from all human subjects. All experiments on human material were approved by the Ethical Committee of University Hospital Leuven, the London–Camberwell St. Giles Research Ethics Committee and the Institutional Review Board (IRB) of Harvard University.
Statistical analysis
Statistics were performed using Graphpad prism 7.01 software (Graphad Software, San Diego, CA) or RStudio (RStudio Team, Boston, MA). Survival and disease onset were analyzed using the log-rank test. Zebrafish aberrant branching were analyzed using logistic regression. Two-way ANOVA was used for neuron and NMJ innervation counts. Zebrafish axonal lengths, mRNA expression levels, protein expression levels and mcm5s2U levels were analyzed by one-way ANOVA. Significance level was defined at 0.05. For multiple comparisons, Tukey’s post-hoc correction was applied. Error bars in graphs are S.E.M. (or mean with 95% confidence interval for affected embryos) of at least 3 independent experiments.
Supplementary Material
Supplementary Material is available at HMG online.
Author Contributions
A.B.A. performed most experiments, analyzed the data and wrote the manuscript. G.J., B.S., S.H., A.J., K.S., I.T., C.E., L.S. and L.R. performed some experiments. R.N. and M.T. are laboratory technicians and performed experiments. A.C. and L.N. provided the knockout mouse. J.R. and A.A.C. provided human samples. R.L., D.C., L.V.D.B., P.V.D. and A.B. supervised some experiments. W.R. supervised and wrote the manuscript. All authors contributed to the final manuscript.
Conflict of Interest statement .None declared.
Funding
This work was supported by the European Research Council under the European’s Seventh Framework Programme (FP7/2007–2013) under the Euro-MOTOR project (Grant agreement No: 259867) and the ERC (grant agreement n° 340429), the Horizon 2020 Programme (H2020-PHC-2014-two-stage; grant agreement number 633413) and an EU Joint Programme-Neurodegenerative Disease Research (JPND) project, www.jpnd.eu (Belgium: FWO G.A001.14JP ND-RiMod-FTD and the Flemish Government initiated Flanders Impulse Program on Networks for Dementia Research (VIND-135043); United Kingdom: Medical Research Council (MR/L501529/1), STRENGTH, and Economic and Social Research Council (ES/L008238/1), ALS-CarE, the Research Foundation Flanders (FWO Flanders, G.0983.14N), the University of Leuven (GOA/11/014), the Interuniversity Attraction Poles program (P7/16) of the Belgian Federal Science Policy Office, the ALS Liga (Belgium), the Association Belge contre les Maladies Neuro-Musculaires (ABMM) and the ‘Opening the Future’ Fund. W.R. was supported through the E. von Behring Chair for Neuromuscular and Neurodegenerative Disorders and the ‘Hart voor ALS’ Fund, KU Leuven. P.V.D. and R.L. are Senior Clinical Investigators of FWO-Flanders. R.L. was supported through ‘Fonds Annie Planckaert-Dewaele’. A.J. and A.A.C. receive salary support from the National Institute for Health Research (NIHR) Biomedical Research Centre at South London and Maudsley NHS Foundation Trust and King’s College London. K.S. is funded by the MDA development award. A.B. was supported by grants from the Swedish Research Council (2016–03949), Karin and Harald Silvanders Foundation and Insamlingsstiftelsen Umeå University. Funding to pay the Open Access publication charges for this article was provided by the European Commission.
Supplementary Material
References
- 1. Robberecht W., Philips T. (2013) The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci., 14, 248–264. [DOI] [PubMed] [Google Scholar]
- 2. Swinnen B., Robberecht W. (2014) The phenotypic variability of amyotrophic lateral sclerosis. Nat. Rev. Neurol., 10, 661–670. [DOI] [PubMed] [Google Scholar]
- 3. Simpson C.L., Lemmens R., Miskiewicz K., Broom W.J., Hansen V.K., van Vught P.W., Landers J.E., Sapp P., Van Den Bosch L., Knight J.. et al. (2009) Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum. Mol. Genet., 18, 472–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. van Blitterswijk M., Mullen B., Wojtas A., Heckman M.G., Diehl N.N., Baker M.C., DeJesus-Hernandez M., Brown P.H., Murray M.E., Hsiung G.Y.. et al. (2014) Genetic modifiers in carriers of repeat expansions in the C9ORF72 gene. Mol. Neurodegener., 9, 38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huang B., Johansson M.J., Bystrom A.S. (2005) An early step in wobble uridine tRNA modification requires the Elongator complex. RNA, 11, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esberg A., Huang B., Johansson M.J., Bystrom A.S. (2006) Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol. Cell, 24, 139–148. [DOI] [PubMed] [Google Scholar]
- 7. Bjork G.R., Huang B., Persson O.P., Bystrom A.S. (2007) A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA, 13, 1245–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen C., Tuck S., Bystrom A.S. (2009) Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet., 5, e1000561.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fernandez-Vazquez J., Vargas-Perez I., Sanso M., Buhne K., Carmona M., Paulo E., Hermand D., Rodriguez-Gabriel M., Ayte J., Leidel S.. et al. (2013) Modification of tRNA(Lys) UUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet., 9, e1003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johansson M.J., Esberg A., Huang B., Bjork G.R., Bystrom A.S. (2008) Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol. Cell Biol., 28, 3301–3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tukenmez H., Xu H., Esberg A., Bystrom A.S. (2015) The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res., 43, 9489–9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karlsborn T., Tukenmez H., Mahmud A.K., Xu F., Xu H., Bystrom A.S. (2014) Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol., 11, 1519–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Selvadurai K., Wang P., Seimetz J., Huang R.H. (2014) Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem.Biol., 10, 810–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Creppe C., Malinouskaya L., Volvert M.L., Gillard M., Close P., Malaise O., Laguesse S., Cornez I., Rahmouni S., Ormenese S.. et al. (2009) Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell, 136, 551–564. [DOI] [PubMed] [Google Scholar]
- 15. Laguesse S., Creppe C., Nedialkova D.D., Prevot P.P., Borgs L., Huysseune S., Franco B., Duysens G., Krusy N., Lee G.. et al. (2015) A dynamic unfolded protein response contributes to the control of cortical neurogenesis. Dev. Cell, 35, 553–567. [DOI] [PubMed] [Google Scholar]
- 16. Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L.. et al. (2001) Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet., 68, 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karlsborn T., Tukenmez H., Chen C., Bystrom A.S. (2014) Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm(5)s(2)U in tRNA. Biochem. Biophys. Res. Commun., 454, 441–445. [DOI] [PubMed] [Google Scholar]
- 18. Greenwood C., Selth L.A., Dirac-Svejstrup A.B., Svejstrup J.Q. (2009) An iron-sulfur cluster domain in Elp3 important for the structural integrity of elongator. J. Biol. Chem., 284, 141–149. [DOI] [PubMed] [Google Scholar]
- 19. Winkler G.S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J.Q. (2002) Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc. Natl. Acad. Sci. U.S.A., 99, 3517–3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pokholok D.K., Hannett N.M., Young R.A. (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell, 9, 799–809. [DOI] [PubMed] [Google Scholar]
- 21. Miśkiewicz K., Jose L.E., Bento-Abreu A., Fislage M., Taes I., Kasprowicz J., Swerts J., Sigrist S., Versées W., Robberecht W.. et al. (2011) ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron, 72, 776–788. [DOI] [PubMed] [Google Scholar]
- 22. Tielens S., Huysseune S., Godin J.D., Chariot A., Malgrange B., Nguyen L. (2016) Elongator controls cortical interneuron migration by regulating actomyosin dynamics. Cell Res., 26, 1131–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lemmens R., Van Hoecke A., Hersmus N., Geelen V., D'Hollander I., Thijs V., Van Den Bosch L., Carmeliet P., Robberecht W. (2007) Overexpression of mutant superoxide dismutase 1 causes a motor axonopathy in the zebrafish. Hum. Mol. Genet., 16, 2359–2365. [DOI] [PubMed] [Google Scholar]
- 24. Swinnen B., Bento-Abreu A., Gendron T.F., Boeynaems S., Bogaert E., Nuyts R., Timmers M., Scheveneels W., Hersmus N., Wang J.. et al. (2018) A zebrafish model for C9orf72 ALS reveals RNA toxicity as a pathogenic mechanism. Acta Neuropathol, doi: 10.1007/s00401-017-1796-5. [DOI] [PubMed] [Google Scholar]
- 25. Stanford W.L., Cohn J.B., Cordes S.P. (2001) Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet., 2, 756–768. [DOI] [PubMed] [Google Scholar]
- 26. Yoo H., Son D., Jang Y.J., Hong K. (2016) Indispensable role for mouse ELP3 in embryonic stem cell maintenance and early development. Biochem. Biophys. Res. Commun., 478, 631–636. [DOI] [PubMed] [Google Scholar]
- 27. Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y.. A role for the elongator complex in zygotic paternal genome demethylation. Nature, 463, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wittschieben B.O., Fellows J., Du W., Stillman D.J., Svejstrup J.Q. (2000) Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J., 19, 3060–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen C., Huang B., Eliasson M., Ryden P., Bystrom A.S. (2011) Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet., 7, e1002258.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nedialkova D.D., Leidel S.A. (2015) Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell, 161, 1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fischer L.R., Culver D.G., Tennant P., Davis A.A., Wang M., Castellano-Sanchez A., Khan J., Polak M.A., Glass J.D. (2004) Amyotrophic lateral sclerosis is a distal axonopathy: evidence in mice and man. Exp. Neurol., 185, 232–240. [DOI] [PubMed] [Google Scholar]
- 32. Gould T.W., Buss R.R., Vinsant S., Prevette D., Sun W., Knudson C.M., Milligan C.E., Oppenheim R.W. (2006) Complete dissociation of motor neuron death from motor dysfunction by Bax deletion in a mouse model of ALS. J. Neurosci., 26, 8774–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young P., Qiu L., Wang D., Zhao S., Gross J., Feng G. (2008) Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat. Neurosci., 11, 721–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin F.J., Shen L., Jang C.W., Falnes P.O., Zhang Y. (2013) Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet., 9, e1003516.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chaverra M., George L., Mergy M., Waller H., Kujawa K., Murnion C., Sharples E., Thorne J., Podgajny N., Grindeland A.. et al. (2017) The familial dysautonomia disease gene, Ikbkap/Elp1, is required in the developing and adult central nervous system. Dis. Model. Mech., 10, 605–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bento-Abreu A., Van Damme P., Van Den Bosch L., Robberecht W. (2010) The neurobiology of amyotrophic lateral sclerosis. Eur. J. Neurosci., 31, 2247–2265. [DOI] [PubMed] [Google Scholar]
- 37. Van der Perren A., Toelen J., Carlon M., Van den Haute C., Coun F., Heeman B., Reumers V., Vandenberghe L.H., Wilson J.M., Debyser Z.. et al. (2011) Efficient and stable transduction of dopaminergic neurons in rat substantia nigra by rAAV 2/1, 2/2, 2/5, 2/6.2, 2/7, 2/8 and 2/9. Gene Ther., 18, 517–527. [DOI] [PubMed] [Google Scholar]
- 38. Sprague J., Clements D., Conlin T., Edwards P., Frazer K., Schaper K., Segerdell E., Song P., Sprunger B., Westerfield M. (2003) The Zebrafish Information Network (ZFIN): the zebrafish model organism database. Nucleic Acids Res., 31, 241–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Elliger S.S., Elliger C.A., Aguilar C.P., Raju N.R., Watson G.L. (1999) Elimination of lysosomal storage in brains of MPS VII mice treated by intrathecal administration of an adeno-associated virus vector. Gene Ther., 6, 1175–1178. [DOI] [PubMed] [Google Scholar]
- 40. Fedorova E., Battini L., Prakash-Cheng A., Marras D., Gusella G.L. (2006) Lentiviral gene delivery to CNS by spinal intrathecal administration to neonatal mice. J. Gene Med., 8, 414–424. [DOI] [PubMed] [Google Scholar]
- 41. Radaelli E., Hermans E., Omodho L., Francis A., Vander Borght S., Marine J.C., van den Oord J., Amant F. (2015) Spontaneous post-transplant disorders in NOD.Cg-Prkdcscid Il2rgtm1Sug/JicTac (NOG) mice engrafted with patient-derived metastatic melanomas. PLoS One, 10, e0124974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bjork G.R., Jacobsson K., Nilsson K., Johansson M.J., Bystrom A.S., Persson O.P. (2001) A primordial tRNA modification required for the evolution of life? EMBO J., 20, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Benoy V., Vanden Berghe P., Jarpe M., Van Damme P., Robberecht W., Van Den Bosch L. (2016) Development of improved HDAC6 inhibitors as pharmacological therapy for axonal Charcot-Marie-Tooth disease. Neurotherapeutics., 14, 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.