Abstract
COSMIC-3D is a comprehensive integration of cancer mutations with protein structure across the human genome and structural proteome, seeking to support the identification and characterisation of protein targets for novel drug design in precision oncology. As an interactive system to explore cancer mutations in three-dimensions, COSMIC-3D is designed to enable a greater understanding of the functional impact of mutations, generate new hypotheses on which mutations are cancer drivers, and provide new opportunities for addressing these mutations pharmaceutically. This combination of genetics, structural proteomics, and drug development, can be best described as “mutation-guided drug design”.
There is a substantial need in precision oncology for treatments directed at specific cancer mutations, to more effectively treat patient populations with specific genomic profiles. Recently, huge gains in targeted and genomic DNA sequencing are driving a growing knowledge of which mutations cause cancer. Yet relative to these genomic knowledge gains, there remains much scope for expansion of the repertoire of precision therapeutic drugs targeting these mutations. Concurrently, the number of publicly available protein structures has been growing exponentially. Both cancer genomic and protein structural data sources individually are great assets for scientists working in the field of oncology drug discovery. However, linking these two important resources enables a much greater understanding of the protein structural aspects of cancer, empowering targeted drug design across cancer proteins.
Here we present COSMIC-3D, a bioinformatics platform that provides greater understanding of the impacts of cancer mutations and how they might be challenged by small-molecule drugs, by placing them in a protein structural context. We have achieved this by combining the rich cancer mutation annotations of COSMIC1 with the 3D human structural proteome data from the Protein Data Bank (PDB)2, using SIFTS3 to translate UniProt sequence coordinates (mapped to COSMIC through sequence alignment) to positions in protein structures. COSMIC-3D currently maps cancer variant data for nearly 9,300 genes to nearly 37,000 protein structures, covering 390 key genes from the Cancer Gene Census4. In total, over 445,000 different missense (non-synonymous) mutations from COSMIC, occurring in total over 736,000 times across tumour samples from 845 disease types, can be located in protein structures, as well as 3,700 in-frame deletions and over 64,000 nonsense mutations.
The COSMIC-3D web interface (see Data Availability, Life Sciences Reporting Summary) combines a WebGL-based interactive 3D structure viewer “NGL”5 (Figs. 1a and 1b) and the “Feature Viewer” sequence viewer (DOI: 10.5281/zenodo.345324) (Fig. 1c). The sequence viewer shows COSMIC mutations, as well as contextual UniProt6 protein annotations such as domains, enzyme active sites, and post-translational modification sites. Users can click on mutations in the sequence viewer to display them in the structure viewer. The structure viewer displays the protein structure and small-molecule ligands, protein-protein or protein-DNA interfaces where available, allowing mutations to be interpreted in the context of these functional binding sites. An additional panel provides further tools for manipulating the 3D structure view, including the ability to load pre-calculated, small-molecule binding site predictions and their associated “druggability” scores, derived from fPocket7.
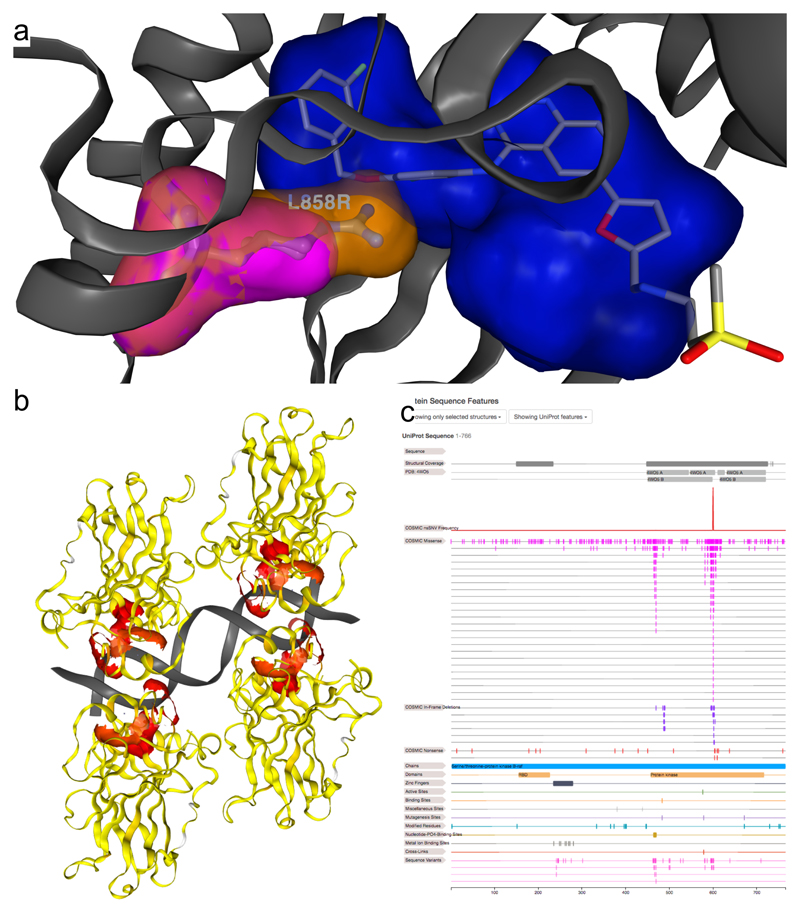
Figure 1.
Individual views of mutation, mutation frequency, and protein sequence views in COSMIC-3D. a) COSMIC-3D structure visualisation of EGFR protein’s kinase domain ATP binding site, showing Leu858 in pink, and an in silico generated model of oncogenic mutant Arg858 in orange. Mutation visualisations are juxtaposed with the highest ranked druggable binding site prediction (blue volume), which accurately encompasses the laptanib binding site (shown in stick representation). Individual missense mutation visualisations can be displayed by clicking on them in the sequence feature viewer. Predicted small molecule binding sites and their druggability can be displayed using the controls in the “Predicted Small-Molecule Binding Sites” section for the active protein structure. PDB: 1XKK. b) TP53 protein DNA binding domain tetramer bound to DNA, visualised with missense mutation recurrence analysis in COSMIC-3D. Recurrence of missense mutations is shown as surfaces coloured on a linear scale, where yellow indicates low recurrence in COSMIC, and red indicates high recurrence. Clearly, the most frequent missense mutations are affecting DNA binding functions. PDB: 4HJE. c) Example of the COSMIC-3D protein sequence feature view for BRAF (NP_004324.2). The 2D sequence view is linked to 3D structure, showing structural coverage of the protein sequence, a graph of missense mutation recurrence, and a “waterfall” of COSMIC mutations which can be clicked on for 3D visualisation. UniProt features add additional context to the structural and functional relevance of COSMIC mutations in proteins.
Individual missense, in-frame deletion, and nonsense somatic cancer mutations are mapped to protein structures and can be located and displayed in COSMIC-3D. For missense mutations, the structure viewer highlights the position of the mutation and visualises the wild-type amino acid, as well as a model of the mutant amino acid, generated using PyMOL (Schrödinger, LLC). This provides the opportunity to generate hypotheses of the impacts of missense mutations on protein structure and function. For example, Fig. 1a shows the COSMIC-3D visualisation of the oncogenic NP_005219.2:p.Leu858Arg mutation in the epidermal growth factor receptor protein (EGFR). The mutation is mapped onto the complex of the EGFR kinase domain bound to lapatinib. The model of the mutant indicates the protrusion of Arg858 into the lapatinib binding site, which also corresponds to the top ranked fPocket binding site prediction. This suggests that the mutant form may sterically interfere with lapatinib binding. Challenges remain in predicting these effects from the wild-type structure alone8, and hypotheses generated should be carefully explored. These challenges include those of protein dynamics; COSMIC-3D provides informative and accessible starting points for assessing local dynamic impacts across the breadth of cancer genomics. In this EGFR example, In vitro studies have shown that EGFR p.Leu858Arg increases the IC50 of lapatinib binding two-fold9, indicating that the mutant form weakens lapatinib binding. This is a well characterised example of how structural data and predictions can generate hypotheses on how mutations may alter the shape of known or predicted drug binding sites.
COSMIC-3D shows the location of recurrent missense mutations on protein structures as 3D “heat-maps” indicating mutation frequency, immediately illustrating their structural and functional contexts. This is intended to highlight key cancer driver mutants such as Arg248 and Arg273 DNA binding site mutants in the TP53 protein (NP_000537.3) (Fig 1b), as well as less well characterised putative targets.
In summary, COSMIC-3D provides a regularly updated system to explore cancer mutations in a protein structural context. The COSMIC-3D “human structural proteome of oncology” aims to empower the drug discovery process with cancer genomics. COSMIC-3D is built to aid the identification of cancer mutations that are cancer drivers of structural and functional importance, and to highlight how these mutations interact with known and predicted drug binding sites. The unique combination of protein structure, mutation location and recurrence, and druggability predictions made accessible in COSMIC-3D presents exciting possibilities for precision medicine by enabling high resolution “mutation-guided drug discovery”.
Acknowledgements
This work was funded by a postdoctoral fellowship awarded to H.C.J. under the “Sustaining Innovation Postdoctoral Training Program” at Astex Pharmaceuticals (H.C.J., H.K.S., M.L.V., S.A.F.). This work was supported by the Wellcome Trust grant [206194]. (H.C.J., S.A.F.). We thank colleagues at the Wellcome Sanger Institute and Astex Pharmaceuticals for helpful comments.
Footnotes
Author Contributions
H.C.J. contributed experimental design, performing the experiments, analysis of the data, and writing the paper. H.K.S., M.L.V., and S.A.F. contributed joint supervision of research, conception and design of the experiments, and writing of the paper.
Competing Financial Interests Statement
COSMIC-3D uses data and systems infrastructure from the COSMIC database group, which is partly funded from Wellcome Trust charitable funds together with sales of licenses to for-profit enterprises. H.K.S. and M.L.V. are employed by Astex Pharmaceuticals.
Data and Code Availability Statement
The datasets generated during and/or analysed during the current study are available via the publicly available COSMIC-3D web platform, https://cancer.sanger.ac.uk/cosmic3d, which will remain available for the foreseeable future.
References
- 1.Forbes SA, et al. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman HM, et al. Curr Opin Struct Biol. 2016;40:17–22. doi: 10.1016/j.sbi.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velankar S, et al. Nucleic Acids Res. 2013;41:D483–9. doi: 10.1093/nar/gks1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futreal PA, et al. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose AS, Hildebrand PW. Nucleic Acids Res. 2015;43:W576–9. doi: 10.1093/nar/gkv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The UniProt Consortium. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Guilloux V, Schmidtke P, Tuffery P. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilmer TM, et al. Cancer Res. 2008;68:571–9. doi: 10.1158/0008-5472.CAN-07-2404. [DOI] [PubMed] [Google Scholar]
- 9.Gazdar AF. Oncogene. 2009;28(Suppl 1):S24–31. doi: 10.1038/onc.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]