Abstract
Interest in the genetic composition of cynomolgus macaques (Macaca fascicularis) has increased due to the rising demand for NHP models in human biomedical research. Significant genetic differences among regional populations of cynomolgus macaques can confound interpretations of research results because they do not solely reflect differences in experimental treatment effects. Therefore, the common origin of cynomolgus macaques used as research subjects should be verified by using region-specific genetic markers to minimize the influence of underlying genetic variation among animals selected as research subjects on phenotypes under study. We compared the effectiveness of 18 short tandem repeat (STR) markers with that of 83 single-nucleotide polymorphism (SNP) markers to differentiate the ancestry of cynomolgus macaques from 6 different populations (Cambodia, Sumatra, Mauritius, Singapore, and the islands of Luzon and Zamboanga in the Philippines). Genetic diversity indices such as allele numbers and expected heterozygosity based on SNP were lower and exhibited lower standard errors than those provided by STR, probably because, unlike STR, most SNP are biallelic and consequently exhibit maximal expected heterozygosity values of 0.50. However, the standard error of estimates of observed heterozygosity based on SNP was higher than that for STR, perhaps reflecting sampling errors. Only 27 SNP were required to match the resolving power of 17 STR to detect population structure, that is, 1.6 SNP:1 STR. Whereas STR only differentiated the Mauritian population from all other populations, SNP detected 4 genetically distinct groups (Cambodia, Singapore-Sumatra, Mauritius, and Zamboanga). SNP are poised to become as valuable as STR for understanding and detecting genetic structure among cynomolgus macaques. Although STR will remain an important tool for cynomolgus macaque population studies, SNP have the potential to become the mainstream marker type.
Abbreviations: FIS, overall inbreeding coefficient; FST, fixation index; HE, expected heterozygosity; HO, observed heterozygosity; mtDNA, mitochondrial DNA; NA, number of alleles; SNP, single-nucleotide polymorphisms; STR, short tandem repeats
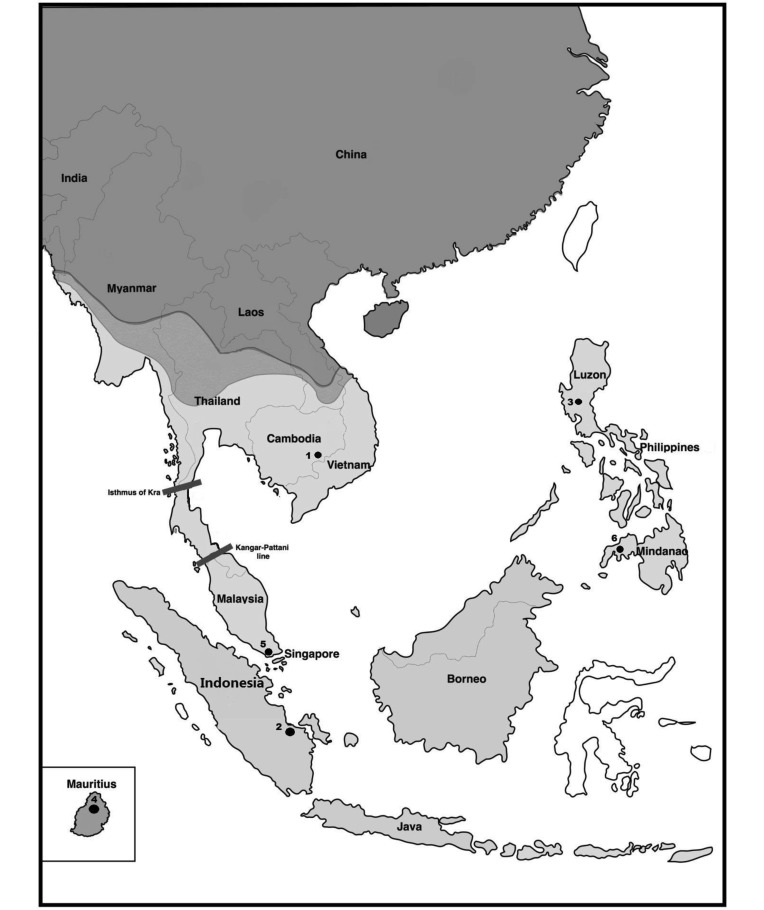
Cynomolgus (or long-tailed) macaques (Macaca fascicularis) are commonly used NHP subjects for biomedical research. Cynomolgus macaques are mainly found in Indonesia, the Philippines, Malaysia, Mauritius, Thailand, Myanmar, and Indochina, including Cambodia, Laos, and Vietnam15,17 (Figure 1). Due to the increasing demand for cynomolgus macaques for biomedical research, more cynomolgus macaques than any other NHP species have been imported into the United States in recent years.4,22,25,43
Figure 1.
Geographic distribution of cynomolgus macaques (M. fascicularis; light gray) primarily is confined to the regions south of the subtropical and temperate geographic range of the rhesus macaque (dark gray), although a region of parapatry occurs in Myanmar, Thailand, and Indochina (medium gray).5 Locations of the cynomolgus macaque populations included in this study are marked with dots and correspond to those in Table 1. Adapted from reference.6
Although some cynomolgus macaques are imported directly to the United States from Mauritius and Indonesia, more than 65% are imported from breeding farms in China,2 a country outside their current natural range. Most cynomolgus macaques in Chinese breeding farms probably originated in Indochina,65 but these farms do not document their origins, and interbreeding of animals from different regions of origin has been reported.28 Genetic evidence also suggests that natural interspecies admixture between cynomolgus and rhesus macaques has occurred in Indochina, Myanmar, and Thailand5,42 (Figure 1). The introgression of rhesus macaque genes into populations of cynomolgus macaques extends well beyond the contact zone between the natural ranges of the 2 species in Indochina in a wide zone of intergradation that ranges southward from 15° to 20° north (of Thailand, Vietnam, and Laos) to southern Thailand and Malaysia.3,5,6,16,29,56,60 The high variability in the level of rhesus introgression inflates diversity parameters of—and genetic differences among—regional populations of Indochinese cynomolgus macaques.6
Previous studies have shown significant genetic differences among regional populations of cynomolgus macaques, with the level of difference between some populations approaching that between some species.2,3,21,29,28,32,33,34,38,54,56,59 Given that phenotypic variation due to these genetic differences can influence responses to experimental treatment effects, thus rendering specific populations inappropriate for particular studies, these differences can confound interpretations of the outcome of experimental research involving cynomolgus macaque subjects from multiple regions of origin.1,9,10,48,50,52,64 For example, the differential responses to Plasmodium knowlesi infection in Philippine, Malaysian, and Mauritian cynomolgus macaques are examples of the importance of knowing the region of origin of research subjects.10,41,48 Because Philippine cynomolgus macaques live near the limit of the geographic range of both the pathogen and its vector and because Mauritian cynomolgus macaques are outside the range of this pathogen and vector, theses NHP exhibit lower rates of infection than those from Malaysia.48,51,64 Unlike cynomolgus macaques from Indonesia, Indochina, or the Philippines, cynomolgus macaques from Mauritius are resistant to symptoms of clinical shigellosis after experimental immunization.10,50 In addition, because alleles contained within the MHC region influence the effectiveness of an individual's immune response to different pathogens, matching subjects in immunologic and infectious disease research for their MHC alleles minimizes the contribution of genetic variance to the phenotypic variance in responses to biomedical treatment effects. For this reason, the Mauritian cynomolgus macaques, which exhibit high frequencies of a relatively few class I MHC alleles,30,36 are desirable subjects for HIV and SIV research.
Because subjects obtained from the entire cynomolgus macaque geographic range are being used in biomedical research, it is important to include provenances throughout this species’ natural range into future population genetic studies.28,54 Consequently, investigators should determine or verify and report the origin and ancestry of the cynomolgus macaques they use as research subjects. The genetic composition of cynomolgus macaques has been studied by using mitochondrial DNA (mtDNA) and microsatellite or short tandem repeat (STR) markers. MtDNA studies have confirmed 2 genetic clusters of cynomolgus macaques in Southeast Asia: a continental population (including Indochina, Thailand, Myanmar, and Peninsular Malaysia north of the Isthmus of Kra), whose members predominantly belong to mtDNA haplogroup Fas154 (that is, the Asian mainland clade39), and an insular population (including Indonesia, Sarawak Malaysia, and the Philippines)21,54,59 belonging predominantly to mtDNA haplogroup Fas254 (that is, the Sundaland clade39).
Other protein polymorphism and mtDNA studies have suggested that Mauritian cynomolgus macaques were introduced from Indonesia by sailors, experienced a severe genetic bottleneck, and have become genetically distinct from other populations of the species,34,47 including those from Indonesia. An analysis using a panel of 24 STR divided cynomolgus macaque populations into 3 genetic clusters: the Philippines (including Luzon, Zamboanga, and Corregidor), Mauritius, and a cluster comprising Sumatra, Cambodia, and Singapore.28,29 In that study, samples from Sumatra and Singapore overlapped those from Cambodia, albeit only slightly, providing a somewhat different clustering than reported in earlier mtDNA studies. Thus, mtDNA and STR together differentiate cynomolgus macaques into 4 different clusters: a continental cluster and the 3 insular clusters of Singapore–Sumatra, the Philippines, and Mauritius.
Although STR are still the mainstream in population genetics, the use of SNP has increased due to its advantages over STR.23 SNP experience a lower false genotyping rate, a much lower rate of mutation (so that shared alleles express identity by descent, rather than only identity by state), and a much greater abundance in the genome and are more suitable for automation and standardization in high-throughput sequencing technologies than STR.18,40,63 However, STR provide higher information content per locus than SNP, because STR are multiallelic whereas the great majority of SNP are only biallelic.26 Therefore, more SNP are required to achieve the same resolution as that provided by fewer STR;47,63 a single STR reportedly is approximately as informative as 1.7 to 5.6 SNP, depending on the characteristics of the marker sets and size of the populations.7,19,20,35,37,49,59 In the current study, we characterized the genetic structure of several regional cynomolgus macaque populations by using panels of 96 SNP and 25 STR and compared the effectiveness of these 2 types of markers in differentiating the ancestry of cynomolgus macaques.
Materials and Methods
The 446 subjects from which the blood samples that we used in this study were acquired were either captured in the wild or recently derived from wild-caught founders maintained in captivity for biomedical research. These samples were obtained directly from in-country breeding farms by Primate Products USA (Immokalee, FL), with all the required CITES and import permits and the greatest possible authentication of the provenance of all samples. Figure 1 illustrates the geographic ranges of the cynomolgus macaques, whereas Table 1 indicates the locations from which the cynomolgus samples originated and the number of samples representing each location. DNA was extracted from the blood samples (QIAamp DNA Blood Mini Kit, Qiagen, Valencia, CA) and then quantified (Qubit dsDNA BR Assay Kit, Applied Biosystems, Foster City, CA); DNA concentrations were adjusted to 80 ng/μL.
Table 1.
Specific sample sites and numbers of cynomolgus macaques used in this study
| No. of samples |
|||
| All subjects |
|||
| Location | STR analysis | SNP analysis | Subjects with both SNP and STR data |
| 1. Cambodia, Indochina | 112 | 23 | 1 |
| 2. Near Palembang in Sumatra, Indonesia | 100 | 38 | 25 |
| 3. Luzon, Philippines | 30 | 23 | 23 |
| 4. Mauritius | 94 | 25 | 0 |
| 5. Singapore | 70 | 25 | 25 |
| 6. Zamboanga on Mindanao, Philippines | 40 | 21 | 21 |
| Total | 446 | 155 | 95 |
Location numbers correspond to those in Figure 1.
To identify SNP for this study, reduced representation libraries of pooled DNA samples from 4 cynomolgus macaques—one each from Sumatra, Cambodia, Zamboanga (in Mindanao, Philippines), and Mauritius—were constructed by using a method previously described for SNP identification in rhesus macaques.61 These analyses generated more than 5,000 SNP that exhibited alleles unique to 1 of the 4 regions. From these geographically unique SNP, a panel of 96 genome-wide ancestry-informative markers was assembled to identify the geographic region of origin of 155 cynomolgus macaques of unknown ancestry. As described previously,65 these SNP then were used to design a custom SNP type assay (Fluidigm, South San Francisco, CA). The assay was run by using the Fluidigm Juno SNP genotyping system at the UC Davis Genome Center DNA Technologies Core; genotypes were assigned by using the Fluidigm SNP Genotyping Analysis software (version 4.1.2).
The STR analysis used in this study has been described elsewhere.29,30,55 For PCR amplification of STR loci, 0.5 to 1.25 μL of DNA extracts from 446 cynomolgus macaque samples were used in each 12.5-μL PCR reaction (67 mM Tris HCl [pH 8.8], 16 mM (NH4)2SO4, 0.01% Tween 20, 0.05 mM each dNTP, 0.2 μM each primer, 1.7 mM MgSO4, 0.025 U/mL Platinum Taq polymerase [Invitrogen, Carlsbad, CA]). The annealing temperatures and extension times of the ‘touch-down’ thermocycler conditions varied for each STR primer pair: 94 °C for 3 min; 60 cycles of 94 °C for 20 s (denaturing), 54 to 62 °C (decreasing 0.1 °C per cycle; annealing), and 72 °C for 45 to 90 s (extension); followed by a final extension at 72 °C for 5 to 60 s. All samples were analyzed on an automated genetic analyzer (ABI 3130xl, Applied Biosystems, Foster City, CA) according to the recommendations of the manufacturer by using the LIZ500 size standard to assign genotypes. Because these markers have been used for assigning parentage to captive cynomolgus macaques without evidence of allelic dropout, we did not routinely confirm genotypes with multiple amplifications. Only STR loci that showed the highest expected heterozygosity or gene diversity estimates in a sample of cynomolgus macaques from several different geographic regions were selected for the present study.29
Because missing data can reduce the power of genetic analyses, only animals and markers with at least 90% complete genotypes were used in subsequent comparisons.8,11 As such, 2 sets of comparative analyses were conducted to evaluate the SNP and STR panels’ ability to differentiate cynomolgus macaque populations; Table 2 presents the SNP and STR markers that were used in each comparison. The first analysis compared the genetic structure and composition of all cynomolgus macaques detected by one or the other of the 2 types of markers while the second analysis only included individuals with both SNP and STR genotypes. Arlequin version 3.5.2.2713 was used to estimate the number of alleles (NA), observed heterozygosity (HO), expected heterozygosity (HE), overall inbreeding coefficient (FIS), fixation index (FST), and pairwise FST for both analyses and marker datasets. FST estimates between pairs of populations were tested for statistical significance by using 1,000,000 genotypic permutations in Arlequin,13 followed by sequential Bonferroni correction for the multiple pairwise population comparisons.45 Because the identities of the individual samples used in the analyses of samples with both SNP and STR genotypes differ from each other and from that for all samples with genotypes for one or the other type of marker, the parameters generated by the 3 different analyses are not strictly comparable.
Table 2.
The combinations of SNP and STR markers that were used in each comparison test
| STR | SNP | |||
| 270o7 | chr1:136655126 | chr11:7015538 | chr14:70139960 | chr2:134110821 |
| 272o12 | chr11:41122854 | chr14:129238251 | chr18:13305801 | chr6:174432781 |
| AGAT007 | chr13:61808312 | chr17:23409587 | chr4:162673570 | chr9:555616 |
| D10s1432 | chr16:58107673 | chr3:55951177 | chr8:21012767 | chr2:132629694 |
| D11s2002 | chr2:72868664 | chr7:93550426 | chr7:110937100 | chr6:167836387 |
| D13s318 | chr7:15073488 | chr8:101877456 | chr3:140215947 | chr6:41508497 |
| D13s765 | chr11: 99104867 | chr2:136013319 | chr1:24843143 | chr13:60894731 |
| D14s306 | chr7:132297042 | chr3:23850940 | chr2:62582885 | chr10:74298352 |
| D16s750 | chr2:181658759 | chr2:54799163 | chr2:79380407 | chr9:87063721 |
| D18s861 | chr14:39287086 | chr16:68291744 | chr2:9107350 | chr11:4110282 |
| D19s255 | chr13:6157050 | chr7:84787844 | chr10:53108031 | chr13:1762849 |
| D1s548 | chr3:61745513 | chr1:66911428 | chr12:72870950 | chr16:57067723 |
| D2s1333 | chr1:160799683 | chr12:53697123 | chr15:11533734 | chr2:42108607 |
| D3s1768 | chr11:57579149 | chr14:28576260 | chr19:36116678 | chr6:93430718 |
| D4s1626 | chr14:100880007 | chr17:86439601 | chr4:35222515 | chr9:8033023 |
| D4s2365 | chr16:74853727 | chr4:156547035 | chr9:48223180 | chr4:135158450 |
| D5s1457 | chr20:75862934 | chr8:10191843 | chr13:11966895 | chr1:162920471 |
| D6s501 | chr7:158143584 | chr11:109373299 | chr6:154596965 | chr9:40160594 |
| D7s1826 | chr3:83236171 | chr4:135158450 | chr19:27552678 | chr1:58733102 |
| D7s794 | chr1:130006021 | chr17:25598362 | chr7:57715344 | chr3:61689424 |
| D8s1106 | chr16:19758927 | chr1939121259 | chr10:73756367 | chr11:90554927 |
| D8s1466 | chr17:4689860 | chr16:71093372 | chr14:82008699 | |
| D9s921 | chr6:6915508 | chr10:8530416 | chr10:57018986 | |
| D9s934 | chr6:86364974 | chr10:52383748 | chr13:10785299 | |
| DXs2506 | chr1:49110258 | chr12:67642116 | chr15:2650820 | |
The markers used in the comparison of all subjects with either SNP or STR data are italicized (Table 3), and markers used in the comparison of animals with both SNP and STR data are bolded (Table 4). The STR have been described elsewhere.29,55 The M. mulatta RheMac2 and M. fascicularis macFas5 reference genomes (https://genome.ucsc.edu/cgi-bin/hgGateway) were used for mapping the chromosome positions of SNP.46,61,65
The program STRUCTURE 2.3.424,44 was used to characterize population structure in both sets of comparative analyses. The simulations were performed with 500,000 iterations after a burn-in period of 100,000 by using the admixture model with a priori location information (LOCPRIOR); LOCPRIOR is used when the locations of origin from which the samples were collected are known. The runs were conducted under the assumption of 1 to K populations, plus 2 to account for potential ancestors in other groups. All STRUCTURE runs were replicated 5 times for each value of K, and the log probability of the data (Ln P[D]) and Δ K12 were used to determine the true number of populations, that is, the K value with the highest Ln P(D) and lowest standard deviation that coincides with the highest Δ K. Additional STRUCTURE analyses were performed to compare the abilities of SNP and STR to detect the population structure and to differentiate regional populations of cynomolgus macaques. Because multiple SNP are required to obtain the same power of resolution as a single STR, the exact number of which varies due to the informativeness of each SNP and STR, STRUCTURE runs were performed on all 83 SNP as well as on 4 sets of SNP randomly selected from all available SNP markers with 90% complete data, that is, 63, 43, 23, and 8 loci. The random selection of SNP was conducted without replacement by using Microsoft Excel (Redmond, WA). Successful assignment by using STRUCTURE was assessed by the proportion of samples for which the proportionate assignment of a subject to its correct population (Q) was 90% or more.
Results
All subjects.
After the removal of markers and subjects with less than 90% data completeness, 381 samples genotyped for 18 of the 25 STR tested and 129 samples genotyped for 83 of the 96 SNP tested remained. The samples were from 5 populations: Cambodia, Sumatra, Mauritius, Singapore, and Zamboanga (Philippines). Luzon (Philippines) was omitted because only 15 of its representatives met the requirement for 90% data completeness for STR. The Luzon sample size was too small for this analysis, whereas the sample sizes of the remaining populations ranged from 37 (Zamboanga) to 103 (Cambodia) subjects with STR data that met the 90% criterion (Table 3).
Table 3.
Population comparison results of all individuals with either SNP or STR data showing number of samples (N), estimates of allele numbers (NA), observed heterozygosity (HO), expected heterozygosity (HE), inbreeding coefficient (FIS), and pairwise FSTvalues averaged across all loci for both marker types and populations
| Cambodia | Sumatra | Mauritius | Singapore | Zamboanga | |
| STR | |||||
| N | 103 | 97 | 84 | 60 | 37 |
| NA | 11.33 ± 4.24 | 10.17 ± 4.40 | 5.44 ± 2.00 | 9.72 ± 3.45 | 6.72 ± 2.76 |
| HO | 0.72 ± 0.11 | 0.71 ± 0.07 | 0.56 ± 0.15 | 0.66 ± 0.12 | 0.64 ± 0.14 |
| HE | 0.78 ± 0.11 | 0.75 ± 0.09 | 0.64 ± 0.12 | 0.77 ± 0.08 | 0.70 ± 0.14 |
| FIS | 0.06 | 0.05 | 0.13 | 0.16 | 0.08 |
| Sumatra | 0.04 | ||||
| Mauritius | 0.14 | 0.11 | |||
| Singapore | 0.04 | 0.04 | 0.13 | ||
| Zamboanga | 0.08 | 0.08 | 0.16 | 0.10 | |
| Mean FST | 0.08 | 0.07 | 0.14 | 0.08 | 0.11 |
| SNP | |||||
| N | 23 | 38 | 23 | 25 | 20 |
| NA | 1.72 ± 0.45 | 1.68 ± 0.47 | 1.46 ± 0.50 | 1.43 ± 0.50 | 1.27 ± 0.44 |
| HO | 0.12 ± 0.22 | 0.14 ± 0.23 | 0.15 ± 0.25 | 0.15 ± 0.26 | 0.10 ± 0.22 |
| HE | 0.20 ± 0.07 | 0.13 ± 0.07 | 0.13 ± 0.09 | 0.13 ± 0.09 | 0.09 ± 0.07 |
| FIS | 0.05 | 0.12 | −0.01 | −0.05 | −0.00 |
| Sumatra | 0.41 | ||||
| Mauritius | 0.49 | 0.24 | |||
| Singapore | 0.39 | 0.07 | 0.20 | ||
| Zamboanga | 0.56 | 0.34 | 0.52 | 0.38 | |
| Mean FST | 0.46 | 0.27 | 0.39 | 0.29 | 0.45 |
Only populations whose members met the 90% data completeness criterion for both SNP and STR were included. Luzon was omitted because too few of its subjects (n = 15) met the 90% data completeness criterion for STR. A total of 83 SNP markers and 18 STR markers met the completeness requirement for inclusion in the analysis. All pairwise FST comparisons (in italics) were statistically significant at the 0.05 level of probability.
The sample numbers and estimates of NA, HO, HE, and FIS averaged across all loci for both marker types and populations for subjects with either STR or SNP data are given in Table 3. Both the means and standard errors of the NA estimates based on SNP were smaller than those generated by STR, reflecting the biallelic and multiallelic nature of SNP and STR, respectively. Not surprisingly, both HO and HE estimates were higher for STR than for SNP. However, although the standard errors of the HE estimates based on both marker types were relatively comparable, those for HO estimates were much higher for SNP than for STR. This pattern might be due to the inflated sampling errors from the lower sample sizes in the SNP analysis (Table 3), because it is evident that when sample numbers are equal, as for the populations whose members were genotyped for both STR and SNP that are compared in Table 4, the NA and HE estimates have much lower standard errors for SNP than for STR.
Table 4.
Population comparison results of all individuals with SNP as well as STR data showing number of samples (N) and estimates of allele numbers (NA), observed heterozygosity (HO), expected heterozygosity (HE), inbreeding coefficient (FIS), and pairwise FSTvalues averaged across all loci for both marker types and populations for subjects with both SNP and STR data
| Luzon | Singapore | Sumatra | Zamboanga | |
| STR | ||||
| N | 15 | 20 | 25 | 17 |
| NA | 2.77 ± 2.50 | 5.94 ± 4.30 | 8.00 ± 5.38 | 5.65 ± 3.01 |
| HO | 0.27 ± 0.24 | 0.51 ± 0.20 | 0.64 ± 0.21 | 0.55 ± 0.24 |
| HE | 0.29 ± 0.28 | 0.69 ± 0.13 | 0.70 ± 0.21 | 0.61 ± 0.21 |
| FIS | 0.13 | 0.28 | 0.10 | 0.13 |
| Singapore | 0.32 | |||
| Sumatra | 0.34 | 0.09 | ||
| Zamboanga | 0.32 | 0.09 | 0.13 | |
| Mean FST | 0.33 | 0.17 | 0.18 | 0.18 |
| SNP | ||||
| NA | 1.11 ± 0.40 | 1.43 ± 0.50 | 1.47 ± 0.50 | 0.44 ± 0.24 |
| HO | 0.07 ± 0.21 | 0.15 ± 0.25 | 0.14 ± 0.22 | 0.10 ± 0.20 |
| HE | 0.05 ± 0.04 | 0.13 ± 0.12 | 0.12 ± 0.12 | 0.10 ± 0.09 |
| FIS | −0.46 | −0.05 | −0.07 | 0.01 |
| Singapore | 0.46 | |||
| Sumatra | 0.40 | 0.07 | ||
| Zamboanga | 0.43 | 0.39 | 0.37 | |
| Mean FST | 0.43 | 0.31 | 0.28 | 0.40 |
Cambodia and Mauritius were omitted from this analysis because these populations lacked sufficient numbers of individuals with both SNP and STR. 83 SNP markers and 17 STR markers met the completeness requirement for inclusion in the analysis. All pairwise FST comparisons (in italics) were statistically significant at the 0.05 level of probability.
The Cambodian population exhibited the highest average number of both SNP and STR alleles per locus (NA = 1.72 ± 0.45 and 11.33 ± 4.24, respectively; Table 3). Mauritius exhibited the lowest NA (5.44 ± 2.00) based on STR, whereas Zamboanga exhibited the lowest NA (1.27 ± 0.44) based on SNP. The estimates of HO and HE based on STR were lowest in Mauritius (0.56 ± 0.15 and 0.64 ± 0.12, respectively) and highest in Cambodia (0.72 ± 0.11 and 0.78 ± 0.11, respectively). Not surprisingly, the estimates of HO and HE from SNP were much lower than those for STR, albeit distributed similarly among populations, ranging from 0.10 ± 0.22 (Zamboanga) to 0.15 ± 0.25 (Mauritius) and 0.09 ± 0.07 (Zamboanga) to 0.20 ± 0.07 (Cambodia), respectively. The differences between corresponding values of HO and HE were not statistically significant at the 0.05 level of probability for any population or marker set.
The population-specific FIS estimates ranged from 0.05 (Sumatra) to 0.16 (Singapore) for STR and –0.05 (Singapore) to 0.12 (Sumatra) for SNP. The overall FIS estimate based on SNP was –0.12 as compared with 0.15 based on STR. Overall FST values based on the panel of SNP were much greater than that of STR (that is, 0.36 compared with 0.20). Table 3 shows the average pairwise FST for each population for both the SNP and STR data. Average genetic differentiation from all other populations was lowest for Sumatra (STR, 0.07; SNP, 0.27) whereas Mauritius and Cambodia had the highest for STR (0.14) and SNP (0.46), respectively.
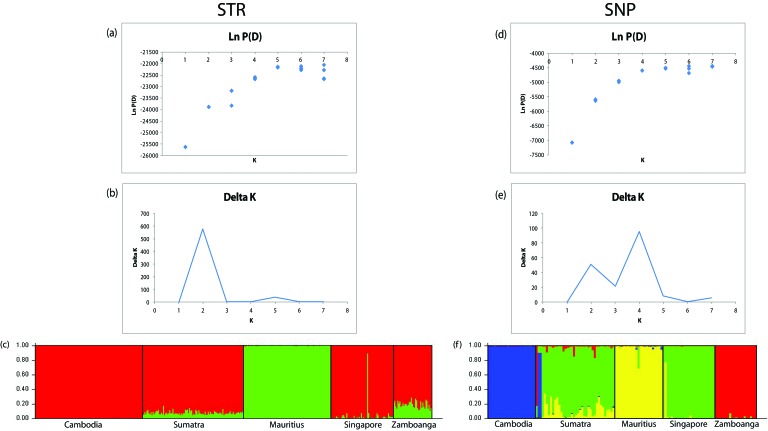
STRUCTURE, Ln P(D), and Δ K results for the STR data suggested that 2 true genetic groups (K = 2) are present among cynomolgus macaque populations, whereas the SNP data yielded 4 true genetic groups (K = 4; Figure 2). With the STR data at K = 2, STRUCTURE was able to differentiate Mauritius from the other populations. For the SNP K = 4 results, STRUCTURE analysis separated Cambodia, Mauritius, and Zamboanga into separate groups and defined a fourth group comprising Singapore and Sumatra.
Figure 2.
Plots for K values from 1 to 8 for all subjects: (A) STR Ln P(D) results and (B) STR Δ K results show 2 possible populations. (C) STR STRUCTURE results for K = 2. Colors represent the proportion of genetic assignment to the respective populations on the y-axis. (D) SNP Ln P(D) results and (E) SNP Δ K results show 4 possible populations. (F) SNP STRUCTURE results for K = 4. The Luzon population was omitted because none of its animals met the 90% data completeness criterion for STR. Diamonds represent the standard deviation of the Ln P(D) estimate.
Subjects with both SNP and STR data.
Although 95 subjects were genotyped for both SNP and STR, only 78 of these samples exhibited at least 90% complete data for the same 17 STR and 83 SNP and represent 5 different populations of origin: Luzon, Singapore, Sumatra, Cambodia, and Zamboanga. Because this analysis only used subjects with both SNP and STR data, all populations had reduced but comparable sample sizes, ranging from 15 (Luzon) to 25 (Sumatra; Table 4). Because only a single subject from Cambodia was included among the 78 samples, it was omitted from this analysis. We also omitted Mauritius from this analysis because its population lacked any subjects with both SNP and STR. The sample numbers and estimates of NA, HO, HE, and FIS averaged across all loci for both marker types and populations for subjects with both SNP and STR data are given in Table 4. The STR data yielded NA estimates ranging from 2.77± 2.50 (Luzon) to 8.00 ± 5.38 (Sumatra), whereas NA estimates for the SNP data ranged from 0.44 ± 0.43 (Zamboanga) to 1.47 ± 0.50 (Sumatra). The estimates of HO and HE for STR ranged from 0.27 ± 0.24 (Luzon) to 0.64 ± 0.21 (Sumatra) and 0.29 ± 0.28 (Luzon) to 0.70 ± 0.21 (Sumatra), respectively. The SNP data exhibited HO estimates ranging from 0.07 ± 0.21 (Luzon) to 0.15 ± 0.25 (Singapore) and HE estimates from 0.05 ± 0.04 (Luzon) to 0.13 ± 0.12 (Singapore). At the 95% confidence level, no statistically significant difference was found between the HO and HE estimates for any populations except that for Singapore based on the STR data.
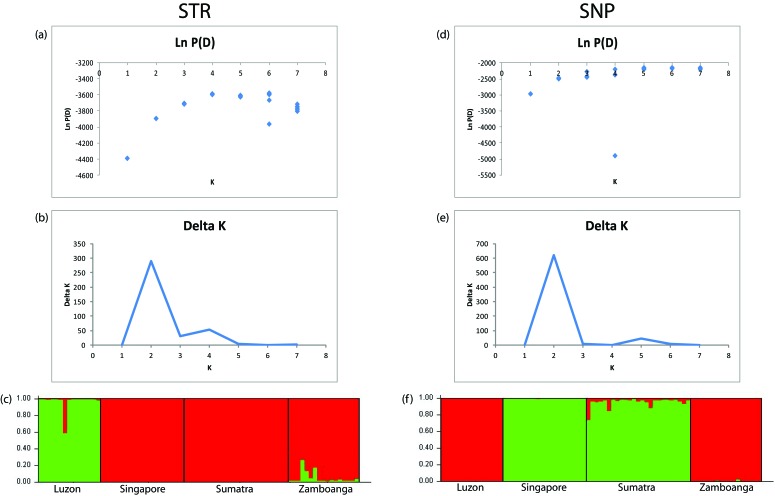
Some disparity between the FIS estimates based on SNP and STR data emerged. Although the STR data exhibited positive FIS estimates for all populations, the range of FIS based on the markers suggested low to moderate levels of inbreeding. The SNP data exhibited negative FIS estimates for all populations except Zamboanga (FIS = 0.01), consistent with an absence of inbreeding. Pairwise FST estimates are shown in Table 4. Average pairwise FST values (that is, average differentiation from all other populations) for STR ranged from 0.17 (Singapore) to 0.33 (Luzon), whereas those for SNP ranged from 0.28 (Sumatra) to 0.43 (Luzon). Therefore, the Luzon population had the highest average pairwise FST estimate according to both SNP and STR results, thus suggesting high divergence from the other 3 populations (Table 4), as has previously been reported.55 The low average pairwise FST of Sumatra and Singapore in addition to their low pairwise FST with each other in both marker sets suggests that the Singaporean and Sumatran populations are closely related.30 This relatedness is also confirmed by the STRUCTURE analysis in Figure 3. The same STRUCTURE analysis revealed an Ln P(D) and Δ K showing the highest probability at 2 (K = 2) for both the SNP and STR data.
Figure 3.
Plots for K values from 1 to 8 for macaques with both SNP and STR data: (A) STR Ln P(D) results and (B) STR Δ K results show 2 populations. (c) STR STRUCTURE results for K = 2, where only 4 samples—1 from Luzon and 3 from Zamboanga—exhibited assignment probabilities to their alleged geographic region of origin lower than 90%. Colors represent the proportion of genetic assignment to the respective populations on the y-axis. (D) SNP Ln P(D) results, where only 3 Sumatran samples exhibited assignment probabilities to their alleged geographic region of origin lower than 90%. (E) SNP Δ K results show 2 populations. (F) SNP STRUCTURE results for K = 2. Diamonds represent the standard deviation of the Ln P(D) estimate. Cambodia and Mauritius were omitted from this analysis because these populations lacked members with both SNP and STR.
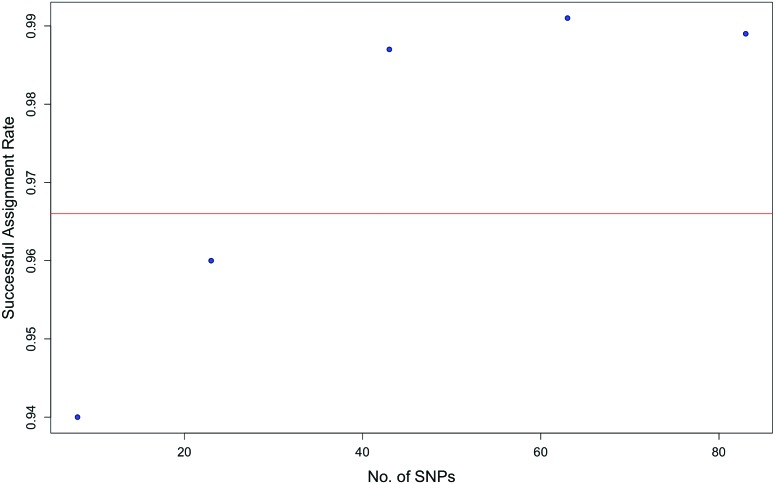
Regarding the fewest SNP that were needed to obtain an equivalent value for accuracy of population assignment (that is, Q value) based on 17 STR, the analyses performed by using 5 sets of SNP—comprising 83, 63, 43, 23 and 8 loci (the last 4 sets comprised SNP that were chosen randomly without replacement)—showed a positive correlation between the number of SNP used and the average rate of successfully assigning a subject to its correct geographic population (Figure 4). As noted earlier, subjects are considered successfully assigned to their correct regional population when their Q value is 90% or more; therefore, according to this analysis, approximately 27 SNP were needed to achieve the same average rate of successful assignment as 17 STR, that is, 96.6% (Figure 4), which translated to approximately 1.6 SNP per STR. This ratio is lower than that reported elsewhere,7,19,20,35,37,58 perhaps because the SNP, but not the STR, that we used here were specifically selected for their ability to differentiate among regional populations of cynomolgus macaques.
Figure 4.
Correlation between number of SNP used and the rate of success in assigning subjects to their respective genetic groups. The horizontal red line represents the average successful assignment rate of 17 STR, that is, approximately 96.6%; 27 SNP were needed to achieve this level of assignment.
Discussion
Previous studies of protein polymorphisms, mtDNA, and STR reported different levels of genetic divisions between insular and mainland cynomolgus macaques and those on the Philippine and Mauritius islands.21,28,32-34,38,59 In the current study, we assessed the genetic composition and structure of cynomolgus macaque populations by using SNP and compared those results with those based on STR.
Given the analyses of both marker types, our results confirmed that genetic differences among cynomolgus macaque populations should be considered in biomedical research. Pairwise FST computations based on both SNP and STR were either similar to or higher than that between Indian and Chinese rhesus macaques as well as that between rhesus and cynomolgus macaques.27,28,30,31 As observed on several islands in Indonesia,33 the elevated differentiation among and reduced variation within insular populations are a consequence of heterogeneity decay and the lack of gene flow due to the presence of significant geographic barriers.28 Conversely, geographic proximity better explains the close genetic relatedness between Singaporean and Sumatran cynomolgus macaque populations.
Unlike a previous study,28 in which SNP-based STRUCTURE and Δ K results indicated 3 true populations (K = 3), our results found 2 true populations (K = 2) in our STR comparison and 4 true populations (K = 4) in our SNP comparison when data from all subjects with either SNP (n = 129) or STR (n = 381) information were used (Figure 2). Our K = 2 STR results only differentiated the Mauritian population from the others but failed to differentiate the Zamboanga population from the Cambodian–Singaporean–Sumatran cluster (Figure 2). In our K = 4 SNP results, Zamboanga and Cambodia populations were separated in addition to Mauritius, leaving Singapore–Sumatra as a fourth population group. Our STRUCTURE analysis of only the 77 samples with both SNP and STR data indicated that only 2 true populations of cynomolgus macaques exist (Figure 3).
The Mauritian population in this study had the lowest NA, HO, and HE according to the STR analyses. Several previous studies have similarly found the Mauritian population to be less genetically diverse than other cynomolgus populations and have attributed this genetic homogeneity to the population's sudden expansion from a small number of founder animals.2,33,45,59,62 In contrast, the Cambodian population exhibited the highest NA, HO, and HE according to STR and, a high degree of differentiation from all other populations based on both marker types, which is likely due to the influence of admixture with rhesus macaques.2,33,57,29,46,51,56,60
Our use of the same subjects to compare the relative powers of STR and the new SNP panel eliminated the influence of variables, such as sample size and individual genetic variance, that could skew comparisons. Although the small sample size clearly influenced the allele numbers, a sample-size-adjusted value is unnecessary for comparing HE and HO, because sample size has the same effect on both data sets. In addition, although increased numbers of equally frequent alleles typically are associated with high estimates of HO and HE, in the current study, the HO and HE estimates were not proportional to the sample sizes of the populations probably because of variation in alternate allele frequencies at each SNP and STR locus.
The lower NA, HO, and HE estimates of the Luzon population as detected by STR suggests the genetic differentiation between Luzon and the other 3 populations. Our SNP results agree with the STR results for the most part, except that our SNP were unable to detect a significant difference between the HO estimates of Luzon and Zamboanga. According to our SNP results, the genetic distance between Luzon and Zamboanga is lower than that between Luzon and Sumatra. The inconsistency between the SNP and STR results regarding Luzon's HO estimate can be explained by the significantly lower inbreeding level that was detected by SNP but not STR. This contradiction suggests that STR tend to overestimate inbreeding levels, perhaps due to allelic homoplasy, or that estimates of HO are more sensitive to sampling error than those of HE. The contradiction might also be attributed in part to differences between the powers of resolution of SNP and STR or the locations of these loci within the macaque genome.
Although the same STR and SNP were used to genotype all subjects in this study, the results of the ‘All subjects’ analysis cannot be compared with those from the ‘Subjects with both SNP and STR data’ evaluation because the criteria for selecting subjects and markers with at least 90% complete genotypes led to subjects with different SNP and STR datasets. Because the earlier analysis compared SNP and STR data from different animals, it was important to compare SNP and STR data from the same animals for which data are reported in Table 2. Sampling errors probably influence the discrepancies between estimates of population genetic metrics from both analyses based on the different sets of SNP and STR.
The comparison of the 2 types of markers shows that population structure affects them differently. In general, the panel of SNP differentiated populations of cynomolgus macaques as well as or better than STR, because SNP revealed higher overall FST values. Whereas previous mtDNA studies were able only to differentiate Sumatran cynomolgus macaques from both Cambodian21,59 and Mauritian34,38,54 cynomolgus macaques and although previous STR studies clustered the species into 3 groups (Mauritius, the Philippines, and Cambodia–Singapore–Sumatra), our current SNP panel distinguished 4 groups (Cambodia, Singapore– Sumatra, Mauritius, and Zamboanga; Figure 2).
Arguably, the more markers used for population discrimination, the greater the resolution of the subject animals’ ancestry. In this study, our results suggest that the SNP are not as discriminating as STR, because multiple SNP are required to achieve the same informative power of a single STR. Therefore, identifying SNP that have great population differentiation power can reduce the number of markers necessary to obtain equivalent STR results. Our results showed that 45 is the optimal number of SNP in our panel for maximizing resources and time efficiency without losing significant discriminating power, although as few as 27 SNP could achieve the same power in detecting population structure as 17 STR in the 4 populations analyzed. Therefore, approximately 1.6 SNP were necessary to provide the same statistical power as a single STR, suggesting that the panel of SNP we used here was more powerful relative to STR than other SNP panels.14,49
Although the current study's focus was limited to 6 source populations—Cambodia, Sumatra, Mauritius, Singapore, and the Philippine populations in Luzon and Zamboanga—some of which represent small island populations, our results underscore the significant genetic subdivision caused by the geographic structure of mainland and maritime Southeast Asia. STR will remain an important tool for cynomolgus population studies, but our findings show that SNP can be valuable for understanding and detecting genetic structure and have potential to move into the mainstream for future population studies.
Acknowledgments
The research described in this article has been reported in George Day's Masters of Science (Forensic Science) thesis, which was submitted to the University of California–Davis Office of Graduate Studies in August 2015. This research has also been presented at the Society of American Primate Veterinarians 40th Annual Workshop, which was held in St Paul, MN, 31 Oct through 3 Nov 2012.
This study was supported by California National Primate Research Center (CNPRC) base grant (no. RR000169-48), ARRA supplement grant RR018144-07 to SK and Nick Lerche (CNPRC), and NIH grants RR005090 and RR025871 to DGS. We thank the staff of the Molecular Anthropology Laboratory at the University of California–Davis for their assistance with sample preparation.
Samples were provided pro bono by Primate Products. Animals involved in this study were managed in compliance with the American Society of Primatologists (ASP) Principles for the Ethical Treatment of Nonhuman Primates and with the National Institutes of Health guidelines prescribing the humane care and use of laboratory animals.
References
- 1.Anderios F, Noorrain A, Vythilingam I. 2010. In vivo study of human Plasmodium knowlesi in Macaca fascicularis. Exp Parasitol 124:181–189. 10.1016/j.exppara.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Bonhomme M, Blancher A, Cuartero S, Chikhi L, Crouau-Roy B. 2008. Origin and number of founders in an introduced insular primate: estimation from nuclear genetic data. Mol Ecol 17:1009–1019. 10.1111/j.1365-294X.2007.03645.x. [DOI] [PubMed] [Google Scholar]
- 3.Bonhomme M, Cuartero S, Blancher A, Crouau-Roy B. 2009. Assessing natural introgression in 2 biomedical model species, the rhesus macaque (Macaca mulatta) and the long-tailed macaque (Macaca fascicularis). J Hered 100:158–169. 10.1093/jhered/esn093. [DOI] [PubMed] [Google Scholar]
- 4.Bowden DM, Smith OA. 1992. Conservationally sound assurance of primate supply and diversity. ILAR J 34:53–56. 10.1093/ilar.34.4.53. [DOI] [Google Scholar]
- 5.Bunlungsup S, Imai H, Hamada Y, Matsudaira K, Malaivijitnond S. 2017. Mitochondrial DNA and 2 Y-chromosome genes of common long-tailed macaques (Macaca fascicularis fascicularis) throughout Thailand and vicinity. Am J Primatol 79:1–13. 10.1002/ajp.22596. [DOI] [PubMed] [Google Scholar]
- 6.Bunlungsup S, Kanthaswamy S, Oldt RF, Smith DG, Houghton P, Hamada Y, Malaivijitnond S. 2017. Genetic analysis of samples from wild populations opens new perspectives on hybridization between long-tailed (Macaca fascicularis) and rhesus macaques (Macaca mulatta). Am J Primatol 79:1–12. 10.1002/ajp.22726. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty R, Stivers DN, Su B, Zhong Y, Budowle B. 1999. The utility of short tandem repeat loci beyond human identification: implications for development of new DNA typing systems. Electrophoresis 20:1682–1696. . [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay B, Garg KM, Ramakrishnan U. 2014. Effect of diversity and missing data on genetic assignment with RAD-Seq markers. BMC Res Notes 7:841 10.1186/1756-0500-7-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin W, Contacos PG, Coatney GR, Kimball HR. 1965. A naturally acquired quotidian-type malaria in man transferable to monkeys. Science 149:865–865. 10.1126/science.149.3686.865. [DOI] [PubMed] [Google Scholar]
- 10.Collins WE, Skinner JC, Broderson JR, Filipski VK, Morris CM, Stanfill PS, Warren M. 1992. Susceptibility of Macaca fascicularis monkeys from Mauritius to different species of Plasmodium. J Parasitol 78:505–511. 10.2307/3283652. [DOI] [PubMed] [Google Scholar]
- 11.de Souto MC, Jaskowiak PA, Costa IG. 2015. Impact of missing data imputation methods on gene expression clustering and classification. BMC Bioinformatics 16:1–9. 10.1186/s12859-015-0494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 13.Excoffier L, Lischer HE. 2010. Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567. 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 14.Fernández ME, Goszczynski DE, Liron JP, Villegas-Castagnasso EE, Carino MH, Ripoli MV, Rogberg-Munoz A, Posik DM, Peral-Garcia P, Giovambattista G. 2013. Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability, and assessment of parentage in an inbred Angus herd. Genet Mol Biol 36:185–191. 10.1590/S1415-47572013000200008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fooden J. 1980. Classification and distribution of living macaques (Macaca lacepede, 1799), p 1–9. In: Lindburg DG. editor. The macaques: studies in ecology, behavior, and evolution. New York (NY): Van Nostrand Reinhold. [Google Scholar]
- 16.Fooden J. 1995. Systematic review of Southeast Asian longtail macaques, Macaca fascicularis (Raffles, 1821). Fieldiana Zoology 81:1–206. [Google Scholar]
- 17.Fooden J. 2000. Systematic review of the rhesus macaque, Macaca mulatta (Zimmerman, 1780). Chicago (IL): Field Museum of Natural History. [Google Scholar]
- 18.Fries R, Durstewitz G. 2001. Digital DNA signatures for animal tagging. Nat Biotechnol 19:508 10.1038/89213. [DOI] [PubMed] [Google Scholar]
- 19.Glaubitz JC, Rhodes OE, Dewoody JA. 2003. Prospects for inferring pairwise relationships with single-nucleotide polymorphisms. Mol Ecol 12:1039–1047. 10.1046/j.1365-294X.2003.01790.x. [DOI] [PubMed] [Google Scholar]
- 20.Goddard KA, Wijsman EM. 2002. Characteristics of genetic markers and maps for cost-effective genome screens using diallelic markers. Genet Epidemiol 22:205–220. 10.1002/gepi.0177. [DOI] [PubMed] [Google Scholar]
- 21.Harihara S, Saitou N, Hirai M, Aoto N, Terao K, Cho F, Honjo S, Omoto K. 1988. Differentiation of mitochondrial DNA types in Macaca fascicularis. Primates 29:117–127. 10.1007/BF02380854. [DOI] [Google Scholar]
- 22.Haus T, Ferguson B, Rogers J, Doxiadis G, Certa U, Rose NJ, Teepe R, Weinbauer GF, Roos C. 2014. Genome typing of nonhuman primate models: implications for biomedical research. Trends Genet 30:482–487. 10.1016/j.tig.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 23.Helyar SJ, Hemmer-Hansen J, Bekkevold D, Taylor MI, Ogden R, Limborg MT, Cariani A, Maes GE, Diopere E, Carvalho GR, Nielsen EE. 2011. Application of SNP for population genetics of nonmodel organisms: new opportunities and challenges. Mol Ecol Resour 11 Suppl 1:123–136. 10.1111/j.1755-0998.2010.02943.x. [DOI] [PubMed] [Google Scholar]
- 24.Hubisz MJ, Falush D, Stephens M, Pritchard JK. 2009. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332. 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Institute for Laboratory Animal Research. 2003. Demands for rhesus monkeys in biomedical research: a workshop report.. ILAR J 44:222–238. 10.1093/ilar.44.3.222. [DOI] [PubMed] [Google Scholar]
- 26.Jorgenson E, Witte JS. 2007. Microsatellite markers for genome-wide association studies. Nat Rev Genet 8:164 doi:10.1038/nrg1962-c2 [DOI] [PubMed] [Google Scholar]
- 27.Kanthaswamy S, Gill L, Satkoski J, Goyal V, Malladi V, Kou A, Basuta K, Sarkisyan L, George D, Smith DG. 2009. The development of a Chinese–Indian hybrid (Chindian) rhesus macaque colony at the California National Primate Research Center (CNPRC) by introgression. J Med Primatol 38:86–96. 10.1111/j.1600-0684.2008.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanthaswamy S, Ng J, Satkoski Trask J, George DA, Kou AJ, Hoffman LN, Doherty TB, Houghton P, Smith DG. 2013. The genetic composition of populations of cynomolgus macaques (Macaca fascicularis) used in biomedical research. J Med Primatol 42:120–131. 10.1111/jmp.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanthaswamy S, Satkoski J, George D, Kou A, Erickson BJ, Smith DG. 2008. Interspecies hybridization and the stratification of nuclear genetic variation of rhesus (Macaca mulatta) and long-tailed macaques (Macaca fascicularis). Int J Primatol 29:1295–1311. 10.1007/s10764-008-9295-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanthaswamy S, Satkoski J, Kou A, Malladi V, Glenn Smith D. 2010. Detecting signatures of inter-regional and inter-specific hybridization among the Chinese rhesus macaque specific pathogen-free (SPF) population using single-nucleotide polymorphic (SNP) markers. J Med Primatol 39:252–265. 10.1111/j.1600-0684.2010.00430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanthaswamy S, Trask JS, Ross CT, Kou A, Houghton P, Smith DG, Lerche N. 2012. A large-scale SNP-based genomic admixture analysis of the captive rhesus macaque colony at the California National Primate Research Center. Am J Primatol 74:747–757. 10.1002/ajp.22025. [DOI] [PubMed] [Google Scholar]
- 32.Kawamoto Y, Ischak TM, Supriatna J. 1984. Genetic variations within and between troops of the crab-eating macaque (Macaca fascicularis) on Sumatra, Java, Bali, Lombok, and Sumbawa, Indonesia. Primates 25:131–159. 10.1007/BF02382387. [DOI] [Google Scholar]
- 33.Kawamoto Y, Kawamoto S, Matsubayashi K, Nozawa K, Watanabe T, Stanley MA, Perwitasari-Farajallah D. 2008. Genetic diversity of longtail macaques (Macaca fascicularis) on the island of Mauritius: an assessment of nuclear and mitochondrial DNA polymorphisms. J Med Primatol 37:45–54. [DOI] [PubMed] [Google Scholar]
- 34.Kondo M, Kawamoto Y, Nozawa K, Matsubayashi K, Watanabe T, Griffiths O, Stanley M-A. 1993. Population genetics of crab-eating macaques (Macaca fascicularis) on the island of Mauritius. Am J Primatol 29:167–182. 10.1002/ajp.1350290303. [DOI] [PubMed] [Google Scholar]
- 35.Krawczak M. 1999. Informativity assessment for biallelic single-nucleotide polymorphisms. Electrophoresis 20:1676–1681. . [DOI] [PubMed] [Google Scholar]
- 36.Krebs KC, Jin Z, Rudersdorf R, Hughes AL, O'Connor DH. 2005. Unusually high-frequency MHC class I alleles in Mauritian origin cynomolgus macaques. J Immunol 175:5230–5239. 10.4049/jimmunol.175.8.5230. [DOI] [PubMed] [Google Scholar]
- 37.Kruglyak L. 1997. The use of a genetic map of biallelic markers in linkage studies. Nat Genet 17:21–24. 10.1038/ng0997-21. [DOI] [PubMed] [Google Scholar]
- 38.Lawler SH, Sussman RW, Taylor LL. 1995. Mitochondrial DNA of the Mauritian macaques (Macaca fascicularis): an example of the founder effect. Am J Phys Anthropol 96:133–141. 10.1002/ajpa.1330960203. [DOI] [PubMed] [Google Scholar]
- 39.Liedigk R, Kolleck J, Boker KO, Meijaard E, Md-Zain BM, Abdul-Latiff MA, Ampeng A, Lakim M, Abdul-Patah P, Tosi AJ, Brameier M, Zinner D, Roos C. 2015. Mitogenomic phylogeny of the common long-tailed macaque (Macaca fascicularis fascicularis). BMC Genomics 16:222 10.1186/s12864-015-1437-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Arias R, Calafell F, Mateu E, Comas D, Andres A, Bertranpetit J. 2001. Sequence variability of a human pseudogene. Genome Res 11:1071–1085. 10.1101/gr.GR-1677RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Migot-Nabias F, Ollomo B, Dubreuil G, Morelli A, Domarle O, Nabias R, Georges AJ, Millet P. 1999. Plasmodium coatneyi: differential clinical and immune responses of 2 populations of Macaca fascicularis from different origins. Exp Parasitol 91:30–39. 10.1006/expr.1999.4342. [DOI] [PubMed] [Google Scholar]
- 42.Osada N, Uno Y, Mineta K, Kameoka Y, Takahashi I, Terao K. 2010. Ancient genome-wide admixture extends beyond the current hybrid zone between Macaca fascicularis and M. mulatta. Mol Ecol 19:2884–2895. 10.1111/j.1365-294X.2010.04687.x. [DOI] [PubMed] [Google Scholar]
- 43.Pavlin BI, Schloegel LM, Daszak P. 2009. Risk of importing zoonotic diseases through wildlife trade, United States. Emerg Infect Dis 15:1721–1726. 10.3201/eid1511.090467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43:223–225. 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 46.Satkoski Trask JA, Garnica WT, Smith DG, Houghton P, Lerche N, Kanthaswamy S. 2013. Single-nucleotide polymorphisms reveal patterns of allele sharing across the species boundary between rhesus (Macaca mulatta) and cynomolgus (M. fascicularis) macaques. Am J Primatol 75:135–144. 10.1002/ajp.22091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN. 2004. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility loci. Am J Hum Genet 75:948–965. 10.1086/425870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmidt LH, Fradkin R, Harrison J, Rossan RN. 1977. Differences in the virulence of Plasmodium knowlesi for Macaca irus (fascicularis) of Philippine and Malayan origins. Am J Trop Med Hyg 26:612–622. 10.4269/ajtmh.1977.26.612. [DOI] [PubMed] [Google Scholar]
- 49.Schopen GC, Bovenhuis H, Visker MH, van Arendonk JA. 2008. Comparison of information content for microsatellites and SNP in poultry and cattle. Anim Genet 39:451–453. 10.1111/j.1365-2052.2008.01736.x. [DOI] [PubMed] [Google Scholar]
- 50.Seekatz AM, Panda A, Rasko DA, Toapanta FR, Eloe-Fadrosh EA, Khan AQ, Liu Z, Shipley ST, Detolla LJ, Sztein MB, Fraser CM. 2013. Differential response of the cynomolgus macaque gut microbiota to Shigella infection. PLoS One 8:1–13. 10.1371/journal.pone.0064212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh B, Daneshvar C. 2013. Human infections and detection of Plasmodium knowlesi. Clin Microbiol Rev 26:165–184. 10.1128/CMR.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ. 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363:1017–1024. 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 53.Smith DG, George D, Kanthaswamy S, McDonough J. 2006. Identification of country of origin and admixture between Indian and Chinese rhesus macaques. Int J Primatol 27:881–898. 10.1007/s10764-006-9026-3. [DOI] [Google Scholar]
- 54.Smith DG, McDonough JW, George DA. 2007. Mitochondrial DNA variation within and among regional populations of longtail macaques (Macaca fascicularis) in relation to other species of the fascicularis group of macaques. Am J Primatol 69:182–198. 10.1002/ajp.20337. [DOI] [PubMed] [Google Scholar]
- 55.Smith DG, Ng J, George D, Trask JS, Houghton P, Singh B, Vilano J, Kanthaswamy S. 2014. A Genetic comparison of 2 alleged subspecies of Philippine cynomolgus macaques. Am J Phys Anthropol 155:136–148. 10.1002/ajpa.22564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevison LS, Kohn MH. 2009. Divergence population genetic analysis of hybridization between rhesus and cynomolgus macaques. Mol Ecol 18:2457–2475. 10.1111/j.1365-294X.2009.04212.x. [DOI] [PubMed] [Google Scholar]
- 57.Street SL, Keyes RC, Grant R, Ferguson B. 2007. Single-nucleotide polymorphisms (SNP) are highly conserved in rhesus (Macaca mulatta) and cynomolgus (Macaca fascicularis) macaques. BMC Genomics 8:1–9. 10.1186/1471-2164-8-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thalamuthu A, Mukhopadhyay I, Ray A, Weeks DE. 2005. A comparison between microsatellite and single-nucleotide polymorphism markers with respect to 2 measures of information content. BMC Genet 6 Suppl 1:S27 10.1186/1471-2156-6-S1-S27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tosi AJ, Coke CS. 2007. Comparative phylogenetics offer new insights into the biogeographic history of Macaca fascicularis and the origin of the Mauritian macaques. Mol Phylogenet Evol 42:498–504. 10.1016/j.ympev.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Tosi AJ, Morales JC, Melnick DJ. 2002. Y-chromosome and mitochondrial markers in Macaca fascicularis indicate introgression with Indochinese M. mulatta and a biogeographic barrier in the Isthmus of Kra. Int J Primatol 23:161–178. 10.1023/A:1013258109954. [DOI] [Google Scholar]
- 61.Trask JS, Garnica WT, Kanthaswamy S, Malhi RS, Smith DG. 2011. 4040 SNP for genomic analysis in the rhesus macaque (Macaca mulatta). Genomics 98:352–358. 10.1016/j.ygeno.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiseman RW, O'Connor DH. 2007. Major histocompatibility complex-defined macaques in transplantation research. Transplant Rev (Orlando) 21:17–25. 10.1016/j.trre.2007.01.001. [DOI] [Google Scholar]
- 63.Xing C, Schumacher FR, Xing G, Lu Q, Wang T, Elston RC. 2005. Comparison of microsatellites, single-nucleotide polymorphisms (SNP) and composite markers derived from SNP in linkage analysis. BMC Genet 6 Suppl 1 :1–5. 10.1186/1471-2156-6-S1-S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, Kadir KA, Quintanilla-Zariñan LF, Villano J, Houghton P, Du H, Singh B, Smith DG. 2016. Distribution and prevalence of malaria parasites among long-tailed macaques (Macaca fascicularis) in regional populations across Southeast Asia. Malar J 15:1–8. 10.1186/s12936-016-1494-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X, Meng Y, Houghton P, Liu M, Kanthaswamy S, Oldt R, Ng J, Trask JS, Huang R, Singh B, Du H, Smith DG. 2017. Ancestry, Plasmodium cynomolgi prevalence, and rhesus macaque admixture in cynomolgus macaques (Macaca fascicularis) bred for export in Chinese breeding farms. J Med Primatol 46:31–41. 10.1111/jmp.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]