Abstract
The recQ-like helicase BLM interacts directly with topoisomerase IIα to regulate chromosome breakage in human cells. We demonstrate that a phosphosite tri-serine cluster (S577/S579/S580) within the BLM topoisomerase IIα-interaction region is required for this function. Enzymatic activities of BLM and topoisomerase IIα are reciprocally stimulated in vitro by ten-fold for topoisomerase IIα decatenation/relaxation activity and three-fold for BLM unwinding of forked DNA duplex substrates. A BLM transgene encoding alanine substitutions of the tri-serine cluster in BLM−/− transfected cells increases micronuclei, DNA double strand breaks and anaphase ultra-fine bridges (UFBs), and decreases cellular co-localization of BLM with topoisomerase IIα. In vitro, these substitutions significantly reduce the topoisomerase IIα−mediated stimulation of BLM unwinding of forked DNA duplexes. Substitution of the tri-serine cluster with aspartic acids to mimic serine phosphorylation reverses these effects in vitro and in vivo. Our findings implicate the modification of this BLM tri-serine cluster in regulating chromosomal stability.
Introduction
Bloom’s syndrome (BS), a genetically determined human chromosome breakage disorder, is caused by loss of function mutations of the recQ-like helicase BLM (1–3). Cells without BLM are characterized by elevated intra- and inter-chromosome recombination, and spontaneous chromosome breakage. Clinical characteristics of BS invariably include short stature, male infertility and an increased risk of cancer. The BLM helicase is an ATP-dependent, 3’-5’ structure-specific helicase vital for DNA replication, transcription and repair (4–6). BLM maintains normal DNA repair functions through its participation in homologous recombination and non-homologous end joining although the exact mechanisms by which it prevents or responds to chromosome breakage are less understood (7–9).
BLM interacts directly with type I topoisomerases (topoisomerase I and topoisomerase IIIα), and type II topoisomerases (topoisomerase IIα) (5,6,10–14). Topoisomerases resolve DNA topological constraints arising from strand separation during replication, replication-repair, recombination and transcription. The most studied complex of BLM with type I topoisomerases is the BTRR complex that consists of BLM, topoisomerase IIIα and the RMI proteins RMI1 and RMI2 and participates in resolving linked intermediates during replication and recombination (12). BLM interacts with topoisomerase IIIα primarily within its N-terminus (amino acids 1–212 of 1417), although an additional region within the extreme C-terminus (amino acids 1266–1417) has been reported (10). BLM also interacts with topoisomerase I to resolve RNA-DNA hybrids during ribosomal DNA transcription (5,6). The interaction of BLM with topoisomerase I reciprocally stimulates respective biochemical activities and most likely facilitates RNA polymerase I transcription in the nucleolus (6). BLM interaction with topoisomerase I is facilitated by amino acids 1332–1349, that also overlap nuclear localization sequences (6,13).
BLM interacts with topoisomerase IIα via amino acids 489–587 (11). This interaction is required for the correction of chromosome breakage, measured by micronuclei formation in transfection experiments in cell lines lacking BLM. Additionally, topoisomerase IIα stimulates BLM helicase activity in vitro using DNA substrates mimicking early homologous recombination structures (11). BLM, in conjunction with the Plk1-interacting checkpoint helicase (PICH), plays a role in the resolution of centromeric chromatin and the recruitment of active topoisomerase IIα to the centromere (12,15,16). Cell lines lacking BLM display excessive numbers of anaphase bridges and lagging chromosomes suggesting a reduced or dysfunctional localization of topoisomerase IIα to the centromere during mitosis (17).
BLM localization and cellular functions are regulated by post-translational modifications in response to cellular stress. These modifications (phosphorylation, ubiquitination and sumoylation) may alter different aspects of its functions, stability, localization to damaged DNA or to PML bodies, and its association with other proteins (18). BLM threonine 99 and 122 are phosphorylated after replicative stress (19,20); phosphorylation of threonine 99 alters the interaction of BLM with topoisomerase III and PML in vivo (21). CHK1 phosphorylation of BLM serine 646 decreases after DNA damage to promote BLM localization to sites of damaged DNA (22,23). BLM also localizes to a class of DAPI-negative/histone-negative anaphase bridges known as ultra-fine bridges (UFBs), as does the Plk1-interacting checkpoint helicase PICH (24,25). UFBs resemble fine, thread-like structures and subsequently are classified into three subtypes dependent on their chromosome ‘anchorage’ origin: telomere/T-UFB, centromere/C-UFB or fragile site/FS-UFB (16,26,27). These subtypes differ in the proteins that mark their ends: FANCD2/FANCI localizes to ends of FS-UFBs while HEC1, an outer kinetochore marker, localizes to C-UFBs (18,28). The DNA structures found within UFBs are not precisely defined but may represent incompletely replicated DNA, hemicatenanes or catenanes. In the G2/M-phase cell cycle transition, sister chromatids are typically connected by hemicatenanes and are catenated at the centromere (29,30). Topoisomerase IIα decatenates these structures to resolve anaphase bridges or UFBs, and prevent chromosome breakage and/or chromosome nondisjunction (14,31). PICH and BLM may collaborate to keep UFBs histone-negative, thus allowing topoisomerase IIα to bind and resolve these aberrant DNA structures (15,16,32). BLM is capable of dissolving hemicatenates between sister chromatids to form non-crossover products (33). Currently, little is known about the regulation and biochemical implications of the BLM/topoisomerase IIα interactions at these structures or at other sites of damaged DNA.
Here, we identify a novel phosphosite tri-serine cluster (S577/S579/S580) within the topoisomerase IIα-interaction domain of BLM that regulates the interaction of BLM and topoisomerase IIα and its subsequent functions in reducing chromosome breakage. Biochemical assays demonstrate that BLM and topoisomerase IIα reciprocally stimulate the enzymatic activity of the other. The increase in BLM activity by topoisomerase IIα is dependent upon the tri-serine cluster, as alanine substitution reduces this stimulation; aspartic acid (phospho-mimic) substitution is similar to that observed with wild-type BLM. In vivo assays monitoring BLM-topoisomerase IIα co-localization, chromosome breakage and UFB formation show that alanine substitution of the serine cluster reduces BLM-topoisomerase IIα co-localization and increases chromosome damage; aspartic acid substitution maintains co-localization and chromosome breakage levels that are similar to wild type BLM. Our studies implicate the tri-serine cluster of BLM in resolving UFBs and reducing subsequent chromosome breakage.
Results
Interaction of BLM and topoisomerase IIα results in reciprocal stimulation of respective biochemical activities
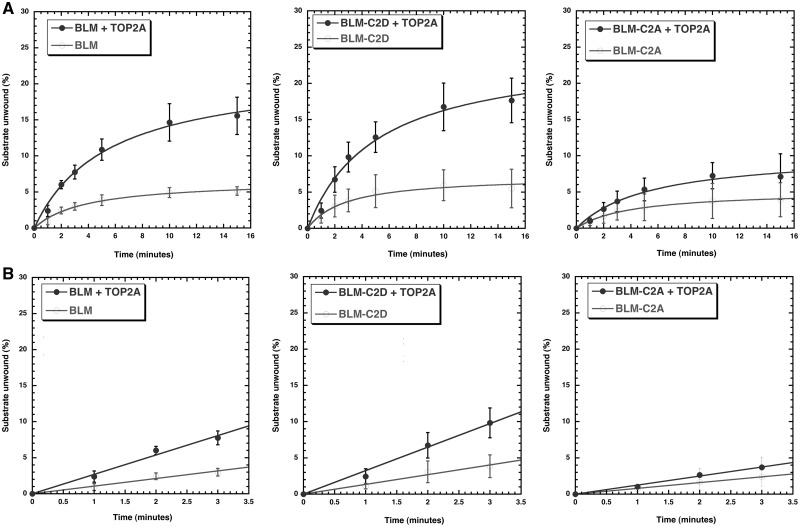
Our previously published work has shown that BLM interacts with topoisomerase IIα via amino acids 489–587 of BLM and that this region is required for topoisomerase IIα-mediated stimulation of BLM helicase activity using short duplex substrates with a 3’ overhang and bubble substrates (11). The experiments presented here were designed to characterize topoisomerase IIα-BLM functions reciprocally. We first determined helicase reaction kinetics and initial rates of unwinding with a forked DNA substrate to model a replication fork in the presence or absence of topoisomerase IIα. The substrates and products of the reaction were resolved on a 10% native PAGE, quantified and the percentage of substrate unwound plotted as a function of time (Fig. 1A and B left panels). Results demonstrate that BLM unwinding follows Michaelis-Menten kinetics and that topoisomerase IIα increases the reaction kinetics with a 3.3-fold increase in the maximum substrate unwound (Fig. 1A left panel, Supplementary Material, Figs S1 and S3A), as well as with a 2.6-fold increase in the initial unwinding rate of BLM (Fig. 1B left panel). Topoisomerase IIα alone is unable to unwind the substrate (Supplementary Material, Fig. S1).
Figure 1.
Topoisomerase IIα stimulates the helicase activity of wild-type BLM and BLM-C2D (phosphomimetic substitutions of S577D, S579D and S580D) by increasing initial rates of unwinding. BLM helicase activities using a forked DNA substrate for BLM, BLM-C2D and BLM-C2A were monitored over time (0, 1, 2, 3, 5, 10 and 15 min) using 0.19 nM BLM (or its mutants) in the absence or presence of 7 nM topoisomerase IIα. Reactions were stopped, products separated on a 10% native PAGE and quantified. Plots of substrate unwound (%) as a function of time were fitted to Michaelis-Menten kinetics. Initial rates of unwinding were calculated from the slope of the curves obtained (using time points 0, 1, 2, 3 and 5 min). (A) (top panels). Kinetics of helicase unwinding in the absence or presence of topoisomerase IIα. (B) (bottom panels). Initial rate of unwinding in the absence or presence of topoisomerase IIα. Phosphomimetic modification of BLM-C2 (S577/S579/S580) is required for topoisomerase IIα-mediated stimulation of BLM helicase activity as the kinetics of helicase unwinding for BLM-C2A in the absence or presence of topoisomerase IIα are similar.
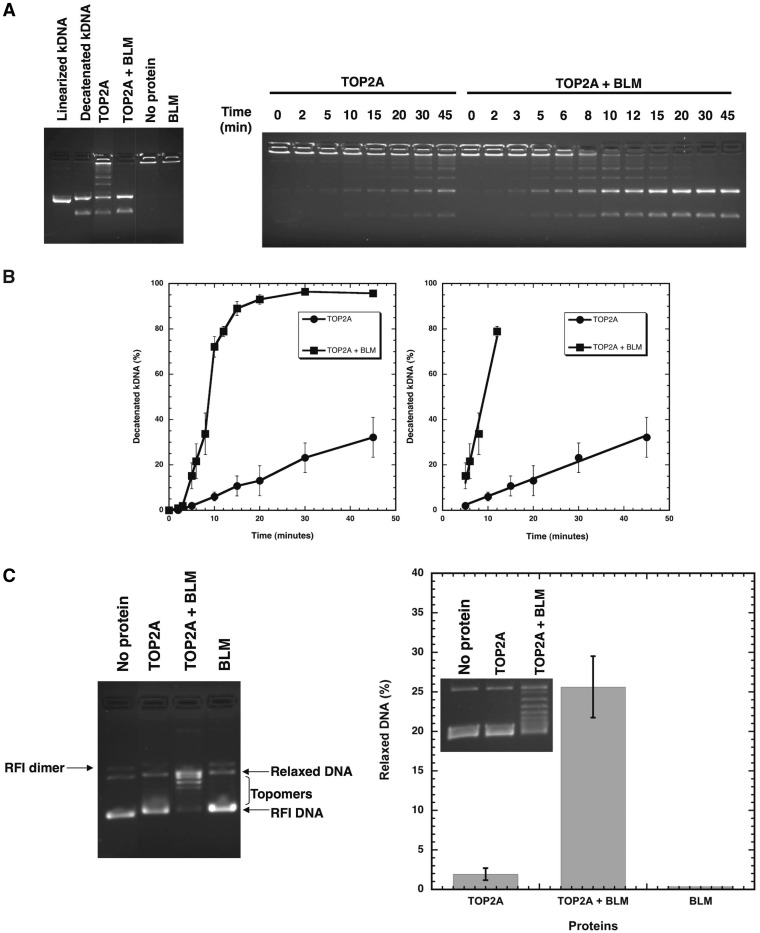
Disentanglement of chromatids during cell division is catalyzed by topoisomerase II. This activity can be biochemically measured in vitro by the ability of an enzyme to decatenate kinetoplast DNA (kDNA) or relax supercoiled DNA. In order to assess if and how the catalytic functions of topoisomerase IIα are affected by BLM, these two activities were monitored in the absence or presence of purified BLM (Fig. 2). BLM alone is unable to decatenate kDNA or relax supercoiled DNA. The concentration of topoisomerase IIα was reduced to observe minimal decatenation of kDNA (Fig. 2A) or relaxation of supercoiled DNA (Fig. 2C). The effects of BLM on the reactions were then monitored. BLM stimulated topoisomerase IIα decatenation activity nearly 10-fold (Fig. 2A and B). Likewise, BLM stimulated the relaxation activity of topoisomerase IIα (Fig. 2C). These results highlight the reciprocal effects of BLM-topoisomerase IIα interaction on enzymatic functions.
Figure 2.
BLM stimulates the decatenation and relaxation activities of topoisomerase IIα. Topoisomerase IIα decatenation activity using kDNA was monitored over time using 0.6 nM TOP2A in the absence or presence of 14nM BLM. Reactions were stopped, products separated on a 1% agarose gel containing ethidium bromide and quantified. (A) Representative gels showing decatenation of kDNA by topoisomerase IIα in the absence or presence of BLM. (B) Plots of decatenated kDNA (%) as a function of time are shown in the graph on the left. The change in rate of decatenation by BLM was calculated from the slope of the linear portion of the reactions and is shown in the graph on the right (9.2% decatenated kDNA/min in the presence of BLM; 0.76% decatenated kDNA/min in the absence of BLM). (C) Topoisomerase IIα relaxation activity with 3kbp supercoiled DNA (RFI) was monitored using 3nM TOP2A in the absence or presence of 27nM BLM. Reactions were stopped, products separated on a 1% agarose gel without ethidium bromide, stained with 0.5µg/ml ethidium bromide for 30 min and topomers monitored by UV. A representative gel is shown. The graph shows relaxation (%) by topoisomerase IIα in the absence or presence of BLM, and by BLM alone; inset shows a representative gel.
A tri-serine cluster S577/S579/S580 within the topoisomerase IIα-interaction domain of BLM is critical for topoisomerase IIα-mediated stimulation of BLM helicase activity
The interaction between BLM and topoisomerase IIα peaks in mitosis, as shown previously by co-localization and co-immunoprecipitation (11,34). BLM is also heavily phosphorylated in M-phase of the cell cycle (18,34). Lastly, ongoing experiments with in vitro expressed and purified BLM protein show BLM mobility shifts suggestive of phosphorylation (Supplementary Material, Fig. 2A). Therefore, we used sequence analysis of the topoisomerase IIα-interaction domain of BLM (amino acids 489–587) to identify five serines organized in two clusters: S517 and S518 in cluster one (C1) and S577, S579 and S580 in cluster two (C2). These serines were evaluated as phosphorylation targets by further informatic analyses and high-resolution mass spectrometry (Supplementary Material, Fig. S2B and C). In order to determine whether these serines contribute to BLM function, serines in each cluster were replaced by either alanine (BLM-C1A, BLM-C2A) or aspartic acid (BLM-C1D, BLM-C2D) using in vitro mutagenesis. Vectors with BLM modifications were generated for protein expression and transfection experiments, and used for biochemical and cytological analyses.
BLM proteins (BLM-C1A, BLM-C1D, BLM-C2A and BLM-C2D) were purified to homogeneity. Protein yields and helicase activities of the BLM proteins were similar to one another (Materials and Methods). Proteins were tested for helicase activity in the presence or absence of topoisomerase IIα (Fig. 1 middle and right panels, Supplementary Material, Fig. S3B). Cluster C1 mutations had no effect on the ability of topoisomerase IIα to stimulate BLM helicase activity (data not shown). However, cluster C2 mutations had a differential effect on the ability of topoisomerase IIα to stimulate BLM helicase activity. BLM-C2D was stimulated by topoisomerase IIα with a 3.4-fold increase in the maximum substrate unwound (Fig. 1A middle panel) as well as a 2.4 fold increase in the initial unwinding rate of BLM-C2D (Fig. 1B middle panel), similar to that seen with wild-type BLM (Fig. 1A and B left panels). Mutation of the C2 serine cluster to alanines (BLM-C2A) significantly reduced topoisomerase IIα stimulation of BLM unwinding activity (Fig. 1 right panels) by more than 2.0-fold in the maximum substrate unwound (Fig. 1A right panel) and in the initial unwinding rate (Fig. 1B right panel). We also tested the ability of the mutants to modulate TOP2A decatenation activity. We did not see a significant difference in the rate of stimulation between the two mutants or in contrast to wild-type but cannot rule out subtle changes due to the fast kinetics of TOP2A stimulation and the rapid jump in decatenation rates between 3 and 5 min (Fig. 2). Co-immunoprecipitation of wild-type BLM or its mutants (C1 or C2) with recombinant topoisomerase IIα also did not reveal significant differences (Supplementary Material, Fig. S2D). However, enzymatic results for BLM helicase activity support the conclusion that the C2 serine cluster plays a critical role in regulating BLM function by topoisomerase IIα.
The tri-serine cluster S577/S579/S580 of BLM is critical for correction of elevated chromosome breakage in cells without BLM
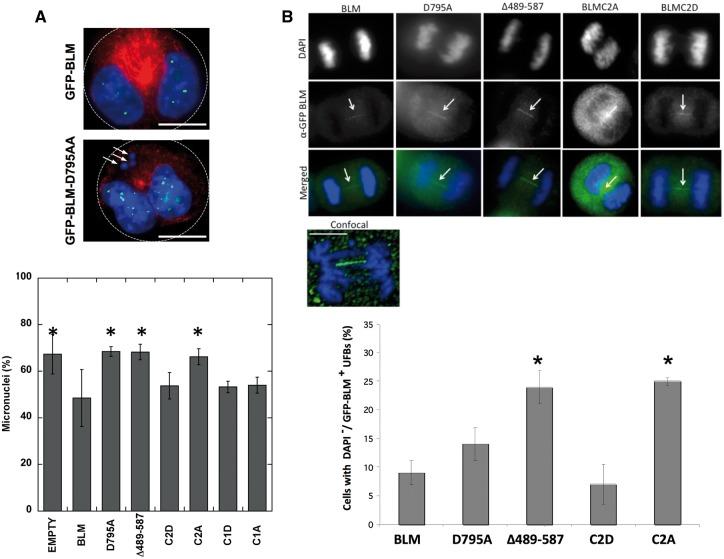
Our biochemical results suggest that serine modifications of BLM at S577, S579 and S580, and the reciprocal enzymatic effects of BLM and topoisomerase IIα may have functional implications for DNA repair. As the interaction of BLM and topoisomerase IIα changes enzymatic efficiency, we asked how it may affect the elevated chromosome breakage characteristic of cells lacking the BLM helicase (11). Micronuclei assays were used to assess whether the biochemical differences between the BLM cluster mutants translated to a cellular phenotype, as these are one of the most sensitive methods for detecting cellular chromosome damage (35). Numerous reports have shown high levels of micronuclei in vivo in those with BS (11,22,36,37). Micronuclei arise from unrepaired DNA double-stranded breaks that generate acentric chromosome fragments that fail to be incorporated in daughter nuclei during mitosis (35,38). BLM protein without the topoisomerase IIα-interaction region (Δ489–587) fails to reduce the number of micronuclei in the BLM−/− cell line GM08505 (11). BLM-C1 and -C2 mutants were tagged with GFP and tested for their ability to reduce the elevated number of micronuclei in GM08505 (BLM−/−) cells. Cells were transfected with pEGFP-empty, pEGFP-BLM, pEGFP-BLM-D795A (helicase-dead mutant), pEGFP-BLM-Δ489–587, Cluster 1 mutants (pEGFP-BLM-C1A, pEGFP-BLM-C1D) or Cluster 2 mutants (pEGFP-BLM-C2A or pEGFP-BLM-C2D) and examined for micronuclei (Fig. 3A).
Figure 3.
Phosphomimetic modification (S577D/S579D/S580D) of BLM or wild-type BLM is required for correction of elevated chromosome breakage and UFB resolution in transfected BLM−/− cells. (A) GMO8505 (BLM−/−) cells were transfected with pEGFP-empty, pEGFP-BLM, pEGFP-BLM-D795A, pEGFP-BLM-Δ489–587, pEGFP-BLM-C1A, pEGFP-BLM-C1D pEGFP-BLM-C2A or pEGFP-BLM-C2D vectors. pEGFP-BLM-D795A encodes a protein without helicase activity; pEGFP-BLM-Δ489–587 encodes a protein without the topoisomerase IIα–interacting region. Cells were treated with cytochalasin B post-transfection to arrest bi-nucleated cells in mitosis. Cells were fixed 18 h post-cytochalasin B treatment and stained with anti-β-tubulin and DAPI for nuclear staining. (Top panel) White arrows point to three micronuclei (MNi) in BLM−/− cells expressing the helicase-dead GFP-BLM-D795A. White dotted lines indicate cell boundaries. Scale bar (white) is 10 μm. (Bottom panel) Micronuclei percentages represent the average number of MNi counted per 100 cells. Four independent experiments counted at least 400 cells in total per vector. pEGFP-empty, pEGFP-BLM-D795A, pEGFP-BLM-Δ489–587 and pEGFP-BLM-C2A failed to reduce MNi frequency significantly compared with pEGFP-BLM. Error bars represent ± SD. Data were analysed by ANOVA; *p < 0.05 was considered significant in comparison to pEGFP-BLM: GFP = 0.0113; pEGFP-BLM-C2A = 0.0239; pEGFP-BLM-D795A = 0.0130; pEGFP-BLM-Δ489–587 = 0.0147. (B) (Top panel) Representative cells in anaphase show GFP-BLM UFBs in GMO8505 (BLM−/−) cells transfected with pEGFP-BLM, pEGFP-BLM-D795A, pEGFP-BLM-Δ489–587, pEGFP-BLM-C2A or pEGFP-BLM-C2D. Scale bar (white) in confocal images is 5 μm. (Bottom panel) Percentages represent the number of cells in anaphase that exhibit GFP-BLM UFBs per 50 GFP-positive cells counted blindly. Two independent experiments counted at least 100 cells in total per vector. Error bars represent ± SD. Data were analysed by Student’s t-test; *p < 0.05 was considered significant in comparison to pGFP-BLM: BLM-C2A = 0.0271; BLM-Δ489–587 = 0.0339.
Transfection of wild-type BLM reduced the percentage of BLM−/− cells containing micronuclei from 67.3 to 48.5% (Fig. 3A, top and bottom panels). Cells transfected with pEGFP-empty, pEGFP-BLM-D795A or pEGFP-BLM-Δ489–587 displayed a high number of cells with at least one micronucleus (67.3–68.5%). Cells transfected with vectors expressing the mutated Cluster 1 (S517/S518) to either alanine (BLM-C1A) or aspartate (BLM-C1D) exhibited recovery comparable to wild-type BLM (54.0%, 53.3%, respectively). Cells transfected with vectors expressing mutated Cluster 2 (S577/S579/S580) exhibited recovery comparable to wild type BLM with the phospho-mimic BLM-C2D only (53.8% compared with 48.5%). The phospho-dead BLM-C2A failed to reduce the percentage of cells containing micronuclei (66.3%, p = 0.0239). Single mutants of each site within this phosphosite cluster (577A/D, 579A/D, 580A/D) behaved similarly to wild-type BLM, as did double mutants (577/579A, 577/580A, 579/580A) (Supplementary Material, Fig. S4). Threonine 581 was also identified as a potential phosphorylation site by the in silico prediction programs (Supplementary Material, Fig. S2B). Interestingly, this site is four amino acids C-terminal to serine 577, a site predicted to be modified by GSK3 kinase, defining serine 581 as a potential pre-phosphorylation priming site (39). Micronuclei studies with cells transfected with pEGFP-BLM577A, pEGFP-BLM577D, pEGFP-BLM581A or pEGFP-BLM581D did not differ from wild-type BLM in correction of chromosome breakage (Supplementary Material, Fig. S4). In total, these results pinpoint the three serines within cluster 2 as important for reducing chromosome damage and imply that phosphorylation of all three serines may be the likely modulator.
The BLM topoisomerase IIα-interaction domain and the S577/S579/S580 tri-serine cluster are required for efficient UFB resolution
BLM localizes to a class of DAPI-negative/histone-negative anaphase bridges known as ultra-fine bridges (UFBs) (18). As BLM−/− cells and those treated with topoisomerase II inhibitors exhibit high numbers of UFBs, we examined the localization of BLM-C2A and -C2D to UFBs. BLM−/− cells were transfected with pEGFP-BLM, pEGFP-BLM-D795A (helicase-dead mutant), pEGFP-BLM-Δ489–587, pEGFP-BLM-C2A or pEGFP-BLM-C2D, and evaluated for UFBs. BLM phospho-mutants BLM-C2A and BLM-C2D localize to UFBs, as do BLM, BLM-Δ489–587 and BLM-D795A proteins (Fig. 3B, top and bottom panels). The percentage of mitotic cells with UFBs was significantly higher in cells expressing BLM-C2A and BLM-Δ489–587 compared with cells expressing wild-type BLM (24% and 25% compared with 9%, p = 0.0271, p = 0.0339) (Fig. 3B, bottom panel). Cells expressing the phospho-mimic BLM-C2D showed similar percentage of mitotic cells with UFBs as cells expressing wild-type BLM (7% versus 9%). Interestingly, increases in DAPI-positive anaphase bridges in cells expressing BLM-C2A or BLMΔ489–587 were not observed compared with BLM-C2A and BLMΔ489–587, highlighting the unique origin of UFBs at anaphase bridges (Supplementary Material, Fig. S5). These data suggest that the BLM topoisomerase IIα-interaction domain and the tri-serine cluster within it are vital for the resolution of UFBs, but are less relevant for anaphase bridges. Although BLM localizes to UFBs, we were unable to demonstrate the localization of topoisomerase IIα to UFBs. Topoisomerase IIα was visible at DAPI-positive anaphase bridges in both normal and BLM−/− cells, suggesting that its localization to anaphase bridges is BLM-independent (Supplementary Material, Fig. S6).
The tri-serine cluster S577/S579/S580 of BLM is critical for BLM co-localization with topoisomerase IIα
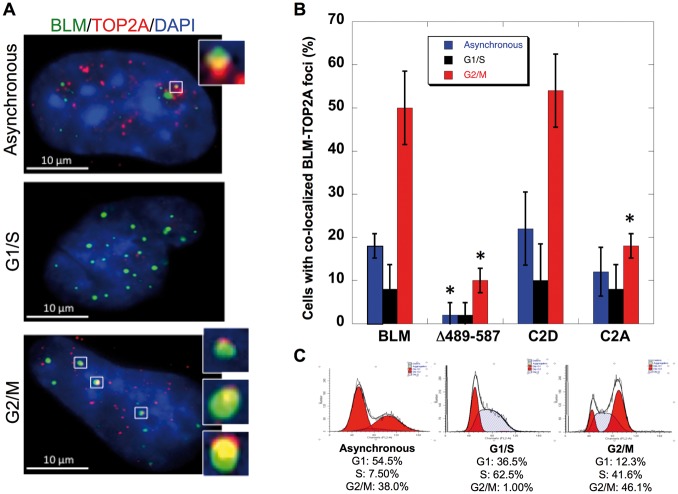
BLM and topoisomerase IIα are minimally expressed during G1-, increase in S- and peak in G2/M-phases of the cell cycle (34,40). Their co-localization is also most prominent in M-phase (11,34). Co-localization of BLM-C2A and BLM-C2D with topoisomerase IIα was examined during G1/S and G2/M (Fig. 4A–C). BLM−/− cells were transfected with pEGFP-BLM, pEGFP-BLM-Δ489–587, pEGFP-BLM-C2A or pEGFP-BLM-C2D. Cells were synchronized using a double-thymidine block and collected 4-h post-release to obtain cells in G1/S or 11-h plus colcemid treatment to obtain cells in G2/M. FACS analysis confirmed synchronization (Fig. 4C). Immunofluorescence studies demonstrated that BLM-C2A is significantly reduced in its co-localization with topoisomerase IIα in G2/M-phase (18% in cells expressing BLM-C2A compared with 50% in cells expressing wild-type BLM, p≤0.0001) (Fig. 4B). The reduction in co-localization of topoisomerase IIα and BLM-C2A is comparable to that in cells expressing the topoisomerase IIα-interaction domain deletion mutant BLM-Δ489–587 in G2/M (10% co-localization). In comparison, cells expressing the phospho-mimic BLM-C2D retain the ability to co-localize BLM-C2D with topoisomerase IIα similarly to cells expressing wild-type BLM (54% in cells expressing BLM-C2D compared with 50% in cells expressing BLM). Localization of wild type and mutant BLM proteins to PML bodies and the nucleolus was examined in order to rule out alteration of cellular localization in considering the effects of serine mutations. BLM-C1A, BLM-C1D, BLM-C2A and BLM-C2D localize to PML bodies and the nucleolus similarly to wild-type BLM (Supplementary Material, Fig. S7). These data suggest that modification of serines 577, 579 and 580 of BLM alters BLM and topoisomerase IIα co-localization in vivo.
Figure 4.
Phosphomimetic modification of BLM-C2 (S577/S579/S580) is required for BLM co-localization with topoisomerase IIα in transfected BLM−/− cells. (A) GMO8505 (BLM−/−) cells were transfected with pEGFP-BLM, pEGFP-BLM-Δ489–587, pEGFP-BLM-C2A or pEGFP-BLM-C2D. Cells were synchronized 24 h post transfection with a double thymidine (Sigma) treatment, fixed and stained for TOP2A with anti-TOP2A (Topogen)/Alexaflor 594 and DAPI. Image inserts show co-localized BLM and TOP2A in asynchronous, G1/S or G2/M cells. Scale bar (white) is 10μm. (B) Cells with at least two GFP-BLM/TOP2A foci were counted as positive for co-localization. A total of 50 GFP-positive cells were counted in two independent and blinded experiments. Percentages of cells with BLM/TOP2A co-localization are represented graphically. Error bars represent ± SD. Data were analysed by Student’s t-test; *p < 0.05 was considered significant in comparison to the percentage of co-localization from cells transfected with pEGFP-BLM in the corresponding cell cycle stage (asynchronous cells, blue; G1/S cells, black; G2/M cells, red): BLMΔ489–587: G2/M≤0.0001; BLMC2A: G2/M≤0.0001; BLMΔ489–587: Asynchronous = 0.0031. (C) Cell cycle distribution profiles and the percentages of cells in cell cycle stages in asynchronous, G1/S and G2S populations.
Discussion
This study has characterized a novel serine cluster (serines 577, 579 and 580) within the topoisomerase IIα-interaction domain of BLM that is necessary for topoisomerase IIα-mediated stimulation of BLM helicase activity in vitro, G2/M-phase cell cycle-specific co-localization with topoisomerase IIα in vivo and maintenance of low levels of chromosome breakage IIα as measured by UFBs and micronuclei formation (Figs 1–4). Phosphomimetic aspartic acid substitutions of all three serines within the cluster are compatible with these functions, while non-phosphorylatable alanine substitutions are not. In contrast, single and double mutants (alanine or aspartic acid substitutions) of each serine within this cluster behaved similarly to wild-type BLM (Supplementary Material, Fig. S4), suggesting that this is a functional cluster of three serines (577/579/580). Decreased co-localization with topoisomerase IIα, and increased UFBs and chromosome breakage occurs in cells transfected with alanine-substituted pEGFP-BLM-C2A. In comparison, cells transfected with the aspartate-substituted, pEGFP-BLM-C2D or wild-type pEGFP-BLM are similar in co-localization of BLM with topoisomerase IIα, UFBs and levels of chromosome breakage.
Phosphosite clusters have been hypothesized to play a functional role in regulating a robust cellular response to phosphorylation, with specific phosphosite combinations determining the rate and duration of various biological responses (41,42). Our experiments argue for expanding the approach of studying single amino acids when evaluating phosphorylation-dependent processes. Our in vitro and cell culture experiments suggest that phosphorylation of the BLM 577/579/580 cluster could facilitate DNA damage repair, likely through its interaction with topoisomerase IIα. Mass-spectrometry approaches to identify modification of these BLM serines were inconsistent most likely due to difficulties in identifying tandem phosphorylation sites, the acidic nature of this protein region and an inopportune trypsin cleavage site.
BLM forms complexes with several protein partners critical for resolving DNA damage. Similar to our previous studies with topoisomerase I, the interaction of BLM with topoisomerase IIα results in the reciprocal stimulation of the respective protein activities (Figs 1 and 2) (6). Their interaction increases the rate of DNA unwinding that is reflected in its total unwinding activity (Fig. 1) and support earlier observations that BLM by itself is a weakly processive helicase with enzymatic activity modulated through its interaction with other proteins to resolve chromosome damage during replication and recombination (6,9,13,43–47). Biochemical experiments demonstrate functional differences in the ability of topoisomerase IIα to stimulate BLM proteins with modifications of the tri-serine cluster; these data correlate with the in vivo studies of micronuclei, DNA damage and UFB.
The cellular endpoints of our experiments (co-localization of BLM with TOP2A, micronuclei recovery and UFB formation) suggest that BLM-C2D behaves similarly to wild-type BLM and that BLM-C2A does not. The endpoints of the biochemical assays also suggest that BLM-C2D behaves similarly to wild-type BLM in helicase stimulation by TOP2A and that BLM-C2A does not. These data support the hypothesis that there is a difference in the ability of the two proteins to functionally interact that depends on the modification of these three serines within the TOP2A-interaction domain of BLM. Two other experimental approaches, co-immunopreciptation (co-IP) of BLM and TOP2A, and decatenation assays, were unable to distinguish between C2D and C2A isoforms of BLM in comparison to wild-type BLM. The negative results from these two assays may be related to the experimental constraints of each approach: both co-IP and decatenation assays were performed with excess protein concentrations. Therefore, any subtle differences in the ability of the BLM isoforms to interact with TOP2A could not be assessed. Our attempts to calculate the apparent interaction coefficient (KD) in vitro were unsuccessful due to variability in the antibody avidity for TOP2A and/or BLM, precluding quantitative analyses. Likewise, the spike in rate of decatenation by TOP2A in the presence of BLM was impossible to slow down with dilutions of protein concentrations. Our results are consistent with the conclusion that the alanine, but not aspartic, substitutions cause a functional defect (in helicase activity stimulation) and perhaps subtle differences in physical interaction with TOP2A that are not detectable in vitro are detectable in vivo, and reflected best in the cellular experiments. It is plausible that the TOP2A-interaction region may not be defined solely by the three amino acid cluster and that this may be contributing to the lack of strong in vitro findings. Our previously published study demonstrated that the interaction domain consists of 99 amino acids (489–587). As explained in Supplementary Material, Fig. S2B, there are several predicted phosphorylation sites that could potentially influence the interaction between TOP2A and BLM. Thus, mutation of S577, S579 and S580 do not necessarily result in an all-or-nothing effect in controlling direct physical interaction, although it clearly affects some biological endpoints measured in our study. It is possible that BLM-TOP2A foci involve other proteins that may be affected by the C2A mutation. The observation of foci with wild-type BLM and BLM-C2D, but not with BLM-C2A, does not necessarily prove a direct physical interaction, as it may also require other proteins to facilitate foci assembly. However, the absence of foci does correlate with the impaired ability of the C2A helicase to be stimulated by TOP2A.
Ultra-fine anaphase bridges (UFBs) are recently discovered histone-negative, DAPI-negative chromatin bridges seen in anaphase due to residual problems in DNA replication or decatenation. UFBs can be centromeric, non-centromeric (at fragile sites) or telomeric in nature, and are reflective of the aberrant (e.g. late-replicating/recombination intermediates) or unresolved (e.g. catenanes) DNA structures at these chromosomal locations. It is not yet clear if the DNA structures within UFBs are homogenous or heterogeneous. Consequently, proteins associated with the different UFBs are diverse but reflective of their putative functions in resolving specific structures (15,16,20,26,48–51). Both BLM and topoisomerase IIα are implicated in the resolution of structures associated with UFBs and a deficiency of either results in an increase in UFBs. Such a role is also consistent with their respective enzymatic activities. Our studies do not address the specific nature or origin of UFBs and hence we cannot ascertain the kind of DNA anomalies that exist at these UFBs. However, the data presented here reinforce a role for the tri-serine cluster within the BLM topoisomerase IIα-interaction domain in resolution of structures associated with UFBs. While TOP2A activity is an important factor in UFB formation, especially in the context of centromeric UFBs (where catenanes are the primary ‘problem’), it is not the only one: defects in replicative repair can also lead to bridges. Our experiments point to the importance of BLM helicase activity and its stimulation by TOP2A to prevent UFB accumulation. We were unable to see topoisomerase IIα localization to the BLM-containing UFBs; however, the topoisomerase IIα-interaction domain of BLM and the tri-serine cluster within the domain were critical for the UFB resolution. We speculate that the observed BLM-containing UFBs primarily represent DNA intermediates specific for BLM, enabling its recruitment. The absence of topoisomerase IIα visualization might reflect a rapid turnover of topoisomerase IIα within the BLM-containing UFBs, the absence of structures that enable recruitment and stabilization of topoisomerase IIα, or non-involvement of topoisomerase IIα for resolving the DNA structures in BLM UFBs. Other proteins may be involved in recruiting topoisomerase IIα and/or BLM to UFBs as in centromeric UFBs where TOPBP1 associates with UFBs independently of BLM and recruits topoisomerase IIα by a direct physical association with topoisomerase IIα (51). DNA structures in centromeric UFBs are DNA catenanes that require TOPBP1 to resolve by recruitment of topoisomerase IIα. BLM is not found in the majority of these UFBs, although a small subset is characterized by partial BLM localization (51). PICH is also a critical protein in recruiting of BLM to centromeric, fragile-site and telomeric UFBs (15,16,26,32,48,50). Some of these UFBs involve topoisomerase IIIα in association with BLM, RMI1 and RMI2 proteins.
Manipulation of the BLM/topoisomerase IIα interaction may make it possible to generate catastrophic genomic instability through very precise targeting of the interaction to enhance the effectiveness of, or replace, some oncolytic chemotherapeutics. Our studies also demonstrate that the investigation of genetically determined chromosome instability in the human can lead to precise mechanistic understanding of how chromosome integrity is maintained.
Materials and Methods
Cell lines and tissue culture
GM08505 cells (BLM−/−) and VA13 (BLM+/+) were obtained from Coriell Cell Repository and cultured in Minimum Essential Medium (Invitrogen) containing 10% fetal bovine serum (Hyclone). All cells were cultured at 37°C and 5% CO2.
Mutagenesis
Mutations of the BLM serines to alanine or aspartic acid were generated by using PCR primers with mutations followed by cloning of PCR products into pYES-BLM and peGFP-BLM. Reactions were transformed into DH5α-competent cells. Sequencing of individual plasmids confirmed desired alterations.
Helicase and topoisomerase IIα assays
BLM proteins were expressed and purified as previously described (5,6,13). Protein expression levels and protein yields were similar for wild-type and mutant BLM proteins. The helicase activity (fmol unwound/min/μM protein) of each mutant BLM protein was similar (e.g. BLM: 3.92; C1A: 3.85; C1D: 3.9; C2A: 3.55; C2D: 3.72). Helicase assays using recombinant TOP2A (purchased from TopoGEN, Buena Vista, CO, USA) and BLM proteins were performed for indicated times at 37°C. Helicase reactions were assembled on ice and started with the addition of BLM in 20 μl reaction volume in buffer containing 50 mM Tris-HCl (pH 8.0), 120 mM NaCl, 10 mM MgCl2, 1 mM ATP, 0.5 mM DTT and 50 μg/ml BSA, using 2 fmole radioactively labeled DNA: DNA forked substrate prepared by annealing the following oligonucleotides (9). Oligonucleotides were purchased from Operon. Oligonucleotide sequences (5’ to 3’ orientation) are
TTTTTTTTTTTTTTTTTAGGGTTAGGGCATGCACTAC GTAGTGCATGCCCTAACCCTAATTTTTTTTTTTTTTT
Reactions were terminated in TOP2A stop buffer (1% Sarkosyl, 5% glycerol, 10 mM EDTA); products were resolved on 10% non-denaturing polyacrylamide gels in 1X TBE. Gels were dried, exposed on a phosphorimager plate and analysed by ImageQuant software on a Typhoon phosphorimager. Bands were quantified, amount of substrate unwound calculated (%) and plotted as a function of time as indicated. Plots were fitted to Michaelis-Menten kinetics using Kaleidagraph software. Standard deviations were calculated from three to five experiments and two independent protein preparations. TOP2A assays were performed as above using either kinetoplast DNA (to monitor decatenation activity) or supercoiled DNA (to monitor relaxation activity). Reactions were analysed on 1% agarose gel as described by the manufacturer (TopoGen).
In silico phosphorylation prediction
Phosphorylation sites, relative solvent accessibility values and conservation of sites were identified using the following six online databases:
KinasePhos2.0: http://kinasephos2.mbc.nctu.edu.tw
PSPP: http://ppsp.biocuckoo.org/aboutPPSP.php
NetPhos2.0: http://www.cbs.dtu.dk/services/NetPhos/
Phosida: http://www.phosida.com
SABLE for RSA values: http://sable.cchmc.org/sable_doc.html
COBALT for conservation: http://www.ncbi.nlm.nih.gov/tools/cobalt/
Immunoprecipitation
Immunoprecipitations were performed using purified proteins at 4°C. Purified BLM, BLM-C1A, BLM-C1D, BLM-C2A or BLM-C2D was incubated with purified TOP2A (Topogen). Two hours of protein incubation was followed by the addition of an anti-BLM antibody or anti-TOP2A antibody and an overnight incubation of the protein-antibody mixture. Protein A/G beads were used to immunoprecipitate the anti-BLM antibody or anti-TOP2A antibody and the associated proteins; goat IgG was used as an isotype-matched negative control. Immunoprecipitated proteins were separated on a 8% SDS-PAGE and detected by western blotting using either anti-BLM or anti-TOP2A antibody as indicated.
Micronuclei analysis
GM08505 cells (BLM−/−) were transfected with Effectene (Qiagen) and cultured for 30-h post-transfection with 5 μg/ml cytochalasin B (Sigma). At 48-h post-transfection, cells were fixed in 4% paraformaldehyde, permeabilized in 0.25% Triton-X-100 in PBS and washed before blocking in 10% normal goat serum. Subsequent staining used 1: 800 anti β-tubulin (Novus) in 1% BSA/0.1% Tween/PBS. Cells were mounted with VectaShield DAPI mounting medium (Vector Labs) onto glass sides (Fisher). Cells were imaged using a Zeiss AxioVert 200M with an attached AxioCam MRm camera. One hundred GFP-positive cells from three independent and blinded experiments were counted for micronuclei. Significance of the micronuclei assays was analysed by ANOVA by comparing each treatment group to the control. p-values < 0.05 were considered significant.
UFB analysis
GM08505 cells (BLM−/−) or VA13 cells (BLM+/+) were transfected with Effectene (Qiagen) and cultured for 24 h. Cells were then treated with 0.1 µg/ml nocodazole (Sigma) for 3 h and incubated in fresh medium for 1 h. Cells were fixed in 4% paraformaldehyde for 15 min at room temperature, permeabilized in 0.25% Triton-X-100 in PBS and washed before blocking in 10% normal goat serum. Subsequent staining used the indicated antibodies in 1% BSA/0.1% Tween/PBS: 1: 1000 anti-GFP (Abcam 290) overnight. Primary antibodies were visualized with fluorescent AlexaFluor (Invitrogen) secondary antibodies. Cells were mounted with VectaShield DAPI mounting medium (Vector Labs) onto glass sides (Fisher) and imaged using a Zeiss AxioVert 200M with an attached AxioCam MRm camera or Olympus Fluoview 1200 with FV-OSRASW software for confocal images. Fifty GFP-positive cells each from each of two independent and blinded experiments were counted. Data were analysed using Student’s t-test; p-values < 0.05 were considered significant.
Cell cycle synchronization and co-localization studies
Cell cycle synchronization used a double-thymidine block and harvest post-release to obtain cells in S- or G2/M-phases. GM08505 cells (BLM−/−) were cultured with 2 mM thymidine (Sigma) for 18 h, washed with PBS and released in fresh media for 9 h; cells were treated a second time with 2 mM thymidine (Sigma) for 17 h. Cells were then washed with PBS and collected at 4 h for G1/S-phase or 11 h for G2/M-phase. Synchronization was confirmed via flow cytometry. Once confirmed, co-localization studies were performed side-by-side. Cells in M-phase were collected following treatment with 40 ng/μl colcemid (Sigma) 6 h prior to harvest. FACS analysis confirmed synchronization. GM08505 cells (BLM−/−) on glass coverslips (Fisher) were washed in PBS and fixed in 4% paraformaldehyde for 15 min at room temperature. Cells were permeabilized in 0.25% Triton-X-100 in PBS and washed before blocking in 10% normal goat serum and subsequent staining with the indicated antibodies in 1% BSA/0.1% Tween/PBS: 1: 1000 anti-B23 (Sigma B0556), 1: 1000 anti-PML (Abcam 53773) or 1: 1500 anti-TOP2A (TopoGen TG2011–1). Cells were washed in 0.1% Tween/PBS; primary antibodies were visualized with fluorescent AlexaFluor (Invitrogen) secondary antibodies. Coverslips were mounted with VectaShield DAPI mounting medium (Vector Labs) onto glass slides (Fisher). Cells were imaged using a Zeiss AxioVert 200M with an attached AxioCam MRm camera. Fifty GFP-positive cells from each of two independent and blinded experiments were counted for co-localization. Data were analysed using Student’s t-test; p-values < 0.05 were considered significant.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We thank Dr. Michael Freitas for sharing his expertise in mass spectrometry and analysis of protein modification, and Dr. Michael McIlhatton for review of the manuscript. We thank The OSU CCC Nucleic Acid Shared Resource, and The OSU Campus Microscopy and Imaging Facility for technical assistance.
Conflict of Interest statement. None declared.
Funding
National Science Foundation [GRFP DGE-0822215 to J.H.], Pelotonia Fellowship Program [A.S.G., J.H.B.], The Bloom’s Syndrome Foundation [J.G.], National Institutes of Health [CA-117898 to J.G.], The Ohio Cancer Research Associates [S.A.] and Concern Foundation [S.A.]. J.G. was supported by Award Number 8UL1TR000090–05, 8KL2TR000112–05, and 8TL1TR000091–05 from the National Center For Advancing Translational Sciences. The work was also supported by The OSU Comprehensive Cancer Center Shared Resources and the National Institutes of Health P30 CA016058.
References
- 1. German J., Archibald R., Bloom D. (1965) Chromosomal breakage in a rare and probably genetically determined syndrome of man. Science, 148, 506–507. [DOI] [PubMed] [Google Scholar]
- 2. German J. (1997) Bloom's syndrome. XX. The first 100 cancers. Cancer Genet. Cytogenet., 93, 100–106. [DOI] [PubMed] [Google Scholar]
- 3. Ellis N.A., Groden J., Ye T.Z., Straughen J., Lennon D.J., Ciocci S., Proytcheva M., German J. (1995) The Bloom's syndrome gene product is homologous to RecQ helicases. Cell, 83, 655–666. [DOI] [PubMed] [Google Scholar]
- 4. Karow J.K., Chakraverty R.K., Hickson I.D. (1997) The Bloom's syndrome gene product is a 3'-5' DNA helicase. J. Biol. Chem., 272, 30611–30614. [DOI] [PubMed] [Google Scholar]
- 5. Grierson P.M., Lillard K., Behbehani G.K., Combs K.A., Bhattacharyya S., Acharya S., Groden J. (2012) BLM helicase facilitates RNA polymerase I-mediated ribosomal RNA transcription. Hum. Mol. Genet., 21, 1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Grierson P.M., Acharya S., Groden J. (2013) Collaborating functions of BLM and DNA topoisomerase I in regulating human rDNA transcription. Mutat. Res., 743, 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Langland G., Elliott J., Li Y., Creaney J., Dixon K., Groden J. (2002) The BLM helicase is necessary for normal DNA double-strand break repair. Cancer Res., 62, 2766–2770. [PubMed] [Google Scholar]
- 8. Chu W.K., Hickson I.D. (2009) RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer, 9, 644–654. [DOI] [PubMed] [Google Scholar]
- 9. Acharya S., Kaul Z., Gocha A.S., Martinez A.R., Harris J., Parvin J.D., Groden J., Huen M.S.-Y. (2014) Association of BLM and BRCA1 during telomere maintenance in ALT cells. PLoS One, 9, e103819. DOI: 10.1371/journal.pone.0103819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu L., Davies S.L., North P.S., Goulaouic H., Riou J.-F., Turley H., Gatter K.C., Hickson I.D. (2000) The Bloom's syndrome gene product interacts with topoisomerase III. J Biol Chem, 27, 9636–9644. [DOI] [PubMed] [Google Scholar]
- 11. Russell B., Bhattacharyya S., Keirsey J., Sandy A., Grierson P., Perchiniak E., Kavecansky J., Acharya S., Groden J. (2011) Chromosome breakage is regulated by the interaction of the BLM helicase and topoisomerase IIα. Cancer Res., 71, 561–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manthei K.A., Keck J.L. (2013) The BLM dissolvasome in DNA replication and repair. Cell. Mol. Life Sci., 70, 4067–4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tangeman L., McIlhatton M.A., Grierson P., Groden J., Acharya S. (2016) Regulation of BLM nucleolar localization. Genes, 7, 69.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pommier Y., Sun Y., Huang S.N., Nitiss J.L. (2016) Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell. Biol., 17, 703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rouzeau S., Cordelières F.P., Buhagiar-Labarchède G., Hurbain I., Onclercq-Delic R., Gemble S., Magnaghi-Jaulin L., Jaulin C., Amor-Guéret M., Sullivan B.A. (2012) syndrome and PICH helicases cooperate with topoisomerase IIα in centromere disjunction before anaphase. PLoS One, 7, e33905. Bloom's DOI: 10.1371/journal.pone.0033905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nielsen C.F., Huttner D., Bizard A.H., Hirano S., Li T.N., Palmai-Pallag T., Bjerregaard V.A., Liu Y., Nigg E.A., Wang L.H.-C., Ian D., Hickson I.D. (2015) PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat. Commun., 6, 8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Payne M., Hickson I.D. (2009) Genomic instability and cancer: lessons from analysis of Bloom's syndrome. Biochem. Soc. Trans., 7, 553–559. [DOI] [PubMed] [Google Scholar]
- 18. Bohm S., Bernstein K.A. (2014) The role of post-translational modifications in fine-tuning BLM helicase function during DNA repair. DNA Repair, 22, 123–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ouyang K.J., Yagle M.K., Matunis M.J., Ellis N.A. (2013) BLM SUMOylation regulates ssDNA accumulation at stalled replication forks. Front. Genet., 4, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang J., Chen J., Gong Z. (2013) TopBP1 controls BLM protein level to maintain genome stability. Mol. Cell, 52, 667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rao V.A., Fan A.M., Meng L., Doe C.F., North P.S., Hickson I.D., Pommier Y. (2005) Phosphorylation of BLM, dissociation from topoisomerase IIIα, and colocalization with γ-H2AX after topoisomerase I-induced replication damage. Mol. Cell. Biol., 25, 8925–8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davies S.L., North P.S., Dart A., Lakin N.D., Hickson I.D. (2004) Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol., 3, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaur S., Modi P., Srivastava V., Mudgal R., Tikoo S., Arora P., Mohanty D., Sengupta S. (2010) CHK1-dependent constitutive phosphorylation of BLM helicase at Serine 646 decreases after DNA damage. Mol. Cancer Res., 8, 1234–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chan K.L., North P.S., Hickson I.D. (2007) BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J., 26, 3397–3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baumann C., Korner R., Hofmann K., Nigg E.A. (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell, 128, 101–114. [DOI] [PubMed] [Google Scholar]
- 26. Liu Y., Nielsen C.F., Yao Q., Hickson I.D. (2014) The origins and processing of ultra fine anaphase DNA bridges. Curr. Opin. Genet. Dev., 26, 1–5. [DOI] [PubMed] [Google Scholar]
- 27. Nera B., Huang H.S., Lai T., Xu L. (2015) Elevated levels of TRF2 induce telomeric ultrafine anaphase bridges and rapid telomere deletions. Nat. Commun., 6, 10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naim V., Rosselli F. (2009) The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat. Cell Bio., 11, 761–768. [DOI] [PubMed] [Google Scholar]
- 29. Lucas I., Hyrien O. (2000) Hemicatenanes form upon inhibition of DNA replication. Nucleic Acids Res., 28, 2187–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Johnson M., Phua H.H., Bennett S.C., Spence J.M., Farr C.J. (2009) Studying vertebrate topoisomerase 2 function using a conditional knockdown system in DT40 cells. Nucleic Acids Res., 37, e98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang L.H., Mayer B., Stemmann O., Nigg E.A. (2010) Centromere DNA decatenation depends on cohesin removal and is required for mammalian cell division. J. Cell Sci., 123, 806–813. [DOI] [PubMed] [Google Scholar]
- 32. Ke Y., Huh J.W., Warrington R., Li B., Wu N., Leng M., Zhang J., Ball H.L., Li B., Yu H. (2011) PICH and BLM limit histone association with anaphase centromeric DNA threads and promote their resolution. embo J., 30, 3309–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chan K.L., Palmai-Pallag T., Ying S., Hickson I.D. (2009) Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat. Cell Biol., 11, 753–760. [DOI] [PubMed] [Google Scholar]
- 34. Dutertre S., Ababou M., Onclercq R., Delic J., Chatton B., Jaulin C., Amor-Gueret M. (2000) Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene, 19, 2731–2738. [DOI] [PubMed] [Google Scholar]
- 35. Rao X., Zhang Y., Yi Q., Hou H., Xu B., Chu L., Huang Y., Zhang W., Fenech M., Shi Q. (2008) Multiple origins of spontaneously arising micronuclei in HeLa cells: direct evidence from long-term live cell imaging. Mutat. Res., 646, 41–49. [DOI] [PubMed] [Google Scholar]
- 36. Rosin M.P., German J. (1985) Evidence for chromosome instability in vivo in Bloom syndrome: increased numbers of micronuclei in exfoliated cells. Hum Genet., 71, 187–191. [DOI] [PubMed] [Google Scholar]
- 37. Froath B., Schmidt-Preuss U., Siemers U., Zollner M., Rudiger H.W. (1984) Heterogenous carriers for Bloom syndrome exhibit a spontaneously increased micronucleus formation in cultured fibroblasts. Hum Genet., 67, 52–55. [DOI] [PubMed] [Google Scholar]
- 38. Bonassi S., Znaor A., Ceppi M., Lando C., Chang W.P., Holland N., Kirsch-Volders M., Zeiger E., Ban S., Barale R.. et al. (2006) An increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of cancer in humans. Carcinogenesis, 28, 625–631. [DOI] [PubMed] [Google Scholar]
- 39. Cohen P., Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol., 2, 769–776. [DOI] [PubMed] [Google Scholar]
- 40. Heck M.M., Hittelman W.N., Earnshaw W.C. (1988) Differential expression of DNA topoisomerase I and II during the eukaryotic cell cycle. Proc. Natl. Acad. Sci., 85, 1086–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gunawardena J. (2005) Multisite protein phosphorylation makes a good threshold but can be a poor switch. Proc. Natl, Acad. Sci., 102, 14617–14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schweiger R., Linial M. (2010) Cooperativity within proximal phosphorylation sites is revealed from large-scale proteomics data. Biol. Direct, 5, 6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brosh R.M. Jr, Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A. (2000) Replication protein A physically interacts with the Bloom’s syndrome protein and stimulates its helicase activity. J. Biol. Chem., 275, 23500–23508. [DOI] [PubMed] [Google Scholar]
- 44. Opresko P.L., von Kobbe C., Laine J.P., Harrigan J., Hickson I.D., Bohr V.A. (2002) Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem., 277, 41110–41119. [DOI] [PubMed] [Google Scholar]
- 45. Opresko P.L., Mason P.A., Podell E.R., Lei M., Hickson I.D., Cech T.R., Bohr V.A. (2005) POT1 stimulates RecQ helicases WRN and BLM to unwind telomeric DNA substrates. J. Biol. Chem., 280, 32069–32080. [DOI] [PubMed] [Google Scholar]
- 46. Wu L. (2007) Role of the BLM helicase in replication fork management. DNA Repair, 6, 936–944. [DOI] [PubMed] [Google Scholar]
- 47. Bizard A.H., Hickson I.D. (2014) The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol., 6, a016477.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chan K.L., Hickson I.D. (2011) New insights into the formation and resolution of ultra-fine anaphase bridges. Semin. Cell Dev. Biol., 22, 906–912. [DOI] [PubMed] [Google Scholar]
- 49. Barefield C., Karlseder J. (2012) The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Res., 40, 7358–7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Biebricher A., Hirano S., Enzlin J.H., Wiechens N., Streicher W, Huttner D., Wang L.H.-C., Nigg E. A., Owen-Hughes T., Liu Y.. et al. (2013) PICH: a DNA translocase specially adapted for processing anaphase bridge DNA. Mol. Cell, 51, 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Broderick R., Nieminuszczy J., Blackford A.N., Winczura A., Niedzwiedz W. (2015) TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat. Commun., 6, 6572.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.