Abstract
Making quick promises of major biomedical breakthroughs based on exciting discoveries at the bench is tempting. But the meandering path from fundamental science to life-saving clinical applications can be fraught with many hurdles. Epigenetics, the study of potentially heritable changes of gene function without modification of the underlying DNA sequence, has dominated the biological research field during the last decade and encountered a large public success. Driven by the unfolding of molecular biology and recent technological progress, the term has evolved significantly and shifted from a conceptual framework to a mechanistic understanding. This shift was accompanied by much hype and raised high hopes that epigenetics might hold both the key to deciphering the molecular underpinning of complex, non-Mendelian diseases and offer novel therapeutic approaches for a large panel of pathologies. However, while exciting reports of biological phenomena involving DNA methylation and histone modifications fill up the scientific literature, the realistic clinical applications of epigenetic medicines remain somewhat blurry. Here, we discuss the state of the art and speculate how epigenetics might contribute to prognostic and therapy approaches in the future.
Keywords: Epigenetics, Epitherapeutics, Medicine

Epigentic – the new “quantum”?
Every scientific sector has its own buzzwords – expressions that ring a bell for everyone and which confer it with an aura of futurist technology mixed with revolutionary insights and solutions for most problems. As much as any obscure procedure in physics becomes credible by the simple addition of the “quantum” particle to it, the term “epigenetics” is the new all-round biological explanation for everything where classical genetics can't go, including the inter-generational transmission of acquired traits, environmental impacts and complex diseases.
Largely fueled by public media excitement, the epigenetic hype has coincided with increasing disappointment from the initial promises of genetics and the decrypting of the human genome. The democratization of sophisticated techniques that render whole-genome studies temporally and financially accessible, has contributed to the uptake of epigenetic approaches. This omnipresent use of the “epigenetics label” has made it increasingly hard to pinpoint its exact definition and technical limitations.
In the midst of an elusive, broad collection of phenomena, partially contradictory definitions and loosely related disciplines, it is sometimes difficult to concentrate on the question if and how epigenetics might one day result in a practical application and if passive observation will give rise to active intervention [1] [Fig. 1].
Fig. 1.
Can we fix the epigenetic landscape? Altered epigenetic modifications are associated with many diseases, but what are the realistic clinical applications of epimedicine?
What are we actually talking about?
As trivial as it might sound, this is quite a legitimate question.
While every scientist and educated layman has a more or less clear notion of epigenetics, pinning down a concrete, palpable definition turns out to be an almost Sisyphean task.
To all appearances, the minimal common denominator of various definitions seems to coalesce around “functionally relevant changes in gene expression that are not due to modifications of the underlying DNA sequence” [2]. Technically, this embraces a substantial amount of biochemical signaling pathways inside a cell, including the binding of any transcriptional modulator to a promoter and subsequent quantitative changes in gene transcription. Most sources agree that these changes in gene function have to happen “via chemical modifications of DNA or histones”, narrowing down the field of action to a finite set of precise biochemical events (Box1).
Box 1. Classical “epigenetic” chromatin marks.
DNA modifications
DNA methylation (5 mC)
-
•
Methyl group added on cytosine or adenine bases
-
•
Mainly studied in CpG context, might be frequent in other sequences
-
•
Leads to gene silencing via DNA compaction, recruitment of transcriptional repressors and exclusion of transcriptional activators
-
•
Initially deposited by de novo methyltransferases guided by sequence, DNA binding proteins, long noncoding RNAs or RNA interference
-
•
Mitotically inherited in a semi-conservative manner via maintenance methyltransferases
-
•
Enzymatic activities responsible for demethylation in mammals are still controversial
DNA hydroxymethylation (5 hmC)
-
•
Found in many mammalian tissues
-
•
Effect on gene expression still relatively unknown
Histone modifications
-
-
Post-translational modifications of specific serine, lysine and arginine residues of the histone amino-terminal tail or histone variants
-
-
Histone-modifying enzymes are recruited by specific DNA-sequences or guided intermediate protein and/or RNA complexes or RNA interference
-
-The replication and inheritance of histone modifications during mitosis is unclear
-
•Acetylation
-
○Associated with transcriptional activation
-
○
-
•Methylation
-
○Mono-, di- or tri-methylation
-
○Either repression- or activation-associated, depending on the targeted residue
-
○
-
•Phosphorylation
-
○Associated with transcriptional activation
-
○
-
•Ubiquitination
-
•Sumoylation
-
○Associated with transcriptional repression
-
○
-
•Histone variants
-
○macroH2A is associated with inactive chromatin
-
○H3.3 might be mitotically heritable and accumulates in active chromatin
-
○
-
•
Are DNA repair and the associated histone variants, like γH2AX, an epigenetic event then [3]? Finally, the real aficionados insist on integrating the notion of meiotic and/or mitotic heritability of epigenetic marks and effects [2], [3], [4], [5]. Yet, heritability remains a major source of confusion, even in its most basic form, cellular division. While the mitotic transmission of DNA methylation via maintenance DNA-methyltransferases is quite well-established and studied, the shuffling and redistribution of histones, histone modifications and variants following cell division remain elusive [3]. Further bias has arisen through the widespread impact of a collection of scientific articles strongly suggesting the repercussion of personal experience and environmental influences on offspring, shifting the attention to meiotic heritability [6], [7]. But so far no mechanistic model can explain how chromatin-encoded information could escape the genome-wide demethylation during the preimplantation stage [3], [4], [8]. Given the dilemma, the NIH Roadmap Epigenomics Mapping Consortium broadened the definition to “also stable, long-term alterations in the transcriptional potential of a cell that are not necessarily heritable”, though no consensus exists on the inferior limit of “long-term” [9].
From that point on, things become gradually confusing. For instance, many publications investigating epigenetic mechanisms also include noncoding RNAs [3], [8], [10]. The latter divide on either side of the arbitrary 200 nucleotides length-limit into small and long noncoding RNAs (lncRNAs), where the first group is further subdivided into a long list of functional classes and the second one mainly unexplored [11].
Most small noncoding RNAs excel indeed in short-term post-transcriptional modulation of gene activity, although not necessarily in an especially durable way nor via chromatin states. Only Piwi-interacting RNAs are said to silence retrotransposons in germ line cells through interaction with methyltransferases [12]. Similarly, microRNAs can prompt the methylation of centromeric sequences through a mechanism termed RNA-directed DNA methylation (RdDM) … in yeast [3], [4], [8]. Its existence in humans is yet speculative [11].
Recent major technical advances in whole-transcriptome sequencing, revealing that the major part of the genome is transcribed but not translated, shifted the interest towards lncRNAs [13].
This led to gigabytes of data and ten thousands of unstudied transcripts. Several specimens are indeed able to recruit chromatin-modifying complexes to specific genomic locations, induce the depositing of epigenetic marks as well as durable changes in gene expression [2], [3], [8] – the X inactivation process un female mammals is without doubt the most impressive example [14].
However, because of several artifact-prone in vitro studies of RNA-protein interactions [15], [16], it was over-hastily assumed that the majority of all long noncoding RNAs acted as guides for chromatin-associated proteins, resolving en passant the mystery of how the adequate subset of binding sites of one protein are chosen among thousands of identical sequences according to one cellular context [10]. A fervid polemic ensued, which disembogued more or less in the temporary consensus that it is way to early to attribute any general function to the overwhelming amount of uncharacterized molecules, and that no, lncRNAs are not epigenetic factors.
In the meantime, more molecules and biochemical events, able either to modify gene expression post-transcriptionally or to perpetuate a modification over cell division and sometimes both, cluster around the gates of epigenetics. In parallel, the definition gradually drifts away from the original center and monopoly of information, the chromatin, now potentially including RNA editing, prions and modifications in non-histone proteins such as microtubules and even organelles or the cellular membrane [10], [17].
We note two fundamental problems here, hampering our quest of a clear definition. One is semantic in nature and results from confusion around the different elements of epigenetics. There are epigenetic effects – heritable changes in gene function for equal DNA-sequences, whatever the underlying mechanism. There are epigenetic marks – covalent modifications of DNA or chromatin proteins. And finally, there are all the putative intermediates, proteins or RNA, leading from mark to effect.
The other problem is the gap in our knowledge. Cases where mark, intermediates and effects are well-characterized are rare and it remains mysterious how the initial marks are established and specific modifications targeted to precise sequences [3].
Journey to the past
But why is such a popular concept difficult to define? A brief journey to the past could shed some light on the current confusion. The original concept of epigenetics can be traced back to the embryologist and philosopher Conrad Waddington in 1942 [18]. Despite the discovery of the basic laws of heredity and chromosomes [3], “genes” were yet an exclusively theoretical principle, incarnated by an elusive material support. Their analysis relied solely on a top-down principle, based on visible phenotypic traits. Genetics and embryology were two completely separate fields – only two years after Waddington's publication, the Avery-MacLeod-McCarty experiment provided concrete proof that DNA, and not proteins, represented the actual support of genetic information [19], [20].
Hence, Waddington's definition is a conceptual, theoretical one. He proposed the epigenotype as the black box between genotype and phenotype, the “complex network of processes and causal mechanisms by which the genes of the genotype bring about phenotypic effects”, and its study as epigenetics, the fusion of epigenesis, a synonym of embryonic development, and genetics [18], [19]. From our “modern” point of view, Waddington's epigenetics incorporated the field of genetic regulation, with all its transcription factors, enhancers, repressors, feedback loops and molecular pathways. By uniting embryology and genetics, Waddington created somehow what we would nowadays call developmental genetics [8], [19].
The following decades brought about an exponential increase in understanding of the molecular nature of genes and their expression, including transcription and translation. Epigenetics however remained devoid of any molecular mechanisms until the mid-seventies, although regularly appealed to in order to substitute for the mysterious “auxiliary mechanisms” that drove the “primary genetic material” into a final phenotype [8]. Then, gradually, molecular evidence started to accumulate.
DNA methylation was discovered in 1950 [8], [21] and in 1969, Griffith and Mahler initially suggested that it might be responsible for memory – literally – as a mechanism to encode long-term memories in brain cells [22]. The idea that DNA methylation could rather influence gene expression was proposed in 1975 [5], [23].
In turn, the interest in the roles of the large panel of histone modifications in the modulation of gene expression is mainly 21st century material, though their identification spreads from 1964 (acetylations, methylations) [24], [25] to 2003 (sumoylations) [26]. The definitive link between histone amino-terminal tail modifications and gene expression regulation was made in the early eighties and the discoveries of other modifications exploded from 1996 on [3]. Concomitant with the new millennium, the terms “histone code” and “epigenetic code” were born [3], [27].
As a consequence, the transition from phenomenon to molecular facts has greatly shifted and narrowed the definition of epigenetics from its initial conceptual nature to a way more mechanistic and molecular one, leading to the opportunistic reinterpretation of the Ancient Greek prefix “epi-“ as “on top of the DNA molecule” [4], [19]. The components have been substituted to the phenomenon they were meant to explain.
The modern vision of epigenetics is a modular one. One brick corresponds to changes in gene expression unrelated to changes in the underlying DNA sequence. One comprises a collection of chemical modifications of DNA and proteins. Another one represents the concept of heritability or durability. Depending on the biological process in question, the different modules are not forcefully a hundred percent compatible; rather a specific selection has to be chosen to match the facts.
Recently, some colleagues have distanced themselves from an overzealous use of the term, preferring concrete terms like chromatin-associated modifications. For the sake of clarity and if not specified otherwise, we will restrict ourselves to “classical” epigenetics, namely DNA methylation and histone marks, for the rest of the review.
Day-to-day epigenetics
The establishment of epigenetic chromatin marks is part and parcel of many fundamental processes during the life of a cell or a multicellular organism. The four most prominent ones are cellular differentiation, genome stability, imprinting and adaption to the environment.
Cellular differentiation
Although the author ignored the exact nature of genes and their relationship with RNA and proteins, Conrad Waddington's famous epigenetic landscape is a visual metaphor for the embryonic development and cellular commitment [28]. He portrayed a totipotent cell rolling down a hill, representing gene networks, epigenotype and environment, leading to the terminally differentiated state of a specific function-related subset of gene expression at the bottom of the mountain [2], [8].
The importance of epigenetic mechanisms during cellular differentiation was further stressed by David Nanney in the late 1950s who hypothesized that “epigenetic systems, regulate the expression of the genetically determined potentialities” [8], [29]. Interestingly, it is Nanney who added the “stability/heritability” compound to Waddington's epigenetic principle, in order to tell the difference from “more trivial, immediately reversible phenotypic mechanisms” [1]. Around 1970, the assumption that all cells of one organism contained the same DNA crystallized into certainty, thanks to somatic nuclear transfer experiments, underlying the need for an interpretation toolbox [3], [23].
The classical epigenetic marks are indeed the felt-tip markers and whiteout of the static DNA instruction manual, stably garnishing during development required genes with active marks, and compacting the chromatin of the undesirable ones with astonishing precision [2], [4]. Ultimately, the simple methylation profile suffices to distinguish a Th1 from a Th2 helper cell [30]. Vice versa, the natural genesis of pluripotent embryonic stem cells in the inner cell mass is preceded by a genome-wide demethylation step, while the artificial reprogramming of terminally differentiated cells into induced pluripotent stem cells (iPS) is promoted by inhibitors of methyltransferases and histone deacetylases, thus demonstrating the amazing plasticity of genetically identical cells [8], [31]. Still, we ignore how the differentiation-related marks are disposed in first place.
Genomic stability
Evolution is a tinkerer and evolution not about building the theoretically optimal construct but about putting up in the best possible way with the opportunities and inconveniences that show up en route [32]. We have to keep in mind that a huge proportion of the genome is of parasitic origin [33]. For example, retrotransposons represent roughly half of the human genome. Although these selfish DNA elements might sometimes lend a hand to evolutionary innovation, the cell has got its hands full bridling centromeres, telomeres and transposable elements in order to warrant the correct attachment of microtubules and to fend off excessive recombination, transposition and insertional mutagenesis [4], [34]. Thus, a stable, mitotically transmissible chemical lock sounds like a convenient tool and the major part of DNA methylation in vertebrates seems indeed to be correlated with repetitive and retroviral sequences [3].
Gene dosage compensation is a prime example of epigenetically orchestrated genomic stability [4]. X chromosome inactivation in female mammals calls on the services of an armada of redundant chromatin modifications – DNA methylation, histone variants and histone tail methylation – as well as their direct and indirect providers: methyltransferases, long noncoding RNAs and the PRC2 complex [4], [14].
Imprinting
Imprinting refers to the monoallelic expression of a gene depending on its parental origin [35], [36]. In other words, only the maternal or paternal version is expressed while the other is extensively silenced. This applies to about 1% of human genes and its putative advantages remain a major evolutionary mystery entwined by many theories [2], [8]. Imprinted loci tend to be growth- and development-associated, such as the best studied example, the Igf2/H19 locus [3], [8], [10]. In females, imprints are a priori placed during folliculogenesis and in males during fetal development and consist concretely in the selective methylation of the DNA and marking of histones with repressive tags [4].
Environmental adaption
| Phenotype = genotype + epigenotype + environment. |
That is the original equation of the epigenetic landscape, thus assigning to the environment an essential role in embryonic development. Nowadays, epigenetics are often discussed in the context of environmental impacts on genomic function and output. A notorious example of an environment-directed and epigenetic-mediated phenomenon is the development of the bee larva into either a worker or a queen, exclusively mediated by the differential methylation of identical genes triggered by diet and available space [37], [38]. Similarly clear-cut examples are difficult to find in humans, although spatial and environmental cues are crucial for early embryogenesis and maturation [39], [40]. The requirement for reactivity and cellular plasticity to adapt to alterations in the physical and chemical environment is clear [2], [41], and the mutation/selection process of DNA too slow. The human organism is equipped with an efficient hormonal signaling system and a horde of molecular sensors, such as heat shock proteins, allowing rapid responses to exterior stimuli. However, these types of adaptions are relatively ephemeral and epigenetic mechanisms have the reputation of perpetuity. Further studies are required to understand how environmental factors might lead to precise long-term modifications in chromatin structure and function [42].
During their lifetime, monozygotic twins drift apart at the scale of their epigenome, an attractive explanation for the development of different phenotypic and behavioral traits, as well as the unequal susceptibility to diseases [43]. What are the possible causes? Fortuity is a by all means a plausible answer. Environmentally directed epigenetic adaption is the answer we want to hear, though. Nevertheless, most studies agree that the critical period of responsiveness to the environment, eventually being translated into long-lasting chromatin modifications with an impact on an individual's life, lies within the embryonic development [18]. Notably, methylation has been shown to be sensitive to the maternal environment [2], [4]. While chromatin marks acquired during this period affect the individual, the epigenetic storage of information to be passed down to progeny has logically to take place during germ cell development, i.e. in utero for females but throughout lifetime in males, albeit the mechanisms are unknown [2], [4], [6], [7].
Epigenetics and disease
As epigenetic chromatin marks play well-established roles in fundamental physiological processes, there is no surprise that their misbehavior can have fatal consequences.
Obvious and well-studied examples are imprinting disorders, in which imprinted genes are aberrantly expressed due to the lack of sufficient silencing via repressive chromatin marks; such as the Prader–Willi, Angelman, Beckwith–Wiedemann and Silver–Russel syndromes, consisting in neurodevelopmental disorders and growth abnormalities [2], [10]. As the establishment of genomic imprints occurs during early embryonic development and could easily be disturbed by environmental factors, this raises some concern about human assisted reproductive technologies [4], [10].
Another scenario with a relatively clear link between cause and consequence arises from mutations of the de novo DNA-methyltransferase DNMT3B in form of the rare Immunodeficiency-Centromere instability-Facial anomalies (ICF) syndrome, characterized by hypomethylation of many loci and aberrant chromosomal configurations [44]. Furthermore, mutations of the methyl-binding protein MECP2 lead to the neurodevelopmental disease X-linked Rett syndrome correlated to lacking transduction of methylation marks into spatial DNA-organization [10].
Changes in the epigenetic profile have been described for numerous pathologies compared to healthy controls, for example cardiovascular disease, mental disorders or amyotrophic lateral sclerosis [45], [46], [47]. In contrast to the previous examples, it remains unclear though how those defective marks arise and to what extend they are the cause or the collateral damage of the disease state.
The most extensively studied pathological case of DNA methylation is without surprise cancer, resulting in the emergence of a separate research field entitled cancer epigenetics [8], [10]. Markert suggested already in 1968 that cancer could be due to gene activity being misprogrammed by epigenetic mechanisms [48]. In 1982, Feinberg and Vogelstein identified the first concrete epigenetic divergence in cancer and described the hypomethylation of a set of genes in several primary tumor types [49]. Both hypomethylation and hypermethylation in many cancer types have since been thoroughly identified, analyzed and made publicly available by the NIH-founded Cancer Genome Atlas (TCGA) research network [50], [51]. These DNA methylation changes bring about a panoply of deleterious secondary effects: hypomethylation unleashes oncogenes, multiresistance genes, pericentromeric satellite sequences or retrotransposable elements, while hypermethylation silences tumor suppressor genes [10].
Yet, pure observation is rapidly unsatisfying and, under financial and publication pressure, the desire to press the handy on/off button made of DNA methylation is omnipresent. The unwritten consensus of the last paragraph of every recent publication dealing with any epigenetic-related phenomenon (and the choice is large, considering the scattering of the notion) is to sell it as a promising biomarker or as a novel therapeutic target [10].
Epimedicine
Like someone trying to learn to play billiard from just watching, there is an intertwined triad in Science conducting the transition from observation to intervention: deducing the rules, predicting the next move and finally playing. By analogy, in the case of epigenetics, they could be renamed epidemiology, prognosis and epitherapy [Fig. 2].
Fig. 2.
The three stages of Science: Observation, Prediction and Intervention.
Guessing the rules: epigenetics and epidemiology
Molecular genetics and the discovery of genetic information revolutionized centuries of empirical and organ-focused medicine in a couple of decades into an era of molecular medicine [10]. The ensuing expectations and speculations climaxed fourteen years ago, when after years of intense labor, the first drafts of the entire sequenced human genome were published [52], [53]. Scientists affirmed that this would “revolutionize the diagnosis, prevention and treatment of most, if not all, human diseases” and many believed that the mutational program of complex diseases would be decrypted and subsequently repaired by genetic therapy [10].
Despite the undeniable achievements of genome-wide association studies (GWAS), a certain feeling of disappointment quickly followed the initial excitement. Fixing genes turned out to be way trickier than expected and suffered both a serious setback and damage to its public image through a few unexpected side effects like the death of Jesse Gelsinger and cases of leukemia after treating X-linked severe combined immunodeficiency using adenoviruses (though Alain Fischer was awarded the well-earned Japan Prize last year) [54], [55]. Moreover, the understanding of complex and non-Mendelian diseases progressed only scarcely – no clear-cut sets of mutations or genetic variants could be attributed to autism or diabetes and even patients with monogenic disorders displayed fluctuating symptoms and disease severity. DNA was obviously not the sole pertinent reference to gain insight into the molecular underpinnings of disease.
The knight in shining armor arrived in the form of epigenetics and suddenly the shortcomings of genetics metamorphosed into the sales pitch for epigenetics – as genes do not hold all the keys to disease, epigenetics will. They were proclaimed the “biggest revolution in biology that is going to forever transform the way we understand genetics, environment, the way the two interact and what causes disease” in 2007 [56]. This sounds strangely familiar, doesn't it?
Epigenetic epidemiology has increasingly focused on the connection between environmental effects and disease phenotypes, including nutrition, chemical and physical influences but also – a novelty compared to genetic studies – social factors [57], [58], [59]. The first cluster of Epigenetic Association Studies (EWAS) was published around 2010 [60], fueled by initial high-impact publications, such as the much-quoted Swedish harvest data putting forward a transgenerational effect of diet mediated by sperm or the link between maternal care and anxiety in rats [7], [61].
Where are the flaws then?
One complication is the logical consequence of the definition dilemma of epigenetics: what exactly should we look for and above all, where should we stop looking? Where is the limit of functional significance and where starts randomness and background noise? Any study can only consider a limited amount of variables, thus the argument that not enough factors were taken into consideration, just like DNA not being sufficient to understand disease, will always apply. DNA methylation of promoter regions, for instance, is currently the widest studied epigenetic phenomenon for technical reasons, however one might argue that they are meaningless as long as non-CpG DNA methylation, histone marks and noncoding RNAs are not taken into account in parallel [60].
Other problems are identical to those encountered by genetic approaches. Just like various sets of mutations can lead to similar disease phenotypes, why would there be a unique set of epimutations corresponding to one symptom? Furthermore, like every nascent scientific domain, epigenetics is currently in a state of technological trial and error. Regarding the state of the art, an exhaustive description and qualitative comparison of current technologies and data analytical approaches for epigenomics can be found in the review by Klaas Mensært and colleagues [62]. As much as the notion, the associated technology is under construction and still imperfect – not enough time has passed yet to inspect reproducibility, agree on foolproof controls and quality-control steps, and rule out artifacts. An armada of different methods is in use and comparing results obtained by different strategies and platforms is difficult, nay impossible [60]. In parallel, the explosion of available data of the last decade, thanks to the advent of high-throughput sequencing technologies and the considerable drop in cost of the latter, has triggered a boom in bioinformatics and analytical methodologies. However, algorithms are as manifold as bench practices and not unconditionally comparable – with the difference that a considerable proportion of biologists is not exactly up-to-date with the rapidly evolving amount of tools and that a reasonable bioinformatic training was only recently integrated into the undergraduate programs.
But EWAS have also their very own set of challenges, different from genetic studies. The main hurdle is the spatio-temporal variability of the epigenome within one individual. DNA is static, identical in all cells of the organism and – modulo some eventual mutations over time – remains also the same during the entire lifetime of the organism. Because of its fundamental role in cell fate, the epigenome in contrast is extremely cell-type specific and because of its plasticity and sensitivity to environmental stimuli, it tends additionally to change over the years [2], [60], [63]. The first corollary is that the precise cell type and cell purity matter. Studying the epigenetic compound of Parkinson's disease using keratinocytes won't make much sense, but getting hold of human dopaminergic neurons of the substantia nigra might be linked to a few inconveniences. Evidently, this impedes as well the comparison between individuals and renders the scaling-up of studies complicated by lack of equivalent material – in cell type, age and maybe environmental context [2], [60]. Traditionally, whole blood samples were collected for genetic cohort studies, like the broadly used 1958 British Birth Cohort Study, yet blood is a heterogeneous tissue and moderately representative for most organs. Nevertheless, this collection has been used to correlate childhood socio-economic status and adult DNA methylation [59].
Moreover, epigenetic epidemiology lacks a fundamental benchmark. Genetics have their hg38 and MM10 – regularly updated “reference genomes” – but there is no absolute reference DNA-methylome to relate to. Considering above-mentioned issues of cell-specificity and variability, a gigantic number of samples would be required to achieve a halfway representative mean of epigenomes and an idea of the degree of inter-individual differences, a methylated equivalent of genetic polymorphisms [2], [60].
Nonetheless, numerous big projects and international collaborations have been launched in order to tackle the task: the European Union Blueprint Consortium is trying to decipher the epigenome of hematopoietic cells, with the goal to furnish about hundred reference epigenomes [64], the NIH Roadmap Epigenomics Mapping Consortium focuses on stem cells and primary ex vivo tissues [9], [65] and the International Human Epigenome Consortium has the modest aim to “understand the extend to which the epigenome has shaped human populations over generations and in response to the environment” by decrypting at least 1000 epigenomes during the next decade [66].
The future will show, how realistic those plans are.
Predicting the moves – a tale of biomarkers
On a daily basis, studies are published introducing a particular noncoding RNA or methylation pattern as the ideal “biomarker” for a specific type of cancer, often without any further investigation of the functions and mechanisms of the candidates. This notion goes hand in hand with the popular concept of “personalized medicine” and the idea that one day, each patient will be treated with a tailor-made cocktail of drugs, corresponding to his precise (epi)genomic profile.
True, each cancer can be considered as unique – an exclusive collection of mutations and epimutations – and the good correlation of thousands of different sets of biomarkers with thousands of disease variations is probably correct. Sequencing large parts of the (epi)genome or even transcriptome of a patient is not that unconceivable either, and might greatly improve the current deficiencies in patient stratification both for therapy and cohort studies. There is one hitch, though. We do not have at our disposal thousands of different treatments or drug combinations. At least, not yet.
Playing the game – epitherapy
The theoretical attractiveness of targeting chromatin modifications lies in their greater reversibility and natural flexibility compared to DNA mutations, making them in principal a more accessible point of attack [2], [4], [10].
Companies aiming to develop epitherapeutic drugs, principally targeting DNA methylation and histone modifications, have sprouted over the last years and the major pharmaceutical concerns branched off substantial financial resources into the creation of novel departments. Most applied research efforts focus currently on cancer therapies and all concrete applications and exciting treatments are restricted for now to cancer treatment. This does not come as a surprise – cancer research benefits historically from a particular background of public attention, economic weight, financial resources and huge quantities of available research material, from cell lines, frozen tumor samples to in vivo models. Hence, the cancer field has always been at the forefront when it comes to the implementation of new technologies and treatment strategies [10].
The purpose of this review is not to present an exhaustive list of available drugs or those at various stages of clinical trials, most of which can be found in the Orange Book of the US Food and Drug Administration (FDA) and various reviews, thus we will content ourselves with a quick, representative overview [67], [68].
Shoot first, ask questions later, is not an unusual approach in the history of drug discovery. 5-azacytidine, alias Vidaza on the market [69], was empirically known for cytotoxic effects on cancer cells since 1968, but its actual role in inhibiting DNA methylation was only established a decade later [68], [69], [70]. Its derivative, 5-aza-2′-deoxycytidine (Decitabine) is used for the treatment of hematological malignancies and similar analogs, such as Zebularine, are at the stage of clinical trials [8], [68]. Similarly, valproic acid was used for nearly half a century to treat neurological diseases like epilepsy under the name of many brands before it turned out to be also a histone deacetylase (HDAC) inhibitor and thus of interest for cancer therapy, where it is currently investigated [71].
After looking, this time on purpose, for modulators of epigenetic mechanisms, the first HDAC inhibitor in form of superanilohydroxamic acid (SAHA, marketed as Vorinostat) was approved by the FDA as a third line treatment against T cell lymphoma [72] [Fig. 3]. Taking into account the considerable delay between the identification of molecules with potential therapeutic applications and their approval as drugs, from proof of mechanism to proof of concept, as well as the high failure rate, most treatment strategies launched by the companies born during the last decade are yet at various stages of (pre)-clinical trials: mainly peptides acting as inhibitors of diverse DNMTs, HATs or HDACs but also some antisense oligonucleotides against the RNA precursors of the latter [68]. The following ten years will without doubt witness an accumulation of epigenetic cancer drugs on the market. However, we will probably have to wait for a couple of more decades before an objective judgment of their usefulness and efficiency becomes pertinent [2].
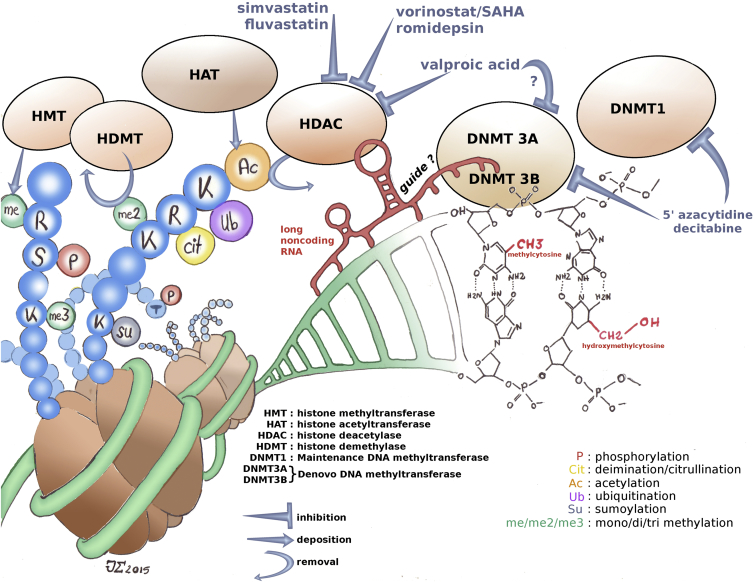
Fig. 3.
Epigenetic chromatin modifications and currently available treatments acting on them. Both DNA and histones can carry many chemical modifications that alter the chromatin state and the expression of genes. They are deposited, altered and removed by a panoply of enzymes. These in turn are regulated by many factors and interact with various molecules, including noncoding RNAs. Several drugs specifically target these enzymes.
To date, the targeting of DNA and histone modifications suffers from the same flaws than any kind of drug administration. The main one is the problem of precise targeting – at the level of the organism, when it comes to aim only at cancer cells but avoid collateral damages in their normal neighbors; and at the level of the chromatin, when it comes to modify the state of only specific genomic loci. While tissue-tropism is vaguely achievable for many drugs, an inhibitor of a methyltransferases will inevitably lower the methylation levels genome-wide. Reaching this degree of precision requires first a way more detailed level of knowledge on how chromatin marks are recruited to specific sequences in specific cellular contexts. It is by all means conceivable to act one day on a long noncoding RNA responsible for guiding a methyltransferase to one locus, for now however, no such concrete approaches exist.
There is an unavoidable gap between the researcher and the clinician. The latter has a somehow more humble approach of the living than the biologist. Empirical trial and error as well as not knowing everything is an acceptable option; and questioning compulsorily all underlying mechanisms is not a necessity. From the clinical point of view, epigenetic cancer therapy has not the slightest pretention to be better, safer or cheaper than existing treatments, rather it is meant to be complementary, to enrich the arsenal. Cancer is a nasty, constantly changing enemy with many resources – having more than one string to one's bow won't be amiss. Most trials don't focus uniquely on epigenetic treatments, by the way, but rather on combinations of different compounds targeting different pathways.
Who is next, on the list of potential targets for epitherapeutics?
Several trials are being conducted in the context of human immunodeficiency virus (HIV) infections [73]. Inflammation maybe, more specifically reversible diseases such as inflammatory bowel disease (IBD), and metabolic diseases linked to tissues presenting a rapid turnover, allowing thus to affect the adult stem cell population.
Conclusions
The big picture
Pointing out that no cellular phenomenon exists independently of others certainly lacks any originality. Yet in the context of the popularity of epigenetics, it might be appropriate to remember that not only the notion is vague, but also that clearly defined components are just another brick in the wall and that one modification may gain functional importance only if it co-occurs with other changes [60].
Since the hype seems to have cooled down across the last five years, the latest trend is integration [2], [3]. Public genome-scale resources call for combining DNA sequence, epigenomic, transcriptomic and proteomic data for each sample [60]. Also, old data are resurrected. To the general puzzlement, many disease-correlated genetic variations locate to noncoding regions and were put on ice. In the light of long noncoding RNA, these variations are reconsidered.
Eu-epigenics
For now, the effect of studies relating the transmission of maternal skills and the link between socio-economic status and suicide, ranges from questionable over inexplicable to fascinating with a taste of Lamarckian theory, which can be considered as rather harmless [8], [61], [74].
Even so, it is a legitimate question, what the long-term effect of epigenetics will be on the human society [2]. Undeniably, genetics have profoundly changed our perception of the interplay of human health and environment. Nowadays, we cover ourselves in sun-cream, fear genetically modified vegetables, and anti-smoking laws and campaigns are omnipresent to the extend that movie posters for Gainsbourg: A Heroic Life were banned from the Parisian metro stations because they featured the eponymous French singer with his hallmark – a cigarette. Maybe our slightly paranoid society will progressively include epigenetic hazard into risk assessments and add a detailed health record of the environmental exposures, diet and social surroundings of early life stages into the health record booklet [2], [42].
Prenatal tests for genomic abnormalities had raised the fear of eugenics and genetic discrimination, as depicted by the 1997 dystopian film Gattaca. In conformity with our current code of ethics, the genetic determinism was properly dismissed in the moral of the story, rehabilitating randomness and free will.
But what kinds of dangers await a new society that believes in the heritability or behavioral programming by parental drug addiction, domestic violence or child abuse? [8], [74].
The cycle of knowledge
Oddly, we currently witness a certain reemergence of the pre-seventies state of mind when it comes to epigenetics, specifically to the issue of inter-generational heritability. During the early post Watson-Crick era, epigenetics were invoked by default when genetics could not provide an explanation any more – a potpourri of phenomena above and beyond genetics [8]. After a brief period of clarity starting in the 1990s, when the definition of epigenetics furtively overlapped the notion of DNA methylation, during the last 15 years, the limits of the term have again moved out of focus by the aggregation of components [1]. And again, epigenetics are the explanation for happenings beyond our current understanding – development and the passing-on of acquired traits between generations.
In an excellent essay, Robert Weinberg, one of the authors of the famous “hallmarks of cancer” illustration, looks back on the history of molecular cancer research. He highlights the eventful journey of research from confusing incomprehension to the glorious years of reductionism and back to overwhelming complexity [75]. A similar scheme seems to apply to (epi)genetics, born into the mystery of embryonic development and heredity, then promoted to the program governing life before drowning in the complexity of omics. Waddington himself was genuinely convinced that “genetics is a way of analyzing an animal into representative units, so that its nature can be indicated by a formula, as we represent a chemical compound by its appropriate symbol”, predicting exactly the phase of reductionism triggered by the first successes of molecular genetics [18].
Due to their popularity (epi)genetics were inevitably bound to deceive, but the powerful comeback of complexity stems also from a profound shift in our way to approach biological questions, after that technological progress unlocked entirely new horizons (high-throughput sequencing just turned 10). In the era of omics – genomics, proteomics, metabolomics – the angle of view on biology has radically changed from focus to global. Instead of investigating one precise object, may it be a gene or a species, the goal is now to provide a description of all components of a system in parallel, assuming that many phenomena make sense only through the study of all their actors.
Any biological process is the result of the coordination of myriads of molecules and individual decisions, and at the very opposite side of reductionism, one might argue that only the knowledge of the spatio-temporal coordinates of every single molecule will allow us to fully understand the process in question [19]. When the ENCODE data was released, some claimed that “possibly every single molecule in the cell is functional” [13].
Maybe in the future, with the increasing resolution and power of data generation, analysis and modeling, all concurrent definitions and mechanisms will fade away and blend in a novel vision of life – the one of Systems Biology.
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Haig D. Commentary: the epidemiology of epigenetics. Int J Epidemiol. 2012;41:13–16. doi: 10.1093/ije/dyr183. [DOI] [PubMed] [Google Scholar]
- 2.Wright R., Saul R.A. Epigenetics and primary care. Pediatrics. 2013;132(Suppl. 3):S216–S223. doi: 10.1542/peds.2013-1032F. [DOI] [PubMed] [Google Scholar]
- 3.Felsenfeld G. A brief history of epigenetics. Cold Spring Harb Perspect Biol. 2014;6:a018200. doi: 10.1101/cshperspect.a018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dupont C., Armant D.R., Brenner C.A. Epigenetics: definition, mechanisms and clinical perspective. Semin Reprod Med. 2009;27:351–357. doi: 10.1055/s-0029-1237423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riggs A.D. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 6.Dias B.G., Ressler K.J. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pembrey M.E., Bygren L.O., Kaati G., Edvinsson S., Northstone K., Sjöström M. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 8.Choudhuri S. From Waddington's epigenetic landscape to small noncoding RNA: some important milestones in the history of epigenetics research. Toxicol Mech Methods. 2011;21:252–274. doi: 10.3109/15376516.2011.559695. [DOI] [PubMed] [Google Scholar]
- 9.http://www.roadmapepigenomics.org/overview.
- 10.Chahwan R., Wontakal S.N., Roa S. The multidimensional nature of epigenetic information and its role in disease. Discov Med. 2011;11:233–243. [PubMed] [Google Scholar]
- 11.Latronico M.V., Condorelli G. Therapeutic applications of noncoding RNAs. Curr Opin Cardiol. 2015;30:213–221. doi: 10.1097/HCO.0000000000000162. [DOI] [PubMed] [Google Scholar]
- 12.Aravin A.A., Bourc’his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ENCyclopedia Of DNA Elements ENCODE, http://www.gemome.gov/10005107.
- 14.Lee J.T. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- 15.Khalil A.M., Guttman M., Huarte M., Garber M., Raj A., Rivea Morales D. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Preer J.R., Jr. Sonneborn and the cytoplasm. Genetics. 2006;172:1373–1377. doi: 10.1093/genetics/172.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waddington C.H. The epigenotype. 1942. Int J Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 19.Gilbert S.F. Commentary: ‘The epigenotype’ by C.H. Waddington. Int J Epidemiol. 2012;41:20–23. doi: 10.1093/ije/dyr186. [DOI] [PubMed] [Google Scholar]
- 20.Avery O.T., Macleod C.M., McCarty M. Studies on the chemical nature of the substances inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyatt G.R. Occurrence of 5-methylcytosine in nucleic acids. Nature. 1950;166:237–238. doi: 10.1038/166237b0. [DOI] [PubMed] [Google Scholar]
- 22.Holliday R., Pugh J.E. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 23.Laskey R.A., Gurdon J.B. Genetic content of adult somatic cells tested by nuclear transplantation from cultured cells. Nature. 1970;228:1332–1334. doi: 10.1038/2281332a0. [DOI] [PubMed] [Google Scholar]
- 24.Murray K. The occurrence of epsilon-N-methyl lysine in histones. Biochemistry. 1964;3:10–15. doi: 10.1021/bi00889a003. [DOI] [PubMed] [Google Scholar]
- 25.Allfrey V.G., Faulkner R., Mirsky A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc Natl Acad Sci USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiio Y., Eisenmann R.N. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci USA. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 28.Waddington C.H. Cambridge University Press; 1940. Organisers and genes. [Google Scholar]
- 29.Nanney D.L. Epigenetic control systems. Proc Natl Acad Sci USA. 1958;44:712–717. doi: 10.1073/pnas.44.7.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson C.B., Rowell E., Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 31.Han J., Sachdev P.S., Sidhu K.S. A combined epigenetic and non-genetic approach for reprogramming human somatic cells. PLoS One. 2010;5:e12297. doi: 10.1371/journal.pone.0012297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 33.Orgel L.E., Crick F.H.C., Selfish D.N.A. The ultimate parasite. Nature. 1980;284:604–607. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 34.Baillie J.K., Barnett M.W., Upton K.R., Gerhardt D.J., Richmond T.A., De Sapio F. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cattanach B.M., Kirk M. Differential activity of maternally and paternally derives chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 36.Skaar D.A., Li Y., bernal A.J., Hoyo C., Murphy S.K., Jirtle R.L. The human imprintome: regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR J. 2012;53:341–358. doi: 10.1093/ilar.53.3-4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patalone S., Hore T.A., Reik W., Sumner S. Shifting behavior: epigenetic reprogramming in eusocial insects. Curr Opin Cell Biol. 2012;24:367–373. doi: 10.1016/j.ceb.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 38.Shi Y.Y., Huang Z.Y., Zeng Z.J., Wang Z.L., Wu X.B., Yan W.Y. Diet and cell size both affect queen-worker differentiation through DNA methylation in honey bees (Apis mellifera, Apidae) PLoS One. 2011;6:e18808. doi: 10.1371/journal.pone.0018808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arrieta M.C., Stiemsma L.T., Amenyogbe N., Brown E.M., Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ralston A., Rossant J. Cdx2 acts downstream of cell polarization to cell-autonomously promote trophectoderm fate in the early mouse embryo. Dev Biol. 2008;313:614–629. doi: 10.1016/j.ydbio.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 41.Wolf C., Linden D.E. Biological pathways to adaptability – interactions between genome, epigenome, nervous system and environment for adaptive behavior. Genes Brain Behav. 2012;11:3–28. doi: 10.1111/j.1601-183X.2011.00752.x. [DOI] [PubMed] [Google Scholar]
- 42.Mirbahai L., Chipman J.K. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposures. Mutat Res Genet Toxicol Environ Mutagen. 2014;764–765:10–17. doi: 10.1016/j.mrgentox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 43.Fraga M.F., Ballestar E., Paz M.F., Ropero S., Setien F., Ballestar M.L. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Velasco G., Walton E.L., Sterlin D., Hédouin S., Nitta H., Ito Y. Germline genes hypomethylation and expression define a molecular signature in peripheral blood ICF patients: implications for diagnosis and etiology. Orphanet J Rare Dis. 2014;9:56. doi: 10.1186/1750-1172-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Udali S., Guarini P., Moruzzi S., Choi S.W., Friso S. Cardiovascular epigenetics: from DNA methylation to microRNAs. Mol Aspects Med. 2013;34:883–901. doi: 10.1016/j.mam.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Kato T., Iwamoto K. Comprehensive DNA methylation and hydroxymethylation analysis in the human brain and its implication in mental disorders. Neuropharmacology. 2014;80:133–139. doi: 10.1016/j.neuropharm.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 47.Martin L.J., Wong M. Aberrant regulation of DNA methylation in amyotrophic lateral sclerosis: a new target of disease mechanisms. Neurotherapeutics. 2013;10:722–733. doi: 10.1007/s13311-013-0205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Markert C.L. Neoplasia: a disease of cell differentiation. Cancer Res. 1968;28:1908–1914. [PubMed] [Google Scholar]
- 49.Feinberg A.P., Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 50.http://cancergenome.nih.gov/.
- 51.Weisenberger D.J. Characterizing DNA methylation alterations from the cancer genome atlas. J Clin Invest. 2014;124:17–23. doi: 10.1172/JCI69740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J. Initial sequencing and analysis of the human genome. Nature. 2001;412:565. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 53.McPherson J.D., Marra M., Hillier L., Waterston R.H., Chinwalla A., Wallis J. A physical map of the human genome. Nature. 2001;409:934–941. doi: 10.1038/35057157. [DOI] [PubMed] [Google Scholar]
- 54.Somia N., Verma I.M. Gene therapy: trials and tribulations. Nat Rev Genet. 2000;1:91–99. doi: 10.1038/35038533. [DOI] [PubMed] [Google Scholar]
- 55.Hacein-Bey-Abina S., von Kalle C., Schmidt M., Le Deist F., Wulffraat N., McIntyre E. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 56.http://www.pbs.org/wgbh/nova/transcripts/3413_genes.html.
- 57.Gronninger E., Weber B., Heil O., Peters N., Stäb F., Wenck H. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Breitling L.P., Yang R., Korn B., Burwinkel B., Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Borghol N., Suderman M., McArdle W., Racine A., Hallett M., Pembrey M. Associations with early-life socio-economic position in adult DNA methylation. Int J Epidemiol. 2012;41:62–74. doi: 10.1093/ije/dyr147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heijmans B.T., Mill J. Commentary: the seven plagues of epigenetic epidemiology. Int J Epidemiol. 2012;41:74–78. doi: 10.1093/ije/dyr225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weaver I.C., Cervoni N., Champagne F.A., D'Alessio A.C., Sharma S., Seckl J.R. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 62.Mensaert K., Denil S., Trooskens G., Van Criekinge W., Thas O., De Meyer T. Next-generation technologies and data analytical approaches for epigenomics. Environ Mol Mutagen. 2014;55:155–170. doi: 10.1002/em.21841. [DOI] [PubMed] [Google Scholar]
- 63.Duarte J.D. Epigenetics primer: why the clinician should care about epigenetics. Pharmacotherapy. 2013;33:1362–1368. doi: 10.1002/phar.1325. [DOI] [PubMed] [Google Scholar]
- 64.http://www.blueprint-epigenome.eu/.
- 65.http://www.genboree.org/epigenomatlas/index.rhtml.
- 66.http://ihec-epigenomes.org/.
- 67.http://www.fda.gov/.
- 68.Heerboth S., Lapinska K., Snyder N., Leary M., Rollinson S., Sarkar S. Use of epigenetic drugs in disease: an overview. Genet Epigenet. 2014;6:9–19. doi: 10.4137/GEG.S12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaminskas E., Farrell A.T., Wang Y.C., Sridhara R., Pazdur R. FDA drug approval summary: azacitidine (s-azacytidine, Vidaza) for injectable suspension. Oncologist. 2005;10:176–182. doi: 10.1634/theoncologist.10-3-176. [DOI] [PubMed] [Google Scholar]
- 70.Jones P.A., Taylor S.M. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1989;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 71.Činčárová L., Zdráhal Z., Fajkus J. New perspectives of valproic acid in clinical practice. Expert Opin Investig Drugs. 2013;22:1535–1547. doi: 10.1517/13543784.2013.853037. [DOI] [PubMed] [Google Scholar]
- 72.Duvic M., Vu J. Update on the treatment of cutaneous T-cell lymphoma (CTCL): focus on vorinostat. Biologics. 2007;1:377–392. [PMC free article] [PubMed] [Google Scholar]
- 73.Ay E., Banati F., Mezei M., Bakos A., Niller H.H., Buzás K. Epigenetics of HIV infection: promising research areas and implications for therapy. AIDS Rev. 2013;15:181–188. [PubMed] [Google Scholar]
- 74.McGowan P.O., Sasaki A., D'Alessio A.C., Dymov S., Labonté B., Szyf M. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weinberg R.A. Coming full circle-from endless complexity to simplicity and back again. Cell. 2014;157:267–271. doi: 10.1016/j.cell.2014.03.004. [DOI] [PubMed] [Google Scholar]