Abstract
Background
Clinical use of next-generation sequencing (NGS) tests has been increasing, but few studies have examined their economic value. Several studies have noted that there are methodological challenges to conducting economic evaluations of NGS tests.
Objective
Our objective was to examine key methodological challenges for conducting economic evaluations of NGS tests, prioritize these challenges for future research, and identify how studies have attempted solutions to address these challenges.
Methods
We identified challenges for economic evaluations of NGS tests using prior literature and expert judgment of the co-authors. We used a modified Delphi assessment to prioritize challenges, based on importance and probability of resolution. Using a structured literature review and article extraction we then assessed whether published economic evaluations had addressed these challenges.
Results
We identified 11 challenges for conducting economic evaluations of NGS tests. The experts identified three challenges as the top priorities for future research: complex model structure, timeframe, and type of analysis and comparators used. Of the 15 published studies included in our literature review, four studies described specific solutions relevant to five of the 11 identified challenges.
Conclusions
Major methodological challenges to economic evaluations of NGS tests remain to be addressed. Our results can be used to guide future research and inform decision-makers on how to prioritize research on the economic assessment of NGS tests
Precis
We examine methodological challenges for conducting economic evaluations of next generation sequencing tests, prioritize these challenges, and identify how studies have applied solutions.
INTRODUCTION
Understanding the economic value of clinical tests that utilize “next-generation sequencing” (NGS) is critical to their appropriate implementation. The use of NGS tests (including multigene panels, whole exome, and whole genome sequencing) has been increasing.(1) However, only a limited number of studies have examined their economic value.(2) Several studies have noted that there are methodological challenges to evaluating NGS tests that may be a barrier to conducting evaluations.(3–12)
Our objective was to examine key methodological challenges for conducting economic evaluations of NGS tests, prioritize these challenges for future research, and identify how studies have attempted solutions. The fundamental key characteristic of NGS tests that complicates their economic evaluation is that, by definition, they simultaneously examine multiple genes and can produce multiple results, each with distinct short and long-term clinical and economic trajectories. In contrast, most economic evaluations examine the value of one test conducted for a specific reason, with one defined result, and with a single trajectory of costs and outcomes, and thus this approach may have to be modified for NGS tests. A previous study noted that researchers need to be “creative” about approaches to evaluating the costs and outcomes of NGS tests.(13) Addressing challenges to conducting economic evaluations can facilitate the ability of researchers to conduct such evaluations as well as increase the clarity and transparency of economic analyses for decision-makers.
METHODS
Overview
We identified challenges for economic evaluations of NGS using prior literature and input from co-authors with expertise in economic methods and NGS. We used a modified Delphi assessment to prioritize these challenges based on their perceived importance and probability of their resolution by methodological consensus. We then used structured literature review and article extraction to assess whether published evaluations had developed and applied solutions to these challenges.
Identifying Challenges for Economic Evaluations of NGS
We developed our list of challenges for economic evaluations of NGS tests in two steps. First, we built on a previous study that defined issues in economic evaluation of personalized medicine more broadly.(14) We then modified the list to include challenges that are particularly relevant to NGS tests, based on studies describing challenges for NGS evaluations.(3–12) Co-authors reviewed the list for accuracy and completeness. We did not restrict the list to only challenges that are unique to NGS, but did focus on those where there was group consensus that NGS testing made them especially challenging. We categorized challenges but we recognize that there is some overlap among them.
Delphi Method
We used the modified Delphi method(15) with the authors who are health economics experts (KP, DM, SW, DR, JB, KC) to rate and rank methodological challenges to economic evaluation of clinical NGS testing. In the first round we described the 11 challenges and asked experts to rate them using the following scales:
-
1)
Importance (4 point rating scale from very important to unimportant, including option to choose “no judgment”)
-
2)
Probability of resolution in the next five years via methodological consensus (5-point rating scale from very probable to very improbable, including option to choose “no judgment”).
Respondents were also asked to provide a written rationale for each of their ratings. After excluding the “no judgment” ratings, we calculated the median scores for both rating scales and selected the top challenges using a threshold median score of three. This threshold corresponded to a rating of “Important” or “Very Important” on the importance scale and “Either Way” (50/50 chance of being resolved), “Probable” (better than a 50% chance of being resolved) or “Very Probable” (almost certain to be resolved) on the probability scale.
The purpose of the second round for the survey was to narrow the list of priority challenges based on the information in the first round. We provided the experts with the subset of challenges that met the above criteria in Round 1 as well as the descriptive rationales for these ratings. We then asked respondents to identify and rank the three top challenges based on their current assessment of importance and probability of resolution and in order of preference for taking action now (1 = most preferred; 3 = less preferred). Respondents provided their rationale for each ranking. We determined the top scoring challenges based on how often each challenge was chosen as either “most preferred” or “preferred”.
Structured Literature Review to Identify Published Economic Evaluations and Their Solutions
We systematically conducted searches of PubMed and Embase to identify economic evaluations of NGS tests. We also used hand searching by reviewing article citations and review articles.
We used ten known relevant articles to identify relevant search terms (16–25) (searches are described in the Appendix). The PubMed search used specific MeSH terms to identify directly relevant articles and title keywords to identify articles not yet indexed. The Embase search was designed to be similar to our PubMed search but was revised to fit Embase terms. We also had to modify searches to capture studies of non-invasive prenatal tests using NGS because of how they were coded.
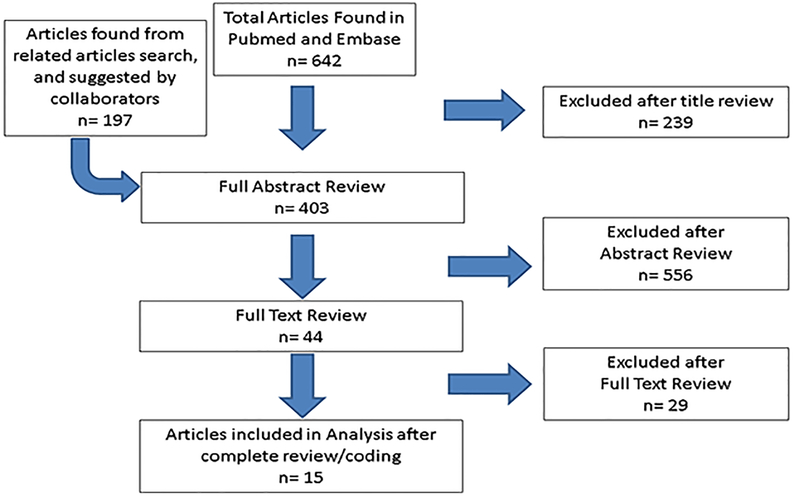
We screened articles by their titles and abstracts, with full text reviewed as necessary (Figure 1). We included studies if they met the following inclusion criteria:
Figure 1:
PRISMA diagram of included and excluded studies.
-
-
Empirical economic evaluation (including cost-effectiveness/cost-benefit/budget impact analyses, but excluding cost/consequence studies that did not calculate a ratio)
-
-
Study of clinical use of next-generation sequencing test (i.e., we did not include gene expression profiling panels or tests of a single gene or gene pairs such as BRCA1/2)
-
-
Published in English
We abstracted study variables using Excel spreadsheets to code study characteristics and solutions used to address challenges (authors KP and PD). Given that our key objective was to identify solutions to challenges rather than simply identify the challenges, we coded studies as follows:
-
(1)
Did the study address any of the identified methodological challenges using a specifically described approach?
-
(2)
If yes, what challenge was addressed and what solution was used?
We then identified how many of the challenges were addressed with specific solutions in the included studies. We did not attempt to define the quality and appropriateness of the methods used by the included studies in terms of whether they identified challenges or not. The challenges were not relevant to all of the studies and thus there was no need for some of the studies to identify challenges or apply solutions. We also did not assess the validity or generalizability of the solutions used.
RESULTS
Challenges for Conducting Economic Evaluations of NGS Tests
We identified 11 challenges, which we grouped into three categories (Table 1).
Table 1:
Challenges Identified for Economic Evaluations of NGS Tests
| Study Questions & Model Structure |
| Complex Model Structure: Modeling multiple pathways, results, & testing uses (as a result of multiple genes being tested). May include modeling potential interactive effects (e.g., of life expectancy across multiple conditions). |
| Timeframe: Modeling upstream (e.g., equipment purchase) and downstream (e.g., recurring testing & storage costs) costs & outcomes specific to NGS when relevant. May include potential savings if doing test up-front with later use of results. |
| Secondary Findings: Incorporating possibility of secondary findings and their impact (positive & negative) when relevant |
| Type of Analysis and Comparators Used: Determining appropriate type of analysis and using approaches other than CEA when relevant; using appropriate comparators that take into account what NGS is being compared to and whether substitution or addition |
| Directly Attributable Outcomes: Identifying costs/outcomes directly attributable to NGS when necessary to parse out |
| Measuring Costs & Outcomes |
| Broad Measures of Patient Outcomes: Quantifying range of outcomes for person being tested when relevant, e.g., measuring personal utility to patients because of psychological benefits from having a diagnosis etc. |
| Broad Measures of Health Outcomes Beyond Person Tested: Modeling individual outcomes beyond person being tested when relevant (e.g., modeling impact on family members) |
| Broad Measures of Societal Outcomes: Modeling impact beyond patient outcomes (e.g., education, employment) |
| Data Aggregation: Aggregating data from multiple sources when necessary to measure NGS impact, e.g., combining data from multiple studies |
| Data Availability & Quality |
| Data Availability Issues: Examining lack of evidence and data variability as relevant to NGS, e.g., prevalence, penetrance, clinical utility, race-specific inputs |
| Statistical Issues: Examining statistical issues as relevant to NGS, e.g., triangulating and integrating data sources, using value of information analysis |
CEA, cost-effectiveness analysis; NGS, next-generation sequencing
-
(1)
Study Questions and Model Structure (Complex Model Structure, Timeframe, Secondary Findings, Type of Analysis and Comparators Used, Directly Attributable Outcomes)
NGS tests can provide multiple results, and they have a much greater likelihood of identifying what are called “variants of unknown significance”, which are variants of a gene that have been identified through genetic testing, but whose significance to the function or health of an organism is not known. They may also generate secondary findings that are unrelated to the original reason for testing. Each of these findings may have distinct clinical trajectories and thus different costs and outcomes, and modeling every possible result and trajectory is often impractical. Secondary findings and variants of unknown significance can have either positive or negative impacts on costs and outcomes. In addition, findings may have interactive effects such that the sum is greater than the parts. For example, knowledge of the patterns of multiple mutations may provide more information than sequential, single-gene testing, thus requiring complex economic models to reflect these interactions. There may also be interactive effects such as the joint impact of multiple outcomes on life expectancy.
Determining the relevant timeframe and costs and outcomes within that timeframe can be particularly complex with NGS tests. There may be upstream costs and outcomes that are incurred prior to testing such as equipment costs downstream costs and outcomes such as data storage costs, variant re-interpretation, and costs as a result of additional testing or work-up due to secondary findings. Of particular relevance is that NGS tests of the individual’s genetic make-up (i.e., germline) may provide information that can be used throughout an individual’s lifetime, and thus costs and outcomes should be appropriately prorated and discounted.
The choice of the type of analysis and relevant comparator(s) can be challenging. NGS tests can be compared to single gene tests, sequential single-gene testing, other types of testing, or no testing. In addition, NGS tests may be simultaneously relevant to multiple conditions (e.g., breast and colorectal cancer, or cancer and heart disease), complicating the determination of the appropriate comparator. Of particular relevance is that NGS tests may substitute for other interventions or may supplement them, which increases the complexity of modeling these tests.
Lastly, it can be challenging to identify which costs and outcomes are directly attributable to NGS versus those that would have occurred anyway. For example, NGS results may suggest cancer screening, which would have been recommended anyway as a preventive measure.
-
(2)
Measuring Costs and Outcomes (Broad Measures of Patient Outcomes/Health Outcomes Beyond Person Tested/Societal Outcomes, Data Aggregation)
NGS tests can produce outcomes that go beyond clinical outcomes for the patient, such as personal utility (personal rationales for and benefits of testing that go beyond clinical outcomes), impacts on family members and impacts beyond individuals on education, employment, etc. Although these effects are not unique to NGS, it has been noted that they may be particularly relevant because of the hereditary nature of genetic diseases and the potential lifetime impacts of testing. Many reviews have noted the challenge of fully capturing the costs and outcomes of NGS tests. For example, testing may end a diagnostic odyssey and thus provide “personal utility” even if it does not change health outcomes. In addition, evaluations of NGS tests may need to aggregate data from multiple studies.
-
(3)
Data Availability and Quality (Data Availability Issues, Statistical Issues)
Data on key variables such as prevalence of mutations, clinical utility of testing, and race-specific variables may be lacking for NGS tests. Evaluation of NGS tests may face data challenges that are more complex than found in other analyses, such as the role of penetrance (the proportion of individuals carrying a particular variant of a gene (the genotype) that also express an associated trait (the phenotype)). Another challenge is that needed data are often not triangulated and integrated so that they can inform economic evaluations. Data may have to be combined from multiple data sources such as provider notes, EHR data, test results reported in PDF format, patient self-report, and other clinics were patients are referred. Lastly, multiple findings also create joint uncertainties that may require complex statistical estimation and may benefit from value of information analyses (i.e., a formal method for quantifying the value of additional evidence).
Priorities for Addressing Challenges
In the first Delphi round, seven challenges (out of 11) scored above the median score of 3 for both importance and probability of resolution (appendix). These challenges were: complex model structure, timeframe, secondary findings, type of analysis and comparators chosen, directly attributable outcomes, data aggregation, and data availability. The experts reassessed the challenges based on the results from Round 1 and chose the following challenges in terms of priority for taking action now: type of analysis and comparator used, complex model structure, and timeframe (Table 2). The experts also explained why they perceived that these challenges were important and feasible to address.
Table 2:
Top Priority Challenges to Address (Modified Delphi results)
| Top Priority Challenges to Address | Expert Working Group Rationales |
|---|---|
| Type of Analysis and Comparators |
Why important?
What’s feasible?
What’s needed?
|
| Complex model structure |
Why important?
What’s feasible?
What’s needed?
|
| Timeframe |
Why important?
What’s feasible?
What’s needed?
|
QALYs, quality adjusted life years; NGS, next-generation sequencing
Notes: We determined the top scoring challenges based on how often each challenge was chosen as either “most preferred” or “preferred”. Each of the above challenges was chosen by two respondents as “most preferred”. For the “preferred” designation, one respondent chose the type of analysis and comparator used, one chose the complex model structure, and two respondents chose timeframe. Given that the three top ranking challenges had similar scores, we did not attempt to further rank them. None of the other challenges were chosen as “most preferred”.
How Studies Have Developed and Applied Solutions to Challenges
We identified 15 studies for inclusion (Table 3). All but one study (Sabatini 2016) were cost-effectiveness analyses. The majority (60%) of the studies were US-based followed by studies from Australia (27%). The studies covered a variety of conditions: 47% were on cancer, 27% were on neurodevelopmental disorders in children, and 20% were on fetal aneuploidies. About half of the studies used intermediate outcome measures (e.g., cost per diagnosis, N=7). Interestingly, despite a concern that NGS technologies are too expensive for health care payers, all the studies except one identified a NGS test scenario that was cost-effective (Doble 2017)
Table 3:
Economic Evaluations of NGS Tests (N=15)
| Author Year | Objective | Country | Disease | Test / Comparators | Outcome measure | Results Summary | Conclusion Summary |
|---|---|---|---|---|---|---|---|
| Bennette 2015 | Clinical/ economic impact of returning IFs | US | Cardiomyopathy, colorectal cancer, healthy individuals with genetic FHx | WGS / not disclosing WGS IFs | cost/QALY |
|
Likely cost-effective for certain populations. Unlikely cost-effective in general population unless NGS <$500. |
| Gallego 2015 | Economic evaluation of NGS panels for CRC | US | Colorectal Cancer | NGS panel / current standard of care | cost/QALY |
|
First-line NGS panel (genes associated with highly penetrant CRCP syndromes + Lynch syndrome genes) cost-effective |
| Kaimal 2015 | Decision-analytic model to assess comprehensive outcomes of prenatal genetic testing strategies among women of varying ages. | US | Fetal aneuploidy | NIPT cell-free DNA / six testing strategies in combination or in sequence | cost/QALY |
|
Multiple marker screening under age 40 most cost-effective option for most women. Age 40+, cell-free DNA as primary screen becomes optimally cost-effective |
| Li 2015 | Is NGS panel (34 genes) for melanoma Tx selection cost-effective? | US | Metastatic Melanoma | NGS panel / Single site BRAF V600 test only | cost/QALY |
|
NGS panel is dominant strategy over single site mutation test strategy (reduced costs and increased QALYs) |
| Walker 2015 | (1) Determine optimum MSS risk cutoff for contingent NIPT (2) Compare cost-effectiveness of optimized contingent NIPT to universal NIPT and conventional MSS | US | Fetal aneuploidy | Universal NIPT cell-free DNA / MSS and optimized contingent NIPT | Cost/ Diagnosis |
|
Most cost-effective policy depended on perspective. Universal NIPT dominated (societal perspective), contingent NIPT dominated (government and payer perspective) |
| Azimi 2016 | Evaluate cost-effectiveness of carrier screening using NGS vs genotyping for 14 recessive disorders for which guidelines recommend screening | US | 14 recessive disorders in carrier screening | NGS panel / genotyping | Cost/ LY gained |
|
NGS-based carrier screening (most prevalent recessive disorders) cost-effective in averting more affected births, creating more LYs gained and reducing annual and lifetime treatments costs as compared with genotyping |
| Fairbrother 2016 | Estimate CEA of fetal aneuploidy screening in general pregnancy population using NIPT vs FTS with serum markers and NT ultrasound | US | Fetal aneuploidy | NIPT cell-free DNA / screening using FTS | Cost/ diagnosis |
|
NIPT in general pregnancy population leads to more prenatal identification of fetal trisomy cases vs FTS and is more economical at NIPT unit cost of $453 |
| Sabatini 2016 | Impact of using targeted gene panel in optimizing care for patients with advanced non small-cell lung cancer, use of targeted gene panel in diagnosis and management of patients with sensorineural hearing loss, and exome sequencing in the diagnosis and management of children with neurodevelopmental disorders of unknown genetic etiology | US | Advanced non small-cell lung cancer, sensorineural hearing loss, and neurodevelopmental disorders of unknown genetic etiology |
Targeted gene panel for three conditions / current standard of care | Cost/ diagnosis; Management, treatment or intervention mix before and after GSP testing |
|
Each model demonstrated value by reducing health care costs or identifying appropriate care pathways, depending on assumptions regarding cost and timing of testing (definition of value differs by clinical scenario). |
| Doble 2017 | Compare use of MTS to select targeted therapy based on tumor genomic profiles to no further testing (w/ chemo or w/ supportive care) in fourth-line treatment of metastatic lung adenocarcinoma | AUS | Metastatic lung adenocarcinoma | MTS / no further testing (chemo or supportive care) | Cost per LY/QALY |
|
MTS not cost-effective. VOI analyses reveal reducing decision uncertainty for cost and resource use parameters, testing parameters and clinical transition probabilities have greatest value |
| Li 2017 | Investigate whether a seven-gene test to identify women who should consider risk-reduction strategies could cost-effectively increase life expectancy |
US | Breast Cancer | 7-gene test (BRCA1, BRCA2, TP53, PTEN, CDH1, STK11, and PALB2). / BRCA1/2 |
cost/QALY |
|
Testing seven breast cancer–associated genes, followed by risk-reduction management starting at either age 40 or 50 years, could cost-effectively improve life expectancy. |
| Saito 2017 | To determine CEA of comprehensive molecular profiling before initiating anti-EGFR therapies for metastatic colorectal cancer | JPY | Metastatic Colorectal Cancer | Comprehensive Molecular Profiling / RAS mutation screening | cost/QALY |
|
Comprehensive screening more cost-effective than RAS screening |
| Schofield 2017 | Evaluate economic value for panel or WES of neuromuscular disease | AUS | neuromuscular disorders | WES and panel / muscle biopsy & protein assays (traditional) | Cost/ additional diagnosis |
|
Panel most cost-effective & WES 2nd most vs. traditional diagnostic pathway |
| Stark 2017 | Evaluation of 3 strategies to include WES in current testing pathway | AUS | Pediatric monogenetic disorders | 1) WES after exhaustive standard investigation, (2) WES to replace some investigations, (3) WES to replace most investigations / standard of care | Cost/ additional diagnosis |
|
Early WES triples diagnostic rate for 1/3 cost per diagnosis |
| Tan 2017 | Investigate impact of WES in sequencing-naive children suspected of having monogenic disorder and evaluate CEA if WES had been available at different time points in diagnostic trajectory | AUS | Monogenic disorders in children | Singleton WES / standard diagnostic pathway (no single gene or panel testing) | Cost/ additional diagnosis |
|
Singleton WES in children with suspected monogenic conditions has high diagnostic yield, and CEA is maximized by early application in the diagnostic pathway. |
| Tsiplova 2017 | Comparison of CMA to WES/WGS in Autism Spectrum Disorder | CAD | Autism Spectrum Disorders | WES, WGS / CMA | Cost/ dx (additional positive finding) |
|
Incremental cost was > CAD$25,000 per additional positive finding if CMA was replaced by newer technology. Future reductions in material and equipment costs and increased understanding of variants will lead to improved value. |
Addl, additional; AUS, Australian; CAD, Canada; CEA, cost-effectiveness analysis; CMA, chromosomal microarray; CRC, colorectal cancer; CRCP, colorectal cancer and polyposis; Dx, diagnosis; FHx, family history; FTS, first trimester combined screening; IFs, incidental findings; JPY, Japan; LY, life year; MSS, maternal serum screening; MTS, multiplex targeted sequencing; NIPT, non-invasive prenatal testing; NT, nuchal translucency; QALY, quality adjusted life years; Tx, treatment; VOI, value on investment; US, United States; WES, whole exome sequencing; WGS, whole genome sequencing; w/, with
Notes:
All studies were CEAs except for Sabatini 2016, which was a cost-impact analysis/budget impact analysis
All studies used the payer perspective except Walker 2015, which used payer, governmental, and societal perspectives.
Of the 11 challenges, six challenges were addressed with specific solutions that were described in four different studies (Table 4). Specific solutions were:
Table 4:
Specific Solutions Applied to Specific Challenges
| CHALLENGES | STUDIES ADDRESSING A SPECIFIC CHALLENGE WITH A SPECIFIC SOLUTION |
|---|---|
| Study Questions & Model Structure | |
| Complex Model Structure |
|
| Timeframe | Studies did not use explicit solutions to address |
| Secondary Findings |
|
| Type of Analysis and Comparators Used |
|
| Directly Attributable Outcomes | Studies did not use explicit solutions to address |
| Measuring Costs & Outcomes | |
| Broad Measures of Patient Outcomes | Studies did not use explicit solutions to address |
| Broad Measures of Health Outcomes Beyond Person Tested | Studies did not use explicit solutions to address |
| Broad Measures of Societal Outcomes | Studies did not use explicit solutions to address |
| Data Aggregation |
|
| Data Availability & Quality | |
| Data Availability Issues | Studies did not use explicit solutions to address |
| Statistical Issues |
|
Note: Challenges are not relevant to all studies.
-
(1)
Bennette et al 2015 (19) addressed the challenges of “Complex Model Structure”, “Secondary Findings”, and “Data Aggregation”. They addressed the modelling complexities introduced by multiple results and conditions and the challenge of modeling secondary findings. Their approach simplified the research question and model to make them manageable and leveraged existing data to make the analyses feasible. They narrowed the research question by modeling three archetypal groups and seven conditions. They also only included genes that were previously defined as having clinical utility rather than all possible secondary findings. They then leveraged existing cost-effectiveness analyses when possible rather than creating their own models.
Bennette et al also addressed the challenge of data aggregation by combining data from multiple studies and creating a composite cost-effectiveness ratio. They multiplied the individual-level estimates for costs and QALYs associated with returning a secondary finding by the expected prevalence of identifying and returning those results to estimate the implications of returning secondary findings at the population level.
-
(2)
Gallego et al 2015 (20) addressed the challenge of “Complex Model Structure” by analyzing hypothetical test scenarios as part of their cost-effectiveness analysis of NGS tests for the diagnosis of colorectal cancer and polyposis symptoms. They noted that tests typically include the most highly penetrant mutations first, but then may expand to include less penetrant mutations. Thus, they analyzed four hypothetical tests in order of increasing effectiveness where each panel was larger than the previous one because of additional, lower prevalence mutations.
-
(3)
Doble et al 2017 (26) addressed the challenge of “Statistical Issues” by using value of information analysis to assess where it would be of greatest value for decision-makers to reduce uncertainty, in their cost-effectiveness analysis of multiplex targeted screening to select targeted therapy for fourth-line treatment of metastatic lung adenocarcinoma. They found that such screening was not cost-effective compared to no testing. However, by using value of information analysis, they determined that additional research to reduce uncertainty may be a worthwhile investment, specifically, that reducing decision uncertainty for cost and resource use parameters, testing parameters, and clinical transition probabilities would have the greatest value.
-
(4)
Sabatini et al 2016 (23) addressed the challenges of “Type of Analysis and Comparators Used” and “Data Aggregation”. They used budget impact analysis, which is a method that has not been as frequently applied to NGS tests or other tests as cost-effectiveness analysis. They also analyzed three different scenarios. By using these approaches, they addressed what they perceived to be the needs of the relevant decision-makers.
Sabatini et al also addressed the challenge of data aggregation by aggregating cost data across laboratories using representative labs and cross-lab comparisons. They noted that one challenge in performing cost analyses for methods with multiple technology platforms and assay steps is the difficulty in determining a representative sample. To address this challenge, several laboratories performing clinical testing that met their definition of a representative laboratory were selected. They also incorporated the full costs of laboratory testing including the costs of bioinformatics and pipeline development, the costs associated with assessing the quality of the run, and the short- and long-term costs of storing data.
We did not find studies that specifically addressed other challenges (Table 4). Some studies mentioned such challenges but did not then attempt to address them with new solutions or with modifications of existing approaches.
DISCUSSION
We identified numerous challenges to conducting economic evaluations of NGS tests, and identified three challenges considered by experts to be the highest priorities for future research. We found that some challenges have been addressed using specific solutions but many challenges have not been addressed and solutions have not been generalized beyond specific studies. Of the three highest priority challenges, we found efforts to apply solutions to two of those challenges but we did not find any studies that have addressed one of the high priority challenges (appropriate timeframes).
Limitations
Our search may have missed relevant studies. As noted in other reviews,(2) the available search terms for identifying NGS panel studies are incomplete. There are no search terms for gene panels or multigene tests and thus we focused on identifying studies of sequencing tests. We also found that studies may be inconsistently coded; for example, Li 2017 (22) was incorrectly coded in PubMed as a “gene expression profiling panel” and thus we located this study using hand searching. To address these limitations, we used a range of data sources (PubMed, Embase, hand-searching) and a range of search terms. The number of studies we included differs from other recent reviews (e.g., Schwarze 2018 (2)) because we focused on multigene panels in addition to WES/WGS tests and we did not include studies only focused on costs.
We cannot ensure that we included all relevant challenges. We thus used a range of sources to identify the most relevant challenges and obtained input from co-authors. Similarly, we also cannot ensure that we identified all solutions used. Our study’s scope did not include determining whether studies should have addressed specific challenges or assess the methodological quality of studies. Instead, we focused on examining what challenges were or were not addressed using solutions. Lastly, we did not assess the appropriateness and adequacy of the identified solutions and other feasible solutions as this was beyond the scope of this study. Future research should obtain additional expert input on the priority challenges to address and potential solutions.
CONCLUSIONS
Although researchers are starting to consider the challenges to conducting economic evaluations of NGS technologies, a great deal more research effort is required to identify and test potential solutions. It would be helpful if future research could further identify viable solutions in addition to examining the solutions already used in published studies. Questions to be addressed include: How generalizable are the identified solutions? What other solutions could be feasible? Can we determine when specific solutions are most relevant? How can economic theory contribute? These questions can be addressed using expert input, case studies, and assessment of ongoing research that is not yet published.
Supplementary Material
Acknowledgements
Michael P. Douglas, MS provided project support, data abstraction, writing support and formatting, and manuscript review. This study was funded by a grant from the National Human Genome Research Institute (R01 HG007063) and consulting agreement with Illumina (no number).
Funding Statements
This study was funded by a grant from the National Human Genome Research Institute (R01 HG007063) and consulting agreement with Illumina (no number).
Footnotes
Financial Disclosures
Dr. Phillips has received honoraria for serving on a scientific advisory panel and is a paid consultant to Illumina. Disclosures have been reviewed by the University of California, San Francisco. One author (K.P.) received consulting fees from Illumina to support the research conducted for this publication. Some authors (K.P., D.R., P.D.) received travel support from Illumina to attend a past working group meeting. Dr. Marshall reports personal fees from Board Membership: Pfizer, personal fees from Consultancy: Optum Insight, personal fees from Consultancy: Research Triangle Institute, personal fees from Consultancy: Roche, personal fees and non-financial support from Consultancy: Novartis, personal fees and non-financial support from Consultancy: Abbvie, personal fees and non-financial support from Consultancy: Janssen, grants from Grants/grants pending, outside the submitted work.
Contributor Information
Kathryn A. Phillips, University of California at San Francisco, Department of Clinical Pharmacy; Center for Translational and Policy Research on Personalized Medicine (TRANSPERS); UCSF Philip R. Lee Institute for Health Policy; and UCSF Helen Diller Family Comprehensive Cancer Center, San Francisco, CA, USA.
Patricia A. Deverka, American Institutes for Research, Chapel Hill, NC, USA.
Deborah A Marshall, Department of Community Health Sciences, University of Calgary, Calgary, Alberta T2N 4Z6, Canada.
Sarah Wordsworth, Nuffield Department of Population Health, Medical Sciences Division, University of Oxford, Oxford, United Kingdom.
Dean A. Regier, Cancer Control BC, School of Population and Public Health, University of British Columbia, Vancouver, BC, Canada.
Kurt D. Christensen, Brigham and Women’s Hospital, Harvard University, Boston, MA, USA.
James Buchanan, Nuffield Department of Population Health, Medical Sciences Division, University of Oxford, Oxford, United Kingdom.
REFERENCES
- 1.Phillips KA, Deverka PA, Hooker GW, et al. Genetic Test Availability And Spending: Where Are We Now? Where Are We Going? Health Aff (Millwood). 2018; 37: 710–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwarze K, Buchanan J, Taylor JC, et al. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018; February 15 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 3.Fugel HJ, Nuijten M, Postma M, et al. Economic Evaluation in Stratified Medicine: Methodological Issues and Challenges. Front Pharmacol. 2016; 7: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annemans L, Redekop K, Payne K. Current methodological issues in the economic assessment of personalized medicine. Value Health. 2013; 16: S20–6. [DOI] [PubMed] [Google Scholar]
- 5.Lu CY. Economic evaluation of whole-genome sequencing in healthy individuals: what can we learn from CEAs of whole-body CT screening? Genet Med. 2016; 18: 103–4. [DOI] [PubMed] [Google Scholar]
- 6.Rogowski W, Payne K, Schnell-Inderst P, et al. Concepts of ‘personalization’ in personalized medicine: implications for economic evaluation. Pharmacoeconomics. 2015; 33: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne K, Eden M, Davison N, et al. Toward health technology assessment of whole-genome sequencing diagnostic tests: challenges and solutions. Per Med. 2017; 14: 235–47. [DOI] [PubMed] [Google Scholar]
- 8.Joosten SE, Retel VP, Coupe VM, et al. Scenario drafting for early technology assessment of next generation sequencing in clinical oncology. BMC Cancer. 2016; 16: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Basu A, Carlson JJ, Veenstra DL. A Framework for Prioritizing Research Investments in Precision Medicine. Med Decis Making. 2016; 36: 567–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payne K, McAllister M, Davies LM. Valuing the economic benefits of complex interventions: when maximising health is not sufficient. Health Econ. 2013; 22: 258–71. [DOI] [PubMed] [Google Scholar]
- 11.Phillips KA, Ladabaum U, Pletcher MJ, et al. Key emerging themes for assessing the cost-effectiveness of reporting incidental findings. Genet Med. 2015; 17: 314–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchanan J, Wordsworth S, Schuh A. Issues surrounding the health economic evaluation of genomic technologies. Pharmacogenomics. 2013; 14: 1833–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen KD, Dukhovny D, Siebert U, et al. Assessing the Costs and Cost-Effectiveness of Genomic Sequencing. J Pers Med. 2015; 5: 470–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Husereau D, Marshall DA, Levy AR, et al. Health technology assessment and personalized medicine: are economic evaluation guidelines sufficient to support decision making? Int J Technol Assess Health Care. 2014; 30: 179–87. [DOI] [PubMed] [Google Scholar]
- 15.Landeta J Current validity of the Delphi method in social sciences. Technological Forecasting & Social Change. 2006; 73: 467–82. [Google Scholar]
- 16.Schofield D, Alam K, Douglas L, et al. Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. npj Genomic Medicine. 2017; 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsiplova K, Zur RM, Marshall CR, et al. A microcosting and cost-consequence analysis of clinical genomic testing strategies in autism spectrum disorder. Genet Med. 2017. [DOI] [PubMed] [Google Scholar]
- 18.Stark Z, Schofield D, Alam K, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017. [DOI] [PubMed] [Google Scholar]
- 19.Bennette CS, Gallego CJ, Burke W, et al. The cost-effectiveness of returning incidental findings from next-generation genomic sequencing. Genet Med. 2015; 17: 587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallego CJ, Shirts BH, Bennette CS, et al. Next-Generation Sequencing Panels for the Diagnosis of Colorectal Cancer and Polyposis Syndromes: A Cost-Effectiveness Analysis. J Clin Oncol. 2015; 33: 2084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Bare LA, Bender RA, et al. Cost Effectiveness of Sequencing 34 Cancer-Associated Genes as an Aid for Treatment Selection in Patients with Metastatic Melanoma. Mol Diagn Ther. 2015; 19: 169–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Arellano AR, Bare LA, et al. A Multigene Test Could Cost-Effectively Help Extend Life Expectancy for Women at Risk of Hereditary Breast Cancer. Value Health. 2017; 20: 547–55. [DOI] [PubMed] [Google Scholar]
- 23.Sabatini LM, Mathews C, Ptak D, et al. Genomic Sequencing Procedure Microcosting Analysis and Health Economic Cost-Impact Analysis: A Report of the Association for Molecular Pathology. J Mol Diagn. 2016; 18: 319–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valencia CA, Husami A, Holle J, et al. Clinical Impact and Cost-Effectiveness of Whole Exome Sequencing as a Diagnostic Tool: A Pediatric Center’s Experience. Front Pediatr. 2015; 3: 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weymann D, Laskin J, Roscoe R, et al. The cost and cost trajectory of whole-genome analysis guiding treatment of patients with advanced cancers. Mol Genet Genomic Med. 2017; 5: 251–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doble B, John T, Thomas D, et al. Cost-effectiveness of precision medicine in the fourth-line treatment of metastatic lung adenocarcinoma: An early decision analytic model of multiplex targeted sequencing. Lung Cancer. 2017; 107: 22–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.