Abstract
Macrophage foam cells store excess cholesterol as cholesteryl esters, which need to be hydrolyzed for cholesterol efflux. We recently reported that silencing expression of carboxylesterase 1 (CES1) in human THP-1 macrophages (CES1KD macrophages) reduced cholesterol uptake and decreased expression of CD36 and scavenger receptor-A in cells loaded with acetylated low-density lipoprotein (acLDL). Here, we report that CES1KD macrophages exhibit reduced transcription of cytochrome P450 27A1 (CYP27A1) in non-loaded and acLDL-loaded cells. Moreover, levels of CYP27A1 protein and its enzymatic product, 27-hydroxycholesterol, were markedly reduced in CES1KD macrophages. Transcription of LXRα and ABCA1 was also decreased in acLDL-loaded CES1KD macrophages, suggesting reduced signaling through PPARγ-CYP27A1-LXRα. Consistent with this, treatment of CES1KD macrophages with agonists for PPARγ, RAR, and/or RAR/RXR partially restored transcription of CYP27A1 and LXRα, and repaired cholesterol influx. Conversely, treatment of control macrophages with antagonists for PPARγ and/or RXR decreased transcription of CYP27A1 and LXRα. Pharmacologic inhibition of CES1 in both wildtype THP-1 cells and primary human macrophages also decreased CYP27A1 transcription. CES1 silencing did not affect transcript levels of PPARγ and RXR in acLDL-loaded macrophages, whereas it did reduce the catabolism of the endocannabinoid 2-arachidonoylglycerol. Finally, the gene expression profile of CES1KD macrophages was similar to that of PPARγ knockdown cells following acLDL exposures, further suggesting a mechanistic link between CES1 and PPARγ. These results are consistent with a model in which abrogation of CES1 function attenuates the CYP27A1-LXRα-ABCA1 signaling axis by depleting endogenous ligands for the nuclear receptors PPARγ, RAR, and/or RXR that regulate cholesterol homeostasis.
Keywords: macrophage, carboxylesterase, peroxisome proliferator-activated receptor gamma, CYP27A1, liver X receptor, 2-arachidonoylglycerol, cholesterol
Introduction
Atherosclerosis is the excess deposition of lipid in the intima of arteries and the underlying pathological process responsible for most cardiovascular disease (1). Macrophages play a central role in atherosclerosis by engulfing oxidized low-density lipoprotein (oxLDL) particles via scavenger receptors, such as scavenger receptor-A (SR-A) and CD36 (2). Scavenger receptor-mediated uptake of modified LDL, unlike uptake of native LDL by LDL receptors, is not controlled by negative feedback regulation. Once internalized, the cholesteryl esters, which are abundant in LDL particles, are hydrolyzed by lysosomal acid lipase to yield free cholesterol (3). This form of cholesterol can be shuttled from macrophages to ApoA1 and high-density lipoprotein (HDL) particles via ATP-binding cassette transporters ABCA1 and ABCG1, respectively (4). Cholesterol effluxed onto ApoA1 and HDL particles is then transported to the liver where it can be metabolized to bile acids and subsequently excreted (5). The process of removing cholesterol from intimal lipid deposits by macrophages and its transport to the liver is referred to as macrophage reverse cholesterol transport (mRCT) and is thought to be responsible for the regression of atherosclerotic plaques (6).
In macrophages, not all free cholesterol generated by lysosomal acid lipase is promptly effluxed from the cell. Because free cholesterol is cytotoxic, it is re-esterified and stored as cholesteryl esters in lipid droplets within the cytoplasm (7). The cholesteryl esters in the droplets are later hydrolyzed by a neutral cholesterol ester hydrolase to again generate free cholesterol for efflux (8). As the plaque progresses and its volume increases, some macrophages take up more cholesterol than they are able to efflux thereby becoming foam cells. As the macrophage’s ability to store the excess cholesterol becomes overwhelmed, the increased free cholesterol content results in endoplasmic reticulum stress-induced apoptosis as well as necrosis, thereby contributing to the accumulation of lipid in the subendothelial space (9). Thus, macrophages appear to have a beneficial role in the early stages of plaque formation, i.e. the removal of cholesterol, whereas at later stages they may enhance plaque formation when their capacity to process cholesterol is overwhelmed.
The two steps thought to be rate limiting in cholesterol removal from atherosclerotic plaques by mRCT are the hydrolysis of cholesteryl esters stored in lipid droplets and ABCA1- and ABCG1-mediated transfer of intracellular cholesterol across the cell membrane to acceptor particles (8). Several candidate enzymes have been proposed to be responsible for the cytoplasmic neutral cholesteryl ester hydrolase activity, including carboxylesterase 1 (CES1) (10), which is a member of the serine hydrolase superfamily and an important xenobiotic detoxication enzyme in the liver (11). With regard to cholesterol metabolism, macrophage-specific transgenic expression of human CES1 in Ldlr−/− mice resulted in reduced atherosclerosis and increased mRCT (12), which is consistent with CES1 having a role in promoting cholesterol efflux from macrophages (13). We have also investigated the role of CES1 in cholesterol efflux from human THP-1 macrophages that were lipid loaded with acLDL (14). Treatment of THP-1 foam cells with either diphenylethane-1,2-dione (benzil), a specific CES inhibitor, or paraoxon, a non-specific CES inhibitor, both resulted in an increased retention of cholesteryl esters following a specified efflux period. This result was consistent with CES1 influencing cholesterol transport through a cytoplasmic neutral cholesteryl ester hydrolase activity. However, a limitation of this study is that benzil and paraoxon may interact with proteins other than CES1 raising the possibility of off-target effects. In addition, both inhibitors were added during the efflux period after the macrophages had been lipid loaded, so their effects (if any) on earlier steps in cholesterol metabolism were not assessed.
To address the issue of off-target effects of benzil and paraoxon, we knocked down the expression of CES1 in THP-1 cells (CES1KD cells) and assessed its impact on cholesterol efflux (15). Surprisingly there was no difference in the fractional rate of cholesterol efflux between CES1KD cells and control cells; however, it was found that CES1 silencing reduced macrophage cholesterol uptake as compared to that in control cells following lipid-loading. Additionally, the transcription and expression of the scavenger receptors SR-A and CD36, the receptors responsible for most of the uptake of acLDL (2), were reduced in CES1KD foam cells as compared to control foam cells. However, the changes in the transcription and expression of SR-A and CD36 caused by CES1 knockdown were not observed in non-lipid-loaded macrophages (i.e. non-foam cells) (15). In addition, a small decrease in ABCA1 expression was observed in CES1KD foam cells, but not in CES1KD macrophages, when compared with the appropriate control cells. Although the mechanism that controls SR-A transcription has not been described, it is known that the transcription of CD36 is regulated by the ligand-activated nuclear receptor PPARγ and its heterodimeric partner RXR (16, 17). Furthermore, the transcription of ABCA1 and ABCG1 is regulated by LXRα/RXR (18–20). This suggested that ablating CES1 expression in THP-1 macrophages and foam cells results in decreased signaling through the nuclear receptor heterodimers PPARγ/RXR and LXRα/RXR. PPARγ and LXR signaling are known to be linked; ligand-activated PPARγ was shown to directly activate the transcription of LXR in macrophages (16, 17). Moreover, Szanto et al. (21) showed that PPARγ could directly regulate the expression of CYP27A1, a cytochrome P450 enzyme that synthesizes the oxysterol 27-hydroxycholesterol (27-OHC), which is an endogenous ligand of LXRα (22). Additionally, RAR/RXR can also trans-activate CYP27A1 expression, suggesting a role for retinoids in the regulation of CYP27A1 as well (21).
In this study, we further investigated the role of CES1 in the regulation of cholesterol homeostasis in human THP-1 macrophages and primary human macrophages. The effect of ablating CES1 activity in both macrophages and foam cells on the transcription of a number of genes important in the regulation of intracellular cholesterol metabolism, including CYP27A1, is reported. We discovered that CES1 silencing profoundly reduced CYP27A1 transcript levels, CYP27A1 protein expression levels, and 27-OHC levels. This result and others reported here are consistent with a mechanism in which abrogation of CES1 expression alters the signaling of PPARγ/RXR and LXRα/RXR. We hypothesize that these effects are caused by the limited availability of CES1-derived endogenous ligands for these nuclear receptors.
Materials and Methods
Cells, Chemicals, and Reagents
Human THP-1 (TIB-202) monocytes, high-glucose RPMI-1640 medium, and gentamicin sulfate solution (50 mg/mL) were purchased from the American Type Culture Collection (ATCC) (Manassas, VA). Primary human monocytes were obtained from Astarte Biologics (Bothell, WA). Low-endotoxin containing fetal bovine serum (FBS) was purchased from Invitrogen (Carlsbad, CA). Acetylated LDL was from Intracel (Bethseda, MD) or Alfa Aesar (Ward Hill, MA). Antibodies against CYP27A1, LXRα, and GAPDH were purchased from Abcam (Cambridge, MA). Phorbol 12-myristate 13-acetate (PMA) was purchased from Sigma (St. Louis, MO), as were selective agonists of RXR (LG100268), RAR (TTNPB), RAR/RXR (9-cis-retinoic acid), and LXR (T0901317). The PPARγ agonist GW1929 and PPARγ antagonist GW9662 were purchased from Cayman (Ann Arbor, MI). The RXR antagonist HX531 was purchased from Fisher Scientific (Pittsburg, PA). The RAR inverse agonist BMS493 was from TOCRIS (Minneapolis, MN). The LXR antagonist GSK2033 was from Axon Medchem (Reston, VA). RIPA lysis buffer and protease inhibitors (PMSF, AEBSF, bestatin, pepstatin A, leupeptin hemisulfate, and aprotinin) were from Santa Cruz Biotechnology (Dallas, TX). Total RNA isolation kits and SYBR-green quantitative real time qRT-PCR master mix were from Qiagen (Valencia, CA). cDNA synthesis reagents were purchased from BioRad Laboratories (Hercules, CA). Primers for RT-PCR consisted of both pre-validated Quantitect primer assays (Qiagen) and self-designed custom oligonucleotide primers were from Invitrogen (Table 1). Lentiviral particles containing CES1, PPARγ, and scrambled control (nonspecific) short hairpin RNA (shRNA) constructs, puromycin hydrochloride, and polybrene were purchased from Santa Cruz Biotechnology. The serine hydrolase activity probes, fluorophosphonate (FP)-biotin and FP-TAMRA, were from Toronto Chemicals and Thermo Pierce, respectively. WWL113 and methyl-β-cyclodextrin (MβCD) were from Sigma.
Table 1.
Primer sequences used for quantitative real-time PCR
| Gene | Forward Sequence | Reverse Sequence | |
|---|---|---|---|
| ABCA1 | 5’-GGGCCTCGTGAAGTATGGAG-3’ | 5’-GCCATCCTAGTGCAAAGAGC-3’ | |
| ABCG1 | 5’-GACAGGGATGCGCATTTCAC-3’ | 5’-GCTGGCATTAGTAACTGTGTCC-3’ | |
| CD36 | 5’-AGGACTTTCCTGCAGAATACCA-3’ | 5’-ACAAGCTCTGGTTCTTATTCACA-3’ | |
| SR-A | 5’-CCTGTGCATTGATGAGAGTGC-3’ | 5’-TGCTCCATACTTCTTTCGTCCT-3’ | |
| NPC1 | 5’-ACTCAGTTACATAGGGCCATCA-3” | 5’-ATCGCTCTTCAGTGGCACAA-3’ | |
| NPC2 | 5’-CCCCGACAGGTTTGTCTTGT-3’ | 5’-ACAGAACCGCAGTCCTTGAA-3’ | |
| STAR | 5’-GAAGGGTGTCATCAGGGCGG-3’ | 5’-TGGCAAAATCCACCTGGGTC-3’ | |
| LXRA | 5’-TCTGGACAGGAAACTGCACC-3’ | 5’-CCGCAGAGTCAGGAGGAATG-3’ | |
| CYP27A1 | 5’-GTGTCTGGCTACCTGCACTT-3’ | 5’-TTGGATGTCGTGTCCACTCC-3’ | |
| CH25H | 5’-ACATCTGGCTTTCCGTGGAG-3’ | 5’-TACGGAGCGAAGTTGCAGTT-3’ | |
| CYP46A1 | 5’-GGCCCAGTTTCCTTCTAGGAC-3’ | 5’-GCGCTCATAGTTGCATTCGG-3’ | |
| PPARγ | 5’-GCCGTGGCCGCAGATTT-3’ | 5’-GGGAGTGGTCTTCCATTACGG-3’ | |
| ALOX15 | 5’-TCAGGTTCCCTTGTTACCGC-3’ | 5’-GTTTCCCCACCGGTACAACT-3’ | |
| PTGS1 | 5’-GCCTCTAACCGTCTGGGAAC-3’ | 5’-AGCATCCCGGCTCCTAAATG-3’ | |
| PTGS2 | 5’-GTTCCACCCGCAGTACAGAA-3’ | 5’-AGGGCTTCAGCATAAAGCGT-3’ | |
| PTGDS | 5’-CCCCCAGGGCTGAGTTAAAG-3’ | 5’-GCACTTATCGGTTTGGGGCA-3’ | |
| PTGES3 | 5’-GCACGTTCATTCTCCGTCCT-3’ | 5’-TCCGCTTTTTCTCTCCGGTC-3’ | |
| TBXAS1 | 5’-GGAACCGACCCGAAAGAGAG-3’ | 5’-AATGTTCTGTGAGCCTCCGC-3’ | |
| CES1 | CES1_2_SG QuantiTect primer assay - QT01155581 | ||
| CES2 | CES2_1_SG QuantiTect primer assay - QT00037443 | ||
| CES3 | CES3_1_SG QuantiTect primer assay - QT00034692 | ||
| PPARα | PPARa_1_SG Quantitect primer assay - QT00017451 | ||
| PPARβ/δ | PPARd_1_SG Quantitect primer assay - QT00078064 | ||
| DAGLβ | DAGLB_1_SG QuantiTect primer assay - QT00074319 | ||
| CB1 | CNR1_2_SG QuantiTect primer assay - QT02305702 | ||
| CB2 | CNR2_1_SG QuantiTect primer assay - QT00012376 | ||
| GAPDH | GAPDH_1_SG QuantiTect primer assay - QT00079247 | ||
General Cell Culture Conditions
Human THP-1 monocytes were cultured in RPMI-1640 medium containing 10% FBS, 2 mM L-glutamine, 10 mM HEPES, 1 mM sodium pyruvate, 4500 mg/L glucose, 1500 mg/L sodium bicarbonate, and 50 μg/ml gentamicin (complete growth medium) and maintained at 37°C in a 1×106 cells/mL with media changes every 48–72 h. THP-1 monocytes were differentiated into macrophages by the addition of PMA (final concentration 100 nM) to complete growth medium for 72 h. Primary human monocytes were plated in complete growth medium and non-adherent cells removed the next day. The adherent cells were cultured for 10 days in complete growth medium. Culture medium was replenished with fresh medium approximately every 3 days.
CES1 and PPARγ expression were individually knocked down (CES1KD and PPARγKD, respectively) in THP-1 monocytes using lentiviral particles containing either a CES1shRNA or a PPARγ shRNA construct, respectively, and subjected to puromycin selection, as described in detail previously for CES1 (23). Control cells were transduced using lentiviral particles containing scrambled shRNA constructs. Abrogation of CES1 expression and activity in THP-1 monocytes was verified by Western blot analysis, real-time PCR, and para-nitrophenyl valerate (pNPV) esterase activity assay. Silencing of PPARγ expression was verified by Western blot analysis and real-time PCR. Control, CES1KD, and PPARγKD monocytes were cultured in complete growth medium with 5 μg/ml puromycin hydrochloride. Only puromycin-resistant cells were used for subsequent experiments. In this report, wildtype cells, control cells, CES1KD cells, and PPARγKD cells refers to THP-1 cells that were non-transduced, transduced with scrambled shRNA, transduced with CES1-specific shRNA, or transduced with PPARγ-specific shRNA respectively.
Activity-Based Protein Profiling (ABPP) of Wildtype, Control, and CES1KD Cell Proteomes
Cell lysates (proteomes) of wildtype, control, and CES1KD cells (monocytes) were prepared in 50 mM Tris-HCl (pH 7.4) buffer by sonication (3 × 10 s, on ice), followed by protein quantitation (BCA reagent, Thermo-Pierce). Proteomes (1 mg/mL of lysate protein) were incubated with FP-TAMRA (2 μM) for 60 min at room temperature. The reactions were quenched by adding 6x SDS-PAGE loading buffer (reducing), followed by heating (90°C, 5 min). After electrophoresis on a 10% PAGE gel, the separated serine hydrolases covalently labeled by the FP-TAMRA probe were visualized using a Typhoon fluorescent gel imager (image shown in grayscale).
The ability of WWL113 (24) to inhibit serine hydrolases in THP-1 wildtype monocytes was evaluated by treating cell lysate proteomes (1 mg protein/mL) with increasing concentrations of WWL113 (0.001–1 μM). After a 30 min preincubation with WWL113, cell proteomes were subsequently treated with FP-biotin (5 μM, 60 min, room temperature) and the reactions were quenched by addition of 6x SDS-PAGE loading buffer (reducing) and heating (90°C, 5 min). Gel-based ABPP was then performed as described previously (11).
In addition to ABPP, the in situ carboxylesterase activity in THP-1 monocytes, following preincubation (30 min, 37°C) with either vehicle (DMSO, 0.1% v/v) or increasing concentrations of WWL113 (0.001–10 μM), was determined using the substrate pNPV as previously described (25). In this assay, intact living cells (105 cells/well) are incubated in PBS containing the inhibitor, then pNPV is added and the extent of its hydrolysis within the live cells is monitored for 10 min at 405 nm in a plate reader.
Quantitative Real-Time PCR Analysis of mRNA Expression
Control, CES1KD, and PPARγ THP-1 monocytes were differentiated into macrophages in complete growth medium with PMA (100 nM) for 72 h. Following differentiation, both control, CES1KD, and PPARγKD macrophages were incubated for either 24 or 48 h in complete growth medium (1% FBS instead of 10% FBS) with and without acLDL (50 μg/mL) and total RNA isolated. In addition, CES1KD macrophages were incubated for 24 h in complete growth medium (1% FBS) with 9-cis-retinoic acid (an RAR/RXR agonist, 1 μM), TTNPB (a specific RAR agonist, 1 μM), or LG100268 (a specific RXR agonist, 100 nM) and total RNA isolated. CES1KD and PPARγKD macrophages were incubated for 48 h in complete growth medium (1% FBS) with GW1929 (a PPARγ agonist, 10 μM). Finally, control macrophages were incubated for 24 h in complete growth medium (1% FBS) with GW9662 (a PPARγ antagonist, 10 μM), HX531 (an RXR antagonist, 100 nM), or BMS493 (a pan RAR inverse agonist, 1 μM) and total RNA collected. Vehicle (DMSO) treatments were also utilized in all experiments. Total RNA was isolated using the RNeasy Plus Mini Kit (Qiagen) according to the manufacturer’s protocol. Recovered RNA was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Waltham, MA) and cDNA was synthesized with an iScript Select cDNA Synthesis Kit (BioRad) using oligo(dT) primers according to the manufacturer’s protocol. Real-time PCR of cDNA products was performed on a Stratagene Mx3005P thermal cycler with Quantifast SYBR Green PCR master mix (Qiagen) using the QuantiTect primer assays or primers detailed in Table 1. The thermocycler program used for all target genes consisted of a five minute hot start at 95°C prior to 40 cycles of 10 s at 95°C, followed by 30 s at 60°C, as recommended by the manufacturer. PCR product quality was assessed via dissociation curve analysis immediately following amplification. Differential expression of target genes was assessed by the ΔΔCt method (26) using GAPDH as the reference gene and results are presented relative to control macrophages that were transduced with lentivirus containing a scrambled shRNA construct.
Western Blot Analysis of CES1, PPARγ, CYP27A1 and LXRα
Whole-cell lysates of control, CES1KD and PPARγKD THP-1 macrophages (2×106 cells per sample), with and without 24-h acLDL loading, were prepared by sonication in RIPA lysis buffer containing protease inhibitors (PMSF, AEBSF, bestatin, pepstatin A, leupeptin hemisulfate, and aprotinin). Thirty μg protein per sample, as determined by bicinchoninic acid assay (Thermo Scientific), was separated on 10% SDS-PAGE gels prior to semidry transfer (20V for 30 min) onto PVDF membranes. Membranes were blocked in 5% (w/v) nonfat dry milk in Tris-buffered saline containing Tween-20 (TBST: 10 mM Tris, 150 mM NaCl, 0.1% Tween-20, pH 7.4) for one h at room temperature and probed for GAPDH (Abcam 37168; final dilution 1:15,000) and CES1 (Abcam 68190; final dilution 1:25000), PPARγKD (Abcam 19481; final dilution 1:500), CYP27A1 (Abcam 126785; final dilution 1:1,000), or LXRα (Abcam 176323; final dilution 1:1,000) overnight at 4°C. After washing with TBST, blots were probed with goat anti-rabbit IgG-HRP (Santa Cruz sc-2030; final dilution 1:15,000) for one h at room temperature. Following final washes, blots were visualized by enhanced chemiluminescence using Thermo Supersignal West Pico ECL reagent. The resulting films were scanned and densitometry analysis was performed using ImageJ v1.49a (NIH).
Biosynthesis of 27-OHC by Monocytes and Macrophages
A 100x working solution of “water-soluble” cholesterol (155 mM MβCD/10 mM cholesterol, 15.5:1 mol:mol ratio) was prepared in water and mixed on a rotator overnight at room temperature. Cholesterol was completely dissolved under these conditions. The cholesterol/MβCD solution was sterile filtered before adding to culture medium. Control and CES1KD monocytes were plated at a density of 2×106 cells per well in a 6-well plate and treated with 100 nM PMA for 48 h to produce macrophages. After washing twice with serum-free medium, the cells were incubated in a 5% CO2 atmosphere at 37°C for 24 h in serum-free medium supplemented with “water-soluble” cholesterol at a final concentration of 0.1 mM cholesterol/1.55 mM methyl-β-cyclodextrin. Negative controls included cells grown in medium that contained 1.55 mM methyl-β-cyclodextrin only. Assuming the Kd for the cholesterol•methyl-β-cyclodextrin complex is the same as that for 2-hydroxypropyl-β-cyclodextrin (Kd = 1.1 mM) (27), the concentration of free cholesterol in the culture medium was estimated to be 40 μM; thus, a sufficiently high concentration of cholesterol was available to the cells to be metabolized. After 24 h, 5β-cholestan-3α-ol (internal standard, 194 nmol) was added to each well as the internal standard. The samples (cells plus medium) were harvested together by scraping into a glass tube and extracted with an equal volume of heptane/isopropanol (3:2, v/v). The layers were separated by centrifugation (5 min, 1,000 x g). The top organic layer was transferred to a new glass tube and the organic extraction step was repeated. The pooled organic extracts were evaporated to dryness under a nitrogen gas stream. Samples were reconstituted in 100 μL of methanol for LC-MS/MS analysis.
LC-MS/MS Analysis of 27-OHC
Samples were analyzed on an Acquity UPLC system (Waters, Milford, MA) coupled to a TSQ Quantum tandem mass spectrometer equipped with an atmospheric pressure chemical ionization (APCI) source (Thermo Fisher Scientific, San Jose, CA). Chromatographic separation was carried out using an Acquity UPLC BEH C18 column (2.1 mm × 100 mm, 1.7 μm) equipped with a precolumn (2.1 mm × 5 mm, 1.7 μm) at 40°C. The mobile phases used were 2 mM ammonium acetate with 0.1% acetic acid (mobile phase A) and methanol with 0.1% acetic acid (mobile phase B). Mobile phase gradient conditions were as follows: hold at 90% A and 10% B for 1 min, linear increase of B to 98% over 16.5 min, hold at 98% B for 4.5 min, then decrease B to 3% in 3 min and re-equilibrate at the initial conditions for 2 min. The overall run time was 27 min and flow rate was 0.2 mL/min. 27-OHC was analyzed in positive ion mode and selected reaction monitoring (SRM). APCI source parameters were as follows: discharge current = 2.0 μA, vaporizer temperature = 450 °C, sheath gas = 30, auxiliary gas = 5 units and capillary temperature = 190 °C. Precursor-to-product ion transitions m/z 385.4 → m/z 161.3 for 27-OHC, and m/z 371.5 → m/z 95.5 for 5β-cholestan-3α-ol (internal standard) were used for quantification. Scan times were 0.1 s per SRM, and the scan width was m/z 0.01. Optimum collision energy and tube-lens conditions were determined for each compound by using autotune software for each analyte by postcolumn infusion of the individual compounds into a 50% A/50% B blend of the mobile phase being pumped at a flow rate of 0.2 mL/min. Xcalibur software was employed for data acquisition and processing. For quantification, calibration standards were prepared using authentic standards ranging in concentration from 5–5,000 nM, and three QC samples (low, medium and high concentration) prepared by adding stock 27-OHC dissolved in methanol to culture medium at 25, 250 and 2500 nM final concentrations (prepared in triplicate). Calibration curves were constructed using 1/x as a weighting factor for 27-OHC with a correlation coefficient (r2) of 0.999. Analyses of the QC samples were 93±8%, 93±8%, and 97±4% of the target concentrations in the low, mid, and high QC samples (mean ± SD), respectively.
Intracellular Total Cholesterol Levels following Treatment with PPARγ/RXR and RAR/RXR agonists and an LXR antagonist
Control and CES1KD macrophages were pretreated for 24 h with DMSO vehicle or GW1929/9cRA (1 μM/10 μM) and GSK2033 (1 μM) in culture medium containing 1% FBS. The culture medium was then removed and replaced with fresh medium containing acLDL (50 μg/mL) and 0.2% w/v BSA. After loading the cells with cholesterol for 24 h, the acLDL-containing medium was removed and the cells washed with 1 x PBS. The cells were then overlaid with equilibration medium (containing 0.2% w/v BSA) and incubated for an additional 18 h. The medium and cells were harvested separately, and cells scraped into 0.1 M potassium phosphate (pH 7.4), 0.05 M NaCl, 5 mM cholic acid, 0.1% Triton X-100 and lysed by sonication. Following brief centrifugation to remove cell debris (1,000 x g, 5 min, 4°C), intracellular total cholesterol and free cholesterol levels in the supernatants were assayed using an Invitrogen Cholesterol Assay kit per the manufacturer’s instructions and normalized on cellular DNA content.
Biosynthesis of 2-AG, Prostaglandins, and Thromboxane B2 by Monocytes and Macrophages
In 6-well plates, control and CES1KD monocytes (3×106 cells/well) in serum-free culture medium were pretreated with vehicle (DMSO, 0.1% v/v) or WWL113 (1 μM) (24) for 30 min, followed by treatment with 100 μM arachidonic acid (AA) to stimulate the cellular production of 2-AG (28). After 60-min incubation with AA, the cells and culture medium were harvested separately by centrifugation. The cell pellet was washed with PBS and suspended in 1.1-mL PBS, followed by removal of a 0.1-mL aliquot for protein determination. The remaining cell suspension and culture medium were each spiked with 2-AG-d8 (270 pmol and 336 pmol, respectively). The cell suspension (in 1-mL PBS) was diluted with 1.5-mL 2:1 (v/v) methanol/water and extracted with ethyl acetate (2 × 2 mL), while the culture medium was diluted with an equal volume of PBS and extracted using C18 SepPak columns. The organic extracts were evaporated to dryness under nitrogen. The residues were reconstituted in 100 μL of 1:1 (v/v) methanol/water and transferred to LC vials for LC−MS/MS analysis (23). For each sample, the amount of 2-AG and in cells and medium were added together and the sum normalized on the total protein content in each well (to control for well-to-well differences in cell numbers).
In a separate experiment using a stable deuterated isotope of 2-AG, wildtype THP-1 macrophages were incubated with 2-AG-d8 (2.5 μM) for either 0 or 4 h in serum-free medium. At each time point, ice-cold methanol (2 mL containing an internal standard 12-[[(cyclohexylamino)carbonyl]amino]-dodecanoic acid, 100 pmol) was added to each dish containing the cells and medium (this lyses the cells and terminates enzymatic reactions), and dishes allowed to sit on ice for 15 min. The lysed cells and medium were then extracted together into ethyl acetate. Levels of AA-d8, PGE2-d7 and PGD2-d7 in the extracts were determined by LC-MS/MS.
Control and CES1KD macrophages were also treated with exogenous 2-AG (10 μM in serum-free medium) for 1 h. Culture medium and cells were then spiked with deuterated internal standards and extracted with ethyl acetate. Extracts were analyzed by LC-MS/MS for prostaglandins and arachidonic acid as previously described (23). In another experiment, wildtype THP-1 macrophages were primed with LPS (1 μg/mL, 4h), then the medium removed and cells gently washed with PBS. Fresh serum-free medium containing 2-AG (10 μM), with or without benzil (50 μM), was overlaid on the cells and incubation continued for 1 h. Culture medium and cells were spiked with deuterated internal standards and extracted into ethyl acetate. Extracts were analyzed by LC-MS/MS for prostaglandins and arachidonic acid (23).
Statistical Analysis
Results are presented as means ± SD. All statistical analyses were performed using either SigmaPlot version 11.0 or SAS 9.3. In some cases, data were transformed (log or square root) to ensure that normality and equal variance assumptions were met. For quantitative RT-PCR gene expression results, data were converted to linearized ΔΔCt values in accordance with the method published by Schmittgen and Livak (26). Statistical comparison between cell types and treatment groups was performed by two-way ANOVA with Student-Newman-Keuls post-hoc testing. For Western blotting and some transcription analyses, statistical comparison between multiple groups was done using one-way ANOVA with Tukey’s post-hoc testing. Comparisons between two groups were analyzed using Student’s t-test. In general, p<0.05 between groups was considered to be statistically different.
Results
Silencing CES1 expression in human THP-1 monocytes/macrophages
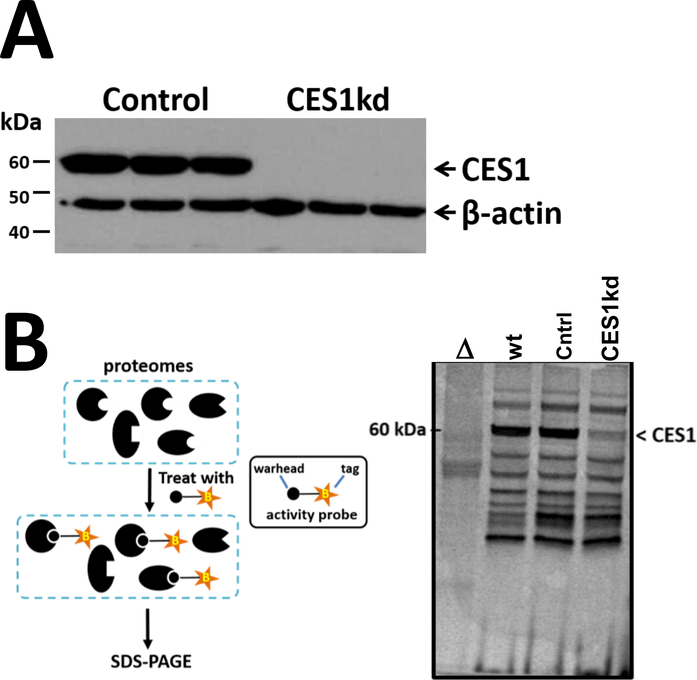
CES1 is one of the most abundant metabolic serine hydrolases in THP-1 macrophages and its activity is potently inhibited by a number of drugs and environmental toxicants, including organophosphorus poisons (Supplementary Figure 1A). However, small molecules that inactivate CES1 often exhibit off-target effects that limit their use in mechanistic experiments. We previously reported, on the basis of reductions in mRNA, protein, and enzymatic activity, the stable ablation of CES1 in human THP-1 cells using a lentivirus encoding CES1 shRNA (15, 23). In the current study, we confirmed and extended these observations to include gel-based ABPP methodology using the fluorescent probe FP-TAMRA, which covalently reacts with the conserved catalytic serine residue in serine hydrolases. Several lines of evidence indicate that CES1 expression is selectively diminished in our CES1KD cells. First, CES1 mRNA levels were markedly lower in the CES1KD cells compared to the control cells, whereas CES2 and CES3 mRNA levels were not significantly altered by CES1 silencing (Supplementary Figure 1B). Relative to CES1 expression, CES2 and CES3 mRNA transcripts are present at very low levels in THP-1 cells (at least 24-fold lower; www.proteinatlas.org), and CES2 protein was previously undetectable by immunoblot analysis in this cell line (Crow et al., 2008). Second, immunoblot analysis indicated that CES1 protein expression was completely ablated in the CES1KD cells (Figure 1A). Third, a prominent band around 60 kDa on the ABPP gel, denoted as CES1, was detected in both wildtype cells and control cells (which had been transduced with scrambled shRNA), but it was markedly diminished in intensity in the CES1KD cells (Figure 1B). Except for CES1, the profiles of serine hydrolase activities detected in the ABPP gel were, in general, qualitatively and quantitatively similar between wildtype, control, and CES1KD cells, suggesting that attenuated CES1 expression did not perturb the overall expression/activities of the other serine hydrolases detected by the ABPP probe.
Figure 1. CES1 silencing in THP-1 cells via lentivirus-mediated siRNA.
(A) Western blot analysis of CES1 in control and CES1KD macrophages. (B) Activity-based protein profiling (ABPP) to examine activities of the serine hydrolases in complex proteomes. Cell lysates were prepared from human THP-1 wildtype, control (scrambled shRNA), and CES1KD (CES1 shRNA) cells. Lysates were treated with the ABPP probe FP-TAMRA (2 μM, 1 h, room temperature) and reactions were stopped by adding reducing SDS-PAGE buffer and incubating at 95°C (5 min). Proteins were separated on SDS-PAGE and the fluorescently-labeled proteins were visualized using a Typhoon fluorescent scanner. Δ, wildtype cell lysate was heat denatured prior to being treated with FP-TAMRA.
Silencing CES1 expression in human THP-1 macrophages alters the transcription of genes involved in cholesterol metabolism
We further discovered that silencing CES1 expression in THP-1 cells had profound effects on gene transcription in both macrophages and foam cells (loaded with acLDL for 48 h), as shown in Figure 2. As expected, CES1 mRNA levels were markedly reduced in the CES1KD macrophages relative to those in control macrophages regardless of acLDL loading status. Strikingly, in macrophages under basal conditions, CES1 silencing caused several genes that encode proteins involved in the metabolism, sensing, and signaling of lipids to be down regulated, including CYP27A1, LXRα, DAGLβ, PPARγ, and RXRα. On the other hand, transcript levels of the scavenger receptors CD36 and SR-A were unchanged in CES1KD macrophages relative to those in control macrophages (<2-fold reduction; Figure 2 and Supplementary Figure 2), as we previously reported (15). CES1 silencing also had no effect on the transcription of ABCA1, ABCG1, CB1, RARα, RARγ, NPC1, and NPC2 in macrophages under basal conditions (Figure 2; NPC1 and NPC2 data not shown).
Figure 2. CES1 silencing alters the transcription of several genes involved in cholesterol metabolism.
mRNA transcript levels for genes associated with cholesterol metabolism in non-acLDL-loaded macrophages (-acLDL) and acLDL-loaded macrophages (+acLDL, 50 μg/mL, 48 h). Data are expressed as relative mRNA levels and the control (-acLDL) was set to 1.0 for each gene (n=3 biological replicates, mean ± SD). * p<0.05 for acLDL-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test). A significant interaction term between cell type and acLDL treatment is denoted in graphs as p<0.05 cell type x acLDL. If not given, no significant interaction was noted between cell type and acLDL treatment.
CES1 silencing in macrophage foam cells also resulted in significant down regulation of multiple genes involved in lipid homeostasis, including CYP27A1, CD36, SR-A, LXRα, ABCA1, DAGLβ, and StAR (Figure 2; StAR data not shown). We previously reported the down regulation of CD36 and SR-A when CES1 was knocked down (15). On the other hand, transcript levels of ABCG1, CB1, PPARγ, RXRα, RARα, RARγ, NPC1, and NPC2 were unaffected by CES1 silencing in foam cells (Figure 2; NPC1 and NPC2 data not shown). Interestingly, the expression of the PPARγ-regulated genes CYP27A1 and LXRα were consistently downregulated in the CES1KD cells relative to those in control cells, regardless of their lipid loading status. The expression of the PPARγ-regulated gene CD36 was also consistently downregulated in the CES1KD cells relative to that of control cells after lipid loading, but was variable when the cells were not lipid loaded. Qualitatively similar results as those shown in Figure 2 were found when macrophages were loaded with acLDL for 24 h instead of 48 h (Supplementary Figure 2). Furthermore, there was a statistically significant cell type x acLDL treatment interaction term for CYP27A1 and CD36 (Figure 2, Supplementary Figure 2), which strongly suggested that the two cell types exhibited a differential PPARγ-mediated induction response toward acLDL. PPARγ (29), RXRα (30), RARα (31), RARγ (31) DAGLβ (32), ABCG1 (33), and CB1 (34) have been reported to exhibit changes in their mRNA levels that roughly correlate with changes in protein expression in various tissues. CD36 and SR-A mRNA transcript levels also correlate with their protein levels in control and CES1KD cells (15). Thus, assessing the mRNA levels of these genes is an appropriate surrogate measure of their protein levels.
Because our data suggested that cellular CES1 activity is linked to downstream PPARγ signaling, this nuclear receptor was silenced in the THP-1 cells to evaluate whether the pattern of gene expression in CES1KD cells mimics that of PPARγKD cells under basal and acLDL-loaded conditions. Knockdown of PPARγ in THP-1 monocytes/macrophages was verified by (i) the marked decrease in its mRNA levels (Figure 3), (ii) Western blot analysis (Figure 3, inset), and (iii) demonstrating that the induction of PPARγ-target genes LXRα and CD36 in control cells treated with a PPARγ agonist (GW1929) was absent in PPARγKD cells (data not shown). We further noted that PPARα and PPARδ expression were not altered by PPARγ silencing (Supplementary Figure 3). The induction patterns of CYP27A1, LXRα, and CD36 in control and PPARγKD macrophages (Figure 3), which had been treated with acLDL for 48 h, were similar to that of control and CES1KD macrophages. Further, a statistically significant cell type x acLDL treatment interaction term for CYP27A1, LXRα, and CD36 was found (Figure 3), which indicates that PPARγKD macrophages do not respond to acLDL in the same way as control macrophages do. These genes were positively induced in control cells by acLDL loading, whereas their induction was blunted in PPARγKD cells, as was also observed in CES1KD cells. Thus the CES1KD cells appeared to phenocopy the behavior of the PPARγKD cells when challenged with acLDL. In support of this, CES1KD macrophages (15) and PPARγKD macrophages (Supplementary Figure 4) both exhibited reduced cholesterol uptake compared to that of control macrophages when exposed to acLDL. This finding is consistent with the reduced amounts of CD36 transcript in both CES1KD and PPARγKD cells compared to that of control cells. In addition, activation of macrophages by LPS challenge resulted in attenuated levels of the lipid mediator PGE2 in both CES1KD and PPARγKD cells compared to that of control cells (Supplementary Figure 5). Together, these results suggest that a mechanistic link exists between CES1 and PPARγ.
Figure 3. PPARγ silencing in macrophages mimics the transcription profile of CES1KD macrophages.
mRNA transcript levels for genes associated with cholesterol metabolism in non-acLDL-loaded macrophages (-acLDL) and acLDL-loaded macrophages (+acLDL, 50 μg/mL, 48 h). Data are expressed as relative mRNA levels and the control (-acLDL) was set to 1.0 for each gene (n=3 biological replicates, mean ± SD). * p<0.05 for acLDL-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test). A significant interaction term between cell type and acLDL treatment is denoted in graphs as p<0.05 cell type x acLDL. If not given, no significant interaction was noted between cell type and acLDL treatment. The western blot (inset) represents control and PPARgKD macrophage cell lysates blotted for PPARg and GAPDH proteins.
Effect of CES1 silencing on CYP27A1-catalyzed biosynthesis of 27-OHC
Of the genes examined, the levels of CYP27A1 transcript were the most dramatically decreased by CES1 silencing (Figure 2). This reduction was confirmed at the protein level, on the basis of Western blots of control and CES1KD macrophage lysates for CYP27A1 protein (Figure 4A). Because CYP27A1 catalyzes the biosynthesis of the oxysterol 27-OHC, an important LXRα agonist, we assessed whether control cells and CES1KD cells differentially biosynthesize this oxysterol. After loading monocytes and macrophages with a water-soluble form of free cholesterol (cyclodextrin-cholesterol), the CES1KD cells produced significantly less 27-OHC than the control cells (Figure 4B), which suggests that CES1 deficiency might alter downstream LXRα signaling pathways. It was estimated that control macrophages produced ~15x more 27-OHC than control monocytes did, which is likely due to the higher relative expression of CYP27A1 in macrophages versus monocytes (Figure 4C). Cells incubated in the absence of exogenous free cholesterol (negative controls) did not produce detectable amounts of 27-OHC in our assay (data not shown).
Figure 4. CYP27A1 protein expression and biosynthetic activity are reduced in CES1 knockdown cells.
(A) Western blot analysis of CYP27A1 in control and CES1KD macrophages. (B) The relative amounts of 27-OHC in control and CES1KD monocytes and macrophages are shown in bar graph. The levels in the control cells were set to 1.0 for monocytes and macrophages. Mass chromatograms for 27-OHC are shown for control and CES1KD macrophage extracts. (C) Western blot analysis of CYP27A1 in THP-1 wildtype monocytes and macrophages. Images represent blots of two lanes from the same gel that were not adjacent to each other, but were processed in the same manner and cut for ease of comparison. There was markedly greater amounts of CYP27A1 in macrophages compared to that of monocytes. Comparable amounts of total protein were loaded in each lane, based on GAPDH loading control. Quantitative data are expressed as the mean ±SD; n=2 for (A), n=3–4 for (B). * p<0.05 CES1KD cells relative to control cells, ** p<0.01 CES1KD cells relative to control cells (Student’s t-test).
Effect of nuclear receptor agonists on the transcription of CYP27A1, LXRα, and CD36 in CES1KD macrophages
Transcription of CYP27A1 and LXRα are regulated in part by the nuclear receptor heterodimers PPARγ/RXR and/or RAR/RXR (21, 35). Therefore, we next examined whether the addition of specific agonists of RXR (LG100268), RAR (TTNPB), PPARγ (GW1929), and RAR/RXR (9-cis-retinoic acid) to CES 1KD macrophages could restore the expression of CYP27A1 and LXRα to levels seen in control macrophages (Figure 5). CD36, whose expression is also regulated in part by PPARγ/RXR, was included as a positive control. Because CYP27A1 and LXRα are components of the PPARγ-CYP27A1-LXRα signaling axis, we also determined the effects of the nuclear receptor agonists on their protein expression. The results for CYP27A1, LXRα, and CD36 are described in turn.
Figure 5. Restoration of CYP27A1, LXRα, and CD36 expression in CES1 knockdown cells by nuclear receptor agonists.
(A) CYP27A1, LXRα, and CD36 transcription in CES1KD macrophages following treatment with selective agonists for RXR (LG100268, 100 nM) and RAR (TTNPB, 1 μM). Western blot analysis of CYP27A1 protein (B) and LXRα protein (C) in vehicle-treated control and CES1KD macrophages, and CES1KD macrophages treated with selective agonists of RXR (LG100268, 100 nM) or RAR (TTNPB, 1 μM). (D and E) CYP27A1, LXRα, and CD36 transcription in CES1KD macrophages following treatment with a specific agonist for PPARγ alone (GW1929, 10 μM) (D) or with GW1929 (10 μM) and 9-cis-RA together (1 μM) (E). (F) Western blot analysis of CYP27A1 in vehicle-treated control and CES1KD macrophages, and CES1KD macrophages treated with GW1929. For panels (A-F), data are expressed as relative mRNA levels or protein levels (n=3 biological replicates, mean ± SD); control cells treated with vehicle were set to 1 for each gene. * p<0.05 CES1KD cells treated with vehicle or agonist vs control cells treated with vehicle; # p<0.05 CES1KD cells treated with agonist vs CES1KD cells treated with vehicle (one-way ANOVA with Tukey’s post-hoc test). (G) Relative total intracellular cholesterol levels in control and CES1KD macrophages pretreated with DMSO vehicle and GW1929/9cRA or GW1929/9cRA/GSK2033. The mass of total cholesterol in control cells + DMSO was 0.47±0.06 μg /μg DNA. See materials and methods for experimental details. Symbols for (G) as follows (one-way ANOVA using Student-Newman-Keuls post-hoc test): * p<0.05 control vs CES1KD cells, # p<0.05 compared to control cells treated with DMSO, & p<0.05 compared to CES1KD cells treated with DMSO. n.s., no significant difference.
CYP27A1
The RAR and PPARγ agonists (TTNPB and GW1929, respectively) both increased CYP27A1 transcription in CES1KD macrophages relative to that of vehicle-treated CES1KD macrophages (Figure 5A and D). Further, the combination of GW1929 and 9-cis-retinoic acid increased CYP27A1 transcription to a greater extent than GW1929 alone (compare Figure 5D to Figure 5E). Moreover, treatment of CES1KD macrophages with either the RAR agonist (TTNPB) or PPARγ agonist (GW1929) could partially (TTNPB, Figure 5B) or almost completely (GW1929, Figure 5F) restore CYP27A1 protein expression to levels seen in control cells. On the other hand, treatment with the RXR agonist LG100268 did not alter CYP27A1 mRNA and protein levels in CES1KD cells (Figure 5A and 5B, respectively).
Next we examined the effects of nuclear receptor antagonists or inverse agonists on CYP27A1 transcription in control cells. Treatment of control macrophages with a PPARγ antagonist (GW9662), an RXR antagonist (HX531), or a pan-RAR inverse agonist (BMS439) caused CYP27A1 mRNA levels to be reduced relative to those in vehicle-treated control cells (Figure 6A-C). Furthermore, the combination of GW9662 and HX531 also reduced CYP27A1 mRNA levels compared to those seen in vehicle-treated control cells (Figure 6D).
Figure 6. Attenuation of CYP27A1 and LXRα transcription in control cells by nuclear receptor antagonists.
CES1, CYP27A1, and LXRα transcription in control macrophages following treatment with selective antagonists for PPARγ (GW9662, 10 μM) (A), RXR (HX531, 100 nM) (B), combined PPARγ and RXR antagonists (GW9662, 10 μM + HX531, 100 nM) (C), and an RAR inverse agonist (BMS493, 1 μM) (D). For each panel, data are expressed as relative mRNA levels (n=3 biological replicates, mean ± SD); control cells treated with vehicle were set to 1 for each gene. * p<0.05 control cells treated with antagonist vs control cells treated with vehicle (Student’s t-test).
Together these results suggested that CES1 has an important role in the regulation of CYP27A1 transcription through PPARγ/RXR and RAR/RXR signaling, and that the transcriptional changes in CYP27A1 are accompanied by corresponding changes in its protein levels.
LXRα
Transcript levels of LXRα were also significantly reduced in CES1KD macrophages as compared to those in control macrophages (Figure 2). Therefore, the ability of specific agonists of PPARγ, RAR, and RXR to restore LXRα transcription and protein expression in CES1KD macrophages was determined (Figure 5A, C, D, and E). Addition of an RXR agonist (LG100268, Figure 5A), PPARγ agonist (GW1929, Figure 5D), or combination of PPARγ agonist and non-selective RAR/RXR agonist (GW1929 and 9-cis-retinoic acid, respectively, Figure 5E) to CES1KD macrophages increased LXRα transcription relative to that of vehicle-treated CES1KD macrophages. Western blot analysis confirmed that the level of LXRα protein was reduced in the CES1KD macrophages as compared to that in control macrophages (Figure 5C). However, treatment of CES1KD cells with either LG100268, TTNPB, or GW1929 did not have a significant effect on LXRα protein expression when compared to vehicle-treated CES1KD cells (Figure 5C; data not shown for GW1929), although the LG100268-treated and GW1929-treated CES1KD cells exhibited significantly greater LXRα mRNA levels relative to that of vehicle-treated CES1KD cells (Figure 5A and Figure 5D, respectively). The RAR agonist (TTNPB) also had no effect on LXRα transcription (Figure 5A).
Although treatment of control macrophages with a selective PPARγ antagonist (GW9662) only slightly affected LXRα mRNA levels as compared to vehicle-treated control cells (Figure 6A), the selective RXR antagonist (HX531), either alone or in combination with GW9662, significantly reduced LXRα transcription levels (Figure 6B,D). The pan-RAR inverse agonist BMS439 also reduced LXRα transcript levels (Figure 6C).
Together these findings suggested that CES1 silencing in THP-1 macrophages can affect LXRα transcriptional regulation. The results indicate that LXRα transcription is partially restored in CES1KD macrophages by both PPARγ and RXR agonists, whereas its transcription in control macrophages could be reduced by PPARγ, RXR, and RAR antagonists.
CD36
The transcript levels of CD36 were only modestly reduced in CES1KD macrophages as compared to those in control macrophages, whereas they were significantly reduced in CES1KD cells versus control cells after acLDL loading (Figure 2 and Supplementary Figure 2). Addition of RXR agonist (LG100268) or PPARγ agonist (GW1929) to CES1KD macrophages increased CD36 transcription relative to that of vehicle-treated CES1KD macrophages (Figure 5A,D). Treatment with a combination of GW1929 and 9-cis-retinoic acid increased CD36 transcript levels approximately 5-fold. On the other hand, the RAR ligand (TTNPB) had no effect on CD36 transcription (Figure 5A). Consistent with a deficit of PPARγ ligands in CES1KD cells, supplementation of CES1KD cells, exposed or not to oxLDL, with the prototypical PPARγ ligand rosiglitazone also restored CD36 expression to control cell levels, whereas it had no effect on the SR-A expression defect (Supplementary Figure 6).
Repair of cholesterol metabolism defect in CES1KD macrophages by pretreatment with PPARγ/RXR and RAR/RXR ligands.
CES1KD macrophages exhibit less cholesterol uptake compared to that of control macrophages following exposure to acLDL (15), which is attributable to reduced CD36 and SR-A expression. This effect on total cholesterol level was confirmed (Figure 5G). Moreover, pretreatment of control and CES1KD macrophages with ligands that activate PPARγ/RXR and RAR/RXR (GW1929 and 9cRA, respectively) prior to cholesterol loading (acLDL) caused significant reductions in intracellular total cholesterol levels (Figure 5G). Despite the fact that acLDL exposure promotes net cholesterol accumulation in cells, the presence of PPARγ/RXR and RAR/RXR ligands will also upregulate the LXR-dependent cholesterol efflux pathway (e.g. induction of the LXR/ABCG1 axis) that counteracts cholesterol influx during the loading period. Thus, in addition to PPARγ/RXR and RAR/RXR ligands, we also added an LXR antagonist (GSK2033) to block the confounding effect of cholesterol efflux caused by upregulated ABCG1-mediated aqueous diffusion of free cholesterol to acLDL acceptors present in the extracellular milieu (36). As depicted in Figure 5G, this approach repaired the cholesterol influx deficit of the CES1KD cells. The level of intracellular total cholesterol in CES1KD cells pretreated with the triple combination of GW1929/9cRA/GSK2033 prior to cholesterol loading was significantly greater than that of CES1KD cells treated with DMSO, whereas it was not statistically different than that of control cells treated with DMSO, indicating that normalization of cholesterol influx had occurred in the CES1KD cells. As expected, control cells pretreated with GW1929/9cRA/GSK2033 exhibited more intracellular total cholesterol compared to that of control cells treated with DMSO (Figure 5G).
Inhibition of CES1 activity in human wildtype THP-1 cells with a small-molecule inhibitor blocks 2-AG catabolism and recapitulates the effects on CYP27A1 transcription seen in CES1KD cells
We next evaluated whether selective inhibition of CES1 activity in wildtype cells using a small molecule [WWL113; (24)] would have the same effect on CYP27A1 transcription as CES1 shRNA did. On the basis of gel-based competitive ABPP (37), we established that WWL113 was a selective and potent inhibitor of CES1 activity in wildtype THP-1 cell proteomes (Figure 7A). The IC50 for WWL113 toward CES1 in THP-1 cell lysates was estimated to be ~0.01 μM. Further, treatment of intact monocytes with WWL113 (1 μM, 1 h) could inhibit ~85% of the pNPV hydrolytic activity in THP-1 cells, establishing that CES1 could be targeted by this inhibitor. Moreover, pretreatment of intact control monocytes with WWL113 (1 μM, 1 h) increased the level of endogenous 2-AG, a lipid mediator and known CES1 substrate (38), compared to that of vehicle-treated control cells upon stimulation with exogenous AA (Figure 7B). We have previously shown that free AA is an effective stimulant of 2-AG biosynthesis in cells via a Nox-dependent mechanism ((28); Supplementary Figure 7). Further, the amount of 2-AG was higher in vehicle-treated CES1KD cells than in vehicle-treated control cells, which was expected since CES1 has a role in regulating 2-AG degradation. There was no difference, however, in 2-AG levels between the vehicle- and WWL113-treated CES1KD cells (Figure 7B), which is also consistent with CES1 having a role in 2-AG catabolism in THP-1 cells. When wildtype THP-1 monocytes were differentiated into macrophages in the presence of WWL113 (1 μM, 48 h), the level of CYP27A1 mRNA was found to be significantly lower in the inhibitor-treated cells than in vehicle-treated cells, whether or not the cells had been exposed to acLDL (Figure 7C). This result is consistent with the substantial downregulation in CYP27A1 transcription seen in the CES1KD cells (Figure 2), although not to the same extent. Furthermore, treatment of primary human macrophages with WWL113 (1 μM, 48 h) also resulted in lower CYP27A1 mRNA levels (Figure 7D).
Figure 7. The pharmacological agent WWL113 is a selective inhibitor of CES1 activity in THP-1 wildtype cells and attenuates CYP27A1 transcription.
(A) Gel-based competitive ABPP using WWL113 (0.001–1 μM final concentration, chemical structure shown at left) and FP-biotin (5 μM) indicated that CES1 was selectively inhibited in THP-1 wildtype cell lysates (ABPP blot shown in middle, CES1 band is indicated). Densitometry of the CES1 band indicated that the IC50 for WWL113 toward CES1 was around 0.01 μM, while WWL113 (1 μM final concentration) could inhibit around 85% of the pNPV hydrolysis activity of intact THP-1 wildtype cells using an in situ assay (25). * p<0.05 WWL113-treated cells vs vehicle-treated cells (control) (Student’s t-test). (B) Control and CES1KD monocytes were pretreated with WWL113 or vehicle (DMSO) for 30 min, followed by the addition of AA (100 μM, 1 h), which stimulates 2-AG production. Data are expressed as relative 2-AG levels (n=3 biological replicates, mean ± SD), * p<0.05 (one-way ANOVA with Student-Newman-Keuls post-hoc test); ns, not significant. (C) THP-1 wildtype monocytes were differentiated into macrophages in the presence of vehicle (DMSO) or WWL113 (1 μM, 48 h). Alternatively, THP-1 wildtype monocytes were differentiated into macrophages in the presence of vehicle (DMSO) or WWL113 (1 μM, 48 h), then exposed to acLDL (50 μg/mL, 48 h) in the absence or presence of WWL113 (1 μM). Total RNA was then isolated and CYP27A1 mRNA levels determined. (D) Primary human monocytes were differentiated for 10 days, followed by treatment with WWL113 (1 μM, 48 h). Total RNA was isolated and CYP27A1 mRNA levels determined. * p<0.05 WWL113-treated cells vs vehicle-treated cells (Student’s t-test).
We previously reported that inactivation of DAGLβ, a 2-AG biosynthetic enzyme, by RHC-80267 decreased prostaglandin synthesis in LPS/ionomycin-stimulated THP-1 macrophages (23). This result was consistent with the attenuated 2-AG and prostaglandin levels in murine peritoneal macrophages when DAGLβ activity was ablated genetically or with a small-molecule inhibitor (39). We extended these findings by treating intact THP-1 wildtype macrophages with deuterium-labeled 2-AG-d8 for 4 h and observed the production of AA-d8, PGE2-d7, and PGD2-d7 (Figure 8A). This demonstrated that 2-AG is a direct source of precursor AA that is subsequently metabolized by macrophages to produce prostaglandins (the conversion of AA-d8 to PGE2-d7 and PGD2-d7 results in the loss of a single deuterium atom). WWL113-treated cells and CES1KD cells both exhibited reduced 2-AG-d8 hydrolytic activity compared to their respective controls (Supplementary Figure 8). Additional evidence that 2-AG levels are regulated by CES1 was noted when control and CES1KD monocytes were differentiated into macrophages for 48 h and steady-state amounts of 2-AG in the cell supernatants were determined (Figure 8B); CES1KD supernatants had ~2.5x more 2-AG than control supernatants. Consistent with the fact that 2-AG hydrolysis provides eicosanoid precursor, when control and CES1KD macrophages were incubated with exogenous 2-AG (10 μM, 1 h) the levels of PGE2 and PGD2 levels in CES1KD cells were lower than those in control cells (Figure 8C), although TxB2 levels were not significantly different. Interestingly, a small molecule inhibitor (benzil) that reversibly inhibits CES1 in THP-1 cells (38) also reduced the levels of 2-AG–derived PGE2, PGD2, and PGF2α when LPS-primed wildtype THP-1 macrophages were treated simultaneously with it and 2-AG (Figure 8D; Wang et al., 2013). On the other hand, benzil had no effect on TxB2 levels (Figure 8D), similar to what was observed in CES1KD cells (Figure 8C).
Figure 8. THP-1 macrophages metabolize 2-AG yielding prostaglandins.
(A) Wildtype THP-1 macrophages were treated with 2-AG-d8 (2.5 μM) for 0 or 4 h in serum-free medium. After 4 h, significant amounts of the hydrolysis product AA-d8 and its COX-derived products, PGE2-d7 and PGD2-d7, were detectable in combined culture medium and cell extracts by LC-MS/MS analysis. (B) THP-1 control and CES1KD monocytes were differentiated into macrophages for 48 h. The culture medium was then removed and extracted for 2AG analysis. OD562, optical density at 562 nm (surrogate measure of cell protein). (C) Control and CES1KD macrophages were treated with exogenous 2-AG (10 μM) for 1 h in serum-free medium. Culture medium and cells were extracted and analyzed by LC-MS/MS for prostaglandins and AA. (D) Wildtype THP-1 macrophages were primed with LPS (1 μg/mL, 4 h). After removal of LPS-containing media followed by cell wash, 2-AG (10 μM) and benzil (50 μM) or vehicle (DMSO, 0.1% v/v) in serum-free medium were overlaid onto the cells. Incubation with compounds proceed for 1 h. Culture medium and cells were extracted and analyzed by LC-MS/MS for prostaglandins and AA. ** p<0.01 in (B) (n=3 biological replicates, mean ± SD); p-values in (C, D) are indicated (n=6 biological replicates, mean ± SD) (Student’s t-test).
Silencing CES1 expression had no effect on LPS-stimulated induction of TNFα and IL-6 (Supplementary Figure 9). This result suggests that LPS-evoked signaling through TLR-4 and NFkB remains intact. Consistent with this notion, expression of TLR-4 was actually somewhat increased in CES1KD cells compared to control cells both at baseline and following LPS treatment. In addition, mRNA levels of the prostaglandin synthetic enzymes PTGS1 (COX1), PTGS2 (COX2), PTGES3, and TBXAS1 were unchanged in CES1KD macrophages compared to controls (Supplementary Figure 10). Only PTGDS expression levels were reduced in the setting of CES1 silencing.
Effect of CES1 silencing on the transcription of cholesterol-25 hydroxylase, CYP46A1, and 15-lipoxygenase
Because knockdown of CES1 expression reduced the transcription of CYP27A1, we also examined the transcription of two other genes that encode oxysterol-synthetic enzymes, namely cholesterol-25 hydroxylase (CH25H), which synthesizes 25-OHC, and cholesterol-24 hydroxylase (CYP46A1), which synthesizes 24-OHC. Similar to 27-OHC, these oxysterols are also ligands for LXRα. Whereas CYP46A1 transcripts in cells were below the limit of detection by our RT-PCR assay, the level of CH25H transcript was slightly greater in CES1KD cells than in control cells (1.6-fold, p=0.008, data not shown).
The effect of CES1 silencing on the transcription of 15-lipoxygenase (ALOX15), an enzyme responsible for the synthesis of hydroxyoctadecadienoic acids (HODEs) and hydroxyeicosotetraenoic acids (HETEs), which are oxygenated metabolites of linoleic acid and AA, respectively, was also examined. The rationale for examining ALOX15 is that HODEs and HETEs are endogenous PPARγ ligands, thus we wondered whether CES1 silencing affected the transcription of ALOX15. Although transcript levels of ALOX15 were above the limit of detection in both control and CES1KD macrophages, they were not statistically different from each other (2.1-fold greater in CES1KD macrophages, but p=0.16, data not shown).
Discussion
The PPARγ-CYP27A1-LXRα signaling axis is a feedforward pathway that regulates the transcription of a number of genes in macrophages involved in restoring cholesterol homeostasis (21). Here we present evidence that the metabolic serine hydrolase CES1 may regulate macrophage cholesterol metabolism in part by signaling through this axis. We found that silencing CES1 expression in THP-1 macrophages resulted in a profound decrease in the transcription and expression of the oxysterol biosynthetic enzyme CYP27A1 and a concomitant decrease in the levels of 27-OHC, an endogenous oxysterol agonist for LXRα. Importantly, the inhibitory effect on CYP27A1 transcription was recapitulated in THP-1 wildtype cells and primary human macrophages treated with a pharmacological agent WWL113 that selectively inhibits CES1 activity. Furthermore, three lines of evidence indicate an important role for PPARγ in the disrupted CYP27A1 expression: (i) CYP27A1 transcription and expression in CES1KD cells could be restored in part by the addition of a PPARγ agonist (GW1929); (ii) CYP27A1 transcription was significantly attenuated in control cells treated with a PPARγ antagonist (GW9662); and (iii) the induction profile of genes (including CYP27A1) in CES1KD cells exposed to acLDL phenocopied that of PPARγKD cells. LXRα and CD36, like CYP27A1, are also regulated by the heterodimeric PPARγ/RXR nuclear receptors (21, 40) and their transcription and expression were reduced by ablating CES1 expression when macrophages were lipid-loaded, although not to the degree that CYP27A1 was. Altogether, these results suggested a mechanistic link between CES1 and PPARγ. It should be noted that CYP27A1 expression was also regulated in part by RAR, because a specific agonist for this nuclear receptor (TTNPB) could partially restore CYP27A1 transcription and expression in CES1KD cells (Figure 5A, B). This finding is also consistent with that of Szanto et al. (21), which suggested that CYP27A1 expression was postively regulated by RAR in addition to PPARγ.
The LXR-regulated ABCA1 transcript levels in CES1KD foam cells were also reduced when compared to those in control foam cells, which indicated that perturbations in PPARγ signaling in CES1KD cells appeared to lead to wide-range reprograming in gene expression, though ABCG1 levels were not reduced. A small reduction in ABCA1 expression, but a larger reduction in ABCG1 expression, was previously found when PPARγ signaling in cells was inhibited by conditional disruption of the PPARγ gene in mice (41). The difference in our results and those previously reported may be due to species differences. Indeed, different responses to PPARγ agonists have been previously described in murine and human macrophages (41, 42). We also noted some variability in CD36 mRNA levels in CES1KD macrophages that were not lipid loaded. CD36 transcriptional regulation is complex and is controlled by transcription factors other than PPARγ/RXR, including Nrf2 (43) and TR4 (44). Subtle changes in signaling through these or other factors might account for the variability in CD36 transcription in CES1KD macrophages under basal conditions. In addition, Zhao et al. (13) reported that stable overexpression of CES1 in THP-1 cells resulted in increased levels of mRNA for SR-A, CD36, ABCA1, and ABCG1 when cells were lipid loaded (CYP27A1 and LXR expression was not assessed), which is the opposite of what we observed for these genes in CES1KD cells. Together, our results suggest that the loss of CES1 expression in THP-1 macrophages attenuates signaling pathways that operate through the ligand-activated nuclear receptors PPARγ/RXR and RAR/RXR. Functionally, this altered signaling results in defective cholesterol influx into the CES1KD macrophages (Figure 9); however, the influx deficit could be repaired by pretreatment of CES1KD macrophages with GW1929 and 9-cis-RA (in the presence of an LXR antagonist to block the confounding effects of the cholesterol efflux pathway) (Figure 5G). This result suggested that supplementing CES1KD cells with ligands that activate PPARγ/RXR and RAR/RXR overcomes the limited availability of endogenous ligands in CES1-deficient macrophages and normalizes cholesterol homeostasis.
Figure 9. Schematic pathway representing findings of this study.
CES1 deficiency leads to reduced cholesterol influx, because of reduced CD36 and SR-A expression, and attenuates the PPARγ/RXR- and RAR/RXR-regulated CYP27A1/27-OHC axis. Black arrows indicate either lipid-protein or lipid-enzyme interactions, whereas purple arrows indicate ligand-dependent nuclear receptor-mediated expression of its target genes. Abbreviations: ABCA1, ATP-binding cassette A1; ABCG1, ATP-binding cassette G1; ATRA, all-trans retinoic acid; CD36, cluster of differentiation 36; CES1, carboxylesterase 1; COX, cyclooxygenase; FAs, fatty acids; HETE, hydroxyeicosatrienoic acid; HODE, hydroxyoctadecadienoic acid; 27-OHC, 27-hydroxycholesterol; 27-COOH-C, 3-hydroxycholest-5-en-26-oic acid; oxLDL, oxidized LDL; LOX, lipoxygenase; 9-c-RA, 9-cis-retinoic acid; PG, prostaglandin.
We next considered how ablation of CES1 activity might mechanistically alter the gene expression profile. CES1 is a notable serine hydrolase because it exhibits a very broad substrate specificity (Bencharit et al., 2006). It is, therefore, feasible that one of its physiological functions is to hydrolyze ester-containing cellular metabolites (pro-ligands) to generate endogenous ligands that affect signaling through the PPARγ/RXR heterodimer. CES1 can metabolize several classes of neutral lipid esters, including cholesteryl esters, glyceryl esters, and retinyl esters (45). For example, CES1 can hydrolyze the monoacylglycerol 2-AG liberating free AA, which is further metabolized in macrophages to prostaglandins (23, 38). Our results directly showed that CES1 can regulate 2-AG levels in THP-1 cells under different conditions (Figures 7B, 8B), and it is known that 2-AG-derived AA is utilized by COX enzymes to produce eicosanoids (39, 46). Consistent with this, CES1KD cells synthesized smaller quantities of several prostaglandins than control cells did upon cellular metabolism of exogenous 2-AG (Figure 8). An exception to this was TxB2 (the stable metabolite of TxA2), which suggests a different pool of AA is used for TxA2/B2 synthesis than for PGE2 and PGD2. In addition, CES1KD macrophages produced less PGE2 than control macrophages when activated by LPS (Supplementary Figure 5). Cytokine expression (TNFα and IL-6), however, remained intact in the LPS-stimulated CES1KD cells (Supplementary Figure 9), which suggested that TLR-4-mediated signaling and downstream NFkB activity are unaffected by CES1 silencing. Levels of COX-1 and COX-2 transcripts were also similar between control and CES1KD macrophages (Supplementary Figure 10). Although the PGD2 synthase transcript (PTGDS) was lower in CES1KD cells compared to control cells, transcript levels of the PGE2 and TxA2 synthases (PTGS3 and TBXAS1, respectively), which make the two most quantitatively important prostaglandins in THP-1 cells, were not different in the two cell types. Therefore, in general, the levels of the prostaglandin synthetic enzymes in CES1KD cells were unaltered compared to control cells, which indicated that the decrease in prostaglandin levels in CES1KD macrophages is caused by a deficit in the provision of AA substrate to COX-1 and COX-2 enzymes. The significance of this is that several COX-derived oxylipins are ligands for PPARγ (47–49), but these oxylipins were depleted in the CES1KD cells when compared to the control cells.
In addition to monoacylglycerols, other neutral lipid esters are substrates of CES1. For example, CES1 hydrolyzes triacylglycerols (50), generating free fatty acids that can undergo additional oxidative biotransformation (51). Furthermore, lipid glyceryl esters containing oxidized fatty acyl moieties are also substrates for CES1. For instance, prostaglandin glyceryl esters are hydrolyzed by both CES1 and CES2 (38), liberating prostaglandins that might act as PPARγ ligands.
The origin of PPARγ ligands in the setting of scavenger receptor-mediated uptake of oxLDL is presumed to be the oxidized fatty acid components of oxidatively-damaged cholesteryl esters and phospholipids (16, 52). Macrophages, which have been lipid loaded with acLDL, respond in a similar way to those lipid loaded with oxLDL, i.e. both loading procedures result in macrophage foam cells. Internalized acLDL likely provides an abundance of substrates for oxidative enzymes, such as COX and LOX, in addition to containing preformed oxidized fatty acyl moieties. Interestingly, cholesterol esters that contain oxidized fatty acyl moieties were reported to be better substrates for neutral cholesterol ester hydrolase than cholesterol esters containing unoxidized fatty acyl moieties (53, 54). This suggests the possibility that CES1 might liberate endogenous ligands for PPARγ by hydrolyzing neutral lipid esters that possess oxidized fatty acyl moieties. More work is required, however, to understand how CES1 in macrophages affects lipid glyceryl ester metabolism and downstream pathways that might furnish endogenous nuclear receptor ligands. These studies are underway in our laboratories.
CES1 may also have an underappreciated role in vitamin A metabolism. Macrophages are exposed to multiple sources of vitamin A, including retinol bound to the retinol-binding protein 4-transthyretin complex (RBP4-TTR), retinoic acid bound to albumin, retinyl esters in VLDL and LDL particles, retinyl esters in chylomicrons, and β-glucuronides of both retinol and retinoic acid (55). The exact role each has in the supply of vitamin A to macrophages is currently uncertain. However, CES1 may increase the availability of ligands for RAR and RXR via the hydrolytic liberation of retinol stored in retinyl esters. Support for this idea comes from the fact that two rat orthologs of human CES1, termed ES10 and ES4, exhibit retinyl palmitate hydrolase activity (56) and are present in large amounts in rat hepatocytes (57). It should be noted, however, that ES10 and ES4 are not expressed in hepatic stellate cells (57), where most vitamin A is stored as retinyl esters. Whether or not human CES1 can hydrolyze retinyl esters has not been determined. Retinol, once released, needs to be converted to retinoic acid to effect gene expression via RAR and/or RXR. A recent study suggested that macrophages in human intestine can biosynthesize retinoic acid from retinol, and that retinoic acid plays an important role in regulating the macrophage phenotype (58). Overall, the roles of retinoids and CES1 in the regulation of human macrophage gene transcription still needs to be explored.
The potential for CES1 to regulate the availability of ligands for PPARγ, RAR, and RXR, and, via these ligand-activated nuclear receptors, to control CYP27A1 expression, might have broader implications (Figure 9). For example, CYP27A1-derived oxysterol, 27-OHC, may be involved in the negative feedback regulation of CES1 activity. We previously described the ability of some oxysterols and polyunsaturated fatty acids to potently inhibit CES1 activity (25). 27-OHC exhibited an apparent inhibitory constant (Ki) for CES1 around 10 nM and showed a significant degree of structural specificity. For instance, 25-OHC, a regioisomer of 27-OHC, had little capacity to inhibit CES1. In addition, several fatty acids could also inhibit CES1 activity, with unsaturated fatty acids being more potent than saturated fatty acids. Interestingly, AA was the most potent fatty acid inhibitor (Ki = 1.7 μM). Further, the PGD2-derived metabolite 15-deoxy-Δ12,14 prostaglandin J2, a known PPARγ ligand (47), was also an inhibitor of CES1. The ability of these lipids to inhibit CES1 suggests a potential self-limiting mechanism whereby CES1 hydrolyzes pro-ligand esters to liberate either preformed PPARγ ligands or fatty acids that are subsequently oxygenated to products that are PPARγ ligands (Figure 9). Release of PPARγ ligands then upregulates CYP27A1 expression resulting in the increased synthesis of 27-OHC (21). Subsequently, fatty acids, released by the hydrolysis of neutral lipid esters, and 27-OHC might directly interact with CES1 and inhibit its enzymatic activity. If such a regulatory pathway is operating in macrophages, ablation of CES1 activity may well affect other PPARγ-mediated processes, e.g. macrophage polarization.
In conclusion, our results are consistent with a model in which CES1 abrogation results in decreased CYP27A1-LXRα signaling by potentially reducing the availability of endogenous ligands for PPARγ, RAR, and/or RXR. We plan to further investigate the role that CES1 plays in macrophage cholesterol homeostasis and its molecular mechanisms, including identification of its endogenous substrates. Finally, it should be pointed out that our interpretation of CES1’s ability to regulate macrophage gene expression does not require it to directly act as a cholesteryl ester hydrolase, a catalytic activity that is intensely debated for CES1 (59–62).
Supplementary Material
Supplementary Figure 1. (A) ABPP-MUDPIT profiling of serine hydrolases in human THP-1 macrophages treated with vehicle or paraoxon (PO). Cells were treated with vehicle (ethanol, 0.1% v/v) or PO (10 μM) at 37°C in serum-free RPMI medium. After 1 h, the cells were harvested, washed with PBS, and processed for ABPP-MUDPIT, as described previously (Wang et al., 2013). Serine hydrolases are denoted by their gene symbols and those inhibited by PO are annotated with red boxes. Data represents the mean ± SD of n=3 biological replicates, * p<0.05 (Student’s t-test). (B) mRNA transcript levels of CES1, CES2, and CES3 in control, CES1KD, and WT THP-1 macrophages. CES1 expression was significantly reduced in CES1KD macrophages relative to that of control and WT macrophages. On the other hand, CES2 and CES3 gene expression were not significantly altered by CES1 knockdown. Data for control and CES1KD macrophages represents the mean ± SD of three independent experiments with quadruplicate replicates each time, whereas data for WT macrophages represents a single experiment with quadruplicate replicates. **p<0.01 (one-way ANOVA).
Supplementary Figure 9. Stimulation of control and CES1KD macrophages with LPS: Effects on cytokine expression. Stimulation of both cell types with LPS (1 μg/mL, 4 h) induced cytokine (TNFα and IL-6) and TLR-4 (only CES1KD cells) mRNA levels. Data represents the mean ± SD of n=3 biological replicates. ** p<0.01 LPS-stimulated control cells vs non-stimulated control cells; # p<0.05, ## p<0.01 LPS-stimulated CES1KD cells vs non-stimulated CES1KD cells; && p<0.01 LPS-stimulated CES1KD cells vs LPS-stimulated control cells; ^ p<0.05 non-stimulated CES1KD cells vs non-stimulated control cells (two-way ANOVA with Student-Newman-Keuls post-hoc test); ns, not significant.
Supplementary Figure 10. CES1 silencing does not alter the transcription of genes involved in eicosanoid biosynthesis. mRNA levels of genes associated with eicosanoid synthesis in control macrophages (-LPS) and LPS-stimulated macrophages (+LPS) were determined. Data are expressed as relative mRNA levels and the control (-LPS) was set to 1.0 for each gene (n=3–4 biological replicates, mean ± SD). * p<0.05 for LPS-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test).
Supplementary Figure 2. CES1 silencing alters the transcription of several genes involved in cholesterol metabolism. mRNA levels of genes associated with cholesterol metabolism in non-acLDL-loaded macrophages (-acLDL) and acLDL-loaded macrophages (+acLDL, 50 μg/mL, 24 h). Data are expressed as relative mRNA levels and the control (-acLDL) was set to 1.0 for each gene (n=3 biological replicates, mean ± SD). * p<0.05 for acLDL-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test). A significant interaction term between cell type and acLDL treatment is denoted in graphs as p<0.05 cell type x acLDL. If not given, no significant interaction was noted between cell type and acLDL treatment.
Supplementary Figure 3. mRNA transcript levels of PPARγ, PPARα, and PPARδ in control and PPARγKD THP-1 macrophages. PPARγ expression was significantly reduced in PPARγKD macrophages relative to that of control macrophages. On the other hand, PPARα (PPARa) and PPARβ/δ (PPARd) gene expression were not significantly altered by PPARγ knockdown. Data represents the mean ± SD of four biological replicates. **p<0.01 (Student’s t-test).
Supplementary Figure 4. Extent of cholesterol loading of control macrophages and PPARγKD macrophages. Macrophages (control and PPARγKD) were exposed to acLDL (50 μg/mL) for 48 h, followed by an overnight equilibration period (no cholesterol acceptors in the culture medium to prevent cholesterol efflux). Cells were then washed with PBS and the levels of cellular cholesterol were determined. Data represents the mean ± SD of n=4–5 biological replicates per group, *p<0.1, **p<0.05 (Student’s t-test).
Supplementary Figure 5. PGE2 biosynthesis in macrophages challenged with LPS. Control, CES1KD, and PPARγKD macrophages were treated with LPS (1 μg/mL, 4 h) or not treated. The levels of PGE2 in extracts of the culture medium and cells were determined by LC-MS/MS and summed together. Data are expressed as relative PGE2 levels and the control (-LPS) was set to 1.0 (n=3 biological replicates, mean ± SD). * p<0.05 for LPS-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test).
Supplementary Figure 6. CD36 and SR-A scavenger receptor transcription in CES1KD macrophages following treatment with a pharmacological agonist for PPARγ (Rosiglitazone, 5 μM) in the absence or presence of oxLDL (25 μg/mL) for 24 h. Data represent the mean ± SD of n=2 biological replicates per group. Rosi vehicle was DMSO.
Supplementary Figure 7. Treatment of THP-1 wildtype monocytes with arachidonic acid stimulates production of 2-AG. Monocytes were treated with the indicated concentrations of AA and the levels of 2-AG in medium were determined by LC-MS/MS analysis. 2-AG levels in untreated monocytes were set to 1. Data represents the mean ± SD of n=3 wells per group.
Supplementary Figure 8. THP-1 wildtype cell lysates pretreated with WWL113 and CES1KD lysates exhibit reduced 2-AG-d8 hydrolysis activities. (A) Wildtype macrophage lysates were pretreated with vehicle (DMSO) and WWL113 (1 μM) for 30 min before incubating with 2-AG-d8 (2.5 μM) for the indicated amounts of time. (B) Control and CES1KD macrophage lysates were incubated with 2-AG-d8 (2.5 μM) for indicated times. Enzymatic reactions were terminated with an equal volume of cold methanol containing CUDA as internal standard. AA-d8 levels were quantified by LC-MS/MS and normalized on the CUDA standard.
Acknowledgments
This work was supported by the National Institutes of Health grant numbers 1R15GM116129–01 and 1R15ES015348–02. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or Mississippi State University.
Funding Statement
Funded by NIH 1R15GM116129–01 and 1R15ES015348–02
Abbreviations:
- AA
arachidonic acid
- ABCA1
ATP binding cassette transporter A1
- ABCG1
ATP binding cassette transporter G1
- acLDL
acetylated LDL
- ALOX15
15-lipoxygenase
- CES1
carboxylesterase 1
- CES1KD
THP-1 cells with CES1 expression knocked down
- COX
cyclooxygenase
- CYP27A1
cytochrome P45027A1
- DAGLβ
diacylglycerol lipase β
- HETEs
hydroxyeicosotetraenoic acids
- HODEs
hydroxyoctadecadienoic acids
- Ki
apparent inhibition constant for inhibitor
- LOX
lipoxygenase
- LXR
liver X receptor
- mRCT
macrophage reverse cholesterol transport
- NPC1 and NPC2
Nieman Pick C1 and C2 proteins
- PPARγ
peroxisome proliferator-activated receptor gamma
- RAR
retinoic acid receptor
- RXR
retinoid X receptor
- StAR
steroidogenic acute regulatory protein
- CB1
cannabinoid receptor 1
- 2-AG
2-arachidonoylglycerol
- 24-OHC
24-hydroxycholesterol
- 25-OHC
25-hydroxycholesterol
- 27-OHC
27-hydroxycholesterol
Footnotes
Declaration of Interests
The authors declare no conflicts of interest
References
- 1.Libby P, Theroux P. Pathophysiology of coronary artery disease. Circulation. 2005;111(25):3481–8. [DOI] [PubMed] [Google Scholar]
- 2.Kunjathoor VV, Febbraio M, Podrez EA, Moore KJ, Andersson L, Koehn S, et al. Scavenger receptors class A-I/II and CD36 are the principal receptors responsible for the uptake of modified low density lipoprotein leading to lipid loading in macrophages. J Biol Chem. 2002;277(51):49982–8. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein JL, Dana SE, Faust JR, Beaudet AL, Brown MS. Role of lysosomal acid lipase in the metabolism of plasma low density lipoprotein. Observations in cultured fibroblasts from a patient with cholesteryl ester storage disease. J Biol Chem. 1975;250(21):8487–95. [PubMed] [Google Scholar]
- 4.Adorni MP, Zimetti F, Billheimer JT, Wang N, Rader DJ, Phillips MC, et al. The roles of different pathways in the release of cholesterol from macrophages. J Lipid Res. 2007;48(11):2453–62. [DOI] [PubMed] [Google Scholar]
- 5.Lewis GF, Rader DJ. New insights into the regulation of HDL metabolism and reverse cholesterol transport. Circ Res. 2005;96(12):1221–32. [DOI] [PubMed] [Google Scholar]
- 6.Rosenson RS, Brewer HB Jr., Davidson WS, Fayad ZA, Fuster V, Goldstein J, et al. Cholesterol efflux and atheroprotection: advancing the concept of reverse cholesterol transport. Circulation. 2012;125(15):1905–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang TY, Chang CC, Lin S, Yu C, Li BL, Miyazaki A. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and −2. Curr Opin Lipidol. 2001;12(3):289–96. [DOI] [PubMed] [Google Scholar]
- 8.Rothblat GH, de la Llera-Moya M, Favari E, Yancey PG, Kellner-Weibel G. Cellular cholesterol flux studies: methodological considerations. Atherosclerosis. 2002;163(1):1–8. [DOI] [PubMed] [Google Scholar]
- 9.Moore KJ, Tabas I. Macrophages in the pathogenesis of atherosclerosis. Cell. 2011;145(3):341–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghosh S Cholesteryl ester hydrolase in human monocyte/macrophage: cloning, sequencing, and expression of full-length cDNA. Physiol Genomics. 2000;2(1):1–8. [DOI] [PubMed] [Google Scholar]
- 11.Ross MK, Borazjani A, Wang R, Crow JA, Xie S. Examination of the carboxylesterase phenotype in human liver. Arch Biochem Biophys. 2012;522(1):44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Song J, Chow WN, St Clair RW, Rudel LL, Ghosh S. Macrophage-specific transgenic expression of cholesteryl ester hydrolase significantly reduces atherosclerosis and lesion necrosis in Ldlr mice. J Clin Invest. 2007;117(10):2983–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao B, Song J, St Clair RW, Ghosh S. Stable overexpression of human macrophage cholesteryl ester hydrolase results in enhanced free cholesterol efflux from human THP1 macrophages. Am J Physiol Cell Physiol. 2007;292(1):C405–12. [DOI] [PubMed] [Google Scholar]
- 14.Crow JA, Middleton BL, Borazjani A, Hatfield MJ, Potter PM, Ross MK. Inhibition of carboxylesterase 1 is associated with cholesteryl ester retention in human THP-1 monocyte/macrophages. Biochim Biophys Acta. 2008;1781(10):643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross MK, Borazjani A, Mangum LC, Wang R, Crow JA. Effects of toxicologically relevant xenobiotics and the lipid-derived electrophile 4-hydroxynonenal on macrophage cholesterol efflux: silencing carboxylesterase 1 has paradoxical effects on cholesterol uptake and efflux. Chem Res Toxicol. 2014;27(10):1743–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93(2):229–40. [DOI] [PubMed] [Google Scholar]
- 17.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93(2):241–52. [DOI] [PubMed] [Google Scholar]
- 18.Costet P, Luo Y, Wang N, Tall AR. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J Biol Chem. 2000;275(36):28240–5. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz K, Lawn RM, Wade DP. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem Biophys Res Commun. 2000;274(3):794–802. [DOI] [PubMed] [Google Scholar]
- 20.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, et al. Control of cellular cholesterol efflux by the nuclear oxysterol receptor LXR alpha. Proc Natl Acad Sci U S A. 2000;97(22):12097–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szanto A, Benko S, Szatmari I, Balint BL, Furtos I, Ruhl R, et al. Transcriptional regulation of human CYP27 integrates retinoid, peroxisome proliferator-activated receptor, and liver X receptor signaling in macrophages. Mol Cell Biol. 2004;24(18):8154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X, Menke JG, Chen Y, Zhou G, MacNaul KL, Wright SD, et al. 27-hydroxycholesterol is an endogenous ligand for liver X receptor in cholesterol-loaded cells. J Biol Chem. 2001;276(42):38378–87. [DOI] [PubMed] [Google Scholar]
- 23.Wang R, Borazjani A, Matthews AT, Mangum LC, Edelmann MJ, Ross MK. Identification of palmitoyl protein thioesterase 1 in human THP1 monocytes and macrophages and characterization of unique biochemical activities for this enzyme. Biochemistry. 2013;52(43):7559–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dominguez E, Galmozzi A, Chang JW, Hsu KL, Pawlak J, Li W, et al. Integrated phenotypic and activity-based profiling links Ces3 to obesity and diabetes. Nat Chem Biol. 2014;10(2):113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crow JA, Herring KL, Xie S, Borazjani A, Potter PM, Ross MK. Inhibition of carboxylesterase activity of THP1 monocytes/macrophages and recombinant human carboxylesterase 1 by oxysterols and fatty acids. Biochim Biophys Acta. 2010;1801(1):31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 27.Shinkyo R, Guengerich FP. Inhibition of human cytochrome P450 3A4 by cholesterol. J Biol Chem. 2011;286(21):18426–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews AT, Lee JH, Borazjani A, Mangum LC, Hou X, Ross MK. Oxyradical stress increases the biosynthesis of 2-arachidonoylglycerol: involvement of NADPH oxidase. Am J Physiol Cell Physiol. 2016;311(6):C960–C74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahmoud AM, Hussein OE, Hozayen WG, Abd El-Twab SM. Methotrexate hepatotoxicity is associated with oxidative stress, and down-regulation of PPARgamma and Nrf2: Protective effect of 18beta-Glycyrrhetinic acid. Chem Biol Interact. 2017;270:59–72. [DOI] [PubMed] [Google Scholar]
- 30.Wnuk A, Rzemieniec J, Litwa E, Lason W, Krzeptowski W, Wojtowicz AK, et al. The Crucial Involvement of Retinoid X Receptors in DDE Neurotoxicity. Neurotox Res. 2016;29(1):155–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchmeyer M, Koufany M, Sebillaud S, Netter P, Jouzeau JY, Bianchi A. All-trans retinoic acid suppresses interleukin-6 expression in interleukin-1-stimulated synovial fibroblasts by inhibition of ERK1/2 pathway independently of RAR activation. Arthritis Res Ther. 2008;10(6):R141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subbanna S, Psychoyos D, Xie S, Basavarajappa BS. Postnatal ethanol exposure alters levels of 2-arachidonylglycerol-metabolizing enzymes and pharmacological inhibition of monoacylglycerol lipase does not cause neurodegeneration in neonatal mice. J Neurochem. 2015;134(2):276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klucken J, Buchler C, Orso E, Kaminski WE, Porsch-Ozcurumez M, Liebisch G, et al. ABCG1 (ABC8), the human homolog of the Drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci U S A. 2000;97(2):817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Notarnicola M, Tutino V, De Nunzio V, Dituri F, Caruso MG, Giannelli G. Dietary omega-3 Polyunsaturated Fatty Acids Inhibit Tumor Growth in Transgenic ApcMin/+ Mice, Correlating with CB1 Receptor Up-Regulation. Int J Mol Sci. 2017;18(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quinn CM, Jessup W, Wong J, Kritharides L, Brown AJ. Expression and regulation of sterol 27-hydroxylase (CYP27A1) in human macrophages: a role for RXR and PPARgamma ligands. Biochem J. 2005;385(Pt 3):823–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phillips MC. Molecular mechanisms of cellular cholesterol efflux. J Biol Chem. 2014;289(35):24020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niphakis MJ, Cravatt BF. Enzyme inhibitor discovery by activity-based protein profiling. Annu Rev Biochem. 2014;83:341–77. [DOI] [PubMed] [Google Scholar]
- 38.Xie S, Borazjani A, Hatfield MJ, Edwards CC, Potter PM, Ross MK. Inactivation of lipid glyceryl ester metabolism in human THP1 monocytes/macrophages by activated organophosphorus insecticides: role of carboxylesterases 1 and 2. Chem Res Toxicol. 2010;23(12):1890–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. DAGLbeta inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8(12):999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, et al. A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis. Mol Cell. 2001;7(1):161–71. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama TE, Sakai S, Lambert G, Nicol CJ, Matsusue K, Pimprale S, et al. Conditional disruption of the peroxisome proliferator-activated receptor gamma gene in mice results in lowered expression of ABCA1, ABCG1, and apoE in macrophages and reduced cholesterol efflux. Mol Cell Biol. 2002;22(8):2607–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chinetti G, Lestavel S, Bocher V, Remaley AT, Neve B, Torra IP, et al. PPAR-alpha and PPAR-gamma activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat Med. 2001;7(1):53–8. [DOI] [PubMed] [Google Scholar]
- 43.Maruyama A, Tsukamoto S, Nishikawa K, Yoshida A, Harada N, Motojima K, et al. Nrf2 regulates the alternative first exons of CD36 in macrophages through specific antioxidant response elements. Arch Biochem Biophys. 2008;477(1):139–45. [DOI] [PubMed] [Google Scholar]
- 44.Xie S, Lee YF, Kim E, Chen LM, Ni J, Fang LY, et al. TR4 nuclear receptor functions as a fatty acid sensor to modulate CD36 expression and foam cell formation. Proc Natl Acad Sci U S A. 2009;106(32):13353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lian J, Nelson R, Lehner R. Carboxylesterases in lipid metabolism: from mouse to human. Protein Cell. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nomura DK, Morrison BE, Blankman JL, Long JZ, Kinsey SG, Marcondes MC, et al. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 2011;334(6057):809–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forman BM, Tontonoz P, Chen J, Brun RP, Spiegelman BM, Evans RM. 15-Deoxy-delta 12, 14-prostaglandin J2 is a ligand for the adipocyte determination factor PPAR gamma. Cell. 1995;83(5):803–12. [DOI] [PubMed] [Google Scholar]
- 48.Kliewer SA, Lenhard JM, Willson TM, Patel I, Morris DC, Lehmann JM. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor gamma and promotes adipocyte differentiation. Cell. 1995;83(5):813–9. [DOI] [PubMed] [Google Scholar]
- 49.Raman P, Kaplan BL, Thompson JT, Vanden Heuvel JP, Kaminski NE. 15-Deoxy-delta12,14-prostaglandin J2-glycerol ester, a putative metabolite of 2-arachidonyl glycerol, activates peroxisome proliferator activated receptor gamma. Mol Pharmacol. 2011;80(1):201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lehner R, Verger R. Purification and characterization of a porcine liver microsomal triacylglycerol hydrolase. Biochemistry. 1997;36(7):1861–8. [DOI] [PubMed] [Google Scholar]
- 51.Schild RL, Schaiff WT, Carlson MG, Cronbach EJ, Nelson DM, Sadovsky Y. The activity of PPAR gamma in primary human trophoblasts is enhanced by oxidized lipids. J Clin Endocrinol Metab. 2002;87(3):1105–10. [DOI] [PubMed] [Google Scholar]
- 52.Salomon RG, Gu X. Critical insights into cardiovascular disease from basic research on the oxidation of phospholipids: the gamma-hydroxyalkenal phospholipid hypothesis. Chem Res Toxicol. 2011;24(11):1791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belkner J, Stender H, Holzhutter HG, Holm C, Kuhn H. Macrophage cholesteryl ester hydrolases and hormone-sensitive lipase prefer specifically oxidized cholesteryl esters as substrates over their non-oxidized counterparts. Biochem J. 2000;352 Pt 1:125–33. [PMC free article] [PubMed] [Google Scholar]
- 54.Weibel GL, Joshi MR, Alexander ET, Zhu P, Blair IA, Rothblat GH. Overexpression of human 15(S)-lipoxygenase-1 in RAW macrophages leads to increased cholesterol mobilization and reverse cholesterol transport. Arteriosclerosis, thrombosis, and vascular biology. 2009;29(6):837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Byrne SM, Blaner WS. Retinol and retinyl esters: biochemistry and physiology. J Lipid Res. 2013;54(7):1731–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanghani SP, Davis WI, Dumaual NG, Mahrenholz A, Bosron WF. Identification of microsomal rat liver carboxylesterases and their activity with retinyl palmitate. Eur J Biochem. 2002;269(18):4387–98. [DOI] [PubMed] [Google Scholar]
- 57.Mello T, Nakatsuka A, Fears S, Davis W, Tsukamoto H, Bosron WF, et al. Expression of carboxylesterase and lipase genes in rat liver cell-types. Biochem Biophys Res Commun. 2008;374(3):460–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sanders TJ, McCarthy NE, Giles EM, Davidson KL, Haltalli ML, Hazell S, et al. Increased production of retinoic acid by intestinal macrophages contributes to their inflammatory phenotype in patients with Crohn’s disease. Gastroenterology. 2014;146(5):1278–88 e1–2. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh S, Zhao B, Bie J, Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascul Pharmacol. 2010;52(1–2):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Igarashi M, Osuga J, Uozaki H, Sekiya M, Nagashima S, Takahashi M, et al. The critical role of neutral cholesterol ester hydrolase 1 in cholesterol removal from human macrophages. Circ Res. 2010;107(11):1387–95. [DOI] [PubMed] [Google Scholar]
- 61.Buchebner M, Pfeifer T, Rathke N, Chandak PG, Lass A, Schreiber R, et al. Cholesteryl ester hydrolase activity is abolished in HSL−/− macrophages but unchanged in macrophages lacking KIAA1363. J Lipid Res. 2010;51(10):2896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sakai K, Igarashi M, Yamamuro D, Ohshiro T, Nagashima S, Takahashi M, et al. Critical role of neutral cholesteryl ester hydrolase 1 in cholesteryl ester hydrolysis in murine macrophages. J Lipid Res. 2014;55(10):2033–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. (A) ABPP-MUDPIT profiling of serine hydrolases in human THP-1 macrophages treated with vehicle or paraoxon (PO). Cells were treated with vehicle (ethanol, 0.1% v/v) or PO (10 μM) at 37°C in serum-free RPMI medium. After 1 h, the cells were harvested, washed with PBS, and processed for ABPP-MUDPIT, as described previously (Wang et al., 2013). Serine hydrolases are denoted by their gene symbols and those inhibited by PO are annotated with red boxes. Data represents the mean ± SD of n=3 biological replicates, * p<0.05 (Student’s t-test). (B) mRNA transcript levels of CES1, CES2, and CES3 in control, CES1KD, and WT THP-1 macrophages. CES1 expression was significantly reduced in CES1KD macrophages relative to that of control and WT macrophages. On the other hand, CES2 and CES3 gene expression were not significantly altered by CES1 knockdown. Data for control and CES1KD macrophages represents the mean ± SD of three independent experiments with quadruplicate replicates each time, whereas data for WT macrophages represents a single experiment with quadruplicate replicates. **p<0.01 (one-way ANOVA).
Supplementary Figure 9. Stimulation of control and CES1KD macrophages with LPS: Effects on cytokine expression. Stimulation of both cell types with LPS (1 μg/mL, 4 h) induced cytokine (TNFα and IL-6) and TLR-4 (only CES1KD cells) mRNA levels. Data represents the mean ± SD of n=3 biological replicates. ** p<0.01 LPS-stimulated control cells vs non-stimulated control cells; # p<0.05, ## p<0.01 LPS-stimulated CES1KD cells vs non-stimulated CES1KD cells; && p<0.01 LPS-stimulated CES1KD cells vs LPS-stimulated control cells; ^ p<0.05 non-stimulated CES1KD cells vs non-stimulated control cells (two-way ANOVA with Student-Newman-Keuls post-hoc test); ns, not significant.
Supplementary Figure 10. CES1 silencing does not alter the transcription of genes involved in eicosanoid biosynthesis. mRNA levels of genes associated with eicosanoid synthesis in control macrophages (-LPS) and LPS-stimulated macrophages (+LPS) were determined. Data are expressed as relative mRNA levels and the control (-LPS) was set to 1.0 for each gene (n=3–4 biological replicates, mean ± SD). * p<0.05 for LPS-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test).
Supplementary Figure 2. CES1 silencing alters the transcription of several genes involved in cholesterol metabolism. mRNA levels of genes associated with cholesterol metabolism in non-acLDL-loaded macrophages (-acLDL) and acLDL-loaded macrophages (+acLDL, 50 μg/mL, 24 h). Data are expressed as relative mRNA levels and the control (-acLDL) was set to 1.0 for each gene (n=3 biological replicates, mean ± SD). * p<0.05 for acLDL-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test). A significant interaction term between cell type and acLDL treatment is denoted in graphs as p<0.05 cell type x acLDL. If not given, no significant interaction was noted between cell type and acLDL treatment.
Supplementary Figure 3. mRNA transcript levels of PPARγ, PPARα, and PPARδ in control and PPARγKD THP-1 macrophages. PPARγ expression was significantly reduced in PPARγKD macrophages relative to that of control macrophages. On the other hand, PPARα (PPARa) and PPARβ/δ (PPARd) gene expression were not significantly altered by PPARγ knockdown. Data represents the mean ± SD of four biological replicates. **p<0.01 (Student’s t-test).
Supplementary Figure 4. Extent of cholesterol loading of control macrophages and PPARγKD macrophages. Macrophages (control and PPARγKD) were exposed to acLDL (50 μg/mL) for 48 h, followed by an overnight equilibration period (no cholesterol acceptors in the culture medium to prevent cholesterol efflux). Cells were then washed with PBS and the levels of cellular cholesterol were determined. Data represents the mean ± SD of n=4–5 biological replicates per group, *p<0.1, **p<0.05 (Student’s t-test).
Supplementary Figure 5. PGE2 biosynthesis in macrophages challenged with LPS. Control, CES1KD, and PPARγKD macrophages were treated with LPS (1 μg/mL, 4 h) or not treated. The levels of PGE2 in extracts of the culture medium and cells were determined by LC-MS/MS and summed together. Data are expressed as relative PGE2 levels and the control (-LPS) was set to 1.0 (n=3 biological replicates, mean ± SD). * p<0.05 for LPS-dependent effects, # p<0.05 for cell type-dependent effects (two-way ANOVA using Student-Newman-Keuls post-hoc test).
Supplementary Figure 6. CD36 and SR-A scavenger receptor transcription in CES1KD macrophages following treatment with a pharmacological agonist for PPARγ (Rosiglitazone, 5 μM) in the absence or presence of oxLDL (25 μg/mL) for 24 h. Data represent the mean ± SD of n=2 biological replicates per group. Rosi vehicle was DMSO.
Supplementary Figure 7. Treatment of THP-1 wildtype monocytes with arachidonic acid stimulates production of 2-AG. Monocytes were treated with the indicated concentrations of AA and the levels of 2-AG in medium were determined by LC-MS/MS analysis. 2-AG levels in untreated monocytes were set to 1. Data represents the mean ± SD of n=3 wells per group.
Supplementary Figure 8. THP-1 wildtype cell lysates pretreated with WWL113 and CES1KD lysates exhibit reduced 2-AG-d8 hydrolysis activities. (A) Wildtype macrophage lysates were pretreated with vehicle (DMSO) and WWL113 (1 μM) for 30 min before incubating with 2-AG-d8 (2.5 μM) for the indicated amounts of time. (B) Control and CES1KD macrophage lysates were incubated with 2-AG-d8 (2.5 μM) for indicated times. Enzymatic reactions were terminated with an equal volume of cold methanol containing CUDA as internal standard. AA-d8 levels were quantified by LC-MS/MS and normalized on the CUDA standard.