Abstract
Back pain is the #1 cause of years lived with disability worldwide, yet surprisingly little is known regarding the biology underlying this symptom. We conducted a genome-wide association study (GWAS) meta-analysis of chronic back pain (CBP). Adults of European ancestry were included from 15 cohorts in the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium, and from the UK Biobank interim data release. CBP cases were defined as those reporting back pain present for ≥3–6 months; non-cases were included as comparisons (“controls”). Each cohort conducted genotyping using commercially available arrays followed by imputation. GWAS used logistic regression models with additive genetic effects, adjusting for age, sex, study-specific covariates, and population substructure. The threshold for genome-wide significance in the fixed-effect inverse-variance weighted meta-analysis was p<5×10−8. Suggestive (p<5×10−7) and genome-wide significant (p<5×10−8) variants were carried forward for replication or further investigation in the remaining UK Biobank participants not included in the discovery sample. The discovery sample comprised 158,025 individuals, including 29,531 CBP cases. A genome-wide significant association was found for the intronic variant rs12310519 in SOX5 (OR 1.08, p = 7.2×10−10). This was subsequently replicated in 283,752 UK Biobank participants not included in the discovery sample, including 50,915 cases (OR 1.06, p = 5.3×10−11), and exceeded genome-wide significance in joint meta-analysis (OR 1.07, p = 4.5×10−19). We found suggestive associations at three other loci in the discovery sample, two of which exceeded genome-wide significance in joint meta-analysis: an intergenic variant, rs7833174, located between CCDC26 and GSDMC (OR 1.05, p = 4.4×10−13), and an intronic variant, rs4384683, in DCC (OR 0.97, p = 2.4×10−10). In this first reported meta-analysis of GWAS for CBP, we identified and replicated a genetic locus associated with CBP (SOX5). We also identified 2 other loci that reached genome-wide significance in a 2-stage joint meta-analysis (CCDC26/GSDMC and DCC).
Author summary
Back pain is the #1 cause of years lived with disability worldwide and one of the most common reasons for health care visits in developed countries, yet surprisingly little is known regarding the biology underlying this symptom. Chronic back pain is the major driver of the societal burden of back pain. Identifying biological pathways involved in chronic back pain through genetic association studies might reveal insights into the underlying mechanisms involved or suggest potential avenues for the development of new treatments. We conducted the first genome-wide association study meta-analysis to examine genetic variants associated with chronic back pain. We identified variants associated with chronic back pain in 158,025 individuals of European ancestry from 16 cohorts in Europe and North America, and replicated our findings in 283,752 UK Biobank participants of European ancestry not included in the discovery sample. Our study identifies three novel genome-wide significant associations with chronic back pain, and suggests possible shared genetic mechanisms with other traits such as cartilage, osteoarthritis, lumbar disc degeneration, depression, and height/vertebral development.
Introduction
Back pain causes more years lived with disability than any other health condition worldwide.[1] Although most adults experience a new (‘acute’) episode of back pain at some point in their lives, the societal burden of back pain is driven by the minority of individuals who fail to recover from such episodes and go on to develop persistent (‘chronic’) back pain.[2] Chronic back pain (CBP) has a number of definitions but is most often considered as back pain of duration ≥3 months in clinical practice, and a duration of ≥6 months is also commonly used in research.[3, 4]
Back pain is moderately heritable. Meta-analysis of 11 twin studies of back pain indicates a heritability of 40%, with a pattern of monozygotic (rMZ = 0.56) and dizygotic (rDZ = 0.28) twin correlations suggesting an additive genetic model (2rDZ = rMZ).[5, 6] Heritability is greater for chronic than for acute back pain.[7] Nevertheless, genetic studies attempting to identify specific genetic markers for CBP have to date been limited to small studies using the candidate gene approach.[8, 9] Although CBP is often attributed to anatomic changes such as intervertebral disc degeneration or disc herniation, such findings have only weak association with CBP, [10, 11] and explain only a small proportion (7–23%) of the genetic influence on back pain[12]. The unexplained genetic contribution to CBP may involve not only spine pathology but also functional predisposition to chronic pain involving higher-order neurologic processes related to the generation and maintenance of pain.[13–15] Furthermore, psychological factors such as depression are widely recognized as important risk factors for CBP.[16] Given the range of processes that might contribute to CBP, the agnostic genome-wide association approach may identify novel genetic variants associated with CBP and provide insights into underlying biological mechanisms not previously considered.
This research was an international collaboration between investigators from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium Musculoskeletal Workgroup[17] and the European Union FP7 project Pain-OMICS (‘Multi-dimensional omics approach to stratification of patients with low back pain’). We conducted a meta-analysis of GWAS of CBP in adults of European ancestry from 16 community- and population-based cohorts, including those from the CHARGE and PainOmics consortia, and the UK Biobank. The aim was to identify novel associations between specific genetic markers and CBP, and elucidate the biological mechanisms underlying this condition.
Results
Study overview
Genome-wide discovery meta-analysis was comprised of adults of European ancestry from 16 cohorts (n = 158,025 including 29,531 CBP cases; Table 1), including 15 CHARGE cohorts and participants from the UK Biobank (UKB) interim data release (UKB1). After quality control, the number of SNPs included in the meta-analysis ranged from 6,205,227 to 9,775,703, depending on the cohort (S1 and S2 Tables). Linkage disequilibrium (LD) score regression (LDsr) was used distinguish polygenicity from potential confounding,[18] using LD scores from European ancestry 1000 Genomes data. The genome-wide significance level was defined as p<5×10−8, and suggestive significance level was defined as p<5×10−7, after using the LDsr intercept as a correction factor. For those SNPs of genome-wide suggestive significance in the discovery phase, replication was conducted in a sample of UKB European ancestry participants (UKB2) who were not part of the interim data release (n = 283,752 subjects, including 50,915 CBP cases), and a joint (discovery-replication) meta-analysis was performed. We then conducted functional characterization of variants and loci achieving genome-wide significance in the joint meta-analysis.
Table 1. Cohorts in meta-analysis of genome-wide association studies of chronic back pain (European ancestry).
| Cohort | Study setting | Country | Sample size | Chronic back pain definition | Prevalence (%) | Age (yr) | BMI (kg/m2) | Women (%) | |
|---|---|---|---|---|---|---|---|---|---|
| Cardiovascular Health Study (CHS) | Community | USA | 2849 | ≥1 month of back pain in consecutive years | 14.2% | Cases (n = 404) | 72.1 ± 5.1 | 27.3 ± 5.0 | 73.3% |
| Controls (n = 2445) | 72.1 ± 5.2 | 26.1 ± 4.3 | 59.6% | ||||||
| Framingham Heart Study[19] | Community | USA | 2673 | ≥6 months of back pain | 21.0% | Cases (n = 561) | 67.7 ± 9.3 | 28.8 ± 5.8 | 62.2% |
| Controls (n = 2112) | 66.4 ± 9.1 | 28.0 ± 52 | 52.9% | ||||||
| Generation Scotland | Population | UK | 5071 | ≥3 months of back pain | 26.0% | Cases (n = 1322) | 54.9 ± 11.8 | 28.1 ± 5.7 | 66.7% |
| Controls (n = 3749) | 52.4 ± 12.7 | 26.4 ± 4.6 | 55.4% | ||||||
| Johnston County Osteoarthritis Project (JoCo) | Population | USA | 480 | ≥6 months of back pain | 38.8% | Cases (n = 186) | 72.0 ± 8.0 | 31.4 ± 6.3 | 65.0% |
| Controls (n = 294) | 73.0 ± 8.0 | 29.3 ± 5.2 | 58.4% | ||||||
| Mr. Os Sweden | |||||||||
| Gothenburg | Population | Sweden | 920 | ≥6 months of back pain | 14.2% | Cases (n = 131) | 75.3 ± 3.2 | 26.7 ± 3.9 | 0% |
| Controls (n = 789) | 75.3 ± 3.2 | 26.1 ± 3.4 | 0% | ||||||
| Malmo | Population | Sweden | 948 | ≥6 months of back pain | 10.8% | Cases (n = 102) | 75.8 ± 3.1 | 27.2 ± 3.7 | 0% |
| Controls (n = 846) | 75.6 ± 3.2 | 26.4 ± 3.6 | 0% | ||||||
| Mr. Os US | Community | USA | 4615 | ≥6 months of back pain | 14.1% | Cases (n = 653) | 74.6 ± 6.1 | 28.0 ± 4.1 | 0% |
| Controls (n = 3962) | 73.9 ± 5.9 | 27.3 ± 3.8 | 0% | ||||||
| Osteoarthritis Initiative (OAI) | Community | USA | 2474 | ≥1 month of back pain in consecutive years | 13.5% | Cases (n = 335) | 61.0 ± 9.1 | 28.9 ± 4.7 | 57.9% |
| Controls (n = 2139) | 61.7 ± 9.1 | 28.1 ± 4.5 | 53.2% | ||||||
| Rotterdam Study (RS) | |||||||||
| RS-1 | Community | Netherlands | 5965 | ≥6 months of back pain | 14.7% | Cases (n = 877) | 69.1 ± 9.2 | 26.7 ± 4.0 | 72.0% |
| Controls (n = 5088) | 70.0 ± 9.4 | 26.2 ± 3.7 | 58.3% | ||||||
| RS-2 | Community | Netherlands | 1566 | ≥6 months of back pain | 36.7% | Cases (n = 574) | 65.3 ± 8.0 | 27.7 ± 4.2 | 66.7% |
| Controls (n = 992) | 64.6 ± 8.0 | 27.3 ± 4.1 | 55.4% | ||||||
| RS-3 | Community | Netherlands | 3019 | ≥6 months of back pain | 38.2% | Cases (n = 1154) | 57.4 ± 6.9 | 28.1 ± 4.8 | 59.5% |
| Controls (n = 1865) | 56.9 ± 6.7 | 27.4 ± 4.5 | 54.3% | ||||||
| Study of Osteoporotic Fractures (SOF) | Community | USA | 3615 | ≥6 months of back pain | 16.3% | Cases (n = 589) | 72.1 ± 5.6 | 27.6 ± 5.3 | 100% |
| Controls (n = 3026) | 71.4 ± 5.2 | 26.6 ± 4.4 | 100% | ||||||
| 10,001 Dalmatians | |||||||||
| Vis | Population | Croatia | 251 | ≥3 months of back pain | 22.3% | Cases (n = 56) | 67.3 ± 13.2 | 27.8 ± 4.4 | 69.6% |
| Controls (n = 195) | 63.7 ± 12.1 | 26.7 ± 4.0 | 52.8% | ||||||
| Korcula | Population | Croatia | 773 | ≥3 months of back pain | 21.2% | Cases (n = 164) | 64.3 ± 12.9 | 28.0 ± 4.3 | 70.7% |
| Controls (n = 609) | 58.0 ± 14.9 | 27.0 ± 41 | 64.0% | ||||||
| UK Biobank | Population | United Kingdom | 120,024 | ≥3 months of back pain | 18.0% | Cases (n = 21,600) | 57.3 ± 7.9 | 28.5 ± 5.4 | 54.0% |
| Controls (n = 98,424) | 57.0 ± 7.9 | 27.4 ± 4.8 | 52.4% | ||||||
| TwinsUK | Population-based twin registry | United Kingdom | 2782 | ≥3 months of back pain | 29.6% | Cases (n = 823) | 56.7 ± 12.6 | 27.4 ± 5.3 | 90.3% |
| Controls (n = 1959) | 54.3 ± 13.9 | 26.0 ± 4.9 | 90.0% | ||||||
| Total of all cohorts | - | - | 158,025 | - | - | Cases (n = 29,531) | - | - | - |
| Controls (n = 128,494) | - | - | - |
Meta-analysis of GWAS of CBP
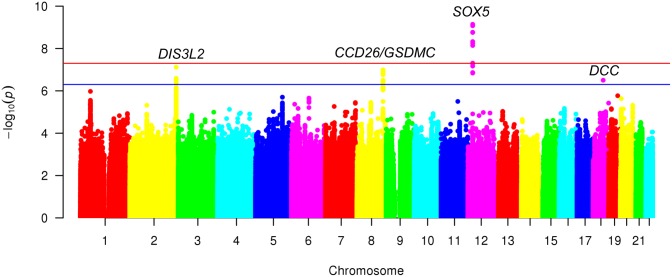
The characteristics of cohorts included in the discovery meta-analysis are shown in Table 1. The mean age of participants in each cohort ranged between 53–76 years. Within cohorts, mean age, BMI, and proportion of females was more often higher among CBP cases than among those without CBP. A quantile-quantile plot comparing the meta-analysis association results with those expected by chance is presented in S1 Fig. The LDsr intercept was 1.007 (standard error 0.006), λ was 1.114, and the LDsr ratio was 0.0581 (standard error 0.053), providing no evidence of inflation of p-values from population stratification. Meta-analysis results are summarized in the Manhattan plot shown in Fig 1.
Fig 1. Manhattan plot for meta-analysis (discovery) of GWAS of chronic back pain (n = 158,025).
GWAS = genome-wide association study. Results use the linkage disequilibrium score regression (LDSR) intercept as a correction factor. Red line depicts genome-wide statistical significance (P <5×10−8). Blue line depicts suggestive significance (P <5×10−7).
A genome-wide significant association (OR 1.08, p = 7.2×10−10) was found for rs12310519 on chromosome 12 in an intronic region of SOX5, with little evidence for heterogeneity (I2 = 0, p = 0.95) (Table 2, S2 Fig). Several other signals were in high LD (r2>0.8) with the top signal (S3 Fig), but none were independently associated with CBP in analyses conditional on rs12310519.
Table 2. Association results for chronic back pain: Meta-analysis (discovery), replication, and joint meta-analysis*.
| Discovery (Meta-analysis of CHARGE and PainOmics cohorts + UKB1)a (n = 158,025) |
Replication (UKB2)b (n = 283,752) |
Joint Meta-Analysis (Discovery-Replication)c (n = 441,777) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP rsID | Chr:Posd | Nearest Gene | Location | Alleles | EAF | OR | SE | p-value | I2 | Het. p-value | OR | SE | p-valueb | OR | SE | p-value |
| rs12310519e | 12:23975219 | SOX5 | intronic | T/C | 0.16 | 1.08 | 0.013 | 7.2 x 10−10 | 0 | 0.95 | 1.06 | 0.009 | 5.3 x 10−11 | 1.07 | 0.008 | 4.5 x 10−19 |
| rs1453867 | 2:232917899 | DIS3L2 | intronic | T/C | 0.65 | 0.95 | 0.010 | 7.7 x 10−8 | 13 | 0.31 | 0.98 | 0.007 | 0.021 | 0.97 | 0.006 | 3.9 x 10−7 |
| rs7833174 | 8:130718772 | CCDC26/GSDMC | intergenic | T/C | 0.77 | 1.06 | 0.011 | 1.0 x 10−7 | 0 | 0.71 | 1.04 | 0.008 | 3.7 x 10−7 | 1.05 | 0.007 | 4.4 x 10−13 |
| rs4384683 | 18:50379032 | DCC | intronic | A/G | 0.54 | 0.95 | 0.009 | 3.2 x 10−7 | 0 | 0.86 | 0.97 | 0.007 | 4.2 x 10−5 | 0.97 | 0.006 | 2.4 x 10−10 |
CHARGE = Cohorts for Heart and Aging Research in Genomic Epidemiology, UKB1 = UK Biobank participants from the interim data release[20], UKB2 = UK Biobank participants not included in the interim data release, chr:pos = chromosome:position, alleles = effect/other, EAF = effect allele frequency OR = odds ratio, het. = heterogeneity
*Top variant at each locus meeting suggestive or genome-wide significance level in discovery stage (p<5.0x10-7).
aAfter genomic control using the LD score regression intercept
bReplication for rs12310519. The threshold for significance in replication of rs12310519 was p<0.05 (0.05/1)
cThe threshold for genome-wide significance in joint analysis was p<5×10−8
dBuild GRCh37/hg19
ers115392701 has merged into rs12310519
No other variants achieved genome-wide significance, but variants in three other loci reached suggestive significance (Table 2, S3 Table, S4–S9 Figs): rs1453867 (OR 0.95, p = 7.7×10−8), located in an intronic region of chromosome 2 within DIS3L2; rs7833174 (OR 1.06, p = 1.0×10−7), located in an intergenic region on chromosome 8 between CCDC26 (a long non-coding RNA) and GSDMC; and rs4384683 (OR 0.95, p = 3.2×10−7), located in an intronic region of chromosome 18 within DCC. In each of these 3 regions, there was no other variant reaching the suggestive significance level in analyses conditional on the lead SNP in the region. Post hoc secondary analyses of the discovery sample showed effects of similar magnitude and direction between the CHARGE cohorts and the UKB interim data release for associations between the lead variants in the top 4 loci and CBP (S4 Table).
We examined these 4 top variants in 283,752 UKB individuals not included in the discovery sample (UKB2), including 50,915 cases (Table 2). For all 4 variants, the direction of association was the same in discovery and replication. The association for rs12310519 in SOX5 replicated in UKB2 (OR 1.06, p = 5.3×10−11), and exceeded genome-wide significance in the joint analysis (OR = 1.07, p = 4.5×10−19). Of the 3 suggestive-significance variants from the discovery stage, rs7833174 at CCDC26/GSDMC (OR 1.05, p = 4.4×10−13) and rs4384683 in DCC (OR 0.97, p = 2.4×10−10) exceeded genome-wide significance in the joint meta-analysis, but rs1453867 in DISL32 (OR 0.98, p = 3.9×10−7) did not (Table 2). Thus, we demonstrate genome-wide significant associations of CBP with loci tagged by rs12310519 (SOX5), rs7833174 (CCDC26/GSDMC), and rs4384683 (DCC), with replication for rs12310519 in SOX5.
Characterization of variants in SOX5, CCDC26/GSDMC, and DCC
Functional characterization followed the same steps for each of the 3 loci that achieved genome-wide significance in the joint meta-analysis. First, we examined cross-phenotype genetic associations between each lead SNP and traits with possible conceptual links to CBP, in look-ups of publicly and privately available GWAS datasets. Where the lead SNP was not present in a dataset, we examined associations with the variant in highest LD with the lead SNP. Second, we annotated lead variants and those in LD (r2≥0.6) for consequences on gene functions (using the combined annotation dependent depletion [CADD] score [21]), potential regulatory functions (using RegulomeDB score[22]), and effects on gene expression (using GTExv6 [23, 24]), and examined whether these variants resided in enhancer regions for selected tissues with connections to the spine or pain processing (using data from the Roadmap Epigenomics Consortium [25, 26]) (Methods, S1 Text).
SOX5
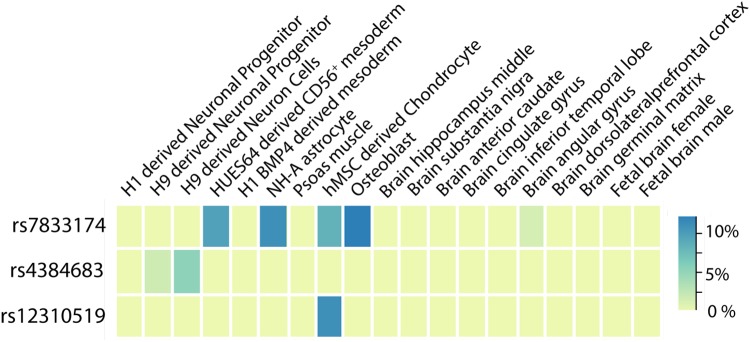
Among CBP-related traits examined, the lead SNP in SOX5, rs12310519, was most strongly associated with imaging-detected lumbar intervertebral disc degeneration (p = 1.1×10−4; S5 Table)[27]. The highest CADD score among SOX5 variants was 10.52 for the lead SNP in the region rs12310519, indicating it is predicted to be among the 10% most deleterious possible substitutions in the human genome (S1 Appendix). However, the overall regulatory potential of these variants was low according to RegulomeDB score (scores of 6 [‘minimal binding evidence’]) (S1 Appendix). There were no meaningful associations with gene expression using GTExv6. The lead SNP rs12310519 and variants in LD (r2>0.6) contained active enhancer marks in chondrogenic cells using Roadmap Epigenomics Consortium data (Fig 2, S1 Appendix).
Fig 2. Significant variants are co-localized with potential gene regulatory markers.
The heatmap depicts the percentage of variants in gene regulatory regions (associated with enhancers/promotors) in LD (r2>0.6) with rs7833174 (CCDC26/GSDMC), rs12310519 (SOX5), and rs4384683 (DCC). We examined epigenetic histone marks in selected cell types/tissues including chondrogenic, bone-related, neuronal and brain cells/tissues; mesodermal cells (related to notochord); and psoas muscle (located proximal to the lumbar spine).
CCDC26/GSDMC
In look-ups of GWAS of selected traits with possible links to CBP (S5 Table), the lead SNP rs7833174 was most strongly associated with height in UKB [28] (p = 1.3×10−59) and hip circumference in UKB [28] (p = 1.8×10−5). The lead SNP rs7833174 was also associated with radiographic hip osteoarthritis[29] (p = 4.9×10−4) (S5 Table). All variants in CCDC26/GSDMC that were suggestively associated with CBP showed cross-phenotypic associations with lumbar microdiscectomy for sciatica in a recent GWAS of Icelandic adults[30] (S6 Table, lowest p = 5.6×10−12). The direction of effect on these other phenotypes was the same as the direction of effect on CBP (i.e. the T allele associated with greater height, greater risk of radiographic hip osteoarthritis, and greater risk of lumbar discectomy for sciatica was also associated with greater CBP risk). The highest CADD score among CBP-associated variants at CCDC26/GSDMC was 18.75 for rs6470778, indicating that this SNP is predicted to be among the 5% most deleterious substitutions in the human genome, and the overall regulatory potential of these variants was substantial according to Regulome DB score (highest RegulomeDB score of 2b [‘likely to affect binding’]) (S1 Appendix). In examination of effects on gene expression using GTExv6, variants in LD with rs7833174 (r2>0.6) were also eQTLs for GSDMC expression in esophageal mucosa, skin, and skeletal muscle (S1 Appendix, p<5×10−8). The lead SNP rs7833174 and variants in LD (r2>0.6) contained active enhancer marks located in regulatory regions for mesodermal cells, astrocytes, chondrogenic cells, and osteoblasts in data from the Roadmap Epigenomics Consortium (Fig 2, S1 Appendix).
DCC
In look-ups of GWAS of selected traits with possible links to CBP (S5 Table), the lead SNP rs4384683 was associated with depressive symptoms[31](p = 5.9×10−4), with the same direction of effect (i.e the A allele was associated with less depressive symptoms and lower CBP risk). The highest CADD score among variants at DCC that were suggestively associated with CBP was 11.21 for rs2116378, indicating that rs2116378 is predicted to be among the 10% most deleterious substitutions in the human genome, but the overall regulatory potential of these variants was low according to Regulome DB score (highest RegulomeDB score of 5) (S1 Appendix). There were no meaningful associations with gene expression using GTExv6. In data from the Roadmap Epigenomics Consortium, the lead SNP rs4384683 and variants in LD (r2>0.6) contained active enhancer marks in H9 human embryonic stem cell-derived neural cells (Fig 2, S1 Appendix).
Secondary analyses to examine interrelationships with height
Post hoc analyses among UKB1 participants from the discovery sample (n = 120,023) indicated that associations with CBP for the lead variant in SOX5 were similar with and without adjustment for height as a covariate, and in conditional analyses accounting for height (S1 Text, S7 Table). Associations with CBP for the lead variant in CCDC26/GSDMC were also similar with and without adjustment for height as a covariate. However, associations with CBP were markedly diminished when conditional on the lead height-associated variant in the region, and associations with height were markedly diminished when conditioned on the lead CBP-associated variant in the region (S7 Table). This suggests that the same functional variant is responsible for association of CCDC26/GSDMC locus with height and CBP, although an alternative explanation is two functional variants in tight LD. Associations with CBP for the lead variant in DCC were similar with and without adjustment for height as a covariate, and in conditional analyses accounting for height (S7 Table).
To examine possible causal effects of height on CBP, we conducted a two-sample Mendelian randomization (MR) analysis using genetic variants associated with standing height in the GIANT consortium as the exposure, and the discovery phase GWAS meta-analysis of CBP as the outcome. Results of the two-sample MR using 326 SNPs and the instruments involved are available in the S2 Appendix. These instruments explained 10.1% of the variance in height, with an average SNP-height F-statistic of 78.5, indicating substantial instrument strength. ORs for CBP were 1.10 per standard deviation increase in height with inverse variance weighted (IVW) regression (p<0.0001). However, there was significant heterogeneity among SNPs (I2 = 0.35; p<0.0001), suggesting horizontal pleiotropy for at least some SNPs (S3 Appendix). Estimates with other MR methods were directionally consistent, and all but MR-Egger regression were statistically significant: an OR for CBP of 1.09 per standard deviation increase in height with MR-Egger regression (p = 0.19); 1.14 per standard deviation increase in height with the weighted median method (p<0.0001), and 1.17 per standard deviation increase in height with the weighted mode method (p = 0.02) (S3 Appendix). The magnitude of MR estimates after excluding 14 outlier SNPs were very similar to the two-sample MR using 326 SNPs, but with substantially less heterogeneity amongst SNPs (I2 = 0.12; p = 0.04), just exceeding nominal significance (S2 and S3 Appendices). MR-Egger intercepts were close to 0 with both the 326 SNP and 312 SNP instruments, and neither were statistically significant, suggesting no strong directional horizontal pleiotropy under the InSIDE (Instrument Strength Independent on Direct Effect) assumption (S3 Appendix).
Secondary analyses to examine for influence of relatedness in UK Biobank
GWAS analyses using linear mixed-effect models in UKB1 yielded associations between the 3 lead SNPs and CBP that were very similar to the original analyses using logistic regression in terms of statistical significance, indicating no meaningful influence on the study results due to relatedness (S8 Table).
Heritability of CBP and genetic correlations
SNP heritability of CBP on the liability scale was 7.6%. Partitioned heritability by functional category using stratified LD score regression showed significant enrichment (p = 0.0004) for regions conserved in mammals, with 2.6% of SNPs explaining 40% of the SNP heritability of CBP, without significant enrichment for other functional categories, including coding regions (S4 Appendix). This pattern of partitioned heritability was broadly similar across cell type groups, including central nervous system (CNS), connective tissue and bone, and skeletal muscle, among others (S4 Appendix). Genetic correlations of nominal significance (range of rg 0.17–0.31, p<0.05) were found with anthropometric traits involving obesity or body fat distribution (waist circumference, hip circumference, waist-hip ratios, overweight/obesity classes, and BMI), but not with height (S9 Table). Larger magnitude nominally significant (p<0.05) genetic correlations were also found with depression-related phenotypes (range of rg 0.46–0.52), self-reported osteoarthritis (rg = 0.63), and ICD-10-defined osteoarthritis (rg = 0.49) phenotypes.
Discussion
This study is the first meta-GWAS of CBP. This collaboration between two international consortia for genomic studies of complex traits in the USA and Europe incorporated data from 16 cohorts and more than 441,000 participants of European ancestry across discovery and replication samples. Our study identifies three novel associations with CBP for loci at SOX5, CCDC26/GSDMC, and DCC.
CBP was most strongly associated with rs12310519 in an intronic region of the SOX5 gene. The SOX genes are a family of transcription factors involved in virtually all phases of embryonic development, and are thought to determine the fate of many cell types. The SOX genes are defined by containing the HMG (‘high mobility group’) box of a gene involved in sex determination called SRY (‘sex determining region’) [32]. SOX5 and SOX6 have overlapping functions and work together in close coordination that is necessary for efficient chondrogenesis [33]. Inactivation of SOX5 leads to minor defects in cartilage and skeletogenesis in mice, whereas SOX5/SOX6 double knockouts have severe chondrodysplasia [34]. Together with SOX9, SOX5 and SOX6 are sometimes referred to as the ‘master chondrogenic SOX trio’ [33, 35]. Prior work indicates an important role for SOX5 in articular cartilage and osteoarthritis [36, 37], and such a role was also supported by our functional annotation showing that rs12310519 (and SNPs in high LD) overlapped with potential regulatory regions for chondrogenic cells. SOX5/6 are also essential for notochord development, and through this role they are critical in the formation of the vertebral column, including the intervertebral discs [33, 35, 38]. Inactivation of SOX5 and/or SOX6 in mice leads to a range of abnormalities in the development of spinal structures [38]. Although variants in SOX5 have not been reported in prior GWAS of limb osteoarthritis (knee, hip, or hand) [39–46], the association of SOX5 with CBP may involve the spinal structures specifically. Consistent with this, we found a cross-phenotypic association for the lead CBP-associated variant in SOX5 with imaging-detected lumbar intervertebral disc degeneration in a prior GWAS meta-analysis [27]. Future GWAS may also be useful to characterize other spine-related phenotypes besides disc degeneration, such as osteoarthritis of the zygapophyseal (‘facet’) joint, the only true synovial joint in the spine [47].
The intergenic variants at CCDC26/GSDMC associated with CBP in the current study were also previously found associated with lumbar microdiscectomy for sciatica due to intervertebral disc herniation.[30] These findings are intriguing, given that lumbar disc herniations (an aspect of lumbar disc degeneration) have long been implicated as a cause of some forms of back pain.[48] Recent studies have concluded that associations between imaging-detected lumbar disc herniation and CBP are of modest magnitude.[10, 11] This might explain the small magnitude association of the top variant at CCDC26/GSDMC with CBP in the current study (OR 1.08 in discovery), in contrast to the larger magnitude association seen with microdiscectomy for sciatica (OR 1.23). Functional characterization of these intergenic variants suggest the likely involvement of the gene GSDMC. GSDMC encodes the protein Gasdermin C, part of the GSDM family of genes that is expressed in epithelial tissues. Although the specific role of GSDMC in lumbar disc herniation and/or sciatica is unclear, GSDMC is associated with differential methylation patterns in osteoarthritis-related cartilage and subchondral bone cartilage, [49, 50] consistent with our findings that variants in LD with rs4384683 were located in potential regulatory regions in chondrocytes and osteoblasts. Our examination of univariate cross-phenotypic genetic associations for CBP-associated variants at CCDC26/GSDMC also suggest pleiotropy with radiographic hip OA for rs6470763.[29] Taken together, these data suggest interconnections between variants at CCDC26/GSDMC and CBP involving cartilage, osteoarthritis, and/or lumbar disc degeneration.
The third significant CBP-associated variant in our study was rs4384683, an intronic variant in the gene DCC (Deleted in Colorectal Carcinoma), which co-localized with regulatory regions in neural embryonic stem cells. DCC encodes a transmembrane protein that is a receptor for netrin-1, an axonal guidance molecule involved in the development of spinal and cortical commissural neurons.[51] Interactions between DCC and netrin-1 are among the best-studied axonal guidance processes, with key roles during development and in adulthood, and they also affect angiogenesis.[52, 53] Increased expression of netrin-1 and DCC occurs in degenerate human intervertebral discs compared to healthy control discs, and in nucleus pulposus compared to annulus fibrosis.[54] Netrin-1/DCC might therefore mediate neurovascular ingrowth into the intervertebral disc, which has long been implicated as a possible mechanism of chronic discogenic back pain.[54, 55] Given the well-known phenotypic correlation between depression and CBP[56], however, another possible explanation for the link between CBP and DCC (suggested by the cross-phenotype association of rs4384683 with depressive symptoms) is pleiotropy. Netrin-1/DCC interactions are also known to play a role in pain processing in the spinal cord in animal models of mechanical allodynia.[52] Taken together, this information suggests various potential mechanisms underlying the association between DCC and CBP, including nociceptive pathways and/or the involvement of mood.
Some epidemiological studies report that greater height confers increased risk of back pain [57–59], although a systematic review found no association.[60] Variants in CCDC26/GSDMC associated with CBP in our meta-analysis were also reported to be associated with height in prior GWAS; hence, post hoc analyses devoted special attention to the role of height in CBP. These region-specific analyses showed that the association of SOX5 and DCC variants with CBP was independent of height; however, CBP- and height-associated variants at CCDC26/GSDMC were tightly linked and could not be disentangled in conditional analyses (S7 Table), indicating that the association of variants in CCDC26/GSDMC with both CBP and height might be explained by biological pleiotropy or mediated pleiotropy (i.e. pleiotropy due to causal effects)[61]. Mendelian randomization analysis, drawing on information from hundreds of genetic markers distributed across the genome, suggested that height may have causal effects on CBP, although with a degree of heterogeneity suggesting horizontal pleiotropy for some SNPs. Such evidence of horizontal pleiotropy is common in MR studies of complex traits,[62] and can be seen even in MR studies of exposure-outcome relationships where causal effects are known.[63] Taken together, our findings suggest a causal component to the relationship between height and CBP, but do not exclude that height and CBP are also linked by biological pleiotropy. Further more advanced studies should be conducted to corroborate our findings. Prior studies demonstrating the vital role of SOX5 in normal vertebrate development [33, 35, 38] are a reminder that measurements of human height used for GWAS may also reflect vertebral column development; if associations with CBP and height are connected via development of the vertebral column, it will be difficult to distinguish pleiotropy and causality using genetic studies alone.
SNP-based heritability in the current study (8%) was considerably lower than estimates from twin studies (~40%). This is a common situation with modern methods of estimating heritability using genotype data, since such estimates reflect only one aspect of narrow-sense heritability captured by the additive genetic components of common variants, excluding the contributions of rare variants, non-additive effects, epistasis, or gene-environment interactions.[64] Similar to what is seen with many other human traits,[65] there was significant enrichment of SNP-based heritability of CBP for genetic regions that are conserved in mammals. Despite the modest heritability of CBP (and other self-reported traits), we found significant and large magnitude genetic correlations between CBP and other phenotypes that may be risk factors for CBP or consequences of CBP, such as depression-, osteoarthritis- and obesity-related traits (but not height). Future GWAS of CBP may benefit from taking these relationships into account, either as covariates, or in multivariate GWAS designs.
A distinguishing feature of the current study as compared to many other GWAS is that the CBP phenotype examined represents a symptom, rather than a disease or a biomarker. Although successful GWAS of self-reported symptoms have been conducted which replicate associations seen with more specific disease phenotypes,[66] our findings highlight potential challenges of GWAS of CBP: despite being one of the largest international studies of CBP ever conducted, our study detected only 3 significant associations with CBP. Still larger sample sizes will be needed in future discovery efforts using this phenotype, or different genetic approaches will be needed. A consequence of the nonspecific nature of the CBP phenotype is that, unlike other musculoskeletal phenotypes such as osteoarthritis, the tissue correlates optimal for conducting functional follow-up studies of findings from CBP GWAS are very unclear. Most animal models for back pain rely on specific mechanisms of inducing pain, such as injuries to the intervertebral disc, zygapophyseal (‘facet’) joint, dorsal root ganglion, or muscle.[67] However, each of these mechanisms likely explain only a certain portion of back pain cases, and do not encompass the important psychosocial aspects of pain and pain reporting that are highly relevant in humans. Despite the importance of psychosocial factors, our meta-GWAS findings are a reminder that structural/anatomic factors involving spinal degeneration, such as disc herniation or osteoarthritis of spinal structures (e.g. facet joints), remain potentially important contributors to the CBP. While our study accounted for age by statistical adjustment, the meta-analysis design including multiple cohorts of older adults may have led to an overrepresentation of genetic variants associated with age-related conditions, such as osteoarthritis. Future GWAS of CBP may benefit from a broader age range of participants, stratification by back pain subtypes, simultaneously studying CBP and spinal degeneration/fracture phenotypes, and examination of interactions between genetic markers for spinal degeneration and markers for pain processing or axonal signaling (including DCC and netrin-1).
Strengths of our study include its multicohort design and large sample size. A potential limitation of our study was heterogeneity of the CBP phenotype used, a consequence of pooling data from numerous cohorts using different definitions. Although this approach helped identify genetic associations shared across CBP subtypes, it might obscure associations pertinent to specific subtypes of back pain. As an example, we examined chronic back pain rather than chronic low back pain, since few of the included cohorts had available question items that isolated the low back region specifically. Given the high agreement between general back pain questions and low back-specific questions,[68] and since mid/upper back pain without concurrent low back pain is uncommon,[69] we expect that our results largely reflect genetic associations with low back pain.[70] Despite phenotype heterogeneity, which would be expected to bias the study towards the null, we successfully identified several associations of statistical significance. Recent efforts to standardize CBP definitions may help to limit phenotype heterogeneity in future meta-GWAS of CBP.[3] Another aspect of the phenotype used in our meta-analysis was that individuals with back pain of less than 3–6 months duration were included as controls. This was done deliberately so as to focus on back pain of chronic duration as the phenotype of interest. That said, GWAS examining back pain of any duration, or analyses excluding those with non-chronic back pain, may find different results. Another possible study limitation was lack of independence in the replication sample of UK Biobank participants from UKB2, given the same study base and methods between the UKB1 and UKB2 subcohorts. Our secondary analyses using linear mixed-effect models demonstrated similar SNP-CBP associations for our top hits when accounting for relatedness within UKB1, but the problem of relatedness across the two subcohorts (UKB1 vs. UKB2) remains. Finally, a limitation of this meta-analysis was that only autosomal variants were analysed, since some included cohorts did not analyze the X chromosome.
In summary, this meta-analysis of GWAS of CBP identified novel genetic associations with CBP at SOX5, CCDC26/GSDMC, and DCC. Analysis of data from other GWAS and functional genomics experiments suggest possible pleiotropic effects of these loci on other traits including cartilage, osteoarthritis, lumbar disc degeneration, depression, and height/vertebral development, and possible causal effects on CBP mediated through height.
Methods
Study design and populations
Discovery meta-analysis included adults of European ancestry from 16 population- and community-based cohorts: Cardiovascular Health Study (CHS), Framingham Heart Study (FHS), Generation Scotland (GS), Johnston County Osteoarthritis Project (JoCo), Osteoporotic Fractures in Men (MrOS) Sweden (MrOS-Gothenburg and MrOS-Malmo), MrOS US, Osteoarthritis Initiative (OAI), Rotterdam Study (RS1, RS2, and RS3), Study of Osteoporotic Fractures (SOF), 10,001 Dalmatians (Vis and Korcula), TwinsUK, and UKB participants from the interim data release[20]. Replication was conducted among UKB European ancestry participants not included in the discovery stage (UKB2), and a joint (discovery-replication) meta-analysis was performed. The separation of analyzed data from UKB into discovery (UKB1) and replication phases (UKB2) reflects the history of this scientific collaboration, in that our initial meta-analysis plan included only the UKB data then available to us and for which we had obtained approval to use (UKB1 [the interim data release]). By the time our meta-analysis was completed, all UKB data had become available; the remainder of UKB data was therefore used for replication. Detailed descriptions of the study cohorts are provided in Table 1 and the Supplemental Methods. This meta-analysis was approved by the Research and Development Committee of VA Puget Sound Health Care System (RDIS 0010, MIRB 00903). Institutional Review Board/Ethics Committee approvals at the individual study sites include those listed in the S2 Text. Written or electronic consent was provided for all studies.
Chronic back pain (CBP) phenotype
There is no “gold-standard” definition for CBP. Consistent with the most commonly accepted clinical and research definitions for CBP [3, 4], CBP cases were defined in this study using one of 3 definitions depending on the cohort (Table 1, S10 Table): 1) ≥3 months of back pain, 2) ≥6 months of back pain, and 3) ≥1 month of back pain in consecutive years (reflecting ≥12 months of back pain). For each cohort, the comparison group (“controls”) was comprised of those who reported not having back pain or reported back pain of insufficient duration to be included as a case. This study used a general definition examining chronic ‘back pain’, as opposed to a more specific chronic ‘low back pain’ definition, due to the fact that most of the included cohorts did not include question items permitting localization of pain to the low back or lumbar region specifically.
Genotyping
Details of genotyping, quality control, imputation methods, and genome-wide analysis for each cohort were study-specific (S1 and S2 Tables). In brief, genotyping was performed using commercially available genome-wide arrays. Imputation of single nucleotide polymorphisms (SNPs) and insertions/deletions (indels) was performed using reference panels from 1000 Genomes phase 1 version 3 or phase 3,[71] or the Haplotype Reference Consortium.[72] Analyses of UKB participants was restricted to the White British ancestry subset who self-report as White, British, and have very similar genetic ancestry backgrounds based on the results of principal components analysis (PCA); further quality control followed recommended practices for UKB[73] (S1 Table).
Statistical analysis
We conducted genome-wide association analyses in each of the 16 cohorts, and subsequent meta-analysis of autosomal SNPs to combine results from all cohorts. Each site conducted GWAS using logistic regression models with additive genetic effects to test for associations between each variant and CBP as a binary trait. These models adjusted for age, sex, study-specific covariates, and population substructure using principal components (S2 Table). Height and body mass index (BMI), calculated as weight in kilograms divided by height in meters squared, were not included as covariates in site-specific GWAS, since these traits might lie along the causal pathway or (in the case of BMI) reflect a consequence of CBP. Harmonization and quality control of GWAS results from each cohort were conducted using the EasyQC software package in the R statistical environment (v3.2.2), using methods described previously.[74] After removal of SNPs with low minor allele frequencies (<0.005 for UKB, <0.03 for Vis, <0.01 for other cohorts) or imputation quality (<0.7 for UKB, <0.6 for other cohorts), deviation from Hardy-Weinberg equilibrium (p < 1 x 10–6), low number of cases (<15) or controls (<15), large absolute values of beta coefficient (≥10), and low minor allele count (≤10), call rate <0.95, the range of SNPs included in the meta-analysis was between 6,205,227 (Croatia-Vis) and 9,775,703 (MrOs-Gothenburg) (S2 Table). Fixed-effect inverse-variance weighted meta-analysis was performed with METAL version 2011-03-25 (http://csg.sph.umich.edu/abecasis/metal/), using the LDsr intercept as a correction factor. The meta-analysis was filtered for variants with fewer than 125,000 informative participants, to ensure that SNP-CBP associations were informed by a plurality of cohorts, and not only the UKB interim data release. For this reason, only variants with MAF<0.01 (SNPs) were included in the meta-analysis. Quality control and meta-analysis were conducted twice, independently of each other, by researchers at the University of Washington (MP and PS) and at PolyOmica (YT, YA, and LCK). The results from the two centers were compared to ensure accuracy. Q-Q and Manhattan plots were generated in R. We conducted conditional and joint (COJO) analysis using summary data (http://cnsgenomics.com/software/gcta/#About) to examine associations conditional on the most significant variant at each locus (S1 Text).
The most highly-associated variants at genome-wide significant loci were subjected to replication among UKB participants not included in the discovery sample (UKB2). Analysis in UKB2 used logistic regression with additive genetic effects, adjusting for age, sex, array, and principal components (S2 Table); significant replication was defined using a Bonferroni-corrected threshold of p<0.05 divided by the number of genome-wide significant loci. The most highly-associated variants at loci with suggestive significance were selected for a joint (discovery-replication) meta-analysis using p<5×10−8 to define genome-wide significance. Further details regarding analysis are provided in S1 Text. A post hoc analysis was conducted to stratify the discovery phase meta-analysis by the CHARGE cohorts (meta-analysis of 15 GWAS) vs. the UKB interim data release cohort (S4 Table).
For genome-wide significant variants, we examined GWAS associations with selected traits with possible links to CBP (anthropometrics, arthritis, depression and depressive symptoms, and imaging-based spinal degeneration) in publicly and privately available GWAS datasets. We conducted functional annotation using FUMA (http://fuma.ctglab.nl). FUMA draws upon multiple publicly available databases, annotating variants for consequences on gene functions using the combined annotation dependent depletion (CADD) score,[21] potential regulatory functions (RegulomeDB score),[22] and effects on gene expression using expression quantitative trait loci (eQTLs) of different tissue types (GTExv6) [23, 24] (S1 Text). The higher the CADD score, the more potentially deleterious is the variant. A CADD score of ≥10 indicates a variant predicted to be among the 10% most deleterious substitutions involving the human genome, a score of ≥20 indicates a variant among the 1% most deleterious, and so forth.[21] We used data from the Roadmap Epigenomics Project to evaluate whether the lead variants at each locus and those in LD (r2>0.6) reside in enhancer regions for selected tissues with possible conceptual connections to the spine via roles in chondrogenesis, vertebral development, muscle, and pain processing in the CNS.[25, 26].
Because two CBP-associated variants were found to be associated with height in prior published GWAS, we conducted post hoc region-specific secondary GWAS analyses accounting for height, among UKB participants from the discovery stage. Further details of functional annotation and secondary analyses are provided in S1 Text. We also conducted a two-sample Mendelian randomization to examine potential causal effects of height on CBP using significant variants associated with standing height from the GIANT consortium as the exposure, and the discovery phase meta-analysis of CBP, using the R package MRbase. We used the inverse-variance weighted regression (IVW) approach as our primary analysis method,[75] and additional analyses with other MR methods (MR-Egger regression, weighted median function, and weighted mode); presenting the results yielded from different MR methods is recommended to demonstrate sensitivity to different patterns of assumption violations.[63, 76] We examined heterogeneity among SNPs using forest plots, funnel plots, heterogeneity statistics, and the MR-Egger intercept test for directional horizontal pleiotropy. Further details of MR methods are provided in the S1 Text.
Given that 30% of UKB participants are related to at least one other person in the cohort, we also conducted post hoc secondary GWAS in UKB1 to examine whether relatedness might have influenced our results. These analyses used linear mixed-effect models (BOLT-LMM), adjusting for age, sex, study-specific covariates, and principal components. The statistical significance of GWAS results for UKB1 using BOLT-LMM were descriptively compared with the original results using logistic regression, for the lead variants achieving suggestive significance in the GWAS meta-analysis.
Finally, we used LDsr of summary-level GWAS results from the discovery stage to estimate heritability due to common autosomal SNPs and genetic correlations.[18] We transformed the observed SNP heritability to the liability scale, in order to make heritability estimates for CBP comparable with traditional heritability estimates from twin studies.[64] We used stratified LDsr to partition heritability across functional categories of the genome, using methods described previously.[65] The threshold for determining the statistical significance of 53 functional categories in partitioning heritability was set at p<9.4 x 10–4 (0.05/53). Further details of methods for partitioning heritability are provided in the S4 Appendix. We used cross-trait LDsr and publicly available meta-GWAS results from LDhub to examine genetic correlations with selected traits with possible links to CBP: anthropometrics (height, waist/hip circumference, BMI, and overweight/obesity), depression and depressive symptoms, osteoarthritis, and rheumatoid arthritis.[77, 78]
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
*LD score regression intercept = 1.0067. GWAS = genome-wide association study.
(PDF)
rs115392701 has merged into rs12310519. Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs115392701 is shown using the colors in the figure legend (rs115392701 has merged into rs12310519).
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs1453867 is shown using the colors in the figure legend.
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs7833174 is shown using the colors in the figure legend.
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs4384683 is shown using the colors in the figure legend.
(PDF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
Acknowledgments
Rotterdam Study: The Rotterdam Study thanks Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera and Marjolein Peters, and Carolina Medina-Gomez, for their help in creating the main GWAS database; and Karol Estrada, and Carolina Medina-Gomez, for the creation and analysis of imputed data. 10,001 Dalmatians: The 10,001 Dalmatians acknowledges the invaluable contributions of the recruitment team in Korcula and the administrative teams in Croatia and Edinburgh.
Data Availability
Meta-GWAS data are available from the dbGaP CHARGE Summary Results site under accession number phs000930, and can be accessed as described in Rich SS, Wang ZY, Sturcke A, Ziyabari L, Feolo M, O'Donnell CJ, et al. Rapid evaluation of phenotypes, SNPs and results through the dbGaP CHARGE Summary Results site. Nat Genet. 2016;48(7):702-3. doi: 10.1038/ng.3582. PubMed PMID: 27350599; PubMed Central PMCID: PMC5787851. The dataset can also be accessed from https://gwasarchive.org under 'Chronic back pain'.
Funding Statement
Infrastructure for the CHARGE Consortium is supported in part by the National Heart, Lung, and Blood Institute grants R01HL105756. This study was supported by the European Commissions’ Seventh Framework Programme funded project PainOmics (Grant agreement n. 602736) Dr. Suri was supported by VA Career Development Award # 1IK2RX001515 from the United States (U.S.) Department of Veterans Affairs Rehabilitation Research and Development (RR&D) Service. Dr. Suri is a Staff Physician at the VA Puget Sound Health Care System. The contents of this work do not represent the views of the U.S. Department of Veterans Affairs or the United States Government. Cardiovascular Health Study: This CHS research was supported by NHLBI contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and NHLBI grants U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, R01HL085251, and R01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the National Institute on Aging (NIA). A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the National Center for Advancing Translational Sciences, CTSI grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Framingham Heart Study: From the Framingham Heart Study of the National Heart Lung and Blood Institute of the National Institutes of Health and Boston University School of Medicine. This work was supported by the National Heart, Lung and Blood Institute's Framingham Heart Study (Contract No. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (Contract No. N02-HL-6-4278), and by grants from the National Institute of Neurological Disorders and Stroke (NS17950) and the National Institute of Aging, (AG08122, AG16495). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Framingham Heart Study or Boston University School of Medicine. Generation Scotland: Generation Scotland (GS) received core funding from the Chief Scientist Office of the Scottish Government Health Directorates CZD/16/6 and the Scottish Funding Council HR03006. Genotyping of samples was carried out by the Genetics Core Laboratory at the Wellcome Trust Clinical Research Facility, Edinburgh, Scotland and was funded by the Medical Research Council UK and the Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” (STRADL) Reference 104036/Z/14/Z). Johnston County Osteoarthritis Project: The JoCo is supported in part by S043, S1734, & S3486 from the CDC/Association of Schools of Public Health; 5-P60-AR30701 & 5-P60-AR49465-03 from NIAMS/NIH; genotyping was supported by Algynomics, Inc. Mr. Os Sweden: MrOS Sweden is supported by the Swedish Research Council, the Swedish Foundation for Strategic Research, the ALF/LUA research grant in Gothenburg, the Lundberg Foundation, the Knut and Alice Wallenberg Foundation, the Torsten Soderberg Foundation, and the Novo Nordisk Foundation, the ALF/FoUU research grant in Skane, Herman Järnhardts, Kocks and Österluds Foundations. Osteoporotic Fractures in Men (Mr Os) US: The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), the National Center for Research Resources (NCRR), and NIH Roadmap for Medical Research under the following grant numbers: U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810, U01 AG042140, U01 AG042143, U01 AG042124, U01 AG042145, U01 AG042139, U01 AG042168, U01 AR066160, UL1 TR000128, and UL1 RR024140. The National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2ARO58973. Osteoarthritis Initiative (OAI): This study was supported by the American Recovery and Reinvestment Act (ARRA) through grant number RC2-AR-058950 from NIAMS/NIH. The OAI is public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the NIH. Additional support was provided by NIH grant P30-DK072488. Rotterdam Study: The Rotterdam Study is supported by the Erasmus Medical Center and Erasmus University, Rotterdam; the Netherlands Organization for Scientific Research (NWO), the Netherlands Organization for Health Research and Development (ZonMw), the Research Institute for Diseases in the Elderly (RIDE), the Ministry of Education, Culture and Science, the Ministry for Health, Welfare and Sports, the European Commission (DG XII), and the Municipality of Rotterdam. The generation and management of GWAS genotype data for the Rotterdam Study (RS I, RS II, RS III) was executed by the Human Genotyping Facility of the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, Rotterdam, The Netherlands. The GWAS datasets are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012), the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) Netherlands Consortium for Healthy Aging (NCHA), project nr. 050-060-810. Study of Osteoporotic Fractures: The Study of Osteoporotic Fractures (SOF) is supported by National Institutes of Health funding. The National Institute on Aging (NIA) provides support under the following grant numbers: R01 AG005407, R01 AR35582, R01 AR35583, R01 AR35584, R01 AG005394, R01 AG027574, and R01 AG027576. The National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) provides funding for the MrOS ancillary study ‘GWAS in MrOS and SOF’ under the grant number RC2ARO58973. 10,001 Dalmatians: Support for field work from University of Split and Zagreb Medical Schools, the Institute for Antropological Research in Zagreb and the Croatian Institute for Public Health. The SNP genotyping for the Vis cohort was performed in the core genotyping laboratory of the Wellcome Trust Clinical Research Facility at the Western General Hospital, Edinburgh, Scotland. The SNP genotyping for the Korcula cohort was performed in Helmholtz Zentrum München, Neuherberg, Germany. The SNP genotyping was performed by AROS Applied Biotechnology, Aarhus, Denmark. The study was funded by the Medical Research Council UK, The Croatian Ministry of Science, Education and Sports (grant 216-1080315-0302), the European Union framework program 6 EUROSPAN project (contract no. LSHG-CT-2006-018947), the Croatian Science Foundation (grant 8875), the Centre for Research Excellence in Personalized Medicine and the Centre of Competencies for Integrative Treatment, Prevention and Rehabilitation using TMS. TwinsUK: TwinsUK receives funding from the Wellcome Trust; European Community’s Seventh Framework Programme (FP7/2007-2013 to Twins UK); the National Institute for Health Research (NIHR) Clinical Research Facility at Guy’s & St Thomas’ NHS Foundation Trust and NIHR Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. UK Biobank: This research has been conducted using the UK Biobank Resource (project # 18219). We are grateful to the UK Biobank participants for making such research possible. Dr. Smith is a Research Scientist at the VA Puget Sound Health Care System. The work of Dr. Tsepilov was partly supported by the Russian Ministry of Science and Education under the 5-100 Excellence Programme. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hoy D, March L, Brooks P, Blyth F, Woolf A, Bain C, et al. The global burden of low back pain: estimates from the Global Burden of Disease 2010 study. Annals of the rheumatic diseases. 2014;73(6):968–74. Epub 2014/03/26. 10.1136/annrheumdis-2013-204428 . [DOI] [PubMed] [Google Scholar]
- 2.Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. The Journal of bone and joint surgery. 2006;88 Suppl 2:21–4. 10.2106/JBJS.E.01273 . [DOI] [PubMed] [Google Scholar]
- 3.Deyo RA, Dworkin SF, Amtmann D, Andersson G, Borenstein D, Carragee E, et al. Report of the NIH Task Force on Research Standards for Chronic Low Back Pain. The spine journal: official journal of the North American Spine Society. 2014;14(8):1375–91. Epub 2014/06/22. 10.1016/j.spinee.2014.05.002 . [DOI] [PubMed] [Google Scholar]
- 4.Meucci RD, Fassa AG, Faria NM. Prevalence of chronic low back pain: systematic review. Rev Saude Publica. 2015;49 10.1590/S0034-8910.2015049005874 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47(7):702–9. 10.1038/ng.3285 . [DOI] [PubMed] [Google Scholar]
- 6.Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, et al. MaTCH (Meta-analysis of Twin Correlations and Heritability) web application 2017 [cited 2017 May 25]. http://match.ctglab.nl/-/specific/plot1.
- 7.Ferreira PH, Beckenkamp P, Maher CG, Hopper JL, Ferreira ML. Nature or nurture in low back pain? Results of a systematic review of studies based on twin samples. Eur J Pain. 2013. Epub 2013/01/22. . [DOI] [PubMed] [Google Scholar]
- 8.Guo TM, Liu M, Zhang YG, Guo WT, Wu SX. Association between Caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res. 2011;52(2):133–8. 10.3109/03008207.2010.487621 . [DOI] [PubMed] [Google Scholar]
- 9.Omair A, Lie BA, Reikeras O, Holden M, Brox JI. Genetic contribution of catechol-O-methyltransferase variants in treatment outcome of low back pain: a prospective genetic association study. BMC Musculoskelet Disord. 2012;13:76 Epub 2012/05/23. 10.1186/1471-2474-13-76 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou D, Samartzis D, Bellabarba C, Patel A, Luk KD, Kisser JM, et al. Degenerative magnetic resonance imaging changes in patients with chronic low back pain: a systematic review. Spine (Phila Pa 1976). 2011;36(21 Suppl):S43–53. Epub 2011/10/05. 10.1097/BRS.0b013e31822ef700 . [DOI] [PubMed] [Google Scholar]
- 11.Endean A, Palmer KT, Coggon D. Potential of magnetic resonance imaging findings to refine case definition for mechanical low back pain in epidemiological studies: a systematic review. Spine (Phila Pa 1976). 2011;36(2):160–9. Epub 2010/08/27. 10.1097/BRS.0b013e3181cd9adb . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battie MC, Videman T, Levalahti E, Gill K, Kaprio J. Heritability of low back pain and the role of disc degeneration. Pain. 2007;131(3):272–80. 10.1016/j.pain.2007.01.010 . [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Raecke R, Niemeier A, Ihle K, Ruether W, May A. Structural brain changes in chronic pain reflect probably neither damage nor atrophy. PLoS One. 2013;8(2):e54475 Epub 2013/02/14. 10.1371/journal.pone.0054475 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baliki MN, Petre B, Torbey S, Herrmann KM, Huang L, Schnitzer TJ, et al. Corticostriatal functional connectivity predicts transition to chronic back pain. Nat Neurosci. 2012;15(8):1117–9. Epub 2012/07/04. 10.1038/nn.3153 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seminowicz DA, Wideman TH, Naso L, Hatami-Khoroushahi Z, Fallatah S, Ware MA, et al. Effective treatment of chronic low back pain in humans reverses abnormal brain anatomy and function. J Neurosci. 2011;31(20):7540–50. Epub 2011/05/20. 10.1523/JNEUROSCI.5280-10.2011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou R, Shekelle P. Will this patient develop persistent disabling low back pain? Jama. 2010;303(13):1295–302. Epub 2010/04/08. 10.1001/jama.2010.344 . [DOI] [PubMed] [Google Scholar]
- 17.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73–80. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291–5. 10.1038/ng.3211 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suri P, Hunter DJ, Rainville J, Kalichman L, Katz JN. Presence and Extent of Severe Facet Joint Osteoarthritis are Associated with Back Pain In Older Adults. Osteoarthritis Cartilage. 2013;21(9):1199–206. 10.1016/j.joca.2013.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.UK Biobank. Genotype imputation and genetic association studies of UK Biobank Interim Data Release, May 2015. 2015.
- 21.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–5. 10.1038/ng.2892 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7. 10.1101/gr.137323.112 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–60. 10.1126/science.1262110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826 10.1038/s41467-017-01261-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–6. 10.1038/nmeth.1906 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–30. 10.1038/nature14248 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams FM, Bansal AT, van Meurs JB, Bell JT, Meulenbelt I, Suri P, et al. Novel genetic variants associated with lumbar disc degeneration in northern Europeans: a meta-analysis of 4600 subjects. Annals of the rheumatic diseases. 2013;72(7):1141–8. 10.1136/annrheumdis-2012-201551 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neale Lab. Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank 2017 [cited 2018 March 25, 2018]. https://docs.google.com/spreadsheets/d/1b3oGI2lUt57BcuHttWaZotQcI0-mBRPyZihz87Ms_No/edit-gid=1209628142.
- 29.Rodriguez-Fontenla C, Calaza M, Evangelou E, Valdes AM, Arden N, Blanco FJ, et al. Assessment of osteoarthritis candidate genes in a meta-analysis of nine genome-wide association studies. Arthritis Rheumatol. 2014;66(4):940–9. 10.1002/art.38300 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjornsdottir G, Benonisdottir S, Sveinbjornsson G, Styrkarsdottir U, Thorleifsson G, Walters GB, et al. Sequence variant at 8q24.21 associates with sciatica caused by lumbar disc herniation. Nat Commun. 2017;8:14265 10.1038/ncomms14265 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48(6):624–33. 10.1038/ng.3552 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hou L, Srivastava Y, Jauch R. Molecular basis for the genome engagement by Sox proteins. Semin Cell Dev Biol. 2017;63:2–12. 10.1016/j.semcdb.2016.08.005 . [DOI] [PubMed] [Google Scholar]
- 33.Liu CF, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43(17):8183–203. 10.1093/nar/gkv688 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smits P, Li P, Mandel J, Zhang Z, Deng JM, Behringer RR, et al. The transcription factors L-Sox5 and Sox6 are essential for cartilage formation. Dev Cell. 2001;1(2):277–90. . [DOI] [PubMed] [Google Scholar]
- 35.Liu CF, Samsa WE, Zhou G, Lefebvre V. Transcriptional control of chondrocyte specification and differentiation. Semin Cell Dev Biol. 2017;62:34–49. 10.1016/j.semcdb.2016.10.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu J, Kang Y, Liao WM, Yu L. MiR-194 regulates chondrogenic differentiation of human adipose-derived stem cells by targeting Sox5. PLoS One. 2012;7(3):e31861 10.1371/journal.pone.0031861 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budd E, de Andres MC, Sanchez-Elsner T, Oreffo ROC. MiR-146b is down-regulated during the chondrogenic differentiation of human bone marrow derived skeletal stem cells and up-regulated in osteoarthritis. Sci Rep. 2017;7:46704 10.1038/srep46704 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smits P, Lefebvre V. Sox5 and Sox6 are required for notochord extracellular matrix sheath formation, notochord cell survival and development of the nucleus pulposus of intervertebral discs. Development. 2003;130(6):1135–48. . [DOI] [PubMed] [Google Scholar]
- 39.arc OC, arc OC, Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380(9844):815–23. 10.1016/S0140-6736(12)60681-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elliott KS, Chapman K, Day-Williams A, Panoutsopoulou K, Southam L, Lindgren CM, et al. Evaluation of the genetic overlap between osteoarthritis with body mass index and height using genome-wide association scan data. Annals of the rheumatic diseases. 2013;72(6):935–41. 10.1136/annrheumdis-2012-202081 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evangelou E, Kerkhof HJ, Styrkarsdottir U, Ntzani EE, Bos SD, Esko T, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Annals of the rheumatic diseases. 2014;73(12):2130–6. 10.1136/annrheumdis-2012-203114 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, Meulenbelt I, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Annals of the rheumatic diseases. 2011;70(2):349–55. 10.1136/ard.2010.132787 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans DS, Cailotto F, Parimi N, Valdes AM, Castano-Betancourt MC, Liu Y, et al. Genome-wide association and functional studies identify a role for IGFBP3 in hip osteoarthritis. Annals of the rheumatic diseases. 2015;74(10):1861–7. 10.1136/annrheumdis-2013-205020 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hollander WD, Boer CG, Hart DJ, Yau MS, Ramos YFM, Metrustry S, et al. Genome-wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Annals of the rheumatic diseases. 2017. 10.1136/annrheumdis-2017-211214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62(2):499–510. Epub 2010/01/30. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yau MS, Yerges-Armstrong LM, Liu Y, Lewis CE, Duggan DJ, Renner JB, et al. Genome-Wide Association Study of Radiographic Knee Osteoarthritis in North American Caucasians. Arthritis Rheumatol. 2017;69(2):343–51. 10.1002/art.39932 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gellhorn AC, Katz JN, Suri P. Osteoarthritis of the spine: the facet joints. Nat Rev Rheumatol. 2013;9(4):216–24. Epub 2012/11/14. 10.1038/nrrheum.2012.199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truumees E. A history of lumbar disc herniation from Hippocrates to the 1990s. Clinical orthopaedics and related research. 2015;473(6):1885–95. 10.1007/s11999-014-3633-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Fukui N, Yahata M, Katsuragawa Y, Tashiro T, Ikegawa S, et al. Genome-wide DNA methylation profile implicates potential cartilage regeneration at the late stage of knee osteoarthritis. Osteoarthritis Cartilage. 2016;24(5):835–43. 10.1016/j.joca.2015.12.013 . [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Fukui N, Yahata M, Katsuragawa Y, Tashiro T, Ikegawa S, et al. Identification of DNA methylation changes associated with disease progression in subchondral bone with site-matched cartilage in knee osteoarthritis. Sci Rep. 2016;6:34460 10.1038/srep34460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finci L, Zhang Y, Meijers R, Wang JH. Signaling mechanism of the netrin-1 receptor DCC in axon guidance. Prog Biophys Mol Biol. 2015;118(3):153–60. 10.1016/j.pbiomolbio.2015.04.001 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu CH, Yuan XC, Gao F, Li HP, Cao J, Liu YS, et al. Netrin-1 Contributes to Myelinated Afferent Fiber Sprouting and Neuropathic Pain. Mol Neurobiol. 2016;53(8):5640–51. 10.1007/s12035-015-9482-x . [DOI] [PubMed] [Google Scholar]
- 53.Dun XP, Parkinson DB. Role of Netrin-1 Signaling in Nerve Regeneration. Int J Mol Sci. 2017;18(3). 10.3390/ijms18030491 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bu G, Hou S, Ren D, Wu Y, Shang W, Huang W. Increased expression of netrin-1 and its deleted in colorectal cancer receptor in human diseased lumbar intervertebral disc compared with autopsy control. Spine (Phila Pa 1976). 2012;37(25):2074–81. 10.1097/BRS.0b013e31825d4ebc . [DOI] [PubMed] [Google Scholar]
- 55.Freemont AJ, Peacock TE, Goupille P, Hoyland JA, O'Brien J, Jayson MI. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet. 1997;350(9072):178–81. . [DOI] [PubMed] [Google Scholar]
- 56.Pinheiro MB, Ferreira ML, Refshauge K, Colodro-Conde L, Carrillo E, Hopper JL, et al. Genetics and the environment affect the relationship between depression and low back pain: a co-twin control study of Spanish twins. Pain. 2015;156(3):496–503. 10.1097/01.j.pain.0000460330.56256.25 . [DOI] [PubMed] [Google Scholar]
- 57.Heuch I, Heuch I, Hagen K, Zwart JA. Association between body height and chronic low back pain: a follow-up in the Nord-Trondelag Health Study. BMJ Open. 2015;5(6):e006983 10.1136/bmjopen-2014-006983 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hershkovich O, Friedlander A, Gordon B, Arzi H, Derazne E, Tzur D, et al. Associations of body mass index and body height with low back pain in 829,791 adolescents. Am J Epidemiol. 2013;178(4):603–9. 10.1093/aje/kwt019 . [DOI] [PubMed] [Google Scholar]
- 59.Smedley J, Egger P, Cooper C, Coggon D. Prospective cohort study of predictors of incident low back pain in nurses. BMJ (Clinical research ed. 1997;314(7089):1225–8. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor JB, Goode AP, George SZ, Cook CE. Incidence and risk factors for first-time incident low back pain: a systematic review and meta-analysis. The spine journal: official journal of the North American Spine Society. 2014;14(10):2299–319. Epub 2014/01/28. 10.1016/j.spinee.2014.01.026 . [DOI] [PubMed] [Google Scholar]
- 61.Solovieff N, Cotsapas C, Lee PH, Purcell SM, Smoller JW. Pleiotropy in complex traits: challenges and strategies. Nat Rev Genet. 2013;14(7):483–95. 10.1038/nrg3461 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8. 10.1038/s41588-018-0099-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7 10.7554/eLife.34408 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zaitlen N, Kraft P. Heritability in the genome-wide association era. Hum Genet. 2012;131(10):1655–64. 10.1007/s00439-012-1199-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228–35. 10.1038/ng.3404 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones SE, Tyrrell J, Wood AR, Beaumont RN, Ruth KS, Tuke MA, et al. Genome-Wide Association Analyses in 128,266 Individuals Identifies New Morningness and Sleep Duration Loci. PLoS Genet. 2016;12(8):e1006125 10.1371/journal.pgen.1006125 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi C, Qiu S, Riester SM, Das V, Zhu B, Wallace AA, et al. Animal models for studying the etiology and treatment of low back pain. J Orthop Res. 2017. 10.1002/jor.23741 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Denard PJ, Holton KF, Miller J, Fink HA, Kado DM, Marshall LM, et al. Back pain, neurogenic symptoms, and physical function in relation to spondylolisthesis among elderly men. The spine journal: official journal of the North American Spine Society. 2010;10(10):865–73. Epub 2010/09/28. 10.1016/j.spinee.2010.07.004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartvigsen J, Nielsen J, Kyvik KO, Fejer R, Vach W, Iachine I, et al. Heritability of spinal pain and consequences of spinal pain: a comprehensive genetic epidemiologic analysis using a population-based sample of 15,328 twins ages 20–71 years. Arthritis and rheumatism. 2009;61(10):1343–51. Epub 2009/10/01. . [DOI] [PubMed] [Google Scholar]
- 70.Suri P, Boyko EJ, Smith NL, Jarvik JG, Williams FM, Jarvik GP, et al. Modifiable risk factors for chronic back pain: insights using the co-twin control design. Spine J. 2017;17(1):4–14. 10.1016/j.spinee.2016.07.533 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. 10.1038/nature15393 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279–83. 10.1038/ng.3643 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. Genome-wide genetic data on ~500,000 UK Biobank participants https://www.biorxiv.org/content/early/2017/07/20/166298: bioRxiv; 2017 [cited 2018 3/25/2018].
- 74.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192–212. 10.1038/nprot.2014.071 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bowden J, Del Greco MF, Minelli C, Davey Smith G, Sheehan NA, Thompson JR. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: the role of the I2 statistic. Int J Epidemiol. 2016;45(6):1961–74. 10.1093/ije/dyw220 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hemani G, Zheng J, Wade KH, Laurin C, Elsworth B, Burgess S, et al. MR-Base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv. 2016.
- 77.Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47(11):1236–41. 10.1038/ng.3406 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272–9. 10.1093/bioinformatics/btw613 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
*LD score regression intercept = 1.0067. GWAS = genome-wide association study.
(PDF)
rs115392701 has merged into rs12310519. Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs115392701 is shown using the colors in the figure legend (rs115392701 has merged into rs12310519).
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs1453867 is shown using the colors in the figure legend.
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs7833174 is shown using the colors in the figure legend.
(PDF)
Point sizes are proportional to inverse variance weights. OR = odds ratio, CI = 95% confidence interval, CHS = Cardiovascular Health Study, FHS = Framingham Heart Study, GenScot = Generation Scotland, JoCo = Johnston County Osteoarthritis Project, MrOs-GBG = Mr. Os Sweden (Gothenburg), MrOs-Malmo = Mr. Os Sweden (Malmo), MrOs-US = Mr. Os United States, OAI = Osteoarthritis Initiative, RS = Rotterdam Study, SOF = Study of Osteoporotic Fractures, UK = United Kingdom, UKB = UK biobank (interim data release).
(PDF)
Association p-values are plotted against genomic location. Negative log of the association p-value is represented on the left-hand y-axis, and recombination rate is displayed on the right-hand y-axis. Genomic location is shown on the x-axis, chr:pos. indicated are GRCh38/hg38. RefSeq genes are indicated in the bottom panel. Linkage disequilibrium r2 relative to the index single nucleotide variant rs4384683 is shown using the colors in the figure legend.
(PDF)
(DOCX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
Data Availability Statement
Meta-GWAS data are available from the dbGaP CHARGE Summary Results site under accession number phs000930, and can be accessed as described in Rich SS, Wang ZY, Sturcke A, Ziyabari L, Feolo M, O'Donnell CJ, et al. Rapid evaluation of phenotypes, SNPs and results through the dbGaP CHARGE Summary Results site. Nat Genet. 2016;48(7):702-3. doi: 10.1038/ng.3582. PubMed PMID: 27350599; PubMed Central PMCID: PMC5787851. The dataset can also be accessed from https://gwasarchive.org under 'Chronic back pain'.