Abstract
The melioidosis bacterium, Burkholderia pseudomallei, is increasingly being recognised as a pathogen in patients with cystic fibrosis (CF). We have recently catalogued genome-wide variation of paired, isogenic B. pseudomallei isolates from seven Australasian CF cases, which were collected between 4 and 55 months apart. Here, we extend this investigation by documenting the transcriptomic changes in B. pseudomallei in five cases. Following growth in an artificial CF sputum medium, four of the five paired isolates exhibited significant differential gene expression (DE) that affected between 32 and 792 genes. The greatest number of DE events was observed between the strains from patient CF9, consistent with the hypermutator status of the latter strain, which is deficient in the DNA mismatch repair protein MutS. Two patient isolates harboured duplications that concomitantly increased expression of the β-lactamase-encoding gene penA, and a 35 kb deletion in another abolished expression of 29 genes. Convergent expression profiles in the chronically-adapted isolates identified two significantly downregulated and 17 significantly upregulated loci, including the resistance-nodulation-division (RND) efflux pump BpeEF–OprC, the quorum-sensing hhqABCDE operon, and a cyanide- and pyocyanin-insensitive cytochrome bd quinol oxidase. These convergent pathoadaptations lead to increased expression of pathways that may suppress competing bacterial and fungal pathogens, and that enhance survival in oxygen-restricted environments, the latter of which may render conventional antibiotics less effective in vivo. Treating chronically adapted B. pseudomallei infections with antibiotics designed to target anaerobic infections, such as the nitroimidazole class of antibiotics, may significantly improve pathogen eradication attempts by exploiting this Achilles heel.
Keywords: bacterial transcriptomics, convergence, RNA-seq, evolution, antibiotic resistance, pathoadaptation
Data Summary
1. Whole-transcriptome sequence data have been deposited into the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) under BioProject accession PRJNA398168 (url – https://www.ncbi.nlm.nih.gov/bioproject/PRJNA398168).
2. The SRA accession number for the raw transcriptomic data of MSHR5654 is SRR6031143 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031143).
3. The SRA accession number for the raw transcriptomic data of MSHR5651 is SRR6031144 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031144).
4. The SRA accession number for the raw transcriptomic data of MSHR5670 is SRR6031145 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031145).
5. The SRA accession number for the raw transcriptomic data of MSHR5662 is SRR6031146 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031146).
6. The SRA accession number for the raw transcriptomic data of MSHR8437 is SRR6031147 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031147).
7. The SRA accession number for the raw transcriptomic data of MSHR8436 is SRR6031148 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031148).
8. The SRA accession number for the raw transcriptomic data of MSHR8440 is SRR6031149 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031149).
9. The SRA accession number for the raw transcriptomic data of MSHR8438 is SRR6031150 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031150).
10. The SRA accession number for the raw transcriptomic data of MSHR8442 is SRR6031151 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031151).
11. The SRA accession number for the raw transcriptomic data of MSHR8441 is SRR6031152 (https://www.ncbi.nlm.nih.gov/sra/?term=SRR6031152).
Impact Statement.
The melioidosis bacterium Burkholderia pseudomallei is a rare but important pathogen in cystic fibrosis (CF). B. pseudomallei infection of the CF airways commonly shifts towards a chronic state, where it becomes difficult to eradicate, leading to accelerated lung decline, respiratory failure and death. Understanding pathogen adaptation to the CF airways is essential for identifying targeted treatment regimens that can successfully slow or halt disease progression. We have recently catalogued the genome-wide evolution of B. pseudomallei in the lungs of seven CF melioidosis patients, which revealed several important adaptations conferring antibiotic resistance, virulence factor attenuation and genetic loss. Here, we extend upon our earlier study by characterising paired B. pseudomallei transcriptomes from five cases using RNA sequencing (RNA-seq). Differential gene expression was identified in four of the five cases, and affected loci involved in antibiotic resistance, quorum sensing, adaptation to microaerobic conditions and inhibition of competing pathogen species. Notably, RNA-seq identified additional within-host evolution signatures not observable with genomic data. Our results provide evidence for parallel evolution of B. pseudomallei in the CF lung, a trait that may be exploited for targeted pathogen eradication and improved patient outcomes.
Introduction
The Gram-negative soil-dwelling bacterium Burkholderia pseudomallei causes melioidosis, an opportunistic tropical infectious disease of humans and animals that has a high fatality rate [1]. B. pseudomallei is found in many tropical and subtropical regions globally, and has been unmasked in temperate and even arid environments following unusually wet weather events [2–4]. Infection occurs following percutaneous inoculation from contaminated soil or water, inhalation, or ingestion. Melioidosis symptoms vary widely due to the ability of B. pseudomallei to infect almost any organ, with pneumonia being the most common presentation [5, 6]. Individuals most at risk of contracting melioidosis include diabetics, those with hazardous alcohol consumption and the immunosuppressed. There has been increasing recognition that people with chronic lung diseases such as cystic fibrosis (CF) are also at a heightened risk [7, 8].
CF is a heritable disorder of the CFTR gene, and defects in CFTR (cystic fibrosis transmembrane conductance regulator) lead to exaggerated airway inflammation, an imbalance in salt regulation in the lungs and pancreas, and a chronic overproduction of thick and sticky mucus in the airways and digestive system [9]. Impaired immunity and mucus clearance encourage infection and subsequent persistence and adaptation of opportunistic bacterial pathogens in the CF lung, leading to the development of bronchiectasis with subsequent progressive pulmonary decline, and ultimately, loss of pulmonary function and death [10].
The most common pathogens of the CF lung are Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and less commonly, Achromobacter xylosoxidans, non-tuberculosis mycobacteria, Stenotrophomonas maltophilia and Burkholderia cepacia complex species [11]. The most common and best-studied CF pathogen is P. aeruginosa, which can adapt to the CF lung environment via various mechanisms. Convergent pathoadaptations in P. aeruginosa include the downregulation or loss of virulence factors and motility-encoding loci, emergence of hypermutators, enhanced antibiotic resistance and immune evasion facilitated by a switch to mucoidy and a biofilm-based lifestyle, and altered expression of other loci enhancing bacterial metabolism and survival within the nutrient-poor CF lung environment [10, 12].
Improving life expectancy for those with CF has led to an increased risk of exposure to B. pseudomallei following travel to melioidosis-endemic regions. Although uncommon, infection of the CF lung by B. pseudomallei has now been documented in at least 25 cases worldwide [13]. Due to low total case numbers, comparatively little is understood about the pathogenic role of B. pseudomallei in CF pulmonary disease. The most common clinical presentation is chronic infection (76 %), which is associated with accelerated lung function decline [13]. This prevalence contrasts with melioidosis in non-CF patients, where chronic infection occurs in only 11 % of cases [6]. To better understand B. pseudomallei pathoadaptation in the CF lung, we recently investigated the genome-wide evolution of isogenic B. pseudomallei strains isolated from seven Australasian CF patients, which were collected between 4 and 55 months apart [14]. Hallmarks of these infections included B. pseudomallei persistence despite multiple eradication attempts, multidrug resistance, mutations in virulence, metabolism and cell wall components, and the first-documented case of hypermutation in B. pseudomallei. In all except one case, multiple single-nucleotide polymorphism (SNP) and insertion-deletion (indel) mutations were identified, with a high rate of nonsynonymous mutations, many of which were predicted to affect protein function [14].
RNA sequencing (RNA-seq) provides a detailed view of the transcriptional landscape in bacterial isolates grown under different conditions or niches [15], and is now a well-established method for examining differential gene expression (DE) in bacterial pathogens [16]. Here, we performed RNA-seq on strains from five of the CF cases that we have recently described [14] to catalogue both within-host and convergent transcriptional evolution during long-term B. pseudomallei infection in the CF lung. Paired isolates representing the initial and the most recent cultures available from each patient were compared. B. pseudomallei cultures were grown in an artificial sputum medium [17, 18] to mimic the conditions found in the CF lung environment.
Methods
CF isolates
The B. pseudomallei strains used in this study are summarised in Table 1. The history and genomic analysis of these cases and strains are detailed elsewhere [14].
Table 1. Summary of the genetic mutations and differentially expressed genes between paired, sequential B. pseudomallei isolates obtained from five CF patients (adapted from [14]).
| Patient | Initial and final isolate IDs | Time between collection (months) | No. of mutational events (Chr I, Chr II) | No. of genes affected by mutations (Chr I, Chr II) | No. of DE genes (Chr I, Chr II) | No. of DE genes (downregulated, upregulated) |
|---|---|---|---|---|---|---|
| CF6 | MSHR5651, MSHR5654 | 27 | 24 (14, 10) | 29 (14, 15) | 229 (69, 160) | 229 (124, 105∗) |
| CF8 | MSHR8436, MSHR8437 | 46 | 12 (7, 5) | 39 (5, 34) | 32 (1, 31) | 32 (32†, 0) |
| CF9 | MSHR5662, MSHR5670 | 55 | 112 | 79 (40, 39) | 792 (381, 411) | 792 (558, 234) |
| CF10 | MSHR8438, MSHR8440 | 10 | 0 | 0 | 0 | 0 |
| CF11 | MSHR8441, MSHR8442 | 14 | 15 | 38 (10, 28) | 169 (62, 107) | 169 (68‡, 101) |
∗Seven of these genes are upregulated due to a 30× duplication affecting these loci in MSHR5654.
†Twenty-nine of these genes are downregulated due to a 35 kb deletion affecting these loci in MSHR8437.
‡Twenty-five of these genes appear as downregulated due to a 10× duplication affecting a 36.7 kb locus in MSHR8441 compared with only a 2× duplication in MSHR8442 [14].
Artificial sputum medium
This medium was made as described elsewhere [17, 18], with modifications detailed here. Antibiotics were not used to maintain media sterility due to concerns that their addition would alter expression profiles. Due to the impracticality of its filtration [19], 1 g porcine stomach mucin (Sigma-Aldrich) dissolved in 40 ml ultrapure water was autoclaved prior to use. All other solutions were sterilised using a 0.22 µM vacuum filter, apart from the UV-irradiated egg yolk emulsion (Oxoid), which was treated aseptically. A stock solution of diethylenetriaminepentaacetic acid (Sigma-Aldrich) was made by dissolving 59.5 mg into 5 ml very basic water (pH=14). CaCl2 was added at a final concentration of 0.22 g l−1 (J. Manos, personal communication). Final concentrations of the components were: 10 g mucin l−1, 1.39 g salmon sperm DNA l−1 (Sigma-Aldrich), 5 g NaCl l−1, 2.2 g KCl l−1, 0.22 g CaCl2 l−1, 5 g casein acid hydrosylate l−1 (Sigma-Aldrich), 10 g BSA l−1 (Roche Diagnostics), 0.005 % diethylenetriaminepentaacetic acid and 0.5 % egg yolk emulsion. Each batch was tested for sterility prior to use by plating 100 µl onto Luria–Bertani (LB) agar (Oxoid) and incubating aerobically for 24 h. pH was tested using an aliquot of the medium to ensure it was within the desired range (pH ~6.5–7). The medium was stored at 4 °C for no longer than 4 weeks prior to use.
Viability counts
Two sets of viability counts were performed for this study. The first was conducted to determine the number of c.f.u. at OD590, which enabled us to standardise the starting number of cells inoculated into the artificial sputum medium. The second was conducted to verify the final concentration of cells across all CF isolates, which enabled us to determine the number of cells for nucleic acid extraction to ensure that approximately equal cell amounts were processed for each pair. The CF isolates were subcultured from glycerol stocks onto LB agar at 37 °C for 24 h. Cells were suspended into PBS followed by spectrophotometric measurement at OD590 in a WPA CO 8000 cell density meter (Biochrom). Tenfold dilutions and plating of cultures onto LB agar were carried out, followed by enumeration at 24 h. Viable counts demonstrated that all CF isolates exhibited a similar cell density when normalised to an OD590=1.0 (range 1.3×108 to 4.9×108 cells). Based on these counts, the starting amount of culture for the CF isolates was standardised to 105 c.f.u. for all subsequent experiments.
Growth curves in artificial sputum medium
To minimize laboratory passage, each culture was again subcultured from the original glycerol stocks onto LB agar at 37 °C for 24 h, followed by a replication of OD590 measurements as determined previously. Based on the viability count data, samples were then diluted to 106 c.f.u. ml−1 in PBS. One hundred microlitres of this suspension (~105 c.f.u.) was used to inoculate 1.9 ml sputum medium, which was aliquoted into 14 ml Nunc round-bottom culture tubes (Thermo Fisher Scientific). Due to biosafety concerns, cells were grown in closed-capped tubes. The cultures were incubated at 37 °C by shaking in an orbital incubator shaker (model BL8500; Bioline) at 50, 200 or 230 r.p.m. for 44 h. Growth curves were obtained by measuring OD590 at regular intervals over this period using un-inoculated sputum medium as the control blank. Shaking at 50 r.p.m. was initially performed to mimic the low-oxygen conditions of the CF lung; however, this speed caused heavy sedimentation of cells and media components, and biofilm formation at the aerobic interface, both of which led to a decrease in optical density values over time and unpredictable, non-uniform growth. Similarly, shaking at 230 r.p.m. was too vigorous for the cells, as observed by inconsistent, non-reproducible viable counts. When shaking at 200 r.p.m., highly reproducible optical density values that correlated with viability counts were obtained (Fig. S1, available with the online version of this article), and the medium did not readily sediment. This speed was therefore used for subsequent experiments, including for RNA harvest. Viability counts were performed at the time of harvest to ensure uniformity of cell concentrations across all isolates.
RNA extraction from isolates grown in artificial sputum medium
The CF strains were grown in duplicate lineages prior to being pooled for extraction, as detailed above. This step was carried out to minimise the chance of rare but significant laboratory-generated mutation(s) affecting gene expression in one of the lineages. In addition, two independent extractions were performed for all isolates, apart from the latter CF11 strain (MSHR8442), where three RNA extractions were carried out. Based on the growth curve analysis, nucleic acids for all cultures were extracted at late log phase (17 h). At the point of harvest, the OD590 of each replicate was measured to ensure that consistent cell density had been obtained prior to combining replicates; final viability counts were also performed. Due to the highly labile nature of bacterial mRNA, two 100 µl aliquots for each strain were immediately placed into 200 µl RNAprotect (Qiagen) and incubated for 5 min to preserve their transcription profiles. Cells were pelleted by centrifugation at 5000 g for 10 min, and the supernatant discarded. Total RNA was extracted using the RNeasy Protect bacteria mini kit (Qiagen). B. pseudomallei cells were lysed following the protocol for genomic DNA extraction [20], with an extended incubation time in proteinase K and lysozyme of 1.5 h. Lysates were loaded onto the RNeasy mini columns and extractions were carried out according to the manufacturer’s instructions, including the recommended on-column DNase I digestion. In our hands, we found this DNase I treatment to be insufficient for removing all contaminating DNA, so extractions for RNA-seq were further treated using a TURBO DNA-free kit (Ambion). For each sample, 35 µl extracted RNA was incubated with 6 U TURBO DNase at 37 °C for 32 min. The remaining RNA was not treated with this second round of DNase; instead, this sample was used as template for PCR contamination screening, as described below. All samples were transferred to clean RNase/DNase-free tubes for downstream processing.
RNA quality control
To verify the removal of DNA from the total RNA extractions, two contamination screens were performed. The first was used to determine the removal of salmon sperm DNA, and the second was to determine the removal of B. pseudomallei DNA. Both the pre- and post-treated RNA samples were used to test for contamination, in duplicate, with the former acting as the positive control. The RNA samples were diluted 1/10 into molecular-grade H2O (Fisher Biotech) prior to PCR. Identification of residual salmon sperm DNA was investigated by targeting the mitochondrial 12S rDNA region of vertebrates [21]. Primers 12 S-6F (5′-CAAACTGGGATTAGATACC-3′) and B-12S-9R (5′-AGAACAGGCTCCTCTAG-3′) were used at a final concentration of 1 µM in a mix containing 1× PCR buffer (Qiagen), 1 U HotStarTaq, 0.2 mM dNTPs, 1 µl template and molecular-grade H2O in a 15 µl total reaction volume. Thermocycling conditions comprised 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 52 °C for 30 s and 72 °C for 30 s, and a final extension at 72 °C for 2 min. Amplicons were detected by agarose gel electrophoresis.
Real-time PCR was used to detect B. pseudomallei DNA contamination. The mmsA (methylmalonate-semialdehyde dehydrogenase) housekeeping gene was targeted using the primers Bp_266152_3012-F1-flap (5′-AATAAATCATAAACGTGAGGCCGGAGATGT-3′) and Bp_266152_ 3012-R1-flap (5′-AATAAATCATAAGACCGACATCACG CACAGC-3′) in combination with a B. pseudomallei-specific TaqMan MGB probe, 266152-T_Bp (5′-VIC-CGGTCTACACGCATGA-3′), as previously described [22], with the following modifications: 0.2 µM probe and 0.4 µM each primer was used, reactions were carried out in a 5 µl total reaction volume, and cycling was performed to 50 cycles.
RNA storage, shipment and RNA-seq
For each sample, 20 µl total RNA was added to an RNAstable tube (Biomatrica), gently mixed with the preservation agent and left to air-dry in a biosafety cabinet for 48 h. Samples were shipped at ambient temperature to Macrogen (Geumcheon-gu, Seoul, Republic of Korea) for RNA-seq. Ribosomal RNA was removed by treatment with the Ribo-Zero rRNA removal kit for bacteria (Epicentre), followed by 100 bp paired-end, stranded library construction using the TruSeq rapid SBS Kit (Illumina). Libraries were sequenced on either the HiSeq2000 or the HiSeq2500 platform (Illumina). All samples were extracted from two separate experiments to account for biological variation, except for MSHR8442, which was extracted thrice. Between 36 and 80 million reads were generated for each sequence, corresponding to between 3.6 and 8.1 billion bp each.
RNA-seq analysis
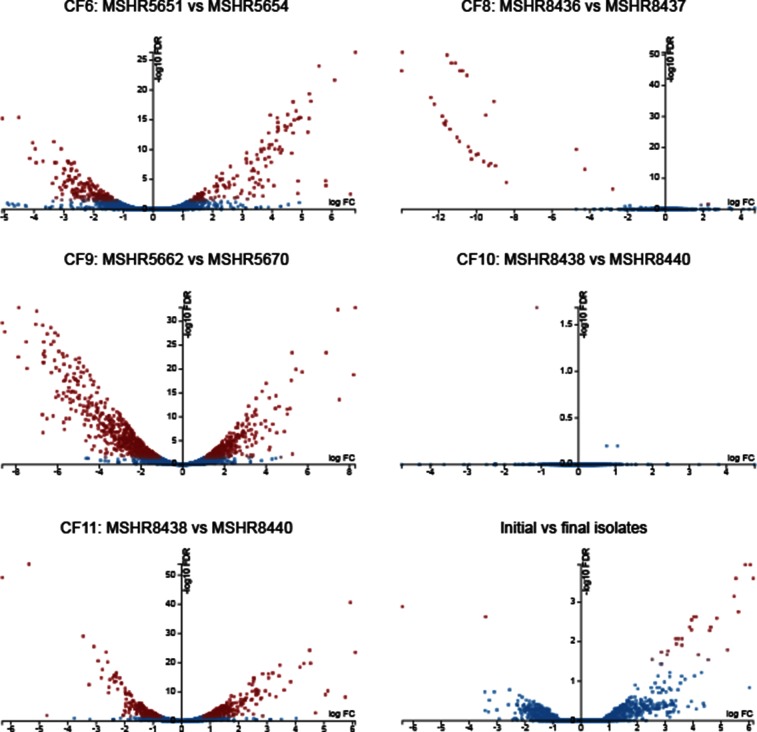
Illumina read filtering was first performed with Trimmomatic v0.33 using the following parameters: TruSeq2-PE adapter removal, leading=3, trailing=3, sliding window=4 : 15 and minimum length=36. Reads were mapped to the prototypic B. pseudomallei K96243 reference genome (RefSeq IDs NC_006350 and NC_006351 for chromosomes 1 and 2, respectively [23]) using Bowtie 2 v2.2.1 [24]. For within-patient analyses, the initial patient genomes were downloaded from the National Center for Biotechnology Information (GenBank references JYBG00000000, JYBH00000000, JYBI00000000, JYBJ00000000 and JYBK00000000 for MSHR5651 [CF6], MSHR8436 [CF8], MSHR5662 [CF9], MSHR8438 [CF10] and MSHR8441 [CF11], respectively) and annotated using Prokka v1.12 [25] with the K96243 proteins as priority (using the ‘−−proteins’ flag) prior to Bowtie 2 v2.3.4 analysis. Transcript quantification was performed with HTSeq (v0.6.1p1) [26] using the intersection non-empty mode and −−stranded=reverse parameters. DE analysis was carried out using the glmFit function of edgeR v3.18.1 [27], implemented in the online Degust 3.1.0 tool (http://degust.erc.monash.edu/). DE loci were visualised using the ‘volcano plot’ function within Degust. Several different groups were compared to determine DE. The first analyses compared initial and latter isolates within CF patients (Table 1) without summing technical replicates (i.e. the RNA-seq data from each independent experiment of a single strain) to identify DE within each patient. To determine convergent DE loci, we summed the reads for each technical replicate prior to analysis and then compared all initial versus all latter isolates, with the latter CF10 isolate excluded due to a lack of DE in this strain and the initial CF11 isolate excluded due to >3 years of infection prior to its isolation [14]. For all analyses, significant DE was defined as a log2 fold change of ≥1.5 and a false discovery rate of ≤0.01. To improve visualisation of DE loci in the volcano plot of the initial and latter comparison (Fig. 1), highly expressed DE loci in only a single strain were omitted.
Fig. 1.
Degust volcano plots showing differentially expressed (DE) genes between paired B. pseudomallei isolates retrieved from five CF lungs, and between initial and latter isolates. Four of the five pairs exhibited DE; CF10, with the shortest time between isolates, exhibited no genetic or significant transcriptomic changes. CF6, CF8, CF9 and CF11 pairs were separated by 229, 32, 792 and 169 DE loci, respectively. The 32 DE loci in CF8 were all downregulated; DE genes in CF6, CF9 and CF11 were downregulated or upregulated. Nineteen loci were differentially expressed between initial and latter isolates, of which 17 were upregulated. Blue, genes with no significant DE; red, genes with significant DE. FDR, false discovery rate; FC, log2 fold change.
Results
DE among CF isogenic pairs
DE was observed in four of the five CF pairs, with only CF10 failing to yield significant transcriptional differences (Fig. 1). We have previously shown that no genetic variants separate the CF10 strains, which had the shortest time between collection of only 10 months [14]. This lack of significant transcriptional differences rules out epigenetic effects (e.g. DNA methylation) on gene expression between the CF10 isolates, at least under the tested growth conditions, and illustrates that RNA-seq is a robust methodology that is not readily prone to false-positive results.
Of the four pairs with significant DE, CF8 had the fewest with 32 loci, followed by CF11, CF6 and CF9 with 169, 229 and 792 DE loci, respectively (Fig. 1, Table 1). These paired isolates were collected 46, 14, 27 and 55 months apart, respectively. There was good correlation between the proportion of DE loci and the genome-wide mutations catalogued between these pairs [14], with 12, 15, 24 and 112 mutational events (i.e. SNPs, indels, deletions or gene duplications) identified in CF8, CF11, CF6 and CF9, respectively (Table 1). The elevated number of mutations seen in CF9 is due to a mutS mutation in the latter strain, which confers a hypermutator phenotype, the first time hypermutation has been described in B. pseudomallei [14]; this in turn contributes to a high number of DE genes. However, when comparing the ratio of DE genes to mutational events, CF11 had the highest proportion of DE genes (11.3), followed by CF6 (9.5), CF9 (7.1) and CF8 (2.7). There was a significant skew towards DE genes located on chromosome II, which contains a lower proportion of housekeeping genes than chromosome I [23]. Despite encoding only 44 % of the genome by size and 41 % of coding sequences, chromosome II loci were significantly overrepresented in the non-hypermutator CF pairs (Pearson’s χ2 test P<0.001), with between 63 and 97 % of the DE genes residing on chromosome II. In CF9, there was a non-significant trend towards chromosome II loci, with 52 % of DE loci located on this chromosome, pointing to the more random nature of mutations in the hypermutator compared with the other cases (Table S1). In addition, to detect any significant DE in the pangenome, we performed DE analysis using the initial strain from each patient as a reference and compared these results with those obtained using K92643 (Tables S1 and S2). On balance, the within-patient analyses were almost identical to those using the K96243 reference, with most discrepancies attributable to differences in gene annotation methods.
Many DE genes are absent in the strain found in chronic-carriage melioidosis patient 314 (P314)
We compared the DE loci in the CF strains to those genes mutated in the strain found in chronic-carriage melioidosis case P314. P314 has the longest B. pseudomallei infection ever documented, and despite multiple eradication attempts, continues to harbour this bacterium in the airways since first being diagnosed in 2000. We have previously shown that the genome of a 139 month isolate from P314, MSHR6686, shows dramatic adaptation to the lung environment, including the loss of 285 kb of chromosome II at four separate locations that collectively encompass 221 genes [28]. When compared with genes lost in MSHR6686, there was a 9, 15, 20 and 97 % overlap in the latter isolates from CF9, CF6, CF11 and CF8, respectively (Table S1). The proportion of downregulated genes varied across this dataset, ranging from 13 % for CF11 to 100 % for CF8, demonstrating that the effect on gene expression at these loci is not unidirectional, with certain overlap loci in fact being upregulated in CF6, CF9 and CF11. Of note, all 29 genes (BPSS1131–BPSS1159) that were lost due to a large deletion in the latter CF8 isolate [14] were also absent in MSHR6686 [28], providing further evidence of their dispensability for long-term B. pseudomallei survival in the mammalian host. As expected, DE showed dramatic downregulation of between 8 and 14 log2 fold (341 to 15 886×) of these loci (Table S1).
DE of surface antigens in the CF6, CF9 and CF11 pairs
B. pseudomallei produces four capsular polysaccharides (CPS-I to -IV) and a lipopolysaccharide (LPS). However, only CPS-I (encoded by BPSL2786–BPSL2810) and LPS (encoded by BPSL1936 and BPSL2672–BPSL2688) are associated with virulence in mammals [29, 30]. Our previous genomic analysis of the CF pairs identified missense mutations affecting the LPS loci wzt (BPSL2681) and rmlA (BPSL2685) in CF6, a missense mutation affecting a putative LPS biosynthesis gene BPSL1119 in CF11 and a CPS-I frameshift mutation affecting wcbA (BPSL2809) in CF11 [14].
Consistent with being under heavy selection during chronic infection, we observed DE of several surface polysaccharide loci in the CF6, CF9 and CF11 pairs, the most dramatic of these being in CF9, with 46 downregulated surface polysaccharide genes (Table S1). The LPS loci wbiE (BPSL2676) and wbiD (BPSL2677) were downregulated by ~1.8 fold (3×) in the latter CF9 isolate. In addition, the poorly characterised LPS biosynthesis-related membrane protein loci BPSS1683–BPSS1685 were downregulated by ~5.9 fold (60×). This isolate also exhibited downregulation of all CPS-I loci (except wcbC) of between 1.7- and 3.1-fold (3 to 8×). In contrast, the latter isolate from CF11 showed upregulation of the CPS-I loci BPSL2793–BPSL2797 (wcbN-wcbM-gmhA-wcbL-wcbK) when compared with its initial isolate, with increases ranging from 2.7 to 6.1-fold (7 to 69×). However, when compared with initial isolates from CF6, CF8, CF9 and CF10, expression of BPSL2793–BPSL2797 in the latter CF11 strain was in fact downregulated (3.4 to 4.5-fold; 11 to 23×). This observation was confirmed as significant downregulation of these loci in the initial CF11 strain (by between 6.1- and 10.1-fold; 69 to 1098×) when compared with all other initial strains, rather than significant upregulation of these CPS-I loci in the latter CF11 strain.
Unlike CPS-I, expression of the CPS-II cluster (BPSS0417–BPSS0429) is induced when grown in water, suggesting that this polysaccharide plays a role in environmental survival [29]. One locus involved in CPS-II biosynthesis, BPSS0425, was downregulated (1.8-fold; 4×) in CF6, and the entire cluster was downregulated in CF9 (range 2.5- to 5.2-fold; 6 to 37×). Conversely, BPSS0417 and BPSS0418 were upregulated in CF11 (~1.6-fold; 3×). However, as with CPS-I, both CF11 strains exhibited significant downregulation of BPSS0417 and BPSS0418 when compared with initial strains from CF6, CF8, CF9 and CF10 (2.0- and 3.1-fold; 4 to 9×). The genes encoding CPS-III (BPSS1825–BPSS1835) were significantly downregulated in the latter isolates from CF6 (2.4- to 3.1-fold; 5 to 9×) and CF9 (5.6- to 7.5-fold; 50 to 178×). Finally, two genes within the CPS-IV cluster (BPSL2769–BPSL2785) were downregulated in CF9 (BPSL2782 and BPSL2785; 1.8- to 2.8-fold; 3 to 7×), but 11 of 17 loci from this cluster were upregulated in CF11 by 1.7- to 5.9-fold (BPSL2769, BPSL2775–BPSL2784). However, unlike CPS-I and CPS-II, this pattern of upregulation in both CF11 isolates was maintained for CPS-IV loci BPSL2769 and BPSL2775–BPSL2781 even when compared with the initial CF6, CF8, CF9 and CF10 isolates (2.0- to 4.0-fold; 4 to 16×).
DE of other virulence-associated loci
In addition to CPS-I and LPS, B. pseudomallei encodes for several other virulence factors that enhance organism survival and replication upon infection or that subvert or disarm host defences. These factors include adhesins, flagella, fimbriae, pili, specialised secretion systems, actin motility proteins, secreted factors and secondary metabolites [30]. Although virulence factors are often critically important during the acute stages of infection, they can become disadvantageous for long-term survival, presumably due their immunogenicity [12, 28]. Consistent with loss-of-virulence as a pathoadaptive mechanism in chronic B. pseudomallei infections, we have previously documented missense mutations affecting the type 3 secretion system 3 (T3SS-3) gene bsaW in CF6, and Burkholderia biofilm factor A-encoding gene, bbfA, and fimbrial protein-encoding BPSL1628 in CF9 [14]. When examining RNA-seq profiles, other virulence genes lacking genetic mutations were found to be significantly downregulated in the latter CF isolates. These loci included three type IV pilus 7 (TFP7) loci (pilR, pilG and pilN; 1.7-fold; 3×), the lysozyme inhibitor-encoding BPSL1057 (3.4-fold; 11×), Burkholderia lethal factor 1 (3.2-fold; 10×), four T3SS-3 loci (bsaS, bsaP, bsaO and bsaN), and 15 flagellum loci in CF9 (mean 2.8-fold; 8×), and a trimeric autotransporter adhesin (bpaC; BPSL1631) in CF11 (1.6-fold; 3×). Of these, the four T3SS-3 loci are also missing in chronic P314 isolates. It is notable that no genetic mutations were identified in these virulence factor loci in the latter CF isolates, highlighting the advantage of using transcriptomics to detect patterns of within-host pathogen adaptation that are not observable with genomic data alone.
High-level trimethoprim/sulfamethoxazole (SXT) resistance in B. pseudomallei involves bpeEF–oprC upregulation
The combination antibiotic SXT, the drug of choice in the eradication phase of melioidosis treatment [31], was administered to patients CF6, CF9 and CF11 during their B. pseudomallei eradication attempts (Table 2). Acquired resistance towards SXT emerged in the latter isolates from CF6 and CF11, and in midpoint isolates from CF9 [14]. The resistance-nodulation-division (RND) efflux pump, BpeEF–OprC, is responsible for widespread trimethoprim resistance in B. pseudomallei and has been implicated in SXT resistance [32]. BpeEF–OprC (BPSS0292–BPSS0294) expression is under the control of two LysR-type regulators, BpeT (BPSS0290) and BpeS (BPSL0731). We, therefore, expected to observe upregulation of bpeEF–oprC in CF strains with elevated SXT minimum inhibitory concentrations (MICs), consistent with defective bpeT or bpeS loci.
Table 2. Summary of clinical aspects and mechanisms of antibiotic resistance in B. pseudomallei isolates retrieved from five CF patients.
Antibiotics highlighted in bold were targeted towards B. pseudomallei eradication. Mutations in red are putative and remain to be functionally characterised. AMC, amoxicillin/clavulanate; AMR, antimicrobial resistance; DOX, doxycycline; F, female; IV Ab, intravenous antibiotic; LT, lung transplant; M, male; MEM, meropenem; na, not applicable; nd, not determined; TET, tetracycline; TN, tobramycin.
| Patient ID | Age at diagnosis (years) [sex] | No. of months between isolates | Treatment | AMR MIC (µg ml−1) | AMR genetic mechanism(s)*,† | DE loci confirming AMR mechanism(s) | Detected co-pathogens‡ | Patient outcome |
|---|---|---|---|---|---|---|---|---|
| CF6 | 21 [M] | 27 | AMC, CAZ, CIP, MEM, SXT | CAZ: ≥256 | PenAC69Y+~30× penA duplication | ↑22× penA | P. aeruginosa, S. aureus, B. cepacia complex | Died (~23 years old) |
| CIP: ≥32 | GyrAY77S | na | ||||||
| SXT: ≥32 | Ptr1R20fs+BpeTT314fs | ↑38-111x bpeEF–oprC | ||||||
| CF8 | 14 [M] | 46 | Nil IV Ab | MEM: 3–4; both isolates | Unknown | Unknown | P. aeruginosa, S. aureus | Unknown |
| CF9§ | 21 [M] | 55 | CAZ, MEM, SXT, TET; LT | DOX: 48 | AmrRL132P+BPSL3085A88fs | nd | P. aeruginosa | Stable despite non-eradication |
| MEM: 4 | AmrRL132P | nd | ||||||
| SXT: ≥32 | MetFN162P+DutN99S+AmrRL132P | nd | ||||||
| CF10 | 25 [F] | 10 | CAZ, MEM, TN | None detected | na | na | P. aeruginosa, S. aureus, B. cenocepacia | B. pseudomallei eradication; died (~34 years old) |
| CF11 | 10 [F] | 14 | CAZ, AMC, TN, MEM, SXT, DOX | SXT: ≥32; both isolates | Ptr1R20-A22ins+BpeSV40I and R247L | ↑40-121× bpeEF–oprC | P. aeruginosa, S. aureus | Died (16 years old) |
| CAZ: 12 | ~10× penA duplication (initial strain); ~2× penA duplication (latter strain) | ↓4× penA in latter strain | ||||||
| DOX: 4–8; both isolates | AmrRΔV62-H223+BPSL3085S130L | ↑6-9× amrAB–oprA | ||||||
| MEM: 3 | AmrRΔV62-H223 | ↑6-9× amrAB–oprA |
*Resistance profiles and mechanisms shown for the latter isolate, unless otherwise specified.
†As reported by Viberg et al. [14].
‡From Geake et al. [13].
§The final isolate from CF9, MSHR5670, was not resistant to any clinically-relevant antibiotics; however, midpoint isolate MSHR5667 (collected 47 months after the initial isolate) was DOX resistant (48 µg ml−1), and all midpoint isolates (MSHR5665, MSHR5666, MSHR5667 and MSHR5669) were SXT resistant (≥32 µg ml−1).
The latter isolate from CF6, which encodes a T314fs mutation in bpeT and is highly resistant towards SXT (MIC≥32 µg ml−1), showed 5.2- to 6.8-fold upregulation of bpeEF–oprC (38 to 111×; Table 2). This isolate also harbours an R20fs mutation in ptr1 (BPSS0039; folM), which encodes a pteridine reductase that is involved in SXT resistance [32]; this frameshift truncates Ptr1 from 267 to 91 residues. Both strains isolated from CF11 are also highly resistant to SXT (MIC≥32 µg ml−1) and encode the BpeS missense variants V40I and R247L (K96243 annotation) when compared with wild-type B. pseudomallei strains [14]. They also encode a three-residue in-frame insertion of R20-A22 in Ptr1 [14, 32]. Because both CF11 strains are SXT resistant, DE was determined by comparison with SXT-sensitive isolates in our dataset. Using this approach, significant upregulation was observed for BpeEF–OprC (5.3- to 6.9-fold; 40 to 121×; Table 2). DE was not observed for ptr1 or other genes involved in the folate biosynthesis pathway in either the CF6 or CF11 isolates (Table S1).
Ceftazidime (CAZ) resistance can occur by upregulation of penA
CAZ is a third-generation cephalosporin antibiotic that is the most commonly recommended therapy for the primary phase of melioidosis treatment [33]. In addition to SXT, the latter isolate from CF6 is highly resistant to CAZ (MIC≥256 µg ml−1) [14]. High-level CAZ resistance is often conferred by a C94Y substitution (C69Y using Ambler’s [34] numbering scheme) in PenA β-lactamase [35–37]. We have recently shown that the latter CF6 strain also harbours a ~30× duplication of a 7.5 kb region that encompasses penA; all 30 copies encode the C69Y variant of this enzyme [14]. Consistent with this duplication event, penA (BPSS0946) expression increased by 4.5-fold (22×) in the latter CF6 strain (Table 2). Six proximal genes (BPSS0945; BPSS0948–BPSS0952) were also upregulated by 3.1- to 4.7-fold (9 to 26×; Table S1). One of these, BPSS0945, is a peptidase and a putative virulence factor that may play a role in multinucleated giant cell formation [38].
A gene duplication event encompassing penA has also been documented in the CF11 isolates. The initial strain showed an elevated MIC towards CAZ (12 µg ml−1), corresponding with a ~10× duplication of a 36.7 kb region that includes penA, whereas the latter strain had a 2× duplication of this region and a wild-type CAZ MIC (2 µg ml−1) [14]. As expected, penA was downregulated by 2.1-fold (4×) in the latter isolate due to five times fewer copies of this gene (Table 2). Downregulation of other genes within the 36.7 kb locus ranged from 1.4- to 3.3-fold (3 to 10×; Table S1).
Increased doxycycline MICs in CF11 are due to amrAB–oprA upregulation and BPSL3085 mutation
Doxycycline was administered to CF11 in combination with SXT as part of a final attempt to eradicate B. pseudomallei [13, 33]. This lengthy administration led to a doxycycline MIC of 4–8 µg ml−1 in the CF11 isolates, both of which were retrieved post-treatment [14].
The RND efflux pump, AmrAB–OprA (BPSL1802–BPSL1804), efficiently effluxes aminoglycoside- and macrolide-class antibiotics [39]. We have recently shown that synergistic mutations affecting both its regulator AmrR (BPSL1805) and an S-adenosyl-l-methionine (SAM)-dependent methyltransferase (BPSL3085) led to doxycycline resistance in an Australian melioidosis case [40]. Consistent with this earlier work, both CF11 strains encode a large deletion in amrR (AmrRΔV62-H223). This mutation results in a 2.6- to 3.2-fold (6 to 9×) upregulation of amrAB–oprA in these isolates (Table 2). In addition, a previously undocumented S130L mutation in BPSL3085 was observed in both strains.
Decreased meropenem susceptibility is generally conferred by amrAB–oprA upregulation
The latter isolate from CF11, a midpoint isolate from CF9 (MSHR5667), and both CF8 isolates possess elevated MICs towards meropenem (3–4 µg ml−1) [14]. Mutations in RND efflux pump regulators, especially AmrR, are responsible for this decreased susceptibility, and importantly, such cases are associated with more refractory treatment and poorer patient outcomes [41]. Alongside its elevated doxycycline MIC, the AmrRΔV62-H223 mutation in the latter CF11 isolate also confers decreased meropenem susceptibility. The midpoint CF9 isolate, MSHR5667, was not examined using RNA-seq or quantitative PCR in our study, but we hypothesise that its AmrRL132P mutation causes ~2–4-fold upregulation of amrAB–oprA in a similar fashion to the CF11 isolates. Curiously, the CF8 isolates had no observed mutations or DE loci involving efflux pumps when compared with wild-type strains, and the basis for the elevated MIC in these isolates remains elusive. Growing these isolates in the presence of low-level meropenem to induce expression prior to RNA-seq/quantitative PCR [42] may be required to identify the transcriptional mechanism/s underpinning this phenotype.
Evidence of convergent DE between early and latter CF isolates
Finally, we performed a comparison of expression profiles from all CF cases to identify a signal of convergent gene expression (pathoadaptation) across early versus latter isolates. To yield the most robust and relevant analysis, we excluded the latter isolate from CF10 due to a lack of DE in this strain, and the initial isolate from CF11, which was retrieved >3 years after infection and had already undergone substantial genetic and transcriptional modifications. Exclusion of both strains was supported by a lack of convergent signal when they were included in the analysis (data not shown). Using these parameters, 17 genes were found to be significantly upregulated, and 2 were significantly downregulated (Table S3). Five (26 %) loci encode for hypothetical proteins with no known function, of which four were upregulated. Among the convergently upregulated genes with known function was the RND efflux pump BpeEF–OprC (4.8- to 6.1-fold; 28 to 69×), the CydAB cytochrome bd quinol oxidase (5.5- to 5.9-fold; 45 to 60×), and the quorum sensing hhqABCDEFG (BPSS0481–BPSS0487) operon (3.4- to 4.1-fold; 11 to 17×). The downregulated locus, BPSS0351, encodes the MerR family transcriptional regulator CueR (3.4-fold; 11×).
Discussion
The melioidosis pathogen, B. pseudomallei, is an uncommon pathogen in CF, with fewer than 30 cases documented worldwide to date. However, it is an important pathogen in CF airways due to its ability to persist despite treatment and its association with accelerating respiratory decline [13]. We have recently performed comparative genomic analysis of isogenic strains collected between 4 and 55 months apart from the airways of the patients in seven of these cases [14]. Here, we sought to further characterise these chronic cases by examining the transcriptomes of five paired B. pseudomallei isolates retrieved between 10 and 55 months apart. Although our sample numbers are modest, our study represents more than one-sixth of globally reported melioidosis cases in CF to date.
Isolates were cultured in an artificial CF sputum medium [18] to mimic their original in vivo environment. Under these conditions, DE was detected in four of the five cases and ranged from 32 to 792 genes, with the hypermutator strain from CF9 contributing the greatest number of DE loci (Table S1). Interestingly, when compared with the number of genetic changes occurring in each isolate pair, the latter isolate from CF11 had a higher proportion of DE loci to mutations (11.3) than CF9 (7.1), demonstrating that hypermutation does not necessarily lead to a similarly high number of transcriptional differences. The one case with no DE, CF10, exhibited no genetic changes (i.e. SNPs, small indels, copy-number variants or large deletions) and had the shortest time between isolate collection at 10 months [14]. All other cases encoded genetic differences between pairs. The DE genes fell into several functional categories (Table S1), reflecting the diversity and versatility of pathoadaptive pathways in B. pseudomallei. Our RNA-seq analysis revealed that many of the DE genes were absent in the chronic P314 strain (range 9–97 %), providing further evidence that these loci are not required for long-term survival in the airways. Perhaps most striking was the observation that nearly one-third (32 %) of DE genes lack a known function, highlighting the relative paucity of functional studies into this important yet under-recognised pathogen.
B. pseudomallei has a ~7.3 Mbp genome that is encoded on two replicons; a ~4.1 Mbp ‘housekeeping’ chromosome I and a ~3.2 Mbp ‘accessory’ chromosome II. The genome of archetypal strain K96243 consists of 3460 and 2395 coding sequences on chromosomes I and II, respectively [23]. There was a greater proportion of DE genes (between 52 and 97 %) on chromosome II in all cases, despite its smaller size and fewer coding sequences. We have previously shown that a greater proportion of mutational events affect chromosome II of B. pseudomallei in a long-term chronic-carriage isolate from P314 [28]. The skew towards DE loci on chromosome II in chronic B. pseudomallei isolates points towards a lesser role for chromosome II loci in bacterial survival and persistence within the human host, which is reflected by the greater degree of reductive evolution affecting this replicon [14, 28]. In their study of the transcriptional landscape of B. pseudomallei, Ooi and co-workers found that only ~28 % of chromosome II genes were expressed under a single condition, compared with ~72 % of chromosome I genes [43]. Taken together, these results support the accessory role of the chromosome II replicon in B. pseudomallei.
Attenuation of immunogenic surface antigens and other virulence factors are hallmarks of chronically persistent infections across many pathogenic bacterial species, including B. pseudomallei [14, 28]. Encoded by the 34.5 kb wcb operon BPSL2786–BPSL2810 [44, 45], the B. pseudomallei CPS-I is a potent virulence determinant that imparts high-level serum resistance and facilitates phagocytic evasion [45]. This capsule is also intact in the equine-adapted B. pseudomallei clone, Burkholderia mallei, where it and has been shown to be essential for its virulence [46, 47]. Our prior genomic analysis identified only a single CF pair with mutated CPS-I in CF11. This frameshift mutation in wcbA results in a truncated protein [14] that likely causes reduced, although not abolished, CPS-I production [48]. The DE analysis provides further evidence of CPS-I inactivation in the CF pairs, with downregulation of all but one of the CPS-I loci in the latter CF9 isolate, and downregulation of wcbN-wcbM-gmhA-wcbL-wcbK in both CF11 isolates. Although CPS-III is not required for virulence, it is noteworthy that this locus was also downregulated in CF9 and CF11, as it has been previously shown that CPS-III expression is tied to that of CPS-I genes [43].
Like CPS-I, the B. pseudomallei LPS is required for capsule biosynthesis, virulence and serum resistance [49]. Its immunogenic outer-membrane component is readily recognised by the host innate immune system [50], which makes LPS a target for inactivation in chronic bacterial infections. We have previously uncovered missense mutations in the LPS wzt and rmlA loci of CF6, and a missense mutation affecting BPSL1119 in CF11 [14]. DE analysis identified additional evidence for reduced or abolished LPS production in a third CF case, CF9, due to the significant downregulation of wbiD, wbiE and BPSS1683–BPSS1685. Prior work has suggested that CPS-I and LPS may be disadvantageous for B. pseudomallei persistence due to their virulence potential and immunogenicity [28]. By examining both genomic and transcriptomic modifications over time, it is now clear that these capsule clusters, in their wild-type form, pose a major issue for successful long-term B. pseudomallei persistence in the CF lung, with the bacterium either mutating or downregulating key genes in CPS-I and LPS pathways. We also observed genetic mutation or transcriptional downregulation of other virulence genes in latter CF strains including TFP7 loci, Burkholderia lethal factor 1, T3SS-3 loci and flagellum loci, suggesting that these loci are similarly detrimental to long-term B. pseudomallei survival in the human host. Importantly, the RNA-seq data identified additional cases of surface antigen and virulence factor abrogation that were not observable with only genomic data. This finding underscores the importance of using both genomic and transcriptomic approaches to identify the functional consequences of within-host evolution of chronic bacterial infections.
SXT is used during the eradication phase of melioidosis treatment and is recommended for post-exposure prophylaxis [51]. We have previously shown that the latter isolate from CF6, and both isolates from CF11, had developed high-level (≥32 µg ml−1) SXT resistance over the course of treatment. These elevated MICs were proposed to be due to mutations within BpeEF–OprC efflux pump regulators (BpeT T314fs in CF6, and BpeS V40I and R247L in CF11) alongside mutations affecting the R20 residue of Ptr1/FolM (R20fs in CF6; R20-A22 duplication in CF11) [14]. Here, we have demonstrated that the efflux pump regulatory mutations cause a dramatic upregulation of bpeEF–oprC in these strains of between 5.2- and 6.9-fold (38 to 121×), mirroring expression levels in bpeS and bpeT laboratory-generated mutants with high-level SXT resistance [32]. Our results confirm those of Podnecky and colleagues [32] showing that upregulation of bpeEF–oprC via BpeS or BpeT dysregulation, together with Ptr1/FolM alteration, leads to a significant increase in SXT MICs that would render this antibiotic ineffective in vivo. RNA-seq is, thus, a useful tool for observing the functional consequences of regulatory mutations that control RND efflux pump expression.
In addition to SXT resistance, the initial isolate from CF11 (12 µg ml−1) and the latter isolate from CF6 (≥256 µg ml−1) are resistant to CAZ. Our prior genomic study showed that CAZ resistance in the initial CF11 strain was due to a 10× duplication of a 36.7 kb region encompassing the β-lactamase gene, penA, the first time that gene duplication has been shown to confer CAZ resistance in B. pseudomallei. In contrast, a 2× duplication of this region in the latter strain did not raise the CAZ MIC above wild-type levels [14]. Similarly, the latter strain from CF6 exhibits a 30× duplication of a 7.5 kb region encompassing penA; however, all 30 copies encode a C69Y missense mutation, which by itself causes high-level (≥256 µg ml−1) CAZ MICs [35]. RNA-seq confirmed the penA duplications, with the 30× variant from CF6 observed as 4.5-fold (22×) upregulation. Similarly, a 2.1-fold (4×) downregulation of penA in the latter strain from CF11 was linked to a 5× greater copy number in the early strain [14]. Thus, RNA-seq provides excellent correlation with gene copy number variation determined from whole-genome sequence coverage data. Taken together, the combined genetic and transcriptional changes affecting antibiotic resistance genes in the CF airways-adapted B. pseudomallei strains illustrates both the intractability of eradicating chronic bacterial infections and the unintended consequences of prolonged antibiotic use in CF treatment.
Although determining within-host transcriptional differences in longitudinal isolates yields valuable insights into the infection dynamics within individual patients, identifying convergent transcriptional changes provides a potential means to predict pathogen behaviour and evolution across multiple CF cases in a relatively straightforward manner. Such predictability could conceivably be exploited to improve the diagnosis or treatment of intractable CF infections, or ideally, to prevent them from progressing in the first place. Therefore, a major objective of this study was to identify evidence of convergence in B. pseudomallei gene expression during its transition to a chronic infection. Despite the small number of CF melioidosis patients available for this study, a signal of convergent pathoadaptation was identified between the initial and latter isolates, with 19 significantly DE loci identified, 17 of which were upregulated (Table S3). This convergence is noteworthy given the large size of the B. pseudomallei genome and the many redundant pathways that could lead to similar adaptive phenotypes, a phenomenon that is well-recognised in P. aeruginosa [52]. One advantage of identifying convergence using transcriptomics rather than genomic data is that it can reveal the transcriptional consequence of multiple genetic mutations; for example, we have observed that multiple missense mutations in the RND efflux pump regulator AmrR lead to the same transcriptional outcome of amrAB–oprA upregulation [41]. As such, RNA-seq data can simplify the identification of convergently expressed loci that can be dysregulated by multiple genetic variants.
The development of antibiotic resistance is a recurring theme in P. aeruginosa isolated from the CF airways [12], and we have recently shown that the same adaptive phenomenon can be observed in the genome of B. pseudomallei in response to prolonged, high-dose antibiotic therapy [14]. It was, therefore, not surprising to identify the convergent upregulation of bpeEF–oprC (4.8- to 6.1-fold; 28 to 69×), which was significantly upregulated in two of the four patients with DE (CF6 and CF11), and which led to SXT resistance as discussed above. The second convergently upregulated locus was the cydAB operon (BPSL0501 and BPSL0502), which encodes cytochrome bd quinol oxidase (5.5- to 5.9-fold; 45 to 60×); this locus was significantly differentially expressed in CF6 and CF9. CydAB is an aerobic terminal oxidase that oxidizes ubiquinol-8 and reduces oxygen to water under oxygen-limiting conditions. This enzyme is better able to scavenge oxygen under microaerobic conditions compared with cytochrome o oxidase, which otherwise predominates as the terminal respiratory enzyme in electron transport-associated energy production [53]. Voggu and colleagues demonstrated that non-pathogenic Staphylococcus species were better able to resist P. aeruginosa antagonism compared with Staphylococcus aureus due to the insensitivity of the non-pathogenic staphylococci cytochrome bd quinol oxidases to the presence of the small respiratory inhibitors hydrogen cyanide and pyocyanin, which are commonly secreted by P. aeruginosa in the CF lung [54]. Thus, it is feasible that the convergent upregulation of cydAB loci represents a defence mechanism employed by B. pseudomallei to counteract the toxic effects of small respiratory inhibitors produced by P. aeruginosa in the CF lung. In support of this hypothesis, P. aeruginosa was co-isolated in all five CF cases examined in this study [14]. Alternatively, cydAB upregulation may simply represent a physiological response to the oxygen-limited environment of the CF airways, as its expression is known to be induced in B. pseudomallei in hypoxic conditions [55]. Under hypoxia, many pathogens including B. pseudomallei become less susceptible to conventional antibiotics, which are typically effective under aerobic conditions, but more susceptible to antibiotics that target anaerobic infections. One example is the nitroimidazole class of antibiotics [55], which reduce their nitro group only under hypoxic conditions, causing DNA strand breakage and bacterial cell death [56]. This phenomenon may explain the difficulties with chronic B. pseudomallei eradication using conventional antibiotics like CAZ and SXT, and raises the exciting but not-yet-tested possibility that nitroimidazoles may be a highly effective therapeutic option for chronic, hypoxia-adapted B. pseudomallei infections such as those adapted to the CF airways.
A third convergently upregulated locus, the quorum sensing operon hhqABCDEFG (3.4- to 4.1-fold; 11× to 17×), is homologous to the B. cepacia complex hmqABCDEFG operon [57]. This operon synthesises a class of compounds known as 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs), the methylated counterparts of 2-alkyl-4(1H)-quinolones (AHQs; also known as HAQs). AHQs were first recognised in P. aeruginosa and are produced by the signalling system pqsABCDE [58]. This cluster produces over 50 different AHQs, and these compounds exhibit diverse biological activities that enable cell-to-cell communication within and between bacterial species and the regulation of various functions including secondary metabolism, virulence, antibacterial activity and biofilm formation [58]. In contrast, little is currently known about the role of HMAQs and AHQs in Burkholderia spp. [57]. The AHQ precursor molecule 2-heptyl-4(1H)-quinolone (HHQ) that is produced by P. aeruginosa actively suppresses the host innate immune response [59], a role that could be shared by B. pseudomallei HHQ. A second possibility is that these compounds impart a competitive advantage in the CF lung environment as HMAQs produced by B. cepacia exhibit antifungal activity [60], so it is feasible that the hmqABCDEFG operon of B. pseudomallei produces similarly potent compounds that can inhibit fungal species from establishing residence in the CF lung. The convergent upregulation of hhqA–hhqG in the B. pseudomallei CF isolates points to a putative role for AHQ-based compounds in B. pseudomallei signalling, immune evasion or competition in the CF lung. More work is needed to elucidate the myriad functions of AHQ compounds in B. pseudomallei, and particularly their role in promoting bacterial persistence in the CF airways.
Of the two convergently downregulated loci, only one, BPSS0351, has an assigned function, although little is known about the role of this gene and its product in B. pseudomallei. This gene encodes CueR (3.4-fold; 11×), a MerR family copper response regulator that is highly sensitive to the presence of copper (Cu) and which regulates the transcription of genes that protect against toxic metal ion concentrations [61, 62]. Cu has a long history as an effective antimicrobial agent due its ability to generate reactive oxygen species, with Cu accumulation in the mammalian host purported to act as an innate immune defence mechanism to restrict pathogen growth [63]. Thus, downregulation of cueR in the latter CF isolates may represent a mechanism for mitigating Cu toxicity in the host, similarly to Escherichia coli [62]. However, there are contradictory reports as to whether Cu levels are elevated in CF sputa [64, 65], and the artificial sputum growth medium does not appear to contain elevated Cu levels [18]. CueR regulates the Cu/silver ATPase CopA and the multicopper oxidase CueO enzymes in E. coli, which correspond to BPSS0224 and BPSL0897 in B. pseudomallei K96243, respectively; however, neither of these genes showed DE in any of the patient pairs. The absence of concomitant increased expression of cueO, which converts periplasmic Cu+ to less toxic Cu2+ in vivo [38], suggests that other enigmatic pressures are responsible for decreasing cueR expression in latter CF isolates. The biological role of these other factors requires further exploration.
We recognise that there are limitations to our study. Growth conditions are known to be an important consideration for mRNA-based investigations due to the alteration of the transcriptome when isolates are grown under different environments or media components [66]. Although our in vitro conditions do not completely mimic the conditions seen in the CF lung, the artificial sputum medium is designed to reflect the nutrient conditions of this environment [18], and our shaking parameters provided a robust way of measuring cellular growth over time while avoiding non-uniform cellular growth, which ensured harvest of B. pseudomallei cultures at the same growth phase (Fig. S1). Additionally, the use of isogenic strain pairs with genomic data [14] enabled us to comprehensively assess the effects of transcriptional adaptation to the CF lung compared with their underlying genetic variants. Our conditions provided transcriptomic data that were consistent with expected expression differences based on genome-wide alterations. Other studies have used artificial sputum media and additional mechanical methods to mimic the CF lung conditions. A rotating wall vessel has been developed to simulate the low fluid shear conditions encountered in CF mucus due to pathological effects of CFTR dysfunction on mucociliary clearance [67], with CF-derived P. aeruginosa isolates demonstrating transcriptional differences depending on shear conditions [19]. The culturing methods for bacterial RNA-seq are a critical consideration in experimental design as they can affect transcriptomic profiles, and the impact of conditions should be considered when comparing transcriptional differences between studies. Another shortcoming is that we only examined five patient pairs due to the relative paucity of melioidosis CF cases worldwide, and only two isolates from each patient due to limited bacterial colony selection and storage at the time of sputum collection and processing. Deeper sampling efforts across a greater number of melioidosis CF patients would be needed to provide greater confidence in our convergent adaptation findings and would allow a more thorough understanding of B. pseudomallei population dynamics and diversity to be attained. Nevertheless, the findings from our study provide important new insights into B. pseudomallei evolution in the CF airways, with many, although not complete, parallels with the common CF pathogens, P. aeruginosa and B. cepacia complex species.
Data bibliography
K96243. Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. NCBI GenBank accession numbers NC_006350.1 and NC_006351.1 (2004).
MSHR5651 (CF6). Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. NCBI GenBank accession number JYBG00000000 (2015).
MSHR8436 (CF8). Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. NCBI GenBank accession number JYBH00000000 (2015).
MSHR5662 (CF9). Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. NCBI GenBank accession number JYBI00000000 (2015).
MSHR8438 (CF10). Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. NCBI GenBank accession number JYBJ00000000 (2015).
MSHR8441 (CF11). Viberg LT, Price EP, Kidd TJ, Bell SC, Currie BJ, Sarovich DS. NCBI GenBank accession number JYBK00000000 (2015).
Funding information
This work was supported by grants 1046812 and 1098337 from the Australian National Health and Medical Research Council. The funder had no role in study design, data collection and interpretation, or the decision to submit the work for publication. E. P. P. was supported by a University of the Sunshine Coast Fellowship, L. T. V. was supported by an Australian Postgraduate Award and Menzies Enhanced Living scholarship, S. C.B . was supported by a Health Research Fellowship from Queensland Health, and D. S. S. was supported by an Advance Queensland Fellowship (award no. AQRF13016-17RD2).
Acknowledgements
We are grateful to Dr Jim Manos (University of Sydney, Sydney, NSW, Australia) for providing advice on the preparation of the artificial sputum medium, Ammar Aziz for helpful conversations regarding RNA-seq analyses, and Jessica Webb, Mark Mayo and Vanessa Theobald (Menzies School of Health Research, Charles Darwin University) for laboratory assistance.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Ethics approval for this study was obtained as previously described [6, 13].
Supplementary Data
Supplementary Data
Supplementary Data
Supplementary Data
Footnotes
Abbreviations: AHQ, 2-alkyl-4(1H)-quinolone; CAZ, ceftazidime; CF, cystic fibrosis; CPS, capsular polysaccharide; DE, differential gene expression; HMAQ, 4-hydroxy-3-methyl-2-alkylquinoline; Indel, insertion-deletion; LPS, lipopolysaccharide; MIC, minimum inhibitory concentration; NCBI, National Center for Biotechnology Information ; RNA-seq, RNA sequencing; RND, resistance-nodulation-division; SNP, single-nucleotide polymorphism; SXT, trimethoprim/sulfamethoxazole; T3SS-3, type 3 secretion system 3.
All supporting data, code and protocols have been provided within the article or through supplementary data files. One supplementary figure and three supplementary tables are available with the online version of this article.
References
- 1.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med. 2012;367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 2.Yip TW, Hewagama S, Mayo M, Price EP, Sarovich DS, et al. Endemic melioidosis in residents of desert region after atypically intense rainfall in central Australia, 2011. Emerg Infect Dis. 2015;21:1038–1040. doi: 10.3201/eid2106.141908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapple SN, Sarovich DS, Holden MT, Peacock SJ, Buller N, et al. Whole-genome sequencing of a quarter-century melioidosis outbreak in temperate Australia uncovers a region of low-prevalence endemicity. Microb Genom. 2016;2:e000067. doi: 10.1099/mgen.0.000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarovich DS, Garin B, de Smet B, Kaestli M, Mayo M, et al. Phylogenomic analysis reveals an Asian origin for African Burkholderia pseudomallei and further supports melioidosis endemicity in Africa. mSphere. 2016;1:e00089-15. doi: 10.1128/mSphere.00089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leelarasamee A, Bovornkitti S. Melioidosis: review and update. Rev Infect Dis. 1989;11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 6.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010;4:e900. doi: 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Carroll MR, Kidd TJ, Coulter C, Smith HV, Rose BR, et al. Burkholderia pseudomallei: another emerging pathogen in cystic fibrosis. Thorax. 2003;58:1087–1091. doi: 10.1136/thorax.58.12.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holland DJ, Wesley A, Drinkovic D, Currie BJ. Cystic fibrosis and Burkholderia pseudomallei infection: an emerging problem? Clin Infect Dis. 2002;35:e138. doi: 10.1086/344447. [DOI] [PubMed] [Google Scholar]
- 9.Amaral MD. Novel personalized therapies for cystic fibrosis: treating the basic defect in all patients. J Intern Med. 2015;277:155–166. doi: 10.1111/joim.12314. [DOI] [PubMed] [Google Scholar]
- 10.Cohen TS, Prince A. Cystic fibrosis: a mucosal immunodeficiency syndrome. Nat Med. 2012;18:509–519. doi: 10.1038/nm.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coutinho HD, Falcão-Silva VS, Gonçalves GF. Pulmonary bacterial pathogens in cystic fibrosis patients and antibiotic therapy: a tool for the health workers. Int Arch Med. 2008;1:24. doi: 10.1186/1755-7682-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winstanley C, O'Brien S, Brockhurst MA. Pseudomonas aeruginosa evolutionary adaptation and diversification in cystic fibrosis chronic lung infections. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geake JB, Reid DW, Currie BJ, Bell SC, Bright-Thomas R, et al. An international, multicentre evaluation and description of Burkholderia pseudomallei infection in cystic fibrosis. BMC Pulm Med. 2015;15:116. doi: 10.1186/s12890-015-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Viberg LT, Sarovich DS, Kidd TJ, Geake JB, Bell SC, et al. Within-host evolution of Burkholderia pseudomallei during chronic infection of seven Australasian cystic fibrosis patients. MBio. 2017;8:e00356-17. doi: 10.1128/mBio.00356-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 16.Creecy JP, Conway T. Quantitative bacterial transcriptomics with RNA-seq. Curr Opin Microbiol. 2015;23:133–140. doi: 10.1016/j.mib.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sriramulu DD, Lünsdorf H, Lam JS, Römling U. Microcolony formation: a novel biofilm model of Pseudomonas aeruginosa for the cystic fibrosis lung. J Med Microbiol. 2005;54:667–676. doi: 10.1099/jmm.0.45969-0. [DOI] [PubMed] [Google Scholar]
- 18.Fung C, Naughton S, Turnbull L, Tingpej P, Rose B, et al. Gene expression of Pseudomonas aeruginosa in a mucin-containing synthetic growth medium mimicking cystic fibrosis lung sputum. J Med Microbiol. 2010;59:1089–1100. doi: 10.1099/jmm.0.019984-0. [DOI] [PubMed] [Google Scholar]
- 19.Dingemans J, Monsieurs P, Yu SH, Crabbé A, Förstner KU, et al. Effect of shear stress on Pseudomonas aeruginosa isolated from the cystic fibrosis lung. MBio. 2016;7:e00813-16. doi: 10.1128/mBio.00813-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Currie BJ, Gal D, Mayo M, Ward L, Godoy D, et al. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect Dis. 2007;7:68. doi: 10.1186/1471-2334-7-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humair PF, Douet V, Morán Cadenas F, Schouls LM, van de Pol I, et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J Med Entomol. 2007;44:869–880. doi: 10.1093/jmedent/44.5.869. [DOI] [PubMed] [Google Scholar]
- 22.Price EP, Dale JL, Cook JM, Sarovich DS, Seymour ML, et al. Development and validation of Burkholderia pseudomallei-specific real-time PCR assays for clinical, environmental or forensic detection applications. PLoS One. 2012;7:e37723. doi: 10.1371/journal.pone.0037723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holden MT, Titball RW, Peacock SJ, Cerdeño-Tárraga AM, Atkins T, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price EP, Sarovich DS, Mayo M, Tuanyok A, Drees KP, et al. Within-host evolution of Burkholderia pseudomallei over a twelve-year chronic carriage infection. MBio. 2013;4:e00388-13. doi: 10.1128/mBio.00388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reckseidler-Zenteno SL, Viteri DF, Moore R, Wong E, Tuanyok A, et al. Characterization of the type III capsular polysaccharide produced by Burkholderia pseudomallei. J Med Microbiol. 2010;59:1403–1414. doi: 10.1099/jmm.0.022202-0. [DOI] [PubMed] [Google Scholar]
- 30.Stone JK, Deshazer D, Brett PJ, Burtnick MN. Melioidosis: molecular aspects of pathogenesis. Expert Rev Anti Infect Ther. 2014;12:1487–1499. doi: 10.1586/14787210.2014.970634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lipsitz R, Garges S, Aurigemma R, Baccam P, Blaney DD, et al. Workshop on treatment of and postexposure prophylaxis for Burkholderia pseudomallei and B. mallei infection, 2010. Emerg Infect Dis. 2012;18:e2. doi: 10.3201/eid1812.120638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podnecky NL, Rhodes KA, Mima T, Drew HR, Chirakul S, et al. Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei: deviation from the norm. MBio. 2017;8:e01357-17. doi: 10.1128/mBio.01357-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Currie BJ. Melioidosis: evolving concepts in epidemiology, pathogenesis, and treatment. Semin Respir Crit Care Med. 2015;36:111–125. doi: 10.1055/s-0034-1398389. [DOI] [PubMed] [Google Scholar]
- 34.Ambler RP, Coulson AF, Frère JM, Ghuysen JM, Joris B, et al. A standard numbering scheme for the class A beta-lactamases. Biochem J. 1991;276:269–270. doi: 10.1042/bj2760269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sam IC, See KH, Puthucheary SD. Variations in ceftazidime and amoxicillin-clavulanate susceptibilities within a clonal infection of Burkholderia pseudomallei. J Clin Microbiol. 2009;47:1556–1558. doi: 10.1128/JCM.01657-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarovich DS, Price EP, von Schulze AT, Cook JM, Mayo M, et al. Characterization of ceftazidime resistance mechanisms in clinical isolates of Burkholderia pseudomallei from Australia. PLoS One. 2012;7:e30789. doi: 10.1371/journal.pone.0030789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rholl DA, Papp-Wallace KM, Tomaras AP, Vasil ML, Bonomo RA, et al. Molecular investigations of PenA-mediated beta-lactam resistance in Burkholderia pseudomallei. Front Microbiol. 2011;2:139. doi: 10.3389/fmicb.2011.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh AP, Lai SC, Nandi T, Chua HH, Ooi WF, et al. Evolutionary analysis of Burkholderia pseudomallei identifies putative novel virulence genes, including a microbial regulator of host cell autophagy. J Bacteriol. 2013;195:5487–5498. doi: 10.1128/JB.00718-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore RA, Deshazer D, Reckseidler S, Weissman A, Woods DE. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 1999;43:465–470. doi: 10.1128/aac.43.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Webb JR, Price EP, Currie BJ, Sarovich DS. Loss of methyltransferase function and increased efflux activity leads to doxycycline resistance in Burkholderia pseudomallei. Antimicrob Agents Chemother. 2017;61:e00268-17. doi: 10.1128/AAC.00268-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarovich DS, Webb JR, Pitman MC, Viberg LT, Mayo M, et al. Raising the stakes: loss of efflux-pump regulation decreases meropenem susceptibility in Burkholderia pseudomallei. Clin Infect Dis. doi: 10.1093/cid/ciy069. doi:10.1093/cid/ciy069 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 42.Webb JR, Price EP, Somprasong N, Schweizer HP, Baird RW, et al. Development and validation of a triplex qPCR assay to detect efflux pump-mediated antibiotic resistance in Burkholderia pseudomallei. bioRxiv. 2018 doi: 10.2217/fmb-2018-0155. doi:10.1101/301960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ooi WF, Ong C, Nandi T, Kreisberg JF, Chua HH, et al. The condition-dependent transcriptional landscape of Burkholderia pseudomallei. PLoS Genet. 2013;9:e1003795. doi: 10.1371/journal.pgen.1003795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reckseidler SL, Deshazer D, Sokol PA, Woods DE. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect Immun. 2001;69:34–44. doi: 10.1128/IAI.69.1.34-44.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reckseidler-Zenteno SL, Devinney R, Woods DE. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect Immun. 2005;73:1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deshazer D, Waag DM, Fritz DL, Woods DE. Identification of a Burkholderia mallei polysaccharide gene cluster by subtractive hybridization and demonstration that the encoded capsule is an essential virulence determinant. Microb Pathog. 2001;30:253–269. doi: 10.1006/mpat.2000.0430. [DOI] [PubMed] [Google Scholar]
- 47.Atkins T, Prior R, Mack K, Russell P, Nelson M, et al. Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J Med Microbiol. 2002;51:539–553. doi: 10.1099/0022-1317-51-7-539. [DOI] [PubMed] [Google Scholar]
- 48.Cuccui J, Milne TS, Harmer N, George AJ, Harding SV, et al. Characterization of the Burkholderia pseudomallei K96243 capsular polysaccharide I coding region. Infect Immun. 2012;80:1209–1221. doi: 10.1128/IAI.05805-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deshazer D, Brett PJ, Woods DE. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol. 1998;30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 50.Tuanyok A, Stone JK, Mayo M, Kaestli M, Gruendike J, et al. The genetic and molecular basis of O-antigenic diversity in Burkholderia pseudomallei lipopolysaccharide. PLoS Negl Trop Dis. 2012;6:e1453. doi: 10.1371/journal.pntd.0001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peacock SJ, Schweizer HP, Dance DA, Smith TL, Gee JE, et al. Management of accidental laboratory exposure to Burkholderia pseudomallei and B. mallei. Emerg Infect Dis. 2008;14:e2. doi: 10.3201/eid1407.071501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol. 2015;10:599–611. doi: 10.2217/fmb.15.3. [DOI] [PubMed] [Google Scholar]
- 53.Cotter PA, Melville SB, Albrecht JA, Gunsalus RP. Aerobic regulation of cytochrome d oxidase (cydAB) operon expression in Escherichia coli: roles of Fnr and ArcA in repression and activation. Mol Microbiol. 1997;25:605–615. doi: 10.1046/j.1365-2958.1997.5031860.x. [DOI] [PubMed] [Google Scholar]
- 54.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, et al. Microevolution of cytochrome bd oxidase in staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol. 2006;188:8079–8086. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamad MA, Austin CR, Stewart AL, Higgins M, Vázquez-Torres A, et al. Adaptation and antibiotic tolerance of anaerobic Burkholderia pseudomallei. Antimicrob Agents Chemother. 2011;55:3313–3323. doi: 10.1128/AAC.00953-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edwards DI. Mechanisms of selective toxicity of metronidazole and other nitroimidazole drugs. Br J Vener Dis. 1980;56:285–290. doi: 10.1136/sti.56.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapalain A, Groleau MC, Le Guillouzer S, Miomandre A, Vial L, et al. Interplay between 4-hydroxy-3-methyl-2-alkylquinoline and N-acyl-homoserine lactone signaling in a Burkholderia cepacia complex clinical strain. Front Microbiol. 2017;8:1021. doi: 10.3389/fmicb.2017.01021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, et al. Functional genetic analysis reveals a 2-alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol. 2006;13:701–710. doi: 10.1016/j.chembiol.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 59.Kim K, Kim YU, Koh BH, Hwang SS, Kim SH, et al. HHQ and PQS, two Pseudomonas aeruginosa quorum-sensing molecules, down-regulate the innate immune responses through the nuclear factor-kappaB pathway. Immunology. 2010;129:578–588. doi: 10.1111/j.1365-2567.2009.03160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kilani-Feki O, Culioli G, Ortalo-Magné A, Zouari N, Blache Y, et al. Environmental Burkholderia cepacia strain Cs5 acting by two analogous alkyl-quinolones and a didecyl-phthalate against a broad spectrum of phytopathogens fungi. Curr Microbiol. 2011;62:1490–1495. doi: 10.1007/s00284-011-9892-6. [DOI] [PubMed] [Google Scholar]
- 61.Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 62.Singh SK, Grass G, Rensing C, Montfort WR. Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol. 2004;186:7815–7817. doi: 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gray RD, Duncan A, Noble D, Imrie M, O'Reilly DS, et al. Sputum trace metals are biomarkers of inflammatory and suppurative lung disease. Chest. 2010;137:635–641. doi: 10.1378/chest.09-1047. [DOI] [PubMed] [Google Scholar]
- 65.Smith DJ, Anderson GJ, Bell SC, Reid DW. Elevated metal concentrations in the CF airway correlate with cellular injury and disease severity. J Cyst Fibros. 2014;13:289–295. doi: 10.1016/j.jcf.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Yoder-Himes DR, Chain PS, Zhu Y, Wurtzel O, Rubin EM, et al. Mapping the Burkholderia cenocepacia niche response via high-throughput sequencing. Proc Natl Acad Sci USA. 2009;106:3976–3981. doi: 10.1073/pnas.0813403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crabbé A, de Boever P, van Houdt R, Moors H, Mergeay M, et al. Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ Microbiol. 2008;10:2098–2110. doi: 10.1111/j.1462-2920.2008.01631.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.