Abstract
Proteomic analysis is an attractive and powerful tool for characterizing the molecular profiles of diseased tissues, such as the vitreous. The complexity of data available for analysis ranges from single (e.g., enzyme-linked immunosorbent assay [ELISA]) to thousands (e.g., mass spectrometry) of proteins, and unlike genomic analysis, which is limited to denoting risk, proteomic methods take snapshots of a diseased vitreous to evaluate ongoing molecular processes in real time. The proteome of diseased ocular tissues was recently characterized, uncovering numerous biomarkers for vitreoretinal diseases and identifying protein targets for approved drugs, allowing for drug repositioning. These biomarkers merit more attention regarding their therapeutic potential and prospective validation, as well as their value as reproducible, sensitive, and specific diagnostic markers
Translational Relevance
Personalized proteomics offers many advantages over alternative precision-health platforms for the diagnosis and treatment of vitreoretinal diseases, including identification of molecular constituents in the diseased tissue that can be targeted by available drugs.
Keywords: personalized proteomics, biomarker, retina, vitreous, drug repositioning, diagnostics
Background—The Precision Health Era
Precision Health aims to tailor medical therapies to each individual patient by taking into account his or her specific genetics, environments, and lifestyle choices. Recently empowered by large sets of molecular and clinical data and high-powered analytics, this concept is changing the field of healthcare, such that we now can customize therapies for each patient. No longer is medical practice confined exclusively to physicians, as basic scientists, engineers, entrepreneurs, healthcare providers, and patients all work together to bring innovative therapies from the laboratory bench to the bedside. Advances in our understanding of the molecular basis of disease are leading to the development of more timely interventions. Precision Health already has transformed the field of cancer and is inspiring a renaissance across multiple medical fields, including ophthalmology. As we move away from symptom-based treatments for blinding eye diseases to a Precision Health approach, we are on the cusp of a new era.
An early achievement of the Precision Health approach is the routine use of cardiac biomarkers for the diagnosis of myocardial infarction (MI). When a patient is admitted for chest pain, for example, MI tops the list of differential diagnoses that must be ruled out. Despite clinical examination, a critical protein biomarker, troponin, is measured routinely as a key to diagnosis and timely intervention. Troponin assays are exquisitely sensitive to the presence of myocardial necrosis and so used to definitively diagnose acute MI.1 Recent advances in Precision Health are exemplified in the personalized treatment of cancer: cellular biomarkers of the tumor microenvironment can help to determine therapeutic approaches and predict prognosis. Robust and reproducible associations have been made between immune gene signatures in tumors and clinical outcomes.2 For example, gene signatures reflecting T- and B-cell specific immune responses have demonstrated positive associations with recurrence-free survival in patients with aggressive subtypes of breast cancer.2–4 Tumors expressing human epidermal growth factor receptor 2 (HER2) are treated with trastuzamab (anti-HER2), while tumors expressing estrogen/progesterone receptors are treated with tamoxifen (selective estrogen receptor modulator).5,6 Similar concepts are now being applied in the context of vitreoretinal diseases.7
In the case of organ-specific diseases, sampling fluid compartments near the diseased tissue (e.g., synovial fluid, urine, cerebral spinal fluid) may be better for diagnosing nonsystemic diseases.8–10 Vitreoretinal diseases often have no equivalent sensitive and specific molecular assay, leaving diagnosis and treatment most often empiric, relying heavily on findings from clinical exams. With appropriate advances, key ophthalmic protein biomarkers similarly could be used routinely to diagnose diseases, such as diabetic retinopathy, retinitis pigmentosa, macular degeneration, proliferative vitreoretinopathy, uveitis, and ocular malignancy.7
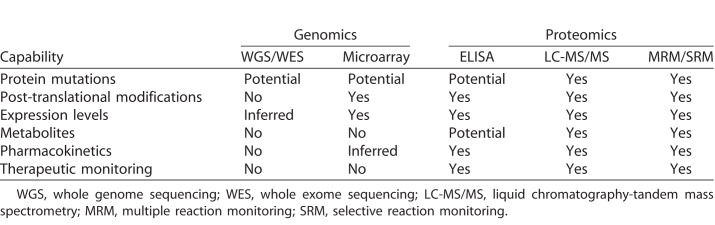
Proteomic analysis of vitreous and aqueous humor is a promising Precision Health strategy for diagnosing and treating numerous complex ophthalmic diseases. Proteomics refers to the large-scale detection of proteins, metabolites, and modifications within a biologic sample. The complexity of proteomic data ranges from the presence or absence of single proteins (e.g., by enzyme-linked immunosorbent assay [ELISA]) to changes in profiles comprised of thousands of proteins (e.g., by mass spectrometry); and while characterizing a single analyte often gives insufficient evidence for making a diagnosis or monitoring complex pathologic processes, the ability to measure multiple proteins or metabolites in real time will enhance our power to diagnose disease or determine the ideal therapeutic regimen. Despite its complexity, mass spectrometry is extremely versatile and offers information on protein expression levels, posttranslational modifications, metabolites, and pharmacokinetics. Precision Health approaches already are using numerous proteomic detection platforms that could be extended immediately for ophthalmic use (Table 1).
Table 1.
Precision Medicine Platforms
Liquid Biopsy Techniques for Ophthalmic Tissues
Retinal biopsy procedures are invasive and carry high rates of visual morbidity, including vitreous hemorrhage and retinal detachment.11 These procedures are uncommon and typically reserved for atypical presentations of ocular inflammation and malignancy.12 Therefore, it is not feasible to routinely biopsy the neurosensory retina. Rather, it is more advantageous to sample the fluid compartment adjacent to the retina to monitor biomarkers. The vitreous humor is an optically-transparent extracellular matrix located in the posterior chamber of the eye, just anterior to the retina (Fig. 1A). Its composition is estimated to be 90% water, with a density that varies depending on anatomic location (i.e., core, cortex, and base).13,14 These characteristics change with age (i.e., from high to low viscosity) and often can be affected by numerous vitreoretinal diseases.13 Damaged retinal cells can release proteins into the vitreous that may remain undetected due to the invasive nature of retinal biopsy procedures.12 Proteomic analysis of the adjacent vitreous may serve as way to indirectly biopsy the diseased retina and identify changes in its proteome.15–19
Figure 1.
Summary of liquid biopsy techniques for ophthalmic tissues: cross sectional image of the human eye. (A) The vitreous is an extracellular matrix that covers the retina, lens, and ciliary body. The vitreous core is biopsied using a 23-gauge needle (depicted) or vitreous cutter and contains native vitreous proteins, systemic protein biomarkers, and retinal biomarkers that can be sampled through proteomic analysis. (B) The aqueous humor, located in the anterior chamber of the eye, is produced by the ciliary body. A 25-gauge needle can be inserted into the anterior chamber at the limbus to sample the aqueous humor for proteomic analysis. Graphical illustrations by Alton Szeto and Vinit Mahajan. Permission to publish granted by original artist.
Previous proteomic analysis of human vitreous from nondiseased postmortem eyes revealed a diverse catalogue of intracellular and extracellular proteins, proteoglycans, and small molecules that originate inside and outside the eye.16 Therefore, changes in the molecular composition of the vitreous can be expected to reflect key pathologic changes during vitreoretinal disease that correlate to disease onset, progression, and response to therapy. Vitreous biopsies frequently are used in the clinical management and diagnosis of intravitreal inflammation, infection, and cancer.14 Proteomic analysis of these liquid biopsies expands their clinical use in the personalized management of patient care.15
Vitreous biopsies can be obtained from living patients in several ways. The least complex method is fine needle aspiration (FNA), where a 23-gauge needle is inserted through the pars plana to manually aspirate small amounts of fluid from the vitreous cavity (Fig. 1A).20,21 We reported that needle biopsies are comparable to vitreous cutter biopsies.14 A prospective case series of patients undergoing this office-based aspiration demonstrated the method to be reproducible and safe with an average of 100 to 200 μL undiluted vitreous obtained in 88% of patients.20 Although this procedure can be done safely under local anesthesia in an outpatient setting, its main limitation often is an inadequate volume of sample for large-scale proteomic studies.14,20,21 A second method involves pars plana vitrectomy (PPV) under local or general anesthesia within the operating room. This technique uses a small, high-speed guillotine called a vitrector to chop and aspirate the vitreous. Although more invasive, PPV ensures adequate sample volume, lower incidence of hypotony, and potentially better sampling of insoluble proteins.14 Previous studies that compared paired samples from 23-gauge FNA and 23-gauge PPV found that both techniques were nearly equivalent with regard to protein concentration, with only minor discrepancies in the relative abundance of certain proteins (as determined by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).14 We anticipated that future studies will address the proteomic quality of samples obtained from small-gauge vitrectomy techniques (e.g., 25- and 27-gauge PPV and microincision vitrectomy), which may be safer and less invasive.
The aqueous humor (AH), produced by the nonpigmented ciliary body epithelium, contains a complex mixture of electrolytes, organic solutes, and proteins that provides nutrition to the avascular tissues of the anterior chamber (Fig. 1B).22 The balance between production and drainage of AH is important in maintaining intraocular pressure and the refractive properties of the eye.22–24 AH typically is sampled before surgical intervention for cataract and contains over 600 nonredundant proteins.22 Changes in the proteomic content of the aqueous humor have been identified in diseases affecting the anterior chamber, including glaucoma and pseudoexfoliation syndrome. Similarly, proteomic studies on AH fluid from patients with macular degeneration and diabetic retinopathy have shown that AH protein content also can be affected in vitreoretinal diseases.25–28 AH fluid can be biopsied in the operating room by inserting a 25-gauge needle into the peripheral cornea at the limbus (Fig. 1B). Using this method, small volumes of AH (up to 100 μL) can be aspirated.24 Despite limitations in sampling AH to characterize vitreoretinal diseases (especially since cytokine profiles of aqueous and vitreous differ significantly29), sampling these tissues may be more beneficial in diseases where vitrectomy surgery is not indicated.
Proper care and handling of surgical specimens is critical to quality control for subsequent proteomic analysis. To ensure tissues are immediately cataloged, processed, and stored, we developed the mobile operating room lab interface (MORLI). The MORLI system has several key components: a mobile operating room cart with a flat, lab bench surface, a computer with secure access to a sample database, a barcode scanner, and drawers with lab supplies for specimen collection (e.g., pipettors, centrifuge, dissecting microscope, cryotubes, and a small liquid nitrogen dewar).30 The MORLI cart allows samples to be processed away from the surgical field. Liquid vitreous samples are collected (via FNA or PPV) and passed to the lab technician who spins down the sample using a microcentrifuge (16,000 × g for 5 minutes at 4°C; to remove cellular debris), transfers the sample to a barcoded cryotube, and flash-freezes it in liquid nitrogen. The corresponding sample barcode is entered into an electronic database for efficient sample logging and retrieval. This biorepository system streamlined our personalized proteomics pipeline for the study of ophthalmic diseases. A similar system could catalog surgical specimens from other tissues.
For ethical reasons, researchers and clinicians do not sample the vitreous of healthy, living patients. Vitrectomy surgery requires a pathologic state, even in noninflammatory conditions, such as idiopathic macular holes and epiretinal membranes. Thus, we derive our control samples from patients with isolated forms of posterior segment pathology, such as idiopathic macular holes (IMHs), which are small, full-thickness retinal disruptions that alter the normal foveal anatomy and lead to severe, unilateral visual distortion.31 Surgical repair of IMHs often involves peeling the internal limiting membrane (ILM) or injecting ocriplasmin into the vitreous cavity to help close the retinal hole.32,33 Another pathology that often serves as our control is a visually significant epiretinal membrane (ERM), which is a thin fibrocellular membrane that forms over the vitreoretinal interface, causing disruption of the normal foveal contour and distorting vision. The exact mechanism for ERM formation is unknown; however, it is believed to be related to cellular changes induced by a posterior vitreous detachment (PVD).34
Although these two conditions are noninflammatory, they likely alter the molecular composition of the vitreous. Likewise, key molecular alterations were detected by proteomic analysis of vitreous from patients undergoing IMH repair, likely representing an underlying pathogenesis driving the formation of macular holes. These include an increased expression of complement pathway effectors and α-2–macroglobulin, a major inducer of Müller cell migration.32 Similarly, analysis of vitreous samples from patients undergoing surgical repair for an ERM identified increased levels of α1-antitrypsin, apolipoprotein-A1, and transthyretin compared to those from IMH vitreous samples.34 As an alternative source of control samples, Wu et al.35 argued that postmortem healthy eyes might be appropriate.35 However, postmortem changes in the vitreous can be reflected in the proteome, confounding the results.36,37 These data reveal how important considerations must be made by the researcher regarding control sample selection for proteomic studies.
Summary of Analytical Methods
Once surgical specimens are properly collected, processed, and stored, their proteomic composition can be analyzed. The choice of an analytical method depends largely on the question being asked. For example, in the case of characterizing an inflammatory disease, the clinician or researcher may wish to focus exclusively on identifying cytokine signaling proteins in a biological sample using multiplex immunoassays. Multiplex immunoassays are a powerful and efficient approach to simultaneously quantifying hundreds of proteins in a biological sample,38 reducing assay costs, time, and the sample volume required for analysis.38
For idiopathic or poorly-characterized diseases, an unbiased approach, like shotgun mass spectrometry (MS), may be more appropriate. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) is a powerful analytical technique that ionizes molecular species and sorts ions based on their mass-to-charge ratio (m/z). This technique is used in a shotgun approach to catalog and quantify the thousands of proteins in a biological sample, which often is performed in tandem with liquid chromatography to separate peptides by size and hydrophobicity before they are ionized. Beforehand, samples are proteolytically digested with trypsin (or another protease) to generate a peptide mixture with less biochemical heterogeneity and simplify the process of protein separation, ionization, and MS characterization.39 Before LC-MS/MS analysis, peptides may be fractionated by strong-cation exchange chromatography (SCX) or isoelectric focusing (IEF).39 However, advances in liquid chromatography, namely ultra-high-pressure liquid chromatography (UPLC), reduce the need to fractionate peptides beforehand.40
Once separated and before entering the mass spectrometer, aqueous phase analytes are ionized to form gas-phase ions. Although many methods exist for this step, biological MS generally uses soft ionization techniques (like electrospray ionization [ESI] and matrix-assisted laser desorption ionization [MALDI]) because they leave large molecules intact.40 Gas-phase ions then are directed toward the mass analyzer, where they are isolated by time (time-of-flight devices; TOF) or space (trap devices) before hitting the detector.39 In the mass spectrometer, isolated peptides produce a fragmentation pattern that yields an individual mass spectrum. Highly advanced bioinformatics algorithms exist that match the thousands of spectra obtained from an LC-MS/MS experiment to known sequences of proteins within large spectral libraries.41–44 One limitation to this approach is that it can only compare sample peptides to those that were previously identified, so advances in search algorithms incorporate simulated proteome-wide spectral libraries, to increase assignments of unique and novel peptides.45
Once peptides are identified, they are quantified using unlabeled and labeled methods. Unlabeled methods include spectral counting or data-independent acquisition (DIA). In DIA MS, peptides within defined m/z windows are fragmented and analyzed without relying on predefined peptides of interest.46 Labeled methods include isotope-coded affinity tags (ICAT), isobaric tags for relative and absolute quantification (iTRAQ), and multiple reaction monitoring (MRM).40 Also referred to as selective reaction monitoring (SRM), MRM is a MS method that detects and quantifies selected target peptides from a complex mixture of proteins.47 Prespecified peptide-precursor ions and their fragments allow for highly-sensitive, reproducible quantification of targeted proteins. This method has advantages over multiplex ELISAs, since it does not rely on antibody quality, and can detect posttranslational modifications (PTMs) and short nucleotide modifications (SNPs) that would otherwise be missed by ELISA (Table 1).47
Vitreous and aqueous, like many serum samples, contain abundant levels of albumin and immunoglobulins,19,48 so these proteins often are depleted before MS analysis so that less abundant proteins can be detected and quantified.49 This process can create false-negative results, however, since many proteins bind to albumin and, therefore, may be depleted during preprocessing.19 Another valuable component of these biopsy fluids is their exosome content. Exosomes are endosome-derived microvesicles released from cells that contain intracellular and membrane-bound proteins, DNA, and RNA.50 Exosomes frequently are isolated from biopsy fluids (e.g., plasma, cerebrospinal fluid [CSF], urine, saliva, synovial fluid, and so forth) and are reported to regulate cellular processes, such as apoptosis, angiogenesis, and inflammation.50–54 Fractionation of liquid biopsy fluids allows for exosome isolation and proteomic detection of their contents.51 Previous studies have identified exosomes in human aqueous humor as well as in the vitreous of uveal melanoma and ERM/IMH patients.55–57 Proteomic analysis of vitreous and aqueous exosomes can expand knowledge of retinal disease pathophysiology and identify novel disease biomarkers. As MS technology advances, sample preprocessing and fractionation may be minimized for proteomic analysis, thereby reducing the number of false-negatives and improving the time from sample collection to analysis.40 Figure 2 summarizes the personalized proteomics pipeline for ophthalmic tissues.
Figure 2.
Personalized proteomics pipeline for precision health in ophthalmology: liquid vitreous biopsies can be obtained in the operating room using a vitreous cutter or 23-gauge needle (left). Vitreous samples can be analyzed for protein content using multiplex ELISA arrays (top row). Custom or commercial antibody arrays quantify protein levels in biological samples using fluorescence or chemiluminescence means. Alternatively, vitreous fluid can be analyzed using a mass spectrometry approach (bottom row). Protein mixtures are digested with trypsin (or another digestive protease) and peptides are extracted with organic solvents. Analytes can be enriched using a variety of affinity chromatography techniques. Chromatography (HPLC, UPLC) is used to separate peptides before ionization and mass acquisition by mass spectrometry (e.g., ESI-MS/MS and MALDI-TOF MS/MS). Highly-advanced algorithms (e.g., MASCOT, OMSSA, and X!Tandem) match the thousands of spectra to known protein sequences and proteins quantified either through unlabeled (e.g., spectral counting or DIA) or labeled methods (e.g., MRM/SRM and iTRAQ). Once protein levels are quantified (either from an ELISA or MS experiment), downstream bioinformatics analysis (right) can help put the identified proteins into the context of the disease.
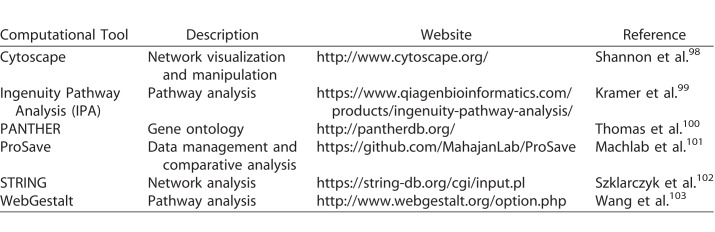
Challenges nevertheless remain when it comes to downstream analysis and management of large LC-MS/MS datasets. Shotgun proteomic experiments can produce data on thousands of proteins, for which meaningful interpretation requires advanced bioinformatics and statistical expertise. Once MS spectra are matched to their respective proteins and quantified, researchers can perform gene ontology as well as pathway and network analysis to interpret the data in a biological or clinical context. This may provide insight into how molecular pathways are affected in diseased tissues. From these data, researchers can study the relevant proteins, their functions, and how they relate to disease onset, timing, severity, and response to therapy. Table 2 summarizes software tools commonly used for bioinformatics analysis of proteomics data. Certain analyses (e.g., Venn diagram, gene ontology, and network analysis) do not incorporate quantitative data (e.g., spectral count or ion abundance), so critical information often is lost. To preserve these important data, we recently developed ProSave, a Java-based program that retrieves quantitative data (e.g., ion abundance or spectral counts) from a curated list of proteins in large proteomics datasets so researchers can derive a better understanding of each protein in a proteomics dataset. For proteomic analysis, development of standardized and user-friendly bioinformatics pipelines will streamline application to routine clinical practice.
Table 2.
Bioinformatics Resources for Personalized Proteomics
Patient Stratification—Proteomics for Biomarker Identification
The proteome represents a network of end products generated from a series of processes related to protein synthesis within a specific cellular environment.58 Biomarkers, on the other hand, are defined by the Biomarkers Definitions Working Group as “a characteristic that is objectively measured and evaluated as an indicator of normal biological processes, pathogenic processes, or pharmacologic responses to a therapeutic intervention.”59 Several intra- and extracellular factors influence the cellular protein profile: (1) early on by the DNA sequence; (2) intermediately by translational, posttranslational, and regulatory steps; and (3) ultimately by the degradative stimuli.58 Thus, proteomic profiles might represent the ultimate biomarkers of cellular status in health and disease.
The discovery of proteomic biomarkers already has improved our understanding of the molecular mechanisms of diseases and should soon become a helpful diagnostic and risk-stratification tool, allowing individualized treatment for safer and more effective therapies. Disease processes alter cellular function and gene expression, such that changes in the protein profile can be used for diagnosis and prognosis.40,60 Normal aqueous humor and vitreous contain endogenously produced proteins that ocular diseases may alter.19,23 Thus, for many eye diseases, the aqueous humor and vitreous may potentially represent readily accessible repositories of proteomic biomarkers.16,61 Unbiased and untargeted proteomic approaches, such as LC-MS/MS, are ideal for identifying biomarkers in aqueous or vitreous biopsies. Even without a priori knowledge of a disease or its causative agent, one can identify novel biomarkers. Such an approach can aid in the systematic understanding of pathophysiology, simply by cataloging upregulated and downregulated proteins in a tissue sample.
Current ophthalmic proteomic studies are investigating the protein profiles related to age-related macular degeneration (AMD), diabetic retinopathy (DR), retinal detachment (RD), proliferative vitreoretinopathy (PVR), uveitis, and ocular cancers.15,61,62 Past proteomic studies from liquid biopsies of patients with vitreoretinal diseases are summarized in Table 3. The methods used in those studies ranged from multiplex ELISA arrays to shotgun MS analysis. For each of the pathologies analyzed, several proteins were found to be up- and downregulated, but further studies are needed to determine how to use this information in clinical practice.
Table 3.
Summary of Proteomic Studies on Liquid Biopsies From Patients With Vitreoretinal Disease as of 2018
Proteomics for the Diagnosis of Idiopathic Uveitis
Uveitis is a family of ocular inflammatory diseases that may involve the iris, ciliary body, vitreous and/or choroid, and it illustrates the potential that personalized proteomics may have to aid in diagnosis. Although often restricted to the eye, uveitis can be an early symptom of debilitating systemic disease with a prevalence of 1 in 4500 people63–67 and should be treated aggressively to prevent significant visual morbidity and blindness. Posterior uveitis involves the choroid and retina, encompassing a group of inflammatory diseases that account for approximately 10% of preventable blindness in the United States.68 This can be caused by infectious agents or systemic inflammatory disease and has high morbidity because the retina is intolerant of immunologic insult. Despite advances in diagnostic procedures, the etiology for over 50% of posterior uveitis cases is not known and, thus, they are labeled as “idiopathic.”68 In certain cases, such as acute retinal necrosis due to a member of the herpes virus family, the inciting agent can be treated directly.69 In most cases, however, the etiologic agent is unknown, and therapy is broadly directed at the inflammatory mediators that cause damage to ocular tissues. Corticosteroids have been the mainstay of uveitis treatment since their introduction in the 1950s, but long-term use results in unfavorable systemic side effects and vision-threatening glaucoma.70,71 Therefore, we need more reliable diagnostic testing to distinguish the various causes of uveitis and guide treatment. Personalized proteomics of vitreous biopsies can aid the diagnosis of these idiopathic cases.
For studies involving noninfectious uveitis, a commonly used disease model is the spontaneous intraocular inflammation observed in horses, known as equine recurrent uveitis (ERU).72 Proteomic studies on ERU tissues have advanced our understanding of the pathogenesis of autoimmune uveitis, and surgical removal of vitreous humor from ERU-affected horses reduces the frequency and severity of relapse.73 This is an important finding that suggests components of the diseased vitreous contribute to the pathogenesis and progression of autoimmune uveitis. Proteomic analysis of vitreous humor from healthy and ERU-affected horses by Deeg et al.74 found that pigment epithelium-derived factor (PEDF) was downregulated in inflamed equine vitreous, while VEGF levels were elevated. PEDF is involved in maintaining the blood/retina barrier (BRB) and regulates neovascularization within the eye.75 Studies on CD4+ T-cells from ERU-affected horses identified formin-like 1 (FMNL1) as a biomarker for inflammatory cell migration in autoimmune uveitis.76 In a cell-culture model of the BRB, treating CD4+ T-cells with monoclonal antibodies to FMNL1 reduced the transmigration rate, suggesting FMNL1 inhibition may delay the progression of inflammatory damage caused by autoimmune uveitis.76 Experimental autoimmune uveitis (EAU) models in mice similarly share many features with human autoimmune uveitis. Immunization with inter-photoreceptor retinol binding protein (IRBP) triggers intraocular inflammation in susceptible mice.77,78 Etiology-specific mouse models for cytomegalovirus (CMV) retinitis and tubercular uveitis also have been developed, highlighting the heterogeneity of immune stimulation seen in human uveitis patients.79–81
Proteomic analysis of vitreous biopsies from uveitis patients has helped identify cytokine signatures for specific forms of infectious and noninfectious uveitis: tumor necrosis factor-α (TNF-α) was implicated in the pathogenesis of juvenile idiopathic arthritis (JIA),82 and IL-6 was identified in patients with Behçet's disease, sarcoidosis, and Fuch's heterochromatic cyclitis (FHC).83 Similarly, interleukin (IL)-17A and IL-10 levels were levated in viral uveitis.84 Nevertheless, these studies only surveyed a limited number of cytokines and etiologies.
Analyses that make use of large-scale proteomic platforms, such as multiplex ELISA arrays or LC-MS/MS, have the potential to differentiate many etiologies of inflammatory eye diseases that otherwise are difficult to diagnose. In a previous study by our group, vitreous biopsies from 15 uveitis patients (three with idiopathic posterior uveitis, four with viral endophthalmitis, one with multifocal choroiditis, one with neovascular inflammatory vitreoretinopathy, two with autoimmune retinopathy, and one with HLA-B27 uveitis) were analyzed by a cytokine array that simultaneously measured the levels of 200 cytokines.15 Differential expression analysis and hierarchical heatmap clustering detected similarities and differences in the cytokine profiles and identified a cytokine signature common to these forms of uveitis (IL-23, PDGFRb, SCF, TIMP-1, TIMP-2, BMP-4, NGF, IGFBP-2, IL-17R, and IL-1RI; Table 3). These data suggest that seemingly different diseases might be targetable and treated by the same therapies.15 More importantly, this could redirect the diagnosis and treatment of a patient who had been previously diagnosed with idiopathic posterior uveitis.15 In a similar study, Kuiper et al.85 used a multiplex ELISA (25 proteins) to analyze 175 aqueous humor samples from four retinal diseases (rhegmatogenous RD, AMD, primary vitreoretinal lymphoma, and idiopathic noninfectious uveitis).85 Three proteins (IL-10, IL-21, and ACE) were further analyzed, using a parsimonious model that could distinguish the four diseases from each other, with 86.7% accuracy.85 This study highlights the potential for proteomic analysis to guide the definitive diagnosis of vitreoretinal diseases.
Proteomics for Drug Repositioning
Drug repositioning is defined as applying approved drugs and compounds towards new indications, which often are rare diseases with few therapeutic options. Ophthalmology is rife with “orphan” diseases (e.g., inherited retinal degenerations and chronic inflammatory diseases) that have small market capitalization, which often present a financial barrier for therapeutic development. The research and development of new drugs often is capital- and time-intensive. When a compound may show therapeutic promise, it may cost upwards of a billion dollars and a further decade of basic research and clinical trials to further develop it into an approved and marketable therapy. Drug repositioning offers a route for clinicians and researchers to circumvent this complex pipeline by using drugs that have standardized, therapeutic doses and well-characterized side-effect profiles.
To identify candidate drugs for repositioning, many current prediction methods make use of genomics-based analyses and retrospective computational methods. For directing treatment of vitreous diseases, personalized proteomics may have more value than personalized genomics because it places more emphasis on biomarkers with therapeutic potential. Using this method, molecular constituents of diseased tissues (e.g., vitreous or aqueous) can be identified and measured. Then, elevated molecular disease effectors can be targeted. Such an approach may be most beneficial in vitreoretinal diseases where nonspecific immunosuppressive medications are the first-line treatments.
PVR is a vision-threatening complication of RD repair characterized by the formation of fibrotic membranes that reopen previously repaired retinal tears and initiate new ones. Its treatment often requires delicate and complex surgery to remove fibrotic membranes, often with poor visual outcomes.86,87 These patients face numerous clinical risk factors, such as prior PVR, longer lasting RD, vitreous or choroidal hemorrhage, and poorer initial visual acuity.86 Despite this, identification of clinical risk factors does little to improve PVR therapy. Corticosteroids and immunosuppressive medications (e.g., 5-fluorourail [5-FU] and daunorubicin) are the mainstay of pharmacologic PVR treatment, but often are ineffective.86
In the case of proteomic analysis, if molecular risk factors can be identified, then they might point to more robust biomarkers and drug targets for precise, patient-specific treatment. Proteomic analysis of vitreous biopsies from patients with early and advanced PVR (grades A-B and C, respectively) suggested key differences in the cellular and molecular profile of the two disease stages: early PVR was characterized by T-cell recruitment and mTOR signaling, whereas the cytokine signatures in the advanced PVR proteome suggested monocyte recruitment.87 This finding strongly suggests that mTOR inhibitors, like intravitreal Sirolimus, would be beneficial in treating PVR. Pathway analysis of differentially expressed proteins in PVR vitreous also suggested why PVR patients may be nonresponsive to glucocorticoids: PVR vitreous contains elevated levels of IL-13, a cytokine shown to make monocytes resistant to glucocorticoids and reduce their suppressive effects on IL-6 production.87,88
The ability of proteomics to guide drug repositioning is exemplified in a prior study by our group, where proteomic analysis of vitreous biopsies successfully directed the repositioning of available drugs for Autosomal Dominant Neovascular Inflammatory Vitreoretinopathy (ADNIV; OMIM 193235) patients.89 ADNIV is a rare, progressive inflammatory intraocular disease caused by mutations in the CAPN5 gene. Before culminating in blindness, ADNIV disease progresses in a series of pathologic stages, characterized by synaptic signaling defects, inflammatory cell infiltration, neovascularization, and intraocular fibrosis.89 Before our study, ADNIV patients were treated with nonspecific immunosuppressive medications, such as oral corticosteroids and infliximab (anti-TNF-α). Our proteomic analysis of ADNIV vitreous revealed that TNF-α levels were normal, explaining why infliximab therapy failed in these patients. The analysis further revealed that the ADNIV vitreous contained abundant levels of VEGF, T-cell proliferative markers, and IL-6. Based on these proteomic data we repositioned bevacizumab (anti-VEGF monoclonal antibody), intravitreal methotrexate (T-cell inhibitor), and tocilizumab (anti-IL-6 monoclonal antibody) and successfully mitigated neovascularization, inflammatory cell infiltration, and persistent fibrosis in these patients.89 Similar strategies can be applied to other diseases where liquid biopsies can be collected to select drug targets and streamline trials.
Future Directions
Historically, physicians have had to use patients' physical signs and symptoms in a constant race to cure poorly-understood diseases. The resulting standardized treatments, often driven by trial and error, cost patients physically and financially. Recent advances in genetics, proteomics, and other molecular sciences are changing the playing field, putting healthcare providers in the lead by allowing them to cure diseases in their earliest stages or even catch them before they appear. Precision Health makes use of big data sets and advanced bioinformatics pipelines to analyze molecular and clinical information to customize patient care. The collaboration between basic scientists, engineers, entrepreneurs, health care providers, and patients is unlocking the causes and prevention of diseases and bringing innovative treatments to the bedside. Molecular diagnoses and targeted treatments hold the key to personalized health that will enable us to live better, longer lives. The significant role Precision Health has had in transforming cancer treatments has inspired changes across the medical fields, including ophthalmology, where ophthalmologists and bench scientists are on the cusp of unraveling the molecular and genetic makeup of blinding eye diseases, moving them away from symptom-based treatment plans to a precision-health approach.
Personalized proteomics offers many advantages over alternative precision-health platforms for the diagnosis and treatment of vitreoretinal diseases. For example, in clinical practice, a patient's genetic profile only denotes risk, which often does little to improve their treatment in the near term. Further, gene expression levels often do not correlate well with protein levels and turnover.90 In contrast, LC-MS/MS analysis can provide information on changes in protein expression levels, posttranslational modifications, metabolites, and response to therapy—information that cannot be ascertained using genomics-based methods (Table 1). Proteomic analysis of vitreous biopsies allows identification of molecular constituents in the diseased tissue that can be targeted by available drugs.15 Approved drugs can be repositioned in real-time to provide precise, personalized therapy. Also, proteomic analysis can point out which drugs to avoid. In the case of ADNIV, when it became apparent that the patient's vitreous contained normal levels of TNF-α, a needless infliximab therapy was halted.89 Similarly, our previous proteomic studies on PVR detected vitreous cytokines that could explain why corticosteroids were ineffective in the advanced, fibrotic stage of the disease.87 Since the retina secretes proteins into the vitreous, proteomic biomarkers may be a more rapid way to monitor therapeutic responses or the success of gene therapy trials than clinical outcomes.
Proteomic analysis is already being applied successfully to other diseases, such as chronic kidney disease (CKD). An MS-based approach used by Good et al.91 analyzed a panel of 273 urinary proteins (named the CKD273 panel) to screen normo-albumineric patients at risk for progression to CKD.91 This panel was validated in several studies in patients with earlier-stage disease, and in larger cohorts, including patients with diabetic kidney disease.92–94 This led to the design of the first urinary proteomics-guided intervention trial, PRIORITY (NCT02040441), in which CKD237-positive patients were randomized to receive spironolactone or placebo.95 The success of this trial led to the support of CKD237 by the Food and Drug Administration (FDA). The success of the PRIORITY trial for CKD highlights the need for prospective validation of candidate proteomic biomarkers in larger patient cohorts. Biomarkers that repeatedly appear in multiple studies and populations are more convincing and, thus, more likely to be reliable indicators of disease risk, progression, and response to therapy.19 Validated biomarkers can then be used in a more routine fashion in the clinical setting. Although numerous proteomic studies identified biomarkers and drug targets for vitreoretinal diseases (Table 3), further analysis and validation is required to determine their role in disease, reproducibility, sensitivity, and specificity.
Finally, although we focused on the vitreous in this review, other ophthalmic tissues can be sampled routinely for proteomic analysis in the clinical setting. Tear fluid, for example, contains complex mixtures of proteins, lipids, and metabolites secreted from the lacrimal gland, cornea, and vascular sources. Absorbent materials (e.g., Schirmer's strips) and microcapillary tubes can be used to sample tear fluid noninvasively (5–10 μL on average) from patients.96 Despite the small sample volume, proteomic analysis has identified close to 2000 unique proteins in human tear fluid and numerous studies identified biomarkers for ocular surface and lacrimal gland diseases.97 We anticipate that advances in small-volume proteomics and safe surgical acquisition of ocular fluids from different anatomical sites will give new insight into the pathophysiology and treatment of eye disease.
Acknowledgments
The authors thank Alton Szeto for anatomic illustrations. Permission to publish was granted by the original artist.
Supported by National Institutes of Health (NIH; Bethesda, MD) Grants R01EY026682, R01EY024665, R01EY025225, R01EY024698, and R21AG050437 (VBM and AGB) and Research to Prevent Blindness (RPB), New York, NY (VBM and AGB), and NIH Grants F30EYE027986 and T32GM007337 (GV). The Barbara & Donald Jonas Laboratory of Regenerative Medicine and Bernard and the Shirlee Brown Glaucoma Laboratory are supported by the NIH Grants 5P30EY019007, R01EY018213, R01EY024698, and R21AG050437, National Cancer Institute Core Grant 5P30CA013696, the RPB Physician-Scientist Award, and unrestricted funds from the RPB. SHT is a member of the RD-CURE Consortium and is supported by the Tistou and Charlotte Kerstan Foundation, the Schneeweiss Stem Cell Fund, New York State (C029572) the Joel Hoffman Fund, the Professor Gertrude Rothschild Stem Cell Foundation, and the Gebroe Family Foundation. The authors alone are responsible for the content and writing of this paper.
Author Contributions: VBM had full access to all data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. Drafting of the manuscript: GV, PHT, TC, GYC, DAM, SHT, AGB, VBM. Critical revision of the manuscript for important intellectual content: SHT, AGB, VBM. Obtained funding: VBM. Administrative, technical, and material support: VBM. Study supervision: VBM.
Disclosure: G. Velez, None; P.H. Tang, None; T. Cabral, None; G.Y. Cho, None; D.A. Machlab, None; S.H. Tsang, None; A.G. Bassuk, None; V.B. Mahajan, None
References
- 1.Daubert MA, Jeremias A. The utility of troponin measurement to detect myocardial infarction: review of the current findings. Vasc Health Risk Manag. 2010;6:691–699. doi: 10.2147/vhrm.s5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gnjatic S, Bronte V, Brunet LR, et al. Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer. 2017;5:44. doi: 10.1186/s40425-017-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagalla S, Chou JW, Willingham MC, et al. Interactions between immunity, proliferation and molecular subtype in breast cancer prognosis. Genome Biol. 2013;14:R34. doi: 10.1186/gb-2013-14-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rody A, Holtrich U, Pusztai L, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt M, Bohm D, von Torne C, et al. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 6.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner TW, Sundstrom JM. A proposal for early and personalized treatment of diabetic retinopathy based on clinical pathophysiology and molecular phenotyping. Vision Res. 2017;139:153–160. doi: 10.1016/j.visres.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar GM, Balaj L, Stott SL, Nahed B, Carter BS. Liquid biopsy for brain tumors. Expert Rev Mol Diagn. 2017;17:943–947. doi: 10.1080/14737159.2017.1374854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Meo A, Bartlett J, Cheng Y, Pasic MD, Yousef GM. Liquid biopsy: a step forward towards precision medicine in urologic malignancies. Mol Cancer. 2017;16:80. doi: 10.1186/s12943-017-0644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosengren S, Firestein GS, Boyle DL. Measurement of inflammatory biomarkers in synovial tissue extracts by enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2003;10:1002–1010. doi: 10.1128/CDLI.10.6.1002-1010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole CJ, Kwan AS, Laidlaw DA, Aylward GW. A new technique of combined retinal and choroidal biopsy. Br J Ophthalmol. 2008;92:1357–1360. doi: 10.1136/bjo.2008.141697. [DOI] [PubMed] [Google Scholar]
- 12.Lim LL, Suhler EB, Rosenbaum JT, Wilson DJ. The role of choroidal and retinal biopsies in the diagnosis and management of atypical presentations of uveitis. Trans Am Ophthalmol Soc. 2005;103:84–91. discussion 91–82. [PMC free article] [PubMed] [Google Scholar]
- 13.Le Goff MM, Bishop PN. Adult vitreous structure and postnatal changes. Eye (Lond) 2008;22:1214–1222. doi: 10.1038/eye.2008.21. [DOI] [PubMed] [Google Scholar]
- 14.Skeie JM, Brown EN, Martinez HD, et al. Proteomic analysis of vitreous biopsy techniques. Retina. 2012;32:2141–2149. doi: 10.1097/IAE.0b013e3182562017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velez G, Roybal CN, Colgan D, Tsang SH, Bassuk AG, Mahajan VB. Precision medicine: personalized proteomics for the diagnosis and treatment of idiopathic inflammatory disease. JAMA Ophthalmol. 2016;134:444–448. doi: 10.1001/jamaophthalmol.2015.5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skeie JM, Roybal CN, Mahajan VB. Proteomic insight into the molecular function of the vitreous. PLoS One. 2015;10:e0127567. doi: 10.1371/journal.pone.0127567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skeie JM, Mahajan VB. Proteomic landscape of the human choroid-retinal pigment epithelial complex. JAMA Ophthalmology. 2014;132:1271–1281. doi: 10.1001/jamaophthalmol.2014.2065. [DOI] [PubMed] [Google Scholar]
- 18.Skeie JM, Mahajan VB. Proteomic interactions in the mouse vitreous-retina complex. PLoS One. 2013;8:e82140. doi: 10.1371/journal.pone.0082140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahajan VB, Skeie JM. Translational vitreous proteomics. Proteomics Clin Appl. 2014;8:204–208. doi: 10.1002/prca.201300062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghodasra DH, Fante R, Gardner TW, et al. Safety and feasibility of quantitative multiplexed cytokine analysis from office-based vitreous aspiration. Invest Ophthalmol Vis Sci. 2016;57:3017–3023. doi: 10.1167/iovs.15-18721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfahler SM, Brandford AN, Glaser BM. A prospective study of in-office diagnostic vitreous sampling in patients with vitreoretinal pathology. Retina. 2009;29:1032–1035. doi: 10.1097/IAE.0b013e3181a2c1eb. [DOI] [PubMed] [Google Scholar]
- 22.Chowdhury UR, Madden BJ, Charlesworth MC, Fautsch MP. Proteome analysis of human aqueous humor. Invest Ophthalmol Vis Sci. 2010;51:4921–4931. doi: 10.1167/iovs.10-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perumal N, Manicam C, Steinicke M, Funke S, Pfeiffer N, Grus FH. Characterization of the human aqueous humour proteome: A comparison of the genders. PLoS One. 2017;12:e0172481. doi: 10.1371/journal.pone.0172481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kliuchnikova AA, Samokhina NI, Illina IY, et al. Human aqueous humor proteome in cataract, glaucoma, and pseudoexfoliation syndrome. Proteomics. 2016;16:1938–1946. doi: 10.1002/pmic.201500423. [DOI] [PubMed] [Google Scholar]
- 25.Kim TW, Kang JW, Ahn J, et al. Proteomic analysis of the aqueous humor in age-related macular degeneration (AMD) patients. J Proteome Res. 2012;11:4034–4043. doi: 10.1021/pr300080s. [DOI] [PubMed] [Google Scholar]
- 26.Yao J, Liu X, Yang Q, et al. Proteomic analysis of the aqueous humor in patients with wet age-related macular degeneration. Proteomics Clin Appl. 2013;7:550–560. doi: 10.1002/prca.201200012. [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi T, Koyama R, Ikeda T, Shimizu A. Catalogue of soluble proteins in the human vitreous humor: comparison between diabetic retinopathy and macular hole. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;776:89–100. doi: 10.1016/s1570-0232(02)00078-8. [DOI] [PubMed] [Google Scholar]
- 28.Chiang SY, Tsai ML, Wang CY, et al. Proteomic analysis and identification of aqueous humor proteins with a pathophysiological role in diabetic retinopathy. J Proteomics. 2012;75:2950–2959. doi: 10.1016/j.jprot.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Ecker SM, Hines JC, Pfahler SM, Glaser BM. Aqueous cytokine and growth factor levels do not reliably reflect those levels found in the vitreous. Mol Vis. 2011;17:2856–2863. [PMC free article] [PubMed] [Google Scholar]
- 30.Skeie JM, Tsang SH, Zande RV, et al. A biorepository for ophthalmic surgical specimens. Proteomics Clin Appl. 2014;8:209–217. doi: 10.1002/prca.201300043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson MW. Improvements in the understanding and treatment of macular hole. Curr Opin Ophthalmol. 2002;13:152–160. doi: 10.1097/00055735-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Zhang P, Zhu M, Zhao Y, et al. A proteomic approach to understanding the pathogenesis of idiopathic macular hole formation. Clin Proteomics. 2017;14:37. doi: 10.1186/s12014-017-9172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steel DH, Lotery AJ. Idiopathic vitreomacular traction and macular hole: a comprehensive review of pathophysiology, diagnosis, and treatment. Eye (Lond) 2013;27(suppl 1):S1–21. doi: 10.1038/eye.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal N, Kofod M, Vorum H, et al. Proteomic analysis of human vitreous associated with idiopathic epiretinal membrane. Acta Ophthalmol. 2013;91:e333–334. doi: 10.1111/aos.12075. [DOI] [PubMed] [Google Scholar]
- 35.Wu CW, Sauter JL, Johnson PK, Chen CD, Olsen TW. Identification and localization of major soluble vitreous proteins in human ocular tissue. Am J Ophthalmol. 2004;137:655–661. doi: 10.1016/j.ajo.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Henke SE, Demarais S. Changes in vitreous humor associated with postmortem interval in rabbits. Am J Vet Res. 1992;53:73–77. [PubMed] [Google Scholar]
- 37.Bishop PN. Structural macromolecules and supramolecular organisation of the vitreous gel. Prog Retin Eye Res. 2000;19:323–344. doi: 10.1016/s1350-9462(99)00016-6. [DOI] [PubMed] [Google Scholar]
- 38.Tighe PJ, Ryder RR, Todd I, Fairclough LC. ELISA in the multiplex era: potentials and pitfalls. Proteomics Clin Appl. 2015;9:406–422. doi: 10.1002/prca.201400130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Fonslow BR, Shan B, Baek MC, Yates JR., III Protein analysis by shotgun/bottom-up proteomics. Chem Rev. 2013;113:2343–2394. doi: 10.1021/cr3003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duarte TT, Spencer CT. Personalized proteomics: the future of precision medicine. Proteomes. 2016;4 doi: 10.3390/proteomes4040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geer LY, Markey SP, Kowalak JA, et al. Open mass spectrometry search algorithm. J Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 42.Bjornson RD, Carriero NJ, Colangelo C, et al. X!!Tandem, an improved method for running X!tandem in parallel on collections of commodity computers. J Proteome Res. 2008;7:293–299. doi: 10.1021/pr0701198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yen CY, Meyer-Arendt K, Eichelberger B, et al. A simulated MS/MS library for spectrum-to-spectrum searching in large scale identification of proteins. Mol Cell Proteomics. 2009;8:857–869. doi: 10.1074/mcp.M800384-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 45.Yen CY, Houel S, Ahn NG, Old WM. Spectrum-to-spectrum searching using a proteome-wide spectral library. Mol Cell Proteomics. 2011. 10:M111 007666. [DOI] [PMC free article] [PubMed]
- 46.Egertson JD, Kuehn A, Merrihew GE, et al. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Q, Chen Z, Zhang S, et al. Multiple and selective reaction monitoring using triple quadrupole mass spectrometer: preclinical large cohort analysis. Methods Mol Biol. 2016;1410:249–264. doi: 10.1007/978-1-4939-3524-6_15. [DOI] [PubMed] [Google Scholar]
- 48.Richardson MR, Price MO, Price FW, et al. Proteomic analysis of human aqueous humor using multidimensional protein identification technology. Mol Vis. 2009;15:2740–2750. [PMC free article] [PubMed] [Google Scholar]
- 49.Colantonio DA, Dunkinson C, Bovenkamp DE, Van Eyk JE. Effective removal of albumin from serum. Proteomics. 2005;5:3831–3835. doi: 10.1002/pmic.200401235. [DOI] [PubMed] [Google Scholar]
- 50.Hegmans JP, Bard MP, Hemmes A, et al. Proteomic analysis of exosomes secreted by human mesothelioma cells. Am J Pathol. 2004;164:1807–1815. doi: 10.1016/S0002-9440(10)63739-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bastos-Amador P, Royo F, Gonzalez E, et al. Proteomic analysis of microvesicles from plasma of healthy donors reveals high individual variability. J Proteomics. 2012;75:3574–3584. doi: 10.1016/j.jprot.2012.03.054. [DOI] [PubMed] [Google Scholar]
- 52.Caby MP, Lankar D, Vincendeau-Scherrer C, Raposo G, Bonnerot C. Exosomal-like vesicles are present in human blood plasma. Int Immunol. 2005;17:879–887. doi: 10.1093/intimm/dxh267. [DOI] [PubMed] [Google Scholar]
- 53.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skriner K, Adolph K, Jungblut PR, Burmester GR. Association of citrullinated proteins with synovial exosomes. Arthritis Rheum. 2006;54:3809–3814. doi: 10.1002/art.22276. [DOI] [PubMed] [Google Scholar]
- 55.Kang GY, Bang JY, Choi AJ, et al. Exosomal proteins in the aqueous humor as novel biomarkers in patients with neovascular age-related macular degeneration. J Proteome Res. 2014;13:581–595. doi: 10.1021/pr400751k. [DOI] [PubMed] [Google Scholar]
- 56.Ragusa M, Barbagallo C, Statello L, et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biol Ther. 2015;16:1387–1396. doi: 10.1080/15384047.2015.1046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Weber SR, Lease J, et al. Liquid biopsy of vitreous reveals an abundant vesicle population consistent with the size and morphology of exosomes. Transl Vis Sci Technol. 2018;7:6. doi: 10.1167/tvst.7.3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams KL. Genomes and proteomes: towards a multidimensional view of biology. Electrophoresis. 1999;20:678–688. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<678::AID-ELPS678>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 59.Biomarkers Definitions Working G. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 60.Kavallaris M, Marshall GM. Proteomics and disease: opportunities and challenges. Med J Aust. 2005;182:575–579. doi: 10.5694/j.1326-5377.2005.tb06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lauwen S, de Jong EK, Lefeber DJ, den Hollander A. Omics biomarkers in ophthalmology. Invest Ophthalmol Vis Sci. 2017;58:BIO88–BIO98. doi: 10.1167/iovs.17-21809. [DOI] [PubMed] [Google Scholar]
- 62.Berry JL, Xu L, Murphree AL, et al. Potential of aqueous humor as a surrogate tumor biopsy for retinoblastoma. JAMA Ophthalmol. 2017;135:1221–1230. doi: 10.1001/jamaophthalmol.2017.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pascolini D, Mariotti SP, Pokharel GP, et al. 2002 global update of available data on visual impairment: a compilation of population-based prevalence studies. Ophthalmic Epidemiol. 2004;11:67–115. doi: 10.1076/opep.11.2.67.28158. [DOI] [PubMed] [Google Scholar]
- 64.Resnikoff S, Pascolini D, Etya'ale D, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–851. [PMC free article] [PubMed] [Google Scholar]
- 65.Jabs DA. Epidemiology of uveitis. Ophthalmic Epidemiol. 2008;15:283–284. doi: 10.1080/09286580802478724. [DOI] [PubMed] [Google Scholar]
- 66.Mattapallil MJ, Sahin A, Silver PB, et al. Common genetic determinants of uveitis shared with other autoimmune disorders. J Immunol. 2008;180:6751–6759. doi: 10.4049/jimmunol.180.10.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pascolini D, Mariotti SP. Global estimates of visual impairment: 2010. Br J Ophthalmol. 2012;96:614–618. doi: 10.1136/bjophthalmol-2011-300539. [DOI] [PubMed] [Google Scholar]
- 68.Lee K, Bajwa A, Freitas-Neto CA, Metzinger JL, Wentworth BA, Foster CS. A comprehensive review and update on the non-biologic treatment of adult noninfectious uveitis: part I. Expert Opin Pharmacother. 2014;15:2141–2154. doi: 10.1517/14656566.2014.948417. [DOI] [PubMed] [Google Scholar]
- 69.Wong RW, Jumper JM, McDonald HR, et al. Emerging concepts in the management of acute retinal necrosis. Br J Ophthalmol. 2013;97:545–552. doi: 10.1136/bjophthalmol-2012-301983. [DOI] [PubMed] [Google Scholar]
- 70.Lee FF, Foster CS. Pharmacotherapy of uveitis. Expert Opin Pharmacother. 2010;11:1135–1146. doi: 10.1517/14656561003713534. [DOI] [PubMed] [Google Scholar]
- 71.Friedman DS, Holbrook JT, Ansari H, et al. Risk of elevated intraocular pressure and glaucoma in patients with uveitis: results of the multicenter uveitis steroid treatment trial. Ophthalmology. 2013;120:1571–1579. doi: 10.1016/j.ophtha.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deeg CA. Ocular immunology in equine recurrent uveitis. Vet Ophthalmol. 2008;11(suppl 1):61–65. doi: 10.1111/j.1463-5224.2008.00625.x. [DOI] [PubMed] [Google Scholar]
- 73.Werry H, Gerhards H. [The surgical therapy of equine recurrent uveitis] Tierarztl Prax. 1992;20:178–186. [PubMed] [Google Scholar]
- 74.Deeg CA, Altmann F, Hauck SM, et al. Down-regulation of pigment epithelium-derived factor in uveitic lesion associates with focal vascular endothelial growth factor expression and breakdown of the blood-retinal barrier. Proteomics. 2007;7:1540–1548. doi: 10.1002/pmic.200600795. [DOI] [PubMed] [Google Scholar]
- 75.Yu H, Chen L, Jiang J. Administration of pigment epithelium-derived factor delivered by adeno-associated virus inhibits blood-retinal barrier breakdown in diabetic rats. Mol Vis. 2010;16:2384–2394. [PMC free article] [PubMed] [Google Scholar]
- 76.Degroote RL, Uhl PB, Amann B, et al. Formin like 1 expression is increased on CD4+ T lymphocytes in spontaneous autoimmune uveitis. J Proteomics. 2017;154:102–108. doi: 10.1016/j.jprot.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 77.Caspi RR, Silver PB, Luger D, et al. Mouse models of experimental autoimmune uveitis. Ophthalmic Res. 2008;40:169–174. doi: 10.1159/000119871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Caspi RR, Roberge FG, Chan CC, et al. A new model of autoimmune disease. Experimental autoimmune uveoretinitis induced in mice with two different retinal antigens. J Immunol. 1988;140:1490–1495. [PubMed] [Google Scholar]
- 79.Dix RD, Cray C, Cousins SW. Mice immunosuppressed by murine retrovirus infection (MAIDS) are susceptible to cytomegalovirus retinitis. Curr Eye Res. 1994;13:587–595. doi: 10.3109/02713689408999892. [DOI] [PubMed] [Google Scholar]
- 80.Rao NA, Albini TA, Kumaradas M, Pinn ML, Fraig MM, Karakousis PC. Experimental ocular tuberculosis in guinea pigs. Arch Ophthalmol. 2009;127:1162–1166. doi: 10.1001/archophthalmol.2009.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bansal S, Barathi VA, Iwata D, Agrawal R. Experimental autoimmune uveitis and other animal models of uveitis: an update. Indian J Ophthalmol. 2015;63:211–218. doi: 10.4103/0301-4738.156914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sen ES, Dick AD, Ramanan AV. Uveitis associated with juvenile idiopathic arthritis. Nat Rev Rheumatol. 2015;11:338–348. doi: 10.1038/nrrheum.2015.20. [DOI] [PubMed] [Google Scholar]
- 83.Ooi KG, Galatowicz G, Calder VL, Lightman SL. Cytokines and chemokines in uveitis: is there a correlation with clinical phenotype? Clin Med Res. 2006;4:294–309. doi: 10.3121/cmr.4.4.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sauer A, Villard O, Creuzot-Garcher C, et al. Intraocular levels of interleukin 17A (IL-17A) and IL-10 as respective determinant markers of toxoplasmosis and viral uveitis. Clin Vaccine Immunol. 2015;22:72–78. doi: 10.1128/CVI.00423-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuiper JJ, Beretta L, Nierkens S, et al. An ocular protein triad can classify four complex retinal diseases. Sci Rep. 2017;7:41595. doi: 10.1038/srep41595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pastor JC, Rojas J, Pastor-Idoate S, Di Lauro S, Gonzalez-Buendia L, Delgado-Tirado S. Proliferative vitreoretinopathy: a new concept of disease pathogenesis and practical consequences. Prog Retin Eye Res. 2016;51:125–155. doi: 10.1016/j.preteyeres.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 87.Roybal CN, Velez G, Toral MA, Tsang SH, Bassuk AG, Mahajan VB. Personalized proteomics in proliferative vitreoretinopathy implicate hematopoietic cell recruitment and mTOR as a therapeutic target. Am J Ophthalmol. 2017;186:152–163. doi: 10.1016/j.ajo.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spahn JD, Szefler SJ, Surs W, Doherty DE, Nimmagadda SR, Leung DY. A novel action of IL-13: induction of diminished monocyte glucocorticoid receptor-binding affinity. J Immunol. 1996;157:2654–2659. [PubMed] [Google Scholar]
- 89.Velez G, Bassuk AG, Colgan D, Tsang SH, Mahajan VB. Therapeutic drug repositioning using personalized proteomics of liquid biopsies. JCI Insight. 2017];2017;2(24):e97818. doi: 10.1172/jci.insight.97818. [published online December 21, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 91.Good DM, Zurbig P, Argiles A, et al. Naturally occurring human urinary peptides for use in diagnosis of chronic kidney disease. Mol Cell Proteomics. 2010;9:2424–2437. doi: 10.1074/mcp.M110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lindhardt M, Persson F, Zurbig P, et al. Urinary proteomics predict onset of microalbuminuria in normoalbuminuric type 2 diabetic patients, a sub-study of the DIRECT-Protect 2 study. Nephrol Dial Transplant. 2017;32:1866–1873. doi: 10.1093/ndt/gfw292. [DOI] [PubMed] [Google Scholar]
- 93.Roscioni SS, de Zeeuw D, Hellemons ME, et al. A urinary peptide biomarker set predicts worsening of albuminuria in type 2 diabetes mellitus. Diabetologia. 2013;56:259–267. doi: 10.1007/s00125-012-2755-2. [DOI] [PubMed] [Google Scholar]
- 94.Schanstra JP, Zurbig P, Alkhalaf A, et al. Diagnosis and prediction of CKD progression by assessment of urinary peptides. J Am Soc Nephrol. 2015;26:1999–2010. doi: 10.1681/ASN.2014050423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lindhardt M, Persson F, Currie G, et al. Proteomic prediction and renin angiotensin aldosterone system inhibition prevention of early diabetic nephropathy in type 2 diabetic patients with normoalbuminuria (PRIORITY): essential study design and rationale of a randomised clinical multicentre trial. BMJ Open. 2016;6:e010310. doi: 10.1136/bmjopen-2015-010310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou L, Beuerman RW. The power of tears: how tear proteomics research could revolutionize the clinic. Expert Rev Proteomics. 2017;14:189–191. doi: 10.1080/14789450.2017.1285703. [DOI] [PubMed] [Google Scholar]
- 97.Chen L, Zhou L, Chan EC, Neo J, Beuerman RW. Characterization of the human tear metabolome by LC-MS/MS. J Proteome Res. 2011;10:4876–4882. doi: 10.1021/pr2004874. [DOI] [PubMed] [Google Scholar]
- 98.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kramer A, Green J, Pollard J, Jr, Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Machlab DA, Velez G, Bassuk AG, Mahajan VB. ProSave: an application for restoring quantitative data to manipulated subsets of proteins [software] 2018 doi: 10.1186/s13029-018-0070-0. Available at: https://github.com/MahajanLab/ProSave. [DOI] [PMC free article] [PubMed]
- 102.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang J, Duncan D, Shi Z, Zhang B. WEB–based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koss MJ, Hoffmann J, Nguyen N, et al. Proteomics of vitreous humor of patients with exudative age-related macular degeneration. PLoS One. 2014;9:e96895. doi: 10.1371/journal.pone.0096895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nobl M, Reich M, Dacheva I, et al. Proteomics of vitreous in neovascular age-related macular degeneration. Exp Eye Res. 2016;146:107–117. doi: 10.1016/j.exer.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 106.Patel S, Ling J, Kim SJ, Schey KL, Rose K, Kuchtey RW. Proteomic analysis of macular fluid associated with advanced glaucomatous excavation. JAMA Ophthalmol. 2016;134:108–110. doi: 10.1001/jamaophthalmol.2015.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sudha D, Kohansal-Nodehi M, Kovuri P, et al. Proteomic profiling of human intraschisis cavity fluid. Clin Proteomics. 2017;14:13. doi: 10.1186/s12014-017-9148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sugioka K, Saito A, Kusaka S, Kuniyoshi K, Shimomura Y. Identification of vitreous proteins in retinopathy of prematurity. Biochem Biophys Res Commun. 2017;488:483–488. doi: 10.1016/j.bbrc.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 109.Naru J, Aggarwal R, Mohanty AK, et al. Identification of differentially expressed proteins in retinoblastoma tumors using mass spectrometry-based comparative proteomic approach. J Proteomics. 2017;159:77–91. doi: 10.1016/j.jprot.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 110.Naru J, Aggarwal R, Singh U, et al. Proteomic analysis of differentially expressed proteins in vitreous humor of patients with retinoblastoma using iTRAQ-coupled ESI-MS/MS approach. Tumour Biol. 2016;37:13915–13926. doi: 10.1007/s13277-016-5162-3. [DOI] [PubMed] [Google Scholar]
- 111.Yang Q, Lu H, Song X, Li S, Wei W. iTRAQ-based proteomics investigation of aqueous humor from patients with Coats' disease. PLoS One. 2016;11:e0158611. doi: 10.1371/journal.pone.0158611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yamane K, Minamoto A, Yamashita H, et al. Proteome analysis of human vitreous proteins. Mol Cell Proteomics. 2003;2:1177–1187. doi: 10.1074/mcp.M300038-MCP200. [DOI] [PubMed] [Google Scholar]
- 113.Kim SJ, Kim S, Park J, et al. Differential expression of vitreous proteins in proliferative diabetic retinopathy. Curr Eye Res. 2006;31:231–240. doi: 10.1080/02713680600557030. [DOI] [PubMed] [Google Scholar]
- 114.Garcia-Ramirez M, Canals F, Hernandez C, et al. Proteomic analysis of human vitreous fluid by fluorescence-based difference gel electrophoresis (DIGE): a new strategy for identifying potential candidates in the pathogenesis of proliferative diabetic retinopathy. Diabetologia. 2007;50:1294–1303. doi: 10.1007/s00125-007-0627-y. [DOI] [PubMed] [Google Scholar]
- 115.Gao BB, Chen X, Timothy N, Aiello LP, Feener EP. Characterization of the vitreous proteome in diabetes without diabetic retinopathy and diabetes with proliferative diabetic retinopathy. J Proteome Res. 2008;7:2516–2525. doi: 10.1021/pr800112g. [DOI] [PubMed] [Google Scholar]
- 116.Takada M, Ban Y, Yamamoto G, et al. Periostin, discovered by nano-flow liquid chromatography and mass spectrometry, is a novel marker of diabetic retinopathy. Biochem Biophys Res Commun. 2010;399:221–226. doi: 10.1016/j.bbrc.2010.07.058. [DOI] [PubMed] [Google Scholar]
- 117.Csosz E, Boross P, Csutak A, et al. Quantitative analysis of proteins in the tear fluid of patients with diabetic retinopathy. J Proteomics. 2012;75:2196–2204. doi: 10.1016/j.jprot.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 118.Velez G, Roybal CN, Binkley E, Bassuk AG, Tsang SH, Mahajan VB. Proteomic Analysis of Elevated Intraocular Pressure with Retinal Detachment. Am J Ophthalmol Case Rep. 2017;5:107–110. doi: 10.1016/j.ajoc.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shitama T, Hayashi H, Noge S, et al. Proteome profiling of vitreoretinal diseases by cluster analysis. Proteomics Clin Appl. 2008;2:1265–1280. doi: 10.1002/prca.200800017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaneko H, Takayama K, Asami T, et al. Cytokine profiling in the sub-silicone oil fluid after vitrectomy surgeries for refractory retinal diseases. Sci Rep. 2017;7:2640. doi: 10.1038/s41598-017-03124-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu J, Peng R, Chen H, Cui C, Ba J. Elucidation of the pathogenic mechanism of rhegmatogenous retinal detachment with proliferative vitreoretinopathy by proteomic analysis. Invest Ophthalmol Vis Sci. 2012;53:8146–8153. doi: 10.1167/iovs.12-10079. [DOI] [PubMed] [Google Scholar]
- 122.Yu J, Liu F, Cui SJ, et al. Vitreous proteomic analysis of proliferative vitreoretinopathy. Proteomics. 2008;8:3667–3678. doi: 10.1002/pmic.200700824. [DOI] [PubMed] [Google Scholar]
- 123.Pollreisz A, Funk M, Breitwieser FP, et al. Quantitative proteomics of aqueous and vitreous fluid from patients with idiopathic epiretinal membranes. Exp Eye Res. 2013;108:48–58. doi: 10.1016/j.exer.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 124.Reich M, Dacheva I, Nobl M, et al. Proteomic analysis of vitreous humor in retinal vein occlusion. PLoS One. 2016;11:e0158001. doi: 10.1371/journal.pone.0158001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dacheva I, Reich M, Nobl M, et al. [Proteome analysis of undiluted vitreous humor in patients with branch retinal vein occlusion] Ophthalmologe. 2018;115:203–215. doi: 10.1007/s00347-017-0469-z. [DOI] [PubMed] [Google Scholar]
- 126.Kalinina Ayuso V, de Boer JH, Byers HL, et al. Intraocular biomarker identification in uveitis associated with juvenile idiopathic arthritis. Invest Ophthalmol Vis Sci. 2013;54:3709–3720. doi: 10.1167/iovs.12-10865. [DOI] [PubMed] [Google Scholar]