Abstract
Polyamine inhibition for cancer therapy is, conceptually, an attractive approach but has yet to meet success in the clinical setting. The aryl hydrocarbon receptor (AHR) is the central transcriptional regulator of the xenobiotic response. Our study revealed that AHR also positively regulates intracellular polyamine production via direct transcriptional activation of 2 genes, ODC1 and AZIN1, which are involved in polyamine biosynthesis and control, respectively. In patients with multiple myeloma (MM), AHR levels were inversely correlated with survival, suggesting that AHR inhibition may be beneficial for the treatment of this disease. We identified clofazimine (CLF), an FDA-approved anti-leprosy drug, as a potent AHR antagonist and a suppressor of polyamine biosynthesis. Experiments in a transgenic model of MM (Vk*Myc mice) and in immunocompromised mice bearing MM cell xenografts revealed high efficacy of CLF comparable to that of bortezomib, a first-in-class proteasome inhibitor used for the treatment of MM. This study identifies a previously unrecognized regulatory axis between AHR and polyamine metabolism and reveals CLF as an inhibitor of AHR and a potentially clinically relevant anti-MM agent.
Keywords: Cell Biology, Oncology
Keywords: Cancer, Polyamines
Introduction
Polyamines are small polycationic molecules that have been implicated in virtually every aspect of eukaryotic cells, including DNA replication, transcription, translation, and proliferation (1–3). Polyamine levels are increased in malignancies, where they contribute to disease progression and metastasis (4–6). The first rate-limiting step in polyamine biosynthesis is the conversion of ornithine to putrescine by the enzyme ornithine decarboxylase (ODC1) (7). ODC1 activity is regulated at multiple levels from transcription to protein degradation, the latter occurring through interaction with the protein ODC1 antizyme (OAZ1-3) (7). The ability of OAZ to interact with and inhibit ODC1 is in turn regulated by the antizyme inhibitors AZIN1-2, thus providing a complex autoregulatory loop (7). With the exception of the transcription factors MYC (8–10) and SP1 (11), little is known about the transcriptional regulation of polyamine biosynthesis. Importantly, although both ODC1 and AZIN1 have been shown to have protumorigenic functions (12–14) and anti-polyamine agents have been under consideration as chemotherapy, none is currently in clinical use (2).
The aryl hydrocarbon receptor (AHR) is a transcription factor that belongs to the ligand-activated nuclear receptor superfamily and has a prominent role in mediating xenobiotic responses (15–17). AHR is sequestered in the cytoplasm by chaperone proteins and, upon ligand binding, translocates into the nucleus, where it dimerizes with the main binding partner AHR nuclear translocator (ARNT) (15–17) to regulate the transcription of genes containing xenobiotic response elements (XREs) within their regulatory regions. Initially considered solely a toxin clearance mediator, AHR has recently been suggested to play a key role in the regulation of several physiological, developmental, and pathological processes (15, 18–24). AHR can act as a tumor suppressor in some settings (25, 26), but an oncogene in others (27–31). However, to date, no specific AHR agonists or antagonists have been approved for use in humans.
Multiple myeloma (MM) is an incurable malignancy of bone marrow–resident plasma cells (32). Despite the progress achieved with the introduction of several agents, including bortezomib (BTZ) (Velcade), the first-in-class proteasome inhibitor (33), MM remains incurable, necessitating the development of novel therapies. Recent studies have linked AHR activation with an increased risk of MM (34–36). Accordingly, a separate study found that polyamine levels are substantially elevated in patients with MM (4), however, the role of AHR or polyamines in MM progression is virtually unknown (37).
Clofazimine (CLF) is a well-tolerated, riminophenazine-based agent approved by the US FDA for the treatment of leprosy and tuberculosis (38, 39). Suppression of transformed phenotypes by CLF has been previously reported to impinge on a variety of pathways and targets (40–44); however, CLF antitumor efficacy has not been evaluated in clinical trials.
Here, we report the identification of AHR as a major transcriptional regulator of polyamine biosynthesis in both normal and transformed cells. Additionally, we provide evidence suggesting that inhibition of AHR by CLF is a clinically relevant antimyeloma strategy.
Results
AHR is a direct transcriptional activator of ODC1 and AZIN1.
ODC1 and AZIN1 have been shown to have protumorigenic functions (12–14), and anti-polyamine agents have long been under consideration as chemotherapeutic agents, although none is currently in clinical use (2). Thus, a therapeutic strategy aimed at the simultaneous inhibition of ODC1 and AZIN1 may prove beneficial. To identify potential transcriptional regulators of ODC1 and AZIN1, human and mouse promoter regions for these genes (from 5,000 bp upstream of the transcription start site [TSS] to the 5′-UTR) were aligned using CONREAL (Conserved Regulatory Elements Anchored Alignment, http://conreal.genomes.nl/) software. A minimal threshold value of 0.9 for the 2 relative scores provided by the program was applied, which resulted in the identification of 38 and 29 transcription factors potentially binding to AZIN1p and ODC1p, respectively, out of which 16 were found to be in common between the 2 genes (Figure 1A). One of these transcription factors was AHR (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI70712DS1). This in silico–based prediction was confirmed by interrogating a second publically available tool, the Gene Promoter Miner (GPMiner, http://gpminer.mbc.nctu.edu.tw/about.htm). To test whether AHR regulates AZIN1 and ODC1, we first used normal human fibroblasts (WI-38) as cells with the least perturbed signaling pathways.
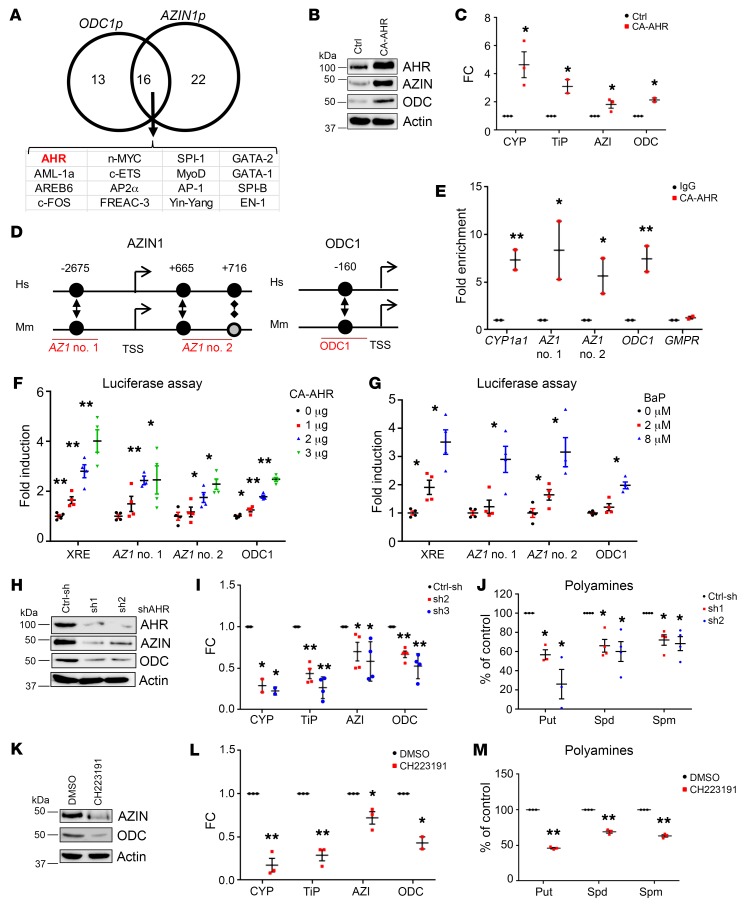
Figure 1. AHR controls ODC1 and AZIN1 transcription.
(A) Predicted transcription factors binding AZIN1 and ODC1 promoters. (B) Extracts from WI-38 cells expressing empty vector control (Ctrl) or CA-AHR probed by immunoblotting for AZIN1 and ODC1. (C) RNA from cells as in B probed in qRT-PCR with the indicated primers and probes. Data represent the average ± SEM of 3 independent experiments performed in triplicate. (D) Schematic of conserved (black circles) or partially conserved (shaded circle) AHR binding sites in the indicated promoters. Hs, Homo sapiens; Mm, Mus musculus. (E) WI-38 DNA was immunoprecipitated with control (IgG) or AHR-specific antibodies and probed in qRT-PCR with primers for the CYP1a1 promoter (positive control), regions in AZIN1 and ODC1 promoters described in D, or GMPR (negative control). Luciferase activity for the AZIN1 and ODC1 promoter regions described in D with increasing amounts of CA-AHR (F) or BaP (G). The XRE-luc plasmid was used as a control. Data represent the average ± SEM of 2 independent experiments performed in duplicate. (H) Cell extracts of WI-38 cells expressing control shRNA (Ctrl-sh) or 2 independent shRNAs against AHR (sh1 and sh2) probed by immunoblotting with the indicated antibodies. (I) RNA from cells as in H probed in qRT-PCR with the indicated primers and probes. Data represent the average ± SEM of 4 independent experiments performed in triplicate. (J) Polyamine content in cells as in H. Data represent the average ± SEM of 4 independent experiments. (K) Extracts of WI-38 cells treated for 2 hours with DMSO or 20 μM CH223191 probed by immunoblotting for AZIN1 and ODC1. (L) RNA from cells as in K probed in qRT-PCR with the indicated primers and probes. Data represent the average ± SEM of 3 independent experiments performed in triplicate. (M) Polyamine content in WI-38 cells treated with CH223191 for 48 hours. Data represent the average ± SEM of 3 independent experiments. *P < 0.05 and **P < 0.001, by 2-tailed Student’s t test. AZI, AZIN1 ; CYP, CYP1a1; ODC, ODC1; TiP, TiPARP; Spd, spermidine; Put, putrescine; Spm, spermine.
Ectopic expression of a constitutively active AHR, CA-AHR, whose transcriptional activity is ligand independent (45), resulted in the induction of both AZIN1 and ODC1 at the protein and mRNA levels (Figure 1, B and C). Two established AHR targets, CYP1a1 (46) and TiPARP (47), were used as positive controls (Figure 1C). Human and mouse promoters for ODC1 and AZIN1 contained XREs that were conserved between species (Figure 1D and Supplemental Figure 1A). We performed ChIP assays using a commercially available antibody against AHR and primers flanking these putative AHR binding sites (Figure 1D), as well as primers flanking the AHR binding site in the CYP1a1 promoter (positive control), or primers flanking a non-XRE promoter (GMPR, negative control). Significant DNA enrichment was detected in AHR- versus IgG-precipitated material, with primers corresponding to the promoters of CYP1a1, AZIN1 (AZ1 no. 1 and AZ1 no. 2), and ODC1, while no enrichment was detected at the negative control promoter (Figure 1E).
XRE-containing regions from AZIN1p or ODC1p were subcloned into pGL3-promoter or pGL3-basic luciferase reporter plasmids, respectively. HEK293FT cells transfected in parallel with the above constructs were either cotransfected with increasing amounts of CA-AHR expression vector (Figure 1F) or treated with the established AHR ligand agonist benzo-a-pyrene (BaP) (16, 48) (Figure 1G). A plasmid expressing the luciferase gene under the control of XRE (XRE-luc) (49) was used as a positive control. Vectors containing either of the AZIN1p sites or the ODC1p site were responsive to AHR activation in a dose-dependent manner, as was seen with the XRE-luc vector. As ODC1 is also a well-established target of MYC (8, 9) and MYC itself has been shown to potentially be a target of AHR (50–54), we extended the sequence of ODC1p in the above construct to include MYC binding elements, either WT or mutated, to abrogate binding. We performed luciferase reporter assays as described above. The presence of MYC binding sites, WT or mutated, did not significantly affect luciferase activity induction, suggesting that MYC does not influence this regulation (Supplemental Figure 1B).
In reciprocal experiments, 2 different shRNAs against AHR (sh1 and sh2) were transduced into WI-38 cells, along with a control shRNA (Ctrl-sh). When compared with control cells, AHR-depleted cells had decreased protein and RNA levels for both AZIN1 and ODC1 (Figure 1, H and I). Importantly, AHR depletion resulted in an approximately 40% to 70% decrease of the 3 major mammalian polyamines putrescine, spermidine, and spermine (Figure 1J).
To test whether pharmacological inhibition of AHR would recapitulate the loss-of-function studies, we treated WI-38 cells with the commercial AHR antagonist CH223191 (55). Consistent with the above data, we observed that treatment with CH223191 suppressed mRNA and protein levels of ODC1 and AZIN1 (Figure 1, K and L) and polyamine amounts (Figure 1M). Together, these data strongly suggest that targeting the AHR may represent a novel anti-polyamine strategy.
CLF is an AHR antagonist.
AHR has long been recognized as an attractive target for therapy (56, 57), and clinical trials for AHR agonists (aminoflavone) (58–61) or antagonists (StemRegenin 1) (62, 63) have been attempted; however, these trials were terminated before completion or withdrawn, partly because of excessive toxicity (64). Thus, we sought to identify small molecules that may be unrecognized AHR antagonists to use as anti-polyamine therapies. We queried the NextBio Pharmaco-Atlas Gene Expression database (www.nextbio.com) for compounds that had resulted in the downregulation of AZIN1 or ODC1 expression, with a score of at least 50 given by the database (range, 0–100), and focused on substances for which the molecular mechanism of action was classified as unknown.
The resulting 23 compounds were further filtered to exclude molecules with known high toxicity (Supplemental Table 1). This limited our set to 10 potential compounds. Three of these agents have been reported in separate high-throughput studies to concomitantly affect the levels of AZIN1 or ODC1 mRNA and classical markers of AHR signaling (CYP1a1, CYP1b1, or TiPARP): CLF, a lipophilic riminophenazine antibiotic used in the treatment of leprosy (65–67); harmol (Har), a β-carboline indoleamine alkaloid found in the plant from the genus Peganum (68); and fenofibrate (FF), a PPARα agonist used to lower cholesterol levels (69, 70).
We treated WI-38 cells with increasing doses of each agent and found that only CLF demonstrated robust reductions in ODC1 and AZIN1 levels in a dose-dependent manner (Figure 2A), which was comparable to the effects of CH223191 (Figure 1K).
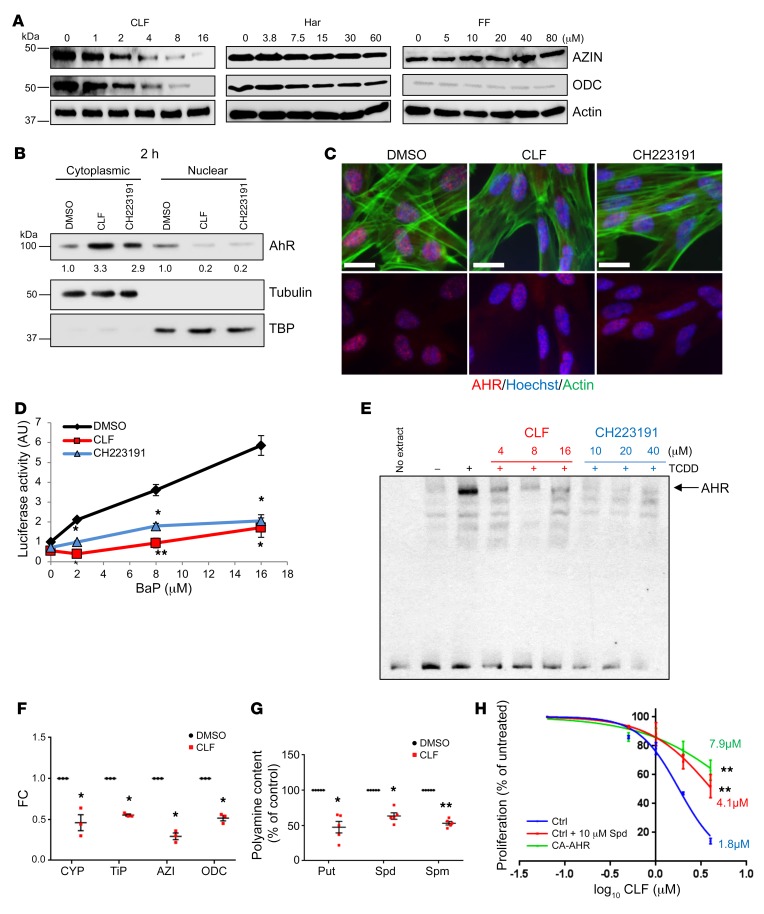
Figure 2. CLF is an AHR antagonist.
(A) Extracts of WI38 cells treated for 2 hours with increasing concentrations of CLF, Har, or FF probed in the immunoblot for AZIN1 and ODC1. (B) Cytoplasmic and nuclear fractions of WI38 cells treated for 2 hours with DMSO, 4 μM CLF, or 20 μM CH223191 resolved in the immunoblot and probed for AHR. Tubulin and TBP were used as positive controls. Quantification of band intensity was performed with ImageJ (NIH). (C) WI38 cells treated as in B and immunostained for AHR (red), DNA (Hoechst-33342, blue), and actin (phalloidin, green). Images are representative of 2 independent experiments. Scale bars: 20 μm. (D) Luciferase activity assay of HEK293FT cells transduced with XRE-luc, with increasing concentrations of BaP and DMSO, 4 μM CLF, or 20 μM CH223191. Data represent the average ± SEM of 2 independent experiments performed in duplicate. (E) Cytosolic fractions of Hepa 1c1c7 cells preincubated with vehicle or 16 nM TCDD and increasing concentrations of CLF or CH223191, and then incubated with XRE-containing biotinylated oligonucleotides and resolved on native gels. Protein-DNA complexes were visualized with a chemiluminescence system. (F) RNA from WI-38 cells treated for 2 hours with DMSO or 4 μM CLF probed in qRT-PCR with the indicated primers and probes. Data represent the average ± SEM of 3 independent experiments performed in triplicate. (G) Polyamine content in WI38 cells treated with 4 μM CLF for 48 hours. Data represent the average ± SEM of 5 independent experiments. *P < 0.05 and **P < 0.001, by 2-tailed Student’s t test. (H) Proliferation of WI38 cells expressing empty vector (Ctrl) or CA-AHR, with increasing concentrations of CLF over time. Control cells were supplemented with 10 μM spermidine and 1 mM aminoguanidine. IC50 values were determined using GraphPad Prism. Data represent the average of 2 experiments performed in quadruplicate. **P < 0.001, by extra sum-of-squares F test using GraphPad Prism.
Immunoblot analysis of WI-38 cells treated with vehicle (DMSO), CH223191, or CLF revealed that treatment with either drug reduced the nuclear content of AHR, promoting its accumulation in the cytoplasm (Figure 2B). We obtained similar results with immunostaining of WI-38 cells treated as above (representative images are shown in Figure 2C).
We transfected HEK293FT cells with the XRE-luc vector and treated them with increasing concentrations of BaP, which resulted in a dose-dependent increase in luciferase activity that was blunted by concomitant treatment with either CH223191 or CLF (Figure 2D), indicating that CLF inhibits AHR signaling.
Next, we used an established EMSA-based approach with cytosolic fractions of mouse hepatoma (Hepa 1c1c7) cells, as under these experimental conditions murine AHR is more stable than the human counterpart (55). Cytosolic extracts were preincubated with the canonical AHR ligand 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (16, 48) in the presence of increasing amounts of CH223191 or CLF, and the ability of AHR to bind DNA was assessed using a biotinylated double-stranded probe containing a single XRE motif. We found that both CH223191 and CLF were able inhibit TCDD-induced AHR-oligonucleotide complex formation in a dose-dependent manner (Figure 2E), suggesting that CLF can prevent the binding of AHR to DNA. At the same time, CLF did not seem to interfere with TCDD-induced AHR binding to its dimerizing partner ARNT, but it reduced basal AHR-ARNT complex formation, as evidenced by co-IP experiments in WI-38 cells (Supplemental Figure 1C). Although the exact mechanisms are unknown, this could be due to an increased retention of AHR in an inactive state through enhanced binding with the hsp90 complex.
CLF treatment reduced, in a dose-dependent manner, the binding of AHR to the promoters of CYP1a1, AZIN1, and ODC1 (Supplemental Figure 1D). Furthermore, we performed ChIP assays in WI-38 control cells, WI-38 cells treated with CLF, or WI-38 cells transduced with AHR-specific shRNAs with antibodies against RNA polymerase II (RNApol II), histone H3 acetylated at lysine 9 (H3K9AC, a marker of actively transcribed chromatin), and histone H3 trimethylated at lysine K27 (H3K27Me3, a marker of repressive chromatin and primers flanking the promoter regions described in Figure 1D). CLF treatment and AHR knockdown (KD) concordantly reduced the ability of RNApol II to bind to these promoters, as compared with the control. Concomitantly, the levels of H3K9AC and H3K27Me3 were decreased and increased, respectively, at the studied promoter regions (Supplemental Figure 1E).
CLF-mediated inhibition of AHR signaling in WI-38 cells decreased the transcription of the AHR targets CYP1a1 and TiPARP as well as AZIN1 and ODC1 (Figure 2F), as was observed with CH223191 treatment (Figure 1K). This was also accompanied by reduced levels of all 3 polyamines (Figure 2G). Loss of the higher polyamines in such a short amount of time suggests that polyamine catabolism may also be increased. The 2 major enzymes of polyamine catabolism are spermine oxidase (SMOX), which converts spermine into spermidine (71, 72), and spermidine/spermine N1-acetyltransferase 1 (SSAT), which acetylates spermine and spermidine, inducing their export from the cell (73, 74). We found that SMOX protein levels were slightly increased by CLF treatment at the 48-hour time point (Supplemental Figure 2A). It is well accepted that SMOX levels and activity are directly proportional (71, 72); thus, these data suggest that oxidation of spermine is slightly increased in response to CLF. The SSAT protein is extremely unstable and cannot be detected by immunoblotting unless stabilized by high amounts of polyamines or, pharmacologically, by selective polyamine analogs (75); thus, as a readout for SSAT levels and activity, we measured the amount of acetylated polyamines in the media. CLF treatment increased the secretion of acetylated polyamines by approximately 2-fold (Supplemental Figure 2B), suggesting that this is another mechanism that contributed to the rapid loss of the increased polyamines.
Polyamine depletion suppresses cell proliferation (3, 6). Thus, we assessed the proliferation of WI-38 cells in response to increasing amounts of CLF and in the presence of CA-AHR overexpression or spermidine supplementation. Both treatments resulted in a 2.3- to 4.4-fold increase in the CLF IC50 (1.8 μM in control vs. 4.1 μM in spermidine and 7.9 μM in CA-AHR; Figure 2H).
The above data, in conjunction with the notion that the CLF structure fits the broad description of AHR ligands (planar, aromatic, and hydrophobic molecules [refs. 16 and 48], and Supplemental Figure 2C), strongly suggest that CLF is an antagonist of AHR and polyamine biosynthesis.
CLF has been previously characterized as an inhibitor of the Kv1.3 channel (40, 76). Since WI-38 cells express detectable amounts of Kv1.3 (Supplemental Figure 2D), we investigated whether Kv1.3 inhibition could account, at least partially, for the observed effects. WI38 cells were treated with the established Kv1.3 inhibitor PSORA-4 (77), which was used in parallel with CLF in previous studies (40, 76). PSORA-4 did not cause any changes in AZIN1 or ODC1 levels (Supplemental Figure 2E), even at a dose of 40 μM, which is double that required for effective inhibition of Kv1.3 in several cell lines (40). Combined with the previous data, these experiments suggest that Kv1.3 inhibition is unlikely to be responsible for the polyamine-inhibiting and growth-suppressing functions of CLF in WI38 cells.
CLF treatment and AHR depletion affect similar pathways.
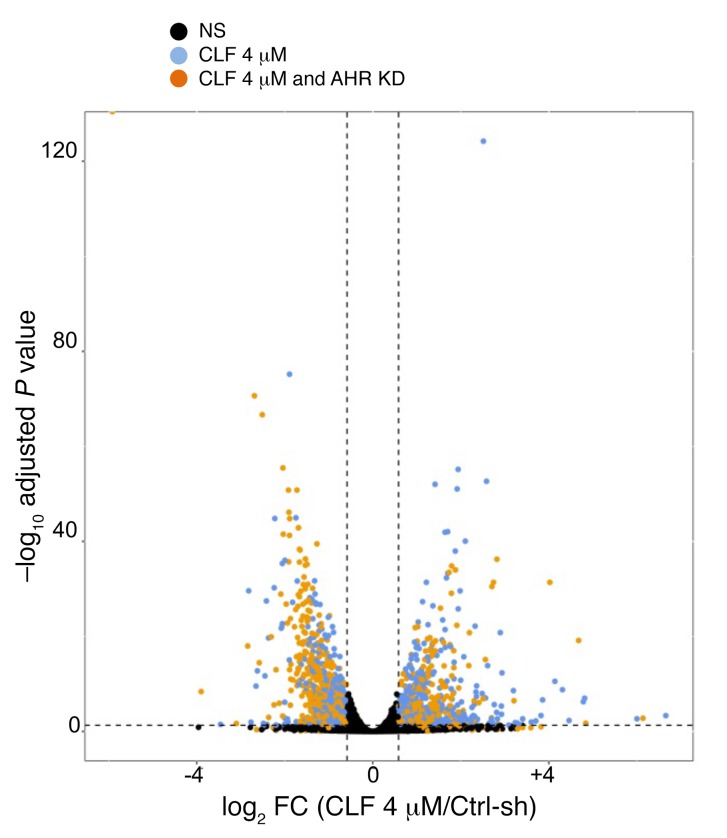
To gain insights into the global transcriptome affected by genetic or pharmacological inhibition of AHR signaling, we performed RNA-sequencing (RAN-seq) in WI-38 cells treated with CLF or transduced with the shAHR constructs described above (Figure 1, H–J). Differential expression (1.5–fold change [FC], adjusted P < 0.05) revealed a strong concordance between genes regulated by individual AHR-targeting shRNAs, suggesting limited off-target binding (Supplemental Figure 2F). Genes regulated by both shRNAs were compared with those coherently affected by CLF treatment. We detected a highly significant overlap between AHR- and CLF-regulated gene sets (Figure 3 and Supplemental Figure 2G), indicating that both treatments affect similar global expression patterns and pathways. This was further confirmed by gene set enrichment analysis (GSEA) of differentially expressed genes (DEGs) using the Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/index.jsp) (78, 79) Gene lists were then cross-referenced for enrichment of Gene Ontology (GO) gene sets for GO biological processes and GO molecular functions. All pathways that were affected by AHR depletion or CLF treatment with a FDR below 0.1 were analyzed, and a strong overlap in the enrichment groups was detected (Supplemental Figure 3 and Supplemental Table 2; hypergeometric test P = 1.73 × 10–86). Altogether, these data indicate that AHR depletion and CLF treatment affect similar pathways and confirm that both of them suppress AZIN1 and ODC1.
Figure 3. CLF and AHR work in overlapping pathways.
Volcano plot of differentially expressed genes in CLF-treated and AHR-KD WI38 cells. A FC of 1.5 and a P < 0.05 threshold were applied. Black dots indicate no significance, blue dots indicate genes that changed significantly only with CLF treatment, and orange dots indicate genes that changed significantly with both CLF treatment and AHR KD.
CLF suppresses AZIN1, ODC1, and polyamine levels in MM.
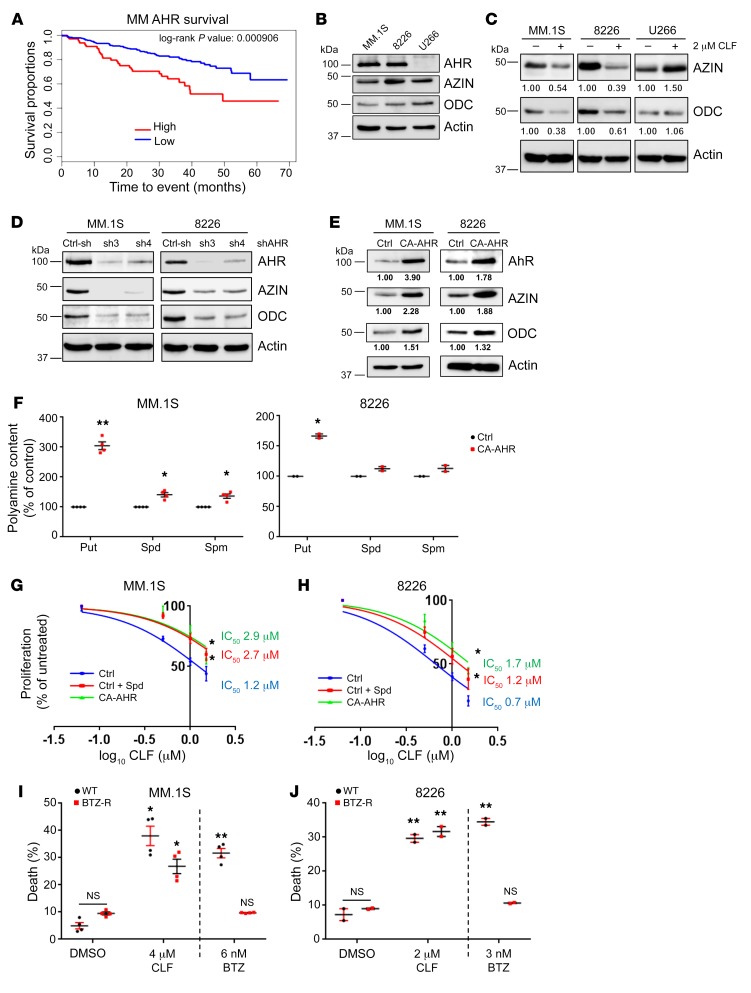
AHR has been demonstrated to possess tumor suppressor (25, 26) or oncogenic (27, 28, 80) functions, depending on the tumor type. We analyzed AHR expression in relation to patient survival in 26 different cancer data sets (see Methods). AHR levels were significantly associated with survival in 5 of the 26 cohorts (Supplemental Figure 4A). In esophageal carcinoma (ESCA) and kidney renal clear-cell carcinoma (KIRC), higher levels of AHR were linked to increased survival rates. In MM, cervical squamous cell carcinoma and cervical and endocervical cancers (CESCs), and glioblastoma multiforme (GBM), AHR levels were instead inversely correlated with survival (Figure 4A and Supplemental Figure 4A). Thus, AHR inhibition in the latter 3 malignancies would potentially be beneficial.
Figure 4. CLF inhibits polyamines in MM cells.
(A) Survival distribution of patients with MM (GEO GSE4581, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi), separated by AHR expression levels. The cohort consisted of 256 patients, 64 of whom were classified as “high AHR” (average expression value = 340.14) and 192 as “low AHR” (average expression value = 27.45). Statistical analysis was performed with a log-rank test. (B) Cell extracts of the human MM cells MM.1S, RPMI-8226, and U266 were probed by immunoblotting with the indicated antibodies. (C) Cell extracts as in B were treated for 2 hours with 2 μM CLF and probed by immunoblotting. Quantification of band intensity was performed with ImageJ. (D) Extracts of MM.1S and 8226 cells were transduced with control shRNA (Ctrl-sh) or 2 independent shRNA against AHR (sh3 and sh4) and probed by immunoblotting with the indicated antibodies. (E) Extracts of MM.1S and 8226 cells transduced with empty vector (Ctrl) or CA-AHR were probed by immunoblotting as in D. Quantification of band intensity was performed with ImageJ. (F) Polyamine content in MM.1S and 8226 cells transduced with control or CA-AHR. Data represent the average ± SEM of 4 (MM1.S) or 2 (RPMI-8226) independent experiments. Cell proliferation of MM.1S (G) or 8226 (H) cells transduced with control or CA-AHR and treated for 48 hours with increasing concentrations of CLF. Control cells were supplemented with 10 μM spermidine and 1 mM aminoguanidine. IC50 values were determined using GraphPad Prism. Viability of MM.1S (I) or RPMI-8226 (J) cells (WT or resistant to BTZ-R) treated with the indicated drugs and doses for 24 hours. Data represent the average ± SEM of 2 experiments performed in duplicate. *P < 0.05 and **P < 0.001 compared with the untreated control, by 2-tailed Student’s t test (F, I, and J) and sum-of-squares F test with GraphPad Prism (G and H).
We decided to test this hypothesis in MM, an incurable malignancy of bone marrow–resident plasma cells (32), as we already gained extensive experience with this tumor type (81–88). Since our previous data revealed that AHR controls polyamine biosynthesis, we decided to assess the degree to which polyamine biosynthesis gene expression correlates with patient survival. To this end, we created a polyamine biosynthesis score by summing the expression levels of the signature enzymes for polyamine biosynthesis (AZIN1, OAZ1, OAZ2, ODC1, SMS, SRM, and AMD1) as previously described for a different gene expression set (89). We analyzed patient survival in relation to this score. Similar to what we observed for AHR (Figure 4A), patients with a high polyamine biosynthesis score had poorer survival compared with patients with a low score (Supplemental Figure 4B), underlining the importance of polyamine biosynthesis in MM pathogenesis.
We selected 3 human MM cell lines (MM.1S, RPMI-8226, and U266) for further studies. All 3 cell lines expressed similar levels of AZIN1 and ODC1, however, unlike MM.1S and RPMI-8226, the U266 cells did not express AHR protein (Figure 4B). Correspondingly, we found that CLF treatment resulted in decreased AZIN1 and ODC1 protein levels in MM.1S and RPMI-8226 cells, but not in AHR-negative U266 cells (Figure 4C). Additionally, while all 3 cell lines expressed Kv1.3 (Supplemental Figure 2D), inhibition of Kv1.3 with PSORA-4 did not change AZIN1 or ODC1 levels in any of the cell lines (Supplemental Figure 4C).
As with WI-38 cells, shRNA-mediated depletion of AHR or overexpression of CA-AHR in MM.1S and RPMI-8226 cells led to, respectively, a reduction or induction of AZIN1 and ODC1 mRNA and protein levels (Figure 4, D and E). In both cell lines, CLF treatment reduced the levels of all 3 polyamines (Supplemental Figure 4C), which was accompanied by a decrease in cell proliferation rates, as was seen with AHR depletion (Supplemental Figure 4E). Ectopic expression of CA-AHR induced all 3 polyamines in MM.1S and RPMI-8226 cells (Figure 4F) and completely (MM.1S) or partially (RPMI-8226) rescued CLF-induced polyamine depletion (Supplemental Figure 4D). CLF-inhibited proliferation in both cell lines was partially rescued by CA-AHR overexpression or spermidine supplementation (as evidenced by a 1.7- to 2.4-fold increase in the CLF IC50 in cells treated with spermidine or overexpressing CA-AHR) (Figure 4, G and H). At the highest concentration, CLF caused a modest increase in cell death, which was rescued by spermidine supplementation or CA-AHR expression (Supplemental Figure 4F). CLF treatment did not cause any change in SMOX levels in either cell line (Supplemental Figure 5A) but did induce acetylated polyamines secretion (Supplemental Figure 5B, measured for 8226 cells).
shRNA-mediated depletion of AHR also caused a modest increase in cell death (from 10% to ~25% to 30%, Supplemental Figure 5C), as was seen with CLF treatment (Supplemental Figure 4F). Importantly, ectopic expression of CA-AHR, AZIN1, or ODC1 had a protective effect (Supplemental Figures 5, D–G).
The CLF/AHR axis was further confirmed in 2 additional AHR-expressing MM cell lines (ARH-77 and KMS-11, Supplemental Figure 5, H–J). Addiction to c-MYC in MM has been previously reported (90, 91). ODC1 is a classical MYC target (8, 9), thus, we decided to test whether AHR regulation would also affect MYC. We treated MM.1S, 8226, and KMS-11 cells with CLF for 4 hours and assessed the levels of MYC and ODC1 (as a control for CLF treatment) by immunoblotting. At this time point, we found that ODC1 levels were substantially depleted by CLF, however MYC levels were unaltered. Furthermore, treatment of MM.1S and 8226 cells with CH223191 did not cause any change in MYC levels, whereas CYP1a1 levels were substantially depleted (Supplemental Figure 5L). These results strongly suggest that AHR effects on ODC1 levels are MYC independent.
To confirm that genetic and pharmacological inhibition of AHR affects overlapping pathways in MM cells as it does in WI38 cells, we performed RNA-seq comparison of MM.1S, depleted of AHR or treated with CLF, and differential expression analysis (FC = 1.5, adjusted P < 0.05). As with WI-38 cells (Figure 3 and Supplemental Figures 2 and 3), the list of genes affected by AHR depletion or CLF treatment in MM cells also significantly overlapped (Supplemental Figure 6, A and B). Unbiased GSEA pathway analysis revealed a strong concordance in the pathways affected by each treatment (Supplemental Figure 7 and Supplemental Table 3; hypergeometric test P = 9.79 × 10–13).
BTZ shows high clinical efficacy in patients with MM; however, the occurrence of drug-resistant relapses is a common event (33). We selected BTZ-resistant (BTZ-R) cell populations, which showed no sensitivity to 6 nM BTZ (MM.1S) or 3 nM BTZ (RPMI-8226), whereas a 24-hour treatment with the same doses caused approximately 30% death of the corresponding parental cells. A dose of CLF that caused approximately 30% death of the parental cells also affected the viability of BTZ-R cells, albeit to a slightly lesser degree (Figure 4, I and J), suggesting that resistance to BTZ does not make MM cells less sensitive to CLF, at least not in vitro.
Thus, in MM cells, AHR functions as an important transcriptional activator of the polyamine biosynthetic pathway, whereas CLF antagonizes this activation.
CLF suppresses MM in vivo.
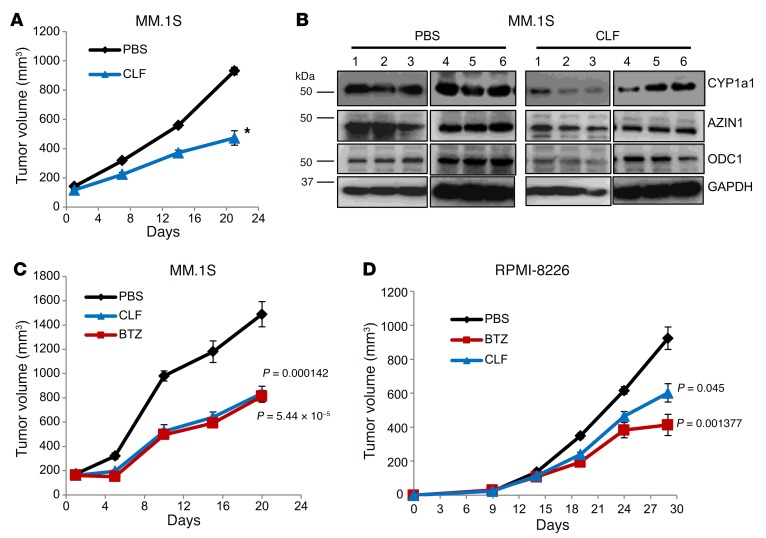
To test whether CLF possesses anti-MM efficacy, we first evaluated its effect on MM cell xenografts growing in immunocompromised mice. MM.1S cells were inoculated in both flanks of SCID mice. When the tumors reached a volume of 100 mm3, the animals were randomized into 2 groups and treated with daily i.p. injections of either vehicle or CLF (10 mg/kg). We detected no signs of toxicity or distress using this CLF treatment regimen, in agreement with previously reported data (40, 41). Tumor volume was measured every 5 days over a 3-week period, at which point the animals in the control group were sacrificed. CLF treatment significantly reduced tumor growth on MM.1S xenografts (Figure 5A and Supplemental Figure 8A). We analyzed 6 tumors per group by immunoblotting and found overall suppression of ODC1 and AZIN1 in the CLF group compared with the control group, as well as downregulation of CYP1a1 expression (Figure 5B). Conversely, we observed that xenografts of MM.1S cells ectopically expressing CA-AHR were resistant to treatment (Supplemental Figure 8B).
Figure 5. CLF inhibits MM xenograft growth.
(A) MM.1S cells were inoculated s.c. into both flanks of 4- to 6-week-old female SCID mice. The animals were randomized into 2 groups (6 animals/group) and treated with daily i.p. injections of vehicle in PBS or CLF (10 mg/kg). Tumor volumes were recorded every 5 days. *P < 0.05, by 2-tailed Student’s t test. (B) Protein extracts from 6 tumors/group from A were resolved by immunoblotting and probed for CYP1a1, AZIN1, or ODC1. Growth of MM.1S (C) or RPMI-8226 (D) cell xenografts was monitored as in A. Animals were randomized into 3 treatment groups: PBS, CLF, or BTZ (1 mg/kg, biweekly i.p. injections). Tumor measurement distributions were tested for normality by Shapiro-Wilk test, and the significance of individual comparisons (treated/untreated) were determined by Student’s t test with Bonferroni’s correction.
To compare the efficacy of CLF and BTZ, MM.1S and RPMI-8226 cells were inoculated into SCID mice, which were randomized for treatment as described above, with the inclusion of a BTZ treatment group as a positive control (1 mg/kg, biweekly i.p. injections) (81, 87). Tumors were measured as described above. Importantly, in both MM1.S- and RPMI-8826–derived xenografts, we found that CLF and BTZ had similar efficacy (Figure 5, C and D).
Next, we evaluated the in vivo effects of CLF in the Vk*MYC mouse model system, an immunocompetent transgenic model in which c-MYC expression is sporadically activated in B cells, causing the spontaneous development of MM with dynamics and features that closely recapitulate those observed in human MM patients (88, 92). Of note, among more than 30 drugs that have been evaluated in clinical trials for MM, 73% of those that induced a response in the Vk*MYC model have been approved for use in the treatment of patients (87, 93)
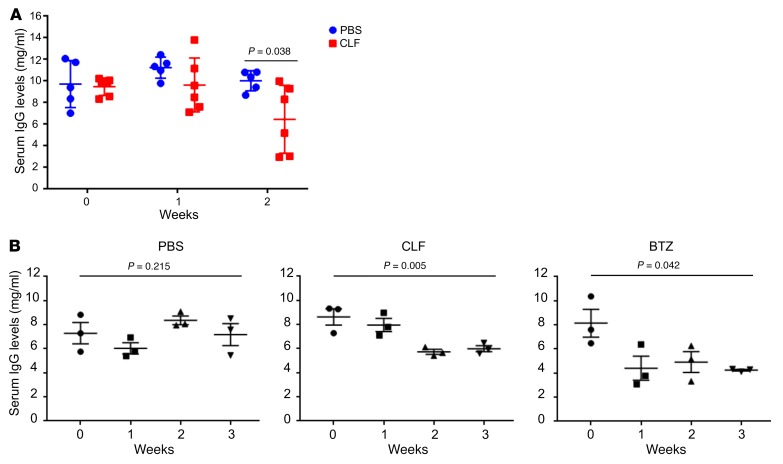
In a pilot experiment, mice with similar disease burdens (as measured by serum IgG secretion levels; see Figure 6A, week 0) were randomized into 1 of 2 groups and treated with either vehicle in PBS or CLF, as described above. Blood was collected every 7 days to assess IgG levels by ELISA. After 2 weeks of treatment, the CLF-treated mice displayed a significantly decreased disease burden (as evidenced by IgG secretion) (Figure 6A).
Figure 6. CLF suppresses MM burden.
(A) 12 Vk*Myc mice with similar disease burdens (day 0) were randomized into 2 groups and treated with vehicle in PBS or CLF (10 mg/kg). Mice were bled once a week, and total murine IgG levels were determined by ELISA. Statistical significance was determined by 2-tailed Student’s t test. (B) Nine Vk*Myc mice with similar disease burdens were randomized into 3 treatment groups: PBS, CLF, or BTZ (1 mg/kg, biweekly i.p. injections). Total murine IgG levels were determined as in A. Statistical significance was determined by 1-way ANOVA.
To further compare the effects of CLF and BTZ, mice with similar disease burdens were selected for a second experiment and randomized into 3 treatment groups (PBS, CLF, or BTZ treatment). Blood was collected every 7 days to assess IgG levels and M-spike by serum protein electrophoresis (SPEP, another indicator of disease burden) (88). As a single agent, we found that CLF was as effective as BTZ at reducing disease burden in mice, as evidenced by both assays (Figure 6B and Supplemental Figure 9).
Cumulatively, our data demonstrate a role for AHR activation in MM proliferation and survival and suggest that CLF could be an efficient therapeutic intervention for MM patients with detectable levels of AHR, setting the foundation for new studies in this area.
Discussion
Polyamines are small cationic molecules that control multiple cellular phenotypes. Cancer cells have been reported to require high polyamine levels (2, 5, 6, 94, 95), thus polyamine inhibition has been suggested as a way to inhibit tumor cells, with minimal effects on normal tissues. Currently, several polyamine-suppressing strategies are being pursued, including the development of polyamine analogs and promoters of polyamine export, although these strategies are not yet used in the clinic (2, 6, 94).
Despite the vast knowledge of the function of polyamines in cells, surprisingly little is known about their transcriptional regulation. The only 2 transcription factors that have been shown to regulate the transcription of ODC1, which encodes for the first rate-limiting enzyme in polyamine biosynthesis, are MYC (8–10) and SP1 (11). Bona fide transcription factors regulating expression of the AZIN1 gene are unknown.
Here, we report that AHR directly controls the transcription of both ODC1 and AZIN1 to support polyamine production (Figure 1). Our findings open the possibility of simultaneous targeting of ODC1 and AZIN1 through AHR inhibition. This 2-point leverage approach could represent a significant therapeutic innovation by reducing the ability of tumor cells to compensate for the reduction in intracellular polyamine. Additionally, while ODC1 inhibition has been previously pursued (i.e., with the irreversible inhibitor D,L-α-difluoromethylornithine [DFMO]) (2, 96, 97), to our knowledge, targeting of AZIN1 has never been attempted before.
The role of AHR in cancer progression appears to be tumor-type specific, as in different malignancies it has been assigned either tumor suppressor (25, 26) or oncogenic (27–31, 80) functions. This duality is also supported by our own analysis of patient databases (Figure 4A and Supplemental Figure 4A). Accordingly, both pharmacological activation and suppression of AHR have been attempted. AHR activation with drugs such as 6-alkyl-1,3,8-trichlorodibenzofuran (6-MCDF) (98) and its derivatives, benzothiazoles (99), and aminoflavones (61) has been tested in malignancies in which AHR acts as a tumor suppressor. In particular, the prodrug of the AHR agonist aminoflavone (AFP464) was included in 2 trials for the treatment of breast cancer (59); however, these trials were terminated before completion or withdrawn.
Conversely, although AHR inhibitors have not been tested as anticancer agents per se, the selective purine analog and AHR antagonist StemRegenin 1 was used in clinical trials to promote the expansion of umbilical cord blood cells for transplantation into hematological malignancies (62, 63). However, these trials were also withdrawn, thus leaving a void in the clinical inhibition of AHR.
Several lines of evidence argue that CLF is a potent and clinically relevant inhibitor of AHR. First, the CLF structure (Supplemental Figure 2C) resembles that of common AHR ligands, which are planar, aromatic, and hydrophobic molecules (16, 48). Second, CLF prevents AHR nuclear translocation (Figure 2, B and C), suppresses ligand-induced AHR-dependent transcription (Figure 2D), and inhibits AHR binding to XRE-containing DNA in vitro (Figure 2E). Consistent with the idea that AHR is a major target of CLF, ectopic expression of CA-AHR was able to rescue MM cells from the antiproliferative effects of CLF (Figure 2H and Figures 4, G and H). Additionally, comparison of RNA-seq profiles showed a significant overlap in pathway regulation via CLF or genetic inhibition of AHR (Supplemental Figures 3 and 7 and Supplemental Tables 2 and 3). As CLF is already being used as an anti-leprosy drug, it could represent the first AHR inhibitor to be fully approved for use in humans.
The potential of CLF as an antitumor drug has been previously explored, and its effects in cultured cells and tumor xenografts have been attributed to its ability to either inhibit the Kv1.3 potassium channel (40–42), interfere with the Wnt signaling pathway (43), or enhance the activity of phospholipase A2 (44). At least some of these effects may be explained by CLF-dependent inhibition of polyamines, as polyamines have been previously shown to inhibit the activity of phospholipases A2 and C (100).
Intriguingly, Durusu et al. (42) have shown that CLF induces cell death in the MM cell line U266, in which we observed no detectable effect, presumably because of the lack of AHR expression. This discrepancy is most likely due to the CLF dosage used in the former study, which was 5 times greater than the one used here (10 μM vs. 2 μM).
Elevated levels of AHR or its constitutive activation were detected in several blood malignancies, including MM (34–36, 101–103). Moreover, publically available gene expression data sets (GSE4581: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) revealed that higher levels of AHR are associated with poor survival of patients with MM (Figure 4A), supporting the hypothesis that AHR may act as a MM oncogene and that its targeting may be beneficial for MM patients.
This hypothesis is also consistent with our published data indicating that MM cells actively generate AHR ligands via induction of the expression of indoleamine 2,3-dioxygenase (IDO) from myeloid DCs in the MM microenvironment (83). Specifically, CD28 receptors expressed on MM cells interact with CD80 and CD86 on DCs to induce IDO, which catalyzes the conversion of tryptophan into kynurenine (a natural ligand for AHR) (104). Both kynurenine-mediated AHR activation and CD28 engagement promote prosurvival signaling in MM cells. Moreover, IDO activation has been shown to induce the suppression of anti-myeloma T cell responses (84, 86, 104). Thus, MM might be particularly dependent on AHR activation for survival.
Vk*MYC mice are an immunocompetent model of spontaneously arising myeloma, in which the actual bone marrow microenvironment is involved in the sustainment of the malignancy, similar to what naturally occurs in the human disease (88, 92). Thus, impairment of AHR signaling, as caused by CLF treatment, may result in both intrinsic and extrinsic effects: inhibition of polyamine biosynthesis and disruption of MM–bone marrow microenvironment interactions, respectively. Additional studies will be required to fully delineate the mechanism of anti-MM activity of CLF.
The role of polyamines in MM has been largely understudied. Recently, Carew et al. (37) targeted MM cell lines with a polyamine analog (CGC-11093), but the drug achieved only modest results as single agent, although it significantly enhanced the action of BTZ. Interestingly, BTZ was able, on its own, to suppress polyamine levels; however, the molecular mechanisms behind this observation have not been fully explored (37).
It is worth noting that the dose used to treat mice in the present study (10 mg/kg) would approximately translate into a dose of 0.87 mg/kg for humans (105), which is well within the FDA recommendations of 100 mg/day and thus supports the feasibility of CLF as a therapy for MM. Most important, unlike existing anti-MM drugs including BTZ, CLF is well tolerated, with minimal side effects in patients (67, 106–110).
In future studies, it will be important to evaluate the role of AHR in the etiology of MM by crossing AHR–/– mice with Vk*MYC mice, as AHR–/– mice are protected from the procarcinogenic effects of several AHR ligands such as TCDD and BaP (56, 111). Additionally, since our data indicated that CLF, at least in vitro, is able to induce cell death in MM cell lines resistant to BTZ (Figure 4, I and J), it will be essential to establish whether CLF can overcome therapy resistance in vivo as well.
Overall, our study builds upon the fundamental discovery of a novel AHR/polyamine biosynthesis signaling pathway and provides actionable insights into an anticancer strategy that opens a new niche for an existing medication.
Methods
Cell lines.
HEK293FT cells were obtained from Clontech (Takara Bio); Phoenix-AMPHO cells were a gift of Andrei Bakin (Roswell Park Comprehensive Cancer Center); human MM MM.1S, RPMI-8226, U266, ARH-77, and KMS-11 cells and human lung fibroblast WI-38 and mouse hepatoma Hepa 1c1c7 cells were obtained from American Type Culture Collection (ATCC). HEK293FT, Phoenix, and WI-38 cells were cultured in DMEM (Invitrogen, Thermo Fisher Scientific) supplemented with 10% FBS, 2 mM glutamine, and penicillin-streptomycin antibiotics. Hepa 1c1c7 cells were cultured in MEM-α without nucleosides (Invitrogen, Thermo Fisher Scientific) supplemented with 10% FBS, 2 mM glutamine and penicillin-streptomycin antibiotics. MM cells were cultured in RPMI (Invitrogen, Thermo Fisher Scientific) supplemented with 10% FBS, 2 mM glutamine, 10 mM HEPES (pH 7.4), nonessential amino acids, 1 mM sodium pyruvate, and penicillin-streptomycin antibiotics. BTZ-R cell lines (MM.1S-BTZ-R and RPMI-8226-BTZ-R) were generated by culturing in the presence of increasing concentrations of BTZ (from 1 nM to 3 to 6 nM) for over 4 months; media and BTZ were replenished every 2 to 3 days. Resistant cells were maintained in media containing BTZ. All cell lines were recently authenticated and verified for being mycoplasma free using a MycoAlert Mycoplasma Detection Kit (Lonza, catalog LT07-318).
Antibodies and other reagents.
Mouse monoclonal antibodies against AHR (A-3; SC-133088), ARNT (A-3; SC-17811), c-MYC (C-33; SC-42), and ODC1 (E-6; SC-398116), as well as normal mouse IgG antibodies (SC-2025), were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against AZIN1 (11548-1-AP) and SMOX (15052-1-AP) and HRP-conjugated β-actin (HRP-60008) and GAPDH (HRP-60004) antibodies were from the Proteintech Group. Mouse monoclonal antibodies against CYP1a1 (MA5-17063) and TBP (MA1-189) were from Thermo Fisher Scientific. Mouse monoclonal antibody against α-tubulin (T6074, clone B-5-1-2) was from Sigma-Aldrich. The ChIP-grade antibodies H3K9ac (ab4441), H3K27me3 (ab6002), and RNAPol-II (ab26721) were from Abcam. Alexa 488–conjugated (green) phalloidin was from Molecular Probes. Alexa 568–conjugated (red) goat anti-mouse antibody was from Invitrogen, Thermo Fisher Scientific. Hoechst-33342 was from Sigma-Aldrich. CLF, PSORA-4, BaP, TCDD, CH223191, and FF were from Sigma-Aldrich, Har was from Santa Cruz Biotechnology, and BTZ was from Selleckchem, and all were dissolved in DMSO. Aminoguanidine and DFMO were from Sigma-Aldrich and dissolved in water; spermidine (0.1 M solution) was from Sigma-Aldrich.
Immunoblotting.
Whole-cell extracts were prepared and analyzed as previously described (81, 112–114). Cytosolic and nuclear fractions were prepared with the Subcellular Protein Fractionation Kit (Thermo Fisher Scientific) according to the manufacturer’s recommendations. Signal quantification was performed with ImageJ (NIH).
Immunoprecipitation assay.
WI-38 cells were treated with drugs for 30 minutes and then lysed on ice. Total lysate (500 μg) was precleared with A/G beads and incubated overnight at 4°C on a rotator in the presence of 10 μg mouse AHR or normal mouse IgG antibodies. A/G beads were added to the extracts for 3 hours, and the bound material was precipitated by centrifugation (800 g, 5 min, 4°C), washed once, and boiled in 2× Laemmli buffer. Equal volumes of IP materials were analyzed by immunoblotting.
EMSA.
Aliquots of Hepa 1c1c7 cells cytosolic extracts were incubated at room temperature (RT) for 2 hours in the presence of vehicle control or 16 nM TCDD and increasing concentrations of CLF or CH223191. Extracts were then incubated for 20 minutes at RT with biotin (Btn) end-labeled oligonucleotides containing a single XRE binding motif and components of the Lightshift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Protein-DNA complexes were resolved on native 6% polyacrylamide gels, transferred onto nylon membranes, and UV crosslinked. Membranes were probed with HRP-conjugated streptavidin and analyzed as above for immunoblotting. EMSA XRE: forward, 5′ Btn-TGAGCTCGGAGTTGCGTGAGAAGAGCCG-3′; EMSA XRE: reverse, 5′ Btn- CGGCTCTTGTCACGCAACTCCGAGCTCA-3′.
Plasmids and infection.
pCMVdeltaR8.2 and pCMV-VSV-G vectors were from Addgene. The pLV-SV4-puro lentiviral vector was obtained from Peter Chumakov (Cleveland Clinic, Cleveland, Ohio, USA). pBabe-puro and the pBabe-puro-CA-AHR vectors were a gift of Andrei Bakin (Roswell Park Comprehensive Cancer Center) and Pedro Fernandez Salguero (Universidad de Extremadura, Badajoz, Spain), respectively. shRNAs against AHR were purchased from Sigma-Aldrich: shAHR no. 1_TRCN0000245285; shAHR no. 2_TRCN000021256; shAHR no. 3_TRCN000021258; shAHR no. 4_TRCN0000021254. Infections were performed as previously described (81, 112–114).
Fluorescence microscopy.
WI-38 cells were grown on coverslips and treated with drugs for 2 hours. Cells were fixed in 4% paraformaldehyde in PBS, permeabilized with 0.05% Triton X-100 in PBS, and blocked in 3% milk in PBS. Cells were incubated for 1 hour with an anti-AHR antibody (1:500 dilution) and for 30 minutes with a fluorescent secondary antibody (1:500) at RT. Actin filaments were stained with phalloidin conjugated with Alexa 488 (green). DNA was visualized with Hoechst-33342. Fluorescence images were captured using a Nikon TE2000-E inverted microscope equipped with a Roper CoolSnap HQ CCD camera and MetaVue software (Molecular Devices).
Quantitative real-time PCR.
Total cellular RNA was isolated using the RNeasy Mini Kit (QIAGEN). cDNA was prepared using a cDNA Reverse Transcription Kit (Invitrogen, Thermo Fisher Scientific). Quantitative real-time PCR (qRT-PCR) was performed on a QS6 Fast Real-Time PCR System (Thermo Fisher Scientific) using either SYBR Green Master Mix or TaqMan Master Mix (Invitrogen, Thermo Fisher Scientific) and the following probes and primers: Hs-AHR Hs00907314_m1; Hs-CYP1a1 Hs01054797_g1; Hs-AZIN1 Hs00210634_m1; Hs-TiPARP, forward, 5′-CTCGTGTTTGAGCTGGTGAA-3′; Hs-TiPARP, reverse, 5′-ACACGTTCATGGCATTCAAA-3′; Hs-ODC1, forward, 5′-TTTTGGACGGGCGAAAGAG-3′; Hs-ODC1, reverse, 5′-TCAAAAACACAGCGGGCATC-3′; Hs-RPS20, forward, 5′-AAGGATACCGGAAAAACACCC-3′; and Hs-RPS20, reverse 5′-TTTACGTTGCGGCTTGTTAGG-3′. PCR data were analyzed using QuantStudio Real-Time PCR Software (Thermo Fisher Scientific).
ChIP.
Interactions between AHR and the AZIN1 and the ODC1 promoters were assessed as previously described (115). The following primers were used for analysis of AHR binding to DNA: AZ1 no. 1, forward, 5′-AGGGTGGTCTTGATCTCTTGAC-3′, reverse, 5′-GGGAGAATTCTGGAGCCATAAAC-3′; AZ1 no. 2, forward, 5′-CTCAGGACCTTCGGGCGG-3′, reverse, 5′-CCCCGCTCCATTCATAACCA-3′; ODC1, forward, 5′-ATCGTGGCTGGTTTGAGCTG-3′, reverse, 5′-TACAGGAGGGACTGACAAAGC-3′; CYP1a1, forward, 5′-CAGGGCTGGGGTCGCAGCGCTTCT-3′, reverse, 5′-GCTACAGCCTACCAGGACTCGGCAG-3′; and GMPR, forward, 5′-AGCAATTCTTCTGCCTCAGC-3′, reverse, 5′-TGGCTAACACAGTGAAACCC-3′.
Dual luciferase reporter assay.
AZIN1 and ODC1 promoter regions containing putative AHR binding sites, as well as the ODC1 promoter including the MYC binding sites (either WT or mutated) were synthesized by Genscript and subcloned into the pGL3 promoter (AZI1 no. 1 and (AZI1 no. 2) or pGL3 basic (ODC1) plasmids (Promega). The XRE-luc plasmid was from Promega. The constructs were mixed with pRLSV40 plasmid expressing the Renilla luciferase gene (Promega). HEK293FT cells were transfected with the plasmid mixtures using LipoD293 (SignaGen Laboratories). Where indicated in the text and figure legend, CA-AHR was added to the plasmid mix, or cells were treated with increasing concentrations of BaP. Forty-eight hours after transfection (24 hours after BaP treatment), firefly luciferase and Renilla signals were detected using a Dual-Luciferase Assay Kit (Promega). Firefly luciferase signals were normalized to corresponding Renilla signals.
Proliferation assay.
WI-38 cells were seeded in 96-well plates and treated with the drugs and doses indicated in the text and figure legend. At the indicated time point, cells were fixed and stained in 0.5% methylene blue in methanol/water (50:50) and solubilized in 1% SDS in PBS. The optical density of solutions was determined using a SpectraMax fluorimeter at 650 nm. The experiments were performed in quadruplicate. MM cells were seeded at 2 × 105 cells/ml in 12-well plates and treated with the indicated drugs and doses. Cell proliferation was assessed by live cell counts with trypan blue exclusion, performed in duplicate.
Ultra-performance liquid chromatography analyses of polyamines.
Ultra-performance liquid chromatographic analyses of polyamines were carried out with adjustments to previously described methods (116). Acetylated polyamines were extracted from media as previously described (116). Alterations were made to the flow rate, gradient, and column as indicated below and as previously described (89). All polyamine measurements were done using an ACQUITY UPLC BEH Shield RP18 (1.7 μm 2.1 × 100 mm) column with a RP18 VanGuard Pre-column (130 Å, 1.7 μm, 2.1 mm × 5 mm) on an ACQUITY UPLC machine (all from Waters) in the Bioanalytics, Metabolomics, and Pharmacokinetics Core Facility at Roswell Park Comprehensive Cancer Center. A constant flow rate was held at 0.17 ml/min. Dancylated polyamines were eluted with a linear gradient from 100% buffer A (55% 10 mM ammonium acetate at pH 4.4 and 45% HPLC grade acetonitrile) to 18% buffer A and 82% buffer B (100% acetonitrile) for 6 minutes, which was then held for 3 minutes. By 10.6 minutes, the conditions returned to 100% buffer A, which also served to equilibrate the column for the next sample.
RNA-seq.
Sequencing libraries were prepared with the TruSeq Stranded mRNA Kit (Illumina) from 500 ng total RNA following the manufacturer’s instructions. PCR-amplified libraries were pooled in an equimolar fashion, loaded into a 75-cycle NextSeq Reagent Cartridge, and single-end sequencing performed on a NextSeq 500 (Illumina) following the manufacturer’s recommended protocol. Genome alignments and feature counting were performed at the Roswell Park Comprehensive Cancer Center’s bioinformatics shared resource. Expression count normalization and differential expression were determined using a standard DESeq2 workflow (117). Overlaps of DEG lists across comparisons were calculated by hypergeometric testing. All analyses were performed using R statistical software, version 3.4.2. Data were deposited in the NCBI’s Gene Expression Omnibus (GEO) database (GEO GSE117160).
Functional annotation of defined gene sets.
Functional annotation for derived gene expression profiles was performed by GSEA using the MSigDB (http://www.broadinstitute.org/gsea/msigdb/index.jsp) (78, 79). Gene lists were cross-referenced for enrichment of GO gene sets specifically for GO biological processes and GO molecular functions. Specifically, GSEA tools were used to assess the enrichment of all Broad Institute Hallmark pathways and curated pathway (KEGG) gene sets available through the MSigDB.
Generation of survival curves stratified on AHR expression.
Data generated by TCGA Research Network (http://cancergenome.nih.gov/) and the GEO data set GSE4581 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi) were used to generate Kaplan-Meier curves. Briefly, within each cohort, patients’ AHR levels were labeled as either low or high as compared with the mean value in each cohort, and survival curves were generated.
Animal studies using a s.c. xenograft model.
MM.1S and 8226 cells (5.0 × 106 cells/flank) were inoculated s.c. into both flanks of 4- to 6-week-old female SCID mice (strain: C.B-Igh-1bIcrTac-Prkdcscid/Ros, bred and maintained by the in-house transgenic mouse facility at the Roswell Park Comprehensive Cancer Center. When tumors reached a volume of 100 mm3, the animals were randomized into 2 groups (6 animals/group) and treated with daily i.p. injections of vehicle in PBS or CLF (10 mg/kg). Tumor volumes were recorded every 5 days, and mice were sacrificed when the tumor volume reached 2 cm3 or when a tumor became ulcerated. No animals were excluded from the study, since all animals developed palpable tumors of approximately 100 mm3 two to three weeks after s.c. inoculation of cells, and none of the animals developed significant morbidity before the end of the study. The animals were coded so that the investigators were blinded to the treatment group until the experiment reached its endpoint.
Animal studies using the Vk*Myc orthotopic model.
Vk*MYC mice were a gift of P.L. Bergsagel (Mayo Clinic, Scottsdale, Arizona, USA) and were housed and bred at the Division of Laboratory Animal Resources (Roswell Park Comprehensive Cancer Center, Buffalo, New York, USA) in a pathogen-free barrier facility. When the animals reached a disease burden of at least 7 mg/ml total IgG, they were randomized into 1 of 3 groups (3 animals/group) and treated with daily i.p. injections of vehicle in PBS or CLF (10 mg/kg), or with biweekly i.p. injections of BTZ (1 mg/kg). Disease burden was measured weekly for 3 consecutive weeks, after which the animals were sacrificed. None of the animals developed significant morbidity before the end of the study, and animals were coded so that the investigators were blinded to the treatment group until the experiment reached its endpoint.
ELISA.
To measure disease burden in the Vk*Myc mouse model, total murine IgG was determined by ELISA (Bethyl Laboratories). Diseased mice were selected on the basis of having total IgG measurements of more than 7 mg/ml. In brief, Nunc MaxiSorp 96-well plates (Thermo Fisher Scientific) were precoated with capture antibody in coating buffer for 1 hour at RT. Serum was diluted 1:100,000 in sample diluent (Bethyl Laboratories) and plated following the manufacturer’s instructions.
Serum protein electrophoresis.
Mice were bled once a week by tail grazing, and sera were analyzed by serum protein electrophoresis (SPEP) as described previously (88).
Statistics.
Each experiment was performed independently at least twice, and the results are expressed, unless otherwise noted, as the average ± SEM of the independent experiments. Statistical analysis was performed using a 2-tailed Student’s t test, 1-way ANOVA, an extra sum-of-squares F test, log-rank test, or a hypergeometric test. A P value of less than 0.05 was considered statistically significant for all analyses.
Study approval.
All experiments in the present studies involving animals were reviewed and approved by the IACUC of the Roswell Park Comprehensive Cancer Center.
Author contributions
ABS, AB, and MAN designed the experiments. ABS and MAN wrote the manuscript. ABS and AB performed most of the experiments and analyzed the data. EEF, SM, KM, BCL, DWW, DHY, ZH, AP, MVR, IIG, KIL, SML, HCA, AMR, MDL, and SRR performed some of the experiments. ESK, AVG, PLB, KPL, and DJS supervised part of the study. PLB provided the Vk*MYC mice. MAN conceived the initial hypothesis and supervised the study. All authors discussed the results and commented on the manuscript.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA220096, CA224434, CA193981, and CA190533 (to MAN); a Ruth L. Kirschstein National Research Service Award (F32CA189622, to ABS); NIH grants CA197996 (to DJS), 1F99CA21245501 (to HCA), and R01AI100157 and R01CA121044 (to KPL); the Jennifer Linscott Tietgen Foundation (to MAN and ESK); and in part by a National Cancer Institute (NCI) Cancer Center Support Grant (P30CA16056, to the Roswell Park Comprehensive Cancer Center, for the Clinical Data Network and the Animal Facility).
Version 1. 09/10/2018
Electronic publication
Version 2. 09/12/2018
Corrected reference 94
Version 3. 10/01/2018
Print issue publication
Footnotes
AB’s present address is: Department of Cancer Immunology and Virology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, Massachusetts, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(10):4682–4696.https://doi.org/10.1172/JCI70712.
See the related Commentary at Targeting the aryl hydrocarbon receptor/polyamine biosynthesis axis of evil for cancer therapy.
Contributor Information
Anna Bianchi-Smiraglia, Email: anna.bianchi-smiraglia@roswellpark.org.
Archis Bagati, Email: archis_bagati@dfci.harvard.edu.
Sudha Moparthy, Email: sudha.moparthy@roswellpark.org.
Dong Hyun Yun, Email: donghyun.yun@roswellpark.org.
Zhannan Han, Email: zhannan.han@roswellpark.org.
Anthony Polechetti, Email: apolechetti@gmail.com.
Matthew V. Roll, Email: matthew.roll@roswellpark.org.
Dominic J. Smiraglia, Email: Dominic.Smiraglia@RoswellPark.org.
References
- 1.Soda K. The mechanisms by which polyamines accelerate tumor spread. J Exp Clin Cancer Res. 2011;30:95. doi: 10.1186/1756-9966-30-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casero RA, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov. 2007;6(5):373–390. doi: 10.1038/nrd2243. [DOI] [PubMed] [Google Scholar]
- 3.Childs AC, Mehta DJ, Gerner EW. Polyamine-dependent gene expression. Cell Mol Life Sci. 2003;60(7):1394–1406. doi: 10.1007/s00018-003-2332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durie BG, Salmon SE, Russell DH. Polyamines as markers of response and disease activity in cancer chemotherapy. Cancer Res. 1977;37(1):214–221. [PubMed] [Google Scholar]
- 5.Murray-Stewart TR, Woster PM, Casero RA. Targeting polyamine metabolism for cancer therapy and prevention. Biochem J. 2016;473(19):2937–2953. doi: 10.1042/BCJ20160383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res. 1988;48(4):759–774. [PubMed] [Google Scholar]
- 7.Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281(21):14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 8.Bello-Fernandez C, Cleveland JL. c-myc transactivates the ornithine decarboxylase gene. Curr Top Microbiol Immunol. 1992;182:445–452. doi: 10.1007/978-3-642-77633-5_56. [DOI] [PubMed] [Google Scholar]
- 9.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A. 1993;90(16):7804–7808. doi: 10.1073/pnas.90.16.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Yosef T, Yanuka O, Halle D, Benvenisty N. Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene. 1998;17(2):165–171. doi: 10.1038/sj.onc.1201939. [DOI] [PubMed] [Google Scholar]
- 11.Kumar AP, Mar PK, Zhao B, Montgomery RL, Kang DC, Butler AP. Regulation of rat ornithine decarboxylase promoter activity by binding of transcription factor Sp1. J Biol Chem. 1995;270(9):4341–4348. doi: 10.1074/jbc.270.9.4341. [DOI] [PubMed] [Google Scholar]
- 12.Olsen RR, Chung I, Zetter BR. Knockdown of antizyme inhibitor decreases prostate tumor growth in vivo. Amino Acids. 2012;42(2–3):549–558. doi: 10.1007/s00726-011-1032-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qiu S, Liu J, Xing F. Antizyme inhibitor 1: a potential carcinogenic molecule. Cancer Sci. 2017;108(2):163–169. doi: 10.1111/cas.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auvinen M, Paasinen A, Andersson LC, Hölttä E. Ornithine decarboxylase activity is critical for cell transformation. Nature. 1992;360(6402):355–358. doi: 10.1038/360355a0. [DOI] [PubMed] [Google Scholar]
- 15.Barouki R, Coumoul X, Fernandez-Salguero PM. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581(19):3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 16.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 17.Poland A, Glover E, Kende AS. Stereospecific, high affinity binding of 2,3,7,8-tetrachlorodibenzo-p-dioxin by hepatic cytosol. Evidence that the binding species is receptor for induction of aryl hydrocarbon hydroxylase. J Biol Chem. 1976;251(16):4936–4946. [PubMed] [Google Scholar]
- 18.Caruso JA, et al. Differential susceptibilities of murine hepatoma 1c1c7 and Tao cells to the lysosomal photosensitizer NPe6: influence of aryl hydrocarbon receptor on lysosomal fragility and protease contents. Mol Pharmacol. 2004;65(4):1016–1028. doi: 10.1124/mol.65.4.1016. [DOI] [PubMed] [Google Scholar]
- 19.Caruso JA, Mathieu PA, Joiakim A, Zhang H, Reiners JJ. Aryl hydrocarbon receptor modulation of tumor necrosis factor-alpha-induced apoptosis and lysosomal disruption in a hepatoma model that is caspase-8-independent. J Biol Chem. 2006;281(16):10954–10967. doi: 10.1074/jbc.M508383200. [DOI] [PubMed] [Google Scholar]
- 20.Mezrich JD, Fechner JH, Zhang X, Johnson BP, Burlingham WJ, Bradfield CA. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J Immunol. 2010;185(6):3190–3198. doi: 10.4049/jimmunol.0903670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Negishi T, et al. Effects of aryl hydrocarbon receptor signaling on the modulation of TH1/TH2 balance. J Immunol. 2005;175(11):7348–7356. doi: 10.4049/jimmunol.175.11.7348. [DOI] [PubMed] [Google Scholar]
- 22.Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Lab Invest. 2009;89(6):695–707. doi: 10.1038/labinvest.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quintana FJ, et al. Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453(7191):65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Zong H, Li S, Zhang D, Zhang L, Xia Q. Activation of aryl hydrocarbon receptor suppresses invasion of esophageal squamous cell carcinoma cell lines. Tumori. 2012;98(1):152–157. doi: 10.1177/030089161209800121. [DOI] [PubMed] [Google Scholar]
- 26.Koliopanos A, et al. Increased arylhydrocarbon receptor expression offers a potential therapeutic target for pancreatic cancer. Oncogene. 2002;21(39):6059–6070. doi: 10.1038/sj.onc.1205633. [DOI] [PubMed] [Google Scholar]
- 27.Gramatzki D, et al. Aryl hydrocarbon receptor inhibition downregulates the TGF-beta/Smad pathway in human glioblastoma cells. Oncogene. 2009;28(28):2593–2605. doi: 10.1038/onc.2009.104. [DOI] [PubMed] [Google Scholar]
- 28.Opitz CA, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478(7368):197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 29.Richmond O, Ghotbaddini M, Allen C, Walker A, Zahir S, Powell JB. The aryl hydrocarbon receptor is constitutively active in advanced prostate cancer cells. PLoS ONE. 2014;9(4):e95058. doi: 10.1371/journal.pone.0095058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ide H, et al. Aryl hydrocarbon receptor signaling involved in the invasiveness of LNCaP cells. Hum Cell. 2017;30(2):133–139. doi: 10.1007/s13577-016-0158-2. [DOI] [PubMed] [Google Scholar]
- 31.DiNatale BC, Smith K, John K, Krishnegowda G, Amin SG, Perdew GH. Ah receptor antagonism represses head and neck tumor cell aggressive phenotype. Mol Cancer Res. 2012;10(10):1369–1379. doi: 10.1158/1541-7786.MCR-12-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351(18):1860–1873. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 33.Field-Smith A, Morgan GJ, Davies FE. Bortezomib (velcadetrade mark) in the treatment of multiple myeloma. Ther Clin Risk Manag. 2006;2(3):271–279. doi: 10.2147/tcrm.2006.2.3.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Consonni D, et al. Mortality in a population exposed to dioxin after the Seveso, Italy, accident in 1976: 25 years of follow-up. Am J Epidemiol. 2008;167(7):847–858. doi: 10.1093/aje/kwm371. [DOI] [PubMed] [Google Scholar]
- 35.Pesatori AC, Consonni D, Rubagotti M, Grillo P, Bertazzi PA. Cancer incidence in the population exposed to dioxin after the “Seveso accident”: twenty years of follow-up. Environ Health. 2009;8:39. doi: 10.1186/1476-069X-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherr DH, Monti S. The role of the aryl hydrocarbon receptor in normal and malignant B cell development. Semin Immunopathol. 2013;35(6):705–716. doi: 10.1007/s00281-013-0390-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carew JS, et al. The novel polyamine analogue CGC-11093 enhances the antimyeloma activity of bortezomib. Cancer Res. 2008;68(12):4783–4790. doi: 10.1158/0008-5472.CAN-07-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gopal M, Padayatchi N, Metcalfe JZ, O’Donnell MR. Systematic review of clofazimine for the treatment of drug-resistant tuberculosis. Int J Tuberc Lung Dis. 2013;17(8):1001–1007. doi: 10.5588/ijtld.12.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CS, Aerts A, Saunderson P, Kawuma J, Kita E, Virmond M. Multidrug therapy for leprosy: a game changer on the path to elimination. Lancet Infect Dis. 2017;17(9):e293–e297. doi: 10.1016/S1473-3099(17)30418-8. [DOI] [PubMed] [Google Scholar]
- 40.Leanza L, et al. Inhibitors of mitochondrial Kv1.3 channels induce Bax/Bak-independent death of cancer cells. EMBO Mol Med. 2012;4(7):577–593. doi: 10.1002/emmm.201200235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zaccagnino A, et al. Tumor-reducing effect of the clinically used drug clofazimine in a SCID mouse model of pancreatic ductal adenocarcinoma. Oncotarget. 2017;8(24):38276–38293. doi: 10.18632/oncotarget.11299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Durusu İZ, et al. Anti-cancer effect of clofazimine as a single agent and in combination with cisplatin on U266 multiple myeloma cell line. Leuk Res. 2017;55:33–40. doi: 10.1016/j.leukres.2017.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Koval AV, et al. Anti-leprosy drug clofazimine inhibits growth of triple-negative breast cancer cells via inhibition of canonical Wnt signaling. Biochem Pharmacol. 2014;87(4):571–578. doi: 10.1016/j.bcp.2013.12.007. [DOI] [PubMed] [Google Scholar]
- 44.Van Rensburg CE, Van Staden AM, Anderson R. The riminophenazine agents clofazimine and B669 inhibit the proliferation of cancer cell lines in vitro by phospholipase A2-mediated oxidative and nonoxidative mechanisms. Cancer Res. 1993;53(2):318–323. [PubMed] [Google Scholar]
- 45.Rico-Leo EM, Alvarez-Barrientos A, Fernandez-Salguero PM. Dioxin receptor expression inhibits basal and transforming growth factor β-induced epithelial-to-mesenchymal transition. J Biol Chem. 2013;288(11):7841–7856. doi: 10.1074/jbc.M112.425009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whitlock JP. Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol. 1999;39:103–125. doi: 10.1146/annurev.pharmtox.39.1.103. [DOI] [PubMed] [Google Scholar]
- 47.Diani-Moore S, et al. Identification of the aryl hydrocarbon receptor target gene TiPARP as a mediator of suppression of hepatic gluconeogenesis by 2,3,7,8-tetrachlorodibenzo-p-dioxin and of nicotinamide as a corrective agent for this effect. J Biol Chem. 2010;285(50):38801–38810. doi: 10.1074/jbc.M110.131573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stejskalova L, Dvorak Z, Pavek P. Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab. 2011;12(2):198–212. doi: 10.2174/138920011795016818. [DOI] [PubMed] [Google Scholar]
- 49.Thomsen JS, Kietz S, Ström A, Gustafsson JA. HES-1, a novel target gene for the aryl hydrocarbon receptor. Mol Pharmacol. 2004;65(1):165–171. doi: 10.1124/mol.65.1.165. [DOI] [PubMed] [Google Scholar]
- 50.Kim DW, Gazourian L, Quadri SA, Romieu-Mourez R, Sherr DH, Sonenshein GE. The RelA NF-kappaB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells. Oncogene. 2000;19(48):5498–5506. doi: 10.1038/sj.onc.1203945. [DOI] [PubMed] [Google Scholar]
- 51.Mandavia C. TCDD-induced activation of aryl hydrocarbon receptor regulates the skin stem cell population. Med Hypotheses. 2015;84(3):204–208. doi: 10.1016/j.mehy.2014.12.023. [DOI] [PubMed] [Google Scholar]
- 52.Dever DP, Opanashuk LA. The aryl hydrocarbon receptor contributes to the proliferation of human medulloblastoma cells. Mol Pharmacol. 2012;81(5):669–678. doi: 10.1124/mol.111.077305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu PY, et al. Aryl hydrocarbon receptor downregulates MYCN expression and promotes cell differentiation of neuroblastoma. PLoS ONE. 2014;9(2):e88795. doi: 10.1371/journal.pone.0088795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang X, et al. The aryl hydrocarbon receptor constitutively represses c-myc transcription in human mammary tumor cells. Oncogene. 2005;24(53):7869–7881. doi: 10.1038/sj.onc.1208938. [DOI] [PubMed] [Google Scholar]
- 55.Kim SH, et al. Novel compound 2-methyl-2H-pyrazole-3-carboxylic acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) prevents 2,3,7,8-TCDD-induced toxicity by antagonizing the aryl hydrocarbon receptor. Mol Pharmacol. 2006;69(6):1871–1878. doi: 10.1124/mol.105.021832. [DOI] [PubMed] [Google Scholar]
- 56.Safe S, Cheng Y, Jin UH. The aryl hydrocarbon receptor (AhR) as a drug target for cancer chemotherapy. Curr Opin Toxicol. 2017;2:24–29. doi: 10.1016/j.cotox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe. Nat Rev Cancer. 2014;14(12):801–814. doi: 10.1038/nrc3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Callero MA, Suárez GV, Luzzani G, Itkin B, Nguyen B, Loaiza-Perez AI. Aryl hydrocarbon receptor activation by aminoflavone: new molecular target for renal cancer treatment. Int J Oncol. 2012;41(1):125–134. doi: 10.3892/ijo.2012.1427. [DOI] [PubMed] [Google Scholar]
- 59.Meng LH, et al. Activation of aminoflavone (NSC 686288) by a sulfotransferase is required for the antiproliferative effect of the drug and for induction of histone gamma-H2AX. Cancer Res. 2006;66(19):9656–9664. doi: 10.1158/0008-5472.CAN-06-0796. [DOI] [PubMed] [Google Scholar]
- 60.Lu YF, Santostefano M, Cunningham BD, Threadgill MD, Safe S. Substituted flavones as aryl hydrocarbon (Ah) receptor agonists and antagonists. Biochem Pharmacol. 1996;51(8):1077–1087. doi: 10.1016/0006-2952(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 61.Loaiza-Pérez AI, et al. Aryl hydrocarbon receptor activation of an antitumor aminoflavone: basis of selective toxicity for MCF-7 breast tumor cells. Mol Cancer Ther. 2004;3(6):715–725. [PubMed] [Google Scholar]
- 62.Wagner JE, et al. Phase I/II Trial of StemRegenin-1 Expanded Umbilical Cord Blood Hematopoietic Stem Cells Supports Testing as a Stand-Alone Graft. Cell Stem Cell. 2016;18(1):144–155. doi: 10.1016/j.stem.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boitano AE, et al. Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells. Science. 2010;329(5997):1345–1348. doi: 10.1126/science.1191536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brinkman AM, et al. Aminoflavone-loaded EGFR-targeted unimolecular micelle nanoparticles exhibit anti-cancer effects in triple negative breast cancer. Biomaterials. 2016;101:20–31. doi: 10.1016/j.biomaterials.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barry VC, et al. A new series of phenazines (rimino-compounds) with high antituberculosis activity. Nature. 1957;179(4568):1013–1015. doi: 10.1038/1791013a0. [DOI] [PubMed] [Google Scholar]
- 66.Barry VC, Conalty ML. The antimycobacterial activity of B 663. Lepr Rev. 1965;36:3–7. doi: 10.5935/0305-7518.19650002. [DOI] [PubMed] [Google Scholar]
- 67.Yawalkar SJ, Vischer W. Lamprene (clofazimine) in leprosy. Basic information. Lepr Rev. 1979;50(2):135–144. doi: 10.5935/0305-7518.19790020. [DOI] [PubMed] [Google Scholar]
- 68.Wong KP, Sourkes TL. Glucuronidation of 3-O-methylnoradrenaline, harmalol and some related compounds. Biochem J. 1968;110(1):99–104. doi: 10.1042/bj1100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gebel T, Arand M, Oesch F. Induction of the peroxisome proliferator activated receptor by fenofibrate in rat liver. FEBS Lett. 1992;309(1):37–40. doi: 10.1016/0014-5793(92)80734-X. [DOI] [PubMed] [Google Scholar]
- 70.Yang LP, Keating GM. Fenofibric acid: in combination therapy in the treatment of mixed dyslipidemia. Am J Cardiovasc Drugs. 2009;9(6):401–409. doi: 10.2165/11203920-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 71.Casero RA, Pegg AE. Polyamine catabolism and disease. Biochem J. 2009;421(3):323–338. doi: 10.1042/BJ20090598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y, Hacker A, Murray-Stewart T, Fleischer JG, Woster PM, Casero RA. Induction of human spermine oxidase SMO(PAOh1) is regulated at the levels of new mRNA synthesis, mRNA stabilization and newly synthesized protein. Biochem J. 2005;386(Pt 3):543–547. doi: 10.1042/BJ20041084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Casero RA, Pegg AE. Spermidine/spermine N1-acetyltransferase--the turning point in polyamine metabolism. FASEB J. 1993;7(8):653–661. doi: 10.1096/fasebj.7.8.8500690. [DOI] [PubMed] [Google Scholar]
- 74.Libby PR, Ganis B, Bergeron RJ, Porter CW. Characterization of human spermidine/spermine N1-acetyltransferase purified from cultured melanoma cells. Arch Biochem Biophys. 1991;284(2):238–244. doi: 10.1016/0003-9861(91)90291-P. [DOI] [PubMed] [Google Scholar]
- 75.Fogel-Petrovic M, Vujcic S, Brown PJ, Haddox MK, Porter CW. Effects of polyamines, polyamine analogs, and inhibitors of protein synthesis on spermidine-spermine N1-acetyltransferase gene expression. Biochemistry. 1996;35(45):14436–14444. doi: 10.1021/bi9612273. [DOI] [PubMed] [Google Scholar]
- 76.Ren YR, et al. Clofazimine inhibits human Kv1.3 potassium channel by perturbing calcium oscillation in T lymphocytes. PLoS ONE. 2008;3(12):e4009. doi: 10.1371/journal.pone.0004009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vennekamp J, et al. Kv1.3-blocking 5-phenylalkoxypsoralens: a new class of immunomodulators. Mol Pharmacol. 2004;65(6):1364–1374. doi: 10.1124/mol.65.6.1364. [DOI] [PubMed] [Google Scholar]
- 78.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ishida M, et al. Activation of aryl hydrocarbon receptor promotes invasion of clear cell renal cell carcinoma and is associated with poor prognosis and cigarette smoke. Int J Cancer. 2015;137(2):299–310. doi: 10.1002/ijc.29398. [DOI] [PubMed] [Google Scholar]
- 81.Fink EE, et al. Mitochondrial thioredoxin reductase regulates major cytotoxicity pathways of proteasome inhibitors in multiple myeloma cells. Leukemia. 2016;30(1):104–111. doi: 10.1038/leu.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mannava S, et al. KLF9 is a novel transcriptional regulator of bortezomib- and LBH589-induced apoptosis in multiple myeloma cells. Blood. 2012;119(6):1450–1458. doi: 10.1182/blood-2011-04-346676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bahlis NJ, et al. CD28-mediated regulation of multiple myeloma cell proliferation and survival. Blood. 2007;109(11):5002–5010. doi: 10.1182/blood-2006-03-012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Murray ME, et al. CD28-mediated pro-survival signaling induces chemotherapeutic resistance in multiple myeloma. Blood. 2014;123(24):3770–3779. doi: 10.1182/blood-2013-10-530964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nair JR, et al. Novel inhibition of PIM2 kinase has significant anti-tumor efficacy in multiple myeloma. Leukemia. 2017;31(8):1715–1726. doi: 10.1038/leu.2016.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nair JR, et al. CD28 expressed on malignant plasma cells induces a prosurvival and immunosuppressive microenvironment. J Immunol. 2011;187(3):1243–1253. doi: 10.4049/jimmunol.1100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chesi M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood. 2012;120(2):376–385. doi: 10.1182/blood-2012-02-412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chesi M, et al. AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell. 2008;13(2):167–180. doi: 10.1016/j.ccr.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Affronti HC, et al. Dietary folate levels alter the kinetics and molecular mechanism of prostate cancer recurrence in the CWR22 model. Oncotarget. 2017;8(61):103758–103774. doi: 10.18632/oncotarget.21911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Holien T, Sundan A. Oncogene addiction to c-MYC in myeloma cells. Oncotarget. 2012;3(8):739–740. doi: 10.18632/oncotarget.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holien T, Våtsveen TK, Hella H, Waage A, Sundan A. Addiction to c-MYC in multiple myeloma. Blood. 2012;120(12):2450–2453. doi: 10.1182/blood-2011-08-371567. [DOI] [PubMed] [Google Scholar]
- 92.Cooke RE, et al. Spontaneous onset and transplant models of the Vk*MYC mouse show immunological sequelae comparable to human multiple myeloma. J Transl Med. 2016;14:259. doi: 10.1186/s12967-016-0994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chesi M, et al. IAP antagonists induce anti-tumor immunity in multiple myeloma. Nat Med. 2016;22(12):1411–1420. doi: 10.1038/nm.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Boncher T, Bi X, Varghese S, Casero RA, Jr, Woster PM. Polyamine-based analogues as biochemical probes and potential therapeutics. Biochem Soc Trans. 2007;35(Pt 2):356–363. doi: 10.1042/BST0350356. [DOI] [PubMed] [Google Scholar]
- 95.Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer. 2004;4(10):781–792. doi: 10.1038/nrc1454. [DOI] [PubMed] [Google Scholar]
- 96.Bey P, Danzin C, Van Dorsselaer V, Mamont P, Jung M, Tardif C. Analogues of ornithine as inhibitors of ornithine decarboxylase. New deductions concerning the topography of the enzyme’s active site. J Med Chem. 1978;21(1):50–55. doi: 10.1021/jm00199a009. [DOI] [PubMed] [Google Scholar]
- 97.Meyskens FL, Gerner EW. Development of difluoromethylornithine (DFMO) as a chemoprevention agent. Clin Cancer Res. 1999;5(5):945–951. [PubMed] [Google Scholar]
- 98.McDougal A, Gupta MS, Morrow D, Ramamoorthy K, Lee JE, Safe SH. Methyl-substituted diindolylmethanes as inhibitors of estrogen-induced growth of T47D cells and mammary tumors in rats. Breast Cancer Res Treat. 2001;66(2):147–157. doi: 10.1023/A:1010608000074. [DOI] [PubMed] [Google Scholar]
- 99.Bradshaw TD, Westwell AD. The development of the antitumour benzothiazole prodrug, Phortress, as a clinical candidate. Curr Med Chem. 2004;11(8):1009–1021. doi: 10.2174/0929867043455530. [DOI] [PubMed] [Google Scholar]
- 100.Sechi AM, Cabrini L, Landi L, Pasquali P, Lenaz G. Inhibition of phospholipase A2 and phospholipase C by polyamines. Arch Biochem Biophys. 1978;186(2):248–254. doi: 10.1016/0003-9861(78)90433-2. [DOI] [PubMed] [Google Scholar]
- 101.Jensen BA, Leeman RJ, Schlezinger JJ, Sherr DH. Aryl hydrocarbon receptor (AhR) agonists suppress interleukin-6 expression by bone marrow stromal cells: an immunotoxicology study. Environ Health. 2003;2(1):16. doi: 10.1186/1476-069X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kashuba EV, et al. Regulation of transactivation function of the aryl hydrocarbon receptor by the Epstein-Barr virus-encoded EBNA-3 protein. J Biol Chem. 2006;281(2):1215–1223. doi: 10.1074/jbc.M509036200. [DOI] [PubMed] [Google Scholar]
- 103.Vogel CF, et al. Pathogenesis of aryl hydrocarbon receptor-mediated development of lymphoma is associated with increased cyclooxygenase-2 expression. Am J Pathol. 2007;171(5):1538–1548. doi: 10.2353/ajpath.2007.070406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Koorella C, Nair JR, Murray ME, Carlson LM, Watkins SK, Lee KP. Novel regulation of CD80/CD86-induced phosphatidylinositol 3-kinase signaling by NOTCH1 protein in interleukin-6 and indoleamine 2,3-dioxygenase production by dendritic cells. J Biol Chem. 2014;289(11):7747–7762. doi: 10.1074/jbc.M113.519686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27–31. doi: 10.4103/0976-0105.177703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dimopoulos MA, et al. Thalidomide and dexamethasone combination for refractory multiple myeloma. Ann Oncol. 2001;12(7):991–995. doi: 10.1023/A:1011132808904. [DOI] [PubMed] [Google Scholar]
- 107.Palumbo A, et al. Bortezomib, doxorubicin and dexamethasone in advanced multiple myeloma. Ann Oncol. 2008;19(6):1160–1165. doi: 10.1093/annonc/mdn018. [DOI] [PubMed] [Google Scholar]
- 108.Chen D, Frezza M, Schmitt S, Kanwar J, Dou QP. Bortezomib as the first proteasome inhibitor anticancer drug: current status and future perspectives. Curr Cancer Drug Targets. 2011;11(3):239–253. doi: 10.2174/156800911794519752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meregalli C. An overview of bortezomib-induced neurotoxicity. Toxics. 2015;3(3):294–303. doi: 10.3390/toxics3030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harvey RD. Incidence and management of adverse events in patients with relapsed and/or refractory multiple myeloma receiving single-agent carfilzomib. Clin Pharmacol. 2014;6:87–96. doi: 10.2147/CPAA.S62512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shimizu Y, et al. Benzo[a]pyrene carcinogenicity is lost in mice lacking the aryl hydrocarbon receptor. Proc Natl Acad Sci U S A. 2000;97(2):779–782. doi: 10.1073/pnas.97.2.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bianchi-Smiraglia A, et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene. 2017;36(1):84–96. doi: 10.1038/onc.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bianchi-Smiraglia A, et al. Pharmacological targeting of guanosine monophosphate synthase suppresses melanoma cell invasion and tumorigenicity. Cell Death Differ. 2015;22(11):1858–1864. doi: 10.1038/cdd.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mannava S, et al. Ribonucleotide reductase and thymidylate synthase or exogenous deoxyribonucleosides reduce DNA damage and senescence caused by C-MYC depletion. Aging (Albany NY) 2012;4(12):917–922. doi: 10.18632/aging.100512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fink EE, et al. Mitochondrial thioredoxin reductase regulates major cytotoxicity pathways of proteasome inhibitors in multiple myeloma cells. Leukemia. 2016;30(1):104–111. doi: 10.1038/leu.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bistulfi G, et al. The essential role of methylthioadenosine phosphorylase in prostate cancer. Oncotarget. 2016;7(12):14380–14393. doi: 10.18632/oncotarget.7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.