Abstract.
Appropriate diagnostic techniques are crucial to global soil-transmitted helminth (STH) control efforts. The recommended Kato–Katz method has low sensitivity in low-transmission settings. Quantitative polymerase chain reaction (qPCR) is a highly sensitive alternative diagnostic option. However, little is known about the variability in qPCR results, and there are few published comparisons between qPCR and other microscopy-based techniques such as sodium nitrate flotation (SNF). Using 865 stool samples collected from 571 individuals, we compared SNF and qPCR in terms of diagnostic sensitivity and infection intensity measurements. In addition, we conducted repeated examinations on a single Necator americanus–positive stool sample over a 6-month period. Results showed good diagnostic agreement between SNF and qPCR for Ascaris spp. (κ = 0.69, P < 0.001), and moderate agreement for hookworm (κ = 0.55, P < 0.001) and Trichuris spp. (κ = 0.50, P < 0.001). Quantitative polymerase chain reaction demonstrated higher sensitivity than SNF for Ascaris spp. (94.1% versus 68.1%) and hookworm (75.7% versus 66.9%) but not for Trichuris spp. (53.1% versus 81.3%), which had very low prevalence. Sodium nitrate flotation and qPCR infection intensity measurements were strongly correlated for Ascaris spp. (ρ = 0.82, P < 0.001) and moderately correlated for hookworm (ρ = 0.58, P < 0.001). Repeated examinations using qPCR showed that N. americanus cycle threshold values decreased significantly at 1 month and remained stable thereafter. Results confirm the high diagnostic sensitivity of qPCR for Ascaris spp. and hookworm, particularly for light-intensity infections, which is ideal for settings approaching transmission elimination. Results support the potential for qPCR to be used as a quantitative assay for STH. Further research is needed in settings where Trichuris trichiura is endemic.
INTRODUCTION
Soil-transmitted helminth (STH) infections—encompassing roundworms (Ascaris lumbricoides), hookworms (Necator americanus, Ancylostoma duodenale, and Ancylostoma ceylanicum), and whipworms (Trichuris trichiura)—are the most prevalent of the neglected tropical diseases, estimated to infect more than a billion people worldwide1 and causing a global disease burden of approximately 3.5 million disability-adjusted life years.2 In recent years, a global focus on the control and elimination of neglected tropical diseases has led to significant scaling up of STH control programs.3 These programs focus on delivering regular deworming treatments to high-risk population groups, mostly children, and aim to reduce morbidity by reducing the proportion of high-intensity infections.4,5 Recently, there has been increasing interest in interrupting STH transmission, through expanding drug administration programs community-wide, such that regular drug treatment is no longer required.6,7
Mapping and monitoring the prevalence and intensity of STH infections in endemic areas is critical for planning mass drug administration programs, assessing program impact, and observing for emerging benzimidazole resistance in humans.8 Diagnostic tests that can accurately and sensitively diagnose infections and classify their intensity are, therefore, essential to the success of mass deworming programs, and represent a crucial component in achieving STH control.
The Kato–Katz technique is most commonly used for STH diagnosis, as per World Health Organization (WHO) guidelines.9 This microscopy-based technique provides a measure of infection intensity in eggs per gram (EPG) of feces, and benefits from being an affordable, relatively simple test that can be carried out in the field.10 A limitation of the Kato–Katz technique is that multiple samples per individual should be examined, given the low sensitivity of a single examination.11 Even when multiple samples are examined, the Kato–Katz technique has poor diagnostic sensitivity in areas of low STH transmission.12 A further limitation is the rapid degradation of hookworm eggs after slide preparation, meaning that examination must occur within 30 minutes of preparation.13
Polymerase chain reaction (PCR)–based techniques are increasingly used to diagnose STH infections. This sensitive molecular approach detects very small quantities of DNA, with previous work demonstrating higher sensitivity compared with the Kato–Katz technique.14–19 Real-time quantitative PCR (qPCR) techniques can also be used to provide quantitative measures of infection intensity.15,19,20 Polymerase chain reaction–determined infection intensity results have demonstrated good correlation with EPG counts obtained using the Kato–Katz technique for both Ascaris spp. and hookworm.15,18,19,21
Despite the increasing use of molecular techniques for STH diagnostics, there are limited published data relating to variability in DNA detection using qPCR. In practice, stool samples are generally preserved for transportation from field sites to a laboratory equipped to perform qPCR, where they are stored before DNA extraction. Therefore, understanding the variability in DNA detection over time is crucial for accurate interpretation of qPCR results.22 A previous report, in which a human stool sample was spiked with N. americanus eggs isolated from hamster stool and preserved for up to 60 days, found that quantitation cycle values (also known as cycle threshold [Ct] values) remained relatively constant over time when stored at 4°C, whereas more substantial variation occurred when samples were stored at 32°C.22 However, no studies have examined samples stored at room temperature (21–22°C), and importantly, none have used samples obtained from infected humans to reflect samples collected during field trials.
Furthermore, there are limited published data comparing qPCR results with other microscopy-based diagnostic methods that may be more sensitive than the Kato–Katz technique. Sodium nitrate flotation (SNF) is a simple technique widely used in veterinary parasitology and has previously demonstrated higher sensitivity for detecting hookworm eggs compared with the Kato–Katz technique.16 Although PCR-based techniques have been shown in two studies to have a higher diagnostic sensitivity than SNF,16,20 there are no published data comparing intensity measurements obtained from SNF and qPCR. A previous attempt to compare quantitative results from these techniques revealed no significant statistical relationship and data were not presented.20
The objectives of this study were 1) to compare the diagnostic performance of SNF and qPCR for STH infections, in terms of both diagnostic sensitivity and infection intensity measurements, using samples from a field trial; and 2) to examine variation in qPCR infection intensity results (Ct values), by conducting repeated analysis on a stool sample positive for N. americanus, preserved in 5% potassium dichromate over a 6-month period.
MATERIALS AND METHODS
Ethics and informed consent.
The research protocol for the field study in which samples were collected for the SNF and qPCR comparison (the [S]WASH-D for Worms pilot study) was approved by the Human Research Ethics Committees at the Australian National University (2015/111) and the Timor-Leste Ministry of Health (2015/196). Written consent was provided by parents of all participating children.
The stool sample used for investigating DNA extraction and PCR variability was obtained from an adult donor at the QIMR Berghofer Medical Research Institute in Brisbane, Australia, who was known to be infected with N. americanus and who provided written informed consent.
Field study area and sample collection.
The stool samples used to compare SNF and qPCR were collected from children attending six primary schools in the Aileu and Manufahi municipalities of Timor-Leste. Samples were collected between May 2015 and June 2016 in the (S)WASH-D for Worms pilot study.23 Children were given labeled stool containers, instructed on how to provide a sample, and requested to collect an early morning sample the following day and bring it to school. Immediately on sample receipt, the research team prepared two aliquots of 2–3 g of stool from each participant. One was preserved in 8 mL of 10% formalin and the other in 5 mL of 5% (w/v) potassium dichromate. The formalin-fixed samples were transported at room temperature to the University of Melbourne, Victoria, Australia, for analysis using SNF. The potassium dichromate–fixed samples were transported at room temperature to the QIMR Berghofer Medical Research Institute, Brisbane, Australia, for analysis using qPCR.
Approximately 5–6 months after baseline sample collection, a single dose of 400 mg albendazole was given to all children under direct observation. Follow-up stool samples were collected and processed using the same procedures 6 months after treatment.
Sodium nitrate flotation procedure.
Formalin-fixed samples were examined at the University of Melbourne, between 1 and 3 months after sample collection, using a technique that has been described previously.16 The samples were centrifuged for 2 minutes at 3,000 × g, formalin poured off, and the sample thoroughly mixed with distilled water. The suspension was then strained through two layers of surgical gauze, poured into a fresh 10 mL centrifuge tube, and centrifuged for 2 minutes at 3,000 × g. The supernatant was carefully removed using a pipette, leaving a fecal pellet between 100 and 300 mg in size. The volume of the fecal pellet was estimated using graduated lines on the centrifuge tube. Sodium nitrate solution at a specific gravity of 1.20 was added to the centrifuge tube and a positive meniscus created. A coverslip was placed over the centrifuge tube for 10 minutes and then transferred to a microscope slide, which was examined in its entirety at ×100 magnification by one of three trained microscopists for STH egg enumeration. Finally, the EPG for each STH was obtained by multiplying the number of eggs counted as required, depending on the size of the filtered fecal pellet examined (e.g., for a 200 mg sample, the number of eggs was multiplied by 5).
One coverslip was examined for most samples; however, for quality control, two coverslips were prepared separately from the same sample and read by individual microscopists for 10% of samples. In these cases, a positive reading by either microscopist was considered a positive result, and when necessary, the two EPG readings were averaged to give a single EPG.
Quantitative PCR procedure.
The potassium dichromate–fixed samples were analyzed at the QIMR Berghofer Medical Research Institute using a multiplex real-time PCR procedure as previously published.20 Following the removal of the preservative, DNA was extracted from stool samples using the PowerSoil DNA Isolation Kit (Mo Bio, Carlsbad, CA) with minor modifications. DNA extractions were carried out between 6 weeks and 6 months after sample collection. The extracted DNA was run in a multiplex real-time PCR reaction that was a quantitative assay for Ascaris spp., N. americanus, Ancylostoma spp., and Trichuris spp.20 A known amount of equine herpesvirus (EHV) plasmid was added as a positive PCR control. Details of all primers and probes are shown in Supplemental Table 1.
The multiplex qPCR assays were run using the Rotor-Gene 6000 (Qiagen, Melbourne, Victoria, Australia), with reactions set up as per previous descriptions.14,20 Cycling conditions consisted of 15 minutes at 95°C followed by 40 cycles of 95°C for 9 seconds and 60°C for 60 seconds. Each qPCR assay returned a Ct value for each STH, representing the cycle number at which a signal exceeding background level was detected. The maximum Ct value considered to represent a positive result was 31 for Ascaris spp. and 35 for all other species, consistent with previous studies.20 For each qPCR assay, two Ct values were generated by performing two reaction runs, and these were averaged to give a single Ct value. The final Ct value for each sample was then converted to a measure of infection intensity in relative fluorescence units (RFU), calculated using the following formula provided by the Rotor-Gene Q software: infection intensity = 10−0.298*Ct + 9.81 RFU.
Assessment of DNA extraction and PCR variability.
Separate from the examination of samples from the field trial, to explore variability in Ct values obtained from qPCR analysis in preserved samples over time, repeated examinations were conducted on a single stool sample. A stool sample was obtained from a donor known to be infected with N. americanus. The sample was homogenized and then separated into aliquots of 1 g each, which were preserved in 5% (w/v) potassium dichromate within several hours of sample collection. The aliquots were stored at room temperature. DNA extraction and subsequent qPCR analysis was performed at five time points: 2 days following preservation, at monthly intervals for 3 months, and then at 6 months after preservation. At each of these time points, three separate aliquots were subjected to triplicate DNA extractions of 200 μg, each of which was examined using the qPCR assay for N. americanus. This gave a total of 15 aliquots and 45 PCR replicates from the original stool sample. Equine herpesvirus was included as an extraction and PCR control.
Statistical analysis.
Diagnostic agreement between SNF and qPCR was examined using Kappa agreement statistics. The Wilcoxon rank sum test was used to compare the Ct values of qPCR-positive samples that were SNF-positive versus SNF-negative. Spearman’s rank correlation coefficients were used to examine the association between PCR intensity and EPG values. The sensitivities of qPCR and SNF were estimated by considering an individual as “true positive” if they had a positive result by either method, creating a diagnostic pseudo-“gold standard.” Specificity was assumed to be 100%.
For the analysis of Ct value variation, a linear mixed model was used to examine the impact of time on Ct values and the variability between and within individual aliquots. Time point was included as a categorical fixed effect and sample number as a random effect. All analyses were conducted using Stata Version 14 (StataCorp, College Station, TX).
RESULTS
Field trial participants.
Diagnostic results for qPCR and SNF were available for 462 children at baseline and 403 at follow-up, for 865 samples in total, from 571 individuals. The mean age of participating children was 9.2 years (range 4–17 years) at baseline and 9.0 years (range 4–17 years) at follow-up. Slightly more than half (52.1%) of the participants were female.
Soil-transmitted helminth prevalence and intensity.
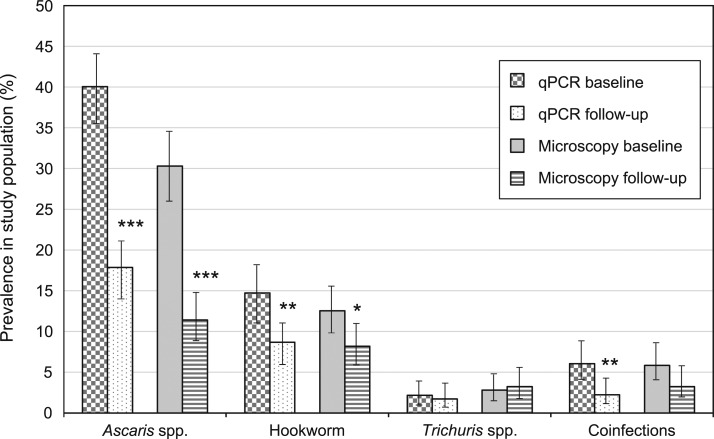
At baseline, Ascaris spp. prevalence was 40.0% (185/462 samples) by qPCR and 30.3% (140/462) by SNF. Hookworm prevalence was 12.6% (58/462) by SNF, whereas by qPCR, 13.9% (64/462) of samples were positive for N. americanus and 0.9% (4/462) for Ancylostoma spp. Prevalence of Trichuris spp. was lower at 2.2% (10/462) by qPCR and 2.8% (13/462) by SNF. Prevalence of all infections was lower at follow-up, apart from Trichuris spp. by SNF (see Table 1 and Figure 1).
Table 1.
Soil-transmitted helminth prevalence and intensity at study time points, as measured by qPCR and SNF
| Prevalence by qPCR (95% CI) | Prevalence by SNF (95% CI) | P value* | Mean cycle threshold value (range) | Mean eggs per gram (range) | |
|---|---|---|---|---|---|
| Baseline (N = 462) | |||||
| Ascaris spp. | 40.0% (35.7–44.6) | 30.3% (26.2–34.7) | 0.002 | 18.3 (9.1–31.0) | 568.2 (4.0–5,244.0) |
| Hookworm | 14.7% (11.8–18.3) | 12.6% (9.8–15.9) | 0.300 | – | 26.6 (2.0–180.0) |
| Necator americanus | 13.9% (11.0–17.3) | – | 25.1 (24.1–26.0) | – | |
| Ancylostoma spp. | 0.9% (0.3–2.3) | – | 22.4 (20.5–24.4) | – | |
| Trichuris spp. | 2.2% (1.2–4.0) | 2.8% (1.6–4.8) | 0.547 | 31.7 (25.7–35.0) | 21.6 (4.4–54.6) |
| Coinfections | 6.1% (4.2–8.7)† | 5.8% (4.0–8.4)‡ | 0.851 | – | – |
| Follow-up (N = 403) | |||||
| Ascaris spp. | 17.9% (14.4–21.9) | 11.4% (8.7–14.9) | 0.010 | 22.6 (11.2–31.0) | 350.8 (2.9–3,373.0) |
| Hookworm | 8.7% (6.3–11.9) | 8.2% (5.9–11.3) | 0.800 | – | 21.2 (2.2–128.6) |
| N. americanus | 8.7% (6.3–11.9) | – | 25.4 (18.6–31.8) | – | |
| Anyclostoma spp. | 0 | – | – | – | |
| Trichuris spp. | 1.7% (0.8–3.6) | 3.2% (1.9–5.5) | 0.174 | 29.7 (21.2–34.1) | 11.1 (2.5–36.7) |
| Coinfections | 2.2% (1.2–4.3)§ | 3.2% (1.9–5.5)¶ | 0.387 | – | – |
CI = confidence interval; qPCR = quantitative polymerase chain reaction; SNF = sodium nitrate flotation.
P values comparing prevalence obtained using SNF and qPCR.
Of 28 coinfections on qPCR at baseline: 21 Ascaris spp. + N. americanus; six Ascaris spp. + Trichuris spp; one Ascaris spp. + Ancylostoma spp.
Of 27 coinfections on SNF at baseline: 19 Ascaris lumbricoides + hookworm, five A. lumbricoides + Trichuris trichiura; two hookworm + T. trichiura; one triple infection.
Of 9 coinfections on qPCR at follow-up: eight Ascaris spp. + N. americanus; one Ascaris spp. + Trichuris spp.
Of 13 coinfections on SNF at follow-up: nine A. lumbricoides + hookworm; three A. lumbricoides + T. trichiura; one triple infection.
Figure 1.
Baseline and follow-up prevalence of each soil-transmitted helminth as measured by quantitative polymerase chain reaction (qPCR) and sodium nitrate flotation (microscopy). Difference between baseline and follow-up prevalence: *P < 0.05, **P < 0.01, ***P < 0.001.
All hookworm and T. trichiura infections diagnosed by SNF at baseline were light-intensity infections according to WHO-defined EPG cut-offs.9 Just 1.4% of baseline A. lumbricoides infections were moderate-intensity infections, whereas the remainder were light-intensity. All infections at follow-up were light-intensity. Quality control results for SNF are shown in Supplemental Table 2.
Diagnostic performance of qPCR and SNF.
As shown in Table 2, there was substantial agreement between qPCR and SNF for diagnosis of Ascaris spp. (κ = 0.69, P < 0.001). Of 186 samples that were positive by SNF, only 16 were negative on qPCR; whereas 87 of 257 qPCR-positive samples were negative on SNF. For hookworm, there was moderate agreement between qPCR and SNF (κ = 0.55, P < 0.001); 45/103 qPCR-positive samples were negative on SNF and 33/91 SNF-positive samples were negative on qPCR. For Trichuris spp., there was moderate agreement between qPCR and SNF (κ = 0.50, P < 0.001), with 6/17 qPCR-positive samples negative on SNF and 15/26 SNF-positive samples negative on qPCR.
Table 2.
Diagnostic agreement of quantitative polymerase chain reaction and sodium nitrate flotation
| PCR result | SNF-positive | SNF-negative | Agreement (%) | Kappa statistic* | P value | |
|---|---|---|---|---|---|---|
| Ascaris spp. | PCR positive | 170 | 87 | 762 (88.1) | 0.6902 | < 0.001 |
| PCR negative | 16 | 592 | ||||
| Hookworm | PCR positive | 58 | 45 | 787 (91.0) | 0.5474 | < 0.001 |
| PCR negative | 33 | 729 | ||||
| Trichuris spp. | PCR positive | 11 | 6 | 844 (97.6) | 0.4997 | < 0.001 |
| PCR negative | 15 | 833 |
PCR = polymerase chain reaction; SNF = sodium nitrate flotation.
Kappa agreement level: < 0.20 Poor; 0.21–0.40 Fair; 0.41–0.60 Moderate; 0.61–0.80 Good; 0.81–1.00 Very good.
For Ascaris spp., the mean Ct value of qPCR-positive samples that were SNF-positive was 15.75 (95% confidence interval (CI): 15.19–16.32), whereas the mean Ct value of qPCR-positive samples that were SNF-negative was significantly higher at 26.78 (95% CI: 25.85–27.61, P < 0.001). Similarly, for hookworm, the mean Ct value was higher for SNF-negative samples (27.62, 95% CI: 26.48–28.76) compared with SNF-positive samples (23.10, 95% CI: 22.41–23.78, P < 0.001).
The sensitivity of qPCR was 94.1% for Ascaris spp. (257/273 samples) and 75.7% for hookworm (103/136 samples), whereas the sensitivity of SNF was lower at 68.1% for Ascaris spp. (186/273 samples) and 66.9% for hookworm (91/136 samples). For Trichuris spp., the sensitivity of qPCR was 53.1% (17/32 samples) and the sensitivity of SNF was higher at 81.3% (26/32 samples).
Comparison of infection intensity.
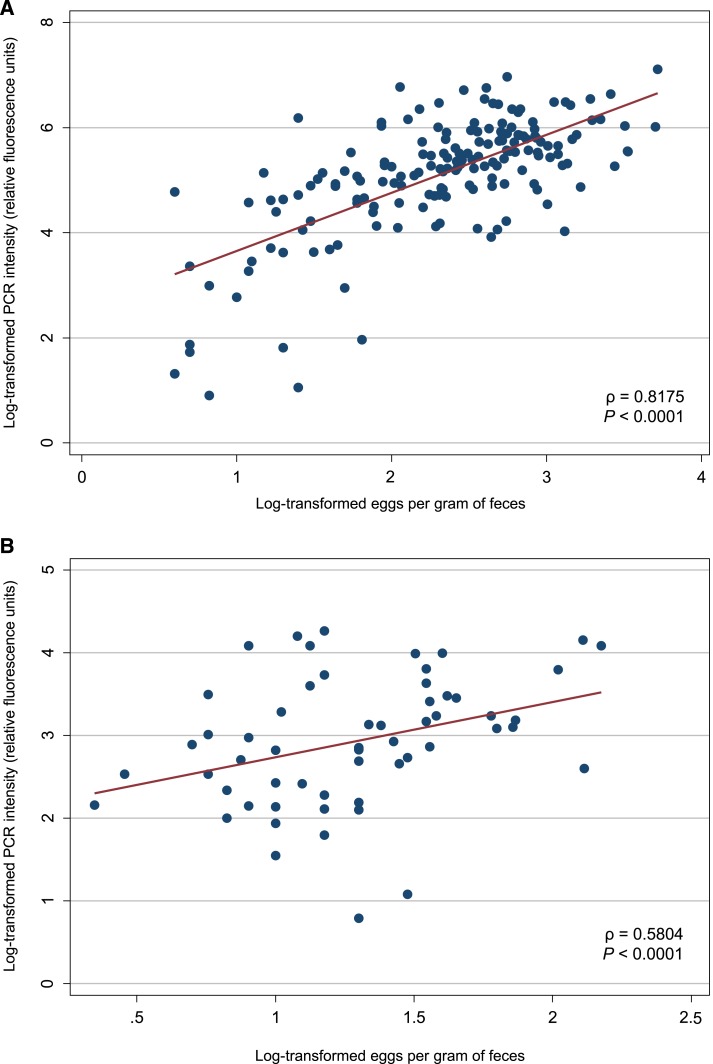
For Ascaris spp., infection intensity values obtained using qPCR were significantly and strongly correlated with EPG values obtained using SNF (ρ = 0.82, P < 0.001). For hookworm, infection intensity values obtained using qPCR were significantly and moderately correlated with EPG values obtained using SNF (ρ = 0.58, P < 0.001). Scatter plots of log-transformed EPG and log-transformed infection intensity for Ascaris spp. and hookworm are shown in Figure 2.
Figure 2.
Scatter plots showing the relationship between infection intensity measured by sodium nitrate flotation (eggs per gram of feces) and quantitative polymerase chain reaction (PCR) (reactive fluorescence units) on universal log10 transformation, for Ascaris spp. (A) and hookworm (B). This figure appears in color at www.ajtmh.org.
Correlations between infection intensity values were stronger at follow-up than baseline for both Ascaris spp. (ρ = 0.85 versus ρ = 0.77) and hookworm (ρ = 0.67 versus ρ = 0.55); see Supplemental Figures 1 and 2. Because of a low number of positive samples, infection intensity comparisons were not performed for Trichuris spp.
Variability in DNA detection.
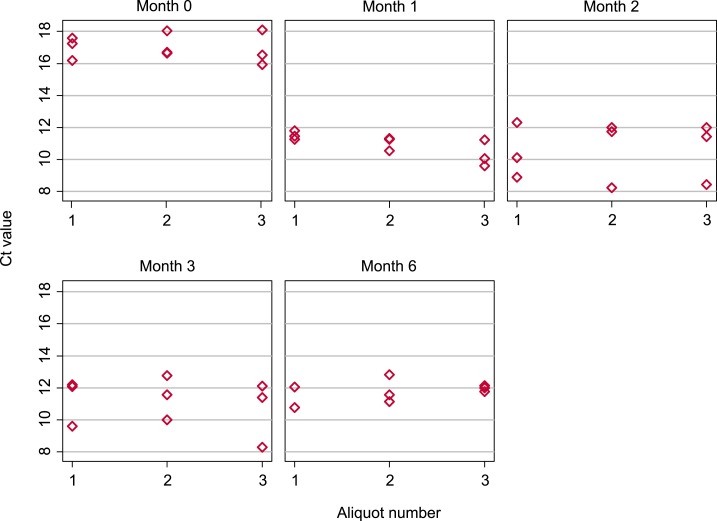
Cycle threshold values obtained for each separate aliquot at each time point are depicted in Figure 3 and summarized in Table 3.
Figure 3.
Box plots showing Necator americanus cycle threshold (Ct) values obtained from individual aliquots measured at five time points following sample preservation. Three aliquots were each subjected to triplicate DNA extractions and quantitative polymerase chain reaction at each time point. This figure appears in color at www.ajtmh.org.
Table 3.
Ct values obtained for Necator americanus and equine herpesvirus from the same stool sample, measured at five time points
| Number of aliquots | Number of PCR replicates | Mean (SD) Ct value | 95% CI for mean Ct value | Minimum Ct value | Maximum Ct value | |
|---|---|---|---|---|---|---|
| N. americanus | ||||||
| Month 0 | 3 | 9 | 17.00 (0.79) | 16.47–17.52 | 15.94 | 18.10 |
| Month 1 | 3 | 9 | 10.95 (0.72) | 10.47–11.43 | 9.62 | 11.80 |
| Month 2 | 3 | 9 | 10.57 (1.67) | 9.45–11.70 | 8.24 | 12.31 |
| Month 3 | 3 | 9 | 11.11 (1.49) | 10.11–12.11 | 8.28 | 12.76 |
| Month 6 | 3 | 8 | 11.78 (0.63) | 11.33–12.23 | 10.77 | 12.83 |
| Equine herpesvirus (positive PCR control) | ||||||
| Month 0 | 3 | 9 | 19.57 (0.31) | 19.37–19.78 | 19.18 | 20.00 |
| Month 1 | 3 | 9 | 18.37 (0.26) | 18.21–18.53 | 18.14 | 18.84 |
| Month 2 | 3 | 9 | 18.78 (0.53) | 18.42–19.14 | 18.30 | 19.93 |
| Month 3 | 3 | 9 | 19.41 (0.63) | 18.99–19.84 | 18.23 | 20.39 |
| Month 6 | 3 | 8 | 18.78 (0.51) | 18.41–19.14 | 18.22 | 19.62 |
CI = confidence interval; Ct = cycle threshold; PCR = polymerase chain reaction.
As shown in Table 3, the mean Ct value for N. americanus obtained from PCR replicates run 2 days after DNA extraction was 17.00 (95% CI: 16.47–17.52), whereas for PCR replicates run at or later than one month, mean Ct values ranged from 10.57 (95% CI: 9.45–11.70) to 11.78 (95% CI: 11.33–12.23). Cycle threshold values for EHV were more consistent across time points, with mean values between 18.37 (95% CI: 18.21–18.53) and 19.57 (95% CI: 19.37–19.78).
Results of the linear mixed model examining the impact of time on N. americanus Ct values and the variability within and between different aliquots are shown in Table 4. These results show that Ct values obtained at month 0 were significantly higher than those obtained at month one (P < 0.001), whereas there were no significant differences between Ct values obtained at month one and any later time point. There was negligible random effects variance for aliquot, reflecting minimal variability between Ct values obtained from separate aliquots at a given time point. On the other hand, the residual variance was higher at 1.18 (95% CI: 0.78–1.79), reflecting greater variability between separate DNA extractions from the same aliquot.
Table 4.
Results of linear mixed model examining impact of time and aliquot on Necator americanus cycle threshold values
| Regression coefficient | 95% CI | P value | |
|---|---|---|---|
| Time point | |||
| Month 0 | 6.05 | 5.04–7.05 | < 0.0001 |
| Month 1 | (Ref) | – | – |
| Month 2 | −0.37 | −1.38 to 0.63 | 0.46 |
| Month 3 | 0.16 | −0.84 to 1.16 | 0.75 |
| Month 6 | 0.83 | −0.20 to 1.87 | 0.11 |
| Random effects variance (95% CI) | |||
| Aliquot | < 0.0001 (< 0.0001 to < 0.0001) | ||
| Residual | 1.18 (0.78–1.79) | ||
CI = confidence interval.
DISCUSSION
We compared the diagnostic performance of SNF and qPCR, both of which have previously demonstrated higher sensitivity compared with the recommended Kato–Katz technique for the diagnosis of STH infections.15,16 Our results confirm that qPCR is more sensitive than SNF for both Ascaris spp. and hookworm.20 For Trichuris spp., qPCR demonstrated lower sensitivity than SNF. Most previous studies comparing qPCR and Kato–Katz have focused on Ascaris spp. and hookworm. Only a small number have compared diagnostic sensitivity for Trichuris spp.; these showed that qPCR had higher sensitivity than Kato–Katz.17,18 Our findings highlight the need for further evaluation of qPCR in settings where T. trichiura is endemic, both before and after mass drug administration. A more sensitive, species-specific PCR assay for T. trichiura may yield improved results.24
Notably, for Ascaris spp. and hookworm, infections that were detected by qPCR but not by SNF were of significantly lighter intensity than those detected by both techniques, confirming the superior performance of qPCR in detecting light-intensity infections.14,15 Stool samples are heterogeneous in terms of helminth egg distribution, and even with thorough mixing during the preparation procedures, there is variability in the number of eggs present in prepared slides. Therefore, light-intensity infections may easily be missed using microscopy-based techniques.
Polymerase chain reaction techniques can detect very small quantities of DNA, which explains their greater sensitivity at lower intensities. However, a number of STH eggs were detected on SNF that were negative by qPCR, particularly for hookworm and Trichuris spp. This may also be explained by heterogeneous egg distribution. For qPCR, an average 200 mg of feces is used, as opposed to 2 g for SNF, which may explain the discrepancy of results. Microscopy-based techniques may also produce false-positive results if strongyle eggs representing non-hookworm genera, for example Oesophagostomum spp. and Trichostrongylus spp.,25,26 or fecal material are mistakenly counted as hookworm eggs.
We demonstrated a strong correlation between intensity results obtained from qPCR and SNF for Ascaris spp. and a moderate correlation for hookworm, which provides additional evidence supporting the potential for qPCR as a quantitative technique for STH diagnosis.15,20 Correlations remained strong at study follow-up, highlighting the capacity of qPCR for evaluating mass drug administration programs at community level. Although qPCR-determined intensity has been shown to correlate with EPG obtained from other microscopy-based techniques,15,18,19,21 this is the first demonstration of such correlation between qPCR and SNF. A previous study found no relationship between these two techniques and also reported lower diagnostic agreement between SNF and qPCR for hookworm, with more infections missed by microscopy.20 This demonstrates the importance of skilled microscopists in performing SNF.
When performing DNA extraction and qPCR on aliquots of the same stool sample, stored at room temperature in 5% potassium dichromate over a 6-month period, N. americanus Ct values decreased significantly between the time of preservation and one month later, and remained relatively stable thereafter. However, results were shown to vary by up to four Ct values at a given time point. This partly reflects heterogeneous egg distribution within stool samples as discussed previously, given the more consistent Ct values obtained for the EHV plasmid positive control. In addition, slight inconsistencies in the volume of stool being added, and in DNA target recovery between replicates, may have contributed to the variability.
The initial decrease in Ct values was likely due to the fact that helminth eggs in the stored stool samples embryonated, causing copy number to exponentially increase. This decrease in Ct values was not seen in a previous study that examined preserved stool samples over a 2-month period.22 However, that study used hookworm-naïve human stool samples, spiked with N. americanus eggs obtained from hamster stool. These eggs may have already embryonated during storage before spiking. To our knowledge, our study is the first to examine variability in DNA detection for STH over time using a stool sample obtained from an infected human, reflecting samples that would be collected in an endemic setting. Our results highlight the importance of time between preservation and DNA extraction on qPCR results, and suggest that to maximize consistency, DNA extraction should occur at least a month after sample preservation because quantitative results appear to stabilize after this time.
Quantitative PCR represents an excellent diagnostic option in scenarios where highly sensitive diagnostic techniques are required to detect light-intensity infections. Such situations include settings approaching STH transmission interruption, or monitoring for STH reemergence following cessation of mass drug administration. Additional benefits of qPCR are its ability to distinguish between hookworm species and its capacity to include assays for other parasitic infections.15,20
Polymerase chain reaction–based techniques require specialized equipment generally available only in central laboratories and are expensive compared with microscopy-based methods. This impacts the feasibility of implementing these techniques in low-resource settings where STH are endemic. However, many low- and middle-income countries are currently undertaking molecular diagnostics in central laboratories. Furthermore, a recent economic evaluation suggests that longer-term programmatic benefits may outweigh the higher cost of novel diagnostic tests,27 particularly in settings approaching STH elimination.
Sodium nitrate flotation represents a less costly alternative diagnostic strategy that could be used in resource-limited settings. This microscopy-based technique, although less sensitive than qPCR, addresses some of the limitations of the Kato–Katz technique, including the need to examine samples within 30 minutes and to examine multiple samples. Our results confirm previous findings that SNF shows good potential as a diagnostic test for STH that could be implemented at scale.16
A limitation of our analysis was that we were unable to convert PCR-determined infection intensity to EPG, and therefore, we could not directly compare infection intensity results obtained using qPCR and SNF. This limits the current ability of qPCR in terms of measuring individual-level infection intensity, and is a crucial area for further research. In particular, determining the relationship between qPCR-determined infection intensity and WHO-defined intensity cut-offs should be prioritized. Additional work is also required to investigate the impact of sample preservation and egg development stage on quantitative results.
A further limitation is that samples were stored in formalin for 1–3 months before analysis using SNF. Diagnostic accuracy of flotation techniques for samples stored in formalin may decrease after 15 days for hookworm infections28; therefore, SNF sensitivity for detecting hookworm may have been suboptimal.
Finally, our analysis of variability in DNA detection used only one stool sample from one individual, who harbored N. americanus only. Further studies should be conducted using multiple samples from multiple infected individuals, including those infected with A. lumbricoides, T. trichiura, A. duodenale, and A. ceylanicum.
In conclusion, our study further highlights the sensitivity of qPCR techniques for detecting light-intensity Ascaris spp. and hookworm infections, and the potential utility of qPCR for determining STH infection intensity. Further research is required to examine the performance of qPCR for detecting Trichuris spp. and to more fully elucidate the ability of qPCR to accurately measure infection intensity. As the global burden of STH decreases and transmission interruption becomes increasingly feasible, ongoing efforts toward incorporating molecular diagnostic methods into STH control efforts should be prioritized.
Supplementary Material
Supplemental tables and figures
Acknowledgments:
We would like to acknowledge Dinh Ng-Nguyen and Harshanie Abeywardena for their work on analyzing the samples via microscopy. We also thank Archie Clements and Darren Gray for their contribution to the (S)WASH-D for Worms pilot study, Salvador Amaral and the study team in Timor-Leste for their efforts in sample collection, and all of the students who participated in the study, their parents, teachers, and community leaders.
Note: Supplemental tables and figures appear at www.ajtmh.org.
Disclaimer: The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Pullan RL, Smith JL, Jasrasaria R, Brooker SJ, 2014. Global numbers of infection and disease burden of soil transmitted helminth infections in 2010. Parasit Vectors 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.GBD 2015 DALYs and HALE Collaborators , 2016. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388: 1603–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uniting to Combat NTDs , 2016. Reaching the Unreached: Fourth Progress Report of the London Declaration. London, United Kingdom: Uniting to Combat NTDs. [Google Scholar]
- 4.WHO , 2012. Eliminating Soil-Transmitted helminthiasis as a Public Health Problem in Children: Progress Report 2001–2010 and Strategic Plan 2011–2020. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 5.WHO , 2017. Preventive Chemotherapy to Control Soil-Transmitted Helminth Infections in at-Risk Population Groups. Geneva, Switzerland: World Health Organization. [PubMed] [Google Scholar]
- 6.Anderson RM, Turner HC, Truscott JE, Hollingsworth TD, Brooker SJ, 2015. Should the goal for the treatment of soil-transmitted helminth (STH) infections be changed from morbidity control in children to community-wide transmission elimination? PLoS Negl Trop Dis 9: e0003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Truscott JE, Hollingsworth TD, Brooker SJ, Anderson RM, 2014. Can chemotherapy alone eliminate the transmission of soil transmitted helminths. Parasit Vectors 7: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCarthy JS, Lustigman S, Yang G-J, Barakat RM, García HH, Sripa B, Willingham AL, Prichard RK, Basáñez M-G, 2012. A research agenda for helminth diseases of humans: diagnostics for control and elimination programmes. PLoS Negl Trop Dis 6: e1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.WHO , 2011. Helminth Control in School Age Children: A Guide for Managers of Control Programmes. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 10.WHO , 1991. Basic Laboratory Methods in Medical Parasitology. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 11.Booth M, Vounatsou P, N’goran E, Tanner M, Utzinger J, 2003. The influence of sampling effort and the performance of the Kato–Katz technique in diagnosing Schistosoma mansoni and hookworm co-infections in rural Côte d’Ivoire. Parasitology 127: 525–531. [DOI] [PubMed] [Google Scholar]
- 12.Nikolay B, Brooker SJ, Pullan RL, 2014. Sensitivity of diagnostic tests for human soil-transmitted helminth infections: a meta-analysis in the absence of a true gold standard. Int J Parasitol 44: 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dacombe R, Crampin A, Floyd S, Randall A, Ndhlovu R, Bickle Q, Fine P, 2007. Time delays between patient and laboratory selectively affect accuracy of helminth diagnosis. Trans R Soc Trop Med Hyg 101: 140–145. [DOI] [PubMed] [Google Scholar]
- 14.Basuni M, Muhi J, Othman N, Verweij JJ, Ahmad M, Miswan N, Rahumatullah A, Aziz FA, Zainudin NS, Noordin R, 2011. A pentaplex real-time polymerase chain reaction assay for detection of four species of soil-transmitted helminths. Am J Trop Med Hyg 84: 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Easton AV, Oliveira RG, O’Connell EM, Kepha S, Mwandawiro CS, Njenga SM, Kihara JH, Mwatele C, Odiere MR, Brooker SJ, 2016. Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: field-based inferences on the impact of mass deworming. Parasit Vectors 9: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inpankaew T, Schär F, Khieu V, Muth S, Dalsgaard A, Marti H, Traub RJ, Odermatt P, 2014. Simple fecal flotation is a superior alternative to quadruple Kato Katz smear examination for the detection of hookworm eggs in human stool. PLoS Negl Trop Dis 8: e3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mationg MLS, et al. 2017. Status of soil-transmitted helminth infections in schoolchildren in Laguna Province, the Philippines: determined by parasitological and molecular diagnostic techniques. PLoS Negl Trop Dis 11: e0006022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejia R, Vicuña Y, Broncano N, Sandoval C, Vaca M, Chico M, Cooper PJ, Nutman TB, 2013. A novel, multi-parallel, real-time polymerase chain reaction approach for eight gastrointestinal parasites provides improved diagnostic capabilities to resource-limited at-risk populations. Am J Trop Med Hyg 88: 1041–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verweij JJ, Brienen EA, Ziem J, Yelifari L, Polderman AM, Van Lieshout L, 2007. Simultaneous detection and quantification of Ancylostoma duodenale, Necator americanus, and Oesophagostomum bifurcum in fecal samples using multiplex real-time PCR. Am J Trop Med Hyg 77: 685–690. [PubMed] [Google Scholar]
- 20.Llewellyn S, Inpankaew T, Nery SV, Gray DJ, Verweij JJ, Clements ACA, Gomes SJ, Traub R, McCarthy JS, 2016. Application of a multiplex quantitative PCR to assess prevalence and intensity of intestinal parasite infections in a controlled clinical trial. PLoS Negl Trop Dis 10: e0004380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knopp S, Salim N, Schindler T, Voules DAK, Rothen J, Lweno O, Mohammed AS, Singo R, Benninghoff M, Nsojo AA, 2014. Diagnostic accuracy of Kato-Katz, FLOTAC, Baermann, and PCR methods for the detection of light-intensity hookworm and Strongyloides stercoralis infections in Tanzania. Am J Trop Med Hyg 90: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaiakovou M, Pilotte N, Baumer B, Grant J, Asbjornsdottir K, Schaer F, Hu Y, Aroian R, Walson J, Williams SA, 2018. A comparative analysis of preservation techniques for the optimal molecular detection of hookworm DNA in a human fecal specimen. PLoS Negl Trop Dis 12: e0006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke NE, Clements ACA, Bryan S, McGown J, Gray D, Nery SV, 2016. Investigating the differential impact of school and community-based integrated control programmes for soil-transmitted helminths in Timor-Leste: the (S)WASH-D for Worms pilot study protocol. Pilot Feasibility Stud 2: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilotte N, Papaiakovou M, Grant JR, Biewert LA, Llewellyn S, McCarthy JS, Williams SA, 2016. Improved PCR-based detection of soil transmitted helminth infections using a next-generation sequencing approach to assay design. PLoS Negl Trop Dis 10: e0004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phosuk I, Intapan PM, Sanpool O, Janwan P, Thanchomnang T, Sawanyawisuth K, Morakote N, Maleewong W, 2013. Molecular evidence of Trichostrongylus colubriformis and Trichostrongylus axei infections in humans from Thailand and Lao PDR. Am J Trop Med Hyg 89: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Verweij JJ, Pit DS, Van Lieshout L, Baeta SM, Dery GD, Gasser RB, Polderman AM, 2001. Determining the prevalence of Oesophagostomum bifurcum and Necator americanus infections using specific PCR amplification of DNA from faecal samples. Trop Med Int Health 6: 726–731. [DOI] [PubMed] [Google Scholar]
- 27.Turner HC, Bettis AA, Dunn JC, Whitton JM, Hollingsworth TD, Fleming FM, Anderson RM, 2017. Economic considerations for moving beyond the Kato–Katz technique for diagnosing intestinal parasites as we move towards elimination. Trends Parasitol 33: 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barda B, et al. 2015. How long can stool samples be fixed for an accurate diagnosis of soil-transmitted helminth infection using mini-FLOTAC? PLoS Negl Trop Dis 9: e0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental tables and figures